Abstract
Oomycetes are eukaryotic microorganisms morphologically similar to but phylogenetically distant from true fungi. Most species in the genus Phytophthora of oomycetes are devastating plant pathogens, causing damages to both agricultural production and natural ecosystems. Tremendous progress has been achieved in recent years in diversity, evolution and lifestyles of oomycete plant pathogens, as well as on the understanding of genetic and molecular basis of oomycete-plant interactions. Phytophthora parasitica is a soilborne pathogen with a wide range of host plants and represents most species in the genus Phytophthora. In this review, we present some recent progress of P. parasitica research by highlighting important features that make it emerge as a model species of oomycete pathogens. The emerged model pathogen will facilitate improved understanding of oomycete biology and pathology that are crucial to the development of novel disease-control strategies and improved disease-control measures.
Keywords: oomycete, Phytophthora parasitica, model pathosystem, tobacco, Arabidopsis thaliana
Introduction
Oomycetes represent a group of eukaryotic microorganisms related to diatoms and brown algae and cause many destructive diseases to plants and animals. Among this group, the genus Phytophthora includes over 100 species (Kroon et al. 2012), and the number is increasing. Phytophthora species, the plant destroyers, have been a great threat to agricultural production and natural ecosystems. A notable example is Phytophthora infestans, the pathogen of the potato and tomato late blight, which triggered the Irish Famine in the 1840s but remains to be a difficult disease to control worldwide (Haas et al. 2009). Also, the recently emerged Phytophthora ramorum caused the Sudden Oak Death in 1990s and severely damaged woodlands in North America and Europe (Grünwald et al. 2012).
Oomycetes have several important characteristics distinct from true fungi (Latijnhouwers et al. 2003). For example, oomycetes are diploid while fungi are haploid, oomycete hyphae are nonseptate and multinucleated while fungi hyphae are septate. Many oomycetes are sterol aux-otrophs. Great differences in cell wall composition between oomycetes (consist mainly of 1,3-b-glucans, some 1,6-b-glucans and 1,4-b-glucans) and fungi (mainly of chitin) are notable (Werner et al. 2002; Latijnhouwers et al. 2003). The majority of fungicides target chitin and sterol synthesis and are ineffective for the control of oomycete diseases.
Understanding the mechanisms of oomycete pathogenicity is essential to develop disease-control measures. Most researches have focused on a few species, particularly P. infestans, Phytophthora sojae and Hyaloperonospora arabidopsidis. P. infestans is a foliar pathogen capable of infecting potato and tomato, P. sojae infects soybean {Glycine max) only. H. arabidopsidis is an obligate biotrophic pathogen of the model plant Arabidopsis thaliana.
Compared with these, Phytophthora parasitica Dastur (syn. Phytophthora nicotianae Breda de Haan) is a typical root pathogen with broad host ranges, being capable of infecting over 72 plant genera (Hickman 1958). With over 100 species in the genus Phytophthora, the features of the pathogen represent majority of Phytophthora species (Kroon et al. 2012). In recent years, many research have focused on this species and extensive molecular genetic tools and genomics resources have been developed. Especially, the pathosystem of compatible interaction (Figure 1) between A. thaliana and P. parasitica has been established. The development of the model pathogen is expected to facilitate accelerated understanding of oomycete pathogenesis, by accessible numerous genetic and genomics resources and associated tools with the model plant A. thaliana, which will ultimately lead to the development of novel disease-control strategies and improved disease-control measures.
Figure 1.

Root infection of Arabidopsis thaliana by Phytophthora parasitica, (a) Heavy colonization of root tissues by P. parasitica. Scale bar = 100 µm. (b) Numerous haustoria-like structures (Ha) developed (arrow). Scale bar = 50 µm.
Phylogeny of Phytophthora parasitica
Oomycetes belong to the kingdom Stramenopila, which also includes brown algae and diatoms. Of all oomycetes, Phytophthora is the best-studied genus. Using a genus-wide phylogeny analysis, 116 Phytophthora species are divided into 10 clades within the genus (Kroon et al. 2012). P. parasitica is classified in the Phytophthora clade 1 and its closest relatives include P. infestans. There are three divisions in this clade (a, b, and c). P. parasitica is singular in this clade, because it could not be placed in one of the subclades based on sequence analysis (Blair et al. 2008; Kroon et al. 2012). The phylogenetic relationships in the genus may serve as an inspiration to help understand more about Phytophthora species.
The biology and life cycle
The typical Phytophthora life cycle includes both asexual and sexual phases. The life cycle of P. parasitica is shown in Figure 2. The hyphae are hyaline and aseptate, sometimes with hyphal swellings. P. parastica produces asexual sporangia, zoospores and chlamydospores. The sporangia are difficult to be released from the hyphae, which is different from the released, airborne sporangia of P. infestans. Zoospores are produced by the sporangia and are wall-less cells with two flagella, which enable them to swim. Chlamydospores are thick-walled, multinucleated asexual spores, usually produced at the tips or in the middle of hyphae. P. parasitica is predominantly het-erothallic, requiring A1 and A2 mating types for the production of oospores (Ko 1981).
Figure 2.
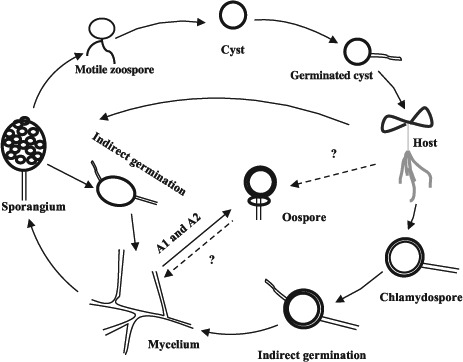
The life cycle of Phytophthora parasitica.
Efficient spore production and dispersal are essential for successful infection (Latijnhouwers et al. 2003). Zoospores are considered to be the major infective agents that initiate plant diseases for most Phytophthora species. Once reached the plant surface, zoospores become immobile cysts, subsequently germinate. P. parasitica infects roots and leaves by producing a specialized structure, appressorium, formed at the tip of germ tubes (Kebdani et al. 2010; Wang et al. 2011). Then, invasive hyphae are developed and haustoria-like structures are formed. At last, abundant sporangia formed on the surface of infected plants. If the environmental condition is unfavorable, chlamydospores are produced from the hyphae. The chlamydospores can survive in soil for several years, and serve as the primary inoculum in the field (Van Jaarsveld 2001). Sexual reproduction is an efficient and important way for the pathogen to produce genetic variation, including potentially large number of various genotypes and pathotypes that enable the pathogen to adapt to unfavorable conditions, particularly the introduction of new resistant genotypes of the host plant. However, to which extent the sexual reproduction play a role in P. parasitica remains unclear.
Genetic manipulation
Genetic manipulation is essential for analysis of gene functions in a given organism. To date, genetic transformation remains difficult for oomycetes, only reported to be successful for few species at variable but low efficiencies compared to true fungi. There are four methods developed for transforming foreign DNA into the genome of Phytophthora. The polyethylene glycol (PEG)-mediated transformation protocol was developed by Judelson et al. (1991), the first description of reliable method for transformation in an oomycete pathogen. Subsequently, the PEG transformation method was successfully used in several other species including P. sojae (Judelson et al. 1993), Saprolegnia monoica (Mort-Bontemps and Fvre 1997), P. parasitica (Bottin et al. 1999), Phytophthora palmivora (van West et al. 1999b), Phytophthora brassicae (Si-Ammour et al. 2003), Pythium aphanidermatum (Mcleod et al. 2008). Alternative transformation procedures were developed, such as particle bombardment in P. infestans (Cvitanich and Judelson 2003), Agrobacterium tumefaciens-mediated transformation in P. infestans (Vijn and Govers 2003) and electroporation of zoospores in Phytophthora capsici (Huitema et al. 2011), for manipulating gene expression levels.
This PEG-mediated transformation method has been successful in P. parasitica (Bottin et al. 1999). The P. parasitica transformants expressing green fluorescent protein (GFP) were used for cytological analysis of the colonization in the host plant tomato (Le Berre et al. 2008; Kebdani et al. 2010) during infection, and for analysis of the potential and application of host-induced gene silencing in P. parasitica (Zhang et al. 2011). The P. parasitica expressing GFP is very stable and can be successfully used for assessment of susceptibility of the host plant to the pathogen (Figure 3).
Figure 3.
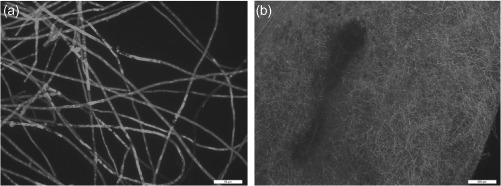
Cytological characterization of Phytophthora parasitica transformant and the infected leaf of Arabidopsis thaliana. (a) Hyphae of P. parasitica transformant expressing GFP. Scale bar = 50 μm. (b) Heavy colonization on leaf tissue of ecotype Col-0. Scale bar = 500 μm.
With the development of genetic transformation method, pathogens were labeled with fluorescent proteins and have successfully been employed to help answer questions about how the pathogens interact with the host plants. For an excellent example, Whisson et al. (2007) used P. infestans transformants expressing GFP and translational fusions of effector Avr3a with the monomeric red fluorescent protein to define that Avr3a is secreted from haustoria and translocated into the host. Ah-Fong and Judelson (2011) labeled various organelles in P. infestans and provided a series of vectors designed for expressing different fluorescent proteins.
Due to diploid nature and lack of homologous recombination-based gene disruption in oomycetes, RNA silencing emerged to be an important approach for downregulation of target genes in Phytophthora. Internuclear gene silencing was reported in P. infestans (Kamoun et al. 1998; van West et al. 1999a). The introductions of sense, antisense, and hairpin constructs were all subsequently confirmed to induce gene silencing, enabling gene function studies in Phytophthora (Ah-Fong et al. 2008). Using inf1 as a target, Ah-Fong et al. (2008) compared three method including PEG treatment of protoplasts, zoospore electroporation, and microprojectile bombardment and they found that hairpin vectors combined with protoplast transformation was the highest to silence genes. Gene silencing has been used to analyze a number of genes in oomycete, including P. parasitica. For example, the suppressed expression of cellulose-binding elicitor lectin (CBEL) in transgenic strains of P. parasitica caused severe impartation in adhesion of the pathogen to the cellophane membrane, differentiation of lobed structures in contact with cellophane, and formation of branched aggregating hyphae (Gaulin et al. 2002). The transformants silenced with PnDLCl in P. parasitica (Narayan et al. 2010) released nonflagellate, nonmotile zoospores from their sporangia. High level (more than 80% reduction) of PnPMA1 silencing in P. parasitica led to the production of nonflagellate and large aberrant zoospores, rapid transition from zoospores to cysts, and a decreased germination rate of cysts, indicating that PnPMAl plays important roles in zoospore development (Zhang et al. 2012).
Genomics
A large-insert bacterial artificial chromosome (BAC) library (Shan and Hardham 2004) using nuclear DNA from P. parasitica was constructed. The library contains 10,752 clones with an average insert size of 90 kb and is free of mitochondrial DNA. The genome size of P. parasitica was estimated to be 95.5 Mb by the analysis using several DNA probes and physical mapping. The BAC library provides important resource for genome analysis of genes of P. parasitica and is useful for the genome reconstruction.
Expressed sequence tags (EST) analysis is a useful approach to get understanding of the basic biology and interaction with host plant of pathogen. Several EST libraries have been generated from different stages including mycelium (Panabières et al. 2005), zoospores (Škalamera et al. 2004), germinated cysts (Shan et al. 2004b), penetration process (Kebdani et al. 2010), late infection stage (Le Berre et al. 2008), and other culture conditions (Rosa et al. 2007) of P. parasitica. The EST resources of different developmental stages of P. parasitica were shown to be important for the identification of specific pathogenicity-related genes, for example, over 300 clones representing 146 unigenes were identified by upregulated expression in germinated cysts (Shan et al. 2004b), more than 400 clones representing 240 genes were shown to be preferentially expressed in zoospores (Škalamera et al. 2004) in the two small EST collections, and 60% of the appressorium-derived sequences were not present in other P. parasitica EST collections (Kebdani et al. 2010). Sequencing of two infection libraries of different stages showed about 9% (147/1689) and 2% (42/2022) of P. parasitica unigenes, respectively, that gave no significant hits in the genome sequences of P. infestans, P. sojae, and P. ramorum (Le Berre et al. 2008; Kebdani et al. 2010). Data from the ESTs of different infection and development stages assist to provide information for future functional analysis and to understand the genetics and physiology of P. parasitica.
The draft genome sequence of P. parasitica has been completed (https://olive.broadinstitute.org/projects/phytophthora_parasitica). The genome sequence of P. parasitica includes about 23,121 predicted genes within the 82-Mb genome compared to 18,178 genes for the 240-Mb genome of P. infestans, 16,988 genes for the 95-Mb genome of P. sojae (Judelson 2012). The number of predicted genes is more than the two narrow host range species P. infestans and P. sojae, implicating possible relation to its capability to infect large number of plant species. The P. parasitica genome project also involves sequencing of multiple isolates isolated from diverse host plants and geographic distant locations, presumably with diverse genetic backgrounds. One of the goals is to make a comparative genomic study to identify genes that determine host range in P. parasitica.
By now, genome sequences have been available for several oomycetes including P. capsici (Lamour et al. 2012), Phytophthora cinnamomi, P. infestans (Haas et al. 2009), P. parasitica, P. ramorum (Tyler et al. 2006), P. sojae (Tyler et al. 2006), Pythium ultimum (Lévesque et al. 2010), H. arabidopsidis (Baxter et al. 2010), Albugo laibachii (Kemen et al. 2011), and Saprolegnia parasitica (Judelson 2012). The sequenced oomycete pathogens genome size is from 37-Mb to 280-Mb and the predicted gene contents range from about 13,000 to 26,000 genes (Judelson 2012). The huge resources of the genome sequences provide more information for answering the questions about the biology, evolution, and pathogenesis of oomycete pathogens. One example is that the sheer number of protein effectors has been uncovered, some of which are crucial for pathogenesis.
The host specificity
P. parasitica is a species complex, capable of infecting numerous plant species, including model plants Nicotiana tabacum and A. thaliana. This allows development of tractable pathosystems for the study of the interaction between P. parasitica and host plants in the laboratory.
P. parasitica caused black shank disease of tobacco worldwide. It can attack all parts of tobacco including roots, stems, and leaves at any growing stages. As its name, the most common symptom of the disease is the black base or shank of the stalk. Infection is usually through the roots. The roots are initially water soaked then rapidly become necrotic. In young plants, the stems become brown to black and the darkening can extend up the stalk several centimeters down into root system, then the plants damp off. The pathogen can directly infect the leaves, brown-to-black or large circular lesions occur following wet weather. The disease can be devastating to tobacco in the greenhouse as well as in the field (Stavely 1979). The cytological observations between P. parasitica and tobacco have also been described (Benhamou and Côté 1992; Séjalon-Delmas et al. 1997; Bottin et al. 1999).
P. parasitica interacts with its host tobacco in a race-cultivar specific manner. Races are defined by the ability of the pathogen to infect various cultivars carrying different resistance genes. The knowledge of resistant resources of tobacco to P. parasitica is limited, mainly including the oligogenic black shank resistance of the cigar-wrapper N. tabacum cv. Florida 301, dominant and monogenic resistance from Nicotiana plumbaginifolia Viv and Nicotiana longiflora Cav, the cigar-type tobacco cv. Beinhart 1000-1, and the flue-cured cultivar Coker 371-Gold (Van Jaarsveld et al. 2002; Antonopoulos et al. 2010).
There are four races (race 0, 1,2, and 3) reported for P. parasitica. Race 0 is defined as being nonpathogenic on N. plumbaginifolia Viv while race 1 is pathogenic (Apple 1962). The evidence indicated that a single dominant gene controlled the resistance to race 0 (Goins and Apple 1970). Race 2 (Lamprecht 1973; Stavely 1979) was defined in South Africa by the differentiated reaction of tobacco ev. Delerest 202, which is resistant to race 2 but susceptible to race 0 and 1. Race 3 (Taylor 1975; McIntyre and Taylor 1978) can overcome resistance in cigar-wrapper tobacco and is tolerant to cold temperature.
The losses of black shank caused by P. parasitica are severe in worldwide tobacco production although many management programs including cultural practices, host resistance, and chemical treatments are deployed. Compared to other methods, the most effective approach to manage black shank is to use cultivars with high levels of resistance to all P. parasitica races. So there is a crying need for new resistance resources for P. parasitica.
The model pathosystem
To better understand the interaction between a pathogen and its host, knowledge obtained from a good model pathosystem is necessary. Compared with the other host plant, A. thaliana is more efficient for accelerated understanding of Phytophthora biology and pathology. The fabulous wealth of Arabidopsis, including its genomic resources, mutant collections, natural ecotypes, and many associated genetic and molecular tools, allowed researchers to obtain fundamental knowledge on understanding molecular and cellular mechanisms in plants interacting with pathogens, notably the pathogen-associated molecular patterns (PAMPs) perception and PAMP-triggered immunity (PTI), nucleotide-binding and leucine-rich repeat domains (NB-LRR) based disease resistance and effector-triggered immunity (ETI), vesicle transport and polarized cellular defense responses, transcriptional output networks, and the interplay between disease resistance and hormone signaling pathways (Nishimura and Dangl 2010).
Attard et al. (2010) and Wang et al. (2011) described the compatible interaction between A. thaliana and P. parasitica, respectively. Cytological characterization showed that both the roots and leaves of Arabidopsis are susceptible to P. paracitica infection, as evidenced by development of water-soaked lesions, extensive pathogen colonization, and formation of abundant haustoria-like structures. The infection process is similar with the natural host, tomato (Le Berre et al. 2008). However, the disease severities were differential, dependent on the ecotypes of A. thaliana and strains of P. parasitica, indicating the presence of natural variation in host specificity between A. thaliana and P. parasitica (Wang et al. 2011). Moreover, the A. thaliana mutants with impaired salicylic acid (SA), jasmonic acid (JA), and ethylene (ET) signaling pathways are more susceptible than the wild type and also the transcripts for marker genes are transient accumulated (Attard et al. 2010). These results suggest that the JA, SA, and ET signaling pathways are all involved in the defense against P. parasitica which is different from three other described Phytophthora species that infect Arabidopsis, including P. brassicae (Roetschi et al. 2001; Schlaeppi et al. 2010), P. cinnamomi (Rookes et al. 2008), and P. capsici (Wang et al. 2013).
The model pathosystem was successfully used to answer interesting questions. For example, Zhang et al. (2011) examined the effect of transgenic expression of dsRNAs on the expression of homologous genes in the invading and colonizing oomycete pathogen. The results suggested that the oomycete pathogens might lack the genetic machinery required for uptake of external silencing signals, in particular dsRNAs, during biotrophic interaction. Diévart et al. (2011) used the pathosystem to analyze the expression of leucine-rich repeat receptor kinases (LRR-RK) genes, and the expression data suggest that oomycete LRR-RKs may play a role in several stages of the oomycete life cycle. Jaouannet et al. (2013) showed that a calreticulin from Meloidogyne incognita (Mi-CRT) plays an important role in infection success. They used the model pathosystem to test the susceptibility of stably transformed A. thaliana plants that express the secreted form of Mi-CRT to P. parasitica. Larroque et al. (2013) investigated the role of CBEL-triggered immunity, which benefit A. thaliana mutants and natural ecotypes, and also the specific interaction between A. thaliana and P. parasitica.
The molecular basis of pathogenesis
The intimate attachment to host cells enables the parasitic pathogen to acquire nutrients easily; however, the relatively conserved components such as PAMPs or danger-associated molecular patterns from pathogens become stimuli of pattern recognition receptors-mediated resistance of plants. The successful pathogen is able to effectively evade or suppress PTI by secretion of a set of effectors, and initiates the disease in the plant. Some effectors could be specifically targeted by resistant proteins in plants and activates ETI (Jones and Dangl 2006).
In recent years, many researches have focused on effectors, which are key virulence factors of pathogens. Effectors are molecules and typically proteins secreted by the pathogen to manipulate host cell structure and function thereby facilitating infection and colonization (Kamoun 2006). Effectors can be classified in two groups according to their subcellular localization. The apoplastic effectors are released into the plant extracellular space and the cytoplasmic effectors are translocated inside the plant cell. Up to now, a number of effectors have been reported (Kamoun 2006), including enzyme inhibitors, small cysteine-rich proteins, Nepl-like family, two large classes of cytoplasmic effectors RXLR and CRN, as well as YxSL motif containing proteins in P. ultimum (Lévesque et al. 2010), and CHXC effectors in Albugo (Kemen et al. 2011). The described effectors of the pathogen play numerous and essential roles in the infection stage (Ali and Bakkeren 2011), such as disarming plant defense enzymes, suppressing host immunity or killing host cells.
The elicitors, like the elicitin gene ParA1 (Kamoun et al. 1993), which are likely PAMP molecules, were identified and well characterized in P. parasitica. Other reported effectors include NEPl-Like protein NPP1 (Fellbrich et al. 2002), and the gene family encoding apoplastic polygalacturonases (Yan and Liou 2005; Wu et al. 2008). One of the apoplastic effectors is CBEL, firstly purified from P. parasitica cell wall. It induces strong defense reactions when infiltrated into leaf tissue of plant species including tobacco and Arabidopsis (Mateos et al. 1997; Séjalon-Delmas et al. 1997; Khatib et al. 2004) and it is necessary for the structure of the hyphal cell wall and attachment to cellulosic substrates such as plant surfaces (Gaulin et al. 2002). CBEL harbors a duplication of two types of domains (Tordai et al. 1999), and it has been detected in many oomycete species (Links et al. 2011).
Recently, a RXLR effector of P. parasitica PSE1, identified in a cDNA library for the penetrating stage of P. parasitica (Kebdani et al. 2010), was proved to favor the pathogen infection by modulating the auxin accumulation during the penetration process (Evangelisti et al. 2013).
Moreover, some RXLR effectors have been proven avirulence functions. Since the first oomycete Avr gene Avr 1b in P. sojae cloned (Shan et al. 2004a), a number of Avr genes have obtained, mainly from species of P. sojae, P. infestans, and H. arabidopsidis (Stassen and Van den Ackerveken 2011). However, little is known about the effector functions in P. parasitica and nothing about the Avr genes of this important pathogen, though host-genotype specificity in P. parasitica is notable.
Many methods have been designed to discover the effectors including Avr proteins, such as positional cloning, bioinformatic prediction, in planta expression, or some methods combined. Understanding what kind of effectors in P. parasitica and how effectors perturb host processes will be the major themes in the study of this pathogen.
Summary
Driven by fundamental questions in oomycete evolution and pathology, P. parasitica emerged to be a model oomycete pathogen for understanding pathogenesis and host-pathogen interaction. The available genetic manipulation, abundant genetic and genomic resources of P. parasitica and its compatible interaction with the model plant A. thaliana will help understand fundamental questions, like the genetic basis of host range in the pathogen, effectors and their roles in pathogenesis, molecular dissection of effector function. The model study of P. parasitica, which represents the majority species of Phytophthora, will accelerate understanding of molecular plant-oomycete interactions and provide insight into novel disease-control strategies.
Acknowledgments
This research was supported by the Natural Science Foundation of China (grant number 31125022) and Chongqing Tobacco Major Program of Integrated Pest management (grant number NY20140401070002) We would like to thank Dr. Biao Gu and Qinhu Wang for the critical reading of the manuscript.
References
- Ah-Fong A, Bormann-Chung CA, Judelson HS. Optimization of transgene-mediated silencing in Phytophthora infestans and its association with small-interfering RNAs. Fungal Genet Biol. 2008;45(8):1197–1205. doi: 10.1016/j.fgb.2008.05.009. [DOI] [PubMed] [Google Scholar]
- Ah-Fong A, Judelson HS. Vectors for fluorescent protein tagging in Phytophthora: tools for functional genomics and cell biology. Fungal Biol. 2011;115(9):882–890. doi: 10.1016/j.funbio.2011.07.001. [DOI] [PubMed] [Google Scholar]
- Ali S, Bakkeren G. Fungal and oomycete effectors-strategies to subdue a host. Can J Plant Pathol. 2011;33(4):425–46. doi: 10.1080/07060661.2011.625448. [DOI] [Google Scholar]
- Antonopoulos DF, Melton T, Mila AL. Effects of chemical control, cultivar resistance, and structure of cultivar root system on black shank incidence of tobacco. Plant Dis. 2010;94(5):613–620. doi: 10.1094/PDIS-94-5-0613. [DOI] [PubMed] [Google Scholar]
- Apple J. Physiological specialization within Phytophthora parasitica var. nicotianae. Phytopathology. 1962;52(4):351–354. [Google Scholar]
- Attard A, Gourgues M, Callemeyn-Torre N, Keller H. The immediate activation of defense responses in Arabidopsis roots is not sufficient to prevent Phytophthora parasitica infection. New Phytol. 2010;187(2):449–460. doi: 10.1111/j.1469-8137.2010.03272.x. [DOI] [PubMed] [Google Scholar]
- Baxter L, Tripathy S, Ishaque N, Boot N, Cabral A, Kemen E, Thines M, Ah-Fong A, Anderson R, Badejoko W, et al. Signatures of adaptation to obligate biotrophy in the Hyaloperonospora arabidopsidis genome. Science. 2010;330(6010):1549–1551. doi: 10.1126/science.1195203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benhamou N, Côté F. Ultrastructure and cytochemistry of pectin and cellulose degradation in tobacco roots infected by Phytophthora parasitica var. nicotianae. Phytopathology. 1992;82(4):468–478. doi: 10.1094/Phyto-82-468. [DOI] [Google Scholar]
- Blair IE, Coffey MD, Park S-Y, Geiser DM, Kang S. A multilocus phylogeny for Phytophthora utilizing markers derived from complete genome sequences. Fungal Genet Biol. 2008;45(3):266–277. doi: 10.1016/j.fgb.2007.10.010. [DOI] [PubMed] [Google Scholar]
- Bottin A, Larche L, Villalba F, Gaulin E, Esquerré-Tugayé MT, Rickauer M. Green fluorescent protein (GFP) as gene expression reporter and vital marker for studying development and microbe-plant interaction in the tobacco pathogen Phytophthora parasitica var. nicotianae. FEMS Microbiol Lett. 1999;176(1):51–56. doi: 10.1111/j.1574-6968.1999.tb13641.x. [DOI] [PubMed] [Google Scholar]
- Cvitanich C, Judelson HS. Stable transformation of the oomycete, Phytophthora infestans, using microprojectile bombardment. Curr Genet. 2003;42(4):228–235. doi: 10.1007/s00294-002-0354-3. [DOI] [PubMed] [Google Scholar]
- Diévart A, Gilbert N, Droc G, Attard A, Gourgues M, Guiderdoni E, Perm C. Leucine-rich repeat receptor kinases are sporadically distributed in eukaryotic genomes. BMC Evol Biol. 2011;11(1):367. doi: 10.1186/1471-2148-11-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evangelisti E, Govetto B, Minet-Kebdani N, Kuhn ML, Attard A, Ponchet M, Panabières F, Gourgues M. The Phytophthora parasitica RXLR effector penetration-specific effector 1 favours Arabidopsis thaliana infection by interfering with auxin physiology. New Phytol. 2013;199(2):476–189. doi: 10.1111/nph.12270. [DOI] [PubMed] [Google Scholar]
- Fellbrich G, Romanski A, Varet A, Blume B, Brunner F, Engelhardt S, Felix G, Kemmerling B, Krzymowska M, Nürnberger T. NPP1, a Phytophthora-associated trigger of plant defense in parsley and arabidopsis. Plant J. 2002;32(3):375–390. doi: 10.1046/j.1365-313X.2002.01454.x. [DOI] [PubMed] [Google Scholar]
- Gaulin E, Jauneau A, Villalba F, Rickauer M, Esquerré-Tugayé M-T BA. The CBEL glycoprotein of Phytophthora parasitica var. nicotianae is involved in cell wall deposition and adhesion to cellulosic substrates. J Cell Sci. 2002;115(23):4565–4575. doi: 10.1242/jcs.00138. [DOI] [PubMed] [Google Scholar]
- Goins RB, Apple J. Inheritance and phenotypic expression of a dominant factor for black shank resistance from Nicotiana plumbaginifolia in a Nicotiana tabacum milieu. Tob Sci. 1970;14:7–11. [Google Scholar]
- Grünwald NJ, Garbelotto M, Goss EM, Heungens K, Prospero S. Emergence of the sudden oak death pathogen Phytophthora ramorum. Trends Microbiol. 2012;20(3):131–138. doi: 10.1016/j.tim.2011.12.006. [DOI] [PubMed] [Google Scholar]
- Haas BJ, Kamoun S, Zody MC, Jiang RH, Handsaker RE, Cano LM, Grabherr M, Kodira CD, Raffaele S, Torto-Alalibo T. Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature. 2009;461(7262):393–398. doi: 10.1038/nature08358. [DOI] [PubMed] [Google Scholar]
- Hickman C. Presidential address: Phytophthora—plant destroyer. Trans Br Mycological Soc. 1958;41(1):1–13. doi: 10.1016/S0007-1536(58)80001-7. [DOI] [Google Scholar]
- Huitema E, Smoker M, Kamoun S, McDowell JM. A straightforward protocol for electro-transformation of Phytophthora capsici zoospores. Plant Immun. 2011;712:129–135. doi: 10.1007/978-1-61737-998-7_11. [DOI] [PubMed] [Google Scholar]
- Jaouannet M, Magliano M, Arguel M, Gourgues M, Evangelisti E, Abad P, Rosso M. The root-knot nematode calreti-culin Mi-CRT is a key effector in plant defense suppression. Mol Plant Microbe Interact. 2013;26(1):97–105. doi: 10.1094/MPMI-05-12-0130-R. [DOI] [PubMed] [Google Scholar]
- Jones JD, Dangl JL. The plant immune system. Nature. 2006;444(7117):323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- Judelson HS. Dynamics and innovations within oomycete genomes: insights into biology, pathology, and evolution. Eukaryotic Cell. 2012;11(11):1304–1312. doi: 10.1128/EC.00155-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judelson HS, Coffey MD, Arredondo FR, Tyler BM. Transformation of the oomycete pathogen Phytophthora megasperma f sp. glycinea occurs by DNA integration into single or multiple chromosomes. Curr Genet. 1993;23(3):211–218. doi: 10.1007/BF00351498. [DOI] [PubMed] [Google Scholar]
- Judelson HS, Tyler BM, Michelmore RW. Transformation of the oomycete pathogen, Phytophthora infestans. Mol Plant Microbe Interact. 1991;4(6):602–607. doi: 10.1094/MPMI-4-602. [DOI] [PubMed] [Google Scholar]
- Kamoun S. A catalogue of the effector secretome of plant pathogenic oomycetes. Annu Rev Phytopathol. 2006;44:41–60. doi: 10.1146/annurev.phyto.44.070505.143436. [DOI] [PubMed] [Google Scholar]
- Kamoun S, Klucher KM, Coffey MD, Tyler BM. A gene encoding A host-specific elicitor protein of Phytophthora parasitica. Mol Plant Microbe Interact. 1993;6(5):573–581. doi: 10.1094/MPMI-6-573. [DOI] [PubMed] [Google Scholar]
- Kamoun S, van West P, Govers F. Quantification of late blight resistance of potato using transgenic Phytophthora infestans expressing β-glucuronidase. Eur J Plant Pathol. 1998;104(5):521–525. doi: 10.1023/A:1008698620906. [DOI] [Google Scholar]
- Kebdani N, Pieuchot L, Deleury E, Panabières F, Le Berre JY, Gourgues M. Cellular and molecular characterization of Phytophthora parasitica appressorium-mediated penetration. New Phytol. 2010;185(1):248–257. doi: 10.1111/j.1469-8137.2009.03048.x. [DOI] [PubMed] [Google Scholar]
- Kemen E, Gardiner A, Schultz-Larsen T, Kemen AC, Balmuth AL, Robert-Seilaniantz A, Bailey K, Holub E, Studholme DJ, MacLean D, et al. Gene gain and loss during evolution of obligate parasitism in the white rust pathogen of Arabidopsis thaliana. Plos Biol. 2011;9(7):e1001094. doi: 10.1371/journal.pbio.1001094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatib M, Lafitte C, Esquerré-Tugayé MT, Bottin A, Rickauer M. The CBEL elicitor of phytophthora parasitica var. nicotianae activates defence in Arabidopsis thaliana via three different signalling pathways. New Phytol. 2004;162(2):501–510. doi: 10.1111/j.1469-8137.2004.01043.x. [DOI] [Google Scholar]
- Ko W. Reversible change of mating type in Phytophthora parasitica. J Gen Microbiol. 1981;125(2):451–154. [Google Scholar]
- Kroon LP, Brouwer H, de Cock AW, Govers F. The genus Phytophthora anno 2012. Phytopathology. 2012;102(4):348–364. doi: 10.1094/PHYTO-01-11-0025. [DOI] [PubMed] [Google Scholar]
- Lamour KH, Mudge J, Gobena D, Hurtado-Gonzales OP, Schmutz J, Kuo A, Miller NA, Rice BJ, Raffaele S, Cano LM, et al. Genome sequencing and mapping reveal loss of heterozygosity as a mechanism for rapid adaptation in the vegetable pathogen Phytophthora capsici. Mol Plant Microbe Interact. 2012;25(10):1350–1360. doi: 10.1094/MPMI-02-12-0028-R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamprecht MP. Breeding flue-cured tobacco resistant to South African black shank (Phytophthora nicotianae (B. de Haan) var. nicotianae) Agroplantae. 1973;5(3):67–72. [Google Scholar]
- Larroque M, Belmas E, Martinez T, Vergnes S, Ladouce N, Lafitte C, Gaulin E, Dumas B. Pathogen-associated molecular pattern-triggered immunity and resistance to the root pathogen Phytophthora parasitica in arabidopsis. J Exp Bot. 2013;64(12):3615–3625. doi: 10.1093/jxb/ert195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latijnhouwers M, De Wit PJ, Govers F. Oomycetes and fungi: similar weaponry to attack plants. Trends Microbiol. 2003;11(10):462–469. doi: 10.1016/j.tim.2003.08.002. [DOI] [PubMed] [Google Scholar]
- Le Berre JY, Engler G, Panabières F. Exploration of the late stages of the tomato-Phytophthora parasitica interactions through histological analysis and generation of expressed sequence tags. New Phytol. 2008;177(2):480–492. doi: 10.1111/j.1469-8137.2007.02269.x. [DOI] [PubMed] [Google Scholar]
- Lévesque CA, Brouwer H, Cano L, Hamilton JP, Holt C, Huitema E, Raffaele S, Robideau GP, Thines M, Win J, et al. Genome sequence of the necrotrophic plant pathogen Pythium ultimum reveals original pathogenicity mechanisms and effector repertoire. Genome Biol. 2010;11(7):R73. doi: 10.1186/gb-2010-11-7-r73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Links MG, Holub E, Jiang RH, Sharpe AG, Hegedus D, Beynon E, Sillito D, Clarke WE, Uzuhashi S, Borhan MH. De novo sequence assembly of Albugo candida reveals a small genome relative to other biotrophic oomycetes. BMC Genomics. 2011;12(1):503. doi: 10.1186/1471-2164-12-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateos FV, Rickauer M, Esquerré-Tugayé M-T. Cloning and characterization of a cDNA encoding an elicitor of Phytophthora parasitica var. nicotianae that shows cellulose-binding and lectin-like activities. Mol Plant Microbe Interact. 1997;10(9):1045–1053. doi: 10.1094/MPMI.1997.10.9.1045. [DOI] [PubMed] [Google Scholar]
- McIntyre JL, Taylor G. Race 3 of Phytophthora parasitica var. nicotianae. Phytopathology. 1978;68(1):35–38. doi: 10.1094/Phyto-68-35. [DOI] [Google Scholar]
- McLeod A, Fry BA, Zuluaga AP, Myers KL, Fry WE. Toward improvements of oomycete transformation protocols. J Eukaryot Microbiol. 2008;55(2):103–109. doi: 10.1111/j.1550-7408.2008.00304.X. [DOI] [PubMed] [Google Scholar]
- Mort-Bontemps M, Fvre M. Transformation of the oomycete Saprolegnia monoïca to hygromycin-b resistance. Curr Genet. 1997;31(3):272–275. doi: 10.1007/s002940050205. [DOI] [PubMed] [Google Scholar]
- Narayan RD, Blackman LM, Shan WX, Hardham AR. Phytophthora nicotianae transformants lacking dynein light chain 1 produce non-flagellate zoospores. Fungal Genet Biol. 2010;47(8):663–671. doi: 10.1016/j.fgb.2010.04.008. [DOI] [PubMed] [Google Scholar]
- Nishimura MT, Dangl JL. Arabidopsis and the plant immune system. Plant J. 2010;61(6):1053–1066. doi: 10.1111/j.1365-313X.2010.04131.X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panabières F, Amselem J, Galiana E, Le Berre J-Y. Gene identification in the oomycete pathogen Phytophthora parasitica during in vitro vegetative growth through expressed sequence tags. Fungal Genet Biol. 2005;42(7):611–623. doi: 10.1016/j.fgb.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Roetschi A, Si-Ammour A, Belbahri L, Mauch F, Mauch-Mani B. Characterization of an Arabidopsis-Phytophthora pathosystem: resistance requires a functional PAD2 gene and is independent of salicylic acid, ethylene and jasmonic acid signalling. Plant J. 2001;28(3):293–305. doi: 10.1046/j.1365-313X.2001.01148.X. [DOI] [PubMed] [Google Scholar]
- Rookes JE, Wright ML, Cahill DM. Elucidation of defence responses and signalling pathways induced in Arabidopsis thaliana following challenge with Phytophthora cinnamomi. Physiol Mol Plant Pathol. 2008;72(4-6):151–161. doi: 10.1016/j.pmpp.2008.08.005. [DOI] [Google Scholar]
- Rosa DD, Campos MA, Targon MLP, Souza AA. Phytophthora parasitica transcriptome, a new concept in the understanding of the citrus gummosis. Genet Mol Biol. 2007;30(3):997–1008. doi: 10.1590/S1415-47572007000500028. [DOI] [Google Scholar]
- Schlaeppi K, Abou-Mansour E, Buchala A, Mauch F. Disease resistance of Arabidopsis to Phytophthora brassicae is established by the sequential action of indole glucosino-lates and camalexin. Plant J. 2010;62(5):840–851. doi: 10.1111/j.1365-313X.2010.04197.X. [DOI] [PubMed] [Google Scholar]
- Séjalon-Delmas N, Mateos FV, Bottin A, Rickauer M, Dargent R, Esquerré-Tugayé M. Purification, elicitor activity, and cell wall localization of a glycoprotein from Phytophthora parasitica var. nicotianae, a fungal pathogen of tobacco. Phytopathology. 1997;87(9):899–909. doi: 10.1094/PHYTO.1997.87.9.899. [DOI] [PubMed] [Google Scholar]
- Shan WX, Cao M, Leung D, Tyler BM. The Avr1b locus of Phytophthora sojae encodes an elicitor and a regulator required for avirulence on soybean plants carrying resistance gene rps1b. Mol Plant Microbe Interact. 2004a;17(4):394–403. doi: 10.1094/MPMI.2004.17.4.394. [DOI] [PubMed] [Google Scholar]
- Shan WX, Hardham AR. Construction of a bacterial artificial chromosome library, determination of genome size, and characterization of an Hsp70 gene family in Phytophthora nicotianae. Fungal Genet Biol. 2004;41(3):369–380. doi: 10.1016/j.fgb.2003.11.012. [DOI] [PubMed] [Google Scholar]
- Shan WX, Marshall J, Hardham A. Gene expression in germinated cysts of Phytophthora nicotianae. Mol Plant Pathol. 2004b;5(4):317–330. doi: 10.1111/j.1364-3703.2004.00231.x. [DOI] [PubMed] [Google Scholar]
- Si-Ammour A, Mauch-Mani B, Mauch F. Quantification of induced resistance against Phytophthora species expressing GFP as a vital marker: beta-aminobutyric acid but not BTH protects potato and Arabidopsis from infection. Mol Plant Pathol. 2003;4(4):237–248. doi: 10.1046/j.1364-3703.2003.00168.x. [DOI] [PubMed] [Google Scholar]
- Škalamera D, Wasson A, Hardham A. Genes expressed in zoospores of Phytophthora nicotianae. Mol Genet Genomics. 2004;270(6):549–557. doi: 10.1007/s00438-003-0946-8. [DOI] [PubMed] [Google Scholar]
- Stassen JH, Van den Ackerveken G. How do oomycete effectors interfere with plant life? Curr Opin Plant Biol. 2011;14(4):407–414. doi: 10.1016/j.pbi.2011.05.002. [DOI] [PubMed] [Google Scholar]
- Stavely JR. Disease resistance in Nicotiana: procedures for experimental use. USDA Tech Bull. 1979;1586:87–110. [Google Scholar]
- Taylor GS. Cold tolerance of Phytophthora parasitica var nicotianae isolated from tobacco in Connecticut. Plant Dis Rep. 1975;59(3):249–252. [Google Scholar]
- Tordai H, Bányai L, Patthy L. The PAN module: the N-terminal domains of plasminogen and hepatocyte growth factor are homologous with the apple domains of the pre-kallikrein family and with a novel domain found in numerous nematode proteins. FEBS Lett. 1999;461(1–2):63–67. doi: 10.1016/S0014-5793(99)01416-7. [DOI] [PubMed] [Google Scholar]
- Tyler BM, Tripathy S, Zhang X, Dehal P, Jiang RH, Aerts A, Arredondo FD, Baxter L, Bensasson D, Beynon JL. Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science. 2006;313(5791):1261–1266. doi: 10.1126/science.1128796. [DOI] [PubMed] [Google Scholar]
- Van Jaarsveld E. Phytophthora nicotianae on tobacco and its control in South Africa [dissertation] Pretoria: University of Pretoria; 2001. [Google Scholar]
- Van Jaarsveld E, Wingfield M, Drenth A. Evaluation of tobacco cultivars for resistance to races of Phytophthora nicotianae in South Africa. J Phytopathol. 2002;150(8-9):456–462. doi: 10.1046/j.1439-0434.2002.00766.x. [DOI] [Google Scholar]
- van West P, Kamoun S, van't Klooster JW, Govers F. Internuclear gene silencing in Phytophthora infestans. Mol Cell. 1999a;3(3):339–348. doi: 10.1016/S1097-2765(00)80461-X. [DOI] [PubMed] [Google Scholar]
- van West P, Reid B, Campbell TA, Sandrock RW, Fry WE, Kamoun S, Gow NA. Green fluorescent protein (GFP) as a reporter gene for the plant pathogenic oomycete Phytophthora palmivora. FEMS Microbiol Lett. 1999b;178(1):71–80. doi: 10.1111/j.1550-7408.2008.00376.x. [DOI] [PubMed] [Google Scholar]
- Vijn I, Govers F. Agrobacterium tumefaciens mediated transformation of the oomycete plant pathogen Phytophthora infestans. Mol Plant Pathol. 2003;4(6):459–467. doi: 10.1046/j.1364-3703.2003.00191.x. [DOI] [PubMed] [Google Scholar]
- Wang Y, Bouwmeester K, van de Mortel JE, Shan WX, Govers F. A novel arabidopsis-oomycete pathosystem: differential interactions with Phytophthora capsici reveal a role for camalexin, indole glucosinolates and salicylic acid in defence. Plant Cell Environ. 2013;36(6):1192–1203. doi: 10.1111/pce.12052. [DOI] [PubMed] [Google Scholar]
- Wang Y, Meng YL, Zhang M, Tong XM, Wang QH, Sun YY, Quan JL, Govers F, Shan WX. Infection of Arabidopsis thaliana by Phytophthora parasitica and identification of variation in host specificity. Mol Plant Pathol. 2011;12(2):187–201. doi: 10.1111/j.1364-3703.2010.00659.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner S, Steiner U, Becher R, Kortekamp A, Zyprian E, Deising HB. Chitin synthesis during in planta growth and asexual propagation of the cellulosic oomycete and obligate biotrophic grapevine pathogen Plasmopara vitícola. FEMS Microbiol Lett. 2002;208(2):169. doi: 10.1111/j.1574-6968.2002.tb11077.x. [DOI] [PubMed] [Google Scholar]
- Whisson SC, Boevink PC, Moleleki L, Avrova AO, Morales JG, Gilroy EM, Armstrong MR, Grouffaud S, van West P, Chapman S, et al. A translocation signal for delivery of oomycete effector proteins into host plant cells. Nature. 2007;450(7166):115–118. doi: 10.1038/nature06203. [DOI] [PubMed] [Google Scholar]
- Wu CH, Yan HZ, Liu LF, Liou RF. Functional characterization of a gene family encoding polygalacturonases in Phytophthora parasitica. Mol Plant Microbe Interact. 2008;21(4):480–489. doi: 10.1094/MPMI-21-4-0480. [DOI] [PubMed] [Google Scholar]
- Yan HZ, Liou RF. Cloning and analysis of pppg1, an inducible endopolygalacturonase gene from the oomycete plant pathogen Phytophthora parasitica. Fungal Genet Biol. 2005;42(4):339–350. doi: 10.1016/j.fgb.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Zhang MX, Meng YL, Wang QH, Liu DD, Quan JL, Hardham AR, Shan WX. PnPMA1, an atypical plasma membrane h+-atpase, is required for zoospore development in Phytophthora parasitica. Fungal Biol. 2012;116(9):1013–1023. doi: 10.1016/j.funbio.2012.07.006. [DOI] [PubMed] [Google Scholar]
- Zhang MX, Wang QH, Xu K, Meng YL, Quan JL, Shan WX. Production of dsRNA sequences in the host plant is not sufficient to initiate gene silencing in the colonizing oomycete pathogen Phytophthora parasitica. Plos One. 2011;6(11):e28114. doi: 10.1371/journal.pone.0028114. [DOI] [PMC free article] [PubMed] [Google Scholar]


