Abstract
In bacteria transcription and translation are linked in time and space. When coupled to RNA polymerase (RNAP), the translating ribosome ensures transcriptional processivity by preventing RNAP backtracking. Recent advances in the field have characterized important linker proteins that bridge the gap between transcription and translation: In particular, the NusE(S10):NusG complex and the NusG homolog, RfaH. The direct link between the moving ribosome and RNAP provides a basis for maintaining genomic integrity while enabling efficient transcription and timely translation of various genes within the bacterial cell.
Introduction
Unlike in eukaryotes, where transcription and translation occur in separate cellular compartments, in bacteria these two processes are spatially and temporally coupled. Both machineries can occupy the same mRNA theoretically facilitating direct physical contact between transcribing RNAPs and translating ribosomes. In fact most genes can initiate translation almost immediately after the ribosome binding site (RBS) emerges from the RNA exit channel of RNAP. However, when translation slows or is stalled, a gap forms between the transcribing RNAP and the trailing ribosome that allows the termination factor Rho to access nascent RNA and prematurely terminate transcription of downstream genes(1). This phenomenon, known as the polarity effect, together with transcription attenuation(2) are two well described events known to result from spatial gaps between RNAP and the trailing ribosome(3).
While the polarity effect and transcription attenuation have been known for decades to be indirect effects of transcription-translation coupling, recent direct links between RNAP and the ribosome have been reported. In this review we will visit some of the recent discoveries that describe direct physical links between the transcribing RNAP and the translating ribosome and the effects of this coupling on transcription processivity and genomic stability in bacteria.
Rate dependencies and genomic stability
During transcription elongation RNAP utilizes a complex Brownian ratchet mechanism to oscillate between productive and inactive (backtracked) states at numerous sites along DNA and RNA(4, 5). Proper balance between these two states determines the rate and fidelity of transcription. Backtracking involves synchronized winding and unwinding of the DNA:RNA hybrid upstream and downstream of the transcription bubble. Backtracking has been implicated in many important processes in bacteria including transcription elongation rates, pausing and genome instability(6) Importantly, if the rate of transcription slows due to backtracking or other pausing events, Rho can terminate transcription owing to the fact that Rho is bound to RNAP throughout the transcription cycle and therefore readily available to do so(7, 8).
The rate of transcription depends on the rate of translation
Unlike RNAP, the ribosome moves through a progressive, unilateral translocation (9). Direct coupling of transcription and translation has been reported both in vivo(10) and in vitro(11). In vivo work has shown that the trailing ribosome “pushes” RNAP, thereby modulating the rate of transcription by preventing backtracking and creating a synchrony of mRNA production and protein synthesis (10, 12) (Figure 1). By inducing expression of a lacZ reporter gene, Proshkin et al. calculated rates for transcription and translation using dot-blot hybridization to detect lacZ transcripts and measuring the subsequent time to β-galactosidase synthesis(10). Despite various growth conditions known to alter the speed of the ribosome, they show that the rate of transcription perfectly matches that of, translation and that both rates depend on codon usage for a particular gene (10). This coupling serves as an energy control mechanism that both limits the number of extra transcripts and prevents premature termination by Rho (Figure 1).
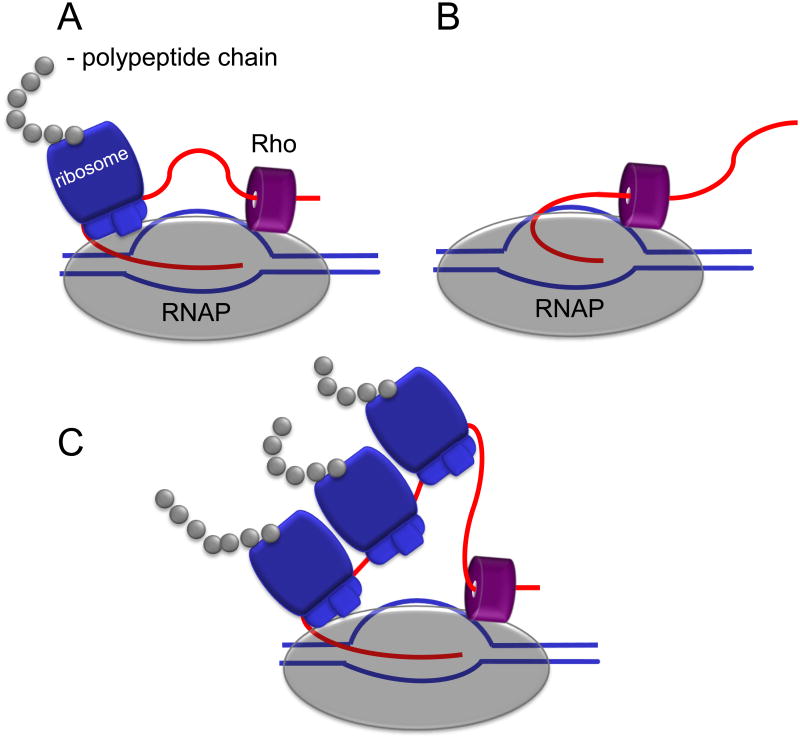
Figure 1. Ribosome coupling and Rho mediated polarity.
Schematics show the ribosome coupled with RNAP to prevent transcription termination by Rho. Translation that begins prior to Rho loading onto RNA is not subject to transcription polarity (A). Rho binds RNAP throughout the transcription cycle and is readily available to terminate transcription in the absence of a ribosome (B)(7). Strong RBS can engage multiple ribosomes to the same nascent transcript, but only the leading one controls the rate of transcription elongation by preventing RNAP backtracking (C)(10).
It also helps RNAP to traverse roadblocks during transcription elongation. RNAP encounters and crosses numerous roadblocks that have been shown to readily halt transcription in vitro. Cooperation between RNAP and ribosomes prevents RNAP backtracking and facilitates displacement of such roadblocks(10). Without this link, RNAP would behave as expected from in vitro studies and backtrack upon encountering nucleoprotein complexes(23). The first 50 nucleotides of transcribed sequences in promoter proximal regions are particularly prone to stalling and backtracking. In fact, in vivo mapping of RNAP on chromosomal DNA showed that 7 out of 34 active promoters harbored stalled RNAP(24); in part because there is little opportunity for ribosomes to bind and push RNAP forward(6).
Recently, Castro-Roa et al. developed a coupled in vitro transcription-translation assay from purified components of both systems to study the interactions between RNAP and the ribosome to exclude contributions from other cellular components. While most pauses studied using this setup behaved as reported in vivo, one of four hairpin-independent RNAP pauses increased in duration in response to a translating ribosome(11). In vitro systems such as these are extremely useful to characterize the nature of the direct physical contact between RNAP and the ribosome; however a purified system excludes effects from the network of interactions bridging the gap between transcription and translation(13) as well as the anti-backtracking and termination factors known to compensate for compromised translation(14). Overall, this in vitro system provides an excellent backdrop for studying the contributions of individual factors on transcription-translation coupling without interference from other cellular components.
The rate of translation depends on secondary mRNA structure
While the rate of transcription depends on the rate of translation, the rate of translation depends on the mRNA fold complexity. Most mRNAs exhibit stable secondary structures within their coding sequence, and the cell uses these secondary structures in a variety of ways to regulate translation. To overcome the kinetic challenge of strand separation, the ribosome behaves as an mRNA helicase, disrupting mRNA duplexes through proteins S3, S4 and S5 of the 30S subunit(15). Unlike traditional helicases, the ribosome unwinds these structures through a two-part mechanism of destabilization at the helical junction and forced strand separation during translocation(16). The speed of translation is therefore directly dependent on the extent and stability of secondary structures formed by the mRNA(17). Therefore, excluding codon usage effects, the rate of transcription can also be influenced by the ribosome in a template RNA dependent way. In fact, translational rates for identical codons differ based on the GC content of mRNA structures at the entry site(16). This suggests that a spatially and temporally close coupling of RNAP and the ribosome that prevents backtracking is necessary for keeping the two processes synchronized despite sequence variability within any individual ORF.
Genomic stability
Actively dividing cells sustain frequent collisions between the replication and transcription machineries since RNAP occupies the same DNA track as the replisome, which moves 20 times more rapidly than RNAP(18). As a conserved feature of genome organization, the majority of essential and ribosomal RNA genes in bacteria are oriented in the same direction as replication(19, 20). This co-orientation results in predominantly co-directional collisions between RNAP and the replisome. Some collisions can disrupt and restart replication(21), while those involving extensive backtracking of RNAPs frequently introduce single- and double-strand breaks (DSBs) into the genome(22) (Figure 2).
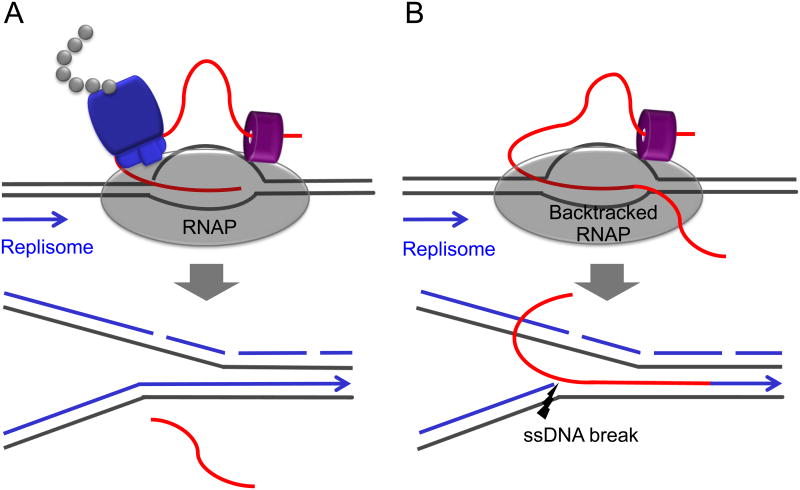
Figure 2. Ribosome coupling and genomic integrity.
Schematics demonstrate the outcomes for co-directional collisions between the replisome and RNAP. Such collisions with non-backtracked RNAP are mostly benign with respect to genomic integrity (A). However, encounters between the replisome and backtracked RNAP can introduce breaks in the DNA leading strand that could eventually lead to DSBs during the next round of replication (B). In this way the ribosome helps to maintain genomic stability by preventing RNAP backtracking (22). Additional factors that deal with backtracked elongation complexes and contribute to genomic stability are termination factors (Rho and Mfd) and transcript cleavage factors (GreA and GreB) (6, 14, 22).
RNAP backtracking links protein synthesis to genome instability and may contribute to stress-driven evolution. Backtracking prone cells exhibit higher mutation rates than wild type cells(22,25) and are critically dependent on DSB repair machineries(22), which are error-prone. In this way the translating ribosome preserves genome integrity through an anti-backtracking mechanism that prevents DSBs(6,22). Since the ribosome is a major sensor of cellular metabolism and stress, its coupling with RNAP provides a mechanistical link between growth conditions, mutagenesis, and cellular adaptation to environmental changes.
Proteins that couple transcription and translation
In bacteria, the NusG family of proteins binds to and alters the translocation properties of RNAP. The most well characterized members in this family of transcriptional regulators are the E. coli proteins, NusG and RfaH. These proteins interface with RNAP through an N-Terminal Domain (NTD) region that surrounds the nucleic acid chains and accelerates transcription(26) (Figure 3). Twenty years ago a possible interaction between NusG and the ribosome was suggested based on genetic data(27); however, it was not until recently that these proteins and their roles in physically coupling transcription to translation was confirmed. Recent work on NusG and RfaH has elucidated novel mechanisms for and expanded the current knowledge on transcription/translation coupling in bacteria.
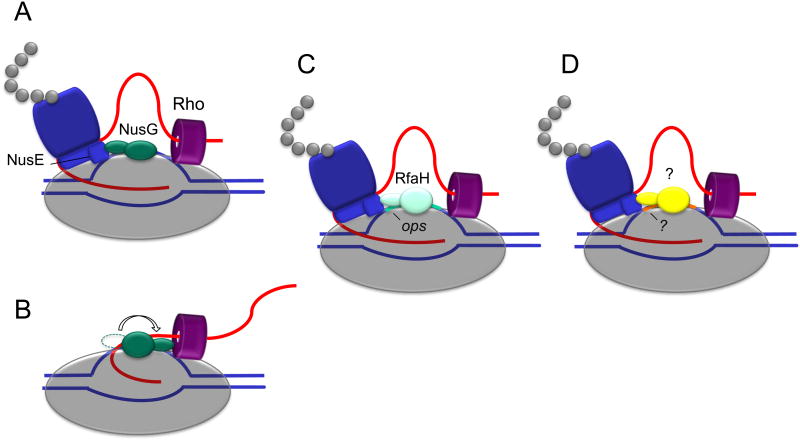
Figure 3. Bridging the ribosome to RNA polymerase.
Schematics demonstrate the known mechanisms for ribosome recruitment to RNAP through the ribosomal subunit NusE (S10). NusG binds to RNAP and complexes with NusE to link the ribosome to RNAP (A)(12). Alternatively, the CTD of NusG can bind Rho-factor, facilitating Rho-mediated termination and precluding binding to NusE (B). The NTD of RfaH binds to RNAP and an ops regulatory site while the CTD of RfaH links RNAP to the ribosome through interactions with NusE (C)(37). Based on the recently identified RfaH fold switching mechanism, it is possible that additional structurally similar factors can recognize specific regulatory elements to draw the ribosome to RNAP and ensure expression of those genes (D).
NusG
NusG is a 20.5kDa protein regulator of RNAP with two separate functional domains. The NusG-NTD is an anti-parallel beta sheet surrounded by three alpha helices that directly interact with RNAP through hydrophobic surface portions(28). In contrast the NusG- C-Terminal Domain (CTD) has a beta barrel fold and binds either Rho-factor for Rho-dependent termination or NusE to couple transcription to translation(12). Crystallographic and NMR studies show that NusG has an overall open conformation, with no contacts formed between the NTD and the CTD(29), hinting at its role as a linker protein through these two functional domains(30).
NusE (S10) is a 11.7kDa protein with dual roles in transcription and translation. NusE binds the transcriptional regulator, NusB, creating a stably folded, heterodimeric complex that can bind to the RNAP elongation complex. NusE is also a component of the 30S ribosomal subunit, the incorporation into which may or may not be dependent on first binding NusB. During transcription, the NusB:NusE complex binds single stranded RNA to prevent premature termination and ensure processive transcription(31). Through this interaction the NusG-CTD is able to directly bind the surface exposed NusE, thereby directly linking the elongating RNAP to the ribosome and preventing Rho-mediated polarity(12, 32). In this way, the NusE:NusG complex physically links the leading ribosome to a waiting RNAP, thereby adjusting transcriptional rate to match the translational needs of the cell.
RfaH
RfaH is a transcriptional activator protein that is homologous to NusG(33). RfaH, is recruited to the elongating RNAP through interactions with an exposed operon polarity suppressor (ops) site on the non-template DNA of horizontally transferred genes. The operon polarity suppressor (ops element) is an 8 bp motif that increases distal gene expression and decreases polarity(34). For years RfaH has been known to be an activator of transcription across a range of genes that assemble and export surface and extracellular components(35). Therefore, RfaH is functionally analogous to NusG for a subset of specific operons that mostly encode extracellular virulence factors.
RfaH is known to share a similar NTD fold with NusG; however, unlike NusG, the full length RfaH exhibits an alpha-helical hairpin in its CTD that folds tightly against the NTD(36), creating a closed conformation. However, recently Artsimovitch, Rosch and colleagues showed that removing the NTD of RfaH or destabilizing interdomain contacts triggers a complete conformational change in the CTD from an all-alpha fold to the NusG-like all-beta fold. In this conformation, the CTD of RfaH efficiently recruits the ribosome through interactions with NusE(37).
Models suggest that the DNA binding motif in the RfaH-NTD includes three amino acids that become obstructed by the CTD when RfaH is in the closed conformation: This creates competition between DNA (open conformation) and CTD (closed conformation) binding to the NTD of RfaH(13). While NusG:NusE interactions are mediated through highly conserved, hydrophobic residues in the CTD(12, 37), the RfaH-NTD residues that contact the ops element are not conserved. This explains the specificity of RfaH for operons containing an ops site(28). The open form of RfaH requires these contacts for efficient expression of ops containing operons that are normally poorly transcribed(37). Transcription across operons containing ops sites is significantly increased by RfaH, which decreases Rho-mediated polarity.
Fold-dependent functional disparities in the RfaH-CTD have important implications for thermodynamically driven folding and binding events that mediate transcription and translation. Changes in fold can present new binding surfaces, stabilize the new structure and extend or change the biological function of the protein(38). Through this elegant transition, RfaH is able to efficiently recruit the ribosome in a template specific way without interfering with housekeeping gene regulation by NusG. RfaH has the unprecedented ability to specifically recognize and bind to a single stranded DNA binding site through a marked structural reorganization that alters the overall conformation from a closed form to an open form. This form shift allows a refolding event to occur that facilitates ribosomal binding that significantly activates translation.
The transcription translation interface
During transcription many factors bind to RNAP to ensure a stable and efficient elongation complex. Owing to crystallization challenges, a high-resolution structure for RNAP from E. coli currently is unavailable. As a result, models are based on structures from the thermophilic bacteria, T. thermophiles, and lack clear structural information in some regions(39). As a result, many domains within RNAP that may be involved in protein interactions have yet to be confidently identified, making it difficult to completely understand functional interactions between the ribosome and RNAP. Current models for transcription translation coupling are subject to change as new structural information emerges(32).
Some suggest a direct interaction whereby the ribosome induces structural changes in the active center of RNAP that serve to inhibit pausing and accelerate transcription(40). Indeed, many ribosomal proteins are known to directly interact with RNAP(41, 42) or localize to transcriptional start sites (43) (Table 1).
Table 1. Proteins that bridge transcription and translation in bacteria.
| Coupling proteins | Name | Function in Transcription | References |
|---|---|---|---|
| Transcription factors | NusG | decreases pausing; links RNAP to ribosome (NusE) | (12, 32) |
|
| |||
| RfaH | decreases ops pause duration; links RNAP to ribosome (NusE) | (37) | |
|
| |||
| Rho | Transcription termination; gene polarity | (1, 7) | |
|
| |||
| Ribosomal proteins | S1 | antitermination; binds RNAP | (41, 44) |
|
| |||
| S4 | antitermination; binds RNAP | (45) | |
|
| |||
| L2 | increases transcription of rRNA genes; binds RNAPα | (46) | |
|
| |||
The largest ribosomal protein, S1, is necessary for translation initiation as well as elongation and purifies in near equimolar amounts with RNAP. Interestingly, purified S1 binds to RNAP and stimulates transcriptional activity from a number of promoters on different DNA templates(44). Similarly, the ribosomal protein S4 directly binds to RNAP and exhibits anti-termination activity similar to that of NusA in vitro(45). The 50S ribosomal protein, L2, also binds the alpha subunits of RNAP and increases transcriptional activity in a promoter-specific way(46).
It has been suggested that the extra-translational roles of ribosomal proteins predate the ribosome itself(41). This easily fits a mechanism by which ribosomes: 1) are attracted to transcription sites through evolutionarily conserved interactions between their protein subunits and RNAP and 2) influence the rate of transcription by preventing backtracking of RNAP and consequently maintaining the integrity and activity of those genes by preventing DSBs and Rho-mediated polarity, respectively.
Conclusion
The link between transcription and translation is an evolutionarily conserved, central feature of bacterial gene expression. The NusG family as well several ribosomal proteins are prime examples of regulatory factors known to directly affect the rate of transcription and physically link RNAP to the ribosome. This coupling prevents RNAP backtracking, maintains genomic integrity, and conserves energy and resource during both transcription and translation. In this way gene expression can be prioritized via ribosomes to be coupled to RNAP preferentially in the most active housekeeping and stress-inducible genes.
Highlights.
In bacteria, the rate of transcription elongation is controlled by the rate of translation
Ribosome coupling prevents RNAP backtracking
Ribosome coupling prevents DNA damage caused by codirectional collisions between backtracked RNAP and the replisome
The NusE:NusG complex links RNAP to the 30S subunit of the ribosome
RfaH adopts a NusG-like fold when it binds to RNAP and can then bind the ribosome through interactions with NusE
Acknowledgments
This work was supported by the NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Richardson JP. Preventing the synthesis of unused transcripts by Rho factor. Cell. 1991;64(6):1047–9. doi: 10.1016/0092-8674(91)90257-y. [DOI] [PubMed] [Google Scholar]
- 2.Yanofsky C. Attenuation in the control of expression of bacterial operons. Nature. 1981;289(5800):751–8. doi: 10.1038/289751a0. [DOI] [PubMed] [Google Scholar]
- 3.Adhya S, Gottesman M. Control of transcription termination. Annu Rev Biochem. 1978;47:967–96. doi: 10.1146/annurev.bi.47.070178.004535. [DOI] [PubMed] [Google Scholar]
- 4.Bar-Nahum G, Epshtein V, Ruckenstein AE, Rafikov R, Mustaev A, Nudler E. A ratchet mechanism of transcription elongation and its control. Cell. 2005;120(2):183–93. doi: 10.1016/j.cell.2004.11.045. [DOI] [PubMed] [Google Scholar]
- 5.Svetlov V, Nudler E. Macromolecular micromovements: how RNA polymerase translocates. Curr Opin Struct Biol. 2009;19(6):701–7. doi: 10.1016/j.sbi.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nudler E. RNA polymerase backtracking in gene regulation and genome instability. Cell. 2012;149(7):1438–45. doi: 10.1016/j.cell.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Epshtein V, Dutta D, Wade J, Nudler E. An allosteric mechanism of Rho-dependent transcription termination. Nature. 2010;463(7278):245–9. doi: 10.1038/nature08669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peters JM, Mooney RA, Kuan PF, Rowland JL, Keles S, Landick R. Rho directs widespread termination of intragenic and stable RNA transcription. Proc Natl Acad Sci U S A. 2009;106(36):15406–11. doi: 10.1073/pnas.0903846106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Steitz TA. A structural understanding of the dynamic ribosome machine. Nat Rev Mol Cell Biol. 2008;9(3):242–53. doi: 10.1038/nrm2352. [DOI] [PubMed] [Google Scholar]
- 10.Proshkin S, Rahmouni AR, Mironov A, Nudler E. Cooperation between translating ribosomes and RNA polymerase in transcription elongation. Science. 2010;328(5977):504–8. doi: 10.1126/science.1184939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *11.Castro-Roa D, Zenkin N. In vitro experimental system for analysis of transcription-translation coupling. Nucleic Acids Res. 2012;40(6):e45. doi: 10.1093/nar/gkr1262. The in vitro assay described here may be used to characterize new transcription-translation coupling factors and their specific role in this process. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burmann BM, Schweimer K, Luo X, Wahl MC, Stitt BL, Gottesman ME, et al. A NusE:NusG complex links transcription and translation. Science. 2010;328(5977):501–4. doi: 10.1126/science.1184953. [DOI] [PubMed] [Google Scholar]
- 13.Svetlov V, Nudler E. Unfolding the bridge between transcription and translation. Cell. 2012;150(2):243–5. doi: 10.1016/j.cell.2012.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Washburn RS, Gottesman ME. Transcription termination maintains chromosome integrity. Proc Natl Acad Sci U S A. 2011;108(2):792–7. doi: 10.1073/pnas.1009564108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takyar S, Hickerson RP, Noller HF. mRNA helicase activity of the ribosome. Cell. 2005;120(1):49–58. doi: 10.1016/j.cell.2004.11.042. [DOI] [PubMed] [Google Scholar]
- 16.Qu X, Wen JD, Lancaster L, Noller HF, Bustamante C, Tinoco I., Jr The ribosome uses two active mechanisms to unwind messenger RNA during translation. Nature. 2011;475(7354):118–21. doi: 10.1038/nature10126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giedroc DP, Cornish PV. Frameshifting RNA pseudoknots: structure and mechanism. Virus Res. 2009;139(2):193–208. doi: 10.1016/j.virusres.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fradkin LG, Kornberg A. Prereplicative complexes of components of DNA polymerase III holoenzyme of Escherichia coli. J Biol Chem. 1992;267(15):10318–22. [PubMed] [Google Scholar]
- 19.Guy L, Roten CA. Genometric analyses of the organization of circular chromosomes: a universal pressure determines the direction of ribosomal RNA genes transcription relative to chromosome replication. Gene. 2004;340(1):45–52. doi: 10.1016/j.gene.2004.06.056. [DOI] [PubMed] [Google Scholar]
- 20.Srivatsan A, Tehranchi A, MacAlpine DM, Wang JD. Co-orientation of replication and transcription preserves genome integrity. PLoS Genet. 2010;6(1):e1000810. doi: 10.1371/journal.pgen.1000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Merrikh H, Machon C, Grainger WH, Grossman AD, Soultanas P. Co-directional replication-transcription conflicts lead to replication restart. Nature. 2011;470(7335):554–7. doi: 10.1038/nature09758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **22.Dutta D, Shatalin K, Epshtein V, Gottesman ME, Nudler E. Linking RNA Polymerase Backtracking to Genome Instability in E.coli. Cell. 2011;146(4):533–543. doi: 10.1016/j.cell.2011.07.034. Here it is reported that co-directional collisions between backtracked RNAP and the replisome induce double-stranded DNA breaks. Genomic integrity is maintained through anti-backtracking mechanisms including coupling of the ribosome to RNAP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Epshtein V, Toulme F, Rahmouni AR, Borukhov S, Nudler E. Transcription through the roadblocks: the role of RNA polymerase cooperation. EMBO J. 2003;22(18):4719–27. doi: 10.1093/emboj/cdg452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hatoum A, Roberts J. Prevalence of RNA polymerase stalling at Escherichia coli promoters after open complex formation. Mol Microbiol. 2008;68(1):17–28. doi: 10.1111/j.1365-2958.2008.06138.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poteete AR. Recombination phenotypes of Escherichia coli greA mutants. BMC Mol Biol. 2011;12:12. doi: 10.1186/1471-2199-12-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sevostyanova A, Belogurov GA, Mooney RA, Landick R, Artsimovitch I. The beta subunit gate loop is required for RNA polymerase modification by RfaH and NusG. Mol Cell. 2011;43(2):253–62. doi: 10.1016/j.molcel.2011.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sullivan SL, Ward DF, Gottesman ME. Effect of Escherichia coli nusG function on lambda N-mediated transcription antitermination. J Bacteriol. 1992;174(4):1339–44. doi: 10.1128/jb.174.4.1339-1344.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Belogurov GA, Mooney RA, Svetlov V, Landick R, Artsimovitch I. Functional specialization of transcription elongation factors. EMBO J. 2009;28(2):112–22. doi: 10.1038/emboj.2008.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *29.Burmann BM, Scheckenhofer U, Schweimer K, Rosch P. Domain interactions of the transcription-translation coupling factor Escherichia coli NusG are intermolecular and transient. Biochem J. 2011;435(3):783–9. doi: 10.1042/BJ20101679. Through NMR paramagnetic relaxation enhancement measurements the authors characterize the two domains of NusG. They conclude that interdomain interaction is unlikely and that NusG, does not require secondary signals or conformational changes to interact with RNAP, unlike its paralogue, RfaH. [DOI] [PubMed] [Google Scholar]
- 30.Mooney RA, Schweimer K, Rosch P, Gottesman M, Landick R. Two structurally independent domains of E. coli NusG create regulatory plasticity via distinct interactions with RNA polymerase and regulators. J Mol Biol. 2009;391(2):341–58. doi: 10.1016/j.jmb.2009.05.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Burmann BM, Luo X, Rosch P, Wahl MC, Gottesman ME. Fine tuning of the E. coli NusB:NusE complex affinity to BoxA RNA is required for processive antitermination. Nucleic Acids Res. 2010;38(1):314–26. doi: 10.1093/nar/gkp736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burmann BM, Rosch P. The role of E. coli Nus-factors in transcription regulation and transcription:translation coupling: From structure to mechanism. Transcription. 2011;2(3):130–134. doi: 10.4161/trns.2.3.15671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bailey MJ, Hughes C, Koronakis V. Increased distal gene transcription by the elongation factor RfaH, a specialized homologue of NusG. Mol Microbiol. 1996;22(4):729–37. doi: 10.1046/j.1365-2958.1996.d01-1726.x. [DOI] [PubMed] [Google Scholar]
- 34.Nieto JM, Bailey MJ, Hughes C, Koronakis V. Suppression of transcription polarity in the Escherichia coli haemolysin operon by a short upstream element shared by polysaccharide and DNA transfer determinants. Mol Microbiol. 1996;19(4):705–13. doi: 10.1046/j.1365-2958.1996.446951.x. [DOI] [PubMed] [Google Scholar]
- 35.Bailey MJ, Hughes C, Koronakis V. RfaH and the ops element, components of a novel system controlling bacterial transcription elongation. Mol Microbiol. 1997;26(5):845–51. doi: 10.1046/j.1365-2958.1997.6432014.x. [DOI] [PubMed] [Google Scholar]
- 36.Belogurov GA, Vassylyeva MN, Svetlov V, Klyuyev S, Grishin NV, Vassylyev DG, et al. Structural basis for converting a general transcription factor into an operon-specific virulence regulator. Mol Cell. 2007;26(1):117–29. doi: 10.1016/j.molcel.2007.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **37.Burmann BM, Knauer SH, Sevostyanova A, Schweimer K, Mooney RA, Landick R, et al. An alpha helix to beta barrel domain switch transforms the transcription factor RfaH into a translation factor. Cell. 2012;150(2):291–303. doi: 10.1016/j.cell.2012.05.042. Using NMR the authors report an unprecidented fold-switch that occurs in the CTD of RfaH. This conformational change allows RfaH to bind both RNAP and the ribosome via NusE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bryan PN, Orban J. Proteins that switch folds. Curr Opin Struct Biol. 2010;20(4):482–8. doi: 10.1016/j.sbi.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Opalka N, Brown J, Lane WJ, Twist KA, Landick R, Asturias FJ, et al. Complete structural model of Escherichia coli RNA polymerase from a hybrid approach. PLoS Biol. 2010;8(9) doi: 10.1371/journal.pbio.1000483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roberts JW. Molecular biology. Syntheses that stay together. Science. 2010;328(5977):436–7. doi: 10.1126/science.1189971. [DOI] [PubMed] [Google Scholar]
- 41.Bhavsar RB, Makley LN, Tsonis PA. The other lives of ribosomal proteins. Hum Genomics. 2010;4(5):327–44. doi: 10.1186/1479-7364-4-5-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Warner JR, McIntosh KB. How common are extraribosomal functions of ribosomal proteins? Mol Cell. 2009;34(1):3–11. doi: 10.1016/j.molcel.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.De S, Brogna S. Are ribosomal proteins present at transcription sites on or off ribosomal subunits? Biochem Soc Trans. 2010;38(6):1543–7. doi: 10.1042/BST0381543. [DOI] [PubMed] [Google Scholar]
- 44.Sukhodolets MV, Garges S. Interaction of Escherichia coli RNA polymerase with the ribosomal protein S1 and the Sm-like ATPase Hfq. Biochemistry. 2003;42(26):8022–34. doi: 10.1021/bi020638i. [DOI] [PubMed] [Google Scholar]
- 45.Torres M, Condon C, Balada JM, Squires C, Squires CL. Ribosomal protein S4 is a transcription factor with properties remarkably similar to NusA, a protein involved in both non-ribosomal and ribosomal RNA antitermination. EMBO J. 2001;20(14):3811–20. doi: 10.1093/emboj/20.14.3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rippa V, Cirulli C, Di Palo B, Doti N, Amoresano A, Duilio A. The ribosomal protein L2 interacts with the RNA polymerase alpha subunit and acts as a transcription modulator in Escherichia coli. J Bacteriol. 2010;192(7):1882–9. doi: 10.1128/JB.01503-09. [DOI] [PMC free article] [PubMed] [Google Scholar]