Abstract
The inducible transcription factor Egr-1, which recognizes a 9-bp target DNA sequence via three zinc-finger domains, rapidly activates particular genes upon cellular stimuli such as neuronal signals and vascular stresses. Here, using the stopped-flow fluorescence method, we measured the target search kinetics of the Egr-1 zinc-finger protein at various ionic strengths between 40 and 400 mM KCl and found the most efficient search at 150 mM KCl. We further investigated the kinetics of intersegment transfer, dissociation, and sliding of this protein on DNA at distinct concentrations of KCl. Our data suggest that Egr-1's kinetic properties are well suited for efficient scanning of chromosomal DNA in vivo. Based on a newly developed theory, we analyzed the origin of the optimal search efficiency at physiological ionic strength. Target association is accelerated by nonspecific binding to nearby sites and subsequent sliding to the target as well as by intersegment transfer. Although these effects are stronger at lower ionic strengths, such conditions also favor trapping of the protein at distant nonspecific sites, decelerating the target association. Our data demonstrate that Egr-1 achieves the optimal search at physiological ionic strength through a compromise between the positive and negative impacts of nonspecific interactions with DNA.
INTRODUCTION
To perform their function, most transcription factors and DNA repair/modifying enzymes must stochastically scan DNA and locate their specific target sites in the vast presence of nonspecific but structurally similar sites. Nonspecific interactions with DNA have both positive and negative impacts on the kinetics of the target search by these proteins. Nonspecific sites near the target serve as an antenna that accelerates target location by the proteins via nonspecific association followed by sliding on DNA (1–10). At the same time, nonspecific sites distant from the target can trap the proteins and decelerate the target search process (6,11). This trapping effect can be particularly strong in the nucleus, where the DNA density is extremely high (∼100 mg/ml) (12). Although ∼80% of chromosomal DNA is occupied by nucleosomes, the total concentration of accessible DNA segments between nucleosomes (average linker length ∼56 bp) (13) is estimated to be ∼0.5 mM. Because this concentration is far higher than the typical apparent affinities of sequence-specific DNA-binding proteins for nonspecific DNA, it is qualitatively obvious that nonspecific DNA effectively traps the proteins and decelerates their target search process. However, the attenuation of search efficiency due to the trapping effect and its relationship to the mechanisms of facilitated target location by these proteins are not well understood quantitatively.
Here we address this issue for the inducible transcription factor Egr-1 (also known as Zif268), which recognizes the 9-base-pair (bp) target sequences GCG(T/G)GGGCG via three zinc-finger domains (14,15). In the brain, Egr-1 is induced by synaptic signals and activates genes for long-term memory formation and consolidation (16,17). In the cardiovascular system, Egr-1 serves as a stress-inducible transcription factor that activates the genes to initiate defense against vascular stress and injury (18,19). Within its short lifetime (∼0.5–1 h) (18), Egr-1 rapidly regulates the target genes, allowing the cells to quickly respond to stimuli. It is therefore important to understand how Egr-1 efficiently scans DNA. Our previous biophysical studies have revealed important features of DNA scanning by Egr-1 (20–23). The nuclear magnetic resonance (NMR) and stopped-flow fluorescence studies demonstrated extremely efficient intersegment transfer of Egr-1 between nonspecific DNA duplexes (20,23). The NMR study showed that Egr-1 scans DNA via two conformationally different states termed the search and recognition modes (23). In the search mode, one zinc finger is locally dissociated from DNA while the other two zinc fingers remain bound to DNA. Our computational and experimental studies have suggested that the search mode facilitates intersegment transfer via intermediates in which a protein molecule transiently bridges two DNA duplexes (21,23).
In the current study, we measure the target search kinetics as a function of ionic strength and demonstrate that Egr-1 exhibits its optimal search efficiency at physiological ionic strength. Varying the ionic strength influences the antenna and trapping effects in completely different manners and therefore allows us to quantitatively assess the interplay between these effects in the target DNA search process. We also introduce a new theoretical method that explicitly investigates the effect of non-specific interactions and intersegment transfer. Analytical expressions for the target search kinetics in terms of the enhancement factors of the antenna effect and intersegment transfer and the attenuation factor of the trapping effect are obtained. Our experimental data and the newly developed theory shed light on Egr-1's properties that are well suited for efficient scanning of DNA in the nucleus.
MATERIALS AND METHODS
Protein and DNA
In this study, we used a protein construct of the Egr-1 DNA-binding domain comprising three zinc fingers (human Egr-1 residues 335–432). For the sake of simplicity, this construct is hereafter referred to as the Egr-1 zinc-finger protein. The protein was expressed in Escherichia coli and purified as described in our previous work (20,22,23). For fluorescence experiments, 33-, 48-, 63-, 88-, 113- and 143-bp probe DNA duplexes with a fluorescein amidite (FAM) attached to the 5′-terminus were prepared using synthetic DNA strands and enzymatic reactions as described (20). The duplexes contain a single Egr-1 target site and share the terminal 33 bp: FAM-AGCGTGGGCGTACCGGTAACTATCGTCTTGAGT-3′ (the Egr-1 target is underlined). Their complete sequences are given in (20). A 28-bp competitor DNA duplex (5′-GTACCGATTGCAGATTCCGAACCTTCAG-3’) containing neither specific nor semi-specific sites was prepared as previously described (20,23).
Stopped-flow fluorescence assay of the target search kinetics
The target search kinetics was measured at 20°C using an ISS PC-1 spectrofluorometer equipped with an Applied Photophysics RX.2000 stopped-flow device as described (20). In this assay, the time courses of the FAM fluorescence were monitored upon mixing two solutions: (i) the Egr-1 zinc-finger protein, and (ii) FAM-labeled probe DNA and 28-bp nonspecific competitor DNA. Hereafter, the total concentrations of the probe DNA, protein and competitor DNA in the reaction mixture are represented by the symbols Dtot, Ptot, and Ctot, respectively. All experimental and theoretical analyses in this study used the conditions Dtot « Ptot « Ctot. Because of these inequalities, the relevant second-order processes occur in a pseudo-first-order manner, which simplifies the kinetic analysis (24). The buffers used were 10 mM Tris·HCl (pH 7.5), 200 nM ZnCl2 and 40–400 mM KCl. Apparent pseudo-first-order rate constants kapp were measured by mono-exponential fitting to the fluorescence time-course data (20). The apparent second-order rate constants ka for the target association were determined from kapp data using various protein concentrations Ptot, typically between 20 and 400 nM (see the Supplementary Materials). This range is biologically relevant, because western blot and DNA association data for nuclear extracts (e.g. 18,19) implies that the maximum level of nuclear Egr-1 in vivo is of order of 10−9–10−7 M. To analyze sliding of Egr-1 on DNA, we measured the rate constants ka using 33-, 48-, 63-, 83-, 113- and 143-bp probe DNA (Dtot = 2.5 nM) and 28-bp competitor DNA (Ctot = 2000 nM). To analyze intersegment transfer between two nonspecific sites on distinct DNA duplexes, we measured the rate constants ka using 113-bp probe DNA (Dtot = 2.5 nM) and 28-bp competitor DNA at various concentrations (Ctot = 1000–16 000 nM) (20).
Analytical expression for the target search kinetics
To analyze the experimental kinetic data, we derived a new general analytical expression for the target search kinetics. The derivation, which is given in the Supplementary Materials, was based on the backward master equations of first-passage probabilities for discrete-state stochastic models (25,26). In our previous study (20), we used the pseudo-first-order rate constant kapp from fluorescence time-course data along with its analytical form that assumes the conditions Dtot « Ptot « Ctot and quasi-equilibrium for nonspecific protein–DNA interactions prior to the equilibrium of target association. In this study, we used the following analytical form of the second-order rate constant ka for protein–target association without assuming quasi-equilibrium:
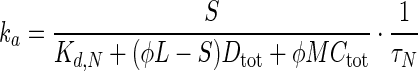 |
(1) |
The parameters in this equation are as follows:
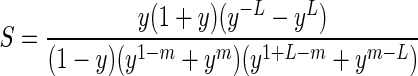 |
(2) |
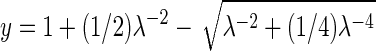 |
(3) |
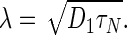 |
(4) |
The parameter λ is the effective sliding length in units of bp (1,26); D1 is the one-dimensional diffusion coefficient for sliding in units of bp2 s−1; Kd,N is the equilibrium constant in molar units for each nonspecific site; L is the total number of binding sites on the probe DNA; M is the total number of binding sites on the competitor DNA; and m is the position of the target (located at the mth site from the edge). The parameter ϕ is the number of possible orientations for each nonspecific site. Due to the structural pseudo-C2 symmetry of double-stranded DNA, ϕ = 2 for proteins that bind as a monomer, and ϕ = 1 for symmetric dimers. When ϕ = 2 is used, the microscopic parameters (e.g. Kd,N, koff,N, kIT,N) are defined for each orientation (20). The parameter τN in Equation (4) is the mean lifetime of a nonspecific complex and is given by
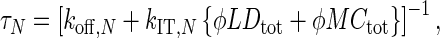 |
(5) |
where koff,N is the dissociation rate constant for a nonspecific complex, and kIT,N is the second-order rate constant for intersegment transfer (also known as direct transfer) between nonspecific sites on two distinct DNA duplexes (20). Although intersegment transfer can occur between two distant sites on the same molecule if the DNA length is significantly longer than the persistence length (i.e. ∼150 bp), such intra-molecular intersegment transfer is not considered here because only relatively short (<150 bp) DNA duplexes are used in our experiments. The annotation of symbols with the subscript ‘N’ indicates that those symbols represent parameters for ‘nonspecific’ interactions. As explained in the Supplementary Materials, the current analytical expression (Equations (1)–(5)) is more general and simpler than the previous expression, while the two expressions provide numerically equivalent results (e.g. Supplementary Figure S-I). In the Discussion section, we also present a physically clearer description of the ka constant in terms of the antenna and trapping effects.
Determination of the parameters for translocation of protein on DNA
The one-dimensional diffusion coefficient D1 and the sliding length λ were determined from the length-dependent ka data by fitting with Equations (1)–(4). The total numbers of binding sites, L for the probe DNA and M for the competitor DNA, were set to A − B + 1, where A is the DNA length in bp, and B is the number of DNA base pairs covered by a protein molecule. Based on structural information about specific and nonspecific DNA complexes of Egr-1 (15,23), B = 9 was used, as previously described (20). Based on the sequences of the probe DNA duplexes, the target position m = 2 was used. Values of ka as a function of L were given as input data, and the parameters D1 and λ were optimized as independent fitting parameters via nonlinear least-squares fitting, in which the sum of the squared differences between observed and calculated ka values was minimized. In this fitting, τN was calculated via Equation (4) with λ and D1 being optimized.
The rate constant kIT,N for intersegment transfer and the rate constant koff,N for dissociation were determined from the dependence of ka data on competitor DNA concentrations by fitting with Equations (1)–(5). In this calculation, values of ka as a function of Ctot were given as input data, and the parameters kIT,N and koff,N were optimized as independent fitting parameters via nonlinear least-squares fitting. The experimentally obtained D1 coefficient was used in this fitting calculation. Simultaneous fitting to the L-dependence and Ctot-dependence data was also conducted using Equations (1)–(5) via optimization of D1, kIT,N, and koff,N as the fitting parameters.
These fitting calculations require values of the Kd,N constant for a nonspecific site (see Equation (1)). Values of Kd,N constants used for nonspecific interactions at 40, 60, 80, 110 and 150 mM KCl were 1.4, 3.6, 4.6, 9.6 and 16 μM, respectively, which were obtained from our previous experimental data (20) in conjunction with the counterion condensation theory (27,28). All nonlinear least-squares fitting calculations were performed with the MATLAB software (MathWorks, Inc.). Some examples of our MATLAB scripts for the fitting calculations are given in the Supplementary Materials.
RESULTS
Target association kinetics at various ionic strengths
Using the stopped-flow fluorescence assay, we measured the target search kinetics for the Egr-1 zinc-finger protein. This assay uses three macromolecular components: the probe DNA, protein and nonspecific competitor DNA (Figure 1A). The probe DNA duplexes contain a target site and a FAM moiety that is tethered to a position near the target. The fluorescence from the FAM probe changes upon binding of the protein to the target site (20). Immediately after a solution of the protein is rapidly mixed with a solution containing the probe DNA and competitor DNA, we began monitoring the time courses of the FAM fluorescence intensity (some example data are shown in Figure 1A). The time courses of fluorescence intensity were mono-exponential. The percentage change from initial to final intensities was virtually the same for the time-course data obtained under solution conditions where complete saturation of binding to the target site is expected at equilibrium (e.g. those shown in Figure 1A). A smaller percentage change was observed under conditions where the saturation level at equilibrium is lower (e.g. at 400 mM KCl). Using the time-course data, the pseudo-first-order rate constants in s−1 units were determined at various concentrations of protein, from which the apparent second-order rate constant ka in M−1 s−1 units was calculated (see Supplementary Figure S-III). Using this assay, we measured the apparent second-order rate constants ka for target association of Egr-1 with the 113-bp probe DNA in the presence of 2 μM nonspecific 28-bp competitor DNA at 40, 60, 80, 110, 150, 190, 230, 300 and 400 mM KCl. Figure 1B shows a logarithmic plot of the ka data as a function of KCl concentration. We found that the ka constant depended strongly on ionic strength in a non-monotonic manner and ranged from 0.055 × 108 to 1.98 × 108 M−1 s−1. Interestingly, the Egr-1 zinc-finger protein exhibited the fastest target search kinetics at 150 mM KCl. Because Mg2+ ions are known to play important roles in some protein–DNA/RNA interactions, we examined the influence of Mg2+ ions on the target search kinetics under the same ionic strength. Keeping 0.5 × [K+] + 2 × [Mg2+] + 0.5 × [Cl−] = 150 mM, we measured the target association kinetics as a function of Mg2+ concentration in a physiological range (0.5–5 mM). The presence of Mg2+ ions caused a slight (up to 23%) increase in the rate constant ka with the maximum ka found at [Mg2+] = 1 mM (Supplementary Figure S-IV). These results indicate that physiological ionic strength renders an optimal condition for target search by the Egr-1 zinc-finger protein.
Figure 1.
Target search kinetics of the Egr-1 zinc-finger protein as a function of ionic strength. (A) Stopped-flow fluorescence assay of the target search kinetics. The fluorescence time-course data shown were obtained using 113-bp probe DNA (Dtot = 2.5 nM), nonspecific 28-bp DNA (Ctot = 2000 nM) and protein (Ptot = 50 nM) at 60, 150 and 300 mM KCl. Concentrations Ptot and Ctot were varied in other measurements. (B) Apparent second-order rate constants ka for target association of the Egr-1 zinc-finger protein in the presence of 2000 nM nonspecific 28-bp DNA. The experiment was performed using 113-bp probe DNA at 40, 60, 80, 110, 150, 190, 230, 300 and 400 mM KCl. Values of ka constant were determined from the pseudo-first-order rate constants for target association at different concentrations of the protein. The solid line shown in the plot is a best-fit curve obtained using Equations (1)–(5) and the counterion condensation theory for the kinetic and thermodynamic parameters (see the main text). The data are shown on a logarithmic scale for each axis. For this panel and for Figures 2 and 3, the error bars represent the standard error of the mean (SEM) estimated from 6–10 replicates. For data points with no error bars, the SEMs were smaller than the size of the symbols.
Ionic-strength dependence of sliding
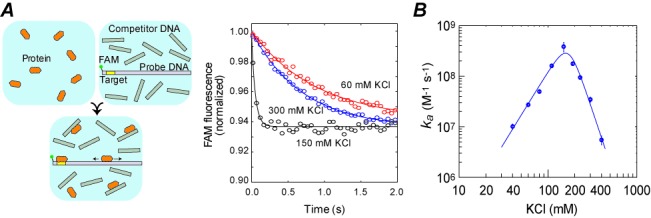
The DNA-length dependence of the target association kinetics provides information on sliding of proteins on DNA (20,29–33). Using the 33-, 48-, 63-, 88-, 113- and 143-bp probe DNA (Dtot = 2.5 nM) and 28-bp nonspecific competitor DNA (Ctot = 2000 nM), we measured the apparent rate constants ka for the Egr-1 zinc-finger protein at various KCl concentrations (Figure 2A). The DNA-length dependence of ka constants for these probe DNA duplexes was almost linear at 40 mM KCl, but more curved and asymptotic at higher ionic strengths. Based on the length-dependence data, we determined the one-dimensional diffusion coefficient D1 (in bp2 s−1; Figure 2B) and the effective sliding length λ (in bp; Figure 2C) as a function of KCl concentration. Although some literature assumed that the D1 coefficient for sliding should be independent of ionic strength (11,34–36), we observed significant ionic-strength dependence of the experimental D1 coefficient for the Egr-1 zinc-finger protein (Figure 2B). Our interpretation of this result is given in the Discussion section. We also found that the sliding length λ depended strongly on ionic strength: for example, λ = 122 ± 2 bp at 40 mM KCl, and λ = 38 ± 3 bp at 150 mM KCl. As noted previously (6,20), the DNA-length dependence of the target association kinetics should be an increasing function with an asymptote for lengths > 2λ. In fact, such asymptotic length dependence was clearly observed (Figure 2A). At ionic strengths as high as 300 mM KCl, no dependence on length was found for the 33–143 bp DNA duplexes (e.g. see ka values in Supplementary Figures S-IIIb and c). Although we cannot determine the sliding length λ in such a case, the absence of length dependence qualitatively suggests a very short sliding length. At 40–150 mM KCl, the length dependence of ka constants for the same set of DNA duplexes was strong enough due to 2λ being comparable to the DNA lengths used in the experiments (Figure 2A and B).
Figure 2.
Ionic-strength dependence of sliding. (A) Length dependence of ka constants at 40, 60, 110 and 150 mM. The horizontal axis represents DNA length in bp. Solid lines are the best-fit curves obtained using Equations (1)–(4). The parameter L, which represents the number of binding sites on the probe DNA, was set to (DNA length in bp) −8 (see the main text). The one-dimensional diffusion coefficient D1 and the effective sliding length λ were optimized in the fitting calculations. Dtot = 2.5 nM and Ctot = 2 μM were used in the measurements. (B) The one-dimensional diffusion coefficient D1 for sliding of the Egr-1 zinc-finger protein as a function of KCl concentration. (C) The effective sliding length λ as a function of ionic strength.
Ionic-strength dependence of intersegment transfer and dissociation
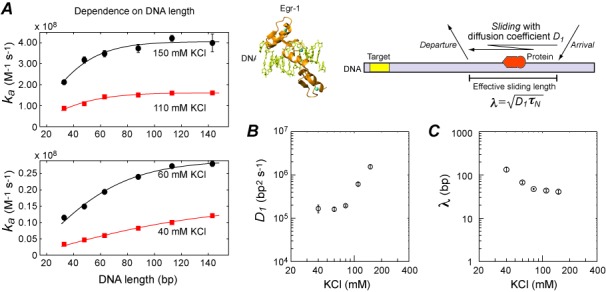
Intersegment transfer is the direct translocation of protein from one DNA duplex to another (without going through the intermediary of free protein) via an intermediate in which a protein molecule transiently bridges two DNA duplexes. The DNA concentration dependence of the translocation kinetics allows us to distinguish intersegment transfer from translocation via dissociation and re-association (20,23,37–43). We varied the concentration of the 28-bp nonspecific competitor DNA between 0.5 and 16 μM and measured the rate constant ka for the association of Egr-1 to the target on the 113-bp probe DNA (Figure 3A). Using the DNA concentration-dependence data, we determined the rate constant kIT,N for intersegment transfer between nonspecific sites of two DNA duplexes and the rate constant koff,N for the dissociation of nonspecific complexes as a function of KCl concentration (Figure 3B and C).
Figure 3.
Ionic-strength dependence of the dissociation and intersegment transfer of the Egr-1 zinc-finger protein. (A) Dependence of the kapp constant on the concentration Ctot of the 28-bp nonspecific DNA. Solid lines are the best-fit curves obtained using a model with intersegment transfer (i.e. Equations (1)–(5)), and dotted lines are the best-fit curves using a model without intersegment transfer (i.e. Equations (1)–(5) with kIT,N = 0). (B) The rate constant koff,N for dissociation from a nonspecific site as a function of KCl concentration. (C) The rate constant kIT,N for intersegment transfer between two nonspecific DNA molecules as a function of KCl concentration.
A surprising finding from this analysis was that the ionic-strength dependence of koff,N is extremely strong; for example, koff,N = 0.20 ± 0.26 s−1 at 40 mM KCl and koff,N = 920 ± 240 s−1 at 150 mM KCl. As shown in Figure 3B, the ionic-strength dependence of koff,N clearly obeys the log–log linear relationship in accordance with the kinetic theory by Lohman et al. (44), which is based on the counterion condensation arguments (27,28). We found that the rate constant kIT,N for intersegment transfer also depended strongly on the ionic strength, but to a lesser degree. Due to this difference, translocation via intersegment transfer is more efficient than translocation via dissociation and re-association at relatively low ionic strengths (i.e. kIT,NϕMCtot > koff,N), whereas the latter can be more efficient (i.e. kIT,NϕMCtot < koff,N) at higher ionic strengths. In fact, for the Ctot-dependence data at low ionic strengths, the kinetic model with intersegment transfer gave a far better fit than the model without intersegment transfer, whereas the two models gave more similar results for the data at high ionic strengths (Figure 2A). At KCl concentrations significantly higher than 150 mM, translocation via dissociation and re-association governs the Ctot-dependence and conceals the influence of intersegment transfer, which precluded us from further analyses.
DISCUSSION
Consideration on the ionic-strength dependent D1 coefficient
Although some previous studies assumed that sliding of proteins on DNA should be virtually independent of ionic strength (11,34–36), our D1 data for Egr-1 (Figure 2B) show significant dependence on ionic strength. Bonnet et al. also showed similar results for the restriction enzyme EcoRV (45). One may interpret that ionic-strength dependence of the experimental D1 data is influenced by ‘hopping’ (i.e. short-range correlated translocation via dissociation and re-association (1)). Because our kinetic model does not discriminate between short-range (i.e. hopping) and long-range translocations via dissociation followed by re-association, kinetics of dissociation in these processes are equally represented by the rate constant koff,N. The D1 measurement can be significantly affected by hopping if the intrinsic dissociation rate constant for hopping is comparable to the first-order rate constant ksl,N for sliding from one site to an adjacent site. To achieve this condition, however, the intrinsic dissociation rate constant for hopping must be >1000-fold larger than the observed dissociation rate constants koff,N (see Figures 2B and 3B; note that the D1 coefficient in bp2 s−1 units is equivalent to ksl,N in s−1 units (20)). Such extremely rapid dissociation only for hopping seems unrealistic, although we cannot completely exclude this possibility. Thus, it is unlikely that the ionic-strength dependence of the D1 data for Egr-1 arises from the influence of hopping.
A more reasonable interpretation of the ionic-strength dependent D1 data is that energy barriers of electrostatic nature are involved in the sliding process. This interpretation is consistent with results of some previous studies. For example, coarse-grained molecular dynamics simulations of the DNA search by proteins also indicated significant dependence of the D1 coefficient on ionic strengths (23,46,47). Besides, if sliding is barrierless diffusion (e.g. ‘movement on an isopotential surface’ as speculated by Winter et al. three decades ago (11)), then the D1 coefficient should be a relatively simple function of the hydrodynamic radius, as is the case for three-dimensional diffusion in solutions. Despite this expectation, data available in the literature for proteins of different sizes show that D1 is virtually independent of molecular size (e.g. 20,35,36,45,48,49). Furthermore, many proteins exhibit sliding on DNA far slower than expected from the Stokes–Einstein relation (as reviewed in (5)). These observations and our ionic-strength dependent D1 data can be explained if sliding involves electrostatic energy barriers, which could be relevant to breakage of ion pairs between protein and DNA (50).
Egr-1's kinetic properties are well suited for efficient scanning of DNA in vivo
Our kinetic data show that the Egr-1 zinc-finger protein exhibits its optimal search efficiency at the physiological ionic strength (i.e. 150 mM KCl). In addition, two other findings in our current study suggest that Egr-1 has characteristics that enable efficient scanning of DNA in the nuclei. First, the effective sliding length for the Egr-1 zinc-finger protein at 150 mM KCl seems to be suitable for scanning accessible regions of chromosomal DNA. According to theoretical work by Mirny et al. (6), proteins can scan DNA most efficiently when 2λ is comparable to the lengths of the linker DNA between nucleosomes. In human cells, linker lengths are widely distributed with an average of ∼56 bp (13). Because our data indicate that 2λ is comparable to these lengths, the sliding and dissociation properties seem suitable for Egr-1 to scan linker DNA in vivo. Second, our data also indicate that the intersegment transfer of Egr-1 is very efficient at physiological ionic strength. The previous study using the coarse-grained molecular dynamics simulations suggested that Egr-1 can conduct intersegment transfer between two DNA duplexes separated by 60 Å (23). Because this separation corresponds to the distance between two DNA ends of a nucleosome particle (51), intersegment transfer could allow Egr-1 to efficiently bypass nucleosome particles and continuously scan DNA (20,23). Thus, our data collectively suggest that the kinetic properties of Egr-1 are well suited to efficiently scan DNA in vivo.
Influences of the antenna and trapping effects and intersegment transfer
Our data show that the target search by the Egr-1 zinc-finger protein is fastest at physiological ionic strength. What is the origin for the optimal kinetic efficiency at physiological ionic strength? To offer a theoretical explanation, we provide a general expression for the kinetics of target location by DNA-binding proteins. This is done by transforming Equation (1) into the following form, which shows how the apparent rate constant ka for the target association is related to the intrinsic association rate constant kon,N ( = koff,N /Kd,N) of each site:
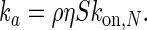 |
(6) |
The parameters ρ and η in this equation are as follows:
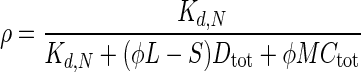 |
(7) |
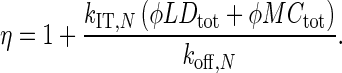 |
(8) |
Equation (6) quantitatively explains how the target association kinetics is influenced by the antenna effect, the trapping effect, and intersegment transfer, which are represented by the parameters S, ρ, and η, respectively. The parameter S given by Equation (2) corresponds to the effective size of the antenna. Nonspecific sites near the target on the same DNA serve as an antenna and make the target association S-fold faster. Because this is equivalent to S sites serving as the target, the term ϕL – S in Equation (7) corresponds to the number of nonspecific sites outside the antenna on the target-containing DNA duplex, and (ϕL – S)Dtot + ϕMCtot corresponds to the net overall concentration of nonspecific sites. Therefore, the parameter ρ represents the fraction of protein molecules that are not trapped by any nonspecific sites during the target search process. If (ϕL – S)Dtot « ϕMCtot (which is usually satisfied), then the parameter ρ becomes independent of S, and the expression becomes equivalent to the expression given by Esadze and Iwahara under the assumption of quasi-equilibrium for nonspecific interactions (20) (see the Supplementary Materials). If the target position is near the middle of the DNA duplex (i.e. m ≈ L/2), the parameter S (Equation (2)) becomes independent of m and can be reduced to
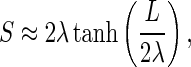 |
(9) |
which corresponds to the expression for the antenna effect given in previous literature (1,6) (see Supplementary Figure S-II). Equation (8) shows that intersegment transfer considerably enhances the target search kinetics (i.e. η » 1) when and only when the pseudo-first-order rate constant for intersegment transfer is substantially larger than the dissociation rate constant of nonspecific complexes. As indicated by Equation (6), the ratio of ka to kon,N is given by ρηS, in which ρ represents an attenuation factor (0 < ρ < 1) due to the trapping effect, η is an enhancement factor (η > 1) due to intersegment transfer, and S is an enhancement factor (1 < S < 2λ) due to the antenna effect.
Optimal target search by Egr-1 at physiological ionic strengths
Based on the above analytical expression of ka, we can now explain the ionic-strength dependence of the target search kinetics in terms of the antenna effect, the trapping effect and intersegment transfer. Using the current experimental data, we calculated the parameters ρ, η and S for the Egr-1 zinc-finger protein as shown in Figure 4. The parameters S and η are decreasing functions of ionic strength, and the antenna effect and intersegment transfer substantially accelerate the target search kinetics at low ionic strengths. In contrast, the parameter ρ is an increasing function of ionic strength, and the trapping effect substantially decelerates the target search kinetics at low ionic strengths. As previously described, the rate constants for electrostatically assisted macromolecular association (such as kon,N) are typically a decreasing function of ionic strength (52,53). The rate constant ka as the product ρηSkon,N (Equation (6)) is therefore maximized at a particular ionic strength as shown in Figure 1B. Our data indicate that the Egr-1 zinc-finger protein achieves its optimal kinetic efficiency at physiological ionic strength through the compromise between the positive and negative impacts of nonspecific interactions with DNA.
Figure 4.
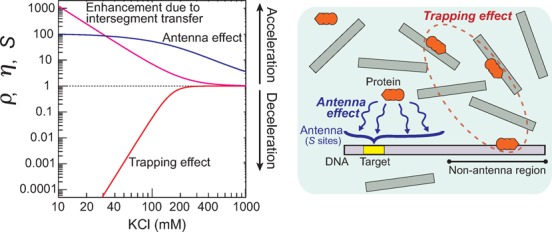
The attenuation factor ρ due to the trapping effect (red), the enhancement factor η due to intersegment transfer (magenta) and the enhancement factor S due to the antenna effect (blue) for the target association kinetics of the Egr-1 zinc-finger protein. These parameters were calculated from the current experimental data on the rate constants koff,N and kIT,N, the one-dimensional diffusion coefficient D1, and the dissociation constant Kd,N for the Egr-1 zinc-finger protein along with Equations (2), (7) and (8). The following conditions were used: Dtot = 2.5 nM; L = 105 sites; Ctot = 2000 nM; M = 20 sites. Ionic-strength dependence represented by 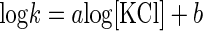 was assumed for koff,N, kIT,N, D1 and Kd,N, and the parameters a and b were calculated from the current experimental data.
was assumed for koff,N, kIT,N, D1 and Kd,N, and the parameters a and b were calculated from the current experimental data.
CONCLUSIONS
In this study, we have demonstrated the optimal search efficiency of the Egr-1 zinc-finger protein at physiological ionic strength and explained the ionic-strength dependence of the target search kinetics in terms of the antenna and trapping effects and intersegment transfer. Our results suggest that Egr-1's kinetic properties in the sliding, dissociation and intersegment transfer processes are well suited for efficient scanning of DNA in vivo. This study provides insight into how the inducible transcription factor Egr-1 rapidly activates the genes in response to cellular stimuli such as synaptic signals and vascular injury. This work also provides a theory that quantitatively explains the interplay between the antenna and trapping effects and intersegment transfer in the target DNA search process. A deeper understanding of this interplay could be helpful in the engineering of transcription factors and DNA repair/modifying enzymes. For example, our theory predicts that protein engineering that simply increases the affinity for DNA will enhance the trapping effect and possibly result in slow target location. Because Egr-1 (Zif268) has been used as a scaffold for zinc-finger technology for artificial gene control, we expect that current and future studies of DNA scanning by Egr-1 at the molecular and atomic levels may facilitate the kinetic improvement of artificial zinc-finger proteins.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We thank Dr Levani Zandarashvili for useful discussion, Dr Yaakov Levy for insights from his computational studies, and Dr Wlodzimierz Bujalowski for helpful advice on our fluorescence experiments.
FUNDING
American Heart Association [12BGIA8960032 to J.I.]; Welch Foundation [C-1559 to A.B.K.]; National Institutes of Health [R01GM105931 to J.I.] (in part). Source of open access funding: R01GM105931 from the National Institutes of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1.Berg O.G., Winter R.B., von Hippel P.H. Diffusion-driven mechanisms of protein translocation on nucleic acids. 1. Models and theory. Biochemistry. 1981;20:6929–6948. doi: 10.1021/bi00527a028. [DOI] [PubMed] [Google Scholar]
- 2.Gorman J., Greene E.C. Visualizing one-dimensional diffusion of proteins along DNA. Nat. Struct. Mol. Biol. 2008;15:768–774. doi: 10.1038/nsmb.1441. [DOI] [PubMed] [Google Scholar]
- 3.Halford S.E. An end to 40 years of mistakes in DNA-protein association kinetics. Biochem. Soc. Trans. 2009;37:343–348. doi: 10.1042/BST0370343. [DOI] [PubMed] [Google Scholar]
- 4.Halford S.E., Marko J.F. How do site-specific DNA-binding proteins find their targets. Nucleic Acids Res. 2004;32:3040–3052. doi: 10.1093/nar/gkh624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kolomeisky A.B. Physics of protein-DNA interactions: mechanisms of facilitated target search. Phys. Chem. Chem. Phys. 2011;13:2088–2095. doi: 10.1039/c0cp01966f. [DOI] [PubMed] [Google Scholar]
- 6.Mirny L., Slutsky M., Wunderlich Z., Tafvizi A., Leith J., Kosmrlj A. How a protein searches for its site on DNA: the mechanism of facilitated diffusion. J. Phys. A: Math. Theor. 2009;42:401335. [Google Scholar]
- 7.Monico C., Capitanio M., Belcastro G., Vanzi F., Pavone F.S. Optical methods to study protein-DNA interactions in vitro and in living cells at the single-molecule level. Int. J. Mol. Sci. 2013;14:3961–3992. doi: 10.3390/ijms14023961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sheinman M., Benichou O., Kafri Y., Voituriez R. Classes of fast and specific search mechanisms for proteins on DNA. Rep. Prog. Phys. 2012;75:026601. doi: 10.1088/0034-4885/75/2/026601. [DOI] [PubMed] [Google Scholar]
- 9.Tafvizi A., Mirny L.A., van Oijen A.M. Dancing on DNA: kinetic aspects of search processes on DNA. Chemphyschem. 2011;12:1481–1489. doi: 10.1002/cphc.201100112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.von Hippel P.H., Berg O.G. Facilitated target location in biological systems. J. Biol. Chem. 1989;264:675–678. [PubMed] [Google Scholar]
- 11.Winter R.B., Berg O.G., von Hippel P.H. Diffusion-driven mechanisms of protein translocation on nucleic acids. 3. The Escherichia coli lac repressor–operator interaction: kinetic measurements and conclusions. Biochemistry. 1981;20:6961–6977. doi: 10.1021/bi00527a030. [DOI] [PubMed] [Google Scholar]
- 12.Lewin B. Genes VII. Oxford: Oxford Univ Press; 2000. [Google Scholar]
- 13.Valouev A., Johnson S.M., Boyd S.D., Smith C.L., Fire A.Z., Sidow A. Determinants of nucleosome organization in primary human cells. Nature. 2011;474:516–520. doi: 10.1038/nature10002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elrod-Erickson M., Rould M.A., Nekludova L., Pabo C.O. Zif268 protein-DNA complex refined at 1.6 Å: a model system for understanding zinc finger-DNA interactions. Structure. 1996;4:1171–1180. doi: 10.1016/s0969-2126(96)00125-6. [DOI] [PubMed] [Google Scholar]
- 15.Pavletich N.P., Pabo C.O. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 Å. Science. 1991;252:809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- 16.Bozon B., Davis S., Laroche S. A requirement for the immediate early gene zif268 in reconsolidation of recognition memory after retrieval. Neuron. 2003;40:695–701. doi: 10.1016/s0896-6273(03)00674-3. [DOI] [PubMed] [Google Scholar]
- 17.Lee J.L., Everitt B.J., Thomas K.L. Independent cellular processes for hippocampal memory consolidation and reconsolidation. Science. 2004;304:839–843. doi: 10.1126/science.1095760. [DOI] [PubMed] [Google Scholar]
- 18.Khachigian L.M., Lindner V., Williams A.J., Collins T. Egr-1-induced endothelial gene expression: a common theme in vascular injury. Science. 1996;271:1427–1431. doi: 10.1126/science.271.5254.1427. [DOI] [PubMed] [Google Scholar]
- 19.Yan S.F., Fujita T., Lu J., Okada K., Shan Zou Y., Mackman N., Pinsky D.J., Stern D.M. Egr-1, a master switch coordinating upregulation of divergent gene families underlying ischemic stress. Nat. Med. 2000;6:1355–1361. doi: 10.1038/82168. [DOI] [PubMed] [Google Scholar]
- 20.Esadze A., Iwahara J. Stopped-flow fluorescence kinetic study of protein sliding and intersegment transfer in the target DNA search process. J. Mol. Biol. 2014;426:230–244. doi: 10.1016/j.jmb.2013.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iwahara J., Levy Y. Speed-stability paradox in DNA-scanning by zinc-finger proteins. Transcription. 2013;4:58–61. doi: 10.4161/trns.23584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takayama Y., Sahu D., Iwahara J. NMR studies of translocation of the Zif268 protein between its target DNA Sites. Biochemistry. 2010;49:7998–8005. doi: 10.1021/bi100962h. [DOI] [PubMed] [Google Scholar]
- 23.Zandarashvili L., Vuzman D., Esadze A., Takayama Y., Sahu D., Levy Y., Iwahara J. Asymmetrical roles of zinc fingers in dynamic DNA-scanning process by the inducible transcription factor Egr-1. Proc. Natl. Acad. Sci. U.S.A. 2012;109:E1724–E1732. doi: 10.1073/pnas.1121500109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pollard T.D., De La Cruz E.M. Take advantage of time in your experiments: a guide to simple, informative kinetics assays. Mol. Biol. Cell. 2013;24:1103–1110. doi: 10.1091/mbc.E13-01-0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Redner S. A Guide to First-Passage Processes. Cambridge: Cambridge University Press; 2001. [Google Scholar]
- 26.Veksler A., Kolomeisky A.B. Speed-selectivity paradox in the protein search for targets on DNA: is it real or not. J. Phys. Chem. B. 2013;117:12695–12701. doi: 10.1021/jp311466f. [DOI] [PubMed] [Google Scholar]
- 27.Manning G.S. Molecular theory of polyelectrolyte solutions with applications to electrostatic properties of polynucleotides. Q. Rev. Biophys. 1978;11:179–246. doi: 10.1017/s0033583500002031. [DOI] [PubMed] [Google Scholar]
- 28.Record M.T., Anderson C.F., Lohman T.M. Thermodynamic analysis of ion effects on binding and conformational equilibria of proteins and nucleic-acids—roles of ion association or release, screening, and ion effects on water activity. Q. Rev. Biophys. 1978;11:103–178. doi: 10.1017/s003358350000202x. [DOI] [PubMed] [Google Scholar]
- 29.Berg O.G., Ehrenberg M. Association kinetics with coupled three- and one-dimensional diffusion. Chain-length dependence of the association rate of specific DNA sites. Biophys. Chem. 1982;15:41–51. doi: 10.1016/0301-4622(82)87015-4. [DOI] [PubMed] [Google Scholar]
- 30.Kim J.G., Takeda Y., Matthews B.W., Anderson W.F. Kinetic studies on Cro repressor-operator DNA interaction. J. Mol. Biol. 1987;196:149–158. doi: 10.1016/0022-2836(87)90517-1. [DOI] [PubMed] [Google Scholar]
- 31.Stanford N.P., Szczelkun M.D., Marko J.F., Halford S.E. One- and three-dimensional pathways for proteins to reach specific DNA sites. EMBO J. 2000;19:6546–6557. doi: 10.1093/emboj/19.23.6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Terry B.J., Jack W.E., Modrich P. Facilitated diffusion during catalysis by EcoRI endonuclease. Nonspecific interactions in EcoRI catalysis. J. Biol. Chem. 1985;260:13130–13137. [PubMed] [Google Scholar]
- 33.Zhou H.X. A model for the mediation of processivity of DNA-targeting proteins by nonspecific binding: dependence on DNA length and presence of obstacles. Biophys. J. 2005;88:1608–1615. doi: 10.1529/biophysj.104.052688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barkley M.D. Salt dependence of the kinetics of the lac repressor-operator interaction: role of nonoperator deoxyribonucleic acid in the association reaction. Biochemistry. 1981;20:3833–3842. doi: 10.1021/bi00516a026. [DOI] [PubMed] [Google Scholar]
- 35.Rau D.C., Sidorova N.Y. Diffusion of the restriction nuclease EcoRI along DNA. J. Mol. Biol. 2010;395:408–416. doi: 10.1016/j.jmb.2009.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tafvizi A., Huang F., Leith J.S., Fersht A.R., Mirny L.A., van Oijen A.M. Tumor suppressor p53 slides on DNA with low friction and high stability. Biophys. J. 2008;95:L01–L03. doi: 10.1529/biophysj.108.134122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bresloff J.L., Crothers D.M. DNA-ethidium reaction kinetics: demonstration of direct ligand transfer between DNA binding sites. J. Mol. Biol. 1975;95:103–123. doi: 10.1016/0022-2836(75)90339-3. [DOI] [PubMed] [Google Scholar]
- 38.Fried M.G., Crothers D.M. Kinetics and mechanism in the reaction of gene regulatory proteins with DNA. J. Mol. Biol. 1984;172:263–282. doi: 10.1016/s0022-2836(84)80026-1. [DOI] [PubMed] [Google Scholar]
- 39.Iwahara J., Clore G.M. Direct observation of enhanced translocation of a homeodomain between DNA cognate sites by NMR exchange spectroscopy. J. Am. Chem. Soc. 2006;128:404–405. doi: 10.1021/ja056786o. [DOI] [PubMed] [Google Scholar]
- 40.Iwahara J., Zweckstetter M., Clore G.M. NMR structural and kinetic characterization of a homeodomain diffusing and hopping on nonspecific DNA. Proc. Natl. Acad. Sci. U.S.A. 2006;103:15062–15067. doi: 10.1073/pnas.0605868103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lieberman B.A., Nordeen S.K. DNA intersegment transfer, how steroid receptors search for a target site. J. Biol. Chem. 1997;272:1061–1068. doi: 10.1074/jbc.272.2.1061. [DOI] [PubMed] [Google Scholar]
- 42.Sidorova N.Y., Scott T., Rau D.C. DNA concentration-dependent dissociation of EcoRI: direct transfer or reaction during hopping. Biophys. J. 2013;104:1296–1303. doi: 10.1016/j.bpj.2013.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takayama Y., Clore G.M. Interplay between minor and major groove-binding transcription factors Sox2 and Oct1 in translocation on DNA studied by paramagnetic and diamagnetic NMR. J. Biol. Chem. 2012;287:14349–14363. doi: 10.1074/jbc.M112.352864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lohman T.M., DeHaseth P.L., Record M.T., Jr Analysis of ion concentration effects of the kinetics of protein-nucleic acid interactions. Application to lac repressor-operator interactions. Biophys. Chem. 1978;8:281–294. doi: 10.1016/0301-4622(78)80011-8. [DOI] [PubMed] [Google Scholar]
- 45.Bonnet I., Biebricher A., Porte P.L., Loverdo C., Benichou O., Voituriez R., Escude C., Wende W., Pingoud A., Desbiolles P. Sliding and jumping of single EcoRV restriction enzymes on non-cognate DNA. Nucleic Acids Res. 2008;36:4118–4127. doi: 10.1093/nar/gkn376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Givaty O., Levy Y. Protein sliding along DNA: dynamics and structural characterization. J. Mol. Biol. 2009;385:1087–1097. doi: 10.1016/j.jmb.2008.11.016. [DOI] [PubMed] [Google Scholar]
- 47.Terakawa T., Kenzaki H., Takada S. p53 searches on DNA by rotation-uncoupled sliding at C-terminal tails and restricted hopping of core domains. J. Am. Chem. Soc. 2012;134:14555–14562. doi: 10.1021/ja305369u. [DOI] [PubMed] [Google Scholar]
- 48.Blainey P.C., van Oijen A.M., Banerjee A., Verdine G.L., Xie X.S. A base-excision DNA-repair protein finds intrahelical lesion bases by fast sliding in contact with DNA. Proc. Natl. Acad. Sci. U.S.A. 2006;103:5752–5757. doi: 10.1073/pnas.0509723103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gorman J., Wang F., Redding S., Plys A.J., Fazio T., Wind S., Alani E.E., Greene E.C. Single-molecule imaging reveals target-search mechanisms during DNA mismatch repair. Proc. Natl. Acad. Sci. U.S.A. 2012;109:E3074–E3083. doi: 10.1073/pnas.1211364109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Anderson K.M., Esadze A., Manoharan M., Brüschweiler R., Gorenstein D.G., Iwahara J. Direct observation of the ion-pair dynamics at a protein-DNA interface by NMR spectroscopy. J. Am. Chem. Soc. 2013;135:3613–3619. doi: 10.1021/ja312314b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Luger K., Mader A.W., Richmond R.K., Sargent D.F., Richmond T.J. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 52.Schreiber G., Fersht A.R. Rapid, electrostatically assisted association of proteins. Nat. Struct. Biol. 1996;3:427–431. doi: 10.1038/nsb0596-427. [DOI] [PubMed] [Google Scholar]
- 53.Vijayakumar M., Wong K.Y., Schreiber G., Fersht A.R., Szabo A., Zhou H.X. Electrostatic enhancement of diffusion-controlled protein-protein association: comparison of theory and experiment on barnase and barstar. J. Mol. Biol. 1998;278:1015–1024. doi: 10.1006/jmbi.1998.1747. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


