Abstract
Semen contains relatively ill-defined regulatory components that likely aid fertilization, but which could also interfere with defense against infection. Each ejaculate contains trillions of exosomes, membrane-enclosed subcellular microvesicles, which have immunosuppressive effects on cells important in the genital mucosa. Exosomes in general are believed to mediate inter-cellular communication, possibly by transferring small RNA molecules. We found that seminal exosome (SE) preparations contain a substantial amount of RNA from 20 to 100 nucleotides (nts) in length. We sequenced 20–40 and 40–100 nt fractions of SE RNA separately from six semen donors. We found various classes of small non-coding RNA, including microRNA (21.7% of the RNA in the 20–40 nt fraction) as well as abundant Y RNAs and tRNAs present in both fractions. Specific RNAs were consistently present in all donors. For example, 10 (of ∼2600 known) microRNAs constituted over 40% of mature microRNA in SE. Additionally, tRNA fragments were strongly enriched for 5’-ends of 18–19 or 30–34 nts in length; such tRNA fragments repress translation. Thus, SE could potentially deliver regulatory signals to the recipient mucosa via transfer of small RNA molecules.
INTRODUCTION
Human semen contains immunosuppressive agents that induce tolerance to paternal antigens and maximize the chances of successful fertilization. This helps explain the low incidence in women of antibodies against sperm and soluble components to semen (1). On the other hand, semen's immunosuppressive properties could also contribute to the prevalence of sexually transmitted infections, which may take advantage of subdued immune responses following exposure to semen (2). For example, the most globally devastating sexually transmitted disease, human immunodeficiency virus (HIV), has proven difficult to be prevented by vaccination, even with vaccine candidates that induced strong anti-HIV immunity in the bloodstream (3,4). This could be partially explained by localized immunosuppression mediated by semen at the time of initial exposure to HIV. Similarly, decreased exposure to semen through the use of condoms helps to clear active human papillomavirus infection (5), consistent with an immunosuppressive function for semen.
Semen is a complex fluid composed of cells and seminal plasma. The immunosuppressive fraction of semen is likely contained in seminal plasma, because isolated washed sperm cells can elicit immunity against paternal antigens, while the plasma alone promotes tolerance to paternal antigens, possibly by redirecting dendritic cells towards a tolerogenic profile (6–8). Seminal plasma contains a high concentration of subcellular lipid-bound microparticles, traditionally termed prostasomes, because the majority of them were thought to derive from epithelial cells in the prostate gland (9). However, these microparticles likely originate from multiple cellular sources in the male genital tract (10) and are morphologically and molecularly consistent with exosomes, so we use the term ‘seminal exosomes’ (SE). SE have already been implicated as immunosuppressive, inhibiting lymphoproliferative responses (2), the activity of phagocytic cells (11) and natural killer cell function (12). Thus, the immunosuppressive properties of seminal plasma appear to reside at least in large part within its exosome fraction.
Exosomes are shed from most cell types and are present in a wide variety of body fluids. They can be taken up by specific target cells via membrane surface proteins, and though exosomes were initially thought to be non-specific waste from cells, evidence that exosomes are important mediators of intercellular communication is rapidly accumulating. Depending on the cell type of origin, exosomes can be involved in both immune stimulation and tolerization. For example, tumor-derived exosomes have been shown to suppress immune responses, promote metastasis, and confer drug resistance to recipient cells in cancer (13–15). In contrast, dendritic cell-derived exosomes loaded with infectious disease antigens can induce immune responses (16–18). The exosomal cargo mediating effects on recipient cells includes cytokines, growth factors and membrane proteins, as well as both messenger and microRNA (19). MicroRNAs (miRNAs) regulate biological functions through degradation or inhibition of specific mRNA targets (20–24). Other small non-coding RNAs carried by exosomes could also act as regulatory elements (25–27). Fragments of transfer RNA (tRNA), for example, can non-specifically inhibit protein translation or function as miRNAs by binding components of the RNA-induced silencing complex (28–30).
We hypothesized that SE, like other exosomes, carry non-coding RNA molecules and that the immunosuppressive effects of SE, and of seminal plasma in general, are at least in part mediated by the activities of these regulatory RNAs. In this study, we found that SE indeed contain substantial amounts of small RNAs (<100 nucleotides [nt]). In contrast to blood plasma and cell culture supernatant, where only a minor fraction of miRNA appears contained within exosomes (31,32), about half of extracellular RNA in seminal plasma purified with the exosomal fraction rather than the exosome-depleted supernatant. We then comprehensively surveyed exosomal small RNAs by deep sequencing, finding that known human miRNAs are carried by SE. SE also contained other potentially immunomodulatory small non-coding RNAs, including tRNAs, Y RNAs and protein-coding mRNA fragments. These findings point to the selective transport and delivery of small RNA molecules in SE as a potentially important mechanism of semen-mediated immunoregulation in the recipient genital mucosa.
MATERIALS AND METHODS
Semen samples
All semen samples were obtained from healthy HIV-negative men at the HIV Vaccine Trials Unit Clinic in Seattle. All protocols were approved by the Institutional Review Boards of the University of Washington and the Fred Hutchinson Cancer Research Center. Written informed consent was obtained from each donor. Donors abstained from sexual activity for > 48 h before semen donation. The entire ejaculates were collected in a sterile container, mixed with 3 ml of Roswell Park Memorial Institute media and kept on ice for <3 h prior to processing.
Isolation of exosomes from semen
Semen samples were allowed to liquefy for 20 min at room temperature. Seminal plasma, containing exosomes, was separated from the cell fraction by centrifugation at 1000 x g for 10 min. Cell debris was removed by subsequent centrifugation at 2400 x g for 30 min followed by 0.45 μm and 0.22 μm syringe filtration (Millex HA). Exosomes were purified from the entire cell- and debris-free seminal plasma (ranging from 0.9 to 4.0 ml in our donors) by ultracentrifugation over a sucrose cushion using a method adapted from Lamparski et al. (33). Up to 2.5 ml of supernatant was added to ultracentrifuge tubes and under-layered with 300 μl of a 20 mM Tris/30% sucrose/deuterium oxide (D2O) cushion (pH 7.4) (Sigma). Samples were ultra-centrifuged at 100 000 x g for 90 min at 4°C in an SW 50 swinging bucket rotor (Beckman). The upper layer was collected and ultracentrifuged again at 100 000 x g for 14 h at 4°C over a 20 mM Tris/25% sucrose/D2O cushion (pH 7.4). The upper layer of exosome-depleted seminal plasma was stored at −80°C. The 30% and 25% sucrose cushions containing the exosome fraction were pooled and brought to 15 ml with Dulbecco's phosphate-buffered saline (PBS). The exosomes were washed by centrifuging at 2400 x g through an Amicon Ultracel 100 kDa cellulose centrifugal filter with 10 ml of PBS and concentrated to a final volume of 425 μl–3.2 ml. Exosomes were stored at −80°C.
Exosome quantification and size determination
Concentration and size distribution of the isolated exosomes were measured by nanoparticle tracking analysis using a Nanosight LM-10 instrument (Nanosight) according to the manufacturer's instructions. In brief, SE samples were vortexed and serially diluted to a final dilution of 1:8000 – 1:80 000 in filtered molecular grade H2O. Blank filtered H2O was run as a negative control. 97 nm ± 3 nm polystyrene latex standards of known concentration (Life technologies) were analyzed along with the diluted exosome solution to validate the instrument. Each sample analysis was conducted for 60 s using Nanosight automatic analysis settings. Samples were evaluated in triplicate and concentration values were averaged.
Western blotting
Total protein concentrations were determined for all SE and SE-depleted plasma samples by Coomassie protein assay (Thermo Scientific) according to the manufacturer's instructions. For western blotting, 40 μg samples of total protein were heated to 95°C for 7 min in 1x DTT-containing sodium dodecyl sulphate (SDS) sample buffer (New England Biolabs) and electrophorized in 4–12% NuPage Bis-Tris-polyacrylamide gel (Novex by Life Technologies), followed by transfer to Immobilon polyvinylidene difluoride membranes (Millipore) and blocking for 1 h or overnight with 5% blotting-grade blocker (Bio-Rad). Monoclonal antibodies used for immunoblotting were anti-Ro/SSA and anti-CD63, both from Santa Cruz Biotechnology, anti-HSP-70 from Enzo Life Sciences and anti-Calnexin from MBL. The secondary antibody was horseradish peroxidase-conjugated goat-anti-mouse IgG (Thermo Pierce). Blots were developed using the Femto ECL chemiluminescent developing kit (Thermo Scientific).
Transmission electron microscopy
Preparation for transmission electron microscopy (EM) analysis was done using the method described by Thery et al. (34), except that an airfuge (Beckman-Coulter) was used to deposit fixed samples onto grids. In brief, SE samples were mixed with an equal volume of 4% PFA and deposited by airfuge onto Formvar/Carbon coated EM grids (Ted Pella). Samples were contrasted and embedded by treatment with uranyl-oxalate solution (Electron Microscopy Services), pH 7, for 5 min, followed by methyl-cellulose-uranyl-acetate (Sigma) on ice for 10 min.
RNAse protection assays and RNA isolation
For RNAse, protease and detergent protection assays, SE and SE-depleted seminal plasma samples were divided into equal fractions, which received: 1) no treatment, 2) RNAse digestion, 3) protease treatment then RNAse digestion, or 4) detergent treatment followed by protease and RNAse digestion. For RNAse treatment, samples were incubated with 12 units/ml of RNAse A and 500 units/ml of RNAse T1 (Ambion RNAse cocktail enzyme mix) for 30 min at 37°C. For protease treatment, samples were digested with pronase (Roche) at 600 μg/ml for 20 min at 40°C. Pronase was inactivated by incubation for 15 min at 80°C. For detergent treatment, samples were incubated with 1% NP-40 detergent for 15 min on ice. Total RNA for RNAse protection assay, sequencing studies or qRT-PCR assays was isolated from SE samples and from SE-depleted seminal plasma using the miRCURYTM RNA Isolation Kit (Exiqon), according to the manufacturer's instructions. All RNA samples were quantified using fluorometry (Qubit RNA Assay Kit, Life Technologies). RNA size distributions were assessed by Agilent Bioanalyzer small RNA chips.
Library generation for small RNA sequencing
Library generation and small RNA sequencing were performed by Ocean Ridge Biosciences. RNA samples for sequencing were digested with RNase-free DNase I and re-purified on RNeasy MinElute columns (Qiagen). Template DNA molecules suitable for cluster generation were prepared from 100 ng of the re-purified RNA samples using the ScriptMiner™ Small RNA-Seq Library Preparation Kit (Epicentre Biotechnologies) according to the manufacturer's instructions, except that following the RNA linker ligation, but before cDNA synthesis, the RNA was fractionated using a 10% acrylamide-urea gel. RNA fragments consisting of inserts of ∼15–40 nt were excised as a lower band, and RNA fragments containing inserts of ∼40–100 nt were excised as an upper band. These two fractions were processed separately through the rest of the library preparation procedure. Following the ScriptMiner protocol, the purified cDNA libraries were electrophorized through a freshly cast 8% native polyacrylamide gel. The library fragments of appropriate size were recovered from the gel by shaking fragments in 1X acrylamide elution buffer (0.01 M Magnesium Acetate, 0.1% SDS, 0.01 M EDTA and 0.5 M Ammonium Acetate) at 37°C at 200 rpm, passage of the eluted cDNA through a 0.45 μm filter and ethanol precipitation. The quality and size distribution of the amplified libraries were confirmed by electrophoresis on Agilent DNA 500 Bioanalyzer microfluidic chips. Libraries were quantified using the KAPA Library Quantification Kit (KK4824, Kapa Biosystems). The libraries were pooled at equimolar concentrations and diluted to 13 pmol prior to loading onto the flow cell of the cBot cluster station (Illumina). The libraries were extended and bridge-amplified to create single sequence clusters using the TruSeq SR Cluster Kit version 3 - HS (Illumina). The flow cell carrying amplified clusters was loaded on the HiSeq 2000 sequencing system (Illumina) and sequenced with 50-bp single-end reads using the TruSeq SBS Kit version 3 - HS (Illumina). Ten percent ΦX174 phage DNA was spiked into all sequencing lanes for sequencer calibration. Real-time image analysis and base calling were performed on the instrument using the HiSeq Sequencing Control Software. CASAVA software version 1.8 (Illumina) was used for de-multiplexing and production of FASTQ sequence files.
Analysis of small RNA sequence data
The FASTX application (www.molecularevolution.org) was used to trim adapter sequences from the 3’-end of the sequence read, discard any sequence of <17 nts after trimming and to collapse identical reads into single entries retaining the read count for each unique sequence. Non-redundant sequences were then aligned to genomic RNA and mRNA sequences of hg19 using Bowtie 2 (www.bowtie-bio.sf.net) (35) and matched sequences were retained and aligned to common and abundant non-coding RNAs (tRNAs, rRNAs, snoRNAs). Remaining sequences were further aligned to known mature and precursor miRNAs using the miRbase 19 database (www.mirbase.org). Additional alignments to a transcript database (Ensembl version 75, http://www.ensembl.org) (36) and the specific piRNA database piRNABank (http://pirnabank.ibab.ac.in/) were also performed (37). Alignment result files were parsed using the R statistical language (http://www.R-project.org) and the Bioconductor project (http://www.bioconductor.org) (38) to generate tables including the read count and annotation information for each entry in the target database. Raw read counts for each RNA class from all samples in the study were then joined in raw data tables. The raw reads were converted to reads per kilobase of target length per million mapped reads (39). All sequences were deposited into the GEO database (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE56076. Calculation of percent of reads by biotype was based on raw read counts of perfectly matched transcripts for each biotype compared to total reads for each library.
Digital droplet PCR
Taqman miRNA assays were used to validate sequencing data on the Biorad digital droplet PCR (ddPCR) platform. miRNA reverse transcription kits and primer probe sets were purchased from Applied Biosystems. ddPCR supermix for probes was purchased from Biorad. During RNA extraction of SE samples for qPCR, 5 μl of 5nM synthetic cel-miR-39 (Qiagen) was added during lysis to normalize extractions and reverse transcription across samples. Reverse transcription was carried out as in (40), starting with 10 ng of RNA. After reverse transcription cDNA was diluted 7-fold with water and 9 μl was used in a 20 μl final PCR reaction volume. Droplets were generated and read using a QX100 droplet digital PCR system (Biorad), according to the manufacturer's instructions.
Analysis of potential mRNA targets of abundant SE-associated miRNAs
Validated immune-related mRNAs targeted by abundant miRNAs in SE were identified using miRTarBase and confirmed by checking original publication references (41).
General statistics
Abundance of RNA biotypes in SE across donors was analyzed using Spearman's correlation coefficient. The difference in RNA content between exosomes and seminal plasma was tested using a paired t test. All statistics were calculated using Prism 6.0 (Graphpad).
RESULTS
Characterization of exosomes from semen
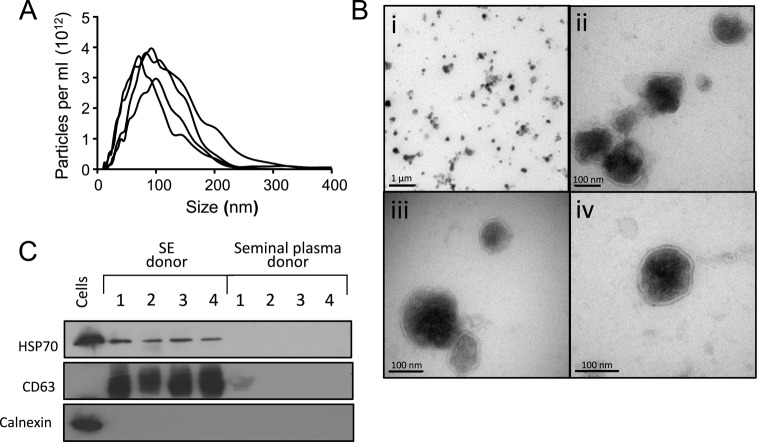
SE isolated from 12 different donors had an average diameter of 93 nm, with >75% of SE between 50 and 200 nm, the characteristic size range of exosomes and small microvesicles (Figure 1A). From these 12 donors, we isolated more than 1012 particles per ejaculate (mean concentration 1.37 × 1013/ml, range 4.7 × 1011–3.12 × 1013 SE/ml). Transmission EM confirmed that our SE samples contained lipid bilayer-bound single and small clumps of particles in the expected size range of exosomes and microvesicles (Figure 1B). Western blotting showed the presence of the universal exosome markers heat shock protein (HSP)-70 and CD63 in the exosomal but not the exosome-depleted seminal plasma fraction. Calnexin was present in cells, but not in either cell-free fraction, indicating that the vesicles are not derived from the endoplasmic reticulum (Figure 1C). These data confirm that the previously described prostasomes are indeed exosomes and microvesicles and that they are present in high concentrations in human semen.
Figure 1.
Microparticles isolated from semen exhibit the characteristic features of exosomes and microvesicles. (A) Representative particle size distribution profiles from four different semen donors. Microparticles were isolated as described in Materials and Methods and their sizes were determined by Nanoparticle Tracking Analysis. (B) Transmission electron micrographs of SE preparation. Panel i—overview of purified microvesicles showing individual exosomes and a few clumps; panels ii–iv—individual exosomes of varying densities with intact lipid bilayers. (C) Western blots on SE and exosome-depleted seminal plasma from four different donors using antibodies against the common exosome markers CD63 and HSP70, and the endoplasmic reticulum marker calnexin. Lysates from HeLa cells were used as a control.
Exosomes in semen carry extracellular RNA
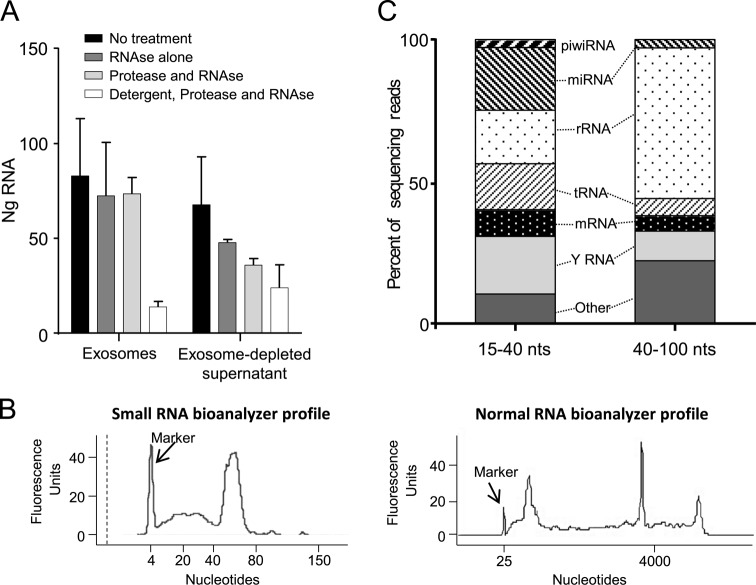
Reports in other systems demonstrate that exosomes can contain RNA and transfer it between cells, delivering regulatory micro and other small RNA molecules that modulate the function of recipient cells (20,22–24,42,43). We found that SE carry an average of 1477 ng of RNA (range 215–3534 ng, n = 8 semen donations) per sample. To test how much RNA in filtered seminal plasma is associated with the exosome fraction, we compared RNA content in three pools of SE, isolated from three separate donors each (nine donors total), with their respective pooled SE-depleted plasmas. Isolated exosomes contained an average of 55% of the total RNA isolated from our samples. The remaining 45% was in the exosome-depleted supernatant (Figure 2A). To show that the SE-associated RNA is carried within intact exosomes, we treated a part of these same samples with either RNAse alone, or with protease followed by RNAse, or with detergent followed by protease then RNAse. More than 88% of RNA in the exosome fractions was protected from combined RNAse/protease digestion. Treatment with detergent to lyse exosomes prior to protease and RNAse treatment resulted in a loss of 83% of the RNA. (Figure 2A). In contrast, 47% of RNA in the seminal plasma fraction was degraded with RNAse/protease treatment alone, confirming that much of this RNA is not protected by vesicles. These data show that a substantial fraction of extracellular RNA in 0.22 μm filtered seminal plasma is exosome-associated and that the majority of SE-associated RNA is enclosed and protected within intact microvesicles or protein complexes.
Figure 2.
SE contain protected small RNAs. (A) The majority of extracellular RNA in semen is protected in exosomes. Total RNA was isolated from SE or exosome-depleted seminal plasma from three pools of three donors each and treated with RNAse alone, with pronase followed by RNAse, or with detergent followed by pronase and RNAse prior to quantification. Equal fractions of each sample type were used for treatment and RNA isolation. Error bars are standard deviations from separate experiments using the three different pools. (B) Small and normal bioanalyzer profiles of SE RNA from one representative donor showing two distinct size fractions of 15–40 nts and 40–100 nts, and enrichment for small RNA. (C) Distribution of RNA biotypes in SE RNA libraries spanning 15–40 nts and 40–100 nts. Segments of the bar indicate the percent of reads attributed to each RNA biotype among all RNA reads that mapped perfectly to known sequences, averaged across six semen donors.
SE contain several small RNA biotypes
Given the extremely high concentration of exosomes in semen and their high RNA content, which suggests biological significance, we analyzed the RNA isolated from SE preparations in six healthy semen donors by next-generation sequencing. A table outlining RNA yields per SE count and volume is provided as Supplemental Table S1. Because a substantial fraction of exosomal RNA was <100 nt in size, and within that size range there were two clearly distinguishable peaks (20–40 nt and 40–100 nt) (Figure 2B), we prepared and sequenced the RNA in two separate size fractions encompassing the two peaks. A total of 195 828 379 raw reads were obtained from the smaller size-fraction libraries, and 212 663 023 reads from the larger. Mapped RNA was analyzed separately by biotype, and total reads for ribosomal RNA, tRNA, miRNA, piwi-RNA, Y RNA and protein-coding mRNA are given in Table 1. The distribution of reads corresponding to the RNA biotypes correlated highly between donors for the larger size-fraction libraries (median Spearman's ρ = 0.96, range 0.89–1.0) (Table 2 and Supplemental Table S2) and less so for the smaller size-fraction libraries (median Spearman ρ = 0.46, range 0.25–0.89) (Table 3 and Supplemental Table S3). Thus, most small RNA in SE mapped to known RNA sequences, and the distribution of reads across the different RNA biotypes was surprisingly consistent between semen donors.
Table 1. Total number of sequencing reads by library.
| Sequencing library | Reads mapped to genome | tRNA reads | miRNA reads | Y RNA reads | Protein-coding reads | Piwi-RNA reads | rRNA reads |
|---|---|---|---|---|---|---|---|
| 40–100 nt libraries | |||||||
| Donor 1 | 12 046 098 | 795 771 | 404 379 | 1 356 599 | 403 392 | 4288 | 6 029 886 |
| Donor 2 | 611 273 | 46 425 | 25 027 | 78 063 | 28 859 | 321 | 296 372 |
| Donor 3 | 10 774 524 | 632 011 | 204 732 | 895 982 | 881 806 | 3448 | 5 663 162 |
| Donor 4 | 13 367 134 | 462 958 | 310 106 | 1 633 277 | 569 234 | 2145 | 7 414 568 |
| Donor 5 | 8 902 958 | 588 513 | 212 253 | 664 106 | 305 079 | 2554 | 5 064 024 |
| Donor 6 | 13 727 563 | 749 751 | 447 043 | 1 303 950 | 1 097 515 | 1873 | 6 835 460 |
| 15–40 nt libraries | |||||||
| Donor 1 | 8 732 973 | 1 744 258 | 2 396 753 | 1 043 526 | 1 021 534 | 411 768 | 1 806 844 |
| Donor 2 | 9 478 197 | 2 292 092 | 1 971 024 | 1 723 312 | 672 676 | 219 097 | 1 438 469 |
| Donor 3 | 10 244 365 | 1 021 183 | 2 355 598 | 3 002 703 | 1 073 212 | 200 640 | 1 399 439 |
| Donor 4 | 9 437 222 | 1 211 062 | 1 850 259 | 3 089 860 | 761 352 | 277 548 | 1 305 193 |
| Donor 5 | 6 821 354 | 1 250 648 | 1 123 228 | 1 076 614 | 385 910 | 155 471 | 1 636 394 |
| Donor 6 | 7 218 397 | 781 771 | 1 668 618 | 861 667 | 899 625 | 165 147 | 1 710 309 |
Table 2. Percent of each RNA type in the larger size-fraction libraries (40–100 nt).
| Donor | 1 | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|---|
| tRNA | 6.61 | 7.59 | 5.87 | 3.46 | 6.61 | 5.46 |
| Ribosomal RNA | 50.06 | 48.48 | 52.56 | 55.47 | 56.88 | 49.79 |
| Y RNA | 11.26 | 12.77 | 8.32 | 12.22 | 7.46 | 9.50 |
| miRNA | 3.36 | 4.09 | 1.90 | 2.32 | 2.38 | 3.26 |
| piwi-RNA | 0.04 | 0.05 | 0.03 | 0.02 | 0.03 | 0.01 |
| Protein coding | 3.35 | 4.72 | 8.18 | 4.26 | 3.43 | 7.99 |
| Other | 25.33 | 22.28 | 23.14 | 22.25 | 23.21 | 23.98 |
Table 3. Percent of each RNA type in the smaller size-fraction libraries (15–40 nt).
| Donor | 1 | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|---|
| tRNA | 19.97 | 24.18 | 9.97 | 12.83 | 18.33 | 10.83 |
| Ribosomal RNA | 20.69 | 15.18 | 13.66 | 13.83 | 23.99 | 23.69 |
| Y RNA | 11.95 | 18.18 | 29.31 | 32.74 | 15.78 | 11.94 |
| miRNA | 27.44 | 20.80 | 22.99 | 19.61 | 16.47 | 23.12 |
| piwi-RNA | 4.72 | 2.31 | 1.96 | 2.94 | 2.28 | 2.29 |
| Protein coding | 11.70 | 7.10 | 10.48 | 8.07 | 5.66 | 12.46 |
| Other | 3.53 | 12.25 | 11.63 | 9.98 | 17.49 | 15.67 |
As expected, the 20–40 nt and 40–100 nt RNA fractions exhibited some notable differences in RNA biotypes (Figure 2C). An average of 21.7% of mappable RNA in the smaller size fractions was identified as miRNA. The larger size fractions contained only 2.89% miRNA. The larger size libraries should have carried only pre-miRNA hairpins, which are ∼70 nt long, and indeed the guide and passenger strands were present in approximately equal abundance in the larger libraries, indicating that the miRNA in the larger size fractions was primarily pre-miRNA. Piwi-RNA, which has a size range of 26–31 nt (44), was present in the smaller RNA fraction (mean 2.75%) and, as expected, was almost undetectable in the larger fractions (0.03%). Full-length tRNA ranges from 73 to 94 nt and therefore was expected in the larger fractions, where tRNA reads accounted for 5.93% of the RNA. Surprisingly, sequences mapping to tRNA were also found in abundance in the smaller libraries, accounting for 16.02% of the sequencing reads, implying that some tRNAs are fragmented in SE. Similarly, Y RNA, the 83–112 nt long non-coding RNA component of the Ro and La ribonucleoprotein complexes (45), unexpectedly accounted for a substantial fraction of the RNA in both size fractions, with the smaller size fractions again holding a higher percentage of Y RNA reads than the larger fractions (mean 19.98% versus 10.25%), implying smaller fragmented Y RNA. Ribosomal RNA constituted 18.6% of the smaller fractions but was the most abundant of all RNA biotypes in the larger fractions (52.2%). The larger fractions also contained more ‘other’ RNA, which consists mainly of retained introns, processed transcripts, signal recognition particle RNA and fragments of larger non-coding RNA (mean for all ‘other’ RNA 23.4% versus 11.8% in the smaller libraries). Protein-coding mRNA fragments were present in both fractions (9.2% for smaller fractions and 5.3% for larger). These results show that SE carry several small RNA biotypes in likely full-length and fragmented forms, which could potentially exert regulatory activities when delivered to target cells.
A few mature miRNA species are highly enriched in SE
The abundance of mature miRNA in SE from all six semen donors encouraged us to look more closely at SE-specific miRNA signatures. Of the 2578 mature human miRNA species identified in miRBase 20 (46–49), only 175 miRNAs were present in all six SE samples (6.8% of all known), and 436 miRNAs in at least one sample (16.9%). However, the miRNAs present in some but not all six donors were at extremely low abundance, never individually representing more than 0.05% of miRNA-specific sequencing reads in any given donor (Supplemental File 1).
The distribution of reads mapping to each of the 175 mature miRNA species common to all six donors correlated highly between donors (median Spearman's ρ = 0. 86, range 0.78–0.92) (Supplemental Table S4). The most abundant miRNA in all donors was let-7b, accounting for a mean of 19.5% of the total miRNA reads (median 20.1%, range 13.7–25.1%) (Figure 3A). Let-7b and the next four most abundant miRNAs (miR-148a, let-7a, miR-375 and miR-99a) together accounted for 33.1% of the total miRNA reads (median 33.0%, range 29.7–37.7%), and adding in the next five most abundant miRNAs covered 41.8% of total miRNA reads (median 42.9%, range 37.9–43.7%) (Figure 3A). We validated the hierarchical miRNA distribution derived from the sequencing reads by quantifying the first through fifth most abundant miRNas (let-7b, miR-148, let-7a, miR-375 and miR-99a) as well as two randomly selected miRNAs with much lower read counts (miR-28 and miR-103), using ddPCR assays with RNA isolated from SE from three additional, unsequenced donors. We confirmed the overall general hierarchical distribution of miRNAs, with the two lower abundant miRNAs by sequencing counts also being much lower by ddPCR copy number counts (on average 15 [miR-103] and 89 [miR-28] fold lower than the five most abundant miRNAs) (Figure 3B). These data demonstrate high consistency of miRNA loading into SE from different healthy semen donors, with only a few select miRNA species being present in abundance.
Figure 3.
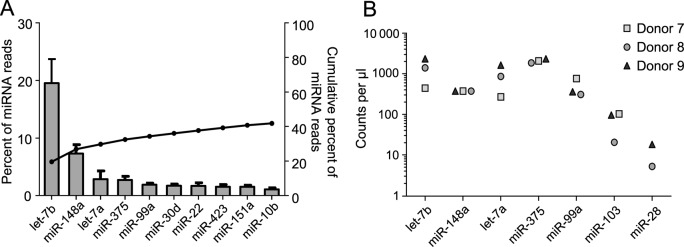
SE contain a few abundant miRNAs. (A) Top 15 most highly expressed unique miRNAs in SE. Left axis and bars: percent of total miRNA reads that mapped to each of the top 15 most abundant miRNAs. Error bars are standard deviation. Right axis and line: cumulative percent of total miRNA reads. (B) ddPCR analysis of miRNAs. RNA was isolated from three additional SE samples that had not been sequenced and assayed for miRNA levels by ddPCR assay. The five highest abundant (by sequencing) and two low abundance miRNAs were tested. Counts per μl of input cDNA are shown for each of the three donors. All reactions were run in triplicate with extremely low standard deviations (error bars always fell within each symbol and are not shown for better visual clarity). miR28 was analyzed in two donors only.
Because only a few miRNA species accounted for the majority of miRNA sequencing reads, we focused on the top 10 miRNAs for our analysis of potential regulatory targets. Within these top 10 miRNAs there was one family with an identical 5’ end seed regions: let-7b/7a, suggesting the possibility that some specific mRNA sequences are targeted by multiple SE-associated miRNAs. The current scientific literature indicates that a number of mRNA sequences that translate to immunity-related proteins could be regulated by miRNAs among the list of the top 10 miRNAs (Table 4) (50–61). Thus, it is feasible that the few abundant miRNAs in SE could influence pathways important for the immunophysiology of the genital tract. Thus, it is feasible that the few abundant miRNAs in SE could influence pathways important for the immunophysiology of the genital tract.
Table 4. Immune related mRNAs are targeted by several of the most common miRNAs in SE.
| miRNAs among the top 10 most abundant SE-associated miRNAs with validated regulatory functions | Immune-related mRNA targets |
|---|---|
| let-7 family | SOC350, CCL750, IL-23R51, CIS52, IL-1353, IL-655, IL-1054,55 |
| miR-148a | CaMKIIa56, HLA-C57, HLA-G58 |
| mir-375 | JAK259, TSLP60 |
| miR-22 | IRF561 |
Y RNA in SE
Y RNAs are evolutionarily conserved small RNAs (83–112 nt in humans) that fold into characteristic stem-loop structures similar to tRNAs. Y RNAs are known as components of the autoantigen ribonucleoprotein complexes targeted by antibodies from patients with Sjogren's syndrome and systemic lupus erythematosus (SLE) (62). Their physiological functions are poorly characterized, though they are reported to be involved in chromosomal DNA replication, in the response to UV damage, and in regulating small RNA biogenesis (63–69). Humans have four types of Y RNAs: hY1, hY3, hY4 and hY5, all of which are present in SE (Figure 4A). Sequencing reads demonstrate the presence of both full-length or ≥50 nt fragments (in the larger size-fractionated libraries) and smaller fragmented Y RNA forms (<50 nts, in the smaller size-fractionated libraries), but the relative distribution of Y RNA types differed between the two libraries. The majority of Y RNA in both libraries was hY4. The larger library contained about 30% hY5, while the smaller library contained almost no hY5. Both libraries contained small amounts of hY1 and hY3 (Figure 4A). The ratios of larger to smaller fragmented Y RNA read counts for the four Y RNA types were extremely consistent between the six donors (Table 5). Many Y RNAs in both size fractions mapped to genomic sequences annotated as pseudogenes and predicted genes (Supplemental File 2). For all four Y RNA types, smaller fragment lengths consistently centered around 30–33 nt (Supplemental Figure S1A). For hY1, hY3 and hY5, >95% of the smaller fragments mapped to the 5’ ends of Y RNA, while for hY4 ∼80% of the smaller fragments mapped to the 5’ end and 20% mapped to the 3’ end (Supplemental Figure S1B). These data indicate that each Y RNA type is loaded into SE in a distinctive manner, implying a selective process.
Figure 4.
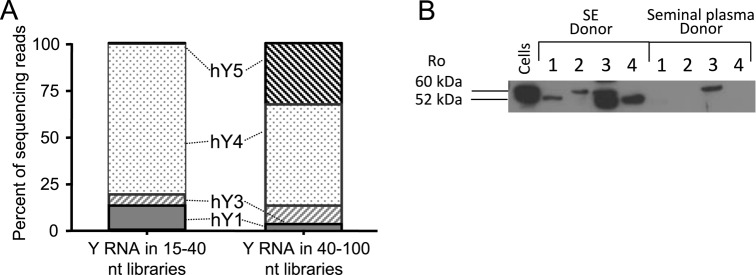
SE carry Y RNA and the binding partner Ro protein. (A) Bar chart of the distribution of Y RNA in larger or smaller libraries by type. Percent reads mapping to each type are averaged across the six donors. (B) Western blot using anti-Ro antibody against SE lysate and exosome-depleted seminal plasma. SE lysates and plasma from four different individual SE samples were tested. HeLa cell lysate served as a positive control.
Table 5. Ratio of full-length to fragmented Y RNA for each donor.
| Donor | 1 | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|---|
| hY1 | 0.29 | 0.18 | 0.16 | 0.29 | 0.26 | 0.21 |
| hY3 | 1.05 | 1.61 | 3.89 | 3.28 | 1.47 | 1.20 |
| hY4 | 0.83 | 0.73 | 0.61 | 0.65 | 0.58 | 0.69 |
| hY5 | 17.32 | 57.32 | 45.62 | 30.85 | 34.00 | 23.51 |
Y RNAs bind to the proteins Ro60 (SSA) and La (SSB) to form the Ro/La ribonucleoprotein complexes, which were first discovered and described as the auto-antigens targeted by antibodies generated in patients with auto-immune diseases. To determine whether such ribonucleoprotein complexes might also be present in SE, we stained SE protein extracts from four semen donors with anti-Ro antibody (Figure 4B). SE carried at least one of the two Ro isoforms (52 kD and 60 kD) in each donor, while only one of the four SE-depleted seminal plasma fractions contained Ro, suggesting that SE carry Ro ribonucleoprotein complexes.
tRNA in SE
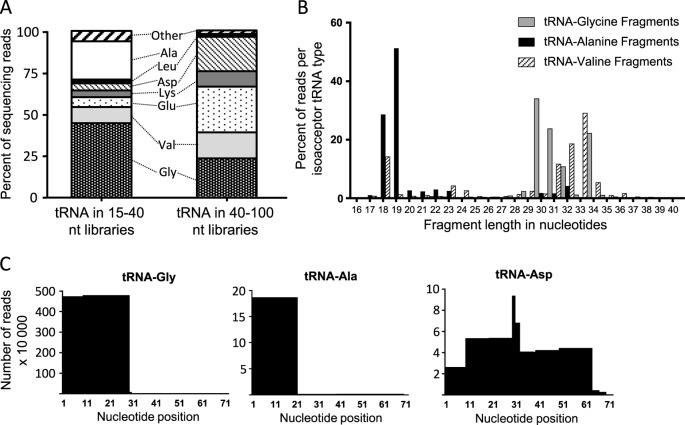
We expected to find tRNAs in the larger sized RNA libraries, since mature tRNAs are ∼72 nt long and have been shown to be carried by exosomes (70). Indeed, tRNA accounted for an average of 5.9% of reads in the larger size fractions (median 6.2%, range 3.5–7.6%) (Figure 2C and Table 2), the sequences of which corresponded to tRNAs with mature ends. Surprisingly, a substantial amount of fragmented tRNA was also present in the smaller size fractions, accounting for 16.0% (median 15.6%, range 10.0–24.2%) of total RNA (Figure 2C and Table 3). tRNA fragments in the smaller library differed in amino acid isoacceptor types from the tRNAs in the larger library (Figure 5A and Table 6), and these differences were highly consistent between donors (median Spearman's ρ = 0.93, range 0.48–1.0, for smaller fragmented tRNA; ρ = 0.95, range 0.90–1.0, for larger tRNA) (Supplemental Tables S5 and S6). For example, tRNA-Gly sequences accounted for 23.0% (median 21.8%, range 20.8–26.9%) of the tRNA in the larger libraries and 44.4% (median 40.8%, range 37.9–58%) of the smaller fragmented tRNA, while tRNA-Ala represented only 0.62% (median 0.62%, range 0.27–0.84%) of tRNA reads in the larger libraries and 23.1% (median 24.8%, range 13.5–30.0%) of smaller fragmented tRNAs. Fragment lengths also varied by isoacceptor type. For example, tRNA-Gly and tRNA-Val fragments were primarily 30–34 nt, while tRNA-Ala fragments were mostly 18–19 nt long (Figure 5B). Regardless of the length, the vast majority of tRNA fragments mapped to the 5’-ends of mature tRNAs, with the exception of a few low-abundance isoacceptor types (Met and Asp), which mapped to more diverse regions including the 3’ ends (Figure 5C). Thus, like miRNA and Y RNA, tRNA types and fragments appear to be selectively loaded into SE.
Figure 5.
SE carry tRNAs. (A) Bar chart of the distribution of tRNA in larger or smaller libraries by amino acid isoacceptor type. Percent reads mapping to each type are averaged across the six donors. (B) Histogram of fragment lengths in nucleotides for tRNA-Gly, tRNA-Ala and tRNA-Val smaller fragments. (C) Sequence coverage of tRNA fragments compared to full-length tRNAs to a resolution of 10 bps. The Y axis indicates the number of reads x 10 000.
Table 6. Ratio of full-length to fragmented tRNA for each donor.
| Donor | 1 | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|---|
| Gly | 0.64 | 0.45 | 0.57 | 0.53 | 0.42 | 0.55 |
| Ala | 0.02 | 0.06 | 0.03 | 0.02 | 0.03 | 0.01 |
| Val | 1.71 | 1.50 | 1.69 | 1.89 | 1.46 | 1.47 |
| Glu | 5.00 | 7.43 | 4.28 | 3.53 | 5.06 | 3.44 |
| Lys | 3.37 | 0.75 | 2.23 | 2.77 | 2.70 | 3.14 |
| Leu | 0.53 | 0.09 | 0.54 | 1.21 | 0.96 | 0.69 |
| Asp | 15.26 | 3.40 | 4.20 | 6.23 | 3.94 | 4.95 |
| Other | 0.27 | 0.97 | 0.15 | 0.24 | 0.25 | 0.17 |
Protein-coding fragments in SE
Though exosomes can transfer mRNAs capable of translation to target cells (20,71), it is thought that the majority of protein-coding RNA in exosomes is fragmented (26). Because we focused on sequencing small RNAs, our analysis does not include full-length mRNA in SE, but we did find protein-coding mRNA fragments of various sizes, accounting for 5.3% of the total sequencing reads in the larger libraries and 9.2% in the smaller (Figure 2C). In contrast to miRNA, Y RNA and tRNA, the distribution of the 80 most abundant mRNA fragments was much more variable between donors (median Spearman's ρ = 0.18 range 0.11–0.22, for the larger size fractions; ρ = 0.20 range 0.16–0.26, for the smaller size fractions) (Supplemental Tables S7 and S8). Only one mRNA fragment, ENST0000360737, corresponding to an uncharacterized protein, was abundant in the libraries (Table 7), accounting for over 50% of the protein-coding reads in the smaller libraries. This transcript also contains the newly described miR-4763, however the fragments carried by SE map to a 24 bp region of the 3’ UTR and not to the miRNA sequence. Other abundant protein-coding fragments are listed in Table 7. The remaining majority of protein-coding fragments in SE each represented <1% of total protein-coding reads. Despite this diversity, the relatively more abundant fragments mapped to only one small segment in the 5’ or 3’ untranslated regions (UTR) of the mRNA (Table 7), rather than randomly across sequences as would be expected if fragments were non-selectively packaged into SE. Of interest, we could not identify any of the mRNA sequences loaded into SE as potential target sequences for the co-loaded miRNAs, indicating that mRNA and miRNA molecules in SE do not associate with each other.
Table 7. mRNA fragments found in SE. Abundance number indicates the mean percent of reads mapping to protein-coding sequences (range). UTR = untranslated region.
| mRNA | Abundance | Location | Number (length) of regions represented |
|---|---|---|---|
| ENST00000360737 | 56.1% (47.4–67.6%) | 3’ UTR | 1 (24 bp) |
| ENST00000335648 | 7.3% (3.8–9.5%) | 5’ UTR | 1 (32 bp) |
| ATAD-5 | 4.6% (2.7–6.0%) | 3’ UTR | 1 (32 bp) |
| NSRP1 | 3.7% (2.6–4.7%) | 5’ UTR | 1 (34 bp) |
| ENST00000284727 | 2.6% (1.2–3.3%) | 3’ UTR | 1 (48 bp) |
DISCUSSION
Sexual intercourse is the primary route of transmission for many infections. Semen, beyond simply carrying pathogens, has well-documented immunosuppressive effects which may influence the infection of new hosts (72). The precise impact of semen on cellular functions important in pathogen response remains poorly understood and is complicated by the fact that unfractionated seminal plasma contains many components, some of which can enhance and others inhibit viral infection of mucosal cells (7,72–78). Thus, the overall role semen plays in infectious disease depends on multiple complex factors in semen itself as well as the interactions of these factors with the specific pathogens and with resident genital tract cells.
Given that exosomes are demonstrated agents of immune information (79), and that small RNAs in exosomes can affect target cells (80), we explored the small RNA content of SE for potential immunomodulatory features. We have established a protocol to isolate SE from semen, which consistently results in 1012 or more purified particles per ejaculate. These particles have the characteristics of exosomes and microvesicles: (i) lipid-bound round shape and size of 40–200 nm, (ii) purify in a density of 1.10–1.27 g/cm3 and (iii) express common exosome markers CD63 and HSP70 and lack expression of the endoplasmic-reticulum marker calnexin (Figure 1 A–C). We were surprised that a substantial fraction of RNA in 0.22 μm filtered seminal plasma was found in the exosomal fraction, because this is in contrast to findings from blood plasma and cell culture supernatant, where the majority of extracellular RNA and particularly miRNA appears independent of exosomes (21–24,31,32). The RNA in SE was particularly enriched for small RNAs ranging in lengths from 20 to 100 nts (Figure 2A and B). By deep sequencing of these small RNAs, we found that their repertoire included miRNAs with known immunomodulatory functions, as well as other small non-coding RNAs representing short, <50 nt fragments and larger, possibly full-length, fragments of >50 nts.
Numerous reports demonstrate that in cell culture conditions, intercellular communication can be mediated through miRNA delivered by exosomes, affecting processes including viral infection, immune responses and cancer progression (19,20). However, most of these experiments were conducted in somewhat artificial conditions and the relevance of this kind of messaging in vivo has yet to be demonstrated. The concentration of individual RNA molecules in each exosome is very small, and whether they can be delivered to the proper location in cells in sufficient quantities to actually regulate cellular targets in vivo remains unclear (19,81). In contrast to exosomes from blood plasma, however, exosomes in semen are delivered at a high concentration to a limited anatomical area—the recipient female genital tract. Our preliminary evidence suggests that phagocytic cells of the genital mucosa preferentially take up exosomes (data not shown). Based on measurement of the size of the vaginal cavity (82), the density of intraepithelial dendritic cells (Langerhans cells) (83) and the high number of SE in each ejaculate it is feasible that phagocytes in the recipient genital tract are exposed to and could take up thousands to tens of thousands of SE per cell, likely above the threshold for triggering physiological effects. Experiments to clarify this hypothesis are in progress in our laboratory.
Based on the observation that all cells studied thus far secrete exosomes and microvesicles (68), it is likely that SE are derived from all tissues in the male genital tract that are connected to the semen-carrying ejaculatory system. In this work we describe in SE the presence of piwi-RNA, a small RNA species expressed primarily in germline cells (84), implying that some SE are derived from spermatozoa. Additionally, polyclonal antisera that were raised against SE suggested a polytopic origin of the microvesicles, because when used for immunostaining the antisera reacted with testes, the epididymis, accessory sex glands and the prostate (10). Thus, since the complex makeup of SE cannot be mimicked by a single cell culture system, we could not compare the small RNA profiles of SE to their cells of origin. However, based on similar sequencing studies comparing exosomes and the cells that make them, it is likely that the RNA content of SE is also distinct from the cells of origin and that small RNAs are specifically packaged and secreted in SE, potentially for the purpose of altering the function of target cells (21,27,70).
miRNA
Mature functional miRNAs of 22 nts are derived from hairpin precursors of ∼70 nts. By sequencing two distinct RNA size fractions, we easily distinguished between mature and pre-miRNA and found that SE contained mature miRNAs and also pre-miRNA. miRNA accounted for ∼21.7% of the total sequencing reads in our smaller size fractions (Figure 2C). Only a small subset of miRNAs are packaged in SE; of 2578 known human miRNAs, 175 mature miRNAs were found in all six semen donors and 434 were detected at greater than the detection threshold in at least one donor. The abundance of specific miRNAs was remarkably consistent between SE isolated from different donors; the correlation coefficient of sequencing reads of the 175 common miRNAs was >0.78 for all donors (Supplemental Table S4). Furthermore, only a few miRNAs accounted for the majority of sequencing reads (Figure 3A). A comparison of sequencing studies of miRNA in exosomes isolated from saliva, breast milk, dendritic cell-T cell co-cultures, human blood plasma, prion-infected cells, glioma cells and trophoblast cultures show that SE share at most 3 of the 10 most abundant miRNAs (21,27,70,85–89). Let-7b is the most common, being an abundant exosomal miRNA in five of these eight studies, with miR-30d found in four of eight. Thus, the profile of abundant miRNAs carried by exosomes varies according to their cell of origin and may represent the different functions of exosomes in various body systems. Accordingly, exosomes in semen exhibited a unique miRNA profile that differed markedly from the profiles found in exosomes of other origins.
Micro and piwi-RNAs in seminal plasma and/or seminal microvesicles have been described in three other papers, primarily with the intent of finding RNA markers for assessing fertility (90–92). Hu et al. (90) focused on RNAs differing between the unfractionated seminal plasma from vasectomized and normal men, making a comparison to our study difficult, though 89% of miRNAs they found were also found in our study. Belleannee et al. (92) used a microarray approach to identify known miRNAs in microvesicles isolated from semen. They report 293 miRNAs detected in SE pooled from normal men; in comparison we detected 175 present in all donors, and another 271 in at least one donor. Microarray and RNAseq technologies have platform-inherent biases and the reliability of miRNA quantification likely differs between these two approaches (93). Nevertheless, we found a notable degree of overlap between the miRNAs they report and those we found; for example, all of our top 10 most abundant miRNAs are within the top 100 ranked miRNAs in their study, and the top 10 from their study are all within our top 85. The third study by Li et al. (91) suggested that in contrast to our study seminal miRNAs are mostly in the supernatant rather than the vesicle-associated fraction of seminal plasma. The authors base this conclusion on only four particular miRNAs tested (detected by qPCR), which represent <0.02% of miRNA reads in our study. This very limited universe of miRNAs studied may explain why their conclusions differed from ours.
The abundant miRNAs in SE could significantly impact the immune function of target cells in the recipient genital mucosa. A number of immune-related mRNAs are targeted by several of the most abundant miRNAs carried by SE (Table 4) and impaired production of any of these proteins in SE-exposed genital leukocytes would alter the normal immune response to pathogens, as some studies have shown. For example, miR-148a was the second most abundant miRNA in SE; it can also be induced by lipopolysaccharide in dendritic cells, where it targets calcium/calmodulin-dependent protein kinase II (CaMKII), leading to reduced MHC II expression and secretion of cytokines, and inhibition of DC-mediated T cell expansion (56). Let-7 family members, also abundant in SE, control the expression of IL-10 and IL-13 in macrophages following pathogenic stimulus, as well as IL-13 in T cells (53,55).
Whether miRNAs can be delivered by SE to genital leukocytes in sufficient quantity to target genes and change cellular function is unknown. However, given the potential for miRNAs delivered by SE to play important roles in regulating immune responses and viral infection in target cells and the selective enrichment of a few miRNAs in SE, this possibility should be studied further.
Y RNA
The presence of putatively full-length and fragmented Y RNA in SE is consistent with other sequencing studies of exosomal small RNAs (70). In a study comparing exosomal small RNA to small RNA profiles in their cells of origin, it was found that Y RNA was enriched in the small RNA fraction of exosomes as compared to cells, suggesting that it is selectively incorporated into exosomes (27). The majority of sequences we identified in the larger libraries were likely full length, because ∼99% of sequences started at the 5’ end and were 50 nts long (the length limit of our sequencing assay). In our exosomal samples hY4 was the most abundant Y RNA, in both larger and smaller sequencing libraries, while hY1 was present mainly as smaller fragments and hY5 mainly as larger or full-length Y RNA (Figure 4A). Sequences that mapped to Y RNA transcripts in Ensembl fell into one of three categories: (i) the four human Y RNAs (hY1, hY2, hY3 and hY5), (ii) pseudogenes from the four human genes and (iii) Y RNAs predicted in the Rfam database (94). We found that 45% of Y RNA in the larger libraries mapped to Y RNA pseudogenes (Supplemental File 2) (95), providing strong evidence that these genes are actually expressed.
In apoptotic cells, a caspase-dependant mechanism generates 5’ Y RNA fragments that are 33 nt in length, mapping to a predicted internal loop (69). Such 33 nt fragments represented the majority of smaller fragments in our data. Y RNA fragments of 33 nt mapping to pseudogenes have also been found as complexes in blood serum (96). Most proteins that bind Y RNAs are involved in alternative splicing and the regulation of translation, thus one function of Y RNAs could be post-transcriptional regulation of gene expression (95). Interestingly, Y RNAs are among the few host RNAs that are packaged along with viral genomes into virions, where it has been suggested that they enhance viral assembly, stability or infectivity (97,98). Otherwise, it is currently unknown what roles Y RNA or Y RNA fragments might play in exosomes, though Nolte-t’-Hoen et al. suggest that they could be involved in sorting regulatory RNAs into exosomes, in stabilizing other RNAs for export, or in guiding other RNAs to specific locations in target cells (27).
We also found that the Y RNA-binding protein Ro is carried by exosomes (Figure 4B), most likely bound to full-length Y RNAs. Ro/La/Y-RNA ribonucleoprotein complexes (RNPs) are the major autoantigenic targets in the autoimmune diseases Sjögren's syndrome and SLE. These complexes have also been found in exosomes secreted by epithelial cells derived from salivary glands, which are the main target organs for autoimmunity in Sjögren's syndrome (99). Sjögren's syndrome and SLE occur nearly 10 times as frequently in women than men, and clinical onset of SLE is most often between 15 and 35 years of age when sexual activity begins and is highest. It is therefore tempting to hypothesize that exposure to RNPs carried by SE plays a role in triggering or maintaining SLE in young women.
tRNA
We found tRNA of full-length or ≥50 nt fragments, as well as numerous smaller tRNA fragments in SE (Figure 5a). tRNA fragments have been described in other types of exosomes (27), as well as in sperm (100,101) and as abundant complexes in blood serum (96). Many studies have demonstrated that tRNA fragmentation is a specific process that depends on cell type and cell state, and tRNA fragments have recently gained attention for their regulatory potential and a possible role in stress response (25,102,103). In mammalian cells, the stress-activated ribonuclease angiogenin cleaves tRNAs in or near the anti-codon internal loop to generate fragments of 30–34 nt, known as tRNA halves (104). The 5’ ends of these tRNA halves inhibit translation by displacing components of the translation initiation complex (105). We find 5’ end tRNA fragments, predominantly 30–34 nt tRNA halves derived from tRNA-Gly and tRNA-Val, consistent with angiogenin cleavage, in SE (Figure 5b). Though only nine different 5’-tRNA fragments were tested, tRNA-Gly halves, which are the most abundant tRNA fragments in SE, were one of the few found to be significant repressors of translation (105). Additionally, smaller tRNA fragments of 19 nt have also been shown to inhibit translation non-specifically (106). These fragments require a GG motif in the 3’ end of the molecule to repress translation, which is present in the 19 nt tRNA-Ala fragments we observe in SE. Thus, multiple tRNA fragments found in SE could globally and non-specifically inhibit translation in cells that take up SE. In addition, some 3’-derived tRNA fragments have been found to be associated with argonaute proteins and thus potentially act as miRNAs. However, we observed very few 3’ tRNA fragments in SE, so tRNA-derived specific translation repression mediated by SE is unlikely.
Protein-coding mRNA fragments
It has recently become apparent that post-transcriptional cleavage of messenger RNA is a regulated and tissue-specific process (107). The mRNA fragments we detected in SE were not individually abundant, with the vast majority of transcripts each accounting for <1% of the total reads attributed to protein-coding RNA (Table 5). In addition to the diversity of different transcripts in SE, mRNA fragments found in SE were diverse across donors, with the lowest correlations of sequencing counts for any RNA biotype analyzed (Supplemental Tables S7 and S8). However, it is unlikely that these RNAs are non-selectively incorporated into SE, because for most transcripts very specific small fragments of mRNA corresponding to only one region were found, rather than fragments spread over entire transcripts (Table 5). We found that all protein-coding fragments that represented over 2% of the reads in this category mapped to untranslated rather than coding regions (Table 5). Small untranslated mRNA fragments have been proposed to play a role in regulating translation of their matching transcripts (26), though whether the particular mRNA fragments in SE function in this way are unknown.
For all of the small RNA classes we detected in SE, the distribution of cleavage fragments was not random between donors, suggesting some mechanism of selective loading or retention. The discrepancy between tRNA and Y RNA types in the larger and smaller libraries in SE further suggests a selective loading process beyond simply reflecting abundance in the cells or origin. For tRNA and Y RNA, 5’ end fragments generated by cleavage in predicted internal loops in the secondary RNA structures appear to be preferentially loaded into SE. The 5’ ends of cleaved nucleic acids are likely to be 5’-monophosphates, while the 5’ ends of 3’ fragments are more likely to be hydroxyl groups (104). miRNAs and piwi-RNAs have 5’ monophosphates as well. Thus the structural predilection for 5’-monophosphate ends could hold clues regarding the mechanism of selective RNA loading into exosomes, occurring either by selection of molecules with 5’ phosphates during the process of loading or merely by providing increased stability and longevity. More research will be required to clarify how small RNA molecules are selectively loaded into exosomes.
In conclusion, we classified the extracellular RNA in human semen, finding the majority was contained and protected within the exosomal fraction. We found that RNA loading into SE is a biologically consistent and highly selective process, with very little variation between different semen donors. Many of the most abundant small RNAs in SE have important regulatory potential for target cells that take up exosomes. Given the rapidly accumulating evidence that exosomes in other systems can deliver regulatory RNAs, and that SE carry a unique and selective profile of RNAs, it will be interesting to study their functional consequences on immune cells in the mucosa in detail. This may lead to a better understanding of how exosomal RNA-mediated regulation contributes to fertilization, but also how it may compromise natural or vaccine-induced immunity to infection.
Accession Number
GEO GSE56076
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
The authors would like to thank the semen donors who contributed to this study. M. Juliana McElrath is head of the clinic where samples were collected. Julie Czartoski, Janine Maenza, Christine Galloway and Gina Braun assisted with collecting samples and with donor recruitment. Thanks to Kim Woodrow from the Department of Bioengineering at the University of Washington and David Willoughby from Ocean Ridge Biosciences for very helpful discussions and comments. We also were assisted by the Electron Microscopy Shared Resource at the Fred Hutchinson Cancer Research Center. The content is solely the responsibility of the authors and does not necessarily represent the official view of the National Institutes of Health or other funders.
FUNDING
R01 [HD051455]; R21 [AI095023]; Royalty Research Grant from the University of Washington (to F.H.); Damon Runyon-Rachleff Innovation Award, US National Institutes of Health Transformative R01 [R01DK085714]; Stand Up To Cancer Innovative Research [SU2C-AACR-IRG1109, all to M.T.]; CFAR Emerging Opportunities Grant [P30 AI027757]; Interdisciplinary Training Grant funded by the National Cancer Institute of the National Institutes of Health [T32CA080416 to L.V.]. Source of Open Access funding: National Institutes of Health R21 [AI095023].
Conflict of interest statement. None declared.
REFERENCES
- 1.Mahdi B.M., Salih W.H., Caitano A.E., Kadhum B.M., Ibrahim D.S. Frequency of antisperm antibodies in infertile women. J. Reprod. Infertil. 2011;12:261–265. [PMC free article] [PubMed] [Google Scholar]
- 2.Kelly R.W., Holland P., Skibinski G., Harrison C., McMillan L., Hargreave T., James K. Extracellular organelles (prostasomes) are immunosuppressive components of human semen. Clin. Exp. Immunol. 1991;86:550–556. doi: 10.1111/j.1365-2249.1991.tb02968.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buchbinder S.P., Mehrotra D.V., Duerr A., Fitzgerald D.W., Mogg R., Li D., Gilbert P.B., Lama J.R., Marmor M., Del Rio C., et al. Efficacy assessment of a cell-mediated immunity HIV-1 vaccine (the Step Study): a double-blind, randomised, placebo-controlled, test-of-concept trial. Lancet. 2008;372:1881–1893. doi: 10.1016/S0140-6736(08)61591-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohen J. More woes for struggling HIV vaccine field. Science. 2013;340 doi: 10.1126/science.340.6133.667. [DOI] [PubMed] [Google Scholar]
- 5.Hogewoning C.J.A., Bleeker M.C.G., van den Brule A.J.C., Voorhorst F.J., Snijders P.J.F., Berkhof J., Westenend P.J., Meijer C.J.L.M. Condom use promotes regression of cervical intraepithelial neoplasia and clearance of human papillomavirus: a randomized clinical trial. Int. J. Cancer. 2003;107:811–816. doi: 10.1002/ijc.11474. [DOI] [PubMed] [Google Scholar]
- 6.Johansson M., Bromfield J.J., Jasper M.J., Robertson S.A. Semen activates the female immune response during early pregnancy in mice. Immunology. 2004;112:290–300. doi: 10.1111/j.1365-2567.2004.01876.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robertson S.A., Guerin L.R., Bromfield J.J., Branson K.M., Ahlström A.C., Care A.S. Seminal fluid drives expansion of the CD4+CD25+ T regulatory cell pool and induces tolerance to paternal alloantigens in mice. Biol. Reprod. 2009;80:1036–1045. doi: 10.1095/biolreprod.108.074658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Remes Lenicov F., Rodriguez Rodrigues C., Sabatte J., Cabrini M., Jancic C., Ostrowski M., Merlotti A., Gonzalez H., Alonso A., Pasqualini R.A., et al. Semen promotes the differentiation of tolerogenic dendritic cells. J. Immunol. 2012;189:4777–4786. doi: 10.4049/jimmunol.1202089. [DOI] [PubMed] [Google Scholar]
- 9.Stegmayr B., Ronquist G. Promotive effect on human sperm progressive motility by prostasomes. Urol. Res. 1982;10:253–257. doi: 10.1007/BF00255932. [DOI] [PubMed] [Google Scholar]
- 10.Renneberg H., Konrad L., Dammshauser I., Seitz J., Aumuller G. Immunohistochemistry of prostasomes from human semen. Prostate. 1997;30:98–106. doi: 10.1002/(sici)1097-0045(19970201)30:2<98::aid-pros5>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 11.Skibinski G., Kelly R.W., Harkiss D., James K. Immunosuppression by human seminal plasma–extracellular organelles (prostasomes) modulate activity of phagocytic cells. Am. J. Reprod. Immunol. 1992;28:97–103. doi: 10.1111/j.1600-0897.1992.tb00767.x. [DOI] [PubMed] [Google Scholar]
- 12.Tarazona R., Delgado E., Guarnizo M.C., Roncero R.G., Morgado S., Sánchez-Correa B., Gordillo J.J., Dejulián J., Casado J.G. Human prostasomes express CD48 and interfere with NK cell function. Immunobiology. 2011;216:41–46. doi: 10.1016/j.imbio.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 13.Taylor D.D., Gercel-Taylor C. Exosomes/microvesicles: mediators of cancer-associated immunosuppressive microenvironments. Semin. Immunopathol. 2011;33:441–454. doi: 10.1007/s00281-010-0234-8. [DOI] [PubMed] [Google Scholar]
- 14.Zhang H.G., Grizzle W.E. Exosomes and cancer: a newly described pathway of immune suppression. Clin. Cancer Res. 2011;17:959–964. doi: 10.1158/1078-0432.CCR-10-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Teng H., Hu M., Yuan L.X., Liu Y., Guo X., Zhang W.J., Jia R.Z. Suppression of inflammation by tumor-derived exosomes: a kind of natural liposome packaged with multifunctional proteins. J. Liposome Res. 2012;22:346–352. doi: 10.3109/08982104.2012.710911. [DOI] [PubMed] [Google Scholar]
- 16.Hosseini H.M., Fooladi A.A., Nourani M.R., Ghanezadeh F. The role of exosomes in infectious diseases. Inflamm. Allergy Drug Targets. 2013;12:29–37. doi: 10.2174/1871528111312010005. [DOI] [PubMed] [Google Scholar]
- 17.Giri P.K., Schorey J.S. Exosomes derived from M. Bovis BCG infected macrophages activate antigen-specific CD4+ and CD8+ T cells in vitro and in vivo. PLoS One. 2008;3:e2461. doi: 10.1371/journal.pone.0002461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hartman Z.C., Wei J., Glass O.K., Guo H., Lei G., Yang X.Y., Osada T., Hobeika A., Delcayre A., Le Pecq J.B., et al. Increasing vaccine potency through exosome antigen targeting. Vaccine. 2011;29:9361–9367. doi: 10.1016/j.vaccine.2011.09.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Turchinovich A., Samatov T.R., Tonevitsky A.G., Burwinkel B. Circulating miRNAs: cell-cell communication function? Front. Genet. 2013;4:119. doi: 10.3389/fgene.2013.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Valadi H., Ekström K., Bossios A., Sjöstrand M., Lee J.J., Lötvall J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 21.Li C.C., Eaton S.A., Young P.E., Lee M., Shuttleworth R., Humphreys D.T., Grau G.E., Combes V., Bebawy M., Gong J. Glioma microvesicles carry selectively packaged coding and noncoding RNAs which alter gene expression in recipient cells. RNA Biol. 2013;10 doi: 10.4161/rna.25281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pegtel D.M., Cosmopoulos K., Thorley-Lawson D.A., van Eijndhoven M.A., Hopmans E.S., Lindenberg J.L., de Gruijl T.D., Wurdinger T., Middeldorp J.M. Functional delivery of viral miRNAs via exosomes. Proc. Natl Acad. Sci. U.S.A. 2010;107:6328–6333. doi: 10.1073/pnas.0914843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Montecalvo A., Larregina a.T., Shufesky W.J., Beer Stolz D., Sullivan M.L.G., Karlsson J.M., Baty C.J., Gibson G.a., Erdos G., Wang Z., et al. Mechanism of transfer of functional microRNAs between mouse dendritic cells via exosomes. Blood. 2012;119:746–766. doi: 10.1182/blood-2011-02-338004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hergenreider E., Heydt S., Treguer K., Boettger T., Horrevoets A.J., Zeiher A.M., Scheffer M.P., Frangakis A.S., Yin X., Mayr M., et al. Atheroprotective communication between endothelial cells and smooth muscle cells through miRNAs. Nat. Cell Biol. 2012;14:249–256. doi: 10.1038/ncb2441. [DOI] [PubMed] [Google Scholar]
- 25.Sobala A., Hutvagner G. Transfer RNA-derived fragments: origins, processing, and functions. Wiley Interdiscip. Rev. RNA. 2011;2:853–862. doi: 10.1002/wrna.96. [DOI] [PubMed] [Google Scholar]
- 26.Batagov A.O., Kurochkin I.V. Exosomes secreted by human cells transport largely mRNA fragments that are enriched in the 3’-untranslated regions. Biol. Direct. 2013;8:12. doi: 10.1186/1745-6150-8-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nolte-'t Hoen E.N., Buermans H.P., Waasdorp M., Stoorvogel W., Wauben M.H., t Hoen P.A. Deep sequencing of RNA from immune cell-derived vesicles uncovers the selective incorporation of small non-coding RNA biotypes with potential regulatory functions. Nucleic Acids Res. 2012;40:9272–9285. doi: 10.1093/nar/gks658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maute R.L., Schneider C., Sumazin P., Holmes A., Califano A., Basso K., Dalla-Favera R. tRNA-derived microRNA modulates proliferation and the DNA damage response and is down-regulated in B cell lymphoma. Proc. Natl Acad. Sci. U.S.A. 2013; 110:1404–1409. doi: 10.1073/pnas.1206761110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cole C., Sobala A., Lu C., Thatcher S.R., Bowman A., Brown J.W., Green P.J., Barton G.J., Hutvagner G. Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. RNA. 2009;15:2147–2160. doi: 10.1261/rna.1738409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loss-Morais G., Waterhouse P.M., Margis R. Description of plant tRNA-derived RNA fragments (tRFs) associated with argonaute and identification of their putative targets. Biol. Direct. 2013;8:6. doi: 10.1186/1745-6150-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arroyo J.D., Chevillet J.R., Kroh E.M., Ruf I.K., Pritchard C.C., Gibson D.F., Mitchell P.S., Bennett C.F., Pogosova-Agadjanyan E.L., Stirewalt D.L., et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl Acad. Sci. U.S.A. 2011;108:5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Turchinovich A., Weiz L., Langheinz A., Burwinkel B. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011;39:7223–7233. doi: 10.1093/nar/gkr254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lamparski H.G., Metha-Damani A., Yao J.Y., Patel S., Hsu D.H., Ruegg C., Le Pecq J.B. Production and characterization of clinical grade exosomes derived from dendritic cells. J. Immunol. Methods. 2002;270:211–226. doi: 10.1016/s0022-1759(02)00330-7. [DOI] [PubMed] [Google Scholar]
- 34.Thery C., Clayton A., Amigorena S., Raposo G. Current Protocols in Cell Biology. Hoboken, NJ: John Wiley & Sons; 2006. pp. 3.22.21–23.22.29. [DOI] [PubMed] [Google Scholar]
- 35.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Flicek P., Amode M.R., Barrell D., Beal K., Billis K., Brent S., Carvalho-Silva D., Clapham P., Coates G., Fitzgerald S., et al. Ensembl 2014. Nucleic Acids Res. 2014;42:D749–D755. doi: 10.1093/nar/gkt1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sai Lakshmi S., Agrawal S. piRNABank: a web resource on classified and clustered Piwi-interacting RNAs. Nucleic Acids Res. 2008;36:D173–D177. doi: 10.1093/nar/gkm696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morgan M., Anders S., Lawrence M., Aboyoun P., Pages H., Gentleman R. ShortRead: a bioconductor package for input, quality assessment and exploration of high-throughput sequence data. Bioinformatics. 2009;25:2607–2608. doi: 10.1093/bioinformatics/btp450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mortazavi A., Williams B.A., McCue K., Schaeffer L., Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 40.Mitchell P.S., Parkin R.K., Kroh E.M., Fritz B.R., Wyman S.K., Pogosova-Agadjanyan E.L., Peterson A., Noteboom J., O’Briant K.C., Allen A., et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl Acad. Sci. U.S.A. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hsu S.D., Tseng Y.T., Shrestha S., Lin Y.L., Khaleel A., Chou C.H., Chu C.F., Huang H.Y., Lin C.M., Ho S.Y., et al. miRTarBase update 2014: an information resource for experimentally validated miRNA-target interactions. Nucleic Acids Res. 2014;42:D78–85. doi: 10.1093/nar/gkt1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Delorme-Axford E., Donker R.B., Mouillet J.F., Chu T., Bayer A., Ouyang Y., Wang T., Stolz D.B., Sarkar S.N., Morelli A.E., et al. Human placental trophoblasts confer viral resistance to recipient cells. Proc. Natl Acad. Sci. U.S.A. 2013;110:12048–12053. doi: 10.1073/pnas.1304718110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Umezu T., Ohyashiki K., Kuroda M., Ohyashiki J.H. Leukemia cell to endothelial cell communication via exosomal miRNAs. Oncogene. 2012;32 doi: 10.1038/onc.2012.295. [DOI] [PubMed] [Google Scholar]
- 44.Yang Q., Hua J., Wang L., Xu B., Zhang H., Ye N., Zhang Z., Yu D., Cooke H.J., Zhang Y., et al. MicroRNA and piRNA profiles in normal human testis detected by next generation sequencing. PLoS One. 2013;8:e66809. doi: 10.1371/journal.pone.0066809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lerner M.R., Boyle J.A., Hardin J.A., Steitz J.A. Two novel classes of small ribonucleoproteins detected by antibodies associated with lupus erythematosus. Science. 1981;211:400–402. doi: 10.1126/science.6164096. [DOI] [PubMed] [Google Scholar]
- 46.Griffiths-Jones S. The microRNA Registry. Nucleic Acids Res. 2004;32:D109–D111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Griffiths-Jones S., Grocock R.J., van Dongen S., Bateman A., Enright A.J. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Griffiths-Jones S., Saini H.K., van Dongen S., Enright A.J. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kozomara A., Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152–D157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bakre A., Mitchell P., Coleman J.K., Jones L.P., Saavedra G., Teng M., Tompkins S.M., Tripp R.A. Respiratory syncytial virus modifies microRNAs regulating host genes that affect virus replication. J. Gen. Virol. 2012;93:2346–2356. doi: 10.1099/vir.0.044255-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li Z., Wu F., Brant S.R., Kwon J.H. IL-23 receptor regulation by Let-7f in human CD4+ memory T cells. J. Immunol. 2011;186:6182–6190. doi: 10.4049/jimmunol.1000917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hu G., Zhou R., Liu J., Gong A.Y., Eischeid A.N., Dittman J.W., Chen X.M. MicroRNA-98 and let-7 confer cholangiocyte expression of cytokine-inducible Src homology 2-containing protein in response to microbial challenge. J. Immunol. 2009;183:1617–1624. doi: 10.4049/jimmunol.0804362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kumar M., Ahmad T., Sharma A., Mabalirajan U., Kulshreshtha A., Agrawal A., Ghosh B. Let-7 microRNA-mediated regulation of IL-13 and allergic airway inflammation. J. Allergy Clin. Immunol. 2011;128 doi: 10.1016/j.jaci.2011.04.034. [DOI] [PubMed] [Google Scholar]
- 54.Swaminathan S., Suzuki K., Seddiki N., Kaplan W., Cowley M.J., Hood C.L., Clancy J.L., Murray D.D., Mendez C., Gelgor L., et al. Differential regulation of the Let-7 family of microRNAs in CD4+ T cells alters IL-10 expression. J. Immunol. 2012;188:6238–6246. doi: 10.4049/jimmunol.1101196. [DOI] [PubMed] [Google Scholar]
- 55.Schulte L.N., Eulalio A., Mollenkopf H.J., Reinhardt R., Vogel J. Analysis of the host microRNA response to Salmonella uncovers the control of major cytokines by the let-7 family. EMBO J. 2011;30:1977–1989. doi: 10.1038/emboj.2011.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu X., Zhan Z., Xu L., Ma F., Li D., Guo Z., Li N., Cao X. MicroRNA-148/152 impair innate response and antigen presentation of TLR-triggered dendritic cells by targeting CaMKIIalpha. J. Immunol. 2010;185:7244–7251. doi: 10.4049/jimmunol.1001573. [DOI] [PubMed] [Google Scholar]
- 57.Kulkarni S., Savan R., Qi Y., Gao X., Yuki Y., Bass S.E., Martin M.P., Hunt P., Deeks S.G., Telenti A., et al. Differential microRNA regulation of HLA-C expression and its association with HIV control. Nature. 2011;472:495–498. doi: 10.1038/nature09914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tan Z., Randall G., Fan J., Camoretti-Mercado B., Brockman-Schneider R., Pan L., Solway J., Gern J.E., Lemanske R.F., Nicolae D., et al. Allele-specific targeting of microRNAs to HLA-G and risk of asthma. Am. J. Hum. Genet. 2007;81:829–834. doi: 10.1086/521200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ding L., Xu Y., Zhang W., Deng Y., Si M., Du Y., Yao H., Liu X., Ke Y., Si J., et al. MiR-375 frequently downregulated in gastric cancer inhibits cell proliferation by targeting JAK2. Cell Res. 2010;20:784–793. doi: 10.1038/cr.2010.79. [DOI] [PubMed] [Google Scholar]
- 60.Biton M., Levin A., Slyper M., Alkalay I., Horwitz E., Mor H., Kredo-Russo S., Avnit-Sagi T., Cojocaru G., Zreik F., et al. Epithelial microRNAs regulate gut mucosal immunity via epithelium-T cell crosstalk. Nat. Immunol. 2011;12:239–246. doi: 10.1038/ni.1994. [DOI] [PubMed] [Google Scholar]
- 61.Polioudakis D., Bhinge A.A., Killion P.J., Lee B.K., Abell N.S., Iyer V.R. A Myc-microRNA network promotes exit from quiescence by suppressing the interferon response and cell-cycle arrest genes. Nucleic Acids Res. 2013;41:2239–2254. doi: 10.1093/nar/gks1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mattioli M., Reichlin M. Heterogeneity of RNA protein antigens reactive with sera of patients with systemic lupus erythematosus. Description of a cytoplasmic nonribosomal antigen. Arthritis Rheum. 1974;17:421–429. doi: 10.1002/art.1780170413. [DOI] [PubMed] [Google Scholar]
- 63.Chen X., Taylor D.W., Fowler C.C., Galan J.E., Wang H.W., Wolin S.L. An RNA degradation machine sculpted by Ro autoantigen and noncoding RNA. Cell. 2013;153:166–177. doi: 10.1016/j.cell.2013.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nicolas F.E., Hall A.E., Csorba T., Turnbull C., Dalmay T. Biogenesis of Y RNA-derived small RNAs is independent of the microRNA pathway. FEBS Lett. 2012;586:1226–1230. doi: 10.1016/j.febslet.2012.03.026. [DOI] [PubMed] [Google Scholar]
- 65.Langley A.R., Chambers H., Christov C.P., Krude T. Ribonucleoprotein particles containing non-coding Y RNAs, Ro60, La and nucleolin are not required for Y RNA function in DNA replication. PLoS One. 2010;5:e13673. doi: 10.1371/journal.pone.0013673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gardiner T.J., Christov C.P., Langley A.R., Krude T. A conserved motif of vertebrate Y RNAs essential for chromosomal DNA replication. RNA. 2009;15:1375–1385. doi: 10.1261/rna.1472009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Krude T., Christov C.P., Hyrien O., Marheineke K. Y RNA functions at the initiation step of mammalian chromosomal DNA replication. J. Cell Sci. 2009;122:2836–2845. doi: 10.1242/jcs.047563. [DOI] [PubMed] [Google Scholar]
- 68.Sim S., Weinberg D.E., Fuchs G., Choi K., Chung J., Wolin S.L. The subcellular distribution of an RNA quality control protein, the Ro autoantigen, is regulated by noncoding Y RNA binding. Mol. Biol. Cell. 2009;20:1555–1564. doi: 10.1091/mbc.E08-11-1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rutjes S.A., van der Heijden A., Utz P.J., van Venrooij W.J., Pruijn G.J. Rapid nucleolytic degradation of the small cytoplasmic Y RNAs during apoptosis. J. Biol. Chem. 1999;274:24799–24807. doi: 10.1074/jbc.274.35.24799. [DOI] [PubMed] [Google Scholar]
- 70.Bellingham S.A., Coleman B.M., Hill A.F. Small RNA deep sequencing reveals a distinct miRNA signature released in exosomes from prion-infected neuronal cells. Nucleic Acids Res. 2012;40:10937–10949. doi: 10.1093/nar/gks832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zomer A., Vendrig T., Hopmans E.S., van Eijndhoven M., Middeldorp J.M., Pegtel D.M. Exosomes: Fit to deliver small RNA. Commun. Integr. Biol. 2010;3:447–450. doi: 10.4161/cib.3.5.12339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sabatté J., Lenicov F.R., Cabrini M., Rodriguez C.R., Ostrowski M., Ceballos A., Amigorena S., Geffner J. The role of semen in sexual transmission of HIV: beyond a carrier for virus particles. Microbes Infect. 2011;13:977–982. doi: 10.1016/j.micinf.2011.06.005. [DOI] [PubMed] [Google Scholar]
- 73.Poiani A. Complexity of seminal fluid: a review. Behav. Ecol. Sociobiol. 2006;60:289–310. [Google Scholar]
- 74.Peters B., Whittall T., Babaahmady K., Gray K., Vaughan R., Lehner T. Effect of heterosexual intercourse on mucosal alloimmunisation and resistance to. Lancet. 2004;363:518–524. doi: 10.1016/S0140-6736(04)15538-4. [DOI] [PubMed] [Google Scholar]
- 75.Stax M.J., van Montfort T., Sprenger R.R., Melchers M., Sanders R.W., van Leeuwen E., Repping S., Pollakis G., Speijer D., Paxton W.A. Mucin 6 in seminal plasma binds DC-SIGN and potently blocks dendritic cell. Virology. 2009;391:203–211. doi: 10.1016/j.virol.2009.06.011. [DOI] [PubMed] [Google Scholar]
- 76.Doncel G.F., Joseph T., Thurman A.R. Role of semen in HIV-1 transmission: inhibitor or facilitator? Am. J. Reprod. Immunol. 2011;65:292–301. doi: 10.1111/j.1600-0897.2010.00931.x. [DOI] [PubMed] [Google Scholar]
- 77.Münch J., Rücker E., Ständker L., Adermann K., Goffinet C., Schindler M., Wildum S., Chinnadurai R., Rajan D., Specht A., et al. Semen-derived amyloid fibrils drastically enhance HIV infection. Cell. 2007;131:1059–1071. doi: 10.1016/j.cell.2007.10.014. [DOI] [PubMed] [Google Scholar]
- 78.Kelly R.W. Immunomodulators in human seminal plasma: a vital protection for spermatozoa in the presence of infection? Int. J. Androl. 1999;22:2–12. doi: 10.1046/j.1365-2605.1999.00142.x. [DOI] [PubMed] [Google Scholar]
- 79.Hwang I. Cell-cell communication via extracellular membrane vesicles and its role in the immune response. Mol. Cells. 2013;36:105–111. doi: 10.1007/s10059-013-0154-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Taylor D.D., Gercel-Taylor C. The origin, function, and diagnostic potential of RNA within extracellular vesicles present in human biological fluids. Front. Genet. 2013;4:142. doi: 10.3389/fgene.2013.00142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Turchinovich A., Weiz L., Burwinkel B. Extracellular miRNAs: the mystery of their origin and function. Trends Biochem. Sci. 2012;37:460–465. doi: 10.1016/j.tibs.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 82.Pendergrass P.B., Belovicz M.W., Reeves C.A. Surface area of the human vagina as measured from vinyl polysiloxane casts. Gynecol. Obstet. Invest. 2003;55:110–113. doi: 10.1159/000070184. [DOI] [PubMed] [Google Scholar]
- 83.Bauer J., Bahmer F.A., Worl J., Neuhuber W., Schuler G., Fartasch M. A strikingly constant ratio exists between Langerhans cells and other epidermal cells in human skin. A stereologic study using the optical disector method and the confocal laser scanning microscope. J. Invest. Dermatol. 2001;116:313–318. doi: 10.1046/j.1523-1747.2001.01247.x. [DOI] [PubMed] [Google Scholar]
- 84.Ishizu H., Siomi H., Siomi M.C. Biology of PIWI-interacting RNAs: new insights into biogenesis and function inside and outside of germlines. Genes Dev. 2012;26:2361–2373. doi: 10.1101/gad.203786.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Munch E.M., Harris R.A., Mohammad M., Benham A.L., Pejerrey S.M., Showalter L., Hu M., Shope C.D., Maningat P.D., Gunaratne P.H., et al. Transcriptome profiling of microRNA by Next-Gen deep sequencing reveals known and novel miRNA species in the lipid fraction of human breast milk. PLoS One. 2013;8:e50564. doi: 10.1371/journal.pone.0050564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ogawa Y., Taketomi Y., Murakami M., Tsujimoto M., Yanoshita R. Small RNA transcriptomes of two types of exosomes in human whole saliva determined by next generation sequencing. Biol. Pharm. Bull. 2013;36:66–75. doi: 10.1248/bpb.b12-00607. [DOI] [PubMed] [Google Scholar]
- 87.Gallo A., Tandon M., Alevizos I., Illei G.G. The majority of microRNAs detectable in serum and saliva is concentrated in exosomes. PloS One. 2012;7:e30679-e30679. doi: 10.1371/journal.pone.0030679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Huang X., Yuan T., Tschannen M., Sun Z., Jacob H., Du M., Liang M., Dittmar R.L., Liu Y., Kohli M., et al. Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genomics. 2013;14:319. doi: 10.1186/1471-2164-14-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Luo S.S., Ishibashi O., Ishikawa G., Ishikawa T., Katayama A., Mishima T., Takizawa T., Shigihara T., Goto T., Izumi A., et al. Human villous trophoblasts express and secrete placenta-specific microRNAs into maternal circulation via exosomes. Biol. Reprod. 2009;81:717–729. doi: 10.1095/biolreprod.108.075481. [DOI] [PubMed] [Google Scholar]
- 90.Hu L., Wu C., Guo C., Li H., Xiong C. Identification of microRNAs predominately derived from testis and epididymis in human seminal plasma. Clin. Biochem. 2013 doi: 10.1016/j.clinbiochem.2013.11.009. in press. [DOI] [PubMed] [Google Scholar]
- 91.Li H., Huang S., Guo C., Guan H., Xiong C. Cell-free seminal mRNA and microRNA exist in different forms. PloS One. 2012;7:e34566–e34566. doi: 10.1371/journal.pone.0034566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Belleannee C., Legare C., Calvo E., Thimon V., Sullivan R. microRNA signature is altered in both human epididymis and seminal microvesicles following vasectomy. Hum. Reprod. 2013;28:1455–1467. doi: 10.1093/humrep/det088. [DOI] [PubMed] [Google Scholar]
- 93.Pritchard C.C., Cheng H.H., Tewari M. MicroRNA profiling: approaches and considerations. Nat. Rev. Genet. 2012;13:358–369. doi: 10.1038/nrg3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Burge S.W., Daub J., Eberhardt R., Tate J., Barquist L., Nawrocki E.P., Eddy S.R., Gardner P.P., Bateman A. Rfam 11.0: 10 years of RNA families. Nucleic Acids Res. 2013;41:D226–D232. doi: 10.1093/nar/gks1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Perreault J., Noel J.F., Briere F., Cousineau B., Lucier J.F., Perreault J.P., Boire G. Retropseudogenes derived from the human Ro/SS-A autoantigen-associated hY RNAs. Nucleic Acids Res. 2005;33:2032–2041. doi: 10.1093/nar/gki504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Dhahbi J.M., Spindler S.R., Atamna H., Boffelli D., Mote P., Martin D.I. 5’ YRNA fragments derived by processing of transcripts from specific YRNA genes and pseudogenes are abundant in human serum and plasma. Physiol. Genomics. 2013;45 doi: 10.1152/physiolgenomics.00129.2013. [DOI] [PubMed] [Google Scholar]
- 97.Garcia E.L., Onafuwa-Nuga A., Sim S., King S.R., Wolin S.L., Telesnitsky A. Packaging of host mY RNAs by murine leukemia virus may occur early in Y RNA biogenesis. J. Virol. 2009;83:12526–12534. doi: 10.1128/JVI.01219-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Onafuwa-Nuga A.A., King S.R., Telesnitsky A. Nonrandom packaging of host RNAs in moloney murine leukemia virus. J. Virol. 2005;79:13528–13537. doi: 10.1128/JVI.79.21.13528-13537.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kapsogeorgou E.K., Abu-Helu R.F., Moutsopoulos H.M., Manoussakis M.N. Salivary gland epithelial cell exosomes: A source of autoantigenic ribonucleoproteins. Arthritis Rheum. 2005;52:1517–1521. doi: 10.1002/art.21005. [DOI] [PubMed] [Google Scholar]
- 100.Kiani J., Rassoulzadegan M. A load of small RNAs in the sperm - how many bits of hereditary information. Cell Res. 2013;23:18–19. doi: 10.1038/cr.2012.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Peng H., Shi J., Zhang Y., Zhang H., Liao S., Li W., Lei L., Han C., Ning L., Cao Y., et al. A novel class of tRNA-derived small RNAs extremely enriched in mature mouse sperm. Cell Res. 2012;22:1609–1612. doi: 10.1038/cr.2012.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hurto R.L. Unexpected functions of tRNA and tRNA processing enzymes. Adv. Exp. Med. Biol. 2011;722:137–155. doi: 10.1007/978-1-4614-0332-6_9. [DOI] [PubMed] [Google Scholar]
- 103.Li Y., Zhou H. tRNAs as regulators in gene expression. Sci. China C Life Sci. 2009;52:245–252. doi: 10.1007/s11427-009-0039-y. [DOI] [PubMed] [Google Scholar]
- 104.Yamasaki S., Ivanov P., Hu G.F., Anderson P. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J. Cell Biol. 2009;185:35–42. doi: 10.1083/jcb.200811106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ivanov P., Emara M.M., Villen J., Gygi S.P., Anderson P. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol. Cell. 2011;43:613–623. doi: 10.1016/j.molcel.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sobala A., Hutvagner G. Small RNAs derived from the 5' end of tRNA can inhibit protein translation in human cells. RNA Biol. 2013;10:553–563. doi: 10.4161/rna.24285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mercer T.R., Dinger M.E., Bracken C.P., Kolle G., Szubert J.M., Korbie D.J., Askarian-Amiri M.E., Gardiner B.B., Goodall G.J., Grimmond S.M., et al. Regulated post-transcriptional RNA cleavage diversifies the eukaryotic transcriptome. Genome Res. 2010;20:1639–1650. doi: 10.1101/gr.112128.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.