Abstract
In mammalian mitochondria, 22 species of tRNAs encoded in mitochondrial DNA play crucial roles in the translation of 13 essential subunits of the respiratory chain complexes involved in oxidative phosphorylation. Following transcription, mitochondrial tRNAs are modified by nuclear-encoded tRNA-modifying enzymes. These modifications are required for the proper functioning of mitochondrial tRNAs (mt tRNAs), and the absence of these modifications can cause pathological consequences. To date, however, the information available about these modifications has been incomplete. To address this issue, we isolated all 22 species of mt tRNAs from bovine liver and comprehensively determined the post-transcriptional modifications in each tRNA by mass spectrometry. Here, we describe the primary structures with post-transcriptional modifications of seven species of mt tRNAs which were previously uncharacterized, and provide revised information regarding base modifications in five other mt tRNAs. In the complete set of bovine mt tRNAs, we found 15 species of modified nucleosides at 118 positions (7.48% of total bases). This result provides insight into the molecular mechanisms underlying the decoding system in mammalian mitochondria and enables prediction of candidate tRNA-modifying enzymes responsible for each modification of mt tRNAs.
INTRODUCTION
Mitochondria, organelles present in most eukaryotic cells, provide the chemical energy required by living cells in the form of adenosine triphosphate (ATP), which is synthesized by the electron transport chain and oxidative phosphorylation (1). Mitochondria contain their own genomic DNA, called mitochondrial (mt)DNA, and unique transcription and translation machinery that converts their genetic information into proteins. In mammals, the mtDNA is a circular double-stranded DNA, ∼16 kilobase pairs (kb) in length, which contains 13 genes encoding essential subunits of the respiratory chain complexes and 24 RNA genes (2 ribosomal RNAs and 22 tRNAs) required for mitochondrial protein synthesis.
The mammalian mitochondrial decoding system differs from the canonical decoding system by its use of four non-universal codons (Table 1) (2): AUA for Met, UGA for Trp and AGR (R = A or G) for Stop. The 60-sense codons are deciphered by 22 species of mitochondrial tRNAs, which constitute the smallest set of tRNAs necessary to translate all sense codons among all kingdoms of life, including other organelle decoding systems. Post-transcriptional modifications at the first letters of tRNA anticodons play a critical role in establishing this minimal decoding system. To reduce the number of tRNA species, each of eight family boxes in mitochondria is decoded by only a single tRNA. The tRNAs responsible for the family boxes frequently have unmodified uridines (U34) at the first (wobble) position of an anticodon. According to Crick's wobble rule (3), U34 can recognize only A and G at the third position of a codon. In the decoding systems of some bacteria and most organelles, U34 can base-pair with any of the four bases by the so-called ‘four-way wobble rule’ (2) or ‘super wobbling’ (4). The conformational flexibility of U34 is thought to enable U:U and U:C pairing. On the other hand, tRNAs responsible for two-codon sets ending in purines (NNR; N = any four nucleotides) have modified uridines (xm5s2U-type) at their wobble positions (5). In general, xm5s2U-type modification restricts conformational flexibility of the wobble base, thereby strengthening recognition of NNR codons and preventing misrecognition of codons ending in pyrimidines (NNY; Y = U and C). We previously discovered that taurine-containing modified uridines in mammalian mt tRNAs are responsible for NNR codon sets: 5-taurinomethyluridine (τm5U) at the wobble position of human and bovine mt tRNALeu(UUR) (6), and 5-taurinomethyl-2-thiouridine (τm5s2U) at the wobble position of human and bovine mt tRNALys (6). Subsequently, τm5s2U was also found at the wobble position of the bovine mt tRNAs for Glu and Gln (7). Determining the proper usage of unmodified or modified U34 in tRNAs would reveal a fundamental principle of the minimal decoding system in mammalian mitochondria.
Table 1. Codon–anticodon pairing in the bovine mitochondrial genetic code.
| Codon | Amino acid (anticodon) | Codon | Amino acid (anticodon) | Codon | Amino acid (anticodon) | Codon | Amino acid (anticodon) |
|---|---|---|---|---|---|---|---|
| UUU | Phe | UCU | UAU | Tyr | UGU | Cys | |
| UUC | (GAA) | UCC | Ser | UAC | (QUA) | UGC | (GCA) |
| UUA | Leu | UCA | (UGA) | UAA | stop | UGA | Trp |
| UUG | (τm5UAA) | UCG | UAG | UGG | (τm5UCA) | ||
| CUU | CCU | CAU | His | CGU | |||
| CUC | Leu | CCC | Pro | CAC | (QUG) | CGC | Arg |
| CUA | (UAG) | CCA | (UGG) | CAA | Gln | CGA | (UCG) |
| CUG | CCG | CAG | (τm5s2UUG) | CGG | |||
| AUU | Ile | ACU | AAU | Asn | AGU | Ser | |
| AUC | (GAU) | ACC | Thr | AAC | (QUU) | AGC | (GCU) |
| AUA | Met | ACA | (UGU) | AAA | Lys | AGA | stop |
| AUG | (f5CAU) | ACG | AAG | (τm5s2UUU) | AGG | ||
| GUU | GCU | GAU | Asp | GGU | |||
| GUC | Val | GCC | Ala | GAC | (QUC) | GGC | Gly |
| GUA | (UAC) | GCA | (UGC) | GAA | Glu | GGA | (UCC) |
| GUG | GCG | GAG | (τm5s2UUC) | GGG |
Non-universal genetic codes are denoted in bold type. AUA: Ile (universal), Met (mitochondria); UGA: stop (universal), Trp (mitochondria); AGA/G: Arg (universal), stop (mitochondria).
5-formylcytidine (f5C), another unique modification in mammalian mitochondria (8), is present at the wobble position of mt tRNAMet. Biochemical studies show that f5C is required for recognition of the non-universal AUA codon, in addition to the canonical AUG codon (9,10). Possible base pairing between f5C and A was demonstrated by a crystallographic study using an f5C-containing anticodon stem-loop bound to the 30S subunit of Thermus thermophilus (11).
Defective mitochondrial translation results in mitochondrial dysfunction, ultimately causing pathological consequences (12). Numerous pathogenic mutations associated with mitochondrial diseases have been found in mtDNA (http://www.mitomap.org/MITOMAP) (13). These pathogenic mutations are maternally inherited. Among over 400 pathogenic mutations compiled to date, ∼200 have been mapped to mt tRNA genes; these mutations diminish biogenesis, stability and function of tRNAs (2,12,14–15). Two major subgroups of mitochondrial encephalomyopathies are caused primarily by point mutations in mt RNAs: mitochondrial encephalopathy, lactic acidosis and stroke-like syndrome (MELAS), caused by a mutation in mt tRNALeu(UUR); and myoclonus epilepsy with ragged-red fibers (MERRF), caused by a mutation in mt tRNALys. Approximately 80% of MELAS patients have an A-to-G mutation at position 3243 (A3243G) (16) in the mt tRNALeu(UUR) gene, and another 10% have a T-to-C mutation at position 3271 (17). MERRF patients have an A8344G mutation in the mt tRNALys gene (18). We previously reported that τm5U and τm5s2U were not present in mutant mt tRNALeu(UUR) harboring the A3243G or T3271C mutation (19), or in mt tRNALys harboring the A8344G mutation (20). We also confirmed the absence of taurine modifications in MELAS patient tissues harboring A3243G, G3244A, T3258C, T3271C or T3291C mutations (21), as well as in MERRF patients harboring the A8344G mutation (22). These pathogenic point mutations are assumed to act as negative determinants of τm5(s2)U biogenesis. In MELAS, the absence of τm5U in mt tRNALeu(UUR) results in a defect in decoding the UUG codon, leading to lower expression of the UUG-rich protein ND6 (23). Similarly, in MERRF the absence of both 5-taurinomethyl and 2-thio groups of τm5s2U in mt tRNALys leads to severe translation failure of both types of AAR codon (24). These observations imply that deficiency in modification of mt tRNA plays a key role in molecular pathogenesis. As mentioned earlier, unmodified U34 can read any of four bases at the third letter of codons in a family box by the four-way wobbling. However, unmodified U34 is only used in tRNAs responsible for family box codons in which at least one G or C is present at the first or second letter of codons. If the codon–anticodon interaction is stabilized by one or two GC pairing at the first two base pairings, U34 is considered to read any of four bases in the family boxes. In the case of mt tRNALeu(UUR) and mt tRNALys, there is no G or C at the first or second letter of their cognate codons. This is the reason why unmodified U34 in the mutant tRNAs cannot decipher cognate codons efficiently, not by expanding their decoding capacity.
Mitochondrial diseases are also caused by pathogenic mutations in nuclear-encoded genes (2), including genes encoding translation factors, aminoacyl-tRNA synthetases, tRNA processing enzymes and tRNA-modifying enzymes. Loss-of-function mutations in these genes hamper the biogenesis and function of mt tRNAs. Several instances of pathogenic point mutants in tRNA-modifying enzymes have been reported to date: mitochondrial myopathy and sideroblastic anemia (MLASA), acute infantile liver failure and hypertrophic cardiomyopathy with lactic acidosis are associated with pathogenic mutations in PUS1 (25), MTU1 (26) and MTO1 (27,28), respectively.
To gain more insight into the molecular basis of the mitochondrial decoding system, as well as the molecular pathogenesis of human diseases caused by deficiencies in mt tRNA modifications, it is necessary to obtain a complete picture of post-transcriptional modifications in all 22 species of mammalian mt tRNAs. To date, 11 species of human or bovine mt tRNAs have been sequenced and their post-transcriptional modifications determined (2). In previous work, we isolated all 22 species of bovine mt tRNAs (29). By analyzing these tRNAs by mass spectrometry, we determined the post-transcriptional modifications of seven species of mitochondrial tRNAs that have never been reported, and provide some revised information regarding the modified bases in five other mitochondrial tRNAs. In total, we identified 15 species of modified nucleosides at 118 positions in 22 species of bovine mt tRNAs. We discuss the basic principles of the mitochondrial decoding system in mammals, and propose candidate tRNA-modifying enzymes whose roles remain to be confirmed experimentally.
MATERIALS AND METHODS
Isolation of individual mitochondrial tRNAs from bovine liver
Bovine liver RNA was prepared as described previously (6,29). Briefly, crude RNA was extracted from buffer-homogenized bovine liver by phenol extraction, and the tRNA fraction was roughly concentrated by anion exchange chromatography. Individual tRNAs were isolated by chaplet column chromatography (29). Seventeen of them were isolated homogeneously based on the polyacrylamide gel electrophoresis analysis. Five mt tRNAs for Leu(UUR), Asn, Thr, Met and Pro were isolated as major bands but had some minor bands. However, the qualities of the isolated tRNAs are sufficient enough to analyze post-transcriptional modifications.
Cyanoethylation of pseudouridine in tRNA
Cyanoethylation of tRNA was performed basically as described (30). Eight micrograms of isolated tRNA dissolved in less than 4 μl of Milli-Q water was added to 30 μl of 41% (v/v) ethanol/1.1 M trimethylammonium acetate (pH 8.6). After addition of 4 μl of acrylonitrile (Wako Pure Chemical Industries), the mixture was incubated at 70°C for 2 h, lyophilized and dissolved in Milli-Q water. The solution was then subjected to RNase digestion and analyzed by RNA mass spectrometry, as described below.
RNA mass spectrometry
RNA fragments digested by RNases were analyzed by mass spectrometry as described previously (31,32). In brief, 2–5 ng of isolated tRNA was digested with RNase T1 (Epicentre) or RNase A (Ambion) and analyzed by an LTQ Orbitrap mass spectrometer (Thermo Scientific) with a nano-electrosprayer connected with a splitless nanoflow high pressure liquid chromatography system (DiNa, KYA Technologies). Alternatively, 0.4–2 μg isolated tRNA was digested with RNase T1 and analyzed on a LCQ DUO ion-trap mass spectrometer with an ESI (electrospray ionization) source (Thermo Finnigan) and HP1100 liquid chromatography system (Agilent Technologies) in negative ion detection mode. Nucleoside analysis was performed as described previously (31). In brief, about 4 μg of isolated tRNA was digested to nucleosides with nuclease P1 (Wako Pure Chemical Industries) and bacterial alkaline phosphatase C75 (Takara bio), and then analyzed on the LCQ DUO ion-trap mass spectrometer with an ESI source and HP1100 liquid chromatography system in positive-ion detection mode. ProMass (Novatia) was used to obtain the uncharged masses of whole tRNAs by deconvolution of the multiply charged mass spectra.
RESULTS
Isolation of 22 species of bovine mt tRNAs
To date, 11 species of bovine mt tRNAs have been described in published work, along with the details of their post-transcriptional modifications: Ser(UCN) (33), Ser(AGY) (34,35), Phe (36), Arg (35), Gly (35), Ile (35), Met (8,37), Val (35), Trp (35), Glu (7) and Gln (7). In addition, sequences of four species of bovine mt tRNAs for Thr, Leu(UUR), Leu(CUN) and Lys have recently been deposited in the tRNA databases as personal communications (38,39). However, some tRNA sequences deposited in the tRNA databases contain unidentified modified bases and mis-annotation of modifications determined by outmoded methods. To accurately define the complete landscape of post-transcriptional modifications of mammalian mt tRNAs, we analyzed all 22 species of mt tRNAs, which we previously isolated from bovine liver by chaplet column chromatography (29).
Mass spectrometric analysis of post-transcriptional modifications in bovine mt tRNAs
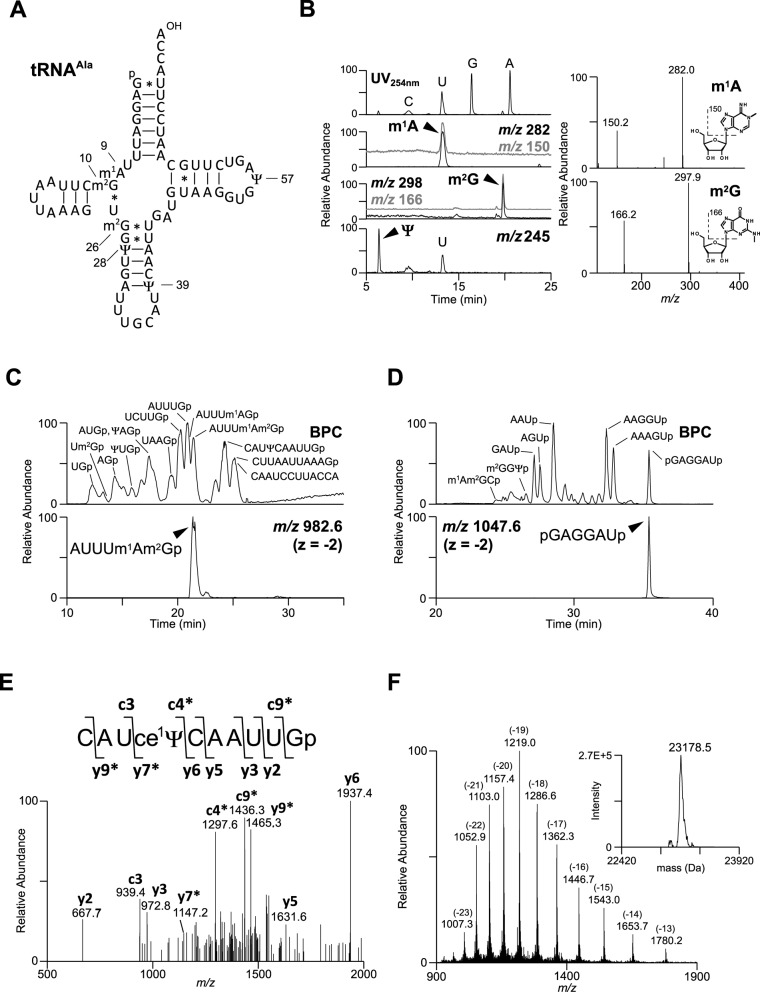
We performed a series of mass spectrometric analyses of each individual mt tRNA to determine its post-transcriptional modifications. As an example, determination of post-transcriptional modifications in mt tRNAAla is depicted in Figure 1A. First, nucleoside analysis by LC/ESI-MS was carried out to determine the composition of modifications, revealing that 1-methyladenosine (m1A), N2-methylguanosine (m2G) and pseudouridine (Ψ) were present in mt tRNAAla (Figure 1B). In parallel, we used capillary LC/nano ESI-MS to analyze RNA fragments generated by digestion with RNase T1 (Figure 1C) or RNase A (Figure 1D). Unmodified fragments were identified by comparing the observed m/z values with the calculated m/z values deduced from the mt tRNAAla gene encoded in bovine mtDNA (GenBank accession number V00654). Nucleotide compositions of internal fragments were calculated as 5′-hydroxyl (5′OH) and 3′-phosphate (3′p). The sequence of each fragment was further analyzed by collision-induced dissociation (CID) (Figure 1E). Because tRNA has 5′-phosphate and 3′-hydroxyl groups, the 5′-terminal fragment pGAGGAUp (MW 2097.2) and the 3′ terminal fragment OH-CAAUCCUUACCA-OH (MW 3697.5) were detected as species with unique molecular masses distinct from those of the internal fragments. Furthermore, RNA fragments containing modified nucleosides could be identified from the deduced molecular mass calculated from the combinations of unmodified and expected modified nucleotides. For example, in the RNase T1 digest of mt tRNAAla, we found a species with m/z 982.627, corresponding to a doubly charged negative ion of dimethylated AUUUAGp. The positions of modifications were determined by CID analysis, which indicated the modified sequence AUUUA(+me)G(+me)p. According to the nucleoside analysis, this species was assigned as AUUUm1Am2Gp, with m1A and m2G at positions 9 and 10, respectively. Similarly, m2G was also found at position 26.
Figure 1.
Mass spectrometric analysis of bovine mt tRNAAla for assignment of post-transcriptional modifications. (A) Secondary structure of bovine mitochondrial tRNAAla with post-transcriptional modifications determined in this study. The position numbers of the modifications are displayed according to the nucleotide numbering system from the tRNA compilation (40). Symbols for modified nucleosides are as follows: m1A, 1-methyladenosine; m2G, N2-methylguanosine and Ψ, pseudouridine. Watson–Crick base pairs are indicated by solid lines, whereas G–U pairs are indicated by asterisks. (B) Nucleoside analysis of bovine mitochondrial tRNAAla. Left, top panel: UV chromatogram at 254 nm of the four major nucleosides (C, U, G and A). Left, lower panels: extracted-ion chromatograms (XIC) for the protonated ion of m1A nucleoside (m/z 282, black line) with its base ion (m/z 150, gray line) (second panel), m2G nucleoside (m/z 298, black line) with its base ion (m/z 166, gray line) (third panel) and Ψ nucleoside (m/z 245, black line) (bottom panel). The XIC for the base ion (20% upper offset) is overlaid on the XIC for the nucleoside ion. Right: mass spectra of m1A and m2G. Cleavage positions for the base-related ions are indicated on the chemical structures. (C) RNA fragment analysis of RNase T1 digests of bovine mitochondrial tRNAAla. Assigned fragments are indicated on the base peak chromatogram (BPC) in the first panel. ‘p’ stands for the terminal phosphate group. The XIC for the doubly charged negative ion of a modification-containing fragment (AUUUm1Am2Gp, m/z 982.6) is indicated in the second panel. Because m2G at position 10 is a partial modification, both AUUUm1Am2Gp and AUUUm1AGp were detected. (D) RNA fragment analysis of RNase A digests of bovine mitochondrial tRNAAla. Assigned fragments are indicated on the BPC. The XIC for the doubly charged negative ion of the 5′-terminal fragment (pGAGGAUp, m/z 1047.6) is indicated in the second panel. (E) A CID spectrum of a cyanoethylated RNA fragment to determine the location of a Ψ site. The doubly charged negative ion of the RNA fragment (m/z 1617.7) shown in the inset was used as the precursor ion for CID. The product ions were assigned according to McLuckey et al. (41). The asterisks in the spectrum denote product ions containing ce1Ψ. (F) Whole mass analysis of intact bovine mt tRNAAla. A series of multiply charged negative ions is shown in the mass spectrum. The charge values are indicated in parentheses. The observed mass obtained by deconvoluting the mass spectra is shown in the inset.
To identify Ψ, a mass-silent modification, each tRNA was treated with acrylonitrile to cyanoethylate Ψ (1-cyanoethyl Ψ; ce1Ψ). The derivatized tRNA was then digested by RNase T1 and subjected to mass spectrometry to detect the cyanoethylated fragments with molecular mass increased by 53 Da. Three cyanoethylated fragments were detected in the RNase T1 digest, and each fragment was further analyzed by CID. As shown in Figure 1E, Ψ at position 39 was identified from the assignment of product ions of CAUce1ΨCAAUUGp. Accordingly, we identified three Ψs at positions 28, 39 and 57. Finally, to confirm the assignment of post-transcriptional modifications, the total molecular mass of mt tRNAAla was measured by deconvoluting the multiply charged negative ions produced by ESI (Figure 1F). The observed mass (23178.5 Da) was fairly close to the calculated mass (23180.6 Da), which corresponds to the base composition of mt tRNAAla (pU27C9A20G16) with three methyl groups. Other tRNAs were basically analyzed using the same procedure as for mt tRNAAla (Supplementary Information and Table S1).
Post-transcriptional modifications in seven bovine mt tRNAs
For 10 of the species of mt tRNAs described in the previous literature, our determinations of modifications yielded results consistent with published findings: Phe, Gly, Leu(CUN), Met, Arg, Ser(UCN), Ser(AGY), Val, Glu and Gln (Supplementary Figure S1).
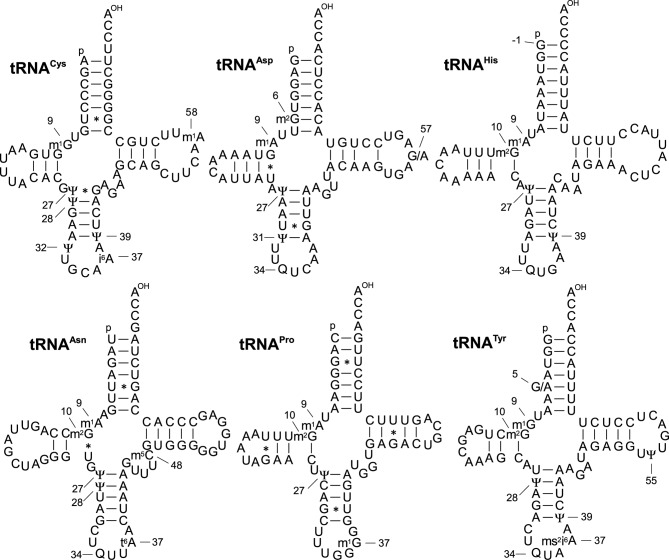
Several of the tRNAs we characterized have not been previously reported or deposited in any databases. We determined the primary structures and modifications for seven such mt tRNAs: Cys, Asp, His, Asn, Pro and Tyr (Figure 2 and Supplementary Table S1) plus the aforementioned mt tRNAAla (Figure 1). We identified queuosine (Q) at the wobble positions of mt tRNAs for Asp, His, Asn and Tyr. In cytosolic tRNAAsp and tRNATyr, Q was glycosylated to mannosyl-Q and galactosyl-Q, respectively; however, Q was not glycosylated in the mt tRNAs. The mt tRNAs for Ala and Pro correspond to family boxes; as expected, unmodified U was present at the wobble position. In mt tRNAHis, we identified a G at position -1 (G-1) (Supplementary Figure S2). This base is likely to be added enzymatically after transcription and 5′-end processing. In mt tRNAAsp, m2G was present at position 6, the first such case to be reported in mammalian mitochondria. At position 37, which is 3′-adjacent to the anticodon, N6-isopentenyladenosine (i6A) and 2-methylthio-N6-isopentenyladenosine (ms2i6A) were present in mt tRNACys and mt tRNATyr, respectively. N6-threonylcarbamoyladenosine (t6A) and m1G were also present in the mt tRNAs for Asn and Pro, respectively. During our analysis, we found two polymorphisms in the sequences of mt tRNAs for Asp and Tyr: in mt tRNAAsp, a mixture of A and G were present at position 57; similarly, A and G were mixed at position 5 in mt tRNATyr (Figure 2). Both sites are encoded as A7356 (H-chain) and A5681 (L-chain) in the bovine mtDNA sequence (GenBank V00654) used as a reference. We speculate that these polymorphisms are the result of mtDNA heteroplasmy in the bovine liver we analyzed. In fact, both sites are encoded as guanosines in the mtDNA sequence of Bos taurus isolate FC3 (GenBank accession number DQ124389.1), suggesting that these sites are polymorphic in populations of healthy individuals.
Figure 2.
Post-transcriptional modifications in six bovine mt tRNAs. Symbols for modified nucleosides are as follows: m1G, 1-methylguanosine; i6A, N6-isopentenyladenosine; Q, queuosine; m5C, 5-methylcytidine; ms2i6A, 2-methylthio-N6-isopentenyladenosine. The ‘G/A’ in tRNAAsp and tRNATyr indicates that both G and A were detected at this position, probably as a result of heteroplasmy in the bovine liver we used in this study.
Full complement of modifications in five bovine mt tRNAs
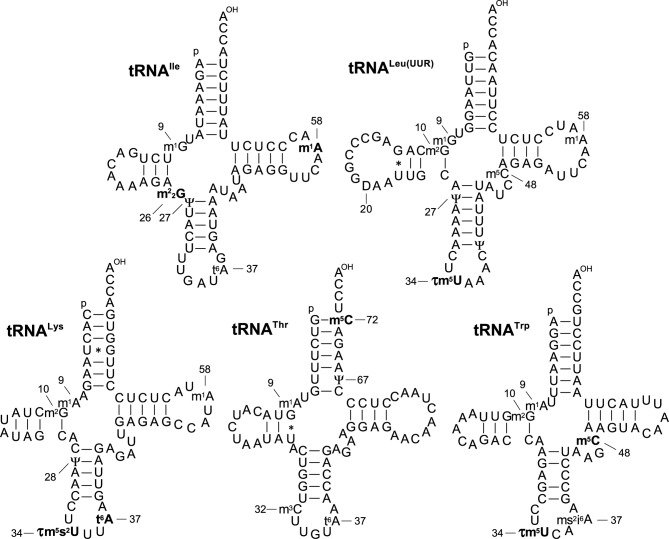
Although sequences and modifications of the remaining five mt tRNAs [Ile, Leu(UUR), Lys, Thr and Trp] have been published or deposited in the database (38,39), we discovered eight previously unreported modifications in these tRNAs (Figure 3): N2, N2-dimethylguanosine (m22G) and m1A at positions 26 and 58 in mt tRNAIle; τm5U at position 34 in mt tRNAs for Leu(UUR) and Trp; τm5s2U and t6A at positions 34 and 37 in mt tRNALys and 5-methylcytidine (m5C) at position 72 in mt tRNAThr and position 48 in mt tRNATrp. The wobble modifications of bovine mt tRNAs for Leu(UUR), Lys and Trp remained unidentified in tRNAdb (38). In a previous study (6), we reported that τm5U and τm5s2U were present in bovine mt tRNALeu(UUR) and mt tRNALys, respectively. To date, however, their full sequences had not been reported. The m5C72 in mt tRNAThr is the first reported instance of this modification in mammalian mitochondria.
Figure 3.
Revised information regarding post-transcriptional modifications of five bovine mt tRNAs. The updated modified bases are represented in bold type. Symbols for modified nucleoside are as follows: m22G, N2, N2-dimethylguanosine; D, dihydrouridine; τm5U, 5-taurinomethyluridine; τm5s2U, 5-taurinomethyl-2-thiouridine; t6A, N6-threonylcarbamoyladenosine; m3C, 3-methylcytidine.
DISCUSSION
Here, we report the post-transcriptional modifications in seven species of bovine mt tRNAs not previously characterized, and eight previously unidentified modified bases in five mt tRNAs whose sequences and modifications were determined in earlier studies. In total, we identified 15 species of modified nucleosides at 118 positions in the complete set of bovine mt tRNAs (Supplementary Table S2), i.e. 7.48% of the bases in these tRNAs are modified. The sites and species of all modifications are summarized in a schematic cloverleaf structure (Figure 4). Notably, all modifications are base modifications; the absence of 2′-O-methylation is a characteristic feature of mt tRNAs.
Figure 4.
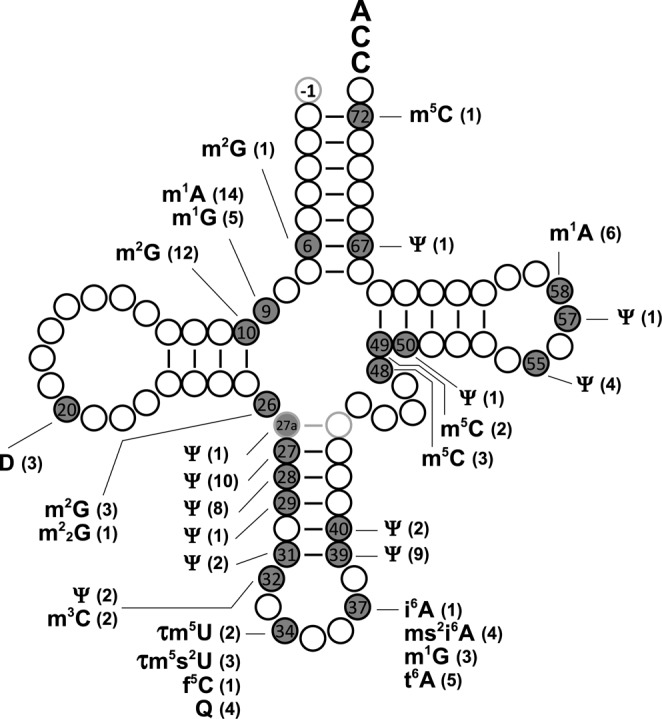
Summary of post-transcriptional modifications in bovine mt tRNAs. Species and numbers of post-transcriptional modifications identified in 22 bovine mt tRNAs are mapped on the schematic secondary structure of tRNA. The modified positions are depicted by gray circles with a symbol indicating each modification. At each position, the number of tRNAs that bear the modification are shown in parenthesis. Positions 27a and 43a, indicated by light gray circles, are unique to mt tRNASer(UCN). G-1 is specific to mt tRNAHis.
Eight mt tRNAs responsible for family boxes had unmodified U at their wobble positions (Table 1), suggesting that the family boxes in mitochondria are decoded by single tRNAs via the four-way wobble rule. For NNR codons, all six mt tRNAs had wobble modifications (Table 1): f5C in mt tRNAMet, τm5U in mt tRNAs for Leu(UUR) and Trp and τm5s2U in mt tRNAs for Lys, Glu and Gln. We consider that these modifications are required for efficient recognition of the cognate codons, as well as to prevent misreading of near-cognate codons. Q was present at the wobble position in four mt tRNAs (Tyr, His, Asn and Asp) responsible for NAY codons (Table 1). Q34 is known to restrict the conformational flexibility of the anticodon loop by making hydrogen bonds between the side chain amine of Q-base and 2′ OH of U33 (42). Q34 enables tRNA to decode NAU efficiently (43). Although the functional role of Q still remains obscure, it is associated with various physiological events in cytoplasmic tRNAs, including cell development and proliferation, neoplastic transformation and translational read-through or frameshift essential for retroviral production (44–46). These facts suggest that the presence of Q in the four mt tRNAs plays a modulatory role in deciphering NAY codons.
Other mt tRNAs responsible for NNY codons [Phe, Ile, Cys, Ser(AGY)] had unmodified G at the wobble position (Table 1). The results clearly reveal a general principle to decipher the minimal decoding system by the base modifications at the wobble positions.
Hypermodifications are frequently found at position 37 of tRNAs. These modifications play important roles in maintaining translational efficiency and integrity (47,48). Four species of base modifications were present at position 37 in 13 mt tRNAs (Figure 4 and Supplementary Figure S1). Among six mt tRNAs responsible for UNN codons, five had i6A or ms2i6A at position 37. t6A37 was present in the mt tRNAs for Ile, Thr, Asn, Lys and Ser(AGY), whereas m1G37 was present in the mt tRNAs for Leu(CUN), Pro and Gln. Last year, we discovered cyclic t6A, which is formed by ATP-dependent dehydration of t6A catalyzed by TcdA, as a bonafide modification at position 37 of tRNAs from bacteria, yeast, plants and protists (49). However, we did not detect any cyclic form of t6A in bovine mt tRNAs, and there is no homolog of TcdA in mammalian genomes.
Loss-of-function mutations in tRNA-modifying enzymes can cause human diseases. Consistent with this idea, large-scale disease-associated exome analyses have identified a number of genes that encode tRNA-modifying enzymes (50,51). Identification of all genes involved in mt tRNA modifications will help us to identify genes and mutations associated with various diseases, especially those linked to mitochondrial dysfunction. Basically, the structure and sequence of human mt tRNAs are similar to those of bovine mt tRNAs. In fact, 81% of total bases in 22 mt tRNAs are conserved between these two mammals. That is why human mt tRNA-modifying enzymes can be predicted based on information of bovine mt tRNAs. So far, nine human genes have been confirmed to be responsible for base modifications in mammalian mt tRNAs, and others have been predicted based on studies of tRNA modifications in model organisms (Table 2). Several of these modifications, and the enzymes that may catalyze their formation, are described in the following paragraphs.
Table 2. List of confirmed and predicted genes responsible for post-transcriptional modifications in mammalian mt tRNAs.
| Position a | tRNA species | Modification b | Confirmed gene(s) in human or mammals | Predicted gene(s) in human |
|---|---|---|---|---|
| 6 | Asp | m2G | THUMPD3 or THUMPD2 (52,53) | |
| 9 | Ala, Asp, Glu, Phe, Gly, His, Lys, Leu(CUN),Asn, Pro, Arg, Thr, Val, Trp | m1A | TRMT10C and SDR5C1 (54) | |
| Cys, Ile, Leu(UUR), Gln, Tyr | m1G | TRMT10C and SDR5C1 (54) | ||
| 10 | Ala, Phe, Gly, His, Lys, Leu(UUR), Leu(CUN), Asn, Pro, Val, Trp, Tyr | m2G | TRMT11 and TRMT112 (55) | |
| 20 | Leu(UUR), Leu(CUN), Ser(UCN) | D | DUS2 (56) | |
| 26 | Ala, Glu, Leu(UUR) | m2G | TRMT1 | |
| Ile | m22G | TRMT1 (57,58) | ||
| 27a c | Ser(UCN) | Ψ | PUS1 | |
| 27 | Cys, Asp, His, Ile, Leu(UUR), Leu(CUN), Asn, Pro, Val, Met | Ψ | PUS1 (25) | |
| 28 | Ala, Cys, Glu, Lys, Leu(CUN), Asn, Ser(UCN), Tyr | Ψ | PUS1 (25) | |
| 29 | Ser(UCN) | Ψ | PUS1 | |
| 31 | Asp, Leu(CUN) | Ψ | RPUSD1, RPUSD2, RPUSD3 or RPUSD4d | |
| 32 | Cys, Val | Ψ | RPUSD1, RPUSD2, RPUSD3 or RPUSD4d | |
| Ser(UCN), Thr | m3C | METTL2B (59,60) | ||
| 34 | Leu(UUR), Trp, Glu, Lys, Gln, | τm5U | GTPBP3 and MTO1 (61,62) | |
| Glu, Lys, Gln | τm5s2U e | MTU1e (63) and NFS1e (64) | ||
| Met | f5C | Unidentified | ||
| Asp, His, Asn, Tyr | Q | hQTRT1 and hQTRTD1 (65,66) | ||
| 37 | Ile, Lys, Asn, Ser(AGY), Thr | t6A | YRDC and QRI7 (OSGEPL1) (67) | |
| Cys, Phe, Ser(UCN), Trp, Tyr | i6A | TRIT1 (68) | ||
| Phe, Ser(UCN), Trp, Tyr | ms2i6Af | CDK5RAP1f (69) | ||
| Leu(CUN), Pro, Gln | m1G | TRMT5 (70) | ||
| 39 | Ala, Cys, Phe, Gly, His, Leu(UUR), Gln, Arg, Tyr | Ψ | PUS3 (71) | |
| 40 | Glu, Gln | Ψ | PUS3d | |
| 48 | Leu(UUR), Asn, Trp | m5C | NSUN2 (72,73) | |
| 49 | Glu, Ser(AGY) | m5C | NSUN2 (72,73) | |
| 50 | Met | Ψ | Unidentified | |
| 55 | Glu, Gln, Ser(UCN), Tyr | Ψ | TRUB2 (74) | |
| 57 | Ala | Ψ | Unidentified | |
| 58 | Cys, Glu, Ile, Lys, Leu(UUR), Ser(UCN) | m1A | TRMT61B (75) | |
| 67 | Thr | Ψ | PUS1d (76) | |
| 72 | Thr | m5C | Unidentified |
aNumbering system for tRNA comes from the tRNAdb compilation (40).
bSymbols for modifications originate from MODOMICS (http://modomics.genesilico.pl/) (39).
cThis position number is unique to mt tRNASer(UCN) (77).
dThese predictions were altered from our previous prediction (2).
eMTU1 and NFS1 are involved in 2-thiolation of τm5s2U34. MTU1 (Mitochondrial tRNA-specific 2-thiouridylase 1) is known as TRMU which originated from bacterial trmU (tRNA methyltransferase U). However, trmU (renamed as mnmA) was found to be a mis-annotation because it is not a tRNA methyltransferase.
fCDK5RAP1 is required for 2-methylthiolation of ms2i6A37.
On the basis of studies of bacterial and yeast mitochondria (63,78), GTPBP3 and MTO1 are predicted to be enzymes involved in τm5U formation; however, to date, no direct evidence in support of this prediction has been published. The GTPBP3–MTO1 complex has been proposed to recognize mt tRNAs for Leu(UUR), Trp, Lys, Glu and Glu. However, these five mt tRNAs share little sequence similarity and no consensus motif. Nonetheless, τm5U formation is sensitive to a single pathogenic point mutation associated with MELAS or MERRF (12), implying strict substrate specificity of the enzyme. To resolve this issue, it will be necessary to perform in vitro reconstitution of τm5U on mt tRNAs using recombinant proteins.
We previously identified MTU1, a thiouridylase responsible for 2-thiolation of τm5s2U in mt tRNAs (63). The sulfur atom comes from Cys, a process mediated by a cysteine desulfurase, NFS1 (64). During bacterial 2-thiouridine formation, several sulfur mediators transfer persulfide sulfur from cysteine desulfurase to thiouridylase (79). To date, however, there is no information available regarding the involvement of such sulfur mediators in the formation of 2-thiouridine in mitochondria.
In mammals, the Q-base is obtained either from the diet or through intestinal microflora (45), because no homologous genes responsible for Q-base biogenesis de novo are encoded in mammalian genome. Mammals possess two tRNA-guanine transglycosylases (TGTases), QTRT1 and QTRTD1, which may transglycosylate the wobble base with the dietary Q-base. According to its subcellular localization, QTRTD1 appears to be the mitochondrial TGTase (65,66). Further studies will be necessary to confirm the identity of the mitochondrial TGTase, as well as the transport pathway by which dietary Q enters the mitochondria. Q is not present in mt tRNAAsp from Morris hepatoma cells (80).
In yeast, t6A37 is synthesized by two enzymes, Sua5p and Qri7p (67). YRDC and OSGEPL1, the human homologs of yeast Sua5p and Qri7p, are presumably involved in t6A formation in mammalian mt tRNAs.
TRIT1 is a tumor suppressor gene, mutations in which are associated with cancer progression. Human TRIT1 was identified as a tRNA isopentenyltransferase (IPTase) homologous to bacterial MiaA and yeast Mod5; the latter is responsible for i6A37 formation in both cytoplasmic and mt tRNAs. Knockdown of TRIT1 in human cells reduced the abundance of i6A in mt tRNASer(UCN) (68). In the present study, we determined that bovine mt tRNAs for Tyr and Cys have i6A37; in total, five mt tRNAs have been identified as potential substrates for TRIT1 in mitochondria.
The human protein CDK5RAP1 is a 2-methylthiolase that is homologous to bacterial MiaB. A CDK5RAP1 variant localizes in mitochondria, and knockdown of CDK5RAP1 results in a reduction of ms2i6A levels (69). Thus, four mt tRNAs [Phe, Ser(UCN), Tyr and Trp] are likely to be modified by CDK5RAP1.
TRMT5, a methyltransferase responsible for m1G37 formation, is predicted to be localized to the mitochondria (70,81), and recombinant TRMT5 can modify in vitro transcribed mt tRNA (70), suggesting that TRMT5 methylates three mt tRNAs: Leu(CUN), Pro and Gln.
Base methylation at position 9 is frequently observed among bovine mt tRNAs. Indeed, 19 mt tRNAs have m1A9 or m1G9 (Figure 4). m1A9 stabilizes the canonical cloverleaf structure of mt tRNAs (82). In addition, m1A9 is indispensable for aminoacylation of nematode mt tRNAs that lack the T-arm. The methyltransferase for m1A9 or m1G9 has been identified as a complex of TRM10C and SDR5C1, both of which are components of mt RNase P (54). The TRM10C–SDR5C1 complex has broad substrate specificity for 19 out of 22 mt tRNA species. Only three mt tRNAs are unmethylated at position 9. Of these, mt tRNAMet has C at this position, and the two mt tRNASer isoacceptors have non-canonical cloverleaf structures: Type I for mt tRNASer(UCN) and Type III for mt tRNASer(AGY) (2).
Ψ is the most abundant modification in bovine mt tRNAs. Among the 22 mt tRNAs, we mapped Ψ at 42 sites, 35 of which reside in the anticodon arm. In humans, there are 13 species of Ψ synthases, each of which has different substrate specificity and introduces Ψ at multiple sites in various RNAs. PUS1 is a human Ψ synthase with broad substrate specificity that introduces Ψ into cytoplasmic tRNAs, mt tRNAs, U2 snRNA and SRA non-coding RNA (25,76,83–84), and it is responsible for Ψ27 and Ψ28 in mt tRNAs (25). We assume that PUS1 is also responsible for Ψ27a and Ψ29 in mt tRNASer(UCN) and Ψ67 in mt tRNAThr (76). Because missense mutations in the PUS1 gene are responsible for the development of MLASA (25), it will be necessary to determine the Ψ sites targeted by PUS1 to gain a deeper understanding of the functional role of this modification, as well as the molecular pathogenesis of MLASA. According to the studies of yeast and Escherichia coli tRNAs, Ψ39 and Ψ40 are likely to be introduced by PUS3 (71), whereas Ψ55 is probably modified by TRUB2 (74). In yeast mitochondria, Ψ31 and Ψ32 are introduced by PUS6 and PUS9, respectively (85,86). Although we previously predicted that either RPUSD2 or RPUSD4 was responsible for Ψ31 and Ψ32 (2), two other homologs, RPUSD1 and RPUSD3, should also be regarded as candidates for these modifications. Regarding Ψ50 and Ψ57, no candidate enzymes can be predicted at present. Biochemical and genetic analyses of Ψ synthase genes will be necessary to determine the enzyme responsible for each Ψ site.
In contrast to cytoplasmic tRNAs, most mammalian mt tRNAs do not have consensus sequences of D- and T-loops, which are conserved among tRNAs in general; consequently, they have lost the canonical D-loop/T-loop interaction. This feature is characteristic of mt tRNAs in mammals (2). In the D-loop, dihydrouridines (D) at positions 16 and 17, which are frequently observed in canonical tRNAs from various sources, are not present in bovine mt tRNAs. Instead, D20 was present in three species of bovine mt tRNAs. Based on a study on yeast tRNA, it is reasonable to predict that human tRNA-dihydrouridine synthase 2 (DUS2) introduces D20 (56). Human DUS2 has been implicated in pulmonary carcinogenesis (87). In the T-loop, Ψ55 and m1A58, both of which are typical modifications in canonical tRNAs, are present in four and six mt tRNAs, respectively. We previously identified human TRMT61B as the methyltransferase responsible for m1A58 in mt tRNAs for Leu(UUR), Ser(UCN) and Lys (75). In addition to these three tRNAs, we also found m1A58 in mt tRNAs for Cys, Glu and Ile. 5-methyluridine (m5U) at position 54 is one of the most common T-loop modifications in canonical tRNAs. m5U54 is present in the human mt tRNAs for Leu(UUR) (19) and Ser(UCN) (88); however, we did not detect m5U54 in any bovine mt tRNAs.
m2G6 is present in bovine mt tRNAAsp. Human mt tRNAAsp does not have m2G6 (89), because this position is replaced by A6. The methyltransferase responsible for m2G6 was identified as Trm14 in Methanocaldococcus jannaschii (52) and TTH1157 in Thermus thermophilus (53). A similarity search using the Trm14 sequence retrieved two human homologs, THUMPD2 and THUMPD3. According to the WoLF P-sort (90) prediction of subcellular localization, the former is likely localized in the cytoplasm and the latter in mitochondria. Although further investigation is necessary, THUMPD3 is the most plausible candidate for the enzyme that introduces m2G6 in mt tRNAAsp, whereas THUMPD2 probably acts on cytoplasmic tRNAs. Although both genes are encoded in the human genome, no m2G6 is present in human mt tRNAAsp, implying the presence of other substrates for this enzyme in human mitochondria.
In this study, we identified G-1 at the 5′ terminus of bovine mt tRNAHis. In general, G-1 is an identity element for aminoacylation by histidyl-tRNA synthetase (91). This is the first reported instance of this base in mammalian mitochondria, although G-1 is present in both yeast and starfish mt tRNAHis (92,93). Judging from the fact that the 5′-adjacent nucleotide of the mt tRNAHis gene in bovine mt DNA is T rather than G (e.g. GenBank V00654), it is likely that G-1 is added enzymatically after 5′-terminal processing by mt RNase P. THG1L is a mammalian guanylyltransferase that adds G-1 at the 5′ terminus of cytoplasmic tRNAHis. Because THG1L is predicted to have a mitochondria-targeting sequence at the N-terminus (94), THG1L is likely to be responsible for G-1 addition of mt tRNAHis.
In summary, we have compiled the first complete picture of post-transcriptional modifications in mammalian mt tRNAs. The results of this study enable a deeper understanding of the molecular mechanisms underpinning the minimal decoding system in mammalian mitochondria, and should help predict the human tRNA-modifying enzymes responsible for each modification in mt tRNAs. The list of tRNA-modifying enzymes serves as a practical landmark that encourages us to identify all genes responsible for tRNA modifications in mammalian mitochondria, and to further investigate human diseases caused by tRNA modification disorders in mitochondria. An important goal for future efforts is the identification of all modifications in human mt tRNAs.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We are grateful to the members of the Suzuki laboratory, in particular, Dr. K. Miyauchi and Dr. A. Nagao, for many insightful discussions. We thank Prof. K. Watanabe (Tokyo University of Pharmacy and Life Sciences) for his abiding encouragement for our mitochondrial tRNA work.
FUNDING
Grants-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Science, Sports, and Culture of Japan [to Takeo S. and Tsutomu S.]; JSPS Fellowship for Japanese Junior Scientists [to Takeo S.].
Conflict of interest statement. None declared.
REFERENCES
- 1.Wallace D.C. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu. Rev. Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Suzuki T., Nagao A., Suzuki T. Human mtochondrial tRNAs: biogenesis, function, structural aspects, and diseases. Annu. Rev. Genet. 2011;45:299–329. doi: 10.1146/annurev-genet-110410-132531. [DOI] [PubMed] [Google Scholar]
- 3.Crick F.H. Codon–anticodon pairing: the wobble hypothesis. J. Mol. Biol. 1966;19:548–555. doi: 10.1016/s0022-2836(66)80022-0. [DOI] [PubMed] [Google Scholar]
- 4.Rogalski M., Karcher D., Bock R. Superwobbling facilitates translation with reduced tRNA sets. Nat. Struct. Mol. Biol. 2008;15:192–198. doi: 10.1038/nsmb.1370. [DOI] [PubMed] [Google Scholar]
- 5.Yokoyama S., Watanabe T., Murao K., Ishikura H., Yamaizumi Z., Nishimura S., Miyazawa T. Molecular mechanism of codon recognition by tRNA species with modified uridine in the first position of the anticodon. Proc. Natl. Acad. Sci. U. S. A. 1985;82:4905–4909. doi: 10.1073/pnas.82.15.4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Suzuki T., Suzuki T., Wada T., Saigo K., Watanabe K. Taurine as a constituent of mitochondrial tRNAs: new insights into the functions of taurine and human mitochondrial diseases. EMBO J. 2002;21:6581–6589. doi: 10.1093/emboj/cdf656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nagao A., Suzuki T., Katoh T., Sakaguchi Y., Suzuki T. Biogenesis of glutaminyl-mt tRNAGln in human mitochondria. Proc. Natl. Acad. Sci. U. S. A. 2009;106:16209–16214. doi: 10.1073/pnas.0907602106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moriya J., Yokogawa T., Wakita K., Ueda T., Nishikawa K., Crain P.F., Hashizume T., Pomerantz S.C., McCloskey J.A., Kawai G., et al. A novel modified nucleoside found at the first position of the anticodon of methionine tRNA from bovine liver mitochondria. Biochemistry. 1994;33:2234–2239. doi: 10.1021/bi00174a033. [DOI] [PubMed] [Google Scholar]
- 9.Takemoto C., Spremulli L.L., Benkowski L.A., Ueda T., Yokogawa T., Watanabe K. Unconventional decoding of the AUA codon as methionine by mitochondrial tRNAMet with the anticodon f5CAU as revealed with a mitochondrial in vitro translation system. Nucleic Acids Res. 2009;37:1616–1627. doi: 10.1093/nar/gkp001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bilbille Y., Gustilo E.M., Harris K.A., Jones C.N., Lusic H., Kaiser R.J., Delaney M.O., Spremulli L.L., Deiters A., Agris P.F. The human mitochondrial tRNAMet: structure/function relationship of a unique modification in the decoding of unconventional codons. J. Mol. Biol. 2011;406:257–274. doi: 10.1016/j.jmb.2010.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cantara W.A., Murphy F.V., Demirci H., Agris P.F. Expanded use of sense codons is regulated by modified cytidines in tRNA. Proc. Natl. Acad. Sci. U. S. A. 2013;110:10964–10969. doi: 10.1073/pnas.1222641110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suzuki T., Nagao A., Suzuki T. Human mitochondrial diseases caused by lack of taurine modification in mitochondrial tRNAs. Wiley Interdiscip Rev. RNA. 2011;2:376–386. doi: 10.1002/wrna.65. [DOI] [PubMed] [Google Scholar]
- 13.Ruiz-Pesini E., Lott M.T., Procaccio V., Poole J.C., Brandon M.C., Mishmar D., Yi C., Kreuziger J., Baldi P., Wallace D.C. An enhanced MITOMAP with a global mtDNA mutational phylogeny. Nucleic Acids Res. 2007;35:D823–D828. doi: 10.1093/nar/gkl927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levinger L., Morl M., Florentz C. Mitochondrial tRNA 3’ end metabolism and human disease. Nucleic Acids Res. 2004;32:5430–5441. doi: 10.1093/nar/gkh884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Belostotsky R., Frishberg Y., Entelis N. Human mitochondrial tRNA quality control in health and disease: a channelling mechanism. RNA Biol. 2012;9:33–39. doi: 10.4161/rna.9.1.18009. [DOI] [PubMed] [Google Scholar]
- 16.Goto Y., Nonaka I., Horai S. A mutation in the tRNA(Leu)(UUR) gene associated with the MELAS subgroup of mitochondrial encephalomyopathies. Nature. 1990;348:651–653. doi: 10.1038/348651a0. [DOI] [PubMed] [Google Scholar]
- 17.Goto Y., Nonaka I., Horai S. A new mtDNA mutation associated with mitochondrial myopathy, encephalopathy, lactic acidosis and stroke-like episodes (MELAS) Biochim. Biophys. Acta. 1991;1097:238–240. doi: 10.1016/0925-4439(91)90042-8. [DOI] [PubMed] [Google Scholar]
- 18.Shoffner J.M., Lott M.T., Lezza A.M., Seibel P., Ballinger S.W., Wallace D.C. Myoclonic epilepsy and ragged-red fiber disease (MERRF) is associated with a mitochondrial DNA tRNA(Lys) mutation. Cell. 1990;61:931–937. doi: 10.1016/0092-8674(90)90059-n. [DOI] [PubMed] [Google Scholar]
- 19.Yasukawa T., Suzuki T., Suzuki T., Ueda T., Ohta S., Watanabe K. Modification defect at anticodon wobble nucleotide of mitochondrial tRNAsLeu(UUR) with pathogenic mutations of mitochondrial myopathy, encephalopathy, lactic acidosis, and stroke-like episodes. J. Biol. Chem. 2000;275:4251–4257. doi: 10.1074/jbc.275.6.4251. [DOI] [PubMed] [Google Scholar]
- 20.Yasukawa T., Suzuki T., Ishii N., Ueda T., Ohta S., Watanabe K. Defect in modification at the anticodon wobble nucleotide of mitochondrial tRNALys with the MERRF encephalomyopathy pathogenic mutation. FEBS Lett. 2000;467:175–178. doi: 10.1016/s0014-5793(00)01145-5. [DOI] [PubMed] [Google Scholar]
- 21.Kirino Y., Goto Y., Campos Y., Arenas J., Suzuki T. Specific correlation between the wobble modification deficiency in mutant tRNAs and the clinical features of a human mitochondrial disease. Proc. Natl. Acad. Sci. U. S. A. 2005;102:7127–7132. doi: 10.1073/pnas.0500563102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yasukawa T., Kirino Y., Ishii N., Holt I.J., Jacobs H.T., Makifuchi T., Fukuhara N., Ohta S., Suzuki T., Watanabe K. Wobble modification deficiency in mutant tRNAs in patients with mitochondrial diseases. FEBS Lett. 2005;579:2948–2952. doi: 10.1016/j.febslet.2005.04.038. [DOI] [PubMed] [Google Scholar]
- 23.Kirino Y., Yasukawa T., Ohta S., Akira S., Ishihara K., Watanabe K., Suzuki T. Codon-specific translational defect caused by a wobble modification deficiency in mutant tRNA from a human mitochondrial disease. Proc. Natl. Acad. Sci. U. S. A. 2004;101:15070–15075. doi: 10.1073/pnas.0405173101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yasukawa T., Suzuki T., Ishii N., Ohta S., Watanabe K. Wobble modification defect in tRNA disturbs codon-anticodon interaction in a mitochondrial disease. EMBO J. 2001;20:4794–4802. doi: 10.1093/emboj/20.17.4794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patton J.R., Bykhovskaya Y., Mengesha E., Bertolotto C., Fischel-Ghodsian N. Mitochondrial myopathy and sideroblastic anemia (MLASA): missense mutation in the pseudouridine synthase 1 (PUS1) gene is associated with the loss of tRNA pseudouridylation. J. Biol. Chem. 2005;280:19823–19828. doi: 10.1074/jbc.M500216200. [DOI] [PubMed] [Google Scholar]
- 26.Zeharia A., Shaag A., Pappo O., Mager-Heckel A.M., Saada A., Beinat M., Karicheva O., Mandel H., Ofek N., Segel R., et al. Acute infantile liver failure due to mutations in the TRMU gene. Am. J. Hum. Genet. 2009;85:401–407. doi: 10.1016/j.ajhg.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ghezzi D., Baruffini E., Haack T.B., Invernizzi F., Melchionda L., Dallabona C., Strom T.M., Parini R., Burlina A.B., Meitinger T., et al. Mutations of the mitochondrial-tRNA modifier MTO1 cause hypertrophic cardiomyopathy and lactic acidosis. Am. J. Hum. Genet. 2012;90:1079–1087. doi: 10.1016/j.ajhg.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baruffini E., Dallabona C., Invernizzi F., Yarham J.W., Melchionda L., Blakely E.L., Lamantea E., Donnini C., Santra S., Vijayaraghavan S., et al. MTO1 mutations are associated with hypertrophic cardiomyopathy and lactic acidosis and cause respiratory chain deficiency in humans and yeast. Hum. Mutat. 2013;34:1501–1509. doi: 10.1002/humu.22393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suzuki T., Suzuki T. Chaplet column chromatography: isolation of a large set of individual RNAs in a single step. Methods Enzymol. 2007;425:231–239. doi: 10.1016/S0076-6879(07)25010-4. [DOI] [PubMed] [Google Scholar]
- 30.Mengel-Jorgensen J., Kirpekar F. Detection of pseudouridine and other modifications in tRNA by cyanoethylation and MALDI mass spectrometry. Nucleic Acids Res. 2002;30:e135. doi: 10.1093/nar/gnf135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suzuki T., Ikeuchi Y., Noma A., Suzuki T., Sakaguchi Y. Mass spectrometric identification and characterization of RNA-modifying enzymes. Methods Enzymol. 2007;425:211–229. doi: 10.1016/S0076-6879(07)25009-8. [DOI] [PubMed] [Google Scholar]
- 32.Soma A., Ikeuchi Y., Kanemasa S., Kobayashi K., Ogasawara N., Ote T., Kato J., Watanabe K., Sekine Y., Suzuki T. An RNA-modifying enzyme that governs both the codon and amino acid specificities of isoleucine tRNA. Mol. Cell. 2003;12:689–698. doi: 10.1016/s1097-2765(03)00346-0. [DOI] [PubMed] [Google Scholar]
- 33.Yokogawa T., Watanabe Y., Kumazawa Y., Ueda T., Hirao I., Miura K., Watanabe K. A novel cloverleaf structure found in mammalian mitochondrial tRNA(Ser) (UCN) Nucleic Acids Res. 1991;19:6101–6105. doi: 10.1093/nar/19.22.6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arcari P., Brownlee G.G. The nucleotide sequence of a small (3S) seryl-tRNA (anticodon GCU) from beef heart mitochondria. Nucleic Acids Res. 1980;8:5207–5212. doi: 10.1093/nar/8.22.5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roe B.A., Wong J.F.H., Chen E.Y., Armstrong P.W. Recombinant DNA: Proceedings of the Third Cleveland Symposium on Macromolecules, Cleveland, Ohio, 22–26 June 1981. 1981. Sequence analysis of mammalian mitochondrial tRNAs in recombinant DNA; pp. 167–176. [Google Scholar]
- 36.Wakita K., Watanabe Y., Yokogawa T., Kumazawa Y., Nakamura S., Ueda T., Watanabe K., Nishikawa K. Higher-order structure of bovine mitochondrial tRNA(Phe) lacking the ‘conserved’ GG and T psi CG sequences as inferred by enzymatic and chemical probing. Nucleic Acids Res. 1994;22:347–353. doi: 10.1093/nar/22.3.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dubin D.T., HsuChen C.C. Sequence and structure of a methionine transfer RNA from mosquito mitochondria. Nucleic Acids Res. 1984;12:4185–4189. doi: 10.1093/nar/12.10.4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Juhling F., Morl M., Hartmann R.K., Sprinzl M., Stadler P.F., Putz J. tRNAdb 2009: compilation of tRNA sequences and tRNA genes. Nucleic Acids Res. 2009;37:D159–D162. doi: 10.1093/nar/gkn772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Machnicka M.A., Milanowska K., Osman Oglou O., Purta E., Kurkowska M., Olchowik A., Januszewski W., Kalinowski S., Dunin-Horkawicz S., Rother K.M., et al. MODOMICS: a database of RNA modification pathways–2013 update. Nucleic Acids Res. 2013;41:D262–D267. doi: 10.1093/nar/gks1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sprinzl M., Vassilenko K.S. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 2005;33:D139–D140. doi: 10.1093/nar/gki012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mcluckey S.A., Vanberkel G.J., Glish G.L. Tandem mass-spectrometry of small, multiply charged oligonucleotides. J. Am. Soc. Mass Spectromet. 1992;3:60–70. doi: 10.1016/1044-0305(92)85019-G. [DOI] [PubMed] [Google Scholar]
- 42.Yokoyama S., Miyazawa T., Iitaka Y., Yamaizumi Z., Kasai H., Nishimura S. Three-dimensional structure of hyper-modified nucleoside Q located in the wobbling position of tRNA. Nature. 1979;282:107–109. doi: 10.1038/282107a0. [DOI] [PubMed] [Google Scholar]
- 43.Meier F., Suter B., Grosjean H., Keith G., Kubli E. Queuosine modification of the wobble base in tRNAHis influences ‘in vivo’ decoding properties. EMBO J. 1985;4:823–827. doi: 10.1002/j.1460-2075.1985.tb03704.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Iwata-Reuyl D., de Crecy-Lagard V. Enzymatic formation of the 7-deazaguanosine hypermodified nucleosides oftRNA. In: Grosjean H., editor. DNA and RNA Modification Enzymes: Structure, Mechanism, Function and Evolution. Austin, TX: Landes Bioscience; 2009. pp. 377–391. [Google Scholar]
- 45.Vinayak M., Pathak C. Queuosine modification of tRNA: its divergent role in cellular machinery. Biosci. Rep. 2010;30:135–148. doi: 10.1042/BSR20090057. [DOI] [PubMed] [Google Scholar]
- 46.Bjork G. Biosynthesis and function of modified nucleosides. In: Soll D., Rajbhandary U.L., editors. t RNA: Structure, Biosynthesis and Function. Washington, DC: American Society for Microbiology; 1995. pp. 165–205. [Google Scholar]
- 47.Suzuki T. Biosynthesis and Function of tRNA Wobble Modifications. In: Grosjean H., editor. Fine-Tuning of RNA Functions by Modification and Editing. Vol. 12. Berlin, Heidelberg: Springer-Verlag; 2005. pp. 23–69. [Google Scholar]
- 48.El Yacoubi B., Bailly M., de Crecy-Lagard V. Biosynthesis and function of posttranscriptional modifications of transfer RNAs. Annu. Rev. Genet. 2012;46:69–95. doi: 10.1146/annurev-genet-110711-155641. [DOI] [PubMed] [Google Scholar]
- 49.Miyauchi K., Kimura S., Suzuki T. A cyclic form of N6-threonylcarbamoyladenosine as a widely distributed tRNA hypermodification. Nat. Chem. Biol. 2013;9:105–111. doi: 10.1038/nchembio.1137. [DOI] [PubMed] [Google Scholar]
- 50.Bamshad M.J., Ng S.B., Bigham A.W., Tabor H.K., Emond M.J., Nickerson D.A., Shendure J. Exome sequencing as a tool for Mendelian disease gene discovery. Nat. Rev. Genet. 2011;12:745–755. doi: 10.1038/nrg3031. [DOI] [PubMed] [Google Scholar]
- 51.Torres A.G., Batlle E., Ribas de Pouplana L. Role of tRNA modifications in human diseases. Trends Mol. Med. 2014 doi: 10.1016/j.molmed.2014.01.008. doi:10.1016/j.molmed.2014.01.008. [DOI] [PubMed] [Google Scholar]
- 52.Menezes S., Gaston K.W., Krivos K.L., Apolinario E.E., Reich N.O., Sowers K.R., Limbach P.A., Perona J.J. Formation of m2G6 in Methanocaldococcus jannaschii tRNA catalyzed by the novel methyltransferase Trm14. Nucleic Acids Res. 2011;39:7641–7655. doi: 10.1093/nar/gkr475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roovers M., Oudjama Y., Fislage M., Bujnicki J.M., Versees W., Droogmans L. The open reading frame TTC1157 of Thermus thermophilus HB27 encodes the methyltransferase forming N(2)-methylguanosine at position 6 in tRNA. RNA. 2012;18:815–824. doi: 10.1261/rna.030411.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vilardo E., Nachbagauer C., Buzet A., Taschner A., Holzmann J., Rossmanith W. A subcomplex of human mitochondrial RNase P is a bifunctional methyltransferase–extensive moonlighting in mitochondrial tRNA biogenesis. Nucleic Acids Res. 2012;40:11583–11593. doi: 10.1093/nar/gks910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Purushothaman S.K., Bujnicki J.M., Grosjean H., Lapeyre B. Trm11p and Trm112p are both required for the formation of 2-methylguanosine at position 10 in yeast tRNA. Mol. Cell Biol. 2005;25:4359–4370. doi: 10.1128/MCB.25.11.4359-4370.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xing F., Hiley S.L., Hughes T.R., Phizicky E.M. The specificities of four yeast dihydrouridine synthases for cytoplasmic tRNAs. J. Biol. Chem. 2004;279:17850–17860. doi: 10.1074/jbc.M401221200. [DOI] [PubMed] [Google Scholar]
- 57.Liu J., Straby K.B. The human tRNA(m(2)(2)G(26))dimethyltransferase: functional expression and characterization of a cloned hTRM1 gene. Nucleic Acids Res. 2000;28:3445–3451. doi: 10.1093/nar/28.18.3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ellis S.R., Morales M.J., Li J.M., Hopper A.K., Martin N.C. Isolation and characterization of the TRM1 locus, a gene essential for the N2,N2-dimethylguanosine modification of both mitochondrial and cytoplasmic tRNA in Saccharomyces cerevisiae. J. Biol. Chem. 1986;261:9703–9709. [PubMed] [Google Scholar]
- 59.Noma A., Yi S., Katoh T., Takai Y., Suzuki T., Suzuki T. Actin-binding protein ABP140 is a methyltransferase for 3-methylcytidine at position 32 of tRNAs in Saccharomyces cerevisiae. RNA. 2011;17:1111–1119. doi: 10.1261/rna.2653411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.D'Silva S., Haider S.J., Phizicky E.M. A domain of the actin binding protein Abp140 is the yeast methyltransferase responsible for 3-methylcytidine modification in the tRNA anti-codon loop. RNA. 2011;17:1100–1110. doi: 10.1261/rna.2652611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li X., Guan M.X. A human mitochondrial GTP binding protein related to tRNA modification may modulate phenotypic expression of the deafness-associated mitochondrial 12S rRNA mutation. Mol. Cell Biol. 2002;22:7701–7711. doi: 10.1128/MCB.22.21.7701-7711.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li X., Li R., Lin X., Guan M.X. Isolation and characterization of the putative nuclear modifier gene MTO1 involved in the pathogenesis of deafness-associated mitochondrial 12 S rRNA A1555G mutation. J. Biol. Chem. 2002;277:27256–27264. doi: 10.1074/jbc.M203267200. [DOI] [PubMed] [Google Scholar]
- 63.Umeda N., Suzuki T., Yukawa M., Ohya Y., Shindo H., Watanabe K., Suzuki T. Mitochondria-specific RNA-modifying enzymes responsible for the biosynthesis of the wobble base in mitochondrial tRNAs. Implications for the molecular pathogenesis of human mitochondrial diseases. J. Biol. Chem. 2005;280:1613–1624. doi: 10.1074/jbc.M409306200. [DOI] [PubMed] [Google Scholar]
- 64.Nakai Y., Umeda N., Suzuki T., Nakai M., Hayashi H., Watanabe K., Kagamiyama H. Yeast Nfs1p is involved in thio-modification of both mitochondrial and cytoplasmic tRNAs. J. Biol. Chem. 2004;279:12363–12368. doi: 10.1074/jbc.M312448200. [DOI] [PubMed] [Google Scholar]
- 65.Boland C., Hayes P., Santa-Maria I., Nishimura S., Kelly V.P. Queuosine formation in eukaryotic tRNA occurs via a mitochondria-localized heteromeric transglycosylase. J. Biol. Chem. 2009;284:18218–18227. doi: 10.1074/jbc.M109.002477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen Y.C., Kelly V.P., Stachura S.V., Garcia G.A. Characterization of the human tRNA-guanine transglycosylase: confirmation of the heterodimeric subunit structure. RNA. 2010;16:958–968. doi: 10.1261/rna.1997610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wan L.C., Mao D.Y., Neculai D., Strecker J., Chiovitti D., Kurinov I., Poda G., Thevakumaran N., Yuan F., Szilard R.K., et al. Reconstitution and characterization of eukaryotic N6-threonylcarbamoylation of tRNA using a minimal enzyme system. Nucleic Acids Res. 2013;41:6332–6346. doi: 10.1093/nar/gkt322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lamichhane T.N., Mattijssen S., Maraia R.J. Human cells have a limited set of tRNA anticodon loop substrates of the tRNA isopentenyltransferase TRIT1 tumor suppressor. Mol. Cell Biol. 2013;33:4900–4908. doi: 10.1128/MCB.01041-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Reiter V., Matschkal D.M., Wagner M., Globisch D., Kneuttinger A.C., Muller M., Carell T. The CDK5 repressor CDK5RAP1 is a methylthiotransferase acting on nuclear and mitochondrial RNA. Nucleic Aacids Res. 2012;40:6235–6240. doi: 10.1093/nar/gks240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Brule H., Elliott M., Redlak M., Zehner Z.E., Holmes W.M. Isolation and characterization of the human tRNA-(N1G37) methyltransferase (TRM5) and comparison to the Escherichia coli TrmD protein. Biochemistry. 2004;43:9243–9255. doi: 10.1021/bi049671q. [DOI] [PubMed] [Google Scholar]
- 71.Lecointe F., Simos G., Sauer A., Hurt E.C., Motorin Y., Grosjean H. Characterization of yeast protein Deg1 as pseudouridine synthase (Pus3) catalyzing the formation of psi 38 and psi 39 in tRNA anticodon loop. J. Biol. Chem. 1998;273:1316–1323. doi: 10.1074/jbc.273.3.1316. [DOI] [PubMed] [Google Scholar]
- 72.Brzezicha B., Schmidt M., Makalowska I., Jarmolowski A., Pienkowska J., Szweykowska-Kulinska Z. Identification of human tRNA:m5C methyltransferase catalysing intron-dependent m5C formation in the first position of the anticodon of the pre-tRNA Leu (CAA) Nucleic Acids Res. 2006;34:6034–6043. doi: 10.1093/nar/gkl765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tuorto F., Liebers R., Musch T., Schaefer M., Hofmann S., Kellner S., Frye M., Helm M., Stoecklin G., Lyko F. RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat. Struct. Mol. Biol. 2012;19:900–905. doi: 10.1038/nsmb.2357. [DOI] [PubMed] [Google Scholar]
- 74.Becker H.F., Motorin Y., Planta R.J., Grosjean H. The yeast gene YNL292w encodes a pseudouridine synthase (Pus4) catalyzing the formation of psi55 in both mitochondrial and cytoplasmic tRNAs. Nucleic Acids Res. 1997;25:4493–4499. doi: 10.1093/nar/25.22.4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chujo T., Suzuki T. Trmt61B is a methyltransferase responsible for 1-methyladenosine at position 58 of human mitochondrial tRNAs. RNA. 2012;18:2269–2276. doi: 10.1261/rna.035600.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Behm-Ansmant I., Massenet S., Immel F., Patton J.R., Motorin Y., Branlant C. A previously unidentified activity of yeast and mouse RNA:pseudouridine synthases 1 (Pus1p) on tRNAs. RNA. 2006;12:1583–1593. doi: 10.1261/rna.100806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Watanabe Y., Kawai G., Yokogawa T., Hayashi N., Kumazawa Y., Ueda T., Nishikawa K., Hirao I., Miura K., Watanabe K. Higher-order structure of bovine mitochondrial tRNA(SerUGA): chemical modification and computer modeling. Nucleic Acids Res. 1994;22:5378–5384. doi: 10.1093/nar/22.24.5378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Armengod M.E., Moukadiri I., Prado S., Ruiz-Partida R., Benitez-Paez A., Villarroya M., Lomas R., Garzon M.J., Martinez-Zamora A., Meseguer S., et al. Enzymology of tRNA modification in the bacterial MnmEG pathway. Biochimie. 2012;94:1510–1520. doi: 10.1016/j.biochi.2012.02.019. [DOI] [PubMed] [Google Scholar]
- 79.Ikeuchi Y., Shigi N., Kato J., Nishimura A., Suzuki T. Mechanistic insights into sulfur relay by multiple sulfur mediators involved in thiouridine biosynthesis at tRNA wobble positions. Mol. Cell. 2006;21:97–108. doi: 10.1016/j.molcel.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 80.Randerath E., Agrawal H.P., Randerath K. Specific lack of the hypermodified nucleoside, queuosine, in hepatoma mitochondrial aspartate transfer RNA and its possible biological significance. Cancer Res. 1984;44:1167–1171. [PubMed] [Google Scholar]
- 81.Lee C., Kramer G., Graham D.E., Appling D.R. Yeast mitochondrial initiator tRNA is methylated at guanosine 37 by the Trm5-encoded tRNA (guanine-N1-)-methyltransferase. J. Biol. Chem. 2007;282:27744–27753. doi: 10.1074/jbc.M704572200. [DOI] [PubMed] [Google Scholar]
- 82.Helm M. Post-transcriptional nucleotide modification and alternative folding of RNA. Nucleic Acids Res. 2006;34:721–733. doi: 10.1093/nar/gkj471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhao X., Patton J.R., Davis S.L., Florence B., Ames S.J., Spanjaard R.A. Regulation of nuclear receptor activity by a pseudouridine synthase through posttranscriptional modification of steroid receptor RNA activator. Mol. Cell. 2004;15:549–558. doi: 10.1016/j.molcel.2004.06.044. [DOI] [PubMed] [Google Scholar]
- 84.Zhao X., Patton J.R., Ghosh S.K., Fischel-Ghodsian N., Shen L., Spanjaard R.A. Pus3p- and Pus1p-dependent pseudouridylation of steroid receptor RNA activator controls a functional switch that regulates nuclear receptor signaling. Mol. Endocrinol. 2007;21:686–699. doi: 10.1210/me.2006-0414. [DOI] [PubMed] [Google Scholar]
- 85.Ansmant I., Motorin Y., Massenet S., Grosjean H., Branlant C. Identification and characterization of the tRNA:Psi 31-synthase (Pus6p) of Saccharomyces cerevisiae. J. Biol. Chem. 2001;276:34934–34940. doi: 10.1074/jbc.M103131200. [DOI] [PubMed] [Google Scholar]
- 86.Behm-Ansmant I., Grosjean H., Massenet S., Motorin Y., Branlant C. Pseudouridylation at position 32 of mitochondrial and cytoplasmic tRNAs requires two distinct enzymes in Saccharomyces cerevisiae. J. Biol. Chem. 2004;279:52998–53006. doi: 10.1074/jbc.M409581200. [DOI] [PubMed] [Google Scholar]
- 87.Kato T., Daigo Y., Hayama S., Ishikawa N., Yamabuki T., Ito T., Miyamoto M., Kondo S., Nakamura Y. A novel human tRNA-dihydrouridine synthase involved in pulmonary carcinogenesis. Cancer Res. 2005;65:5638–5646. doi: 10.1158/0008-5472.CAN-05-0600. [DOI] [PubMed] [Google Scholar]
- 88.Toompuu M., Yasukawa T., Suzuki T., Hakkinen T., Spelbrink J.N., Watanabe K., Jacobs H.T. The 7472insC mitochondrial DNA mutation impairs the synthesis and extent of aminoacylation of tRNASer(UCN) but not its structure or rate of turnover. J. Biol. Chem. 2002;277:22240–22250. doi: 10.1074/jbc.M200338200. [DOI] [PubMed] [Google Scholar]
- 89.Messmer M., Putz J., Suzuki T., Suzuki T., Sauter C., Sissler M., Catherine F. Tertiary network in mammalian mitochondrial tRNAAsp revealed by solution probing and phylogeny. Nucleic Acids Res. 2009;37:6881–6895. doi: 10.1093/nar/gkp697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Horton P., Park K.J., Obayashi T., Fujita N., Harada H., Adams-Collier C.J., Nakai K. WoLF PSORT: protein localization predictor. Nucleic Acids Res. 2007;35:W585–W587. doi: 10.1093/nar/gkm259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Giege R., Sissler M., Florentz C. Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Res. 1998;26:5017–5035. doi: 10.1093/nar/26.22.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sibler A.P., Martin R.P., Dirheimer G. The nucleotide sequence of yeast mitochondrial histidine-tRNA. FEBS Lett. 1979;107:182–186. doi: 10.1016/0014-5793(79)80491-3. [DOI] [PubMed] [Google Scholar]
- 93.Tomita K., Ueda T., Watanabe K. The presence of pseudouridine in the anticodon alters the genetic code: a possible mechanism for assignment of the AAA lysine codon as asparagine in echinoderm mitochondria. Nucleic Acids Res. 1999;27:1683–1689. doi: 10.1093/nar/27.7.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jackman J.E., Gott J.M., Gray M.W. Doing it in reverse: 3’-to-5’ polymerization by the Thg1 superfamily. RNA. 2012;18:886–899. doi: 10.1261/rna.032300.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.