Abstract
Humans have exceptionally long lifespans compared with other mammals. However, our longevity evolved when our ancestors had two copies of the apolipoprotein E (APOE) ε4 allele, a genotype that leads to a high risk of Alzheimer’s disease (AD), cardiovascular disease, and increased mortality. How did human aging evolve within this genetic constraint? Drawing from neuroscience, anthropology, and brain-imaging research, we propose the hypothesis that the evolution of increased physical activity approximately 2 million years ago served to reduce the amyloid plaque and vascular burden of APOE ε4, relaxing genetic constraints on aging. This multidisciplinary approach links human evolution with health and provides a complementary perspective on aging and neurodegenerative disease that may help identify key mechanisms and targets for intervention.
Keywords: aging, aerobic fitness, apolipoprotein, Alzheimer’s disease, dementia, vitamin D
The human lifespan from an evolutionary perspective
Humans live longer than any other primate, and are unique in having a prolonged postreproductive lifespan [1–4]. In humans living more traditional lifestyles (e.g., hunter-gatherers), despite having a life expectancy of under 40 (driven mainly by high infant and juvenile mortality), the average modal adult life span is approximately 72 years of age (with a range of 68–78; [5]). By comparison, our closest living relatives, chimpanzees, have a modal lifespan of 15 years and a life expectancy of approximately 30 years of age if they survive until puberty [6]. Most hypotheses for the evolution of human longevity (e.g., the ‘Grandmother hypothesis’) are centered on the ability of postreproductive individuals (grandparents) to aid their offspring and their offspring’s children, thereby improving their own reproductive success [2–4,7] (Box 1). In all cases, to contribute substantially to the well being and survival of younger kin, the evolution of the long human lifespan likely required older individuals to maintain high levels of both physical and cognitive health.
Box 1. Hypotheses for the evolution of the human lifespan.
The evolution of the long human lifespan and, in particular, the long postreproductive lifespan of females, poses a vexing question. Why would natural selection act to prolong the lifespan if individuals are living beyond the point where they can reproduce, or living to ages where reproductive rates are greatly diminished? Recent work suggests that, by aiding offspring of reproductive age in provisioning grandchildren, older men and women can greatly increase their own reproductive success by both contributing food to existing grandchildren and by increasing reproductive rates of their own children [2,3,109]. Two important hypotheses, the Grandmother Hypothesis (GH) and the Embodied Capital Hypothesis (ECH), present models for the evolution of the long human lifespan.
Williams [110] and later Hawkes and colleagues [3,111] suggested that the evolution of a long lifespan is the result of selection acting on grandmothers and their ability to provide resources for their kin. A key aspect of the GH is that, as climates changed at around the time of the evolution of the genus Homo, food became more difficult to find and extract [111]. The required foraging effort increased, and mothers who were pregnant or lactating may have had difficulty providing for their own offspring. Grandmothers, living beyond their reproductive years and without young offspring to care for, could provide increased foraging effort for their daughter’s children and, therefore, improve their own fitness [111]. This hypothesis is supported by ethnographic data from hunter-gatherer groups (primarily the Hadza of northern Tanzania) that show that older females provide a large proportion of food calories to their offspring and to the offspring of their offspring [3,111]. Provisioning is especially important to newly weaned children just after their mothers give birth to a sibling, because the mother’s resources are diverted to the newborn. This evolutionary scenario suggests that females who maintained their vigor past their reproductive senescence would have improved reproductive fitness through their daughters’ offspring and, thus, selection would favor their increased lifespan [111].
Kaplan and colleagues hypothesized that increased longevity is the result of investment in ‘embodied capital’ (EC: somatic tissue, such as the brain; and functional capacity, such as cognitive complexity) [2,109]. In the EC hypothesis, selection acts to extend the lifespan when early life is dominated by learning, and delayed maturation for increased learning pays off with increased reproductive fitness [2]. Thus, early life investment in brain tissue and cognitive complexity in humans (e.g., learning to hunt, finding food on the landscape, creating tools, etc.) pays off if lifespan is increased, and the return on EC investment occurs through intergenerational transfers of resources. This model links knowledge gained during growth to the evolution of the long lifespan and, again, highlights the importance of cognition in the evolution of long postreproductive life [109].
Thus, major explanations for the evolution of increased lifespan, especially increased postreproductive lifespan in females, require not only long lives, but also physical and cognitive health late in life. Foraging efforts to provision grandchildren require aerobic fitness because hunter-gatherers must trek long distances each day [112]. Given that foraging by hunter-gatherers also requires extensive experience, memory, and skill [109], intergenerational provisioning also requires the maintenance of cognitive function well past reproductive senescence.
Although many hypotheses address why human longevity evolved (e.g., [2–4]), how the long human lifespan evolved remains an open question. In a series of classic articles, Finch, Sapolsky, and Stanford [1,8–10] argued that the evolutionary origins of the long human lifespan are tied to the interaction between diet and genotype. Specifically, carriers of the ε4 allele of the APOE gene (responsible for lipid transport) have higher levels of total cholesterol and accumulation of atherosclerotic plaques in arteries, leading to increased risks of cardiovascular disease and stroke, as well as dementia and AD [11]. The APOE ε2 and ε3 alleles confer reduced risks of these diseases of aging relative to the ε4 allele, and are relatively recent additions to the human genome, with the ε3 and ε2 allele clade having evolved by 200 000 years ago [11]. Today, prevalence of these alleles varies around the world, but in most populations, ε3 is found in the highest frequency (mean = 78.3%; range: 8.5–98.0%), followed by ε4 (mean = 14.5%; range: 0–49.0%), and ε2 (mean = 6.4%; range: 0–37.5%) [12]. Finch and colleagues argue that humans’ exceptionally long lifespans are a product, in part, of the evolution of the ε3 allele, especially because diets shifted to include more meat and increased dietary fat and cholesterol later during our evolutionary history [10].
However, evidence suggests that increases in the human lifespan began as early as 1.8 million years ago [13], when our ancestors were likely homozygous for APOE ε4. To understand how the long human lifespan evolved within this constraint, it is important to view lifespan evolution within the context of aging outcomes that we see in living humans. Variation in human aging today is often expressed as a continuum that ranges from successful aging, having a long lifespan with high levels of cognitive and physical function, to pathological aging with impaired cognition and diminished physical capacities that can lead to dementia and relatively increased mortality [14,15]. If longevity evolved in humans to, in part, allow grandparents to aid offspring in raising grandchildren, then we must understand how our ancestors aged successfully despite being homozygous for APOE ε4. Recent studies suggest that lifestyle factors, specifically aerobic exercise, can have a positive impact on the aging brain, as well as physical longevity, especially in carriers of the ε4 allele [16–18]. Here, we argue that aerobic physical activity had an important role in the evolution of the human lifespan before the origins of the ε2 and ε3 alleles. Our review of the evolutionary links between physical activity, brain aging, and longevity are especially timely given the recent intense research focus on the use of exercise to reduce risks of developing disorders such as cardiovascular disease, AD, and other age-related dementias, including vascular dementia [18]. By providing the context of human evolutionary history, we hope that this review will serve as a foundation to further advance multidisciplinary efforts toward unraveling the mechanisms behind exercise-induced neuroprotection and increased longevity with successful aging, while helping to identify those who may benefit most from exercise interventions.
APOE functions and risks
APOE is a protein that circulates in the plasma and is present in the central nervous system, helping to regulate cholesterol and lipid metabolism, as well as aid cellular reparative processes [19,20]. The APOE gene, located on chromosome 19, is polymorphic, with three isoforms that differentially affect the affinity of the APOE protein for lipoprotein particles and the binding of the protein to low-density lipoprotein (LDL) receptors [11,21]. In addition, nearby single nucleotide polymorphisms (SNPs) are associated with disease states [22]. The presence of the ε4 allele tends to increase total cholesterol levels compared with the homozygous ε3 condition, whereas carriers of the ε2 allele have lower total cholesterol levels due to a reduced binding affinity for LDL [23]. Based on these functional differences, researchers have found carrying the ε4 allele leads to increased risk of coronary artery disease (CAD; up to 40% higher) due to higher circulating cholesterol levels and atherosclerotic plaque build-up in peripheral arteries [23,24], sometimes manifesting early in life [25,26].
Carriers of the ε4 allele also have a well-known higher risk of developing AD, dementia, and increased cognitive decline [27–34]. As the most well-established susceptibility gene for late-onset AD, APOE ε4 homozygotes have an increased risk of developing AD [35]. Although risk varies across populations (Table 1), in general each ε4 allele exerts a dose-related earlier onset of AD [36,37]. Interestingly, in two populations with high ε4 frequencies [Nigerians and Kenyans (Kenyan data are not shown in Table 1 due to lack of data for specific genotypes)], ε4 is not strongly associated with increased risk of AD or CAD [38–41]. Although it is not clear why there is a lack of association between ε4 and AD in these groups, future work is needed with these regional populations to unravel whether environmental or behavioral interactions (e.g., physical activity) affect the expression of this genetic risk factor.
Table 1.
Worldwide sample of odds ratios for Alzheimer’s diseasea
| Continent | Country | n | Odds ratio ε3/ε4 relative to referent (95% confidence interval) |
Odds ratio ε4/ε4 relative to referent (95% confidence interval) |
Refs |
|---|---|---|---|---|---|
| Africa | Nigeria | 582 | 1.33 (0.83–2.13) | 1.68 (0.73–3.65) | [39] |
| Tunisia | 129 | 2.9 (1.27–6.62) | 5.4 (1.35–21.48) | [113] | |
| Asia | Iran | 234 | 3.2 (1.6–6.5) | 12.75 (1.6–104) | [114] |
| Korea | 336 | 2.9 (1.7–5.2) | 24.7 (2.8–214.5) | [115] | |
| Japan | 1541 | 3.9 (1.9–8.0) | 21.8 (8.6–55.3) | [116] | |
| Europe | France | 1447 | 2.2 (1.5–3.5) | 11.2 (4.0–31.6) | [117] |
| Norway | 937 | 4.2 (2.98–5.89) | 12.9 (6.64–26.68) | [118] | |
| North America | US Caucasians (clinical) | 6305 | 3.2 (2.8–3.8) | 14.9 (10.8–20.6) | [119] |
| US Caucasians | 4858 | 2.7 (2.2–3.2) | 12.5 (8.8–17.7) | [119] | |
| US African-Americans | 474 | 1.1 (0.7–1.8) | 5.7 (2.3–14.1) | [119] | |
| US Hispanics | 528 | 2.2 (1.3–3.4) | 2.2 (0.7–6.7) | [119] | |
| US Japanese | 2313 | 5.6 (3.9–8.0) | 33.1 (13.6–80.5) | [119] | |
| South America | Brazil | 241 | 3.22 (1.73–6.01) | 9.57 (2.07–44.13) | [120] |
| Chile | 282 | 2.4 (1.3–4.5) | 12.8 (3.9–47.6) | [121] |
Odds ratios (ORs) for each genotype relative to referent genotype: ε3/ε3. This sample is not an exhaustive list but is meant to show the range in variation of ORs around the world. All studies are case–control samples with the exception of [39], which was a community-based longitudinal study design. It is noteworthy that the ORs calculated from data in [39] show non-significant associations with APOE e4 heterozygotes and homozygotes (95% confidence intervals include 1.0). Future work is needed to demonstrate the reproducibility of these findings in other samples from this region and to determine whether the low risk is due to gene × environment and/or lifestyle interactions, or other factors.
There remains disagreement over the specific mechanisms by which the ε4 allele increases the risk of AD and age-related cognitive decline. The relation between the ε4 allele and multiple pathological impacts, including on amyloid deposition, synaptogenesis, mitochondrial function, and phosphorylation of tau, have been suggested (reviewed in [42,43]), but with the most notable effects related to increased accumulation and reduced clearance of amyloid β-peptide (Aβ), marked by the formation of extracellular neuritic plaques whose build-up in the brain is one of the key pathological features of AD [44].
Brain-imaging studies have also detailed ε4-related effects associated with AD and dementia that begin before the onset of overt clinical symptoms. In this case, reductions in cerebral glucose metabolism revealed by positron emission tomography (PET) imaging in young to late middle-aged adults have been most consistently observed in brain areas typically affected in AD, including parietotemporal and frontal brain regions, along with indications of increased fibrillar amyloid deposition with PET [45–48]. Using magnetic resonance imaging (MRI) scans in young to late middle-aged and older adults, alterations in regional cerebral activity during cognitive task performance and brain network connectivity, as well as reductions and declines in structural gray matter volume and cortical thickness, have also been reported in asymptomatic ε4 carriers compared with noncarriers [49–58]. However, these MRI structural, functional, and network connectivity effects have been less consistently observed across different-aged samples and methods [54,57,59–61].
Given that an APOE genotype increases the risk of developing diseases that can contribute to both mid-life and late-life mortality, the presence of a single ε4 allele helps to explain a significant amount of lifespan variation in humans [62,63]. Homozygous ε4 carriers have an even greater reduction in life expectancy compared with individuals with other genotypes, including carriers of only one ε4 allele (6.4 years younger than ε3/ε4 heterozygotes) [1,64]. APOE ε4 is also considered a frailty gene, reducing the probability of successful aging even with a long lifespan [65]. Thus, the presence of the ε4 allele should affect the ability of individuals late in life to contribute to the health and well being of their offspring and grandchildren [66], and understanding the evolutionary history of the APOE gene will help us clarify its role in the evolution of the uniquely long human lifespan.
APOE evolution
Based on a comprehensive analysis of haplotype diversity, Fullerton et al. [11] showed that ε4 is the ancestral APOE allele in humans, and the divergence of the ε4 and the ε2 and ε3 clades occurred between 200 000 and 300 000 years ago. Even after the evolution of other alleles, ε4 remained present in our ancestors, given that McIntosh et al. [67] showed that the Denisovan hominin (30 000–80 000-years old) appears to have had the ε4 allele. Although direct measures of lipid binding have not been assessed, living chimpanzees have a form of the APOE gene that is believed to be functionally similar to the human APOE ε3 allele based on amino acid predictions, and the chimpanzee APOE amino acid sequence is monomorphic [67], suggesting the ε4 allele was an evolutionary novelty during human evolution (Box 2).
Box 2. Why did APOE ε4 evolve?
Our chimpanzee-like ancestors had a monomorphic APOE allele that is thought to be functionally similar to humans’ ε3 allele [67]. Thus, the ε4 allele likely evolved in response to selection pressures early in our evolutionary lineage. Some have suggested that APOE ε4 evolved in response to shifts in diet (‘a thrifty gene’), allowing for fat accumulation when nutrition access fluctuated [1,12,122]. Others suggest the ε4 allele evolved due to its enhanced inflammatory response, which may reflect an adaptation to highly infectious environments [1,8]. We highlight a third possibility that both accounts for why ε4 evolved early in human evolution and also explains the current observed differences in latitude-dependent worldwide distribution of the allele [12].
Heubbe et al. [19] suggested that the ε4 allele leads to greater intestinal absorption and renal reuptake of vitamin D. In mice with APOE ε4-targeted replacement, vitamin D levels were significantly higher than in APOE ε2 and APOE ε3 mice [19]. In addition, in a human sample, ε4 carriers had significantly higher levels of vitamin D compared with ε2 and ε3 carriers (13–25% higher) [19]. Selection likely acts strongly to regulate vitamin D status due to the farranging deleterious effects of low levels of vitamin D, including impaired reproductive function, increased risk of infections, rickets, and cardiovascular disease [123].
Based on the benefits of ε4 to support higher vitamin D levels, we suggest a possible evolutionary scenario that can account for both the evolution of ε4 from our last common ancestor with chimpanzees, and the current latitudinal gradient of ε4 prevalence [12]. Beginning approximately 2 million years ago, our ancestors moved into more open environments and likely lost body hair to improve thermoregulation through sweating [107] (Figure 1, main text). At this time, melanin-rich skin evolved to protect the subdermis from the damaging effects of ultraviolet (UV) radiation, especially on blood folate levels, which are essential to fetal development [124]. However, melanization also inhibits vitamin D synthesis [124]. Thus, the original evolution of the ε4 allele from the isoform present in our last common ancestor with chimpanzees may have allowed for highly melanized skin to protect blood folate, and still permit the absorption of vitamin D. Human migrations north into Europe and Asia, likely between 200 000 and 500 000 years ago, led to reduced levels of UV radiation and lighter skin colors [124]. For a given dose of UVB radiation, lighter skin color leads to relatively higher levels of vitamin D synthesis compared with darker skin colors [125]. Thus, with the evolution of lower levels of melanin in the skin, it is possible that selection for vitamin D synthesis and uptake was relaxed, allowing for the evolution of APOE ε2 and ε3 isoforms. Although this, and other possible hypotheses for the early evolution of the ε4 allele have been suggested, it is important to note that our overall conjecture that the evolution of increased aerobic activity relaxed a constraint on aging in our ancestors having the ε4 allele does not specifically depend on the vitamin D or other explanations for the early evolution of the ε4 allele.
Researchers have proposed several possible scenarios for the evolution of the ε4 allele [1,8,19], including a recent suggestion that the ε4 allele evolved to protect against vitamin D deficiency (Box 2). However, with the evolution of the ε3 and ε2 alleles, the ε4 allele became less prevalent. Today, the frequency of the ε4 allele in human populations around the world generally follows a U-shaped latitudinal gradient, with the highest frequencies (up to approximately 40–50% of the population) in equatorial and high latitudes, and lower frequencies in middle latitudes ([12], but see [68] for additional factors that contribute to worldwide variation).
This evolutionary history of the APOE gene presents a vexing paradox. As described in more detail below, data suggest that lifespans began to increase before the evolution of ε2 and ε3. In this context, how did selection generate longer human lifespans when all individuals had two copies of the deleterious ε4 allele? We argue that exercise and aerobic physical activity played an important role in the evolution of the long human lifespan by reducing the deleterious effects of ε4 on the development of AD and CAD. Support for this hypothesis requires evidence of: (i) an interaction between behavior and genotype, such that increased aerobic activity reduces deleterious effects of APOE ε4 on diseases of aging; (ii) increased physical activity during human evolution before the evolution of the ε2 and ε3 alleles; and (iii) increases in human lifespan following the evolution of increased physical activity. Below, we consider each of these three elements in support of our overall hypothesis that aerobic physical activity had an essential role in the evolution of human aging and longevity.
Interaction between physical activity and APOE genotype
Recent work suggests that exercise and physical activity interact with the APOE genotype to mediate the effects of the ε4 allele on CAD. In a cross-sectional study, ε4 carriers showed a significant protective effect of high-intensity activity, with athletic ε2, ε3, and ε4 carriers having similar blood lipid profiles and sedentary ε4 carriers showing significantly high levels of lipid risk factors for CAD [69]. A similar result was found in a cross-sectional study of middle-aged highly fit and age-matched sedentary groups, where inactive ε4 carriers had elevated risk factors of CAD, including higher circulating LDL levels compared with physically active ε4 carriers, and compared with ε3 carriers regardless of aerobic activity levels [70]. Thus, physical activity seems to diminish CAD risks in APOE ε4 carriers to ε4 noncarrier levels, whereas sedentary lifestyles may exacerbate CAD risk factors among ε4 carriers.
In addition, exercise appears to improve brain aging in individuals carrying the ε4 allele. Longitudinal studies suggest that physical inactivity leads to increased risk of developing dementia or AD in APOE ε4 carriers [71]. For example, whereas mid-life exercise generally reduces the risk of developing AD or dementia in all subjects, carriers of the ε4 allele who participated in higher amounts of mid-life leisure-time exercise had greater protection against dementia or AD approximately 20 years later compared with ε4 noncarriers [16,71]. Odds ratios for cognitive decline are significantly higher among ε4 carriers who exercised for less than 1 h per day over 3 years compared with ε4 carriers who exercised for more than 1 h per day [17]. In some studies, engagement in physical activity seems to reduce risks of cognitive decline in ε4 carriers only [72], whereas others have found that physical activity reduces risk of AD in ε4 carriers, but has a similar reduction in dementia risk for all genotypes [73]. Although two studies have not shown a specific effect of exercise on development of AD or dementia in ε4 carriers [74,75], methodological limitations with these studies have been highlighted, reducing their relevance in addressing the question of a genotype–activity interaction, including limitations in the quality and extent of physical activity assessment [18,73]. Thus, in studies with highquality measures of physical activity, there is clear and growing support that engagement in physical exercise has a protective effect for APOE ε4 carriers.
Brain function as measured by functional MRI also interacts with genetic risk for AD and exercise status [18]. For example, in a test of semantic memory (famous name discrimination task), researchers found a significant interaction between physical activity participation and APOE genotype, with highly physically active ε4 carriers showing higher levels of semantic memory activation across several brain regions compared with those with lower genetic risk, or less active subjects [18,76]. APOE ε4 carriers with high levels of aerobic fitness (measured by maximum aerobic capacity) had greater glucose uptake in the temporal lobe [measured by 18-fluoro-deoxyglucose (FDG)-PET] following a working memory task compared with ε4 noncarriers [77]. APOE ε4 carriers who were physically active also had a lower probability of cognitive decline compared with less active ε4 carriers [72]. Finally, in homozygous ε4 women, cardiovascular fitness was positively correlated with performance on a series of cognitive tests [78].
Although the mechanisms behind the protective effects of exercise on pathological brain aging remain unclear with indications of enhanced neurogenesis [79], recent work indicates that exercise and physical activity may reduce amyloid deposition in the brain, a key marker of AD pathology [80,81]. In a study of 201 cognitively normal adults, Head et al. [81] found an interaction between ε4 status and exercise engagement for [11C]PiB PET binding (amyloid imaging), with sedentary ε4 carriers having significantly higher binding compared with active noncarriers. A similar recent study of 116 cognitively normal adults aged 60–95 found that higher levels of physical activity were associated with lower amyloid brain loads in ε4 carriers [80].
To our knowledge, no studies have yet reported on the potential interaction between APOE genotype, physical activity, and brain structure. However, several studies have found evidence that, without taking genotype into account, physical activity and exercise can affect the volume of brain structures in both cognitively normal older adults, and in individuals with AD or dementia. For example, greater aerobic fitness in cross-sectional studies of older adults is associated with increased white matter tract integrity in the frontal lobes, temporal lobes, and the uncinate fasciculus and cingulum [82,83], larger gray matter volumes in temporal, parietal, and inferior frontal areas [84–87], and increased cortical, hippocampal, and whole-brain volume [88–90]. Intervention studies in aging cohorts found that several months of aerobic exercise can increase gray matter volume in the prefrontal cortex, lateral temporal lobe, and the hippocampus [85,91,92]. Finally, in adults in the early stages of AD, aerobically fit subjects have larger total gray matter volumes, and greater volumes of the medial temporal lobe compared with less fit subjects [88,93].
Thus, there is growing evidence that physical activity, exercise, and aerobic fitness significantly reduce CAD risk and improve cognitive aging and biomarkers of AD pathology in APOE ε4 carriers. Given the observed differences in brain function, structure, and connectivity, as well as reports showing the preferential build-up of Aβ in adult ε4 carriers, it is possible that early or lifelong physical activity increases clearance of Aβ potentially through improved sleep (e.g., [94]) or other mechanisms, and enhances brain resilience through neuroprotective processes, such as increased perfusion and neurogenesis [18]. This hypothesis may provide an explanation for how the long human lifespan evolved despite the early high prevalence of the ε4 allele.
The evolutionary history of human exercise and longevity
There is a great deal of evidence suggesting that aerobic exercise became an increasingly important element of the human lifestyle beginning approximately 1.8 million years ago. With the evolution of Homo erectus, our ancestors began a new lifestyle oriented around a combination of meat eating and gathering [95,96]. This lifestyle contrasts with the more sedentary ape-like lifestyles of the earlier hominin genus, Australopithecus, who are reconstructed to have ranged much shorter distances than H. erectus [96,97]. Hunter-gatherers cover greater distances in single-day foraging bouts than other living primates, and these treks require high levels of cardiovascular endurance [98,99]. In addition to long foraging distances, endurance running (ER) is thought to be an important component of this early hunting and gathering lifestyle [98,100,101]. There is evidence that a change in body shape during the evolution of the genus Homo is consistent with skeletal adaptations for ER, and differs from the more ape-like body shape of earlier hominins [98,102]. Traits that improve ER performance, but seem to have little effect on walking performance, include increased semicircular canal radii [102], increased joint surface areas in the lower, but not upper, limbs [98], and the possible convergent evolution of many skeletal and neurobiological traits that improve endurance performance in mammals and humans [98,100,101,103–105]. Thus, beginning with the evolution of H. erectus, our ancestors became endurance athletes, regularly engaging in high levels of aerobic physical activity [97].
We suggest this shift to higher levels of physical activity during human evolutionary history relaxed APOE-related constraints on lifespan as far back as 1.8 million years ago (Figure 1; Box 3). Evidence for an early evolution of longevity comes mainly from studies of life-history schedules in fossil taxa and is based on the strong links between the timing of developmental milestones and maximum lifespan across mammals (slow development occurs in association with long lifespan; [106]). Growth and development data from H. erectus (timing of third molar eruption and growth of body and brain size) suggest a lifespan of 60 years [106,107], a major jump over earlier species (e.g., species of Australopithecus) with estimated lifespans of 30 or 40 years. Additionally, this longer lifespan for Homo erectus is likely an underestimate, because this method predicts a lifespan of 66 years old for our own species [106].
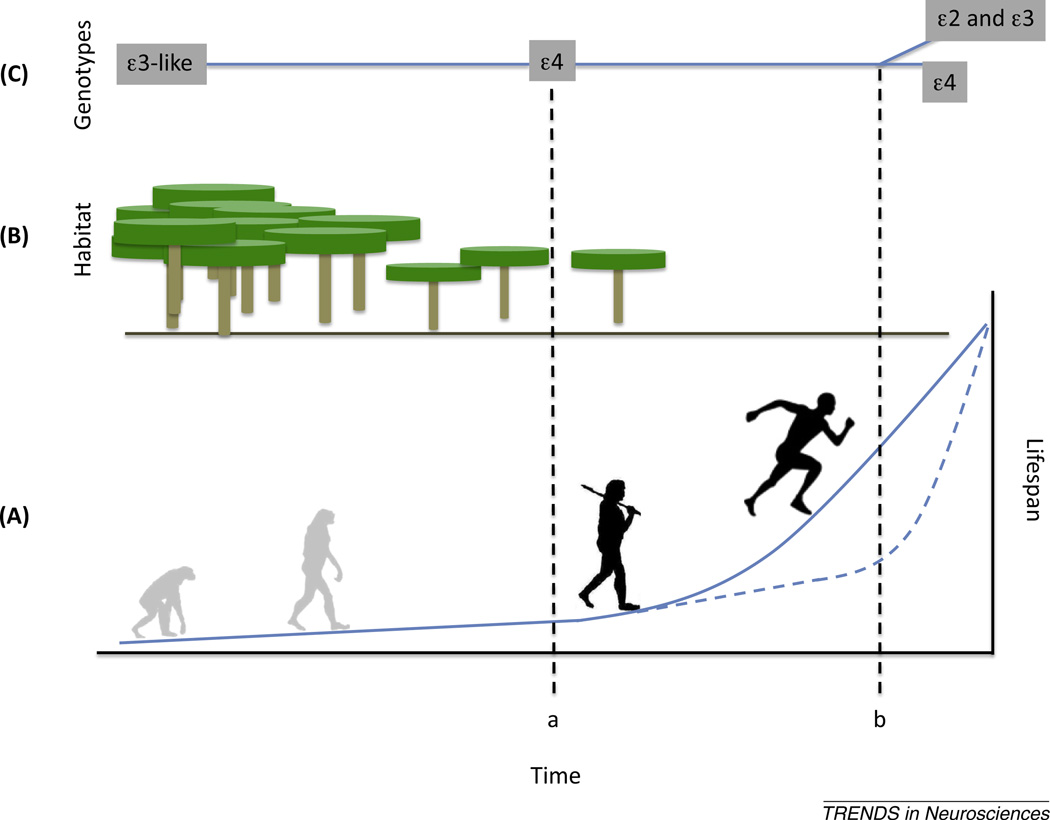
Figure 1.
Hypothesized relation between physical activity, apolipoprotein E (APOE) evolution, and human longevity. A conceptual time-line showing the relation between changes in lifespan reflecting both physical and cognitive health and physical activity (A), habitat (B), and APOE gene evolution (C) in human ancestors. Early in our evolutionary history, human ancestors lived in heavily forested environments and engaged in little aerobic activity. As habitats became more open in Africa (approximately 2 million years ago; time a), a loss of fur combined with increased melanin content in the skin may have led to the evolution of the APOE ε4 allele (Box 1). An increase in aerobic physical activity under these conditions would have allowed an increase in lifespan before the evolution of less deleterious APOE alleles (ε2 and ε3; time b). The solid blue line in (A) shows the change in human lifespan suggested by paleolife-history reconstructions. An alternative lifespan evolution, with a more recent prominent increase in longevity suggested by some interpretations of paleodemographic data [108], is shown as the dashed blue line in (A).
Box 3. Life expectancy versus lifespan and the evolution of human longevity.
A popular view of our Paleolithic ancestors is one of a Hobbesian nasty, brutish, and short life, with individuals living to old age only recently [5]. This model of the human lifespan is often based on studies of life expectancy at birth (Le), and data showing that Le increased dramatically over the past two centuries [5]. However, Le is often driven by high levels of infant and adolescent mortality, and on its own, this variable does not provide much information about the evolutionary timing of the long human lifespan. One way to better understand how and when old age evolved is to explore lifespan in modern humans living lives similar in many ways to those of our hunter-gatherer ancestors.
In living hunter-gatherer groups, and in a pre-industrial European population (18th-century Sweden), Le varies between 21 and 37 years, and is driven mainly by high mortality rates for infants and juveniles (approximately 57% of individuals lives to age 15) [5]. Thus, Gurven and Kaplan [5] examined the modal adult life span of hunter-gatherer populations as a way to estimate the natural age of death for humans. Among living hunter-gatherer groups, modal age of death ranges from 68 to 78 years, compared with a modal age of death of 85 years in the USA, and 15 years in wild chimpanzees [5]. In traditionally living human groups, most deaths in the total population, and in the population over the age of 60, are due to infectious illness or injury rather than to a degenerative disease [5].
Thus, in humans living lifestyles similar to those of our ancestors, modal lifespans are longer than that of our closest relatives, the chimpanzee. Although debates over when these long lifespans evolved continue, data from both paleodemography and life-history theory suggest that a shift towards longer lives occurred sometime during the evolution of the genus Homo [3,108]. Lifespans of living hunter-gatherers support this notion that human longevity may have evolved early in our lineage, possibly with the transition to the hunter-gatherer lifestyle that marks the origins of Homo erectus 1.8 million years ago.
Paleodemographic studies based on coarse age-estimates of individual fossils suggest a shift towards the presence of larger numbers of older individuals with the evolution H. erectus, and an even larger number of older individuals with the evolution of modern H. sapiens [108]. However, these analyses must be read with caution, given the difficulty in estimating age in the fossil record and the possible influences of postmortem processes on preservation of fossil remains (e.g., death assemblages do not necessarily reflect age structures of the populations due to preservation biases, including differential effects of burial and erosion on skeletons of different aged-individuals and effects of predation that can vary by age-class [13]). Despite some uncertainty over the timing of old age, the early evolution of longevity (approximately 1.8 million years ago) with H. erectus is most consistent with hypotheses for the evolution of the postreproductive lifespan that link successful aging to the origins of hunting and gathering [2–4,13]. If analyses of paleolife-history schedules are correct, as suggested by the fossil record, then the long postreproductive lifespan of humans began to evolve in concert with the shift towards higher aerobic activity in H. erectus, when the only available APOE allele was ε4. Thus, our review of evidence that exercise and physical activity can moderate the effects of APOE status on brain aging suggests that physical activity played an important role in relaxing disease-related constraints on the evolution of humans’ uniquely long lifespans.
Implications for research on cognitive aging, longevity, and health
Understanding how the long human lifespan evolved in the context of the deleterious effects of the APOE homozygous ε4 genotype may have important implications for current research efforts focused on enhancing cognitive aging, longevity, and the prevention of neurodegenerative disease in older populations. It is well established that human aging today is characterized by substantial heterogeneity, often expressed as a continuum that can extend from successful to pathological aging [14,15]. Such individual differences can present a significant obstacle to identifying universally effective treatments and prevention therapies for age-related decline and neurodegenerative disease. Our hypothesis suggests that increased selection for exercise and physical activity served to ameliorate the effects of APOE ε4, supporting the evolution of longer human lifespans and achieving the potential for successful advanced aging. Thus, changes induced by modern-day environmental constraints and human behavior may have led to greater vulnerability to the effects of APOE ε4 in subgroups of elderly in which high levels of physical activity throughout life are no longer required. Gaining a better understanding of the evolutionary origins linking the benefits of physical activity with the presence of the APOE ε4 allele may help in clarifying the underlying mechanisms and key molecular pathways that can serve to modify the deleterious effects of the ε4 allele and potentially help in elucidating targets for interventions that may be effective for subgroups and general aging populations today. It is also expected that this multidisciplinary approach linking together evolutionary origins for common genetic risk factors, behaviors, and their impacts on health provides a complementary perspective on aging and disease that may uniquely contribute to our understanding of other human disease processes and their links to potential interventions for disease prevention.
Concluding remarks
Here, we have developed a hypothesis for the evolution of the human lifespan that shows how increases in aerobic activity during our transition from a low-activity, sedentary, ape-like lifestyle, to a high-activity hunter-gather lifestyle served to relax constraints on aging imposed by the deleterious homozygous APOE ε4 genotype. This increased lifespan, and high levels of function in the elderly, would have enabled older adults to assist younger kin, reinforcing the selective benefits of old age [2,4,13]. The later evolution of ε3 and ε2 alleles (Box 2), may have paved the way for further increases in lifespan evident in Upper Paleolithic human populations and modern humans [1,9,10].
These studies provide an evolutionary context that suggests that diseases such as CAD, AD and other age-related dementias, may be due, in part, to the mismatch between our genetic heritage and our modern environment. We believe that the evolutionary hypothesis detailed here sheds important light on current ideas for prevention of CAD and AD. The genetic risk imposed by the ε4 allele may be particularly high among individuals leading sedentary lifestyles, with physical activity equalizing disease risk among all genotypes. Thus, our evolutionary heritage may help us tailor preventative interventions for individuals who have ancestral genotypes. These conclusions are important for populations around the world, where ε4 allele frequencies can vary from <5% to nearly 50% [12]. Without a continued lifestyle of high physical activity, regions of the world where the ε4 allele remains highly prevalent (e.g., equatorial Africa with ε4 frequencies approaching 50%) may experience a substantial increase in CAD, AD, and dementia with the increasing globalization of sedentary lifestyles. In the end, our understanding of human evolutionary history can help inform on how we should proceed from both a local and a global health perspective today, suggesting future research directions leading to public health policies to support successful aging in older populations worldwide (Box 4).
Box 4. Outstanding questions.
Although we suggest that our evolutionary history links physical activity with successful aging and longevity, more interdisciplinary research is needed to further address the many important, related questions that remain. Such future research has the potential to provide a more complete evolutionary context for understanding the human lifespan today, may help to advance our knowledge of the underlying biological mechanisms contributing to the individual differences observed during aging, and may help guide the development of effective approaches for the treatment and prevention of age-related diseases for different aging populations. Currently unresolved questions that may help in achieving these research goals include:
Do other genes that show an association with both exercise and cognitive aging [e.g., fat mass and obesity-associated (FTO) and brain-derived neurotrophic factor (BDNF)] have a similar evolutionary history to that of APOE?
How does physical activity influence the development of brain structure, function, and connectivity for ε4 carriers and noncarriers over the lifespan?
How does physical activity exert its beneficial effects in the context of APOE ε4 status to reduce the risk of CAD and AD?
Does APOE status interact with other genes to influence successful aging and longevity?
How do different amounts of exercise and at different points in the lifespan (e.g, young adult versus midlife versus old age) impact aging outcomes?
Acknowledgments
The authors would like to acknowledge support from the National Institute on Aging (AG025526 and AG19610), the National Science Foundation (NSF BCS 0820270), the Wenner Gren Foundation, the state of Arizona and Arizona Department of Health Services, Arizona Advanced Research Institute for Biomedical Imaging, and the Evelyn F. McKnight Brain Institute.
References
- 1.Finch CE. Evolution of the human lifespan, past, present, and future: phases in the evolution of human life expectancy in relation to the inflammatory load. Proc. Am. Philos. Soc. 2012;156:9. [PubMed] [Google Scholar]
- 2.Gurven M, et al. How long does it take to become a proficient hunter? Implications for the evolution of extended development and long life span. J. Hum. Evol. 2006;51:454–470. doi: 10.1016/j.jhevol.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 3.Hawkes K, et al. Grandmothering, menopause, and the evolution of human life histories. Proc. Natl. Acad. Sci. U.S.A. 1998;95:1336–1339. doi: 10.1073/pnas.95.3.1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marlowe F. The patriarch hypothesis. Hum. Nat. 2000;11:27–42. doi: 10.1007/s12110-000-1001-7. [DOI] [PubMed] [Google Scholar]
- 5.Gurven M, Kaplan H. Longevity among hunter-gatherers: a cross-cultural examination. Popul. Dev. Rev. 2007;33:321–365. [Google Scholar]
- 6.Hill K, et al. Mortality rates among wild chimpanzees. J. Hum. Evol. 2001;40:437–450. doi: 10.1006/jhev.2001.0469. [DOI] [PubMed] [Google Scholar]
- 7.Kim PS, et al. Increased longevity evolves from grandmothering. Proc. R. Soc. B: Biol. Sci. 2012;279:4880–4884. doi: 10.1098/rspb.2012.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Finch CE. Evolution of the human lifespan and diseases of aging: roles of infection, inflammation, and nutrition. Proc. Natl. Acad. Sci. U.S.A. 2010;107:1718–1724. doi: 10.1073/pnas.0909606106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Finch CE, Sapolsky RM. The evolution of Alzheimer disease, the reproductive schedule, and apoE isoforms. Neurobiol. Aging. 1999;20:407. doi: 10.1016/s0197-4580(99)00053-6. [DOI] [PubMed] [Google Scholar]
- 10.Finch CE, Stanford CB. Meat-adaptive genes and the evolution of slower aging in humans. Q. Rev. Biol. 2004;79:3–50. doi: 10.1086/381662. [DOI] [PubMed] [Google Scholar]
- 11.Fullerton SM, et al. Apolipoprotein E variation at the sequence haplotype level: implications for the origin and maintenance of a major human polymorphism. Am. J. Hum. Genet. 2000;67:881–900. doi: 10.1086/303070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eisenberg DT, et al. Worldwide allele frequencies of the human apolipoprotein E gene: climate, local adaptations, and evolutionary history. Am. J. Phys. Anthropol. 2010;143:100–111. doi: 10.1002/ajpa.21298. [DOI] [PubMed] [Google Scholar]
- 13.Hawkes K, O’Connell J. How old is human longevity? J. Hum. Evol. 2005;49:650–653. doi: 10.1016/j.jhevol.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 14.Alexander GE, et al. Characterizing cognitive aging in humans with links to animal models. Front. Aging Neurosci. 2012;4:21. doi: 10.3389/fnagi.2012.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Daffner KR. Promoting successful cognitive aging: a comprehensive review. J. Alzheimers Dis. 2010;19:1101–1122. doi: 10.3233/JAD-2010-1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rovio S, et al. Leisure-time physical activity at midlife and the risk of dementia and Alzheimer’s disease. Lancet Neurol. 2005;4:705–711. doi: 10.1016/S1474-4422(05)70198-8. [DOI] [PubMed] [Google Scholar]
- 17.Schuit AJ, et al. Physical activity and cognitive decline, the role of the apolipoprotein e4 allele. Med. Sci. Sports Exerc. 2001;33:772–777. doi: 10.1097/00005768-200105000-00015. [DOI] [PubMed] [Google Scholar]
- 18.Smith JC, et al. Physical activity and brain function in older adults at increased risk for Alzheimer’s disease. Brain Sci. 2013;3:54–83. doi: 10.3390/brainsci3010054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huebbe P, et al. APOE ε4 is associated with higher vitamin D levels in targeted replacement mice and humans. FASEB J. 2011;25:3262–3270. doi: 10.1096/fj.11-180935. [DOI] [PubMed] [Google Scholar]
- 20.Mahley RW, Huang Y. Apolipoprotein e sets the stage: response to injury triggers neuropathology. Neuron. 2012;76:871–885. doi: 10.1016/j.neuron.2012.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hui DY, et al. Defective hepatic lipoprotein receptor binding of B-very low density lipoporetins from type III hyperlipoproteinemic patients. J. Biol. Chem. 1984;259:860–869. [PubMed] [Google Scholar]
- 22.Martin ER, et al. SNPing Away at complex diseases: analysis of single-nucleotide polymorphisms around APOE Alzheimer disease. Am. J. Hum. Genet. 2000;67:383–394. doi: 10.1086/303003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eichner JE, et al. Apolipoprotein E polymorphism and cardiovascular disease: a HuGE review. Am. J. Epidemiol. 2002;155:487–495. doi: 10.1093/aje/155.6.487. [DOI] [PubMed] [Google Scholar]
- 24.Stengård JH, et al. An ecological study of association between coronary heart disease mortality rates in men and the relative frequencies of common allelic variations in the gene coding for apolipoprotein E. Hum. Genet. 1998;103:234–241. doi: 10.1007/s004390050811. [DOI] [PubMed] [Google Scholar]
- 25.Hixson J. Apolipoprotein E polymorphisms affect atherosclerosis in young males. Pathobiological Determinants of Atherosclerosis in Youth (PDAY) Research Group. Arterioscler. Thromb. Vasc. Biol. 1991;11:1237–1244. doi: 10.1161/01.atv.11.5.1237. [DOI] [PubMed] [Google Scholar]
- 26.Ilveskoski E, et al. Age-dependent association of apolipoprotein E genotype with coronary and aortic atherosclerosis in middle-aged men: an autopsy study. Circulation. 1999;100:608–613. doi: 10.1161/01.cir.100.6.608. [DOI] [PubMed] [Google Scholar]
- 27.Alexander GE, et al. Relation of age and apolipoprotein E to cognitive function in Down syndrome adults. Neuroreport. 1997;8:1835–1840. doi: 10.1097/00001756-199705260-00009. [DOI] [PubMed] [Google Scholar]
- 28.Caselli RJ, et al. Longitudinal modeling of age-related memory decline and the APOE ε4 effect. N. Engl. J. Med. 2009;361:255–263. doi: 10.1056/NEJMoa0809437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Caselli RJ, et al. Longitudinal changes in cognition and behavior in asymptomatic carriers of the APOE e4 allele. Neurology. 2004;62:1990–1995. doi: 10.1212/01.wnl.0000129533.26544.bf. [DOI] [PubMed] [Google Scholar]
- 30.Mortensen EL, Høgh P. A gender difference in the association between APOE genotype and age-related cognitive decline. Neurology. 2001;57:89–95. doi: 10.1212/wnl.57.1.89. [DOI] [PubMed] [Google Scholar]
- 31.Roses M, Allen D. Apolipoprotein E alleles as risk factors in Alzheimer’s disease. Annu. Rev. Med. 1996;47:387–400. doi: 10.1146/annurev.med.47.1.387. [DOI] [PubMed] [Google Scholar]
- 32.Saunders A, et al. Apolipoprotein E ∈ 4 allele distributions in late-onset Alzheimer’s disease and in other amyloid-forming diseases. Lancet. 1993;342:710–711. doi: 10.1016/0140-6736(93)91709-u. [DOI] [PubMed] [Google Scholar]
- 33.Smith JD. Apolipoproteins and aging: emerging mechanisms. Ageing Res. Rev. 2002;1:345–365. doi: 10.1016/s1568-1637(02)00005-3. [DOI] [PubMed] [Google Scholar]
- 34.Strittmatter WJ, et al. Apolipoprotein E: high-avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc. Natl. Acad. Sci. U.S.A. 1993;90:1977–1981. doi: 10.1073/pnas.90.5.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Corder E, et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science. 1993;261:921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 36.Duijn Cv, et al. Apolipoprotein E4 allele in a population-based study of early-onset Alzheimer’s disease. Nat. Genet. 1994;7:74–78. doi: 10.1038/ng0594-74. [DOI] [PubMed] [Google Scholar]
- 37.Nunomura A, et al. Apolipoprotein E polymorphism and susceptibility to early-and late-onset sporadic Alzheimer’s disease in Hokkaido, the northern part of Japan. Neurosci. Lett. 1996;206:17–20. doi: 10.1016/0304-3940(96)12415-0. [DOI] [PubMed] [Google Scholar]
- 38.Chen C-H, et al. A comparative study to screen dementia and APOE genotypes in an ageing East African population. Neurobiol. Aging. 2010;31:732–740. doi: 10.1016/j.neurobiolaging.2008.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gureje O, et al. APOE ε4 is not associated with Alzheimer’s Disease in elderly Nigerians. Ann. Neurol. 2006;59:182–185. doi: 10.1002/ana.20694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kalaria R, et al. Evaluation of risk factors for Alzheimer’s disease in elderly east Africans. Brain Res. Bull. 1997;44:573–577. doi: 10.1016/s0361-9230(97)00310-9. [DOI] [PubMed] [Google Scholar]
- 41.Kalaria RN, et al. Alzheimer’s disease and vascular dementia in developing countries: prevalence, management, and risk factors. Lancet Neurol. 2008;7:812–826. doi: 10.1016/S1474-4422(08)70169-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang Y. Aβ-independent roles of apolipoprotein E4 in the pathogenesis of Alzheimer’s disease. Trends Mol. Med. 2010;16:287–294. doi: 10.1016/j.molmed.2010.04.004. [DOI] [PubMed] [Google Scholar]
- 43.Kim J, et al. The role of apolipoprotein E in Alzheimer’s disease. Neuron. 2009;63:287–303. doi: 10.1016/j.neuron.2009.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cedazo-Mínguez A. Apolipoprotein E and Alzheimer’s disease: molecular mechanisms and therapeutic opportunities. J. Cell. Mol. Med. 2007;11:1227–1238. doi: 10.1111/j.1582-4934.2007.00130.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reiman EM, et al. Functional brain abnormalities in young adults at genetic risk for late-onset Alzheimer’s dementia. Proc. Natl. Acad. Sci. U.S.A. 2004;101:284–289. doi: 10.1073/pnas.2635903100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reiman EM, et al. Correlations between apolipoprotein E ε4 gene dose and brain-imaging measurements of regional hypometabolism. Proc. Natl. Acad. Sci. U.S.A. 2005;102:8299–8302. doi: 10.1073/pnas.0500579102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reiman EM, et al. Fibrillar amyloid-β burden in cognitively normal people at 3 levels of genetic risk for Alzheimer’s disease. Proc. Natl. Acad. Sci. U.S.A. 2009;106:6820–6825. doi: 10.1073/pnas.0900345106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Small GW, et al. Apolipoprotein E type 4 allele and cerebral glucose metabolism in relatives at risk for familial Alzheimer disease. JAMA. 1995;273:942–947. [PubMed] [Google Scholar]
- 49.Alexander GE, et al. Gray matter network associated with risk for Alzheimer’s disease in young to middle-aged adults. Neurobiol. Aging. 2012;33:2723–2732. doi: 10.1016/j.neurobiolaging.2012.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bookheimer SY, et al. Patterns of brain activation in people at risk for Alzheimer’s disease. N. Engl. J. Med. 2000;343:450–456. doi: 10.1056/NEJM200008173430701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Burggren AC, et al. Reduced cortical thickness in hippocampal subregions among cognitively normal apolipoprotein E e4 carriers. Neuroimage. 2008;41:1177–1183. doi: 10.1016/j.neuroimage.2008.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen K, et al. Correlations between apolipoprotein E ε4 gene dose and whole brain atrophy rates. Am. J. Psychiatry. 2007;164:916–921. doi: 10.1176/ajp.2007.164.6.916. [DOI] [PubMed] [Google Scholar]
- 53.Dennis NA, et al. Temporal lobe functional activity and connectivity in young adult APOE ε4 carriers. Alzheimers Dement. 2010;6:303–311. doi: 10.1016/j.jalz.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Filippini N, et al. Distinct patterns of brain activity in young carriers of the APOE-ε4 allele. Proc. Natl. Acad. Sci. U.S.A. 2009;106:7209–7214. doi: 10.1073/pnas.0811879106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mueller SG, et al. Selective effect of Apo e4 on CA3 and dentate in normal aging and Alzheimer’s disease using high resolution MRI at 4 T. Neuroimage. 2008;42:42–48. doi: 10.1016/j.neuroimage.2008.04.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shaw P, et al. Cortical morphology in children and adolescents with different apolipoprotein E gene polymorphisms: an observational study. Lancet Neurol. 2007;6:494–500. doi: 10.1016/S1474-4422(07)70106-0. [DOI] [PubMed] [Google Scholar]
- 57.Trivedi MA, et al. Reduced hippocampal activation during episodic encoding in middle-aged individuals at genetic risk of Alzheimer’s disease: a cross-sectional study. BMC Med. 2006;4:1. doi: 10.1186/1741-7015-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wishart H, et al. Regional brain atrophy in cognitively intact adults with a single APOE ε4 allele. Neurology. 2006;67:1221–1224. doi: 10.1212/01.wnl.0000238079.00472.3a. [DOI] [PubMed] [Google Scholar]
- 59.Mondadori CR, et al. Better memory and neural efficiency in young apolipoprotein E ε4 carriers. Cereb. Cortex. 2007;17:1934–1947. doi: 10.1093/cercor/bhl103. [DOI] [PubMed] [Google Scholar]
- 60.Reiman EM, et al. Hippocampal volumes in cognitively normal persons at genetic risk for Alzheimer’s disease. Ann. Neurol. 1998;44:288–291. doi: 10.1002/ana.410440226. [DOI] [PubMed] [Google Scholar]
- 61.Schmidt H, et al. Apolipoprotein E4allele in the normal elderly: neuropsychologic and brain MRI correlates. Clin. Genet. 1996;50:293–299. doi: 10.1111/j.1399-0004.1996.tb02377.x. [DOI] [PubMed] [Google Scholar]
- 62.Ewbank DC. The APOE gene and differences in life expectancy in Europe. J. Gerontol. A: Biol. Sci. Med. Sci. 2004;59:B16–B20. doi: 10.1093/gerona/59.1.b16. [DOI] [PubMed] [Google Scholar]
- 63.Jacobsen R, et al. Increased effect of the ApoE gene on survival at advanced age in healthy and long-lived Danes: two nationwide cohort studies. Aging Cell. 2010;9:1004–1009. doi: 10.1111/j.1474-9726.2010.00626.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kulminski AM, et al. Trade-off in the effects of the apolipoprotein E polymorphism on the ages at onset of CVD and cancer influences human lifespan. Aging Cell. 2011;10:533–541. doi: 10.1111/j.1474-9726.2011.00689.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Christensen K, et al. The quest for genetic determinants of human longevity: challenges and insights. Nat. Rev. Genet. 2006;7:436–448. doi: 10.1038/nrg1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hawkes K. How grandmother effects plus individual variation in frailty shape fertility and mortality: guidance from human–chimpanzee comparisons. Proc. Natl. Acad. Sci. U.S.A. 2010;107:8977–8984. doi: 10.1073/pnas.0914627107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McIntosh AM, et al. The Apolipoprotein E (APOE) gene appears functionally monomorphic in chimpanzees (Pan troglodytes) PLoS ONE. 2012;7:e47760. doi: 10.1371/journal.pone.0047760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Singh P, et al. APOE distribution in world populations with new data from India and the UK. Ann. Hum. Biol. 2006;33:279–308. doi: 10.1080/03014460600594513. [DOI] [PubMed] [Google Scholar]
- 69.Bernstein MS, et al. Physical activity may modulate effects of ApoE genotype on lipid profile. Arterioscler. Thromb. Vasc. Biol. 2002;22:133–140. doi: 10.1161/hq0102.101819. [DOI] [PubMed] [Google Scholar]
- 70.Pisciotta L, et al. Physical activity modulates effects of some genetic polymorphisms affecting cardiovascular risk in men aged over 40 years. Nutr. Metab. Cardiovasc. Dis. 2003;13:202–210. doi: 10.1016/s0939-4753(03)80012-1. [DOI] [PubMed] [Google Scholar]
- 71.Kivipelto M, et al. Apolipoprotein E ε4 magnifies lifestyle risks for dementia: a population-based study. J. Cell. Mol. Med. 2008;12:2762–2771. doi: 10.1111/j.1582-4934.2008.00296.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Woodard JL, et al. Lifestyle and genetic contributions to cognitive decline and hippocampal integrity in healthy aging. Curr. Alzheimer Res. 2012;9:436. doi: 10.2174/156720512800492477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Luck T, et al. Apolipoprotein E epsilon 4 genotype and a physically active lifestyle in late life: analysis of gene–environment interaction for the risk of dementia and Alzheimer’s disease dementia. Psychol. Med. 2013;44:1319–1329. doi: 10.1017/S0033291713001918. [DOI] [PubMed] [Google Scholar]
- 74.Lindsay J, et al. Risk factors for Alzheimer’s disease: a prospective analysis from the Canadian Study of Health and Aging. Am. J. Epidemiol. 2002;156:445–453. doi: 10.1093/aje/kwf074. [DOI] [PubMed] [Google Scholar]
- 75.Podewils LJ, et al. Physical activity, APOE genotype, and dementia risk: findings from the Cardiovascular Health Cognition Study. Am. J. Epidemiol. 2005;161:639–651. doi: 10.1093/aje/kwi092. [DOI] [PubMed] [Google Scholar]
- 76.Smith JC, et al. Interactive effects of physical activity and APOE-ε4 on BOLD semantic memory activation in healthy elders. Neuroimage. 2011;54:635–644. doi: 10.1016/j.neuroimage.2010.07.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Deeny SP, et al. Cardiovascular fitness is associated with altered cortical glucose metabolism during working memory in ε4 carriers. Alzheimers Dement. 2012;8:352–356. doi: 10.1016/j.jalz.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Etnier JL, et al. Cognitive performance in older women relative to ApoE-e4 genotype and aerobic fitness. Med. Sci. Sports Exerc. 2007;39:199–207. doi: 10.1249/01.mss.0000239399.85955.5e. [DOI] [PubMed] [Google Scholar]
- 79.Vukovic J, et al. Microglia modulate hippocampal neural precursor activity in response to exercise and aging. J. Neurosci. 2012;32:6435–6443. doi: 10.1523/JNEUROSCI.5925-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Brown B, et al. Physical activity and amyloid-β plasma and brain levels: results from the Australian Imaging, Biomarkers and Lifestyle Study of Ageing. Mol. Psychiatry. 2013;18:875–881. doi: 10.1038/mp.2012.107. [DOI] [PubMed] [Google Scholar]
- 81.Head D, et al. Exercise engagement as a moderator of the effects of APOE genotype on amyloid deposition. Arch. Neurol. 2012;69:636–643. doi: 10.1001/archneurol.2011.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Marks BL, et al. Role of aerobic fitness and aging on cerebral white matter integrity. Ann. N. Y. Acad. Sci. 2007;1097:171–174. doi: 10.1196/annals.1379.022. [DOI] [PubMed] [Google Scholar]
- 83.Voss MW, et al. The influence of aerobic fitness on cerebral white matter integrity and cognitive function in older adults: results of a one-year exercise intervention. Hum. Brain Mapp. 2012;34:2972–2985. doi: 10.1002/hbm.22119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bugg JM, Head D. Exercise moderates age-related atrophy of the medial temporal lobe. Neurobiol. Aging. 2011;32:506–514. doi: 10.1016/j.neurobiolaging.2009.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Erickson KI, et al. Physical activity, brain plasticity, and Alzheimer’s disease. Arch. Med. Res. 2012;43:615–621. doi: 10.1016/j.arcmed.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Floel A, et al. Physical activity and memory functions: are neurotrophins and cerebral gray matter volume the missing link? Neuroimage. 2010;49:2756–2763. doi: 10.1016/j.neuroimage.2009.10.043. [DOI] [PubMed] [Google Scholar]
- 87.Gordon BA, et al. Neuroanatomical correlates of aging, cardiopulmonary fitness level, and education. Psychophysiology. 2008;45:825–838. doi: 10.1111/j.1469-8986.2008.00676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Burns JM, et al. Cardiorespiratory fitness and brain atrophy in early Alzheimer disease. Neurology. 2008;71:210–216. doi: 10.1212/01.wnl.0000317094.86209.cb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Erickson KI, et al. Interactive effects of fitness and hormone treatment on brain health in postmenopausal women. Neurobiol. Aging. 2007;28:179–185. doi: 10.1016/j.neurobiolaging.2005.11.016. [DOI] [PubMed] [Google Scholar]
- 90.Erickson KI, Kramer AF. Aerobic exercise effects on cognitive and neural plasticity in older adults. Br. J. Sports Med. 2009;43:22–24. doi: 10.1136/bjsm.2008.052498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Erickson KI, et al. Physical activity predicts gray matter volume in late adulthood: The Cardiovascular Health Study. Neurology. 2010;75:1415–1422. doi: 10.1212/WNL.0b013e3181f88359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Erickson KI, et al. Exercise training increases size of hippocampus and improves memory. Proc. Natl. Acad. Sci. U.S.A. 2011;108:3017–3022. doi: 10.1073/pnas.1015950108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Honea R, et al. Cardiorespiratory fitness and preserved medial temporal lobe volume in Alzheimer’s disease. Alzheimer Dis. Assoc. Disord. 2009;23:188. doi: 10.1097/WAD.0b013e31819cb8a2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Spira AP, et al. Self-reported sleep and β-amyloid deposition in community-dwelling older adults. JAMA Neurol. 2013;70:1537–1543. doi: 10.1001/jamaneurol.2013.4258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Anton SC. Natural history of Homo erectus. Yrbk. Phys. Anthropol. 2003;46:126–169. doi: 10.1002/ajpa.10399. [DOI] [PubMed] [Google Scholar]
- 96.Anton SC, et al. An ecomorphological model of the initial hominid dispersal from Africa. J. Hum. Evol. 2002;43:773–785. doi: 10.1006/jhev.2002.0602. [DOI] [PubMed] [Google Scholar]
- 97.Lieberman DE, et al. Brains versus brawn and the evolution of Homo. In: Grine F, Leakey REF, editors. The Origin of Homo. Plenum; 2009. pp. 77–92. [Google Scholar]
- 98.Bramble DM, Lieberman DE. Endurance running and the evolution of Homo . Nature. 2004;432:345–352. doi: 10.1038/nature03052. [DOI] [PubMed] [Google Scholar]
- 99.Malina RM, Little BB. Physical activity: the present in the context of the past. Am. J. Hum. Biol. 2008;20:373–391. doi: 10.1002/ajhb.20772. [DOI] [PubMed] [Google Scholar]
- 100.Carrier DR. The energetic paradox of human running and hominid evolution. Curr. Anthropol. 1984;25:483–495. [Google Scholar]
- 101.Raichlen DA, et al. Calcaneus length determines running economy: implications for endurance running performance in modern humans and Neandertals. J. Hum. Evol. 2011;60:299–308. doi: 10.1016/j.jhevol.2010.11.002. [DOI] [PubMed] [Google Scholar]
- 102.Spoor F, et al. The bony labyrinth of Neanderthals. J. Hum. Evol. 2003;44:141–165. doi: 10.1016/s0047-2484(02)00166-5. [DOI] [PubMed] [Google Scholar]
- 103.Raichlen DA, et al. Wired to run: exercise-induced endocannabinoid signaling in humans and cursorial mammals with implications for the ‘runner’s high’. J. Exp. Biol. 2012;215:1331–1336. doi: 10.1242/jeb.063677. [DOI] [PubMed] [Google Scholar]
- 104.Raichlen DA, Gordon AD. Relationship between exercise capacity and brain size in mammals. PLoS ONE. 2011;6:e20601. doi: 10.1371/journal.pone.0020601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Raichlen DA, Polk JD. Linking brains and brawn: exercise and the evolution of human neurobiology. Proc. R. Soc. B: Biol. Sci. 2013;280:20122250. doi: 10.1098/rspb.2012.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Smith BH. Dental development and the evolution of life history in Hominidae. Am. J. Phys. Anthropol. 1991;86:157–174. [Google Scholar]
- 107.Hemmer H. Estimation of basic life history data of fossil hominoids. In: Henke HCW, Tattersall I, editors. Handbook of Paleoanthropology. Springer; 2007. pp. 587–619. [Google Scholar]
- 108.Caspari R, Lee S-H. Older age becomes common late in human evolution. Proc. Natl. Acad. Sci. U.S.A. 2004;101:10895–10900. doi: 10.1073/pnas.0402857101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kaplan H, et al. A theory of human life history evolution: diet, intelligence, and longevity. Evol. Anthropol. 2000;9:156–185. [Google Scholar]
- 110.Williams GC. Pleiotropy, natural selectio, and the evolution of senescence. Evolution. 1957;11:398–411. [Google Scholar]
- 111.Hawkes K. Grandmothers and the evolution of human longevity. Am. J. Hum. Biol. 2003;15:380–400. doi: 10.1002/ajhb.10156. [DOI] [PubMed] [Google Scholar]
- 112.Pontzer H, et al. Hunter-gatherer energetics and human obesity. PLoS ONE. 2012;7:e40503. doi: 10.1371/journal.pone.0040503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Rassas AA, et al. High APOE epsilon 4 allele frequencies associated with Alzheimer disease in a Tunisian population. Neurol. Sci. 2012;33:33–37. doi: 10.1007/s10072-011-0663-8. [DOI] [PubMed] [Google Scholar]
- 114.Raygani AV, et al. Association between apolipoprotein E polymorphism and Alzheimer disease in Tehran, Iran. Neurosci. Lett. 2005;375:1–6. doi: 10.1016/j.neulet.2004.10.073. [DOI] [PubMed] [Google Scholar]
- 115.Kim KW, et al. Association between apolipoprotein E polymorphism and Alzheimer’s disease in Koreans. Neurosci. Lett. 1999;277:145–148. doi: 10.1016/s0304-3940(99)00867-8. [DOI] [PubMed] [Google Scholar]
- 116.Bertram L, et al. Systematic meta-analyses of Alzheimer disease genetic association studies: the AlzGene database. Nat. Genet. 2007;39:17–23. doi: 10.1038/ng1934. [DOI] [PubMed] [Google Scholar]
- 117.Bickeböller H, et al. Apolipoprotein E and Alzheimer disease: genotype-specific risks by age and sex. Am. J. Hum. Genet. 1997;60:439. [PMC free article] [PubMed] [Google Scholar]
- 118.Sando SB, et al. APOE ε4 lowers age at onset and is a high risk factor for Alzheimer’s disease: a case control study from central Norway. BMC Neurol. 2008;8:9. doi: 10.1186/1471-2377-8-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Farrar L, et al. Effects of age, sex and ethnicity on the association between apolipoprotein E genotype and Alzheimer’s disease: a meta-analysis. JAMA. 1997;278:1349–1356. [PubMed] [Google Scholar]
- 120.Bahia VS, et al. Polymorphisms of APOE and LRP genes in Brazilian individuals with Alzheimer disease. Alzheimer Dis. Assoc. Disord. 2008;22:61–65. doi: 10.1097/WAD.0b013e31815a9da7. [DOI] [PubMed] [Google Scholar]
- 121.Quiroga P, et al. Apolipoprotein E polymorphism in elderly Chilean people with Alzheimer’s disease. Neuroepidemiology. 1998;18:48–52. doi: 10.1159/000026195. [DOI] [PubMed] [Google Scholar]
- 122.Corbo R, Scacchi R. Apolipoprotein E (APOE) allele distribution in the world. Is APOE* 4 a ‘thrifty’ allele? Ann. Hum. Genet. 1999;63:301–310. doi: 10.1046/j.1469-1809.1999.6340301.x. [DOI] [PubMed] [Google Scholar]
- 123.Yuen A, Jablonski N. Vitamin D: in the evolution of human skin colour. Med. Hypotheses. 2010;74:39–44. doi: 10.1016/j.mehy.2009.08.007. [DOI] [PubMed] [Google Scholar]
- 124.Jablonski NG, Chaplin G. The evolution of human skin coloration. J. Hum. Evol. 2000;39:57–106. doi: 10.1006/jhev.2000.0403. [DOI] [PubMed] [Google Scholar]
- 125.Armas LA, et al. Ultraviolet-B radiation increases serum 25-hydroxyvitamin D levels: the effect of UVB dose and skin color. J. Am. Acad. Dermatol. 2007;57:588–593. doi: 10.1016/j.jaad.2007.03.004. [DOI] [PubMed] [Google Scholar]