Abstract
The transcription factor nuclear factor κB (NF-κB) rapidly reprograms gene expression in response to various stimuli, and its activity is regulated by several posttranslational modifications, including phosphorylation, methylation, and acetylation. The addition of O-linked β-N-acetylglucosamine (a process known as O-GlcNAcylation) is an abundant posttranslational modification that is enhanced in conditions such as hyperglycemia and cellular stress. We report that the NF-κB subunit c-Rel is modified and activated by O-GlcNAcylation. We identified serine 350 as the site of O-GlcNAcylation, which was required for the DNA binding and transactivation functions of c-Rel. Blocking the O-GlcNAcylation of this residue abrogated c-Rel–mediated expression of the cytokine-encoding genes IL2, IFNG, and CSF2 in response to T cell receptor (TCR) activation, whereas increasing the extent of O-GlcNAcylation of cellular proteins enhanced the expression of these genes. TCR- or tumor necrosis factor (TNF)–induced expression of other NF-κB target genes, such as NFKBIA (which encodes IκBα) and TNFAIP3 (which encodes A20), occurred independently of the O-GlcNAcylation of c-Rel. Our findings suggest a stimulus-specific role for hyperglycemia-induced O-GlcNAcylation of c-Rel in promoting T cell–mediated autoimmunity in conditions such as type 1 diabetes by enhancing the production of T helper cell cytokines.
INTRODUCTION
Nuclear factor κB (NF-κB) is a pleiotropic, evolutionarily conserved transcription factor family with multiple roles in cell survival, development, apoptosis, immunity, and inflammatory responses (1). The NF-κB family is composed of five monomers that function in dimeric pairs: p65 (RelA), RelB, c-Rel, p50 (and its precursor p105), and p52 (and its precursor p100). NF-κB family members are preformed proteins that reside, in unstimulated cells, mostly in the cytoplasm bound to inhibitory proteins of the inhibitor of κB (IκB) family. In general, activation occurs through signal-induced phosphorylation and proteasome-mediated degradation of IκB, which releases bound NF-κB, enabling its translocation to the nucleus where it activates transcription (2). Because the NF-κB family members are preformed proteins, their initial activation and activity is often regulated by posttranslational modifications rather than by induction of their synthesis. All NF-κB members are regulated by phosphorylation and ubiquitylation (3, 4). The p105, p100, and p65 subunits are also regulated by acetylation. Modifications of NF-κB proteins by S-nitrosylation, oxidation, nitration, and alkylation are also known, although the physiological roles of the individual modifications remain poorly defined (3).
O-GlcNAc glycosylation (O-GlcNAcylation) is an essential and abundant form of intracellular posttranslational protein modification. It involves attachment of the monosaccharide N-acetyl-d-glucosamine (GlcNAc) to serine or threonine residues in nuclear and cytoplasmic proteins of multicellular eukaryotes (5, 6). O-GlcNAcylation is a dynamic, reversible process akin to phosphorylation, with the addition and removal of O-GlcNAc catalyzed by O-GlcNAc transferase (OGT) and O-GlcNAcase (OGA), respectively (6). Several lines of evidence suggest a reciprocal relationship between O-GlcNAc and phosphate, which may occupy the same or adjacent serine or threonine residues in proteins or may occur simultaneously at different sites in a protein (6, 7). O-GlcNAcylation plays an important role in many key cellular processes, including cell cycle regulation (8, 9), transcription and translation (6), protein stability and turnover (10), cellular stress (11), and gene expression (6, 12). Lack of O-GlcNAcylation is embryonically lethal, as is seen in OGT-deficient mice, and O-GlcNAcylation is essential for T cell function (13, 14).
The extent of O-GlcNAcylation of proteins is increased under hyperglycemic conditions, and O-GlcNAcylation enhances NF-κB–dependent transcription (15). Both O-GlcNAcylation and NF-κB activation are associated with experimental and clinical diabetes (16–19). Growing evidence supports a pivotal role for O-GlcNAcylation in the activation of NF-κB in B cells and T cells (20). Regulation of the NF-κB subunit p65 by O-GlcNAcylation has received some attention, with identification of the sites of modification and of their role in influencing p65 function (21, 22). Although both NF-κB and O-GlcNAcylation were discovered more than 25 years ago, the role of this posttranslational modification in regulating NF-κB subunits other than p65 remains largely uncharacterized. Moreover, relatively few studies have pinpointed the site of modification on proteins or established the site-specific functions for O-GlcNAc. So far, less than 10% of the known O-GlcNAcylated proteins have had their modification sites identified (6).
While studying the glucose dependency of various cell types, we found that a high glucose concentration selectively promoted the survival of potentially self-reactive T cells by increasing the expression of several NF-κB–dependent genes that encode antiapoptotic proteins (23). Because hyperglycemia also enhances the extent of O-GlcNAcylation of proteins, we examined the O-GlcNAcylation status of all of the NF-κB family proteins in lymphocytes under hyperglycemic conditions. Here, we showed that c-Rel is the major O-GlcNAcylated NF-κB subunit in lymphocytes, and that enhancement of its O-GlcNAcylation increased its transcriptional activity. We identified serine 350 (Ser350) as the site of O-GlcNAcylation in c-Rel by collision-induced dissociation mass spectrometry (CID-MS) and CID-MS after BEMAD [β-elimination followed by Michael addition with 2-aminoethanethiol (2-AET)] derivatization and site-directed mutagenesis. Mutation of Ser350 completely blocked the O-GlcNAcylation of c-Rel, compromising its DNA binding and transactivation in cells in response to stimulation of the T cell receptor (TCR). Our study links the physiological functions of c-Rel and O-GlcNAcylation in T cells, and it suggests a previously unknown and potentially pathologic role of O-GlcNAcylation: promoting autoimmunity by enhancing the production of cytokines by helper T cells through modulation of c-Rel function.
RESULTS
The NF-κB protein c-Rel is modified by O-GlcNAcylation
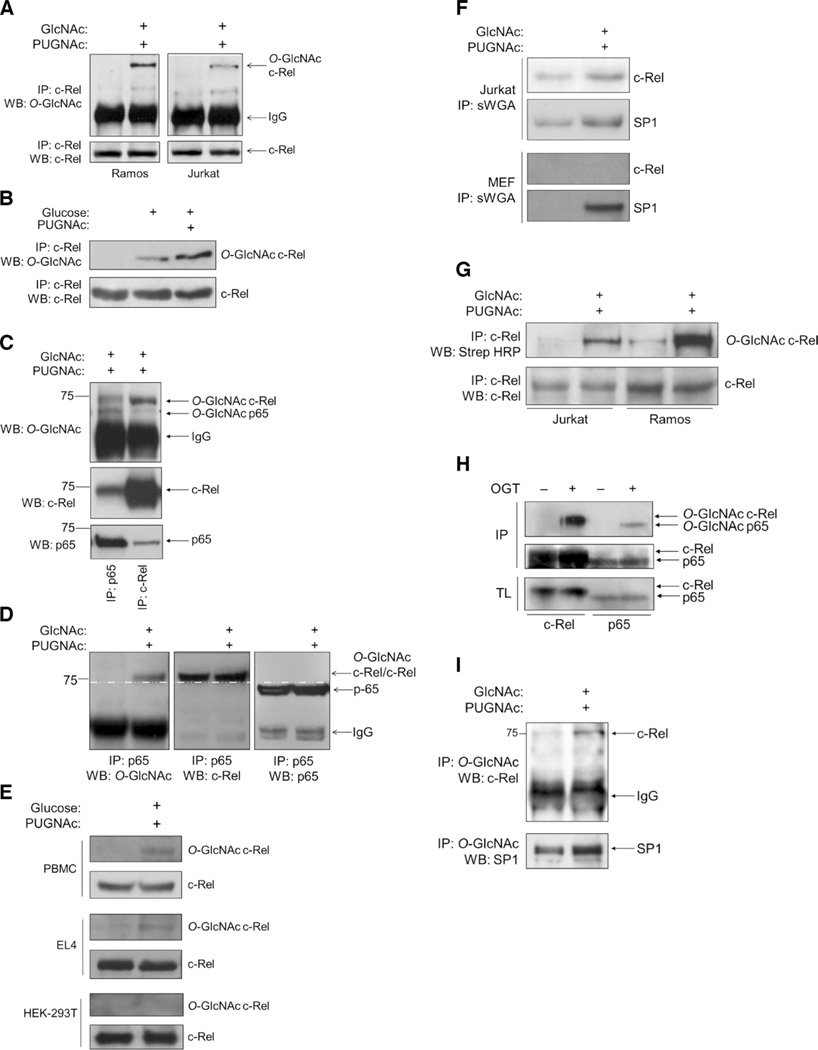
We examined NF-κB proteins in various cellular systems for evidence of increased O-GlcNAcylation, and we found that c-Rel was a major O-GlcNAcylated NF-κB protein in both B (Ramos) and T (Jurkat) lymphoblastoid cell lines (Fig. 1A). To enrich the O-GlcNAcylated pool of proteins, we exposed cells to GlcNAc, a precursor of GlcNAcylation, and to O-(2-acetamido-2-deoxy-d-glucopyranosylidene)amino-N-phenylcarbamate (PUGNAc) (24), a potent inhibitor of the enzyme OGA, which catalyzes the cleavage of O-GlcNAc from serine and threonine residues of modified proteins. Inhibition of OGA increases the pool of O-GlcNAcylated proteins in cells. We also detected the O-GlcNAcylation of c-Rel in cells exposed to a pathophysiologically relevant hyperglycemic concentration of glucose (30 mM), and the addition of PUGNAc further enhanced the extent of O-GlcNAcylation of c-Rel (Fig. 1B). Under similar conditions, c-Rel was also O-GlcNAcylated in primary mouse splenocytes (Fig. 1C). As previously reported (15, 20–22), we also observed the O-GlcNAcylation of the NF-κB protein p65 in primary splenocytes (Fig. 1C). O-GlcNAcylated c-Rel existed in a complex with p65 in the cytoplasm, as indicated by their reciprocal coimmunoprecipitation (Fig. 1, C and D). Unlike phosphorylation, O-GlcNAcylation does not change the electrophoretic mobility of modified proteins (6). The O-GlcNAcylated band in p65 immunoprecipitates appeared to have a greater molecular mass than that of p65, suggesting that the coimmunoprecipitated c-Rel had undergone O-GlcNAcylation (Fig. 1D). Furthermore, after depletion of c-Rel from the lysates, reimmunoprecipitated p65 failed to show evidence of O-GlcNAcylation (fig. S1A), suggesting that c-Rel was the major O-GlcNAcylated protein in lymphocytes.
Fig. 1. The NF-κB subunit c-Rel is modified by O-GlcNAcylation.
(A) Ramos cells and Jurkat cells (50 × 106 each) were left untreated or were treated with 4 mM GlcNAc and 100 µM PUGNAc for 12 hours. Samples were subjected to immunoprecipitation (IP) with an anti–c-Rel antibody and were analyzed by Western blotting (WB) with an antibody against O-GlcNAc (RL2). (B) Jurkat cells were left untreated or were treated with 100 µM PUGNAc for 12 hours and 30 mM glucose for 3 hours or with glucose alone. Samples were analyzed as described in (A). (C) Splenocytes (75 × 106) from C57BL/6 mice were treated with 4 mM GlcNAc and 100 µM PUGNAc for 8 hours, subjected to immunoprecipitation with antibodies against p65 or c-Rel, and analyzed by Western blotting with antibodies against the indicated proteins. (D) Ramos cells were treated with 4 mM GlcNAc and 100 µM PUGNAc for 12 hours and then were subjected to immunoprecipitation with anti-p65 antibody. O-GlcNAcylation of the 75-kD protein was detected with the anti–O-GlcNAc (RL2) antibody (left). The blot was reincubated with antibodies against c-Rel and p65. c-Rel coimmunoprecipitated with p65 (middle). The p65 protein ran with a lower molecular mass than did the O-GlcNAcylated protein and c-Rel (75 kD) (right). The white dashed line running across all three blots indicates separation between c-Rel and p65. (E) Human PBMCs were activated with phytohemagglutinin for 48 hours. EL4 cells (50 × 106) and HEK 293T cells (10 × 106) were cultured overnight in medium containing 5 mM glucose. Cells were treated and analyzed as described in (B). (F) Jurkat cells (30 × 106) and MEFs (10 × 106) were left untreated or were treated as described in (B) and then were subjected to immunoprecipitation with succinylated WGA (sWGA) to pull down O-GlcNAcylated proteins. Samples were analyzed by Western blotting with antibodies against c-Rel and SP1, which was used as a positive control for O-GlcNAcylation and sWGA immunoprecipitation. (G) Chemoenzymatic O-GlcNAc detection assay. Jurkat cells (30 × 106) and Ramos cells (30 × 106) were left untreated or were treated with 4 mM GlcNAc and 100 µM PUGNAc for 8 hours and then were subjected to chemoenzymatic labeling and immunoprecipitation with anti–c-Rel antibody. Samples were then analyzed by Western blotting as indicated. (H) Baculoviral expression of c-Rel or p65 in insect cells in the absence or presence of OGT. Cell lysates were subjected to immunoprecipitation with antibodies against c-Rel or p65 and then were analyzed by Western blotting with an anti–O-GlcNAc antibody. Total cell lysates were analyzed by Western blotting with anti–c-Rel and anti-p65 antibodies. (I) Reverse immunoprecipitation of O-GlcNAcylated c-Rel. Jurkat cells were treated as described in (A). Anti–O-GlcNAc immunoprecipitates were analyzed by Western blotting with anti–c-Rel and anti-SP1 antibodies. Western blots in (A) through (I) are representative of three independent experiments.
We also found the O-GlcNAcylation of c-Rel in human peripheral blood mononuclear cells (PBMCs), as well as in the glucose-dependent mouse T cell line EL4 (Fig. 1E) (23); however, we did not observe O-GlcNAcylation of endogenous c-Rel in human embryonic kidney (HEK) 293T cells, an epithelial cell line, or in mouse embryonic fibroblasts (MEFs) (Fig. 1, E and F), even when global O-GlcNAcylation in these cells was enhanced (fig. S1B). We found that c-Rel in Jurkat cells bound to the O-GlcNAc–binding lectin wheat germ agglutinin (WGA), and its reactivity to the lectin increased in the presence of GlcNAc and PUGNAc (Fig. 1F). Furthermore, a chemoenzymatic method to biotinylate and detect O-GlcNAcylated proteins showed O-GlcNAcylated c-Rel in Ramos cells and Jurkat cells (Fig. 1G). We found that both c-Rel and p65 were O-GlcNAcylated in a heterologous baculoviral expression system when coexpressed with OGT (Fig. 1H). Finally, we precipitated c-Rel from GlcNAc- and PUGNAc-treated cells with an anti–O-GlcNAc antibody (Fig. 1I).
c-Rel is O-GlcNAcylated at Ser350 in the Rel inhibitory domain
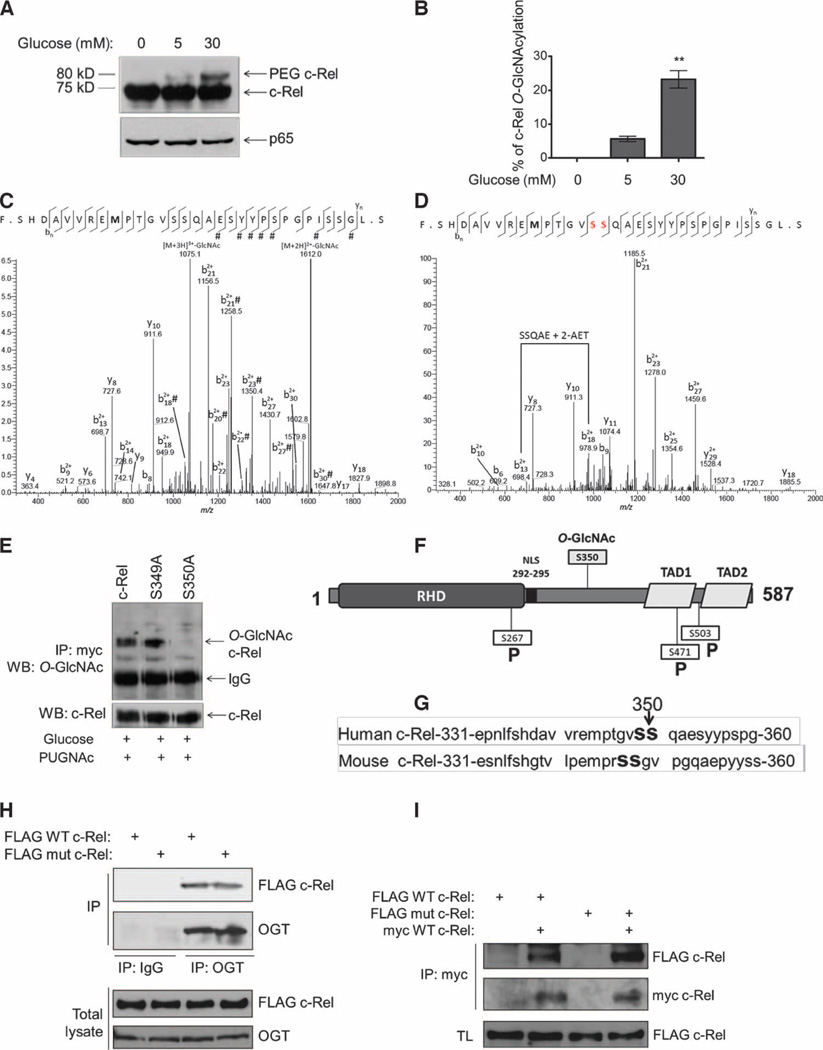
To identify the stoichiometry of c-Rel O-GlcNAcylation, we used a chemoenzymatic approach to label O-GlcNAc residues with a 5-kD polyethylene glycol 5000 (PEG 5000) mass tag (25). This approach increases the molecular mass of the glycoprotein in 5-kD increments, depending on the number of O-GlcNAc moieties that are attached to the protein. We found that human c-Rel showed a shift of 5 kD in Jurkat cells and Ramos cells after treatment with PUGNAc and high concentrations of glucose, which suggests that modified c-Rel was predominantly mono-O-GlcNAcylated (Fig. 2A). We saw an increased proportion of O-GlcNAcylated c-Rel in cells in response to 30 mM glucose compared to that in cells treated with 5 mM glucose, suggesting that the O-GlcNAcylation of c-Rel was both dynamic and inducible (Fig. 2A). We quantified the stoichiometry of O-GlcNAcylated c-Rel in cells in 30 mM glucose by densitometry and found that about 20% of the total c-Rel protein was modified with O-GlcNAc (Fig. 2B).
Fig. 2. c-Rel is O-GlcNAcylated at Ser350.
(A) Ramos cells (10 × 106) were cultured overnight in RPMI medium without glucose and then were treated for 3 hours with 5 or 30 mM glucose and 100 µM PUGNAc. Cells were lysed by boiling in buffer containing 1% SDS and complete protease inhibitor cocktail, and 100 µg of total protein was chemoenzymatically labeled with a 5-kD PEG mass tag and analyzed by Western blotting with antibodies against c-Rel and p65. The PEG–c-Rel band represents the O-GlcNAc–modified subpopulation of c-Rel. Data are representative of four independent experiments. (B) Percentage of c-Rel O-GlcNAcylation. Data are means ± SEM from three independent experiments. **P < 0.01 comparing 30 to 5 mM. (C and D) CID-MS site mapping of the O-GlcNAcylation of c-Rel. (C) Fragment ions were calculated and assigned as unmodified by GlcNAc because of facile neutral loss of GlcNAc in CID except where indicated by “#.” GlcNAc modification was localized to the N-terminal 18 amino acid residues. (D) Digested c-Rel was derivatized by BEMAD with 2-AET and analyzed by nanoLCMS with CID. The methionine in both spectrums (bold) is oxidized. (E) HEK 293T cells were transfected with plasmids encoding myc-tagged wild-type (WT) c-Rel or the indicated c-Rel mutants. Cells were cultured overnight in medium containing 5 mM glucose. Twenty-four hours after transfection, cells were treated with 30 mM glucose and 100 µM PUGNAc for 3 hours, subjected to immunoprecipitation with anti-myc antibody, and analyzed by Western blotting with antibodies against the indicated proteins. (F) Representation of the structure of c-Rel. RHD, Rel homology domain; NLS, nuclear localization signal; RID, Rel inhibitory domain; TAD, transactivation domain; P, phosphorylation sites. (G) Sequences of human and mouse c-Rel with the O-GlcNAcylation site indicated. (H) HEK 293T cells were transfected with plasmids encoding FLAG-tagged WT or S350A mutant c-Rel. Cell lysates were subjected to immunoprecipitation with an anti-OGT antibody or with rabbit immunoglobulin G (IgG) as a negative control, and samples were analyzed by Western blotting with antibodies against the FLAG tag or OGT. Total lysates were also analyzed by Western blotting with the same antibodies. (I) HEK 293T cells were transfected with plasmids encoding the indicated constructs. Cell lysates were subjected to immunoprecipitation with anti-myc antibody and were analyzed by Western blotting with an anti-FLAG antibody to examine c-Rel dimer formation. Total lysates (TL) were also analyzed by Western blotting with anti-FLAG antibody. Western blotting data in (E), (H), and (I) are representative of three independent experiments.
To map the site of O-GlcNAcylation, we immunoprecipitated endogenous c-Rel from Jurkat cells or Ramos cells treated with a high concentration of glucose (30 mM) and PUGNAc (fig. S2), and we analyzed the chymotrypsin digest and the BEMAD-derivatized digests by nano–liquid chromatography–tandem mass spectrometry (nanoLC-MS/MS). O-GlcNAc residues are often labile during CID-MS. BEMAD derivatization replaces the labile carbon-oxygen linkage of the O-GlcNAc modification with a more stable carbon-sulfur linkage that can be identified by MS. Together, these experiments mapped O-GlcNAcylation to Ser349 or Ser350 of human c-Rel (Fig. 2, C and D). We separately mutated each of these serines to alanines, expressed the mutant c-Rel proteins in transfected cells, and examined their O-GlcNAcylation under hyperglycemic conditions. Unlike endogenous c-Rel, exogenous c-Rel was O-GlcNAcylated in HEK 293T cells. Although the c-Rel S349A mutant was O-GlcNAcylated to the same extent as wild-type c-Rel, the S350A mutation completely blocked O-GlcNAcylation (Fig. 2E). Ser350 in c-Rel is located in its Rel inhibitory domain (RID) (Fig. 2F) (26). Both Ser349 and Ser350 are also conserved in mouse c-Rel (Fig. 2G). The location of Ser350 in c-Rel corresponds to the location of the O-GlcNAcylation site in p65 (Thr352) (21), suggesting conserved accessibility of this region for the enzyme OGT. Thus, consistent with the PEG labeling data (Fig. 2A), we identified a single serine residue in c-Rel, Ser350, as the O-GlcNAcylation site.
OGT is capable of forming complexes with its substrates (27).We found that the lack of O-GlcNAcylation of the c-Rel S350A mutant was not because of its inability to bind to OGT, because overexpressed wild-type c-Rel and the S350A c-Rel mutant bound to endogenous OGT to similar extents (Fig. 2H). Moreover, the S350A mutation did not interfere with the ability of c-Rel to form dimers, indicating its competence for performing the protein-protein interactions that are crucial for NF-κB function (Fig. 2I).
Mutation of Ser350 to alanine blocks the O-GlcNAcylation and transactivation of c-Rel but not its TCR-dependent phosphorylation
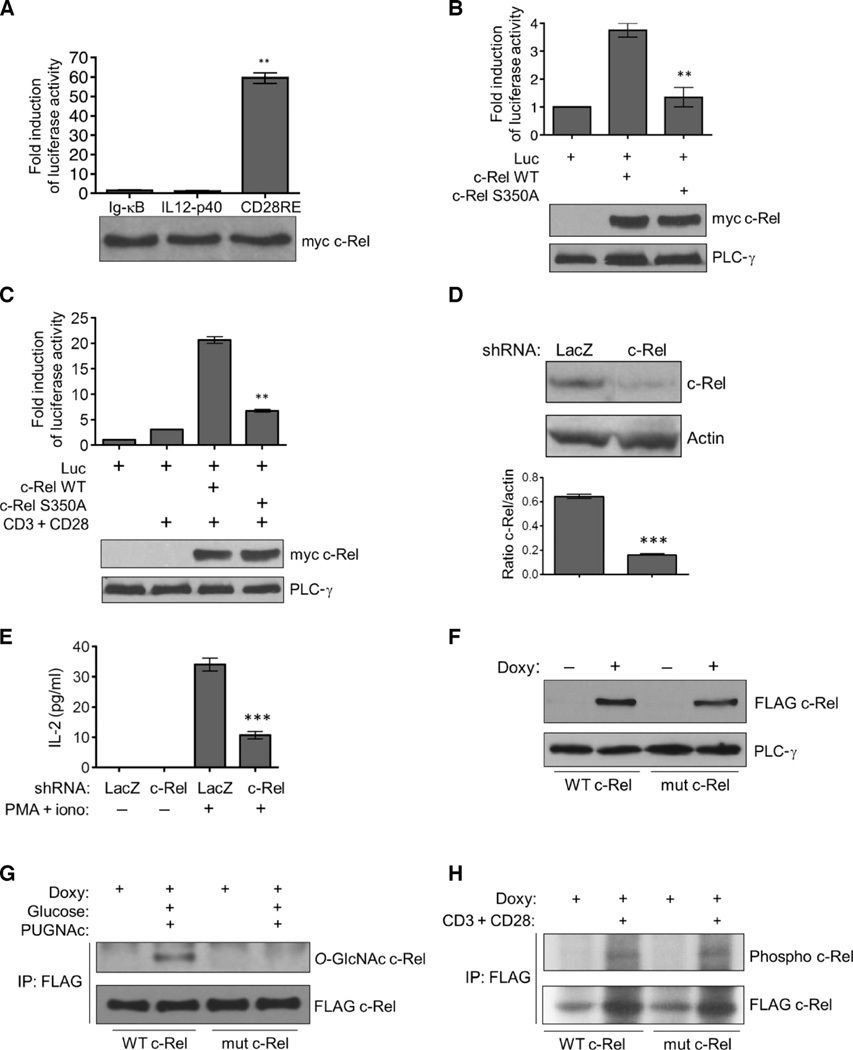
To investigate the function of the O-GlcNAcylation of c-Rel, we initially assayed the transactivation ability of overexpressed c-Rel. We found that a CD28-responsive DNA element–driven luciferase reporter gave a greater response to c-Rel–induced activation than did immunoglobulin κB (Ig-κB) or IL12-p40-κB site-driven luciferase reporters (Fig. 3A). On this basis, we studied c-Rel function by costimulating Jurkat cells with anti-CD3 and anti-CD28 antibodies. We found that the S350A mutant c-Rel had greatly reduced transactivation potential compared to that of wild-type c-Rel when increased in abundance (Fig. 3B) or when cells were costimulated by anti-CD3 and anti-CD28 antibodies (Fig. 3C).
Fig. 3. Mutating the O-GlcNAcylation site blocks the activity, but not the phosphorylation, of c-Rel.
(A) HEK 293T cells were transfected with plasmid encoding myc-tagged WT c-Rel together with the indicated luciferase reporter plasmids. Luciferase activity was assessed with a dual luciferase assay system. **P < 0.01 for the CD28RE reporter compared to the Ig-κB or IL12-p40 reporters. (B and C) Jurkat cells were transfected with plasmids encoding myc-tagged WT c-Rel or the S350Amutant c-Rel together with the CD28RE luciferase reporter plasmid (Luc). (C) Twenty-four hours after transfection, cells were stimulated with anti-CD3 and anti-CD28 antibodies for 4 hours. (B and C) Luciferase activity was assessed as described in (A). **P < 0.01, when comparing the S350A mutant c-Rel to WT c-Rel. (A to C) Data in the bar graphs are means ± SEM of three independent experiments. Bottom: Western blotting analysis of total cell lysates with antibodies against the myc tag or phospholipase C-γ (PLC-γ). Blots are representative of three independent experiments. (D) shRNA-mediated knockdown of endogenous c-Rel in Jurkat T-REx cells. Bottom: Densitometric quantification of c-Rel amounts relative to those of actin. Data are means ± SEM from three independent experiments; the Western blots are from one representative experiment. ***P < 0.001. (E) Jurkat T-REx cells were treated with PMA (50 ng/ml) and ionomycin (250 ng/ml) for 16 hours, and the amounts of IL-2 secreted into the culture medium were determined by enzyme-linked immunosorbent assay (ELISA). Data are means ± SEM from three independent experiments. ***P < 0.001. (F) Jurkat T-REx cell clones were treated with doxycycline (1 µg/ml) for 22 hours, and the production of WT and mutant c-Rel was determined by Western blotting analysis with an anti-FLAG antibody. PLC-γ was used as a loading control. (G) Jurkat T-REx cells (75 × 106) expressing WT and mutant c-Rel were incubated with doxycycline, then treated as described in Fig. 1B, and subjected to immunoprecipitation and Western blotting analysis with antibodies against the indicated proteins. (H) Cellular phosphorylation of WT and S350A mutant c-Rel. FLAG-tagged c-Rel proteins were immunoprecipitated from 75 × 106 Jurkat T-REx cells and analyzed by autoradiography (top) and Western blotting (bottom). Data in (F) to (H) are representative of three independent experiments.
We attempted to study the S350A mutant c-Rel by creating a cell line that constitutively overexpresses wild-type or mutant c-Rel. But these efforts were plagued by the fact that the increased amounts of c-Rel in transfected cells led to cell death after a few days, consistent with previous reports (28, 29). To circumvent this problem, we expressed c-Rel in Jurkat T-REx cells with a tetracycline-inducible system. We first suppressed endogenous c-Rel through the lentiviral delivery of a short hairpin RNA (shRNA) that targeted the 3′ noncoding region of c-Rel, which then enabled the cells to tolerate increased amounts of exogenous c-Rel, and enabled us to analyze the function of the inducibly expressed wild-type and mutant protein with minimum interference from the endogenous protein. We achieved about 75% knockdown of endogenous c-Rel abundance (Fig. 3D) and observed a substantial reduction in c-Rel–dependent interleukin-2 (IL-2) production in response to stimulation of the cells with phorbol 12-myristate 13-acetate (PMA) and ionomycin (Fig. 3E). We then generated stable cell clones that, under the control of a tetracycline-regulated promoter, could inducibly express FLAG-tagged wild-type c-Rel or the S350A mutant c-Rel, which was not subject to O-GlcNAcylation. Addition of the tetracycline analog doxycycline induced the production of detectable amounts of FLAG-tagged wild-type and mutant c-Rel proteins in these clones after 20 to 24 hours (Fig. 3F).We analyzed multiple cell clones to exclude clonal variation and to verify the consistency of inducible c-Rel production and the responses of the cells to costimulation with anti-CD3 and anti-CD28 antibodies. Consistent with the results of our overexpression experiments, we found that the doxycycline-induced wild-type c-Rel was O-GlcNAcylated, whereas the S350A mutant c-Rel protein, although it was present at a similar amount to that of the wild-type protein, showed a complete lack of O-GlcNAcylation (Fig. 3G).
On the basis of findings from previous studies, c-Rel Ser350 is not thought to be phosphorylated (3). We stimulated the phosphorylation of c-Rel by treating cells with anti-CD3 and anti-CD28 antibodies, conditions that cause an increase in c-Rel abundance (30, 31), as well as the extent of its phosphorylation (32). However, we could not detect any difference in the overall phosphorylation extent of doxycycline-induced wild-type or mutant c-Rel proteins when we performed an orthophosphate labeling assay (Fig. 3H).
The S350A mutation compromises the DNA binding ability of c-Rel
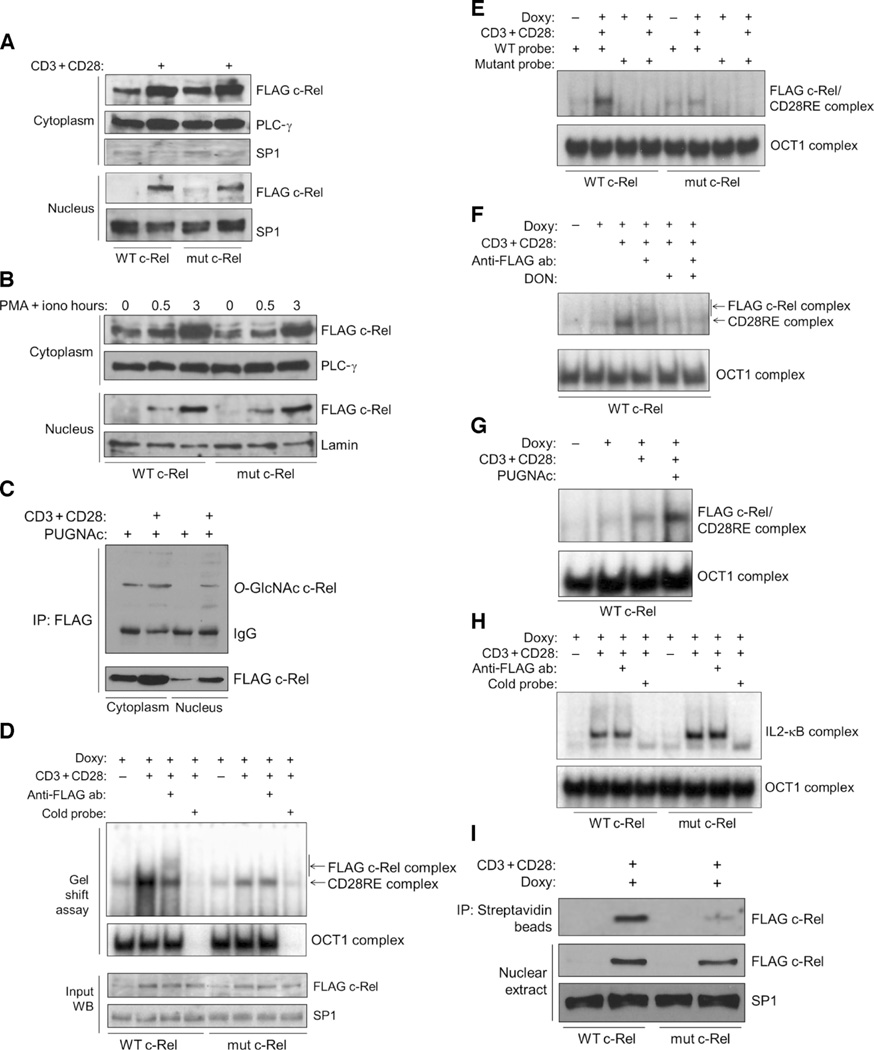
To study nuclear translocation, we induced c-Rel protein in Jurkat T-REx cell clones and stimulated these cells with anti-CD3 and anti-CD28 antibodies. We then prepared and analyzed cytoplasmic and nuclear extracts of the cells and found comparable translocation of wild-type and mutant c-Rel proteins into the nucleus (Fig. 4A). We observed similar results in experiments in which we transiently transfected mouse EL4 T cells with plasmids encoding wild-type or S350A mutant c-Rel and then stimulated the nuclear translocation of these proteins by treating the cells with anti-CD3 and anti-CD28 antibodies (fig. S3). Treatment of the Jurkat T-REx cell clones with PMA and ionomycin, which activates the same downstream signaling components that are activated in response to costimulation with anti-CD3 and anti-CD28, also resulted in comparable nuclear translocation of wild-type and S350A mutant c-Rel (Fig. 4B). As previously reported (30–32), we also observed an increase in the amounts of c-Rel proteins after stimulation of cells with anti-CD3 and anti-CD28 or with PMA and ionomycin, and this response occurred in cells expressing either wild-type or the S350A mutant c-Rel (Figs. 3H and 4, A to C). We also found inducibly expressed FLAG-tagged O-GlcNAcylated c-Rel in the nuclei of Jurkat cells in response to costimulation with anti-CD3 and anti-CD28, which was suggestive of a nuclear role for the modified c-Rel (Fig. 4C).
Fig. 4. Mutation of the O-GlcNAcylation site of c-Rel does not affect its nuclear translocation but impairs its binding to the CD28RE.
(A) Jurkat T-REx cell clones were treated with doxycycline for 22 hours to induce production of WT c-Rel or S350A mutant c-Rel and then were left untreated or were treated with anti-CD3 and anti-CD28 antibodies for 3 hours. Cytoplasmic and nuclear extracts were analyzed with antibodies against the indicated proteins. PLC-γ and SP1 were used as loading controls for the cytoplasmic and nuclear fractions, respectively. (B) Jurkat T-REx cell clones were treated with doxycycline for 22 hours to induce production of WT or mutant c-Rel and then were left untreated or were treated with PMA and ionomycin for the indicated times. Cytoplasmic and nuclear extracts were analyzed with antibodies against the indicated proteins. (C) Jurkat T-REx cells (75 × 106) expressing WT c-Rel were treated as described in (A). PUGNAc (100 µM) was added during the last 12 hours of doxycycline treatment. FLAG-tagged c-Rel was immunoprecipitated from cytoplasmic and nuclear extracts, and samples were analyzed by Western blotting with anti–O-GlcNAc and anti-FLAG antibodies. (D) Jurkat T-REx cell clones expressing WT c-Rel or mutant c-Rel were treated as described in (A), and nuclear extracts were analyzed by EMSA for binding to a CD28RE probe. OCT1 was used as a control for normalization of the nuclear extracts, and its specificity was determined by competition with unlabeled OCT1 probe. Bottom: Western blotting analysis of the input amounts of FLAG-tagged WT and mutant c-Rel. (E) Nuclear extracts from the samples shown in (D) were incubated with a mutated CD28RE probe as a specificity control for binding. (F) Jurkat T-REx cells expressing WT c-Rel were treated overnight with 50 µM DON. Other treatments and EMSA analyses were performed as described in (D). (G) Cells were treated overnight with 100 µM PUGNAc in culture medium containing 5 mM glucose. Other treatments and EMSA analyses were performed as described in (D). (H) Cells were treated as described in (D), and binding to the IL2-κB probe was examined by EMSA. (I) In vitro oligonucleotide binding assay. Top: Binding of WT and mutant c-Rel to the CD28RE probe. Bottom: Amounts of the respective proteins in the samples. Data in (A) to (I) are representative of three to five independent experiments. PLC-γ, lamin, and SP1 were used as loading controls.
Because the nuclear translocation of the S350A mutant c-Rel was intact, we next examined its DNA binding properties in an attempt to account for its compromised transactivation ability. With an electrophoretic mobility shift assay (EMSA), we observed that nuclear extracts from cells expressing FLAG-tagged wild-type c-Rel showed enhanced binding to the CD28RE probe (33) upon costimulation with anti-CD3 and anti-CD28 antibodies (Fig. 4D). The addition of an anti-FLAG antibody to the binding reaction predominantly blocked DNA-protein binding and led to a supershift in a portion of the complex, implying that FLAG-tagged c-Rel was bound to the DNA. In contrast, nuclear extracts from cells with the S350A mutant c-Rel mutant showed substantially less DNA binding upon costimulation with anti-CD3 and anti-CD28, and they lacked a shifted complex in the presence of the anti-FLAG antibody, as determined by EMSA experiments (Fig. 4D). We could compete both wild-type and mutant c-Rel complexes with a 100-fold excess of unlabeled probe. We used the DNA binding of OCT1, which could be competed with an OCT1 probe, as a control for the gel shift assays. The observed differences in the DNA binding abilities of wild-type and S350A mutant c-Rel were not a result of differences in the amount of the relevant proteins in the nuclear extracts (Fig. 4D, bottom). Mutations in the CD28RE region completely abolished the binding of the c-Rel protein complexes to the probe, indicating the specificity of the interaction (Fig. 4E). Similar to the situation in cells expressing the S350A mutant c-Rel, treatment of cells with 6-diazo-5-oxo-l-norleucine (DON), which inhibits O-GlcNAcylation (34), decreased the extent of CD28RE binding in nuclear extracts from cells expressing wild-type c-Rel upon costimulation with anti-CD3 and anti-CD28 antibodies (Fig. 4F). Conversely, enhancement of O-GlcNAcylation by treating cells with PUGNAc (Fig. 4G) or culturing cells with an increased concentration of glucose (fig. S4A) substantially increased the extent of binding of c-Rel to the CD28RE DNA probe.
Maximal induction of IL2 expression depends on multiple transcription factors, including c-Rel, p65, activating protein 1 (AP-1), and nuclear factor of activated T cells (NFAT) (35). To be certain that mutation of the O-GlcNAcylation site of c-Rel specifically affected its binding to the AT-rich CD28RE region and not to the other, GC-rich NF-κB site in the IL2 promoter, we studied DNA binding with a probe for the p65-binding region (IL2-κB) of the IL2 promoter (36). Anti-CD3– and anti-CD28–inducible IL2-κB–DNA–protein complexes were formed from samples of cells expressing either wild-type or mutant c-Rel, with a modest increase in the extent of DNA binding of the latter (Fig. 4H). An anti-FLAG antibody did not supershift these DNA-bound complexes, indicating the absence of FLAG–c-Rel in these complexes (Fig. 4H). This result also demonstrates that TCR activation efficiently stimulates both wild-type and mutant c-Rel–expressing cell lines. To further validate that the S350A mutant c-Rel had a defect in its DNA binding ability, we performed an in vitro oligonucleotide pulldown assay. Consistent with the gel shift assays, we found that the S350A mutant c-Rel showed markedly reduced binding to a biotinylated CD28RE probe compared to that of the wild-type c-Rel (Fig. 4I). In addition, we found that the binding of p65 to the CD28RE probe occurred similarly in cell lines expressing either wild-type or mutant c-Rel (fig. S4B).
The O-GlcNAcylation of c-Rel induces the expression of a subclass of TCR-induced genes
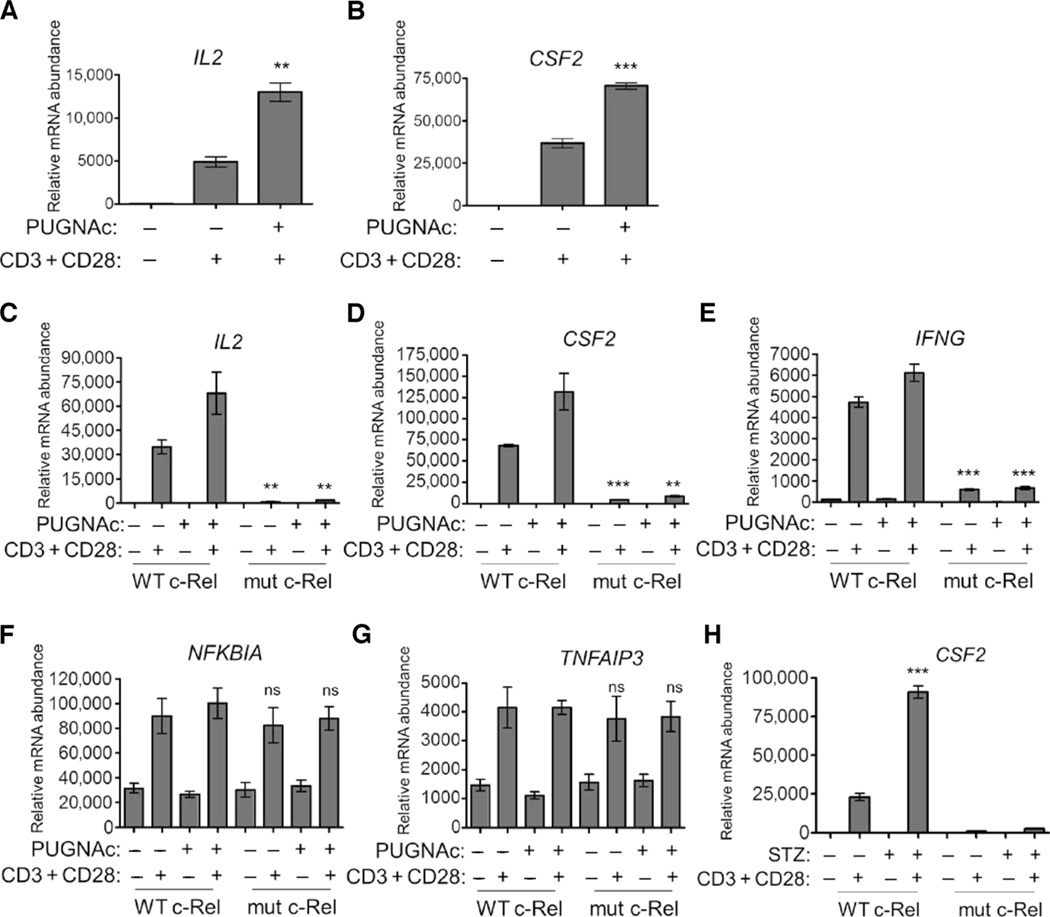
TCR signaling mediates autoimmune T cell responses in hyperglycemic conditions such as diabetes (37). We studied the costimulation of T cells with anti-CD3 and anti-CD28 antibodies, conditions that mimic the TCR activation that induces the expression of several c-Rel–dependent genes, including IL2, IFNG, and CSF2 (26, 38, 39). In experiments with c-Rel–deficient T cells, we confirmed that these genes were indeed dependent on c-Rel for their expression (fig. S5A). We observed substantially potentiated anti-CD3– and anti-CD28–induced expression of IL2 and CSF2 in Jurkat cells after exposure to PUGNAc compared to that in cells that were not treated with PUGNAc (Fig. 5, A and B), substantiating the requirement of O-GlcNAcylation for c-Rel transactivation (Fig. 3, B and C).
Fig. 5. O-GlcNAcylation of c-Rel is required for TCR-induced, CD28RE-dependent gene expression.
(A and B) PUGNAc increases anti-CD3– and anti-CD28–induced, c-Rel–dependent gene expression in Jurkat cells. Cells were treated with 100 µM PUGNAc for 12 hours and then were treated with plate-bound anti-CD3 and anti-CD28 antibodies (each at 2 µg/ml) for 3 hours. Samples were then analyzed by quantitative real-time polymerase chain reaction (PCR) to determine the abundances of (A) IL2 and (B) CSF2 mRNAs relative to that of transferrin receptor (TFRC) mRNA. **P < 0.01, ***P < 0.001, when comparing PUGNAc-treated to untreated samples. (C to G) Jurkat T-REx cell clones were incubated with doxycycline for 22 hours to induce production of WT c-Rel or S350A mutant c-Rel, left untreated or treated with PUGNAc overnight, and then were left untreated or were treated with anti-CD3 and anti-CD28 antibodies for 3 hours. Samples were then analyzed by quantitative real-time PCR to determine the abundances of (C) IL2, (D) CSF2, (E) IFNG, (F) NFKBIA, and (G) TNFAIP3 mRNAs relative to that of TFRC mRNA. **P < 0.01, ***P < 0.001, when comparing cells expressing WT and mutant c-Rel under similar treatments. ns, not significant. (H) Jurkat T-REx cell clones expressing WT c-Rel or mutant c-Rel were treated overnight with 2 mM STZ and then were treated as described in (C). ***P < 0.001, when comparing STZ-treated samples to untreated samples. Data in (A) to (H) are means ± SEM of triplicate samples and are representative of three independent experiments, with similar results.
Consistent with the impaired DNA binding of the S350A mutant c-Rel (Fig. 4, D, E, and I), mutation of the O-GlcNAcylation site of c-Rel markedly suppressed anti-CD3– and anti-CD28–induced expression of IL2, CSF2, and IFNG (Fig. 5, C to E). However, other NF-κB–dependent genes, including NFKBIA and TNFAIP3, which have potential c-Rel–binding sites in their promoters (40) but are mainly induced by p65, were expressed to similar extents in cells containing wild-type or mutant c-Rel (Fig. 5, F and G). This finding indicates that TCR signaling and NF-κB activation are intact in cells expressing the S350A mutant c-Rel. PUGNAc substantially potentiated the expression of c-Rel–dependent genes only in wild-type cells, emphasizing the role of O-GlcNAcylation in this transactivation process (Fig. 5, C to E). Further confirming the effect of PUGNAc, streptozotocin (STZ), another OGA inhibitor that promotes O-GlcNAcylation (Fig. 5H) (41), and high glucose (fig. S5B) also enhanced the anti-CD3– and anti-CD28–induced, c-Rel–dependent expression of IL2, CSF2, or both. The S350A mutant c-Rel did not appear to exhibit any dominant-negative effect, because it did not affect the basal expression of several c-Rel–dependent genes (fig. S6, A to D).
Neither the S350A mutation of c-Rel nor PUGNAc-enhanced O-GlcNAcylation alters TNF-induced gene expression
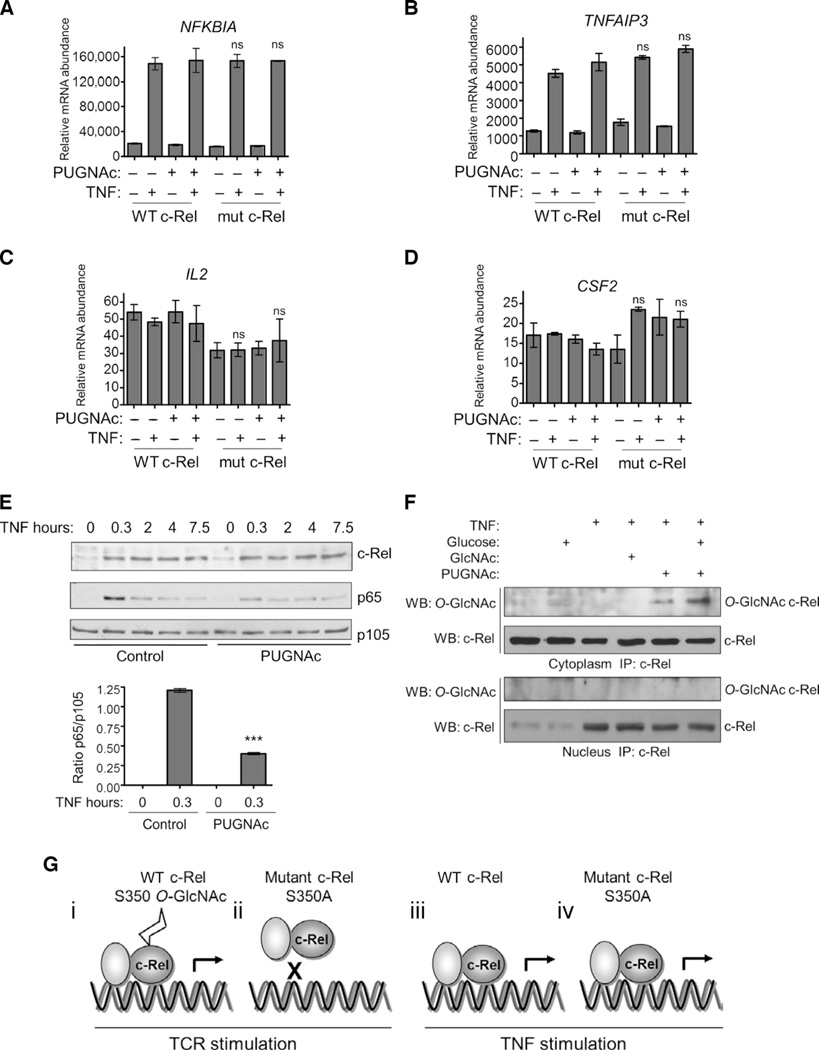
TNF stimulates the phosphorylation of c-Rel and induces its transactivation function (42). We studied the role of the enhanced O-GlcNAcylation of c-Rel and the effect of the S350A mutation on TNF-induced, NF-κB–dependent gene expression. Neither the S350A mutation nor PUGNAc affected the TNF-induced expression of NFKBIA or TNFAIP3 (Fig. 6, A and B). In addition, TNF did not enhance the expression of IL2 or CSF2 in these cells (Fig. 6, C and D). Consistent with these observations, we found that PUGNAc, which enhances the general extent of O-GlcNAcylation within the cell, had no effect on the TNF-dependent nuclear translocation of c-Rel, although it decreased the amount of p65 that translocated to the nucleus in response to TNF (Fig. 6E). Further, TNF did not stimulate the translocation of O-GlcNAcylated c-Rel to the nucleus (Fig. 6F and fig. S7). These results suggest that TNF induces the expression of NFKBIA and TNFAIP3 independently of c-Rel in Jurkat T cells or that the induction of these genes in response to TNF is mediated by c-Rel that is not GlcNAcylated. Because p65 takes a dominant and redundant role in NF-κB–dependent gene expression in response to TNF, the ability to independently define TNF-induced, c-Rel–dependent genes remains a challenge. Thus, c-Rel and its O-GlcNAcylation are stimulus-specific and seem relevant in TCR-induced, but not TNF-induced, expression of selected genes (Fig. 6G).
Fig. 6. TNF-induced gene expression and c-Rel activation are independent of c-Rel O-GlcNAcylation.
(A to D) Jurkat T-REx cell clones were incubated with doxycycline for 22 hours to induce production of WT c-Rel or mutant c-Rel, left untreated or treated with PUGNAc overnight, and then were left untreated or were treated with TNF (100 ng/ml) for 3 hours. Samples were then analyzed by quantitative real-time PCR to determine the abundances of (A) NFKBIA, (B) TNFAIP3, (C) IL2, and (D) CSF2 mRNAs relative to that of TFRC mRNA. Data are means ± SEM of triplicate samples and are representative of three independent experiments with similar results. ns, not significant. (E) Increased O-GlcNAcylation does not affect TNF-dependent c-Rel activation, but decreases p65 activation. Jurkat cells (30 × 106) were cultured overnight in medium containing 5 mM glucose and were treated with 100 µM PUGNAc as indicated. Cells were then treated with 4 mM GlcNAc and 30 mM glucose for 3 hours, and TNF was applied for the last 20 min of the treatment. Cytoplasmic and nuclear extracts were prepared, subjected to immunoprecipitation with antibodies against c-Rel, and then analyzed by Western blotting with antibodies specific for O-GlcNAc and c-Rel. Data are representative of three independent experiments. Western blotting analysis of p105 abundance confirmed uniform loading. Bottom: Densitometric quantification of the amount of nuclear p65 relative to that of p105 after 20 min of stimulation with TNF. Data are means ± SEM from three independent experiments. ***P < 0.001, when comparing PUGNAc-treated to control samples. (F) Jurkat cells (40 × 106) were cultured overnight in medium containing 5 mM glucose and then were treated with 4 mM GlcNAc and 30 mM glucose in the absence or presence of PUGNAc for 3 hours. TNF was applied for the last 20 min of the treatment. Cytoplasmic and nuclear extracts were prepared, subjected to immunoprecipitation with antibody against c-Rel, and analyzed by Western blotting with antibodies specific for O-GlcNAc and c-Rel. Blots in (E) and (F) are representative of three independent experiments. (G) Hypothetical schematic models of the regulation of the DNA binding and transactivation functions of c-Rel by O-GlcNAcylation. (i) Stimulation of the TCR induces the O-GlcNAcylation of c-Rel at Ser350, the binding of c-Rel to target DNA, and the induction of gene expression. (ii) The S350A mutation blocks the TCR-induced binding of c-Rel to DNA and inhibits gene expression. (iii and iv) The S350A mutation of c-Rel has no effect on TNF-induced gene expression.
DISCUSSION
Here, we identified a single site of O-GlcNAcylation, Ser350, in the NF-κB protein c-Rel. O-GlcNAcylation substantially stimulated c-Rel–dependent transactivation and was required for the expression of several genes after TCR costimulation. We delineated the mechanism by which c-Rel was regulated by O-GlcNAc modification, showing that Ser350 was required for the binding of c-Rel to DNA, and also how the extent of O-GlcNAcylation of c-Rel modulated this process.
Our results show that O-GlcNAcylation of c-Rel is required for TCR-induced expression of IL2, CSF2, and IFNG. All of these genes contain the CD28RE in their promoters (33), suggesting a critical role of O-GlcNAcylated c-Rel in mediating CD28RE-dependent transcription. We did not observe any effect of mutating the O-GlcNAcylation site of c-Rel or of treating cells with PUGNAc on TCR- or TNF-induced expression of TNFAIP3 and NFKBIA, genes that are thought to contain c-Rel–binding sites in their promoters (40); however, it has not been shown unambiguously that these genes are indeed dependent on c-Rel. Additionally, we found that increased O-GlcNAcylation did not affect the TNF-dependent nuclear translocation of c-Rel, and conversely, TNF had no substantial effect on glucose- or GlcNAc-induced O-GlcNAcylation of c-Rel (Fig. 6F and fig. S7). These observations suggest that TNF might stimulate the transactivation of c-Rel independently of its O-GlcNAcylation status. We found that TCR stimulation mediated transactivation of c-Rel through both O-GlcNAcylation–dependent and O-GlcNAcylation–independent pathways. Although both the TCR and TNF signaling stimulated the nuclear translocation of c-Rel, only TCR signaling resulted in the presence of O-GlcNAcylated c-Rel in the nucleus. This might depend on a TCRdependent cofactor that binds to O-GlcNAcylated c-Rel in the cytoplasm to facilitate its nuclear translocation. Alternatively, c-Rel may become O-GlcNAcylated in the nucleus after TCR activation. These results suggest a stimulus-specific role of O-GlcNAcylation in modulating c-Rel function.
Both OGT- and c-Rel–deficient mice show impaired T helper 1 (TH1)–type responses (14, 43). Inhibition of OGT in Jurkat cells causes impaired anti-CD3– and anti-CD28–dependent NF-κB activation and IL2 expression (20). These findings suggest an overlap in the function of O-GlcNAcylation and NF-κB. Golks et al. (20) showed that OGT is critical for the activation of NF-κB in lymphocytes, and they suggested that p65 was the O-GlcNAcylated NF-κB protein. Our results show that c-Rel is the major O-GlcNAcylated protein in lymphocytes and that it exists in a complex with p65 (Fig. 1, C and D). In addition to p65-bound c-Rel, it is possible that the c-Rel homodimer and a c-Rel–p50 heterodimer are also O-GlcNAcylated. This could be tested in cells devoid of p65 or p50.
The CD28RE region exhibits high-affinity binding to c-Rel homodimers. Complexes containing p65 and p50 also bind, albeit weakly, to this region (44, 45). We found that the binding of p65 to the CD28RE region occurred independently of c-Rel binding (fig. S4B), suggesting the presence of distinct c-Rel– and p65-containing complexes bound to this region. Thus, although both c-Rel and p65 are capable of binding to the CD28RE region, it seems likely that only c-Rel homodimers or c-Rel–p50 heterodimers are involved in CD28RE-dependent transactivation. Consistent with this notion, anti-CD3– and anti-CD28–induced transcription of IL2 occurs independently of p65 (46). Because we found that O-GlcNAcylated c-Rel was mostly in the cytoplasm in complex with p65, it is plausible that modified c-Rel may sequester p65 in the cytoplasm and thus dampen p65-dependent transcription. Supporting this notion, we found that the extent of DNA binding by p65 was modestly enhanced in cells with a mutant c-Rel, and that the PUGNAc-mediated increase in O-GlcNAcylation decreased the extent of the TNF-dependent nuclear translocation of p65 (Figs. 4H and 6E). Consistent with our data, others have also reported that PUGNAc decreases the p65-mediated induction of some NF-κB–dependent genes (47). In contrast, high concentrations of glucose and glucosamine increase the extent of p65 O-GlcNAcylation and enhance the NF-κB–dependent transcription of target genes (15, 21). The differences between these studies may be due to the use of different cell types or different types of NF-κB–activating stimuli, which may dictate the functional outcome of NF-κB O-GlcNAcylation.
An increase in the extent of O-GlcNAcylation of p65 decreases its affinity for IκB and facilitates the nuclear translocation of p65 (21). However, we did not see any marked differences in the nuclear translocation of either wild-type c-Rel or the S350A mutant c-Rel, making it unlikely that O-GlcNAcylation modifies the binding of c-Rel to IκB. Ser350 is well-separated from the Rel homology domain (RHD), which is required for DNA binding and dimerization (44, 48). Whether O-GlcNAcylation of Ser350 causes any structural alteration of the DNA binding domain and how this might enhance the DNA binding property of c-Rel are important questions to be addressed in future studies. O-GlcNAcylation facilitates the DNA binding of other transcription factors, such as Pdx-1 (49) and YY1 (50). Similarly, our results suggest that O-GlcNAcylation regulates the NF-κB protein c-Rel by facilitating its DNA binding and subsequent transactivation. We also reported a similar role for methylation of p65 in promoting its DNA binding property (51). In addition, acetylation of p65 enhances its affinity for DNA (52). These findings suggest that posttranslational modifications play a major role in inducing NF-κB–DNA interactions.
Our discovery of the O-GlcNAcylation of c-Rel adds yet another layer of complexity to the regulation of NF-κB. c-Rel is crucial for lymphocyte function and has important roles in T cell–mediated immune responses and autoimmunity. Similarly, the process of O-GlcNAcylation is also important for T cell function. Our study reveals important crosstalk between sugar metabolism, NF-κB signaling, and T cell function that have implications for autoimmunity, diabetes, and cancer, conditions in which dysregulation of both NF-κB signaling and O-GlcNAcylation are implicated. c-Rel is particularly important for the development and proliferation of immunosuppressive regulatory T cells (Tregs) (53). Understanding how hyperglycemia and the O-GlcNAcylation of c-Rel affect Tregs and autoimmunity is an important area of future research, and our investigations in this direction are in progress. High glucose–induced O-GlcNAcylation of c-Rel and its enhanced activation might act as a positive regulator of autoimmune diabetes. The increased expression of the c-Rel–dependent genes IL2 and IFNG may enhance TH1 cell function and cell-mediated cytotoxicity, accelerating the destruction of pancreatic β cells. Therefore, targeting the O-GlcNAcylation of c-Rel may prove to be an effective tool to suppress autoreactive T cells and thereby suppress autoimmunity.
MATERIALS AND METHODS
Reagents and antibodies
Glucose, GlcNAc, STZ, DON, PMA, ionomycin, blasticidin, doxycycline, and polyinosinic-polycytidylic acid [poly(I:C)] were purchased from Sigma. Zeocin was obtained from Invivogen. PUGNAc was from Toronto Research Chemicals. Recombinant TNF protein was purchased from PeproTech. [32P]Orthophosphate and [γ-32P]adenosine 5′-triphosphate (ATP) were obtained from PerkinElmer Radiochemicals. The IL-2 ELISA kit and antibodies against mouse CD3 (145-2C11) and CD28 (37.51) were from BioLegend. Antibodies against human CD3 (OKT3) and CD28 (clone 28.2) were obtained from eBioscience. Antibodies against c-Rel, p65, SP1, OGT, β-actin, PLC-γ, and lamin were from Santa Cruz Biotechnology. The anti-FLAG antibody was from Sigma. The anti–O-GlcNAc antibodies RL2 and CTD 110.6 were from Abcam and Covance, respectively.
Cells
Jurkat, Jurkat T-REx, Ramos, EL4,MEFs, and HEK 293T cells were grown as previously described (23). Human PBMCs were purchased from AllCells. The tetracycline-inducible Jurkat T-REx cells were from Invitrogen.
Mice
c-Rel–deficient mice were provided by H.-C. Liou (Weill Medical College of Cornell University, New York). C57BL/6 mice were from a colony maintained in-house. Mice were housed and handled in accordance with the National Institutes of Health (NIH) guidelines under protocols approved by the Institutional Animal Care and Use Committee. All mice used in this study were 6 to 10 weeks old, unless otherwise indicated.
Plasmids
The complementary DNAs (cDNAs) encoding human c-Rel and its mutants were cloned into the pCMX vector, which encodes a myc tag, for transient expression in transfected cells. The S349A and S350A mutants of c-Rel were generated by PCR-based site-directed mutagenesis. For inducible expression experiments, cDNAs encoding wild-type and mutant c-Rel containing the FLAG tag were cloned into the pcDNA4 vector (Invitrogen). For baculoviral expression, cDNAs encoding c-Rel, p65, and OGT were cloned into the vector pVL1393 (AB Vector).
Orthophosphate labeling
Cellular phosphorylation assays were performed as described previously (54). Briefly, Jurkat T-REx cells (75 × 106) were treated with doxycycline to induce expression of the vectors encoding wild-type and mutant c-Rel, starved for 90 min in phosphate-free medium, and stimulated with anti-CD3 and anti-CD28 antibodies for 3 hours in the presence of radioactive orthophosphate (0.2 mCi/ml). Cells were harvested, washed with phosphate-free medium, and lysed. FLAG-tagged c-Rel was immunoprecipitated with an anti-FLAG antibody, samples were transferred onto membranes, and phosphate incorporation was assessed by autoradiography.
In vitro chemoenzymatic O-GlcNAc labeling assay
Jurkat or Ramos cells (30 × 106) were treated for 8 hours with 4 mM GlcNAc and 100 µM PUGNAc. Cells were lysed in a buffer containing 20 mM Hepes (pH 7.9) and 1% SDS. Proteins were precipitated with methanol/chloroform/water, and labeling reactions were performed with the Click-IT O-GlcNAc enzymatic labeling system (Invitrogen) according to the manufacturer’s instructions. Biotin-labeled samples were precipitated with methanol/chloroform/water, resuspended in buffer containing 50 mM tris-HCl (pH 8.0) and 1% SDS, and analyzed by Western blotting with a streptavidin–horseradish peroxidase conjugate.
WGA binding assay
Jurkat cells (30 × 106) and MEFs (10 × 106) were cultured overnight in RPMI medium containing 5 mM glucose with or without 4 mM GlcNAc and 100 µM PUGNAc. Cells were lysed in lysis buffer [50 mM Hepes (pH 7.9), 150 mM NaCl, 1% Triton X-100, 50 µM PUGNAc, and EDTA-free protease inhibitor cocktail]. The lysate was precleared with protein A/G beads (Santa Cruz Biotechnology) and then incubated with succinylated WGA (Vector Labs) for 4 hours at 4°C. WGA also binds to sialic acid, and we used succinylated WGA to limit its binding to sialic acid and increase its specificity for O-GlcNAc. The immunoprecipitate was resolved by SDS–polyacrylamide gel electrophoresis, and we performed Western blotting analysis to detect c-Rel (and SP1 as a control) with specific antibodies.
Baculoviral expression
Baculovirus was produced by transfecting Sf9 insect cells with the pVL1393 vector encoding the gene of interest with Lipofectamine reagent (Invitrogen). For co-infections, High Five insect cells were infected with OGT-expressing virus and either c-Rel– or p65-expressing viruses. Cells were harvested 60 hours after infection and lysed on ice for 30 min in lysis buffer [11 mM sodium phosphate (pH 7.4), 150 mM NaCl, 0.1% SDS, 0.5% sodium deoxycholate, 1% Triton X-100, 50 µM PUGNAc, and 1× complete protease inhibitor cocktail]. Proteins were immunoprecipitated from the lysates and were detected by Western blotting analysis with an anti–O-GlcNAc antibody (RL2).
In vitro PEG labeling
Jurkat or Ramos cells (10 × 106) were cultured overnight in RPMI medium without glucose. Cells were treated for 3 hours with 5 or 30 mM glucose and 100 µM PUGNAc and then lysed by boiling in buffer containing 1% SDS and complete protease inhibitor cocktail (Roche), and 100 µg of total protein was chemoenzymatically labeled with a 5-kD PEG mass tag as described previously (25). The labeled lysates were resolved on an 8% polyacrylamide gel and analyzed by Western blotting. The stoichiometry of O-GlcNAcylation was determined by taking the ratio of the densitometric value for the PEG–c-Rel band and dividing it by the sum of the values for the bands corresponding to PEG–c-Rel and c-Rel.
Mass spectrometry
Jurkat or Ramos cells (4.5 × 108) were cultured overnight in medium containing 5 mM glucose. Cells were stimulated for 3 hours with 30 mM glucose and 100 µM PUGNAc, lysed, subjected to immunoprecipitation with an anti–c-Rel antibody, and resolved on a 4 to 12% bis-tris gel in Mops buffer. The band corresponding to c-Rel was excised, manually digested with chymotrypsin, and analyzed by nanoLC-MS with collision-induced dissociation (CID) and electron-transfer dissociation (ETD) fragmentation; data were analyzed as previously described, but with modifications (7). Peptides were separated with a 120-min gradient, and duplicate analyses were performed on an LTQ Orbitrap XL for accurate mass determination of precursor ions. A portion of the digested peptides was subjected to BEMAD (55). After 2 hours at 50°C, the reaction was quenched with acetic acid, and organic solvents were removed by vacuum centrifugation. The sample was analyzed by nanoLC-MS on an LTQ Orbitrap XL as described earlier. Data were searched with Mascot software, allowing for the possibility that peptides were modified by the BEMAD reagent 2-AET at serines or threonines.
shRNAs and lentiviral transduction
shRNAs specific for the 3′ noncoding region of human c-Rel and for LacZ (as a nonspecific control) in a microRNA-155–based expression cassette were synthesized and then amplified by PCR with primers containing restriction sites. Primers used for the c-Rel shRNA expression cassette were as follows: CTGGAGGCTTGCTGAAGGCTGTATGCTG-(TTCGCTATGTCCAAAGTTGTA)-gttttggccactgactgac-(TACAACTTGACATAGCGAA)-CAGGACACAAGGCCTGTTACTAGCACTCACATGGAACAAATGGCCCA. Primers used for the LacZ shRNA expression cassette were as follows: CTGGAGGCTTGCTGAAGGCTGTATGCTG-(TACATCGGGCAAATAATATCG)-gttttggccactgactgac-(CGATATTATGCCCGATGTA)-CAGGACACAAGGCCTGTTACTAGCACTCACATGGAACAAATGGCCCA. The 5′ and 3′ linker sequences are represented in bold uppercase letters, the loop is in lowercase italics, and the shRNA sequence is underlined inside parentheses. These cassettes were cloned into the pHAGE6 lentiviral vector, and viruses were produced by transfecting HEK 293T cells as described previously (56). To select transduced cells, cDNA encoding the extracellular domain of human CD271 was placed downstream of the shRNA cassette and was separated by an internal ribosome entry site (IRES) sequence. Virus was concentrated 100-fold by ultracentrifugation and resuspended in RPMI medium, and 50 µl of virus was used to infect 200 × 103 Jurkat cells per well in six-well plates in the presence of polybrene (5 µg/ml). Cells were spun at 1200g for 90 min at 30°C to facilitate viral adherence to the cell surface, supernatants were discarded, and cells were cultured in fresh medium. Cells were analyzed for shRNA-mediated knockdown of c-Rel 72 to 96 hours after infection.
Magnetic-activated cell sorting and fluorescence-activated cell sorting
Jurkat cells transduced with lentivirus expressing c-Rel–specific shRNA were selected on the basis of their cell-surface expression of CD271 by magnetic sorting with anti-CD271 microbeads (Miltenyi Biotec) according to the manufacturer’s instructions. Cells were blocked with Fc block reagent (anti-mouse CD16 and CD32 antibodies, BD Biosciences), and CD271 expression was confirmed by fluorescence-activated cell sorting with fluorescein isothiocyanate–conjugated anti-CD271 antibody (Miltenyi Biotec) and analyzed with a FACSCalibur flow cytometer (BD Biosciences) and FlowJo software (TreeStar).
In vitro activation of cells with anti-CD3 and anti-CD28 antibodies
Jurkat cells were cultured at 1.5 × 106 cells/ml in six-well plates or 10-cm dishes in the presence of doxycycline (1 µg/ml) for 22 hours to induce the production of FLAG-tagged c-Rel. Cells were stimulated for the times indicated in the figure legends with plate-bound anti-CD3 and anti-CD28 antibodies (each at 2 µg/ml).
Immunoprecipitations and Western blotting
Total cell lysates were prepared, and immunoprecipitations and Western blotting analysis were performed as described previously (54).
Quantitative real-time PCR analysis
DNA-free RNA and cDNA were prepared as previously described (54). Quantitative real-time PCR with cDNA corresponding to 20 to 60 ng of total RNA and TaqMan Gene Expression Assays (Applied Biosystems) was performed in a real-time PCR machine (Realplex, Eppendorf). The expression of individual genes of interest was normalized to the expression of the gene encoding the TFRC for human genes and UBE2D2 for mouse genes.
Electrophoretic mobility shift assays
Nuclear protein extracts were prepared as described previously (57). Nuclear extracts corresponding to 4 to 7 µg of protein were tested for DNA binding with probes containing the c-Rel–binding region of CD28RE and the p65-binding region of IL2. The OCT1 probe was used as an internal control for normalization of the nuclear extracts. In the probe containing the CD28RE-NF-IL2B AP-1 region, the AP-1–binding site was mutated as previously described to restrict protein binding to only the CD28RE site in our assays (33). Primer sequences are as follows. CD28RE-AP mutant probe: forward, 5′-tcgagTTTAAAGAAATTCCAAATCAACATCAg; reverse, 5′-tcgacTGATGTTGATTTGGAATTTCTTTAAAc; the c-Rel–binding site is shown in bold and is underlined. CD28RE mutant-AP mutant probe: forward, 5′-tcgagTTTAAAGACCTCGAAAAGCAACATCAg; reverse, 5′-tcgacTGATGTTGCTTTTCGAGGTCTTTAAAc. IL2-κB probe: forward, 5′-CCAAGAGGGATTTCACTAAATCC; reverse, 5′-GGATTTAGGTGAAATCCCTCTTGG; the p65-binding site is shown in bold and is underlined. OCT1 probe: forward, 5′-TGTCGAATGCAAATCACTAGAA; reverse, 5′-TTCTAGTGATTTGCATTCGACA. Lysates were incubated with [γ-32P]ATP–labeled probe in binding buffer [10 mM tris-Cl (pH 7.5), 50 mM NaCl, 4% glycerol, 1 mM MgCl2, 0.1 mM EDTA, poly(I:C) (50 µg/ml), 50 µM PUGNAc, 0.1 mM dithiothreitol (DTT)] for 20 min at room temperature. Antibody supershift assays to identify the DNA binding of FLAG-tagged c-Rel were performed on ice for 30 min with 1 µg of anti-FLAG antibody (M2). Samples were treated with a 100-fold excess of unlabeled probe for competition to determine the specificity of binding. Samples were resolved by electrophoresis through 5% nondenaturing polyacrylamide gels and analyzed by autoradiography.
Oligonucleotide pulldown assays
Cells expressing wild-type or mutant c-Rel (40 × 106) were cultured overnight in RPMI medium containing doxycycline (1 µg/ml) for 22 hours. Cells were then stimulated with plate-bound anti-CD3 and anti-CD28 antibodies for 3 hours. Cytoplasmic extracts were prepared as described previously (33). Nuclear extracts were prepared in lysis buffer [20 mM Hepes (pH 7.9), 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 1 mM DTT, 50 µM PUGNAc, 0.1% NP-40, with protease inhibitor cocktail] and diluted in the same buffer without NaCl to reach a final concentration of 150 mM NaCl. Lysates containing about 200 µg of nuclear proteins were incubated with 10 µg of poly(I:C) and 1 µg of annealed biotinylated oligonucleotides containing the CD28RE region at 4°C in a rotator. Oligonucleotide sequences used were as follows: biotinylated forward, 5′-Biosg-TCGAGTTTAAAGAAATTCCAAATCAACATCAG; reverse, 5′-TCGACTGATGTTGATTTGGAATTTCTTTAAAC. After 1 hour, streptavidin-agarose beads were added to the mixture for an additional 3 hours. The beads were washed thrice in buffer containing 250 mM NaCl and then were analyzed by Western blotting.
Statistical analysis
Statistical differences in gene expression were analyzed with a two-tailed Student’s unpaired t test with Prism software (GraphPad). Data are presented as means ± SEM, unless otherwise indicated.
Supplementary Material
Acknowledgments
We express our deep and sincere gratitude to the late N. Sharon for invaluable advice and discussions. We thank A. Weiss for the CD28RE luciferase reporter plasmid; A. Balazs for the lentiviral expression plasmids; the Caltech animal facility; the Caltech protein expression center for baculoviral expression; and J. Rexach, R. Parameswaran, C.-K. Ea, L. Yang, and other members of the Baltimore laboratory for insightful discussions.
Funding: This work was initially supported by NIH grant 2R01 GM039458 to D.B., 2RO1 GM084724 to L.C.H.-W., and later by a Mizutani Foundation for Glycoscience grant to P.R.
Footnotes
SUPPLEMENTARY MATERIALS
www.sciencesignaling.org/cgi/content/full/6/290/ra75/DC1
Fig. S1. The NF-κB protein c-Rel is modified by O-GlcNAcylation.
Fig. S2. Coomassie blue staining of immunoprecipitated c-Rel.
Fig. S3. Mutation of the O-GlcNAcylation site of c-Rel does not affect its nuclear translocation.
Fig. S4. Glucose enhances the DNA binding ability of c-Rel, and expression of the S350A mutant c-Rel does not affect the DNA binding ability of p65.
Fig. S5. IL2, IFNG, and CSF2 are c-Rel–dependent genes.
Fig. S6. Mutant c-Rel does not have a dominant-negative effect on the function of wild-type c-Rel.
Fig. S7. TNF-induced activation of c-Rel is independent of its O-GlcNAcylation.
Author contributions: P.R. conceived the project and designed and performed the experiments; P.M.C. performed the PEG labeling; D.E.M. and E.C.P. performed the mass spectrometry; P.R., P.M.C., L.C.H.-W., and D.B. analyzed and interpreted the data; and P.R. and D.B. wrote, and all of the authors edited, the manuscript.
Competing interests: A patent was filed for the targeting of c-Rel O-GlcNAcylation (File No. CIT-6437-P).
REFERENCES AND NOTES
- 1.Baltimore D. NF-κB is 25. Nat. Immunol. 2011;12:683–685. doi: 10.1038/ni.2072. [DOI] [PubMed] [Google Scholar]
- 2.Hayden MS, Ghosh S. NF-κB in immunobiology. Cell Res. 2011;21:223–244. doi: 10.1038/cr.2011.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perkins ND. Post-translational modifications regulating the activity and function of the nuclear factor kappa B pathway. Oncogene. 2006;25:6717–6730. doi: 10.1038/sj.onc.1209937. [DOI] [PubMed] [Google Scholar]
- 4.Leidner J, Palkowitsch L, Marienfeld U, Fischer D, Marienfeld R. Identification of lysine residues critical for the transcriptional activity and polyubiquitination of the NF-κB family member RelB. Biochem. J. 2008;416:117–127. doi: 10.1042/BJ20080432. [DOI] [PubMed] [Google Scholar]
- 5.Hart GW, Housley MP, Slawson C. Cycling of O-linked β-N-acetylglucosamine on nucleocytoplasmic proteins. Nature. 2007;446:1017–1022. doi: 10.1038/nature05815. [DOI] [PubMed] [Google Scholar]
- 6.Hart GW, Slawson C, Ramirez-Correa G, Lagerlof O. Cross talk between O-GlcNAcylation and phosphorylation: Roles in signaling, transcription, and chronic disease. Ann. Rev. Biochem. 2011;80:825–858. doi: 10.1146/annurev-biochem-060608-102511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rexach JE, Clark PM, Mason DE, Neve RL, Peters EC, Hsieh-Wilson LC. Dynamic O-GlcNAc modification regulates CREB-mediated gene expression and memory formation. Nat. Chem. Biol. 2012;8:253–261. doi: 10.1038/nchembio.770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Slawson C, Zachara NE, Vosseller K, Cheung WD, Lane MD, Hart GW. Perturbations in O-linked β-N-acetylglucosamine protein modification cause severe defects in mitotic progression and cytokinesis. J. Biol. Chem. 2005;280:32944–32956. doi: 10.1074/jbc.M503396200. [DOI] [PubMed] [Google Scholar]
- 9.Wang Z, Udeshi ND, Slawson C, Compton PD, Sakabe K, Cheung WD, Shabanowitz J, Hunt DF, Hart GW. Extensive crosstalk between O-GlcNAcylation and phosphorylation regulates cytokinesis. Sci. Signal. 2010;3 doi: 10.1126/scisignal.2000526. ra2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang WH, Kim JE, Nam HW, Ju JW, Kim HS, Kim YS, Cho JW. Modification of p53 with O-linked N-acetylglucosamine regulates p53 activity and stability. Nat. Cell Biol. 2006;8:1074–1083. doi: 10.1038/ncb1470. [DOI] [PubMed] [Google Scholar]
- 11.Chatham JC, Marchase RB. Protein O-GlcNAcylation: A critical regulator of the cellular response to stress. Curr. Signal Transduct. Ther. 2010;5:49–59. doi: 10.2174/157436210790226492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanover JA, Krause MW, Love DC. Bittersweet memories: Linking metabolism to epigenetics through O-GlcNAcylation. Nat. Rev. Mol. Cell Biol. 2012;13:312–321. doi: 10.1038/nrm3334. [DOI] [PubMed] [Google Scholar]
- 13.Shafi R, Iyer SP, Ellies LG, O’Donnell N, Marek KW, Chui D, Hart GW, Marth JD. The O-GlcNAc transferase gene resides on the X chromosome and is essential for embryonic stem cell viability and mouse ontogeny. Proc. Natl. Acad. Sci. U.S.A. 2000;97:5735–5739. doi: 10.1073/pnas.100471497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O’Donnell N, Zachara NE, Hart GW, Marth JD. Ogt-dependent X-chromosome-linked protein glycosylation is a requisite modification in somatic cell function and embryo viability. Mol. Cell. Biol. 2004;24:1680–1690. doi: 10.1128/MCB.24.4.1680-1690.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.James LR, Tang D, Ingram A, Ly H, Thai K, Cai L, Scholey JW. Flux through the hexosamine pathway is a determinant of nuclear factor κB-dependent promoter activation. Diabetes. 2002;51:1146–1156. doi: 10.2337/diabetes.51.4.1146. [DOI] [PubMed] [Google Scholar]
- 16.Yerneni KK, Bai W, Khan BV, Medford RM, Natarajan R. Hyperglycemia-induced activation of nuclear transcription factor κB in vascular smooth muscle cells. Diabetes. 1999;48:855–864. doi: 10.2337/diabetes.48.4.855. [DOI] [PubMed] [Google Scholar]
- 17.Yuan M, Konstantopoulos N, Lee J, Hansen L, Li ZW, Karin M, Shoelson SE. Reversal of obesity- and diet-induced insulin resistance with salicylates or targeted disruption of Ikkβ. Science. 2001;293:1673–1677. doi: 10.1126/science.1061620. [DOI] [PubMed] [Google Scholar]
- 18.Lamhamedi-Cherradi SE, Zheng S, Hilliard BA, Xu L, Sun J, Alsheadat S, Liou HC, Chen YH. Transcriptional regulation of type I diabetes by NF-κB. J. Immunol. 2003;171:4886–4892. doi: 10.4049/jimmunol.171.9.4886. [DOI] [PubMed] [Google Scholar]
- 19.Zhao Y, Krishnamurthy B, Mollah ZU, Kay TW, Thomas HE. NF-κB in type 1 diabetes. Inflamm. Allergy Drug Targets. 2011;10:208–217. doi: 10.2174/187152811795564046. [DOI] [PubMed] [Google Scholar]
- 20.Golks A, Tran TT, Goetschy JF, Guerini D. Requirement for O-linked N-acetylglucosaminyltransferase in lymphocytes activation. EMBO J. 2007;26:4368–4379. doi: 10.1038/sj.emboj.7601845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang WH, Park SY, Nam HW, Kimdo H, Kang JG, Kang ES, Kim YS, Lee HC, Kim KS, Cho JW. NFκB activation is associated with its O-GlcNAcylation state under hyperglycemic conditions. Proc. Natl. Acad. Sci. U.S.A. 2008;105:17345–17350. doi: 10.1073/pnas.0806198105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Allison DF, Wamsley JJ, Kumar M, Li D, Gray LG, Hart GW, Jones DR, Mayo MW. Modification of RelA by O-linked N-acetylglucosamine links glucose metabolism to NF-κB acetylation and transcription. Proc. Natl. Acad. Sci. U.S.A. 2012;109:16888–16893. doi: 10.1073/pnas.1208468109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramakrishnan P, Kahn DA, Baltimore D. Anti-apoptotic effect of hyperglycemia can allow survival of potentially autoreactive T cells. Cell Death Differ. 2011;18:690–699. doi: 10.1038/cdd.2010.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haltiwanger RS, Grove K, Philipsberg GA. Modulation of O-linked N-acetylglucosamine levels on nuclear and cytoplasmic proteins in vivo using the peptide O-GlcNAc-β-N-acetylglucosaminidase inhibitor O-(2-acetamido-2-deoxy-d-glucopyranosylidene)amino-N-phenylcarbamate. J. Biol. Chem. 1998;273:3611–3617. doi: 10.1074/jbc.273.6.3611. [DOI] [PubMed] [Google Scholar]
- 25.Rexach JE, Rogers CJ, Yu SH, Tao J, Sun YE, Hsieh-Wilson LC. Quantification of O-glycosylation stoichiometry and dynamics using resolvable mass tags. Nat. Chem. Biol. 2010;6:645–651. doi: 10.1038/nchembio.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gilmore TD, Gerondakis S. The c-Rel transcription factor in development and disease. Genes Cancer. 2011;2:695–711. doi: 10.1177/1947601911421925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Slawson C, Hart GW. O-GlcNAc signalling: Implications for cancer cell biology. Nat. Rev. Cancer. 2011;11:678–684. doi: 10.1038/nrc3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abbadie C, Kabrun N, Bouali F, Smardova J, Stéhelin D, Vandenbunder B, Enrietto PJ. High levels of c-rel expression are associated with programmed cell death in the developing avian embryo and in bone marrow cells in vitro. Cell. 1993;75:899–912. doi: 10.1016/0092-8674(93)90534-w. [DOI] [PubMed] [Google Scholar]
- 29.Gerondakis S, Grumont R, Gugasyan R, Wong L, Isomura I, Ho W, Banerjee A. Unravelling the complexities of the NF-κB signalling pathway using mouse knockout and transgenic models. Oncogene. 2006;25:6781–6799. doi: 10.1038/sj.onc.1209944. [DOI] [PubMed] [Google Scholar]
- 30.Venkataraman L, Burakoff SJ, Sen R. FK506 inhibits antigen receptor–mediated induction of c-rel in B and T lymphoid cells. J. Exp. Med. 1995;181:1091–1099. doi: 10.1084/jem.181.3.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nolz JC, Fernandez-Zapico ME, Billadeau DD. TCR/CD28-stimulated actin dynamics are required for NFAT1-mediated transcription of c-rel leading to CD28 response element activation. J. Immunol. 2007;179:1104–1112. doi: 10.4049/jimmunol.179.2.1104. [DOI] [PubMed] [Google Scholar]
- 32.Bryan RG, Li Y, Lai JH, Van M, Rice NR, Rich RR, Tan TH. Effect of CD28 signal transduction on c-Rel in human peripheral blood T cells. Mol. Cell. Biol. 1994;14:7933–7942. doi: 10.1128/mcb.14.12.7933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shapiro VS, Truitt KE, Imboden JB, Weiss A. CD28 mediates transcriptional upregulation of the interleukin-2 (IL-2) promoter through a composite element containing the CD28RE and NF-IL-2B AP-1 sites. Mol. Cell. Biol. 1997;17:4051–4058. doi: 10.1128/mcb.17.7.4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Housley MP, Rodgers JT, Udeshi ND, Kelly TJ, Shabanowitz J, Hunt DF, Puigserver P, Hart GW. O-GlcNAc regulates FoxO activation in response to glucose. J. Biol. Chem. 2008;283:16283–16292. doi: 10.1074/jbc.M802240200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bunting K, Wang J, Shannon MF. Control of interleukin-2 gene transcription: A paradigm for inducible, tissue-specific gene expression. Vitam. Horm. 2006;74:105–145. doi: 10.1016/S0083-6729(06)74005-5. [DOI] [PubMed] [Google Scholar]
- 36.Kang SM, Tran AC, Grilli M, Lenardo MJ. NF-kappa B subunit regulation in nontransformed CD4+ T lymphocytes. Science. 1992;256:1452–1456. doi: 10.1126/science.1604322. [DOI] [PubMed] [Google Scholar]
- 37.Herold KC. Achieving antigen-specific immune regulation. J. Clin. Invest. 2004;113:346–349. doi: 10.1172/JCI20963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Köntgen F, Grumont RJ, Strasser A, Metcalf D, Li R, Tarlinton D, Gerondakis S. Mice lacking the c-rel proto-oncogene exhibit defects in lymphocyte proliferation, humoral immunity, and interleukin-2 expression. Genes Dev. 1995;9:1965–1977. doi: 10.1101/gad.9.16.1965. [DOI] [PubMed] [Google Scholar]
- 39.Hilliard BA, Mason N, Xu L, Sun J, Lamhamedi-Cherradi SE, Liou HC, Hunter C, Chen YH. Critical roles of c-Rel in autoimmune inflammation and helper T cell differentiation. J. Clin. Invest. 2002;110:843–850. doi: 10.1172/JCI15254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bunting K, Rao S, Hardy K, Woltring D, Denyer GS, Wang J, Gerondakis S, Shannon MF. Genome-wide analysis of gene expression in T cells to identify targets of the NF-κB transcription factor c-Rel. J. Immunol. 2007;178:7097–7109. doi: 10.4049/jimmunol.178.11.7097. [DOI] [PubMed] [Google Scholar]
- 41.Roos MD, Xie W, Su K, Clark JA, Yang X, Chin E, Paterson AJ, Kudlow JE. Streptozotocin, an analog of N-acetylglucosamine, blocks the removal of O-GlcNAc from intracellular proteins. Proc. Assoc. Am. Physicians. 1998;110:422–432. [PubMed] [Google Scholar]
- 42.Martin AG, San-Antonio B, Fresno M. Regulation of nuclear factor κB transactivation. Implication of phosphatidylinositol 3-kinase and protein kinase C ζ in c-Rel activation by tumor necrosis factor α. J. Biol. Chem. 2001;276:15840–15849. doi: 10.1074/jbc.M011313200. [DOI] [PubMed] [Google Scholar]
- 43.Liou HC, Hsia CY. Distinctions between c-Rel and other NF-κB proteins in immunity and disease. Bioessays. 2003;25:767–780. doi: 10.1002/bies.10306. [DOI] [PubMed] [Google Scholar]
- 44.Huang DB, Chen YQ, Ruetsche M, Phelps CB, Ghosh G. X-ray crystal structure of proto-oncogene product c-Rel bound to the CD28 response element of IL-2. Structure. 2001;9:669–678. doi: 10.1016/s0969-2126(01)00635-9. [DOI] [PubMed] [Google Scholar]
- 45.Kane LP, Lin J, Weiss A. It’s all Relative: NF-κB and CD28 costimulation of T-cell activation. Trends Immunol. 2002;23:413–420. doi: 10.1016/s1471-4906(02)02264-0. [DOI] [PubMed] [Google Scholar]
- 46.Doi TS, Takahashi T, Taguchi O, Azuma T, Obata Y. NF-κB RelA-deficient lymphocytes: Normal development of T cells and B cells, impaired production of IgA and IgG1 and reduced proliferative responses. J. Exp. Med. 1997;185:953–961. doi: 10.1084/jem.185.5.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xing D, Gong K, Feng W, Nozell SE, Chen YF, Chatham JC, Oparil S. O-GlcNAc modification of NFκB p65 inhibits TNF-α-induced inflammatory mediator expression in rat aortic smooth muscle cells. PLoS One. 2011;6:e24021. doi: 10.1371/journal.pone.0024021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Müller CW, Harrison SC. The structure of the NF-κB p50:DNA-complex: A starting point for analyzing the Rel family. FEBS Lett. 1995;369:113–117. doi: 10.1016/0014-5793(95)00541-g. [DOI] [PubMed] [Google Scholar]
- 49.Gao Y, Miyazaki J, Hart GW. The transcription factor PDX-1 is post-translationally modified by O-linked N-acetylglucosamine and this modification is correlated with its DNA binding activity and insulin secretion in min6 β-cells. Arch. Biochem. Biophys. 2003;415:155–163. doi: 10.1016/s0003-9861(03)00234-0. [DOI] [PubMed] [Google Scholar]
- 50.Hiromura M, Choi CH, Sabourin NA, Jones H, Bachvarov D, Usheva A. YY1 is regulated by O-linked N-acetylglucosaminylation (O-GlcNAcylation) J. Biol. Chem. 2003;278:14046–14052. doi: 10.1074/jbc.M300789200. [DOI] [PubMed] [Google Scholar]
- 51.Ea CK, Baltimore D. Regulation of NF-κB activity through lysine monomethylation of p65. Proc. Natl. Acad. Sci. U.S.A. 2009;106:18972–18977. doi: 10.1073/pnas.0910439106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen LF, Mu Y, Greene WC. Acetylation of RelA at discrete sites regulates distinct nuclear functions of NF-κB. EMBO J. 2002;21:6539–6548. doi: 10.1093/emboj/cdf660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Isomura I, Palmer S, Grumont RJ, Bunting K, Hoyne G, Wilkinson N, Banerjee A, Proietto A, Gugasyan R, Wu L, McNally A, Steptoe RJ, Thomas R, Shannon MF, Gerondakis S. c-Rel is required for the development of thymic Foxp3+ CD4 regulatory T cells. J. Exp. Med. 2009;206:3001–3014. doi: 10.1084/jem.20091411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ramakrishnan P, Baltimore D. Sam68 is required for both NF-κB activation and apoptosis signaling by the TNF receptor. Mol. Cell. 2011;43:167–179. doi: 10.1016/j.molcel.2011.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wells L, Vosseller K, Cole RN, Cronshaw JM, Matunis MJ, Hart GW. Mapping sites of O-GlcNAc modification using affinity tags for serine and threonine post-translational modifications. Mol. Cell. Proteomics. 2002;1:791–804. doi: 10.1074/mcp.m200048-mcp200. [DOI] [PubMed] [Google Scholar]
- 56.O’Connell RM, Balazs AB, Rao DS, Kivork C, Yang L, Baltimore D. Lentiviral vector delivery of human interleukin-7 (hIL-7) to human immune system (HIS) mice expands T lymphocyte populations. PLoS One. 2010;5:e12009. doi: 10.1371/journal.pone.0012009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schreiber E, Matthias P, Müller MM, Schaffner W. Rapid detection of octamer binding proteins with “mini-extracts,” prepared from a small number of cells. Nucleic Acids Res. 1989;17:6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.