Abstract
CRISPR/Cas9 -mediated DNA cleavage (CCMDC) is becoming increasingly used for efficient genome engineering. Proto-spacer adjacent motif (PAM) adjacent to target sequence is one of the key components in the design of CCMDC strategies. It has been reported that NAG sequences are the predominant non-canonical PAM for CCMDC at the human EMX locus, but it is not clear whether it is universal at other loci. In the present study, we attempted to use a GFP-reporter system to comprehensively and quantitatively test the efficiency of CCMDC with non-canonical PAMs in human cells. The initial results indicated that the effectiveness of NGA PAM for CCMDC is much higher than that of other 14 PAMs including NAG. Then we further designed another three pairs of NGG, NGA and NAG PAMs at different locations in the GFP gene and investigated the corresponding DNA cleavage efficiency. We observed that one group of NGA PAMs have a relatively higher DNA cleavage efficiency, while the other groups have lower efficiency, compared with the corresponding NAG PAMs. Our study clearly demonstrates that NAG may not be the universally predominant non-canonical PAM for CCMDC in human cells. These findings raise more concerns over off-target effects in CRISPR/Cas9-mediated genome engineering.
CRISPR (Clustered Regularly Interspaced Short Palindrome Repeats)/Cas9-mediated targeted genome engineering technologies have a broad range of research and medical applications1,2,3. The process is based on a natural bacterial immune defense system identified in Streptococcus pyogenes, and originally included three minimal components; the CRISPR-associated nuclease Cas9 (SpCas9), a specificity-determining CRISPR RNA (crRNA), and an auxiliary trans-activating crRNA (tracrRNA). In further developments, a chimeric single guide RNA (sgRNA) was generated by the fusion of crRNA and tracrRNA duplexes, which mimics the natural crRNA-tracrRNA hybrid.
The Cas9 nuclease is targeted to specific genomic loci by a specific 20 nucleotide guide sequence. Target sites must include a protospacer adjacent motif (PAM) at the 3′ end adjacent to the 20-base-pair target site; Different Cas9 use different PAMs for the target sites4. As to the Streptococcus pyogenes Cas9, the PAM sequence is NGG, which is the most widely used in customized CRISPR/Cas9-mediated DNA cleavage (CCMDC).
Although the NGG rule is generally accepted by the Cas9 research community, it was recently reported that NAG is the predominant non-canonical PAM for CCMDC in human cells5. Since it is important to minimize off-target effects in CCMDC strategies even with the double-nickase Cas9 strategy6, here we sought to investigate and compare different PAMs for CRISPR/Cas9-mediated DNA cleavage in human cells.
Results
Generation of a GFP-reporter system and optimization of genome editing conditions
We first generated a GFP-reporter system (Figure 1A) in HEK-293 cells by lentiviral transduction and selection with puromycin. Further studies were performed with cells isolated from a single puromycin-resistant colony, 293-SC1, which we used for further experiments. The copy number of GFP gene in 293-SC1 were measured by Q-PCR (Supplementary methods) and its results (Figure S1) indicated there is only one GFP gene copy per cell.
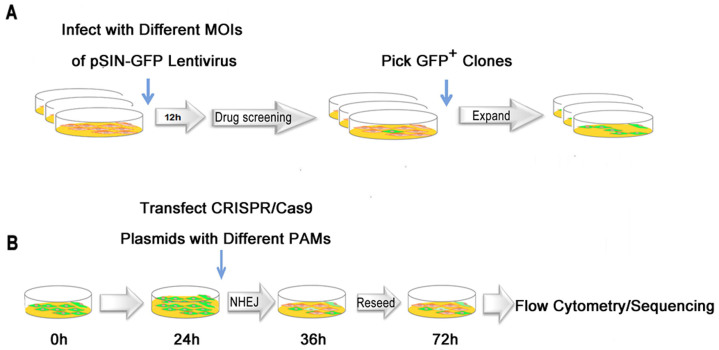
Figure 1. Generation of a GFP-reporter system and its application for CRISPR/Cas9-mediated DNA cleavage.
(A) HEK-293 cells expressing GFP were generated by transduction with lentivirus at different MOIs and selection with puromycin. Single colony were picked and expanded to obtain a homogenous cell population. The single colony isolated was named 293-SC1. (B) An illustration of the GFP-reporter system for assessing CRISPR/Cas9-mediated DNA cleavage in human cells.
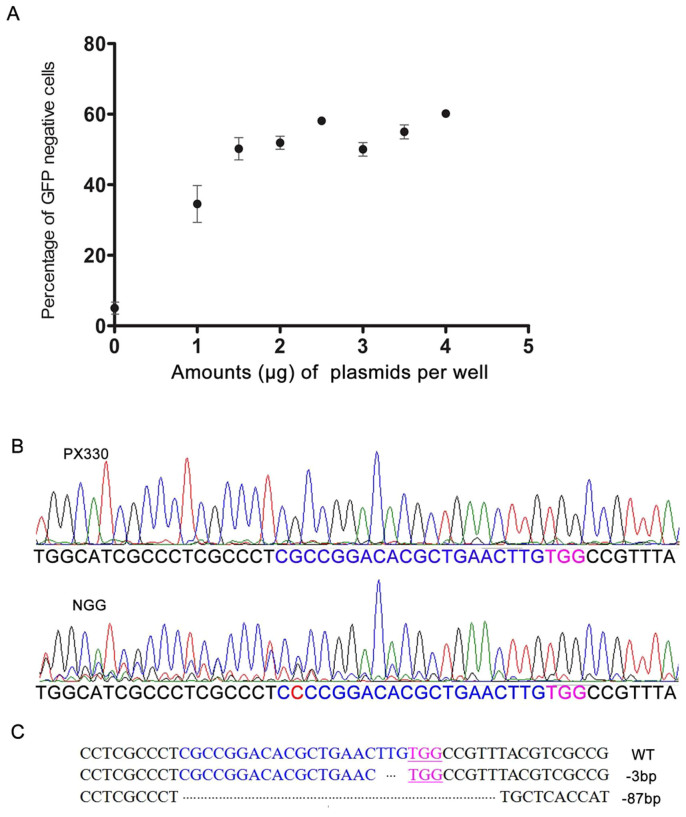
Cas9 can be programmed to induce DNA double strand breaks (DSBs) at specific genomic loci through a synthetic sgRNA, which when targeted to coding regions of genes can create frameshift indel mutations that result in a loss-of-function allele. We took advantage of the GFP-reporter system because it is easily detectable by flow cytometry. We designed and tested NGG PAM for CCMDC using the GFP-reporter system (Figure 1B). The plasmids for the CRISPR/Cas9 expression system were transfected into 293-SC1 cells and the level of effective genome editing was measured by identifying GFP-negative cells by flow cytometry. We found that transfection of 1.5 μg plasmid per well of a six-well plate was the lowest amount that could lead to maximum levels of CCMDC of approximately 51% efficiency (from 41% to 56%, Figures 2 and 3). DNA sequence chromatograms (Figure 2B) confirmed the occurrence of non-homologous end joining (NHEJ) in these cells. Two GFP-negative colonies were sequenced, revealing two deletion mutations in the GFP gene (Figure 2C). This further supported the existence of an NGG PAM for CCMDC. We rationalize that some NHEJ mutations from CCMDC do not lead to frame-shift mutations that deplete GFP, so the numbers of cells with NHEJ mutations from CCMDC was in reality greater than the 51% of cells identified as GFP negative.
Figure 2. Optimization of transfection conditions of CRISPR/Cas9 plasmid to inactivate GFP.
(A) 293-SC1 cells were transfected with different amounts of CRISPR/Cas9 plasmid. The cells were saturated with 1.5 μg of CRISPR/Cas9 plasmids. With 1.5 μg plasmids/well, approximately 51% of cells had inactivated GFP expression. (B) DNA sequence chromatograms of cells transfected with CRISPR/Cas9 are in a mass, compared with controls. (C) DNA sequence analysis of single GFP-negative colonies. WT, −3 bp, −87 bp represent wild-type, or with 3 bp deletion and 87 bp deletion of GFP gene, respectively.
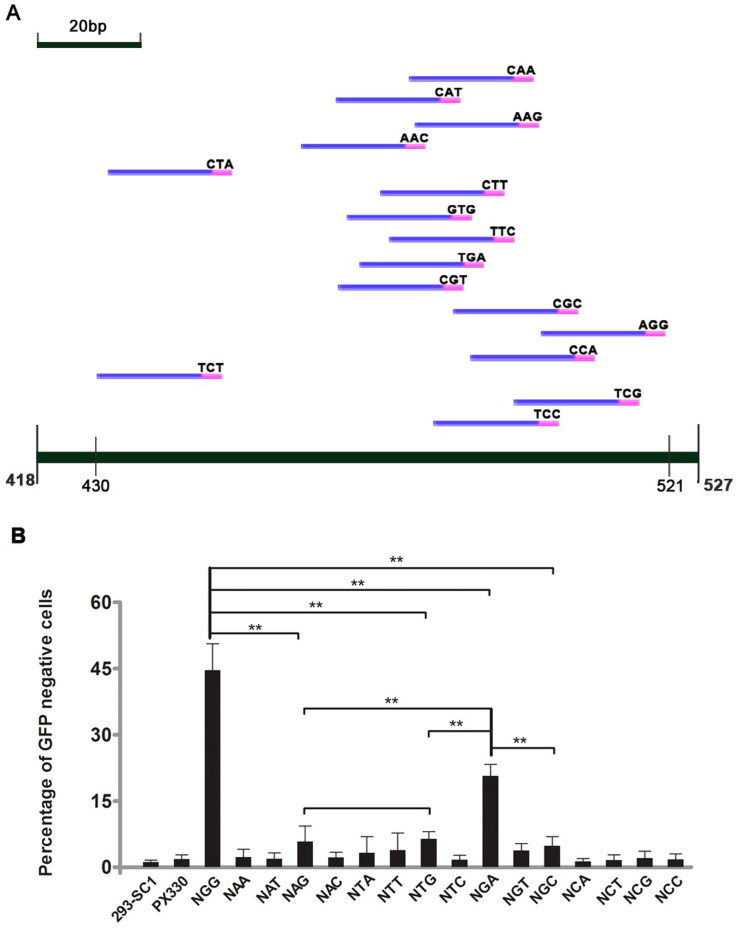
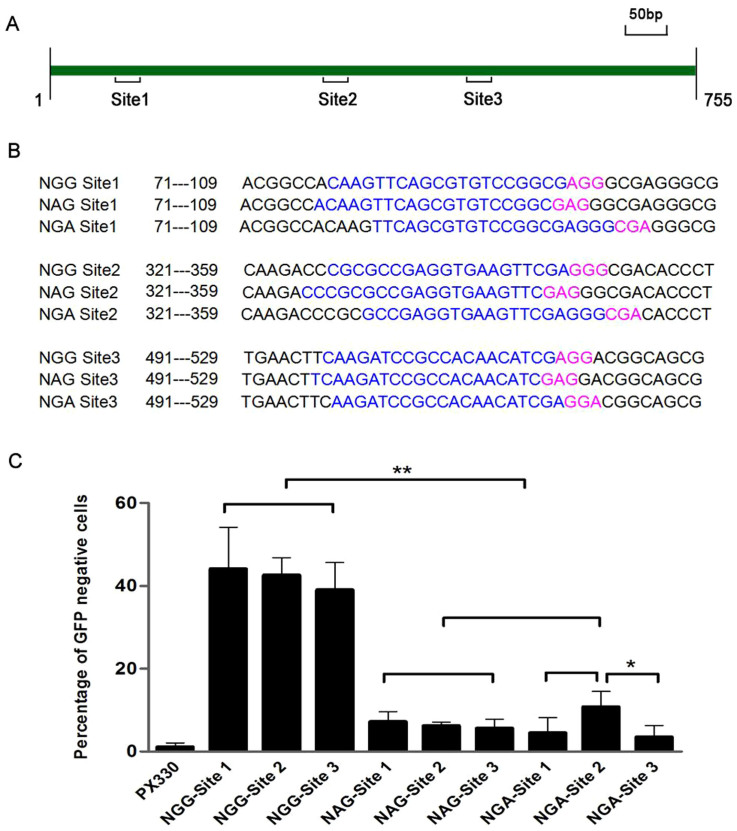
Figure 3. Effectiveness of different PAMs for CRISPR/Cas9-mediated NHEJ to inactivate GFP.
(A) Schematic diagram of targeted sites with different PAMs in GFP gene. The targeted sites with different PAMs were designed at this 90-bp window to minimize the locus bias. Targeted sites and PAMs sequences are in blue and pink, respectively. The targeting sequences were show in Table S1, S2. (B) Among these 16 PAMs (nNN), NGA PAM has the relative highest level of CRISPR/Cas9 mediated DNA cleavage except NGG. Paired sample T test method was used for analyze the data. Significant difference is as follows: **p < 0.01.
N(NN) panel CCMDC efficiency
We used the optimized transfection conditions to examine CCMDC efficiency in a panel of 16 N(NN) sites. We surprisingly found that the efficiency of NGA for CCMDC is much higher than that of other PAMs except NGG (Figure 3 and 4), including NAG, which was previously reported as the predominant non-canonical PAM at human EMX locus5. Specially, NGG, NGA and NAG PAM for CCMDC average efficiencies were 48%, 16% and 4%, respectively. This result was confirmed by transfecting SC1 cells with a targeting NGA PAM for CCMDC, which showed that more GFP negative cells were observed compared with other PAMs except NGG (Figure 3B). We picked GFP-negative colonies from NGA PAM panel and sequencing the whole GFP gene. The sequencing results of one GFP-negative colony showed that it has an 11-bp deletion mutation, including one nucleotide (A) of PAM and 10-bp PAM following sequence, which leads to frame-shift mutation of GFP gene (Figure S2). It further supported the existence of an NGA PAM for CCMDC at GFP locus.
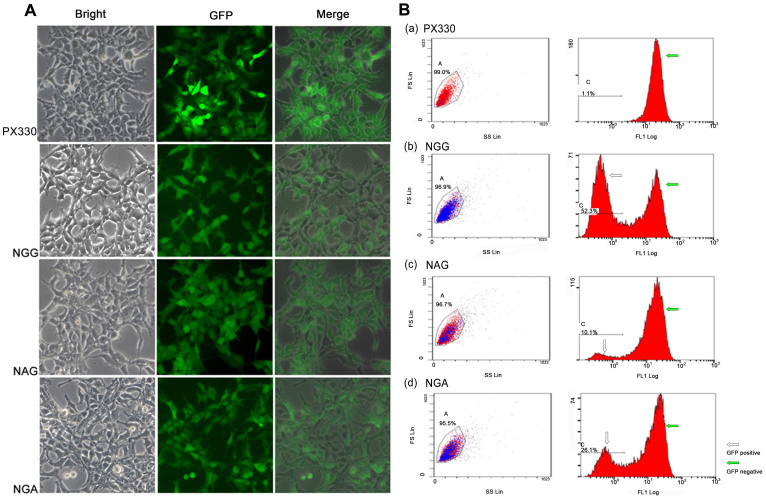
Figure 4. CRISPR/Cas9-mediated cleavage of DNA sequences to inactivate GFP.
Cleavage efficiency of different PAMs of CRISPR/Cas9 from high to low were NGG, NGA and NAG, respectively, as shown by fluorescence microscopy (A) and flow cytometry (B).
Efficiency of additional NGG/NAG/NGA PAMs in CCMDC
We designed a further three pairs of targeting oligonucleotides with NGG, NGA and NAG PAMs at different locations in the GFP gene (Figure 5A) and investigated the corresponding DNA cleavage efficiency (Figure 5A, B). Not surprisingly, all three NGG PAMs still have robust DNA cleavage efficiency. It is reasonable that site 3 of NGG PAMs have slightly lower DNA cleavage because its location is much closer to the stop code of GFP gene, which will results in a relative longer GFP gene with frameshift indel mutations to maintain its activity. We observed that one group of NGA PAMs still have higher DNA cleavage efficiency, while the other two groups of NGA PAMs have lower DNA cleavage efficiency, compared with that of NAG PAM (Figure 5C). Because we selected the same site for the design of guide sequences with different PAMs for these three pairs to avoid DNA cleavage bias, this cleavage should be comparable to measure the efficiency of CRISPR/Cas9-mediated GFP inactivation. Taken together, our study clearly demonstrates that NAG may not be the universally predominant non-canonical PAM for CCMDC in human cells, which is not consistent with the current literature.
Figure 5. NAG/NGA PAM CRISPR/Cas9-mediated NHEJ to inactivate GFP.
(A) Schematic diagram of targeted sites with NAG/NGA PAMs in GFP gene. (B) The targeting sequences are in blue and NAG/NGA PAMs sequences are in pink. (C) One group of NGA sites still has higher DNA cleavage efficiency, while another two groups of NGA sites have lower DNA cleavage efficiency, compared with that of NAG PAM. Paired sample T test method was used for analyze the data. Significant differences are as follows: *p < 0.05, **p < 0.01.
Discussion
The CRISPR/Cas9 recombination system from bacteria has been recently applied to genome engineering in different species, including Drosophila7, C. elegans8, zebrafish9,10, mouse11, rat12, and human13,14,15. Off-target effects are still a major issue for the application for the CRISPR/Cas9-mediated genome engineering. Several studies have reported off-target effects in genome manipulation both in vitro and in vivo5,12,16,17.
CRISPR/Cas9 includes two key components; the sgRNA guide sequence and PAMs. Off-target effects, in principle, will be determined by these two factors. For the guide sequence off-target effects are relatively clear, and can be detected at loci that vary at up to five nucleotides from the target sequence17,18,19. In contrast it is not clear whether PAMs will affect CCMDC. We designed this study to further test this. In the process of this work, one group reported that NAG is the predominant non-canonical PAM for CCMDC4.
Enzymatic specificity and activity are often highly dependent on reaction conditions, which at high enzyme concentrations might amplify off-target activity4. To avoid this, we firstly optimized the CRISPR/Cas9-mediated DNA cleavage using a GFP-reporter and used the minimum amount of CRISPR/Cas9 expression plasmid needed (Figure 2).
We examined the specificity of PAMs for CCMDC in human cells with a GFP-reporter system. Surprisingly, we found some NGA PAMs have relatively high CCMDC (up to 16%), while others have low levels of CCMDC. We speculate that other factors, including neighboring sequence or the guide sequence itself, may affect the different PAMs used in CCMDC. Our finding that significant off-target mutagenesis can be induced by CRISPR/Cas9 with non-NGG PAMs in three sites of the GFP gene in human cells, has important implications for the future extensive use of this genome-engineering platform. To avoid potential ‘off-target’ genomic sequences, the guide sequence for cas9 cleavage should not be followed by a PAMs with either 5′-NGG or 5′-NGA sequence. Meanwhile, it would be useful to screen and identify specific Cas9 mutants, which are optimized for genome engineering to minimize the off-target effects.
Recently, a new strategy using double nickase (DN) has been proposed5, but the presence of off-target effects due to CAS9/gRNA may still exist. Further studies are needed to provide detailed insights into the mechanism of off-target effects in DN-mediated genome engineering.
The most recent progress of using CRISPR/Cas9 to knock-out the genes at the genome level for dissection corresponding functions will be of great interest to the whole basic medical research society3. While off-target has to be highlighted and then taken into account for the careful interpretation of results from these studies, especially on human cells, because off-target on animals or plants could be minimized through outcrossing.
In summary, we have comprehensively and quantitatively examined the specificity of PAMs for CCMDC in human cells. Our findings support the idea that NAG is not the universal predominant non-canonical PAM for CCMDC in human cells. Also, for the first time, we showed that NGA PAM of CRISPR/Cas9 has a relatively high DNA cleavage efficiency. These findings raise more concerns over the design of CRISPR/Cas9 strategies for genome engineering to minimize off-target effects.
Methods
Plasmids and DNA analysis
The lentiviral vector plasmid pSIN-GFP containing a GFP gene, IRES and Puromycin gene, was generated from pSIN-EF2-Lin28-Puro (obtained from Addgene; ID 16580) using EcoR I and BamH I restriction enzyme sites. CRISPR/Cas9 plasmids were constructed as described online (http://www.genome-engineering.org/crispr/). The oligonucleotide sequences used are summarized in Tables S1, S2 and S3. Plasmid DNA and genomic DNA were isolated by standard techniques. The DNA sequencing confirmed the desired specific sequence in the constructs.
Cells and cell culture
HEK-293 cells were obtained from ATCC (CAT#CRL-1573), and grown at 37°C in 5% CO2 in Dulbecco's modified Eagle's medium (Life Technologies, Carlsbad, CA), 10% heat-inactivated fetal bovine serum, penicillin, and streptomycin.
HEK-293 cells expressing GFP were generated by transduction with lentivirus at serial dilution and selection with puromycin (0.9 μg/ml) until all cells in control dishes had detached (6 to 8 days). Drug-resistant single colonies of transduced HEK-293 cells were isolated and named 293-SC1. To maintain GFP expression, the medium for 293-SC1 culture included puromycin.
Lentiviral vector preparation
Helper-free lentivirus vector preparations of pSIN-GFP were made by transient transfection of HEK-293 cells with pSIN-GFP and helper plasmids. The culture supernatants containing the viral particles were collected and stored at -80 degree until use.
Nuclease assay for gene edit
The protocol used for nuclease assays is illustrated in Figure 1B. Briefly, 5 × 105 293-SC1 cells were seeded in six-well plates on day 1, and transfected with CRISPR/Cas plasmids by the calcium-phosphate precipitation method on day 2. The transfected 293-SC1 cells were treated with trypsin and replated in a six-well plate on day 3. Cells were harvested for flow cytometry and genomic DNAs isolation on day 4. GFP-negative colonies were marked and picked under a microscope using a pipette. The GFP sequence flanking the CRISPR target site was PCR amplified, and products were purified and then sequenced on an ABI PRISM 3730 DNA Sequencer (Sequencing primers are shown in Table S3).
Author Contributions
F.G. conceived the idea, Y.L.Z., X.L.G., F.Y.Y., L.P.Z., J.Y.Z. and X.F.T. performed the experiments, Z.B.J., J.Q. and F.G. performed data analyses, and F.G. wrote the manuscript. All authors have read and approved the final manuscript.
Supplementary Material
Supplementary File
Acknowledgments
The authors wish to thank Dr. Zhang,Feng (Massachusetts Institute of Technology, Cambridge, MA,USA) for providing CRISPR-Cas9 plasmids and comments. The authors also wish to acknowledge the comments by Dr. Xu,Ren-he (University of Connecticut Health Center, Farmington, CT, USA). This work was supported by grants from the Chinese National Program on Key Basic Research Project (973 Program, 2013CB967502, F.G.), the Natural Science Foundation of China (81201181/H1818, F.G.), Zhejiang provincial & Ministry of Health research fund for medical sciences (WKJ2013-2-023, F.G.), and Wenzhou Medical University PI starting grant (QTJ12011, F.G.).
References
- Ding Q. et al. Enhanced efficiency of human pluripotent stem cell genome editing through replacing TALENs with CRISPRs. Cell Stem Cell 12, 393–4 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi L. S. et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152, 1173–83 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O. et al. Genome-Scale CRISPR-Cas9 Knockout Screening in Human Cells. Science (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esvelt K. M. et al. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat Methods 10, 1116–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu P. D. et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol 31, 827–32 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran F. A. et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 154, 1380–9 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z. et al. Highly efficient genome modifications mediated by CRISPR/Cas9 in Drosophila. Genetics 195, 289–91 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedland A. E. et al. Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat Methods 10, 741–3 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jao L. E., Wente S. R. & Chen W. Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc Natl Acad Sci U S A 110, 13904–9 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang W. Y. et al. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol 31, 227–9 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H. et al. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell 154, 1370–9 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D. et al. Heritable gene targeting in the mouse and rat using a CRISPR-Cas system. Nat Biotechnol 31, 681–3 (2013). [DOI] [PubMed] [Google Scholar]
- Cong L. et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–23 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P. et al. RNA-guided human genome engineering via Cas9. Science 339, 823–6 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Z. et al. Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis. Proc Natl Acad Sci U S A 110, 15644–9 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y. et al. Correction of a Genetic Disease in Mouse via Use of CRISPR-Cas9. Cell Stem Cell 13, 659–62 (2013). [DOI] [PubMed] [Google Scholar]
- Pattanayak V. et al. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat Biotechnol 31, 839–43 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y. et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol 31, 822–6 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii W., Kawasaki K., Sugiura K. & Naito K. Efficient generation of large-scale genome-modified mice using gRNA and CAS9 endonuclease. Nucleic Acids Res 41, e187 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary File