Conspectus
DNA performs a vital function as a carrier of genetic code, but in the field of nanotechnology, DNA molecules can catalyze chemical reactions in the cell, that is, DNAzymes, or bind with target-specific ligands, that is, aptamers. These functional DNAs with different modifications have been developed for sensing, imaging, and therapeutic systems. Thus, functional DNAs hold great promise for future applications in nanotechnology and bioanalysis. However, these functional DNAs face challenges, especially in the field of biomedicine. For example, functional DNAs typically require the use of cationic transfection reagents to realize cellular uptake. Such reagents enter the cells, increasing the difficulty of performing bioassays in vivo and potentially damaging the cell’s nucleus. To address this obstacle, nanomaterials, such as metallic, carbon, silica, or magnetic materials, have been utilized as DNA carriers or assistants. In this Account, we describe selected examples of functional DNA-containing nanomaterials and their applications from our recent research and those of others. As models, we have chosen to highlight DNA/nanomaterial complexes consisting of gold nanoparticles, graphene oxides, and aptamer–micelles, and we illustrate the potential of such complexes in biosensing, imaging, and medical diagnostics.
Under proper conditions, multiple ligand–receptor interactions, decreased steric hindrance, and increased surface roughness can be achieved from a high density of DNA that is bound to the surface of nanomaterials, resulting in a higher affinity for complementary DNA and other targets. In addition, this high density of DNA causes a high local salt concentration and negative charge density, which can prevent DNA degradation. For example, DNAzymes assembled on gold nanoparticles can effectively catalyze chemical reactions even in living cells. And it has been confirmed that DNA–nanomaterial complexes can enter cells more easily than free single-stranded DNA.
Nanomaterials can be designed and synthesized in needed sizes and shapes, and they possess unique chemical and physical properties, which make them useful as DNA carriers or assistants, excellent signal reporters, transducers, and amplifiers. When nanomaterials are combined with functional DNAs to create novel assay platforms, highly sensitive biosensing and high-resolution imaging result. For example, gold nanoparticles and graphene oxides can quench fluorescence efficiently to achieve low background and effectively increase the signal-to-background ratio. Meanwhile, gold nanoparticles themselves can be colorimetric reporters because of their different optical absorptions between monodispersion and aggregation.
DNA self-assembled nanomaterials contain several properties of both DNA and nanomaterials. Compared with DNA–nanomaterial complexes, DNA self-assembled nanomaterials more closely resemble living beings, and therefore they have lower cytotoxicity at high concentrations. Functional DNA self-assemblies also have high density of DNA for multivalent reaction and three-dimensional nanostructures for cell uptake. Now and in the future, we envision the use of DNA bases in making designer molecules for many challenging applications confronting chemists. With the further development of artificial DNA bases using smart organic synthesis, DNA macromolecules based on elegant molecular assembly approaches are expected to achieve great diversity, additional versatility, and advanced functions.
1. Introduction
Traditionally, DNA is a carrier of genetic information, but functional DNAs are also able to bind specifically with various targets (DNA aptamers) and possess catalytic activity (DNAzymes) with the ability to regulate gene expression.1 Such functional DNAs can be designed and selected in vitro, enabling them to play important roles in biological analysis and clinical diagnostics.
Aptamers are single-stranded oligonucleotides that possess high stability and high affinity and selectivity for specific targets. Aptamers can be generated through a technology termed “systematic evolution of ligands by exponential enrichment” (SELEX). Immense combinatorial libraries that contain trillions of different sequences are used to select different aptamers toward a variety of targets, including metal ions, metabolites, proteins, and even whole cells.2 The chemical nature of aptamers is based on the nucleic acid molecule, affording many distinct advantages in bioapplications, such as small size, nontoxicity, ease of synthesis and chemical modification with various functional groups, and facile surface immobilization.3 In addition, aptamers can be synthesized in vitro through mature solid state synthesis technology, eliminating the need for an animal source. All of these unique properties suggest that aptamers are superior to other molecular recognizers, including antibodies. In particular, these unique properties make aptamers new recognition elements in novel biosensors, termed aptasensors, which can be designed by various sensing strategies suited for almost any kind of target.4,5 The flexibility offered by aptamers also improves innovative biochemical applications through imaging.
DNAzymes, also called catalytic DNA or deoxyribozymes, are also functional DNA molecules selected in vitro. They can catalyze different kinds of chemical and biological reactions, including cleavage of nucleic acid substrates,6 ligation,7 phosphorylation8 and porphyrin metalation.9 The active sites of DNAzymes can distinguish substrates at the atomic level by short DNA strands. Meanwhile, their backbones are negatively charged and have a certain flexibility that exposes their bases to the outside, achieving, in turn, highly efficient catalysis. Thus, in many sensing platforms, DNAzymes have been implemented for the amplified detection of nucleic acids10 and metal ions.6,11−13
Despite the utility of SELEX, the discovery of new functional DNAs is a time-consuming process. As single-stranded oligonucleotides, the ability of DNAs to change conformation remains limited, even though functional DNAs, including aptamers and DNAzymes, require the presentation of many different conformations in order to generate signals that can be controlled and monitored. Furthermore, functional DNAs are negatively charged molecules having low binding affinity with the cell membrane, which complicates entry into living cells14,15 and thus limits applications in biochemical assays and medical diagnostics. In the past few years, different reformational nanomaterials, such as metallic, carbon, silica, and magnetic nanomaterials, have been developed to address these problems. Such nanomaterials enhance the use of DNA in nucleic acids-based biochemical assays since they possess composition-dependent chemical and physical properties, including unique optical, electronic, magnetic, and catalytic properties, and they can be designed and synthesized in unique sizes and shapes. As a consequence, nanomaterials are able to act as outstanding signal reporters, transducers, and amplifiers. Meanwhile, nanomaterials generally have a large surface area and can be chemically modified, leading to high loading efficiency of cargos, such as DNAs, drugs, or other molecules, to carry out different functions.16−18 Furthermore, the unusual biocompatibility and overall structural robustness of nanomaterials make them powerful candidates as delivery nanovehicles able to protect nucleic acids against nuclease degradation.19
In this Account, we will review the achievements made by our group and others in the application of functional DNA-containing nanomaterials in bioanalysis and biomedicine.
2. The Assembly of Functional DNA and Nanomaterials
Functional DNAs can be simply assembled on a given nanomaterial using methods such as covalent binding or π–π stacking. Generally, the nanoarchitecture of this complex contains a nanomaterial core and a monolayer of functional DNA. In order to take full advantage of the properties of functional DNA, analytic conditions need to be optimized in order to reduce steric hindrance and nonspecific adsorption. For example, the introduction of spacers, such as short DNA, bovine serum albumin (BSA), and poly(ethylene glycol) (PEG), can separate functional DNA and cover the blank surfaces of nanomaterials. Systems based on functional DNA-nanomaterial complexes not only possess the properties of both the functional DNA and the nanomaterial but also generate several new features, such as high affinity between the functional DNA–nanomaterial complex and target, strong degradation resistance, and high cellular uptake. Such properties can play a key role in biomedical applications. For instance, highly efficient self-delivery into cancer cells is the first requirement for exploring the intracellular environment. One study recently demonstrated that DNA with a 3D nanostructure could enter cells more easily than linear DNA strands.14 In addition, the formation of a 3D DNA/nanomaterial structure via self-assembly results in a nanoarchitecture that possesses the properties of both nanomaterials and DNA at the same time.
2.1. Gold Nanomaterials
As the most common and stable metallic nanomaterials, gold nanoparticles (AuNPs) have attracted much attention over the past decade due to their many features and properties. First, AuNPs can be synthesized in a straightforward manner, and their size and shape can be tuned by changing the parameters during synthesis. Second, AuNPs possess size-related electronic and optical properties that make them superquenchers and good colorimetric reporters. Third, AuNPs exhibit good biocompatibility, high intracellular stability, high DNA-loading capacity, and easy surface modification. Specifically, the strong interaction between thiol and gold provides an easy-to-handle and low-cost approach for AuNP modification.
Functional DNA-containing nanostructures based on the incorporation of AuNPs can result in a synergism between the functional DNAs and AuNPs. For example, the large surface area of AuNPs can accommodate a high density of DNA bound to the NP surface, leading to a higher binding strength for complementary targets than that of single DNA strands of the same sequence,20 conditions that are favorable for enhancing the efficiency of catalytic reactions for DNAzyme cleavage. AuNP–aptamer conjugates can increase affinity to target analytes through multiple ligand–receptor interactions, increased surface roughness, and ligand density.21 Meanwhile, the high local salt concentration and negatively charged surface of DNA-assembled AuNPs can enhance the degradation resistance of DNA (Figure 1).22 Taking advantage of the high density of DNA on AuNPs, DNA–AuNP conjugates gain more cellular uptake.23 Recently, the Mirkin group14 demonstrated that three-dimensional nanostructures of DNA, termed spherical nucleic acids (SNAs), promote intracellular transport and possess rapid cellular uptake kinetics. As shown in Figure 2, oligonucleotide dimensionality has a significant influence on endocytosis. High affinity with biotargets and facile cellular uptake of DNA-assembled AuNPs make them ideal candidates for biomedical applications.
Figure 1.
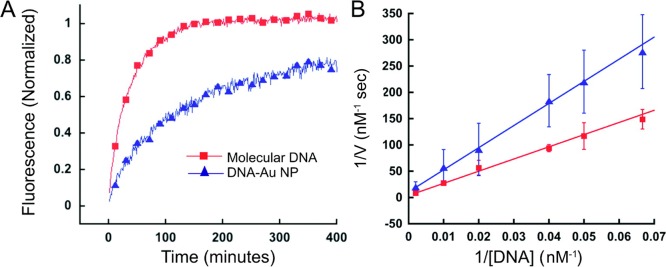
Comparison of the degradation rates of molecular DNA and DNA–AuNP systems. (A) Fluorescence-based progress curves of the enzyme-catalyzed reaction as a function of time. (B) Double reciprocal (Lineweaver–Burk) plot of the initial degradation velocity as a function of DNA-duplex concentration, which is used to calculate the kinetic parameters of the reaction. Reproduced with permission from ref (22). Copyright 2008 American Chemical Society.
Figure 2.
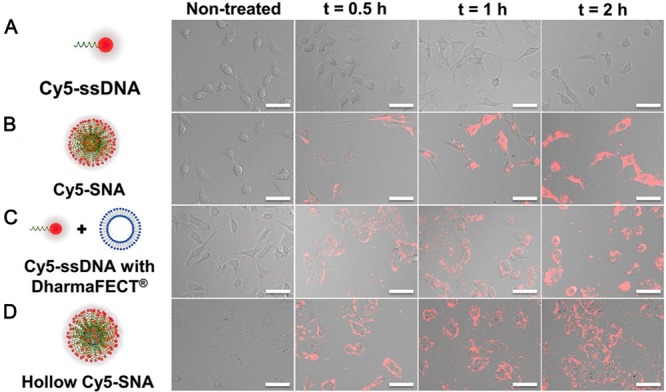
Effect of oligonucleotide dimensionality on endocytosis. (A) Cy5–ssDNAs cannot enter C166 cells in amounts detectable by confocal microscopy. (B) However, with their 3D architecture, Cy5–SNAs that contain the same total DNA concentration can readily enter cells, starting from binding to the cell membrane 30 min after incubation to populating the cytosol 2 h after incubation. (C) Encapsulation of Cy5–ssDNAs with DharmaFECT, a conventional cationic transfection agent, led to the tracing of the cell border by Cy5–ssDNAs without significant intracellular accumulation. (D) Hollow Cy5–SNAs enter cells with kinetics and degree comparable to Cy5–SNAs. Reproduced with permission from ref (14). Copyright 2013 National Academy of Sciences.
2.2. Carbon Nanomaterials
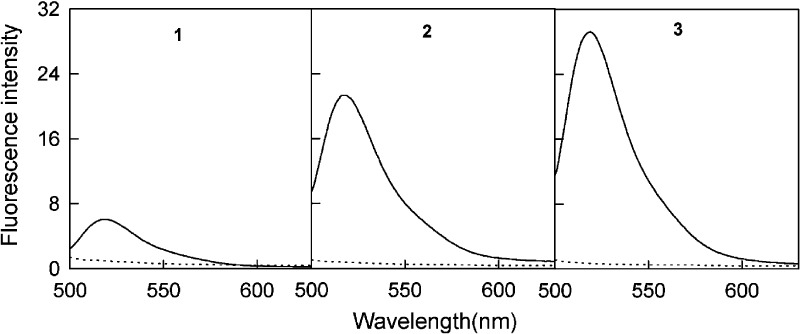
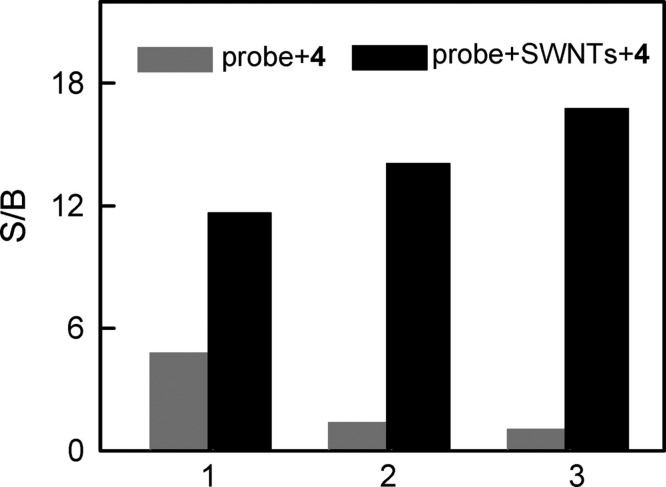
Single-walled carbon nanotubes (SWNTs) and graphene oxide (GO) have powerful quenching capability for organic dyes. A variety of complexes, including DNA strands, can be adsorbed noncovalently onto the surface of SWNTs and GO by virtue of π–π stacking. Thus, traditional molecular beacon-based fluorescent systems can be prepared.24,25 For SWNTs, in 2008 the Yang group reported a novel self-assembled SWNT–oligonucleotide complex as an efficient molecular beacon (MB) and demonstrated that it could be utilized in recognizing and detecting specific DNA sequences in a single step in homogeneous solution.24 They first tested three different nucleic-acid detection methods based on (i) a conventional MB (1), (ii) the self-assembled carbon-nanotube complex of 2 (2–SWNT), and (iii) the self-assembled carbon-nanotube complex of 3 (3–SWNT) (Table 1). As shown in Figure 3, it can be seen that the SWNTs provided further fluorescence quenching for MB 1 but more than 98% quenching for concentrations of 2, which ranged from 50 to 200 nM in their experiment, proving that ssDNA bound tightly on SWNTs. The experimental results summarized in Figure 4 showed a rather large variation in S/B in these assays because of the different background signals of the probes. In the same year, the Yang group constructed a new fluorescent sensor based on a ssDNA–SWNT complex for the detection of both DNA and protein. Human R-thrombin (Tmb) binding aptamers were self-assembled on SWNTs to detect Tmb in a fluorescent assay.25 All these studies illustrate that the SWNT–oligonucleotide complexes are excellent and universal fluorescent probes for bioanalysis.
Table 1. Designs of Probes and Target Oligonucleotides.
| type | sequence |
|---|---|
| FAM-labeled-MB (1)a | 5′-Dabcyl-CCTAGCTCTAAATCACTATGGTCGCGCTAGG-FAM-3′ |
| FAM-labeled-HP (2)b | 5′-CCTAGCTCTAAATCACTATGGTCGCGCTAGG-FAM-3′ |
| FAM-labeled-LN (3)c | 5′-CCTAGCTCTAAATCACTATGGTCGCCGATCC-FAM-3′ |
| pc-DNA (4)d | 5′-GCGACCATAGTGATTTAGA-3′ |
| sm-DNA(5)e | 5′-GCGACCATACTGATTTAGA-3′ |
Molecular beacon.
Hairpin-structured probe.
Linear probe.
Perfectly complementary target.
Single-base mismatched target.
Reproduced with permission from ref (24). Copyright 2008 American Chemical Society.
Figure 3.
Fluorescence emission spectra of 1, 2, and 3 in the absence (continuous lines) and the presence (dotted lines) of SWNTs. The concentrations of 1–3 were 50 nM, and excitation wavelength was at 480 nm. Reproduced with permission from ref (24). Copyright 2008 American Chemical Society.
Figure 4.
Comparisons of the signal-to-background ratio (S/B) of the fluorescent oligonucleotides generated by their perfectly complementary target 4. Gray bar: S/B of 1–3 generated by 6-fold excess of 4 in the absence of SWNTs. Black bar: S/B of 1–3 generated by 6-fold excess of 4 in the presence of SWNTs. The concentrations of 1–3 were 50 nM. Excitation was at 480 nm, and emission was monitored at 520 nm. Reproduced with permission from ref (24). Copyright 2008 American Chemical Society.
Graphene oxide (GO), a water-soluble derivative of graphene, possesses advantages such as easy synthesis, good water dispersibility, and facile surface modification. Most sensing and imaging systems with DNA/GO complexes are based on the fact that ssDNA with different lengths and conformations can exhibit different affinities for GO. For fluorescent assays, a rational design based on DNA–GO complexes can achieve very low background fluorescence. Moreover, several studies have suggested that GO can be an excellent vehicle and can also protect DNA on its surface, making GO favorable for efficient delivery of aptamers into living cells to carry out in situ detection.26
2.3. Magnetic Nanomaterials
In recent years, researchers have developed various types of magnetic nanoparticles (MNPs). By investigating their properties and behaviors, the application of MNPs in many important areas can be improved, including drug delivery, hyperthermia, magnetic resonance imaging (MRI), tissue engineering and repair, biosensing, biochemical separation, and bioanalysis. One of the most outstanding aspects of magnetic nanoparticles is their high-throughput separation capabilities, which can be combined with aptamers to realize the capture and concentration of cancer cells in one assay. To accomplish this, researchers often integrate MNPs with other nanoparticles to construct multifunctional systems for different applications. Most MNP systems utilize inorganic nanocrystals ranging from metals and alloys to metal oxides as their magnetic cores. In order to ensure their tolerance and biocompatibility, as well as specific localization at the biological target site, the surface of MNPs can be coated with a few atomic layers of a metal oxide, such as silica, or inorganic metal, such as gold, making MNPs suitable for further functionalization by the attachment of various bioactive molecules.
2.4. Functional DNA Self-Assembled Nanomaterials
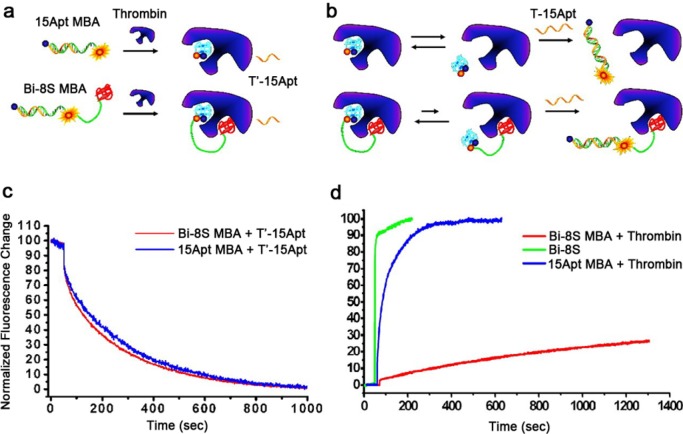
DNA is naturally water-soluble and biocompatible. Moreover, it is relatively simple to synthesize DNA with a commercial synthesizer. Recently, DNA has emerged as a favorable material for constructing DNA nanostructures with promising applications in biomedicine and biotechnology. For functional DNAs, multivalent interaction can result in better affinity and selectivity in contrast to monovalent interaction in the design of high-performance ligands. In 2008, we assembled two thrombin-binding aptamers together, and the aptamer assembly proved to be a high-performance inhibitor for treating various diseases related to blood clotting disorders under optimized conditions.27 Bivalent interaction of the aptamer assembly with thrombin, which increases overall binding affinity, is proposed as the mechanism for the enhanced inhibition. The binding affinity is directly related to kinetic parameters, such as kon′ and koff. As demonstrated in Figure 5, the increased thrombin inhibition potency of the aptamer assembly originates from the kinetic changes caused by cooperative binding.
Figure 5.
Comparison of binding kinetics. (a) Cartoon to describe kon′ measurement. (b) Cartoon to describe koff measurement. (c) Real-time fluorescence signal change of kon′ measurement. After thrombin was added, each sample mixture showed fluorescence decay. The decreasing rate was comparable in both cases. According to the calculation of the initial reaction rate, Bi-8S (bivalent nucleic acid candidate, Bi-xS, where x is the number of spacer bases linking two aptamers) exhibited a kon 1.2 times faster than that of 15Apt, a thrombin aptamer 15 bases long. (d) Real-time fluorescence signal change of koff′ measurement. Free 15Apt molecular beacon aptamer (MBA, green line) showed very rapid hybridization kinetics with its target DNA. Thrombin-bound 15Apt MBA (blue) showed slower hybridization kinetics compared with the free form. Interestingly, thrombin-bound Bi-8S MBA (red) showed a dissociation rate that was 51.7 times slower. The ka of the 15Apt domain of Bi-8S is about 62 times stronger than that of free 15Apt. Reproduced with permission from ref (27). Copyright 2008 National Academy of Sciences.
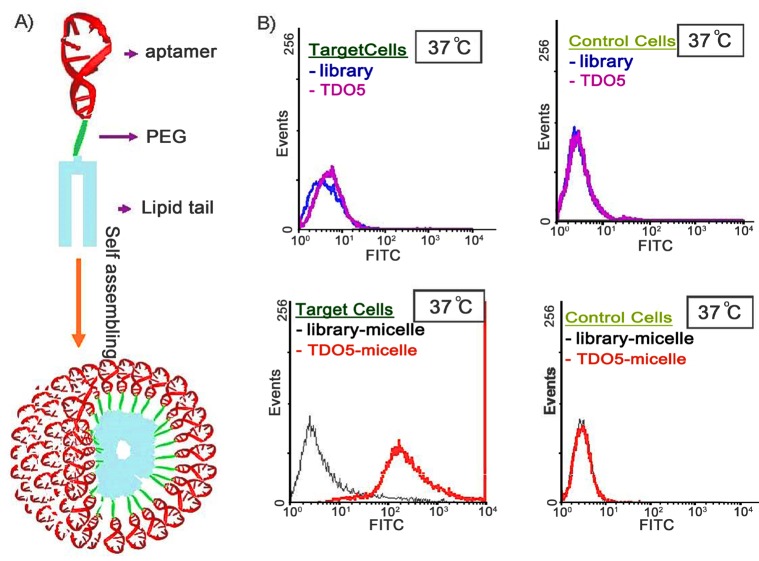
Inspired by amphiphilic block copolymers, which can self-assemble into different morphologies, the copolymer that contains a hydrophilic DNA segment and a hydrophobic organic polymer unit can form a DNA micelle under certain conditions. Compared with DNA-conjugated nanoparticles, DNA micelles have no inorganic cores, which would be cytotoxic at high concentrations, and the time required to synthesize DNA micelles can generally be abbreviated. In order to endow DNA micelles with more applicable properties and functions, we chose an aptamer to replace general DNA and conjugated it with a hydrophobic lipid tail. In 2010, we demonstrated that the aptamers in aptamer–micelle conjugates could still recognize their specific targets.28 Figure 6 shows that aptamer TDO5 was unable to bind with Ramos cells at physiological temperature. However, the TDO5–micelle conjugate displayed high affinity and selectivity for its target Ramos cells, as a result of densely packed aptamers that could enhance affinity for the target.
Figure 6.
(A) Schematic illustration of aptamer–micelle formation. (B) Flow cytometric assay to monitor the binding of free TDO5 (250 nM) with Ramos cells (target cells) and HL60 (control cells) at 37 °C for 5 min. The blue and black curves represent the background binding of unselected DNA library or library–micelle. The purple and red curves represent the binding of TDO5 or TDO5–micelle. Reproduced with permission from ref (28). Copyright 2010 National Academy of Sciences.
Instead of conventionally used short DNA, long DNA building blocks generated via rolling circle replication (RCR) can also form nanoarchitectures, such as DNA nanoflowers (NFs). RCR is an isothermal enzymatic reaction involving the replication of many circular genomic DNAs. These assemblies display higher biostability than complexes composed of many different DNA strands with sophisticated designs. The dense DNA packaging in NFs presumably affords the ability to resist nuclease degradation, denaturation, or dissociation at extremely low concentration.
3. Applications in Biosensing, Imaging, Drug Delivery, and Therapy
Because of the advantages of functional DNA-containing nanomaterials, as discussed above, multifunctional systems can be constructed to improve biomedical applications, including biosensing, imaging, drug delivery, and clinical therapy. For example, in fluorescent assays, aptamers can serve as recognition moieties and control the distance between quenchers, such as AuNPs and GO, and fluorophores via their conformational changes. Thus, two types of fluorescent sensors can be designed. If fluorescence is quenched in the absence of target but restored in the presence of target, the sensor is termed “turn-on”.14,25,26 On the other hand, if fluorescence is activated in the absence of targets but quenched in their presence, the sensor is termed “turn-off”. Fluorescence “turn-off” sensors may report false positive results caused by other quenchers in practical samples and are undesirable for practical analytical applications.
3.1. Functional DNA-Assembled Gold Nanomaterials
AuNP–DNA conjugates are stable in serum and can enter cells without transfection reagents. Based on their special electronic properties, AuNPs display “superquenching” ability for fluorescence via long-range resonance energy transfer.29 The Mirkin group reported aptamer–AuNP complexes, termed aptamer nanoflares, and detected intracellular ATP levels.30 DNA polymers assembled on AuNPs can be variously designed, for example by labeling with imaging fluorescent tags or the simultaneous loading of recognition elements and anticancer drugs.31 The AuNP–DNA conjugates show high stability and good biocompatibility, and the size of the complex can be controlled by changing the length of the self-assembled DNA biopolymer shell, which might provide a new and highly effective means for transporting cargos. Recently, the Lu group developed a novel DNAzyme–gold nanoparticle probe, which, for the first time, could be successfully applied to detect target analytes in living cells.32 The 39E DNAzyme, which has exceptional selectivity and sensitivity for the uranyl ion (UO22+), was chosen as an initial demonstration, and AuNP was chosen to be the carrier for cellular delivery of the DNAzyme. The assembly strategy of this novel system is shown in Figure 7. In the presence of uranyl, cleavage of the fluorophore-labeled substrate strand is catalyzed by DNAzyme. The shorter product strand labeled with Cy3 is released, and fluorescence is simultaneously recovered. Subsequently, this DNAzyme–AuNP probe was demonstrated to easily enter cells and serve as a metal ion sensor in the cellular environment.
Figure 7.
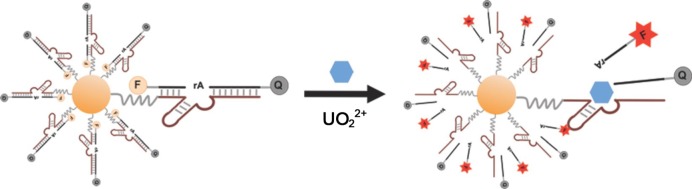
Design of a fluorescent DNAzyme immobilized on AuNPs as a selective probe of uranyl inside live cells. Reproduced with permission from ref (32). Copyright 2013 American Chemical Society.
Owing to their remarkable optical properties, AuNPs can also be used as brilliant colorimetric reporters.33 In terms of color changes resulting from the aggregation of AuNPs, aptamer-assembled AuNPs have provided more efficient biosensors for proteins and cancer cells. We have developed a colorimetric assay for the direct detection of diseased cells based on cancer cell aptamer-conjugated AuNPs (ACGNPs).34 Figure 8 shows ACGNPs bound to target cells with high specificity, despite the occurrence of some nonspecific binding and nonbinding, neither of which could dramatically alter the color of the assay solution as a consequence of the negligible increase in absorption and scattering of ACGNPs. These results implied that this colorimetric approach was very sensitive for the detection of target cancer cells and that ACGNPs could provide a direct visualization of cancer cells by assembling on a cell membrane surface to induce spectral changes.
Figure 8.

Schematic representation of the ACGNP-based colorimetric assay. Reproduced with permission from ref (34). Copyright 2008 American Chemical Society.
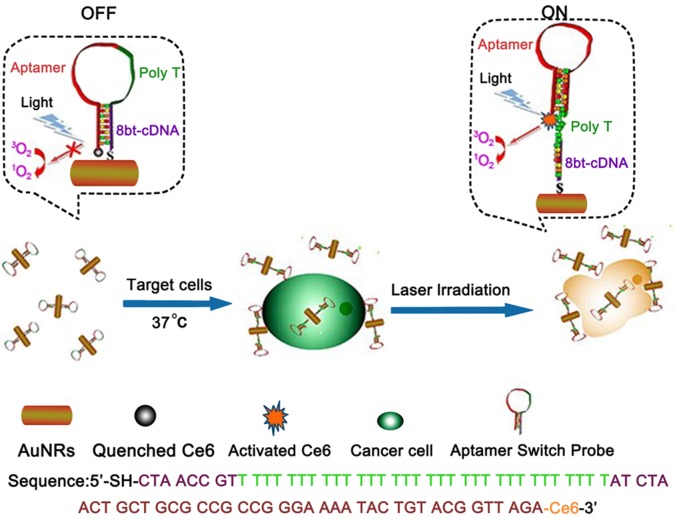
By change of the aspect ratio, gold nanoparticles may become nanorods, which present strong absorption in the near-infrared region. Taking advantage of this phenomenon, gold nanorods (AuNRs) are emerging as efficient photothermal therapy (PTT) nanomaterials.35−37 Compared with photothermal therapy alone, a combinatorial PTT/photodynamic therapy (PDT) approach can enhance therapeutic efficacy.36 Figure 9 shows the integration of DNA–AuNRs with a photosensitizer for use in multimodal cancer therapy. In order to manipulate the quenching and recovery of photosensitizer fluorescence, the aptamer, poly T, and 8bt-cDNA were designed for the probes, which were assembled on AuNRs, thus achieving controlled singlet oxygen generation (SOG) for PDT. The strategy of utilizing a highly selective aptamer combined with the synergistic effect of PTT and PDT promises to be a more efficient therapeutic regimen against cancer cells than nonspecific methods using either PTT or PDT alone.
Figure 9.
Schematic representation of aptamer switch probe (ASP)–photosensitizer–AuNRs for PTT and PDT. Reproduced with permission from ref (36). Copyright 2012 American Chemical Society.
3.2. Functional DNA-Assembled Carbon Nanomaterials
Among the various properties of single-walled carbon nanotubes (SWNT) and GO, the construction of sensitive systems for biosensing and imaging takes advantage of their excellent fluorescence quenching. In 2011, we demonstrated that SWNTs could be an excellent nanoquencher for lanthanide ion compounds.38 We first demonstrated the feasibility of this hypothesis and then constructed a label-free and time-resolved luminescence sensing platform for the detection of protein in complex biological fluids. The sensing approach was based on the noncovalent assembly of SWNTs and aptamers, the quenching ability of the SWNT complex for rare-earth chelates, and the restoration of the luminescence signal in the presence of target. Furthermore, the long lifetime of Eu3+ luminescence made it favorable for discriminating background signal in complex biological samples. With such unique properties, this time-resolved luminescence assay approach could achieve a low limit of detection under conditions of high autofluorescence without sample pretreatment.
An intracellular ATP probe based on an aptamer/graphene oxide nanocomplex has been used to monitor graphene in living cells and probe ATP in situ.26 However, because of the fetal bovine serum (FBS) used in culture medium and the presence of intracellular proteins, some nonspecific desorption can occur. To overcome this problem, an ATP aptamer molecular beacon (AAMB) has been used to substitute for the ATP aptamer, as reported by the Li group. We have also constructed an internal reference platform for a semiquantitative assay for intracellular ATP imaging.
3.3. Functional DNA-Assembled Magnetic Nanomaterials
MNPs can enhance the magnetic resonance (MR) signal of protons from surrounding water molecules.39 Aggregation of aptamer-conjugated MNPs is caused by their binding to target cells, which induces the coupling of magnetic spin moments and generates strong local magnetic fields. Such strong local magnetic fields lead to inhomogeneities that accelerate the spin-dephasing of adjacent water protons. In response, the spin–spin relaxation time (T2) of the surrounding water protons correspondingly decreases. Thus, aggregation of MNPs can be detected by ΔT2 changes. Capitalizing on this phenomenon, we have developed magnetic relaxation switches (MRSw) for the detection of specific targets. Figure 10 illustrates how aptamer-conjugated magnetic nanoparticles (ACMNPs) can reversibly self-assemble when binding to the same targets.40 The dense packing of aptamers on ACMNPs creates a multivalent effect for target binding that results in high sensitivity and specificity of this system in complex biological systems, including serum, plasma, and whole blood. Furthermore, by use of an array of ACMNPs, recognition patterns were generated for multiple types of cancer cells, thus creating a cellular molecular profile that allows not only the identification of cancer cells, but also the ability to differentiate between cancer and normal cells.
Figure 10.
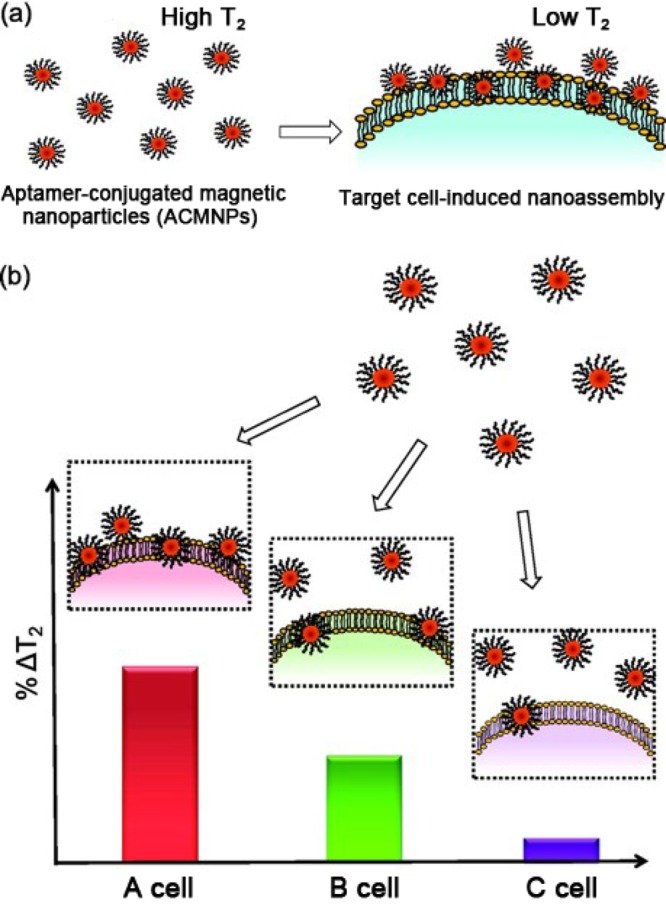
Schematic illustration of a magnetic nanosensor used for cancer cell detection and pattern recognition. (a) The magnetic nanoparticles conjugated with aptamers have highly specific binding to their target cells. Without target cells, ACMNPs are well dispersed, resulting in a high T2 of surrounding water protons. The addition of target cells leads to the aggregation of magnetic nanoparticles, decreasing the T2 of adjacent water protons. (b) Distinct recognition patterns generated for various cell lines with different receptor expression levels using the magnetic nanosensor. The cell line with the most abundant (A cell) receptors gives the largest ΔT2, followed by the cell line with the medium number of receptors (B cell), and the smallest ΔT2 was obtained for the cell line with the lowest receptor expression level (C cell). Reproduced with permission from ref (40). Copyright 2012 American Chemical Society.
MNPs can be useful alternatives to porous silica nanoparticles as nanovehicles for drug delivery.41 By means of acid etching, a porous hollow magnetite nanoparticle (PHMNP) can be generated and loaded with the anticancer drug doxorubicin (DOX) in its hollow interior as a carrier. In this system, aptamers were used as targeting moieties. However, instead of straightforward modification onto the surface of PHMNPs, a heterobiofunctional PEG ligand with a catechol group on one end and a carboxyl group on the other end served as a linker assembled onto the surface of PHMNPs for conjugation with special aptamers. This nanostructure was named as a smart multifunctional nanostructure (SMN) and was successfully utilized for targeted chemotherapy and magnetic resonance imaging (MRI). The acidic environment of lysosomes facilitated the release of DOX from the acid-labile pores of SMNs, and the release of DOX enabled efficient killing of target cancer cells.
3.4. Functional DNA Self-Assembled Nanomaterials
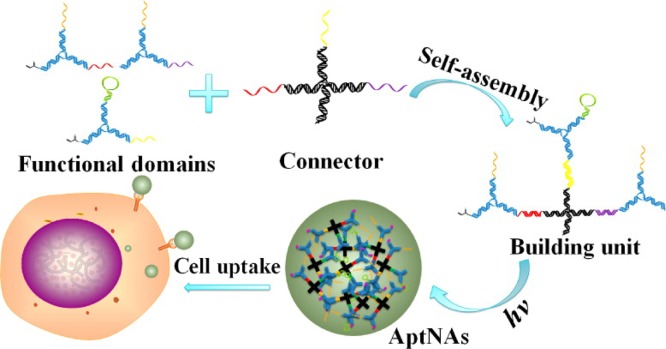
Functional DNA self-assembled nanomaterials, as distinct from the above-mentioned complexes, need no inorganic scaffold to form a 3D nanostructure. Combining DNA hybridization with a cross-linking reaction, we constructed a multifunctional and programmable aptamer nanoassembly structure.42 In Figure 11, an X-shaped core connector came from predesigned base-pair hybridization of four single-stranded DNAs, and three Y-shaped DNA functional domains were designed to link with the core connector. Targeting aptamers and acrydite-groups could be incorporated in different domains, as well as intercalated anticancer drugs, therapeutic antisense oligonucleotides, or other functional groups. Thus, each nanoassembly building unit was composed of one core connector and three functional domains. The acrydite-modified building units were further photo-cross-linked to form different nanostructures with controllable diameters. These nanostructures exhibited several remarkable features: facile modular design, facile assembly and preparation, high programmability, excellent biostability and biocompatibility, and selective recognition and transportation.
Figure 11.
Schematic illustration of the multifunctional self-assembled nanoassembly building units and photo-cross-linked nanoassembly structure. Reproduced with permission from ref (42). Copyright 2013 American Chemical Society.
Without Watson–Crick base-pairing, DNA can also grow together to form elongated DNA building blocks via liquid crystallization and dense packaging. Primer, circular template, and DNA polymerase participate in a DNA replication reaction to generate elongated DNA. Consequently, DNA NFs based on RCR completely consisted of deoxynucleotides, and their sizes were readily tunable over a wide range by simply adjusting such parameters as assembly time and template sequences. The circular template in our study was integrated with an antisense oligonucleotide aptamer sequence and drug loading sites. The resultant multifunctional NFs can be valuable tools for the recognition of targets and the simultaneous delivery of dyes and drugs.43
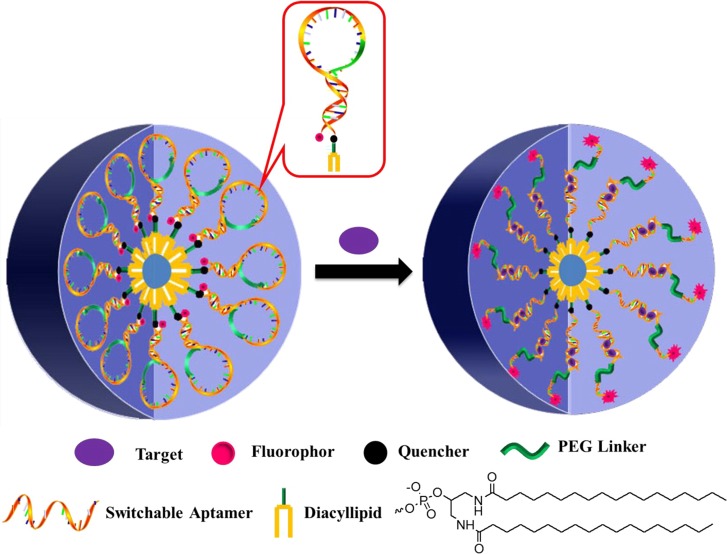
Functional DNA–micelles contain both hydrophilic and hydrophobic segments that self-assemble under certain conditions. Compared with aptamer-assembled AuNP and aptamer-GO complexes, respectively, the synthesis of aptamer–micelles is less time-consuming and produces fewer false positive signals. Recently, we reported an aptamer–micelle system for intracellular molecule detection.44 An aptamer was designed as a molecular beacon (MB) able to quench and restore fluorescence, as shown in Figure 12. This MB was modified with a lipid tail to form the nanostructure of micelle flares. In the presence of ATP, the conformation of the MB containing the ATP aptamer was altered, leading to the restoration of fluorescence. Therefore, intracellular detection and imaging of ATP could be achieved in real time.
Figure 12.
Working principle of switchable aptamer–micelle flares. Reproduced with permission from ref (44). Copyright 2013 American Chemical Society.
4. Future Perspectives
Combining the unique properties of both DNAs and such classical nanomaterials as AuNPs or GOs, functional DNA-containing nanomaterials can serve as smart biosensing, imaging, and therapy systems. Recent advances in functional DNA nanotechnology have shown the versatility and tremendous potential of functional DNA–nanomaterial complexes. However, some challenges lie ahead. For example, compared with buffer solutions, the intracellular environment is obviously much more complex. As such, nanomaterials used as scaffolding may adsorb some nontarget organic molecules or biomolecules in living cells, resulting in false signals. To address this issue, existing functional DNA-containing nanomaterials require more artful modifications to optimize their 3D nanostructures and surfaces, for example, by introducing spacers into the scaffold and changing the charge on the nanomaterial’s surface.
Meanwhile, researchers are working on the development of composite nanomaterials that possess remarkable properties, but with very low biological toxicity, for various bioassays. From a natural perspective, DNA self-assemblies, such as DNA–micelles and DNA nanoflowers, may be ideal platforms for intracellular assays. Moreover, by smart organic synthesis, artificial DNA bases would enrich this field with innovative properties and can also give rise to novel choreographed approaches of functional DNA self-assembly. Multimodality and clinical practicality are trends in the development of functional DNA-containing nanomaterial systems of the future, with more attention on improving their sensing capabilities for environmental monitoring, medical diagnostics, and therapeutics.
Biographies
Hao Liang is a student in the Department of Chemistry at Hunan University of Sciences. She obtained her B.S. (2011) in Chemistry from Hunan University, China. Currently, she is a Ph.D. candidate majoring in Analytical Chemistry in the Department of Chemistry at Hunan University. Her research interests include constructing novel functional nucleic acid-conjugated nanostructures for bioassay and bioimaging development.
Xiao-Bing Zhang is a professor in the Department of Chemistry at Hunan University of Sciences. He completed his B.S. in 1993 and Ph.D. in 2001, both in chemistry from Hunan University. He worked at the Ecole Normale Superieure de Lyon (France) and the Royal Institute of Technology (Sweden) as a postdoctoral fellow from 2003 to 2005. He served as an invited professor at ENS de Lyon in 2008 and as a visiting professor at the University of Illinois at Urbana–Champaign in 2009. Professor Zhang’s research interests concern fluorescent chemosensors and functional DNA-based biosensors.
Yifan Lv is a student in the Department of Chemistry at Hunan University of Sciences. He obtained his B.S. (2012) in Chemistry from Hunan University, China. He is a Ph.D. candidate majoring in Analytical Chemistry in the Department of Chemistry at Hunan University. His research interests include constructing novel logic gates and circuits based on nucleic acids.
Liang Gong is a student in the Department of Chemistry at the Hunan University of Sciences. She obtained her B.S. (2011) in Chemistry from Hunan Normal University, China. Currently, she is a Ph.D. candidate majoring in Analytical Chemistry in the Department of Chemistry at Hunan University. Her research interests include constructing novel nanomaterials for biosensor and drug delivery development.
Ruowen Wang is an associate researcher in the Collaborative Innovation Center for Chemistry and Molecular Medicine at Hunan University. He earned his Ph.D. in organic chemistry from the Shanghai Institute of Organic Chemistry, CAS, in 2006 and worked as a postdoctoral researcher on artificial DNA at the University of Pittsburgh. From July 2009 to June 2013, he worked at the Shanghai Institute of Organic Chemistry as an associate research professor. His research interests include bioorganic chemistry, molecular biology, and medicinal chemistry.
Xiaoyan Zhu is a student in the Department of Chemistry at the Hunan University of Sciences. She obtained her B.S. (2010) in Chemistry from Huanggang Normal University, China. She obtained her M.S. (2013) in Chemistry from Tianjin University, China. Currently, she is a Ph.D. candidate majoring in Analytical Chemistry in the Department of Chemistry at Hunan University. Her research interests include constructing novel artificial nucleic acid bases.
Ronghua Yang is a professor in the Department of Chemistry at Hunan University of Sciences. He earned his Ph.D. in 2000 in chemistry from Hunan University. He worked at Beijing University as a postdoctoral fellow from 2001 to 2003. He served as an associate professor at Beijing University in 2003 and as a visiting professor at the University of Florida in 2006. His research interests concern fluorescent chemosensors and their applications in food security and environmental analysis.
Weihong Tan is V. T. and Louis Jackson Professor of Chemistry and Physiology at the University of Florida. He obtained his B.S. from Hunan Normal University, his M.S. from the Chinese Academy of Sciences, and his Ph.D. in physical chemistry in 1993 from the University of Michigan at Ann Arbor. His research interests include chemical biology, bioanalytical chemistry, bionanotechnology, and biomedical engineering. Recently, his group developed cell-SELEX for the generation and application of aptamers as molecular tools to elucidate the molecular foundation of biological and pathological processes.
This work is supported by grants awarded by the National Institutes of Health (Grants GM079359 and CA133086). This work is also supported by the National Key Scientific Program of China (Grant 2011CB911000), NSFC (Grants 21325520, 21327009, 21221003, J1210040, 21177036, and 21135001), National Instrumentation Program (Grant 2011YQ030124), the Ministry of Education of China (Grant 20100161110011), and Hunan Provincial Natural Science Foundation (Grant 11JJ1002).
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
References
- Teller C.; Willner I. Functional nucleic acid nanostructures and DNA machines. Curr. Opin. Biotechnol. 2010, 21, 376–391. [DOI] [PubMed] [Google Scholar]
- Ellington A. D.; Szostak J. W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. [DOI] [PubMed] [Google Scholar]
- Liu J.; Cao Z.; Lu Y. Functional nucleic acid sensors. Chem. Rev. 2009, 109, 1948–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willner I.; Zayats M. Electronic aptamer-based sensors. Angew. Chem., Int. Ed. 2007, 46, 6408–6418. [DOI] [PubMed] [Google Scholar]
- Eltzov E.; Cosnier S.; Marks R. S. Biosensors based on combined optical and electrochemical transduction for molecular diagnostics. Expert Rev. Mol. Diagn. 2011, 11, 533–546. [DOI] [PubMed] [Google Scholar]
- Faulhammer D.; Famulok M. The Ca2+ ion as a cofactor for a novel RNA-cleaving deoxyribozyme. Angew. Chem., Int. Ed. 1996, 35, 2837–2841. [Google Scholar]
- Cuenoud B.; Szostak J. W. A DNA metalloenzyme with DNA ligase activity. Nature 1995, 375, 611–614. [DOI] [PubMed] [Google Scholar]
- Li Y.; Breaker R. R. Phosphorylating DNA with DNA. Proc. Natl. Acad. Sci. U.S.A. 1999, 96, 2746–2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.; Sen D. A catalytic DNA for porphyrin metallation. Nat. Struct. Mol. Biol. 1996, 3, 743–747. [DOI] [PubMed] [Google Scholar]
- Lu C. H.; Wang F.; Willner I. Zn2+-ligation DNAzyme-driven enzymatic and nonenzymatic cascades for the amplified detection of DNA. J. Am. Chem. Soc. 2012, 134, 10651–10658. [DOI] [PubMed] [Google Scholar]
- Liu J.; Brown A. K.; Meng X.; Cropek D. M.; Istok J. D.; Watson D. B.; Lu Y. A catalytic beacon sensor for uranium with parts-per-trillion sensitivity and million fold selectivity. Proc. Natl. Acad. Sci. U.S.A. 2007, 104, 2056–2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei S. H.; Liu Z.; Brennan J. D.; Li Y. An efficient RNA-cleaving DNA enzyme that synchronizes catalysis with fluorescence signaling. J. Am. Chem. Soc. 2003, 125, 412–420. [DOI] [PubMed] [Google Scholar]
- Santoro S. W.; Joyce G. F.; Sakthivel K.; Gramatikova S.; Barbas C. F. RNA cleavage by a DNA enzyme with extended chemical functionality. J. Am. Chem. Soc. 2000, 122, 2433–2439. [DOI] [PubMed] [Google Scholar]
- Choi C. H.; Hao L.; Narayan S. P.; Auyeung E.; Mirkin C. A. Mechanism for the endocytosis of spherical nucleic acid nanoparticle conjugates. Proc. Natl. Acad. Sci. U.S.A. 2013, 110, 7625–7630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosi N. L.; Giljohann D. A.; Thaxton C. S.; Lytton-Jean A. K.; Han M. S.; Mirkin C. A. Oligonucleotide-modified gold nanoparticles for intracellular gene regulation. Science 2006, 312, 1027–1030. [DOI] [PubMed] [Google Scholar]
- Pavlov V.; Xiao Y.; Shlyahovsky B.; Willner I. Aptamer-functionalized Au nanoparticles for the amplified optical detection of thrombin. J. Am. Chem. Soc. 2004, 126, 11768–11769. [DOI] [PubMed] [Google Scholar]
- Slowing I. I.; Vivero-Escoto J. L.; Wu C. W.; Lin V. S. Y. Mesoporous silica nanoparticles as controlled release drug delivery and gene transfection carriers. Adv. Drug Delivery Rev. 2008, 60, 1278–1288. [DOI] [PubMed] [Google Scholar]
- Lu A. H.; Salabas E. L.; Schüth F. Magnetic nanoparticles: Synthesis, protection, functionalization, and application. Angew. Chem., Int. Ed. 2007, 46, 1222–1244. [DOI] [PubMed] [Google Scholar]
- Gao L.; Cui Y.; He Q.; Yang Y.; Fei J.; Li J. Selective recognition of co-assembled thrombin aptamer and docetaxel on mesoporous silica nanoparticles against tumor cell proliferation. Chem.—Eur. J. 2011, 17, 13170–13174. [DOI] [PubMed] [Google Scholar]
- Lytton-Jean A. K.; Mirkin C. A. A thermodynamic investigation into the binding properties of DNA functionalized gold nanoparticle probes and molecular fluorophore probes. J. Am. Chem. Soc. 2005, 127, 12754–12755. [DOI] [PubMed] [Google Scholar]
- Sheng W.; Chen T.; Tan W.; Fan Z. H. Multivalent DNA nanospheres for enhanced capture of cancer cells in microfluidic devices. ACS Nano 2013, 7, 7067–7076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seferos D. S.; Prigodich A. E.; Giljohann D. A.; Patel P. C.; Mirkin C. A. Polyvalent DNA nanoparticle conjugates stabilize nucleic acids. Nano Lett. 2009, 9, 308–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giljohann D. A.; Seferos D. S.; Patel P. C.; Millstone J. E.; Rosi N. L.; Mirkin C. A. Oligonucleotide loading determines cellular uptake of DNA-modified gold nanoparticles. Nano Lett. 2007, 7, 3818–3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R.; Jin J.; Chen Y.; Shao N.; Kang H.; Xiao Z.; Tang Z.; Wu Y.; Zhu Z.; Tan W. Carbon nanotube-quenched fluorescent oligonucleotides: Probes that fluoresce upon hybridization. J. Am. Chem. Soc. 2008, 130, 8351–8358. [DOI] [PubMed] [Google Scholar]
- Yang R.; Tang Z.; Yan J.; Kang H.; Kim Y.; Zhu Z.; Tan W. Noncovalent assembly of carbon nanotubes and single-stranded DNA: An effective sensing platform for probing biomolecular interactions. Anal. Chem. 2008, 80, 7408–7413. [DOI] [PubMed] [Google Scholar]
- Wang Y.; Li Z.; Hu D.; Lin C.-T.; Li J.; Lin Y. Aptamer/graphene oxide nanocomplex for in situ molecular probing in living cells. J. Am. Chem. Soc. 2010, 132, 9274–9276. [DOI] [PubMed] [Google Scholar]
- Kim Y.; Cao Z.; Tan W. Molecular assembly for high-performance bivalent nucleic acid inhibitor. Proc. Natl. Acad. Sci. U.S.A. 2008, 105, 5664–5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y.; Sefah K.; Liu H.; Wang R.; Tan W. DNA aptamer–micelle as an efficient detection/delivery vehicle toward cancer cells. Proc. Natl. Acad. Sci. U.S.A. 2010, 107, 5–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan C.; Wang S.; Hong J. W.; Bazan G. C.; Plaxco K. W.; Heeger A. J. Beyond superquenching: Hyper-efficient energy transfer from conjugated polymers to gold nanoparticles. Proc. Natl. Acad. Sci. U.S.A. 2003, 100, 6297–6301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng D.; Seferos D. S.; Giljohann D. A.; Patel P. C.; Mirkin C. A. Aptamer nano-flares for molecular detection in living cells. Nano Lett. 2009, 9, 3258–3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J.; Zhu G.; Li Y.; Li C.; You M.; Chen T.; Song E.; Yang R.; Tan W. A spherical nucleic acid platform based on self-assembled DNA biopolymer for high-performance cancer therapy. ACS Nano 2013, 7, 6545–6554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P.; Hwang K.; Lan T.; Lu Y. A DNAzyme-gold nanoparticle probe for uranyl ion in living cells. J. Am. Chem. Soc. 2013, 135, 5254–5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava S.; Frankamp B. L.; Rotello V. M. Controlled plasmon resonance of gold nanoparticles self-assembled with PAMAM dendrimers. Chem. Mater. 2005, 17, 487–490. [Google Scholar]
- Medley C. D.; Smith J. E.; Tang Z.; Wu Y.; Bamrungsap S.; Tan W. Gold nanoparticle-based colorimetric assay for the direct detection of cancerous cells. Anal. Chem. 2008, 80, 1067–1072. [DOI] [PubMed] [Google Scholar]
- Wang J.; Sefah K.; Altman M. B.; Chen T.; You M.; Zhao Z.; Huang C. Z.; Tan W. Aptamer-conjugated nanorods for targeted photothermal therapy of prostate cancer stem cells. Chem.—Asian J. 2013, 8, 2417–2422. [DOI] [PubMed] [Google Scholar]
- Wang J.; Zhu G.; You M.; Song E.; Shukoor M. I.; Zhang K.; Altman M. B.; Chen Y.; Zhu Z.; Huang C. Z. Assembly of aptamer switch probes and photosensitizer on gold nanorods for targeted photothermal and photodynamic cancer therapy. ACS Nano 2012, 6, 5070–5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasun E.; Gulbakan B.; Ocsoy I.; Yuan Q.; Shukoor M. I.; Li C.; Tan W. Enrichment and detection of rare proteins with aptamer-conjugated gold nanorods. Anal. Chem. 2012, 84, 6008–6015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang X.; Yu R.; Jin J.; Li J.; Yang R.; Tan W.; Yuan J. New strategy for label-free and time-resolved luminescent assay of protein: conjugate Eu3+ complex and aptamer-wrapped carbon nanotubes. Anal. Chem. 2011, 83, 782–789. [DOI] [PubMed] [Google Scholar]
- Weissleder R.; Moore A.; Mahmood U.; Bhorade R.; Benveniste H.; Chiocca E. A.; Basilion J. P. In vivo magnetic resonance imaging of transgene expression. Nat. Med. 2000, 6, 351–354. [DOI] [PubMed] [Google Scholar]
- Bamrungsap S.; Chen T.; Shukoor M. I.; Chen Z.; Sefah K.; Chen Y.; Tan W. Pattern recognition of cancer cells using aptamer-conjugated magnetic nanoparticles. ACS Nano 2012, 6, 3974–3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T.; Shukoor M. I.; Wang R.; Zhao Z.; Yuan Q.; Bamrungsap S.; Xiong X.; Tan W. Smart multifunctional nanostructure for targeted cancer chemotherapy and magnetic resonance imaging. ACS Nano 2011, 5, 7866–7873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.; Han D.; Chen T.; Peng L.; Zhu G.; You M.; Qiu L.; Sefah K.; Zhang X.; Tan W. Building a multifunctional aptamer-based DNA nanoassembly for targeted cancer therapy. J. Am. Chem. Soc. 2013, 135, 18644–18650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G.; Hu R.; Zhao Z.; Chen Z.; Zhang X.; Tan W. Noncanonical self-assembly of multifunctional DNA nanoflowers for biomedical applications. J. Am. Chem. Soc. 2013, 135, 16438–16445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.; Chen T.; Han D.; You M.; Peng L.; Cansiz S.; Zhu G.; Li C.; Xiong X.; Jimenez E. Engineering of switchable aptamer micelle flares for molecular imaging in living cells. ACS Nano 2013, 7, 5724–5731. [DOI] [PMC free article] [PubMed] [Google Scholar]