SUMMARY
Mitochondrial fission requires recruitment of dynamin-related protein 1 (Drp1) to the mitochondrial surface and activation of its GTP-dependent scission function. The Drp1 receptors MiD49 and MiD51 recruit Drp1 to facilitate mitochondrial fission, but their mechanism of action is poorly understood. Using X-ray crystallography, we demonstrate that MiD51 contains a nucleotidyl transferase domain that binds ADP with high affinity. MiD51 recruits Drp1 via a surface loop that functions independently of ADP binding. However, in the absence of nucleotide binding, the recruited Drp1 cannot be activated for fission. Purified MiD51 strongly inhibits Drp1 assembly and GTP hydrolysis in the absence of ADP. Addition of ADP relieves this inhibition and promotes Drp1 assembly into spirals with enhanced GTP hydrolysis. Our results reveal ADP as an essential cofactor for MiD51 during mitochondrial fission.
INTRODUCTION
Mitochondrial dynamics requires a balance between the opposing processes of fusion and fission and serves to protect mitochondrial function (Chan, 2012; Westermann, 2010; Youle and van der Bliek, 2012). Disruption of either process causes midgestational lethality in mice and neurologic disease in humans (Chen et al., 2003; Ishihara et al., 2009; Wakabayashi et al., 2009).
Drp1, a dynamin-related GTPase, is the central player in mitochondrial fission (Bleazard et al., 1999; Labrousse et al., 1999; Sesaki and Jensen, 1999; Smirnova et al., 2001). A pool of Drp1 exists in the cytosol and can be recruited to the surface of mitochondria, where it assembles into highly ordered structures that wrap around mitochondrial tubules. Assembly of Drp1 increases its GTP hydrolysis activity, leading to conformational changes that mediate constriction and scission of the mitochondrial tubule (Ingerman et al., 2005; Mears et al., 2011). In mammals, four proteins on the mitochondrial outer membrane act as Drp1 receptors: Fis1, Mff, MiD49, and MiD51 (Gandre-Babbe and van der Bliek, 2008; Palmer et al., 2011; Stojanovski et al., 2004; Yoon et al., 2003; Zhao et al., 2011). Overexpression of Mff causes mitochondrial fragmentation (Otera et al., 2010), and depletion of Mff causes mitochondrial elongation (Gandre-Babbe and van der Bliek, 2008). Fis1 behaves similarly, but the degree of mitochondrial elongation upon depletion depends on the study (Losón et al., 2013; Otera et al., 2010; Yoon et al., 2003; Yu et al., 2005).
MiD49 and MiD51 appear to operate in a more regulated manner. As Drp1 receptors, overexpression of either causes increased recruitment of Drp1 to the mitochondrial surface. Paradoxically, this increased recruitment is not normally accompanied by increased fission rates. The mitochondria instead dramatically elongate, indicating inhibition of the recruited Drp1 (Liu et al., 2013; Palmer et al., 2013; Palmer et al., 2011; Zhao et al., 2011). Cells treated with carbonyl cyanide 3-chloro-phenylhydrazone (CCCP) causes rapid activation of fission and mitochondrial fragmentation, with a greater effect on MiD51 than on MiD49 (Losón et al., 2013).
To address the structural biology of MiD51 function, we used X-ray crystallography to solve the atomic structure of the cytosolic domain of MiD51. Structural and biophysical analyses indicate that MiD51 contains a variant nucleotidyl transferase fold that binds ADP with high affinity. Although ADP binding is dispensable for Drp1 recruitment, it is essential for activation of Drp1 in vitro and in vivo. These results identify ADP as an essential cofactor for MiD51 function.
RESULTS
MiD51 Has a Nucleotidyl Transferase Domain that Binds ADP
Like other Drp1 receptors on mitochondria, MiD51 is an integral outer membrane protein, with most of the protein exposed to the cytosol. Initial attempts to crystallize the entire cytoplasmic portion of mouse MiD51 failed to yield promising hits. Secondary structure prediction indicated that the membrane-proximal region is likely to be disordered (Figure 1A). Limited trypsin proteolysis of the cytosolic domain produced a stable product (Figure S1A available online). N-terminal peptide sequencing combined with mass spectrometry revealed that proteolysis removed almost all the predicted unstructured region, with the C terminus of MiD51 remaining intact (Figure S1B). Using a recombinant form of this shorter polypeptide (MiD51Δ1–133), we readily obtained crystals that diffract to high resolution and solved the structure by multiwavelength anomalous diffraction (MAD) analysis (Table 1) of a selenomethionine-substituted crystal. The native structure was solved at a resolution of 2.2 Å (Table 1; Figure 1B). Model building and refinement produced a final structure with excellent stereochemistry, with an Rfree of 27.4% and an Rwork of 21.9%.
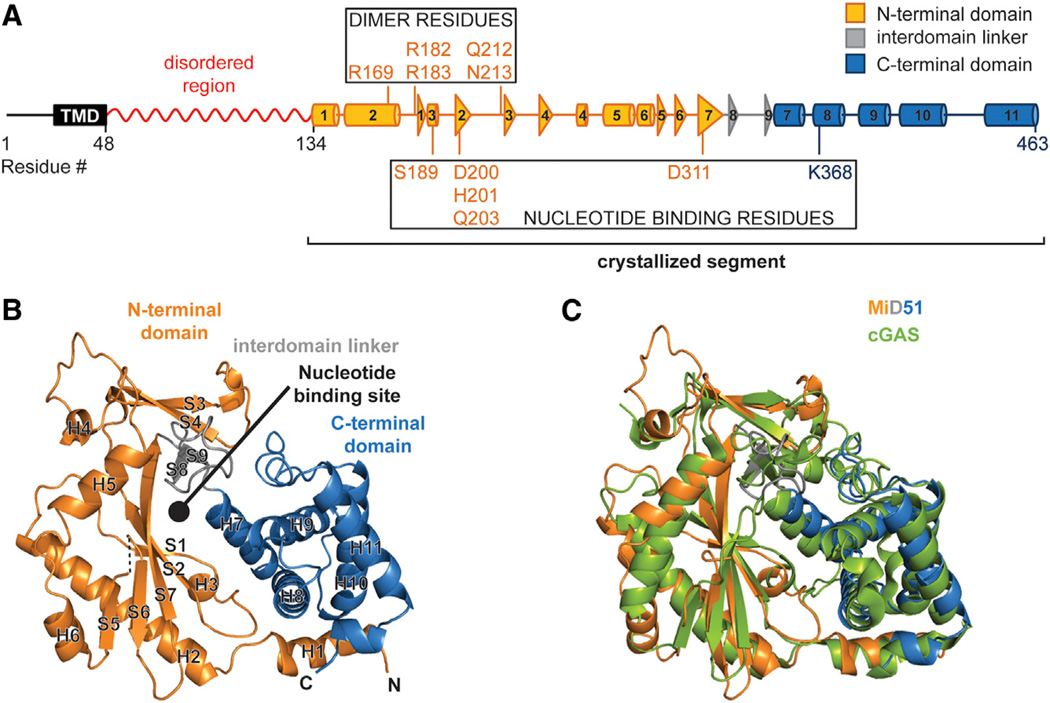
Figure 1. The Cytosolic Region of MiD51 Has a Nucleotidyl Transferase Domain.
(A) Schematic of MiD51. The region determined by X-ray crystallography is indicated and color-coded as in (B). The red squiggle indicates a region predicted to have low probability of regular secondary structure. Boxes highlight residues important for nucleotide binding and the dimer interface. TMD, transmembrane domain. Cylinders represent α-helical segments and triangles strand segments. α helices and β strands from the crystallized segment are numbered.
(B) Ribbon representation of mouse MiD51Δ1–133. Orange, N-terminal domain; gray, interdomain linker; blue, C-terminal domain. α helices and strands are numbered according to (A). Dashed lines denote residues missing from the model. N and C denote the N and C termini. The circle indicates the predicted nucleotide-binding pocket.
(C) Structural overlay of MiD51 (colored as in B)and cyclic GMP-AMP synthase (cGAS) (green).
Table 1.
Data Collection and Refinement Statistics
| Data | Se-Met | Native | ADP Bound | H201A Mutant | CDM Mutant |
|---|---|---|---|---|---|
| Space group | P 1 21 1 | P 1 21 1 | P 1 21 1 | P 1 21 1 | P 21 21 21 |
| Unit cell (Å, °) | 81.42, 79.22, 103.17, 90, 97.86, 90 | 91.07, 78.59, 102.31, 90, 96.61, 90 | 62.07, 80.81, 65.38, 90, 105.74, 90 | 82.43, 79.15, 103.45, 90, 98.04, 90 | 63.68, 67.1, 79.41, 90, 90, 90 |
| Number of molecules (in ASU) | 4 | 4 | 2 | 4 | 1 |
| Resolution (Å) | 39.4–2.6 (2.74–2.60)a | 39.3–2.2 (2.32–2.2) | 38.43–2.0 (2.11–2.0) | 39.58–2.0 (2.11–2.0) | 34.17–2.0 (2.11–2.0) |
| Rmerge (%)b | 7.9 (60.5) | 7.2 (64.0) | 5.7 (37.1) | 6.2 (51.5) | 7.3 (65.2) |
| Completeness (%) | 95.8 (95.9) | 96.1 (95.4) | 97.7 (97.4) | 98.1 (98.7) | 98 (99.8) |
| Mean I/σ | 16.4 (3.4) | 10.5 (2.3) | 16.6 (3.4) | 12.8 (2.4) | 17.1 (2.8) |
| Number of measured reflections | 272,481 (39,317) | 200,830 (28,326) | 141,942 (20,295) | 282,928 (41,589) | 116,233 (17,069) |
| Number of unique reflections | 38,509 (5,598) | 70,056 (10,157) | 41,061 (5,957) | 87,318 (12,772) | 23,493 (3,379) |
| Redundancy | 7.1 (7.0) | 2.9 (2.8) | 3.5 (3.4) | 3.2 (3.3) | 4.9 (5.1) |
| Rwork (%)c | 20.2 | 21.9 | 17.8 | 17.7 | 17.7 |
| Rfree (%) | 25.7 | 27.4 | 22.5 | 21.9 | 21.8 |
| Average B-factor (Å)2 | 44.8 | 56.4 | 30.4 | 31.0 | 26.1 |
| Rmsd from ideal values | |||||
| Bonds (Å) | 0.010 | 0.009 | 0.009 | 0.009 | 0.008 |
| Angle (°) | 1.37 | 1.39 | 1.312 | 1.289 | 1.155 |
| Ramachandran statistics (%) | |||||
| Favored | 97.7 | 97.3 | 96.9 | 97.8 | 97.2 |
| Allowed | 2.1 | 2.5 | 3.1 | 2.1 | 2.8 |
| Outliers | 0.2 | 0.2 | 0 | 0.1 | 0 |
Values in parentheses are for the highest-resolution shell.
Rmerge = ∑∑i|Ii – < I > |/∑ < I >, where < I > is the mean intensity of N reflections with intensities Ii and common indices h, k, and I.
Rwork = ∑∑hkl‖Fobs|– kFcal‖/∑hkl |Fobs|, where Fobs and Fcal are the observed and calculated structure factors, respectively.
MiD51Δ1–133 has a compact, globular structure consisting of two α-helical regions separated by a central β strand region (Figure 1B). To find structurally related proteins, we used the DALI server to search the MiD51 structure against structures deposited in the Protein Data Bank (PDB). Proteins belonging to the nucleotidyl transferase (NTase) family had the highest Z scores. Cyclic GMP-AMP synthase (PDB 4K99; Gao et al., 2013) showed the greatest similarity, with a Z score of 27.7 and a root-mean-square deviation (rmsd) of 2.4 Å for backbone atoms (Figure 1C). NTase proteins typically catalyze the polymerization of nucleic acids from triphosphate nucleotides. They bind nucleotide triphosphates in the cleft located between the central β sheet and the C-terminal α-helical segment (Kristiansen et al., 2011). Intriguingly, the corresponding region of MiD51Δ1–133 contained additional electron density, suggesting partial occupancy by a ligand (see Experimental Procedures). Structural alignments show that residues canonically used for nucleotide binding are partially conserved in MiD51. These same residues are more divergent in MiD49 (Figure S1C).
To identify potential MiD51 ligands, we used a fluorescence-based thermal shift assay to screen a broad panel of nucleotides (Figures S2A–S2D). Contrary to what would be expected for a canonical NTase, MiD51Δ1–133 shows no stabilization when incubated with NTPs, except for a modest stabilization with ATP. Instead, MiD51Δ1–133 shows substantial stabilization with ADP (4–6 degrees) and modest stabilization with GDP. No potential ligands were identified for MiD49 (Figure S2E).
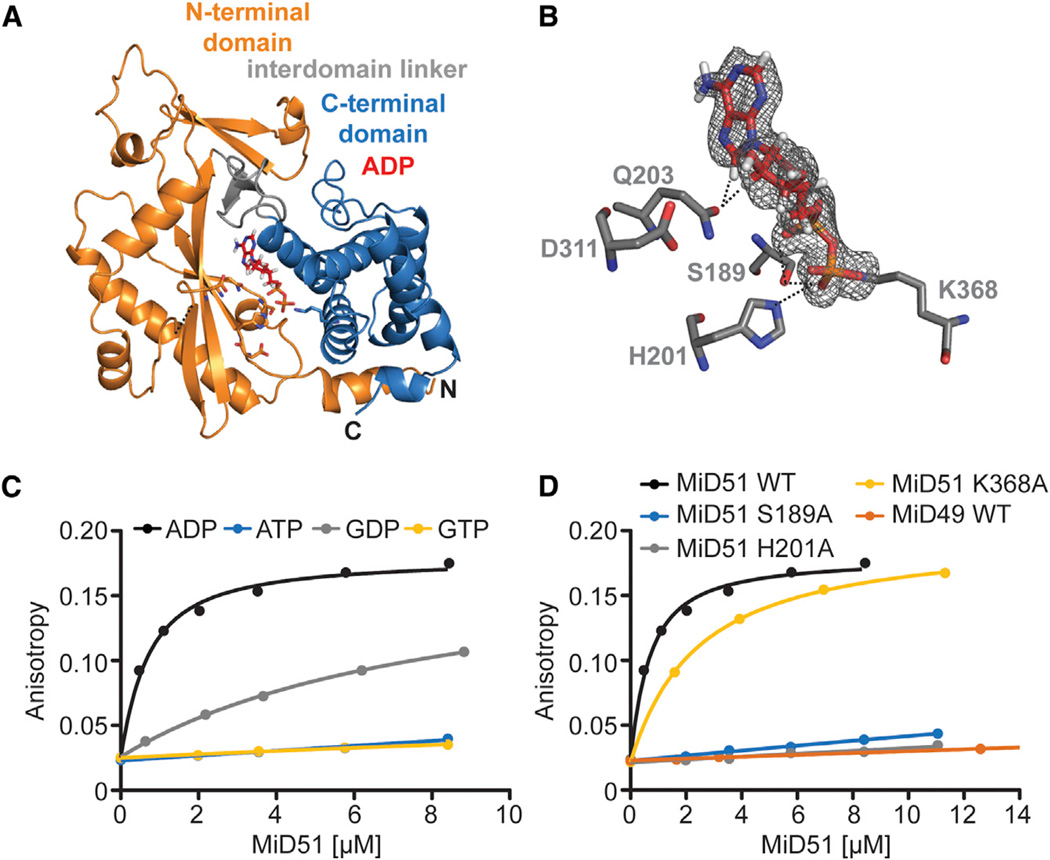
Because MiD51 showed the greatest stabilization with ADP, we performed crystallization trials in the presence of ADP and determined the structure of MiD51Δ1–133 bound to ADP at a resolution of 2.0 Å(Table 1; Figures 2A and 2B). NTases use a triad of negatively charged residues (either Asp or Glu) from the central β sheet to coordinate two Mg2+ ions that buffer the negative charges of the β and γ phosphate groups of NTPs (Kristiansen et al., 2011). Our structure revealed that MiD51 uses a related triad of residues (H201, Q203, and D311) in the central β sheet, but two of these residues (H201 and Q203) directly contact ADP and do not coordinate Mg2+. H201 forms a hydrogen bond with the β-phosphate group of ADP, and Q203 has hydro-phobic interactions with carbons in the ribose and base rings (Figure 2B). Like other NTases, a serine (S189) from the N-terminal domain and a lysine (K368) from the C-terminal domain help to coordinate the negative charge from the phosphate groups of the nucleotide. Specifically, S189 forms hydrogen bonds with both the α- and β-phosphates of ADP, and K368 forms a salt bridge with the β phosphate.
Figure 2. MiD51 Binds ADP with High Affinity.
(A) Ribbon representation of mouse MiD51Δ1–133 bound to ADP.
(B) Details of the nucleotide-binding pocket. The 2 Fo-Fc map for ADP (red) is contoured at 1.2 σ. Key residues are depicted, and interactions with the ADP are denoted with black dashed lines.
(C) Binding of MiD51 to ADP. Equilibrium titrations were performed with MANT-labeled nucleotides to determine the affinity of MiD51Δ1–133 for MANT-ATP (no binding), MANT-GTP (no binding), MANT-ADP (Kd = 0.5 µM ± 0.008 µM) and MANT-GDP (8.0 µM ± 0.9 µM).
(D) Mutational analysis of residues structurally implicated in ADP binding. Note that MiD51 S189A, H201A, and MiD49 do not bind MANT-ADP. MiD51 K368A has a Kd of 2.6 µM ± 0.4 µM.
To measure the binding affinities of MiD51 to nucleotides, we monitored changes in the fluorescence anisotropy of 2′/3′-O-(N-methyl-anthraniloyl) (MANT)-labeled nucleotides upon incubation with MiD51 (Rome et al., 2013). MiD51 has the highest affinity for ADP (Kd of 0.5 µM ± 0.008 µM). We found no detectable binding to ATP, despite its ability to modestly stabilize MiD51 in the thermal shift assay. MiD51 binds guanine-based nucleotides more weakly, but still exhibits a strong preference for GDP (Kd = 8 µM ± 0.9 µM) relative to GTP (not detectable) (Figure 2C). Binding to ADP does not require Mg2+. Mutation of the residues structurally implicated in nucleotide binding (H201, S189, and K368) greatly reduces the affinity of MiD51 for ADP. Mutations H201A and S189A abolish nearly all nucleotide binding, whereas K368A (Kd = 2.6 µM ± 0.4 µM) causes a 5-fold reduction (Figure 2D). Taken together, these results show that MiD51 has a nucleotidyl transferase domain but differs substantially from other proteins with this motif. It uses a variant set of nucleotide binding residues to directly contact ADP rather than using a Mg2+ cofactor to bind a nucleotide triphosphate. MiD49 does not bind ADP (Figure 2D), and we have not identified an alternative ligand. With MiD51, we have not detected nucleotide transfer reactions with ATP or GTP (data not shown), suggesting that although MiD51 has an NTase fold, it may not possess enzymatic activity.
MiD51 Forms a Dimer through Electrostatic Interactions in the N-Terminal α-Helical Segment
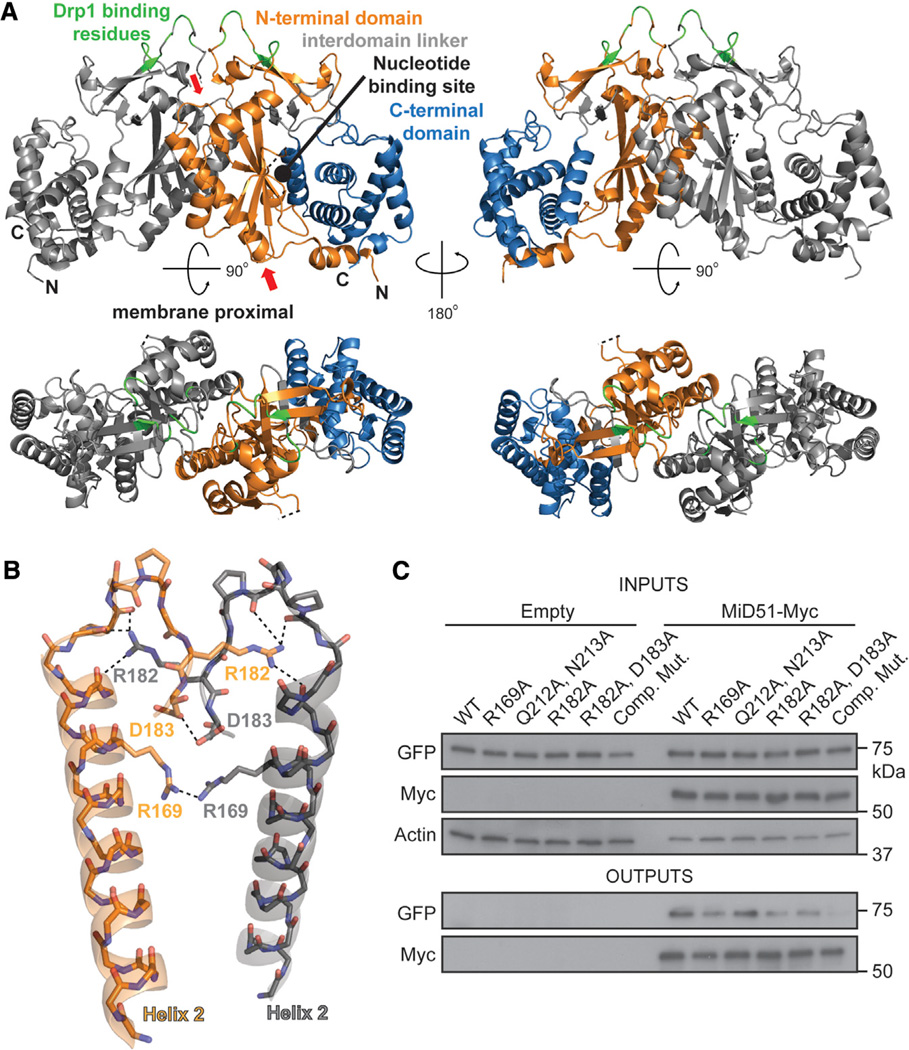
MiD51Δ1–133 crystallizes as a dimer (Figure 3) using the N-terminal α-helical segments from two monomers as an interaction interface. The interface is largely composed of electrostatic interactions between charged residues from a loop, and the interface has a buried surface area of 1,184.1 Å2 (Figure 3B). Additionally, the guanidinium groups from a pair of Arg169 residues from α-helix number 2 adopt a planar stacking conformation (Neves et al., 2012), and their interaction is bridged through hydrogen bonds with a sulfate group. The interaction surface of MiD51 is distinct from that of other NTase family members known to dimerize. To determine the biological relevance of this interface, we designed a series of mutants to disrupt the dimer interface and tested their ability to self-associate in a coimmunoprecipitation assay (Figure 3C). Compared to wild-type MiD51, several mutants showed a moderately reduced ability to self-associate. A mutant (hereafter referred to as the “compound dimer mutant”) combining five substitutions in the dimer interface showed a severe ablation of self-association.
Figure 3. MiD51 Forms a Dimer through Electrostatic Interactions.
(A) MiD51 crystallizes as a dimer through an interface found in the N-terminal domain. The two molecules are differentially colored gray (left) and as in Figure 1B (right). In the bottom figure, the dimer has been rotated to view the interface from the top. The right panels are a 2-fold axis rotation. Red arrows indicate the α helix and loop shown in (B). Drp1 binding residues are colored green.
(B) Detailed view of residues important for the dimer interface. Interactions are indicated with dashed lines. The color scheme is the same as in (A). Electrostatic interactions were determined using the PDBePISA server. R169 self-associates by hydrogen bonding with a sulfate ion (not depicted).
(C) Mutational analysis of dimer formation. MiD51-GFP mutants were cotransfected with either empty vector or Myc tagged MiD51 constructs. For each reaction, the same mutant was used for both the GFP and Myc tagged constructs. Cells were treated with a reversible crosslinker and solubilized before immunoprecipitation against the Myc epitope. Top: expression of MiD51-GFP and MiD51-Myc in cell lysates. Actin is a loading control. Bottom: anti-Myc immunoprecipitates analyzed for MiD51-GFP. Comp Mut, compound dimerization mutant.
MiD51 Does Not Undergo a Significant Structural Change upon ADP Binding or Dimerization
Interestingly, the structures of MiD51 in its apo form (crystallized without added ADP) and in the ADP complex are identical (Figure S3A). This observation suggests that MiD51 does not undergo a conformational change upon ADP binding. However, this interpretation is complicated by the possibility of low levels of ADP being present in our apo MiD51 crystals. To address this issue, we crystallized the MiD51 H201A mutant, which is incapable of nucleotide binding. Upon structure determination, we found that the H201A mutant has no significant structural changes compared to the ADP-bound form of MiD51 (Figures S3B and S3D). Importantly, the H201A mutant had no apparent electron density in the putative nucleotide-binding pocket. These results indicate that, although ADP stabilizes MiD51, it does not induce a conformational change. Moreover, we solved the structure of the compound dimer mutant (Figure S3C) and found that this monomer is identical to the monomers within the dimeric structures of wild-type MiD51 and H201A (Figure S3D).
Identification of a Surface Loop Necessary for Drp1 Binding
We tested whether ADP binding or dimerization of MiD51 is necessary for its recruitment of Drp1 to mitochondria. MiD51 mutants defective for either function are fully active in recruiting Drp1 puncta to mitochondria (Figure S4A). Furthermore, the MiD51 mutants coimmunoprecipitated with Drp1 from cells as efficiently as wild-type (Figure S4B). These results clearly indicate that Drp1 binding is independent of ADP binding or dimer formation.
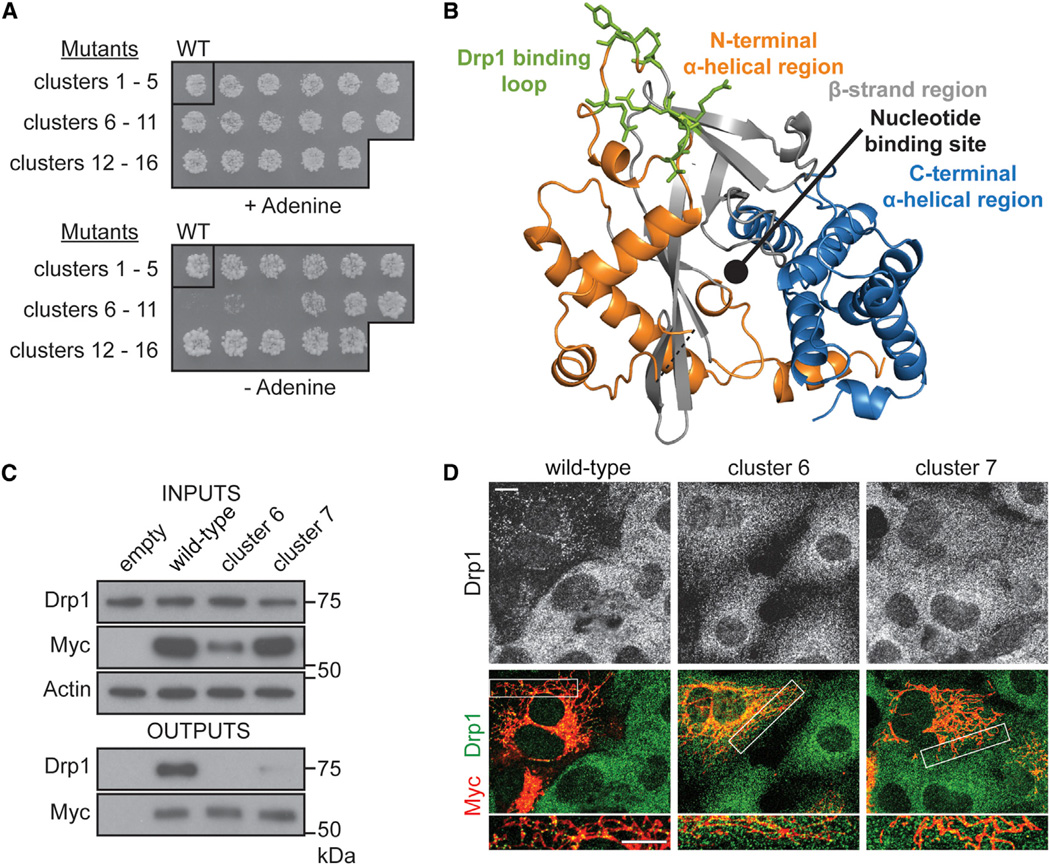
To determine the Drp1 binding surface on MiD51, we designed 16 mutants, each containing a cluster of three to four point mutations, to systematically sample the solvent-exposed surface of MiD51 (Figures S4C and S4D). As assessed by yeast two-hybrid analysis, mutants 6 and 8 show nearly complete loss of Drp1 binding, and mutant 7 shows substantially reduced binding (Figure 4A). Remarkably, all three clusters converge onto a single, exposed loop on the top surface of MiD51 (green residues, Figure 4B). The residues within this loop are well conserved between MiD51 and MiD49 (Figures S1C and S1D). We expressed mutants 6 and 7 in 293T cells and found that neither can coimmunoprecipitate Drp1 (Figure 4C). Mutant 6, but not 7, shows reduced expression. Furthermore, these mutants are defective in recruiting Drp1 puncta to mitochondria, even though they localize properly to mitochondria (Figure 4D).
Figure 4. MiD51 Uses a Surface Loop to Bind Drp1.
(A) A yeast two-hybrid screen to identify MiD51 regions necessary for Drp1 binding. The top panel shows the diploid selection plate, and the bottom panel shows the interaction selection plate. Three mutational clusters (6, 7, and 8) perturb MiD51 binding to Drp1.
(B) The three clusters localize to a loop at the top surface of MiD51. Mutated residues are colored green and depicted as sticks.
(C) Analysis of MiD51-Drp1 binding in 293T cells. Wild-type or mutant MiD51-Myc was coexpressed with mouse Drp1 and Myc-immunoprecipitates were analyzed for Drp1. Top: expression of MiD51-Myc and Drp1 in cell lysates. Actin is a loading control. Bottom: anti-Myc immunoprecipitate analyzed for Drp1 levels. Loading of immunoprecipitates was normalized to Myc levels.
(D) MiD51 cluster mutants 6 and 7 fail to rescue mitochondrial Drp1 recruitment in Fis1/Mff null cells. Drp1 was visualized with anti-Drp1 immunofluorescence, and transfected cells were Myc positive. Fis1/Mff null cells are used because they have severely reduced recruitment of Drp1 to mitochondria, allowing the recruitment activity of MiD51 to be readily assessed (Losón et al., 2013).
Inhibition of Respiration Stimulates Mitochondrial Fission by MiD51
Cells overexpressing MiD51 have severely elongated and interconnected mitochondria (Palmer et al., 2011; Zhao et al., 2011). We previously showed that overexpressed MiD51 recruits Drp1 to the mitochondrial outer membrane in an inactive form (Losón et al., 2013). Treatment of MiD51-overexpressing cells with CCCP (a proton ionophore that dissipates the mitochondrial membrane potential) activates the recruited Drp1 and promotes rapid mitochondrial fragmentation independently of Fis1 or Mff (Losón et al., 2013).
We found that treatment with antimycin A (an inhibitor of complex III of the electron transport chain) also induces rapid mitochondrial fission in MiD51-overexpressing cells (Figures 5A and 5B). This fragmentation occurs independently of Fis1 and Mff, because it occurs in mouse embryonic fibroblasts (MEFs) deleted for Fis1 and Mff (Figures 5C and 5D). Mitochondrial fragmentation induced by antimycin A is specific to MiD51-expressing cells and does not occur with MiD49-expressing cells (Figure 5B). CCCP treatment causes enhanced processing of OPA1 (Ishihara et al., 2006), but antimycin A treatment does not (Figure S5D).
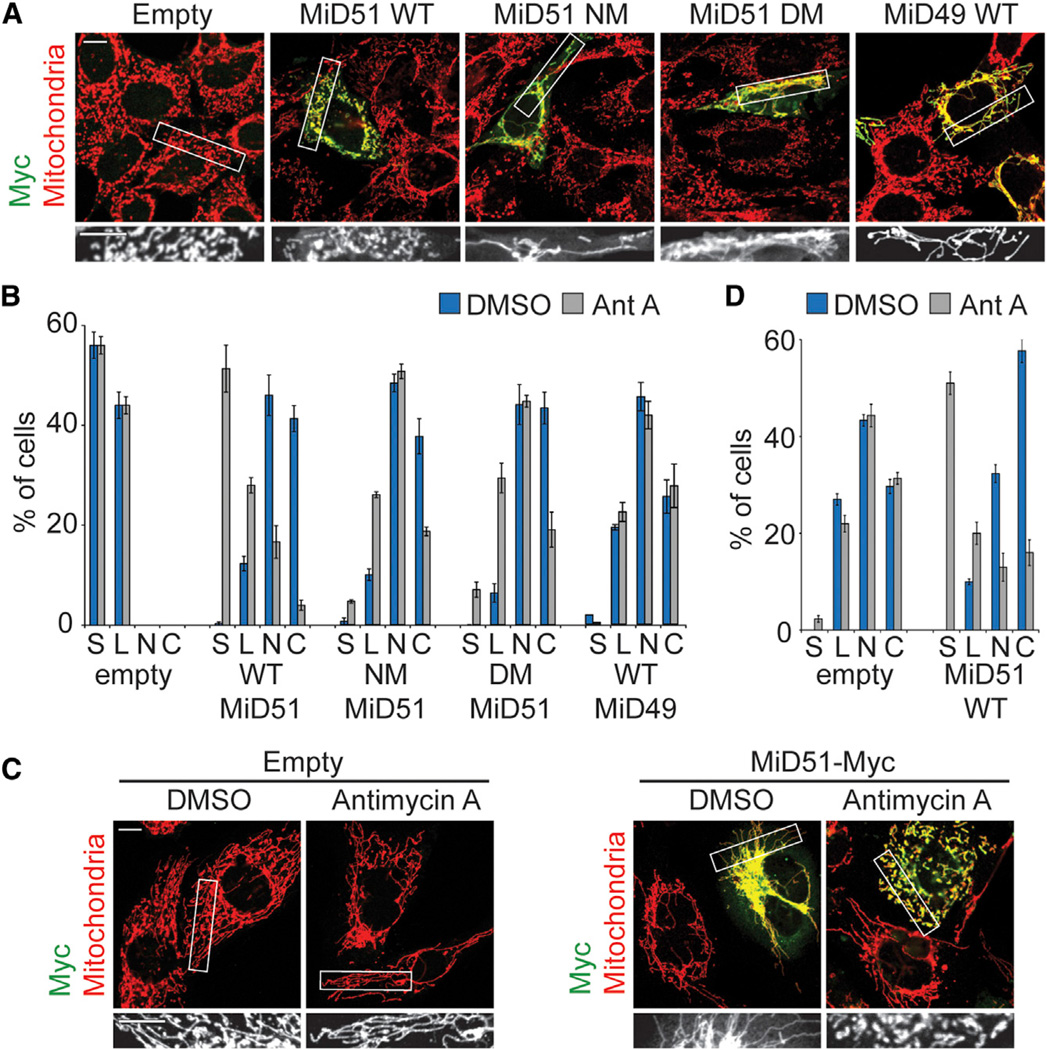
Figure 5. ADP Binding and Dimerization Are Required for the Fission Activity of MiD51.
(A) ADP and dimerization mutants localize to mitochondria but are defective for mitochondrial fission. Wild-type MEFs were transfected with empty vector or MiD51-Myc constructs and treated with antimycin A. NM, nucleotide-binding site mutant (S189A); DM, compound dimer mutant.
(B) Quantification of results in (A). Mitochondrial morphologies were scored as described previously (Losón et al., 2013).
(C) Mitochondrial morphology in Fis1/Mff null MEFs transfected with empty vector or MiD51-Myc and treated with vehicle (DMSO) or antimycin A. Transfected cells were visualized with anti-Myc immunofluorescence, and mitochondria were highlighted with anti-Tom20 immunofluorescence.
(D) Quantification of results in (C).
In (B) and (D), data are averages from three independent experiments ±SD. Mitochondrial morphology scoring: S, short; L, long; N, net-like; C, collapsed. Scale bars, 10 µm. Regions within the white boxes are shown in greater magnification below.
These observations allowed us to use CCCP or antimycin A treatment to assess the mitochondrial fission activity of MiD51 mutants. When expressed in MEFs, MiD51 mutants defective in ADP binding or dimerization promote Drp1 recruitment (Figures S4A and S4B) and robust elongation of mitochondria (Figures S5A–S5C). However, upon addition of CCCP or antimycin A, all the nucleotide-binding mutants show little or no stimulation of mitochondrial fission. For the dimerization mutants, the fission defects correlate well with the dimerization defect. The Q212A/N213A mutant, which has normal dimerization, stimulates fission as well as wild-type. The compound dimer mutant has the most severe fission defect among the dimer mutants, and the rest have intermediate defects. Taken together, these results indicate that MiD51 mitochondrial fission activity, but not Drp1 recruitment, requires ADP binding and dimerization.
ADP Binding to MiD51 Stimulates Drp1 Oligomerization
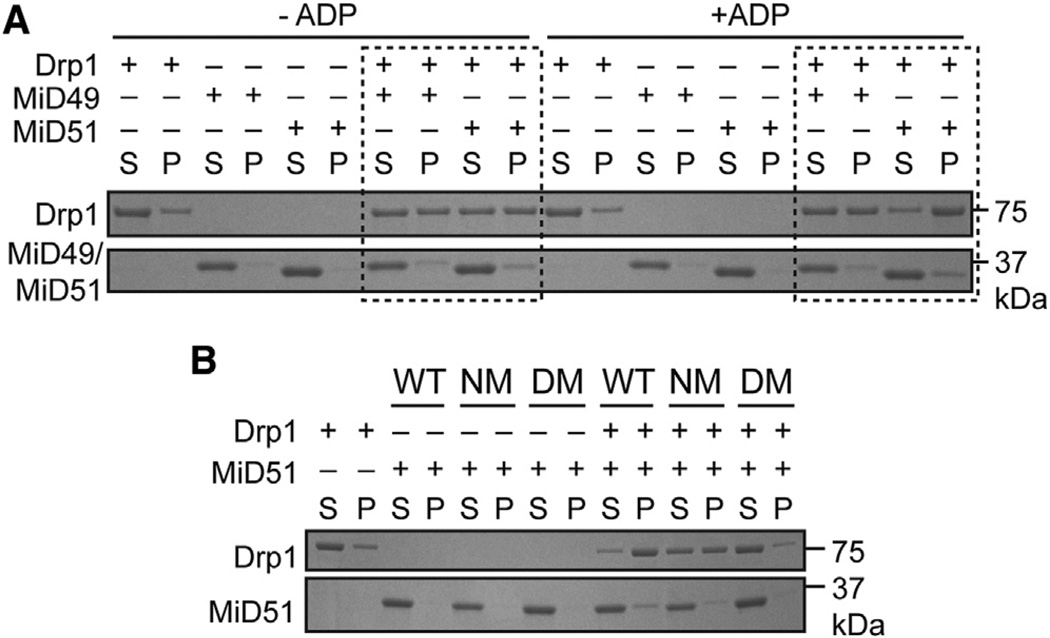
Like dynamin (Ferguson and De Camilli, 2012; Schmid and Frolov, 2011), Drp1 and its yeast homolog dynamin 1 (Dnm1) polymerize into ordered oligomers (Fröhlich et al., 2013; Ingerman et al., 2005; Mears et al., 2011) that have enhanced GTP hydrolysis activity. To test whether MiD49 or MiD51 can regulate Drp1 oligomerization, we performed sedimentation experiments using recombinant Drp1 and either recombinant MiD49 or MiD51. In the absence of ADP, both MiD49Δ1–124 and MiD51Δ1–133 moderately stimulate Drp1 sedimentation in the presence of GTPγS (a nonhydrolyzable form of GTP). Remarkably, MiD51Δ1–133, but not MiD49Δ1–124, further stimulates Drp1 sedimentation upon addition of ADP (Figure 6A). In contrast, the MiD51 nucleotide-binding mutant S189A is incapable of enhancing Drp1 sedimentation upon addition of ADP. The compound dimer mutant also does not promote any sedimentation of Drp1 in the presence of ADP (Figure 6B). These results demonstrate that both MiD49 and MiD51 can basally promote Drp1 oligomerization. This function of MiD51 requires dimerization and is enhanced by ADP binding.
Figure 6. ADP Promotes Drp1 Oligomerization by MiD51.
(A) Effect of MiD51 and ADP on Drp1 sedimentation. Recombinant mouse MiD49Δ1–124 or MiD51Δ1–133 was incubated with recombinant mouse Drp1 in the presence of GTPγS with or without ADP. Reactions were incubated for 30 min, and Drp1 oligomers were sedimented at 100,000 × g. Equivalent volumes of the supernatant (S) and pellet (P) were loaded and resolved by SDS-PAGE. Dashed boxes indicate key comparison groups.
(B) Analysis of MiD51 mutants on Drp1 sedimentation. The MiD51 nucleotide binding mutant (NM, S189A) and the compound dimer mutant (DM) were assessed for their ability to facilitate Drp1 sedimentation in the presence of GTPγS and ADP.
Binding of ADP to MiD51 Stimulates Assembly of Drp1 Tubules and GTP Hydrolysis
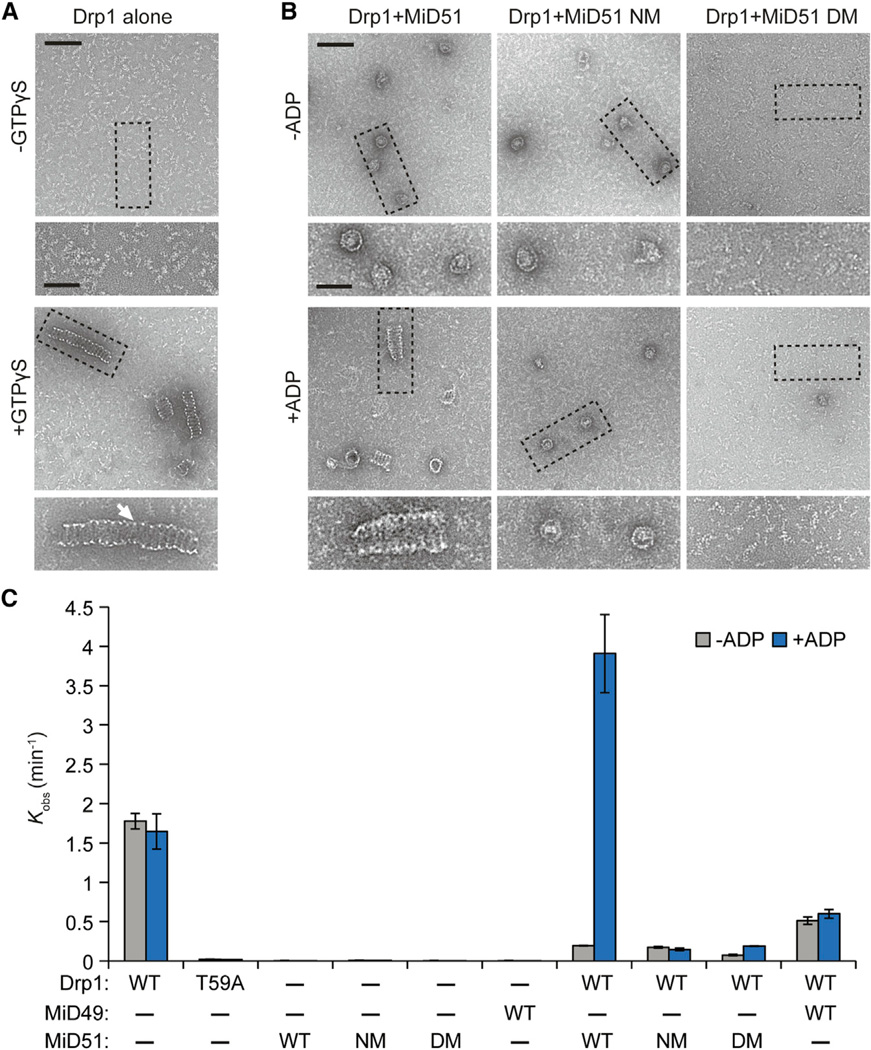
We used negative stain transmission electron microscopy to examine the effects of MiD51 on Drp1 oligomeric structures at high resolution. In the presence of GTPγS, recombinant Drp1 forms tubules with a repeating spiral pattern (Figure 7A). Interestingly, in samples incubated with MiD51 in the absence of ADP, these tubules are replaced with short, ring-like structures (Figure 7B). In the presence of both MiD51 and ADP, Drp1 forms spiral tubules that are slightly wider than those formed from Drp1 alone. The nucleotide-binding mutant S189A is completely defective in stimulation of tubule formation by ADP. Moreover, the compound dimerization mutant prevents any Drp1 assembly with or without ADP (Figure 7B). Our ultrastructural analysis demonstrates that MiD51 without ADP inhibits formation of Drp1 tubules and promotes a different form of Drp1 assembly. ADP relieves this inhibition and stimulates Drp1 tubule formation in a dimerization-dependent manner.
Figure 7. MiD51 and ADP Promote Drp1 Spiral Assembly and GTP Hydrolysis.
(A and B) Negative stain transmission electron microscopy of Drp1 oligomers. Images are representative of three independent experiments. (A) Recombinant mouse Drp1 forms regular spiral-tubular structures in the presence of a nonhydrolyzable analog of GTP, GTPγS. White arrow highlights the thin edge of spiral-tubular structures. (B) Effects on Drp1 spiral-tubular formation upon addition of MiD51 proteins with and without ADP. GTPγS was present in all reactions. Dashed boxes indicate inset boundaries. Scale bar, 100 nm; inset scale bar, 50 nm.
(C) Effect of MiD51 and ADP on GTP hydrolysis by Drp1. Initial GTP hydrolysis rates were measured with the indicated proteins, with or without ADP. Reactions were performed at saturating GTP (1 mM) with 150 mM NaCl. NM, nucleotide-binding site mutant (S189A); DM, compound dimer mutant. T59A is a Drp1 catalytic mutant. Results are the average of three independent experiments ±SD.
We further determined how the structural states of Drp1 correlate with its GTP hydrolysis activity (Figure 7C). Under physiological salt concentration and saturating GTP (1 mM), recombinant Drp1 hydrolyzes approximately 1.5 molecules of GTP per minute. Like other dynamins (Praefcke and McMahon, 2004), mutation of a putative catalytic threonine (T59) abolishes GTP hydrolysis (Figure 7C). Incubation of Drp1 with MiD51Δ1–133 causes a dramatic suppression of GTP hydrolysis. Remarkably, addition of ADP to the reaction relieves this inhibition, resulting in a 20-fold stimulation of Drp1 GTP hydrolysis between the ADP-free and the ADP-containing reactions. The S189A and compound dimer mutants similarly inhibited GTP hydrolysis in the absence of ADP, but addition of ADP had no stimulatory effect. Analogously, MiD49Δ1–124 inhibited the GTPase activity of Drp1 and did not respond to ADP. Thus, both apo-MiD49 and apo-MiD51 inhibit the GTPase activity of Drp1, but in the case of MiD51, ADP binding relieves this inhibition and promotes GTP hydrolysis.
DISCUSSION
Our results indicate that MiD51 has a variant nucleotidyl transferase domain that binds ADP instead of a nucleotide triphosphate. A previous bioinformatics study using sensitive protein-fold recognition algorithms greatly expanded the number of putative nucleotidyl transferase family members, suggesting that this family may have diverse functions in the cell. This study also suggested that MiD49 and MiD51 may be distant members of the NTase family (Kuchta et al., 2009). Our results raise the possibility that some noncanonical members of the NTase family may not catalyze nucleotide transfer but instead use metabolites as cofactors. Based on their sequence similarity, MiD49 is expected to be structurally similar to MiD51. MiD49 has also been shown to stimulate Drp1 assembly (Koirala et al., 2013). However, it does not bind ADP and shows only partial conservation in the key residues used by MiD51 for nucleotide binding (Figure S1C). In future studies, it will be important to determine whether MiD49 indeed binds another cofactor.
Our results identify ADP as an essential cofactor for MiD51. In the absence of ADP binding, MiD51 can still recruit Drp1 via a binding loop we have identified by mutational analysis.In dimeric MiD51, the adjacent binding loops form an extended interface on the top surface, which would be expected to protrude furthest from the mitochondrial outer membrane (Figure 3A). Although capable of recruiting Drp1, nucleotide-binding mutants of MiD51 are unable to activate mitochondrial fission in cells. In vitro, apo-MiD51 strongly suppresses the assembly of Drp1 into spirals and its GTP hydrolysis activity. Upon addition of ADP, suppression of both activities is relieved. Drp1 spirals are tailored to the diameter of mitochondrial tubules and are thought to wrap around mitochondria to mediate constriction (Ingerman et al., 2005). Drp1 spirals also have enhanced GTP hydrolysis, which is critical for mitochondrial fission (Mears et al., 2011).
Rather than inducing a conformational change, ADP appears to act as a structural cofactor that stabilizes the folding of MiD51. Our results favor a model in which ADP binding is necessary to stabilize MiD51 so that Drp1 can assemble properly. Dimerization of MiD51 is also necessary for mitochondrial fission, and further work will be necessary to understand whether there is a relationship between ADP binding and dimerization. Because of the unexpected role for ADP, it will also be interesting to explore the possibility that cellular metabolism, by regulating ADP levels, can impact MiD51 function.
EXPERIMENTAL PROCEDURES
Materials
Antibody sources included Drp1 (BD Biosciences), Tom20 (Santa Cruz), MiD51 (also known as SMCR7L; Thermo Pierce), Actin (Millipore), Myc (mouse monoclonal 9E10 from Covance, and rabbit polyclonal from Sigma-Aldrich), and OPA1 (in-house mouse monoclonal). CCCP (Sigma-Aldrich) was used at 50 µM. Antimycin A (Sigma-Aldrich) was used at 10 µM in cell culture experiments. Cells were grown in LabTek chambered glass slides (Nunc) for fixed cell imaging. Dithiobis (succinimidyl propionate) (DSP) was purchased from Thermo-Pierce. MANT-labeled nucleotides were from Jena Biosciences.
Recombinant Protein Production, Purification, and Crystallization
Recombinant mouse MiD49Δ1–51, MiD49Δ1–124, MiD51Δ1–48, MiD51Δ1–133, and Drp1 variant 2 were produced in Rosetta (DE3) BL21 E. coli (Invitrogen). In a typical purification, 1 I of terrific broth containing 100 µg/ml ampicillin and 50 µg/ml chloramphenicol was grown at 37°C to an optical density 600 (OD600) of 1.5. Cultures were cooled on ice for 30 min and induced with 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) overnight at 16°C. The cells were harvested and stored at −80°C. A total of 10 g of cells expressing MiD proteins were lysed in 50 ml glutathione S-transferase (GST) buffer (50 mM HEPES, 300 mM NaCl, 10% glycerol, and 2 mM dithio-threitol [DTT] [pH 7.4]) using sonication. A total of 10 g of cells expressing Drp1 were lysed in 50 ml Hisx6 buffer (50 mM KH2PO4, 300 mM NaCl, 5 mM imidazole, and 10% glycerol [pH 8.0]) using sonication. Lysates were cleared by centrifugation at 43,000 × g for 30 min at 4°C. GST-tagged MiD49 and MiD51 proteins were captured with glutathione Sepharose (GE Healthcare) and washed with GST buffer. Hisx6-tagged Drp1 was captured with Ni-NTA Sepharose (QIAGEN) and washed with Hisx6 wash buffer (20 mM Tris, 300 mM NaCl, 40 mM imidazole, 10% sucrose, 10% glycerol, pH 8.0). Beads for MiD49 and MiD51 protein purification were exchanged into protease buffer (50 mM HEPES, 150 mM NaCl, and 2 mM DTT [pH 7.4]). PreScission Protease (80 units, GE Healthcare) was incubated for 20 hr at 4°C with continuous end-over-end mixing. The eluted protein was further purified by size exclusion on a Hi-Load Superdex 200 16/ 60 column (GE Healthcare) pre-equilibrated with GST column buffer (50 mM HEPES, 150 mM NaCl, and 2 mM DTT [pH 7.4]) and driven by an AKTA Purifier (Amersham). Eluted Drp1 protein was further purified by size exclusion on a Hi-load Superdex 200 16/60 column pre-equilibrated with Hisx6 column buffer (10 mM Tris, 100 mM NaCl, 2 mM DTT, and 1 mM EDTA [pH 8.0]) and driven by an AKTA Purifier. Drp1 protein was additionally purified by binding to a Hi-Load HiTrapQ column (GE Healthcare) pre-equilibrated with Hisx6 column buffer and driven by an AKTA Purifier. The protein was then eluted with a NaCl gradient. Peak fractions were collected and concentrated to approximately 1 mM for MiD49 and MiD51 proteins and 0.5 mM for Drp1 proteins using Amicon Ultra-15 concentrators (Millipore) with a molecular weight cutoff of 30 kDa. Proteins were flash-frozen in liquid nitrogen and stored at −80°C.
Selenomethionine-labeled MiD51Δ1–133 was produced by growing cells in M9 minimal media at 37°C to OD600 of 1.0 and then incubating with amino acids (lysine, phenylalanine, and threonine at 100 mg/l; isoleucine, leucine, and valine at 50 mg/l; and selenomethionine at 60 mg/l) for 15 min, cooling cells on ice for 30 min, and finally inducing with 1 mM IPTG overnight at 16°C. Protein purification was carried out as described above.
Limited proteolysis of MiD51Δ1–48 was performed with 1 µg/µl recombinant protein and 0.001 µg/µl trypsin (Promega) at 4°C. Time points were taken by diluting aliquots in Laemmli buffer (25 mM Tris, 10% glycerol, 1% SDS, 0.01% bromophenol blue, and 2% β-mercaptoethanol [pH 6.8]) and boiling samples immediately.
Crystallization, Data Collection, and Structure Determination
Crystallization trials by hanging drop-vapor diffusion at room temperature identified a condition (100 mM HEPES [pH 7.0], 200 mM LiSO4, 20% polyethylene glycol (PEG) 3,350 [w/v]] that yielded rod-shaped crystals for wild-type, selenomethionine-substituted, and H201A mutant proteins. The compound dimer mutant crystallized in a differential condition [200 mM NaH2PO4, 20% PEG 3,350 (w/v)]. Diffraction data were collected from vitrified crystals on beamline 12-2 at the Stanford Synchrotron Radiation Light-source. All data were processed with XDS (Kabsch, 2010) and merged using SCALA (Evans, 2006) as implemented in CCP4 (Bailey, 1994). Selenomethionine-substituted MiD51Δ1–133 was used for phasing. Using intensity data at 2.6 Å from three wavelengths, all four selenium sites were located by PHENIX (Adams et al., 2002). After solvent flattening and density modification in PHENIX, the map revealed clear density for the protein. Manual model building in COOT (Emsley and Cowtan, 2004) using the 2.6 Å experimental map generated a starting model. Refinement was carried out using PHENIX, with an initial round of rigid body refinement followed by a round of simulated annealing. The selenomethionine-substituted crystal is nonisomorphic to the native crystal. However, molecular replacement in PHENIX with the 2.2 Å native data set produced an excellent map, and refinement produced density for most of the side chains. After a few rounds of manual model building and refinement with translation/libration/screw obtained from the TLSMD server (Painter and Merritt, 2006), the Rwork converged to 21.9% and the Rfree to 27.4% for the apo structure. For the ADP-bound structure, Rwork was 17.8% and Rfree was 22.5%. Rwork was 17.7% and Rfree was 21.9% for the H201A mutant structure, and Rwork was 17.7% and Rfree was 21.8% for the compound dimer mutant structure. After refinement of the native model, significant density was present in the binding cleft. ADP was fitted into this density, and the final model and refinement statistics were produced with ADP in the binding cleft. With the exception of a stretch of three prolines (residues 290–291), the final models include residues 134–463 of MiD51 and have excellent stereochemistry with no/few Ramachandran outliers, as assessed by MOLPROBITY (Davis et al., 2007). To determine the structure of the MiD51-ADP complex, MiD51Δ1–133 was mixed with 10 mM ADP and 5 mM MgCl2 and incubated on ice for 1 hr before setting crystallization trials. Molecular replacement in PHENIX for a 2.0 Å MiD51-ADP complex data set produced an excellent map, and refinement produced clear density for the nucleoside and phosphate groups of ADP.
Fluorescence Anisotropy and Determination of Kd Values
Fluorescence measurements were performed using a Fluorolog-3–22 spectro-fluorometer (Jobin Yvon) at 25°C (50 mM HEPES, 150 mM NaCl, 10 mM MgCl2, and 2 mM DTT [pH 7.5]). Samples were excited at 355 nm, and fluorescence emission was monitored at 448 nm. For titrations, MANT-nucleotide was held constant at 300 nM and the protein concentration was varied as indicated. To determine Kd values, observed anisotropy values (Aobsd) were plotted as a function of MiD concentration and fit to Equation 1
| (Equation 1) |
in which A0 is the anisotropy value of free MANT-nucleotide, A1 is the anisotropy when MANT-nucleotide is bound to protein, co is the concentration of total MANT-nucleotide, and Kd is the equilibrium dissociation constant of MiD for MANT-nucleotide.
Immunofluorescence and Imaging
For immunofluorescence, cells were fixed in 4% formaldehyde for 10 min at 37°C, permeabilized with 0.1% Triton X-100 at room temperature, and incubated with antibodies in 5% fetal calf serum in PBS. Bound antibody was visualized with Alex Fluor-conjugated secondary antibodies (Life Technologies). Scoring of mitochondrial network morphology was performed blind to genotype and treatment. All quantifications were done in triplicate, and 100 cells were scored per experiment.
All fluorescence imaging was performed using a Plan-Apochromat 63×/1.4 oil objective on a Zeiss LSM 710 confocal microscope driven by Zen 2009 software (Carl Zeiss). Image cropping and global adjustments to brightness and contrast were performed using Photoshop (Adobe).
Cell Culture
All cell lines were cultured in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum and supplemented with 100 U/ml penicillin and 100 µg/ml streptomycin. Treatment with 50 µM CCCP occurred for 1 hr, and treatment with 10 µM antimycin A occurred for 2 hr.
Cloning and Transfection
MiD49, MiD51, and Drp1 variant 2 transcripts were amplified from an MEF cDNA library using PCR. MiD49 and MiD51 were cloned into the XhoI and BamHI sites of a pcDNA3.1(–) plasmid containing a C-terminal 4xMyc tag and were also cloned into the XhoI and BamHI sites of pEGFP-N2 (Invitrogen) for C-terminal GFP tagging. For the generation of an MiD51-Myc constitutively expressing cell line, MiD51 was cloned into the NotI and EcoRI sites of the retroviral vector pQCXIP (Invitrogen). The entire open reading frames were confirmed by DNA sequencing. For recombinant protein expression in bacteria, mouse MiD49Δ1–51, MiD49Δ1–124, MiD51Δ1–48, and MiD51Δ1–133 were cloned into the BamHI and XhoI sites of plasmid pGEX6P1 (GE Healthcare). Mouse Drp1 variant 2 was cloned into the NdeI and BamHI sites of a modified pET21b vector (Novagen). All mutants for MiD51 and Drp1 were constructed by PCR using oligonucleotides encoding mutations.
Plasmids were transfected using Lipofectamine 2000 (Invitrogen). Cells transfected with plasmids were assessed 24 hr posttransfection. MiD49/51-Myc-positive cells were visualized with Myc immunofluorescence and mitochondria were visualized with Tom20 immunofluorescence.
Immunoprecipitation
To assess MiD51 dimerization or MiD51 interaction with Drp1, mouse MiD51-Myc was cotransfected with either mouse MiD51-GFP or mouse Drp1 into 293T cells growing in 35 mm plates. Then 24 hr posttransfection, cells were trypsinized, washed once with PBS, and crosslinked with 250 µM DSP in PBS for 20 min at room temperature. Crosslinker was quenched by the addition of Tris (pH 7.4) to a 100 mM final concentration, and cells were washed once with PBS containing 100 mM Tris (pH 7.4). Cells were lysed in immunoprecipitation (IP) buffer (1% Triton X-100, 150 mM NaCl, 25 mM Tris-HCl, and 1 mM EDTA [pH 7.4]) and lysates were cleared with a 21,000 × g spin at 4°C for 10 min. Immunoprecipitations were performed in IP buffer, and immune complexes were captured with protein A/G agarose (Thermo-Pierce). Beads were washed with IP buffer and cross-linker reversed by boiling samples in Laemmli buffer containing 5% β-mercaptoethanol.
Drp1 Sedimentation Assay
A total of 5 µM Drp1 was incubated in sedimentation assay buffer (10 mM HEPES, 110 mM NaCl, 2 mM MgCl2, and 1 mM GTPγS [pH 7.0]) for 30 min at 25°C. Where indicated, MiD49 or MiD51 protein was added to 10 µM and ADP to 100 µM. The reactions were centrifuged at 100,000 × g (TLA100.3 rotor) in a Beckman Optima MAX Ultracentrifuge for 20 min at 4°C. Supernatants were removed, and pellet fractions were resuspended in an equal volume of buffer. Supernatant and pellet fractions were resolved on gradient (4%-20%) SDS/PAGE gels and stained with Coomassie brilliant blue dye.
GTPase Assay
All reactions were performed in GTPase assay buffer (10 mM HEPES, 150 mM NaCl, 2 mM MgCl2, and 1 mM GTP [pH 7.0]) at 37°C with 50 nM [γ-32P]-GTP (MP Biomedicals). Drp1 proteins were used at 3 µM, and MiD49 and MiD51 proteins were used at 15 µM. Where indicated, ADP was added to 100 µM. Reactions were quenched in 0.75 M potassium phosphate (pH 3.3), resolved by polyethylenimine cellulose thin-layer chromatography in 1 M formic acid/0.5 M LiCl, and quantified by autoradiography. Initial rates were derived from a linear fit to the initial stage of reactions, in which <40% [γ-32P]-GTP had been hydrolyzed.
Electron Microscopy
For negative-stain electron microscopy (EM), carbon-coated copper grids were glow discharged for 60 s. A total of 3 µl of sample was added to the surface and then blotted and stained with 2% uranyl acetate. Images were acquired using an FEI Tecnai T12 electron microscope equipped with a LaB6 filament and operated at 120 kV. Magnifications of 15,000× and 42,000× were recorded on a Gatan CCD. Samples for EM were prepared by incubating 2.5 µM Drp1 alone or with 12.5 µM MiD51 in reaction buffer (10 mM HEPES, 150 mM NaCl, 2 mM MgCl2, 1 mM GTPγS, pH 7.0) for 2 hr at 25°C. Where indicated, ADP was added to 100 µM. Samples were diluted 5-fold in reaction buffer before application to grids.
Thermal Shift Assay
Sypro Orange (Sigma-Aldrich) was used at 1× to report protein unfolding. Fluorescence measurements were taken in a Bio-Rad CFX96 thermal cycler using the FRET mode and performed in assay buffer (50 mM HEPES, 150 mM NaCl, 1 mM DTT, and 2 mM MgCl2 [pH 7.5]). MiD49Δ1–51 and MiD51Δ1–133 were used at 1 µM.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Meera Rao for assistance with the GTPase assay and Alasdair McDowell for guidance with EM. We thank the Beckman Foundation at Caltech for support of the EM resource and the Gordon and Betty Moore Foundation and Augoron Institute for support of the Grant Jensen lab microscopy center. We acknowledge the Gordon and Betty Moore Foundation, the Beckman Institute, and the Sanofi-Aventis Bioengineering Research Program at Caltech for their generous support of the Molecular Observatory at Caltech. Operations at SSRL are supported by the US Department of Energy and the National Institutes of Health (NIH). This work was supported by a grant from the NIH (GM062967). O.C.L. was supported by an R. L. Kirschstein National Research Service Award (5F31GM089327) and an American Physiological Society William Townsend Porter predoctoral fellowship.
Footnotes
ACCESSION NUMBERS
The Protein Data Bank accession numbers for the crystal structures reported in this paper are 4OAF (native structure), 4OAG (bound to ADP), 4OAH (H201A mutant structure), and 4OAI (dimer mutant structure).
SUPPLEMENTAL INFORMATION
Supplemental Information includes five figures and can be found with this article online at http://dx.doi.org/10.1016/j.str.2014.01.001.
REFERENCES
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- Bailey S Collaborative Computational Project. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994 Number;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Bleazard W, McCaffery JM, King EJ, Bale S, Mozdy A, Tieu Q, Nunnari J, Shaw JM. The dynamin-related GTPase Dnm1 regulates mitochondrial fission in yeast. Nat. Cell Biol. 1999;1:298–304. doi: 10.1038/13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan DC. Fusion and fission: interlinked processes critical for mito-chondrial health. Annu. Rev. Genet. 2012;46:265–287. doi: 10.1146/annurev-genet-110410-132529. [DOI] [PubMed] [Google Scholar]
- Chen H, Detmer SA, Ewald AJ, Griffin EE, Fraser SE, Chan DC. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J. Cell Biol. 2003;160:189–200. doi: 10.1083/jcb.200211046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, 3rd, Snoeyink J, Richardson JS, Richardson DC. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. (Web Server issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Evans P. Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- Ferguson SM, De Camilli P. Dynamin, a membrane-remodelling GTPase. Nat. Rev. Mol. Cell Biol. 2012;13:75–88. doi: 10.1038/nrm3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fröhlich C, Grabiger S, Schwefel D, Faelber K, Rosenbaum E, Mears J, Rocks O, Daumke O. Structural insights into oligomerization and mitochondrial remodelling of dynamin 1-like protein. EMBO J. 2013;32:1280–1292. doi: 10.1038/emboj.2013.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandre-Babbe S, van der, Bliek AM. The novel tail-anchored membrane protein Mff controls mitochondrial and peroxisomal fission in mammalian cells. Mol. Biol. Cell. 2008;19:2402–2412. doi: 10.1091/mbc.E07-12-1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P, Ascano M, Wu Y, Barchet W, Gaffney BL, Zillinger T, Serganov AA, Liu Y, Jones RA, Hartmann G, et al. Cyclic [G(2′,5′)pA(3′,5′)p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase. Cell. 2013;153:1094–1107. doi: 10.1016/j.cell.2013.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingerman E, Perkins EM, Marino M, Mears JA, McCaffery JM, Hinshaw JE, Nunnari J. Dnm1 forms spirals that are structurally tailored to fit mitochondria. J. Cell Biol. 2005;170:1021–1027. doi: 10.1083/jcb.200506078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara N, Fujita Y, Oka T, Mihara K. Regulation of mitochondrial morphology through proteolytic cleavage of OPA1. EMBO J. 2006;25:2966–2977. doi: 10.1038/sj.emboj.7601184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara N, Nomura M, Jofuku A, Kato H, Suzuki SO, Masuda K, Otera H, Nakanishi Y, Nonaka I, Goto Y, et al. Mitochondrial fission factor Drp1 is essential for embryonic development and synapse formation in mice. Nat. Cell Biol. 2009;11:958–966. doi: 10.1038/ncb1907. [DOI] [PubMed] [Google Scholar]
- Kabsch W. Xds. Acta Crystallogr. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koirala S, Guo Q, Kalia R, Bui HT, Eckert DM, Frost A, Shaw JM. Interchangeable adaptors regulate mitochondrial dynamin assembly for membrane scission. Proc. Natl. Acad. Sci. USA. 2013;110:E1342–E1351. doi: 10.1073/pnas.1300855110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristiansen H, Gad HH, Eskildsen-Larsen S, Despres P, Hartmann R. The oligoadenylate synthetase family: an ancient protein family with multiple antiviral activities. J Interferon Cytokine Res. 2011;31:41–47. doi: 10.1089/jir.2010.0107. [DOI] [PubMed] [Google Scholar]
- Kuchta K, Knizewski L, Wyrwicz LS, Rychlewski L, Ginalski K. Comprehensive classification of nucleotidyltransferase fold proteins: identification of novel families and their representatives in human. Nucleic Acids Res. 2009;37:7701–7714. doi: 10.1093/nar/gkp854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labrousse AM, Zappaterra MD, Rube DA, van der Bliek AM. C. elegans dynamin-related protein DRP-1 controls severing of the mitochondrial outer membrane. Mol. Cell. 1999;4:815–826. doi: 10.1016/s1097-2765(00)80391-3. [DOI] [PubMed] [Google Scholar]
- Liu T, Yu R, Jin SB, Han L, Lendahl U, Zhao J, Nistér M. The mitochondrial elongation factors MIEF1 and MIEF2 exert partially distinct functions in mitochondrial dynamics. Exp. Cell Res. 2013;319:2893–2904. doi: 10.1016/j.yexcr.2013.07.010. [DOI] [PubMed] [Google Scholar]
- Losón OC, Song ZY, Chen HC, Chan DC. Fis1, Mff, MiD49, and MiD51 mediate Drp1 recruitment in mitochondrial fission. Mol. Biol. Cell. 2013;24:659–667. doi: 10.1091/mbc.E12-10-0721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mears JA, Lackner LL, Fang S, Ingerman E, Nunnari J, Hinshaw JE. Conformational changes in Dnm1 support a contractile mechanism for mitochondrial fission. Nat. Struct. Mol. Biol. 2011;18:20–26. doi: 10.1038/nsmb.1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neves MA, Yeager M, Abagyan R. Unusual arginine formations in protein function and assembly: rings, strings, and stacks. J. Phys. Chem. B. 2012;116:7006–7013. doi: 10.1021/jp3009699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otera H, Wang C, Cleland MM, Setoguchi K, Yokota S, Youle RJ, Mihara K. Mff is an essential factor for mitochondrial recruitment of Drp1 during mitochondrial fission in mammalian cells. J. Cell Biol. 2010;191:1141–1158. doi: 10.1083/jcb.201007152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Painter J, Merritt EA. TLSMD web server for the generation of multi-group TLS models. J. Appl. Cryst. 2006;39:109–111. [Google Scholar]
- Palmer CS, Osellame LD, Laine D, Koutsopoulos OS, Frazier AE, Ryan MT. MiD49 and MiD51, new components of the mitochondrial fission machinery. EMBO Rep. 2011;12:565–573. doi: 10.1038/embor.2011.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer CS, Elgass KD, Parton RG, Osellame LD, Stojanovski D, Ryan MT. Adaptor proteins MiD49 and MiD51 can act independently of Mff and Fis1 in Drp1 recruitment and are specific for mitochondrial fission. J. Biol. Chem. 2013;288:27584–27593. doi: 10.1074/jbc.M113.479873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praefcke GJ, McMahon HT. The dynamin superfamily: universal membrane tubulation and fission molecules? Nat. Rev. Mol. Cell Biol. 2004;5:133–147. doi: 10.1038/nrm1313. [DOI] [PubMed] [Google Scholar]
- Rome ME, Rao M, Clemons WM, Shan SO. Precise timing of ATPase activation drives targeting of tail-anchored proteins. Proc. Natl. Acad. Sci. USA. 2013;110:7666–7671. doi: 10.1073/pnas.1222054110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid SL, Frolov VA. Dynamin: functional design of a membrane fission catalyst. Annu. Rev. Cell Dev. Biol. 2011;27:79–105. doi: 10.1146/annurev-cellbio-100109-104016. [DOI] [PubMed] [Google Scholar]
- Sesaki H, Jensen RE. Division versus fusion: Dnm1p and Fzo1p antagonistically regulate mitochondrial shape. J. Cell Biol. 1999;147:699–706. doi: 10.1083/jcb.147.4.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova E, Griparic L, Shurland DL, van der Bliek AM. Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol. Biol. Cell. 2001;12:2245–2256. doi: 10.1091/mbc.12.8.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojanovski D, Koutsopoulos OS, Okamoto K, Ryan MT. Levels of human Fis1 at the mitochondrial outer membrane regulate mitochondrial morphology. J. Cell Sci. 2004;117:1201–1210. doi: 10.1242/jcs.01058. [DOI] [PubMed] [Google Scholar]
- Wakabayashi J, Zhang Z, Wakabayashi N, Tamura Y, Fukaya M, Kensler TW, Iijima M, Sesaki H. The dynamin-related GTPase Drp1 is required for embryonic and brain development in mice. J. Cell Biol. 2009;186:805–816. doi: 10.1083/jcb.200903065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann B. Mitochondrial fusion and fission in cell life and death. Nat. Rev. Mol. Cell Biol. 2010;11:872–884. doi: 10.1038/nrm3013. [DOI] [PubMed] [Google Scholar]
- Yoon Y, Krueger EW, Oswald BJ, McNiven MA. The mitochondrial protein hFis1 regulates mitochondrial fission inmammalian cells through an interaction with the dynamin-like protein DLP1. Mol. Cell. Biol. 2003;23:5409–5420. doi: 10.1128/MCB.23.15.5409-5420.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youle RJ, van der Bliek AM. Mitochondrial fission, fusion, and stress. Science. 2012;337:1062–1065. doi: 10.1126/science.1219855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu T, Fox RJ, Burwell LS, Yoon Y. Regulation of mitochondrial fission and apoptosis by the mitochondrial outer membrane protein hFis1. J. Cell Sci. 2005;118:4141–4151. doi: 10.1242/jcs.02537. [DOI] [PubMed] [Google Scholar]
- Zhao J, Liu T, Jin SB, Wang XM, Qu MQ, Uhlén P, Tomilin N, Shupliakov O, Lendahl U, Nistér M. Human MIEF1 recruits Drp1 to mitochondrial outer membranes and promotes mitochondrial fusion rather than fission. EMBO J. 2011;30:2762–2778. doi: 10.1038/emboj.2011.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.