Abstract
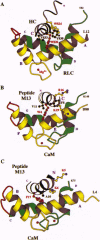
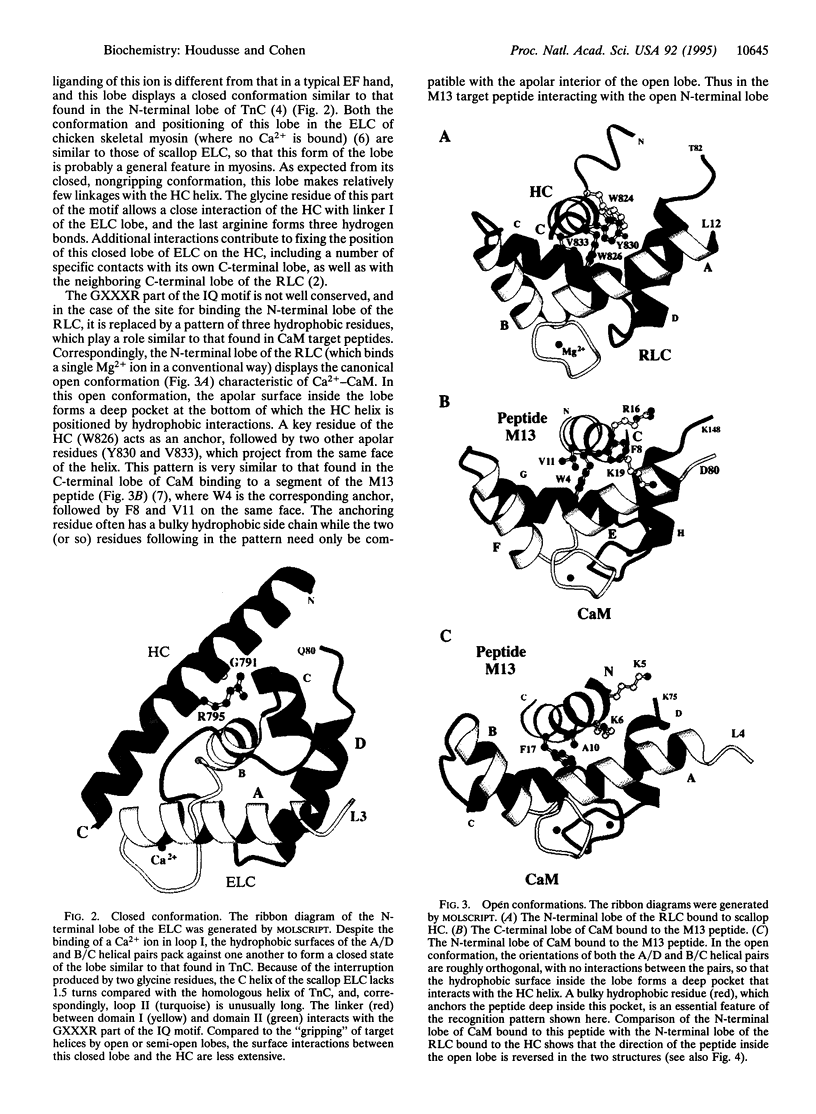
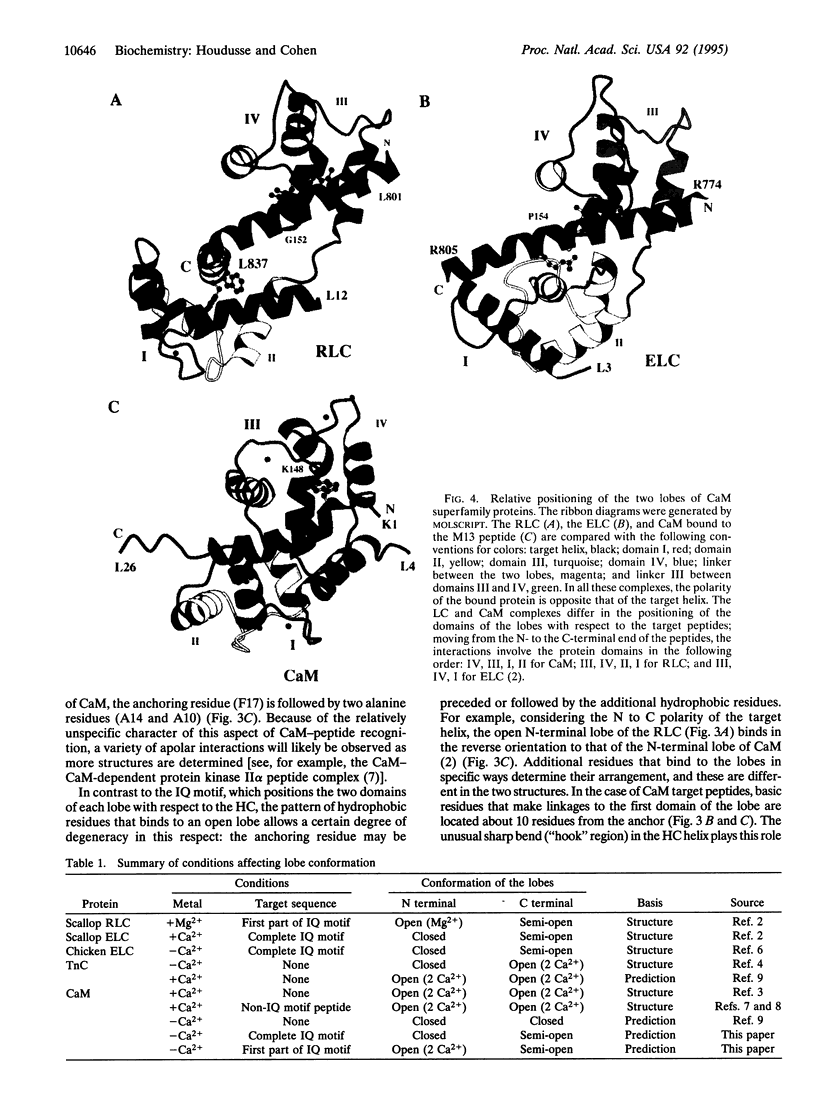
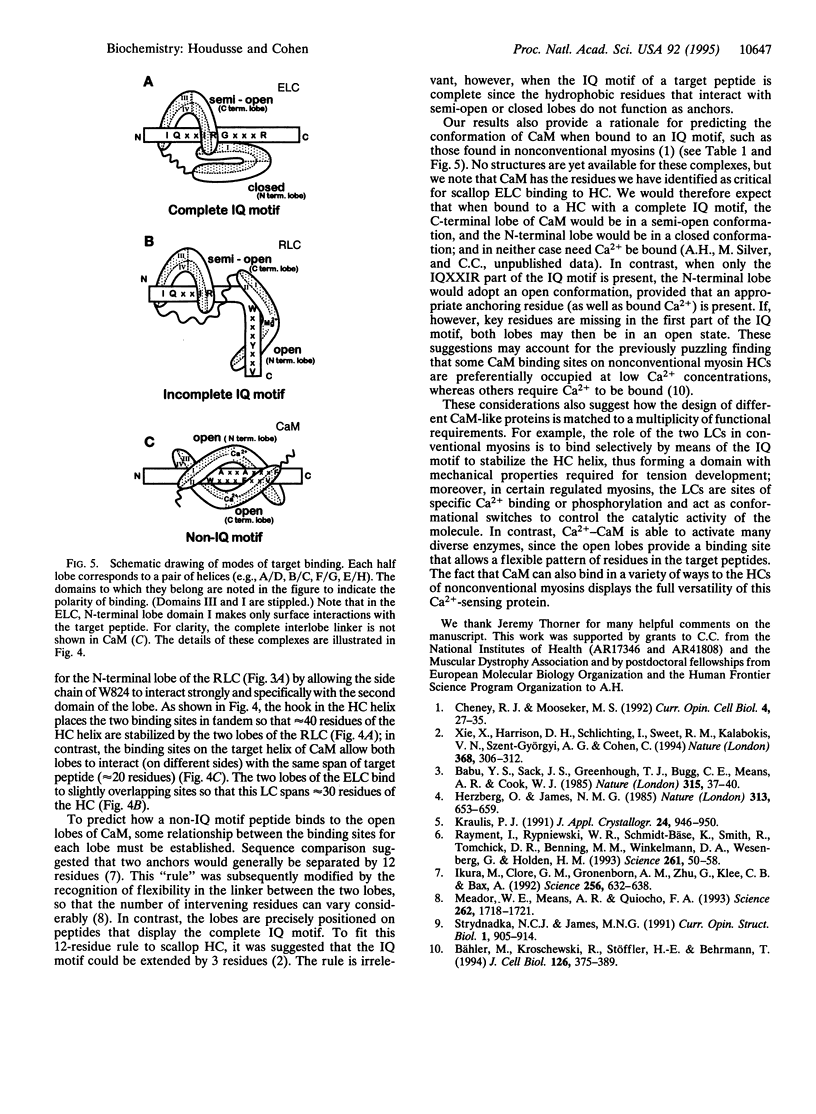
Some of the rules for how members of the calmodulin (CaM) superfamily bind to target peptides are revealed by the crystal structure of the regulatory domain of scallop myosin. The structure shows that the IQ motif of the heavy chain in this invertebrate myosin imposes constraints on both the positioning and conformation of the individual lobes of the light chains. In contrast, analysis of the contact residues in the targets bound by Ca(2+)-CaM reveals how the structure of CaM accommodates a broader range of sequences consonant with this protein's functional diversity.
Full text
PDF
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Babu Y. S., Sack J. S., Greenhough T. J., Bugg C. E., Means A. R., Cook W. J. Three-dimensional structure of calmodulin. Nature. 1985 May 2;315(6014):37–40. doi: 10.1038/315037a0. [DOI] [PubMed] [Google Scholar]
- Bähler M., Kroschewski R., Stöffler H. E., Behrmann T. Rat myr 4 defines a novel subclass of myosin I: identification, distribution, localization, and mapping of calmodulin-binding sites with differential calcium sensitivity. J Cell Biol. 1994 Jul;126(2):375–389. doi: 10.1083/jcb.126.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheney R. E., Mooseker M. S. Unconventional myosins. Curr Opin Cell Biol. 1992 Feb;4(1):27–35. doi: 10.1016/0955-0674(92)90055-h. [DOI] [PubMed] [Google Scholar]
- Herzberg O., James M. N. Structure of the calcium regulatory muscle protein troponin-C at 2.8 A resolution. Nature. 1985 Feb 21;313(6004):653–659. doi: 10.1038/313653a0. [DOI] [PubMed] [Google Scholar]
- Ikura M., Clore G. M., Gronenborn A. M., Zhu G., Klee C. B., Bax A. Solution structure of a calmodulin-target peptide complex by multidimensional NMR. Science. 1992 May 1;256(5057):632–638. doi: 10.1126/science.1585175. [DOI] [PubMed] [Google Scholar]
- Meador W. E., Means A. R., Quiocho F. A. Modulation of calmodulin plasticity in molecular recognition on the basis of x-ray structures. Science. 1993 Dec 10;262(5140):1718–1721. doi: 10.1126/science.8259515. [DOI] [PubMed] [Google Scholar]
- Rayment I., Rypniewski W. R., Schmidt-Bäse K., Smith R., Tomchick D. R., Benning M. M., Winkelmann D. A., Wesenberg G., Holden H. M. Three-dimensional structure of myosin subfragment-1: a molecular motor. Science. 1993 Jul 2;261(5117):50–58. doi: 10.1126/science.8316857. [DOI] [PubMed] [Google Scholar]
- Xie X., Harrison D. H., Schlichting I., Sweet R. M., Kalabokis V. N., Szent-Györgyi A. G., Cohen C. Structure of the regulatory domain of scallop myosin at 2.8 A resolution. Nature. 1994 Mar 24;368(6469):306–312. doi: 10.1038/368306a0. [DOI] [PubMed] [Google Scholar]