Abstract
U12-type introns are a rare class of introns in the genomes of diverse eukaryotes. In the human genome, they number over 700. A subset of these introns has been shown to be spliced at a slower rate compared to the major U2-type introns. This suggests a rate-limiting regulatory function for the minor spliceosome in the processing of transcripts containing U12-type introns. However, both the generality of slower splicing and the subsequent fate of partially processed pre-mRNAs remained unknown. Here, we present a global analysis of the nuclear retention of transcripts containing U12-type introns and provide evidence for the nuclear decay of such transcripts in human cells. Using SOLiD RNA sequencing technology, we find that, in normal cells, U12-type introns are on average 2-fold more retained than the surrounding U2-type introns. Furthermore, we find that knockdown of RRP41 and DIS3 subunits of the exosome stabilizes an overlapping set of U12-type introns. RRP41 knockdown leads to slower decay kinetics of U12-type introns and globally upregulates the retention of U12-type, but not U2-type, introns. Our results indicate that U12-type introns are spliced less efficiently and are targeted by the exosome. These characteristics support their role in the regulation of cellular mRNA levels.
INTRODUCTION
Precursor messenger RNAs (pre-mRNAs), transcribed from the genomic DNA, undergo extensive processing prior to their export to the cytoplasm as mature messenger ribonucleoprotein particles (mRNPs). The processing steps include 5' capping, pre-mRNA splicing and polyadenylation. A large amount of this processing is now known to occur co-transcriptionally (1). At the same time, each step of mRNA processing is subject to quality control in order to achieve a high fidelity in relaying genetic information. Nuclear quality-control pathways are triggered following a delay or accumulation of a particular processing intermediate, and the transcript in question will be either retained at the site of transcription, anchored at the nuclear pore or completely degraded by nuclear exonucleases (2). Such a degrade-when-delayed mechanism has been dubbed kinetic competition; in other words, the kinetic rates of processing reactions versus degradation reactions will determine the overall fate of a particular transcript (3,4).
Quality control of splicing has been studied extensively both in yeast and mammalian cells where the RNA exosome and, more recently, the 5'→3' exonuclease Rat1/XRN2 (5,6) have been implicated in this process. The exosome is an exoribonuclease complex that is involved in biogenesis, degradation and surveillance of many RNA species, including processing of ribosomal RNA, small nuclear and nucleolar RNAs, mRNA turnover and surveillance of aberrant RNAs, and it is present both in the nucleus and in the cytoplasm (7). It consists of a core of nine subunits (called in human CSL4, RRP4, RRP40, RRP41, RRP46, MTR3, RRP42, OIP2 and PM/SCL-75) forming a barrel-shape structure, which is catalytically inactive in eukaryotes, and associated catalytic subunits, which can include RRP6 (or PM/SCL-100), DIS3 or DIS3 homologs (8–12).
A diverse group of eukaryotes harbors two types of spliceosomal introns in their genomes. These introns, termed as U2- and U12-type introns (or major and minor introns), are removed by specific U2- or U12-dependent spliceosomes, respectively. The minor spliceosome contains four unique snRNA components, U11, U12, U4atac and U6atac, and shares the U5 snRNA with the major spliceosome (13). Moreover, in addition to more than a hundred protein components that are shared with the major spliceosome, there are seven unique protein components in the minor spliceosome, all located in the U11/U12 di-snRNP that functions in the recognition of the U12-type introns (14–17). The expression levels of several of the unique minor spliceosome components, namely U11-48K and U11/U12-65K proteins and the U6atac snRNA, have been shown to be tightly regulated in evolutionarily distant organisms (18–20), suggesting that precise control of spliceosome component levels may be important for cellular survival or organismal development (21,22). Mutations in the specific snRNA or protein components of the minor spliceosome have also been associated with two pediatric diseases, microcephalic osteodysplastic primordial dwarfism type 1/Taybi-Linder syndrome (MOPD1/TALS) (23–25) and Isolated familial growth hormone deficiency (IGHD) (26).
Reverse transcriptase-polymerase chain reaction (RT-PCR) investigations of individual genes containing U12-type introns have reported elevated levels of unspliced minor introns compared to the major introns within the same genes in cellular mRNA pools at the steady state (27,28), suggesting a model in which slow splicing of U12-type introns would provide a rate-limiting control for the expression of genes containing U12-type introns (27,29). Consistent with observed increased intron retention levels, quantitative RT-PCR analysis of newly synthesized pre-mRNAs revealed ∼2-fold longer excision kinetics for U12-introns compared to major introns (30). Furthermore, conversion of a U12-type intron to U2-type (27) led to increased protein production, suggesting the rate-limiting control would not only affect mRNA but also protein levels. The rate-limiting regulation model has been recently further reinforced by findings showing that the efficiency of minor splicing can be regulated through the levels U6atac snRNA, which in turn responds to p38MAPK pathway activation (20).
In this study, we have investigated the retention of U12-type introns at the transcriptome level using RNAseq analysis of nuclear RNAs. Consistent with earlier single-gene investigations, we found that on average unspliced U12-type introns are present at ∼2-fold higher level in the nuclear fraction compared to their neighboring U2-type introns. Furthermore, we asked if the unspliced U12-type introns would be the targets of the nuclear quality-control pathways by knocking down two separate components, RRP41 and DIS3, of the exosome complex known to be involved in nuclear RNA turnover and quality control. Our RNAseq analysis, in particular of the RRP41 knockdown, revealed global stabilization of unspliced U12-type introns, in striking contrast to U2-type introns which showed significantly lower proportion of intron retention events, of which only a few were further stabilized by the exosome knockdown. Similar contrast was observed in our kinetic analyses of intron decay in RRP41 knockdown cells, which revealed that U12-type introns show significantly slower decay kinetics and display elevated baseline levels compared to the U2-type introns. Together, our data provide evidence of a transcriptome-wide retention of U12-type introns through poor splicing and their targeting by nuclear quality-control mechanisms.
MATERIALS AND METHODS
Cell culture
All cell lines were maintained in Dulbecco's modified Eagle's medium supplemented with FBS, penicillin and streptomycin. HEp-2 and HEK293 cells were used for small-scale experiments, whereas RNAseq samples were collected from HEp-2 cells only. HEp-2 cells were chosen for their consistent performance both in siRNA transfection experiments and in the subsequent nuclear and cytoplasmic fractionations. Transfections were performed using Oligofectamine or Lipofectamine RNAiMAX following manufacturer's recommendations. Forty eight hours after transfection, cells were harvested by trypsinization and each sample was divided into RNA and protein preparations. Trizol reagent was used for RNA extraction. For protein analysis, the cells were suspended in lysis buffer (25 mM Tris-HCl pH 7.5, 100 mM KCl, 1 mM EDTA, 0.5 mM DTT, protease inhibitor cocktail), sonicated 3 × 30 s using Bioruptor sonicator and cleared by 10 min 13 000 rpm centrifugation in an Eppendorf centrifuge. Rescue cotransfections were performed by transfecting siRNAs first with Lipofectamine RNAiMAX, followed by plasmid transfection after 24 h with Lipofectamine 2000, and cell harvesting 24 h after the second transfection. mRNA decay experiments were performed 48 h after knockdown (control and RRP41 siRNA treated cells) using 100 μM DRB (5,6-dichloro-1-β-D-ribofuranosylbenzimidazole) and harvested directly into Trizol at indicated time points.
Cellular fractionation
Cells were trypsinized, washed once with cold phosphate buffered saline, resuspended in hypotonic buffer (10 mM HEPES-KOH pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT, protease inhibitors) and incubated on ice for 10 min. Then cells were collected by centrifugation, resuspended in hypotonic buffer and lysed with a Kontes tight pestle (B) douncer. Lysis was monitored under the microscope using Trypan blue staining. The lysate was layered on 1 M sucrose in hypotonic buffer and centrifuged 2300 g 30 min at 4°C, resulting in separation of the nuclear pellet from the cytoplasmic fraction at the top of sucrose cushion. Nuclei were washed once with hypotonic buffer containing 1 M sucrose. Nuclear samples were subsequently sonicated and insoluble material was cleared by centrifugation as described above for total cell lysate. The quality of fractionation was monitored on western blots with nuclear (topoisomerase I) and cytoplasmic (eIF2α) markers.
siRNAs
siRNAs were purchased from Eurogentec and Sigma-Aldrich. Exosome and control siRNA sequences are listed in Supplementary Table S2. siRNAs targeting XRN2 and DCP2 have been described by Davidson et al. (6).
Plasmids
The open reading frame of RRP41 was cloned into the pCI-neo vector, as described previously (31), which allows the expression of the N-terminally VSV-tagged RRP41 protein (VSV-RRP41). Two silent point mutations in the coding sequence of RRP41 (corresponding to the siRNA target site) were introduced with the QuikChange Site-Directed Mutagenesis Kit (Agilent Technologies) using the manufacturer's recommendations and primers listed in Supplementary Table S3. More specifically, this procedure modified the codons encoding Q126 and V127 from CAG to CAA and GTG to GTA, respectively.
Antibodies
RRP41 rabbit polyclonal antibodies are described elsewhere (32). DIS3 antibodies were purchased from Abnova (cat. no. H00022894-A01), XRN2 and DCP2 antibodies from Bethyl laboratories (A301-103A and A302-597A) and VSV-tag antibodies from Abcam (ab1874).
RT-PCR and northern blotting
Complementary DNA (cDNA) was synthesized using AMV reverse transcriptase (Promega) and random hexamer primers. RT-PCR primers for PCR (agarose gel analysis) have been described in (16). The gels were scanned with Fuji LAS-3000 CCD camera and images were quantified using AIDA image analyzer software (Raytest). Intensities were normalized according to the lengths of the PCR products. LightCycler 480 DNA SYBR Green I master mix and LightCycler 480 (Roche) were used in all RT-qPCR analyses. Quantitative PCR (qPCR) primers are listed in Supplementary Table S3. For steady-state analysis, each U12-type intron signal was normalized to a U2-type exon–exon signal in the same transcript, and in decay experiments, U12 and U2 intron signal was normalized to a U6 snRNA amplicon, using primers described in (33). All RT-qPCR samples were assayed in triplicates. Northern blotting and probes were as described in (19).
RNA sequencing and analysis
Sequencing libraries were constructed from nuclear and cytoplasmic RNA preparations of HEp-2 cells using SOLiD Total RNA-Seq Kit (Applied Biosystems). Briefly, library construction contained the following steps: ribosomal RNA removal, fragmentation using RNaseIII, reverse transcription, amplification (14 cycles) and size selection (70–150 bp). Sequencing was performed on SOLiD 4. Initial read mapping to the human reference genome (hg19) was performed using SOLiD Bioscope V1.3, followed by removal of PCR duplicates. The paired reads mapping to U12-intron containing genes were normalized to the length of each intron and exon and were also normalized to the number of the paired reads mapped to the intron/exon and its neighboring introns and exons to obtain the corresponding normalized FPKM (Fragments Per Kilobase per Million mapped fragments) values.
In order to avoid biases caused by misannotation and alternative splicing, all intron and exon coordinates of the multiple transcript annotations of the U12-intron containing genes were collapsed and the genes with changes in their U12 intron coordinates were disregarded. Differentially retained introns (U12 and U2-type introns) were identified using the Bioconductor package edgeR 2.4.6 (34) using common dispersion value of 0.02 and P-value cutoff 0.05 for the intron. Intron–exon junction read pairs or individual reads were identified based on their shared mapping to both intronic and exonic sequences. Intron–exon junction levels were normalized by the summation of the intron–exon junction levels across the transcripts. Intron retention factor is the relative retention of the U12 introns (FPKM of the U12 introns) to the retention of their 5' and 3' neighbor introns (the mean of their FPKM).
For the global U2 intron retention analysis, mRNA annotations were extracted from RefSeq. Reads mapped to a single transcript and reads mapped to multiple transcripts were counted separately. For each intron/exon, the number of the reads that were mapped to introns/exons of multiple transcripts was divided uniformly and added to their single mapped reads count. Analysis related to alternative splicing was done using most recent (2013) MISO alternative splicing annotation files (35) downloaded from http://genes.mit.edu/burgelab/miso/docs/annotation.html#id5.
RESULTS
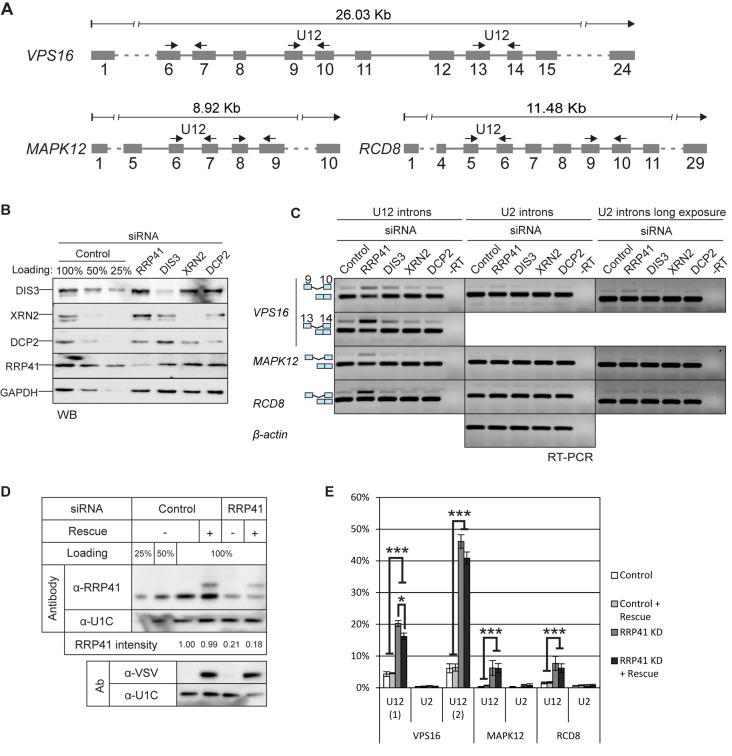
Previous RT-PCR investigations have documented an elevated level of unspliced U12-type introns in the cellular mRNA pool at the steady state (27,36). Given that pre-mRNAs containing unspliced U12-type introns are not exported to the cytoplasm (37), we asked whether nuclear quality-control mechanisms would target mRNAs containing unspliced U12-type introns. We knocked down two exosome subunits in HEp-2 cells, the RRP41 subunit that is one of the core components of the exosome and DIS3, which is one of the catalytic subunits. In both cases, we obtained good knockdown levels as verified by western blotting (Figure 1B). We then assessed the U12-type intron retention levels of three genes, VPS16, MAPK12 and RCD8, using primers in the exons flanking the U12-type introns (Figure 1A). These genes were chosen as they were previously shown to contain moderate to high level unspliced U12-type introns in the steady-state cellular mRNA pool (16). As a control, we analyzed U2-type intron retention levels from the same transcripts, but observed no stabilization of these introns in any of the knockdowns (Figure 1C).
Figure 1.
U12-intron-containing transcripts are stabilized by the exosome knockdown. (A) Exon/intron structures of human VPS16, MAPK12 and RCD8 genes and positions of the primers used in RT-PCR experiments (arrows). (B) Western blot illustrating the depletion of RRP41, DIS3, XRN2 and DCP2 upon siRNA-mediated knockdown. Cell lysates from cells treated with control siRNA were loaded in the indicated amounts. (C) RT-PCR analysis of the three transcripts described in panel (A) and beta-actin after knockdown of RRP41, DIS3, XRN2 or DCP2. The positions of the transcripts (i.e. mRNA versus pre-mRNA) are indicated on the left. U2 panels are shown in two versions, in the middle panel contrast settings are the same as in the U12 panel; in the rightmost panel the same gel is shown with darker settings to reveal the weak U2 intron signals. (D) Expression level of VSV-epitope-tagged RRP41 containing silent mutations at the siRNA target site, analyzed by western blotting using a polyclonal anti-RRP41 antibody and anti-VSV tag antibody. The relative intensity of the endogenous RRP41 signal (normalized to U1C signal) is shown below the panels. (E) Quantification of intron retention levels upon RRP41 knockdown and with or without RRP41 rescue. VPS16, MAPK12 and RCD8 spliced and unspliced signals in agarose gels containing separated RT-PCR products were quantified (*P < 0.05 and ***P < 0.001 with Student's t-test).
Consistent with a hypothesis that the nuclear exosome is involved in the degradation of transcripts containing U12-type introns, we observed a notable stabilization of the retained U12-type introns (2- to 8-fold for RRP41 knockdown and 2- to 4-fold for DIS3 knockdown), but not of the U2 control introns in the same transcripts, nor in the beta-actin gene used as an independent control (Figure 1C). Interestingly, even though knockdown of either exosome subunit seemed to stabilize unspliced U12-type introns, the effect was generally more pronounced for RRP41 knockdown. A possible explanation for the stronger effect is the codepletion of several other exosome subunits, including core exosome components and RRP6 catalytic subunit (Supplementary Figure S1) upon RRP41 depletion, as shown earlier (38). We also tested the other exosome catalytic subunits, of which RRP6 (also known as PM/SCL-100) showed a small, but not repeatable stabilizing effect on U12-type intron retention that varied from experiment to experiment with our test gene set, while the effects of DIS3L1 and DIS3L2 were always negative (Supplementary Figure S1). We also knocked down XRN2 and DCP2, which function in the 5'→3' degradation pathway and have recently been shown to co-transcriptionally monitor and degrade aberrant pre-mRNAs containing unspliced introns (6). Neither knockdown had any effect on either U2- or U12-type intron retention of the tested VPS16, MAPK12 or RCD8 transcripts (Figure 1B and C).
As the exosome is known to be involved in snRNA processing and sn/snoRNA transcription termination (39–42) and the U6atac snRNA has been linked to regulation of the splicing efficiency of U12-type introns (20), we tested whether exosome knockdown causes aberrant processing or destabilization snRNAs of the U12-dependent spliceosome. However, our northern blot analysis of the snRNAs did not reveal any effect on the levels or processing (Supplementary Figure S2A). Neither were the levels of mRNAs encoding the minor spliceosome specific proteins altered in our subsequent RNAseq data set analysis (Supplementary Figure S2D). Finally, we carried out a rescue experiment of the knockdown by transfecting an RRP41 construct with silent point mutations in the siRNA target site after the knockdown. With a moderate level of VSV-RRP41 expression, we were able to obtain a partial rescue of the levels of unspliced U12-type introns (Figure 1D and E). Taken together, these results suggest a role for the DIS3-exosome in the degradation of U12-type intron-containing pre-mRNA processing intermediates.
Global analysis of U12-type intron retention
The above results showed that at least a subset of the more slowly spliced U12-type intron-containing mRNA precursors is degraded by the nuclear exosome. To ask if this is a general property of U12-type intron pre-mRNAs, we carried out a global transcriptome analysis of HEp-2 cells treated with RRP41, DIS3 or control siRNAs targeting green fluorescent protein (GFP). We verified knockdown levels by western blotting (DIS3: 83%; RRP41: 81%; Figure 2A). To increase the relative abundance of mRNAs with unspliced introns, we enriched nuclear RNA by separating nuclei from cytoplasm (Figure 2B). Based on the quantification of the western blots, we observed very low levels (<10%) of cytoplasmic contamination in the nuclear fraction and virtually no nuclear contamination in the cytoplasmic fraction (Figure 2B). The resulting RNA fractions were rRNA depleted and randomly fragmented after which directional sequencing libraries were constructed by ligation of SOLiD sequencing primers to RNA fragments, followed by cDNA synthesis, amplification, size selection and SOLiD sequencing (see Supplementary Figure S3 for overall strategy).
Figure 2.
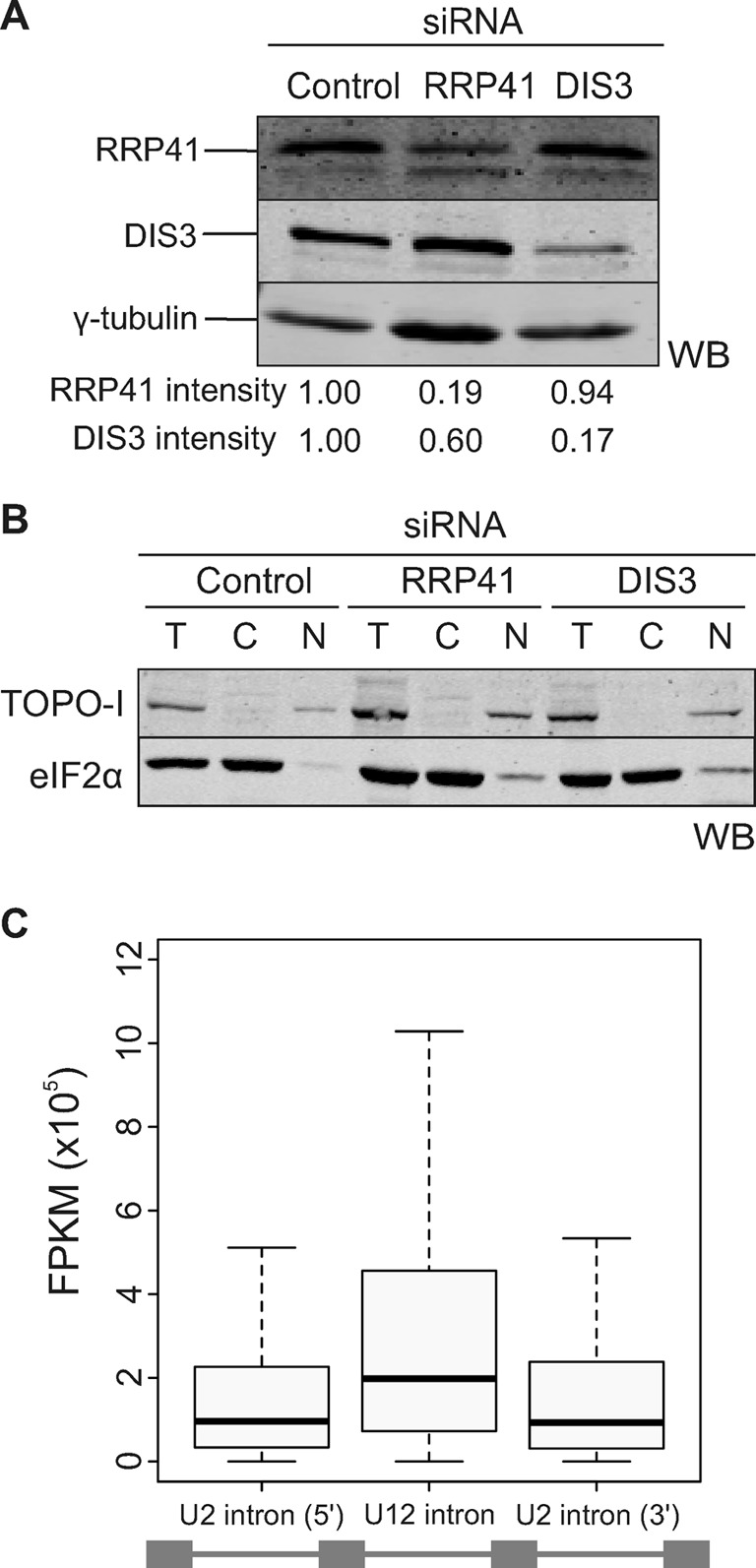
Global analysis of U12-type intron retention. (A) Western blot showing the depletion levels of RRP41 and DIS3 after knockdown. Intensities of the individual bands (shown below each lane) were quantified using the Odyssey Imaging System (LI-COR) and normalized to the γ-tubulin signal. (B) Western blot of nuclear and cytoplasmic fractions of cells after knockdown. Topoisomerase I (TOPO-I) was used as a nuclear marker and eIF2α as a cytoplasmic marker. T, total cell lysate; C, cytoplasmic fraction; N, nuclear fraction. (C) Distribution of normalized read values in U12-type introns in comparison with the flanking U2-type introns, as indicated below the figure, in the nuclear fraction of control siRNA treated cells. The whiskers represent 1.5 times the interquartile range.
We mapped the reads from both nuclear and cytoplasmic fractions to genes containing U12-type introns, followed by removal of PCR duplicates (see Table 1 for overall read statistics). Normalized FPKM values were calculated for each exon and intron within the gene set as described in the Materials and Methods section.
Table 1. RNAseq statistics.
| Sequencing platform | SOLiD 4 |
|---|---|
| Reads size | 50–35 |
| Fragment size | ∼70–150 bps |
| Overall readsa | 865 136 419 |
| Reads mapped to genome | 354 850 544 |
| Reads mapped to U12 genes | 26 861 817 |
| Unique reads mapped to: | |
| Introns of U12 genes | 7 187 277 |
| Exons of U12 genes | 1 788 277 |
| U12 introns | 683 051 |
| U2 introns of U12 genes | 6 468 221 |
| Flanking exons of U12 introns | 167 648 |
aTotal of all samples.
We detected the RNAseq reads from 544 U12-type introns, covering 78% of all known human U12-type introns (43). We first compared the global retention of U12-type introns. FPKM values from U12-type introns were compared to the values obtained from the neighboring U2-type introns. As seen in Figure 2C, we found that the retention levels of the U12-type introns were on average two times higher than their close U2-type introns. Similar results were also observed with RRP41 and DIS3 knockdown samples, but with a further increased contrast between U12- and U2-type introns (see below). To exclude the possibility that the above result would be caused by stabilization of excised introns in our RNA pool, we separately examined exon–intron junction reads (Supplementary Figure S4). Although the number of such reads was limited in our data set due to the short read length, the general result, i.e. ≥2-fold elevated retention of U12-type introns in comparison to neighboring U2-type introns, was also seen with all the three exon–intron junction read data sets (control, RRP41 and DIS3 knockdowns), suggesting that our results with intron-specific reads reflect the stabilization of transcripts containing unspliced U12-type introns, and not the stabilization of spliced intron lariats. This interpretation is further supported by our initial RT-PCR observations using intron–exon spanning primers (see Figure 1) and by the validation of the most stabilized introns (see below), which all showed elevated U12-type intron signals compared to the U2-type introns used as controls. Finally, to investigate if a subset of our results could be explained by possible intron-encoded stable RNAs (such as miRNAs, snoRNAs or other non-coding RNAs) residing within the U12-type introns, we also examined the distribution of annotated RNA and protein coding genes within the U12-type introns. Interestingly, U12-type introns in the human genome appear to be completely devoid of annotated ncRNA genes. Taken together, our transcriptome-wide results indicate increased levels of transcripts containing unspliced U12-type introns in the nuclear RNA pool, substantiating the previous observations with individual U12-type intron-containing transcripts.
Exosome knockdown leads to stabilization of U12-type intron containing pre-mRNAs
We investigated transcriptome-wide effects of the exosome subunit knockdowns on U12-type intron retention by identifying introns that were differentially retained in either knockdown using a P-value cutoff of 0.05 (34). Similarly as with the total U12-type intron data set, the stabilized introns also showed higher retention levels compared to the neighboring U2-type introns (cf. Figures 2C and 3D). Of the 119 introns meeting the statistical criteria, 80% were derived from RRP41 knockdown. Majority of those were stabilized (Figure 3A and C) and displayed on average 2-fold higher retention levels compared to the control knockdown (Figure 3D). This set includes introns in PSMC4, KIFAP and IFT80 that have been identified earlier [(30); see Supplementary Table S1 (36)]. With the DIS3 knockdown the number of statistically significant introns was significantly smaller and the ratio between stabilized and destabilized U12-type introns more even, with a slight excess of destabilized U12-type introns (Figure 3B and C). Nevertheless, there was a statistically significant overlap between the two knockdowns for both the stabilized and destabilized U12-type introns (P = 1.74E−10 and P = 0.0122, respectively, using hypergeometric test).
Figure 3.
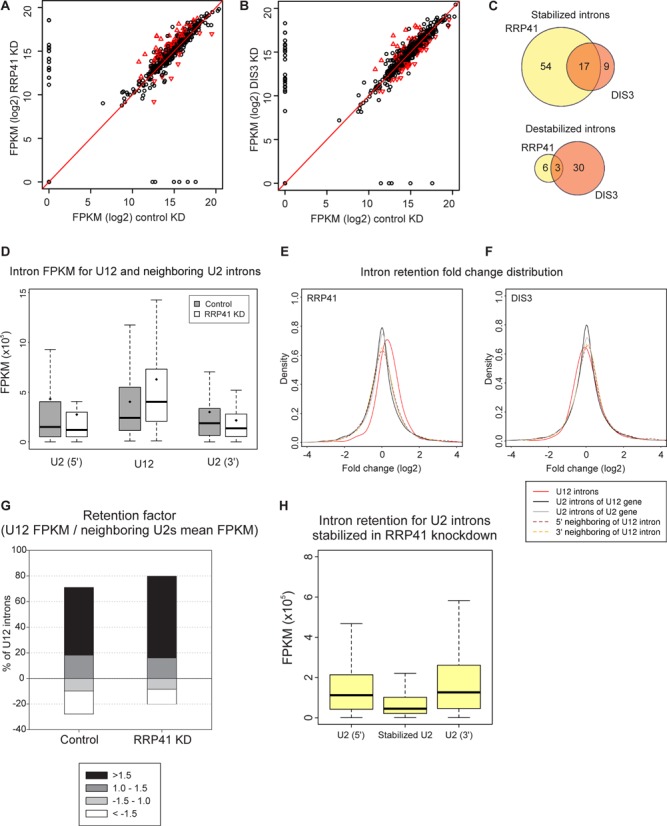
Exosome knockdown increases retention of U12-type introns. (A) Pairwise comparison of U12 intron retention values in the control sample versus the RRP41 knockdown (KD) sample. Black circles: introns that are not significantly changed; red triangles: introns that are significantly changed in knockdown versus control. (B) As in panel (A), DIS3 knockdown versus control knockdown. (C) Venn diagram of significantly stabilized or destabilized U12-type introns upon RRP41 and DIS3 knockdown. (D) Intron FPKM values for the stabilized U12 introns and their neighboring U2 introns (when comparing RRP41 KD to control). Gray boxes: control; white boxes: RRP41 knockdown. The black line in the box represents the median, diamond represents the mean and whiskers represent 1.5 times the interquartile range. (E) Fold change density plot for the retention of introns in RRP41 versus control knockdown. Red line: all the detected U12-type introns; black line: all the U2-type introns in genes containing U12-type introns; gray line: randomly sampled U2-type introns in genes lacking U12-type introns; brown dashed line: U2-type introns located upstream of U12-type introns; yellow dashed line: U2-type introns located downstream of U12-type introns. (F) As in panel (E), for DIS3 knockdown. (G) Intron retention factor analysis of U12 introns in the control and RRP41 samples. For the calculation of intron retention factor, the U12 intron FPKM value was divided by the mean FPKM values obtained from the surrounding introns. The obtained retention factors were used to divide the U12-type intron in four classes as indicated. For illustrative purposes, the retention values that are below 1 (i.e. U2-type intron signals are higher than U12-type intron signals) have been shown as negative values: >1.5-fold retention; 1 to 1.5-fold retention; −1.5 to 1-fold retention and <−1.5-fold retention. (H) Analysis of U2-type introns in the control nuclear knockdown data set using the introns that show statistically significant stabilization after RRP41 knockdown. Distribution of normalized read values in the U2-type intron stabilized in RRP41 knockdown as compared to the flanking U2-type introns as indicated.
A closer examination of the data set revealed a more widespread effect upon RRP41 knockdown. Significantly, the entire group of U12-type introns, comprising also the majority of those above the statistical cutoff, has been stabilized as a result of the RRP41 depletion as indicated by the off-diagonal shift seen in Figure 3A and more clearly in a frequency plot (Figure 3E; see also Supplementary Figure S7). This is also seen as an increased number of stabilized U12-type introns (Figure 3G). While the DIS3 knockdown does not lead to similar prominent off-diagonal global changes (Figure 3B and F), a pairwise comparison of both the RRP41 and DIS3 data at intron level (Supplementary Figure S5) indicates a similar response of U12-type intron retention, but the magnitude of change with DIS3 is less pronounced than with the RRP41 knockdown (slope 0.607).
Importantly, the global shift observed with RRP41 knockdown was statistically highly significant (P < 2.2E−16, Jonckheere trend test) and was not observed with the U2-type introns located immediately up- or downstream of the U12-type introns (Figure 3F) or in the group of all the U2-type introns in the same genes. Similarly, random sampling of U2-type introns from the genes with similar expression levels as those carrying U12-type introns also returned a profile centered at 0-fold change (Figure 3E). We conclude that U12-type introns are preferably stabilized upon exosome subunit knockdown and that the effect of stabilization with RRP41 subunit is more prominent compared to the DIS3 subunit. In many cases the effect with U12-type introns is small, but it is still distinguishable in our global analysis, while the largest effects are about 4-fold. Furthermore, it suggests that in these transcripts U12-type introns remain unspliced whereas U2-type introns have already been excised.
To ask if the differential responses of the U12-type introns to exosome subunit depletion can be explained by differences in sequence characteristics, we compared the statistically significant and unchanged introns. The analysis included intron length, position (first, last or middle location), distance to 3' end of the gene, intron subtype (AT-AC vs. GT-AG) and the strength of splice sites. For splice site analysis, we also generated sequence logos for the 5'ss, branchpoint and 3'ss of statistically stabilized and unchanged introns for visual examination (Supplementary Figure S6). Finally, we carried out k-mer analysis searching for over-represented or under-represented 6-mers in the stabilized and destabilized U12 introns and their flanking exons using Fisher's exact test. In none of the analyses did we detect any significant correlation that would explain the differences between statistically stabilized and unchanged introns. Our negative results suggest either that there is no clear-cut classification between the two groups, but perhaps rather a continuum of the responses as suggested by the results in Figure 3E. Alternatively, there may also be multiple signals and pathways resulting the same exosome response, which make identification difficult from a relatively small set used in our analyses.
To ask if we could detect U2-type introns responding in the same way to exosome knockdown as U12-type introns, we analyzed the entire human U2-type intron complement to identify U2-type introns showing statistically significant stabilization in our edgeR analysis. Strikingly, out of 200.033 unique introns analyzed we identified only 0.6% (1208 introns) that showed stabilization after knockdown while the percentage is 14.7% for U12-type introns using the same statistical cutoff. A further analysis of the retention profile of these introns in the control cells revealed that they displayed a lower retention compared to the neighboring U2-type introns unlike the U12-type introns that were more retained in the control cells (cf. Figures 3H and 2C). Further selection to identify introns that show a similar retention pattern as U12-type introns in Figure 2C and respond to RRP41 knockdown returned only 86 introns. Our analysis of the 1208 U2-type introns that are stabilized by RRP41 knockdown or the group of 86 introns that show higher retention levels in control cells and further stabilization after RRP41 knockdown did not return statistically significant correlations using the same sequence and position characteristics as we used in the U12-type intron analysis above. However, within the group of 86 U2-type introns 30 are listed as introns related to exon-skipping events (see Supplementary Table S5) in alternative splicing database. This result is statistically significant (P < 0.01, Fisher's exact test) and is interesting because alternative splicing events have been reported to often occur post-transcriptionally (44–46).
Together, our analysis with U2-type introns emphasizes differences between the two spliceosomes with U12-type intron being affected significantly more by the exosome knockdown than the U2-type introns. Interestingly, a small subset of alternatively spliced U2-type introns showed increased retention in control cells and further stabilization upon RRP41 knockdown similarly as the U12-type introns.
Kinetic differences between U12- and U2-type intron decay
To ask if there are differences in the kinetics of mRNA decay between the two intron types, we investigated a subset of U12-type introns identified from the RNAseq analysis. First we used RT-qPCR using total RNA preparations from three independent knockdown reactions to confirm the U12-type intron retention with primers that span exon–intron junctions (Figure 4A). Of the top eight U12-type introns in the RRP41 knockdown list showing elevated intron retention (Supplementary Table S1), seven out of eight were validated in RT-qPCR, all showing typically more than 3-fold higher levels than in control knockdown (Figure 4B), even though the exact stabilization values varied somewhat between the individual knockdown reactions (see Figure 4C). Five of these introns were also showing higher retention levels in the RNAseq data of the DIS3 knockdown, and only one of these (RABL2B) was validated by RT-qPCR (Figure 4B). This may reflect that DIS3 knockdown effect size is smaller, and while it is seen in the nuclear enriched RNAseq dataset, it may be more easily missed in the total RNA samples that were used in the RT-qPCR (compare also to Figure 1C).
Figure 4.
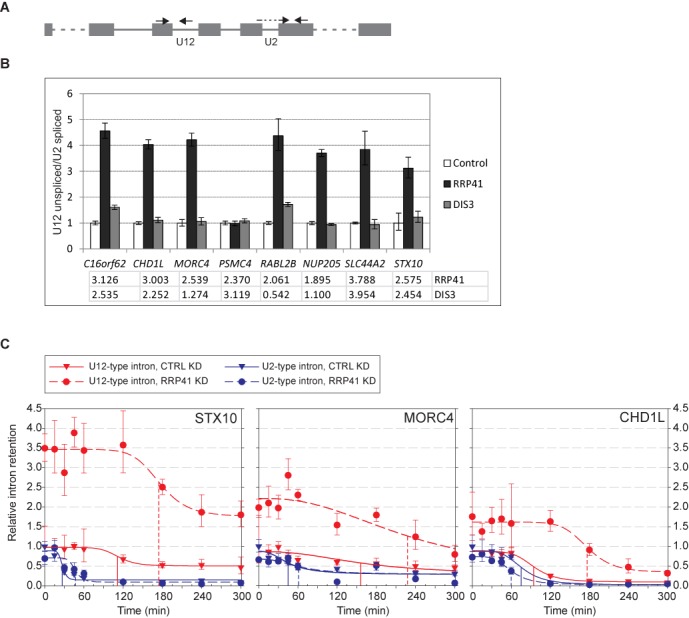
The effect of RRP41 knockdown on U12- and U2-type intron nuclear decay. (A) Schematic of the RT-qPCR primer setup. In each case, the intron retention is measured using a forward primer in the upstream exon and a reverse primer in the intron. The obtained value is normalized with a U2-type exon–exon junction value from the same transcript as shown in the figure. (B) RT-qPCR quantification of U12 intron retention in control, RRP41 knockdown and DIS3 knockdown samples. Total RNA was used as starting material. U12-type intron retention in control sample was set at 1. Error bars represent standard deviation of three replicates. Numbers under the chart represent intron retention fold change values in RNAseq data. (C) RT-qPCR quantification of intron retention in total RNA following transcription inhibition by DRB. DRB was added on the cells at t = 0 and RNA samples were harvested in Trizol at indicated time points. U2-type intron values are shown in blue and the U12-type intron values in red, control experiments in triangles and knockdown in circles. All values were normalized to a U6 snRNA amplicon. U2- and U12-type intron data were scaled independently so that control reaction t = 0 time point was set to 1.0. The data were fitted to a four-parameter logistic equation and the resulting curve fits for each data set are shown. Control and RRP41 knockdown fits are indicated with the solid and dotted lines, respectively. Inferred half-lives are indicated by drop lines for each intron and condition.
Subsequently we selected three qPCR-validated genes (see Figure 4B), STX10, MORC4 and CHD1L, that have RRP41 knockdown-stabilized U12-type introns in either 5' end, in the middle of the transcript or 3' end, respectively. We investigated pre-mRNA decay kinetics between the U12-type intron and a U2-type intron in the same gene following inhibition of transcription initiation. We treated control and knockdown cells with DRB, which inhibits CDK7 phosphorylation, and thus prevents transcription initiation, but does not affect ongoing elongation or pre-mRNA processing (30). Therefore, the decline of the pre-mRNA signal is a combination of both pre-mRNA splicing and nuclear decay. As seen in Figure 4C, there is a clear accumulation of transcripts containing unspliced U12-type, but not U2-type introns, upon RRP41 knockdown. Following DRB treatment, the U12-type intron levels decline after a gene-specific lag period; however, the lag period does not correlate with estimated elongation time after DRB addition, as all three transcripts are expected to complete transcription within ∼15 min of incubation start, calculated with an elongation rate of 3.8 kb/min as in (30). The lag period was clearly longer for U12-type introns than U2-type introns. RRP41 knockdown led to a further, ∼2-fold, extension of the lag period with the U12-type introns in CHDL1 and STX10 genes. Using the data we estimated the half-lives of the U2- and U12-type intron containing pre-mRNAs. With U2-type introns the half-lives varied between ∼30 min and 75 min with little differences between the control and RRP41 knockdown. In comparison, with the U12-type introns half-lives were clearly increased with each gene in the RRP41 knockdown, but the exact numbers were difficult to estimate because of very high baseline levels of the U12-type introns even after extended incubation in the presence of DRB. Nevertheless, conservative estimates indicate ∼180–210-min minimal half-life after RRP41 knockdown, which is 1.5 to 2-fold longer than in control cells.
Together, the half-life measurements indicate clear differences between the two intron types. The slow decline of the U12-type pre-mRNA signal in particularly in the RRP41 knockdown cells supports the model in which the nuclear exosome is involved in the decay of transcripts containing unspliced U12-type introns.
DISCUSSION
In this study, we have investigated the nuclear retention of transcripts containing unspliced U12-type introns at the whole transcriptome level using RNAseq methods. On average, we find ∼2-fold higher levels of unspliced U12-type introns compared to the neighboring U2-type introns on the same transcript in human cells. Additionally, the nuclear levels of a subset of unspliced U12-type introns are further increased after the knockdown of exosome subunits. In particular, depletion of the core subunit RRP41 leads to stabilization of large number of U12-type introns that display significantly delayed and inefficient decay kinetics, suggesting that transcripts containing unspliced U12-type introns are actively degraded by the exosome complex. Thus our results provide the first transcriptome-level evidence that the inefficient splicing and accumulation of unspliced introns is a general property of most U12-type introns. Furthermore, we show that nuclear exosome, possibly together with the other nuclear processing mechanisms, targets such transcripts and prevents their accumulation in the nucleus.
Our results on minor intron retention are consistent with previous single-gene observations in mammalian and Drosophila cells that have similarly reported 2 to 4-fold higher levels of unspliced U12-type introns in cellular steady-state RNA pools but extend these results to transcriptome-wide level (27,36). Furthermore, we observed selective stabilization of majority of U12-type introns, but only a very small subset of U2-type introns by the nuclear exosome depletion. Thus our results provide now an experimental evidence that links the slower splicing of U12-type introns that has been observed both in vivo (30) and in vitro (47,48), and the proposed role of these introns as rate-limiting controls that regulate the formation of fully processed mRNAs (20,27). Nuclear degradation has been implicated with earlier experiments using a splicing reporter with a U12-type intron which show increased pre-mRNA degradation (27) and after an antisense oligonucleotide blocking of U6atac snRNA that leads to downregulation of all minor-intron containing genes (20), but in those earlier studies the nuclear components involved in pre-mRNA degradation had not been identified.
The underlying mechanisms behind the slower splicing or elevated intron retention levels are not fully understood. However, recent work has provided evidence that U6atac snRNA, a component of the catalytic core of the minor spliceosome, could be a molecular switch that regulates the efficiency of splicing of U12-type introns (20). Alternatively or additionally, the splicing efficiency could be regulated by limiting the abundance of the key protein components of the minor spliceosome, for example via the negative feedback loop that controls the cellular homeostasis of the U11/U12 di-snRNP. This regulatory system destabilizes the transcripts of the U11-48K and U11/U12-65K proteins, which are necessary for intron recognition (19). In both cases the regulatory pathways respond to external factors, such as MAPK signalling and hnRNP H protein levels (18,20).
Analogous results have also been observed in a number of experiments with U2-type introns. For example, defects in spliceosome components or splicing factors (5), inhibition of splicing by spliceostatin A (6), defunct splice sites (49) and changes in nuclear retention (50) have all been shown to induce degradation of incompletely spliced transcripts. Thus, any deviation in the kinetics of pre-mRNA splicing that prevents nuclear export also leads to nuclear decay as suggested by the kinetic proofreading hypothesis (3,4). In yeast, a similar nuclear pathway controlling expression of a set of genes has been described. There, it has been shown that mRNAs containing slowly spliced introns are degraded and that this response requires PAB2 and RRP6 (51). Together with the yeast study, the U12-type intron findings presented here support the conclusion that this is a widespread mechanism for gene regulation.
Since alternative splicing has been reported to favor post-transcriptional processing (44–46), introns related to alternative splicing events are potential candidates to display similar retention and exosome-related degradation patterns as U12-type introns. Intriguingly, our query to such U2-type introns revealed an enrichment of introns related to exon-skipping events in the nuclear compartment. However, given that our retrieved group of introns represents a minor subpopulation of all exon-skipping events (or alternative splicing in general), this is unlikely to be a general characteristic of exon-skipping introns.
Recent transcriptome-wide work has indicated that the exosome targets more than half of the yeast intron-containing genes. (40). Similarly, crosslinking and cDNA sequencing (CRAC) experiments in yeast revealed that DIS3 binds to intron-containing pre-mRNAs (41). The observation that so many transcripts are subjected to degradation pathways suggests that mis-processing events are very common, but in the case of U12-type intron-containing transcripts, and others, it can also be a mechanism for regulation. Excess production of transcripts that are constitutively degraded by the exosome can be used to quickly activate gene expression patterns in a switch-like manner, as shown in fission yeast meiotic gene expression (52), response to environmental stress (51), and a similar mechanism has been suggested for U12-type introns recently (20).
The effect with RRP41 knockdown on retention of U12-type intron-containing transcripts led mostly to stabilization of the pre-mRNAs. In addition to the statistically significant effect on individual transcripts, the U12-type intron-containing transcripts were also stabilized as a group. The most likely explanation for such a strong effect is that depletion of the RRP41 destabilizes the core of the exosome, causing co-depletion of the other core subunits, similarly as observed with mutations inactivating Rrp41, which inhibit the activity of the whole exosome complex (7,53). In contrast, with the DIS3 knockdown the overall stabilization of the U12-type intron-containing transcripts was not detected at all (Figure 3F) and many introns were in fact destabilized. Why this would be the case is somewhat unclear; however, it may be speculated that the knockdown of DIS3 may cause a disturbance in the ratio of active exosome subunits or in their relative association with the core exosome. Thus, overall exonucleolytic activity may increase upon the knockdown of one active subunit, as the others may compensate (R. Staals and G. Pruijn, manuscript in preparation). Nevertheless, of the introns showing a significant stabilization effect 17 (out of 80) were shared between the two knockdowns.
A possible alternative hypothesis to explain our data on U12-type intron stabilization is that exosome (or the RRP41 subunit) is itself involved in the splicing of U12-type introns. However, our data argue against this possibility as the knockdown of neither exosome component leads to reduction of U12-type spliced reads in the nucleus (exon–exon junction reads; see Supplementary Figure S2B) or reduction of mRNA levels in the cytoplasm (Supplementary Figure S2C). Nor are the minor spliceosome components affected by exosome knockdown (neither snRNAs as detected on northern blot nor the mRNA levels of the protein components specific to the minor spliceosome, as detected in RNAseq data; Supplementary Figure S2A and D). Finally, earlier mass spectrometry analyses on proteins interacting with the exosome components have failed to detect minor spliceosome components, and, conversely, analysis of minor spliceosome protein composition has failed to detect exosome components (12,17,54).
Given that only a subset of U12-type intron-containing genes showed a strong response to either exosome knockdowns, we sought for potential characteristics from the sequence of the introns themselves and the surrounding exons that would explain differential stabilization. However, none of characteristics tested, including intron length, position within the gene, intron subtype, splice site strength, expression level of the transcript, enrichment or absence of sequence elements within the intron or the surrounding exons, showed any correlation with the observed stabilization effect. The most likely explanation for the lack of enriched signals is that many different sequences and interactions can result in a similar outcome or that our strongest responding introns represent only a subset of introns which all share the similar characteristics but with varying strengths as suggested by the data in Figure 3E.
In summary, we provide here transcriptome-wide evidence that the overall intron retention is higher in the steady state for U12-type introns compared to U2-type introns. Furthermore, our data support a role for the nuclear exosome, among its other tasks, in the quality control of pre-mRNAs and in targeting such transcripts that have retained unspliced U12-type introns. We postulate that U12-type introns are needed for the proper regulation of the genes that they reside in and that the mechanism of regulation relies on surveillance of nuclear processing, suggesting that the minor spliceosome is essential for the regulatory networks of eukaryotic cells.
ACCESSION NUMBER
RNAseq raw read data and processed intron analyses are available at NCBI GEO database under accession number GSE52539.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We thank Marja-Leena Peltonen for technical assistance during this work and the members of both Frilander and Pruijn laboratories for stimulating discussions. We thank Finnish IT Center for Science Ltd (CSC) and Finnish Institute for Molecular Medicine (FIMM) for computing resources.
Footnotes
The authors wish it to be known that, in their opinion, the first two authors should be regarded as Joint First Authors.
FUNDING
Academy of Finland [140087 to M.J.F.]; Sigrid Jusélius Foundation [to M.J.F.]; Helsinki Graduate School in Biotechnology and Molecular Biology [to E.H.N.]; Viikki Doctoral Programme in Molecular Biosciences [to A.O.]; Orion-Farmos Foundation [to E.H.N.]. Funding for open access charge: Academy of Finland [140087 to M.J.F.].
Conflict of interest statement. None declared.
REFERENCES
- 1.Moore M.J., Proudfoot N.J. Pre-mRNA processing reaches back totranscription and ahead to translation. Cell. 2009;136:688–700. doi: 10.1016/j.cell.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 2.Schmid M., Jensen T.H. Nuclear quality control of RNA polymerase II transcripts. Wiley Interdiscip. Rev. RNA. 2010;1:474–485. doi: 10.1002/wrna.24. [DOI] [PubMed] [Google Scholar]
- 3.Burgess S.M., Guthrie C. Beat the clock: paradigms for NTPases in the maintenance of biological fidelity. Trends Biochem. Sci. 1993;18:381–384. doi: 10.1016/0968-0004(93)90094-4. [DOI] [PubMed] [Google Scholar]
- 4.Doma M.K., Parker R. RNA quality control in eukaryotes. Cell. 2007;131:660–668. doi: 10.1016/j.cell.2007.10.041. [DOI] [PubMed] [Google Scholar]
- 5.Bousquet-Antonelli C., Presutti C., Tollervey D. Identification of a regulated pathway for nuclear pre-mRNA turnover. Cell. 2000;102:765–775. doi: 10.1016/s0092-8674(00)00065-9. [DOI] [PubMed] [Google Scholar]
- 6.Davidson L., Kerr A., West S. Co-transcriptional degradation of aberrant pre-mRNA by Xrn2. EMBO J. 2012;31:2566–2578. doi: 10.1038/emboj.2012.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Dijk E.L., Schilders G., Pruijn G.J. Human cell growth requires a functional cytoplasmic exosome, which is involved in various mRNA decay pathways. RNA. 2007;13:1027–1035. doi: 10.1261/rna.575107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dziembowski A., Lorentzen E., Conti E., Seraphin B. A single subunit, Dis3, is essentially responsible for yeast exosome core activity. Nat. Struct. Mol. Biol. 2007;14:15–22. doi: 10.1038/nsmb1184. [DOI] [PubMed] [Google Scholar]
- 9.Makino D.L., Baumgartner M., Conti E. Crystal structure of an RNA-bound 11-subunit eukaryotic exosome complex. Nature. 2013;495:70–75. doi: 10.1038/nature11870. [DOI] [PubMed] [Google Scholar]
- 10.Malecki M., Viegas S.C., Carneiro T., Golik P., Dressaire C., Ferreira M.G., Arraiano C.M. The exoribonuclease Dis3L2 defines a novel eukaryotic RNA degradation pathway. EMBO J. 2013;32:1842–1854. doi: 10.1038/emboj.2013.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Staals R.H., Bronkhorst A.W., Schilders G., Slomovic S., Schuster G., Heck A.J., Raijmakers R., Pruijn G.J. Dis3-like 1: a novel exoribonuclease associated with the human exosome. EMBO J. 2010;29:2358–2367. doi: 10.1038/emboj.2010.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tomecki R., Kristiansen M.S., Lykke-Andersen S., Chlebowski A., Larsen K.M., Szczesny R.J., Drazkowska K., Pastula A., Andersen J.S., Stepien P.P., et al. The human core exosome interacts with differentially localized processive RNases: hDIS3 and hDIS3L. EMBO J. 2010;29:2342–2357. doi: 10.1038/emboj.2010.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Turunen J.J., Niemelä E.H., Verma B., Frilander M.J. The significant other: splicing by the minor spliceosome. Wiley Interdiscip. Rev. RNA. 2013;4:61–76. doi: 10.1002/wrna.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frilander M.J., Steitz J.A. Initial recognition of U12-dependent introns requires both U11/5’ splice-site and U12/branchpoint interactions. Genes Dev. 1999;13:851–863. doi: 10.1101/gad.13.7.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schneider C., Will C.L., Makarova O.V., Makarov E.M., Lührmann R. Human U4/U6.U5 and U4atac/U6atac.U5 tri-snRNPs exhibit similar protein compositions. Mol. Cell. Biol. 2002;22:3219–3229. doi: 10.1128/MCB.22.10.3219-3229.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Turunen J.J., Will C.L., Grote M., Lührmann R., Frilander M.J. The U11–48K protein contacts the 5’ splice site of U12-type introns and the U11–59K protein. Mol. Cell. Biol. 2008;28:3548–3560. doi: 10.1128/MCB.01928-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Will C.L., Schneider C., Hossbach M., Urlaub H., Rauhut R., Elbashir S., Tuschl T., Lührmann R. The human 18S U11/U12 snRNP contains a set of novel proteins not found in the U2-dependent spliceosome. RNA. 2004;10:929–941. doi: 10.1261/rna.7320604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Turunen J.J., Verma B., Nyman T.A., Frilander M.J. HnRNPH1/H2, U1 snRNP and U11 snRNP co-operate to regulate the stability of the U11–48K pre-mRNA. RNA. 2013;19:380–389. doi: 10.1261/rna.036715.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Verbeeren J., Niemelä E.H., Turunen J.J., Will C.L., Ravantti J.J., Lührmann R., Frilander M.J. An ancient mechanism for splicing control: U11 snRNP as an activator of alternative splicing. Mol. Cell. 2010;37:821–833. doi: 10.1016/j.molcel.2010.02.014. [DOI] [PubMed] [Google Scholar]
- 20.Younis I., Dittmar K., Wang W., Foley S.W., Berg M.G., Hu K.Y., Wei Z., Wan L., Dreyfuss G. Minor introns are embedded molecular switches regulated by highly unstable U6atac snRNA. eLife. 2013;2:e00780. doi: 10.7554/eLife.00780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Markmiller S., Cloonan N., Lardelli R.M., Doggett K., Keightley M.-C., Boglev Y., Trotter A.J., Ng A.Y., Wilkins S.J., Verkade H., et al. Minor class splicing shapes the zebrafish transcriptome during development. Proc. Natl. Acad. Sci. U.S.A. 2014;111:3062–3067. doi: 10.1073/pnas.1305536111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Otake L.R., Scamborova P., Hashimoto C., Steitz J.A. The divergent U12-type spliceosome is required for pre-mRNA splicing and is essential for development in Drosophila. Mol. Cell. 2002;9:439–446. doi: 10.1016/s1097-2765(02)00441-0. [DOI] [PubMed] [Google Scholar]
- 23.Pessa H.K.J., Frilander M.J. Minor splicing, disrupted. Science. 2011;332:184–185. doi: 10.1126/science.1205503. [DOI] [PubMed] [Google Scholar]
- 24.He H., Liyanarachchi S., Akagi K., Nagy R., Li J., Dietrich R.C., Li W., Sebastian N., Wen B., Xin B., et al. Mutations in U4atac snRNA, a component of the minor spliceosome, in the developmental disorder MOPD I. Science. 2011;332:238–240. doi: 10.1126/science.1200587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Edery P., Marcaillou C., Sahbatou M., Labalme A., Chastang J., Touraine R., Tubacher E., Senni F., Bober M.B., Nampoothiri S., et al. Association of TALS developmental disorder with defect in minor splicing component U4atac snRNA. Science. 2011;332:240–243. doi: 10.1126/science.1202205. [DOI] [PubMed] [Google Scholar]
- 26.Argente J., Flores R., Gutiérrez-Arumí A., Verma B., Martos-Moreno G.A., Cuscó I., Oghabian A., Chowen J.A., Frilander M.J., Pérez-Jurado L.A. Defective minor spliceosome mRNA processing results in isolated familial growth hormone deficiency. EMBO Mol. Med. 2014;6:299–306. doi: 10.1002/emmm.201303573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patel A.A., McCarthy M., Steitz J.A. The splicing of U12-type introns can be a rate-limiting step in gene expression. EMBO J. 2002;21:3804–3815. doi: 10.1093/emboj/cdf297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pessa H.K.J., Greco D., Kvist J., Wahlström G., Heino T.I., Auvinen P., Frilander M.J. Gene expression profiling of U12-type spliceosome mutant Drosophila reveals widespread changes in metabolic pathways. PLoS ONE. 2010;5:e13215. doi: 10.1371/journal.pone.0013215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patel A.A., Steitz J.A. Splicing double: insights from the second spliceosome. Nat. Rev. Mol. Cell Biol. 2003;4:960–970. doi: 10.1038/nrm1259. [DOI] [PubMed] [Google Scholar]
- 30.Singh J., Padgett R.A. Rates of in situ transcription and splicing in large human genes. Nat. Struct. Mol. Biol. 2009;16:1128–1133. doi: 10.1038/nsmb.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raijmakers R., Egberts W.V., van Venrooij W.J., Pruijn G.J. Protein-protein interactions between human exosome components support the assembly of RNase PH-type subunits into a six-membered PNPase-like ring. J. Mol. Biol. 2002;323:653–663. doi: 10.1016/s0022-2836(02)00947-6. [DOI] [PubMed] [Google Scholar]
- 32.Brouwer R., Allmang C., Raijmakers R., van Aarssen Y., Egberts W.V., Petfalski E., van Venrooij W.J., Tollervey D., Pruijn G.J.M. Three novel components of the human exosome. J. Biol. Chem. 2001;276:6177–6184. doi: 10.1074/jbc.M007603200. [DOI] [PubMed] [Google Scholar]
- 33.Galiveti C.R., Rozhdestvensky T.S., Brosius J., Lehrach H., Konthur Z. Application of housekeeping npcRNAs for quantitative expression analysis of human transcriptome by real-time PCR. RNA. 2010;16:450–461. doi: 10.1261/rna.1755810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Katz Y., Wang E.T., Airoldi E.M., Burge C.B. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat. Methods. 2010;7:1009–1015. doi: 10.1038/nmeth.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pessa H., Ruokolainen A., Frilander M.J. The abundance of the spliceosomal snRNPs is not limiting the splicing of U12-type introns. RNA. 2006;12:1883–1892. doi: 10.1261/rna.213906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Friend K., Kolev N.G., Shu M.-D., Steitz J.A. Minor-class splicing occurs in the nucleus of the Xenopus oocyte. RNA. 2008;14:1459–1462. doi: 10.1261/rna.1119708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kammler S., Lykke-Andersen S., Jensen T.H. The RNA exosome component hRrp6 is a target for 5-fluorouracil in human cells. Mol. Cancer Res. 2008;6:990–995. doi: 10.1158/1541-7786.MCR-07-2217. [DOI] [PubMed] [Google Scholar]
- 39.Allmang C., Petfalski E., Podtelejnikov A., Mann M., Tollervey D., Mitchell P. The yeast exosome and human PM-Scl are related complexes of 3’ –> 5’ exonucleases. Genes Dev. 1999;13:2148–2158. doi: 10.1101/gad.13.16.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gudipati R.K., Xu Z., Lebreton A., Seraphin B., Steinmetz L.M., Jacquier A., Libri D. Extensive degradation of RNA precursors by the exosome in wild-type cells. Mol. Cell. 2012;48:409–421. doi: 10.1016/j.molcel.2012.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schneider C., Kudla G., Wlotzka W., Tuck A., Tollervey D. Transcriptome-wide analysis of exosome targets. Mol. Cell. 2012;48:422–433. doi: 10.1016/j.molcel.2012.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Hoof A., Lennertz P., Parker R. Yeast exosome mutants accumulate 3’-extended polyadenylated forms of U4 small nuclear RNA and small nucleolar RNAs. Mol. Cell. Biol. 2000;20:441–452. doi: 10.1128/mcb.20.2.441-452.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alioto T.S. U12DB: a database of orthologous U12-type spliceosomal introns. Nucleic Acids Res. 2007;35:D110–D115. doi: 10.1093/nar/gkl796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tilgner H., Knowles D.G., Johnson R., Davis C.A., Chakrabortty S., Djebali S., Curado J., Snyder M., Gingeras T.R., Guigó R. Deep sequencing of subcellular RNA fractions shows splicing to be predominantly co-transcriptional in the human genome but inefficient for lncRNAs. Genome Res. 2012;22:1616–1625. doi: 10.1101/gr.134445.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Khodor Y.L., Menet J.S., Tolan M., Rosbash M. Cotranscriptional splicing efficiency differs dramatically between Drosophila and mouse. RNA. 2012;18:2174–2186. doi: 10.1261/rna.034090.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vargas Diana Y., Shah K., Batish M., Levandoski M., Sinha S., Salvatore A.E. Marras, Schedl P., Tyagi S. Single-molecule imaging of transcriptionally coupled and uncoupled splicing. Cell. 2011;147:1054–1065. doi: 10.1016/j.cell.2011.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tarn W.-Y., Steitz J.A. A novel spliceosome containing U11, U12 and U5 snRNPs excises a minor class (AT-AC) intron in vitro. Cell. 1996;84:801–811. doi: 10.1016/s0092-8674(00)81057-0. [DOI] [PubMed] [Google Scholar]
- 48.Wu Q., Krainer A.R. U1-mediated exon definition interactions between AT-AC and GT-AG introns. Science. 1996;274:1005–1008. doi: 10.1126/science.274.5289.1005. [DOI] [PubMed] [Google Scholar]
- 49.Eberle A.B., Hessle V., Helbig R., Dantoft W., Gimber N., Visa N. Splice-site mutations cause Rrp6-mediated nuclear retention of the unspliced RNAs and transcriptional down-regulation of the splicing-defective genes. PLoS ONE. 2010;5:e11540. doi: 10.1371/journal.pone.0011540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sayani S., Chanfreau G.F. Sequential RNA degradation pathways provide a fail-safe mechanism to limit the accumulation of unspliced transcripts in Saccharomyces cerevisiae. RNA. 2012;18:1563–1572. doi: 10.1261/rna.033779.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lemieux C., Marguerat S., Lafontaine J., Barbezier N., Bahler J., Bachand F. A Pre-mRNA degradation pathway that selectively targets intron-containing genes requires the nuclear poly(A)-binding protein. Mol. Cell. 2011;44:108–119. doi: 10.1016/j.molcel.2011.06.035. [DOI] [PubMed] [Google Scholar]
- 52.McPheeters D.S., Cremona N., Sunder S., Chen H.M., Averbeck N., Leatherwood J., Wise J.A. A complex gene regulatory mechanism that operates at the nexus of multiple RNA processing decisions. Nat. Struct. Mol. Biol. 2009;16:255–264. doi: 10.1038/nsmb.1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Drazkowska K., Tomecki R., Stodus K., Kowalska K., Czarnocki-Cieciura M., Dziembowski A. The RNA exosome complex central channel controls both exonuclease and endonuclease Dis3 activities in vivo and in vitro. Nucleic Acids Res. 2013;41:3845–3858. doi: 10.1093/nar/gkt060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen C.Y., Gherzi R., Ong S.E., Chan E.L., Raijmakers R., Pruijn G.J., Stoecklin G., Moroni C., Mann M., Karin M. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell. 2001;107:451–464. doi: 10.1016/s0092-8674(01)00578-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.