Abstract
CorA is the major transport system responsible for Mg2+ uptake in bacteria and can functionally substitute for its homologue Mrs2p in the yeast inner mitochondrial membrane. Although several CorA crystal structures are available, the molecular mechanism of Mg2+ uptake remains to be established. Here we use EPR spectroscopy, electrophysiology and molecular dynamic simulations to show that CorA is regulated by cytoplasmic Mg2+ acting as a ligand and elucidate the basic conformational rearrangements responsible for Mg2+-dependent gating. Mg2+ unbinding at the divalent cation sensor triggers a conformational change that leads to the inward motion of the stalk helix, which propagates to the pore forming transmembrane helix TM1. Helical tilting and rotation in TM1 generates an iris-like motion that increases the diameter of the permeation pathway, triggering ion conduction. This work establishes the molecular basis of a Mg2+-driven negative feedback loop in CorA as the key physiological event controlling Mg2+ uptake and homeostasis in prokaryotes.
Introduction
Magnesium (Mg2+) is the most abundant divalent cation in biology1 and is essential to all living cells as it participates in a myriad of key physiological and biochemical processes, from enzymatic activity to genomic stability. In bacteria, Mg2+ homeostasis is carried out by three molecularly distinct translocation systems, MgtA/B, MgtE and CorA2. CorA belongs to the GMN family and has been proposed to be one of the major Mg2+ uptake pathway3. Since the discovery of the CorA gene4, the pioneering work of Maguire and colleagues using in vivo radiotracer measurement have provided the functional and genetic basis for its role in bacteria5, 6, 7. The structures of CorA from Thermotoga maritima offered the first structural template to understand Mg2+ permeation and transport8, 9, 10. The GMN family is characterized by relatively low sequence conservation and several reports have suggested conflicting transport mechanisms for TmCorA11, 12. However, electrophysiological data shows that CorA-driven Mg2+ translocation involves a channel like mechanism where the inwardly-biased electrochemical gradient serves as the main driving force for Mg2+ permeation13.
So far, the available TmCorA structures have shown the presence of a density commensurate with a divalent cation at the GMN signature sequence and another ion density located in the cytoplasmic end of the pore14, 15. This leaves a long (≈ 30 Å) narrow pathway without apparent coordination sites for permeating ions, providing no obvious insights in regards to the molecular basis of Mg2+− selectivity and permeation for TmCorA and other members of the GMN family. Molecular dynamic simulations and electrostatic calculations have suggested that the present set of divalent-bound crystal structures, represents a non-conductive state16, 17. A recent structure of a distant TmCorA homolog from M. jannaschii has revealed the first structural glimpses into the determinants of Mg2+ selectivity. An electron density asymmetrically positioned in the outer mouth of the pore has been interpreted as Mg2+ with its first water shell18. Interestingly, this new structure points to a putative role for residue Asn314 at the GMN signature sequence in selectivity. A recent TmCorA structure shows a similar density coordinated by the same residue triad, suggesting that Mg2+ coordination by the GMN signature sequence is a common motif within the family14. Using a combination of electrophysiology and mutagenesis, we have shown that the GMN motif likely participates in the formation of a high affinity Mg2+ binding site that defines CorA selectivity via a knock-in knock-out repulsion mechanism 13
Indeed, as derived from the present set of structures, such a long and seemingly inhospitable pore (devoid of ions) has been considered as one of the key arguments suggesting that widespread conformational rearrangements must take place to allow Mg2+ permeation16, 17. However, attempts to stabilize the conductive conformation of CorA by eliminating divalent ions in the crystallizations matrix have been unsuccessful, and the quest for a conductive conformation remains ongoing.
Here, we have used functional and spectroscopic approaches to address two fundamental questions regarding CorA-driven Mg2+ translocation: (i) the nature and location of the gate that opens the permeation pathway, (ii) the molecular movements that underlie Mg2+-driven gating. Using a large set of cysteine mutants targeted to the two transmembrane segments (TM) and the entire stalk helix we show that binding of Mg2+ to the intracellular divalent cation sensor triggers channel closing. Comparison of individual EPR parameters from the complete set of spin-labeled mutant provided information on probe dynamics, solvent accessibility and inter-subunit distances in the conformationally-trapped states. These data, combined with existing crystal structures and computational approaches were used to depict the structural rearrangements associated with Mg2+-dependent gating and to generate three-dimensional models of CorA in its putative conductive conformation.
Results
CorA is gated by intracellular Mg2+
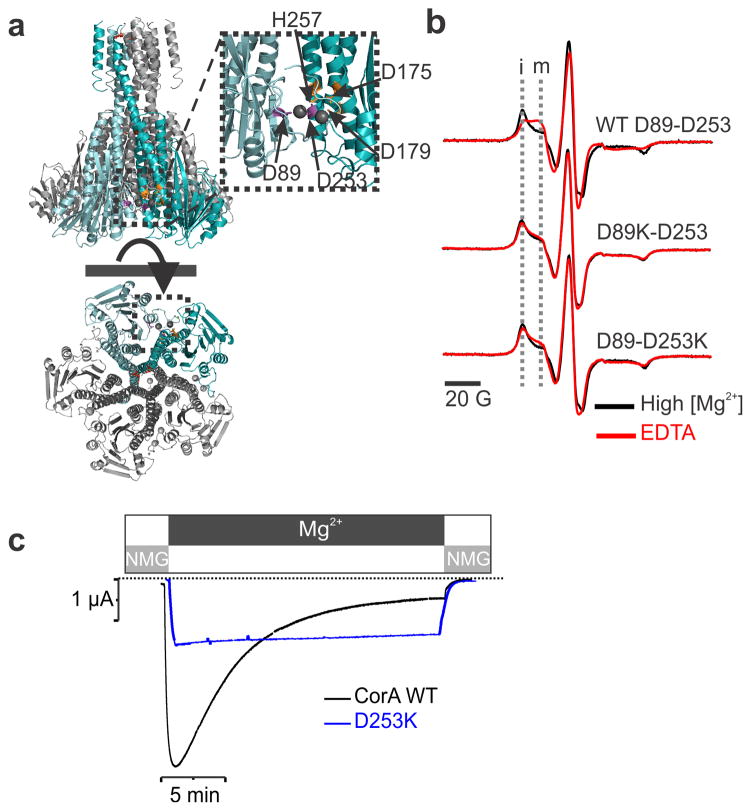
In vivo transport measurements of CorA show that shortly after a rapid Mg2+ uptake, its intracellular concentration remains steady, suggesting that the activity of CorA might be self-regulated19. Further, in vitro studies have revealed a Mg2+-dependent protease susceptibility, a clear indication that Mg2+ translocation through CorA must involve substantial structural rearrangements10, 14, 20. We have generated over-expression constructs that produce large TmCorA-catalyzed ionic currents in Xenopus oocytes13. This Mg2+ inward current peaks within a few seconds (a reflection of the speed of the solution exchange) and spontaneously decays over the course of 15–20 min to a small (less than 5 % of peak) steady state current (Fig. 1a). The decay time constants are correlated with the current intensity and the nature of the permeant ion, leading us to suggest that internal Mg2+ likely serves a dual role as both permeant ion and gating ligand for CorA (Fig. 2). At high intracellular Mg2+ concentration (>5 mM), CorA-catalyzed currents are small or non-existent, but as the intracellular concentration drops below 1–2 mM channel opening is triggered, supporting robust Mg2+ inward currents. This was directly confirmed by using a cut-open oocyte setup where the local Mg2+ concentration is tightly buffered and constantly perfused through a cannula positioned close to the membrane (Fig. 1b). Under those conditions, the inward current is abolished as a consequence of increasing internal Mg2+ concentration with an apparent Mg2+ affinity of 2.4 mM, which also corresponds to the physiological concentration in bacteria (2–3 mM)21. These conformational transitions are best fitted with the Hill number of 2 (nH = 2) and suggest a positive cooperativity for the Mg2+-driven gating transition (Fig. 1f).
Figure 1. CorA is gated by intracellular Mg2+.
a. TmCorA specific Mg2+ currents are recorded on a perfused Two Electrode Voltage Clamp setup. The representative traces are recorded at a constant −60mV for a Xenopus oocyte injected with mRNA coding for TmCorA, and an oocyte injected with water (control). The currents are recorded with constant external perfusion changing from a divalent free solution (N-Methyl Glucamine) to a 20 mM Mg2+ buffer. The inset represents a confocal microscopy image of an oocyte injected with the YFP-TmCorA fusion construct. b. The internal perfusion cut open oocyte Voltage clamp setup used for this experiment is schematized in the upper panel. The representative trace was shown in the lower panel for each internal buffer tested c. Schematic representation of the cysteine scan used for this work. The covered sequence is shown, black red, green and blue. d. Statistical representation of the probe mobility parameter for the different secondary structure elements. Box are the 25–75% with the median as horizontal line, the little square show the mean values, and the asterisk show the 1–99% using the same color code as for panel c. e. CW-EPR spectra recorded for the Q260C-spin labeled mutant in presence of various Mg2+ concentration. f. Titration plot of the normalized Mg2+ dependent currents superimposed with Mg2+ conformational transition recorded by CW-EPR. Both plots were fitted by a Hill equation. The inset is a magnification of the low concentration range.
Figure 2. Divalent cations are both charge carriers and gating factors.
a, Representative traces of Mg2+ currents are recorded in CorA expressing Xenopus laevis oocyte clamped at −60mV. Right trace; Mg2+ (20 mM) current traces are in black and the Co2+ (20 mM) current is in Red. b, Correlation between peak currents inhibition time constant (single exponential decay fit). The results are plotted in Black and red for Mg2+ and Co2+ as charge carriers respectively. c. Low field region of X-band CW EPR spectra of CorA spin labeled at position 260C titrated by a Co2+ and Mg2+ are shown. The resulting titration plots are displayed using the same color code for the ions. A magnification of the lower concentration range is displayed in the inset. The plots were fitted with a Hill equation and the apparent affinity is 2.4 mM for Mg2+ and 0.29 mM for Co2+. d. I/V curve of excised patch inside in the inside out configuration when the cytoplasmic domain is bathed in presence of divalent chelating compound or a 10 mM concentration of Mg2+.
We have investigated the conformation of liposome-reconstituted CorA in two different experimental conditions: without Mg2+ (apo form) and at saturating Mg2+ concentrations (20 mM). Continuous Wave EPR spectroscopy (CW-EPR) was used to determine the spectral properties of for more than 100 spin-labeled cysteine mutants (Fig. 1c). Fig. 1d reports the global change in probe mobility ( )22 in four different regions of CorA, clearly showing widespread structural rearrangement associated with the presence of Mg2+. From this large data set, position 260 displays one of the largest spectral changes between the apo and Mg2+-bound forms of CorA, making it an ideal reporter to establish the Mg2+ dependence of the conformational changes in CorA (Fig. 1d–e). Assessed from the spin label mobility, the overall Mg2+ dependence of the structural changes is similar to that obtained for the inhibitory gating measured by perfused cut-open oocyte voltage clamp (Fig. 1f). Equivalent conformational transitions were observed in the absence of a lipid bilayer, implying that the Mg2+-dependent structural changes are self-contained, with no direct participation of the lipid bilayer (Supplementary Fig. 1). We conclude that the Mg2+-driven deactivation of Mg2+ currents (Fig. 1a,b) and the Mg2+-induced conformational transitions observed by EPR (Fig. 1e) are a reflection of the very same molecular events that underlie Mg2+ dependent gating in CorA.
This conclusion is further supported by several pieces of information that relate these time-dependent current changes to a fundamental property of CorA gating. First, when Mg2+ current are increased by variation of the CorA expression levels, a concomitant acceleration in Mg2+ current decay is observed (Fig. 2a,b). Second, other divalent cations like Co2+ are not only able to permeate CorA but also induce current decay with much faster kinetics than those obtained with Mg2+ as charge carrier (Fig. 2a,b). Structural rearrangements can be followed as a function of Mg2+ or Co2+ concentrations and are best fitted by a classical Hill binding model with an apparent affinity for Mg2+ of K0.5 = 2.4 mM and for Co2+ of K0.5 = 0.2 mM, both showing positive cooperativity (nH = 2) (Fig. 2c). The higher Co2+ affinity detected by spectroscopy is consistent with the faster rate of inward current decays (Fig. 2a,b) and with previous Co2+ binding experiments, but is in the toxicity range of this cation which seems incompatible with a role of CorA as a regulator of Co2+ homeostasis as previously proposed11. Third, excised patch clamp experiments show that exposure of the intracellular domain of CorA to mM concentrations of Mg2+ leads to a reduction and eventually, the complete inhibition of the CorA-driven Mg2+ currents. However, pre-exposing the cytoplasmic side to EDTA under the same experimental conditions consistently enhances Mg2+ current amplitudes (Fig. 2d). We interpreted these phenomena to be the result of a self-regulatory mechanism, where an increase in the local Mg2+ concentration on the intracellular side of the membrane saturates a CorA Mg2+-binding site, ultimately triggering channel closure. Under this scheme, the apo form of CorA is conductive and Mg2+ binding triggers channel deactivation and stabilizes the non-conductive (closed) state of the channel. This mechanism where the charge carrier being also the gating factor fully supports the suggestion that Mg2+ homeostasis in bacteria takes place through a Mg2+ negative feedback loop2, 10, 20.
The divalent cation sensor
The crystal structures of TmCorA obtained in a high concentration of divalent ions (Mg2+, Co2+, Ca2+) point to the likely location of the divalent cation sensor8, 9, 10. A clear electron density was unequivocally assigned to divalent ions coordinated between two aspartate residues, Asp253 from one subunit, and Asp89 from its adjacent subunit8 (Fig. 3a). Mutating these residues alters the Mg2+ dependent protease digestion susceptibility, implying that these residues might be involved in magnesium sensing10. Using both spectroscopic and functional approaches, we find that disrupting the contribution of individual components of this putative site dramatically alters Mg2+ binding. Replacing Asp253 or Asp89 by a Lysine abolishes the Mg2+ dependent conformational transition observed at position Asn260 (near the divalent binding site) by EPR spectroscopy (Fig. 3b). Also, given the similarities with the EPR spectra of D89K-D253, D89-D253K and Mg2+-bound WT CorA (again at position Asn260), the conformational equilibrium of the divalent binding site mutant seems to be biased towards the closed state. This observation is supported by a moderate loss of function phenotype in a cell complementation assay20. Based on this observation, we argue that the conformational motions monitored at position 260 directly correlate with the presence of Mg2+ at the divalent binding site. Oocytes injected with the D253K construct can still generate a CorA-specific Mg2+ inward current, but in contrast to the WT channel, they do not exhibit a time-dependent decay (Fig. 3c) and thus facilitating permeation studies 13. This result is the most direct demonstration that the interacting pair Asp253-Asp89 acts as an important component of the divalent cation sensor in CorA. This divalent sensor presumably probes the intracellular concentration of Mg2+ and triggers an allosteric conformational wave that leads to channel closure as Mg2+ reaches above its physiological level. Interestingly, MgtE also seems to operate with a self-regulated mechanism despite any structural nor sequence similarity to CorA23, 24.
Figure 3. The divalent cation sensor.
a. Close up view of the putative regulatory Mg2+ binding domain revealed by the crystal structure. b. EPR spectra of the Q260C-spin labeled mutant recorded in presence of 20 mM Mg2+ shown in black and in absence of divalent cation shown in red. The experiments were repeated in the background of the WT protein, D89K and D253K mutation. The immobile (i) and mobile (m) components of the low field spectra are highlighted by two vertical dash line. c. Representative macroscopic current traces recorded for WT CorA and D253K are shown in black and blue respectively.
Gating Involves Expansion of the Intracellular Cavity
To gain further insight into the CorA gating mechanism, we carried out a complete characterization of the environmental properties of spin labels attached to cysteine residues located along the long stalk helix (residues 246 to 293), the pore forming TM1 helix (residues 294 to 314), the extracellular loop (residues 315 to 325) and the lipid facing TM2 helix (residues 326 to 348). We analyzed three structural parameters from CW-EPR spectral properties: probe mobility ( ) which reflects the conformational flexibility22 (larger values imply higher motional freedom); collision with NiEDDA, an indicator of water exposure; and collision with O2, which monitors lipid exposure25, 26 (Fig. 4a). Measurements were carried out in high Mg2+, which should favor the closed conformation, and in the nominal absence of Mg2+ (or other divalent ions), which should populate the open state of CorA.
Figure 4. Probe environmental parameters and structural rearrangement associated with gating.
a. Probe mobility, NiEDDA accessibility and O2 accessibly values were plotted against the residue number for two experimental conditions: 20 mM Mg2+ and No Mg2+. The values for sample recorded in presence of Mg2+ (closed state) are shown in gray closed circle whereas the values obtained for CorA in its open conformation are shown as colored open circles. b. The differential mobility values between the open and the closed conformation were color coded from red to blue and mapped on the crystal structure of CorA. For clarity purposes only the stalk helix and Tm1 are shown and one subunit has been removed. Similar representation is shown on the right side for the differential values in NiEDDA colliding agent. c. differential O2 accessibility are color scaled and mapped on TM1 (left panel) and TM2 (right panel). Only 2 subunits are shown for clarity purposes.
Comparing solvent accessibility and probe mobility revealed a series of dramatic Mg2+-driven structural changes in CorA (Fig. 4b). In the Apo form, residues from the stalk helix forming the inner cavity of the intracellular funnel show increased mobility and NiEDDA accessibility (ΠNiEDDA), particularly for residues Gly274, Leu280, Val283 and Thr287. Differences in ΠNiEDDA between the apo and Mg2+-bound states mapped onto the crystal structure of TmCorA (Closed state) (Fig. 4b) show that the major increases in ΠNiEDDA and take place inside the funnel cavity (arrows). These results point to a widening of the inner funnel cavity as Mg2+ is removed from its regulatory domain. Consistent with the fairly large cavity expansion, the sharp increase in probe mobility seems to occur in both faces of the helix as a result of reduced helical packing in the open (apo) conformation. Given the size of NiEDDA compared to a water molecule, we anticipate that the functionally-relevant magnitudes of the conformational transitions extend further beyond the mapped increases and represent a widespread reorientation of the permeation pathway as a whole. This is supported by significant changes in the O2 accessibility (ΠO2) along the transmembrane helices (Fig. 4d). The relatively large increase in ΠO2 profile along pore facing residue in TM1 indicates a relatively reorientation of this Helix (Fig. 4c). These changes are compatible with the unbending of helix TM1 ultimately leading to an increase in the diameter of the permeation pathway. The very high ΠO2 values reported for TM2 indicate that the intimate contact between TM1 and TM2 are disrupted in the open conformation. Conformational changes were also observed for TM1 as evaluated from the collision to the DOGS-NTA[Ni(II)] lipid, a collisional agent that reports on interfacial accessibility (Supplementary Fig. 2). This strongly supports a sizable and widespread structural rearrangement upon Mg2+-driven gating that appears to be larger than the relatively modest radial and lateral tilts of the stalk helix α7 recently reported14.
Inner motion of the stalk helix leads to pore opening
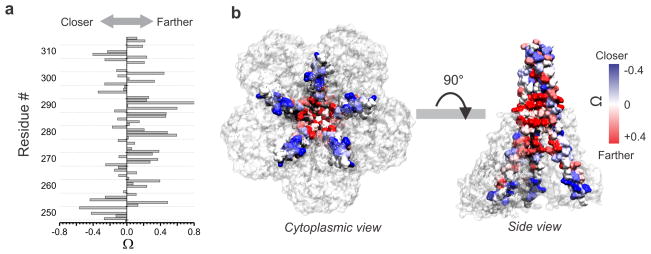
As expected from a radially symmetric homopentameric protein, line shape analyses of CorA EPR spectra reveal the presence of strong spin-spin interaction at residues facing the fivefold symmetry axis along the pore17. Any movement that alters the intersubunit proximity would generate changes in the spin-spin coupling of spatially close residues (< ~15 Å). Because of the complications due to the distance relations in a pentameric geometry, we have estimated the intersubunit relative proximity from the ratio of the normalized amplitude of the spectra in the open and closed conformation (the Ω parameter adapted from27, see Methods section). Negative Ω values indicate that the labeled positions are moving closer to the fivefold axis of symmetry while positive Ω values reflect motion away from the symmetry axis. Plotting the Ω values vs. the residue sequence revealed that residues at the tip of the C-terminal end of the stalk helix move closer to the symmetry axis as the channel opens (Fig. 5a,b). Beyond residue Asp270, the vast majority of the positions move further away from the center, indicating an expansion of the cavity and permeation path (Fig. 5b). In the closed conformation structure we and others have shown that permeation of a point charge through CorA is limited by several energy barriers along the permeation pathway16, 17. Any conformational change triggering CorA opening must lower the amplitude of these barriers. Considering the presence of a kink in the stalk helix at position Gly273 and a pivot point, it is conceivable that a closure of the tip of the stalk helix would translate into an expansion of the funnel and the permeation pathway (Supplementary Fig. 3). Dipolar coupling at positions flanking the constricted permeation pathway clearly indicate the dilation of the permeation pathway throughout its length, supporting the idea of an open channel (Supplementary Fig. 4).
Figure 5. Change in intersubunit proximities in the Stalk and in the pore forming TM1 helices.
a. The bars represent the Ω parameter, which reports on the relative inter-subunit proximity change when the channel goes from the closed to the open state. Ω is defined by the ratio of the intensities of the central resonance line of the EPR spectra for the same sample in presence of 20 mM Mg2+ and in absence of Mg2+ (see Methods section) b. Ω values are color scaled and mapped on the CorA crystal structure (closed conformation). Two views are presented and for clarity purposes TM2 has been removed and on the side view one subunit has been removed so the pore facing residues are visible.
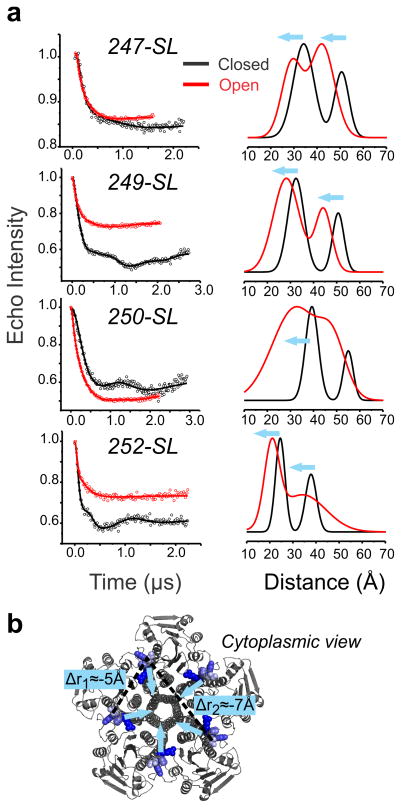
This type of structural rearrangement is incompatible with a classical transporter-like mechanism where the protein alters its conformation from an inward facing to outward facing conformation by means of a rocking motion. This interpretation is also supported by the anomalous mole fraction effect (AMFE) observed in Ni2+/ Mg2+ mixtures, indicating that CorA pore is multi-occupied13. Our results are also consistent with molecular dynamic simulations data suggesting an iris-like mechanism of opening16. To quantify the amplitude of the motion described above, we carried out double electron-electron resonance (DEER) spectroscopy experiments to calculate the average distances and distance distributions of inter-probe distances28 under different Mg2+ concentrations. Recently, we have shown that using a symmetry-based constraints fit we can accurately measure distance distributions on homomeric proteins29. Labeling positions at residues Thr247, Pro249, Tyr250 and Arg252 were selected to be within the working distance range of the DEER technique (20–70 Å). Figure 6 shows that for the four positions at the end of the stalk helix, both the short adjacent and the long diagonal distance in the pentamer decrease around ~5Å when the open state is populated (Supplementary Table 1). These measurements describe relatively large closure movement of the stalk helix upon gating (Fig. 6b). The larger amplitude of the distance distribution in the open channel is compatible with an increase of protein dynamics which could interfere with the crystallization of this conductive conformation14. Such a movement is likely to dampen away from the hinge point and is expected to produce sufficient torque to propagate into the permeation pathway (TM1), generating the relatively large pore widening suggested above on the basis of the changes in environmental parameters (Fig. 4).
Figure 6. Quantification of the stalk helix inward motion by DEER spectroscopy.
a. Background corrected dipolar evolutions are shown as open circles for four different mutants. The fitted data are represented as solid lines and the corresponding distance distributions are illustrated using the same color code. b. The labeled positions are shown in blue on the cytoplasmic view of CorA crystal structures. Two dotted line represent the adjacent <r1> and the diagonal <r2> components of the distance distribution and the blue arrows schematize the inferred motion.
A Model of CorA in its open conformation
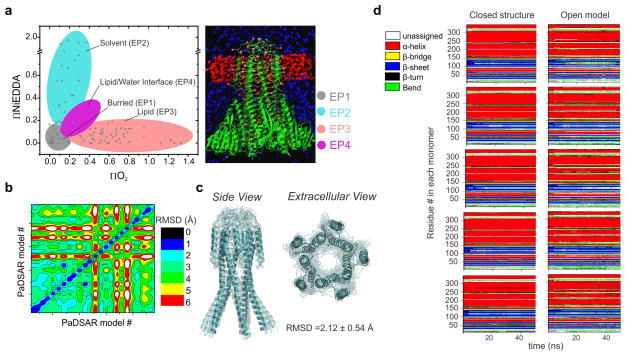
We have taken advantage of the present set of structural data as constraints to build three-dimensional models of CorA in its open conformation. Accessibility data together with intersubunit distance changes were applied to the existing “closed” conformation structure (Mg2+-bound) by means of restrained molecular dynamic (MD) simulation using solvent accessibility as a soft constraint30 (Fig. 7, Supplementary Fig. 6,7). Model generation was limited to the regions of CorA from which experimental data is available, including the long stalk helix, the transmembrane helices and the extracellular loop. The accessibility restraints were enforced through Lennard-Jones-like interactions between a pseudoatom representation of the spin-label and environmental probe-surrounding particles30 (Fig. 7a). Intersubunit Cβ-Cβ distances estimated from DEER experiments, together with the accessibility data were used to generate multiple models by varying the distance constraint boundary, increasing MD refining cycles and replicating the pentamer structure from individual subunits (Supplementary Table 2). After validation, the top 25 models were subsequently refined using the accessibility restraints (Fig 7b, Supplementary Fig. 7). After a second round of refinement, the top 10 models show good structural convergence with a root mean square difference (RMSD) of 2.12±0.54 Å of each other (Fig. 7c). The stability of the top ranked structure was evaluated by an all-atom MD simulation in a fully hydrated phospholipid bilayer. After 5ns of the simulation, the stalk and inner helices are stable around 3–5Å RMSD from the starting structure for the whole simulated time (25 ns) (Supplementary Fig. 8). From the simulations, few transient changes in secondary structure of the open model are observed but, the overall secondary pattern of the open model is similar to that of the closed structure (Fig. 7d). This suggests that the open model is a plausible structure for CorA in its open conformation.
Figure 7. Structural models building and validation for CorA in its open conformation.
a. PaDSAR translates solvent accessibility into structural restraints30 (for details see Methods section and Supplementary Fig. 6). The pseudoatom probes are classified in four categories according to their EPR environmental parameters (left panel) and the starting structure (right panel) is equilibrated into a slab of O2 molecules, representing the lipid bilayer (red spheres). NiEDDA relaxing agent is represented by blue spheres. b. Hundreds of models were generated and evaluated according to their agreement with experimental data (see supplementary Fig. 7). The 25 best models candidates were selected for a second round of PaDSAR refinement and the pairwise Cα-RMSD matrix of the 25 PaDSAR models are color scaled and coded on the contour plot. c. An ensemble of ten models show strong converging structures with backbone RMSD of 2.1 Å ± 0.5 for the regions where EPR data were obtained. d. An all-atoms MD simulations of closed and open CorA embedded in a hydrated lipid bilayer was performed. Secondary structure elements were assigned according to DSSP algorithm52. Stability of the closed and open structures secondary structure elements was followed during the time course of the unrestrained molecular dynamic simulation. The residue run from 6 to 349 are colored coded depending on their secondary structure as followed: α-helix (red), stand in β-sheet (blue), β-bridge (yellow), β-turn (black), coil (green), unassigned (white).
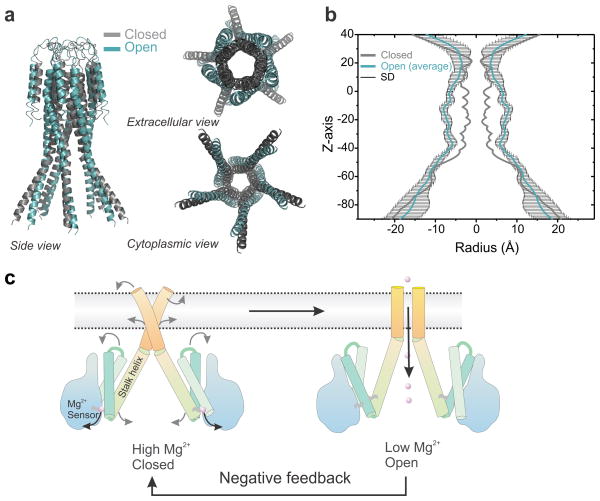
Superimposing the two CorA permeation pathway models offers a glimpse of the nature and extent of the conformational transition taking place during Mg2+-driven gating (Fig. 8). The tips of the stalk helix come together towards the axis of symmetry like the nozzle of a jet-engine. After a kink, this motion translates into an expansion of the cavity and the mouth of the pore opens up with a motion reminiscent of an iris of a camera. These conformational transitions are best appreciated by observing the linear interpolation between the two states and illustrates the negative feedback mechanism first anticipated by Maguire and colleagues (Supplementary movie). Given the low resolution of our spectroscopic approach we choose to analyze the pore radius profile of our ten best-ranked models (Fig. 8b). The comparison of the two conformational states clearly indicates a pore expansion throughout the length of the permeation pathway. Interestingly, the expansion is relatively modest on the upper part of the pathway around the signature sequence residues, consistent with their putative participation in Mg2+ selectivity14, 18. The pore dilation is larger (+5Å) for the most constricted region of the pore, especially for the hydrophobic gates formed by Met292 and Leu294. These increases in the pore radius profile are consistent with expectations for an open channel and suggest that our average model represents a plausible structure for the conductive state of CorA. Poisson-Boltzmann electrostatic calculation performed on our ten best ranked models shows that the large energy barriers impeding ion conduction in the closed state are largely alleviated (Supplementary Fig. 9). Altogether, our results support an explicit molecular mechanism of magnesium regulation in prokaryotes where CorA acts as a Mg2+-deactivated Mg2+-selective channel as part of an homeostatic negative feedback loop (Fig. 8c).
Figure 8. Mg2+ dependent Gating mechanism of CorA.
a. The open model generated by restrained molecular dynamic simulation is shown in teal and the closed state crystal structure is shown as grey cartoon. Only the positions measured by EPR are shown. b. The pore radius profile was calculated for our ten best ranked open models using the program HOLE40. The average value is plotted in teal color and the standard deviation values are plotted in black. The profile for the closed structure is plotted in grey color for comparison. c. Cartoon summarizing the structural rearrangements associated with the Mg2+ gating process regulating closing and opening of CorA. When the intracellular [Mg2+] drops below 2 mM, Mg2+ leaves its regulatory binding site, triggering a closing motion of the stalk helix. This motion transduce after a kink in a widening of the permeation pathway switching the conformation to a conductive state.
Methods
Molecular Biology and Xenopus laevis oocyte overexpression
CorA from Thermotoga maritima was sub-cloned in the pbstA vector optimized for oocyte expression31 using the unique restriction sites NcoI and Xba1 and the PCR primers 5′-AAAAACCATGGAGGAAAAGAGGCTGTCTGCT and 5′-AAAAATCTAGATTAGAGCCACTTCTTTTTCTTGAA. For expression assay only, the same gene was cloned in C-Terminal fusion with Venus, a brighter version of the fluorescent protein YFP32 using the primers 5′-AAAAAACTAGTATGGGCAGCAGCCATCATCATCAT and 5′-AAAAATCTAGATTAGAGCCACTTCTTTTTCTTGAA and unique restriction site SpeI and XbaI. Expression was monitored after 24 and 48 hours of injection by confocal microscopy on an Olympus DSU Spinning Disk Confocal microscope on oocyte injected with constructs and with blank as control. Mutations were introduced by PCR using mismatch mutagenic primers as described by the Stratagene Quick Change kit. All mutant plasmid were amplified in E.Coli XL-1 Blue Strain and purified by Mini or Midi Prep kit (Qiagen) and verified by whole gene sequencing. 10 μg of plasmid-DNA were linearized by overnight Not1 digestion, verified by 1% Agarose electrophoresis and purified by the QIAquick PCR purification (Qiagen) following the suppliers recommendation. 1 μg of purified linear DNA was used as template for in vitro translation (Ambion) and after purification the RNA were aliquoted and kept frozen at −80°C. Oocyte were harvested from survival surgery on adult frog according to standard protocol and our animal care procedure # 71475 and injected with 50 ng of mRNA.
Electrophysiology
Electrophysiology measurements were performed on custom made Two Electrodes Voltage Clamp (TEVC) setups according to basic standards procedure33. The external solutions were isosmotic and typically composed by Hepes/N-Methyl-D-Gluconamide (NMG), 10 mM, NMG-Methanesulfonic Acid (MSA), 30 mM NMG-Cl, 20 mM MgCl2. The cut-open oocyte preparation was used in experiments requiring control of intracellular [Mg2+]. This consisted of a Vaseline-gap oocyte chamber following the original design of Perozo et al. 34 where an internal perfusion cannula runs through the internal and guard compartments using a small stainless steel needle (33 gauge) connected to a syringe pump. After mounting the oocyte in the chamber, a micro needle was inserted midway through the bottom of the oocyte and perfusion was begun with internal solution buffered with a mixture of EDTA/EGTA. Internal Mg2+ was successively increased from 0 to 2, 4, 40 and 60 mM by changing the internal perfusion solution. The internal perfusion rate was constant at 10 μL/h, and 10–15 min was required for equilibration at each new internal [Mg2+].
Sample preparation and CW-EPR spectroscopy
Cysteine mutants were expressed in E.Coli BL21DE3-pLysS strain as previously described17. Immediately after elution from the Co2+ column, the purified protein were reacted with a 10 molar excess of spin label for 30 min on ice, followed by second addition of 10 molar probe excess and reacted for an additional 30 min. The reaction was stopped by addition of 1000 fold excess of L-Cysteine and the excess probe was removed by Gel filtration chromatography. Reconstitution was carried out by destabilizing the pre-formed POPC-POPG 3:1 liposomes with a saturating concentration of Triton™ X-100 (0.2 w/w ratio) before adding labeled protein to the liposome suspension in a 1/1500 protein/lipid molar ratio. Detergent was removed by successive addition of Bio-Beads SM-2 (Bio-Rad) and proteoliposomes were harvested by ultracentrifugation at 200,000g and resuspended into 200 μL. Samples were split into two and were equilibrated with a buffer containing either 1 mM EDTA or 20 mM MgCl2. Continuous-wave EPR spectroscopic measurements were performed at room temperature on a Bruker EMX X-band spectrometer equipped with a dielectric resonator (ER 4123D). Spin labeled samples were loaded into a gas-permeable TPX plastic capillary and spectra were recorded at 2.0 mW incident power, 100 kHz modulation frequency with 1.0 G of modulation amplitude. The motional freedom of the spin label was quantified as a peak-to-peak first-derivative width of the central resonance line ( )22. Solvent exposure was measured form power-saturation experiments in which the vertical peak-to-peak amplitude of the central line of the EPR spectra is measured as a function of increasing incident microwave power25, 26. Prior and during the recording of the spectra the sample are equilibrated with N2, Air (20% O2) or with 25 mM NiEDDA (Ni[II]ethylenediaminediacetic acid) in presence of N2. Accessibility parameter Π were calculated as described in a previous report17. High accessibility of the spin probe to O2 is witness by a High value of the ΠO2 and is diagnostic of a membrane exposed residue whereas high NiEDDA accessibility (ΠNiEDDA) is diagnostic of a water exposed residue. The relative probe proximity change was determined by calculating the Ω parameter. The Ω parameter is defined by the ratio of the intensities of the central resonance line M = 0 for the same sample exposed in two different experimental conditions as followed: where is the amplitude of M = 0 measured in absence of Mg2+ (open conformation) and Ā is the amplitude of M = 0 measured in presence of 20 mM Mg2+ (closed conformation). We shifted the axis with the operator −1 to scale the values for negative/positive values instead of below/above 1.
DEER Experiments
Samples concentration was empirically optimized to 75 μM of CorA pentamer and were loaded into a quartz capillary (Vitrocom) and equilibrated at 80 K under a flow of liquid N2 using an Oxford cryostat. A standard four-pulse DEER sequence was conducted on a Bruker Elexsys 580 EPR spectrometer equipped with a 3 mm split-ring resonator. The four-pulse DEER sequence was set with pulses of 16 ns (90°) and 32 ns (180°), and evolution times were typically set to 1800–2500 ns depending on distance and signal quality. Refocused echo intensity evolutions were recorded, and these phase-corrected signals were background-corrected assuming a homogeneous 3D distribution (Supplementary Fig. 5). The distance distribution P(r) was recovered from DEER measurements by a model fit of a two-component Rice3D mixture with distance ratio constraint using 1.578<k<1.645 as determined from our previous study29. Analysis was performed on a modified version of DeerAnalysis 2011 program35. Each time domain dipolar evolution data set was preprocessed using tools provided by DeerAnalysis to correct for experimental phase errors and to separate the intramolecular distances from the intermolecular background contribution. The background was assumed as a homogeneous distribution in three dimensions, appropriate for spin-labeled membrane proteins in detergent micelles.
Molecular modeling
The starting structure of CorA was taken from our previous work17. As mentioned previously, this model of the closed state of CorA was derived from the crystal structure (PDB 2UIB) with remodeling of the unresolved loop (residues 315 to 325). The structure of CorA is a homopentamer with 344 amino acid residues per monomer starting from residues 6 to 349. For the calculation of the CorA open model, we employed our previously developed method called PaDSAR (pseudoatom driven solvent accessibility refinement), restrained molecular dynamics using EPR accessibility restraints30. For the present work, the PaDSAR method has been incorporated in CHARMM c35b636. Two sources of experimentally-based restraints used in PaDSAR are the interprobe distances and solvent accessibility data. The detailed method is described below. Analyses of intersubunit DEER distances and the proximity profile of the open and closed states of CorA allows one to estimate a range of distance changes of the labeled positions at the backbone level. In the structure calculation, distance restraints (ΔrCβ) were defined as a distance difference between intersubunit Cβ-positions of the labeled residues measured between the candidate open-model and the closed-state x-ray structure as follow:
| (1) |
Where i is the residue number, of which the experimentally derived distance accounted for the following residue pairs: 247–247′, 249-249′, 250-250′ and 252-252′ (the prime is used to denote different subunits). As a pentameric protein, the “short” and “long” distance restraints were used to keep five subunits arranged about a fivefold symmetry axis. The short distance defined an intersubunit Cβ-Cβ separation of the residue between a subunit and its first-adjacent subunit whilst the long distance defined that of the residue between a subunit and its second adjacent subunit. From experimental data, the short and long distance restraints were set to −5±δÅ and −7±δÅ, respectively. The negative value of distance indicates the residue pair of an expected open model is getting closer with respect to that of the closed-state structure. The δ values were used to define the upper and lower bound. A list of the intersubunit Cβ-Cβ distance restraints used in the calculations is given in Supplementary Table 2. The accessibility restraints used in PaDSAR were derived from the experimental ΠO2 and ΠNiEDDA accessibility data of more than 100 spin-labeled (SL) CorA mutants. The SL-positions covered a broad region of the protein domain including the long stalk helix (residue 246 to 293), the pore forming TM1 helix (residues 294 to 314), the extracellular loop (residue 315 to 325) and the lipid facing TM2 helix (residue 326 to 348). The restraints are described in terms of the Lennard-Jones-like interactions between various types of spin probe pseudoatoms attached to the Cα-atom of protein backbone and the surrounding environment pseudoatoms presented by OXY (O2), NIC (NiEDDA) or PROT (amino acid residue). In the calculation, four types of the spin pseudoatoms (Fig. 7, Supplementary Fig. 6) have first been assigned to individual SL-residue of CorA before the PaDSAR calculation. The assignment was on the basis of residue-based surface exposure from an analysis of ΠO2, ΠNiEDDA and ΔHo−1 profiles. An initial configuration of modeling system comprising of the starting structure of the CorA pentamer attached with the 83 assigned spin pseudoatoms and several hundreds of OXY and NIC pseudoatoms is illustrated in Fig. 7a. The modeling of the CorA open structure may be divided into two main PaDSAR runs. The first run was carried out using the delta distance (ΔrCB) and accessibility restraints imposed on the starting structure as described in previous section. The protein was modeled with the united-atom CHARMM PARAM19 force field37. During the calculation, we restrained the N-terminal domain (residue 6 to 156) and secondary structures including residue 165 to 199, 204 to 237, 243 to 310 (stalk and TM1 helices) and 327 to 347 (TM2 helix). OXY and NIC pseudoatoms move within a defined boundary using the Miscellaneous Mean Field Potential (MMFP) in CHARMM38. OXY pseudoatoms were allowed moving in the membrane region whereas NIC pseudoatoms distribute outside the membrane but within the defined boundary (Supplementary Fig. 6). We varied the upper and lower bound of distance restraints (δ = 1 and 2) and increased MD refining cycles to generate a set of model candidates. However, it appears that these model candidates tend to lose its fivefold symmetry of the pentamer. Therefore, we took an individual subunit of all the resulting models and replicated their own pentamer structure. As a result, hundreds of model candidates were generated, and subsequently evaluated by considering with the following experimental-based validation: 1) the restraint penalty (Prstrnt) and distance restraint violation (viol) 39, and 2) a consensus change between the evaluated model and the intersubunit proximity (Ω). For the former validation, Prstrnt is given as
| (2) |
| (3) |
Where and are the upper and lower distance restraints, and krstrnt is an arbitrary constant value. The latter validation was made by identifying the closer or further Cβ-Cβ distance of individual SL-residues (the short distance or 1st adjacent subunit) with respect to the closed-state conformation and comparing with the positive or negative signs of Ω parameters (Figure 5). The best-fit 25 models selected based on an averaged fit criterion were subjected to structure refinement of the PaDSAR run. In the second run, the top 25 models were refined using the accessibility restraint only. Similar protocol of PaDSAR refinement described above, except no distance restraint was imposed at this step. In addition, PaDSAR treated each model as a single subunit, and refined it using the image facility within the CHARMM program to maintain the fivefold symmetry of the CorA pentamer. In addition to the model validation described previously, inspections of the resulting structures obtained in the second run include structure convergence (RMSD), the HOLE plot40, the accessibility-mapped molecular surface and all-atom MD simulations.
Model validation by Poisson-Boltzmann energy calculation of ion translocation
Model quality was assessed by calculating the solvation free energy profile of ion translocation along the permeation pathway. We employed Poisson-Boltzmann (PB) continuum method to compute free-energy barrier for Mg2+ translocation through the CorA tranmembrane pore of the closed and open conformations. The PB calculation was performed using Adaptive Poisson-Boltzmann Solver (APBS)41. The program PDB2PQR was used to add hydrogen atoms and assign atomic charges and radii to each generated configuration42, 43. Partial atomic charges and radii of the protein were taken from the CHARMM parameter sets44. The CorA protein was oriented with its symmetry axis coinciding with the z-axis. The sequential focusing multigrid algorithm in solving PB equation consists of three resolution maps: 300×300×300 Å3 for coarse, 200×200×200 Å3 for medium, and 100×100×100 Å3 for fine resolutions. Grid points of 161×161×161 were employed to all maps. The implicit membrane hydrophobic slab was defined between z = ±12 Å. This corresponds to the thickness of hydrophobic slab of 24 Å. The dielectric constants for protein, membrane and water were 2, 2 and 80, respectively45. Ionic strength was 0.15 M with coulomb charge +1 and −1, and radius 2.0 Å. Water probe radius was 1.4 Å.
The solvation free energy profile of Mg2+ translocation through the TmCor pore was computed by systematically varying Mg2+ positions from z = +20 Å (periplasmic region) to z = −40 Å (cytoplasmic region) with 1 Å steps along the z-axis. The program APBSmem was used to set up and to monitor the membrane region46. For each configuration, electrostatic contribution to the solvation free energy was computed twice, one for implicit aqueous model and one for water-membrane model, giving rise to ΔGelec,water and ΔGelec,membrane, respectively. The solvation free enrgy (ΔΔGelec) that is the electrostatic free energy difference of the proteins embedded in the two solvent environments was obtained according to Eq.(4)
| (4) |
Model validation by explicit solvent molecular dynamics simulation
An obtained model was subsequently inserted into a pre-equilibrated lipid bilayer, made up of ~480 molecules of 1-palmitoyl-2-oleoyl-sn-glycerol-3-phosphatidylcholine (POPC) embedded in ~64,000 TIP3P waters. Side-chain ionization states that were expected at pH = 7 were assigned based on pKa calculations using PROPKA47. Sodium and chloride counter ions were added to neutralize charges on the system using VMD’s Autoionize plugin48. Total number of atoms in the simulation is ~300,000 atoms. The MD simulations were carried out using NAMD 2.8b2 with the all-atoms CHARMM22 and CHARMM27 force fields for protein, lipid, and ion49. The TIP3P models were used for waters. At this stage, the pseudoatoms present in the model were removed; no pseudoatom restraints were included during the simulation with explicit solvent models. To reduce finite-size effects, periodic boundary conditions were used in molecular dynamics simulations with a periodic dimension of 144×144×155 Å3. The calculation of long-rang electrostatic interactions in periodic membrane-protein system employed the particle mesh Ewald (PME) summation via fast Fourier transform (FFT). The PME grid size was set to at least 1 cell per Angstrom in each dimension. A distance cut-off of 12 Å was employed with a switching distance set at 10 Å. The non-iterative SETTLE algorithm was used to keep waters rigid. Rigid bonds between the hydrogen and the heavy atom to which it is attached were enabled. The simulations were performed at constant pressure of 1 atm and constant temperature of 300K. Langevin dynamics was used to keep a constant temperature of the system with Langevin damping coefficient of 1 ps−1. The pressure of the system was controlled using a Nośe-Hoove Langevin Piston. Energy minimizations and restrained MD simulations were employed to relax geometric, angle, torsional and other structural strains of the model systems. During the first restrained MD run, atoms for protein and lipid head groups were restrained to their initial positions using harmonic constraints with a force constant of 1 kcal.mol−1.Å−2. In the subsequent run, protein atoms were restrained, leaving the rest of the system to be relaxed. Finally, all positional restraints were removed and an equilibration run was performed. MD simulations were performed for 25 ns with the time step of 2 fs. The configurations and velocities were stored every 2 ps. The programs VMD, WORDOM50, DSSP51, APBS41, 46 were used for assessment of protein structure stability, RMSD and visualization.
Supplementary Material
Acknowledgments
O.D thanks Prof. Luis G. Cuello for enlightening discussion throughout the course of this work. The authors are thankful to Perozo lab members for critical reading of the manuscript. This work was supported by NIH Grant GM088406. RES560530217-HR provides grant support to PS.
Footnotes
Author Contributions. O.D and E.P designed and conceived the project and wrote the paper. O.D. performed experiments and analyzed the data. P.S performed the modeling and molecular dynamic simulations. F.B. provided guidance and experimental advice. All authors discussed the results and participated in MS preparation.
Competing financial interests. The authors declare no competing financial interests.
References
- 1.Romani A. Cellular magnesium homeostasis. Archives of biochemistry and biophysics. 2011;512:1–23. doi: 10.1016/j.abb.2011.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maguire ME. Magnesium transporters: properties, regulation and structure. Front Biosci. 2006;11:3149–3163. doi: 10.2741/2039. [DOI] [PubMed] [Google Scholar]
- 3.Knoop V, Groth-Malonek M, Gebert M, Eifler K, Weyand K. Transport of magnesium and other divalent cations: evolution of the 2-TM-GxN proteins in the MIT superfamily. Mol Genet Genomics. 2005;274:205–216. doi: 10.1007/s00438-005-0011-x. [DOI] [PubMed] [Google Scholar]
- 4.Nelson DL, Kennedy EP. Magnesium transport in Escherichia coli. Inhibition by cobaltous ion. The Journal of biological chemistry. 1971;246:3042–3049. [PubMed] [Google Scholar]
- 5.Maguire ME. The structure of CorA: a Mg(2+)-selective channel. Curr Opin Struct Biol. 2006;16:432–438. doi: 10.1016/j.sbi.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 6.Snavely MD, Florer JB, Miller CG, Maguire ME. Magnesium transport in Salmonella typhimurium: 28Mg2+ transport by the CorA, MgtA, and MgtB systems. J Bacteriol. 1989;171:4761–4766. doi: 10.1128/jb.171.9.4761-4766.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Snavely MD, Florer JB, Miller CG, Maguire ME. Magnesium transport in Salmonella typhimurium: expression of cloned genes for three distinct Mg2+ transport systems. J Bacteriol. 1989;171:4752–4760. doi: 10.1128/jb.171.9.4752-4760.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eshaghi S, Niegowski D, Kohl A, Martinez Molina D, Lesley SA, Nordlund P. Crystal structure of a divalent metal ion transporter CorA at 2.9 angstrom resolution. Science. 2006;313:354–357. doi: 10.1126/science.1127121. [DOI] [PubMed] [Google Scholar]
- 9.Lunin VV, et al. Crystal structure of the CorA Mg2+ transporter. Nature. 2006;440:833–837. doi: 10.1038/nature04642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Payandeh J, Pai EF. A structural basis for Mg2+ homeostasis and the CorA translocation cycle. EMBO J. 2006;25:3762–3773. doi: 10.1038/sj.emboj.7601269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xia Y, Lundback AK, Sahaf N, Nordlund G, Brzezinski P, Eshaghi S. Co2+ selectivity of Thermotoga maritima CorA and its inability to regulate Mg2+ homeostasis present a new class of CorA proteins. J Biol Chem. 2011;286:16525–16532. doi: 10.1074/jbc.M111.222166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Niegowski D, Eshaghi S. The CorA family: structure and function revisited. Cell Mol Life Sci. 2007;64:2564–2574. doi: 10.1007/s00018-007-7174-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dalmas O, Sandtner W, Medovoy D, Frezza L, Bezanilla F, Perozo E. A repulsion mechanism explains magnesium permeation and selectivity in CorA. Proc Natl Acad Sci U S A. 2014 doi: 10.1073/pnas.1319054111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pfoh R, Li A, Chakrabarti N, Payandeh J, Pomès R, Pai E. Structural asymmetry in the magnesium channel CorA points to sequential allosteric regulation. Proc Natl Acad Sci U S A. 2012;109:18809–18814. doi: 10.1073/pnas.1209018109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nordin N, et al. Exploring the structure and function of Thermotoga maritima CorA reveals the mechanism of gating and ion selectivity in Co2+/Mg2+ transport. Biochem J. 2013;451:365–374. doi: 10.1042/BJ20121745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chakrabarti N, Neale C, Payandeh J, Pai EF, Pomes R. An iris-like mechanism of pore dilation in the CorA magnesium transport system. Biophys J. 2010;98:784–792. doi: 10.1016/j.bpj.2009.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dalmas O, Cuello LG, Jogini V, Cortes DM, Roux B, Perozo E. Structural dynamics of the magnesium-bound conformation of CorA in a lipid bilayer. Structure. 2010;18:868–878. doi: 10.1016/j.str.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guskov A, et al. Structural insights into the mechanisms of Mg2+ uptake, transport, and gating by CorA. Proc Natl Acad Sci U S A. 2012;109:18459–18464. doi: 10.1073/pnas.1210076109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Froschauer EM, Kolisek M, Dieterich F, Schweigel M, Schweyen RJ. Fluorescence measurements of free [Mg2+] by use of mag-fura 2 in Salmonella enterica. FEMS Microbiol Lett. 2004;237:49–55. doi: 10.1016/j.femsle.2004.06.013. [DOI] [PubMed] [Google Scholar]
- 20.Payandeh J, Li C, Ramjeesingh M, Poduch E, Bear CE, Pai EF. Probing structure-function relationships and gating mechanisms in the CorA Mg2+ transport system. J Biol Chem. 2008;283:11721–11733. doi: 10.1074/jbc.M707889200. [DOI] [PubMed] [Google Scholar]
- 21.Alatossava T, Jutte H, Kuhn A, Kellenberger E. Manipulation of intracellular magnesium content in polymyxin B nonapeptide-sensitized Escherichia coli by ionophore A23187. J Bacteriol. 1985;162:413–419. doi: 10.1128/jb.162.1.413-419.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McHaourab HS, Lietzow MA, Hideg K, Hubbell WL. Motion of spin-labeled side chains in T4 lysozyme. Correlation with protein structure and dynamics. Biochemistry. 1996;35:7692–7704. doi: 10.1021/bi960482k. [DOI] [PubMed] [Google Scholar]
- 23.Hattori M, et al. Mg(2+)-dependent gating of bacterial MgtE channel underlies Mg(2+) homeostasis. The EMBO journal. 2009;28:3602–3612. doi: 10.1038/emboj.2009.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hattori M, Tanaka Y, Fukai S, Ishitani R, Nureki O. Crystal structure of the MgtE Mg2+ transporter. Nature. 2007;448:1072–1075. doi: 10.1038/nature06093. [DOI] [PubMed] [Google Scholar]
- 25.Altenbach C, Froncisz W, Hyde JS, Hubbell WL. Conformation of spin-labeled melittin at membrane surfaces investigated by pulse saturation recovery and continuous wave power saturation electron paramagnetic resonance. Biophys J. 1989;56:1183–1191. doi: 10.1016/S0006-3495(89)82765-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Farahbakhsh ZT, Altenbach C, Hubbell WL. Spin labeled cysteines as sensors for protein-lipid interaction and conformation in rhodopsin. Photochem Photobiol. 1992;56:1019–1033. doi: 10.1111/j.1751-1097.1992.tb09725.x. [DOI] [PubMed] [Google Scholar]
- 27.Perozo E, Cortes DM, Cuello LG. Structural rearrangements underlying K+-channel activation gating. Science. 1999;285:73–78. doi: 10.1126/science.285.5424.73. [DOI] [PubMed] [Google Scholar]
- 28.Jeschke G, Polyhach Y. Distance measurements on spin-labelled biomacromolecules by pulsed electron paramagnetic resonance. Phys Chem Chem Phys. 2007;9:1895–1910. doi: 10.1039/b614920k. [DOI] [PubMed] [Google Scholar]
- 29.Dalmas O, Hyde HC, Hulse RE, Perozo E. Symmetry-Constrained Analysis of Pulsed Double Electron-Electron Resonance (DEER) Spectroscopy Reveals the Dynamic Nature of the KcsA Activation Gate. J Am Chem Soc. 2012;134:16360–16369. doi: 10.1021/ja3069038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sompornpisut P, Roux B, Perozo E. Structural refinement of membrane proteins by restrained molecular dynamics and solvent accessibility data. Biophys J. 2008;95:5349–5361. doi: 10.1529/biophysj.108.142984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Starace DM, Stefani E, Bezanilla F. Voltage-dependent proton transport by the voltage sensor of the Shaker K+ channel. Neuron. 1997;19:1319–1327. doi: 10.1016/s0896-6273(00)80422-5. [DOI] [PubMed] [Google Scholar]
- 32.Nagai T, Ibata K, Park ES, Kubota M, Mikoshiba K, Miyawaki A. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat Biotechnol. 2002;20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- 33.Dascal N. Voltage clamp recordings from Xenopus oocytes. Curr Protoc Neurosci. 2001;Chapter 6(Unit 6):12. doi: 10.1002/0471142301.ns0612s10. [DOI] [PubMed] [Google Scholar]
- 34.Perozo E, Papazian DM, Stefani E, Bezanilla F. Gating currents in Shaker K+ channels. Implications for activation and inactivation models. Biophys J. 1992;62:160–168. doi: 10.1016/S0006-3495(92)81802-7. discussion 169–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jeschke G, et al. DeerAnalysis2006 - a comprehensive software package for analyzing pulsed ELDOR data. Appl Magn Reson. 2006;30:473–498. [Google Scholar]
- 36.Brooks BR, et al. CHARMM: the biomolecular simulation program. J Comput Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neria E, Fischer S, Karplus M. Simulation of activation free energies in molecular systems. J Chem Phys. 1996;105:1902–1921. [Google Scholar]
- 38.Beglov D, Roux B. Finite Representation of an Infinite Bulk System - Solvent Boundary Potential for Computer-Simulations. J Chem Phys. 1994;100:9050–9063. [Google Scholar]
- 39.Sompornpisut P, Liu YS, Perozo E. Calculation of rigid-body conformational changes using restraint-driven Cartesian transformations. Biophys J. 2001;81:2530–2546. doi: 10.1016/S0006-3495(01)75898-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smart OS, Neduvelil JG, Wang X, Wallace BA, Sansom MS. HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J Mol Graph. 1996;14:354–360. 376. doi: 10.1016/s0263-7855(97)00009-x. [DOI] [PubMed] [Google Scholar]
- 41.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dolinsky TJ, et al. PDB2PQR: expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 2007;35:W522–525. doi: 10.1093/nar/gkm276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dolinsky TJ, Nielsen JE, McCammon JA, Baker NA. PDB2PQR: an automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 2004;32:W665–667. doi: 10.1093/nar/gkh381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sitkoff D, Sharp KA, Honig B. Accurate Calculation of Hydration Free-Energies Using Macroscopic Solvent Models. J Phys Chem-Us. 1994;98:1978–1988. [Google Scholar]
- 45.Grabe M, Lecar H, Jan YN, Jan LY. A quantitative assessment of models for voltage-dependent gating of ion channels. Proc Natl Acad Sci U S A. 2004;101:17640–17645. doi: 10.1073/pnas.0408116101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Callenberg KM, Choudhary OP, de Forest GL, Gohara DW, Baker NA, Grabe M. APBSmem: a graphical interface for electrostatic calculations at the membrane. PLoS One. 2010;5 doi: 10.1371/journal.pone.0012722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li H, Robertson AD, Jensen JH. Very fast empirical prediction and rationalization of protein pKa values. Proteins. 2005;61:704–721. doi: 10.1002/prot.20660. [DOI] [PubMed] [Google Scholar]
- 48.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33–38. 27–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 49.Feller SE, MacKerell AD. An improved empirical potential energy function for molecular simulations of phospholipids. J Phys Chem B. 2000;104:7510–7515. [Google Scholar]
- 50.Seeber M, et al. Wordom: A User-Friendly Program for the Analysis of Molecular Structures, Trajectories, and Free Energy Surfaces. Journal of Computational Chemistry. 2011;32:1183–1194. doi: 10.1002/jcc.21688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 52.Joosten RP, et al. A series of PDB related databases for everyday needs. Nucleic Acids Res. 2011;39:D411–419. doi: 10.1093/nar/gkq1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.