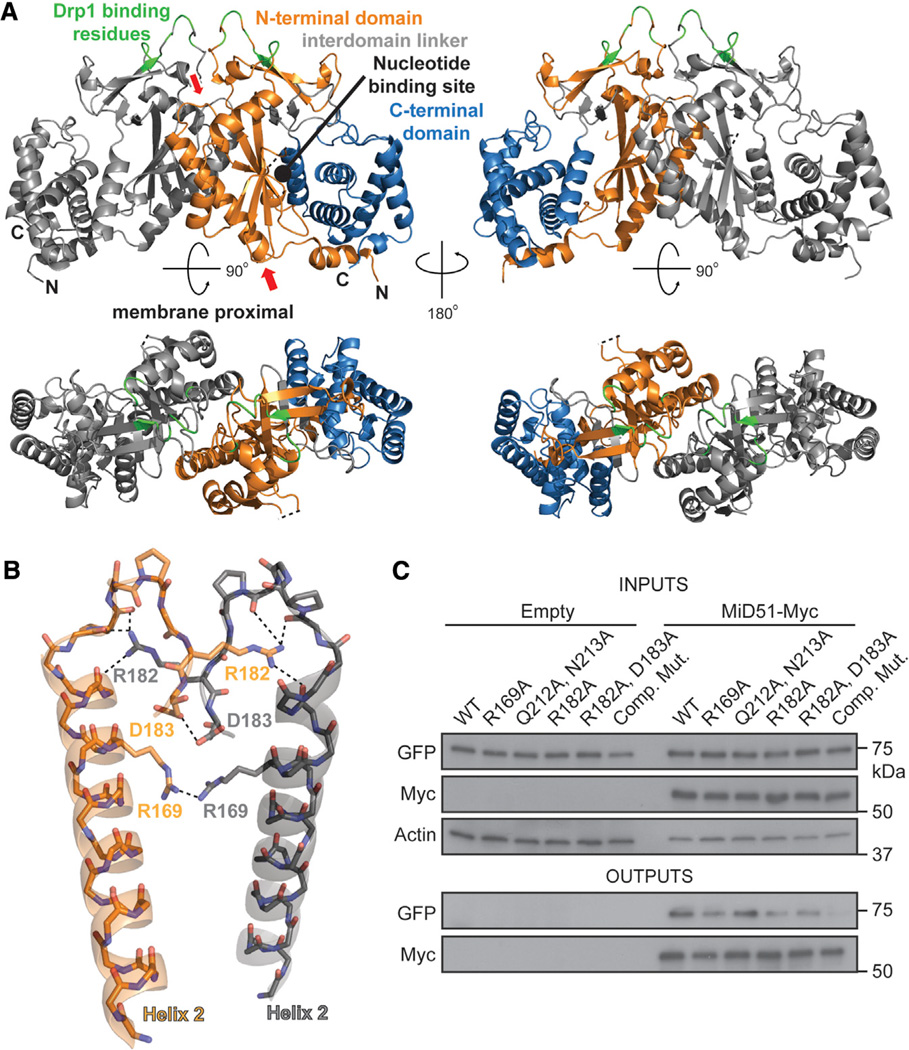
Figure 3. MiD51 Forms a Dimer through Electrostatic Interactions.
(A) MiD51 crystallizes as a dimer through an interface found in the N-terminal domain. The two molecules are differentially colored gray (left) and as in Figure 1B (right). In the bottom figure, the dimer has been rotated to view the interface from the top. The right panels are a 2-fold axis rotation. Red arrows indicate the α helix and loop shown in (B). Drp1 binding residues are colored green.
(B) Detailed view of residues important for the dimer interface. Interactions are indicated with dashed lines. The color scheme is the same as in (A). Electrostatic interactions were determined using the PDBePISA server. R169 self-associates by hydrogen bonding with a sulfate ion (not depicted).
(C) Mutational analysis of dimer formation. MiD51-GFP mutants were cotransfected with either empty vector or Myc tagged MiD51 constructs. For each reaction, the same mutant was used for both the GFP and Myc tagged constructs. Cells were treated with a reversible crosslinker and solubilized before immunoprecipitation against the Myc epitope. Top: expression of MiD51-GFP and MiD51-Myc in cell lysates. Actin is a loading control. Bottom: anti-Myc immunoprecipitates analyzed for MiD51-GFP. Comp Mut, compound dimerization mutant.