Abstract
Our understanding of the pervasive involvement of small RNAs in regulating diverse biological processes has been greatly augmented by recent application of deep-sequencing technologies to small RNA across diverse eukaryotes. We review the currently-known small RNA classes and place them in context of the reconstructed evolutionary history of the RNAi protein machinery. This synthesis indicates the earliest versions of eukaryotic RNAi systems likely utilized small RNA processed from three types of precursors: 1) sense-antisense transcriptional products, 2) genome-encoded, imperfectly-complementary hairpin sequences, and 3) larger non-coding RNA precursor sequences. Structural dissection of PIWI proteins along with recent discovery of novel families (including Med13 of the Mediator complex) suggest that emergence of a distinct architecture with the N-terminal domains (also occurring separately fused to endoDNases in prokaryotes) formed via duplication of an ancestral unit was key to their recruitment as primary RNAi effectors and use of small RNAs of certain preferred lengths. Prokaryotic PIWI proteins are typically components of several RNA-directed DNA restriction or CRISPR/Cas systems. However, eukaryotic versions appear to have emerged from a subset that evolved RNA-directed RNA interference. They were recruited alongside RNaseIII domains and RdRP domains, also from prokaryotic systems, to form the core eukaryotic RNAi system. Like certain regulatory systems, RNAi diversified into two distinct but linked arms concomitant with eukaryotic nucleo-cytoplasmic compartmentalization. Subsequent elaboration of RNAi proceeded via diversification of the core protein machinery through lineage-specific expansions and recruitment of new components from prokaryotes (nucleases and small RNA-modifying enzymes), allowing for diversification of associating small RNAs.
As the reality of pervasive transcription of the genome across the three superkingdoms of life becomes more and more widely-accepted [1–6], efforts to characterize the non-protein coding component of the transcriptome have intensified. Among non-coding transcripts, distinct classes of small RNAs have been among the first to be characterized, a process accelerated by the development of second-generation deep sequencers well-suited to the task of sequencing cellular small RNA fractions [7]. In the wake of this veritable avalanche of discoveries, attempts to classify small RNAs at times can seem as varying as the number of researchers investigating them. These classifications have mainly relied on some combination of length, genome context, shared functional traits, phylogenetic patterns, and structural features of the small RNAs [8–14]. This somewhat ad hoc approach to classification largely reflects the inherent difficulties in formulating a natural evolutionary classification due to the lower constraints on sequences and structures of these RNAs and the possibility of convergent evolution of functionally similar RNAs. This starkly contrasts the situation in the protein universe or even other more structurally constrained small/mid-sized RNA molecules such as riboswitches and tRNAs [15–17].
While the relationships between different small RNAs can be murky, particularly across large phylogenetic distances, these molecules have emerged as key players in a large range of core biological processes across the tree of life. A few notable, experimentally-tested roles for various classes of small RNA include: 1) translational repression in bacteria [18]; 2) guidance of pseudouridylation and methylation during ribosomal RNA maturation in eukaryotes and archaea [19]; and 3) immunity against viruses and other invasive nucleic acid elements in all the three superkingdoms of life [20, 21]. The roles of small RNAs are particularly well-studied in eukaryotes, currently implicated in interacting and overlapping processes such as: 1)post-transcriptional gene silencing [22]; 2) histone modification, DNA modification and chromatin dynamics [23, 24]; 3) germline maintenance [25]; and 4) pre-mRNA splicing [24].
One large group of eukaryotic small RNA classes can be unified on the basis of their role in pathways that might be generally termed RNA interference (RNAi). These RNAi systems rely on a conserved set of proteins, with the central player being a member of the PIWI/Argonaute superfamily (hereinafter PIWI superfamily), which contains a conserved domain of the RNase H fold. The PIWI protein acts as the basic platform for RNAi systems [26] by providing the scaffold for the interaction between a substrate small RNA strand, often termed the “guide strand”, and a complementary polynucleotide target strand (this pairing of protein and RNA components is referred to as the RNA-induced silencing complex, or RISC). Additional core components of the RNAi systems are those involved in generation of the mature small RNAs that are utilized by the PIWI superfamily proteins. Other than the PIWI proteins, these include an endoribonuclease domain of the RNaseIII superfamily proteins (e.g. Dicer) and in several cases an RNA-dependent RNA polymerase (RdRp) that acts as an amplificatory component by synthesizing more small RNAs.
The broad contours of the evolution of the protein components of RNAi, along with reviews of specific classes of small RNAs, have been previously discussed on several occasions [27–29]. However, a flurry of recent investigations is beginning to clarify and vastly expand the scope of PIWI-bound small RNA cargo across eukaryotes. These findings enable a more complete assessment of the natural history of the RNAi system in terms of both RNA and protein components. To this end, we present an overview of classes of small RNA associated with RNAi. We then synthesize these observations with a comprehensive reconstruction of both the core and ancillary components of the RNAi protein machinery in light of new genomic data while attempting to highlight currently underappreciated avenues of research relating to small RNA and RNAi systems.
CLASSES OF SMALL RNA ASSOCIATING WITH PIWI
RNAi-related literature is rife with reports identifying distinct classes of small RNA. Caution must be applied when interpreting these findings; reports relying exclusively on computational methods exploring kinship with known classes of PIWI domain-associating small RNAs can reach poorly-supported conclusions (example: [30]). On the other hand, certain reports exclude the presence of particular small RNA classes based on sequencing with limited depth/coverage [31]. Likewise, the presence of a class of small RNAs in a dataset does not necessarily imply physical association with a PIWI/AGO member and active participation in an RNAi-like pathway. To address this issue, recent reports have focused on identifying PIWI-bound small RNA through direct PIWI immunoprecipitation –sequencing (IP-seq) and/or confirming association with experiments targeting specific members of a small RNA class. While deep IP-seq datasets are also limited by potential detection of spurious associations (particularly in the absence of a negative-control IP-seq dataset), they tend to offer the most accurate picture of the RNAi-functional small RNA classes to date. Table 1 provides an overview of key deep-sequencing datasets (both IP-seq and whole cell fractionation small RNA-seq) while Table 2 provides a summary of known PIWI-associating small RNA classes across diverse eukaryotic lineages. Figure 1 provides an overlay of the information from Table 2 onto the evolutionary history of the components of the RNAi system, while Figure 2 provides a generalized overview of pathways which generate the classes of small RNAs outlined in Table 2. The following subsections briefly summarize individual classes of small RNA currently known to associate with PIWI proteins.
Table 1.
Selected small RNA deep sequencing in diverse eukaryotic lineages.
| Organism | Clade | Type of dataset | References |
|---|---|---|---|
| Human | animal vertebrate | IP-seq/small RNA-seq | [84, 265–267] |
| mouse | animal vertebrate | IP-seq/small RNA-seq | [47, 268, 269] |
| Drosophila | animal insect | IP-seq/small RNA-seq | [270–272] |
| C. elegans | animal nematode | IP-seq/small RNA-seq | [119, 273] |
| Saccharomyces castellii | budding yeast | small RNA-seq | [167] |
| Saccharomyces cerevisiae | budding yeast | small RNA-seq | [167] |
| Schizosaccharomyces pombe | fission yeast | IP-seq | [51] |
| Magnaporthe oryzae | filamentous fungus | small RNA-seq | [85] |
| Arabidopsis thaliana | plant | IP-seq/small RNA-seq | [50, 274–276] |
| Chlamydomonas reinhardtii | algal plant | small RNA-seq | [277] |
| Entamoeba histolytica | amoebozoan | IP-seq | [60] |
| Dictyostelium discoideum | amoebozoan mycetozoa (slime mold) | small RNA-seq | [40] |
| Toxoplasma gondii | apicomplexa | small RNA-seq | [39] |
| Tetrahymena thermophila | chromalveolate ciliate | IP-seq/small RNA-seq | [59] |
| Trypanosoma brucei | kinetoplastid | IP-seq | [52] |
| Trypanosoma cruzi | kinetoplastid | small RNA-seq | [173] |
| Trichomonas vaginalis | parabasalid | small RNA-seq | [37, 164] |
| Giardia lamblia | diplomonad | small RNA-seq | [37, 164] |
Table 2.
Broad overview of PIWI-associating RNA classes, from the perspective of small RNA genome source.
| small RNA genome source | general small RNA “class”1 | phyletic spread2 | functional role(s)3 |
|---|---|---|---|
| genomically-encoded, independently-transcribed hairpins | imperfectly-complementary hairpins | Giardia, Trichomonas | at least one example is linked to PTGS [79] |
| genomically-encoded, independently-transcribed hairpins | miRNA | plants, animals, T. gondii*, D. discoideum* | PTGS |
| genomically-encoded, non- independently-transcribed double-stranded sequences | miRtrons | animals | PTGS |
| genomically-encoded, non- independently-transcribed double-stranded sequences | Endo-siRNA (including esiRNAs and social RNA∧) | animals, plants, fungi/yeast, T. brucei∧ | PTGS, viral resistance |
| double-stranded sequences derived from s-as transcription | Endo-siRNAs, proximal promoter/transcriptional unit-derived small RNA3, and small RNA toxins/effectors∧ | animals, plants, fungi/yeast, slime mold, T. thermophila, E. histolytica, T. gondii∧, T. brucei, Giardia | PTGS, retrotransposon silencing, co-transcriptional regulation, polII positioning |
| double-stranded sequences derived from single-stranded transcription4 | endo-siRNAs (including qiRNAs, priRNAs, meiotic silencing RNAs, nat-siRNAs, plant heterochromatic siRNAs, ta-siRNAs, 22G-RNAs) | C. elegans, plants, some fungi | regulation of chromatin dynamics, germline surveillance |
| double-stranded sequences derived from s-as transcription | splice site-associated RNA | animals, E. histolytica∧ | regulation of splicing |
| double-stranded sequences derived from s-as transcription | scan RNAs (scnRNAs) | ciliates | DNA elimination |
| double-stranded sequences derived from s-as transcription | DNA damage-induced small RNA | animals, plants | recruitment of DSB repair proteins, clearance of DSB- proximal transcribed byproducts |
| single-stranded sequences | piRNAs | animals | germline surveillance, regulation of chromatin dynamics |
| ncRNA precursor | sdRNAs (snoRNA-derived) | animals, plants, fungi, Giardia | PTGS |
| ncRNA precursor | tRF (includes 3′ tRF-CCA, 3′ tRF-U, and 5′ tRF classes) (tRNA-derived) | animals, plants, Magnaporthe oryzae*, amoebozoans*, ciliates, T. gondii* | PTGS, rRNA processing |
| ncRNA precursor | svRNAs (vault RNA-derived) | animals | PTGS |
| ncRNA precursor | other (including potential snRNA, RMRP, and rRNA- derived small RNAs) | animals | RMRP fragments likely involved in PTGS [105], otherwise unknown |
small RNA classes have often been labeled according to functional roles. Rendering the functional role secondary to the source potentially enables better delineation of the evolutionary descent of small RNA classes.
symbol meanings: *, association with PIWI inferred from small RNA-seq; ∧, inclusion pending further investigation.
abbreviation: PTGS, post-transcriptional gene silencing
further research is needed to resolve differences or overlap between s-as gene body-derived small RNAs involved in regulating gene expression (as “endo-siRNAs”) and those involved in regulation of chromatin dynamics/polII positioning.
classes of small RNA derived from gene bodies or repeat regions which form a duplex by adding the complementary strand after transcription via RdRP activity instead of independently transcribing the opposite strand.
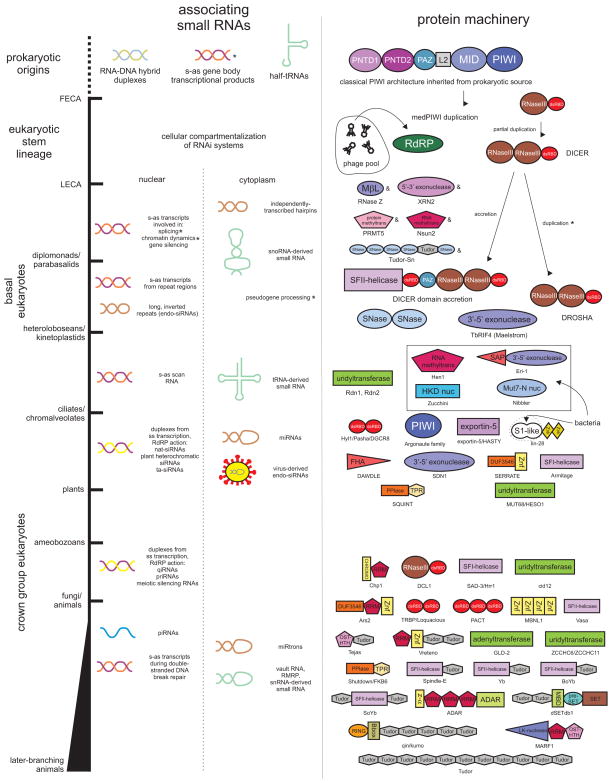
Figure 1.
Temporal diagram depicting emergence of RNA substrates (left column) and core and ancillary protein domains (right column) comprising RNAi systems against the divergence of major eukaryotic lineages labeled to the left. RNAi substrates are divided according to potential for acting in the nuclear and cytoplasmic compartments. Protein domains are depicted by labeled polygonal shapes and are not drawn to scale. Names of the proteins with the depicted architectures are provided below the domains. Inferred origins for several proteins are depicted where appropriate. Affixed asterisks indicate uncertainty in terms of timing of the origins of a RNA substrate or associating protein. Affixed ampersands are indicative of a protein with a deep phylogenetic origin but potential associations with RNAi pathways in early lineages remains unexplored. Abbreviations: PNTD1, PIWI N-terminal domain 1; PNDT2, PIWI N-terminal domain 2, L2, Linker-2 domain; SNase, Staphylcoccal nuclease; nuc, nuclease; Znk, zinc knuckle; FHA, Forkhead-associated; PPlase, peptidyl prolyl-cis/trans-isomerase; Znf, zinc finger; dsRBD, double-stranded RNA-binding domain; HTH, helix-turn-helix; RdRP, RNA-dependent RNA polymerase.
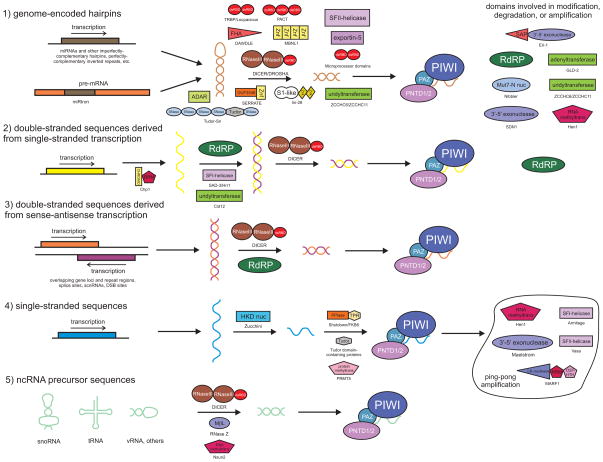
Figure 2.
General overview of the biogenesis of various classes of small RNA. Extremely generalized biogenesis pathways for the classes of small RNA listed in Table 2 are provided for reference. Classes of small RNA cargo are depicted and labelled numerically on the far left. Domains catalyzing or providing assistance in various lineages at steps in the pathway are shown in relevant locations. For the temporal timing of the emergence of such domains in eukaryotic evolution, please refer to Figure 1. Domains contributing to post-transcriptional modification, degradation, or amplification of small RNAs are shown to the left of each pathway. For ease in comparison, coloring of RNA and protein domains matches Figure 1. Proteins/domains which participate in RNAi pathways but cannot yet be clearly assigned to any specific pathway are not included. Core domains are labelled in white lettering.
miRNAs and other small RNA derived from imperfectly complementary hairpins
Perhaps the most studied classes of small RNA are plant and animal microRNAs (miRNAs). miRNAs are generated via the successive action of RNaseIII domain-containing enzymes on double-stranded, imperfectly complementary hairpins directly transcribed from the genome by RNA polII or contained in spliceosomal introns of other genes (miRNA variants known as mirtrons observed thus far both in animals and plants [32, 33]) (Figs. 1,2). miRNAs are typically involved in post-transcriptional gene silencing through binding of the PIWI-miRNA complex to complementary stretches of nucleotides in the 3′ UTR of mRNA transcripts. miRNAs have been the subject of extensive and exhaustive reviews (see [22, 34, 35]), including discussions on the divergent vs. convergent origin divide for plant and animal miRNAs [27, 36].
Perhaps more so than other small RNA classes, the potential presence of “miRNA-like” small RNAs in other branches of the eukaryotic tree of life has been the subject of debate since the first reports of the miRNA system in plants and animals (Table 2). Small RNA sequencing complemented in some instances by single-gene studies have uncovered support for possible “miRNA-like” hairpin precursors in the diplomonad Giardia lamblia [37, 38], the parabasalid Trichomonas vaginalis [37], the apicomplexan Toxoplasma gondii [39], and the slime mold Dictyostelium discoideum [40]. The hairpins observed in T. gondii are of particular interest due to their observed affinity in terms of structural and thermodynamic properties with mammalian miRNA [39]. It is crucial to point out that while most current literature often describes imperfectly complementary hairpin sequences in basal eukaryotes as “miRNA-like”, this does not necessarily imply a common origin for all such “miRNA-like” sequences; these could be products of the lineage-specific, convergent emergence of hairpin precursors. Indeed, several features of the currently known “miRNA-like” hairpins in diverse eukaryotes, particularly in Giardia and Trichomonas, such as length, relative location of the derived transcript on the hairpin, potential 3′ and 5′ modifications, and possible underlying steps in biogenesis pathways show differences with canonical plant and animal miRNAs. Nevertheless, all examples of such systems across eukaryotes (Table 2) might be viewed as variations on a generalized theme of processing of imperfect RNA hairpin precursors.
The core RNAi protein apparatus used in processing these imperfect hairpins therefore can be seen as a preadaptation allowing fixation of independent precursor RNAs in conjunction with other selective pressures on multiple occasions in eukaryotic evolution. Of particular importance were the selective advantages accrued from the deployment of these miRNA-like regulators as an additional level of control, over and beyond the conserved transcriptional and chromatin-based regulatory systems [41]. As gene product control mechanisms come under strong selection due to lineage-specific adaptations [42] [43], it is likely that the corresponding miRNA-like repertoire was constantly subject to turnover with new precursor hairpins being selected and old ones being partially or entirely lost. A similar phenomenon has been reported in the case of the evolution of another major class of regulatory molecules in eukaryotes, transcription factors [44]. Under this scenario the conserved animal and plant miRNAs likely represent versions that were “institutionalized” as crucial gene regulators owing to the unique adaptations in their respective lineages.
siRNAs from diverse sources
Short interfering RNA (siRNA) is a collective label affixed to a class of small RNAs typically processed from multiple distinct sources but sharing a common feature, i.e., processing from perfect or nearly-perfect complementary sequences (Table 2, Fig. 2). Sources of endogenous siRNAs (endo-siRNAs) (distinct from exogenous siRNAs referring to synthetically designed siRNAs for laboratory and therapeutic use) can be grouped into three categories: 1) genome-encoded complementary double-stranded sequences including long non-coding RNA (ncRNA) loci (for example, the esiRNAs in Drosophila[45] and potentially the so-called “social RNA” in C. elegans[311]), independently-folding units in some RNA transcripts like mRNA [46], inverted repeat sequences [47], and double-stranded RNA (dsRNA) viral genomes [48, 49]. 2) Double-stranded sequences forming in the wake of sense-antisense (s-as) transcription at convergent protein-coding loci [47, 50, 51] and retrotransposed elements [45, 52, 314]. 3) Amplification of single-stranded RNA (ssRNA) substrates via the action of RdRP producing suitable dsRNA substrates.
Of these the first category is found across the so-called crown group eukaryotes including plants, fungi/yeast, animals, including recent discoveries of such in vertebrates [47, 53–55]. Such siRNAs are possibly present in the kinetoplastid Trypanosoma brucei [52]. The second category has been observed widely in eukaryotes. In addition to a well-characterized presence in animals, plants, fungi, and amoebozoans, the basal eukaryotic diplomonad Giardia lamblia processes double-stranded substrates derived from sense-antisense (s-as) transcription at variant-specific surface protein (VSP) family gene loci. Association of the resulting small RNAs with Giardia PIWI results in silencing of expression at all but one VSP locus which is selected as the surface-presented antigen [56]. PIWI IP-seq also revealed s-as transcription giving rise to a highly-expressed class of endo-siRNAs in the ciliate Tetrahymena thermophila [57–59]. Remarkably, these small RNA are similarly found antisense to lineage-specific, uncharacterized protein-coding gene families, a scenario paralleling s-as transcription at VSP family genes in Giardia. Smaller-scale PIWI IP-seq in the amoebozoan Entamoeba histolytica is also strongly suggestive of endo-siRNA generation from sites of s-as transcription [60], although notably the E. histolytica genome does not encode for any known RNaseIII-like enzymes involved in RNAi. Finally, small RNAs derived from s-as transcription are observed in T. brucei [52] (Table 2). The kinetoplastid genome is unusual among eukaryotes due to its organization into transcribed long directional clusters containing consecutive protein-coding genes. The edges of these clusters are often characterized by convergent transcription and appear to represent hotspots for small RNA transcript generation [52]. The third mechanism, observed in organisms such as C. elegans [61, 62], plants [63, 64] and some fungi, is dependent upon the presence of the RdRP proteins and include the well-studied trans-acting (ta-) and natural cis-antisense transcripts-associating (nat-) siRNAs [65] (Table 2). These siRNAs seem to be largely involved in regulation of chromatin dynamics at protein-coding loci (see below, Figs. 1,2).
Little research on small RNAs has been performed in other basal eukaryotic lineages including the heteroloboseans (Naegleria) and parabasalids (Trichomonas) and later-branching chromalveolate lineages like apicomplexa and stramenopiles. However, they are phylogenetically bracketed by Giardia, kinetoplastid, ciliate, amoebozoan, plant, animal, and fungal lineages, which use sense-antisense siRNA; hence, is possible these lineages also produce such s-as small RNA transcripts (Fig. 1). Thus, based on current experimental evidence, s-as transcription appears to be the phylogenetically most widespread source for RNAi-related small RNA going back to the last eukaryotic common ancestor (LECA), an observation with deeper implications discussed throughout the remainder of this article. Functions of small RNAs derived from s-as transcription appear to extend beyond the classical endo-siRNA roles of post-transcriptional gene or repeat silencing to include several forms of co-transcriptional and other more specialized forms of regulation; these roles are discussed in subsections below. Research in Giardia and T. thermophila suggests that the silencing of lineage-specific gene families via s-as transcriptional endo-siRNA-associated with PIWI proteins might be more widespread than previously thought. This observation is of particular interest in the context of regulation of lineage-specifically expanded multigene families which are present throughout eukaryotes, including several parasite antigens [66]. Possible parallels can also be seen in surface receptor protein silencing via small RNA generation from s-as transcription in natural killer (NK) cells in vertebrates [67]. It should be noted that while these small RNA transcripts were classified as piRNA-like by the authors on the basis of transcript length and the presence of a protective 3′ modification [67], there are multiple arguments against classifying these small RNAs as canonical piRNAs: 1) piRNAs are not known to be generated by s-as transcription; 2) several classes of small RNAs which are not piRNAs have been characterized with similar transcript lengths in a range of eukaryotes; 3) 3′ modifications are not restricted to piRNAs. As such, these transcripts appear more akin to endo-siRNA-like transcripts generated by s-as transcription.
piRNA
piRNAs constitute a class of small RNA involved in germline maintenance [25] (Table 2). On binding PIWI proteins, the piRNA, acting as a guide strand, associates with transposable elements (TEs) resulting in TE-silencing and also influences heterochromatin dynamics by mediating DNA cytosine methylation [68]. piRNAs are typically observed in clusters on the genome, co-localizing with regions heavily-concentrated with transposable elements [69]. piRNAs are amplified via the “ping-pong” mechanism following single-stranded transcription (Figs. 1,2), requiring the action of a series of nucleases and helicases on the ribonucleoprotein complex formed after initial association of the piRNA with the PIWI protein [70]. Notably, the piRNA biogenesis pathway lacks RNaseIII-like nucleases. piRNA signatures have been detected in sponges and cnidarians, suggesting piRNAs were already present in the common ancestor of metazoans [71] (Fig. 1). While the expression of piRNAs has long-thought to be restricted to germline cells, increasing evidence suggests that piRNAs may also function in select somatic cell types in vertebrates including neurons [72, 73] and pancreatic tissue [74], suggesting a more generalized role for piRNAs in animal cell differentiation.
snoRNA fragments
Early deep-sequencing small RNA profiles identified classes of sequences mapping to well-characterized classes of longer ncRNA loci [53] (Table 2, Figs. 1,2). Several features of these small RNAs, including conserved processing site locations, nucleotide biases, and the predicted double-stranded character of the small RNA-derived region in the transcribed ncRNA suggested that instead of representing decay products, a conserved functional role, possibly linked to RNAi, existed for these small RNA [53]. Early PIWI IP-seq studies detected a highly-expressed small nucleolar RNA (snoRNA) fragment; detailed characterization confirmed an RNAi-related role for this snoRNA fragment in post-transcriptional gene silencing similar to miRNAs [75]. Targeted investigation of snoRNA-derived RNA (sdRNA) indicated sdRNAs were involved in post-transcriptional gene silencing across fungi, animals, and plants [76]. The characterization of sdRNAs associated with PIWI proteins in Giardia [77, 78] has shown that at least some, like the endo-siRNAs described above, appear to function in the context of VSP transcript silencing [79]. Given this distribution, it is likely that this class of small RNAs is an ancient, conserved processing system that was present in the LECA (Fig. 1). sdRNA-like sequences were also observed in T. brucei, however, due to the randomly-distributed locations for the recovered fragments on the snoRNA precursors, the authors speculated that these may represent non-specific PIWI associations of degradation products [52].
tRNA fragments
In addition to the sdRNA class, recent research has uncovered PIWI-associated small RNAs derived from tRNAs (Table 2, Figs. 1,2). Several distinct tRNA fragment (tRF) small RNA classes have been identified based on location within the precursor tRNA transcript. At least two of these classes are experimentally-confirmed to associate with PIWI proteins, the 3′ tRF-CCA class (derived from the 3′ end of the mature tRNA transcript) and the 3′ tRF-U class (derived from the 3′ end of the precursor tRNA transcript) [80–82]. These two classes have been found to associate with PIWI domains in animals [80–82] and at least the former in plants [83]. Some evidence indicates the 5′ tRF class additionally associates with the PIWI domain in both animals and plants [83, 84]. Fragments derived from tRNA falling within the length range typically associated with PIWI cargo have also been observed in Magnaporthe oryzae, amoebozoans, the apicomplexan T. gondii, and T. thermophila (Fig. 1) [39, 40, 85]. Notably, in T. thermophila, the Twi12 PIWI protein was found to be predominantly loaded with 3′ CCA tRNA cargo [86], starkly contrasting plant and animal PIWI domains where tRFs comprise a small fraction of the total small RNA PIWI load. While accumulating evidence implicates plant and animal tRF-PIWI complexes in post-transcriptional gene silencing [80, 81, 83, 87], in T. thermophila tRF-Twi12 appears to be involved in a drastically distinct function: assembly into a complex with the nuclear 5′–3′ exonuclease Xrn2 [88], activating it for rRNA pre-processing and degradation of improperly processed rRNA transcripts [89–91]. Similar to snoRNA, tRNA fragments observed in T. brucei PIWI IP-seq were claimed to be a non-specific association [52]. While this manuscript was under review, a small RNA PIWI-IP seq dataset from the bacterium Rhodobacter sphaeroides was published (see discussion below) [92]. Perhaps the most surprising finding in this dataset was the association of half-tRNA sequences (~45nt) cleaved at the anticodon with PIWI. Due primarily to sequencer length constraints, most small RNA PIWI-IP deep seq datasets to this point have only captured small RNA with lengths up to 30nt. However, the presence of a substantial, ancestral half-length tRNA sequence population in eukaryotic cellular small RNA pools has been established [53, 93–95], indicating that half-tRNAs could represent an ancient PIWI-associating substrate and could be more widespread in eukaryotes than currently known. Intriguingly, half-tRNAs have been linked to regulation of gene expression through association with the RNAse Z nuclease [96–98], suggesting further investigation into potential functional roles for half-tRNA-PIWI complexes in gene silencing pathways could be warranted.
Other fragments from larger ncRNA precursors
Classical PIWI proteins associate with several additional small RNAs processed from larger ncRNA precursors (Table 2, Figs. 1,2). The vault RNA (vRNA) gives rise to a set of small vault RNA-derived RNAs (svRNAs), one member of this class associates with a classical PIWI protein to post-transcriptionally regulate expression of the CYP3A4 gene [99]. The vRNA is associated with the vault which is an enigmatic nucleoprotein organelle found in several eukaryotes. Its primary protein components are the MVP, with conserved EF-hand domains, the vault-polyADP ribose polymerase, and the TEP1 protein which contains an N-terminal RNA-binding ROT/TROVE domain, a central NACHT NTPase module and a C-terminal WD40 beta propeller. The vRNA probably binds the ROT/TROVE domain of the latter protein. Two prevailing functional themes currently under investigation link the vRNA to 1) drug resistance [99, 100] and 2) neuronal differentiation [101, 102]. Poor sequence conservation of vRNA complicates efforts to trace its phylogenetic depth: while it is found in the mycetozoan slime mold Dictyostelium as well as in vertebrates, it appears to have been lost in many animal lineages and appears to be absent in other crown eukaryotic lineages including plants and fungi [103]. svRNA functionality through association with the RNAi pathway may therefore represent the recent colonization of a functional niche in vertebrates, consistent with the differential gene expression patterns observed for svRNAs [84] and the terminal cell types within which vRNA and derived svRNAs are thought to function.
IP-seq experiments have also revealed the presence of small RNAs derived from hairpin-like regions in small nuclear RNA (snRNA) and ribosomal RNA (rRNA) precursors associating with PIWI/AGO [84, 104]; however, little is known about potential functional roles for these small RNA-PIWI complexes and thus far they have been identified only in vertebrate IP-seq datasets. In the ascomycete Neurospora crassa, qiRNAs are transcribed from rDNA loci as ssRNA and subsequently amplified by RdRP activity before entering the RNAi pathway [65]; a mechanism distinct from the dsRNA-cleaving mechanism used to generate the ncRNA-derived small RNA classes described above (see below, Table 2). A recent report describes at least two fragments generated by DICER-dependent processing of double-stranded regions in the RNA component of mitochondrial RNA processing endoribonuclease P (RMRP) which appear to be involved in RNAi-mediated post-transcriptional gene silencing. Disruption of their expression is linked to Cartilage Hair Hypoplasia syndrome [105]. A related report claims the telomerase reverse transcriptase (TERT) complexes with RMRP to provide a substrate for RNAi in cancer lines; independent confirmation awaits this observation [106].
More generally, the repeated extraction of small RNA sequences for PIWI cargo from larger ncRNA sequences can again be viewed as the result of the preadaptions conferred by the core RNAi protein apparatus to process such substrates. Like the class of transcribed hairpin sequences described above, it seems likely that the selective advantage accrued from the deployment of ncRNA-derived small RNA in lineage-specific gene silencing contexts led to fixation of such small RNAs in several terminal lineages. The lack of known conservation of any single ncRNA precursor across a wide range of eukaryotes can thus again be understood through the lens of repertoire turnover: new ncRNA precursors are selected with old ones being partially or entirely lost.
Proximal promoter and transcriptional unit-derived small RNA
The control exerted by synthetic, exogenously-introduced antigene or small activating RNAs (agRNAs, saRNAs) designed to be complementary to promoter regions on gene activation and repression following PIWI association have been discussed elsewhere [107, 108]; we will focus here on PIWI association with endogenous sequences derived from proximal promoter and protein-coding gene body regions. Small RNA classes derived from divergent transcription at transcriptional start sites have been described as transcription-initiated RNAs (tiRNAs) and transcription start site-associated RNAs (TSSa) in two separate reports, and are likely derived from polII transcriptional “backtracking” events [109, 110]. Most reviews list these as distinct classes of promoter region-derived RNAs; however, similar median lengths and distribution of the small RNA start sites around transcriptional start sites indicate tiRNAs and TSSa RNAs are two descriptors for the same class of RNA. The primary difference in the tiRNA and TSSa studies is that the latter investigated targeted enrichment of promoter-derived small RNA transcripts at specific genes and observed that the dominant small RNAs derived from these loci tended towards a longer length distribution (~20–90nt) than evidenced by small RNA deep sequencing (limited to a length of ~30nt at the time), while the former work did not investigate such enrichment. Regardless, researchers immediately recognized the presence of small RNAs in the range of ~20nt as attractive potential cargo for PIWI domains. Initial reports did not recover TSSa/tiRNAs in a Drosophila PIWI IP-seq dataset [109]; however, a subsequent report suggested at least a subset of tiRNAs may associate with human AGO proteins [84]. The relationship between these smaller RNAs and much longer divergent transcripts at transcriptional start sites [111–113] remains unclear; however, similarities in the distributions of start sites could suggest a relationship between these two RNA populations with the former representing abortive or nuclease-processed versions of the latter.
Small RNA sequences associated with PIWI proteins that map along the length of protein-coding loci in the genome have also been reported. One such example is seen at heat shock loci in Drosophila; these sequences are enriched during heat shock but are also observed under normal conditions across the complete set of Drosophila promoters [114]. Association of these small RNAs with a classical PIWI protein in Drosophila positions polII and contributes to transcriptional control at active loci. While the biogenesis of these Drosophila small RNAs remains unexplored, the involvement of the DICER enzyme and the observed antisense transcription start sites originating from points downstream of the 3′ end of the sense transcript [114] suggests biogenesis occurs via a s-as transcriptional intermediate. Given these observations, exploration of a potential link between the ISWI SWI2/SNF2 ATPase, which enables a switch to antisense transcription [115], RNAi pathways, and gene expression may be of interest, particularly in organisms lacking canonical RdRP activity.
While their functions appear to be distinct, the s-as transcriptional mechanism generating these small RNAs parallels the production of the gene-silencing “endo-siRNAs” described above in Giardia and T. thermophila. s-as transcriptional products derived from gene bodies and associating with PIWI proteins have also been observed in the plant [50], fission yeast [51], amoebozoan [60], and kinetoplastid lineages [52] (Table 2). Additionally, in the apicomplexan T. gondii many sequences are reported as mapping to protein coding regions; it is unclear whether the authors distinguished between those mapping to sense and anti-sense strands [39]. While these small RNAs might have endo-siRNA-like functions, their generally wider distribution across the genome and lower levels of expression could point to roles in co-transcriptional regulation, perhaps involving regulation of chromatin dynamics or polII positioning as in Drosophila [52, 114].
Typical production of RNAi-associating small RNAs from gene bodies appears to entail: 1) s-as transcription at the generating locus, 2) re-association of complementary s-as transcripts, 3) nuclease processing to generate short duplexes, 4) loading of a single strand from the duplex onto PIWI, and, when encoded by the genome, 5) amplification of the substrate via RdRP activity. In some lineages including plants, fission yeast, and C. elegans, variations on this biogenesis mechanism have emerged (Table 2, Figs. 1,2); however, these small RNAs can be functionally unified with the endo-siRNA-like molecules based on probable shared roles in triggering heterochromatin formation and transcriptional inactivation [63, 116]. In many plants, this process is accomplished through the plant-specific polIV and polV RNA polymerases. We were unable to detect polIV or polV homologs in algae, suggesting these polymerase versions are not an ancestral feature of the whole plant clade. Mosses such as Physcomitrella contain fully-differentiated polIV and polV homologs. Thus, polIV likely emerged at the base of land plant evolution through polII gene duplication with polV emerging after subsequent polIV duplication [117]. In plants containing these homologs, polIV generates transcripts at gene loci which are in turn subject to RdRP activity, generating long s-as duplex transcripts which are then processed into smaller duplexes by DICER activity and loaded onto a PIWI protein [63]. PolV concomitantly generates long transcripts at the same loci which are targeted by the PolIV/RdRP-generated complementary sequences associating with PIWI, resulting in recruitment of chromatin modifying factors and ultimately transcriptional repression. In fission yeast, RdRP acts on small transcript degradation fragments associating with a PIWI protein (known as primal RNAs or priRNAs) [116]; meiotic silencing small RNAs in N. crassa are transcribed from unpaired stretches of DNA which are, similar to priRNAs, amplified by RdRP activity and ultimately loaded onto PIWI [118]. In C. elegans, specific mRNA transcripts are targeted for endo-siRNA production via terminal RdRP activity and subsequent DICER processing [119], a subset of these small RNAs is linked to chromatin modification-induced silencing at gene loci [120, 121]. These lineage-specific mechanistic variations are likely to represent secondary innovations from a broader theme involving silencing through s-as transcription for reinforcement of particular regulatory interactions.
Despite mechanistic and functional shifts, two overriding implications can be derived from combining the preceding observations: 1) s-as transcription at gene bodies represents an ancestral source of RNA substrates for PIWI protein association and 2) small RNA derived from s-as transcription for incorporation into RNAi pathways has an ancient eukaryotic functional role in the regulation of chromatin dynamics, a possibility first raised over a decade ago based on the strong concordance in phyletic patterns between RNAi components and chromatin modifying factors [122].
An added wrinkle relevant to this discussion is the recent discovery of a novel, inactive PIWI family in the Med13 protein (MedPIWI) [123], a component of the CDK8 subcomplex which functions as the negative regulator of the Mediator transcriptional coactivator complex. This domain is found across all major eukaryotic lineages with the exception of the kinetoplastids and is predicted to bind small RNAs, triggering a conformational change in the subcomplex resulting in allosteric inhibition of Mediator-polII association and subsequent transcriptional activation. While the MedPIWI-associating small RNA substrate is unknown, either of the above classes of small RNAs represents possible candidate binding partners.
Splice site-associated small RNA
Small RNAs derived from intronic regions have been identified as associating with the classical PIWI module of AGO proteins by IP-seq experiments in humans and Drosophila [24, 124, 125] (Table 2, Fig. 1). Splice site-associated RNA-PIWI complexes appear to play a crucial role in recruitment of the splicing machinery, affecting decisions during alternative splicing [24]. While the biogenesis of these small RNAs has yet to be determined definitively, several clues have been presented: 1) the PIWI module appears to preferentially associate with small RNAs transcribed in the sense direction [24, 124]. 2) targeted studies of the CD44 gene locus indicated that the strongest enrichment for splice-site associated RNAs overlapped with the site of a known small RNA transcribed antisense to CD44 [24]. 3) DICER is required for their processing. These three observations strongly suggest splice site-associated RNAs are also a product of convergent s-as transcription yielding duplex RNA processed by RNaseIII activity.
While the complete extent of the deployment of RNAi in regulation of splicing has not yet been extensively analyzed outside of animals, enrichment of small RNAs at splice sites has been previously reported in at least one amoebozoan [60] (Table 2, Fig. 1). Also, as mentioned above, many basal lineages have reported PIWI association with small RNAs derived from s-as transcription along protein-coding gene bodies. This, taken with 1) the observation that RNAi components strongly co-occur with splicing machinery components across eukaryotic lineages [122] and 2) the documented links between the introduction of introns and the initiation of eukaryogenesis [126, 127] suggests investigation into potential ancestral roles for RNAi in regulation of splicing could provide substantial insight into the evolution and functional diversification of RNAi systems (Fig. 1).
scanRNAs and DNA elimination/genome unscrambling in ciliates
Ciliates contain two nuclei in the same cell: the micronucleus and its derived macronucleus which is formed through the process of DNA elimination of specific sequences present in the micronuclear genome followed by extensive endoreplication [128]. In certain ciliates like T. thermophila, a class of small RNAs, the scan RNAs (scnRNAs), guides this process via association with a PIWI protein (Table 2, Fig. 1) [128, 129]. scnRNAs are generated via s-as transcription likely occurring along the complete length of the micronuclear genome followed by DICER processing. Thus, in terms of their biogenesis, scnRNAs be viewed as a specialized, ciliate-specific class of small RNAs arising from s-as transcription (Table 2). During conjugation, loaded scnRNA-PIWI complexes are imported into the parental macronucleus. By processes not yet fully-understood, association between these complexes with transcribed RNA results in degradation of scnRNAs which bind to cognate complementary sequences. The remaining loaded PIWI proteins contain only scnRNA which matches portions of the micronucleus excised in the parental macronucleus. These scnRNA-PIWI complexes migrate from the pyknotic parental macronucleus to the newly-forming macronucleus initiated by micronucleus duplication. scnRNA again base pairs with nascent ncRNA transcripts during macronucleus transcription, resulting in recruitment of the histone methyltransferase Ezl1p and H3K9 methylation, which in turn acts as a marker for genome elimination [130].
All ciliates studied to this point appear to utilize scnRNAs transcribed from the micronucleus in some form of DNA elimination, including the oligohymenophorean ciliates Tetrahymena [128, 129] and Paramecium [131] and the spirotrich ciliates Stylonychia and Oxytricha [132]. Spirotrichs are likely to harbour a more complex DNA elimination pathway due to the high proportion of genome sequence eliminated in the macronucleus (up to 95%) and the size of many of the eliminated sequences (as small as 10nt, unlikely to be efficiently targeted by ~25nt scnRNAs) [128]. Additionally, spirotrichal macronuclear genomes undergo “unscrambling”, a process which restores the order of genes observed in the parental macronucleus in newly-formed macronuclei which are scrambled during frequent rearrangement events occurring during micronuclear meiosis. Unscrambling is mediated not by small ncRNA, but by long ncRNA transcribed from the parental macronucleus and imported into the new macronucleus wherein it appears to impose the “correct” gene order as read by the long ncRNA [133].
From a broader perspective, the linkage between RNAi pathways and transmission of epigenetic information is by no means unique to ciliates. As described above and elsewhere, small RNAs and long ncRNAs contribute to and direct chromatin modifications in a variety of contexts. However, one striking feature of ciliate scnRNA and ncRNA usage is that the epigenetic information is passed to progeny not through the formation of chromatin marks but in the transmission of the RNA itself as proposed in the case of the plant-specific phenomenon of paramutation [134]; the formation of chromatin marks in ciliate DNA elimination acts as transient guides directing removal of marked DNA segments. In some ways this ciliate-specific process parallels trans-, rather than cis-based epigenetic transfer, as is seen in ncRNA-mediated imprinting in animals genomes.
DNA damage-induced small RNA
Recent studies have demonstrated small RNAs are generated at double stranded break (DSB) sites of DNA damage in humans, Drosophila, and plants. The common denominator in these events is the activity of polII and DICER homologs, which is supplemented by DROSHA and RdRP activity in humans and plants, respectively [135–137] (Table 2, Figs. 1,2). In at least plants and Drosophila, these small RNAs are loaded onto classical PIWI proteins and the resulting complexes appear to contribute to DSB repair by either recruiting DSB repair proteins to breakage sites [135] or by binding to and clearing transcribed products from the repair region [136]. The generation of these small RNAs occurs either through processing of duplexes formed via convergent s-as transcription around the breakage site [136, 137] or, in plants, through the activity of RdRP on ssRNA generated at the site [135].
The aforementioned qiRNAs in fungi are also transcribed in response to DNA damage, but in contrast to the cis-acting effects of the above-described small RNAs, these appear to enable a trans response as DNA damage stimulates the generation of qiRNA at rDNA loci. Complements of these transcripts are then transcribed by RdRP action, cleaved by DICER, and loaded onto PIWI; this association then appears to globally inhibit protein synthesis by a mechanism which remains unclear [65, 138].
Potential prokaryotic PIWI substrates
Two distinct PIWI families are present in prokaryotes, the prokaryotic homologs of the classical eukaryotic PIWI/Argonaute family (pPIWI) [29, 139] and the recently-discovered pPIWI-RE family [123]. Biochemical studies indicate that several pPIWI proteins have a preference for DNA-RNA hybrids, unlike the eukaryotic versions that have been primarily shown to bind dsRNA substrates [140, 141]. Consistent with this, pPIWI-RE is predicted to function as an RNA-dependent restriction system in prokaryotes as part of a mobile three-gene operon. In this system, the pPIWI-RE domain is predicted to bind/process DNA-RNA hybrid R-loops formed by invasive plasmids and/or phages during replication in cooperation with the R-loop-specific DinG-type helicase encoded by one of the three operon genes [123]. pPIWI-RE loaded with R-loop-derived small RNA sequences are then predicted to initiate cleavage of invasive DNA by recruiting the third protein encoded by these operons, a restriction endonuclease fold DNase. The substrates for the classical pPIWI proteins are poorly characterized; it has been suggested on the basis of genome associations that pPIWI proteins might act as components of prokaryotic systems involved in defense against invasive DNA [142]. A more recent examination of the operonic associations of pPIWI proteins reveals that nearly all major lineages of pPIWI domains are combined in operons with a diverse range of endoDNases (Further Resources). These versions could very well participate in plasmid/phage restriction systems in a manner similar to the pPIWI-RE family with RNA guides to target restriction by the coupled DNases.
While this manuscript was under review, the first description of the nucleotide content associating with a bacterial PIWI protein (from Rhodobacter sphaeroides) was published [92]. The results support much of the above-outlined experimental and computational research in this area: two major classes of nucleotide sequences in addition to the aforementioned half-tRNAs (see above) were observed associating with PIWI: 1) a class of 16–19nt small RNAs predominantly antisense to genic regions and 2) a class of 21–25nt small DNAs sense to genic regions. The two classes of sequences map to the same genome regions, indicating that the duplex initially loaded into the PIWI protein, as predicted by biochemical studies [140, 141], is a DNA-RNA hybrid duplex. Consequently, the authors named the classes the DNA-interacting RNA (diRNA) and RNA-interacting DNA (riDNA) [92]. diRNA appears to associate much more frequently than riDNA, suggesting a bias towards retention of the RNA of the duplex as the guide strand. Such bias is not a novel concept, the structural thermodynamic determinants governing duplex orientation during loading and subsequent guide strand selection has been well-studied in eukaryotes [143, 144]. The authors speculate that the source of the diRNA is likely to be RNA degradation fragments. We agree on this possibility, but question the assertion that these degradation fragments are loaded as single strands onto the PIWI protein. Parallels drawn to piRNA are unconvincing; from an evolutionary standpoint the poorly-understood single-stranded piRNA loading step represents a highly-specialized, lineage-specific adaptation present only in the animal lineage and is therefore extremely unlikely to represent an ancestral form of PIWI loading. Considering the discovery of the pPIWI-RE domain and its accompanying conserved operonic associations, we can instead propose that RNA fragments in Rhodobacter and prokaryotes with related PIWI domains (see below, Further Resources) initially form R-loop-like hybrid duplexes with DNA which in turn are recognized and bound by PIWI as a hybrid duplex. PIWI binding then recruits the strongly associated endonuclease domain (see below) that cleaves the regions surrounding the site of diRNA-PIWI binding. After cleavage, one strand of the PIWI-duplex is unwound and the resulting PIWI-diRNA or less-frequent PIWI-riDNA complex would be free to re-associate with another stretch of invasive DNA/RNA to effect a continued silencing. As such R-loops are more likely to be formed at sites of invasive DNA replication [145], this also provides an explanation for the capacity, described in the recent paper, of the R. sphaeroides PIWI domain to discriminate between invasive DNA species and “self” DNA. Such targeting explains why diRNA production is dependent on PIWI expression[92]; PIWI association is upstream of the DNA endonuclease activity (see below for further discussion on the evolution of PIWI proteins).
A subset of pPIWI proteins, distinct from R. sphaeroides and closely related pPIWI proteins, is encoded as part of a novel CRISPR/Cas system. In this case we predict that they are likely to use the small CRISPR RNAs as potential substrates. Yet another distinct subset of pPIWI proteins with the closest affinities to the eukaryotic PIWI proteins does not seem to show any conserved operonic association with any other genes. Given the extensive utilization of s-as transcription as material for PIWI cargo in eukaryotes, this group of pPIWI domains could recognize s-as transcription at overlapping coding regions, recently observed as a rampant occurrence through the use of RNA-seq technologies in diverse prokaryotic lineages [146]. The persistent role observed across classical PIWI domains in regulation of transcription along with the recent discovery of the MedPIWI module [123] suggests that in the eukaryotic stem lineage itself the PIWI domains already had distinct roles in transcription regulation. Hence it is conceivable that pPIWI domains which potentially recognize s-as RNA products might also have an as-yet uncharacterized role in transcription regulation, marking a shift from roles in defence against invasive DNA.
EVOLUTION OF PROTEIN COMPONENTS IN RNAI PATHWAYS IN RELATION TO ASSOCIATED SMALL RNAS
While several overviews of the natural history of the protein components of the RNAi pathway have been previously published [27, 28, 147, 148], until recently it has not been possible to include a comprehensive account of the RNA components of the pathway in this analysis. However, with the publishing of several RNA IP-seq and total cell small RNA-seq datasets from diverse eukaryotic lineages (Table 1), we can begin to consider the contributions of the RNA components of the system and analyze its evolutionary trajectories in comparison to the proteins. Moreover, recent genomic and structural data have thrown considerable new light on the origin of the key protein components of the eukaryotic RNAi system. The core eukaryotic protein apparatus for RNAi in many of its various manifestations consists of the PIWI domain, an RNaseIII (Dicer-like) domain, and the RdRP domain. This section will first discuss the prokaryotic origins of these core eukaryotic RNAi components and then move into discussions on innovations occurring after the core system was in place. In addition to information contained in Figure 1, Figure 3 provides an overview of expansion and loss of core protein RNAi components.
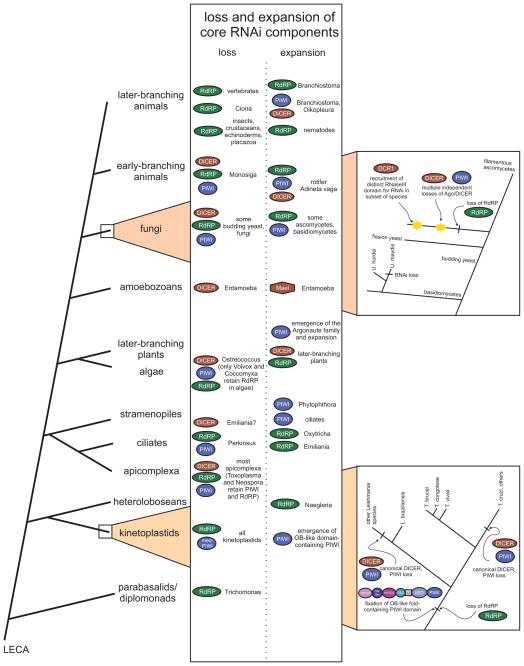
Figure 3.
Loss and expansion events of core RNAi proteins during eukaryotic evolution. Core RNAi protein components undergoing loss or extensive expansion are listed and labeled with specific affected lineages in the center columns, plotted against major eukaryotic evolutionary transitions to the left. Boxes to the far right provide more detailed resolution of loss and gain events in the kinetoplastid (bottom) and fungal/yeast lineages (top). Abbreviations: Mael, Maelstrom.
The anatomy and origin of the PIWI superfamily, the heart of the eukaryotic RNAi system
The PIWI superfamily proteins are the primary players in the eukaryotic RNAi system. Indeed, there is evidence that they can function in RNAi systems independently of the other core components, namely the DICER-like and RdRP enzymes. In the piRNA system, different versions of the PIWI superfamily function with distinct, non-RNaseIII nucleases both in piRNA production and as effectors of piRNAs [25]. In E. histolytica and T. gondii, PIWI proteins bind a variety of small RNAs which appear to be the end products of complex processing pathways [39, 60] despite the apparent absence of genomically-encoded RNaseIII enzymes. Finally, a DICER-independent miRNA pathway also exists in which a catalytically active PIWI domain cleaves the precursor hairpin independent of RNaseIII to yield mature miRNA [149]. While this pathway was once thought to be restricted to a single miRNA-like hairpin substrate, it now appears that a range of precursors can be processed [150, 151]. In most archaea with pPIWI proteins there is no RNaseIII encoded in the genome. Furthermore, there is neither direct physical evidence nor contextual support from comparative genomics for associations between RNase III-like and RdRP enzymes and PIWI proteins in bacteria. This suggests that basic versions of RNAi pathways could have emerged with just a nuclease-active PIWI protein.
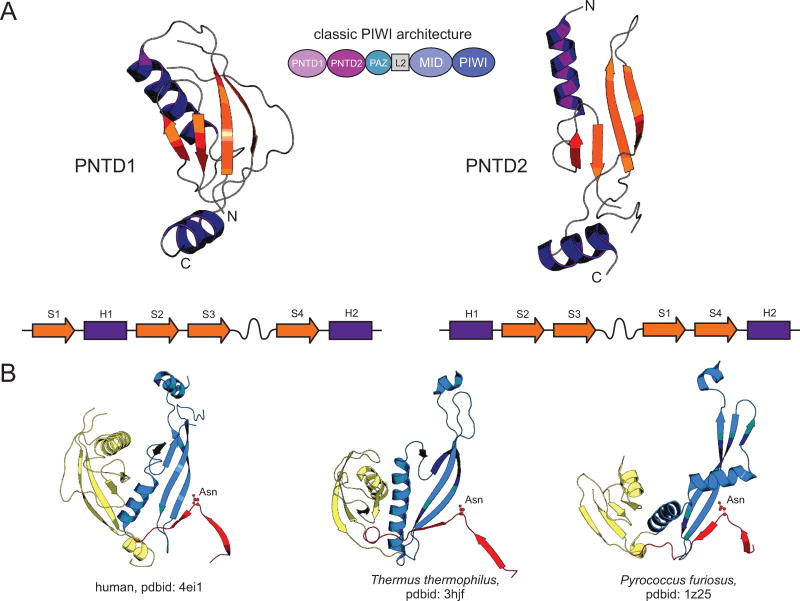
Recent structure-function studies on the PIWI proteins have elucidated how they might accomplish several distinct actions as part of their function. Eukaryotic PIWI proteins are comprised of the following domains: 1) a dyad of PIWI-N-terminal domains (PNTD1 and PTND2; Figs. 4, 5A). These two domains have arisen through duplication followed by a circular permutation at the N-terminus of one of the copies from an ancestral domain with 4 strands and two helices (Fig. 4). The N-terminal extension to the PNTD1 and PNTD2 domains contains an extended region with a highly conserved asparagine (Further Resources). The boundaries of these two domains have been inaccurately established in several studies resulting in two inappropriately-defined segments termed the N-terminal and Linker-1 domains. In the kinetoplastid-specific PIWI proteins a RNA-binding OB-fold domain of the NOB superfamily [152] is inserted between PNTD1 and PNTD2. 2) These domains are followed by PAZ, which is a RNA-binding domain adopting a SH3-like fold and plays an important role in recognition of the 3′end of the guide strand. 3) A conserved linker region (typically termed Linker-2). 4) The α/β sandwich MID domain which specifically binds the 5′ end of the guide strand. 5) The RNaseH fold domain which binds the target strand, and if active, uses its metal-dependent RNaseH active site to cleave target and passenger strands (Figs. 4,5A). This architecture of the PIWI proteins along with the distinctive domains for the recognition of the termini of the guide strands appears to have been the primary evolutionary constraint for the characteristic modal length of the small RNAs deployed in RNAi. Recent studies have shown that the PNTD1 and PNTD2 domains form an extended channel for binding of the duplex substrate and also play a key role in melting the passenger strand (i.e. the complementary strand in the dsRNA precursor of miRNA)-guide strand duplex by actively prying them apart to facilitate cleavage of the former. It is also supposed to play a similar role during cleavage of the target strand [153, 154]. This suggests the PNTD1/2 domain dyad acts as an inbuilt and ancestral switch allowing the RNaseH domain of the PIWI proteins to catalyze cleavage only when the former domains establish an appropriate conformation of the nucleic-acid protein complex. Furthermore, the PNTD1/2 domain dyad facilitates disengagement by preventing duplex propagation.
Figure 4.
Structural anatomy of PIWI N-terminal domain 1 (PNTD1) and PIWI N-terminal domain 2 (PNTD2). (A) Cartoon rendering and topological diagrams of the PNTD1 and PNTD2 domains. PNTD2 has previously been referred to as the Linker-1 domain, however, as these renderings make clear, the PNTD2 domain emerged via duplication and circular permutation from the PNTD1 domain, forming a single N-terminal module which is evolutionary present across all three superkingdoms of Life and is found in architectures outside of the well-studied classical PIWI architecture depicted in the center of the figure (see also Fig. 5A). β-strands are colored in orange, α-helices are colored in purple, and extended loop regions are colored in grey. N- and C-termini are labeled with “N” and “C”, respectively. (B) Cartoon renderings of the PNTD1/PNTD2 dyad along with the N-terminal leader region containing the well-conserved asparagine residue. Three protein representatives from the three superkingdoms of Life are depicted, labeled by species and pdbid at the bottom. PNTD1 is colored in yellow, PNTD2 is colored in blue, and the leader region is colored in red. The asparagine residue is rendered as a ball and stick, colored in red, and labeled as “Asn”.
Figure 5.
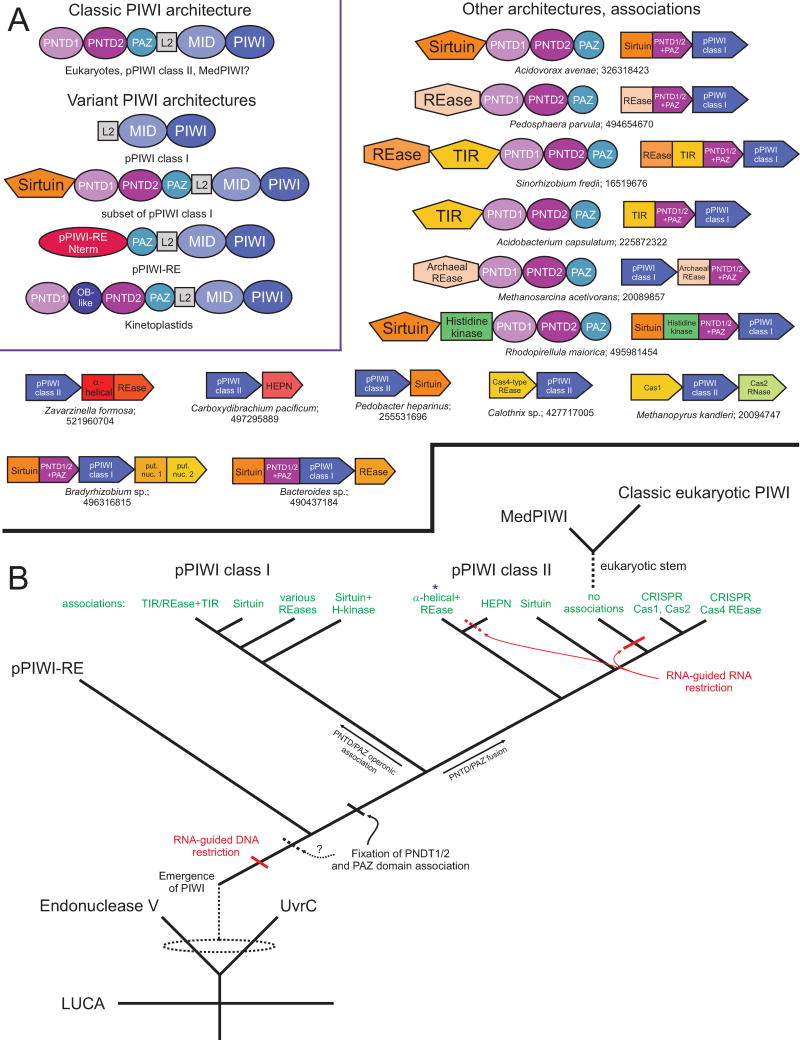
Genome associations and evolutionary history of the PIWI domain. (A) Domain architectures and conserved gene neighborhoods involving the PIWI domain. The inset, boxed in purple, depicts all known domain architectural themes for the PIWI domain with the phylogenies and/or PIWI families which contain these architectures provided below. Individual domains are depicted as labeled, boxed polygons. Outside of the inset, architectures and conserved gene neighborhoods containing PIWI domains and various nucleases in prokaryotes are provided. Depicted gene neighborhoods are single representatives of a cluster of sequences with the same, conserved neighborhood (for complete lists, see Further Resources). Conserved neighborhoods and domain architectures are labeled by species name and gene identifier (gi) number, separated by a semicolon. (B) Major events in the evolutionary history of the PIWI domain. Emergence of the PIWI domain from an Endonuclease V/UvrC ancestor, the likely cluster of pPIWI domains from which the eukaryotic PIWI versions emerged, and the higher-order relationships of individual PIWI families are depicted. Related groups of pPIWI class I and class II proteins are labeled in green according to the genome associations observed in (A). The group containing the recently-published PIWI-IP seq dataset from R. sphaeroides (α-helical+REase) is denoted with a blue asterisk. Predicted functional shifts for PIWI-centered systems are labeled in red. Dashed lines indicate uncertainty in terms of the origins of a lineage or timing of an event. Abbreviations: REase, Restriction endonuclease; put. nuc., putative nuclease; H-kinase, histidine kinase.
The PIWI RNaseH fold domain is most closely related to the Endonuclease V (EndoV) family of endoDNases, which is found in the EndoV and UvrC proteins [155]. While pPIWI proteins show a sporadic distribution in a relatively small group of prokaryotes, the UvrC proteins are highly conserved across bacteria and EndoV proteins across eukaryotes and archaea suggesting that an ancestral EndoV-like domain was probably already present in the last universal common ancestor. Hence, the PIWI RNaseH fold domain could have been derived from such an EndoV-like precursor originally involved in DNA repair in one of the prokaryotic superkingdoms (Fig. 5B). The pPIWI proteins, in particular the recent discovery of the pPIWI-RE clade of the PIWI superfamily, further clarifies the evolution of the characteristic domain architecture and the ancestral role of the PIWI superfamily. Sequence comparisons suggest that the pPIWI-RE clade was the first to branch off, with the remaining pPIWI proteins falling into two major clades, pPIWI class-I and class-II, in phylogenetic trees. Eukaryotic PIWI proteins emerge from within the class-II pPIWI clade (Fig. 4B). By the time of the divergence of the pPIWI and pPIWI-RE proteins, the PIWI proteins had already acquired the distinctive combination of the MID and the RNaseH domains and had probably added the distinctive N-terminal domains in addition [156]. Genome context analysis reveals that, like pPIWI-RE, most members of both classes are encoded in conserved operons alongside one or more several distinct endoDNases (Fig. 5, Further Resources): 1) 7 distinct families of nucleases of the restriction endonuclease (REase) fold; 2) a distinctive family of Sirtuin domains, which is comparable to those associated with the HerA-like DNA-pumping ATPases and is predicted to function as a DNase rather than as a deacylase [157]; 3) a distinct clade of TIR domain protein which were previously found in Restriction-Modification systems [158] and are also predicted to function as DNases. These observations, when superimposed on the relationships of the PIWI proteins (Fig. 5B), suggest that the ancestral function of the PIWI domain was in RNA-guided restriction of DNA. The report of the nucleotide content bound to a pPIWI domain in R. sphaeroides published while this manuscript was under review supports this contention [145]; the pPIWI system in R. sphaeroides belongs to the class-II pPIWI clade (Fig. 5B).
A major subset of the class-I pPIWI proteins are typically distinguished by the absence of the N-terminal PNTD1/2 and PAZ domains. However, in these cases these domains are found fused to the C-termini of the different endoDNase operonic partners with which the pPIWI is combined (Fig. 5A). A subset of these PNTD1/2 and PAZ domains in the case of those pPIWIs associated with Sirtuins were previously erroneously described as the analog of PAZ (aPAZ) domains [142]. This combination of the PNTD1/2 domains with the endoDNase domains, separately from the catalytic core of the class-I pPIWI proteins, suggests that they probably play a role in preventing deployment of the DNase component of the system until it joins the guide RNA-bound pPIWI protein and the PNTD1/2 facilitate proper target-guide pairing by virtue of their role in prying apart duplexes. This separation of the N-terminal domains is not observed in the class-II pPIWI clade, though a few of them might have entirely lost those domains (Fig. 5). This suggests that coupling with the DNA restriction component proceeds through distinct mechanisms in the two pPIWI clades (see above for more discussion on pPIWI-associating substrates). Certain class-II pPIWI proteins constitute a distinctive CRISPR/Cas system in which they are combined with the spacer-integration components Cas1 and Cas2 (Fig. 5). Since these systems lack the typical CRISPR/Cas processing RNases like Cas6, the class-II pPIWI proteins are likely to play a role in possibly processing and utilizing CRISPR RNAs for targeting invasive nucleic acids. Certain class-II pPIWI proteins are combined in operons with the endoRNase HEPN domains (Fig. 5) [159]. These along with the other class-II versions which show no linkages to endoDNases, including those close to the origin of the eukaryotic PIWI proteins, might be involved in purely RNA-guided RNA-targeting activities (Fig. 5). Thus, the situation in the pPIWI systems with numerous DNA-targeting versions alongside few RNA-targeting ones is comparable to the CRISPR/Cas systems, where most are RNA-guided DNA-targeting versions (such as the Cas9-containing type-II, Type-U, Type-I and many Type-IIIs) along with a few that target RNA (type III-B) [20, 160]. Eukaryotes, in contrast, appear to have built their systems primarily upon the RNA-targeting PIWI systems acquired early in their evolution from prokaryotes.
Coalescence of the eukaryotic RNAi core apparatus: the RdRP and RNaseIII domains
Like the PIWI superfamily, both the eukaryotic RdRP and the RNaseIII domains ultimately emerged from prokaryotic precursors. Studies on early-branching eukaryotic lineages and the shared phyletic distribution patterns of RNaseIII, PIWI domains, and RdRP suggest that they had come to together as a functional unit in the LECA [27–29]. Of these there is no evidence that the prokaryotic precursors of the RdRP had any direct role at all in RNAi-like processes. Instead the eukaryotic RdRPs evolved from enzymes that functioned as DdRPs in the transcription of certain bacteriophages, such as SPBC2, or primer-synthesis of certain distinctive bacterial mobile elements [125, 126]. However, it is possible that the transcripts generated by these DdRPs were used as substrates by prokaryotic phage-restriction systems with PIWI proteins. Thus, in eukaryotes there appears to have been a shift to using RNA templates as opposed a DNA one, paralleling the RNA-targeting specificity of the eukaryotic PIWI proteins. Unlike their prokaryotic precursors, upon acquisition the RdRPs have been inherited vertically throughout eukaryotic evolution.
Studies on the bacterial type II CRISPR/Cas systems have shown that the regular bacterial RNaseIII used in cellular dsRNA processing works in conjunction with the Cas9 protein (a RNaseH fold nuclease like PIWI) in dicing CRISPR transcripts in a complex with a complementary small RNA called the tracrRNA [161]. This suggests that RNaseIII enzymes might have generically functioned with multiple, evolutionarily distinct systems that involved dsRNA processing even in prokaryotes. Evolution of the eukaryotic RNaseIII enzymes has been unclear because previous sequence-based phylogenetic analyses have not been able to unequivocally confirm relationships between DICERs characterized in basal eukaryotes and those found in the crown group [148]. This is exacerbated by the high divergence of RNAi-active RNaseIII superfamily nucleases that have recently been identified in select kinetoplastids including T. brucei, Trypanosoma congolese, Trypanosoma vivax (but not Trypanosoma cruzi) [162], and Leishmania braziliensis (AMB, LA personal observations).
In an effort to resolve this issue, we exhaustively collected individual RNaseIII domains across prokaryotes and eukaryotes and generated phylogenetic trees from their sequences. The following main features of these trees are relevant to the early evolution of eukaryotic RNaseIII proteins: 1) all eukaryotic RNaseIII domains appear to be of common origin. 2) At least two copies of the domain can be traced to the LECA, both arranged in tandem in a single protein [163]. 3) The tandem RNaseIII domain copies observed in basal eukaryotes are closely related to each other and form a monophyletic assemblage, while the tandem repeats found in proteins belonging to crown eukaryotic lineages have diverged considerably. 4) The two repeats are followed by a single dsRBD domain, consistent with the canonical architecture observed in bacteria (RNaseIII+dsRBD). We can thus speculate the RNaseIII domain was acquired from an ancestral bacterial source with the dsRBD domain intact and subsequently underwent a partial duplication of only the RNaseIII domain. Sequence specialization between the two domains likely did not begin until the emergence of the kinetoplastid and chromalveolate lineages, which also coincided with the emergence of the mitochondrial MRPL44-like protein which can be traced to kinetoplastids and contains a single inactive RNaseIII domain and likely emerged via duplication and inactivation of a single copy of the more ancestral tandem RNAseIII protein. This tandem version might have also undergone a N-terminal fusion to the 5′ end RNA-binding PAZ domain as suggested by the architecture of the version from the basal eukaryote Giardia. Several additional domains associated with these core domains early in eukaryotic evolution. A member of the superfamily II (SF-II) helicases appears to have fused with the core DICER domains prior to the divergence of the heterolobosean Naegleria lineage (Fig. 1).
Early evolution of the small RNA component in RNAi systems
In attempting to reconstruct the ancestral small RNA substrate of RNAi systems, here we consider 1) known/probable RNAs associating with basal eukaryotic PIWI domains and 2) distribution of classes of RNA substrates across all eukaryotic lineages; the more widely appropriated an RNA substrate, the more likely it is to be ancestral. While keeping in mind that research on PIWI-RNA cargo in basal eukaryotes is still in its infancy, current findings suggest these RNAi pathways are capable of incorporating 1) hairpin-derived small RNA in both Giardia and Trichomonas [37, 38, 164], 2) dsRNA derived from convergent transcription as confirmed experimentally in Giardia and possibly in Trichomonas based on observed small RNA-mapping to gene loci [37, 56, 164], and 3) small RNA derived from double-stranded regions of longer classes of ncRNA such as snoRNAs whose derivatives are observed in Giardia [77, 78]. These three classes also appear to be the most broadly-distributed classes of small RNA associating with PIWI across diverse eukaryotic lineages (Table 2). Of the three, the most widely-distributed substrates appearing in every major lineage are dsRNA duplexes arising from convergent s-as transcription through a largely-conserved common mechanism. This is a particularly attractive candidate for an ancestral substrate given the widespread presence of overlapping s-as transcription in prokaryotic genomes, possibly reflective of conditions in the stem eukaryotic lineage [146].
While imperfectly-complementary hairpin processing and product association with PIWI from genome-encoded loci appears to be present in the basal eukaryotes Giardia and Trichomonas, as well as the apicomplexan T. gondii, plant, and fungi/animal lineages, evidence for these products is currently lacking elsewhere. Processing of larger ncRNA classes into small RNAs for RNAi pathways shows a widespread yet patchy distribution. ncRNA processing into PIWI cargo has not yet been observed in Trichomonas, kinetoplastids, or fungi, although tRNA fragments have recently been reported in Magnaporthe [85] but not yet investigated for PIWI-binding potential. Of the ncRNA-derived small RNA classes, tRNA-derived fragments are most commonly reported, notably present in amoebozoans, ciliates, and apicomplexa although they appear to be absent in Giardia and kinetoplastids, while snoRNA fragments are perhaps the most ancient form of processing given their presence in the basal Giardia [77]. Of additional potential relevance is the observation of half-tRNAs associating with a pPIWI domain [145]. Thus, products of imperfectly-complementary hairpins (miRNA-like) and larger ncRNA precursors could have been small RNA partners of the RNAi machinery in early eukaryotes; however, the functions of these RNA classes appear to have undergone considerable lineage-specific tinkering.
Later gene duplications and component specialization in the core RNAi machinery
The core protein components of the RNAi machinery display an extensive evolutionary history of lineage-specific duplications (Fig. 3). Diversification of PIWI proteins through duplication began right in the eukaryotic stem lineage, where it bifurcated into the classical PIWI family and the MedPIWI family (Figs. 1,5B), which was incorporated as an integral component of the CDK8 subcomplex of the Mediator complex [123]. A much later duplication of the PIWI proteins gave rise to the distinct Argonaute (AGO) subfamily in the common ancestor of animal, fungi, and plants [28] (Fig. 2). In animals, the PIWI “proper” clade became tightly functionally-linked with the emerging piRNA class of small RNAs and the nuage complex comprised of several proteins with RNA-binding and methylated arginine-binding domains involved in protection of germline integrity. The AGO clade retained roles typically attributed to the ancestral PIWI proteins in other eukaryotic lineages. Several lineage-specific duplications have occurred in PIWI/AGO proteins (Fig. 2). These duplication events can be accompanied by specialization of the small RNA substrates binding the PIWI protein with notable examples observed in T. thermophila, Oikopleura (a tunicate with an otherwise reduced genome), Adineta vaga (a bdelloid rotifer which is an asexual animal), and C. elegans [147, 165] (Fig. 2). On other occasions duplication events have given rise to apparently functionally redundant PIWI proteins, as in the mammalian set of four AGO proteins [166].
Similarly, duplications have played a major role in the evolution of the RNaseIII superfamily. A second version of the tandem RNaseIII repeat-containing protein emerged possibly as early as the heterolobosean lineage. The first version, showing strongest affinity to the single ortholog present in basal lineages like Giardia and Trichomonas, is related to the classical DICER-like enzymes from other eukaryotes. The second, faster-evolving version includes the DROSHA-like enzymes known in animals for their intermediary role in miRNA production. While it has long been assumed that DROSHA homologs are restricted to animals, we contend on the basis of the sequence profile comparisons and phylogenetic analyses outlined above that DROSHA homologs exist across many major eukaryotic lineages including the plants (as typified by the previously uncharacterized Arabidopsis thaliana AT4G37510 protein—see Further Resources), chromalveolates, and possibly the heterolobosean Naegleria. The most parsimonious evolutionary scenario for the emergence of the DROSHA-containing clade is simple duplication of the DICER-like protein of basal eukaryotes described above. This is consistent with the general dearth of N-terminal domain fusions (e.g. the helicase domains) observed in both basal DICER-like enzymes and members of the DROSHA-containing clade. However, we cannot rule out the possibility of DROSHA-like enzymes emerging first in the common ancestor of animals and plants, with lateral transfers to heteroloboseans and chromalveolates. The discovery of DROSHA-like homologs in plants also raises questions about their function. In animals, DROSHA processes the pre-miRNA from the pri-miRNA transcript; in plants, however, this step, along with the ensuing step processing the dsRNA hairpin from the pre-miRNA, are catalyzed by successive action of the DICER enzymes. Given that the tandem RNaseIII-like architecture in eukaryotes to this point has exclusively been involved in RNAi pathways, we predict the plant DROSHA-like enzymes have an as-yet-undetermined role in a plant RNAi pathway.
In kinetoplastids, the tandem RNaseIII architecture-containing protein has duplicated on at least one occasion and is currently known from T. brucei, Trypanosoma congolese, Trypanosoma vivax [162], and Leishmania braziliensis (AMB, LA personal observations). The rapid divergence of the kinetoplastid RNaseIII repeats in these proteins contributes to a lack of confidence in assessing their affinities, although the second RNaseIII repeat shows affinity for the DICER-like clade suggesting a possible vertical inheritance from the basal tandem RNaseIII domain-containing versions followed by a particularly rapid divergence of the first RNaseIII repeat in kinetoplastids (Fig. 3). RNaseIII domains belonging to two distinct proteins containing the tandem architecture from the amoebozoan Acanthamoeba, Dictyostelium, and Polysphondylium organisms similarly cluster together largely outside of the major clades. The first and second RNaseIII repeats from both versions cluster together to the exclusion of other RNaseIII domains, suggesting the independent emergence of the tandem RNaseIII architecture in these organisms. Fungi contain several distinct RNaseIII domain proteins. While the loss of the canonical DICER in budding yeast has been well-documented, several ascomycetes and basidiomycetes have retained canonical DICER-like enzymes. In addition, most fungi contain inactive Yml15-like enzymes belonging to the MRLP44-like clade. This clade, despite a sporadic distribution in early-branching eukaryotic lineages, appears to have been fixed in the common ancestor of plants, animals, and fungi. Rnt1p-like enzymes represent a group of active single RNaseIII domain proteins in fungi along with their sister group, the so-called DCR1-like enzymes linked to RNAi activity in some budding yeasts [167–169]. Our analyses suggest that the Rnt1p-, DCR1-, and pac1 (a group of snoRNA-processing enzymes primarily observed in fission yeast)-like enzymes share a monophyletic origin, suggesting late emergence from a more ancestral active RNaseIII domain. Retention of a C-terminal dsRBD domain and their monophyly with other eukaryotic RNaseIII domains points to the origin of these fungal proteins from the tandem domain-containing version accompanied by loss of the N-terminal RNaseIII domain (Figs. 1,3).
Duplication of the RdRPs at various points in eukaryotic evolution (e.g. in plants and in A. vaga) appear to have been frequently accompanied by a concomitant proliferation and specialization of a DICER/RdRP homolog with specific small RNA substrates [170] (Fig. 3). Of note in this regard is A. vaga which possesses an expansion of RdRPs (20 copies) and DICERs (8 copies) [165].
Gene loss in the core RNAi machinery
The RdRP enzyme is observed to be frequently lost despite retention of DICER and PIWI/AGO (e.g. independently in insects and vertebrates) (Fig. 3). Loss of RdRP does not appear to coincide with a decrease in either the diversity of small RNA substrates found associating with PIWI/AGO or in the number/variety of distinct functional contexts occupied by RNAi, at least in well-studied lineages. In addition to isolated component loss, the entire or partial complement of core RNAi components has been lost in several fungal lineages, with the best-known example being the complete loss of the RNAi machinery in several budding yeast lineages [171] and in the fungus Ustilago maydis [172]. In the budding yeasts like S. cerevisiae the loss of the core RNAi system is accompanied by extensive loss of spliceosomal introns and spliceosomal components. Similarly, several kinetoplastids including Trypanosoma cruzi, Leishmania major, Leishmania infantum, Leishmania donovani, and Leishmania mexicana as well as most apicomplexa have respectively partially or entirely lost the complement of core RNAi components, although notably all kinetoplastids contain at least one PIWI domain with the characteristic insertion of the NOB domain (Figs. 3,5A).
Loss and re-emergence of core RNAi components in closely-related organisms from both fungi and kinetoplastids can be instructive in understanding the adaptability of the system and its relation to the small RNA substrate (Fig. 3). IP-seq has been performed on classical PIWI proteins for RNAi-active T. brucei [52] and small RNA-seq on T. cruzi, which contains only the distinctive kinetoplastid-type PIWI protein with the NOB domain (Figs. 3,5A) [173]. Similarly, small RNA-seq has also been performed in RNAi-active Saccharomyces castellii (now known as Naumovozyma castellii) and the RNAi-deficient Saccharomyces cerevisiae that lacks even a PIWI protein [167]. In both pairs of organisms, RNA profiles of the RNAi-deficient organisms appear to primarily consist of degradation products derived from rRNA, tRNA, repeats, and coding regions. Interestingly, consistent with the presence of a PIWI protein in T. cruzi, at least a portion of the sequences derived from snRNA, snoRNA, and coding regions are found in the length range typical of AGO/PIWI-associating small RNA (~20–23nt) [173]. Hence, it is conceivable that kinetoplastids other than T. brucei, while lacking conventional RNAi, possess some lineage-specific specialized version that uses their distinctive version of the PIWI protein. These profiles contrast drastically with those from organisms with active RNAi. In T. brucei the PIWI-IP profile reveals association with several distinct classes of small RNA including small RNAs derived from s-as retrotransposon transcription, satellite-like repeats, convergently-transcribed protein coding regions, and inverted repeats [52]. The small RNA population of S. castellii and related budding yeasts, which appear to have reused a member of the Rntp1/DCR1/Pac1 radiation for endonuclease activity to “reconstitute” an active RNAi system, also contain a rich assortment of sequences similar to other eukaryotes including those derived from s-as, inverted repeats, and other ncRNA sources [167] (Fig. 2). Thus, it appears the re-emergence of a functional RNAi system results in generation of similar small RNA substrates, pointing again to the inherent predisposition of the system to specific classes of precursor substrates.
Evolution of ancillary RNAi machinery and specialized classes of small RNA
The above-described core machinery is in large part sufficient to process substrates for RNAi from double-stranded sequences derived from s-as transcription, simple genome-encoded hairpins, and regions from larger ncRNA species. Consistent with this, little to no elaboration to the core machinery is typically observed in the processing of these substrates across eukaryotes (Fig. 2). Instead, elaboration of the core machinery appears to have occurred primarily in two contexts: 1) the emergence of novel classes of small RNA substrates, e.g. piRNAs and 2) the “institutionalization” and subsequent rapid expansion of particular classes of small RNA substrates, with examples including the animal and plant miRNA pathways. An unusual elaboration is observed in the case of the so-called “social RNA”, where a RNA substrate import apparatus centered on the ceramidase domain-containing transmembrane SID-1 protein appears to have been superimposed on the core machinery [311–313].
Recruitment of further RNase domains to the RNAi system
One of the earliest elaborations on the core RNAi machinery appears to have occurred in the kinetoplastid lineage. In addition to core components, RNAi in T. brucei was recently discovered to depend on the TbRIF4 and TbRIF5 genes, both of which are present only in kinetoplastid lineages which retained the active RNAi pathway [174]. TbRIF4 contains an active Maelstrom-like RNase H fold 3′->5′ exonuclease domain and assists in processing RNAs bound by the guide strand of the T. brucei PIWI domain [174]. We also propose a similar role for the Maelstrom-like nucleases in processing small RNAs in Entamoeba, which lacks a member of the RNaseIII superfamily (Fig. 3). While Maelstrom-like nucleases in many later-branching eukaryotic lineages are inactive, experimental evidence indicates they retain roles related RNAi, namely in piRNA biogenesis and transposon silencing in the germline [175]. Inactive Maelstrom-like domains are predicted to contribute to these functions through a RNA-binding role [176] which in turn regulates cytosine methylation of DNA, perhaps requiring their N-terminal HMG domain (Fig. 1) [177]. While the initial study identifying TbRIF5 did not observe any conserved regions in the protein, we identify for the first time here the presence of two tandem, N-terminal copies of the Staphylococcus nuclease (SNase) domain which appear most closely related to SNase domains found in the Tudor-SN/P100 proteins (AMB, LA, personal observations) (Further Resources) [178]. Tudor-SN proteins have previously been linked to RNAi systems, involved in degradation of hyper-edited inosine-containing miRNA precursors [179, 180]. Tudor-SN is an innovation traceable to the LECA (also present in kinetoplastids); suggesting TbRIF5 likely emerged via at least partial duplication of Tudor-SN in the kinetoplastid lineage. While we currently do not observe a Tudor domain in TbRIF5, there remain several uncharacterized globular regions C-terminal to the SNase domains that could harbour divergent copies of Tudor or additional SNase domains. The observed physical association between the T. brucei tandem RNaseIII domain-containing (DICER-like) protein and TbRIF5 in T. brucei [174], suggests that the SNase domains of TbRIF5 contribute to the processing of dsRNA targets.
In course of eukaryotic evolution several other nucleases appear to have been recruited to the RNAi machinery. The HKD-type nuclease found in the Zucchini protein and the Mut7-N (Nibbler) nuclease of the PIN fold were first experimentally identified as RNAi components in Drosophila [181–183] and Caenorhabditis [184], respectively (see below). They emerged at least prior to the divergence of the alveolates, plants, animals and fungi from their common ancestor, but have been lost in several lineages. The Drosophila squash protein was originally claimed to be a nuclease [181] but analysis using sequence profiles indicates that it is a low-complexity protein limited to arthropods. In addition, two distinct nucleases were recruited for regulation of miRNA following maturation: the RNase H fold 3′-to-5′ exonuclease SDN1 in plants [185] and the 5′-to-3′ exonuclease XRN2 in animals [186]. XRN2 is highly conserved and likely to have been present in the LECA with a primary role in rRNA processing; further investigation into potential roles in RNAi for XRN2 in other eukaryotes might be of interest, particularly in light of its linkage to RNAi systems in T. thermophila (Fig. 1) [88]. Similarly, the Eri-1 nuclease, which combines an N-terminal SAP domain with a 3′-to-5′ exonuclease domain has been found to have a role in both 5.8S rRNA processing and regulation of RNAi [187]. In the latter processes it has been implicated in both the maturation of certain siRNAs and negative regulation of silencing by degradation of small RNAs [188]. Bacterial orthologs of Eri-1 are often found in a conserved gene-neighborhood with genes for tRNAs and rRNAs, suggesting that it plays a role in the processing of these RNAs. The LK-nuclease domain, a member of the 5′->3′ exonuclease superfamily [189], is fused to C-terminal RRM and OST-HTH RNA-binding domains [189, 190] in the MARF1 protein and its homologs. Recently MARF1 has been implicated as a female germ cell analog to the piRNA-PIWI complex in control of retrotransposon silencing in oocytes [191]. This nuclease domain is likely to regulate processing of as-yet-unidentified small RNAs in this process. Both the Eri-1-like and LK-nuclease domains are widespread in bacteria but largely absent in archaea. Thus, they appear to have been transferred to eukaryotes at different points from bacteria: prior to the divergence of kinetoplastids (Eri-1) and in the common ancestor of plants and animals (LK-nuclease). The metallo-β-lactamase (MβL) fold-containing RNase Z protein has been implicated in DICER-independent processing of tRNA in animals during production of 3′ tRF-U small RNAs [81]. RNase Z belongs to a higher-order assemblage in the MβL fold which includes two other families which act as endonucleases [192], the DNA-cleaving SNM1 family involved in DNA repair [193] and the RNA-cleaving CPSF family which cleaves the 3′ cleavage signal in pre-mRNA transcripts [194]. Two paralogous versions of RNase Z are present in eukaryotes with at least one of these versions traceable to the LECA; however, the version which appears to be implicated in small RNA generation is traceable only to Naegleria, stramenopiles, and possibly some apicomplexan lineages, suggesting it emerged from the more ancestral version later in eukaryotic evolution.
Recruitment of enzymes modifying small RNAs to the RNAi system
RNA-modifying enzymes such as methylases and nucleotidyltransferases play a major role in maturation, processing, and stability of certain RNA components of the RNAi system. The distinctive methylation of 2′ OH at the 3′end of specific small RNAs is catalyzed by the HEN1 RNA methylases [195]. In animals this modification is an integral part of the maturation of piRNAs. In plants, degradation of miRNAs is mediated by competition between degradation-blocking HEN1 RNA methylation [196, 197] and degradation-inducing polyuridylation by the nucleotidyltransferase MUT68/HESO1 [198–200]. In ciliates, Hen1 methylates the 3′ terminus of scnRNA during biogenesis, contributing to scnRNA stability [195]. Hen1 appears to have been first recruited to RNAi machinery via HGT from a bacterial source, where it appears to regulate RNA repair (Fig. 1) [201]. The above observations suggest that HEN1’s recruitment in eukaryotes was followed by its deployment in modifying small RNAs from different limbs of the RNAi system in different eukaryotic lineages. However, given the role of both animal piRNAs and ciliate scnRNA in suppressing gene disruption via transposons or selfish DNA, it is conceivable that modification by HEN1 is a fairly ancient aspect of RNAi-dependent protection of genes from selfish elements [202] (Fig. 1). Nsun2 is an additional methyltransferase containing the same fold as Hen1 recently observed in position-specific methylation of vault RNA as a prerequisite for svRNA generation [203]. Nsun2 is a tRNA-modifying enzyme traceable to the LECA, suggesting it acquired an additional functionality in vault RNA processing following the fixation of the vault RNA after emergence of the crown group (Figs. 1,2).
The other major modification, namely 3′ addition of nucleotides, is catalyzed by the TRF polymerase family of 3′ nucleotidyltransferases of the DNA polymerase β (DNA pol-β) superfamily [204] (Table 3, Figs. 1,2). The TRF family of polymerases is specifically related to the poly(A) polymerase, also catalyzing 3′ template-free homopolymerization of particular nucleotides. In animals, the GLD-2 subfamily of TRF-like nucleotidyltransferases adenylates mature miRNAs which stabilizes miR-122 in liver cells [205] and appears to generally act to reduce affinity for AGO proteins [206]. The ZCCHC6/ZCCH611 nucleotidyltransferases were recently shown to catalyze the monouridylation of pre-let-7 miRNAs, a step required for maturation [207]; however, in the presence of the lin28 cofactor (see below) this subfamily polyuridylates pre-let-7 miRNAs, apparently blocking interaction with DICER and leading to decay [208]. They also uridylate mature miRNAs, modulating their effectiveness [209, 210] and perhaps facilitating their degradation similar to the plant MUT68/HESO1 uridylyltransferases [198–200]. MUT68/HESO1 uridylyltransferases are also implicated in 3′ modification of RISC-cleaved fragments [198, 211]. In Tetrahymena, the uridylyltransferases Rdn1 and Rdn2 form distinct complexes with the RdRP to mediate processing/amplification of distinct groups of 23–25nt RNAs [212]. Related TRF nucleotidyltransferases also appear to be involved in highly selective modification of other subsets of RNA transcripts in animals; TUT1 is implicated in polyadenylation of specific mRNAs [213] and polyuridylation of specific snRNA [214] transcripts in the nucleus while MTPAP polyadenylates mitochondrial transcripts [215].
Table 3.
Classification of TRF4/TRF5-like family of nucleotidyltransferases
| Subfamily | Phyletic distribution | Prominent members1 | Known functions | Comments |
|---|---|---|---|---|
| classical TRF4/5 | all eukaryotes | TRF4 (Scer), TRF5 (Scer), cid14 (Spom), DmTRF4-1 (Dmel), DmTRF4-2 (Dmel), GLD-4 (Cele), ncPAP1 (Tbru) | snRNA polyadenylation [278], U14 snoRNA, U5 snRNA, pre- ribosomal rRNA degradation [279], intergenic transcript degradation [280], hypomodified tRNA degradation [281, 282], etc. | PAPD7-like and PAPD5-like assemblages formed via gene duplication event occurring in TRF4 during chordate evolution; Scer TRF4 and TRF5, meanwhile, result from an Scer-specific gene duplication event |
| **PAPD7-like | jawed vertebrates | PAPD7/POLS/TRF4- 1/LAK1/POLK/TRF4 (Hsap) | - | |
| **PAPD5-like | jawed vertebrates and Xenopus | PAPD5/TRF4-2(Hsap) | snoRNA adenylation [283], miRNA modification [206, 218] | |
| TUT1 | animals | TUT1/PAPD2/TUTase/U6- TUTase/STAR-PAP/Hs5 (Hsap) | Regulation of selected mRNAs [213, 284], U6 snRNA uridylyl transferase [214], miRNA modification [218] | TUT1 & MTPAP subfamilies form a monophyletic higher- order assemblage |
| MTPAP | animals | MTPAP/PAPD1/Hs4 (Hsap) | mt-mRNA polyA tail extension [215, 285], miRNA modification [218] | |
| ZCCHC6/ZCCHC11 repeat 2 subfamily | crown group eukaryotes | ZCCHC6/PAPD6/Hs2 (Hsap), ZCCHC11/PAPD3 (Hsap), cid- 1(Cele), pup-2 (Cele), cid11 (Spom), cid13 (Spom), cid1 (Spom) | checkpoint control [286, 287], mRNA polyuridylation [288], targets ribonucleotide reductase mRNA [289], histone mRNA uridylation [290], pre-miRNA polyuridylation [227, 290, 291] miRNA uridylation [207, 209, 210] | Repeat 1 subfamily associated with repeat 2 subfamily before divergence of nematodes. cid11, cid13 are closely related to the exclusion of other sequences, suggesting late gene duplication event in S. pombe. Repeat 2 subfamily and GLD-2/PAPD4 likely share a common ancestor which originally emerged as single copy in Naegleria or chromalveolate lineages. |
| ZCCHC6/ZCCHC11 repeat 1 subfamily | chordates, Strongylocentrotus, C. elegans | ZCCHC6/PAPD6/Hs2 (Hsap), ZCCHC11/PAPD3 (Hsap), cid-1 (Cele) | see above | |
| GLD-2/PAPD4 | animals | GLD-2 (Cele), PAPD4/Hs1 (Hsap), XGLD-2 (Xlae), mut-2 (Cele) | Cytoplasmic mRNA polyadenylation [292–294], miRNA adenylation [205], transposon silencing and RNAi [295, 296] | |
| basal ZCCHC6/ZCCHC11 repeat 2-like, GLD- 2/PAPD4-like subfamily | Naegleria, ciliates, stramenopiles, slime molds, possible apicomplexan representation | Rdn1 (Tthe), Rdn2 (Tthe) | endo-siRNA production [212, 297] | |
| AT3G56320-like2 | plants, Giardia, kinetoplastids, stramenopiles, Naegleria | AT3G56320 (Atha), KPAP2 (Tbru) | mitochondrial mRNA editing [298] | |
| RET1-like | kinetoplastids | RET1 (Ltar), RET2 (Ltar), TbTUT3(Tbru), TbTUT4 (Tbru), KPAP1 (Tbru) | mRNA, gRNA uridylation, mitochondrial mRNA editing [299–303], mitochondrial transcript polyadenylation [304] | Monophyletic kinetoplastid- specific expansion |
| MEE44-like2 | plants, slime molds, stramenopiles | MEE44 (Atha) | - | Arabidopsis deletion linked to embryonic arrest [305] |
| MUT68-like | plants | MUT68 (Crei) | adenylation of RISC-cleaved mRNA fragments [211], miRNA and siRNA uridylation [198] | |
| MEAT1-like | kinetoplastids | MEAT1 (Tbru) | mitochondrial mRNA editing [306] | |
| ncPAP2-like | kinetoplastids | ncPAP2 (Tbru) | nuclear adenylation [307] | |
| monkey king | insects | mkg (Dmel), mkg-p(Dmel) | - | previously described [308] |
| CG1091-like2 | insects | CG1091 (Dmel) | - | |
| PFD1045c-like2 | apicomplexa | PFD1045c (Pfal) | - | PFD1045c was identified as an erythrocyte membrane- associated antigen [217] |
| Paramecium expansion2 | Paramecium | -- | - | Paramecium lineage-specific expansion |
Several members of the TRF4/TRF5 family notably found in S. pombe and C. elegans failed to reliably fall within a clear, monophyletic assemblage, including the functionally characterized, telomeric repeat-adenylating cid12 [309, 310].
Indicates newly-characterized subfamily
differentiated lineages emerging after a duplication event split the classical TRF4/5 subfamily late in animal evolution.
A detailed analysis performed on the TRF polymerase family revealed two members of the family can be confidently traced to LECA, one belonging to the classical TRF4/5 subfamily found across eukaryotes and involved in nuclear RNA surveillance which tags various non-coding RNA through 3′ polyadenylation for degradation by the exosome [216] and another belonging to the functionally uncharacterized AT3G56320-like subfamily found in Giardia, kinetoplastids, Naegleria, chromalveolates, and plants (Table 3). Extensive diversification in the TRF family occurred in kinetoplastids: in addition to retaining members of the aforementioned subfamilies, additional kinetoplastid-specific subfamilies emerged, including the RET1-like, MEAT1-like, and ncPAP2-like subfamilies, which play a wide range of roles in the cell (Table 3). Phylogenetic analysis indicates the large RET1-like clade, which includes the TbTUT3, TbTUT4, and KPAP1 paralogs in addition to RET1 and RET2, might be derived from early duplication of a member of the AT3G56320-like clade. Despite this diversification, to this point no kinetoplastid TRF members have been linked to RNAi; however, they could be involved in the distinctive RNA editing of kinetoplastids, which involves additions of polyuridine tracts. The ciliate Rdn1 and Rdn2[251] cluster together with other representatives from ciliates and several alveolates, stramenopiles, and Naegleria along with the GLD-2/PAPD4-like and ZCCHC6/ZCCHC11 repeat 2-like clades. The ZCCHC6/ZCCHC11 repeat 2-like clade, present across crown group eukaryotes, appears to retain the 3′ RNA uridylation observed in more basal versions typified by the ciliate enzymes. In animals the ancestral ZCCHC6/ZCCHC11 catalytic domain underwent an intra-protein duplication resulting in the repeat 1-like subfamily (Table 3). This version with polymerase domains further duplicated spawning the paralogous ZCCHC6 and ZCCHC11 clades early in animal evolution. Orthologs of these enzymes underwent additional, distinct duplications in certain lineages, most notably the Cid11 and Cid13 paralogs in S. pombe and a duplication at the base of fungi giving rise to a set of paralogs including S. pombe Cid1 (Table 3). The GLD-2/PAPD4-like family emerged later in animals, likely concomitant with the apparent functional shift to 3′ RNA adenylation. The extensive linkage of this assemblage of clades (Table 3) to different steps in RNAi systems (see below) suggests a possible ancestral role in modifying RNA substrates of RNAi pathways. However, both the GLD-2/PAPD4-like and ZCCHC6/ZCCHC11-like subfamilies have been linked to additional roles in cytoplasmic mRNA 3′ modification suggesting considerable substrate promiscuity (Table 3). Additionally, we identify three uncharacterized T. thermophila homologs clustering tightly with Rdn1 and Rdn2, exploration of the functions of these homologs could shed further light on the potential ancestral role for this assemblage of TRF subfamilies. Two additional clades, prototyped by TUT1 and MTPAP, form a monophyletic, higher-order assemblage which emerged at the base of animals but diverged into distinct clades early in animal evolution, sometime after early-branching animal lineages like Nematostella and Trichoplax which contain only single representative copies of the assemblage (Table 3). Several additional lineage-specific clades are seen in the TRF family including the plant-specific MUT68-like clade, the uncharacterized MEE44-like clade, the uncharacterized insect-specific clades Monkey King and CG1091, and the apicomplexan-specific PFD1045c (Table 3). PFD1045c was earlier linked to host antigen expression in Plasmodium falciparum [217]. In light of the recently-uncovered role for s-as transcription in regulation of the surface antigen var gene family in P. falciparum [66] and noted cases of TRF family-mediated 3′ addition to hairpin/duplex RNA, it might be worth investigating potential roles for PFD1045c-mediated degradation of repressed var transcripts.
Thus, it appears that on at least two occasions TRF family polymerases have been specifically recruited to RNAi activities, once at the base of the GLD-2/PAPD4-ZCCHC6/ZCCHC11 assemblage and once in the plant MUT68-like subfamily. Other RNAi-related roles observed for the family in the classical TRF, MTPAP, and TUT1 subfamilies likely represent secondary functional acquisitions, consistent with the restricted number and cell-type specificity of observed substrates [206, 218]. Thus, the current data point to the accretion of HEN1 methylases occurring relatively late, i.e. prior to divergence of chromalveolates and the crown group, whereas nucleotidyltransferases might have been recruited earlier (Figs. 1,2). Currently modification of pri-miRNAs (e.g. pri-miR-142, the precursor of miRNA-142) by deamination of adenines to inosines by the dsRNA-specific ADAR enzymes has been reported in vertebrates; this modification counters processing by Drosha and routes the pri-miRNA for degradation by Tudor-SN [179]. The ADAR enzymes emerged first in metazoans via duplication of the pan-eukaryotic tRNA adenine deaminase Tad1 and modify both mRNAs and miRNAs (Figs. 1,2). it remains to be seen whether Tad1 might intersect with the RNAi system in other eukaryotes: recently a clade of predicted cytidine deaminases fused to Tudor domains has been reported in stramenopiles which might have independently been recruited for modification of small RNAs in this clade [219].
RNA-binding proteins and other ancillary components of the RNAi system
Above discussion regarding the recruitment of processing and modifying enzymes indicates that in the crown group clade (the common ancestor of animal and plants and all their descendants), many major innovations are centered on the miRNA system. Analysis of other ancillary components also points in a similar direction, with comparable roles emerging in plants and animals: the dual dsRBD-containing Hyl1/Pasha/DGCR8 protein which binds pri-miRNAs and the exportin-5/HASTY protein which exports pre-miRNAs to the cytoplasm have been found in both plants and animals but are lost in fungi (Fig. 1). Several of these proteins appear to have been recruited as regulatory factors at different steps in miRNA biogenesis; these globally regulate miRNAs or function in narrower contexts regulating specific miRNAs or miRNA families. Some examples include the C2H2-Zn-finger-containing Ars2/SERRATE protein which widely stabilizes pri-miRNAs and augments their processing to miRNAs in plants and animals [220–223] (Fig. 1). Likewise the FHA domain-containing DAWDLE protein, which globally stabilizes pri-miRNAs [173], points to a common mechanism involving regulation of miRNA maturation via protein serine/threonine phosphorylation in both plants and animals. The lin-28 protein which combines a S1-like and dual zinc knuckle RNA-binding domains inhibits DICER processing of particular miRNAs in conjunction with 3′ terminal polyuridylation [224–227] (Fig. 1). While lin-28 has been implicated in a miRNA-related role in both plants and animals, highly-divergent copies appear to be present in earlier-branching lineages such as ciliates and kinetoplastids, where they are implicated in RNA editing [228]. This suggests that a Lin-28 like protein might have been acquired earlier from bacteria, where it has been shown to function as a mRNA chaperone and transcription anti-terminator that destabilizes mRNA hairpins [229] (Figs. 1,2). In contrast, other proteins associated with the miRNA system show a more limited phyletic pattern: the RRM domain-containing proteins TRBP/Loquacious and PACT, which associate with DICER and contribute to miRNA substrate and cleavage specificity, appear to be animal- and vertebrate-specific, respectively [230]. The animal RNA-binding zinc finger domain-containing MBNL1 protein binds to and regulates expression of pre-miR-1 in muscle cells [231]. In plants the SQUINT protein with a peptidyl prolyl-cis/trans-isomerase cyclophilin and a TPR domain appears to be a protein folding chaperone that augments AGO activity [232] (Fig. 1).
The above discussion points to a rather extensive regulatory infrastructure being in place prior to the divergence of animals and plants, which could have acted as a pre-adaptation facilitating emergence of the numerous miRNAs in these lineages. It has been proposed that miRNAs with physiologically important targets originate with low expression levels. As deleterious off-target sites are purified over time the expression and relative functional importance of the miRNA is believed to increase [233]. Indeed, the above-outlined regulatory infrastructure could have provided sufficient robustness to limit the negative consequences of off-target interactions by means of multiple control steps even as the miRNA-system evolved greater specificity. In fungi, the concerted loss of this system appears to have precluded the use of miRNAs. Nevertheless, the remnant RNAi systems in fungi appear to have undergone further elaboration by addition of distinctive ancillary proteins including the SF1-helicase SAD-3/Hrr1, the CHROMO+RRM domain-containing Chp1, and the nucleotidyltransferase Cid12 [65] (Figs. 1,2).
The other major recruitment of ancillary proteins concerns the interface between the RNAi machinery and chromatin dynamics. This link appears to have emerged as an offshoot of ancestral transcriptional regulation by PIWI family proteins in the nucleus. Evidence from ciliates suggests that relatively early in eukaryotic evolution the RNAi-chromatin interface was already regulating generation of epigenetic marks by histone methylation. This also appears to have marked the emergence of the precursors of the nuage-like RNP complexes, which expanded further in animals with the emergence of piRNAs as major defenders of the set aside germline. The initial phase of the evolution of the precursors of nuage-like complexes appears to have been accompanied by the recruitment of the HKD nuclease Zucchini [181], the SF-I helicase-containing Armitage (also found in plants) [234–236] and possibly the 3′-5′ nuclease domain-containing Maelstrom [175]. Another key innovation in this phase was the expansion of the predicted dsRNA-binding OST-HTH [190] and Tudor domain proteins which bind methylated arginines common in RNA-binding proteins generated by the PRMT family of methylases. In animals there is evidence for the apparent burgeoning of this system as suggested by several distinct components identified over the past two decades: 1) SF-II RNA helicases like Vasa [237], 2) the chaperones Hsp90 and Shutdown/FKB6 with a peptidyl prolyl-cis/trans-isomerase and TPR domains [238, 239], 3) and an expansion of Tudor domain fusion proteins, such as Tudor [240], the insect-specific Krimper [241], SF-II helicase-fused Spindle-E [237, 241], Yb, BoYb and SoYb [234–236], the dsRBD-containing Vret present in at least insects and sea urchins [242], the RING and B-box containing qin/Kumo proteins [243, 244], and the histone methyltransferase SET domain-fused SETDB1 [245]. In addition, RNAi systems of both animals and plants interface DNA modification by DNA cytosine methylases which generate 5mC. In this context it is of interest to note that a SM-like domain is found at the N-terminus of many DNMT1-like methylases [246]. In prokaryotes, SM domains bind several specialized small RNAs, effecting regulation of mRNA stability and ribosome association [247, 248]. Hence, the possibility of direct interactions between small RNAs and DNMT1 in eukaryotes remains a matter of interest.
General Conclusions
Characterization of the RNAi system has been a principal driving force in the discovery of a veritable “RNA world” which practically remained out of sight even as little as 15 years ago. The characterization of these RNAs has led to an appreciation of their extensive involvement with several major biological processes. This has also had a major biotechnological impact with RNAi-based technologies rapidly occupying the center-stage in molecular biology for several model systems. However, several basic biological questions concerning this system are only recently coming into greater focus.
First among these is the functional relevance of the RNAi system in the greater hierarchy of regulatory mechanisms comprised of direct transcription control, chromatin level control, splicing, and RNAi-independent regulation of transcript maturation and stability. Some workers have expressed a rather maximalist view of the role of the RNAi system: 1) based on the role of the system in countering killer plasmids and other selfish elements some have speculated that organismal lineages cannot survive over long evolutionary times without such systems [249]. 2) Others have suggested that emergence of complex nervous systems in vertebrates relates to the proliferation of miRNAs [249, 250]. 3) Yet others have proposed a role for the RNAi system in emergence of organizational complexity in animal and plants [71]. However, actual evidence for such maximalist positions remains equivocal at best. Studies on regulatory networks have suggested that a distributed architecture with regulatory backups and division of the regulatory “responsibility” among several separate systems, each with a small individual effect, helps minimize the risk of failure [251–253]. Elaboration of the RNAi system in eukaryotes supports this principle as it adds a level across which regulatory “responsibility” can be distributed and also provides a backup for other systems. However, there still appears to be a hierarchy among the eukaryotic control systems – while very few eukaryotic lineages apparently lack specific transcription factors, there are several lineages that have partly or entirely lost their RNAi system. For example, the blood cell-infecting apicomplexans have thrived without a functional RNAi system for at least 300 million years. On the other hand, successive sister groups of vertebrates like the tunicate Ciona and Branchiostoma respectively possess reduced and elaborate RNAi systems, yet they display comparable organizational complexity (Fig. 2). Thus, even though a fairly well-developed RNAi system can be reconstructed as being already present in the LECA, it appears to have been repeatedly lost or reduced in several lineages. However, practically all these lineages never had a paucity of transcription factors (barring kinetoplastids) or chromatin-level regulators, suggesting that as a regulatory system RNAi is probably lower in the hierarchy than the former two nuclear systems. The exact reasons for loss or diminution of RNAi remain unclear, particularly because of the presence of sister species with similar lifestyles that differ in its presence or absence (e.g. Naumovozyma and Saccharomyces or T. cruzi and T. brucei). However, the converse issue, i.e., the strong tendency to retain RNAi in certain lineages (e.g. plants) might relate to the entanglement of the system with regulation of important genes. Indeed, the presence of conserved miRNAs across animals supports this possibility for the animal miRNA system.
The second major question regarding the natural history of the RNAi system is that of it being an apparently unique derived feature of eukaryotes. This is particularly interesting because prokaryotes possess their own widespread, well-elaborated RNA-based interference or restriction system, namely the CRISPR/Cas system; in addition to the less frequent pPIWI-dependent systems. One extreme proposal could be that there was a RNAi system already present in the last universal common ancestor but the protein components associated with it have diverged extensively. Indeed such a situation is seen in the case of the ribozyme RNaseP, where the RNA component is conserved across the three superkingdoms of life, but its protein components are markedly distinct between bacteria and the archaea-eukaryotic lineage. However, a comparison of the RNA and protein components and processing mechanisms of the eukaryotic and prokaryotic RNA-based interference systems does not currently support a unified origin for them in their common ancestor. This raises the question as to why the widespread prokaryotic CRISPR/Cas system was never acquired by eukaryotes. A possible answer for this comes from the observation that eukaryotes also lost several other multigene defense systems such as the Restriction-Modification systems (along with their recently described RNA-targeting components), the Pgl system and toxin-antitoxin systems [159]. All these systems are themselves mobile, selfish elements that appear to depend on strong genomic linkage (i.e. existence of operons) for the physical assembly of their products and neutralization of their toxic components via the linkage of transcription and translation in prokaryotes. The emergence of the nucleus in eukaryotes, with the resulting breakdown of transcription-translation coupling rendered such systems incapable of survival due to the potential danger of the toxic restriction components to the cell. Indeed, expressions of CRISPR/Cas systems in eukaryotes with appropriate RNA guides, e.g. type-II systems, introduce double strand breaks in DNA with serious mutagenic consequences [254, 255]. The eukaryotic RNAi system appears to have been rebuilt by elaboration around a core formed by the simpler prokaryotic pPIWI-based systems, specifically those that did not have strong operonic linkages with DNA-targeting components.
This process appears to have proceeded via: 1) accretion of several components from unrelated prokaryotic RNA processing and other systems and 2) drawing on components (e.g. helicases and RNA-binding domains) from the parallel, massive expansion of RNA-processing systems related to processes such as splicing and polyadenylation that were emerging or elaborating in eukaryotes. Among the proteins drawn from unrelated prokaryotic systems are the RdRPs which appear to have been derived from among DdRPs of distinct phages and selfish elements that were involved in transcription or production of primers. Likewise, the RNases, namely, RNaseIII, Eri-1-like, Mut7-C, HKD-type Zucchini-like, LK, and SNase, the methylase Hen1, and the RNA-binding protein Lin-28 all appear to have been picked up from general RNA-processing and modifying systems in prokaryotes. To date none of these, barring the general role of RNaseIII proteins in processing Type-II CRISPR/Cas RNAs [256], have been implicated specifically in a prokaryotic RNAi-like process. Thus, it appears that their generic ability to handle RNA substrates similar to those generated in the RNAi system led to their recruitment. We can state with some confidence that ancestral eukaryotic RNAi system had proteins with two tandem RNaseIII domains, a PIWI module, the RdRP module, at least a single dsRBD, at least one PAZ RNA-binding domain, and the tandem PIWI N-terminal repeat domains. This probably operated on dsRNA products formed by overlapping s-as transcription, likely initially at protein coding genes and later at other non-coding and repeat sequences as these began to expand in eukaryotic genomes. Hence, the origin of the RNAi system of eukaryotes from ancestral prokaryotic components appears to be rather distinct from the origin of the Ubiquitin (Ub) system, which also coevally elaborated in the eukaryotes [257–260]. In the latter case the core components had already developed functional linkages and physical interactions in the prokaryotes themselves and these interactions were inherited and retained as is in the eukaryotes [261–263]. The evolution of the RNAi system instead more closely resembles that of the eukaryotic chromatin modification system wherein numerous components were picked up from functionally unrelated prokaryotic systems, particularly those involved in genomic conflicts, and assembled in to an entirely distinct regulatory system in eukaryotes [246, 264]. However, just as in the case of the Ub-system, the eukaryotic nucleus appears to have facilitated the evolution of compartmentalized versions of the RNAi system, with distinct limbs acting in the nucleus and cytoplasm [257]. Nuclear RNAi systems are primarily concerned with pre-transcriptional regulation through interface with chromatin modification systems, regulation of polII, and direct interactions with nascent transcripts via splicing regulation and silencing, while cytoplasmic RNAi systems are primarily involved in post-transcriptional gene regulation (Fig. 1). The small RNA substrates participating in these RNAi systems are largely derived from a pool of RNA sources consisting of independently-transcribed, genome-encoded hairpins, double-stranded structures in larger ncRNA precursors such as snoRNA, tRNA, invasive RNA, and vRNA, and, most prominently, double-stranded duplexes formed via s-as transcription. Incorporation of small RNA into RNAi systems from sources outside of this pool appears to have occurred only on rare occasions. However, the ability to appropriate such substrates from this pool regardless of context likely contributed to the expansion of functional roles for RNAi systems outside of the ancestral role in gene regulation, with examples including DNA repair in the crown group and exonuclease activation in T. thermophila.
In conclusion, further studies on both the protein components and RNA cargo via deep-sequencing of the immunoprecipitated PIWI proteins from diverse prokaryotes and eukaryotes are likely to yield an even-clearer picture of the evolution of the RNAi systems. Further clarification of the evolutionary principles governing the emergence and diversification of eukaryotic RNAi systems, such as those discussed here, might help in the design of synthetic versions of RNAi-like systems.
Acknowledgments
AMB and LA’s research is supported by the Intramural Research Program of the NIH, National Library of Medicine.
Footnotes
Further Resources
Please see the following for the further resources mentioned in the text: ftp://ftp.ncbi.nih.gov/pub/aravind/RNAi/RNAi_review.html
Contributor Information
Alexander Maxwell Burroughs, Email: burrough@ncbi.nlm.nih.gov, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health.
Yoshinari Ando, Department of Biochemistry and Molecular Biology, Bloomberg School of Public Health, Johns Hopkins University.
L Aravind, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health.
References
- 1.Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 2.Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 3.Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermuller J, Hofacker IL, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 5.Berretta J, Morillon A. Pervasive transcription constitutes a new level of eukaryotic genome regulation. EMBO reports. 2009;10:973–982. doi: 10.1038/embor.2009.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Venters BJ, Pugh BF. Genomic organization of human transcription initiation complexes. Nature. 2013 doi: 10.1038/nature12535. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 7.Ando Y, Burroughs AM, Kawano M, de Hoon MJL, Hayashizaki Y. Targeted Methods to Improve Small RNA Profiles Generated by Deep Sequencing. In: Mallick B, Ghosh Z, editors. Regulatory RNAs. Springer; Berlin Heidelberg: 2012. pp. 253–271. [Google Scholar]
- 8.Zhang X, Rossi JJ. Phylogenetic comparison of small RNA-triggered transcriptional gene silencing. The Journal of biological chemistry. 2011;286:29443–29448. doi: 10.1074/jbc.R111.276378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li L, Liu Y. Diverse small non-coding RNAs in RNA interference pathways. Methods Mol Biol. 2011;764:169–182. doi: 10.1007/978-1-61779-188-8_11. [DOI] [PubMed] [Google Scholar]
- 10.Holley CL, Topkara VK. An introduction to small non-coding RNAs: miRNA and snoRNA. Cardiovascular drugs and therapy/sponsored by the International Society of Cardiovascular Pharmacotherapy. 2011;25:151–159. doi: 10.1007/s10557-011-6290-z. [DOI] [PubMed] [Google Scholar]
- 11.Joshua-Tor L, Hannon GJ. Ancestral roles of small RNAs: an Ago-centric perspective. Cold Spring Harbor perspectives in biology. 2011;3:a003772. doi: 10.1101/cshperspect.a003772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choudhuri S. Lesser known relatives of miRNA. Biochemical and biophysical research communications. 2009;388:177–180. doi: 10.1016/j.bbrc.2009.08.039. [DOI] [PubMed] [Google Scholar]
- 13.Farazi TA, Juranek SA, Tuschl T. The growing catalog of small RNAs and their association with distinct Argonaute/Piwi family members. Development. 2008;135:1201–1214. doi: 10.1242/dev.005629. [DOI] [PubMed] [Google Scholar]
- 14.Kim VN. Small RNAs: classification, biogenesis, and function. Molecules and cells. 2005;19:1–15. [PubMed] [Google Scholar]
- 15.Andreeva A, Howorth D, Chandonia JM, Brenner SE, Hubbard TJ, Chothia C, Murzin AG. Data growth and its impact on the SCOP database: new developments. Nucleic acids research. 2008;36:D419–425. doi: 10.1093/nar/gkm993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sillitoe I, Cuff AL, Dessailly BH, Dawson NL, Furnham N, Lee D, Lees JG, Lewis TE, Studer RA, Rentzsch R, et al. New functional families (FunFams) in CATH to improve the mapping of conserved functional sites to 3D structures. Nucleic acids research. 2013;41:D490–498. doi: 10.1093/nar/gks1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, et al. The Pfam protein families database. Nucleic acids research. 2012;40:D290–301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Desnoyers G, Bouchard MP, Masse E. New insights into small RNA-dependent translational regulation in prokaryotes. Trends in genetics: TIG. 2013;29:92–98. doi: 10.1016/j.tig.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 19.Watkins NJ, Bohnsack MT. The box C/D and H/ACA snoRNPs: key players in the modification, processing and the dynamic folding of ribosomal RNA. Wiley interdisciplinary reviews RNA. 2012;3:397–414. doi: 10.1002/wrna.117. [DOI] [PubMed] [Google Scholar]
- 20.Makarova KS, Aravind L, Wolf YI, Koonin EV. Unification of Cas protein families and a simple scenario for the origin and evolution of CRISPR-Cas systems. Biology direct. 2011;6:38. doi: 10.1186/1745-6150-6-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aliyari R, Ding SW. RNA-based viral immunity initiated by the Dicer family of host immune receptors. Immunological reviews. 2009;227:176–188. doi: 10.1111/j.1600-065X.2008.00722.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nature reviews Genetics. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 23.Cam HP. Roles of RNAi in chromatin regulation and epigenetic inheritance. Epigenomics. 2010;2:613–626. doi: 10.2217/epi.10.46. [DOI] [PubMed] [Google Scholar]
- 24.Ameyar-Zazoua M, Rachez C, Souidi M, Robin P, Fritsch L, Young R, Morozova N, Fenouil R, Descostes N, Andrau JC, et al. Argonaute proteins couple chromatin silencing to alternative splicing. Nature structural & molecular biology. 2012;19:998–1004. doi: 10.1038/nsmb.2373. [DOI] [PubMed] [Google Scholar]
- 25.Ishizu H, Siomi H, Siomi MC. Biology of PIWI-interacting RNAs. new insights into biogenesis and function inside and outside of germlines. Genes & development. 2012;26:2361–2373. doi: 10.1101/gad.203786.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 27.Shabalina SA, Koonin EV. Origins and evolution of eukaryotic RNA interference. Trends in ecology & evolution. 2008;23:578–587. doi: 10.1016/j.tree.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muljo SA, Kanellopoulou C, Aravind L. MicroRNA targeting in mammalian genomes. genes and mechanisms. Wiley interdisciplinary reviews Systems biology and medicine. 2010;2:148–161. doi: 10.1002/wsbm.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anantharaman V, Koonin EV, Aravind L. Comparative genomics and evolution of proteins involved in RNA metabolism. Nucleic acids research. 2002;30:1427–1464. doi: 10.1093/nar/30.7.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mallick B, Ghosh Z, Chakrabarti J. MicroRNA switches in Trypanosoma brucei. Biochemical and biophysical research communications. 2008;372:459–463. doi: 10.1016/j.bbrc.2008.05.084. [DOI] [PubMed] [Google Scholar]
- 31.Rathjen T, Nicol C, McConkey G, Dalmay T. Analysis of short RNAs in the malaria parasite and its red blood cell host. FEBS letters. 2006;580:5185–5188. doi: 10.1016/j.febslet.2006.08.063. [DOI] [PubMed] [Google Scholar]
- 32.Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC. Mammalian mirtron genes. Molecular cell. 2007;28:328–336. doi: 10.1016/j.molcel.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell. 2007;130:89–100. doi: 10.1016/j.cell.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leung AK, Sharp PA. Function and localization of microRNAs in mammalian cells. Cold Spring Harbor symposia on quantitative biology. 2006;71:29–38. doi: 10.1101/sqb.2006.71.049. [DOI] [PubMed] [Google Scholar]
- 35.Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nature reviews Genetics. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Axtell MJ, Westholm JO, Lai EC. Vive la difference: biogenesis and evolution of microRNAs in plants and animals. Genome biology. 2011;12:221. doi: 10.1186/gb-2011-12-4-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen XS, Collins LJ, Biggs PJ, Penny D. High throughput genome-wide survey of small RNAs from the parasitic protists Giardia intestinalis and Trichomonas vaginalis. Genome biology and evolution. 2009;1:165–175. doi: 10.1093/gbe/evp017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saraiya AA, Li W, Wang CC. A microRNA derived from an apparent canonical biogenesis pathway regulates variant surface protein gene expression in Giardia lamblia. RNA. 2011;17:2152–2164. doi: 10.1261/rna.028118.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Braun L, Cannella D, Ortet P, Barakat M, Sautel CF, Kieffer S, Garin J, Bastien O, Voinnet O, Hakimi MA. A complex small RNA repertoire is generated by a plant/fungal-like machinery and effected by a metazoan-like Argonaute in the single-cell human parasite Toxoplasma gondii. PLoS pathogens. 2010;6:e1000920. doi: 10.1371/journal.ppat.1000920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Avesson L, Reimegard J, Wagner EG, Soderbom F. MicroRNAs in Amoebozoa: deep sequencing of the small RNA population in the social amoeba Dictyostelium discoideum reveals developmentally regulated microRNAs. RNA. 2012;18:1771–1782. doi: 10.1261/rna.033175.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schulze SR, Wallrath LL. Gene regulation by chromatin structure: paradigms established in Drosophila melanogaster. Annual review of entomology. 2007;52:171–192. doi: 10.1146/annurev.ento.51.110104.151007. [DOI] [PubMed] [Google Scholar]
- 42.Ohno S. Evolution by gene duplication. Berlin: Springer-Verlag; 1970. [Google Scholar]
- 43.Demuth JP, Hahn MW. The life and death of gene families. BioEssays: news and reviews in molecular, cellular and developmental biology. 2009;31:29–39. doi: 10.1002/bies.080085. [DOI] [PubMed] [Google Scholar]
- 44.Aravind L, Anantharaman V, Venancio TM. Apprehending multicellularity: regulatory networks, genomics, and evolution. Birth defects research Part C, Embryo today: reviews. 2009;87:143–164. doi: 10.1002/bdrc.20153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kawamura Y, Saito K, Kin T, Ono Y, Asai K, Sunohara T, Okada TN, Siomi MC, Siomi H. Drosophila endogenous small RNAs bind to Argonaute 2 in somatic cells. Nature. 2008;453:793–797. doi: 10.1038/nature06938. [DOI] [PubMed] [Google Scholar]
- 46.Ghildiyal M, Seitz H, Horwich MD, Li C, Du T, Lee S, Xu J, Kittler EL, Zapp ML, Weng Z, Zamore PD. Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science. 2008;320:1077–1081. doi: 10.1126/science.1157396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Polikepahad S, Corry DB. Profiling of T helper cell-derived small RNAs reveals unique antisense transcripts and differential association of miRNAs with argonaute proteins 1 and 2. Nucleic acids research. 2013;41:1164–1177. doi: 10.1093/nar/gks1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mourrain P, Beclin C, Elmayan T, Feuerbach F, Godon C, Morel JB, Jouette D, Lacombe AM, Nikic S, Picault N, et al. Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell. 2000;101:533–542. doi: 10.1016/s0092-8674(00)80863-6. [DOI] [PubMed] [Google Scholar]
- 49.Dalmay T, Hamilton A, Rudd S, Angell S, Baulcombe DC. An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell. 2000;101:543–553. doi: 10.1016/s0092-8674(00)80864-8. [DOI] [PubMed] [Google Scholar]
- 50.Mi S, Cai T, Hu Y, Chen Y, Hodges E, Ni F, Wu L, Li S, Zhou H, Long C, et al. Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell. 2008;133:116–127. doi: 10.1016/j.cell.2008.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Buhler M, Spies N, Bartel DP, Moazed D. TRAMP-mediated RNA surveillance prevents spurious entry of RNAs into the Schizosaccharomyces pombe siRNA pathway. Nature structural & molecular biology. 2008;15:1015–1023. doi: 10.1038/nsmb.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tschudi C, Shi H, Franklin JB, Ullu E. Small interfering RNA-producing loci in the ancient parasitic eukaryote Trypanosoma brucei. BMC genomics. 2012;13:427. doi: 10.1186/1471-2164-13-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kawaji H, Nakamura M, Takahashi Y, Sandelin A, Katayama S, Fukuda S, Daub CO, Kai C, Kawai J, Yasuda J, et al. Hidden layers of human small RNAs. BMC genomics. 2008;9:157. doi: 10.1186/1471-2164-9-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smalheiser NR, Lugli G, Thimmapuram J, Cook EH, Larson J. Endogenous siRNAs and noncoding RNA-derived small RNAs are expressed in adult mouse hippocampus and are up-regulated in olfactory discrimination training. RNA. 2011;17:166–181. doi: 10.1261/rna.2123811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yi R, Pasolli HA, Landthaler M, Hafner M, Ojo T, Sheridan R, Sander C, O’Carroll D, Stoffel M, Tuschl T, Fuchs E. DGCR8-dependent microRNA biogenesis is essential for skin development. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:498–502. doi: 10.1073/pnas.0810766105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Prucca CG, Slavin I, Quiroga R, Elias EV, Rivero FD, Saura A, Carranza PG, Lujan HD. Antigenic variation in Giardia lamblia is regulated by RNA interference. Nature. 2008;456:750–754. doi: 10.1038/nature07585. [DOI] [PubMed] [Google Scholar]
- 57.Howard-Till RA, Yao MC. Induction of gene silencing by hairpin RNA expression in Tetrahymena thermophila reveals a second small RNA pathway. Molecular and cellular biology. 2006;26:8731–8742. doi: 10.1128/MCB.01430-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee SR, Collins K. Two classes of endogenous small RNAs in Tetrahymena thermophila. Genes & development. 2006;20:28–33. doi: 10.1101/gad.1377006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Couvillion MT, Lee SR, Hogstad B, Malone CD, Tonkin LA, Sachidanandam R, Hannon GJ, Collins K. Sequence, biogenesis, and function of diverse small RNA classes bound to the Piwi family proteins of Tetrahymena thermophila. Genes & development. 2009;23:2016–2032. doi: 10.1101/gad.1821209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang H, Ehrenkaufer GM, Hall N, Singh U. Small RNA pyrosequencing in the protozoan parasite Entamoeba histolytica reveals strain-specific small RNAs that target virulence genes. BMC genomics. 2013;14:53. doi: 10.1186/1471-2164-14-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pak J, Fire A. Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science. 2007;315:241–244. doi: 10.1126/science.1132839. [DOI] [PubMed] [Google Scholar]
- 62.Sijen T, Steiner FA, Thijssen KL, Plasterk RH. Secondary siRNAs result from unprimed RNA synthesis and form a distinct class. Science. 2007;315:244–247. doi: 10.1126/science.1136699. [DOI] [PubMed] [Google Scholar]
- 63.Haag JR, Pikaard CS. Multisubunit RNA polymerases IV and V: purveyors of non-coding RNA for plant gene silencing. Nature reviews Molecular cell biology. 2011;12:483–492. doi: 10.1038/nrm3152. [DOI] [PubMed] [Google Scholar]
- 64.Xie Z, Qi X. Diverse small RNA-directed silencing pathways in plants. Biochimica et biophysica acta. 2008;1779:720–724. doi: 10.1016/j.bbagrm.2008.02.009. [DOI] [PubMed] [Google Scholar]
- 65.Chang SS, Zhang Z, Liu Y. RNA interference pathways in fungi: mechanisms and functions. Annual review of microbiology. 2012;66:305–323. doi: 10.1146/annurev-micro-092611-150138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jiang L, Mu J, Zhang Q, Ni T, Srinivasan P, Rayavara K, Yang W, Turner L, Lavstsen T, Theander TG, et al. PfSETvs methylation of histone H3K36 represses virulence genes in Plasmodium falciparum. Nature. 2013 doi: 10.1038/nature12361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cichocki F, Lenvik T, Sharma N, Yun G, Anderson SK, Miller JS. Cutting edge: KIR antisense transcripts are processed into a 28-base PIWI-like RNA in human NK cells. J Immunol. 2010;185:2009–2012. doi: 10.4049/jimmunol.1000855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Watanabe T, Tomizawa S, Mitsuya K, Totoki Y, Yamamoto Y, Kuramochi-Miyagawa S, Iida N, Hoki Y, Murphy PJ, Toyoda A, et al. Role for piRNAs and noncoding RNA in de novo DNA methylation of the imprinted mouse Rasgrf1 locus. Science. 2011;332:848–852. doi: 10.1126/science.1203919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 70.Olovnikov I, Aravin AA, Fejes Toth K. Small RNA in the nucleus: the RNA-chromatin ping-pong. Current opinion in genetics & development. 2012;22:164–171. doi: 10.1016/j.gde.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Grimson A, Srivastava M, Fahey B, Woodcroft BJ, Chiang HR, King N, Degnan BM, Rokhsar DS, Bartel DP. Early origins and evolution of microRNAs and Piwi-interacting RNAs in animals. Nature. 2008;455:1193–1197. doi: 10.1038/nature07415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rajasethupathy P, Antonov I, Sheridan R, Frey S, Sander C, Tuschl T, Kandel ER. A role for neuronal piRNAs in the epigenetic control of memory-related synaptic plasticity. Cell. 2012;149:693–707. doi: 10.1016/j.cell.2012.02.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lee EJ, Banerjee S, Zhou H, Jammalamadaka A, Arcila M, Manjunath BS, Kosik KS. Identification of piRNAs in the central nervous system. RNA. 2011;17:1090–1099. doi: 10.1261/rna.2565011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yan Z, Hu HY, Jiang X, Maierhofer V, Neb E, He L, Hu Y, Hu H, Li N, Chen W, Khaitovich P. Widespread expression of piRNA-like molecules in somatic tissues. Nucleic acids research. 2011;39:6596–6607. doi: 10.1093/nar/gkr298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ender C, Krek A, Friedlander MR, Beitzinger M, Weinmann L, Chen W, Pfeffer S, Rajewsky N, Meister G. A human snoRNA with microRNA-like functions. Molecular cell. 2008;32:519–528. doi: 10.1016/j.molcel.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 76.Taft RJ, Glazov EA, Lassmann T, Hayashizaki Y, Carninci P, Mattick JS. Small RNAs derived from snoRNAs. RNA. 2009;15:1233–1240. doi: 10.1261/rna.1528909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Saraiya AA, Wang CC. snoRNA, a novel precursor of microRNA in Giardia lamblia. PLoS pathogens. 2008;4:e1000224. doi: 10.1371/journal.ppat.1000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li W, Saraiya AA, Wang CC. Gene regulation in Giardia lambia involves a putative microRNA derived from a small nucleolar RNA. PLoS neglected tropical diseases. 2011;5:e1338. doi: 10.1371/journal.pntd.0001338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li W, Saraiya AA, Wang CC. The profile of snoRNA-derived microRNAs that regulate expression of variant surface proteins in Giardia lamblia. Cellular microbiology. 2012;14:1455–1473. doi: 10.1111/j.1462-5822.2012.01811.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lee YS, Shibata Y, Malhotra A, Dutta A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs) Genes & development. 2009;23:2639–2649. doi: 10.1101/gad.1837609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Haussecker D, Huang Y, Lau A, Parameswaran P, Fire AZ, Kay MA. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA. 2010;16:673–695. doi: 10.1261/rna.2000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cole C, Sobala A, Lu C, Thatcher SR, Bowman A, Brown JW, Green PJ, Barton GJ, Hutvagner G. Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. RNA. 2009;15:2147–2160. doi: 10.1261/rna.1738409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Loss-Morais G, Waterhouse PM, Margis R. Description of plant tRNA-derived RNA fragments (tRFs) associated with argonaute and identification of their putative targets. Biology direct. 2013;8:6. doi: 10.1186/1745-6150-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Burroughs AM, Ando Y, de Hoon MJ, Tomaru Y, Suzuki H, Hayashizaki Y, Daub CO. Deep-sequencing of human Argonaute-associated small RNAs provides insight into miRNA sorting and reveals Argonaute association with RNA fragments of diverse origin. RNA biology. 2011;8:158–177. doi: 10.4161/rna.8.1.14300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nunes CC, Gowda M, Sailsbery J, Xue M, Chen F, Brown DE, Oh Y, Mitchell TK, Dean RA. Diverse and tissue-enriched small RNAs in the plant pathogenic fungus, Magnaporthe oryzae. BMC genomics. 2011;12:288. doi: 10.1186/1471-2164-12-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Couvillion MT, Sachidanandam R, Collins K. A growth-essential Tetrahymena Piwi protein carries tRNA fragment cargo. Genes & development. 2010;24:2742–2747. doi: 10.1101/gad.1996210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Maute RL, Schneider C, Sumazin P, Holmes A, Califano A, Basso K, Dalla-Favera R. tRNA-derived microRNA modulates proliferation and the DNA damage response and is down-regulated in B cell lymphoma. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:1404–1409. doi: 10.1073/pnas.1206761110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Couvillion MT, Bounova G, Purdom E, Speed TP, Collins K. A Tetrahymena Piwi bound to mature tRNA 3′ fragments activates the exonuclease Xrn2 for RNA processing in the nucleus. Molecular cell. 2012;48:509–520. doi: 10.1016/j.molcel.2012.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Henry Y, Wood H, Morrissey JP, Petfalski E, Kearsey S, Tollervey D. The 5′ end of yeast 5. 8S rRNA is generated by exonucleases from an upstream cleavage site. The EMBO journal. 1994;13:2452–2463. doi: 10.1002/j.1460-2075.1994.tb06530.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Geerlings TH, Vos JC, Raue HA. The final step in the formation of 25S rRNA in Saccharomyces cerevisiae is performed by 5′-->3′ exonucleases. RNA. 2000;6:1698–1703. doi: 10.1017/s1355838200001540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang M, Pestov DG. 5′-end surveillance by Xrn2 acts as a shared mechanism for mammalian pre-rRNA maturation and decay. Nucleic acids research. 2011;39:1811–1822. doi: 10.1093/nar/gkq1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Olovnikov I, Chan K, Sachidanandam R, Newman DK, Aravin AA. Bacterial argonaute samples the transcriptome to identify foreign DNA. Molecular cell. 2013;51:594–605. doi: 10.1016/j.molcel.2013.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Li Y, Luo J, Zhou H, Liao JY, Ma LM, Chen YQ, Qu LH. Stress-induced tRNA-derived RNAs: a novel class of small RNAs in the primitive eukaryote Giardia lamblia. Nucleic acids research. 2008;36:6048–6055. doi: 10.1093/nar/gkn596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fu H, Feng J, Liu Q, Sun F, Tie Y, Zhu J, Xing R, Sun Z, Zheng X. Stress induces tRNA cleavage by angiogenin in mammalian cells. FEBS letters. 2009;583:437–442. doi: 10.1016/j.febslet.2008.12.043. [DOI] [PubMed] [Google Scholar]
- 95.Saikia M, Krokowski D, Guan BJ, Ivanov P, Parisien M, Hu GF, Anderson P, Pan T, Hatzoglou M. Genome-wide identification and quantitative analysis of cleaved tRNA fragments induced by cellular stress. The Journal of biological chemistry. 2012;287:42708–42725. doi: 10.1074/jbc.M112.371799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Elbarbary RA, Takaku H, Uchiumi N, Tamiya H, Abe M, Takahashi M, Nishida H, Nashimoto M. Modulation of gene expression by human cytosolic tRNase Z(L) through 5′-half-tRNA. PloS one. 2009;4:e5908. doi: 10.1371/journal.pone.0005908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Elbarbary RA, Takaku H, Uchiumi N, Tamiya H, Abe M, Nishida H, Nashimoto M. Human cytosolic tRNase ZL can downregulate gene expression through miRNA. FEBS letters. 2009;583:3241–3246. doi: 10.1016/j.febslet.2009.09.015. [DOI] [PubMed] [Google Scholar]
- 98.Ivanov P, Emara MM, Villen J, Gygi SP, Anderson P. Angiogenin-induced tRNA fragments inhibit translation initiation. Molecular cell. 2011;43:613–623. doi: 10.1016/j.molcel.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Persson H, Kvist A, Vallon-Christersson J, Medstrand P, Borg A, Rovira C. The non-coding RNA of the multidrug resistance-linked vault particle encodes multiple regulatory small RNAs. Nature cell biology. 2009;11:1268–1271. doi: 10.1038/ncb1972. [DOI] [PubMed] [Google Scholar]
- 100.Mossink MH, van Zon A, Scheper RJ, Sonneveld P, Wiemer EA. Vaults: a ribonucleoprotein particle involved in drug resistance? Oncogene. 2003;22:7458–7467. doi: 10.1038/sj.onc.1206947. [DOI] [PubMed] [Google Scholar]
- 101.Minones-Moyano E, Friedlander MR, Pallares J, Kagerbauer B, Porta S, Escaramis G, Ferrer I, Estivill X, Marti E. Upregulation of a small vault RNA (svtRNA2–1a) is an early event in Parkinson disease and induces neuronal dysfunction. RNA biology. 2013:10. doi: 10.4161/rna.24813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Skreka K, Schafferer S, Nat IR, Zywicki M, Salti A, Apostolova G, Griehl M, Rederstorff M, Dechant G, Huttenhofer A. Identification of differentially expressed non-coding RNAs in embryonic stem cell neural differentiation. Nucleic acids research. 2012;40:6001–6015. doi: 10.1093/nar/gks311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Stadler PF, Chen JJ, Hackermuller J, Hoffmann S, Horn F, Khaitovich P, Kretzschmar AK, Mosig A, Prohaska SJ, Qi X, et al. Evolution of vault RNAs. Molecular biology and evolution. 2009;26:1975–1991. doi: 10.1093/molbev/msp112. [DOI] [PubMed] [Google Scholar]
- 104.Wei H, Zhou B, Zhang F, Tu Y, Hu Y, Zhang B, Zhai Q. Profiling and identification of small rDNA-derived RNAs and their potential biological functions. PloS one. 2013;8:e56842. doi: 10.1371/journal.pone.0056842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Rogler LE, Kosmyna B, Moskowitz D, Bebawee R, Rahimzadeh J, Kutchko K, Laederach A, Notarangelo LD, Giliani S, Bouhassira E, et al. Small RNAs derived from lncRNA RNase MRP have gene-silencing activity relevant to human Cartilage Hair Hypoplasia. Human molecular genetics. 2013 doi: 10.1093/hmg/ddt427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Maida Y, Yasukawa M, Furuuchi M, Lassmann T, Possemato R, Okamoto N, Kasim V, Hayashizaki Y, Hahn WC, Masutomi K. An RNA-dependent RNA polymerase formed by TERT and the RMRP RNA. Nature. 2009;461:230–235. doi: 10.1038/nature08283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Portnoy V, Huang V, Place RF, Li LC. Small RNA and transcriptional upregulation. Wiley interdisciplinary reviews RNA. 2011;2:748–760. doi: 10.1002/wrna.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gagnon KT, Corey DR. Argonaute and the nuclear RNAs: new pathways for RNA-mediated control of gene expression. Nucleic acid therapeutics. 2012;22:3–16. doi: 10.1089/nat.2011.0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Taft RJ, Glazov EA, Cloonan N, Simons C, Stephen S, Faulkner GJ, Lassmann T, Forrest AR, Grimmond SM, Schroder K, et al. Tiny RNAs associated with transcription start sites in animals. Nature genetics. 2009;41:572–578. doi: 10.1038/ng.312. [DOI] [PubMed] [Google Scholar]
- 110.Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA, Young RA, Sharp PA. Divergent transcription from active promoters. Science. 2008;322:1849–1851. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Preker P, Nielsen J, Kammler S, Lykke-Andersen S, Christensen MS, Mapendano CK, Schierup MH, Jensen TH. RNA exosome depletion reveals transcription upstream of active human promoters. Science. 2008;322:1851–1854. doi: 10.1126/science.1164096. [DOI] [PubMed] [Google Scholar]
- 113.Almada AE, Wu X, Kriz AJ, Burge CB, Sharp PA. Promoter directionality is controlled by U1 snRNP and polyadenylation signals. Nature. 2013 doi: 10.1038/nature12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Cernilogar FM, Onorati MC, Kothe GO, Burroughs AM, Parsi KM, Breiling A, Lo Sardo F, Saxena A, Miyoshi K, Siomi H, et al. Chromatin-associated RNA interference components contribute to transcriptional regulation in Drosophila. Nature. 2011;480:391–395. doi: 10.1038/nature10492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Whitehouse I, Rando OJ, Delrow J, Tsukiyama T. Chromatin remodelling at promoters suppresses antisense transcription. Nature. 2007;450:1031–1035. doi: 10.1038/nature06391. [DOI] [PubMed] [Google Scholar]
- 116.Halic M, Moazed D. Dicer-independent primal RNAs trigger RNAi and heterochromatin formation. Cell. 2010;140:504–516. doi: 10.1016/j.cell.2010.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Tucker SL, Reece J, Ream TS, Pikaard CS. Evolutionary history of plant multisubunit RNA polymerases IV and V: subunit origins via genome-wide and segmental gene duplications, retrotransposition, and lineage-specific subfunctionalization. Cold Spring Harbor symposia on quantitative biology. 2010;75:285–297. doi: 10.1101/sqb.2010.75.037. [DOI] [PubMed] [Google Scholar]
- 118.Shiu PK, Raju NB, Zickler D, Metzenberg RL. Meiotic silencing by unpaired DNA. Cell. 2001;107:905–916. doi: 10.1016/s0092-8674(01)00609-2. [DOI] [PubMed] [Google Scholar]
- 119.Gu W, Shirayama M, Conte D, Jr, Vasale J, Batista PJ, Claycomb JM, Moresco JJ, Youngman EM, Keys J, Stoltz MJ, et al. Distinct argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline. Molecular cell. 2009;36:231–244. doi: 10.1016/j.molcel.2009.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gu SG, Pak J, Guang S, Maniar JM, Kennedy S, Fire A. Amplification of siRNA in Caenorhabditis elegans generates a transgenerational sequence-targeted histone H3 lysine 9 methylation footprint. Nature genetics. 2012;44:157–164. doi: 10.1038/ng.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Burton NO, Burkhart KB, Kennedy S. Nuclear RNAi maintains heritable gene silencing in Caenorhabditis elegans. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:19683–19688. doi: 10.1073/pnas.1113310108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Aravind L, Watanabe H, Lipman DJ, Koonin EV. Lineage-specific loss and divergence of functionally linked genes in eukaryotes. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:11319–11324. doi: 10.1073/pnas.200346997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Iyer LM, Zhang D, Maxwell Burroughs A, Aravind L. Computational identification of novel biochemical systems involved in oxidation, glycosylation and other complex modifications of bases in DNA. Nucleic acids research. 2013;41:7635–7655. doi: 10.1093/nar/gkt573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Valen E, Preker P, Andersen PR, Zhao X, Chen Y, Ender C, Dueck A, Meister G, Sandelin A, Jensen TH. Biogenic mechanisms and utilization of small RNAs derived from human protein-coding genes. Nature structural & molecular biology. 2011;18:1075–1082. doi: 10.1038/nsmb.2091. [DOI] [PubMed] [Google Scholar]
- 125.Taliaferro JM, Aspden JL, Bradley T, Marwha D, Blanchette M, Rio DC. Two new and distinct roles for Drosophila Argonaute-2 in the nucleus: alternative pre-mRNA splicing and transcriptional repression. Genes & development. 2013;27:378–389. doi: 10.1101/gad.210708.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Rogozin IB, Carmel L, Csuros M, Koonin EV. Origin and evolution of spliceosomal introns. Biology direct. 2012;7:11. doi: 10.1186/1745-6150-7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Koonin EV. The origin of introns and their role in eukaryogenesis: a compromise solution to the introns-early versus introns-late debate? Biology direct. 2006;1:22. doi: 10.1186/1745-6150-1-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Mochizuki K. DNA rearrangements directed by non-coding RNAs in ciliates. Wiley interdisciplinary reviews RNA. 2010;1:376–387. doi: 10.1002/wrna.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Mochizuki K, Fine NA, Fujisawa T, Gorovsky MA. Analysis of a piwi-related gene implicates small RNAs in genome rearrangement in tetrahymena. Cell. 2002;110:689–699. doi: 10.1016/s0092-8674(02)00909-1. [DOI] [PubMed] [Google Scholar]
- 130.Kataoka K, Mochizuki K. Programmed DNA elimination in Tetrahymena: a small RNA-mediated genome surveillance mechanism. Advances in experimental medicine and biology. 2011;722:156–173. doi: 10.1007/978-1-4614-0332-6_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lepere G, Nowacki M, Serrano V, Gout JF, Guglielmi G, Duharcourt S, Meyer E. Silencing-associated and meiosis-specific small RNA pathways in Paramecium tetraurelia. Nucleic acids research. 2009;37:903–915. doi: 10.1093/nar/gkn1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Juranek SA, Rupprecht S, Postberg J, Lipps HJ. snRNA and heterochromatin formation are involved in DNA excision during macronuclear development in stichotrichous ciliates. Eukaryotic cell. 2005;4:1934–1941. doi: 10.1128/EC.4.11.1934-1941.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Nowacki M, Vijayan V, Zhou Y, Schotanus K, Doak TG, Landweber LF. RNA-mediated epigenetic programming of a genome-rearrangement pathway. Nature. 2008;451:153–158. doi: 10.1038/nature06452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Brzeski J, Brzeska K. The maze of paramutation: a rough guide to the puzzling epigenetics of paramutation. Wiley interdisciplinary reviews RNA. 2011;2:863–874. doi: 10.1002/wrna.97. [DOI] [PubMed] [Google Scholar]
- 135.Wei W, Ba Z, Gao M, Wu Y, Ma Y, Amiard S, White CI, Rendtlew Danielsen JM, Yang YG, Qi Y. A role for small RNAs in DNA double-strand break repair. Cell. 2012;149:101–112. doi: 10.1016/j.cell.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 136.Michalik KM, Bottcher R, Forstemann K. A small RNA response at DNA ends in Drosophila. Nucleic acids research. 2012;40:9596–9603. doi: 10.1093/nar/gks711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Francia S, Michelini F, Saxena A, Tang D, de Hoon M, Anelli V, Mione M, Carninci P, d’Adda di Fagagna F. Site-specific DICER and DROSHA RNA products control the DNA-damage response. Nature. 2012;488:231–235. doi: 10.1038/nature11179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Lee HC, Chang SS, Choudhary S, Aalto AP, Maiti M, Bamford DH, Liu Y. qiRNA is a new type of small interfering RNA induced by DNA damage. Nature. 2009;459:274–277. doi: 10.1038/nature08041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Cerutti L, Mian N, Bateman A. Domains in gene silencing and cell differentiation proteins: the novel PAZ domain and redefinition of the Piwi domain. Trends in biochemical sciences. 2000;25:481–482. doi: 10.1016/s0968-0004(00)01641-8. [DOI] [PubMed] [Google Scholar]
- 140.Ma JB, Yuan YR, Meister G, Pei Y, Tuschl T, Patel DJ. Structural basis for 5′-end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature. 2005;434:666–670. doi: 10.1038/nature03514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Yuan YR, Pei Y, Ma JB, Kuryavyi V, Zhadina M, Meister G, Chen HY, Dauter Z, Tuschl T, Patel DJ. Crystal structure of A. aeolicus argonaute, a site-specific DNA-guided endoribonuclease, provides insights into RISC-mediated mRNA cleavage. Molecular cell. 2005;19:405–419. doi: 10.1016/j.molcel.2005.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Makarova KS, Wolf YI, van der Oost J, Koonin EV. Prokaryotic homologs of Argonaute proteins are predicted to function as key components of a novel system of defense against mobile genetic elements. Biology direct. 2009;4:29. doi: 10.1186/1745-6150-4-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kawamata T, Seitz H, Tomari Y. Structural determinants of miRNAs for RISC loading and slicer-independent unwinding. Nature structural & molecular biology. 2009;16:953–960. doi: 10.1038/nsmb.1630. [DOI] [PubMed] [Google Scholar]
- 144.Yoda M, Kawamata T, Paroo Z, Ye X, Iwasaki S, Liu Q, Tomari Y. ATP-dependent human RISC assembly pathways. Nature structural & molecular biology. 2010;17:17–23. doi: 10.1038/nsmb.1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Aguilera A, Garcia-Muse T. R loops: from transcription byproducts to threats to genome stability. Molecular cell. 2012;46:115–124. doi: 10.1016/j.molcel.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 146.Lasa I, Toledo-Arana A, Gingeras TR. An effort to make sense of antisense transcription in bacteria. RNA biology. 2012;9:1039–1044. doi: 10.4161/rna.21167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Hock J, Meister G. The Argonaute protein family. Genome biology. 2008;9:210. doi: 10.1186/gb-2008-9-2-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Cerutti H, Casas-Mollano JA. On the origin and functions of RNA-mediated silencing: from protists to man. Current genetics. 2006;50:81–99. doi: 10.1007/s00294-006-0078-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Yang JS, Lai EC. Dicer-independent, Ago2-mediated microRNA biogenesis in vertebrates. Cell Cycle. 2010;9:4455–4460. doi: 10.4161/cc.9.22.13958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Burroughs AM, Kawano M, Ando Y, Daub CO, Hayashizaki Y. pre-miRNA profiles obtained through application of locked nucleic acids and deep sequencing reveals complex 5′/3′ arm variation including concomitant cleavage and polyuridylation patterns. Nucleic acids research. 2012;40:1424–1437. doi: 10.1093/nar/gkr903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Yang JS, Maurin T, Lai EC. Functional parameters of Dicer-independent microRNA biogenesis. RNA. 2012;18:945–957. doi: 10.1261/rna.032938.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Koonin EV, Wolf YI, Aravind L. Protein fold recognition using sequence profiles and its application in structural genomics. Advances in protein chemistry. 2000;54:245–275. doi: 10.1016/s0065-3233(00)54008-x. [DOI] [PubMed] [Google Scholar]
- 153.Kwak PB, Tomari Y. The N domain of Argonaute drives duplex unwinding during RISC assembly. Nature structural & molecular biology. 2012;19:145–151. doi: 10.1038/nsmb.2232. [DOI] [PubMed] [Google Scholar]
- 154.Schurmann N, Trabuco LG, Bender C, Russell RB, Grimm D. Molecular dissection of human Argonaute proteins by DNA shuffling. Nature structural & molecular biology. 2013;20:818–826. doi: 10.1038/nsmb.2607. [DOI] [PubMed] [Google Scholar]
- 155.Aravind L, Walker DR, Koonin EV. Conserved domains in DNA repair proteins and evolution of repair systems. Nucleic acids research. 1999;27:1223–1242. doi: 10.1093/nar/27.5.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Sasaki HM, Tomari Y. The true core of RNA silencing revealed. Nature structural & molecular biology. 2012;19:657–660. doi: 10.1038/nsmb.2302. [DOI] [PubMed] [Google Scholar]
- 157.Iyer LM, Makarova KS, Koonin EV, Aravind L. Comparative genomics of the FtsK-HerA superfamily of pumping ATPases: implications for the origins of chromosome segregation, cell division and viral capsid packaging. Nucleic acids research. 2004;32:5260–5279. doi: 10.1093/nar/gkh828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Iyer LM, Abhiman S, Aravind L. MutL homologs in restriction-modification systems and the origin of eukaryotic MORC ATPases. Biology direct. 2008;3:8. doi: 10.1186/1745-6150-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Anantharaman V, Makarova KS, Burroughs AM, Koonin EV, Aravind L. Comprehensive analysis of the HEPN superfamily: identification of novel roles in intra-genomic conflicts, defense, pathogenesis and RNA processing. Biology direct. 2013;8:15. doi: 10.1186/1745-6150-8-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Reeks J, Graham S, Anderson L, Liu H, White MF, Naismith JH. Structure of the archaeal Cascade subunit Csa5: Relating the small subunits of CRISPR effector complexes. RNA biology. 2013;10:762–769. doi: 10.4161/rna.23854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Shi H, Tschudi C, Ullu E. An unusual Dicer-like1 protein fuels the RNA interference pathway in Trypanosoma brucei. RNA. 2006;12:2063–2072. doi: 10.1261/rna.246906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Koc EC, Burkhart W, Blackburn K, Moyer MB, Schlatzer DM, Moseley A, Spremulli LL. The large subunit of the mammalian mitochondrial ribosome. Analysis of the complement of ribosomal proteins present. The Journal of biological chemistry. 2001;276:43958–43969. doi: 10.1074/jbc.M106510200. [DOI] [PubMed] [Google Scholar]
- 164.Huang PJ, Lin WC, Chen SC, Lin YH, Sun CH, Lyu PC, Tang P. Identification of putative miRNAs from the deep-branching unicellular flagellates. Genomics. 2012;99:101–107. doi: 10.1016/j.ygeno.2011.11.002. [DOI] [PubMed] [Google Scholar]
- 165.Flot JF, Hespeels B, Li X, Noel B, Arkhipova I, Danchin EG, Hejnol A, Henrissat B, Koszul R, Aury JM, et al. Genomic evidence for ameiotic evolution in the bdelloid rotifer Adineta vaga. Nature. 2013 doi: 10.1038/nature12326. [DOI] [PubMed] [Google Scholar]
- 166.Su H, Trombly MI, Chen J, Wang X. Essential and overlapping functions for mammalian Argonautes in microRNA silencing. Genes & development. 2009;23:304–317. doi: 10.1101/gad.1749809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Drinnenberg IA, Weinberg DE, Xie KT, Mower JP, Wolfe KH, Fink GR, Bartel DP. RNAi in budding yeast. Science. 2009;326:544–550. doi: 10.1126/science.1176945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Bernstein DA, Vyas VK, Weinberg DE, Drinnenberg IA, Bartel DP, Fink GR. Candida albicans Dicer (CaDcr1) is required for efficient ribosomal and spliceosomal RNA maturation. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:523–528. doi: 10.1073/pnas.1118859109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Bernstein DA, Vyas VK, Fink GR. Genes come and go: the evolutionarily plastic path of budding yeast RNase III enzymes. RNA biology. 2012;9:1123–1128. doi: 10.4161/rna.21360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Zong J, Yao X, Yin J, Zhang D, Ma H. Evolution of the RNA-dependent RNA polymerase (RdRP) genes: duplications and possible losses before and after the divergence of major eukaryotic groups. Gene. 2009;447:29–39. doi: 10.1016/j.gene.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 171.Nakayashiki H, Kadotani N, Mayama S. Evolution and diversification of RNA silencing proteins in fungi. Journal of molecular evolution. 2006;63:127–135. doi: 10.1007/s00239-005-0257-2. [DOI] [PubMed] [Google Scholar]
- 172.Laurie JD, Linning R, Bakkeren G. Hallmarks of RNA silencing are found in the smut fungus Ustilago hordei but not in its close relative Ustilago maydis. Current genetics. 2008;53:49–58. doi: 10.1007/s00294-007-0165-7. [DOI] [PubMed] [Google Scholar]
- 173.Franzen O, Arner E, Ferella M, Nilsson D, Respuela P, Carninci P, Hayashizaki Y, Aslund L, Andersson B, Daub CO. The short non-coding transcriptome of the protozoan parasite Trypanosoma cruzi. PLoS neglected tropical diseases. 2011;5:e1283. doi: 10.1371/journal.pntd.0001283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Barnes RL, Shi H, Kolev NG, Tschudi C, Ullu E. Comparative genomics reveals two novel RNAi factors in Trypanosoma brucei and provides insight into the core machinery. PLoS pathogens. 2012;8:e1002678. doi: 10.1371/journal.ppat.1002678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Soper SF, van der Heijden GW, Hardiman TC, Goodheart M, Martin SL, de Boer P, Bortvin A. Mouse maelstrom, a component of nuage, is essential for spermatogenesis and transposon repression in meiosis. Developmental cell. 2008;15:285–297. doi: 10.1016/j.devcel.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Zhang D, Xiong H, Shan J, Xia X, Trudeau VL. Functional insight into Maelstrom in the germline piRNA pathway: a unique domain homologous to the DnaQ-H 3′–5′ exonuclease, its lineage-specific expansion/loss and evolutionarily active site switch. Biology direct. 2008;3:48. doi: 10.1186/1745-6150-3-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Iyer LM, Tahiliani M, Rao A, Aravind L. Prediction of novel families of enzymes involved in oxidative and other complex modifications of bases in nucleic acids. Cell Cycle. 2009;8:1698–1710. doi: 10.4161/cc.8.11.8580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Ponting CP. P100, a transcriptional coactivator, is a human homologue of staphylococcal nuclease. Protein science: a publication of the Protein Society. 1997;6:459–463. doi: 10.1002/pro.5560060224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Yang W, Chendrimada TP, Wang Q, Higuchi M, Seeburg PH, Shiekhattar R, Nishikura K. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nature structural & molecular biology. 2006;13:13–21. doi: 10.1038/nsmb1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Li CL, Yang WZ, Chen YP, Yuan HS. Structural and functional insights into human Tudor-SN, a key component linking RNA interference and editing. Nucleic acids research. 2008;36:3579–3589. doi: 10.1093/nar/gkn236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Pane A, Wehr K, Schupbach T. zucchini and squash encode two putative nucleases required for rasiRNA production in the Drosophila germline. Developmental cell. 2007;12:851–862. doi: 10.1016/j.devcel.2007.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Ipsaro JJ, Haase AD, Knott SR, Joshua-Tor L, Hannon GJ. The structural biochemistry of Zucchini implicates it as a nuclease in piRNA biogenesis. Nature. 2012;491:279–283. doi: 10.1038/nature11502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Nishimasu H, Ishizu H, Saito K, Fukuhara S, Kamatani MK, Bonnefond L, Matsumoto N, Nishizawa T, Nakanaga K, Aoki J, et al. Structure and function of Zucchini endoribonuclease in piRNA biogenesis. Nature. 2012;491:284–287. doi: 10.1038/nature11509. [DOI] [PubMed] [Google Scholar]
- 184.Ketting RF, Haverkamp TH, van Luenen HG, Plasterk RH. Mut-7 of C. elegans, required for transposon silencing and RNA interference, is a homolog of Werner syndrome helicase and RNaseD. Cell. 1999;99:133–141. doi: 10.1016/s0092-8674(00)81645-1. [DOI] [PubMed] [Google Scholar]
- 185.Ramachandran V, Chen X. Degradation of microRNAs by a family of exoribonucleases in Arabidopsis. Science. 2008;321:1490–1492. doi: 10.1126/science.1163728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Chatterjee S, Grosshans H. Active turnover modulates mature microRNA activity in Caenorhabditis elegans. Nature. 2009;461:546–549. doi: 10.1038/nature08349. [DOI] [PubMed] [Google Scholar]
- 187.Ansel KM, Pastor WA, Rath N, Lapan AD, Glasmacher E, Wolf C, Smith LC, Papadopoulou N, Lamperti ED, Tahiliani M, et al. Mouse Eri1 interacts with the ribosome and catalyzes 5. 8S rRNA processing. Nature structural & molecular biology. 2008;15:523–530. doi: 10.1038/nsmb.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Gabel HW, Ruvkun G. The exonuclease ERI-1 has a conserved dual role in 5. 8S rRNA processing and RNAi. Nature structural & molecular biology. 2008;15:531–533. doi: 10.1038/nsmb.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Anantharaman V, Zhang D, Aravind L. OST-HTH: a novel predicted RNA-binding domain. Biology direct. 2010;5:13. doi: 10.1186/1745-6150-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Callebaut I, Mornon JP. LOTUS, a new domain associated with small RNA pathways in the germline. Bioinformatics. 2010;26:1140–1144. doi: 10.1093/bioinformatics/btq122. [DOI] [PubMed] [Google Scholar]
- 191.Su YQ, Sun F, Handel MA, Schimenti JC, Eppig JJ. Meiosis arrest female 1 (MARF1) has nuage-like function in mammalian oocytes. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:18653–18660. doi: 10.1073/pnas.1216904109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Aravind L. An evolutionary classification of the metallo-beta-lactamase fold proteins. In silico biology. 1999;1:69–91. [PubMed] [Google Scholar]
- 193.Li X, Moses RE. The beta-lactamase motif in Snm1 is required for repair of DNA double-strand breaks caused by interstrand crosslinks in S. cerevisiae. DNA repair. 2003;2:121–129. doi: 10.1016/s1568-7864(02)00192-1. [DOI] [PubMed] [Google Scholar]
- 194.Murthy KG, Manley JL. The 160-kD subunit of human cleavage-polyadenylation specificity factor coordinates pre-mRNA 3′-end formation. Genes & development. 1995;9:2672–2683. doi: 10.1101/gad.9.21.2672. [DOI] [PubMed] [Google Scholar]
- 195.Kurth HM, Mochizuki K. 2′-O-methylation stabilizes Piwi-associated small RNAs and ensures DNA elimination in Tetrahymena. RNA. 2009;15:675–685. doi: 10.1261/rna.1455509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Li J, Yang Z, Yu B, Liu J, Chen X. Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis. Current biology: CB. 2005;15:1501–1507. doi: 10.1016/j.cub.2005.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Yu B, Yang Z, Li J, Minakhina S, Yang M, Padgett RW, Steward R, Chen X. Methylation as a crucial step in plant microRNA biogenesis. Science. 2005;307:932–935. doi: 10.1126/science.1107130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Ibrahim F, Rymarquis LA, Kim EJ, Becker J, Balassa E, Green PJ, Cerutti H. Uridylation of mature miRNAs and siRNAs by the MUT68 nucleotidyltransferase promotes their degradation in Chlamydomonas. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:3906–3911. doi: 10.1073/pnas.0912632107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Ren G, Chen X, Yu B. Uridylation of miRNAs by hen1 suppressor1 in Arabidopsis. Current biology: CB. 2012;22:695–700. doi: 10.1016/j.cub.2012.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Zhao Y, Yu Y, Zhai J, Ramachandran V, Dinh TT, Meyers BC, Mo B, Chen X. The Arabidopsis nucleotidyl transferase HESO1 uridylates unmethylated small RNAs to trigger their degradation. Current biology: CB. 2012;22:689–694. doi: 10.1016/j.cub.2012.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Huang RH. Unique 2′-O-methylation by Hen1 in eukaryotic RNA interference and bacterial RNA repair. Biochemistry. 2012;51:4087–4095. doi: 10.1021/bi300497x. [DOI] [PubMed] [Google Scholar]
- 202.Gao S, Liu Y. Intercepting noncoding messages between germline and soma. Genes & development. 2012;26:1774–1779. doi: 10.1101/gad.199992.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Hussain S, Sajini AA, Blanco S, Dietmann S, Lombard P, Sugimoto Y, Paramor M, Gleeson JG, Odom DT, Ule J, Frye M. NSun2-Mediated Cytosine-5 Methylation of Vault Noncoding RNA Determines Its Processing into Regulatory Small RNAs. Cell reports. 2013;4:255–261. doi: 10.1016/j.celrep.2013.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Aravind L, Koonin EV. DNA polymerase beta-like nucleotidyltransferase superfamily: identification of three new families, classification and evolutionary history. Nucleic acids research. 1999;27:1609–1618. doi: 10.1093/nar/27.7.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Katoh T, Sakaguchi Y, Miyauchi K, Suzuki T, Kashiwabara S, Baba T. Selective stabilization of mammalian microRNAs by 3′ adenylation mediated by the cytoplasmic poly(A) polymerase GLD-2. Genes & development. 2009;23:433–438. doi: 10.1101/gad.1761509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Burroughs AM, Ando Y, de Hoon MJ, Tomaru Y, Nishibu T, Ukekawa R, Funakoshi T, Kurokawa T, Suzuki H, Hayashizaki Y, Daub CO. A comprehensive survey of 3′ animal miRNA modification events and a possible role for 3′ adenylation in modulating miRNA targeting effectiveness. Genome research. 2010;20:1398–1410. doi: 10.1101/gr.106054.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Heo I, Ha M, Lim J, Yoon MJ, Park JE, Kwon SC, Chang H, Kim VN. Mono-uridylation of pre-microRNA as a key step in the biogenesis of group II let-7 microRNAs. Cell. 2012;151:521–532. doi: 10.1016/j.cell.2012.09.022. [DOI] [PubMed] [Google Scholar]
- 208.Heo I, Joo C, Kim YK, Ha M, Yoon MJ, Cho J, Yeom KH, Han J, Kim VN. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009;138:696–708. doi: 10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 209.Jones MR, Blahna MT, Kozlowski E, Matsuura KY, Ferrari JD, Morris SA, Powers JT, Daley GQ, Quinton LJ, Mizgerd JP. Zcchc11 uridylates mature miRNAs to enhance neonatal IGF-1 expression, growth, and survival. PLoS genetics. 2012;8:e1003105. doi: 10.1371/journal.pgen.1003105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Jones MR, Quinton LJ, Blahna MT, Neilson JR, Fu S, Ivanov AR, Wolf DA, Mizgerd JP. Zcchc11-dependent uridylation of microRNA directs cytokine expression. Nature cell biology. 2009;11:1157–1163. doi: 10.1038/ncb1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Ibrahim F, Rohr J, Jeong WJ, Hesson J, Cerutti H. Untemplated oligoadenylation promotes degradation of RISC-cleaved transcripts. Science. 2006;314:1893. doi: 10.1126/science.1135268. [DOI] [PubMed] [Google Scholar]
- 212.Talsky KB, Collins K. Strand-asymmetric endogenous Tetrahymena small RNA production requires a previously uncharacterized uridylyltransferase protein partner. RNA. 2012;18:1553–1562. doi: 10.1261/rna.033530.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Mellman DL, Gonzales ML, Song C, Barlow CA, Wang P, Kendziorski C, Anderson RA. A PtdIns4,5P2-regulated nuclear poly(A) polymerase controls expression of select mRNAs. Nature. 2008;451:1013–1017. doi: 10.1038/nature06666. [DOI] [PubMed] [Google Scholar]
- 214.Trippe R, Sandrock B, Benecke BJ. A highly specific terminal uridylyl transferase modifies the 3′-end of U6 small nuclear RNA. Nucleic acids research. 1998;26:3119–3126. doi: 10.1093/nar/26.13.3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Tomecki R, Dmochowska A, Gewartowski K, Dziembowski A, Stepien PP. Identification of a novel human nuclear-encoded mitochondrial poly(A) polymerase. Nucleic acids research. 2004;32:6001–6014. doi: 10.1093/nar/gkh923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Anderson JT, Wang X. Nuclear RNA surveillance: no sign of substrates tailing off. Critical reviews in biochemistry and molecular biology. 2009;44:16–24. doi: 10.1080/10409230802640218. [DOI] [PubMed] [Google Scholar]
- 217.Kun J, Hesselbach J, Schreiber M, Scherf A, Gysin J, Mattei D, Pereira da Silva L, Muller-Hill B. Cloning and expression of genomic DNA sequences coding for putative erythrocyte membrane-associated antigens of Plasmodium falciparum. Research in immunology. 1991;142:199–210. doi: 10.1016/0923-2494(91)90059-r. [DOI] [PubMed] [Google Scholar]
- 218.Wyman SK, Knouf EC, Parkin RK, Fritz BR, Lin DW, Dennis LM, Krouse MA, Webster PJ, Tewari M. Post-transcriptional generation of miRNA variants by multiple nucleotidyl transferases contributes to miRNA transcriptome complexity. Genome research. 2011;21:1450–1461. doi: 10.1101/gr.118059.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Iyer LM, Zhang D, Rogozin IB, Aravind L. Evolution of the deaminase fold and multiple origins of eukaryotic editing and mutagenic nucleic acid deaminases from bacterial toxin systems. Nucleic acids research. 2011;39:9473–9497. doi: 10.1093/nar/gkr691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Grigg SP, Canales C, Hay A, Tsiantis M. SERRATE coordinates shoot meristem function and leaf axial patterning in Arabidopsis. Nature. 2005;437:1022–1026. doi: 10.1038/nature04052. [DOI] [PubMed] [Google Scholar]
- 221.Yang L, Liu Z, Lu F, Dong A, Huang H. SERRATE is a novel nuclear regulator in primary microRNA processing in Arabidopsis. The Plant journal: for cell and molecular biology. 2006;47:841–850. doi: 10.1111/j.1365-313X.2006.02835.x. [DOI] [PubMed] [Google Scholar]
- 222.Gruber JJ, Zatechka DS, Sabin LR, Yong J, Lum JJ, Kong M, Zong WX, Zhang Z, Lau CK, Rawlings J, et al. Ars2 links the nuclear cap-binding complex to RNA interference and cell proliferation. Cell. 2009;138:328–339. doi: 10.1016/j.cell.2009.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Sabin LR, Zhou R, Gruber JJ, Lukinova N, Bambina S, Berman A, Lau CK, Thompson CB, Cherry S. Ars2 regulates both miRNA- and siRNA- dependent silencing and suppresses RNA virus infection in Drosophila. Cell. 2009;138:340–351. doi: 10.1016/j.cell.2009.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Heo I, Joo C, Cho J, Ha M, Han J, Kim VN. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Molecular cell. 2008;32:276–284. doi: 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- 225.Newman MA, Thomson JM, Hammond SM. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA. 2008;14:1539–1549. doi: 10.1261/rna.1155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Hagan JP, Piskounova E, Gregory RI. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nature structural & molecular biology. 2009;16:1021–1025. doi: 10.1038/nsmb.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Miller MM, Halbig K, Cruz-Reyes J, Read LK. RBP16 stimulates trypanosome RNA editing in vitro at an early step in the editing reaction. RNA. 2006;12:1292–1303. doi: 10.1261/rna.2331506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Bae W, Xia B, Inouye M, Severinov K. Escherichia coli CspA-family RNA chaperones are transcription antiterminators. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:7784–7789. doi: 10.1073/pnas.97.14.7784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Lee HY, Zhou K, Smith AM, Noland CL, Doudna JA. Differential roles of human Dicer-binding proteins TRBP and PACT in small RNA processing. Nucleic acids research. 2013 doi: 10.1093/nar/gkt361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Rau F, Freyermuth F, Fugier C, Villemin JP, Fischer MC, Jost B, Dembele D, Gourdon G, Nicole A, Duboc D, et al. Misregulation of miR-1 processing is associated with heart defects in myotonic dystrophy. Nature structural & molecular biology. 2011;18:840–845. doi: 10.1038/nsmb.2067. [DOI] [PubMed] [Google Scholar]
- 232.Smith MR, Willmann MR, Wu G, Berardini TZ, Moller B, Weijers D, Poethig RS. Cyclophilin 40 is required for microRNA activity in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:5424–5429. doi: 10.1073/pnas.0812729106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Chen K, Rajewsky N. The evolution of gene regulation by transcription factors and microRNAs. Nature reviews Genetics. 2007;8:93–103. doi: 10.1038/nrg1990. [DOI] [PubMed] [Google Scholar]
- 234.Olivieri D, Sykora MM, Sachidanandam R, Mechtler K, Brennecke J. An in vivo RNAi assay identifies major genetic and cellular requirements for primary piRNA biogenesis in Drosophila. The EMBO journal. 2010;29:3301–3317. doi: 10.1038/emboj.2010.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Saito K, Ishizu H, Komai M, Kotani H, Kawamura Y, Nishida KM, Siomi H, Siomi MC. Roles for the Yb body components Armitage and Yb in primary piRNA biogenesis in Drosophila. Genes & development. 2010;24:2493–2498. doi: 10.1101/gad.1989510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Handler D, Olivieri D, Novatchkova M, Gruber FS, Meixner K, Mechtler K, Stark A, Sachidanandam R, Brennecke J. A systematic analysis of Drosophila TUDOR domain-containing proteins identifies Vreteno and the Tdrd12 family as essential primary piRNA pathway factors. The EMBO journal. 2011;30:3977–3993. doi: 10.1038/emboj.2011.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Patil VS, Kai T. Repression of retroelements in Drosophila germline via piRNA pathway by the Tudor domain protein Tejas. Current biology: CB. 2010;20:724–730. doi: 10.1016/j.cub.2010.02.046. [DOI] [PubMed] [Google Scholar]
- 238.Preall JB, Czech B, Guzzardo PM, Muerdter F, Hannon GJ. shutdown is a component of the Drosophila piRNA biogenesis machinery. RNA. 2012;18:1446–1457. doi: 10.1261/rna.034405.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Olivieri D, Senti KA, Subramanian S, Sachidanandam R, Brennecke J. The cochaperone shutdown defines a group of biogenesis factors essential for all piRNA populations in Drosophila. Molecular cell. 2012;47:954–969. doi: 10.1016/j.molcel.2012.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Nishida KM, Okada TN, Kawamura T, Mituyama T, Kawamura Y, Inagaki S, Huang H, Chen D, Kodama T, Siomi H, Siomi MC. Functional involvement of Tudor and dPRMT5 in the piRNA processing pathway in Drosophila germlines. The EMBO journal. 2009;28:3820–3831. doi: 10.1038/emboj.2009.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Lim AK, Kai T. Unique germ-line organelle, nuage, functions to repress selfish genetic elements in Drosophila melanogaster. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:6714–6719. doi: 10.1073/pnas.0701920104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Zamparini AL, Davis MY, Malone CD, Vieira E, Zavadil J, Sachidanandam R, Hannon GJ, Lehmann R. Vreteno, a gonad-specific protein, is essential for germline development and primary piRNA biogenesis in Drosophila. Development. 2011;138:4039–4050. doi: 10.1242/dev.069187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Zhang Z, Xu J, Koppetsch BS, Wang J, Tipping C, Ma S, Weng Z, Theurkauf WE, Zamore PD. Heterotypic piRNA Ping-Pong requires qin, a protein with both E3 ligase and Tudor domains. Molecular cell. 2011;44:572–584. doi: 10.1016/j.molcel.2011.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Anand A, Kai T. The tudor domain protein kumo is required to assemble the nuage and to generate germline piRNAs in Drosophila. The EMBO journal. 2012;31:870–882. doi: 10.1038/emboj.2011.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Rangan P, Malone CD, Navarro C, Newbold SP, Hayes PS, Sachidanandam R, Hannon GJ, Lehmann R. piRNA production requires heterochromatin formation in Drosophila. Current biology: CB. 2011;21:1373–1379. doi: 10.1016/j.cub.2011.06.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Iyer LM, Abhiman S, Aravind L. Natural history of eukaryotic DNA methylation systems. Progress in molecular biology and translational science. 2011;101:25–104. doi: 10.1016/B978-0-12-387685-0.00002-0. [DOI] [PubMed] [Google Scholar]
- 247.Wassarman KM, Repoila F, Rosenow C, Storz G, Gottesman S. Identification of novel small RNAs using comparative genomics and microarrays. Genes & development. 2001;15:1637–1651. doi: 10.1101/gad.901001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Anantharaman V, Aravind L. Novel conserved domains in proteins with predicted roles in eukaryotic cell-cycle regulation, decapping and RNA stability. BMC genomics. 2004;5:45. doi: 10.1186/1471-2164-5-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Aravin AA, Hannon GJ, Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007;318:761–764. doi: 10.1126/science.1146484. [DOI] [PubMed] [Google Scholar]
- 250.Technau U. Evolutionary biology: Small regulatory RNAs pitch in. Nature. 2008;455:1184–1185. doi: 10.1038/4551184a. [DOI] [PubMed] [Google Scholar]
- 251.Venancio TM, Aravind L. Reconstructing prokaryotic transcriptional regulatory networks: lessons from actinobacteria. Journal of biology. 2009;8:29. doi: 10.1186/jbiol132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Venancio TM, Balaji S, Iyer LM, Aravind L. Reconstructing the ubiquitin network: cross-talk with other systems and identification of novel functions. Genome biology. 2009;10:R33. doi: 10.1186/gb-2009-10-3-r33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Balaji S, Iyer LM, Babu MM, Aravind L. Comparison of transcription regulatory interactions inferred from high-throughput methods: what do they reveal? Trends in genetics: TIG. 2008;24:319–323. doi: 10.1016/j.tig.2008.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J. RNA-programmed genome editing in human cells. eLife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Burroughs AM, Balaji S, Iyer LM, Aravind L. Small but versatile: the extraordinary functional and structural diversity of the beta-grasp fold. Biology direct. 2007;2:18. doi: 10.1186/1745-6150-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Burroughs AM, Iyer LM, Aravind L. Natural history of the E1-like superfamily: implication for adenylation, sulfur transfer, and ubiquitin conjugation. Proteins. 2009;75:895–910. doi: 10.1002/prot.22298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Burroughs AM, Jaffee M, Iyer LM, Aravind L. Anatomy of the E2 ligase fold: implications for enzymology and evolution of ubiquitin/Ub-like protein conjugation. Journal of structural biology. 2008;162:205–218. doi: 10.1016/j.jsb.2007.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Grau-Bove X, Sebe-Pedros A, Ruiz-Trillo I. A genomic survey of HECT ubiquitin ligases in eukaryotes reveals independent expansions of the HECT system in several lineages. Genome biology and evolution. 2013;5:833–847. doi: 10.1093/gbe/evt052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Burroughs AM, Iyer LM, Aravind L. Functional diversification of the RING finger and other binuclear treble clef domains in prokaryotes and the early evolution of the ubiquitin system. Molecular bioSystems. 2011;7:2261–2277. doi: 10.1039/c1mb05061c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Iyer LM, Burroughs AM, Aravind L. The prokaryotic antecedents of the ubiquitin-signaling system and the early evolution of ubiquitin-like beta-grasp domains. Genome biology. 2006;7:R60. doi: 10.1186/gb-2006-7-7-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Nunoura T, Takaki Y, Kakuta J, Nishi S, Sugahara J, Kazama H, Chee GJ, Hattori M, Kanai A, Atomi H, et al. Insights into the evolution of Archaea and eukaryotic protein modifier systems revealed by the genome of a novel archaeal group. Nucleic acids research. 2011;39:3204–3223. doi: 10.1093/nar/gkq1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Aravind L, Abhiman S, Iyer LM. Natural history of the eukaryotic chromatin protein methylation system. Progress in molecular biology and translational science. 2011;101:105–176. doi: 10.1016/B978-0-12-387685-0.00004-4. [DOI] [PubMed] [Google Scholar]
- 265.Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jr, Jungkamp AC, Munschauer M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Azuma-Mukai A, Oguri H, Mituyama T, Qian ZR, Asai K, Siomi H, Siomi MC. Characterization of endogenous human Argonautes and their miRNA partners in RNA silencing. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:7964–7969. doi: 10.1073/pnas.0800334105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Vourekas A, Zheng Q, Alexiou P, Maragkakis M, Kirino Y, Gregory BD, Mourelatos Z. Mili and Miwi target RNA repertoire reveals piRNA biogenesis and function of Miwi in spermiogenesis. Nature structural & molecular biology. 2012;19:773–781. doi: 10.1038/nsmb.2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Leung AK, Young AG, Bhutkar A, Zheng GX, Bosson AD, Nielsen CB, Sharp PA. Genome-wide identification of Ago2 binding sites from mouse embryonic stem cells with and without mature microRNAs. Nature structural & molecular biology. 2011;18:237–244. doi: 10.1038/nsmb.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Ghildiyal M, Xu J, Seitz H, Weng Z, Zamore PD. Sorting of Drosophila small silencing RNAs partitions microRNA* strands into the RNA interference pathway. RNA. 2010;16:43–56. doi: 10.1261/rna.1972910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Czech B, Zhou R, Erlich Y, Brennecke J, Binari R, Villalta C, Gordon A, Perrimon N, Hannon GJ. Hierarchical rules for Argonaute loading in Drosophila. Molecular cell. 2009;36:445–456. doi: 10.1016/j.molcel.2009.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Czech B, Malone CD, Zhou R, Stark A, Schlingeheyde C, Dus M, Perrimon N, Kellis M, Wohlschlegel JA, Sachidanandam R, et al. An endogenous small interfering RNA pathway in Drosophila. Nature. 2008;453:798–802. doi: 10.1038/nature07007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Claycomb JM, Batista PJ, Pang KM, Gu W, Vasale JJ, van Wolfswinkel JC, Chaves DA, Shirayama M, Mitani S, Ketting RF, et al. The Argonaute CSR-1 and its 22G-RNA cofactors are required for holocentric chromosome segregation. Cell. 2009;139:123–134. doi: 10.1016/j.cell.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Wang H, Zhang X, Liu J, Kiba T, Woo J, Ojo T, Hafner M, Tuschl T, Chua NH, Wang XJ. Deep sequencing of small RNAs specifically associated with Arabidopsis AGO1 and AGO4 uncovers new AGO functions. The Plant journal: for cell and molecular biology. 2011;67:292–304. doi: 10.1111/j.1365-313X.2011.04594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Takeda A, Iwasaki S, Watanabe T, Utsumi M, Watanabe Y. The mechanism selecting the guide strand from small RNA duplexes is different among argonaute proteins. Plant & cell physiology. 2008;49:493–500. doi: 10.1093/pcp/pcn043. [DOI] [PubMed] [Google Scholar]
- 276.Wang XB, Jovel J, Udomporn P, Wang Y, Wu Q, Li WX, Gasciolli V, Vaucheret H, Ding SW. The 21-nucleotide, but not 22-nucleotide, viral secondary small interfering RNAs direct potent antiviral defense by two cooperative argonautes in Arabidopsis thaliana. The Plant cell. 2011;23:1625–1638. doi: 10.1105/tpc.110.082305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Zhao T, Li G, Mi S, Li S, Hannon GJ, Wang XJ, Qi Y. A complex system of small RNAs in the unicellular green alga Chlamydomonas reinhardtii. Genes & development. 2007;21:1190–1203. doi: 10.1101/gad.1543507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Nakamura R, Takeuchi R, Takata K, Shimanouchi K, Abe Y, Kanai Y, Ruike T, Ihara A, Sakaguchi K. TRF4 is involved in polyadenylation of snRNAs in Drosophila melanogaster. Molecular and cellular biology. 2008;28:6620–6631. doi: 10.1128/MCB.00448-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, Tollervey D. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell. 2005;121:713–724. doi: 10.1016/j.cell.2005.04.029. [DOI] [PubMed] [Google Scholar]
- 280.Wyers F, Rougemaille M, Badis G, Rousselle JC, Dufour ME, Boulay J, Regnault B, Devaux F, Namane A, Seraphin B, et al. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell. 2005;121:725–737. doi: 10.1016/j.cell.2005.04.030. [DOI] [PubMed] [Google Scholar]
- 281.Kadaba S, Krueger A, Trice T, Krecic AM, Hinnebusch AG, Anderson J. Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes & development. 2004;18:1227–1240. doi: 10.1101/gad.1183804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Vanacova S, Wolf J, Martin G, Blank D, Dettwiler S, Friedlein A, Langen H, Keith G, Keller W. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS biology. 2005;3:e189. doi: 10.1371/journal.pbio.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Berndt H, Harnisch C, Rammelt C, Stohr N, Zirkel A, Dohm JC, Himmelbauer H, Tavanez JP, Huttelmaier S, Wahle E. Maturation of mammalian H/ACA box snoRNAs: PAPD5-dependent adenylation and PARN-dependent trimming. RNA. 2012;18:958–972. doi: 10.1261/rna.032292.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Laishram RS, Anderson RA. The poly A polymerase Star-PAP controls 3′-end cleavage by promoting CPSF interaction and specificity toward the pre-mRNA. The EMBO journal. 2010;29:4132–4145. doi: 10.1038/emboj.2010.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Nagaike T, Suzuki T, Katoh T, Ueda T. Human mitochondrial mRNAs are stabilized with polyadenylation regulated by mitochondria-specific poly(A) polymerase and polynucleotide phosphorylase. The Journal of biological chemistry. 2005;280:19721–19727. doi: 10.1074/jbc.M500804200. [DOI] [PubMed] [Google Scholar]
- 286.Olsen A, Vantipalli MC, Lithgow GJ. Checkpoint proteins control survival of the postmitotic cells in Caenorhabditis elegans. Science. 2006;312:1381–1385. doi: 10.1126/science.1124981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Read RL, Martinho RG, Wang SW, Carr AM, Norbury CJ. Cytoplasmic poly(A) polymerases mediate cellular responses to S phase arrest. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:12079–12084. doi: 10.1073/pnas.192467799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Rissland OS, Mikulasova A, Norbury CJ. Efficient RNA polyuridylation by noncanonical poly(A) polymerases. Molecular and cellular biology. 2007;27:3612–3624. doi: 10.1128/MCB.02209-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Saitoh S, Chabes A, McDonald WH, Thelander L, Yates JR, Russell P. Cid13 is a cytoplasmic poly(A) polymerase that regulates ribonucleotide reductase mRNA. Cell. 2002;109:563–573. doi: 10.1016/s0092-8674(02)00753-5. [DOI] [PubMed] [Google Scholar]
- 290.Schmidt MJ, West S, Norbury CJ. The human cytoplasmic RNA terminal U-transferase ZCCHC11 targets histone mRNAs for degradation. RNA. 2011;17:39–44. doi: 10.1261/rna.2252511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Thornton JE, Chang HM, Piskounova E, Gregory RI. Lin28-mediated control of let-7 microRNA expression by alternative TUTases Zcchc11 (TUT4) and Zcchc6 (TUT7) RNA. 2012;18:1875–1885. doi: 10.1261/rna.034538.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Wang L, Eckmann CR, Kadyk LC, Wickens M, Kimble J. A regulatory cytoplasmic poly(A) polymerase in Caenorhabditis elegans. Nature. 2002;419:312–316. doi: 10.1038/nature01039. [DOI] [PubMed] [Google Scholar]
- 293.Suh N, Jedamzik B, Eckmann CR, Wickens M, Kimble J. The GLD-2 poly(A) polymerase activates gld-1 mRNA in the Caenorhabditis elegans germ line. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:15108–15112. doi: 10.1073/pnas.0607050103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Barnard DC, Ryan K, Manley JL, Richter JD. Symplekin and xGLD-2 are required for CPEB-mediated cytoplasmic polyadenylation. Cell. 2004;119:641–651. doi: 10.1016/j.cell.2004.10.029. [DOI] [PubMed] [Google Scholar]
- 295.Ketting RF, Plasterk RH. A genetic link between co-suppression and RNA interference in C. elegans. Nature. 2000;404:296–298. doi: 10.1038/35005113. [DOI] [PubMed] [Google Scholar]
- 296.Collins J, Saari B, Anderson P. Activation of a transposable element in the germ line but not the soma of Caenorhabditis elegans. Nature. 1987;328:726–728. doi: 10.1038/328726a0. [DOI] [PubMed] [Google Scholar]
- 297.Lee SR, Talsky KB, Collins K. A single RNA-dependent RNA polymerase assembles with mutually exclusive nucleotidyl transferase subunits to direct different pathways of small RNA biogenesis. RNA. 2009;15:1363–1374. doi: 10.1261/rna.1630309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Kao CY, Read LK. Targeted depletion of a mitochondrial nucleotidyltransferase suggests the presence of multiple enzymes that polymerize mRNA 3′ tails in Trypanosoma brucei mitochondria. Molecular and biochemical parasitology. 2007;154:158–169. doi: 10.1016/j.molbiopara.2007.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Blum B, Simpson L. Guide RNAs in kinetoplastid mitochondria have a nonencoded 3′ oligo(U) tail involved in recognition of the preedited region. Cell. 1990;62:391–397. doi: 10.1016/0092-8674(90)90375-o. [DOI] [PubMed] [Google Scholar]
- 300.McManus MT, Adler BK, Pollard VW, Hajduk SL. Trypanosoma brucei guide RNA poly(U) tail formation is stabilized by cognate mRNA. Molecular and cellular biology. 2000;20:883–891. doi: 10.1128/mcb.20.3.883-891.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Aphasizhev R, Aphasizheva I, Simpson L. Multiple terminal uridylyltransferases of trypanosomes. FEBS letters. 2004;572:15–18. doi: 10.1016/j.febslet.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 302.Stagno J, Aphasizheva I, Rosengarth A, Luecke H, Aphasizhev R. UTP-bound and Apo structures of a minimal RNA uridylyltransferase. Journal of molecular biology. 2007;366:882–899. doi: 10.1016/j.jmb.2006.11.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Stagno J, Aphasizheva I, Aphasizhev R, Luecke H. Dual role of the RNA substrate in selectivity and catalysis by terminal uridylyl transferases. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:14634–14639. doi: 10.1073/pnas.0704259104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Etheridge RD, Aphasizheva I, Gershon PD, Aphasizhev R. 3′ adenylation determines mRNA abundance and monitors completion of RNA editing in T. brucei mitochondria. The EMBO journal. 2008;27:1596–1608. doi: 10.1038/emboj.2008.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Pagnussat GC, Yu HJ, Ngo QA, Rajani S, Mayalagu S, Johnson CS, Capron A, Xie LF, Ye D, Sundaresan V. Genetic and molecular identification of genes required for female gametophyte development and function in Arabidopsis. Development. 2005;132:603–614. doi: 10.1242/dev.01595. [DOI] [PubMed] [Google Scholar]
- 306.Aphasizheva I, Ringpis GE, Weng J, Gershon PD, Lathrop RH, Aphasizhev R. Novel TUTase associates with an editosome-like complex in mitochondria of Trypanosoma brucei. RNA. 2009;15:1322–1337. doi: 10.1261/rna.1538809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Etheridge RD, Clemens DM, Gershon PD, Aphasizhev R. Identification and characterization of nuclear non-canonical poly(A) polymerases from Trypanosoma brucei. Molecular and biochemical parasitology. 2009;164:66–73. doi: 10.1016/j.molbiopara.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Wang W, Yu H, Long M. Duplication-degeneration as a mechanism of gene fission and the origin of new genes in Drosophila species. Nature genetics. 2004;36:523–527. doi: 10.1038/ng1338. [DOI] [PubMed] [Google Scholar]
- 309.Win TZ, Stevenson AL, Wang SW. Fission yeast Cid12 has dual functions in chromosome segregation and checkpoint control. Molecular and cellular biology. 2006;26:4435–4447. doi: 10.1128/MCB.02205-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Bah A, Wischnewski H, Shchepachev V, Azzalin CM. The telomeric transcriptome of Schizosaccharomyces pombe. Nucleic acids research. 2012;40:2995–3005. doi: 10.1093/nar/gkr1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.McEwan DL, Weisman AS, Hunter CP. Uptake of extracellular double-stranded RNA by SID-2. Mol Cell. 2012;47:746–754. doi: 10.1016/j.molcel.2012.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Sarkies P, Miska EA. Is there social RNA? Science. 2013;341:467–468. doi: 10.1126/science.1243175. [DOI] [PubMed] [Google Scholar]
- 313.Pei J, Millay DP, Olson EN, Grishin NV. CREST--a large and diverse superfamily of putative transmembrane hydrolases. Biol Direct. 2011;6:37. doi: 10.1186/1745-6150-6-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Weiberg A, Wang M, Lin F, Zhao H, Zhang Z, Kaloshian I, Huang H, Jin H. Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science. 2013;342:118–123. doi: 10.1126/science.1239705. [DOI] [PMC free article] [PubMed] [Google Scholar]