Abstract
Recent studies have suggested that IL-18 −607C/A and −137G/C polymorphisms may be associated with the risk of allergic disease; however, individually published results are inconclusive. Therefore, we performed a meta-analysis to clarify whether IL-18 −607C/A and −137G/C polymorphisms were associated with the risk of allergic disease. A total of 21 studies including 5,331 cases and 9,658 controls were involved in this meta-analysis. In the overall analysis and the subgroup analysis according to ethnicity, we did not find significant association between IL-18 −607C/A or −137G/C polymorphism and the risk of allergic disease (all P > 0.05). However, in a stratified analysis by type of allergic disease, our results indicated that IL-18 −607C/A polymorphism was associated with a significantly decreased risk of allergic asthma in heterozygous comparison and IL-18 −137G/C was associated with a significantly decreased risk of allergic dermatitis in recessive model and homozygous comparison. In the stratified analysis by source of control, IL-18−607C/A showed significantly reduced risk in population-based subgroup, and for IL-18 −137G/C only significantly decreased risk was found in the hospital-based subgroup. Our meta-analysis suggests that IL-18 −607C/A and −137G/C polymorphisms may be protective factors for the risk of allergic asthma and allergic dermatitis, respectively.
1. Introduction
People with allergic disorders such as allergic dermatitis (AD), allergic rhinitis (AR), allergic asthma (AA), and food allergy can experience acute signs and symptoms of disease within minutes of exposure to the associated allergens [1]. To date, allergic diseases are causes of tremendous morbidity, and the rise in allergic disease is fast becoming a major global health issue [2, 3]. However, the reasons and mechanisms are not well understood. Accumulating evidence demonstrated that allergic diseases are complex genetic diseases resulting from the effect of multiple genetic and interacting environmental factors on their pathophysiology. The nature of the individual genes that have been identified as susceptibility factors for allergic disease has been comprehensively reviewed elsewhere, and the list of these genetic factors is likely to expand considerably in the future with the recent advent of genome-wide association approaches [4–6].
IL-18, formerly called interferon- (IFN-) γ-inducing factor, is a novel cytokine belonging to the IL-1 family and is produced by a wide range of immune cells, such as monocytes, activated macrophages, and Kupffer cells [7–9]. IL-18 has become recognized as an important regulator of innate and acquired immune responses and is expressed at sites of chronic inflammatory disease [10]. To date, accumulating evidence demonstrated that IL-18 synergizes with IL-12 to induce IFN-γ production and promote Th1 responses [11, 12]. However, IL-18 alone can also induce Th2 cytokine, IL-13, and promote Th2 responses [13, 14]. Indeed, abnormal imbalance of Th1 and Th2 functions contributes to the pathogeneses of allergic disorders [15–17]. IL-18 was also involved in allergic inflammatory reactions by indirectly inducing B cell isotype switching to IgE and by inducing the production of Th2 cytokines such as IL-4 and IL-13 [18]. In addition, IL-18 is generally considered to be involved in the Th1-mediated immune response and inhibits IgE synthesis, usually by acting synergistically with IL-12 [19]. Taken together, these studies show that IL-18 may be having both allergy-promoting and antiallergic functions. More recently, positive relationship has been found between IL-18 levels in the lesion or circulation and allergic diseases, such as AA, AR, and AD [20]. However, IL-18 was also raised in patients with other diseases in which dominant Th1 cells play a key role for immunity [21–24]. These findings confirm the evidence of an association between IL-18 gene and allergic disease but remain controversial.
IL-18 gene is located on chromosome 11q22 and contains many functional polymorphisms in the promoter region. Many studies show that the variations in IL-18 gene promoter are able to influence IL-18 production and activity, especially the IL-18 gene −607C/A (rs1946518) and −137G/C (rs187238) polymorphisms [25]. Giedraitis et al. reported that IL-18 −607C/A can alter a cAMP-responsive element binding site and result in a decrease of IL-18 transcription [26]. The IL-18 −137G/C polymorphism leads to a G/C amino acid substitution at position −137 in the promoter region of the IL-18 gene, which could alter the IL-18 promoter activity [27, 28]. Recent studies have suggested that IL-18 polymorphisms may be associated with the risk of allergic disease; however, individually published results are inconclusive due to small sample sizes. Therefore, we performed a meta-analysis of all eligible studies to clarify whether these two polymorphisms of IL-18 gene were associated with the risk of allergic disease, which may promote our understanding of the exact role of IL-18 in the development of allergic disease.
2. Material and Methods
2.1. Search Strategy
A systematic search was performed in PubMed, Web of Science, Science Direct, and Chinese National Knowledge Infrastructure (CNKI) databases to identify all the studies on the association between IL-18 −607C/A and/or −137G/C polymorphisms and allergic disease susceptibility (last search updated to November 2013). The search strategies were based on combinations of the following key words: “allergy or allergic disease or allergic disorder” and “IL-18 or −607C/A or −137G/C or rs1946518 or rs187238” and “polymorphism or variant or mutation or genotype or SNP.” There was no limit on time period, sample size, population, or language for minimizing potential publication bias.
2.2. Selection Criteria
Studies consistent with the following criteria were included in the meta-analysis: (1) case-control studies focus on the associations between IL-18 promoter polymorphisms and the risk of allergic disease; (2) all patients meet the diagnostic criteria for allergic disease; (3) sufficient published data for estimating the odds ratio (OR) and their corresponding 95% confidence interval (95% CI). The exclusion criteria of the meta-analysis were (1) studies with duplicate data; (2) studies with incomplete data; and (3) reviews, abstracts, and meta-analysis. In the case of several articles from the same study group, the most complete and recent results were used.
2.3. Data Extraction
Two authors independently extracted the data from the included studies (Cheng and Hao). For each eligible study, the following information was extracted: first author's name, year of publication, country, ethnicity, type of allergic disease, numbers of cases and controls, source of controls (hospital-based controls or population-based controls), genotyping method, and Hardy-Weinberg equilibrium (HWE). In case of discrepancies, a consensus on each item was reached among the authors.
2.4. Statistical Analysis
The ORs with 95% CIs for genotypes were used to evaluate the strength of the association between IL-18 −607C/A and −137G/C polymorphisms and the risk of allergic disease. The pooled ORs were calculated for allele model (mutation [M] allele versus wild [W] allele), dominant model (WM + MM versus WW), recessive model (MM versus WM + WW), homozygote comparison (MM versus WW), and heterozygote comparison (WM versus WW), respectively. The significance of the combined ORs was determined by a Z-test and two-sided P value <0.05 was considered significant.
Chi square-based Q-test and the I 2 statistic were performed to evaluate possible heterogeneity (P < 0.10 and I 2 > 50% indicated evidence of heterogeneity). A random-effects model or fixed-effects model was used to calculate pooled OR in the presence or absence of heterogeneity, respectively. Subgroup analyses were conducted according to the type of allergic disease, ethnicity, and source of control. Sensitivity analysis was carried out by deleting one single study each time to examine the influence of individual data set on the pooled ORs. Publication bias was evaluated with Begg's funnel plot [29] and Egger's regression method [30], and P < 0.05 was considered representative of statistically significant publication bias. Data analyses were performed using Stata 11.0 (StataCorp, College Station, TX) and Review Manager software 5.0 (Oxford, England).
3. Results
3.1. Characteristics of the Eligible Studies
As shown in Figure 1, a total of 66 potentially relevant articles were identified from PubMed, Web of Science, Science Direct, and CNKI databases using different combinations of key terms. After reading the titles and abstracts, we excluded 38 articles that assessed unrelated polymorphisms, were not case-control studies, were conducted in cell lines, and were performed in animal model. After reading the full texts of the remaining 28 articles regarding the association between IL-18 polymorphisms and allergic disease, seven articles were excluded, 1 for meta-analysis related to IL-18 promoter polymorphism and asthma risk, 4 with incomplete data (no available genotype frequency), 1 not related to IL-18 polymorphism, and 1 for overlapping data. Finally, a total of 21 articles were identified for data extraction and assessment, including 5,331 cases and 9,658 controls. The studies identified and their main characteristics were summarized in Table 1. Of the 21 case-control studies included, an array of allergic diseases including AA [31–40], AD [41–45], AR [10, 46], drug allergy [47, 48], and other types [49, 50] was involved. Of all studies included, 12 studies were conducted in Asian populations [31, 33, 36, 37, 39, 40, 43–48], while 9 studies were performed in Caucasian populations [10, 32, 34, 35, 38, 41, 42, 49, 50]. There were 16 studies concerning IL-18 −607C/A [10, 31, 32, 35–40, 44–50] and 19 studies concerning −137G/C [10, 31–37, 39–45, 47–49]. The genotype distributions among the controls of all studies were in agreement with HWE, except 2 studies for −607C/A [38, 46] and 1 study for −137G/C [33].
Figure 1.
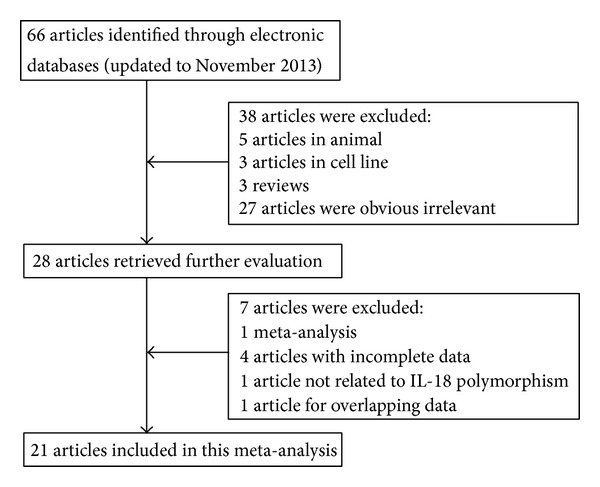
Flow diagram of studies included and excluded in the present meta-analysis.
Table 1.
Characteristics of studies included in the present meta-analysis.
| Studies | Year | Country | Ethnicity | Type | Source of controls | Sample size | SNP studied | Genotyping method | HWE |
|---|---|---|---|---|---|---|---|---|---|
| Harada et al. [31] | 2009 | Japan | Asian | AA | PB | 453/719 | −607C/A; −137G/C |
Taqman | 0.29; 0.48 |
| Heinzmann et al. [32] | 2004 | German | Caucasian | AA | PB | 230/269 | −607C/A; −137G/C |
Taqman | 0.68; 0.17 |
| Birbian et al. [33] | 2013 | Indian | Asian | AA | PB | 410/414 | −137G/C | ARMS-PCR | <0.001 |
| Yang et al. [40] | 2009 | China | Asian | AA | PB | 102/100 | −607C/A −137G/C |
PCR-SSP | 0.08; 0.74 |
| Imboden et al. [34] | 2006 | Swiss | Caucasian | AA | HB | 530/5204 | −137G/C | Taqman | 0.37 |
| Lachheb et al. [38] | 2008 | Tunisia | Caucasian | AA | HB | 105/112 | −607C/A | PCR-RFLP | 0.002 |
| Pawlik et al. [35] | 2007 | Poland | Caucasian | AA | PB | 231/305 | −607C/A; −137G/C |
Allele-specific PCR | 0.75; 0.08 |
| Sebelova et al. [10] | 2007 | Czech | Caucasian | AR | HB | 539/312 | −607C/A; −137G/C |
PCR-RFLP | 0.06; 0.80 |
| Shin et al. [37] | 2005 | Korea | Asian | AA | PB | 438/149 | −607C/A; −137G/C |
PCR-sequencing | 0.14; 0.36 |
| Wu et al. [36] | 2012 | China | Asian | AA | PB | 120/120 | −607C/A; −137G/C |
PCR-SSP | 0.05; 0.40 |
| Chen [39] | 2008 | China | Asian | AA | HB | 82/78 | −607C/A; −137G/C |
PCR-SSP | 0.66; 0.89 |
| Ibrahim et al. [41] | 2012 | Egypt | Caucasian | AD | HB | 25/25 | −137G/C | PCR-RFLP | 0.10 |
| Trzeciak et al. [42] | 2010 | Poland | Caucasian | AD | PB | 67/46 | −137G/C | ARMS-PCR | 0.07 |
| Luo et al. [45] | 2008 | China | Asian | AD | PB | 82/100 | −607C/A; −137G/C |
PCR-SSP | 0.62; 0.76 |
| Izakovicova et al. [49] | 2010 | Czech | Caucasian | Allergic disorder | PB | 633/325 | −607C/A; −137G/C |
Taqman | 0.09; 0.71 |
| Lee et al. [46] | 2006 | China | Asian | AR | HB | 160/166 | −607C/A | PCR-RFLP | <0.001 |
| Ming et al. [47] | 2011 | China | Asian | Drug allergy | HB | 606/614 | −607C/A; −137G/C |
PCR-sequencing | 0.07; 0.09 |
| Osawa et al. [44] | 2007 | Japan | Asian | AD | PB | 21/100 | −607C/A; −137G/C |
PCR-sequencing | 0.88; 0.48 |
| Kim et al. [48] | 2011 | Korea | Asian | Drug allergy | PB | 275/196 | −607C/A; −137G/C |
SNaPshot | 0.84; 0.17 |
| Kato et al. [43] | 2009 | Japan | Asian | AD | HB | 160/104 | −137G/C | PCR-RFLP | 0.11 |
| Torres et al. [50] | 2010 | Spain | Caucasian | Henoch-Schönlein purpura | PB | 62/200 | −607C/A; −137G/C |
Taqman | 0.05; 0.10 |
AA, allergic asthma; AD, allergic dermatitis; AR, allergic rhinitis; PB, population-based controls; HB, hospital-based controls; HWE, Hardy-Weinberg equilibrium; PCR, polymerase chain reaction; RFLP, restriction fragment length polymorphism; SSP, sequence-specific primers.
3.2. Quantitative Analysis
A summary of associations between IL-18 −607C/A and −137G/C polymorphisms and allergic disease risk was shown in Table 2.
Table 2.
Meta-analysis of IL-18 −607C/A and −137G/C polymorphisms with risk of allergic disease.
| Allele model | Dominant model | Recessive model | Homozygous comparison | Heterozygous comparison | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| OR [95% CI] | P | OR [95% CI] | P | OR [95% CI] | P | OR [95% CI] | P | OR [95% CI] | P | |
| IL-18 −607C/A | A allele versus C allele | AA + AC versus CC | AA versus CC + AC | AA versus CC | AC versus CC | |||||
| Overall | 1.03 [0.91–1.15] | 0.67 | 1.02 [0.85–1.22] | 0.86 | 1.07 [0.88–1.28] | 0.51 | 1.01 [0.77–1.33] | 0.94 | 1.00 [0.83–1.21] | 0.98 |
| Type of disease | ||||||||||
| AA | 0.98 [0.89–1.08] | 0.72 | 0.87 [0.74–1.03] | 0.10 | 1.08 [0.82–1.43] | 0.58 | 0.88 [0.61–1.27] | 0.48 | 0.82 [0.69–0.98] | 0.03 |
| AD | 1.15 [0.81–1.64] | 0.44 | 1.39 [0.80–2.41] | 0.24 | 1.01 [0.55–1.87] | 0.98 | 1.27 [0.61–2.65] | 0.53 | 1.42 [0.79–2.56] | 0.24 |
| AR | 1.05 [0.88–1.24] | 0.60 | 1.36 [0.55–3.34] | 0.51 | 1.00 [0.73–1.36] | 0.98 | 1.21 [0.84–1.75] | 0.30 | 1.49 [0.47–4.78] | 0.50 |
| Other types | 1.02 [0.74–1.43] | 0.89 | 1.02 [0.66–1.60] | 0.92 | 1.09 [0.67–1.78] | 0.72 | 1.09 [0.55–2.18] | 0.80 | 1.03 [0.70–1.51] | 0.88 |
| Ethnicity | ||||||||||
| Asian | 1.04 [0.88–1.24] | 0.61 | 1.10 [0.82–1.46] | 0.53 | 1.02 [0.80–1.31] | 0.85 | 0.96 [0.64–1.45] | 0.86 | 1.12 [0.82–1.52] | 0.47 |
| Caucasian | 0.97 [0.88–1.08] | 0.57 | 0.90 [0.77–1.04] | 0.15 | 1.14 [0.83–1.56] | 0.42 | 1.02 [0.82–1.27] | 0.86 | 0.86 [0.73–1.01] | 0.06 |
| Source of control | ||||||||||
| PB | 0.93 [0.86–1.01] | 0.11 | 0.86 [0.76–0.98] | 0.03 | 0.97 [0.85–1.11] | 0.69 | 0.80 [0.68–0.95] | 0.009 | 0.85 [0.74–0.98] | 0.03 |
| HB | 1.27 [1.03–1.57] | 0.03 | 1.36 [0.94–1.96] | 0.10 | 1.46 [0.97–2.21] | 0.07 | 1.75 [1.14–2.68] | 0.01 | 1.27 [0.84–1.94] | 0.26 |
|
| ||||||||||
| IL-18 −137G/C | C allele versus G allele | CC + CG versus GG | CC versus CG + GG | CC versus GG | CG versus GG | |||||
| Overall | 0.93 [0.80–1.07] | 0.31 | 0.97 [0.82–1.14] | 0.70 | 0.77 [0.58–1.02] | 0.07 | 0.76 [0.55–1.04] | 0.08 | 1.02 [0.88–1.18] | 0.78 |
| Type of disease | ||||||||||
| AA | 0.94 [0.86–1.03] | 0.21 | 0.97 [0.87–1.08] | 0.63 | 0.80 [0.63–1.02] | 0.07 | 0.80 [0.63–1.02] | 0.07 | 1.00 [0.90–1.12] | 0.95 |
| AD | 0.66 [0.30–1.46] | 0.30 | 0.70 [0.29–1.71] | 0.44 | 0.30 [0.15–0.60] | <0.001 | 0.26 [0.12–0.53] | <0.001 | 0.84 [0.38–1.82] | 0.65 |
| AR | 0.93 [0.75–1.16] | 0.51 | 0.94 [0.71–1.25] | 0.68 | 0.81 [0.48–1.37] | 0.43 | 0.80 [0.47–1.37] | 0.42 | 0.97 [0.73–1.30] | 0.85 |
| Other types | 0.96 [0.61–1.53] | 0.87 | 1.00 [0.60–1.66] | 0.99 | 0.71 [0.47–1.09] | 0.12 | 0.70 [0.46–1.08] | 0.11 | 1.05 [0.66–1.68] | 0.84 |
| Ethnicity | ||||||||||
| Asian | 1.05 [0.85–1.30] | 0.64 | 1.12 [0.89–1.39] | 0.34 | 0.78 [0.54–1.12] | 0.18 | 0.81 [0.56–1.16] | 0.25 | 1.15 [0.93–1.43] | 0.20 |
| Caucasian | 0.80 [0.66–0.98] | 0.03 | 0.83 [0.68–1.02] | 0.08 | 0.71 [0.49–1.02] | 0.06 | 0.66 [0.43–1.00] | 0.05 | 0.93 [0.83–1.05] | 0.24 |
| Source of control | ||||||||||
| PB | 0.87 [0.73–1.05] | 0.14 | 0.91 [0.76–1.10] | 0.35 | 0.76 [0.51–1.11] | 0.15 | 0.73 [0.48–1.12] | 0.15 | 0.98 [0.87–1.10] | 0.72 |
| HB | 1.04 [0.77–1.39] | 0.81 | 1.09 [0.79–1.51] | 0.60 | 0.72 [0.54–0.96] | 0.03 | 0.72 [0.53–0.97] | 0.03 | 1.14 [0.85–1.54] | 0.38 |
AA, allergic asthma; AD, allergic dermatitis; AR, allergic rhinitis; PB, population-based control; HB, hospital-based control.
For IL-18 −607C/A polymorphism, a total of 4,089 cases and 3,840 controls were included in the meta-analysis. There was significant between-study heterogeneity among all the genetic models (allele model: I 2 = 63%; dominant model: I 2 = 63%; recessive model: I 2 = 57%; homozygous comparison: I 2 = 71%; heterozygous comparison: I 2 = 62%). Therefore, random-effect model was applied to synthesize the data. In the overall analysis, we did not find significant association between IL-18 −607C/A polymorphisms and the risk of allergic disease under all models (A allele versus C allele: OR = 1.03, 95% CI = 0.91–1.15, P = 0.67; AA + AC versus CC: OR = 1.02, 95% CI = 0.85–1.22, P = 0.86; AA versus AC + CC: OR = 1.07, 95% CI = 0.88–1.28, P = 0.51; AA versus CC: OR = 1.01, 95% CI = 0.77–1.33, P = 0.94; AC versus CC: OR = 1.00, 95% = 0.83–1.21, P = 0.98, Figure 2). In the subgroup analysis by the type of allergic disease, we only found significant association between IL-18 −607C/A polymorphism and AA under heterozygous comparison (AC versus CC: OR = 0.82, 95% CI = 0.69–0.98, P = 0.03). In a stratified analysis by ethnicity, there was no significant association between IL-18 −607C/A polymorphism and the risk of allergic disease (all P > 0.05 under all models) in Asian and Caucasian populations. In a stratified analysis by source of control, IL-18 −607C/A showed significantly reduced risk in dominant model (OR = 0.86, 95% CI = 0.76–0.98, P = 0.03), homozygous comparison (OR = 0.80, 95% CI = 0.68–0.95, P = 0.009), and heterozygous comparison (OR = 0.85, 95% CI = 0.74–0.98, P = 0.03) in population-based subgroup but showed increased risk in allele model (OR = 1.27, 95% CI = 1.03–1.57, P = 0.03) and homozygous comparison (OR = 1.75, 95% CI = 1.14–2.68, P = 0.01) in hospital-based subgroup.
Figure 2.
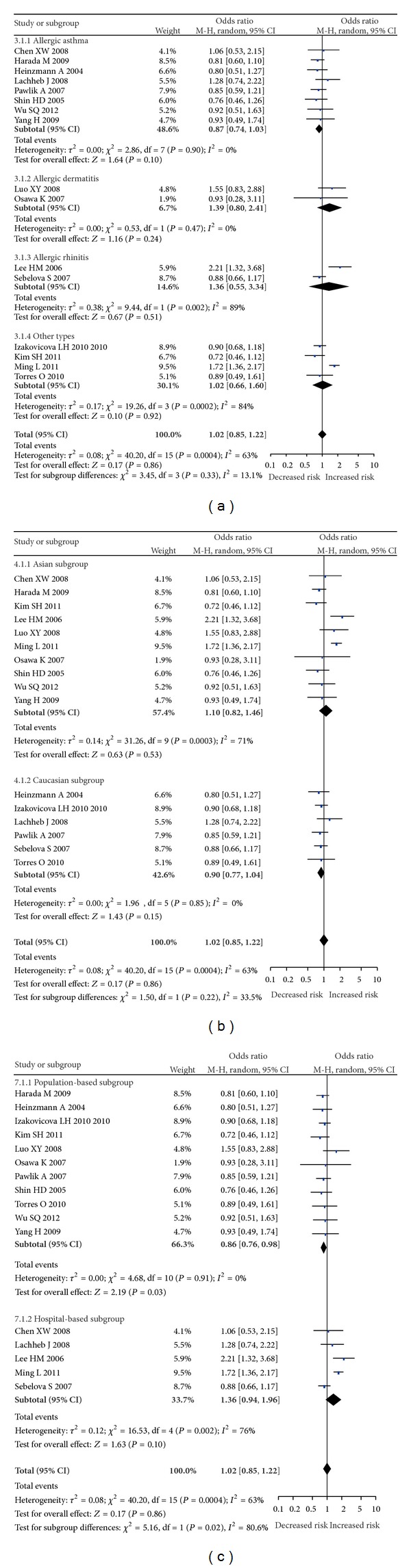
Forest plots for IL-18 −607C/A polymorphism and allergic disease risk under dominant model. (a) Subgroup analysis by type of disease. (b) Subgroup analysis by ethnicity. (c) Subgroup analysis by source of control.
For IL-18 −137G/C polymorphism, a total of 5,067 cases and 9,379 controls were included in the meta-analysis. We found significant between-study heterogeneity under allele model (I 2 = 73%), dominant model (I 2 = 67%), and heterozygous comparison (I 2 = 56%) and the random-effects model was used, whereas the fixed-effect model was used in recessive model (I 2 = 39%) and homozygous comparison (I 2 = 46%). In the overall analysis, we did not find significant association between IL-18 −137G/C polymorphism and the risk of allergic disease under all models (C allele versus G allele: OR = 0.93, 95% CI = 0.80–1.07, P = 0.31; CC + CG versus GG: OR = 0.97, 95% CI = 0.82–1.14, P = 0.70; CC versus CG + GG: OR = 0.77, 95% CI = 0.58–1.02, P = 0.07; CC versus GG: OR = 0.76, 95% CI = 0.55–1.04, P = 0.08; CG versus GG: OR = 1.02, 95% CI = 0.88–1.18, P = 0.78, Figure 3). When the data were stratified by the type of allergic disease, a significant association between IL-18 −137G/C polymorphism and AD was found under recessive model (CC versus CG + GG: OR = 0.30, 95% CI = 0.15–0.60, P < 0.001) and homozygous comparison (CC versus GG: OR = 0.26, 95% CI = 0.12–0.53, P < 0.001). In the subgroup analysis by ethnicity, the results suggested that IL-18 −137G/C polymorphism was not associated with the risk of allergic disease (all P > 0.05 under all models) in Asian and Caucasian populations. In a stratified analysis by source of control, only significantly decreased risk was found in the hospital-based subgroup under recessive model (OR = 0.72, 95% CI = 0.54–0.96, P = 0.03) and homozygous comparison (OR = 0.72, 95% CI = 0.53–0.97, P = 0.03).
Figure 3.
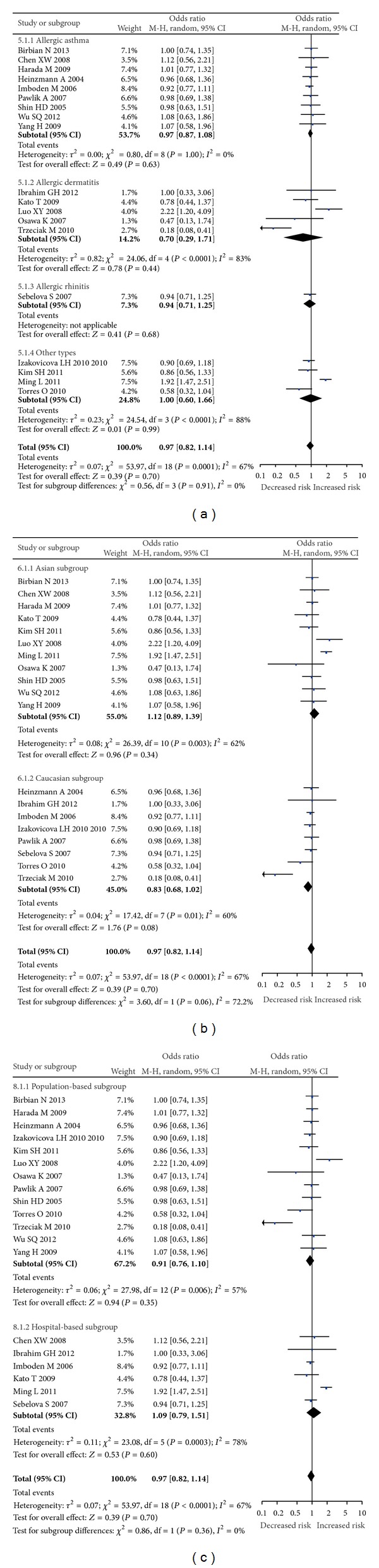
Forest plots for IL-18 −137G/C polymorphism and allergic disease risk under dominant model. (a) Subgroup analysis by types of disease. (b) Subgroup analysis by ethnicity. (c) Subgroup analysis by source of control.
3.3. Sensitivity Analysis
In order to assess the reliability of our results, we performed a sensitivity analysis by sequentially excluding individual study. Statistically similar results were obtained after sequentially excluding each study, suggesting the stability of this meta-analysis.
3.4. Publication Bias
Potential publication bias in this meta-analysis was estimated using Begg's funnel plot and Egger's test. The shape of funnel plot revealed the evidence of funnel plot symmetry for IL-18 −607C/A and −137G/C polymorphisms (Figure 4), and Egger's test provided statistical evidence which identified the absence of publication bias (IL-18 −607C/A, P = 0.741; IL-18 −137G/C, P = 0.438).
Figure 4.
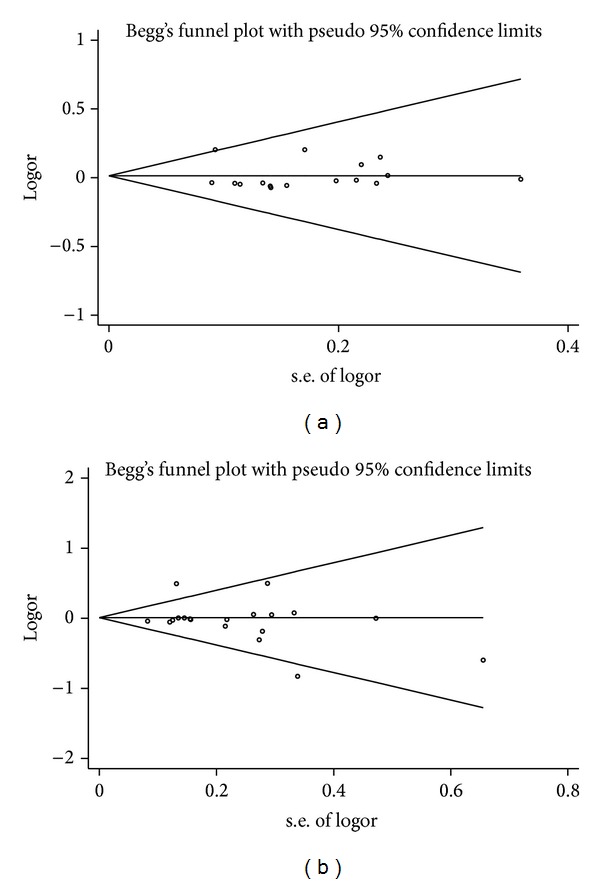
Publication bias represented by Begg's funnel plot for the association between IL-18 polymorphisms and the risk of allergic disease under the dominant model. (a) IL-18 −607C/A polymorphism; (b) IL-18 −137G/C polymorphism.
4. Discussion
The enormous health importance of allergy disease has stimulated much work aimed at identifying susceptibility genes. Investigation into common genetic variation in the human genome has highlighted the contribution of genetics to etiology and pathogenesis of allergic diseases. However, findings are not always consistent. Considering a single study may lack statistical power to provide compelling evidence as a result of small sample size and clinical heterogeneities, we performed this meta-analysis by combining all eligible publications to derive an accurate assessment of the association between IL-18 polymorphisms and the risk of allergic disease.
IL-18, belonging to the IL-1 superfamily, is a pleiotropic proinflammatory cytokine, which functions as a crucial regulator of IgE production through balancing the TH1-cell- and TH2-cell-mediated immune responses. A surfeit of IL-18 has been found in patients with allergic diseases, including AA, AD, and AR, in which a predominance of Th1 cells is significant [9, 20, 51]. Yoshimoto et al. suggested that IL-18 may be critical in regulation of IgE production in vivo, providing a potential therapeutic target for allergic disorders [52]. Therefore, the functions of IL-18 are very heterogeneous and complicated. In principle, IL-18 enhances the IL-12-driven Th1 immune responses, but it can also stimulate Th2 immune responses in the absence of IL-12 [53]. All of these findings suggest that IL-18 plays important roles in both antiallergic and allergy-promoting effects. Currently, IL-18 genetic polymorphisms have been postulated to be implicated in the development of allergic disease. Large quantities of evidence support the hypothesis that IL-18 genetic polymorphisms may cause the abnormal expression of IL-18, which is responsible for mediating the T-cell regulation, thereby leading to the pathological development of allergic disease.
In the present meta-analysis, including 5,331 cases and 9,658 controls, IL-18 −607C/A and −137G/C polymorphisms were not found to be associated with the risk of allergic disease in the overall analysis. The conclusion of this meta-analysis is different from the previous meta-analysis [54]. As we knew, this meta-analysis is the latest one and has the largest sample size. So the result of our meta-analysis is more likely convincing.
Subgroup analyses according to ethnicity revealed similar results, suggesting that the environment they lived in did not play an obvious role in the association between IL-18 polymorphisms and the risk of allergic disease. However, in a stratified analysis by types of allergic disease, our results indicated that IL-18 −607C/A polymorphism was associated with a significantly decreased risk of AA in heterozygous comparison, and IL-18 −137G/C was associated with a significantly decreased risk of AD in recessive model and homozygous comparison. As mentioned above, IL-18 gene promoter polymorphisms at positions −607C/A and −137G/C were associated with their transcription activity. Low promoter activity was observed for the −607A and −137C alleles, whereas higher promoter activity was observed for the −607C and −137G alleles in these positions [54]. Our meta-analysis showed that IL-18 −607C/A and −137G/C polymorphisms might be involved in the etiology of allergic disease, revealing a significant protective role or decreased risk towards AA and AD, respectively. Allergic disease is a multifactorial disease influenced by interactions of multiple susceptibility genes and environmental factors, and there will not be a single gene or single environmental factor that has a large effect on allergic disease susceptibility. However, our results should be interpreted with much caution. Only eight case-control studies for AA and five for AD were included in this meta-analysis, which might reduce statistical power to get a reliable result. In subgroup analysis, according to the source of control, significantly decreased risk was found in the population-based subgroup for IL-18 −607C/A under three models (dominant model, homozygous comparison, and heterozygous comparison), but significant increased risk was found in hospital-based subgroup under two models (allele model and homozygous comparison). As for IL-18 −137G/C, only significantly decreased risk was found in the hospital-based subgroup under recessive model and homozygous comparison. The reason may be that the hospital-based controls have a high risk of producing unreliable results because hospital-based controls may not always be strictly healthy individuals and hospital-based controls may have other diseases and also been given the corresponding drugs which exerted a confounding effect on the risk for allergic disease. Thus, we cannot completely exclude the possibility that a true genetic effect was overestimated, and the results of hospital-based studies should be explained with caution. Therefore, a proper and representative population-based high quality study is of great value in case-control studies.
There are several limitations in this meta-analysis that should be considered. First, there is a significant heterogeneity in studies, which may influence the interpretation of the results. Second, in subgroup analysis by ethnicity, the included studies were only Asians and Caucasians. More studies containing the full range of possible ethnic differences in genotype association studies are needed to avoid selection bias. Third, only published data was collected, leading to possible publication bias in this meta-analysis. Forth, it is well acknowledged that many other factors, such as gene-gene or gene-environment interactions, may affect the risk of allergic disease. Determining whether or not these factors influence the results of this meta-analysis would need further investigation.
5. Conclusion
In conclusion, our meta-analysis provided a more precise estimation based on larger sample size compared with the individual studies. Our pooled results demonstrated no significant association between IL-18 polymorphisms and allergic disease. The subgroup analysis indicated that IL-18 −607C/A and −137G/C polymorphisms may be a protective factor for the risk of AA and AD, respectively. In the future, more large-scale studies should be carried out to confirm or refute the relationship between IL-18 polymorphisms and the risk of allergic disease.
Acknowledgment
This work has been supported by Grants from the National Natural Science Foundation of China (no. 81301835).
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- 1.Galli SJ, Tsai M. IgE and mast cells in allergic disease. Nature Medicine. 2012;18(5):693–704. doi: 10.1038/nm.2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frischmeyer-Guerrerio PA, Guerrerio AL, Oswald G, et al. Allergy: TGFβ receptor mutations impose a strong predisposition for human allergic disease. Science Translational Medicine. 2013;5(195) doi: 10.1126/scitranslmed.3006448.195ra94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asher MI, Montefort S, Björkstén B, et al. Worldwide time trends in the prevalence of symptoms of asthma, allergic rhinoconjunctivitis, and eczema in childhood: ISAAC Phases One and Three repeat multicountry cross-sectional surveys. The Lancet. 2006;368(9537):733–743. doi: 10.1016/S0140-6736(06)69283-0. [DOI] [PubMed] [Google Scholar]
- 4.Vercelli D. Discovering susceptibility genes for asthma and allergy. Nature Reviews Immunology. 2008;8(3):169–182. doi: 10.1038/nri2257. [DOI] [PubMed] [Google Scholar]
- 5.Ober C, Hoffjan S. Asthma genetics 2006: the long and winding road to gene discovery. Genes and Immunity. 2006;7(2):95–100. doi: 10.1038/sj.gene.6364284. [DOI] [PubMed] [Google Scholar]
- 6.Holloway JW, Yang IA, Holgate ST. Genetics of allergic disease. Journal of Allergy and Clinical Immunology. 2010;125(2):S81–S94. doi: 10.1016/j.jaci.2009.10.071. [DOI] [PubMed] [Google Scholar]
- 7.Dinarello CA. IL-18: a T(H1)-inducing, proinflammatory cytokine and new member of the IL-1 family. Journal of Allergy and Clinical Immunology. 1999;103(1 I):11–24. doi: 10.1016/s0091-6749(99)70518-x. [DOI] [PubMed] [Google Scholar]
- 8.Krakowiak A, Walusiak J, Krawczyk P, et al. IL-18 levels in nasal lavage after inhalatory challenge test with flour in bakers diagnosed with occupational asthma. International Journal of Occupational Medicine and Environmental Health. 2008;21(2):165–172. doi: 10.2478/v10001-008-0015-y. [DOI] [PubMed] [Google Scholar]
- 9.Wong CK, Ho CY, Ko FWS, et al. Proinflammatory cytokines (IL-17, IL-6, IL-18 and IL-12) and Th cytokines (IFN-γ, IL-4, IL-10 and IL-13) in patients with allergic asthma. Clinical and Experimental Immunology. 2001;125(2):177–183. doi: 10.1046/j.1365-2249.2001.01602.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sebelova S, Izakovicova-Holla L, Stejskalova A, Schüller M, Znojil V, Vasku A. Interleukin-18 and its three gene polymorphisms relating to allergic rhinitis. Journal of Human Genetics. 2007;52(2):152–158. doi: 10.1007/s10038-006-0093-2. [DOI] [PubMed] [Google Scholar]
- 11.Fukao T, Matsuda S, Koyasu S. Synergistic effects of IL-4 and IL-18 on IL-12-dependent IFN-γ production by dendritic cells. Journal of Immunology. 2000;164(1):64–71. doi: 10.4049/jimmunol.164.1.64. [DOI] [PubMed] [Google Scholar]
- 12.Yoshimoto T, Okamura H, Tagawa Y-I, Iwakura Y, Nakanishi K. Interleukin 18 together with interleukin 12 inhibits IgE production by induction of interferon-γ production from activated B cells. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(8):3948–3953. doi: 10.1073/pnas.94.8.3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoshino T, Yagita H, Ortaldo JR, Wiltrout RH, Young HA. In vivo administration of IL-18 can induce IgE production through Th2 cytokine induction and up-regulation of CD40 ligand (CD154) expression on CD4+ T cells. European Journal of Immunology. 2000;30(7):1998–2006. doi: 10.1002/1521-4141(200007)30:7<1998::AID-IMMU1998>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 14.Hoshino T, Wiltrout RH, Young HA. IL-18 is a potent coinducer of IL-13 in NK and T cells: a new potential role for IL-18 in modulating the immune response. Journal of Immunology. 1999;162(9):5070–5077. [PubMed] [Google Scholar]
- 15.Shi Y-H, Shi G-C, Wan H-Y, et al. Coexistence of Th1/Th2 and Th17/Treg imbalances in patients with allergic asthma. Chinese Medical Journal. 2011;124(13):1951–1956. [PubMed] [Google Scholar]
- 16.Yoshida A, Kohchi C, Inagawa H, Nishizawa T, Soma G-I. Improvement of allergic dermatitis via regulation of the Th1/Th2 immune system balance by macrophages activated with lipopolysaccharide derived from Pantoea agglomerans (IP-PA1) Anticancer Research. 2009;29(11):4867–4870. [PubMed] [Google Scholar]
- 17.Herberth G, Daegelmann C, Weber A, et al. Association of neuropeptides with Th1/Th2 balance and allergic sensitization in children. Clinical and Experimental Allergy. 2006;36(11):1408–1416. doi: 10.1111/j.1365-2222.2006.02576.x. [DOI] [PubMed] [Google Scholar]
- 18.Tanaka H, Miyazaki N, Oashi K, et al. IL-18 might reflect disease activity in mild and moderate asthma exacerbation. Journal of Allergy and Clinical Immunology. 2001;107(2):331–336. doi: 10.1067/mai.2001.112275. [DOI] [PubMed] [Google Scholar]
- 19.Kim E, Lee J-E, Namkung J-H, et al. Association of the single-nucleotide polymorphism and haplotype of the interleukin 18 gene with atopic dermatitis in Koreans. Clinical and Experimental Allergy. 2007;37(6):865–871. doi: 10.1111/j.1365-2222.2007.02717.x. [DOI] [PubMed] [Google Scholar]
- 20.Tanaka T, Tsutsui H, Yoshimoto T, et al. Interleukin-18 is elevated in the sera from patients with atopic dermatitis and from atopic dermatitis model mice, NC/Nga. International Archives of Allergy and Immunology. 2001;125(3):236–240. doi: 10.1159/000053821. [DOI] [PubMed] [Google Scholar]
- 21.Lee C-C, Lin W-Y, Wan L, et al. Association of interleukin-18 gene polymorphism with asthma in Chinese patients. Journal of Clinical Laboratory Analysis. 2008;22(1):39–44. doi: 10.1002/jcla.20218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamada G, Shijubo N, Shigehara K, Okamura H, Kurimoto M, Abe S. Increased levels of circulating interleukin-18 in patients with advanced tuberculosis. American Journal of Respiratory and Critical Care Medicine. 2000;161(6):1786–1789. doi: 10.1164/ajrccm.161.6.9911054. [DOI] [PubMed] [Google Scholar]
- 23.Shigehara K, Shijubo N, Ohmichi M, et al. Increased levels of interleukin-18 in patients with pulmonary sarcoidosis. American Journal of Respiratory and Critical Care Medicine. 2000;162(5):1979–1982. doi: 10.1164/ajrccm.162.5.9911113. [DOI] [PubMed] [Google Scholar]
- 24.Kanai T, Watanabe M, Okazawa A, et al. Interleukin 18 is a potent proliferative factor for intestinal mucosal lymphocytes in Crohn’s disease. Gastroenterology. 2000;119(6):1514–1523. doi: 10.1053/gast.2000.20260. [DOI] [PubMed] [Google Scholar]
- 25.Yang X, Qiu MT, Hu JW, et al. Association of interleukin-18 gene promoter -607 C>A and -137G>C polymorphisms with cancer risk: a meta-analysis of 26 studies. PLoS ONE. 2013;8(9) doi: 10.1371/journal.pone.0073671.e73671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Giedraitis V, He B, Huang W-X, Hillert J. Cloning and mutation analysis of the human IL-18 promoter: a possible role of polymorphisms in expression regulation. Journal of Neuroimmunology. 2001;112(1-2):146–152. doi: 10.1016/s0165-5728(00)00407-0. [DOI] [PubMed] [Google Scholar]
- 27.Liang XH, Cheung W, Heng CK, Wang DY. Reduced transcriptional activity in individuals with IL-18 gene variants detected from functional but not association study. Biochemical and Biophysical Research Communications. 2005;338(2):736–741. doi: 10.1016/j.bbrc.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 28.Arimitsu J, Hirano T, Higa S, et al. IL-18 gene polymorphisms affect IL-18 production capability by monocytes. Biochemical and Biophysical Research Communications. 2006;342(4):1413–1416. doi: 10.1016/j.bbrc.2006.02.096. [DOI] [PubMed] [Google Scholar]
- 29.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50(4):1088–1101. [PubMed] [Google Scholar]
- 30.Egger M, Smith GD, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. British Medical Journal. 1997;315(7109):629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harada M, Obara K, Hirota T, et al. A functional polymorphism in IL-18 is associated with severity of bronchial asthma. American Journal of Respiratory and Critical Care Medicine. 2009;180(11):1048–1055. doi: 10.1164/rccm.200905-0652OC. [DOI] [PubMed] [Google Scholar]
- 32.Heinzmann A, Gerhold K, Ganter K, et al. Association study of polymorphisms within interleukin-18 in juvenile idiopathic arthritis and bronchial asthma. Allergy. 2004;59(8):845–849. doi: 10.1111/j.1398-9995.2004.00538.x. [DOI] [PubMed] [Google Scholar]
- 33.Birbian N, Singh J, Jindal SK. Protective role of IL-18 -137G/C polymorphism in a North Indian population with asthma: a pilot study. Cytokine. 2013;61(1):188–193. doi: 10.1016/j.cyto.2012.09.015. [DOI] [PubMed] [Google Scholar]
- 34.Imboden M, Nicod L, Nieters A, et al. The common G-allele of interleukin-18 single-nucleotide polymorphism is a genetic risk factor for atopic asthma. The SAPALDIA cohort study. Clinical and Experimental Allergy. 2006;36(2):211–218. doi: 10.1111/j.1365-2222.2006.02424.x. [DOI] [PubMed] [Google Scholar]
- 35.Pawlik A, Kaminski M, Kuśnierczyk P, et al. Interleukin-18 promoter polymorphism in patients with atopic asthma. Tissue Antigens. 2007;70(4):314–318. doi: 10.1111/j.1399-0039.2007.00908.x. [DOI] [PubMed] [Google Scholar]
- 36.Wu S-Q, Liang W, Wang G-L, Li L-Y, Wang D-L, Chen C. Polymorphisms of the IL-18 promoter and bronchial asthma. Molecular Medicine Reports. 2012;6(6):1385–1388. doi: 10.3892/mmr.2012.1098. [DOI] [PubMed] [Google Scholar]
- 37.Shin HD, Kim LH, Park BL, et al. Association of interleukin 18 (IL18) polymorphisms with specific IgE levels to mite allergens among asthmatic patients. Allergy. 2005;60(7):900–906. doi: 10.1111/j.1398-9995.2005.00619.x. [DOI] [PubMed] [Google Scholar]
- 38.Lachheb J, Chelbi H, Ammar J, Hamzaoui K, Hamzaoui A. Promoter polymorphism of the IL-18 gene is associated with atopic asthma in Tunisian children. International Journal of Immunogenetics. 2008;35(1):63–68. doi: 10.1111/j.1744-313X.2007.00738.x. [DOI] [PubMed] [Google Scholar]
- 39.Chen XW. The Association between IL-16 and IL-18 Polymorphisms and the Risk of Asthma. Nanchang, China: Nanchang Medical College; 2008. [Google Scholar]
- 40.Yang H, Liu XN, Chen XW, Liu YL. Association of polymorphisms in interleukin218 gene promoter region with asthma in Jiangxi Han population. Chinese Journal of Immunology. 2009;25(6):524–527. [Google Scholar]
- 41.Ibrahim GH, ElTabbakh MT, Gomaa AHA, Mohamed EA. Interleukin-18 gene polymorphisms in Egyptian patients with allergic diseases. American Journal of Rhinology and Allergy. 2012;26(5):385–389. doi: 10.2500/ajra.2012.26.3806. [DOI] [PubMed] [Google Scholar]
- 42.Trzeciak M, Gleń J, Roszkiewicz J, Nedoszytko B. Association of single nucleotide polymorphism of interleukin-18 with atopic dermatitis. Journal of the European Academy of Dermatology and Venereology. 2010;24(1):78–79. doi: 10.1111/j.1468-3083.2009.03272.x. [DOI] [PubMed] [Google Scholar]
- 43.Kato T, Tsunemi Y, Saeki H, et al. Interferon-18 gene polymorphism -137 G/C is associated with susceptibility to psoriasis vulgaris but not with atopic dermatitis in Japanese patients. Journal of Dermatological Science. 2009;53(2):162–163. doi: 10.1016/j.jdermsci.2008.08.016. [DOI] [PubMed] [Google Scholar]
- 44.Osawa K, Etoh T, Ariyoshi N, et al. Relationship between Kaposi’s varicelliform eruption in Japanese patients with atopic dermatitis treated with tacrolimus ointment and genetic polymorphisms in the IL-18 gene promoter region. Journal of Dermatology. 2007;34(8):531–536. doi: 10.1111/j.1346-8138.2007.00326.x. [DOI] [PubMed] [Google Scholar]
- 45.Luo XY, Jiang LP, Liu W, Wang H. Relationship between interleukin-18 gene promoter polymorphism and atopic dermatitis in children. Chinese Journal of Dermatology. 2008;41(7):447–450. [Google Scholar]
- 46.Lee H-M, Park SA, Chung S-W, et al. Interleukin-18/-607 gene polymorphism in allergic rhinitis. International Journal of Pediatric Otorhinolaryngology. 2006;70(6):1085–1088. doi: 10.1016/j.ijporl.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 47.Ming L, Wen Q, Qiao H-L, Dong Z-M. Interleukin-18 and IL18 -607A/C and -137G/C gene polymorphisms in patients with penicillin allergy. Journal of International Medical Research. 2011;39(2):388–398. doi: 10.1177/147323001103900206. [DOI] [PubMed] [Google Scholar]
- 48.Kim S-H, Son JK, Yang E-M, Kim J-E, Park H-S. A functional promoter polymorphism of the human IL18 gene is associated with aspirin-induced urticaria. British Journal of Dermatology. 2011;165(5):976–984. doi: 10.1111/j.1365-2133.2011.10467.x. [DOI] [PubMed] [Google Scholar]
- 49.Izakovicova Holla L, Hrdlič ková B, Schüller M, et al. Haplotype analysis of the interleukin-18 gene in Czech patients with allergic disorders. Human Immunology. 2010;71(6):592–597. doi: 10.1016/j.humimm.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 50.Torres O, Palomino-Morales R, Miranda-Filloy JA, Vazquez-Rodriguez TR, Martin J, Gonzalez-Gay MA. IL18 gene polymorphisms in Henoch-Schönlein purpura. Clinical and Experimental Rheumatology. 2010;28(1):p. S114. [PubMed] [Google Scholar]
- 51.Verhaeghe B, Gevaert P, Holtappels G, et al. Up-regulation of IL-18 in allergic rhinitis. Allergy. 2002;57(9):825–830. doi: 10.1034/j.1398-9995.2002.23539.x. [DOI] [PubMed] [Google Scholar]
- 52.Yoshimoto T, Mizutani H, Tsutsui H, et al. IL-I 8 induction of IgE: dependence on CD4+ T cells, IL-4 and STAT6. Nature Immunology. 2000;1(2):132–137. doi: 10.1038/77811. [DOI] [PubMed] [Google Scholar]
- 53.Nakanishi K, Yoshimoto T, Tsutsui H, Okamura H. Interleukin-18 regulates both Th1 and Th2 responses. Annual Review of Immunology. 2001;19:423–474. doi: 10.1146/annurev.immunol.19.1.423. [DOI] [PubMed] [Google Scholar]
- 54.Ma Y, Zhang B, Tang R-K, Liu Y, Peng G-G. Interleukin-18 promoter polymorphism and asthma risk: a meta-analysis. Molecular Biology Reports. 2012;39(2):1371–1376. doi: 10.1007/s11033-011-0871-6. [DOI] [PubMed] [Google Scholar]


