Abstract
Background
Staphylococcus epidermidis is a pathogen that is frequently encountered in the hospital environment. Healthcare workers (HCWs) can serve as a reservoir for the transmission of S. epidermidis to patients.
Methods
The aim of this study was to compare and identify differences between S. epidermidis isolated from 20 patients with catheter-related bloodstream infections (CRBSIs) and from the hands of 42 HCWs in the same hospital in terms of antimicrobial resistance, biofilm production, presence of the intercellular adhesion (ica) operon and genetic diversity (pulsed field gel electrophoresis (PFGE), multilocus sequence typing (MLST) and staphylococcal cassette chromosome (SCC) mec typing).
Results
S. epidermidis isolates that caused CRBSI were resistant to significantly more non-betalactam drugs than were isolates collected from HCWs. Among the 43 mecA positive isolates (26 from HCWs), the most frequent SCCmec type was type IV (44%). The ica operon was significantly more prevalent in CRBSI isolates than in HCWs (P < 0.05). Weak in vitro biofilm production seemed to correlate with the absence of the ica operon regardless of the commensal or pathogenic origin of the isolate. The 62 isolates showed high diversity in their PFGE patterns divided into 37 different types: 19 harbored only by the CRBSI isolates and 6 shared by the clinical and HCW isolates. MLST revealed a total of ten different sequence types (ST). ST2 was limited to CRBSI-specific PFGE types while the “mixed” PFGE types were ST5, ST16, ST88 and ST153.
Conclusion
One third of CRBSI episodes were due to isolates belonging to PFGE types that were also found on the hands of HCWs, suggesting that HCW serve as a reservoir for oxacillin resistance and transmission to patients. However, S. epidermidis ST2, mecA-positive and icaA-positive isolates, which caused the majority of clinically severe CRBSI, were not recovered from the HCW’s hands.
Keywords: Staphylococcus epidermidis, Catheter-related bloodstream infection, Molecular epidemiology, PFGE, MLST
Introduction
Catheter-related bloodstream infection (CRBSI) is one of the most common healthcare-associated infections [1], and coagulase-negative staphylococci, predominantly Staphylococcus epidermidis, are the most common pathogens involved. A key factor in the pathogenesis of S. epidermidis CRBSI is its ability to form a biofilm, which is mediated by the production of an intercellular polysaccharide adhesin (PIA) encoded by an accessory gene cluster called the intercellular adhesion (ica) operon [2].
In a recent study, the genotypes of 33 S. epidermidis isolates from blood cultures of patients with nosocomial CRBSI have been investigated and compared with 33 commensal S. epidermidis isolates from healthy students [3]. The two populations were found to be genetically different using pulsed field gel electrophoresis (PFGE), a typing method known to provide reliable short-term epidemiological data: only 23% of the 37 PFGE types observed were common to both CRBSIs and commensal isolates. Furthermore, clinically severe CRBSIs were due to multidrug-resistant ica positive isolates. Using multilocus sequence typing (MLST), a method used to study long-term global epidemiology, these isolates were found to belong to two closely related genetic lineages (sequence type (ST)2 and ST54), which were not found among the healthy volunteers (HVs). Similarly, Muldrew et al. demonstrated the presence of a predominant and persistent clone among central line isolates from a specific ward. They showed that this clone was not found among S. epidermidis isolates collected from a control group living in the community [4]. These data suggest that particular lineages are transmitted within the hospital, which, by combining resistance and virulence, can cause more severe disease. However, controversy remains over the source of these bacteria isolated from central venous catheters. Cross-transmission of these pathogens might occur directly between patients in contact with one another, indirectly through contamination from the environment or through the contaminated hands of healthcare workers (HCWs). Indeed, acquisition of antibiotic-resistant flora such as methicillin-resistant S. epidermidis (MRSE) is an occupational hazard for HCWs. Such flora can serve as a reservoir for transmission to the environment, other staff, and ultimately to patients. Several studies have demonstrated that HCWs carry S. epidermidis that belong to the same clones that cause infections in their respective wards [5-7].
In this study we analyzed the antimicrobial drug resistance and molecular epidemiology of S. epidermidis that resulted in CRBSIs and commensal S. epidermidis collected from HCWs working in the same hospital over a close period.
We also studied the correlation between in vitro biofilm formation and the presence of the ica operon in this collection and on a set of commensal S. epidermidis isolates from HVs and from CRBSIs, which were previously collected [3]. The aim was to compare and identify differences between the S. epidermidis isolates causing CRBSI and commensal isolates from HCWs and HVs in terms of genetic relatedness, resistance to antibiotics, presence of the ica operon, and in vitro biofilm formation.
Methods
During two days in September 2012, 51 HCWs volunteered to participate in this study. They worked in the ten wards of the Erasme University Hospital that present the majority of reported CRBSIs cases, such as the intensive care department and gastroenterology department.
At least five HCWs per unit were sampled. Samples were obtained by taking fingerprints from the dominant hand directly onto mannitol salt agar plates (Becton-Dickinson, Heidelberg, Germany) at the end of the morning from HCWs that have already worked for several hours. Plates were incubated at 35°C in an aerobic atmosphere for two days, and a maximum of four mannitol non-fermentative colonies were randomly picked and sub-cultured onto Columbia blood agar for 18–24 hours at 35°C in an aerobic atmosphere. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDITOF-MS) was used to identify the bacteria present in each specimen. A maximum of one isolate per volunteer (the first one identified as S. epidermidis) was selected for the study.
The clinical strains originated from blood cultures of patients with CRBSI and were identified through the Erasme University Hospital Infection Control Unit database from January 2011 to September 2012. Among the 56 CRBSIs episodes identified, 20 episodes due to S. epidermidis (at least two blood cultures; one episode per patient) and treated with antibiotics were included. The patients were located on various wards, including the intensive care unit (n = 10), gastroenterology (n = 7), nephrology (n = 1), neurology (n = 1), and thoracic surgery (n = 1) departments. All of the 20 episodes of CRBSI except one were hospital-acquired and were distributed through non-tunneled central venous catheters (n = 16), peripheral arterial catheters (n = 2), peripheral venous catheter (n =1) and tunneled central venous catheter (n = 1).
Definitions
Confirmation of a S. epidermidis-BSI requires that the patient presents with fever (temperature above 38°C) or hypothermia (temperature below 35°C) and/or hypotension and at least two pairs of blood cultures positive for S. epidermidis collected at different times. We defined S. epidermidis-CRBSI as a primary BSI with a catheter positive for S. epidermidis presenting an identical susceptibility profile to the blood cultures. Clinically, the condition was considered severe if the patient presented with severe sepsis or septic shock when blood samples were collected for culture. Multidrug resistance (MDR) was defined as resistance to at least 5 of the 12 antibiotics tested in the study.
Bacterial isolate identification (ID) and susceptibility testing
Isolates, conserved at −80°C in pure glycerin, were sub-cultured onto Columbia blood agar for 18–24 hours at 35°C in an aerobic atmosphere. ID was confirmed by MALDITOF-MS. Susceptibility to 16 antimicrobial agents (penicillin, cefoxitin -used as marker for the detection of methicillin resistance-, erythromycin, clindamycin, levofloxacin, gentamicin, kanamycin, tobramycin, fusidic acid, minocycline, rifampin, trimethoprim-sulfamethoxazole, mupirocin, vancomycin, linezolid, and tigecycline) was determined by Vitek 2® using the antibiotic susceptibility P-610 card.
Bacterial DNA was extracted as described by Ünal et al. [8], and subsequently all isolates were tested for the presence of the mecA gene as previously described [9]. The presence of the antibiotic resistance genes aadC, aacA-aphD, aph3, erm (A, B and C), and msrA was determined using PCR on all kanamycin, tobramycin, gentamycin and erythromycin non-susceptible isolates [10,11]. Additionally, all isolates were tested for the presence of icaA to assess the presence of the ica operon [12].
Molecular typing techniques
For all of the isolates, SmaI genomic DNA restriction fragments were separated by PFGE as previously described [13]. As established by the previous criteria [14], patterns differing by less than 79% (corresponding to less than seven band differences) were considered to belong to the same type (represented by a letter). Arbitrarily, PFGE types represented by a single isolate were called “singletons”, while those represented by three or more isolates were considered “epidemic types.”
MLST was performed as described by Thomas et al. [15] on a randomly selected isolate from each PFGE type represented by more than one isolate. SCCmec typing was performed on all isolates based on determination of the ccr and mec gene complex using M-PCR1 and M-PCR2 as previously described [16].
Comparative analysis of the isolates from CRBSIs, HVs and HCWs
We compared a total of 53 CRBSI isolates (20 collected from the present study and 33 collected previously [3]) to commensal S. epidermidis isolated from the 42 HCWs from the present study and from the 33 HVs of our previous study [3] in terms of genetic relatedness, resistance to antibiotics, and biofilm formation.
Study of biofilm formation with the crystal violet staining method on 128 S. epidermidis isolates
An overnight growth culture in brain heart infusion (BHI) liquid was adjusted to a final OD600 of 1.00 ± 0.05 by adding sterile BHI medium. This OD-adjusted suspension was then diluted 250-fold to obtain the initial bacterial suspension (IBS). The wells of a 96-well microplate were inoculated with 200 μL of the IBS and incubated at 35°C in a humid atmosphere. A control well was inoculated with sterile BHI medium. Each isolate was evaluated using six samples. The medium was removed and the wells were washed three times with sterile distilled water. The wells were air dried for 45 min, and the adherent cells were stained with a 1/1000 solution of crystal violet. After 45 min, the excess crystal violet was removed and the wells were washed five times with 300 μL sterile distilled water. The dye was dissolved with 200 μL of a 33% acetic acid solution, and the absorbance of each well was read at 540 nm in a microplate reader (Synergy HT, BioTek), as described by Stepanovic et al. [17].
Confidentiality and ethics committee approval
The demographic, clinical, microbiological and molecular epidemiology data were collected anonymously. The ethics committee of the Erasme Hospital approved the protocol before the beginning of the study (No. P2012/208).
Data analysis
Stata data analysis and statistical software (version 12.0; StataCorp LP: College Station, Texas, USA) were used for all data analyses. Categorical analyses were carried out using Fisher’s exact test or the chi-square test, as appropriate. The Mann–Whitney U test or Kruskal-Wallis test was used for comparison between groups of non-normally distributed variables, as appropriate. Statistical significance was set at a two-sided P-value of < 0.05.
Results
A total of 62 S. epidermidis isolates (42 from HCWs and 20 from CRBSIs) were collected. All of the isolates were confirmed as S. epidermidis by MALDITOF-MS analysis. Overall, S. epidermidis was recovered from the hands of 82% of the HCWs (42 out of 51).
Resistance
The resistance results are shown in Table 1. The majority of isolates (69%) were resistant to methicillin, and there were no significant difference in the presence of the mecA gene between the HCW and CRBSI isolates. S. epidermidis isolates from patients with CRBSI were significantly more resistant to all non-betalactam drugs, except for erythromycin and fusidic acid, than the S. epidermidis isolates collected from HCWs. A higher percentage of mupirocin-resistant strains was found among isolates from CRBSIs in comparison with HCWs (P = 0.013).
Table 1.
Antimicrobial resistance profiles and resistance (R)-encoding genes of S. epidermidis isolates collected from healthcare workers (HCWs; n =42) vs. S. epidermidis isolates causing catheter-related bloodstream infections (CRBSIs; n = 20)
| Antimicrobial | Encoding gene | HCWs (% R) | CRBSIs (% R) | P-value |
|---|---|---|---|---|
|
Penicillin |
|
37 (88%) |
20 (100%) |
0.165 |
|
Methicillin (cefoxitin) |
|
26 (62%) |
17 (85%) |
0.065 |
| |
mecA |
26 |
17 |
0.065 |
|
Erythromycin |
|
26 (62%) |
15 (75%) |
0.308 |
| |
ermC |
11 |
12 |
0.009 |
| |
msrA |
13 |
0 |
0.006 |
| |
ermA |
1 |
3 |
0.034 |
|
Clindamycin |
|
14 [6]* (33%) |
15 [5]* (75%) |
0.002 |
|
Levofloxacin |
|
10 (24%) |
14 (70%) |
< 0.001 |
|
Fusidic acid |
|
19 (45%) |
13 (65%) |
0.146 |
|
Trimethoprim/sulfamethoxazole |
|
4 (10%) |
11 (55%) |
< 0.001 |
|
Aminoglycosides§ (at least one) |
|
10 (24%) |
13 (65%) |
0.002 |
|
Kanamycin |
|
9 (21%) |
13 (65%) |
< 0.001 |
|
Tobramycin |
|
6 (14%) |
13 (65%) |
< 0.001 |
|
Gentamicin |
|
5 (12%) |
10 (50%) |
0.003 |
| |
aadC |
6 |
11 |
< 0.001 |
| |
aacA-aphD |
5 |
10 |
0.003 |
| |
aph3 |
3 |
1 |
1.000 |
|
Mupirocin |
|
4 (10%) |
8 (40%) |
0.013 |
|
Rifampin |
|
0 (0%) |
5 (25%) |
0.002 |
| Median sum of resistance to non-beta-lactams (range) | 2 (0–5) | 5 (0–8) | < 0.001 |
Legend:
No resistance to minocycline, vancomycin, linezolid, and tigecycline was observed.
§The three aminoglycosides were regarded as a whole, and isolates resistant to at least one of the three drugs were considered resistant to aminoglycosides.
*For clindamycin, the values in square brackets represent the number of isolates with a clindamycin-inducible resistant phenotype.
When we assayed the genes that encode these resistance profiles, 23 erythromycin-resistant isolates had the ermC gene, whereas 13 isolates (only from HCWs) were msrA positive. Constitutive phenotypes were harbored by isolates with the ermC (n = 13) and ermA (n = 3) genes. When present, aminoglycoside resistance was mainly mediated by aadC (n = 17) alone or in combination with aacA-aphD (n = 10).
Comparative resistance data from CRBSI, HCW and HV isolates
A total of 128 isolates were analyzed, classified into three groups: HCWs (n = 42), CRBSIs (n = 53) and HVs (n = 33). Among the 12 antibiotics tested, which included the betalactams, the median sum of resistances observed for HCWs, HVs and CRBSIs were 4, 1, and 6, respectively. The difference in these values was statistically significant (P < 0.001). The difference in resistance to methicillin among the three sub-populations was also statistically significant (P < 0.001).
Molecular epidemiology
PFGE
The 62 isolates produced a broad range of restriction patterns, which were distributed into 37 different PFGE types (see Table 2). Twenty-three PFGE types were represented by a single isolate (singletons). Eight PFGE types (of which six singletons) were harbored by CRBSI isolates only. Twenty-three PFGE types (17 singletons) were harbored by a total of 34 HCW isolates. Six PFGE types were shared by CRBSI and HCW isolates (n = 17).
Table 2.
PFGE types and Sequence types (ST) distribution of S. epidermidis isolates collected from health care workers (HCW) and catheter-related bloodstream infections (CRBSI)
| |
|
N° of isolates |
|
|---|---|---|---|
| PFGE type | ST | HCW | CRBSI |
|
Y
|
ST5
|
3
|
3
|
|
I |
ST22 |
5 |
0 |
|
ZB |
ST33 |
4 |
0 |
|
ZH |
ST2 |
0 |
3 |
|
ZV
|
ST5
|
1
|
2
|
| R |
ST88 |
2 |
0 |
| YA |
New single locus variant of ST370 |
2 |
0 |
| YI |
ST16 |
2 |
0 |
| ZO |
ST130 |
2 |
0 |
| ZL |
ST2 |
0 |
2 |
|
ZU |
ST5 |
1 |
1 |
|
F |
ST153 |
1 |
1 |
|
YM |
ST16 |
1 |
1 |
|
ZR |
ST88 |
1 |
1 |
| Singletons |
ND |
17 |
6 |
| Total | 42 | 20 | |
Legend:
ND not done.
Mixed PFGE types are identified by bold type.
Epidemic PFGE types are identified by italic type.
MLST
Fourteen isolates were typed by MLST (one randomly selected per PFGE type composed of more than one isolate). MLST revealed a total of ten different sequence types. ST2 was limited to CRBSI-specific PFGE types while the “mixed” PFGE types were ST5, ST16, ST88 and ST153. HCW-specific PFGE types were ST22, ST33, ST88, ST130 and a new ST single locus variant (SLV) ST370.
SCCmec type and presence of the ica operon
Among the 43 mecA positive isolates (26 HCWs), nine different combinations of ccr and mec complexes were found. The most frequent type was SCCmec type IV (n = 19). Up to 12 isolates carried composite SCCmec types, amplifying more than one ccr gene complex. Eleven isolates were unable to be SCCmec typed because of the absence of amplification for one of the two complexes.
We observed icaA positive isolates in both HCWs and patients with a ratio of 50% (10 out of 20) for CRBSI isolates and 24% (10 out of 42) for HCW isolates (P = 0.04).
Comparative molecular epidemiology of CRBSI, HCW and HV isolates
Among the 128 isolates, 64 different PFGE types were found, which included 41 singletons. Sixty-five isolates (51%) belonged to one of the 12 epidemic PFGE types that were identified. Distribution of epidemic S. epidermidis PFGE types is shown in Table 3. Four epidemic PFGE types, representing a total of 24 isolates (19%), were found among the three epidemiological groups (group 1). Few group 1 isolates were mecA positive, but all were icaA negative. Three epidemic PFGE types, representing 20 isolates (16%), were only found in the hospital environment (group 2). All group 2 isolates were mecA positive, but only five isolates were icaA positive. Three epidemic PFGE types (12 isolates, 9%) were exclusively found among CRBSIs (“CRBSI-specific” PFGE types, group 3). All group 3 isolates were mecA and icaA positive and possessed MDR. One epidemic PFGE type (five isolates) was found among both HCWs and HVs, and one epidemic PFGE was found (four isolates) among only HCWs (“commensal-specific” epidemic PFGE types, group 4). Both of these group 4 epidemic PFGE types were mecA and icaA negative.
Table 3.
Distribution of epidemic S. epidermidis PFGE types (≥3 isolates) and related characteristics
|
Group 1
|
Group 2
|
Group 3
|
Group 4
|
P- value | |
|---|---|---|---|---|---|
| Common to the three epidemiological groups (CRBSIs-HCWs-HVs) | Specific to hospital environment (CRBSIs-HCWs) | Specific to CRBSIs | Commensal (HCWs, HCWs-HVs) | ||
|
N° isolates |
24 |
20 |
12 |
9 |
NA |
|
Epidemic PFGE types (N° isolates) |
F (6), ZO (4), R (11), N (3) |
I (6), ZV (13), Y (11) |
ZK (3), ZL (5), ZH (4) |
ZB (4), E (5) |
NA |
|
MLST |
ST153, ST130, ST89, ST59 |
ST22, ST5 |
ST2 |
ST33, ST73 |
NA |
|
N°
mec
A positive isolates |
6 |
20 |
12 |
1 |
‹0.001 |
|
N°
ica
A positive isolates |
1 |
5 |
12 |
0 |
‹0.001 |
| Median sum of resistances (range) | 2 (0–6) | 5 (2–8) | 8 (5–8) | 1 (1–4) | ‹0.001 |
Legend:
NA not available.
In vitro biofilm production and correlation with the presence of the icaA gene
One hundred and twenty-seven isolates were tested for the ability to form biofilms (one CRBSI isolate was lost). Isolates were distributed in three populations according to the importance of the biomass formation: one population with absorbance values less than 1.5, one with values between 1.5 and 3, and one population with absorbance values greater than 3 (Figure 1).
Figure 1.
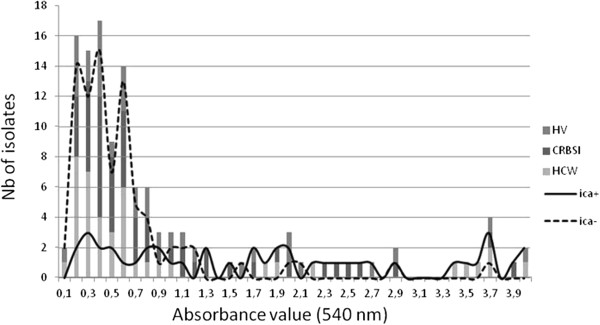
Biofilm production and correlation with the presence of the ica A in S . epidermidis population among HVs (healthy volunteers), CRBSIs (catheter-related bloodstream infections) and HCWs (healthcare workers).
Seventy-nine of the icaA negative isolates (95%) presented an absorbance value that was less than 1.5, but one of the ten strong producers (absorbance value >3) was icaA negative, suggesting a strong but not absolute correlation between the presence of the icaA gene and the capacity to form a strong biofilm.
Strong biofilm producers were mainly icaA positive commensal isolates (six from HCWs and two from HVs), but not all icaA positive isolates (especially CRBSI isolates) produced strong biofilms. These isolates were evenly distributed among the three populations (Figure 1).
Clinical data
Among the 53 CRBSI S. epidermidis isolates, 13 were associated with severe clinical symptoms, all were mec positive (P = 0.047), and all were resistant to a median of seven antibiotics (including beta-lactams), compared to resistance to only four antibiotics for the non-severe group (P = 0.03). All but one (12 out of 13) were icaA positive. Among the 11 isolates MLST typed, only two STs (single locus variants of one another) were found: ST2 (n = 8) and ST54 (n = 3).
Discussion
In a previous study, we compared a set of S. epidermidis isolates responsible for CRBSI to a set of commensal S. epidermidis isolated from HVs [3]. Although almost one-quarter of CRBSI episodes were due to isolates from PFGE types that were also found to colonize the healthy students, all severe CRBSI episodes were caused by isolates belonging to “specific” PFGE types that were not found in the community. These latter isolates shared the additional characteristics of being MDR, icaA and mecA positive and belonged to MLST ST2 or ST54 (single locus variant of ST2). To understand the role of HCWs in the complex process of CRBSI, it was important to study the relatedness of S. epidermidis isolates from the hospital environment. The prevalence of staphylococci recovered from the HCW’s hands (82%) was similar to that reported by Cimiotti et al. [18]. We found that the S. epidermidis isolated from CRBSIs were significantly resistant to more antibiotics than the isolates from HCWs. For example, a higher percentage of mupirocin-resistant strains was found among isolates from CRBSIs in comparison with HCWs. The resistance of these strains could be partly due to the use of topical mupirocin calcium ointment for the eradication of nasal carriage of methicillin-resistant S. aureus in hospitalized patients. Conversely, resistant S. epidermidis isolates and especially MRSE were significantly more prevalent in HCWs than in the HV population we previously studied [3]. These findings confirm that the resistances of HCW S. epidermidis isolates reflect an adaptation of the HCW flora to the pressure caused by the use of antibiotics in the hospital environment, which is where colonization likely occurred. Indeed, it was previously shown that new graduate nurses acquire antibiotic-resistant staphylococci over time [18]. Conversely, Hira et al. found that characteristics of coagulase-negative staphylococci isolates from healthcare personnel changed after a period of absence from the hospital with a replacement of “hospital” strains with “community” strains [6].
Twenty-six percent (14 out of 53) of all CRBSI episodes (2006–2012) were due to isolates belonging to PFGE types that were also found on the hands of the HCWs but not among the HVs. These isolates were resistant to oxacillin and to a median of five non-beta-lactam antibiotics, but very few were icaA positive. They belonged to a limited number of genetic lineages (mainly ST5 and ST22), confirming that HCWs were not only colonized by hospital-adapted S. epidermidis clones, but they also served as a reservoir and vector for transmission to patients.
ST 73 has been previously described in isolates obtained from hospital employees or families [19]; however, in our study, five isolates belonging to the genotype PFGE type E and identified as ST73 were obtained from one HCW and 4 HVs with no contact with hospital settings.
Few CRBSI episodes (15%) were due to isolates belonging to PFGE types that were common to all the three epidemiological groups (HVs, HCWs and CRBSI patients). These isolates belonged to various genetic lineages: the vast majority was oxacillin susceptible, and all were icaA negative. Finally, the majority (57%) of CRBSI episodes, including all the severe CRBSI episodes caused by ST2/ST54 icaA and mecA positive MDR isolates were due to isolates belonging to PFGE types that were neither found on the skin of HV nor on the hands of HCWs.
These results should be interpreted with caution. For example, we did not check whether the nurses were newly employed at the hospital, nor did record the turnover of the nursing staff. Moreover, chlorhexidine solutions, which are used for hand hygiene, may select specific genotypes in the hospital environment, especially on the hands of HCWs [20]. Other factors could also play a role in the transmission of these particular clones. For example, environmental contamination of MRSE has been previously documented in the literature [21]. In particular, air can become contaminated with MRSE [22]: in a recent study by Botelho et al., identical clones of S. epidermidis were recovered from both patient and indoor air samples, and some airborne isolates displayed virulence profiles and levels of biofilm accumulation similar to those found in patient isolates [23].
S. epidermidis CRBSI characteristically involves biofilm formation, which is the most important factor involved in its pathogenesis [2]. In the present study, approximately 95% of icaA negative isolates presented only weak biofilm production while 90% of the strong biofilm producers were icaA positive. PIA production does not seem to be of universal importance for biofilm formation because PIA-independent biofilm formation has been demonstrated [24]. Furthermore, some isolates from biofilm-associated infections do not harbor the ica genes [25]. In these ica negative strains, biofilm formation was mediated by proteins such as the accumulation-associated protein Aap [26] or biofilm-associated protein Bap/Bhp [27]. It is now known that an insertion sequence element called IS256 is actively involved in the modulation of biofilm formation through reversible insertion into the ica operon and its regulatory genes [28]. This insertion sequence occurs more frequently in MDR and ica-positive isolates [29]. This “phase variation” phenomenon may explain why half of our ST2/ST54 icaA and mecA positive MDR isolates were not strong in vitro biofilm producers.
ST2 (n = 12) isolates were exclusively found among CRBSIs and were responsible for the majority of clinically severe CRBSIs. This widespread ST, identified by Miragaia et al. [30], seems to be a highly successful strain because most of the nosocomial infections that occur worldwide are due to ST2 isolates. In a recent study, Du et al. examined 120 clinical S. epidermidis isolates and 204 commensal isolates in parallel (92 from HCWs and 112 from HVs) [31]. As in the present study, the ST2 isolates, almost exclusively responsible for the catheter-related infections were not recovered from HVs, were MDR and icaA positive; however, they were also IS256-positive. The authors postulated that the combination of these two genetic elements (ica operon and IS256) with antibiotic resistance leads to the success of ST2 in the hospital environment and among device-related infections.
Our study presents several limitations. First, the sample size was small (n = 62) and limited to a single hospital. Additionally, the sampling of HCWs (2012) was not at the exact same time compared with that for patients with CRBSI (2011–2012) because CRBSIs due to S. epidermidis are rare events. Furthermore, only one S. epidermidis strain per patient and per HCW was studied, which limits the interpretation of our results given the frequent polyclonality of S. epidermidis infection and colonization.
Conclusions
S. epidermidis isolates from the hospital environment (CRBSIs and HCWs) were resistant to more antibiotics than S. epidermidis colonizing HVs. Almost one third of CRBSI episodes were due to isolates belonging to PFGE types that were also found on the hands of HCWs, but not among HVs, suggesting that HCWs serve as a reservoir for transmission to patients. However, nosocomial CRBSIs, particularly clinically severe CRBSIs were mainly due to mecA positive, icaA positive S. epidermidis isolates belonging to PFGE types and MLST sequence types (ST2 and ST54) that were not found on the skin of HVs or on the hands of HCWs. These findings suggest that these S. epidermidis genotypes, through a combination of virulence and resistance, are particularly adapted to survive in the hospital environment and cause severe catheter-related infections.
Abbreviations
CRBSI: Catheter-related bloodstream infection; HCW: Healthcare worker; HV: Healthy volunteer; ica: Intercellular adhesion; ID: Identification; MALDITOF-MS: Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry; MDR: Multidrug resistance; MLST: Multilocus sequence typing; MRSE: Methicillin-resistant Staphylococcus epidermidis; PFGE: Pulsed field gel electrophoresis; PIA: Intercellular polysaccharide adhesin; SCCmec: Staphylococcal cassette chromosome mec; ST: Sequence type.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
SC and MH designed the study and drafted the manuscript. SC, BB, MH and OD analyzed and interpreted the data. AD, CNo. and CNa. performed the laboratory work, including resistance tests, molecular biology and biofilm formation study. BB helped with statistical testing and manuscript preparation. BB and MH supervised the manuscript. All authors read and approved the final manuscript.
Contributor Information
Soraya Cherifi, Email: soraya.cherifi@chu-brugmann.be.
Baudouin Byl, Email: baudouin.byl@erasme.ulb.ac.be.
Ariane Deplano, Email: ariane.deplano@erasme.ulb.ac.be.
Carole Nagant, Email: carole.nagant@erasme.ulb.ac.be.
Claire Nonhoff, Email: claire.nonhoff@erasme.ulb.ac.be.
Olivier Denis, Email: odenis@ulb.ac.be.
Marie Hallin, Email: marie_hallin@stpierre-bru.be.
Acknowledgements
This project was conducted with support from the Brugmann Foundation and from the Fonds David and Alice Van Buuren.
References
- World Health Organization. Preventing bloodstream infections from central line venous catheters. 2012. Available online at: http://www.who.int/patientsafety/implementation/bsi/en/index.html.
- Fey PD, Olson ME. Current concepts in biofilm formation of Staphylococcus epidermidis. Future Microbiol. 2010;5:917–933. doi: 10.2217/fmb.10.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherifi S, Byl B, Deplano A, Nonhoff C, Denis O, Hallin M. Comparative epidemiology of staphylococcus epidermidis isolates from patients with catheter-related bacteremia and from healthy volunteers. J Clin Microbiol. 2013;51:1541–1547. doi: 10.1128/JCM.03378-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muldrew KL, Tang Y-W, Li H, Stratton CW. Clonal dissemination of staphylococcus epidermidis in an oncology ward. J Clin Microbiol. 2008;46:3391–3396. doi: 10.1128/JCM.00115-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liakopoulos V, Petriniki E, Efthimiadi G. Clonal relatedness of methicillin-resistant coagulase-negative staphylococci in the haemodialysis unit of a single university center in Greece. Nephrol Dial Transpl. 2008;23:2599–2603. doi: 10.1093/ndt/gfn101. [DOI] [PubMed] [Google Scholar]
- Hira V, Sluijter M, Wil HF, Goessens WFH, Oh A, de Groot R, Hermans PWM, Kornelisse RF. Coagulase-negative staphylococcal skin carriage among neonatal intensive care unit personnel: from population to infection. J Clin Microbiol. 2010;48:3876–3881. doi: 10.1128/JCM.00967-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milisavljevic V, Wu F, Cimmoti J, Haas J, Della-Latta P, Larson E, Saiman L. Genetic relatedness of staphylococcus epidermidis from infected infants and staff in the neonatal intensive care unit. Am J Infect Control. 2005;33:341–347. doi: 10.1016/j.ajic.2005.02.003. [DOI] [PubMed] [Google Scholar]
- Ünal S, Hoskins J, Flokowitsch JE, Wu CY, Preston DA, Skatrud PL. Detection of methicillin-resistant staphylococci by using the polymerase chain reaction. J Clin Microbiol. 1992;30:1685–1691. doi: 10.1128/jcm.30.7.1685-1691.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maes N, Magdalena J, Rottiers S, De Gheldre Y, Struelens MJ. Evaluation of a triplex PCR assay to discriminate staphylococcus aureus from coagulase-negative staphylococci and determine methicillin resistance from blood cultures. J Clin Microbiol. 2002;40:1514–1517. doi: 10.1128/JCM.40.4.1514-1517.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhoof R, Godard C, Content J, Nyssen HJ, Hannecart-Pokorni E. Detection by polymerase chain reaction of genes encoding aminoglycoside-modifying enzymes in methicillin-resistant staphylococcus aureus isolates of epidemic phage types. Belgian study group of hospital infections (GDEPIH/GOSPIZ) J Med Microbiol. 1994;41:282–290. doi: 10.1099/00222615-41-4-282. [DOI] [PubMed] [Google Scholar]
- Lina G, Quaglia A, Reverdy ME, Leclercq R, Vandenesch F, Etienne J. Distribution of genes encoding resistance to macrolides, lincosamides, and streptogramins among staphylococci. Antimicrob Agents Chemother. 1999;43:1062–1066. doi: 10.1128/aac.43.5.1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozitskaya S, Olson ME, Fey PD, Witte W, Ohlsen K, Ziebuhr W. Clonal analysis of staphylococcus epidermidis isolates carrying or lacking biofilm-mediating genes by multilocus sequence typing. J Clin Microbiol. 2005;43:4751–4757. doi: 10.1128/JCM.43.9.4751-4757.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deplano A, Witte W, van Leeuwen WJ, Brun Y, Struelens MJ. Clonal dissemination of epidemic methicillin-resistant staphylococcus aureus in Belgium and neighboring countries. Clin Microbiol Infect. 2000;6:239–245. doi: 10.1046/j.1469-0691.2000.00064.x. [DOI] [PubMed] [Google Scholar]
- Miragaia M, Carriço JA, Thomas JC, Couto I, Enright MC, de Lencastre H. Comparison of molecular typing methods for characterization of staphylococcus epidermidis: proposal for clone definition. J Clin Microbiol. 2008;46:118–129. doi: 10.1128/JCM.01685-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JC, Vargas MR, Miragaia M, Peacock SJ, Archer GL, Enright MC. Improved multilocus sequence typing scheme for staphylococcus epidermidis. J Clin Microbiol. 2007;45:616–619. doi: 10.1128/JCM.01934-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo Y, Ito T, Ma XX, Watanabe S, Kreiswirth BN, Etienne J, Hiramatsu K. Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob Agents Chemother. 2007;51:264–274. doi: 10.1128/AAC.00165-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanovic S, Vukovic D, Dakic I, Savic B, Svabic-Vlahovic M. A modified microtiter-plate test for quantification of staphylococcal biofilm formation. J Microbiol Methods. 2000;40:175–179. doi: 10.1016/S0167-7012(00)00122-6. [DOI] [PubMed] [Google Scholar]
- Cimiotti JP, Wu F, Della-Latta P, Nesin M, Larson E. Emergence of resistant staphylococci on the hands of new graduate nurses. Infect Control Hosp Epidemiol. 2004;25:431–435. doi: 10.1086/502418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widerström M, Wiström J, Ek E, Edebro H, Monsen T. Near absence of methicillin-resistance and pronounced genetic diversity among staphylococcus epidermidis isolated from healthy persons in northern Sweden. APMIS. 2011;119:505–512. doi: 10.1111/j.1600-0463.2011.02757.x. [DOI] [PubMed] [Google Scholar]
- Milisavljevic V, Tran LP, Batmalle C, Bootsma HJ. Benzyl alcohol and ethanol can enhance the pathogenic potential of clinical staphylococcus epidermidis strains. Am J Infect Control. 2008;36:552–558. doi: 10.1016/j.ajic.2007.10.025. [DOI] [PubMed] [Google Scholar]
- Kelly S, Collins J, Maguire M, Gowing C, Flanagan M, Donnelly M, Murphy PG. An outbreak of colonization with linezolid-resistant staphylococcus epidermidis in an intensive therapy unit. J Antimicrob Chemother. 2008;61:901–907. doi: 10.1093/jac/dkn043. [DOI] [PubMed] [Google Scholar]
- Lis DO, Pacha JZ, Idzik D. Methicillin resistance of airborne coagulase-negative staphylococci in homes of persons having contact with a hospital environment. Am J Infect Control. 2009;37:177–182. doi: 10.1016/j.ajic.2008.09.013. [DOI] [PubMed] [Google Scholar]
- Botelho AM, Nunes ZD, Asensi MD, Gomes MZ, Fracalanzza SE, Figueiredo AM. Characterization of coagulase-negative staphylococci isolated from hospital indoor air and a comparative analysis between airborne and inpatient isolates of staphylococcus epidermidis. J Med Microbiol. 2012;61:1136–1145. doi: 10.1099/jmm.0.035931-0. [DOI] [PubMed] [Google Scholar]
- Rohde H, Burdelski C, Bartscht K, Hussain M, Buck F, Horstkotte MA, Knobloch JK, Heilmann C, Herrmann M, Mack D. Induction of Staphylococcus epidermidis biofilm formation via proteolytic processing of the accumulation-associated protein by staphylococcal and host proteases. Mol Microbiol. 2005;55:1883–1895. doi: 10.1111/j.1365-2958.2005.04515.x. [DOI] [PubMed] [Google Scholar]
- Arciola CR, Campoccia D, Baldassarri L, Donati ME, Pirini V, Gamberini S, Montanaro L. Detection of biofilm formation in Staphylococcus epidermidis from implant infections. Comparison of a PCR-method that recognizes the presence of ica genes with two classic phenotypic methods. J Biomed Mater Res A. 2006;76:425–430. doi: 10.1002/jbm.a.30552. [DOI] [PubMed] [Google Scholar]
- Rohde H, Burandt EC, Siemssen N, Frommelt L, Burdelski C, Wurster S, Scherpe S, Davies AP, Harris LG, Horstkotte MA, Knobloch JK, Ragunath C, Kaplan JB, Mack D. Polysaccharide intercellular adhesion or protein factors in biofilm accumulation of Staphylococcus epidermidis and Staphylococcus aureus isolated from prosthetic hip and knee joint infections. Biomaterials. 2007;28:1711–1720. doi: 10.1016/j.biomaterials.2006.11.046. [DOI] [PubMed] [Google Scholar]
- Tormo MA, Knecht E, Götz F, Lasa I, Penades JR. Bap-dependant biofilm formation by pathogenic species of staphylococcus: evidence of horizontal gene transfer? Microbiology. 2005;151:2465–2475. doi: 10.1099/mic.0.27865-0. [DOI] [PubMed] [Google Scholar]
- Valle J, Vergara-Irigaray M, Merino N, Penadés JR, Lasa I. SigmaB regulates IS256-mediated staphylococcus aureus biofilm phenotypic variation. J Bacteriol. 2007;189:2886–2896. doi: 10.1128/JB.01767-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozitskaya S, Cho SH, Dietrich K, Marre R, Naber K, Ziebuhr W. The bacterial insertion sequence element IS256 occurs preferentially in nosocomial staphylococcus epidermidis isolates: association with biofilm formation and resistance to aminoglycosides. Infect Immun. 2004;72:1210–1215. doi: 10.1128/IAI.72.2.1210-1215.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miragaia M, Thomas JC, Couto I, Enright MC, de Lencastre H. Inferring a population structure for staphylococcus epidermidis from multilocus sequence typing data. J Bacteriol. 2007;189:2540–2552. doi: 10.1128/JB.01484-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du X, Zhu Y, Song Y, Li T, Luo T, Sun G, Yang C, Cao C, Lu Y, Li M. Molecular analysis of staphylococcus epidermidis strains isolated from community and hospital environments in China. PLoS One. 2013;8:e62742. doi: 10.1371/journal.pone.0062742. [DOI] [PMC free article] [PubMed] [Google Scholar]


