Abstract
Identification of recent HIV infection within populations is a public health priority for accurate estimation of HIV incidence rates and transmitted drug resistance. Determining HIV incidence rates by prospective follow-up of HIV-uninfected individuals is challenging and serological assays have important limitations. HIV diversity within an infected host increases with duration of infection. In this analysis, we explore a simple bioinformatics approach to assess viral diversity by determining the percentage of ambiguous base calls in sequences derived from standard genotyping of HIV-1 protease and reverse transcriptase. Sequences from 691 recently infected (≤1 year) and chronically infected (>1 year) individuals from Sweden, Vietnam and Ethiopia were analyzed for ambiguity. A significant difference (p <0.0001) in the proportion of ambiguous bases was observed between sequences from individuals with recent and chronic infection in both HIV-1 subtype B and non-B infection, consistent with previous studies. In our analysis, a cutoff of <0.47% ambiguous base calls identified recent infection with a sensitivity and specificity of 88.8% and 74.6% respectively. 1,728 protease and reverse transcriptase sequences from 36 surveys of transmitted HIV drug resistance performed following World Health Organization guidance were analyzed for ambiguity. The 0.47% ambiguity cutoff was applied and survey sequences were classified as likely derived from recently or chronically infected individuals. 71% of patients were classified as likely to have been infected within one year of genotyping but results varied considerably amongst surveys. This bioinformatics approach may provide supporting population-level information to identify recent infection but its application is limited by infection with more than one viral variant, decreasing viral diversity in advanced disease and technical aspects of population based sequencing. Standardization of sequencing techniques and base calling and the addition of other parameters such as CD4 cell count may address some of the technical limitations and increase the usefulness of the approach.
Keywords: HIV, viral diversity, ambiguity, incidence, resistance, bioinformatics
1. Introduction
Identification of recent HIV infection within populations is a public health priority both for accurate estimation of HIV incidence and estimation of levels of transmitted drug resistant (TDR) HIV (Bennett et al., 2008a; Sexton et al., 2012). The gold standard for determining HIV incidence rates has been prospective follow-up of HIV-uninfected people. However, this approach is expensive, time-consuming and logistically challenging, especially in resource-limited settings. Moreover, this type of study has inherent biases, which may lead to lower observed HIV incidence rates than in the source population of interest. To address these challenges, laboratory testing methods have been developed to calculate HIV incidence using cross-sectional studies. These laboratory methods and testing algorithms rely on the ability to distinguish recently infected from chronically infected individuals using biomarkers such as antibody levels or avidity as indicators of recent infection (Busch et al., 2010; Mastro et al., 2010). BED capture enzyme immunoassays (BED) have proven useful in calculating incidence rates but misclassify a proportion of long-term infections as recent; therefore, false recent rates must be calculated separately in each population studied. Host and virological factors such as HIV subtype are known to influence BED performance making their successful application in populations infected with HIV-1 non-B or mixed subtypes a challenge (Parekh et al., 2011; Sexton et al., 2012); thus limiting this approach in Africa and Asia, where HIV-1 non-B subtypes and mixed subtype epidemics predominate.
Current HIV incidence research is focused on standardizing approaches to evaluating incidence assays and building consensus on statistical methods to interpret results. This research aims to define false recent rates within different populations and develop new and improved incidence assays or multi-test recent infection testing algorithms (RITAs) with lower false recent rates and which are less susceptible to host factors.
The need to estimate incident infection and document levels of TDR in recently infected populations is especially important in low- and middle-income countries to support HIV prevention and antiretroviral therapy program planning. Limited laboratory infrastructure and funding in resource limited settings as well as sensitivity and specificity concerns limit the use of RITAs. Therefore, the World Health Organization (WHO) has proposed surrogate epidemiological definitions to use when performing surveillance of TDR for the purpose of creating cohorts with individuals likely to be recently infected.
From 2004 – 2012, WHO TDR survey guidance recommended use of a truncated sequential sampling method to categorize the prevalence of TDR to each relevant drug class as <5% (low), 5–15% (moderate) or >15% (high), using ≤47 specimens from individuals consecutively diagnosed with HIV in defined geographic regions during the survey period (Bennett et al., 2008b; Myatt and Bennett, 2008). All sequencing of HIV-1 pol for surveys following WHO guidance is performed at WHO-designated drug resistance genotyping laboratories using in-house or commercial methods. All WHO-designated laboratories have undergone a rigorous vetting process and participate in an external quality assurance scheme (WHO, 2008). Drug resistance detected by genotyping is defined using the WHO surveillance drug resistance mutations list (Bennett et al., 2009). TDR surveys are frequently performed in primigravid women age less than 25 years at time of HIV-diagnosis (Bennett et al., 2008a; Myatt and Bennett, 2008; WHO, 2012). Age and gravidity criteria are intended to minimize inclusion of women with previous antiretroviral drug exposures, including drugs provided as part of prevention of mother to child transmission initiatives, thus maximizing the likelihood that detected drug resistance was truly transmitted. In settings of low HIV prevalence or concentrated epidemics, surveillance of HIV TDR has often been conducted at voluntary counseling and testing (VCT) sites using specimens from newly diagnosed individuals age less than 25 years and never pregnant, if female.
A complementary approach to antibody assays and surrogate epidemiological definitions of recent infection, which has been proposed, is assessment of the level of HIV genetic diversity observed with sequencing. Studies measuring viral diversity among sequences generated by single genome sequencing (SGS) conclude that most individuals are infected with a single HIV founder virus from which subsequent diversity arises (Abrahams et al., 2009; Karlsson et al., 1998; Kearney et al., 2009; Keele et al., 2008; Salazar-Gonzalez et al., 2008). Viral diversity increases in a linear fashion in early infection, then reaches a plateau and may eventually decrease in advanced disease (Kearney et al., 2009; Shankarappa et al., 1999). Viral diversity in individual patients is reflected in the proportion of ambiguous bases observed in HIV-1 pol sequences obtained from population based genotyping with increasing diversity seen with increased duration of infection (Kouyos et al., 2011). Ambiguous bases appear in sequences derived by population based sequencing due to the simultaneous detection of more than one nucleotide at the same position of the genome. Viral diversity has been used to estimate time since infection in HIV-1, mainly subtype B infection (Abrahams et al., 2009; Kouyos et al., 2011; Ragonnet-Cronin et al., 2012). In an analysis of the Swiss Cohort study, a linear increase in viral diversity of 0.2 % ambiguous bases per year was observed during the first 8 years of infection (Kouyos et al., 2011).
An inexpensive and simple bioinformatics approach estimating the likely duration of HIV infection using the proportion of ambiguous bases in HIV-1 pol may be a useful measure to strengthen epidemiological surrogate definitions of recent HIV infection in surveys assessing TDR. The aim of our study was to evaluate the feasibility of this approach in HIV-1 non-B subtypes, calculate a sequence ambiguity cutoff between recent and chronic HIV infection, and apply this cutoff to sequences generated from TDR surveys in order to evaluate the utility of this algorithm in providing supportive evidence of recent HIV infection.
2. Materials and methods
2.1 Reference sequences used to differentiate recent from chronic infection
The Swedish InfCareHIV national database and clinical decision support tool (Dalgaard et al., 2012) includes information on all Swedish residents with known HIV infection (prior to 2007 patients diagnosed at some small clinics were not included). The database contains demographic and clinical information such as treatment history, genotyping result and viral sequences when available. As of November 2010, the time of data abstraction, 8,225 patients were registered in InfCareHIV and 3,843 HIV-1 pol sequences from treatment naïve patients were available. Based on seroconversion intervals, clinical data and CD4 cell counts a total of 518 HIV-1 protease and reverse transcriptase sequences were abstracted from individuals as described below. The shortest sequence was 852 base pairs and the longest 1055 base pairs.
Patients with recent infection (≤1 year), subtype B (n=171) and non-B (n=79), were selected based on clinical diagnosis of primary HIV infection or diagnosis due to contact tracing and no indication of earlier infection, or by documented seroconversion within 365 days prior to sampling for sequencing purposes. Chronic infection in patients with subtype B (n=126) was defined as documented seroconversion >1 year before the specimen was obtained for sequencing. Few patients with HIV-1 non-B infection and seroconversion >1 year prior to sequencing were available in InfCareHIV; therefore, a surrogate definition of CD4 ≤200 cells/mm3 at time of diagnosis was used to identify individuals chronically infected with HIV-1 non-B (n=142). This threshold is supported in a recent publication by Lodi et al. (2011) demonstrating that the median time from seroconversion to CD4 ≤200 cells/mm3in a cohort of 18,495 treatment naïve individuals was 7.93 years (7.76–8.09).
The sequences abstracted from the Swedish InfCareHIV database for this study had been generated between 1993–2010. Sequencing and base identification was performed at Karolinska University Hospital, Stockholm, Sweden, Gothenburg University Hospital, Gothenburg, Sweden or the Swedish Institute for Infectious Disease Control, Stockholm, Sweden per established in-house protocols (1993–1999) (Birk and Sönnerborg, 1998; Karlsson et al., 2012)o commercial methods (2000–2002: Trugene HIV-1 Genotyping kit, Visible Genetics, Inc., Toronto, Canada; 2003–2010: ViroSeq HIV-1 genotyping kit version 2, Celera Diagnostics, Alameda, California, USA and ABI PRISM 3100 genetic analyzer, Applied Biosystems, Foster City, California, USA).
The patient characteristics of sequences included from Swedish InfCareHIV are shown in Table 1.
Table 1.
Characteristics and HIV-1 subtypes of patients with recent and chronic infection abstracted from the Swedish InfCareHIV database.
| Recent B | Recent non-B | Chronic B | Chronic non-B | ||
|---|---|---|---|---|---|
| Number of patients | 171 | 79 | 126 | 142 | |
| HIV-1 subtype | A1 | 9 (11%) | 17 (12%) | ||
| B | 171 (100%) | 126 (100%) | |||
| C | 13 (16%) | 47 (33%) | |||
| D | 2 (3%) | 1 (1%) | |||
| F | 3 (2%) | ||||
| G | 5 (6%) | 4 (3%) | |||
| CRF01AE | 33 (42%) | 36 (25%) | |||
| CRF02AG | 19 (13%) | ||||
| CRF06CPX | 3 (2%) | ||||
| CRF12BF | 1 (1%) | ||||
| CRF24BG | 6 (8%) | ||||
| unassigned | 10 (13%) | 12 (8%) | |||
| Patient characteristics | |||||
| Sex | Male | 163 (95%) | 61 (77%) | 110 (87%) | 66 (46%) |
| Female | 8 (5%) | 18 (23%) | 16 (13%) | 76 (54%) | |
| Origin | African | 3 (2%) | 13 (16%) | 4 (3%) | 74 (52%) |
| Caucasian | 144 (84%) | 55 (70%) | 97 (77%) | 30 (21%) | |
| Asian | 8 (5%) | 4 (5%) | 4 (3%) | 27 (19%) | |
| Latin American | 12 (7%) | 2 (3%) | 8 (6%) | 1 (1%) | |
| Other/unknown | 4 (2%) | 5 (6%) | 13 (11%) | 10 (7%) | |
| Transmission mode | MSM | 146 (85%) | 23 (29%) | 94 (75%) | 5 (4%) |
| heterosexual | 12 (7%) | 35 (44%) | 16 (13%) | 108 (76%) | |
| IDU | 13 (8%) | 18 (23%) | 13 (10%) | 3 (2%) | |
| Mother-to-child | 1 (1%) | 3 (2%) | |||
| Other/unknown | 3 (4%) | 2 (2%) | 23 (16%) | ||
MSM, men having sex with men; IDU, intravenous drug user
To augment the analysis, additional HIV-1 protease and reverse transcriptase sequences from chronically infected treatment naïve Ethiopian patients with HIV-1 subtype C (n=110), and Vietnamese patients with HIV-1 subtype CRF01_AE (n=63) initiating antiretroviral therapy at Karolinska Institutet collaborating centers in Ethiopia and Vietnam were analyzed. Although seroconversion dates for these cohorts were unknown, clinically advanced disease at time for sampling assured that they had been infected >1 year. The Vietnamese cohort had a median CD4 count of 56 cells/mm3 (IQR: 26–163) and the Ethiopian cohort 100 cells/mm3 (IQR: 56–144). No sequences from recently infected Ethiopian or Vietnamese patients were available. Sequencing of HIV-1 pol from these two cohorts was performed at the Karolinska Institutet using identical in-house protocols for both the Vietnamese (Bontell et al., 2011) and the Ethiopian cohort (unpublished data).
In total, 691 protease and reverse transcriptase sequences from 250 recently and 441 chronically infected patients were analyzed. HIV-1 subtype distribution is shown in Table 2.
Table 2.
HIV-1 subtypes in study material.
| HIV-1 subtype | Number in reference material | Number in TDR surveys |
|---|---|---|
| A1 | 26 (3.8%) | 66 (3.8%) |
| B | 297 (43.0%) | 111 (6.4%) |
| C | 170 (24.6%) | 531 (30.7%) |
| D | 3 (0.4%) | 27 (1.6%) |
| F | 3 (0.4%) | 3 (0.2%) |
| G | 9 (1.3%) | 60 (3.5%) |
| J | 3 (0.2%) | |
| CRF01AE | 132 (19.1%) | 363 (21.0%) |
| CRF02AG | 19 (2.7%) | 28 (1.6%) |
| CRF06CPX | 3 (0.4%) | 34 (2.0%) |
| CRF07BC | 251 (14.5%) | |
| CRF08BC | 35 (2.0%) | |
| CRF24_BG | 6 (0.9%) | |
| CRF12_BF | 1 (0.1%) | |
| CRF11CPX | 11 (0.6%) | |
| CRF13CPX | 2 (0.1%) | |
| CRF18CPX | 1 (0.1%) | |
| CRF25CPX | 1 (0.1%) | |
| CRF37CPX | 2 (0.1%) | |
| unassigned | 22 (3.2%) | 199 (11.5%) |
| total | 691 | 1728 |
Reference material from Swedish InfCareHIV and Ethiopian and Vietnamese cohorts.
2.2 Sequences from surveys of transmitted HIV drug resistance
1,728 HIV-1 protease and reverse transcriptase sequences from 36 surveys of TDR conducted according to WHO guidelines in Africa, Asia and Latin America during 2005–2009 were included (WHO, 2012). Sixteen of the surveys (mainly from Africa) enrolled primigravid women and 20 surveys (mainly from Asia) enrolled VCT attendees. All sequences were derived from plasma or serum specimens and base calling was performed at WHO-designated laboratories (WHO, 2010). The data were deidentified for country of origin and each survey was labeled with a number between 1 and 36. Surveys using dried blood spots (DBS) as the specimen type were excluded to avoid DNA contribution, which may have affected observed ambiguity.
HIV-1 subtypes for TDR survey sequences are shown in Table 2.
2.3 Analysis methods
HIV-1 pol sequences covering the protease and reverse transcriptase genes were aligned and trimmed using MEGA 4.0 and 5.0 (Tamura et al., 2007; Tamura et al., 2011) and the REGA HIV-1 Subtyping Tool was used to identify subtype (Alcantara et al., 2009; de Oliveira et al., 2005). The percentage of ambiguous base calls (R, Y, K, M, S, W, B, D, H, V or N) in each sequence was calculated using BioEdit (Hall, 1999) and Microsoft Excel. The percentages of ambiguities in sequences derived from recently infected patients (≤1 year) from the InfCareHIV database were compared to the percentages of ambiguities in chronically infected patients (>1 year) abstracted from the InfCareHIV database and from chronically infected cohorts from Vietnam and Ethiopia using unpaired t-test. (GraphPad: http://www.graphpad.com/quickcalcs/ttest1/). The results were confirmed by bootstrap analysis, where individual ambiguity results were randomly picked with replacement for 200 rounds for recently and chronically infected patients respectively. The difference between means of the two groups obtained with bootstrapping was calculated using unpaired t-test function in GraphPad online application. The non-parametric Mann-Whitney test was used for subanalyses assessing recent versus chronic infection within the smaller groups of subtypes A1, C and CRF01_AE (GraphPad InStat version 3.1a for Macintosh, GraphPad Software, San Diego California USA, www.graphpad.com).
A Perl script program was used to calculate the sensitivity and specificity of all possible ambiguity cutoffs. Results of this sensitivity analysis were used to select the cutoff that maximized the specificity while maintaining sensitivity when differentiating sequences from patients with recent (≤1 year) versus chronic infection (>1 year). This ambiguity cutoff was subsequently applied to 1,728 HIV-1 protease and reverse transcriptase sequences from 36 surveys of TDR.
3. Results
3.1 HIV-1 pol sequence ambiguity distinguishes recent from chronic infection in subtype B and non-B
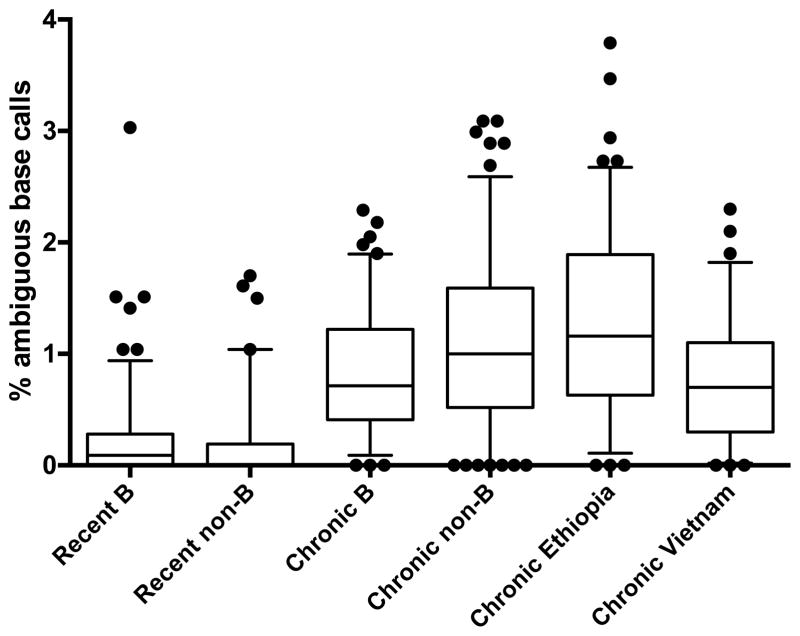
In order to investigate whether the frequency of ambiguous base calls observed in HIV-1 pol sequences could be used to distinguish recent from chronic HIV infection using subtype B and non-B sequences, we first analyzed ambiguity in a set of sequences from patients with known duration of infection abstracted from the InfCareHIV database and sequences from Ethiopian and Vietnamese patients with known chronic infection. Sequences from patients with known recent or known chronic infection formed a set of reference sequences. Sequences from individuals with known recent infection (n=250) had a significantly lower frequency of ambiguous base calls when compared to sequences from those with chronic infection (n=441) (p <0.0001; unpaired t-test). Since the t-test was compromised by the non-normal distribution of the data, results were confirmed with bootstrap analysis, which documented statistical significance (p <0.0001). This finding was consistent across all available subtypes among Swedish InfCareHIV patients: subtype A1 (n=26; p<0.0002), C (n=60; p<0.0005) and CRF01_AE (n=69; p<0.0001) with the Mann-Whitney test and B (n=297; p<0.0001) by bootstrap analysis. Ambiguity observed in sequences from our recently and chronically infected reference population is shown in Figure 1.
Figure 1.
Ambiguity analysis of reference sequences from recent and chronic infection. All sequences from recent infections were abstracted from Swedish InfCareHIV. Sequences from individuals with chronic infection were abstracted from Swedish InfCareHIV and from research cohorts in Vietnam and Ethiopia. The ends of the whiskers mark the 5th and 95th percentile. Outliers are marked by dots.
No significant difference in viral diversity due to subtype was seen in recently infected patients. For the two recently infected reference populations the median values were 0.09% (subtype B) and 0.00% (subtype non-B) respectively (range 0–3.03%). In contrast, wide variation in viral diversity amongst the four populations of chronically infected patients was observed [median 0.72% (subtype B from InfCareHIV), 1.00% (subtype non-B from InfCareHIV), 1.16% (Ethiopia) and 0.70% (Vietnam) ambiguous base calls observed respectively; overall range 0–7.28%]. Each population of chronically infected individuals contained a low number (2–5%) of sequences with no ambiguous bases.
A cutoff of <0.47% ambiguous bases was found to best discriminate recent from chronic infection amongst sequences from the reference population (see section 2.3). This cutoff detected recent HIV-infection with a sensitivity of 88.8% and a specificity of 74.6%. The sensitivity and specificity of the cutoffs <0.45%, <0.47% and <0.5% is shown in Table 3.
Table 3.
Sensitivity and specificity of cutoffs for recent HIV-infection in recently and chronically infected reference populations.
| Cutoff | Sensitivity | Specificity |
|---|---|---|
| 0.45 % | 84.0 % | 75.7 % |
| 0.47 % | 88.8 % | 74.6 % |
| 0.50 % | 88.8 % | 73.5 % |
Cutoff measuring the percentage of ambiguous bases in pol
3.2 Ambiguity in recent infection and mode of transmission
In our reference material of recently infected patients (Table 1) we did not detect a difference in percentage of ambiguous base calls due to mode of transmission in subtype B (n=171) [heterosexual transmission (n=12, median 0.09, range 0–0.30), men having sex with men (MSM) (n=146, median 0.05, range 0–3.03), injection drug users (IDU) (n=13, median 0.09, range 0–1.41)] or in other subtypes (n=76) [heterosexual transmission (n=35, median 0, range 0–1.70), MSM (n=23, median 0, range 0–0.47), IDU (n=18, median 0, range 0–0.85)].
3.3 Surveys of transmitted HIV drug resistance performed following WHO guidance
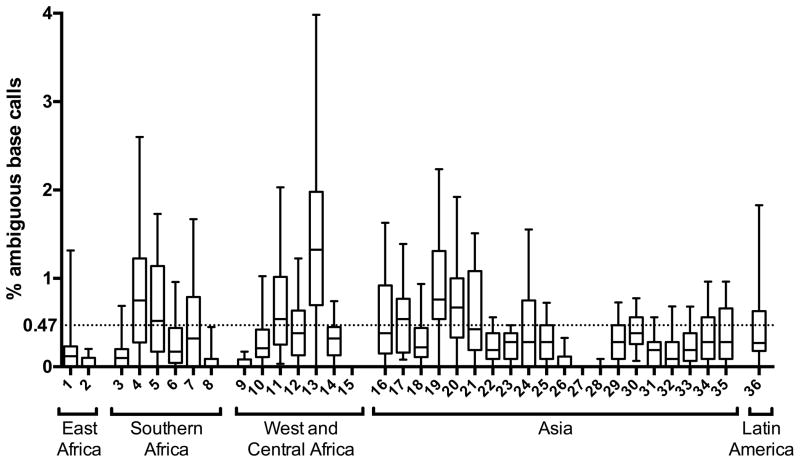
The cutoff of 0.47% ambiguous bases established using reference sequences was applied to 1,728 HIV-1 pol sequences obtained from 36 TDR surveys in order to classify them as likely having originated from individuals with recent (≤1 year) or chronic (> 1 year) HIV infection.
1,233 and 495 sequences had <0.47% and >0.47% ambiguous base calls, respectively. The majority, 1,233 (71%) of individuals, included in TDR surveys were classified by this cutoff as likely to have been recently infected (within one year at time of sampling). The other 495 (29%) individuals included in TDR surveys were classified as likely having chronic infection. Notably, the proportion of individuals classified as having recent infection by the ambiguity cutoff varied considerably amongst surveys (range 22–100%). Pregnant women (873 sequences) and VCT attendees (855 sequences) were classified as recently infected in 73% and 69% of cases respectively. The results of this analysis are shown in Figure 2 with each survey represented by a number from 1 to 36. The number of patients and the HIV-1 subtypes in each survey, the type of survey, and the proportion of sequences in each survey classified as coming from an individual classified as recently infected are shown in supplementary Table 4.
Figure 2.
Ambiguity analysis results from surveys of transmitted HIV drug resistance conducted according to WHO guidance. Each survey is numbered from 1 to 36 and presented by WHO subregion. The ends of the whiskers mark the 5th and 95th percentile. Outliers are omitted for clarity. The cutoff for recent infection (<0.47%) is depicted by the dotted line.
4. Discussion
The identification of recent HIV infection within populations is a public health priority both for accurate estimation of HIV incidence and levels of transmitted HIV drug resistance. A variety of serologic approaches are available to detect recent HIV infection and current assays measure relative levels of HIV specific and total IgG antibodies. Limitations of these BED assays impair their use especially in resource-limited settings and in populations infected with non-subtype B.
Our study explores the application of a bioinformatics algorithm based on increasing HIV viral diversity within individuals over time as a method to support differentiation of sequences as being from recently or chronically infected populations. Use of sequencing data already obtained for resistance testing makes the approach cost-effective and raises the possibility of its integration into existing surveillance protocols. Our study aimed to evaluate this approach further in HIV-1 subtype non-B virus since most surveys of TDR are performed in settings where non-B virus predominates. Our data demonstrate the feasibility of differentiating between sequences from individuals with recent (≤1 year) and chronic (> 1 year) infection, a finding that was independent of HIV subtype. Our observation is consistent with published data from both the Swiss Zurich Primary HIV Infection Study (mainly subtype B) (Kouyos et al., 2011) and the Canadian HIV Strain and Drug Resistance Surveillance Program (only subtype B) (Ragonnet-Cronin et al., 2012). Therefore, assessment of sequence ambiguity and application of cutoffs of viral diversity may provide useful evidence when attempting to enrich HIV-1 sequence sets with individuals likely to have been recently infected.
Kouyos et al. (2011) suggest a cutoff of <0.5% to distinguish recent (≤1 year) from chronic (> 1 year) HIV-1 infection and Ragonnet-Cronin et al. (2012) suggest a cutoff of <0.45% to differentiate duration of subtype B infections as <6 or >6 months. Using our reference material to distinguish recent (≤1 year) from chronic (> 1 year) infection, the cutoff that had the best balance of sensitivity and specificity was <0.47%. The cutoffs are nearly identical, supporting the universality of this approach. The slight discrepancies may be due to technical differences in PCR assays, sequencing and base calling. Discrepancies in the true duration of infections included in the studies may also have affected the calculated cutoffs.
We identified several limitations to our approach. In optimizing the cutoff we only achieved a sensitivity of 88.8% and a specificity of 74.6%. Our model assumes that all patients are infected with one founder virus that subsequently gives rise to multiple quasispecies, yielding more ambiguous bases within a sequence derived by population based sequencing as the duration of infection increases. Amongst recently infected patients in our reference population, 13% had an ambiguity index of ≥0.47%, impairing the sensitivity of the approach. Studies show that a minority of patients are initially infected with more than one viral variant, causing more genetic variation in early infection; thus limiting the value of this approach. The proportion of early HIV-1 infections with more than one documented variant range from 14% (Kearney et al., 2009), 22% (Abrahams et al., 2009), 24% (Keele et al., 2008) to 33% (Salazar-Gonzalez et al., 2008). In the Swiss Zurich Primary HIV Infection Study, 18% (24/130) of patients genotyped within a month from infection had >0.68% ambiguous bases in pol, suggesting infection with more than one founder virus. Swiss data also suggest that there may be more genetic diversity in early infection amongst patients infected via an intravenous route compared to infection acquired sexually (Kouyos et al., 2011). We found no difference in ambiguity amongst recently infected patients due to mode of acquisition of HIV in our study (limited by a much smaller study material) and neither did Ragonnet-Cronin et al (2012). Further studies of viral diversity in recently infected IDUs might be indicated to evaluate the feasibility of this measure of recent infection within this subgroup.
The specificity of applying a cutoff of sequence ambiguity to differentiate recent from chronic infection is likely reduced by decreased viral diversity, which may occur in advanced disease (Karlsson, 1999; Shankarappa et al., 1999). Decreasing diversity amongst patients with advanced disease may lead to low proportions of ambiguous bases amongst sequences from patients with failing immune response. In our reference material, wide variation in ambiguity was seen within and among the four cohorts of chronically infected individuals. The presence of sequences with a low proportion of ambiguous bases may represent individuals with advanced disease. Other explanations include poor sequence quality or operator error leading to underestimation of mixed bases.
The difference between the Ethiopian subtype C (median 1.16%) and Vietnamese subtype CRF01_AE (median 0.70%) chronic cohorts may relate to more advanced disease in the Vietnamese (median CD4 count 56 cells/mm3) than in the Ethiopian cohort (median CD4 count 100 cells/mm3). However, there is a possibility that differences in viral diversity between the cohorts related to subtype and/or host factors affect the results.
Due to the retrospective nature of this analysis, original chromatograms were not available for realignment using an automated tool with standardized parameters. Technical differences resulting from different PCR primers or protocols, sequencing methods, DNA contribution and subjectivity in manual assignment of double peaks as ambiguous bases during the interpretation of chromatograms must be considered as possible sources of bias in this analysis. Nonetheless, we demonstrate that this approach is feasible when using sequences generated using different techniques from different laboratories. Thus, our results together with recent publications (Kouyos et al., 2011; Ragonnet-Cronin et al., 2012) suggest that measurement of sequence ambiguity to differentiate recent from chronic infection may be performed in a context where complete standardization of methods is not feasible. Performance of this analytical method may be augmented by analyzing only ambiguity in certain variable parts of the pol gene as suggested by Ragonnet-Cronin et al. (2012) in an analysis of HIV-1 subtype B sequences. Analysis of ambiguity in variable parts of pol merits study with sequences from all common HIV-1 subtypes. Sequences derived by standard base calling are required for this analysis to minimize random error, which may have greater effect on ambiguity when using short sequences.
In applying the ambiguity cutoff to sequences from TDR surveys following WHO survey guidance, we suggest a possible context where this approach may provide useful population-level information to strengthen surrogate definitions currently used to identify individuals with likely recent HIV infection. Notably, our results suggest that the vast majority of patients (71%) included in TDR surveys were likely to be recently infected (within 1 year); however, in some surveys the majority (up to 78%) had levels of diversity above the 0.47 % cutoff. Possible explanations include differences in age of sexual debut between populations and incorrect application of WHO TDR survey inclusion criteria in the field. The high frequency of super infections with another HIV-strain in some populations (Chohan et al., 2005; Grobler et al., 2004; Powell et al., 2009; Redd et al., 2011; Vidal et al., 2012) is another possible explanation to these results. It should be considered that the approach may be less sensitive in populations where early re-infection is common. Whether the surveys included primigravid women or VCT attendees did not affect the results when all surveys were compared.
The assessment of ambiguous base calls as a method to differentiate chronic versus recent HIV-1 infection holds promise; however, results should be treated cautiously and future research using sequences aligned by standardized base calling software should be performed. Even under optimal technical circumstances, inherent limitations of the approach suggest that it should not be used to assess the duration of infection of individual patients, but rather to contribute to population-level information on HIV-incidence and TDR.
The specificity of the algorithm can be improved if it is used in concert with clinical, epidemiological or laboratory parameters, such as CD4 cell count, that further minimize inclusion of individuals with advanced disease.
Supplementary Material
Acknowledgments
We would like to thank the WHO Internship programme and Karolinska University Laboratory for the opportunity to start this study and collaboration during a WHO Internship in 2010.
We are grateful to Dr Neil Parkin, Data First Consulting, Belmont, CA, USA for his help in making the figures for this paper.
Financial support: EU FP7 Marie Curie ERG (Global ART #246599) and NIH3K233AIO74423-053(MRJ).
Contributor Information
Emmi Andersson, Email: emmi.andersson@gmail.com.
Wei Shao, Email: shaow@mail.nih.gov.
Irene Bontell, Email: irene.bontell@ki.se.
Fatim Cham, Email: qualabs@gmail.com.
Do Duy Cuong, Email: doduy.cuong@gmail.com.
Amogne Wondwossen, Email: wonamogne@yahoo.com.
Lynn Morris, Email: lynnm@nicd.ac.za.
Gillian Hunt, Email: GillianH@nicd.ac.za.
Anders Sönnerborg, Email: Anders.Sonnerborg@ki.se.
Silvia Bertagnolio, Email: bertagnolios@who.int.
Frank Maldarelli, Email: fmalli@mail.nih.gov.
Michael R Jordan, Email: mjordan@tuftsmedicalcenter.org.
References
- Abrahams M, Anderson J, Giorgi E, Seoighe C, Mlisana K, Ping L, Athreya G, Treurnicht F, Keele B, Wood N, Salazar-Gonzalez J, Bhattacharya T, Chu H, Hoffman I, Galvin S, Mapanje C, Kazembe P, Thebus R, Fiscus S, Hide W, Cohen M, Karim S, Haynes B, Shaw G, Hahn B, Korber B, Swanstrom R, Williamson C C.A.I.S.T.C.f.H.-A.V.I Consortium. Quantitating the Multiplicity of Infection with Human Immunodeficiency Virus Type 1 Subtype C Reveals a Non-Poisson Distribution of Transmitted Variants. J Virol. 2009;83:3556–3567. doi: 10.1128/JVI.02132-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcantara L, Cassol S, Libin P, Deforche K, Pybus O, Van Ranst M, Galvao-Castro B, Vandamme A-M, de Oliveira T. A Standardized Framework for Accurate, High-throughput Genotyping of Recombinant and Non-recombinant Viral Sequences. Nucleic Acids Res. 2009:W634–642. doi: 10.1093/nar/gkp455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett D, Bertagnolio S, Sutherland D, Gilks C. The World Health organization’s global strategy for prevention and assessment of HIV drug resistance. Antivir Ther. 2008a;13(Suppl 2):1–13. [PubMed] [Google Scholar]
- Bennett DE, Camacho RJ, Otelea D, Kuritzkes DR, Fleury H, Kiuchi M, Heneine W, Kantor R, Jordan MR, Schapiro JM, Vandamme AM, Sandstrom P, Boucher CA, van de Vijver D, Rhee SY, Liu TF, Pillay D, Shafer RW. Drug resistance mutations for surveillance of transmitted HIV-1 drug-resistance: 2009 update. PLoS One. 2009;4:e4724. doi: 10.1371/journal.pone.0004724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett DE, Myatt M, Bertagnolio S, Sutherland D, Gilks CF. Recommendations for surveillance of transmitted HIV drug resistance in countries scaling up antiretroviral treatment. Antivir Ther. 2008b;13(Suppl 2):25–36. [PubMed] [Google Scholar]
- Birk M, Sönnerborg A. Variations in the human immunodeficiency virus type 1 pol gene associated with reduced sensitivity to antiretroviral drugs in treatment naive patients. AIDS. 1998;12:2369–2375. doi: 10.1097/00002030-199818000-00005. [DOI] [PubMed] [Google Scholar]
- Bontell I, Cuong D, Agneskog E, Diwan V, Larsson M, Sönnerborg A. Transmitted drug resistance and phylogenetic analysis of HIV CRF01_AE in Northern Vietnam. Infect Genet Evol. 2011;12:448–452. doi: 10.1016/j.meegid.2011.04.034. [DOI] [PubMed] [Google Scholar]
- Busch M, Pilcher C, Mastro T, Kaldor J, Vercauteren G, Rodriguez W, Rousseau C, Rehle T, Welte A, Averill M, Garcia Calleja J W.W.G.o.H.I Assays. Beyond detuning: 10 years of progress and new challenges in the development and application of assays for HIV incidence estimation. AIDS. 2010;24:2763–2771. doi: 10.1097/QAD.0b013e32833f1142. [DOI] [PubMed] [Google Scholar]
- Chohan B, Lavreys L, Rainwater S, Overbaugh J. Evidence for frequent reinfection with human immunodeficiency virus type 1 of a different subtype. J Virol. 2005;79:10701–10708. doi: 10.1128/JVI.79.16.10701-10708.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalgaard L, Søgaard O, Jensen-Fangel S, Larsen C, Sönnerborg A, Østergaard L. Use of InfCare HIV to identify and characterize suboptimally treated HIV patients at a Danish HIV clinic: a cross-sectional cohort study. Scand J Infect Dis. 2012:108–114. doi: 10.3109/00365548.2011.616223. [DOI] [PubMed] [Google Scholar]
- de Oliveira T, Deforche K, Cassol S, Salminem M, Paraskevis D, Seebregts C, Snoeck J, van Rensburg E, Wensing A, van de Vijver D, Boucher C, Camacho R, Vandamme AM. An Automated Genotyping System for Analysis of HIV-1 and other Microbial Sequences. Bioinformatics. 2005;21:3797–3800. doi: 10.1093/bioinformatics/bti607. [DOI] [PubMed] [Google Scholar]
- Grobler J, Gray C, Rademeyer C, Seoighe C, Ramjee G, Karim S, Morris L, Williamson C. Incidence of HIV-1 dual infection and its association with increased viral load set point in a cohort of HIV-1 subtype C-infected female sex workers. J Infect Dis. 2004;190:1355–1359. doi: 10.1086/423940. [DOI] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999:95–98. [Google Scholar]
- Karlsson Reappearance of Founder Virus Sequence in human Immunodeficiency Virus Type 1-Infected Patients. J Virol. 1999;73:6191–6196. doi: 10.1128/jvi.73.7.6191-6196.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson A, Björkman P, Bratt G, Ekvall H, Gisslén M, Sönnerborg A, Mild M, Albert J. Low prevalence of transmitted drug resistance in patients newly diagnosed with HIV-1 infection in Sweden 2003–2010. PLoS One. 2012;7:e33484. doi: 10.1371/journal.pone.0033484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearney M, Maldarelli F, Shao W, Margolick JB, Daar ES, Mellors JW, Rao V, Coffin JM, Palmer S. Human immunodeficiency virus type 1 population genetics and adaptation in newly infected individuals. J Virol. 2009;83:2715–2727. doi: 10.1128/JVI.01960-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keele BF, Giorgi EE, Salazar-Gonzalez JF, Decker JM, Pham KT, Salazar MG, Sun C, Grayson T, Wang S, Li H, Wei X, Jiang C, Kirchherr JL, Gao F, Anderson JA, Ping LH, Swanstrom R, Tomaras GD, Blattner WA, Goepfert PA, Kilby JM, Saag MS, Delwart EL, Busch MP, Cohen MS, Montefiori DC, Haynes BF, Gaschen B, Athreya GS, Lee HY, Wood N, Seoighe C, Perelson AS, Bhattacharya T, Korber BT, Hahn BH, Shaw GM. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci U S A. 2008;105:7552–7557. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouyos RD, von Wyl V, Yerly S, Boni J, Rieder P, Joos B, Taffe P, Shah C, Burgisser P, Klimkait T, Weber R, Hirschel B, Cavassini M, Rauch A, Battegay M, Vernazza PL, Bernasconi E, Ledergerber B, Bonhoeffer S, Gunthard HF. Ambiguous nucleotide calls from population-based sequencing of HIV-1 are a marker for viral diversity and the age of infection. Clin Infect Dis. 2011;52:532–539. doi: 10.1093/cid/ciq164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodi S, Phillips A, Touloumi G, Geskus R, Meyer L, Thiébaut R, Pantazis N, Amo J, Johnson A, Babiker A, Porter K EuroCoord CCi. Time From Human Immunodeficiency Virus Seroconversion to Reaching CD4+ Cell Count Thresholds <200, <350, and <500 Cells/mm3: Assessment of Need Following Changes in Treatment Guidelines. Clin Infect Dis. 2011;53:817–825. doi: 10.1093/cid/cir494. [DOI] [PubMed] [Google Scholar]
- Mastro T, Kim A, Hallett T, Rehle T, Welte A, Laeyendecker O, Oluoch T, Garcia-Calleja J. Estimating HIV incidence in populations using tests for recent infection: Issues, challenges and the way forward. J HIV AIDS Surveill Epidemiol. 2010;2:1–14. [PMC free article] [PubMed] [Google Scholar]
- Myatt M, Bennett D. A novel sequential sampling technique for the surveillance of transmitted HIV drug resistance by cross sectional survey for use in low resource settings. Antivir Ther. 2008;13(Suppl 2):37–48. [PubMed] [Google Scholar]
- Parekh B, Hanson D, Hargrove J, Branson B, Green T, Dobbs T, Constantine N, Overbaugh J, McDougal J. Determination of Mean Recency Period for Estimation of HIV Type 1 Incidence with the BED-Capture EIA in Persons Infected with Diverse Subtypes. AIDS Res Hum Retroviruses. 2011;27:265–273. doi: 10.1089/aid.2010.0159. [DOI] [PubMed] [Google Scholar]
- Powell R, Urbanski M, Burda S, Kinge T, Nyambi P. High frequency of HIV-1 dual infections among HIV-positive individuals in Cameroon, West Central Africa. J Acquir Immune Defic Syndr. 2009;50:84–92. doi: 10.1097/QAI.0b013e31818d5a40. [DOI] [PubMed] [Google Scholar]
- Ragonnet-Cronin M, Aris-Brosou S, Joanisse I, Merks H, Vallée D, Caminiti K, Rekart M, Krajden M, Cook D, Kim J, Malloch L, Sandstrom P, Brooks J. Genetic Diversity as a Marker for Timing Infection in HIV-Infected Patients: Evaluation of a 6-Month Window and Comparison With BED. J Infect Dis. 2012;206:756–764. doi: 10.1093/infdis/jis411. [DOI] [PubMed] [Google Scholar]
- Redd A, Collinson-Streng A, Martens C, Ricklefs S, Mullis C, Manucci J, Tobian A, Selig E, Laeyendecker O, Sewankambo N, Gray R, Serwadda D, Wawer M, Porcella S, Quinn T Program RHS. Identification of HIV superinfection in seroconcordant couples in Rakai, Uganda, by use of next-generation deep sequencing. J Clin Microbiol. 2011;49:2859–2867. doi: 10.1128/JCM.00804-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar-Gonzalez JF, Bailes E, Pham KT, Salazar MG, Guffey MB, Keele BF, Derdeyn CA, Farmer P, Hunter E, Allen S, Manigart O, Mulenga J, Anderson JA, Swanstrom R, Haynes BF, Athreya GS, Korber BT, Sharp PM, Shaw GM, Hahn BH. Deciphering human immunodeficiency virus type 1 transmission and early envelope diversification by single-genome amplification and sequencing. J Virol. 2008;82:3952–3970. doi: 10.1128/JVI.02660-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton C, Costenbader E, Vinh D, Chen P, Hoang T, Lan N, Feldblum P, Kim A, Giang le T. Correlation of prospective and cross-sectional measures of HIV type 1 incidence in a higher-risk cohort in Ho Chi Minh City, Vietnam. AIDS Res Hum Retroviruses. 2012;28:866–873. doi: 10.1089/aid.2011.0221. [DOI] [PubMed] [Google Scholar]
- Shankarappa R, Margolick JB, Gange SJ, Rodrigo AG, Upchurch D, Farzadegan H, Gupta P, Rinaldo CR, Learn GH, He X, Huang XL, Mullins JI. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J Virol. 1999;73:10489–10502. doi: 10.1128/jvi.73.12.10489-10502.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei MSK. MEGA5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance and Maximum Parsimony Methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. World Health Organization/HIVresNet drug resistance laboratory strategy. Antiviral Therapy. 2008;13(Suppl 2):49–58. [PubMed] [Google Scholar]
- WHO. World Health Organization/HIV/ResNet HIV drug resistance laboratory strategy. World Health Organization; Geneva, Switzerland: 2010. [Last accessed 29 October, 2012]. July 2010 Update. Available at: http://www.who.int/hiv/pub/drugresistance/hiv_reslab_strategy.pdf. [Google Scholar]
- WHO. WHO HIV drug resistance report 2012. World Health Organization; Geneva, Switzerland: 2012. [Google Scholar]
- Vidal N, Diop H, Montavon C, Butel C, Bosch S, Ngole E, Touré-Kane C, Mboup S, Delaporte E, Peeters M. A novel multiregion hybridization assay reveals high frequency of dual inter-subtype infections among HIV-positive individuals in Cameroon, West Central Africa. Infect Genet Evol. 2012;14C:73–82. doi: 10.1016/j.meegid.2012.11.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.