Abstract
Microglia are myeloid cells of the central nervous system (CNS) that participate both in normal CNS function and disease. We investigated the molecular signature of microglia and identified 239 genes and 8 microRNAs that were uniquely or highly expressed in microglia vs. myeloid and other immune cells. Out of 239 genes, 106 were enriched in microglia as compared to astrocytes, oligodendrocytes and neurons. This microglia signature was not observed in microglial lines or in monocytes recruited to the CNS and was also observed in human microglia. Based on this signature, we found a crucial role for TGF-β in microglial biology that included: 1) the requirement of TGF-β for the in vitro development of microglia that express the microglial molecular signature characteristic of adult microglia; and 2) the absence of microglia in CNS TGF-β1 deficient mice. Our results identify a unique microglial signature that is dependent on TGF-β signaling which provides insights into microglial biology and the possibility of targeting microglia for the treatment of CNS disease.
INTRODUCTION
Microglia are resident myeloid cells of the CNS and participate both in normal CNS function and in the progression and resolution of disease. If a unique microglial gene and microRNA signature were identified, it would provide the basis to both understand microglia biology and to modulate microglia for the treatment of CNS diseases. Related to this, the investigation of microglia has been complicated by controversy and nomenclature disputes1–3 and a challenge to investigators has been the development of markers that distinguish microglia from hematogenous infiltrating macrophages which have identical morphologies2.
Recent studies suggest that resident microglia represent a unique, indigenous cell population in the brain. Specifically, it has been shown that adult microglia derive from primitive macrophages4 in a Myb-independent manner5 via PU.1 and IRF8 dependent pathways6. This lineage is mainly regulated by CSFR14 and its ligand, IL-347. In addition, it has been reported that in the experimental autoimmune encephalomyelitis (EAE) model, infiltrating monocytes do not contribute to the residual microglial pool8 and that microglia can be distinguished from monocytes using red-green mice in which microglia and monocyte-derived macrophages are labeled with CX3CR1 (GFP) and CCR2 (RFP) respectively9. Thus, there is a resident pool of microglia that is separate from peripheral myeloid cells that infiltrate the nervous system.
We embarked on a series of investigations to identify unique biological features of microglial cells using two approaches: 1) gene and microRNA array analysis and 2) quantitative proteomic analysis. We used these two approaches to profile murine CNS-derived adult microglia vs. splenic Ly6C monocyte subsets and other immune cell types. These investigations have led to the identification of a unique TGF-β dependent microglial signature in mice, features of which are also observed in human microglial cells.
RESULTS
Identification of a unique microglial signature
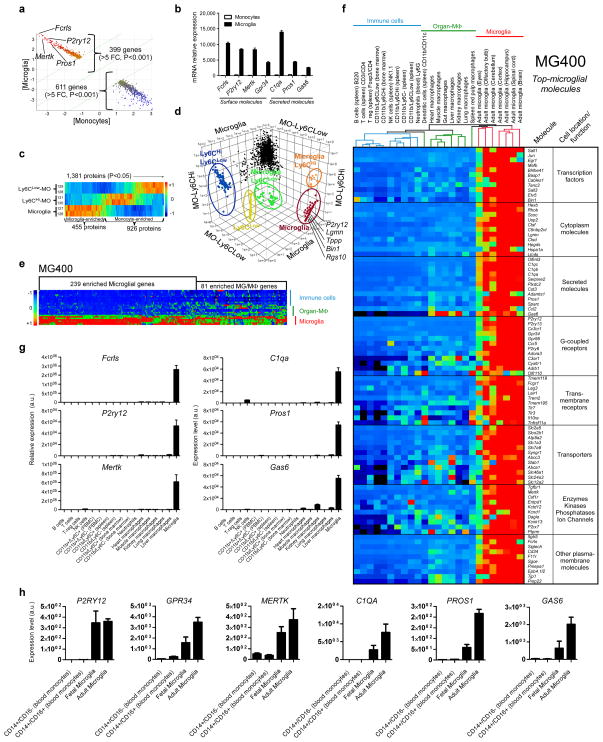
To identify a unique microglia signature we performed gene profiling (Source data Fig. 1) and quantitative mass spectrometry analysis (Supplementary Fig. 1 and Source data Fig. 1) of CD11b+CD45Low microglia isolated from the CNS and CD11b+Ly6C+ monocyte subsets isolated from the spleen of naïve adult mice. We chose Ly6C+ monocytes as this subset is known to be recruited to the CNS in association with inflammation10–12 and it was our goal to identify unique microglial signatures. Gene array identified 1572 genes that were enriched in microglia (Source data Fig. 1). Fig. 1a shows a scatterplot of 399 microglia enriched genes vs. 611 monocyte enriched genes with a greater than 5-fold difference (P<0.001). We highlight four highly expressed microglial genes in the scatterplot: Fcrls, P2ry12, Mertk and Pros1. Fig. 1b demonstrates the normalized intensity of both the surface and secreted microglia specific molecules. Following this we performed quantitative mass spectrometry analysis (tandem mass tagging, TMT) of microglial cells vs. splenic Ly6CHi and Ly6CLow subsets. Mass spectrometry identified a total of 1991 proteins (Supplementary Fig. 1a,b and Source data Fig. 1). As shown in Fig. 1c, we found that 1381 of these proteins were differentially expressed between microglia and Ly6C subsets. 455 of these proteins were enriched in microglia and 926 proteins (greater than 2-fold difference) were enriched in Ly6C monocytes (Source data Fig. 1). Fig. 1d is a three dimensional scatterplot based on these 1381 proteins which shows a proteomic signature of microglia vs. Ly6C subsets in which we highlight five highly expressed microglial proteins in the scatterplot: P2ry12, Lgmn, Tppp, Bin1 and Rgs10. In addition to these 5 proteins, we identified a total of 74 proteins which were uniquely expressed in microglia (Supplementary Fig. 1c and Source data Fig. 1). In addition, out of 103 microglial enriched proteins above 5 fold as compared with Ly6CLow and Ly6CHi monocytes (Supplementary Fig. 1d), 64 of the proteins were also identified in gene profiling of microglia (Source data Fig. 1).
Figure 1. Identification of a microglia signature by gene expression and quantitative mass spectrometry.
(a) AffyExon1 genearray expression profile of adult mouse microglia and splenic Ly6C monocytes (biological triplicates) identified 399 genes in microglia vs. 611 genes in monocytes (>5 fold, P<0.001, Student’s t test, 2-tailed) (see Source data – Figure 1). (b) Gene expression of microglial molecules. Bars show mean normalized intensity ± s.e.m. (n = 3). (c) Heatmap of 1,381 mass spectrometry identified proteins differentially expressed between microglia and Ly6C subsets (ANOVA, P<0.05) (biological duplicates) (see Source data – Figure 1). 455 of these proteins were enriched in microglia and 926 proteins in Ly6C monocytes (see Source data – Figure 1). (d) 3D-scatter plot based on the 1,381 differentially expressed proteins in microglia and monocytes. (e) Heatmap of microglia vs. F4/80+CD11b+ macrophages and immune cells using the MG400 chip. MG, microglia; MΦ, macrophages (see Source data – Figure 1). (f) Heatmap and hierarchical clustering of microglia, macrophages and immune cells analyzed with the MG400 chip (see Source data – Figure 1). Results were log-transformed, normalized and centered, and populations and genes were clustered by Pearson correlation. Data are representative of three different experiments with microglia pooled from 15 mice, macrophages from 10 mice and immune cells pooled from 5 mice. (g) qPCR analysis of identified microglial genes in different cells. Expression levels were normalized to Gapdh (n = 2). Bars show mean ±s.e.m. Shown is one representative of three individual experiments. (h) qPCR validation of the selected 6 microglial genes in both fetal and adult human microglia and human blood-derived monocytes. Expression levels were normalized to Gapdh (n = 2). Bars show mean ± s.e.m.
In order to investigate the degree to which the microglial molecules we identified were expressed in other immune cell types and organ specific macrophages, we constructed a Nanostring chip that contained 354 microglial enriched genes, 40 inflammation-related genes we previously found affected in microglia in SOD1 mice12 and 6 housekeeping genes. We termed this chip MG400 (Source data Fig. 1). 254 genes were chosen from 1572 genes and 100 proteins were chosen from 455 proteins (Source data Fig. 1), all of which were specifically or highly expressed in adult mouse microglia. Fig. 1e shows a heatmap analysis of microglia vs. organ specific macrophages and immune cells using the MG400 chip. We found 239 genes which were specifically expressed by microglia and 81 genes which were expressed by both microglia and organ specific F4/80+CD11b+ macrophages (Source data Fig. 1). A heatmap and hierarchical clustering of microglia, organ specific macrophages and immune cell populations analyzed with the MG400 chip shows the top microglial molecules grouped according to cell localization and function (Fig. 1f and Source data Fig. 1).
We then performed quantitative RT-PCR (qPCR) analysis of 6 selected microglial genes to validate their expression in different cell populations (Fig. 1g). We found minimal if any expression of these 6 microglial genes in other immune cells or organ-specific macrophages. Finally, in order to determine whether there was similar expression of these genes in human microglia, we performed qPCR in both fetal and adult human CNS-derived microglia and human blood-derived CD14+CD16− and CD14+CD16+ monocytes which are analogous to murine Ly6CHi and Ly6CLow subsets13. As shown in Fig. 1h, we found that P2ry12, Gpr34, Mertk, C1qa, Pros1 and Gas6 are highly or uniquely expressed in human microglia. Of note, there is no human orthologous gene of Fcrls in humans, the most highly expressed gene in murine microglia. In summary, as shown in Fig. 1f, we identified genes (e.g., Fcrls, P2ry12, Tmem119, Olfml3, Hexb, Tgfbr1, Gpr34 and Sall1), which appear to be expressed in microglia as we did not detect them in immune cells, Ly6C monocytes or organ specific macrophages. Recent work demonstrated that all tissue resident macrophages are different to an extent yet cluster together when compared to monocytes and DCs and that spleen red pulp macrophages were closely associated with microglia14. Consistent with this, we found that red pulp macrophages are the tissue resident macrophages closest to microglia (Fig. 1f and Source data Fig. 1). In addition, we found that genes related to the TAM system (e.g. Mertk, Pros1 and Gas6) were highly expressed in microglia (Fig. 1g). It is well known that TAM family receptors/ligands are expressed in macrophages15. Furthermore, we found increased microglial expression of C1q, Csfr1 and Cd34 genes, which are known to be expressed on tissue resident macrophages14.
Thus, it appears that there are common features between CNS resident microglia and tissue macrophages. Our findings are consistent with reports that macrophage progenitors develop from the primitive ectoderm of the yolk sac4 and give rise to tissue macrophages that do not have a monocytic progenitor16,17.
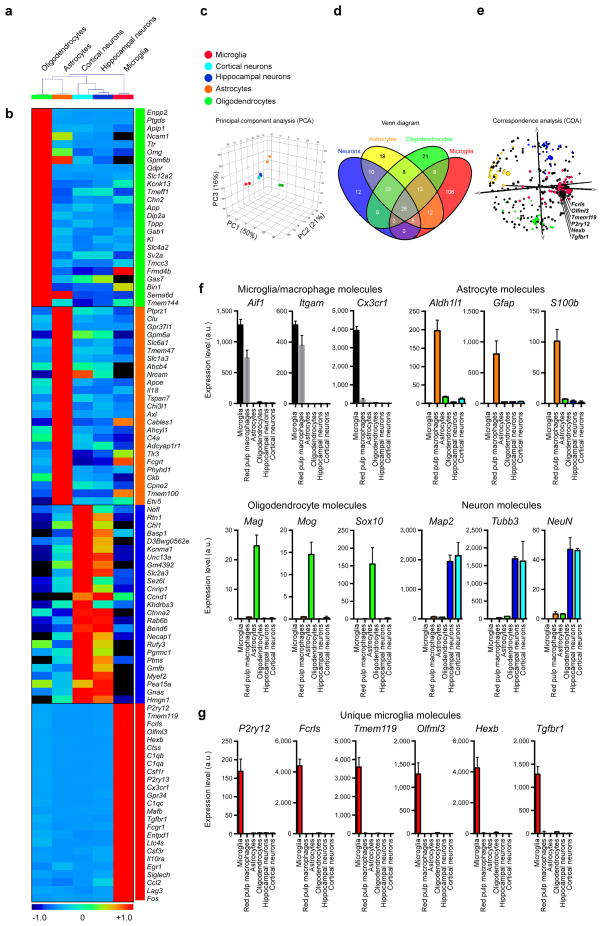
Next, we investigated whether the microglial genes we identified were expressed in CNS cells including astrocytes, oligodendrocytes and neurons by profiling these CNS cells with the MG400 microglial chip (Source data Fig. 2). A dendogram based on unsupervised hierarchical clustering of CNS cell types demonstrates that microglia are different from CNS cells (Fig. 2a). Fig. 2b shows a heatmap of the 25-top specific genes in microglia and the other CNS cells (Supplementary Table 1). Principal component analysis (PCA) based on the MG400 gene expression profile shows that microglia are different than other CNS cells (Fig. 2c). We compared the identified molecular signature of detected genes across all four populations and found 106 genes which were specifically expressed in microglia (Fig. 2d). Correspondence analysis (COA) shows the 106 microglial specific genes (Source data Fig. 2). We highlight six highly expressed microglial genes in the COA plot: Fcrls, Olfml3, Tmeme119, P2ry12, Hexb and Tgfbr1 (Fig. 2e). The purity of CNS cells types was established by qPCR analysis using their known unique markers (Fig. 2f). qPCR analysis demonstrated that the microglia unique genes Fcrls, Olfml3, Tmeme119, P2ry12, Hexb and Tgfbr1 were absent in oligodendrocytes, astrocytes and neurons (Fig. 2g).
Figure 2. MG400 profile in microglia vs. astrocytes, oligodendrocytes and neurons.
(a) Dendogram of unsupervised hierarchical clustering (Pearson correlation; average linkage) of biological duplicates for FCRLS+ adult microglia (n = 5 mice), Glt-EGFP+ adult astrocytes (n = 9 mice), adult oligodendrocytes (n = 5 mice) and primary postnatal hippocampal and cortical neurons (see Source data – Figure 2). Individual cell types are identified as follows: green, oligodendrocytes; orange, astrocytes; cyan, cortical neurons; dark-blue, hippocampal neurons and red, microglia. (b) Heatmap of top 25 enriched genes in each cell type based on hierarchical clustering. Each lane represents the average expression value of two biological duplicates per cell type. (c) Principle component analysis based on MG400 expression for CNS cells. (d) MG400 profile of detected genes (>100 mRNA transcripts) in microglia, astrocytes, oligodendrocytes and neurons. Venn diagram displays unique and intersecting genes among cell types. (e) Correspondence analysis of samples (large spheres) and genes (small spheres). (f) qPCR analysis of CNS cell type specific genes for each population. (g) qPCR analysis of microglial unique genes (P2ry12, Fcrls, Tmem119, Olfml3, Hexb and Tgfbr1) as compared to spleen red pulp macrophages and CNS cell types. Expression levels were normalized to Gapdh (n = 3). Bars show mean ± s.e.m. Shown is one of two individual experiments.
In addition, we analyzed a published database of identified genes in oligodendrocytes, astrocytes and neurons18. We compared the identified molecular signature of genes with expression >3-fold across all four populations and found 152 genes which were specifically expressed in microglia (Supplementary Table 2 and Source data – Supplementary Table 2). Consistent with our results (Fig. 2), P2ry12, Fcrls, Tmem119, Olfml3, Hexb and Tgfbr1 were identified as unique microglial genes. Thus, these unique microglial genes may serve as targets to investigate microglia biology.
In order to better understand the physiological function of microglia based on our identified molecular signature, we performed Ingenuity pathway analysis (IPA). We found that the top microglial functions in naïve adult microglia were related to nervous system development and were not observed in monocytes (Supplementary Fig. 2). The genes related to this pathway are shown in Supplementary Fig. 3. GeneGo analysis identified PU.1 as the top transcriptional factor differentially expressed in microglia (Supplementary Table 3 and Supplementary Fig. 4), which confirms previous observations that PU.1 deficient mice do not develop myeloid cells19,20. Finally, canonical pathway (Supplementary Fig. 5) and upstream regulator analysis (Supplementary Table 4) revealed that TGF-β was among the most affected molecules in microglia (Supplementary Fig. 6). These results are consistent with our findings presented below that TGF-β is important for development and maintenance of microglia both in vitro and in vivo.
Identification of a microglial microRNA signature
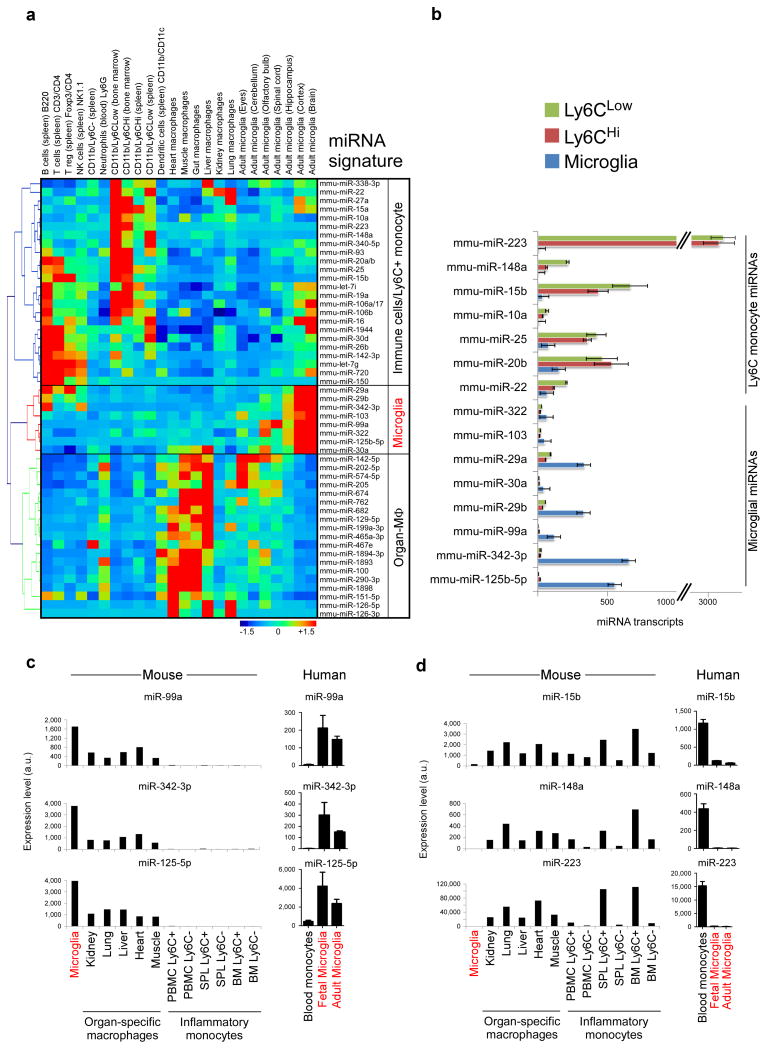
To further characterize microglia, we used a Nanostring based miRNA chip that contains 600 microRNAs to profile microglia vs. immune cells (Source data Fig. 3). Fig. 3a shows a heatmap and hierarchical clustering of mouse microglia, organ specific macrophages and immune cell populations. We found 8 microRNAs that were highly expressed in microglia. Three of these (miR-125b-5p, miR-342-3p, miR-99a) were specifically expressed in microglia as compared to other immune cell types. In addition, we identified 24 microRNAs that were highly expressed in Ly6C+ inflammatory monocytes and immune cells. Three of these (miR-223, miR-148a, miR-15b) were specifically expressed in Ly6C monocyte subsets. In Fig. 3b we show miRNA transcript copies of unique or highly expressed microRNAs in microglia and Ly6C monocyte subsets. To validate these findings, we performed qPCR analysis of microglial miRNAs (miR-125b-5p, miR-99a, miR-342-3p) and inflammatory Ly6C+ miRNAs (miR-15b, miR-148a, miR-223) and found a similar expression pattern that we observed by Nanostring (Fig. 3c,d). In order to determine whether there was similar expression of microglial and inflammatory monocyte miRNAs in humans, we performed qPCR in both fetal and adult CNS-derived human microglia and human blood-derived CD14+CD16− and CD14+CD16+ monocytes (analogues of Ly6CHi and Ly6CLow subsets). As shown in Fig. 3c,d, we found that miR-99a, miR-125-5p and miR-342-3p were highly expressed in human microglia. Consistent with our results in mice, we also found that miR-15b, miR-148a and miR-223 were highly expressed in human monocytes with miR-223 the most selectively expressed miRNA.
Figure 3. Identification of a miRNA microglia signature.
(a) Heatmap and hierarchical clustering of differentially expressed miRNAs in microglia, organ specific macrophages and immune cell populations based on 600 miRNA nCounter chip (see Source data – Figure 3). (b) miRNA transcript copies of highly expressed microRNAs in microglia and Ly6C monocyte subsets. Bars show mean normalized intensity ± s.e.m. of miRNA transcripts per 100 ng of total RNA (n = 2). (c) qPCR validation of microglial miRNAs (miR-99a, miR-342-3p miR-125b-5p) and (d) inflammatory Ly6C+ miRNAs (miR-15b, miR-148a, miR-223) in Ly6C monocytes, organ-specific macrophages and human fetal and adult microglia. Bars show mean normalized intensity ± s.e.m. (n = 3). miRNA expression level was normalized against U6 miRNA using ΔCt. One representative of two individual experiments is shown.
In order to investigate whether the microglial miRNAs we identified were expressed in CNS cells including astrocytes, oligodendrocytes and neurons we profiled these cells with the Nanostring miRNA chip (Source data - Supplementary Fig. 7). We found that miR-342-3p was enriched in microglia, whereas both miR-125b-5p and miR-99a were highly expressed in microglia, astrocytes and neurons but not in oligodendrocytes (Supplementary Fig. 7).
Identification of unique microglial surface molecules
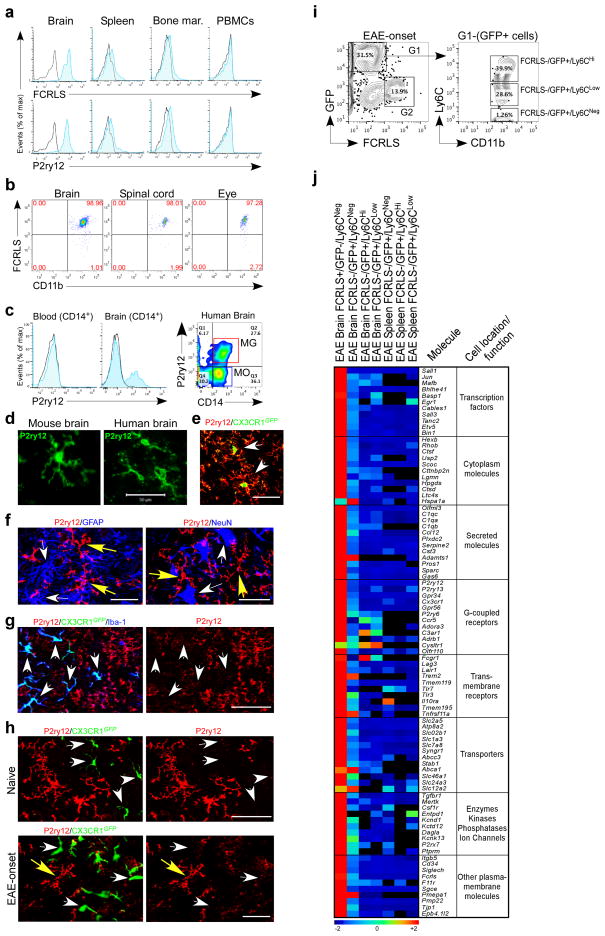
We identified P2ry12 and Fcrls as unique microglial genes. We thus studied the surface expression of these molecules by both FACS and immunohistochemistry. To study P2ry12, we obtained a polyclonal rabbit anti-P2ry12 antibody previously described to regulate microglial activation21 and generated ourselves a rat anti-P2ry12 antibody. To study FCRLS, we constructed an expression vector to generate an FCRLS-Fc fusion protein and immunized rats to produce an anti-FCRLS mAb. FACS analysis shows that anti-FCRLS and anti-P2ry12 antibodies stain adult microglia isolated by conventional methods22 from naïve murine brain but did not stain CD11b-gated myeloid cells from murine spleen, bone marrow and peripheral blood (Fig. 4a). In addition, as shown in Fig. 4b, anti-FCRLS stained >97% of total CD11b-gated cells in the brain, spinal cord and eye. FACS analysis of human cells showed that anti-P2ry12 stained a subpopulation of CD14+ myeloid cells isolated from fresh post-mortem brain but did not stain peripheral blood CD14+ monocytes (Fig. 4c). No staining of human cells was observed with anti-FCRLS. Of note, the gene for FCRLS is not expressed in humans.
Figure 4. Surface expression of microglial molecules and molecular signature of recruited monocytes during EAE.
(a) FACS analysis of CD11b-gated cells stained with FCRLS and P2ry12 Abs. Black line represents isotype control. Data are representative of three or more replicates. (b) Surface expression of FCRLS in murine brain (n = 6), spinal cord (n = 5) and eye (n = 4) CD11b-gated cells. (c) FACS analysis of P2ry12 expression in human peripheral blood and brain isolated mononuclear cells (n = 3). (d) Immunohistochemistry of cells with microglial morphology stained with anti-P2ry12 Ab in murine (n = 6) and human brain (n = 2). (e) GFP+ microglia in CX3CR1GFP transgenic mouse co-stained with anti-P2ry12 Ab (arrows). (f) Left panel shows staining with P2ry12 and GFAP (astrocyte marker); right panel shows staining with P2ry12 and NeuN (neuronal marker). Yellow arrows point to P2ry12+ microglia. (g) Rat anti-P2ry12 antibody stained Iba-1+GFP− microglia in naïve CX3CR1GFP/+ chimeric mouse but not recruited Iba-1+GFP+ monocytes (arrows). (h) Confocal images of spinal cord from naïve and EAE onset. Resident GFP− microglia are P2ry12 positive. Recruited GFP+ monocytes are P2ry12 negative. Scale bar, 50 μm. (i) FACS sorting of FCRLS+ microglia from EAE chimeric mice at disease onset. Ly6C is expressed by FCRLS−GFP+CD11b+ recruited monocytes (G1). Ly6C is not expressed by FCRLS+GFP−CD11b+ microglia (G2). FACS histogram represents a pool of 5 mice. (j) Heatmap of top microglial genes in sorted microglia, recruited monocytes from brain (n = 6) and splenic monocyte (n = 6) subsets from EAE chimeric mice at disease onset as analyzed by MG400 chip (see Source data – Figure 4). Data represent two independent experiments.
Immunohistochemical staining of both murine and human brain with anti-P2ry12 antibody identified cells with a microglia morphology (Fig. 4d). In addition, the P2ry12 signal co-localized on CX3CR1GFP microglia in which GFP is expressed in resident microglia17,23 (Fig. 4e). The P2ry12 signal was not observed on GFAP+ astrocytes or NeuN+ neurons (Fig. 4f) or peripheral tissue resident macrophages in splenic red and white pulp, lung, kidney, liver and skin (Supplementary Fig. 8a). Staining of murine microglia with a polyclonal anti-FCRLS antibody was observed by FACS (Supplementary Fig. 8b) and immunohistochemistry (Supplementary Fig. 8c,d). As shown in Fig. 4g, rat anti-P2ry12 antibody stained Iba-1+GFP− resident microglia in naïve CX3CR1GFP/+ chimeric mice but not recruited Iba-1+GFP+ monocytes. To investigate whether anti-P2ry12 Ab recognized recruited monocytes under inflammatory conditions, we induced EAE in CX3CR1GFP/+ chimeric mice. As shown in Fig. 4h recruited GFP+ monocytes do not express P2ry12 and resident GFP− microglia are positive for P2ry12 in both naïve and EAE mice.
Recruited monocytes do not acquire the microglia signature
Although the lineage relationship between microglia and macrophages is clear4,5, a major question is whether or not circulating monocytes and/or myeloid progenitor cells can acquire a unique molecular microglia signature and restore microglial function in the healthy CNS and disease.
To address this question, we investigated whether Ly6C+ recruited monocytes acquire the microglial signature during neuroinflammation. Thus, we sorted FCRLS+CD11b+GFP− resident microglia and recruited Ly6C+CD11b+GFP+ monocytes from EAE chimeric mice (Fig. 4i). We found that recruited GFP+Ly6CHi, GFP+Ly6CLow and GFP+Ly6CNeg monocyte subsets did not acquire the microglia signature during EAE and FCRLS+CD11b+GFP− resident microglia express the microglial signature as defined by the MG400 chip (Fig. 4j and Source data - Figure 4). Thus, microglia and recruited monocytes maintain their own molecular signature during neuroinflammation.
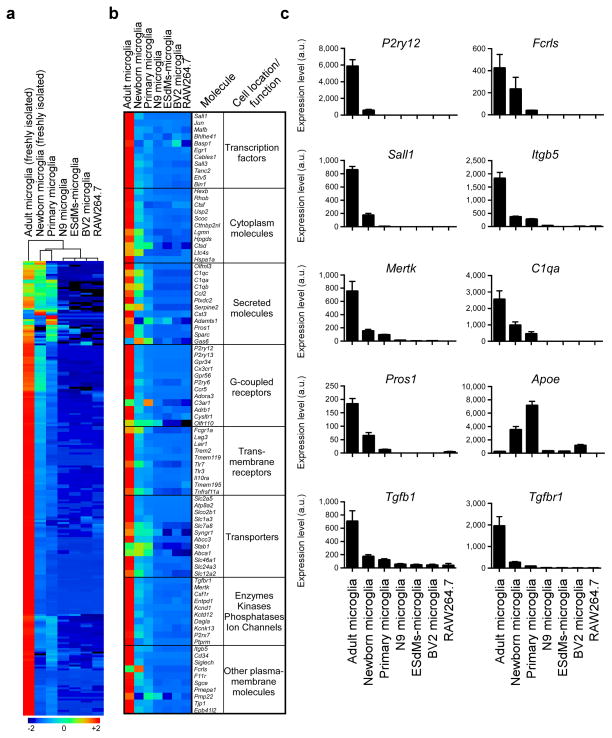
Microglia cell lines do not express a microglia signature
Investigators have utilized microglial cell lines to investigate microglial biology in vitro. We thus compared the adult murine microglia signature we identified to microglial cell lines and other microglial preparations to determine the degree to which they expressed a microglial signature. We investigated newborn microglia (P1), primary cultured newborn microglia (P1-P2), microglial cell lines (N9, BV2), embryonic stem cell microglia (ESdMs)24 and RAW264.7 macrophages. We found that none of these cells expressed the adult microglia signature we identified. Hierarchical clustering demonstrated that freshly sorted newborn and cultured primary microglia were closest to adult microglia (Fig. 5a and Source data Fig. 5). Fig. 5b compares expression of the top microglial genes we found to be expressed in adult microglia in vivo to the cell lines we tested. We then performed qPCR analysis of 10 selected microglial genes to validate their expression in different microglial cell lines. We found minimal if any expression of these 10 microglial genes in microglial cell lines. Of note, Apoe was highly expressed in newborn and primary microglia and had minimal expression in adult microglia (Fig. 5c).
Figure 5. Microglia signature is not present in microglial cell lines.
(a) MG400 expression profile of adult microglia (n = 10), newborn microglia (P1) (n = 30), primary cultured newborn microglia (P1-P2), microglial cell lines (N9, BV2), embryonic stem cell microglia (ESdMs) and RAW264.7 macrophages (see Source data – Figure 5). Data are representative of two different experiments. (b) Heatmap of top microglial genes. One representative of two individual experiments is shown. (c) qPCR analysis of 10 selected microglia genes. Gene expression level was normalized against Gapdh using ΔCt (n = 2). Bars show mean ± s.e.m. Shown is one representative of three individual experiments.
To investigate when during the course of development microglia acquire a microglia signature, we profiled embryonic (E10.5 and E12.5), post-natal (P4, P21 and P30) and adult microglia (2 months). Heatmap hierarchical cluster analysis showed that microglia expressed the lowest level of microglial genes between embryonic (E10.5, E 12.5) and postnatal P4 stages (Supplementary Fig. 9 and Source data - Supplementary Fig. 9). The highest expression of microglial genes occurred in the second period, between P21 and 2 months of age (Supplementary Fig. 9a). Supplementary Fig. 9b shows expression of the top microglial genes and Supplementary Fig. 10c shows TaqMan confirmation of 10 selected genes. Of note, Apoe expression was downregulated in microglia during development and expression of Apoe reciprocally correlated with the induction of microglial genes (Supplementary Fig. 9c).
TGF-β is required for development of microglia in vitro
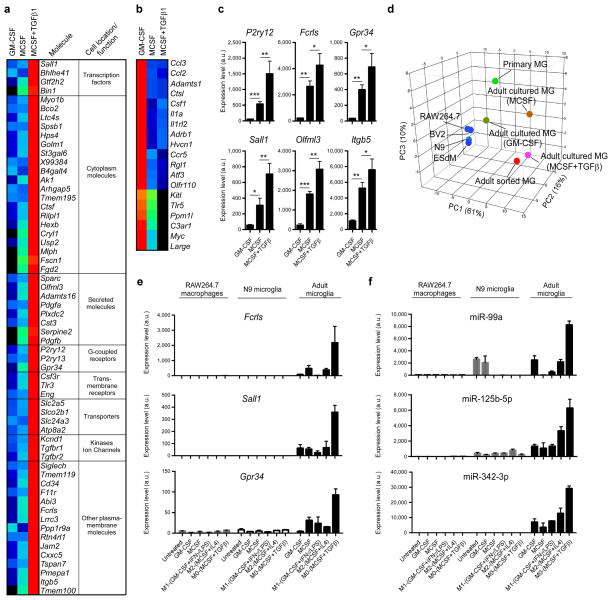
Using the microglia signature we identified, we investigated factors responsible for the growth and development of microglia in vitro. We cultured adult microglia in the presence of GM-CSF or MCSF with or without the addition of TGF-β1, which we identified above to be a major upstream regulator in microglia (Supplementary Fig. 5 and Supplementary Table 4). As shown in Fig. 6a, TGF-β1 plus MCSF induced the expression of 60 microglial genes above two fold as compared to microglia cultured in the presence of MCSF only and 108 microglial genes as compared to microglia cultured in the presence of GM-CSF (Source data Fig. 6). We termed these microglia ‘M0’ as their phenotype resembles freshly isolated adult microglia. Adult microglia do not survive in the presence of TGF-β1 alone and no effect was observed when TGF-β1 was added to GM-CSF. 19 genes, mostly related to pro-inflammatory responses including Ccl2 and Ccl3, were suppressed when cultured with MCSF or MCSF+TGF-β1 (Fig. 6b). The expression of 6 selected microglial genes in MCSF+TGF-β1 cultured microglia was confirmed by qPCR (Fig. 6c). Principal component analysis of cells analyzed with the MG400 chip showed that adult cultured microglia in the presence of MCSF+TGF-β1 most closely resemble freshly sorted adult microglia (Fig. 6d). Of note, MCSF+TGF-β1 did not induce the expression of microglial genes (Fig. 6e) or microglial miRNAs (Fig. 6f) in either the N9 microglia cell line or the RAW264.7 macrophage cell line. Consistent with our findings, others have reported that in vitro, TGF-β1 signaling induces a quiescent microglial phenotype25 and promotes resting microglial properties26
Figure 6. Role of TGF-β in the development of microglia in vitro.
(a and b) MG400 analysis of (a) upregulated microglial genes and (b) downregulated microglial genes in cultured adult microglia in the presence of MCSF and TGF-β1 in comparison to adult microglia cultured in the presence of GM-CSF or MCSF alone (see Source data – Figure 6). One representative of three individual experiments is shown. (c) qPCR analysis of 6 selected microglial genes in cultured adult microglia in the presence of MCSF and TGF-β1. Gene expression level was normalized against Gapdh using ΔCt (n = 6, each biological triplicate consisted of two wells per treatment). Bars show mean normalized intensity ± s.e.m. *P<0.05, **P<0.01, ***P<0.001, F2,15=5.829; 1-Way ANOVA followed by Dunnett’s multiple-comparison post-hoc test. (d) Principal component analysis (PCA) of different cell populations based on MG400 expression profile. (e and f) qPCR analysis of Fcrls, Sall1 and Gpr-34 gene expression (e) and miR-99a, miR-125b-5p and miR-342-3p expression (f) in RAW264.7 macrophages, N9 microglia cell line and adult microglia cultures. Cells were untreated or cultured in the presence of MCSF, GM-CSF, or polarized for 48h to M0 (MCSF+TGF-β1), M1 (GM-CSF+IFNγ+LPS) or M2 (MCSF+IL4) phenotypes. RAW264.7 macrophages and N9 cells survive without MCSF or GM-CFS, whereas adult microglia require either MCSF or GM-CSF. Gene expression level was normalized against Gapdh using ΔCt (n = 3). miRNA expression level was normalized against U6 miRNA using ΔCt (n = 3). Bars show mean ± s.e.m. Shown is one of two individual experiments.
In addition, we investigated the MG400 profile in GM-CSF cultured microglia with LPS/IFN-γ or MCSF+IL4, which are known to polarize M1 and M2 microglia, respectively27 (Supplementary Fig. 10). We found that M1 microglia express a high level of inflammatory-related transcripts including Ccl2, Ccl3, Ccl5, whereas M2 microglia express genes including IGF-1 (Supplementary Fig. 10a,b), which play a role in tissue development and neural cell renewal28. In order to better understand the physiological function of M0 vs. M1/M2 polarized microglia, we performed IPA analysis based on identified affected microglial genes. We found that microglial functions in M0-polarized microglia were related to nervous system development and were not observed in M1 or M2 polarized microglia (Supplementary Fig. 10c). In addition, we found that the top upstream regulators in M0 microglia were related to TGF-β1 signaling (Supplementary Fig. 10d). It should be noted however, that the regulators of M1/M2 phenotypes are not found in the intact CNS and that freshly isolated microglia from naive or diseased mice do not demonstrate these expression profiles.
TGF-β plays a critical role in microglia development in vivo
Investigation of TGF-β1 in the adult mouse brain is limited since TGF-β1−/− mice develop a lethal autoinflammatory syndrome shortly after birth29,30 which is linked to a T-cell specific deficiency of TGF-β31. We previously generated a TGF-β1-Tg mouse in which TGF-β is linked to the IL-2 promoter (IL2TGF-β1-Tg) and T cells transiently overexpress TGF-β after TCR stimulation32. We crossed this IL2TGF-β1-Tg mouse with TGF-β1−/− mice. Intercrossing these mice generated IL2TGF-β1-Tg-TGF-β1−/− mice, which bear the transgenic Tgfβ1 gene, but lack both endogenous TGF-β1 alleles. In IL2TGF-β1-Tg-TGF-β1−/− mice, the expression of TGF-β1 is thus limited to T-lymphocytes. We found that IL2TGF-β1-Tg-TGF-β1−/− mice were protected from lethal inflammation, survived into adulthood and were indistinguishable from IL2TGF-β1-Tg-TGF-β1+/− littermates at the age of 4–10 weeks with no signs of inflammation or organ dysfunction. However, when we investigated the CNS of these mice, we found that these animals did not express TGF-β1 in the brain or spinal cord and we found no TGF-β1-producing T-cells in the CNS of those animals33. These animals have defects in extracellular glutamate homeostasis and synaptic plasticity33 and we have termed these animals CNS-TGFβ1−/− mice. These mice thus provided the unique opportunity to investigate the effects of CNS TGF-β1 deficiency on microglia.
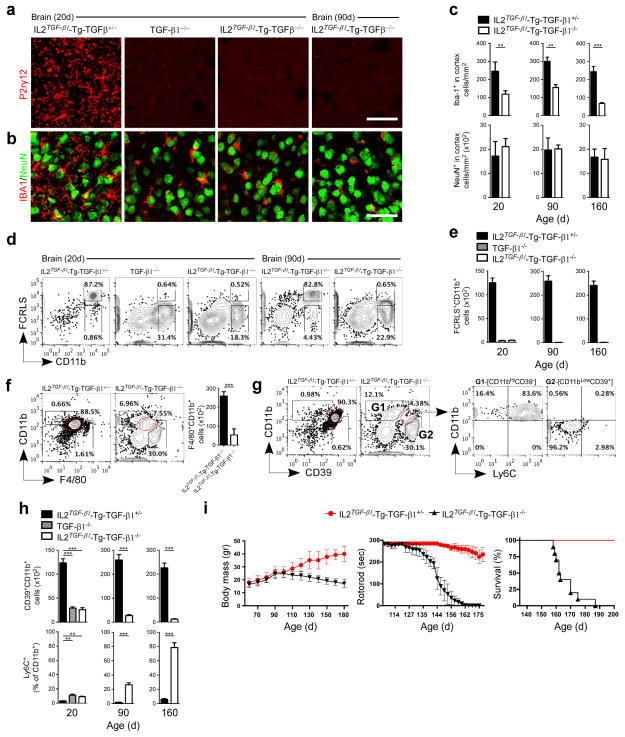
We found a loss of microglial cells in the CNS of adult CNS-TGFβ1−/− mice as measured by immunohistochemistry with anti-P2ry12 antibody at 20 and 90 days of age and in TGF-β1−/− mice just prior to death at 20 days (Fig. 7a). Iba-1 staining (Fig. 7b) and quantitative analysis of Iba-1+ myeloid cells demonstrated a reduced number of Iba-1+ cells in CNS-TGFβ1−/− mice (Fig. 7c) and these cells had a round morphology without ramified processes as compared to wild type mice (Fig. 7b). We did not observe neuronal loss in either CNS-TGFβ1−/− or TGF-β1−/− mice (Fig. 7b,c). FACS analysis with anti-FCRLS antibody at 20 and 90 days of age (Fig. 7d) and quantitative analysis of FCRLS+CD11b+ cells at 20, 90 and 160 days showed that CNS-TGFβ1−/− mice had loss of FCRLS+ microglia (Fig. 7e). In addition, CD11b+CD45Low microglia were lost in CNS-TGFβ1−/− mice (Supplementary Fig. 11a). We also found increased apoptosis of CD39+CD11b+ cells at 20, 90 and 160 days of age (Supplementary Fig. 11b,c). We then investigated the expression of F4/80 at 90 days in CNS-TGFβ1−/− mice; F4/80 stains organ specific macrophages and microglia5,34. We found that there was significant reduction of F4/80+CD11b+ cells as compared to wild type mice and that the level of CD11b expression was reduced in CNS-TGFβ1−/− mice (Fig. 7f). We previously reported that CD39 is expressed on adult microglia and can distinguish recruited Ly6C+ monocytes from resident microglia12. In CNS-TGFβ1−/− mice, we found that CD39+CD11bLow cells were decreased in an identical fashion to F4/80+CD11bLow cells (Fig. 7f,g). Associated with this, we found an increase in CD39−CD11bHi cells which express Ly6C (Fig. 7g) and which represent peripherally infiltrating monocytes9,12. Quantitative analysis showed significant reduction of CD39+CD11b+ cells and a reciprocal increase in Ly6C+CD11b+ cells at 20, 90 and 160 days in CNS-TGFβ1−/− mice (Fig. 7h). Of note, it has been reported that endogenous TGF-β1 is essential for normal murine Langerhans cell development35 and TGF-β1 acts directly on Langerhans cells through an autocrine/paracrine loop36. However, we did not find differences in peripheral organ macrophages, dendritic cells (DCs) or Langerhans cells (LCs) in CNS-TGFβ1−/− mice (Supplementary Fig. 12), consistent with the fact that loss of TGF-β1 is restricted to the CNS.
Figure 7. Loss of microglia in CNS-TGF-β1−/− mice.
(a and b) Immunohistochemistry of brain stained with (a) anti-P2ry12 (microglia) and (b) anti-Iba-1 (myeloid cells) and anti-NeuN (neurons) at 20 and 90 days of age (IL2TGF-β1-Tg-TGF-β1+/− and IL2TGF-β1-Tg-TGF-β1−/−) (n = 6) and 20d (TGF-β1−/−) (n = 5) mice. Scale bar, 50 μm. (c) Quantification of Iba-1+ and NeuN+ cells. Data represent mean ± s.e.m. at 20 days **P<0.01, t=13.51; 90 days **P<0.01, t=4.41; and 160 days ***P<0.001, t=11.29. Student’s t test, 2-tailed. (d) Representative FACS analysis of brain-derived mononuclear cells stained for CD11b and FCRLS among CD45+ cells at 20 (n = 5) and 90 (n = 6) days. (e) Quantification of FCRLS+CD11b+ cells at 20 (n = 6), 90 (n = 5) and 160 (n = 5) days. Data represent mean ± s.e.m. (f and g) FACS plots show the percentage and total cell number ± s.e.m. of (f) F4/80+CD11b+ cells (***P<0.001, t=15.60 Student’s t test, 2-tailed) and (g) the percentage of CD39+CD11b+ cells among CD45+ cells at 90 days of age in IL2TGF-β1-Tg-TGF-β1+/− (n = 5) and IL2TGF-β1-Tg-TGF-β1−/− (n = 6) mice. (h) Quantification of CD39+CD11b+ cells among CD45+ cells and Ly6C+ cells as percentage of CD11b+ cells at 20 (n = 6), 90 (n = 5) and 160 (n = 5) days. Data represent mean ± s.e.m. For CD39+CD11b+ cells ***P<0.001, F2,13=12.34; 1-Way ANOVA followed by Dunnett’s multiple-comparison post-hoc test for comparison at 20d and ***P<0.001, t= 20.20 and ***P<0.001, t=26.36 Student’s t test, 2-tailed for comparison at 90d and 160d, respectively. (i) Body mass, rotorod performance and survival in IL2TGF-β1-Tg-TGF-β1+/− and IL2TGF-β1-Tg-TGF-β1−/− mice (n = 10/group).
It is known that, in mice, the earliest developmental stage at which seeding of cells in the brain with myeloid features occurs at E8.5/E9.037. Subsequent reports showed that microglia arise early during development from progenitors in the embryonic yolk sac4,38,39. Thus, primitive macrophages exit the yolk sac blood islands at the onset of circulation and colonize the neuroepithelium from E9.5 to give rise to microglia. The blood bran barrier begins to form at E13.5 and may isolate the developing brain from the contribution of fetal liver hematopoiesis40. We found no difference in CD39+CD11b+ cells between wild type and CNS-TGFβ1−/− mice at E10.5 (Supplementary Fig. 13). However, at E14.5 we found a significant reduction in CD39+CD11b+ cells (from 82.7% to 14.7%) associated with reduced surface expression of CD11b. In addition, at E14.5 we found an increase in CD39−CD11b+ cells (from 6.8% to 44.5%). This same pattern was observed in newborn brain (P1). At 160 days of age we found that CD39+CD11bLow cells were lost in CNS-TGFβ1−/− mice and this was associated with a concomitant increase in CD39−CD11b+Ly6C+ cells (Fig. 7h and Supplementary Fig. 13a). We did not find a change in the number of F4/80+CD11b+ primitive macrophages in the yolk sac at E10.5 (Supplementary Fig. 13b,c).
CNS-TGFβ1−/− mice were normal until they reached 100–120 days of age when they developed motor abnormalities (Supplementary Movie 1) as measured by decreased body weight and reduced rotorod performance (Fig. 7j). These mice steadily progressed to paralysis and death by 23–25 weeks of age. We also observed microglia loss in the spinal cord of CNS-TGFβ1−/− mice; the number of neurons were not affected (Supplementary Fig. 14). Thus, apart from the physiologic changes described previously33, the only morphological changes in the brains of CNS-TGFβ1−/− mice were loss of microglia.
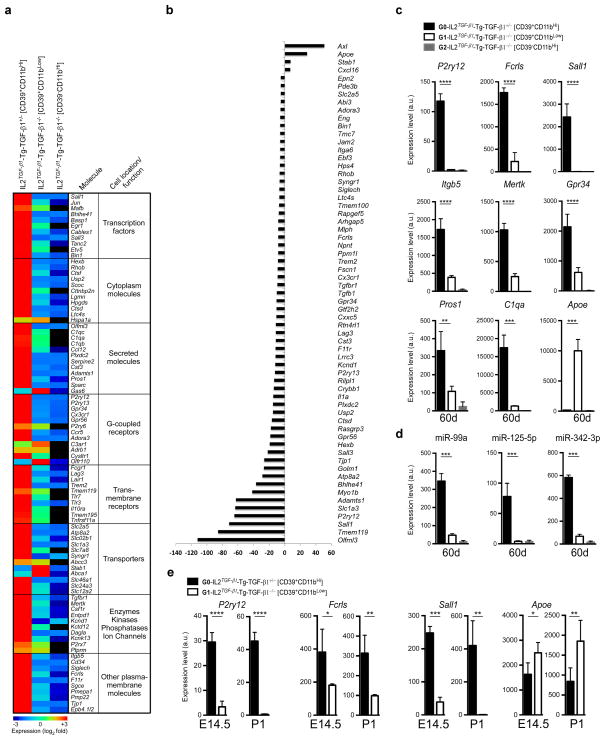
In order to determine whether the remaining CD39+Ly6C−CD11bLow cells in the CNS of CNS-TGFβ1−/− mice were resident microglia which express the microglial molecular signature, we profiled them using the MG400 chip. We sorted CD39+Ly6C−CD11b+ by FACS from both CNS-TGFβ1−/− and WT mice. We found that at 60 days of age, CD39+Ly6C−CD11b+ cells from CNS-TGFβ1−/− mice only expressed low levels of microglial genes and recruited CD39−CD11bHi cells did not express microglial genes (Fig. 8a,b and Source data Fig. 8). qPCR analysis demonstrated that microglial genes were absent (Fig. 8c). Of note, the expression of Apoe was markedly increased in CD39+Ly6C−CD11b+ cells from CNS-TGFβ1−/− mice (Fig. 8c). The microglia specific miRNAs miR-99a, miR-125-5p, and miR-342-3p were markedly decreased in CD39+Ly6C−CD11b+ cells (Fig. 8d). We then investigated the expression of microglial genes during development in CNS-TGFβ1−/− mice. We found that P2ry12, Sall1 and Fcrls were markedly decreased in microglia from CNS-TGFβ1−/− mice at E14.5 and that P2ry12 and Sall1 were undetectable at P1. Of note, there was increased expression of Apoe at both E14.5 and P1 (Fig. 8e). IPA analysis of canonical pathway and upstream regulator analysis (Supplementary Fig. 5 and Supplementary Table 4, respectively) based on the expression of microglial genes (Fig. 8a) revealed that TGF-β pathway and downstream microglial molecules, which we identified as the most affected molecules in microglia, were suppressed in CNS-TGFβ1−/− mice (Supplementary Figure 15). In summary, lack of TGF-β1 affected microglial development beginning at E14.5 but did not affect microglial progenitors at the E10.5 stage.
Figure 8. Molecular microglial signature in CNS-TGF-β1 deficient mice.
(a) MG400 profile of brain CD39+CD11b+ cells in IL2TGF-β1-Tg-TGF-β1+/− (n = 3) and IL2TGF-β1-Tg-TGF-β1−/− mice at 60d of age (n = 3) (see Source data – Figure 8). Heatmap of top microglial genes is shown. Results were log-transformed, normalized (to the mean expression of zero across samples) and centered. (b) Affected microglial genes in CD39+CD11b+ cells from and IL2TGF-β1-Tg-TGF-β1−/− mice as compared to IL2TGF-β1-Tg-TGF-β1+/− mice showing at least >5-fold difference. (c and d) qPCR validation of (c) 9 selected microglial genes and (d) 3 miRNAs at 60 days of age. Expression levels were normalized to Gapdh. Data represent mean ± s.e.m. (n = 3 mice per group) for P2ry12, ****P<0.0001, F2,9=335.5; Fcrls, ****P<0.0001, F2,9=221.5; Sall1, ****P<0.0001, F2,9=69.4; Itgb5, ****P<0.0001, F2,9=102.0; Mertk, ****P<0.0001, F2,9=234.8; Gpr34, ****P<0.0001, F2,9=71.5; Pros1, ***P=0.002, F2,9=24.2; C1qa, ****P<0.0001, F2,9=93.4; Apoe, ****P<0.0001, F2,9=110.8; 1-Way ANOVA followed by Dunnett’s multiple-comparison post-hoc test. (e) qPCR validation of 4 selected microglial genes at E14.5 (n = 4) and P1 (n = 5) mice. Data represent mean ± s.e.m. For P2ry12 at E14.5 ****P<0.0001, t=11.91; at P1 ****P<0.0001, t=18.48; For Fcrls at E14.5 *P=0.035, t=3.66; at P1 **P<0.006, t=5.26; For Sall1 at E14.5 ****P<0.0001, t=18.38; at P1 **P=0.0034, t=6.24; For Apoe at E14.5 *P=0.0221, t=2.97; at P1 **P=0.0088, t=3.62; (Student’s t test, unpaired).
DISCUSSION
Using a multifaceted approach we identified a unique genetic and microRNA signature for both mouse and human microglia cells that distinguishes microglia in the CNS from peripheral monocytes and other immune cells and from other cells of the nervous system. The plasticity of cell populations remains a major biologic feature of cell lineages including T cells and neurons. A similar plasticity exists in microglia that are clearly more diverse than an M1/M2 classification. Based on the microglial molecular signature we identified, we were able to define the plasticity of microglia and the factors in vivo and in vitro that affect their biology.
We identified TGF-β1 as a major differentiation factor for microglia both in vitro and in vivo. Consistent with this, we found loss of microglia in mice deficient for TGF-β1 in the CNS. Although there was no overt neuronal loss in CNS-TGF-β1−/− mice, these mice developed late onset motor dysfunction. We have previously reported that these mice have defects related to glutamate recycling and synaptic plasticity33. Consistent with this, deficits in microglia function have been reported to contribute to synaptic abnormalities which may be present in neuro-developmental disorders41. CNS-TGFβ−/− mice appear to show a healthy behavior phenotype throughout much of their adult life. This observation raises the possibility that other cells assume some of the functions of resident microglia at earlier stages of life such as synaptic pruning during early brain development. Detailed neuromorphological and behavioral analysis will be required to determine the extent to which abnormalities of microglia alter neurodevelopment and define which cell types might compensate.
TGF-β and their receptors are expressed at low levels by both neurons and glial cells42,43, and their expression is upregulated in response to a wide range of neuronal insults and during aging44,45. We observed high expression of Tgfb1 and the Tgfbr1 in microglia (Fig. 1f). Numerous studies have demonstrated that TGF-β1 exerts a protective effect against various neuronal insults46.
Tgfb1−/− animals die at 3 weeks of age of a severe multifocal autoinflammatory disorder30 and it has been reported that the loss of TGF-β1 in these mice leads to increased neuronal cell death and microgliosis in the brain47. In these studies, microglia were identified as F4/80+ cells which does not distinguish resident microglia from infiltrating macrophages. Using our microglial specific markers and molecular signature, we demonstrate that there is loss of microglia as opposed to microgliosis, and this loss is associated with infiltration of Ly6C positive monocytes into the brain.
Because we identified genes related to neuronal function both in cultured as well as ex vivo adult microglia, the presence of these genes does not simply represent extracellular absorption as the microglial genes we identified were upregulated by exogenous TGF-β1 in vitro. In addition to the known innate immune function of microglia, IPA identified neuronal and development function as the most significantly expressed pathways in microglia. Our results are consistent with previous studies on the role of microglial in the development and function of the nervous system41,48–50.
We found that microglia have different phenotypes that are linked to development. Thus, embryonic and early post-natal microglia acquire the features of adult microglia after post-natal day four, possibly as newly-proliferated microglial cells begin to express differentiated functions. Commonly studied microglia cell lines and primary microglia do not express the adult microglial molecular signature and do not express the signature after culture with TGF-β1. The identification of a molecular microglial signature can now be used to determine the features which account for microglia CNS homeostatic function as well as neurotoxic vs. protective properties. We also found that organ-resident macrophages in the gut and liver are “microglia-like” as they express Gas6 and Mertk and these genes may have a role in maintaining tissue homeostasis which is consistent with a recent transcriptional profiling of murine tissue macrophages14.
In summary, we have identified a TGFβ-1 dependent microglial signature and structures on the surface of microglia that provide the ability to distinguish microglia from infiltrating myeloid cells in the CNS. The genetic and microRNA signature we observed in mice was also found in human microglia. These findings provide new insights into microglial biology and the possibility of targeting microglia for the treatment of CNS disease.
METHODS
Animals
C57BL6 males and females, B6.Cg-Tg were obtained from Jaxmice laboratories. Adult male and female heterozygous TGF-β1−/+ mice were kindly provided by Sharon M. Wahl (National Institutes of Health). These mice were kept on C57BL6/Sv129 background. On the C57BL6/Sv129 background, 60–80% of (statistically expected) homozygous TGF-β1−/− conceptuses reached parturition as described29. C57BL/6 IL2-transgenic mice that express TGF-β1 under the control of an IL2 promoter (termed IL2TGF-β1-Tg-TGF-β1+/+) were generated in our laboratory and are described elsewhere32. Those mice were crossed with TGF-β1−/+ animals to obtain IL2TGF-β1-Tg-TGF-β1+/− mice. IL2TGF-β1-Tg-TGF-β1+/− mice do not show any clinical or pathological abnormalities and breed well. Progeny mice were intercrossed at least 6 generations prior to all experimental procedures, and IL2TGF-β1-Tg-TGF-β1−/− mice were generated from crossing heterozygous IL2TGF-β1-Tg-TGF-β1+/− mice33. All experiments were performed in mice with a mixed inbred genetic background (75% C57BL6 and 25% Sv129). Because TGF-β1−/− mice are not viable on a 100% C57BL6 background29, we refrained from backcrossing them. Heterozygous IL2TGF-β1-Tg-TGF-β1+/− mice were used as a littermate control mice for IL2TGF-β1-Tg-TGF-β1−/− mice. All mice were housed with sterile food and water ad libitum. Animals were euthanized through CO2 inhalation. The Institutional Animal Care and Use Committee (IACUC) at Harvard Medical School approved all experimental procedures involving animals.
Generation of chimeric mice
Recipient lethally irradiated (950 Rad) 30 day old C57BL/6 mice were transplanted with donor syngeneic BM cells from CX3CR1-GFP+/−-WT mice. In these chimeras, peripheral monocytes are distinguishable from microglia by FACS12. We determined the percentage of chimerism 8 weeks after transplant by examining cells from the spleen. We found that 95–97% of F4/80+CD11b+ cells in the spleen were GFP+. In chimeric naïve or EAE mice, mononuclear cells stained for the monocyte marker CD11b could be easily subdivided into three distinct subsets: 1) CD11b+GFP−FCRLS+ microglia and 2) CD11b+GFP+FCRLS− peripheral macrophages (Fig. 4i).
Generation of anti-P2ry12 and anti-FCRLS antibodies
P2ry12 polyclonal antibody was generated in rats. Adult Lewis rats were vaccinated (ip) five times (two weeks apart) with a synthetic peptide corresponding to the mouse P2Y12 C terminus (NH2-Cys-Gly-Thr-Asn-Lys-Lys-Lys-Gly-Gln-Glu-Gly-Gly-Glu-Pro-Ser-Glu-Glu-Thr-Pro-Met-OH; Anaspec)21. The first vaccination was given with Complete Freund’s Adjuvant (CFA) and then next 3 injections with Incomplete Freund’s Adjuvant (IFA) combined with the synthetic P2ry12 peptide ip to generate rat antibodies. For the 1st injection, the peptide was conjugated to Keyhole Limpet Hemocyanin (KLH) plus (CFA) followed by a 2nd injection with 50μg of the synthetic peptide conjugated to BSA + Incomplete Freund’s Adjuvant (IFA) and then a 3rd injection with 50μg of a synthetic peptide conjugated to OVA + IFA. All injections were given ip. The above injections were given 2 weeks apart. A 4th injection was given two weeks later with 50μg of the synthetic peptide conjugated to KLH plus Titermax (200μl). The 5th injection was given with 50μg of the synthetic peptide conjugated to BSA. First three injections were given at a 1:1 ratio with CFA or IFA. CFA, IFA and Titermax were purchased from Sigma. P2ry12 antibody was affinity purified using a Sulfolink coupling gel (Pierce) to immobilize the antigenic peptide.
In order to generate FCRLS mAb, we constructed an expression vector pFUSE-hIgG2-FC2 (InvivoGen) to generate FCRLS-Fc fused protein. Adult Lewis rats were vaccinated as described above and identified positive FCRLS 16 oligoclones were subclonned to produce FCRLS mAbs. 4G11 FCRLS clone was used in this study.
Mouse microglia isolation and sorting
Mononuclear cells were directly isolated from C57BL6 embryos (E10.5, E12.5), newborn (P4) and adult (P21, P30 and 2m) mice. Mice were transcardially perfused with ice-cold phosphate-buffer saline (PBS), spinal cords and brains separately dissected. Single cell suspensions were prepared and centrifuged over a 37%/70% discontinuous Percoll gradient (GE Healthcare), mononuclear cells isolated from the interface, and total cell count determined. Isolated cells were labeled with FCRLS mAb to specifically sort resident microglia followed by total RNA isolation and gene/miRNA expression profiling. Resident microglia were sorted with combination of FRCLS and CD11b antibodies.
Cell suspension preparation
Cell suspensions were made from brain12 and other tissues as described51 before analysis by flow cytometry. Yolk sac from mouse embryos was dissected as described52 and yolk sac cell suspensions were made as described4. Skin cell suspensions were isolated as previously described53 and analyzed by flow Cytometry. In brief, mouse ears were split in two dorsal and ventral parts and incubated for 1h in HBSS containing 2.4 mg/ml Dispase (working activity 1.7 U/mg; Invitrogen) to allow for separation of dermal and epidermal sheets. Epidermal and dermal sheets were then cut into small pieces, incubated fir 2h in HBSS containing 1% FBS and 0.2 mg/ml collagenase type IV (working activity of 770 U/mg; Sigma-Aldrich), and then passed through a 19G syringe to obtain homogenous cell suspension. Lung, liver, kidney and spleen cells were isolated, homogenized and incubated for 1h in 10% FBS HBSS containing 0.2 mg/ml collagenase type IV (working activity 0f 770 U/mg), and passed through a 19G syringe to obtain homogeneous cell suspension. Cell suspensions were then run either through a room temperature NycoPrep gradient 1.077 (Axis-Shield; for lung and kidney) and a room temperature isotonic Percoll (GE Healthcare; for liver) centrifugation to enrich the percentage of hematopoietic cells, as per manufacturer’s instructions.
Flow Cytometry
For fluorescent-activated cell sorting (FACS) analysis, mice were transcardially perfused with 50ml 0.01 M PBS and their brains were homogenized using a Teflon-stick homogenizer. Mononuclear cells were separated through Percoll (GE Healthcare Life Sciences) gradient centrifugation. Cells were pre-blocked with anti-CD16/CD32 Fc Block (clone 2.4G2, BD Biosciences, 5 μg ml−1, validated in ref. 12) and stained on ice for 30 min with combinations of CD45-PerCP (clone 30-F11, BD Biosciences, 2 μg ml−1, validated in ref. 12), CD11C-PeCy7 (clone N418, eBioscience, 5 μg ml−1, validated in ref. 7), Ly6C-FITC (clone AL-21, BD Biosciences, 5 μg ml−1, validated in ref. 12), CD11b-PeCy™7 (clone M1/70, BD Biosciences, 2 μg ml−1, validated in ref. 12), CD11b-PE (clone M1/70, BD Biosciences, 2 μg ml−1, validated in ref. 12), CD3 PE (clone 145-2C11; BD Bioscience, 1 μg ml−1, validated in ref. 54), CD4-FITC (clone RM4-5, BD Biosciences, 5 μg ml−1, validated in ref. 55) and rat IgG isotype controls (clone R35-38, BD Biosciences, 2 μg ml−1, validated in ref. 12). FCRLS (clone 4G11, newly generated in this study) mAb was detected with goat anti-rat IgG conjugated to APC (clone Poly4054, Biolegend, 0.7 μg ml−1, validated in ref. 12). Tissue macrophages, dendritic cells and Langerhans cells were analyzed with combinations of MHCII (I-A)-APC-Cy7 (clone M5/114.15.2, eBioscience, 2 μg ml−1, validated in ref. 7), F4/80-PE (clone Cl:A3-1, AbD Serotec, 1 μg ml−1, validated in ref. 7), CD11C-PeCy7 (as described before), CD45-PerCp (as described before), CD3-PE (as described before), CD19-PE (clone eBiofD3; eBioscience; 0.5 μg ml−1, validated in ref. 54), NK1.1-PE (clone PK-136, eBioscience, 2 μg ml−1, validated in ref. 54), CD11b-PeCy7 (as described before) or CD11b-PacBlu (clone M1/70, eBioscience, 2 μg ml−1, validated in ref. 12), CD103-APC (clone 2E7, eBioscience, 5 μg ml−1, validated in ref. 7), CD39-APC (clone 5F2, eBioscience, 2 μg ml−1, validated in ref. 12) or CD39 and Ly6C previously produced in our laboratory (clone 5E12, validated in ref. 12) and Ly6C (clone 6C3, Millipore, 1 μg ml−1, validated in ref. 12). Resting red-pulp macrophages from the spleen were sorted after nonenzymatic disaggregation of the spleen and were identified as F4/80hi cells that lacked B220 but had high expression of CD11c and MHC class II14. A combination of AnnexinV-PE or -FITC-conjugated and 7AAD-PerCP was used to stain apoptotic and necrotic cells, respectively (BD Biosciences). Appropriate antibody IgG isotype controls (BD Biosciences) were used for all staining. FACS analysis was performed on a LSR machine (BD Biosciences), and data analyzed with FlowJo Software (TreeStar). Cell sorting was performed using a Becton Dickinson FACSARIA cell sorter.
Primary newborn mouse microglial culture
Brains from neonatal (P0–P1) C57BL6J mice were stripped of their meninges and minced with scissors under a dissecting microscope (Stemi DV4; Zeiss) in Leibovitz-15 medium (Biological Industries). After trypsinization (0.5% trypsin for 10 minutes at 37°C, 5% CO2), the tissue was triturated. The cell suspension was washed in culture medium for glial cells (DMEM supplemented with 10% FCS [Sigma-Aldrich], 1mM l-glutamine, 1mM sodium pyruvate, 100U ml−1 penicillin, and 100mg ml−1 streptomycin) and cultured at 37°C, 5% CO2 in 75-cm2 Falcon tissue-culture flasks (BD Biosciences) coated with poly-d-lysine (PDL; 10mg ml−1; Sigma-Aldrich) in borate buffer (2.37g borax and 1.55g boric acid dissolved in 500 ml sterile water, pH 8.4) for 1 hour, then rinsed thoroughly with sterile, glass-distilled water. Half of the medium was changed after 6 hours in culture and every second day thereafter, starting on day 2, for a total culture time of 10–14 days. MG were shaken off the primary mixed brain glial cell cultures (150 rpm, 37°C, 6 hours) with maximum yields between days 10 and 14, seeded (105 cells ml−1) onto PDL-pretreated 24-well plates (1ml/well; Corning Incorporated), and grown in culture medium for MG (RPMI-1640 medium; Sigma-Aldrich; supplemented with 10% FCS, 1mM l-glutamine, 1mM sodium pyruvate, 50mM β-mercaptoethanol, 100U ml−1 penicillin, and 100mg ml−1 streptomycin). The cells were allowed to adhere to the surface of a PDL-coated culture flask for 1 hour at 37°C, 5% CO2, and nonadherent cells were rinsed off.
Primary adult mouse microglial culture
Adult microglia were isolated from C57BL/6 mice at age 6–10 weeks from brains, spinal cords or eyes. Tissue was homogenized using a Teflon-stick homogenizer. Mononuclear cells were separated through Percoll (GE Healthcare Life Sciences) 37%/70% gradient centrifugation (1200rpm, room temperature, with minimal acceleration and no brake using a swinging bucket rotor). Mononuclear cells were isolated from the interface. Cells were stained on ice for 20 min with anti-FCRLS mAb at 1:200 dilution in blocking buffer containing 0.25% BSA (Sigma-Aldrich) in HBSS (Invitrogen). Cell sorting was performed using Becton Dickinson FACSARIA cell sorter. Sorted microglia were cultured 6-well plate (1.2×105 cells/well in 3ml) or 12-well plate (6.5×104 cells/well in 2ml) in poly-D-lysine coated plates (BD Biosciences), and grown in microglia culture medium (DMEM/F-12 Glutamax; Invitrogen; supplemented with 10% FCS, 100U ml−1 penicillin, 100mg ml−1 streptomycin and mouse recombinant carrier free MCSF 10ng/ml (R&D Systems) or mouse recombinant carrier free GM-CSF 10ng ml−1 (R&D Systems) at 37°C, 5% CO2. Cells were cultured for at least 5 days without changing media before treatment with additional cytokines.
Generation of M0- M1- M2 adult microglia cultures
Adult primary microglia were isolated as described above. M1 and M2 microglia were polarized as described elsewhere27. To generate M0 microglia, sorted adult primary microglia were cultured in microglia culture medium described above containing mouse recombinant carrier free MCSF 10ng ml−1 (R&D Systems) and 50ng/ml human recombinant TGFβ1 (Miltenyi Biotec) for 5 days.
Human Microglia isolation and cell culture
Human fetal CNS tissue was obtained from either a human fetal tissue repository (Albert Einstein College of Medicine, Bronx, NY), or Novogenix Laboratories (Torrance, CA), following Canadian Institutes of Health Research guidelines. Human fetal microglia were isolated by mincing tissue and treating with DNase and trypsin as described previously27. Cells were then passed through a nylon mesh column and plated at 7×106 cells ml−1 in DMEM containing 5% FBS. After 10–14 days in culture, floating microglia were removed and plated at 105 cells ml−1. Human adult microglia were isolated from temporal lobe brain tissue from patients undergoing surgery for non tumor-related intractable epilepsy. All tissue used was outside of the suspected focal site and cells were isolated as described previously. Tissue was treated with DNase and trypsin, passed through a nylon mesh and cell suspension was separated on a 30% Percoll gradient. Cells were then washed, and plated at 105 cells ml−1 in MEM containing 5% FBS. All studies using human cells received institutional review board approval.
Isolation of astrocytes
12- to 16-week-old adult BAC-Glt1-EGFP reporter mice were anesthetized and perfused with ice-cold Hank’s Balanced Salt Solution (HBSS). Whole brains were immediately extracted, minced, and digested with 20 unit/ml papain and 0.005% DNase in Earle’s Balanced Salt Solution (EBSS)(Worthington, Lakewood, NJ) for 90 minutes at 37°C with 5% CO2. The tissues were then triturated with 5ml pipette and centrifuged at 300 x g for 6 minutes. Cell pellets were resuspended in low ovomucoid protease inhibitor solution (1mg/ml ovomucoid inhibitor, 1mg/ml albumin, 0.005% DNase in EBSS), overlaid on top of high inhibitor solution (10mg/ml ovomucoid inhibitor, 10mg/ml albumin in EBSS) and centrifuged at 300 x g for 6 minutes. Pelleted cells were resuspended in 10ml 30% Percoll (GE Healthcare, Piscataway, NJ) with 1% BSA and centrifuged at 300 x g for 25 min to remove myelin. Cells were then pre-blocked with anti-CD16/CD32 (Fc Block, BD Biosciences), and stained on ice for 30 min with anti-CD11b-PE-Cy7™. EGFP+/CD11b− astrocytes were collected by sorting using a Becton Dickinson FACSARIA cell sorter.
Isolation of oligodendrocytes
For PLP-eGFP positive cell sorting, PLP-eGFP mice (C57/BL6 background)56 were perfused with cold Hank’s balanced salt solution (1xHBSS). The brains were dissected and spinal cords were flushed with cold 1xHBSS. Tissues were minced into small pieces and digested with papain (30U/ml, Roche) and DNase I (10ug/ml, Sigma, St. Louis, MO) for 30 min at 37°C. The samples were dissociated by sequentially triturating for 10 times through 18G and 23G needle syringes. For pelleting, 1xHBSS containing 20% FCS was added. The cell pellets were suspended in 1xHBSS and separated in a discontinuous 30%/70% Percoll gradient57. Cells were collected from the 30%/70% interphase and washed in the cell sorting buffer (1xHBSS, 1%BSA, 1ug/ml DNase, 2mM EDTA). The cells were stained with CD45-APC antibody (Biolegend, San Diego, CA) as described previously57 and sorted by fluorescence activated cell sorting (FACS) (BD FACSAria III, Becton, Dickinson and Company). PLP-eGFP+ cells were uniformly CD45-negative.
Isolation of cortical and hippocampal neurons
Primary cortical and hippocampal neurons were prepared from embryos at age E16.5. Cerebral cortices and hippocampi were isolated and freed from meninges. Tissues were first digested with 0.25% trypsin in Hank’s balanced salt solution (HBSS) for 15 min at 37°C, then washed three times with HBSS and triturated with fire-polished glass pipettes until single cells were obtained. Cell suspension was then filtered through a 70 and a 40 μm cell strainer and subjected to a spin at 1000g for a few minutes. Supernatant was removed and cell pellet was resuspended with 10 ml of HBSS. Cell density was then determined with a hemocytometer and cells were seeded at different densities according to the experimental design and need. We used Dulbecco’s modified Eagle’s media (DMEM) supplemented with 10% FBS for the initial plating, and the medium was changed to Neurobasal supplemented with 1× B27 (Invitrogen) in 3 h. Half medium was changed every 3 days.
RNA isolation
Total RNA was extracted using mirVanaTM miRNA isolation kit (Ambion) according to the manufacturer’s protocol.
Real-time PCR
Total RNA (20–40ng) was used in 20–40μl of reverse transcription reaction (high-capacity cDNA Reverse Transcription Kit; Applied Biosystems) and 3ng RNA in 5μL reverse transcription reaction with specific miRNA probes (Applied Biosystems). qPCR reactions were performed in duplicates or triplicates. mRNA or miRNAs levels were normalized relative to U6 or GAPDH, respectively, by the formula 2^(−ΔCt), where ΔCt = CtmiR-X-CtU6 or GAPDH. All data are mean of duplicates, and the standard errors of mean were calculated between duplicates or triplicates. Real time PCR reaction was performed using Vii7 (Applied Biosystems). All qRT-PCRs were performed in duplicate or triplicate, and the data are presented as mean ± standard error of mean (s.e.m).
Human brain specimens
Fresh human brain was obtained from Massachusetts General Hospital pathology department within 5h from time of death. Tissue was used for immunohistochemical and FACS analysis (see Fig. 4c,d).
Microarray processing and analysis
Microarray analysis was performed by genetic core facility at Harvard. Briefly, total RNA was isolated from adult brain sorted adult CD11b+/CD45Low microglia (30 mice per sample, in biological triplicates) and splenic CD11b+/Ly6C+ monocytes (20 mice per sample, in biological triplicates). Cells were isolated from C57Bl6 mice (8–10 weeks old, pooled 1:1 females and males). RNA quality and integrity were assessed using Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA) with all samples having high-quality RNA [RNA integrity number (RIN) = 8.7–10]. Hybridization was performed with Affymetrix GeneChip® Mouse Exon 1.0 ST arrays (Affymetrix, Inc, Santa Clara, CA). Briefly, 200ng of total RNA from each sample were reverse transcribed to cDNA, followed by overnight in vitro transcription to generate cRNA, which was reverse transcribed, and the 5.5μg of sense cDNA were fragmented and labeled. The quality of cDNA and fragmented cDNA was assessed in the Agilent bioanalyzer. Microarrays were hybridized, washed, stained, and scanned according to the protocol described in manual from Affymetrix (v.4). To minimize technical (nonbiological) variability among arrays, densitometry values between arrays were normalized using the Robust Multichip Average function and further transformed to the logarithmic scale (log2). Statistically significant differences between groups were determined using GeneSpring (v.12.5) to identify the most robustly differentially expressed genes between adult brain microglia and splenic monocytes. Functional annotations were carried out using the Ingenuity Pathway Analysis (IPA; Ingenuity Systems, Redwood City, CA, http://www.ingenuity.com), in which gene symbols and fold changes of the up- and downregulated genes were imported (see Supplementary Table 1). Hierarchical cluster analysis was performed to see how data aggregate and a heatmap was generated with pluripotency genes. All data analysis was performed using GeneSpring v.12.5.
Mass spectrometry analysis: cell culture, harvest, and lysis
CD11b+/CD45Low microglia (15 pooled mouse brains in biological duplicates) and splenic Ly6C+/CD11b+ monocytes (10 pooled spleens in biological duplicates) were sorted and mechanically lysed with a mechanical homogenizer with ice-cold lysis buffer composed of 50mM TRIS pH8.5, 50mM β-glycerophosphate 1mM sodium orthovanadate, 1mM PMSF and EDTA free protease inhibitor cocktail (Promega) in 8M Urea. The lysate was centrifuged at 10,000 x g for 20min to remove cell debris.
Mass spectrometry analysis: Sample preparation, protein digest, and peptide TMT-labeling
Protein content was measured using a BCA assay (Thermo Scientific, Rockford, IL); disulfide bonds were reduced with 5mM DTT and cysteine residues alkylated with iodoacetamide (14 mM) essentially as previously described58. Protein lysates were cleaned up by methanol-chloroform precipitation and digested overnight with LysC (Wako, Japan) in a 1/100 enzyme/protein ratio in 2 M urea and 25 mM Tris-HCl, pH 8.5. The digest was acidified with 10% formic acid (FA) to a pH of ~ 2–3 and subjected to C18 solid-phase extraction (SPE) (Sep-Pak, Waters, Milford, MA). Isobaric labeling of the digested peptides was accomplished with 6-plex tandem mass tag (TMT) reagents (Thermo Fisher Scientific, Rockford, IL). Reagents, 0.8 mg, were dissolved in 40μl acetonitrile (ACN) and 1/4 of the solution were added to 100 μg of peptides dissolved in 100 μl of 200mM HEPES, pH 8.5. After 1hr (RT), the reaction was quenched by adding 8 μl of 5 % hydroxylamine. Labeled peptides (all six reagents (126–131) were combined and acidified prior to C18 SPE on Sep-Pak cartridges (50 mg).
Mass spectrometry analysis: Strong cation exchange (SCX) chromatography sample fractionation
Orthoganol sample fractionation was performed using strong cation exchange (SCX) chromatography as previously described58. Briefly, the labeled peptides were solubilized in SCX buffer A (7 mM KH2PO4, pH 2.7, 30% ACN) and separated via a 4.6 × 200 mm polysulfoethyl A HPLC column (particle size, 5μm; pore size, 200 Å; PolyLC, Columbia, MD). The separation was performed using applying a dual two-buffer (SCX A and B) gradient from 0 to 50% SCX buffer B (7 mM KH2PO4, 350 mM KCl, pH 2.7, 30% ACN) in 47 min at a flow rate of 1.0 ml min−1, followed by 50 to 100% SCX buffer A to buffer B in 4.5 min using an Agilent 1100 quaternary pump outfitted with a degasser and a photodiode array detector (PDA) (Thermo Scientific, San Jose, CA). Samples were collected in ~40 s increments into a 96-well plate, and dried under vacuum. Fractions were then redissolved with 5% FA/5% acetonitrile and combined into a total of 20 samples, which were then desalted by C18 StageTips (3M Empore), and dried under vacuum.
Mass spectrometry analysis: Liquid chromatography separation and tandem mass spectrometry (LC-MS/MS)
All 20 LC-MS/MS experiments were performed on a LTQ Orbitrap Velos (Thermo Fischer) equipped with a Famos autosampler (LC Packings, Sunnyvale, CA) and an Agilent 1200 binary HPLC pump (Agilent Technologies, Santa Clara, CA). Peptides were separated onto a 100 μm I.D. microcapillary column packed first with approximately 1 cm of Magic C4 resin (5μm, 100 Å, Michrom Bioresources, Auburn, CA) followed by ~25 cm of Maccel C18AQ resin (1.8 μm, 200 Å, Nest Group, Southborough, MA). Separation was achieved through applying a gradient from 7 to 35% ACN in 0.5% FA over 160min at ~250 nl min−1. Electrospray ionization was enabled through applying a voltage of 1.8 kV using an inert gold electrode via a PEEK junction at the end of the microcapillary column. The LTQ Orbitrap Velos was operated in data-dependent mode for the MS methods59. The MS survey scan was performed in the Orbitrap in the range of 400–1500 m/z at a resolution of 3×104, followed by the selection of the ten most intense ions (TOP10) for CID-MS2 fragmentation in the ion trap using a precursor isolation width window of 2 m/z, AGC setting of 2000, and a maximum ion accumulation of 150 ms. Singly charged ion species were not subjected to CID fragmentation. Normalized collision energy was set to 35% and an activation time of 20ms. Ions within a 10ppm m/z window around ions selected for MS2 were excluded from further selection for fragmentation for 120 s. Directly following each MS2 event, the most intense fragment ion in an m/z range between 110–160% of the precursor m/z was selected for HCD-MS3. The fragment ion isolation width was set to 4 m/z, AGC was set to 20,000, the maximum ion time was 250 ms, normalized collision energy was set to 60% and an activation time of 50 ms for each MS3 scan.
Mass spectrometry analysis: Data processing and spectra assignment
A compendium of in-house developed software was used to convert mass spectrometric data (Raw file) to the mzXML format, as well as to correct monoisotopic m/z measurements and erroneous assignments of peptide charge state. Assignment of MS/MS spectra was performed using the Sequest algorithm by searching the data against a protein sequence database including all entries from the mouse IPI database (version 3.62; ~56,000 entries) and other known contaminants such as human keratins and its reverse decoy component (reversed order)60. Sequest searches were performed using a 20ppm precursor ion tolerance and requiring each peptides N-/C- termini to adhere with Ly6C protease specificity, while allowing up to two missed cleavages. Six-plex TMT tags on peptide N termini and lysine residues (+229.162932 Da) and carbamidomethylation of cysteine residues (+57.02146 Da) were set as static modifications while methionine oxidation (+15.99492 Da) was set as variable modification. A MS2 spectra assignment false discovery rate (FDR) of less than 1% was achieved by applying the target-decoy database search strategy60. Filtering was performed using a in-house linear discrimination analysis method to create one combined filter parameter from the following peptide ion and MS2 spectra metrics: Sequest parameters XCorr and ΔCn, peptide ion mass accuracy and charge state, in-solution charge of peptide, peptide length and mis-cleavages. Linear discrimination scores were used to assign probabilities to each MS2 spectrum for being assigned correctly and these probabilities were further used to filter the dataset with an MS2 spectra assignment FDR of smaller than a 1% at the protein level58.
Mass spectrometry analysis: Determination of TMT reporter ion intensities and quantitative data analysis
For quantification, a 0.03 m/z window centered on the theoretical m/z value of each the six reporter ions and the intensity of the signal closest to the theoretical m/z value was recorded. Reporter ion intensities were further denormalized based on their ion accumulation time for each MS2 or MS3 spectrum and adjusted based on the overlap of isotopic envelopes of all reporter ions (as determined by the manufacturer). The total signal intensity across all peptides quantified was summed for each TMT channel, and all intensity values were adjusted to account for potentially uneven TMT labeling and/or sample handling variance.
Data normalization and analysis
Heatmaps were generated employing Multi Experiment Viewer v 4.8. Expression values were transformed using the mean and standard deviation of the row (gene) of the matrix to which the value belongs, using the following formula: Value = [(Value) – Mean(Row)]/[Standard deviation(Row)]. Hierarchical clustering analysis was performed using Pearson correlation (average linkage clustering method). Principle component analysis (PCA) and 3D scatterplot were performed with JMP v.10 (SAS Institute, Cary, NC). Correspondence analysis (COA) was performed in MeV. Venn diagrams were used to compare gene lists (http://bioinfogp.cnb.csic.es/tools/venny/index.html).
GeneGo Biological Pathway Analysis
Brain microglia and splenic monocyte gene expression profile was compared using Affymetrix GeneChip® Mouse Exon 1.0 ST arrays and analyzed by GeneSpring (v.12.5) as described above. Differentially regulated transcripts were analyzed using GeneGO Metacore pathway analysis (GeneGO, St. Joseph, MI; genego.com). A set of identified 399 microglial genes and 611 monocyte genes (see Fig. 1a and Supplementary Table 1) were imported into MetaCore to build an analysis of functional ontologies including GeneGo process, GeneGo disease process, canonical pathway maps, and networks.
IPA (Ingenuity®) analysis
Data were analyzed through the use of IPA (Ingenuity® Systems, www.Ingenuity.com). Differentially expressed genes (with corresponding fold-changes and p values) were incorporated in canonical pathways and bio-functions and were used to generate biological networks. Uploaded dataset for analysis were filtered using the following cutoff definitions: 5-fold change, P<0.01). IPA provides the most comprehensive, validated knowledgebase of interactions between biomolecules including miRNA. Further, they also provide comprehensive annotation of different functional and pathway enrichment along with the ability to present this knowledge in the form of a network of interaction.
MG400 chip design
The MG400 chip was designed using on quantitative NanoString nCounter platform. Genearray analyses and mass spectrometry quantitative TMT-analysis, described above, were used to identify 254 genes and 100 proteins which were specifically or highly expressed in adult mouse microglia. In addition, the MG400 chip contains 40 inflammation-related genes which were significantly affected in EAE, APP/PS1 and SOD1 mice (see Supplementary Table 6).
Immunohistochemistry
Transverse sections of the spinal cord or coronal sections of the brain (30μm) were treated with a permeabilization/blocking solution containing 10% FCS, 2% bovine serum albumin, 1% glycine, and 0.05% Triton X-100 (Sigma-Aldrich). Primary antibodies were applied for 1 hour in a humidified chamber at room temperature. Sections were blocked for 1 hour with blocking solution. The tissue shown in Fig. 4d was stained with a rabbit anti-P2ry12 pAb (1:200, validated in ref. 21). The tissue shown in Fig. 4e–h was stained in combination with the rat anti-P2ry12 pAb (1:2000, newly generated in this study) we generated plus mouse anti-NeuN (1:500; Millipore, validated in ref. 12), rabbit anti-Iba1 (1:500; Wako, cat. no. 019-19741, validated in ref. 12) or mouse anti-GFAP (1:300; BD Pharmingen, clone 2E1, validated in ref. 33). Antibodies were diluted in PBS containing 0.05% Triton X-100, 0.1% Tween 20, and 2% horse serum. Sections were incubated with the primary antibody for 12 hours at 4°C, washed with PBS, and incubated with the secondary antibodies in PBS for 1 hour at room temperature while protected from light. Secondary antibodies used for both immunocytochemistry and immunohistochemistry were donkey anti-rat or donkey anti-sheep Cy3, donkey anti-rabbit Cy5; donkey anti-mouse FITC. All antibodies were purchased from Jackson ImmunoResearch Laboratories Inc. and used at a dilution of 1:250–1:500. Control sections (not treated with primary antibody) were used to distinguish specific from nonspecific staining or autofluorescent components. Sections were then washed with PBS and coverslipped in polyvinyl alcohol with diazabicylo-octane anti-fading agent or ProLong® Gold antifade reagent with DAPI (Invitrogen).
Quantitative NanoString nCounter miRNA/gene expression analysis
Nanostring nCounter technology (www.nanostring.com) allows expression analysis of multiple genes (up to 800 genes) from a single sample. We performed nCounter multiplexed target profiling of 179 inflammation genes which consist of genes differentially expressed during inflammation and immune responses, nCounter 578 miRNA (see complete list of genes and miRNAs at www.nanostring.com) and 400 microglial transcripts (MG400, see MG400 chip design). 100ng per samples of total RNA were used in nCounter analysis according to the manufacturer’s suggested protocol.
Statistical analysis
For all statistical analyses, data distribution was assumed to be normal, but this was not formally tested. Unless otherwise indicated, data are presented as means ± s.e.m. and two-tailed Student’s t-tests (unpaired) or ANOVA multiple comparison tests were used to assess statistical significance (*P<0.05, **P<0.01, ***P<0.001) and calculated with GraphPad Prism 6 software (La Jolla, CA). No statistical methods were used to predetermine sample sizes, but our sample sizes are similar to those reported in previous publication12. Data collection and analysis were performed blind to the conditions of the experiments. Also, data for each experiment were collected and processed randomly and animals were assigned to various experimental groups randomly as well. All n and P values and statistical tests are indicated in figure legends. All errors bars represent s.e.m., and mean ± s.e.m is plotted for all graphs except were noted.
Supplementary Material
Acknowledgments
We thank Dr. David Julius for providing polyclonal anti-P2ry12 antibody and N. Kassam for support in antibody generation. Dr. Anna Krichevsky and Dr. Andus Wong Hon-Kit for providing neurons and Lisa Spangler for technical assistance for oligodendrocyte isolation. We thank Deneen Kozoriz for the FACS sorting. This work was supported by NIH Grant AG027437, NIH Transformative Grant AG-043975, a grant from the Amyotrophic Lateral Sclerosis Association, a Thome Foundation AMD grant, and philanthropic support. We thank Prize4Life for providing SOD1 mice.
Footnotes
Accession codes. The gene and miRNA data have been deposited in Gene Expression Omnibus (GEO) database under the ID code GSE48579 and protein data has been deposited in PeptideAtlas under the ID code PASS00352.
Contributions
O.B. and H.L.W. conceived the study, designed experiments and wrote the paper. O.B., A.J.L., G.G, T.K., B.D., R.C., P.M.W., C.E.D. and Z.F. performed experiments. M.P.J. and S.P.G performed mass spectrometry experiments. C.S.M. and J.P.A. performed human microglia studies. Z.C., J.D.R. and LL., R.M.R. performed CNS cell isolation studies. All authors were participants in the discussion of results, determination conclusions and review of the manuscript.
Competing financial interests
The authors declare no competing financial interests.
Supplementary information includes fifteen figures, nine tables, one movie and ten source datasets.
References
- 1.Prinz M, Priller J, Sisodia SS, Ransohoff RM. Heterogeneity of CNS myeloid cells and their roles in neurodegeneration. Nat Neurosci. 2011;14:1227–1235. doi: 10.1038/nn.2923. [DOI] [PubMed] [Google Scholar]
- 2.Ransohoff RM, Cardona AE. The myeloid cells of the central nervous system parenchyma. Nature. 2010;468:253–262. doi: 10.1038/nature09615. [DOI] [PubMed] [Google Scholar]
- 3.Saijo K, Glass CK. Microglial cell origin and phenotypes in health and disease. Nat Rev Immunol. 2011;11:775–787. doi: 10.1038/nri3086. [DOI] [PubMed] [Google Scholar]
- 4.Ginhoux F, et al. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science. 2010;330:841–845. doi: 10.1126/science.1194637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schulz C, et al. A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science. 2012;336:86–90. doi: 10.1126/science.1219179. [DOI] [PubMed] [Google Scholar]
- 6.Kierdorf K, et al. Microglia emerge from erythromyeloid precursors via Pu.1- and Irf8-dependent pathways. Nat Neurosci. 2013 doi: 10.1038/nn.3318. [DOI] [PubMed] [Google Scholar]
- 7.Greter M, et al. Stroma-derived interleukin-34 controls the development and maintenance of langerhans cells and the maintenance of microglia. Immunity. 2012;37:1050–1060. doi: 10.1016/j.immuni.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ajami B, Bennett JL, Krieger C, McNagny KM, Rossi FM. Infiltrating monocytes trigger EAE progression, but do not contribute to the resident microglia pool. Nature neuroscience. 2011;14:1142–1149. doi: 10.1038/nn.2887. [DOI] [PubMed] [Google Scholar]
- 9.Saederup N, et al. Selective chemokine receptor usage by central nervous system myeloid cells in CCR2-red fluorescent protein knock-in mice. PLoS ONE. 2010;5:e13693. doi: 10.1371/journal.pone.0013693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.King IL, Dickendesher TL, Segal BM. Circulating Ly-6C+ myeloid precursors migrate to the CNS and play a pathogenic role during autoimmune demyelinating disease. Blood. 2009;113:3190–3197. doi: 10.1182/blood-2008-07-168575. blood-2008-07-168575 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dimitrijevic OB, Stamatovic SM, Keep RF, Andjelkovic AV. Absence of the chemokine receptor CCR2 protects against cerebral ischemia/reperfusion injury in mice. Stroke. 2007;38:1345–1353. doi: 10.1161/01.STR.0000259709.16654.8f. 01.STR.0000259709.16654.8f [pii] [DOI] [PubMed] [Google Scholar]
- 12.Butovsky O, et al. Modulating inflammatory monocytes with a unique microRNA gene signature ameliorates murine ALS. J Clin Invest. 2012;122:3063–3087. doi: 10.1172/JCI62636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geissmann F, Jung S, Littman DR. Blood monocytes consist of two principal subsets with distinct migratory properties. Immunity. 2003;19:71–82. doi: 10.1016/s1074-7613(03)00174-2. S1074761303001742 [pii] [DOI] [PubMed] [Google Scholar]
- 14.Gautier EL, et al. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat Immunol. 2012;13:1118–1128. doi: 10.1038/ni.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rothlin CV, Ghosh S, Zuniga EI, Oldstone MB, Lemke G. TAM receptors are pleiotropic inhibitors of the innate immune response. Cell. 2007;131:1124–1136. doi: 10.1016/j.cell.2007.10.034. [DOI] [PubMed] [Google Scholar]
- 16.Hashimoto D, et al. Tissue-resident macrophages self-maintain locally throughout adult life with minimal contribution from circulating monocytes. Immunity. 2013;38:792–804. doi: 10.1016/j.immuni.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yona S, et al. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity. 2013;38:79–91. doi: 10.1016/j.immuni.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cahoy JD, et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci. 2008;28:264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scott EW, et al. PU.1 functions in a cell-autonomous manner to control the differentiation of multipotential lymphoid-myeloid progenitors. Immunity. 1997;6:437–447. doi: 10.1016/s1074-7613(00)80287-3. [DOI] [PubMed] [Google Scholar]
- 20.Beers DR, et al. Wild-type microglia extend survival in PU.1 knockout mice with familial amyotrophic lateral sclerosis. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:16021–16026. doi: 10.1073/pnas.0607423103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haynes SE, et al. The P2Y12 receptor regulates microglial activation by extracellular nucleotides. Nature neuroscience. 2006;9:1512–1519. doi: 10.1038/nn1805. [DOI] [PubMed] [Google Scholar]
- 22.Cardona AE, Huang D, Sasse ME, Ransohoff RM. Isolation of murine microglial cells for RNA analysis or flow cytometry. Nat Protoc. 2006;1:1947–1951. doi: 10.1038/nprot.2006.327. nprot.2006.327 [pii] [DOI] [PubMed] [Google Scholar]
- 23.Jung S, et al. Analysis of fractalkine receptor CX(3)CR1 function by targeted deletion and green fluorescent protein reporter gene insertion. Mol Cell Biol. 2000;20:4106–4114. doi: 10.1128/mcb.20.11.4106-4114.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beutner C, Roy K, Linnartz B, Napoli I, Neumann H. Generation of microglial cells from mouse embryonic stem cells. Nature protocols. 2010;5:1481–1494. doi: 10.1038/nprot.2010.90. [DOI] [PubMed] [Google Scholar]
- 25.Abutbul S, et al. TGF-beta signaling through SMAD2/3 induces the quiescent microglial phenotype within the CNS environment. Glia. 2012;60:1160–1171. doi: 10.1002/glia.22343. [DOI] [PubMed] [Google Scholar]
- 26.Schilling T, Nitsch R, Heinemann U, Haas D, Eder C. Astrocyte-released cytokines induce ramification and outward K+ channel expression in microglia via distinct signalling pathways. Eur J Neurosci. 2001;14:463–473. doi: 10.1046/j.0953-816x.2001.01661.x. [DOI] [PubMed] [Google Scholar]
- 27.Durafourt BA, et al. Comparison of polarization properties of human adult microglia and blood-derived macrophages. Glia. 2012;60:717–727. doi: 10.1002/glia.22298. [DOI] [PubMed] [Google Scholar]
- 28.Butovsky O, et al. Induction and blockage of oligodendrogenesis by differently activated microglia in an animal model of multiple sclerosis. J Clin Invest. 2006;116:905–915. doi: 10.1172/JCI26836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kulkarni AB, et al. Transforming growth factor beta 1 null mutation in mice causes excessive inflammatory response and early death. Proc Natl Acad Sci U S A. 1993;90:770–774. doi: 10.1073/pnas.90.2.770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shull MM, et al. Targeted disruption of the mouse transforming growth factor-beta 1 gene results in multifocal inflammatory disease. Nature. 1992;359:693–699. doi: 10.1038/359693a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gorelik L, Flavell RA. Transforming growth factor-beta in T-cell biology. Nat Rev Immunol. 2002;2:46–53. doi: 10.1038/nri704. [DOI] [PubMed] [Google Scholar]
- 32.Carrier Y, Yuan J, Kuchroo VK, Weiner HL. Th3 cells in peripheral tolerance II TGF-beta-transgenic Th3 cells rescue IL-2-deficient mice from autoimmunity. J Immunol. 2007;178:172–178. doi: 10.4049/jimmunol.178.1.172. 178/1/172 [pii] [DOI] [PubMed] [Google Scholar]
- 33.Koeglsperger T, et al. Impaired glutamate recycling and GluN2B-mediated neuronal calcium overload in mice lacking TGF-beta1 in the CNS. Glia. 2013;61:985–1002. doi: 10.1002/glia.22490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perry VH, Hume DA, Gordon S. Immunohistochemical localization of macrophages and microglia in the adult and developing mouse brain. Neuroscience. 1985;15:313–326. doi: 10.1016/0306-4522(85)90215-5. [DOI] [PubMed] [Google Scholar]
- 35.Borkowski TA, Letterio JJ, Farr AG, Udey MC. A role for endogenous transforming growth factor beta 1 in Langerhans cell biology: the skin of transforming growth factor beta 1 null mice is devoid of epidermal Langerhans cells. J Exp Med. 1996;184:2417–2422. doi: 10.1084/jem.184.6.2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaplan DH, et al. Autocrine/paracrine TGFbeta1 is required for the development of epidermal Langerhans cells. J Exp Med. 2007;204:2545–2552. doi: 10.1084/jem.20071401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alliot F, Godin I, Pessac B. Microglia derive from progenitors, originating from the yolk sac, and which proliferate in the brain. Brain research. Developmental brain research. 1999;117:145–152. doi: 10.1016/s0165-3806(99)00113-3. [DOI] [PubMed] [Google Scholar]
- 38.Herbomel P, Thisse B, Thisse C. Zebrafish early macrophages colonize cephalic mesenchyme and developing brain, retina, and epidermis through a M-CSF receptor-dependent invasive process. Dev Biol. 2001;238:274–288. doi: 10.1006/dbio.2001.0393. [DOI] [PubMed] [Google Scholar]
- 39.Chan WY, Kohsaka S, Rezaie P. The origin and cell lineage of microglia: new concepts. Brain research reviews. 2007;53:344–354. doi: 10.1016/j.brainresrev.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 40.Ginhoux F, Lim S, Hoeffel G, Low D, Huber T. Origin and differentiation of microglia. Frontiers in cellular neuroscience. 2013;7:45. doi: 10.3389/fncel.2013.00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paolicelli RC, et al. Synaptic pruning by microglia is necessary for normal brain development. Science. 2011;333:1456–1458. doi: 10.1126/science.1202529. [DOI] [PubMed] [Google Scholar]
- 42.Flanders KC, et al. Localization and actions of transforming growth factor-beta s in the embryonic nervous system. Development. 1991;113:183–191. doi: 10.1242/dev.113.1.183. [DOI] [PubMed] [Google Scholar]
- 43.Hamby ME, Hewett JA, Hewett SJ. Smad3-dependent signaling underlies the TGF-beta1-mediated enhancement in astrocytic iNOS expression. Glia. 2010;58:1282–1291. doi: 10.1002/glia.21005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Henrich-Noack P, Prehn JH, Krieglstein JT. GF-beta 1 protects hippocampal neurons against degeneration caused by transient global ischemia. Dose-response relationship and potential neuroprotective mechanisms. Stroke. 1996;27:1609–1614. doi: 10.1161/01.str.27.9.1609. discussion 1615. [DOI] [PubMed] [Google Scholar]
- 45.Wiessner C, et al. Expression of transforming growth factor-beta 1 and interleukin-1 beta mRNA in rat brain following transient forebrain ischemia. Acta Neuropathol. 1993;86:439–446. doi: 10.1007/BF00228578. [DOI] [PubMed] [Google Scholar]
- 46.Flanders KC, Ren RF, Lippa CF. Transforming growth factor-betas in neurodegenerative disease. Prog Neurobiol. 1998;54:71–85. doi: 10.1016/s0301-0082(97)00066-x. [DOI] [PubMed] [Google Scholar]
- 47.Brionne TC, Tesseur I, Masliah E, Wyss-Coray T. Loss of TGF-beta 1 leads to increased neuronal cell death and microgliosis in mouse brain. Neuron. 2003;40:1133–1145. doi: 10.1016/s0896-6273(03)00766-9. [DOI] [PubMed] [Google Scholar]
- 48.Wake H, Moorhouse AJ, Jinno S, Kohsaka S, Nabekura J. Resting microglia directly monitor the functional state of synapses in vivo and determine the fate of ischemic terminals. J Neurosci. 2009;29:3974–3980. doi: 10.1523/JNEUROSCI.4363-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saijo K, et al. A Nurr1/CoREST pathway in microglia and astrocytes protects dopaminergic neurons from inflammation-induced death. Cell. 2009;137:47–59. doi: 10.1016/j.cell.2009.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peri F, Nusslein-Volhard C. Live imaging of neuronal degradation by microglia reveals a role for v0-ATPase a1 in phagosomal fusion in vivo. Cell. 2008;133:916–927. doi: 10.1016/j.cell.2008.04.037. [DOI] [PubMed] [Google Scholar]
- 51.Ginhoux F, et al. The origin and development of nonlymphoid tissue CD103+ DCs. J Exp Med. 2009;206:3115–3130. doi: 10.1084/jem.20091756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gekas C, Rhodes KE, Mikkola HK. Isolation and visualization of mouse placental hematopoietic stem cells. Current protocols in stem cell biology. 2008;Chapter 2(Unit 2A 8 1–2A 8 14) doi: 10.1002/9780470151808.sc02a08s6. [DOI] [PubMed] [Google Scholar]
- 53.Ginhoux F, et al. Blood-derived dermal langerin+ dendritic cells survey the skin in the steady state. J Exp Med. 2007;204:3133–3146. doi: 10.1084/jem.20071733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Coquery CM, Loo W, Buszko M, Lannigan J, Erickson LD. Optimized protocol for the isolation of spleen-resident murine neutrophils. Cytometry. Part A: the journal of the International Society for Analytical Cytology. 2012;81:806–814. doi: 10.1002/cyto.a.22096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weigert O, et al. CD4+Foxp3+ regulatory T cells prolong drug-induced disease remission in (NZBxNZW) F1 lupus mice. Arthritis Res Ther. 2013;15:R35. doi: 10.1186/ar4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mallon BS, Shick HE, Kidd GJ, Macklin WB. Proteolipid promoter activity distinguishes two populations of NG2-positive cells throughout neonatal cortical development. J Neurosci. 2002;22:876–885. doi: 10.1523/JNEUROSCI.22-03-00876.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu L, et al. CXCR2-positive neutrophils are essential for cuprizone-induced demyelination: relevance to multiple sclerosis. Nat Neurosci. 2010;13:319–326. doi: 10.1038/nn.2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huttlin EL, et al. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell. 2010;143:1174–1189. doi: 10.1016/j.cell.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ting L, Rad R, Gygi SP, Haas W. MS3 eliminates ratio distortion in isobaric multiplexed quantitative proteomics. Nat Methods. 2011;8:937–940. doi: 10.1038/nmeth.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.