Abstract
Ribbon synapses in the retina lack the t-SNARE (target-soluble N-ethylmaleimide-sensitive factor attachment protein receptor) syntaxin 1A that is found in conventional synapses of the nervous system, but instead contain the related isoform syntaxin 3B. Previous studies have demonstrated that syntaxin 3B is essential for synaptic vesicle exocytosis in ribbon synapses, but syntaxin 3B is less efficient than syntaxin 1A in binding the t-SNARE protein SNAP-25 and catalyzing vesicle fusion. We demonstrate here that syntaxin 3B is localized mainly on the presynaptic membrane of retinal ribbon synapses and that a subset of syntaxin 3B is localized in close proximity to the synaptic ribbon. We show further, that syntaxin 3B can be phosphorylated by the Ca2+/calmodulin-dependent protein kinase II (CaMKII). We determine that the phosphorylation site is located close to the N-terminus at T14. Syntaxin 3B with a phosphomimetic mutation (T14E) had a stronger binding affinity for SNAP-25 compared with wild type syntaxin 3B. We propose that phosphorylation of syntaxin 3B by CaMKII can modulate the assembly of the SNARE complex in ribbon synapses of the retina, and might regulate the exocytosis of synaptic vesicles in ribbon synapses.
Keywords: Ribbon synapse, retina, SNARE complex, SNAP-25, Munc18
Introduction
Ribbon synapses are specialized synapses that are found in photoreceptors and bipolar cells of the retina, hair cells of the inner ear, and pinealocytes of the pineal gland (Matthews and Fuchs, 2010). They contain a specialized plasma membrane attached structure called the synaptic ribbon (Mercer and Thoreson, 2011). The synaptic ribbon has synaptic vesicles tethered to it and the majority of the synaptic vesicle exocytosis in ribbon synapses is thought to occur in close proximity to the synaptic ribbon. Besides releasing neurotransmitters via fusion of individual synaptic vesicles with the plasma membrane, as conventional synapses do, ribbon synapses are also thought to release neurotransmitters via compound fusion of multiple synaptic vesicles (Matthews and Sterling, 2008; He et al., 2009).
Release of neurotransmitters via synaptic vesicle exocytosis in conventional synapses is dependent on the interaction between a set of so called v-SNARE (vesicular soluble N-ethylmaleimide-sensitive factor attachment protein receptor) proteins, localized on the synaptic vesicle and plasma membrane localized t-SNARE proteins. These specialized proteins interact via specialized coiled coil domains, the so-called SNARE domains to form a very stable complex that is thought to be essential for vesicle fusion.
In conventional synapses, synaptic vesicle fusion is dependent on the synaptic vesicle-associated v-SNARE protein synaptobrevin 2 (also called VAMP2) and the plasma membrane associated t-SNARE proteins syntaxin 1A or 1B and SNAP-25 (Jahn and Fasshauer, 2012; Rizo and Südhof, 2012). Ribbon synapses in the retina also contain SNAP-25 and synaptobrevin2/VAMP2, but they do not contain syntaxin 1A or 1B (Morgans et al., 1996). Instead, they contain the related isoform syntaxin 3B (Curtis et al., 2008).
Different syntaxin iso-proteins are encoded by a large gene family of syntaxin genes and are thought to mediate membrane fusion in different membrane trafficking processes. Syntaxin proteins have a characteristic domain structure consisting of (from N to C-terminus) a short N-terminal sequence called the N-peptide, three coiled domains forming the so called Habc domain, a flexible linker region, the SNARE domain and a C-terminal transmembrane spanning domain. Syntaxins are thought to exist in two conformations, the closed conformation where the three subdomains of the Habc domain form an intra-molecular parallel helical complex with the SNARE domain and an open conformation where the SNARE domain is free and can interact with SNAP-25 (Dulubova et al., 1999).
Syntaxin 3B is one of the two major transcripts (syntaxin 3A and B) of the syntaxin 3 gene that are generated by differential splicing. The N-terminal part of the two proteins up to the first half of the SNARE domain is identical between syntaxin 3A and 3B, but the second half of the SNARE domains and the TMD differ significantly. The syntaxin 3B transcript is expressed at high levels in the mouse retina, but only at very low levels in brain, kidney or liver. In contrast syntaxin 3A is expressed at high levels in the kidney and at low levels in the brain but no expression is detectable in the retina using sensitive RT-PCR analysis (Curtis et al., 2008). Syntaxin 3B has previously been shown to be enriched in synaptic ribbon synapses (Morgans et al., 1996; Sherry et al., 2006) but its subcellular localization is not known.
Syntaxin 3A is thought to be involved in the trafficking of transport vesicles from the trans-golgi network to the apical membrane of non-neuronal cells (Sharma et al., 2006). In contrast syntaxin 3B has been shown to be important for synaptic vesicle exocytosis in retinal bipolar cells indicating a specific role for syntaxin 3B in ribbon synapses (Curtis et al., 2010). Furthermore, Syntaxin 3B, SNAP-25 and synaptobrevin can catalyze SNARE mediated vesicle fusion in an in vitro fusion assay (Curtis et al., 2008) but in comparison to syntaxin 1A, syntaxin 3B has a lower affinity to bind SNAP-25 and is less efficient in catalyzing vesicle fusion. However, the molecular basis for the reduced affinity of syntaxin 3B to SNAP-25 is unknown.
Syntaxin 1A and syntaxin 3A can be phosphorylated by different kinases. Syntaxin 1A is phosphorylated by Casein kinase II at serine 14 and the phosphorylation affects the binding of syntaxin 1A to synaptotagmin and the localization of syntaxin 1A in neurons (Risinger and Bennett, 1999; Foletti et al., 2000). In contrast syntaxin 3A is not a substrate for Casein kinase II but can be phosphorylated by Calcium/calmodulin dependent kinase II (CaMKII) (Risinger and Bennett, 1999). It is not known if syntaxin 3B can also be phosphorylated by CaMKII, what the function of such a phosphorylation could be and at what position the phosphorylation occurs.
Experimental procedures
Materials
Molecular biology reagents were obtained from New England Biolabs (Beverly, MA, U.S.A.), the vector expressing His6-SNAP-25b from pET-15b was a gift from Dr. James McNew (Rice University, TX, U.S.A.), recombinant alpha-CaMKII and calmodulin were gifts from Dr. M. Neal Waxham (University of Texas Medical School at Houston, TX, U.S.A.), Glutathione sepharose beads was obtained from Amersham Biosciences (Piscataway, New Jersey, U.S.A.), Ni-NTA agarose was from Qiagen (Hilden, Germany). Anti-SNAP-25 monoclonal antibody (Cl 71.1) was obtained from Synaptic Systems (Göttingen, Germany), monoclonal antibodies against CtBP2 and Munc18 were obtained from BD Biosciences (San Jose, CA, U.S.A.) and against PSD-95 (7E3-1B8) from Pierce/Thermo Fisher Scientific (Waltham, MA). Polyclonal rabbit antibody against glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (NB100-56875) was obtained from Novus Biologicals (Littleton, CO).
Animals
All animal procedures conformed to National Institutes of Health (NIH) guidelines and were approved by the Animal Welfare Committee of the University of Texas Health Science Center at Houston.
Antibody generation
Peptides from the N-terminus of syntaxin 3B KDRLEQLKAKQLTQDDC (UT478) and phosphopeptide KAKQL[pT]QDDDTC (UT649) with added cysteines (bolded) were synthesized by Biosyn (Levisville, TX) and peptides where coupled via the cysteine to maleimide-activated keyhole limpet hemocyanin(KLH) (Pierce, Rockford, IL). Antibodies were generated in rabbits by Cocalico Biologicals Inc. (Reamstown, PA). The rabbit sera were affinity purified using the immunizing peptide fixed to agarose with the Sulfolink® kit (Pierce, Rockford, IL) as described (Janz et al 1999) with the exception of phospho-antibody that was eluted with 3M MgCl2 and further purified by passing over a peptide column containing the unphospho-peptide (KAKQLTQDDDTC).
Plasmid construction
A mouse EST clone (pCMV-syntaxin 3B, accession No. BC024844, IMAGE clone No. 5357204) coding for full-length syntaxin 3B was used as a template to generate syntaxin 3B expression constructs by PCR. The GST-syntaxin 3B fusion construct (pGEX-STX 3B) was generated by PCR amplification of the sequence coding for the cytoplasmic domain without the transmembrane domain (residues 2–264) and subcloning into the BamHI and EcoRI site of pGEX-KG vector. The GST-syntaxin 3B truncated mutation construct (pGEX-STX3B SNARE) was generated by PCR amplification of the sequence coding for the SNARE domain (residues 186-264) and subcloning into the BamHI and EcoRI site of pGEX-KG vector. Syntaxin 3B point mutant constructs were generated by PCR using Pfu polymerase with full-length syntaxin 3B as template. PCR products were cloned into the BamHI and EcoRI site of pGEX-KG, and mutations were verified by DNA sequencing.
Expression and purification of recombinant proteins
Glutathione S-transferase (GST) fusion proteins of the full cytoplasmic domain of mouse syntaxin 1A, mouse syntaxin 3A, and mouse syntaxin 3B as well as its mutations T14A, T14E, T81A, S145A, and S187A, were expressed in Escherichia coli BL21 cells and purified with gluthathione-sepharose beads. His6-SNAP-25 was expressed in BL21 cells and purified with Ni-NTA agarose.
GST pulldown assays
Mouse retinas were isolated and homogenized in 10 volumes buffer A (20 mM Hepes-NaOH, pH 7.4, with protease inhibitors (Complete (Roche)). After homogenization, an equal amount of buffer B (20 mM Hepes-NaOH, pH 7.4, 0.2 M NaCl, 2% Triton X-100) was added, and the homogenate was incubated at 4°C with rotation for 30 minutes. The homogenate was then centrifuged for 1 hour at 20,000 rpm at 4°C in a JA-20 rotor and the supernatant was used for binding experiments. The protein concentration in the supernatant was adjusted by dilution to 0.1 mg/ml. One milliliter of the extract was mixed with the glutathione-sepharose beads for each binding reaction. The samples were incubated overnight at 4°C, with rotation. After overnight incubation, the samples were centrifuged at 3,000 rpm for 30 seconds. The supernatant was removed, and the beads were washed six times with cold wash buffer (20 mM Hepes-NaOH, pH 7.4, 0.1 M NaCl, 1% Triton X-100). After the washes, SDS sample buffer was added, and the samples were analyzed by SDS-PAGE and western blotting.
Phosphorylation reactions
Phosphorylation reactions contained 12 μg/ml CaMKII in a total volume of 50 μl with the following reaction buffers: 25 mM HEPES, pH 7.5, 10 mM MgCl2, 50 mM KCl, 4 mM CaCl2, 2 μM calmodulin, and 0.4 mM dithiothreitol. Samples were preincubated for 1 min at 30°C before addition of 5 μl of [γ-32P] ATP (200 mM; 40 mCi/ml). The reactions were continued for 10 min at 30°C and then terminated by adding 10 μl of 4x SDS-PAGE sample buffer. The samples were separated by SDS-PAGE, transferred onto nitrocellulose and analyzed by autoradiography.
Western blotting
The following primary antibodies were used for western blotting: monoclonal antibodies against SNAP25 (Cl 71.1) 1:2,000 (Synaptic Systems, Göttingen, Germany), Munc18 (clone 31) 1:2,000 (BD Transduction Labs, San Jose, CA) and PSD-95 (7E3-1B8) 1:2000 (Pierce/Thermo Fisher Scientific, Waltham, MA); polyclonal antibodies against (GAPDH) (NB100-56875) 1:500 (Novus Biologicals, Littleton, CO), syntaxin 3 (UT478) 1:500, phospho-syntaxin 3 (UT649) 1:500. For non-quantitative analysis, peroxidase-labeled secondary antibodies (Zymed, South San Francisco, CA) were used with a chemiluminescence detection system (ECL Plus Kit, Amersham Biosciences, Piscataway, NJ). For quantitative analysis, the blots were probed with Alexa Fluor 488-labeled secondary antibodies (Molecular Probes, Eugene, OR) and the signals were detected by a Typhoon 9400 imager (Amersham Biosciences) and were analyzed quantitatively using ImageJ. Blocking peptides where used at a concentration of 10μg/ml.
Isolation of Bipolar Cell of Retina
Bipolar cells were isolated from 2- to 6-month-old C57Bl6/J mice by mechanical trituration after enzymatic digestion using methods similar to those previously described (Zhou et al., 2006). The resulting cell suspension was plated onto glass coverslips coated with 0.1 mg/ml poly-D-lysine. Rod bipolar cells were identified based on their characteristic morphology using previously described criteria (Zhou et al., 2006).
Immunocytochemistry and confocal microscopy
Fixed retinal sections and dissociated cells were incubated with primary antibodies diluted in 3% normal goat serum (NGS), 1% bovine serum albumin (BSA), 0.5% Triton X-100, 0.05% NaN3, and 0.1 M Phosphate buffer (PB) overnight at 4°C. Whole-mount preparations were incubated in primary antibody for 2 days at 4°C. Primary antibodies used were: purified antibodies against syntaxin 3 (UT478) (1:400 dilution) and phosphosyntaxin 3 (UT 649) (1:500 dilution), monoclonal antibodies against CtBP2/ribeye, (1:500 dilution) (BD Transduction Laboratories, San Jose, CA), monoclonal antibodies against SV2 (1:500 dilution) (Developmental Studies Hybridoma Bank at University of Iowa), and SNAP-25 (Cl 71.1) (1:200 dilution) (Synaptic Systems, Göttingen, Germany). Blocking peptides where used at a concentration of 10μg/ml. For fluorescence labeling, sections or cells were incubated with a mixture of Alexa-tagged anti-mouse (Alexa 488) and anti-rabbit (Alexa 568) immunoglobulin secondary antibodies (Molecular Probes, Eugene, OR) at 1:400 dilutions for 1 h at room temperature in a light-protected environment. Sections or cells were rinsed and mounted on a slide with Vectashield mounting medium for fluorescence (H-1000, Vector Laboratories, Burlingame, CA). Immunolabeled specimens were scanned with a 0.2-μm step size on a Zeiss Laser Scanning Microscope 510 Meta (Carl Zeiss, Inc., Thornwood, NY). Confocal images were analyzed using the Zeiss LSM 510 proprietary software (Zeiss, Oberkochen, Germany). Intensity levels and brightness/contrast were adjusted in Adobe Photoshop CS v.10.0 (Adobe Systems, San Jose, CA).
Subcellular fractionation of retina
A subcellular fractionation of retina was performed using a modification of a method developed for brain tissue (Huttner et al., 1983; Janz et al., 1998). Four mouse retinas were homogenized in 500 ul buffer 1 (0.32M sucrose, 10mM HEPES-NaOH, pH 7.4 with Complete™ (Roche) protease inhibitors (without EDTA)) with 10 strokes at 900 rpm in a glass-teflon homogenizer. The homogenate was centrifuged at 1,000 rpm for 5 minutes in Eppendorf centrifuge yielding pellet P1. The supernatant (S1) was centrifuged again at 10,000 rpm for 10 minutes. The pellet was resuspended in 1ml buffer 1 and centrifuged again in an Eppendorf centrifuge at 10,000 rpm for 15 minutes yielding pellet P2 (crude synaptosomal pellet) and supernatant S2. The crude synaptosomes (P2) were resuspended in 100μl buffer 1 and lysed hypo-osmotically by dilution with 900μl of 5mM HEPES buffer, homogenization with 10 strokes at 900 rpm in a glass-Teflon homogenizer, shaking at 4°C for 15 minutes and a second homogenization again with 10 strokes at 900 rpm in the glass-Teflon homogenizer. The lysed synaptosomes were then centrifuged at 14000 rpm in an Eppendorf centrifuge yielding a synaptic plasma-membrane pellet (LP1). The supernantant (LS1) is centrifuged 1 hour at 60000 rpm in a TLA-120 rotor to yield the synaptic vesicle-enriched pellet (LP2) and supernatant (LS2). Protein concentration of all the fractions was measured using the Biorad Protein assay and adjusted to the same concentration by diluting with 5 mM Hepes-NaOH lysis buffer. Subcellular fractions were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) and western blot.
RESULTS
Syntaxin 3B protein expression
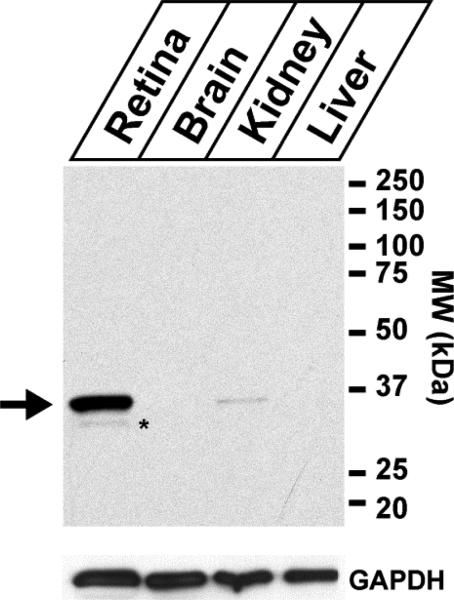
The expression of syntaxin 3B in different tissues has previously only been studied on the mRNA level. In order to analyze the distribution of syntaxin 3B protein in different tissues, an antibody directed against the N-terminus of syntaxin 3 was generated and purified by using affinity chromatography with the immunizing peptide. The immunizing peptide is located in an area that is identical between two main splice forms of the syntaxin 3 gene (syntaxin 3A and 3B), so the antibody does not distinguish between these two isoforms. The expression of syntaxin 3 protein in different tissues from mouse was then analyzed by western blot analysis (figure 1). A strong protein band of the predicted molecular weight of syntaxin 3B (33 kDa) was observed in retina extract. A weaker band of approximately the same size was observed in the kidney tissue. No protein was detectable under these conditions in the liver or brain extracts. The absence of any reactive protein of the size of syntaxin 1A or 1B (about 33 kDa), which are expressed at high levels in brain (Bennett et al., 1993), confirms that the antibody used here does not cross-react with syntaxin 1A or 1B.
Figure 1. Analysis of the expression of syntaxin 3 protein in different tissues.
Top panel: Tissue extracts from retina, brain, kidney and liver where separated by SDS-PAGE and analyzed by western blotting using the syntaxin 3 antibody. The predicted position of syntaxin 3B (33kDa) is labeled with an arrow. A weak band that migrates slightly lower and probably represents a breakdown product is labeled with asterisk. The analysis shows that syntaxin 3 is highly expressed in retina and weakly expressed in the kidney. No expression of syntaxin 3 protein was detectable in brain or liver under these conditions. Bottom panel: The expression of the ubiquitously expressed marker Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was analyzed to confirm equal protein concentrations in the tissue extracts.
The expression pattern observed for the syntaxin 3 protein in the different tissue extracts is consistent with the expression of the syntaxin 3 mRNA (Curtis et al., 2008). Only syntaxin 3B mRNA is expressed in mouse retina (Curtis et al., 2008), therefore protein in retina reacted with the antibodies corresponds to syntaxin 3B. The identity of the protein recognized by our antibody in the retina was confirmed as syntaxin 3B by purifying the 33 kDa band using immuno-precipitation from bovine retina extract, and analyzing the purified band by MS fingerprinting analysis (data not shown). Therefore the protein identified in the following experiment by the syntaxin 3 antibodies in the mouse retina will be referred to as syntaxin 3B.
Syntaxin 3B localization in the retina
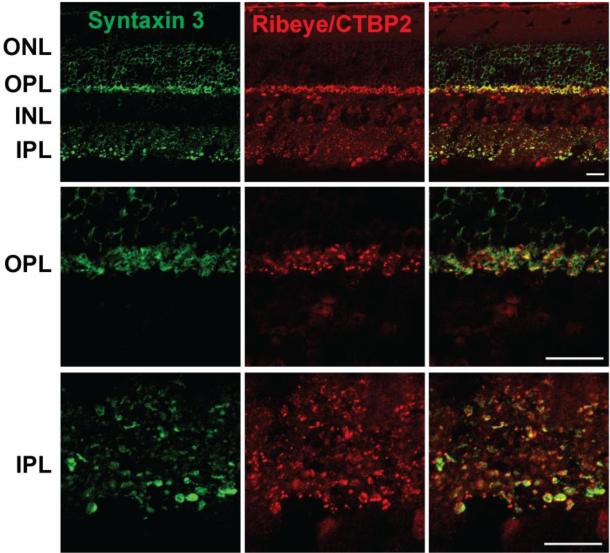
To investigate the localization of syntaxin 3B in retina, mouse retina sections and whole mounted mouse retinas were double labeled with the syntaxin 3 antibody and an antibody that recognizes a common domain found in the nuclear protein CTBP2 (C-terminal-binding protein 2) and in ribeye, a marker for ribbon synaptic terminals (Schmitz et al., 2000). Syntaxin 3B labeling in retina cross-sections showed strong labeling of the inner and outer plexiform layers (IPL and OPL), the main synaptic layers in the retina, consistent with previous studies (Morgans et al., 1996; Sherry et al., 2006). In the OPL, strong syntaxin 3B labeling is seen in photoreceptor terminals, identified by the staining of ribeye (figure 2). In the inner plexiform layer (IPL), the innermost portion of the IPL, corresponding to the synaptic terminals of ribbon synapses, showed strong syntaxin 3B and ribeye labeling.
Figure 2. Syntaxin 3B is found in synaptic layers of the mouse retina.
Top panel: Retina sections were labeled with antibodies directed against syntaxin 3 and the ribbon synapses and nuclear markers Ribeye/CTPB2. The synaptic layers (inner plexiform layer (IPL) and outer plexiform layer (OPL)) show strong syntaxin 3B labeling. INL, Inner nuclear layer, ONL, outer nuclear layer; GCL, ganglion cell layer.
Bottom panels: Higher magnifications of the OPl and IPL regions.
Scale bar = 20 μm.
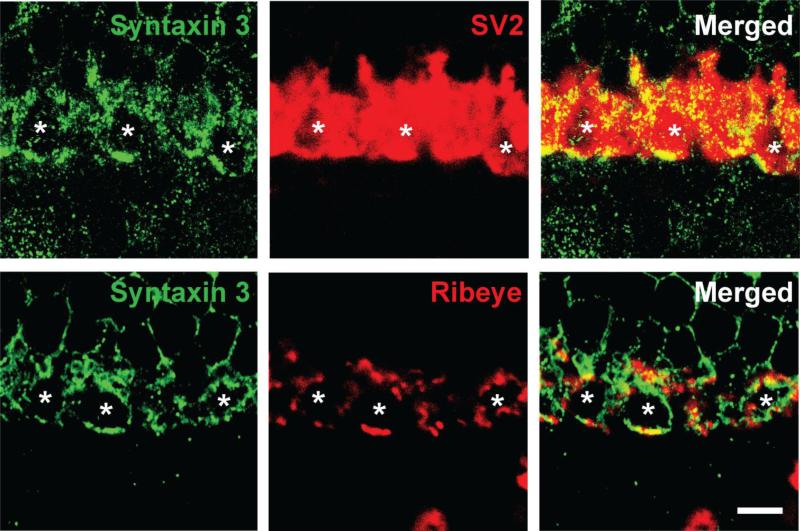
Next we analyzed the localization of syntaxin 3B in photoreceptor terminals using high resolution confocal microscopy and double labeling with the synaptic vesicle marker SV2 and ribeye (figure 3). This experiment reveals that syntaxin 3B in the OPL is mainly located at the synaptic plasma membrane of the photoreceptor terminals. While some syntaxin 3B is found in close proximity to the synaptic ribbons (as identified by the ribeye antibody) the majority of the syntaxin 3B is found in the extra ribbon area on the membrane of the terminals. As mentioned before, there is considerable evidence that synaptic vesicles in ribbon synapses can undergo compound fusion that would involve inter-vesicle fusion. Such a process would likely require the presence of t-SNARE proteins such as syntaxin 3B on the synaptic vesicles. However, the majority of syntaxin 3B was found at the plasma membrane and there was very little co-localization detectable between the synaptic vesicle marker SV2 and syntaxin 3B.
Figure 3. Syntaxin 3B in photoreceptor terminals partially colocalizes with ribeye.
Retinal sections where double labeled with antibodies against syntaxin 3 and ribeye/CTBP2 or the synaptic vesicle marker SV2. Synaptic terminals are marked with asterisks. Some syntaxin 3B is colocalized with ribeye but a large fraction occupies ribeye free parts of the plasma membrane. Scale bar = 5 μm.
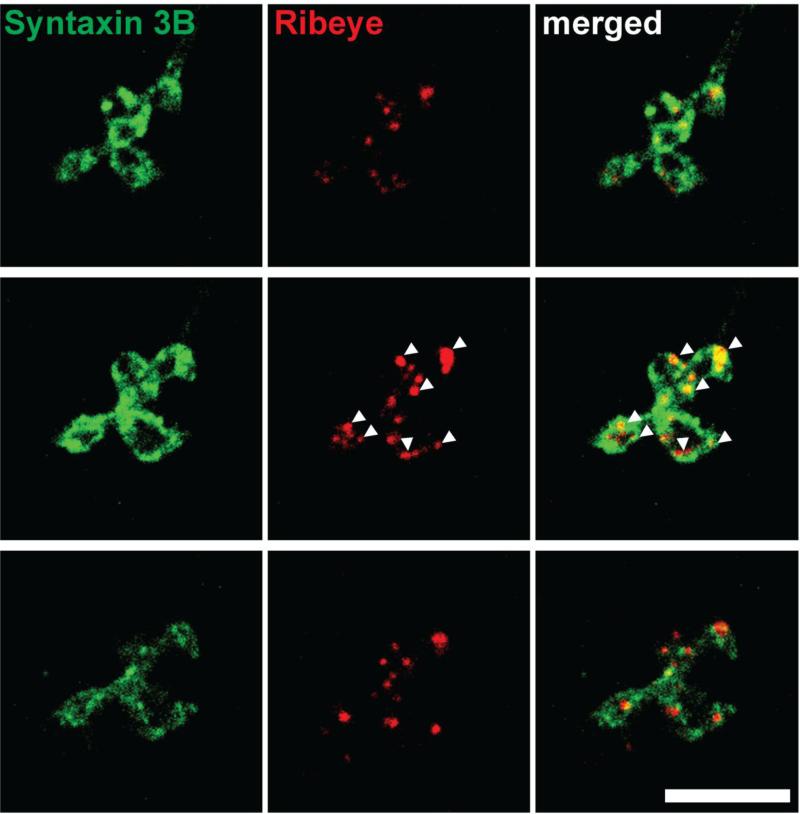
In another set of experiments we investigated the subcellular localization of syntaxin 3B in retina bipolar cells. Retinas were dissociated and isolated bipolar cells were double labeled with antibodies against syntaxin 3B and CtBP2/ribeye (Fig. 4) and analyzed by confocal microscopy. The terminals of the isolated bipolar cells showed strong labeling for syntaxin 3B on the synaptic plasma membrane. Similar to the localization in the photoreceptor terminals, syntaxin 3B was not restricted to the proximity of the synaptic ribbons. These findings indicate that the preferential fusion of synaptic vesicles at the proximity of the synaptic ribbons in retinal ribbon synapses is not caused by a restricted distribution of syntaxin 3B at the plasma membrane.
Figure 4. Syntaxin 3B is localized on the plasma membrane of bipolar cell terminals.
Consecutive optical sections of isolated retinal bipolar cells terminals were double labeled for syntaxin 3B and ribeye/CTBP2. Arrowheads mark the position of individual synaptic ribbons. Scale bar = 5 μ m.
Sub cellular distribution of syntaxin 3B
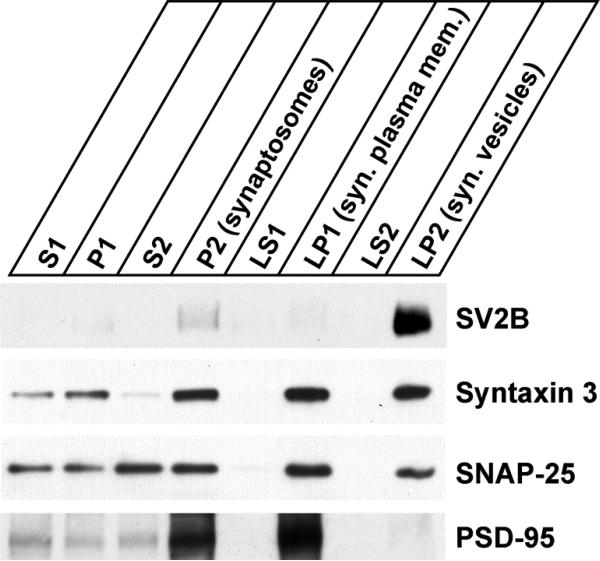
In order to confirm the localization of syntaxin 3B on the synaptic plasma membrane of ribbon synapses using an independent technique, we performed a subcellular fractionation of mouse retinas. Synaptosomes were prepared from mouse retinas by differential centrifugation using a modification of techniques developed for brain tissue (Huttner et al. 1983, Janz et al. 1998). The synaptosomes (P2) were then hypoosmotically lysed and further fractionated into soluble fractions containing soluble presynaptic proteins (synaptosol) (LS1 and LS2), synaptic plasma membrane fraction (LP1) and synaptic vesicle fraction (LP2). The distribution of syntaxin 3B, SNAP-25, SV2B, a marker protein for synaptic vesicles in ribbon synapses of the retina (Wang et al. 2003) and the postsynaptic density marker PSD-95 in the different fractions was then analyzed by SDS-PAGE and western blotting (figure 5). As expected, the synaptic vesicle marker SV2B was highly enriched in the synaptic vesicle fraction (LP2) and almost undetectable in the other synaptic fractions (LS1, LS2 and LP1). In contrast the postsynaptic density protein PSD-95 was highly enriched in the synaptic plasma membrane fraction (LP1) but not detectable in the other synaptic fractions (LS1, LS2 and LP2). This demonstrates the efficient separation of the different subcellular compartments, especially of the synaptic plasma membrane and the synaptic vesicle fractions. SNAP-25 was enriched in the synaptic plasma membrane fraction (LP1), but was also present in the synaptic vesicle fraction. This is consistent with the localization of SNAP-25 in conventional synapses where the majority is found on the synaptic plasma membrane but a sub-fraction is present on the synaptic vesicles (Takamori et al. 2006). Syntaxin 3B was enriched in the synaptic plasma membrane fraction (LP1) but was also found in the synaptic vesicle fraction (LP2), similar to SNAP-25. These subcellular fractionation experiments confirm the localization of syntaxin 3B to the synaptic plasma membrane in ribbon synapses but also suggest that a significant fraction of syntaxin 3B is found on synaptic vesicles. This finding differs from the previous immuno-staining results that indicated only very little co-localization between the synaptic vesicle marker SV2 and syntaxin 3B in ribbon synaptic terminals of the retina and is likely due to differences in the sensitivity of the two experiments.
Figure 5. Syntaxin 3B is associated with the synaptic plasma membrane.
Subcellular fractionation of mouse retina was performed to analyze the distribution of syntaxin 3B. Synaptosomes (P2) were isolated from mouse retina by differential centrifugation. The synaptosomes were then hypo-osmotically lysed and separated into synaptic plasma membrane fraction (LP1), synaptic vesicle fraction (LP2) and soluble presynaptic protein fractions (LS1 and LS2). Equal protein amounts were analyzed by SDS-PAGE and western blotting using specific antibodies directed against SV2B (ribbon synaptic vesicle marker), syntaxin 3, SNAP-25 and PSD-95 (postsynaptic density protein).
Reduced SNAP-25 binding is caused by the N-terminus of syntaxin 3B
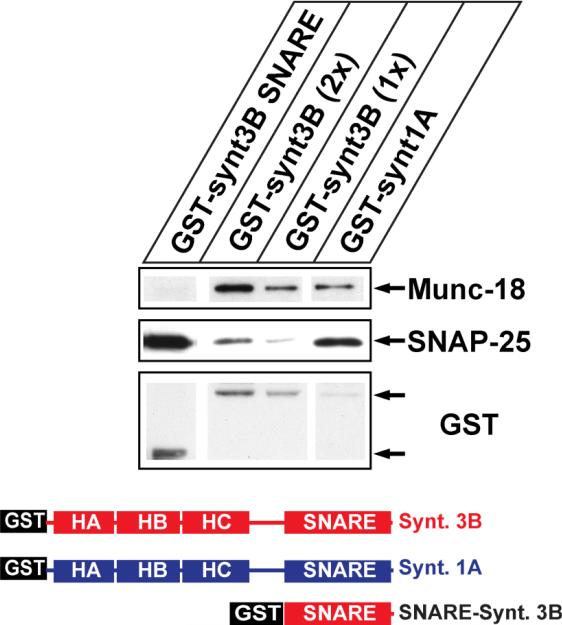
As mentioned previously, syntaxin 3B binds SNAP-25 less efficiently than syntaxin 1A (Curtis et al. 2008) despite the fact that the SNARE domains of syntaxin 1A and syntaxin 3B are very similar. In contrast, the N-terminal domains of syntaxin 3B and syntaxin 1A are much less homologous. This makes it likely that the differences between the N-terminal domains of syntaxin 3B and syntaxin 1A could be responsible for the reduced binding of SNAP-25, possibly by keeping syntaxin 3B in a more closed conformation. In order to analyze the role of the N-terminal domain of syntaxin 3B in SNAP-25 binding, we generated a GST fusion construct that contains only the SNARE domain of syntaxin 3B without the N-terminus. The binding of SNAP-25 from brain extract to the SNARE domain construct in comparison to constructs containing the full cytoplasmatic domains of syntaxin 3B and syntaxin1A was analyzed using GST-pulldown assays with protein extract from mouse brain (figure 6). The experiment demonstrated that the syntaxin 3B SNARE domain binds SNAP-25 with stronger affinity than the full length syntaxin 3B, confirming the hypothesis that the N-terminal part of syntaxin 3B has a negative effect on the SNAP-25 binding.
Figure 6. The N-terminal domain reduces binding of syntaxin 3B to SNAP25.
SNAP-25 and Munc18 binding to GST fusion proteins of syntaxins 1A and 3B without transmembrane domains and the isolated syntaxin 3B SNARE domain was analyzed using pulldown assays with retina extract followed by SDS-PAGE and western blotting using SNAP-25 antibodies and GST antibodies (loading control). The structure of the recombinant proteins is depicted below. Two different amounts of GST-syntaxin 3B protein were loaded to allow a better comparison. Recombinant GST fusion proteins of the predicted size are marked with arrows.
Syntaxin 3B is phosphorylated at T14 by CaMKII
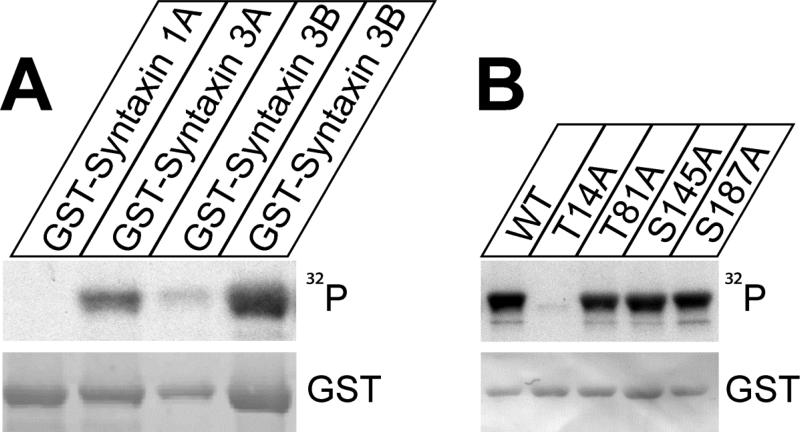
Previous studies have demonstrated that syntaxin 1A is substrate of casein kinase whereas syntaxin 3A is substrate of CaMKII (Risinger and Bennett, 1999). While the exact phosphorylation site in syntaxin 3A has not been determined it was previously shown that the major CaMKII phosphorylation site in syntaxin3A is located in the N-terminal region of the protein (aminoacids 4-187) (Riesinger and Bennet, 1999). Since this area is identical between syntaxin 3A and 3B we tested if syntaxin 3B can also be phosphorylated by CaMKII. Fusion proteins of GST and the cytoplasmic domains of syntaxin 1A, 3A and 3B were in vitro phosphorylated by CaMKII in the presence of Ɣ-32P-ATP. The reaction products were then resolved by SDS-PAGE, transferred to nitrocellulose membranes and analyzed by autoradiography to detect 32P incorporation and by immunoblot with GST monoclonal antibodies to control for protein amounts (figure 7A). The experiment showed that syntaxin 3B was phosphorylated with a similar efficiency as syntaxin 3A. In contrast syntaxin 1A was not phosphorylated by CaMKII under these conditions. This experiment confirms that syntaxin 3B is an efficient substrate of CaMKII.
Figure 7. Syntaxin 3B is phosphorylated by CaMKII at T14.
A. GST-syntaxin fusion proteins were in vitro phosphorylated by CaMKII and Ɣ -32P ATP, separated by SDS-PAGE and analyzed by autoradiography (32P) and immunoblotting with GST antibodies. Two different amounts of syntaxin 3B were loaded to allow better comparison with the other proteins.
B: Wildtype GST-syntaxin 3B as well as mutated proteins were phosphorylated and analyzed as in A. Wild type syntaxin 3B and mutant proteins GST-syntaxin 3B T81A, S145A, and S187A were phosphorylated similarly, while the mutant protein GST-syntaxin 3B T14A was not phosphorylated, demonstrating that T14 is the phosphorylation site in syntaxin 3B.
To identify potential phosphorylation site of syntaxin 3B for CaMKII the protein sequence of mouse syntaxin 3B was analyzed for potential CaMKII phosphorylation sites within syntaxin 3B. Several potential sites were identified and four sites that are not conserved between syntaxin 3B and syntaxin 1A (T14, T81, S145, and S187) were selected for site directed mutagenesis to alanine. Four corresponding GST fusion proteins (GST-Syntaxin T14A, T81A, S145A, and S187A) were generated and tested for their capacity to be phosphorylated by CaMKII (figure 7B). The experiment revealed that only the T14A mutation effectively eliminated phosphorylation by CaMKII, whereas the other mutations had no effect. Therefore, T14 was identified as the CaMKII phosphorylation site in syntaxin 3B.
A phosphomimetic mutation of syntaxin 3B increases the affinity to SNAP25
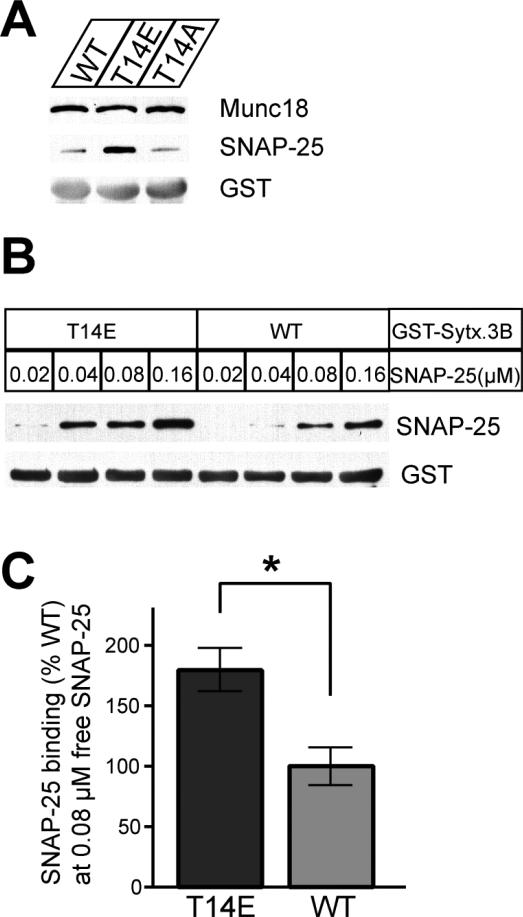
Syntaxin 3B has a lower binding affinity to SNAP-25 than syntaxin1A and is less efficient in catalyzing SNARE mediated in vitro fusion of vesicles (Curtis et al. 2008), but seems to efficiently catalyze vesicle fusion in ribbon synapses in vivo (Curtis et al 2010). This discrepancy between in vitro and in vivo activity suggests that extra factors are required that allow syntaxin 3B to function efficiently in ribbon synapses. Therefore, we decided to investigate if the phosphorylation of syntaxin 3B could modulate the efficiency of syntaxin 3B binding to SNAP-25. We generated a phosphomimetic mutation (T14E) and a control mutation (T14A) in GST-syntaxin 3B (without the transmembrane domain) to test the binding to native SNAP-25 and Munc18 from mouse retina extract in GST pulldown assays (figure 8). The experiment revealed that GST-syntaxin3B with the phosphomimetic mutation bound more SNAP25 than the wildtype protein or the T14A mutation. In contrast, neither the phosphomimetic T14E mutation nor the T14A mutation changed the binding of GST-syntaxin 3B to munc18 from retina extract.
Figure 8. A phosphomimetic mutation of syntaxin 3B (T14E) increases the binding affinity to SNAP25.
A. GST pulldown assays with retina extract were used to investigate the binding of SNAP-25 and Munc18 to wildtype GST-syntaxin 3B (WT), a phosphomimetic mutant (T14E) and a control mutant (T14A). The pulldown samples were analyzed by SDS-PAGE and immunoblotting with antibodies against Munc18, SNAP-25 and GST. The phosphomimetic T14E mutant protein had a much higher affinity to SNAP-25 than the wildtype or the T14A mutant protein but Munc18 was bound with similar efficiency by all three proteins. B. GST pulldown experiments were performed to directly test the binding of recombinant SNAP-25 to wildtype (WT) or mutant (T14E) GST-syntaxin 3B. The concentration of SNAP-25 in the assay is given above the lanes. The phosphomimetic T14E mutant protein bound recombinant SNAP-25 with a higher affinity than GST-syntaxin 3B. C. Multiple pulldown experiments were performed using recombinant SNAP-25 (0.08 μM) and the amount of bound SNAP-25 was quantified. Results were normalized to the amount bound by the wildtype protein. The T14E mutant protein showed a significantly higher affinity for recombinant SNAP-25 than the wild type protein. (WT: 100.0 ± 15.97%; T14E: 179.5 ± 18.63% (+/- SEM, n=3, p-value= 0.0317 (unpaired two tailed t test)).
We next investigated the binding properties of syntaxin 3B phosphomimetic T14E mutation with recombinant SNAP-25 to investigate if the changed binding affinity of syntaxin 3B is an endogenous property of syntaxin 3B that does not depend on other proteins. Different amounts of recombinant SNAP-25 were incubated with GST-syntaxin 3B wild type and T14E mutation. The pulldown samples were analyzed by immunoblotting with SNAP-25 monoclonal antibodies. The membrane was also probed with an antibody to GST to ensure that the amount of the recombinant proteins in each pulldown was comparable. The phosphomimetic T14E mutation bound more SNAP-25 than GST-syntaxin 3B wild type at each corresponding concentrations of SNAP-25 (figure 8B). To quantify the change in the binding properties of syntaxin 3B with recombinant SNAP-25 quantitatively, recombinant SNAP-25 at a concentration that represented about the half maximal binding affinity (0.08μM) was incubated with GST-syntaxin 3B wild type and T14E mutation respectively. The pulldown samples were analyzed by immunoblotting with SNAP-25 monoclonal antibodies and fluorescence-labeled secondary antibodies. The phosphomimetic T14E mutant showed a significantly stronger affinity for recombinant SNAP-25 than GST-syntaxin 3B wild type protein (figure 8C).
Syntaxin 3B in the mouse retina is phosphorylated at T14
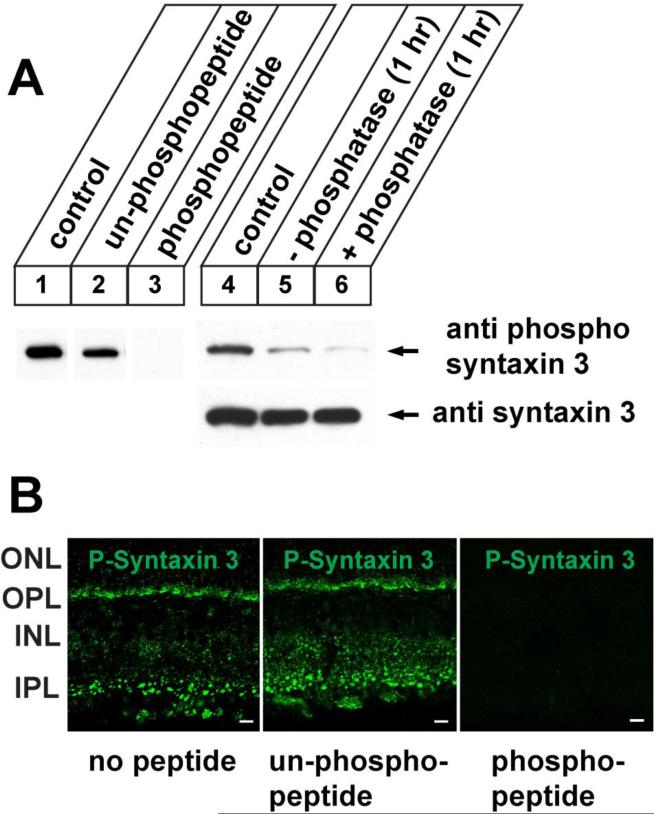
We have so far demonstrated using recombinant proteins that syntaxin 3B can be phosphorylated in vitro by CaMKII at T14. This raises the question if this phosphorylation also happens in vivo in the mammalian retina. In order to test this question we generated a phospho-specific peptide antibody against the T14 site of syntaxin 3. The antibody was affinity purified using the immunizing phospho-peptide and not phospho-specific antibodies were removed by absorption with the corresponding unphosphorylated peptide. The antibody was then tested on western blots of mouse retina extract and shown to react with a protein of the predicted mobility of syntaxin 3B (figure 9, lane1). The specificity of the antibody was confirmed by blocking experiments using the immunizing phospho-peptide or the corresponding unphosphorylated peptide. In a second set of experiments we confirmed that the antibody recognizes a phosphoprotein by incubating retina extract for 1 hour at 370C in the presence and absence of alkaline phosphatase. The reactivity of the antibody with the predicted phospho-syntaxin 3B band was already greatly reduced after incubation without additional phosphatases, indicating that endogenous phosphatases in the extract are already sufficient to dephosphorylate syntaxin 3B. The addition of alkaline phosphatase reduced the observed signal even further. Blotting of the samples with the syntaxin 3B antibody confirmed that the observed reduction in phospho-syntaxin 3B was not caused by degradation of the protein. These experiments confirm that syntaxin 3B in the retina is phosphorylated at T14. Next we investigated the distribution of the phosphorylated syntaxin 3B in the retina by immunolabeling mouse retina sections with the phospho-specific antibody and secondary fluorescent antibodies. In order to confirm the specificity of the labeling, blocking experiments with the phosphorylated peptide used for the generation of the antibody or the corresponding unphosphorylated peptide were performed. The antibody specifically labeled the synaptic layers in the retina (IPL and OPL) demonstrating that the phosphorylated syntaxin 3B is found in retinal ribbon synapses.
Figure 9. Syntaxin 3B in the mouse retina is phosphorylated at T14.
A. Left panel (1-3): Western blot analysis of mouse retina extract using a peptide antibody directed against a phosphopeptide derived from the N-terminus of syntaxin 3B detected a band of the predicted size of syntaxin 3B (1). Addition of the immunizing phospopeptide to the western blot blocked the signal (lane 3), whereas the corresponding unphosphorylated peptide had only a weak effect on the phospo-syntaxin 3B signal (lane 2). Right panel (4-6): Retina extract was incubated for 1 hr at 370C in the presence (6) or absence (5) of alkaline phosphatase and analyzed by western blotting using the phospho-syntaxin 3 antibody. Unincubated extract was used as a control (4). Stability of syntaxin 3B protein during the incubation was confirmed by blotting with the syntaxin 3 antibody.
B. Retina sections were labeled with the phospho-syntaxin 3 antibody with or without blocking antibodies as in A. Strong labeling of the ribbon synapse containing layers (IPL and OPL) with the phospho-specific antibodies indicates that syntaxin 3B in ribbon synapses is phosphorylated at T14. Scale bar = 20μm.
Discussion
Structural consequence of the phosphorylation of syntaxin 3B
What is the molecular basis for the reduced binding of SNAP-25 by syntaxin 3B compared to Syntaxin 1A and the phosphorylated syntaxin 3B?
As mentioned before, the regulatory Habc domain causes syntaxin 1A to exist in two different confirmations: open and closed (Dulubova et al., 1999). Only the open configuration can bind SNAP-25 and a mutation that keeps syntaxin 1B, in a constitutive open conformation has been shown to lead to epileptic seizures and altered synaptic transmission in mutant mice (Gerber et al., 2008), indicating that the balance between the open and closed forms can regulate the efficiency of synaptic vesicle exocytosis. We show here that removing of the N-terminal domain of syntaxin 3B causes the truncated protein to have an increased SNAP-25 binding affinity. This demonstrates that the regulatory N-terminus and not the SNARE domain cause the reduced SNAP-25 binding affinity of syntaxin 3B. The most plausible explanation for this finding is that the N-terminal domain causes syntaxin 3B to be mainly in the closed conformation. The phosphomimetic mutation of syntaxin 3B increases the binding to SNAP-25 without the requirement of other cofactors, indicating that the phosphorylation changes syntaxin 3B into a more open conformation as depicted in figure 10. This model is also consistent with the fact that the binding of Munc18 from retina extracts to GST syntaxin 3B is not affected by the phosphomimetic mutation since munc18 is able to bind to the closed syntaxin as well as to the full SNARE complex (Dulubova et al., 2007).
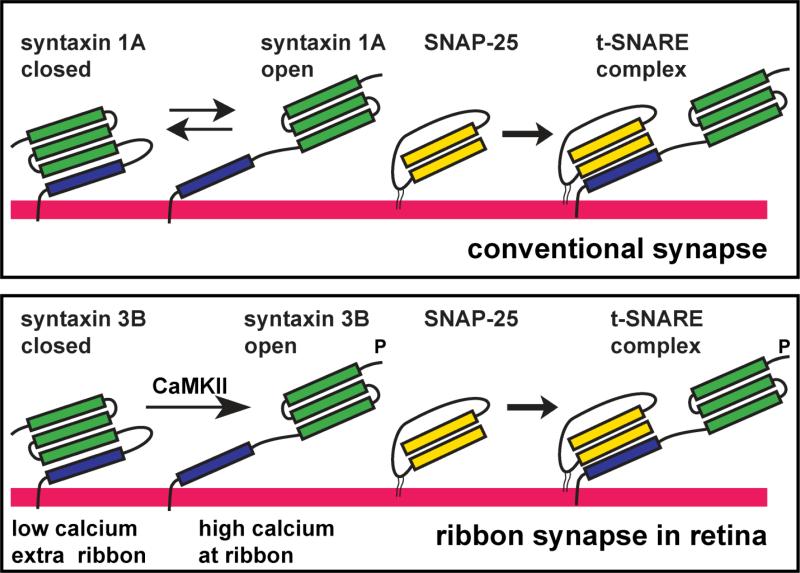
Figure 10. Model of the regulation of syntaxin 3B in ribbon synapses of the retina.
In conventional synapses syntaxin 1A exists in two conformations: “open” and “closed”. The “closed” conformation has the Habc domain (green) folding back on the SNARE domain (blue) and cannot bind to SNAP-25 to form a t-SNARE complex. We propose that syntaxin 3B in ribbon synapses of the retina exists mainly in the closed conformation. Activation of CaMKII by elevated calcium leads to the phosporylation of syntaxin 3B in the proximity of the ribbon. The phosphorylation of syntaxin 3B stabilizes the open conformation that can then efficiently form a t-SNARE complex and increase synaptic vesicle exocytosis.
Phosphorylation of syntaxin 3B can regulate the efficiency of neurotransmitter release in ribbon synapses
We demonstrate here that the syntaxin isoform found at ribbon synapses of the retina, syntaxin 3B, is a substrate for CaMKII and that a phosphomimetic mutation at the CaMKII phosphorylation site T14 increases the binding of syntaxin 3B to SNAP-25. In contrast the syntaxin isoform found in conventional synapses syntaxin 1A cannot be phosphorylated by CaMKII. What could be the functional relevance of the phosphorylation of syntaxin 3B by CaMKII in ribbon synapses? Different studies have shown that ribbon synapses are enriched in CaMKII (Bronstein et al., 1988; Ullrich and Südhof, 1994) and CaMKII has also recently been identified in a ribbon associated complex of proteins (Uthaiah and Hudspeth, 2010; Kantardzhieva et al., 2012). CaMKII has been implicated in regulating synaptic signal transmission in conventional synapses on the presynaptic (reviewed in Wang, 2008) and post synaptic side (reviewed in Lisman et al., 2012). On the presynaptic side one of the main substrate of CamKII is thought to be synapsin 1 (Chi et al., 2003; Benfenati et al., 1992; Kennedy et al., 1983). However, while CaMKII has been shown to be present in ribbon synapses of the retina (Ullrich and Sudhof, 1994;Bronstein et al., 1988), synapsin1 has been shown to be absent from ribbon synapses in the retina (Mandell et al., 1990; Ullrich and Südhof, 1994; von Kriegstein et al., 1999). These findings make syntaxin 3B an attractive candidate for a target of CaMKII phosphorylation in ribbon synapses of the retina.
What could be the biological function of the phosphorylation of syntaxin 3B by CaMKII? In contrast to conventional synapses, that release neurotransmitters at a relatively constant rate in response to an depolarizing action potential, ribbon synapses release neurotransmitters with a wide range from releasing the content of a single vesicle to a constitutive release of several 100 vesicles per second (Baden et al., 2013) as response to graded changes in membrane. As a consequence ribbon synapses require neurotransmitter release machinery that is able to adapt to a wide range of release speeds. The presence of a specialized syntaxin isoform in ribbon synapses that has a lower efficiency to catalyze synaptic vesicle fusion in the un-phosphorylated state and is able to catalyze vesicle fusion at a higher rate when activated via calcium dependent phosphorylation by CaMKII would allow fine tuning the release machinery for different release rates.
Spatial restriction of vesicle fusion to the proximity of the synaptic ribbon
Synaptic vesicle fusion in ribbon synapse of the retina is thought to occur mainly at the base of the synaptic ribbon (Heidelberger et al., 2005). CaMKII is part of a ribbon associated complex of proteins (Uthaiah and Hudspeth, 2010; Kantardzhieva et al., 2012) and presynaptic CaMKII is thought to be activated by the influx of Ca2+ through voltage dependent Ca2+ channels during synaptic activity (Gorelick et al., 1988). In ribbon synapses the majority of the channel mediated Ca2+ influx is located to the proximity of the synaptic ribbon probably due to a clustering of voltage dependent Ca2+ channels at the ribbon (Matthews and Fuchs, 2010). The activation of syntaxin 3B by CaMKII and its interaction with SNAP-25 to form a t-SNARE complex would therefore be dependent on activity and spatial proximity to the synaptic ribbon (see model in figure 10). Since the majority of synaptic vesicle fusion happens at the base of the ribbon it is plausible that the restriction of t-SNARE complexes to the base of the ribbon allows the preservation of a reserve pool of free syntaxin 3B and SNAP-25 that can be recruited to the synaptic ribbon in an activity dependent manner. Such a mechanism would be essential for the properly regulated release of neurotransmitters in the retina. Future studies evaluating the ultrastructural distribution of phosphorylated syntaxin 3B in ribbon synapses will be required to test this hypothesis.
Conclusions
We show here that syntaxin 3B can be phosphorylated by CaMKII and have identified the phosphorylation site in syntaxin 3B. Furthermore, a mutant form of syntaxin3B that mimics the phosphorylation by CaMKII binds SNAP-25 more efficiently than the wildtype. Based on these findings we propose a model how synaptic vesicle exocytosis in ribbon synapses is modulated by syntaxin 3B and CaMKII (figure 10).
Acknowledgments
This work was supported by grants R01 EY12128 (to R.H.) and R01 EY016452 (to R.J.) from the NEI/NIH. We thank the confocal core facilities at UT Houston Medical School supported by NIH grants S10 RR022531 and P30 EY010608. We would like to thank Drs. James McNew, Neal Waxham and John O'Brien for gifts of reagents and helpful discussions and Nataliia Bogdanova for excellent technical assistance.
Abbreviations
- CaMKII
Calcium/calmodulin-dependent protein kinase II
- GST
Glutathione S-transferase
- SNARE
solubleN-ethylmaleimide-sensitive factor attachment protein receptor
- GAPDH
Glyceraldehyde 3-phosphate dehydrogenase
- CTBP2
C-terminal binding protein 2
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Bibliography
- Baden T, Euler T, Weckstrom M, Lagnado L. Spikes and ribbon synapses in early vision. Trends Neurosci. 2013;36:480–488. doi: 10.1016/j.tins.2013.04.006. [DOI] [PubMed] [Google Scholar]
- Benfenati F, Valtorta F, Rubenstein JL, Gorelick FS, Greengard P, Czernik AJ. Synaptic vesicle-associated Ca2+/calmodulin-dependent protein kinase II is a binding protein for synapsin I. Nature. 1992;359:417–420. doi: 10.1038/359417a0. [DOI] [PubMed] [Google Scholar]
- Bennett MK, Garcia-Arraras JE, Elferink LA, Peterson K, Fleming AM, Hazuka CD, Scheller RH. The syntaxin family of vesicular transport receptors. Cell. 1993;74:863–873. doi: 10.1016/0092-8674(93)90466-4. [DOI] [PubMed] [Google Scholar]
- Bronstein JM, Wasterlain CG, Bok D, Lasher R, Farber DB. Localization of retinal calmodulin kinase. Exp Eye Res. 1988;47:391–402. doi: 10.1016/0014-4835(88)90050-4. [DOI] [PubMed] [Google Scholar]
- Chi P, Greengard P, Ryan TA. Synaptic vesicle mobilization is regulated by distinct synapsin I phosphorylation pathways at different frequencies. Neuron. 2003;38:69–78. doi: 10.1016/s0896-6273(03)00151-x. [DOI] [PubMed] [Google Scholar]
- Curtis L, Datta P, Liu X, Bogdanova N, Heidelberger R, Janz R. Syntaxin 3B is essential for the exocytosis of synaptic vesicles in ribbon synapses of the retina. Neuroscience. 2010;166:832–841. doi: 10.1016/j.neuroscience.2009.12.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis LB, Doneske B, Liu X, Thaller C, McNew JA, Janz R. Syntaxin 3b is a t-SNARE specific for ribbon synapses of the retina. J Comp Neurol. 2008;510:550–559. doi: 10.1002/cne.21806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulubova I, Khvotchev M, Liu S, Huryeva I, Südhof TC, Rizo J. Munc18-1 binds directly to the neuronal SNARE complex. Proc Natl Acad Sci U S A. 2007;104:2697–2702. doi: 10.1073/pnas.0611318104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulubova I, Sugita S, Hill S, Hosaka M, Fernandez I, Südhof TC, Rizo J. A conformational switch in syntaxin during exocytosis: role of munc18. EMBO J. 1999;18:4372–4382. doi: 10.1093/emboj/18.16.4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foletti DL, Lin R, Finley MA, Scheller RH. Phosphorylated syntaxin 1 is localized to discrete domains along a subset of axons. J Neurosci. 2000;20:4535–4544. doi: 10.1523/JNEUROSCI.20-12-04535.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber SH, Rah JC, Min SW, Liu X, de WH, Dulubova I, Meyer AC, Rizo J, Arancillo M, Hammer RE, Verhage M, Rosenmund C, Südhof TC. Conformational switch of syntaxin-1 controls synaptic vesicle fusion. Science. 2008;321:1507–1510. doi: 10.1126/science.1163174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorelick FS, Wang JK, Lai Y, Nairn AC, Greengard P. Autophosphorylation and activation of Ca2+/calmodulin-dependent protein kinase II in intact nerve terminals. J Biol Chem. 1988;263:17209–17212. [PubMed] [Google Scholar]
- He L, Xue L, Xu J, McNeil BD, Bai L, Melicoff E, Adachi R, Wu LG. Compound vesicle fusion increases quantal size and potentiates synaptic transmission. Nature. 2009;459:93–97. doi: 10.1038/nature07860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidelberger R, Thoreson WB, Witkovsky P. Synaptic transmission at retinal ribbon synapses. Prog Retin Eye Res. 2005;24:682–720. doi: 10.1016/j.preteyeres.2005.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttner WB, Schiebler W, Greengard P, De CP. Synapsin I (protein I), a nerve terminal-specific phosphoprotein. III. Its association with synaptic vesicles studied in a highly purified synaptic vesicle preparation. J Cell Biol. 1983;96:1374–1388. doi: 10.1083/jcb.96.5.1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn R, Fasshauer D. Molecular machines governing exocytosis of synaptic vesicles. Nature. 2012;490:201–207. doi: 10.1038/nature11320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janz R, Hofmann K, Südhof TC. SVOP, an evolutionarily conserved synaptic vesicle protein, suggests novel transport functions of synaptic vesicles. J Neurosci. 1998;18:9269–9281. doi: 10.1523/JNEUROSCI.18-22-09269.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantardzhieva A, Peppi M, Lane WS, Sewell WF. Protein composition of immunoprecipitated synaptic ribbons. J Proteome Res. 2012;11:1163–1174. doi: 10.1021/pr2008972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy MB, McGuinness T, Greengard P. A calcium/calmodulin-dependent protein kinase from mammalian brain that phosphorylates Synapsin I: partial purification and characterization. J Neurosci. 1983;3:818–831. doi: 10.1523/JNEUROSCI.03-04-00818.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisman J, Yasuda R, Raghavachari S. Mechanisms of CaMKII action in long-term potentiation. Nat Rev Neurosci. 2012;13:169–182. doi: 10.1038/nrn3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandell JW, Townes-Anderson E, Czernik AJ, Cameron R, Greengard P, De CP. Synapsins in the vertebrate retina: absence from ribbon synapses and heterogeneous distribution among conventional synapses. Neuron. 1990;5:19–33. doi: 10.1016/0896-6273(90)90030-j. [DOI] [PubMed] [Google Scholar]
- Matthews G, Fuchs P. The diverse roles of ribbon synapses in sensory neurotransmission. Nat Rev Neurosci. 2010;11:812–822. doi: 10.1038/nrn2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews G, Sterling P. Evidence that vesicles undergo compound fusion on the synaptic ribbon. J Neurosci. 2008;28:5403–5411. doi: 10.1523/JNEUROSCI.0935-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer AJ, Thoreson WB. The dynamic architecture of photoreceptor ribbon synapses: cytoskeletal, extracellular matrix, and intramembrane proteins. Vis Neurosci. 2011;28:453–471. doi: 10.1017/S0952523811000356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgans CW, Brandstatter JH, Kellerman J, Betz H, Wassle H. A SNARE complex containing syntaxin 3 is present in ribbon synapses of the retina. J Neurosci. 1996;16:6713–6721. doi: 10.1523/JNEUROSCI.16-21-06713.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risinger C, Bennett MK. Differential phosphorylation of syntaxin and synaptosome-associated protein of 25 kDa (SNAP-25) isoforms. J Neurochem. 1999;72:614–624. doi: 10.1046/j.1471-4159.1999.0720614.x. [DOI] [PubMed] [Google Scholar]
- Rizo J, Südhof TC. The membrane fusion enigma: SNAREs, Sec1/Munc18 proteins, and their accomplices--guilty as charged? Annu Rev Cell Dev Biol. 2012;28:279–308. doi: 10.1146/annurev-cellbio-101011-155818. [DOI] [PubMed] [Google Scholar]
- Schmitz F, Königstorfer A, Südhof TC. RIBEYE, a component of synaptic ribbons: a protein's journey through evolution provides insight into synaptic ribbon function. Neuron. 2000;28:857–872. doi: 10.1016/s0896-6273(00)00159-8. [DOI] [PubMed] [Google Scholar]
- Sharma N, Low SH, Misra S, Pallavi B, Weimbs T. Apical targeting of syntaxin 3 is essential for epithelial cell polarity. J Cell Biol. 2006;173:937–948. doi: 10.1083/jcb.200603132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry DM, Mitchell R, Standifer KM, du PB. Distribution of plasma membrane-associated syntaxins 1 through 4 indicates distinct trafficking functions in the synaptic layers of the mouse retina. BMC Neurosci. 2006;7:54. doi: 10.1186/1471-2202-7-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamori S, Holt M, Stenius K, Lemke EA, Grønborg M, Riedel D, Urlaub H, Schenck S, Brügger B, Ringler P, Müller SA, Rammner B, Gräter F, Hub JS, De Groot BL, Mieskes G, Moriyama Y, Klingauf J, Grubmüller H, Heuser J, Wieland F, Jahn R. Molecular anatomy of a trafficking organelle. Cell. 2006;127:831–846. doi: 10.1016/j.cell.2006.10.030. [DOI] [PubMed] [Google Scholar]
- Ullrich B, Südhof TC. Distribution of synaptic markers in the retina: implications for synaptic vesicle traffic in ribbon synapses. J Physiol Paris. 1994;88:249–257. doi: 10.1016/0928-4257(94)90088-4. [DOI] [PubMed] [Google Scholar]
- Uthaiah RC, Hudspeth AJ. Molecular anatomy of the hair cell's ribbon synapse. J Neurosci. 2010;30:12387–12399. doi: 10.1523/JNEUROSCI.1014-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Kriegstein K, Schmitz F, Link E, Südhof TC. Distribution of synaptic vesicle proteins in the mammalian retina identifies obligatory and facultative components of ribbon synapses. Eur J Neurosci. 1999;11:1335–1348. doi: 10.1046/j.1460-9568.1999.00542.x. [DOI] [PubMed] [Google Scholar]
- Wang MM, Janz R, Belizaire R, Frishman LJ, Sherry DM. Differential distribution and developmental expression of synaptic vesicle protein 2 isoforms in the mouse retina. J Comp Neurol. 2003;460:106–122. doi: 10.1002/cne.10636. [DOI] [PubMed] [Google Scholar]
- Wang ZW. Regulation of synaptic transmission by presynaptic CaMKII and BK channels. Mol Neurobiol. 2008;38:153–166. doi: 10.1007/s12035-008-8039-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou ZY, Wan QF, Thakur P, Heidelberger R. Capacitance measurements in the mouse rod bipolar cell identify a pool of releasable synaptic vesicles. J Neurophysiol. 2006;96:2539–2548. doi: 10.1152/jn.00688.2006. [DOI] [PubMed] [Google Scholar]