Abstract
The relationship between protein dynamics and function is a subject of considerable contemporary interest. Although protein motions are frequently observed during ligand binding and release steps, the contribution of protein motions to the catalysis of bond making/breaking processes is more difficult to probe and verify. Here, we show how the quantum mechanical hydrogen tunneling associated with enzymatic C–H bond cleavage provides a unique window into the necessity of protein dynamics for achieving optimal catalysis. Experimental findings support a hierarchy of thermodynamically equilibrated motions that control the H-donor and -acceptor distance and active-site electrostatics, creating an ensemble of conformations suitable for H-tunneling. A possible extension of this view to methyl transfer and other catalyzed reactions is also presented. The impact of understanding these dynamics on the conceptual framework for enzyme activity, inhibitor/drug design, and biomimetic catalyst design is likely to be substantial.
Keywords: enzyme catalysis, hydrogen tunneling, protein dynamics
INTRODUCTION
Rarely can the trajectory of a scientific discipline be traced to an anecdotal comment by an eminent researcher. This appears to be the case for enzymology, following Linus Pauling’s proposal that the enormous rate accelerations achieved by enzymes might arise from a preferential binding interaction between an enzyme and its substrate at the activated complex, commonly referred to as enhanced transition-state (TS) binding (1). This concept is the standard textbook explanation for the origin of enzyme catalysis and played a central role in enzyme-targeted drug design for several decades.
With the availability of many X-ray structures for enzymes and other proteins, examining protein-ligand interactions in great detail has been possible. This has raised the question of how the small changes in bond lengths, angles, and charges that are anticipated upon formation of a TS could account for the requisite differentiation in binding between the ground state and TS. An upper limit of 1025-fold has been estimated from experimental rate differences between enzymatic and solution reactions, and the true limit may be much higher (2). As an important test of the enhanced TS stabilization explanation, catalytic antibodies were generated in response to small, stable molecules designed to resemble the activated complex. Testing of these antibodies for activity with the corresponding substrate(s) generally led to fairly small rate enhancements relative to the corresponding reaction in solution. The reasons for this observation are complex: imperfect TS analogs used as haptens and the absence of sufficient active-site functional groups, together with an inflexible protein-fold architecture that is not well suited to catalysis (3). An apparent limiting component in the catalytic antibody design strategy was a focus on a rigid TS structure and omission of the dynamics needed to reach that state. The latter feature (i.e., the link of protein dynamics to function) has emerged as a central issue in enzymology. The resulting viewpoints regarding the role of protein dynamics are not yet universally agreed upon, although there is growing consensus about the key concepts.
Major distinguishing features of enzymes are their need to bring substrates from the solution to the active site and their need to release their final products to the bulk solvent while catalyzing chemical transformation. This implies two types of flexibility. The first enables the enzyme to accommodate a range of bound species (reactant, intermediates, TSs, and product), and the second is the conformational adjustments that bring new functional groups into the active site (such as loop closures subsequent to entry of the substrate into the active site) (4–6). There has been considerable discussion in the recent literature that focuses attention on conformational selection by substrates upon binding (7, 8), in contrast to the earlier proposal of induced fit, in which new conformational states are achieved only after binding (9, 10). Conformational selection implies a conformational landscape whose equilibrium position is shifted upon ligand binding. As increasingly emphasized, the role of conformational selection does not stop at the level of formation of the enzyme-substrate complex (E-S) but continues throughout the catalytic cycle, with only some fraction of protein conformers achieving substates that are optimal for the bond making and breaking steps of catalysis. One of the major challenges has been to obtain experimental evidence that demonstrates a link between such conformational sampling and enzymatic bond cleavage events. In this context, the study of H-tunneling has played a major role. This is because tunneling implies the ability of an H-donor and -acceptor to achieve short internuclear distances, focusing attention on a dynamic control of barrier width, in contrast to the classical focus on the reduction in barrier height as the origin of catalysis. Clarifying several concepts at the outset is useful (see sidebar, Clarification of Concepts Regarding Protein Dynamics).
The newer experimental observations and concepts relate back to the original proposal by Pauling. Both views rely on many active-site contacts and interactions between the enzyme and substrate. However, in contrast to the earlier focus on static protein structures and TS stabilization, the newer theories attribute the achievement of optimal catalytic states to a dynamical sampling within the protein population that transiently achieves reactant states.
EMPIRICAL DEVIATIONS AND RESULTING THEORETICAL MODELS FOR TUNNELING
Historical Context
Modern enzymology has relied heavily on the use of isotope effects to determine the nature of rate-limiting steps and, later, TS structure (17–19). The theory of isotope effects developed after World War II primarily focused on a semiclassical approximation that included changes in force constants (F) between the ground state and TS together with an impact of the reduced mass (µ) at the reacting bond on the frequency (ν) of the vibrating/reacting bond (Equation 1) (20):
| 1 |
Beginning in the 1980s, investigators noted experimental observations that could not be explained in these seemingly simple terms and introduced the possibility of quantum mechanical (QM) tunneling. The early tests for the contribution of H-tunneling in enzyme reactions relied heavily on the measurement of kinetic hydrogen isotope effects (KIEs) (21–23). The benefits of incorporating isotopes of hydrogen [protium, deuterium (D), and tritium (T)] into a stable, nonexchanging position of a substrate led to a focus on C–H bond cleavage via hydride, proton, and hydrogen atom transfer. The semiclassical approximation provided a set of expectations regarding the empirical rate ratios (i.e., kH/kD, kH/kT, and/or kD/kT), and these included an upper limit to the size of the KIE; a reasonable, well-defined interrelationship among multiple KIEs (e.g., the magnitude of kH/kT in relation to kD/kT); the temperature dependence of the KIE (24); and its pressure dependence (Table 1) (25). In direct contrast to the predictions in Table 1, accumulated data over more than 20 years now indicate consistent deviations in a direction indicative of QM tunneling.
Table 1.
Deviations from semiclassical predictions for KIEs
| Parameter | Predictiona | Observations | References |
|---|---|---|---|
| Primary H/D KIE | <7 | >100 | 149, 150 |
| Secondary hydrogen KIE | <Equilibrium limit | >Equilibrium limit | 21, 22 |
| Interrelationship of secondary KIEs | Determined by mass of isotope in labeled bond(s) | Secondary KIE is suppressed for D-transfer. | 23, 58, 143 |
| Temperature dependence of KIE | Extrapolated KIE close to unity at high temperature | Extrapolated KIE at high temperature can assume any value up to the experimentally observed one. | 31, 32, 38 |
| Pressure dependence of KIE | No impact | Variable | 56 |
Reference 24.
Tunneling
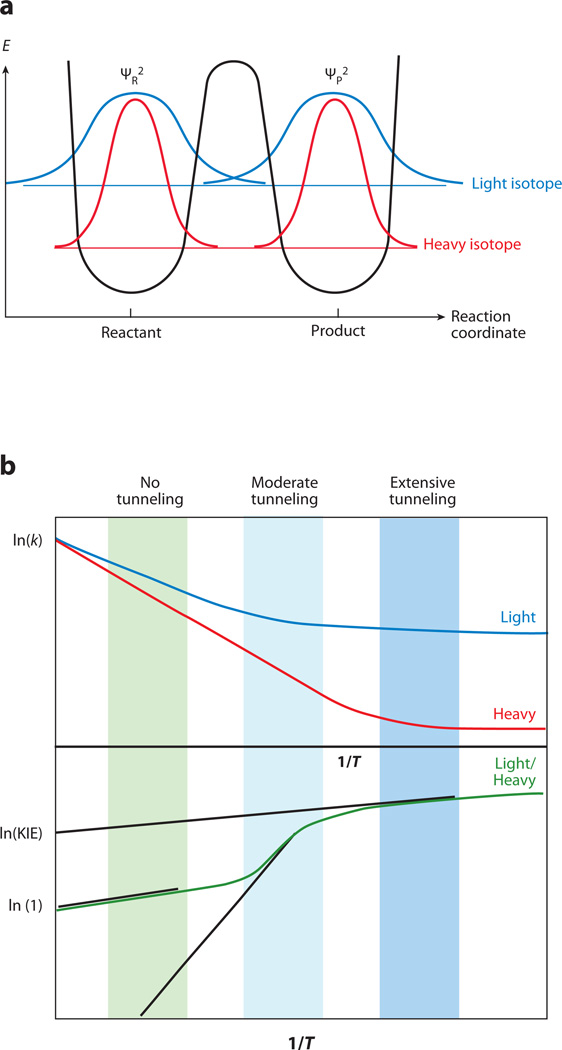
QM tunneling is a phenomenon in which a particle crosses a barrier between reactants and products owing to its wave-like nature. Tunneling occurs when the probability of finding a particle in the reactant well overlaps with the probability of finding the particle in the product well (Figure 1). The mass dependence of tunneling predicts that the lightest isotope can tunnel over longer distances (broader barriers) than heavier isotopes (Figure 1a).
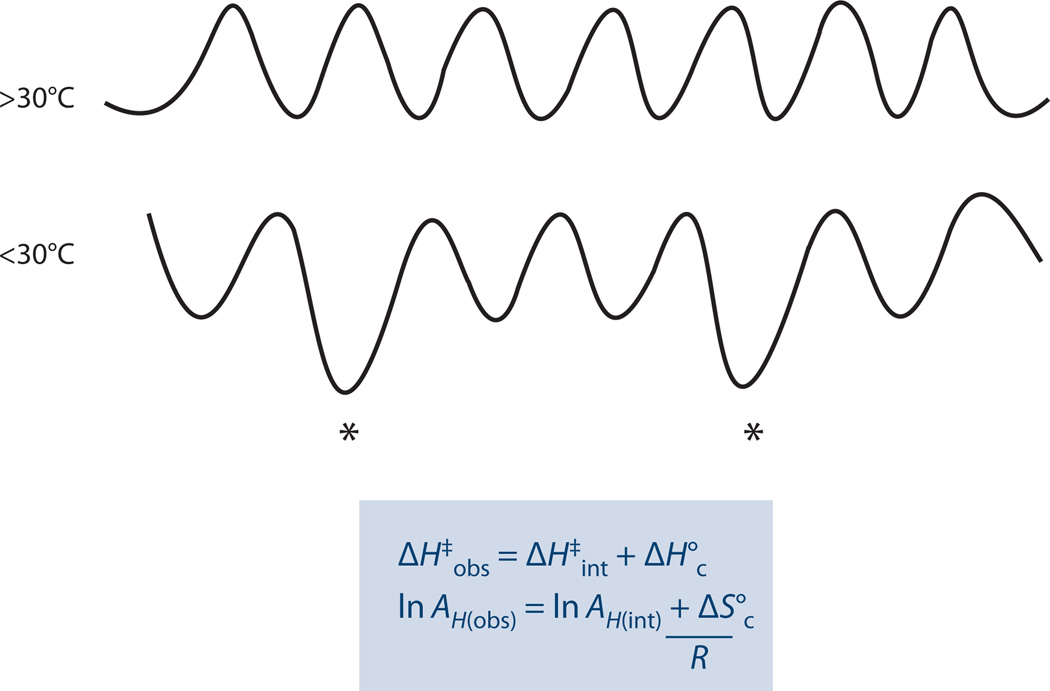
Figure 1.
(a) Illustration of a ground-state quantum mechanical tunneling for light and heavy isotopes. Reprinted with permission from Acc. Chem. Res. Copyright © 1998, American Chemical Society (26). (b) An Arrhenius plot of a hydrogen transfer applicable to a semiclassical transition-state (TS) model with a tunneling correction. Arrhenius plot of reaction rates for two isotopes, light (blue line) and heavy (red line). Arrhenius plot for their kinetic hydrogen isotope effects (KIEs) ( green line). The KIE on the Arrhenius pre-exponential factor (A) is an intercept of a tangent (black line) to a curve at the experimental temperature range. Highlighted regions indicate three distinct systems: no tunneling contribution, in which Al/Ah is close to unity; moderate tunneling contribution, resulting in Al/Ah smaller than 1; and extensive tunneling contribution, in which Al/Ah is larger than unity. Reproduced with permission from Chem. Biol. Copyright © 1999, American Chemical Society (59). Abbreviations: E, energy; P, product; R, reactant; ψ, wave function probability.
Tunneling as a correction to transition-state theory
An early model for H-tunneling, developed by Bell (27), introduced a multiplier term (Q) to the semiclassical rate constant. Q represents the probability that a particle will move through an inverse parabolic barrier (Equation 2):
| 2 |
where kSC is the semiclassical rate (no tunneling), W represents the energy of the particle, k is the Boltzmann constant, and T is the absolute temperature. Barrier penetration occurs below the classical TS, and its effect is predicted to be the most significant for the lightest isotope.
The temperature dependence of the rate can be written in the usual manner, leading to pre-exponential and exponential terms that vary depending on the extent of barrier penetration and the isotope being transferred:
| 3 |
where A is the Arrhenius prefactor, e is the exponent (natural base), Ea is the energy of activation, and h and l represent a heavy and light isotope, respectively. R is the gas constant and T is absolute temperature. For the majority of reactions in the condensed phase (including enzyme reactions), only a narrow experimental temperature range is available (0–100°C), such that plots of ln(k) versus 1/T often appear linear with slopes exponentially proportional to Ea (Figure 1b, moderate tunneling regime). This analysis provides a means to explain inflated KIEs arising from inflated isotopic differences in the energy of activation, ΔEa, and can explain inverse KIEs upon extrapolation of rate data to infinite temperature (i.e., an apparent Al/Ah less than unity). The behavior changes in the limit of very low temperatures, at which tunneling provides the only route to product formation, producing temperature-independent rates, enormously inflated KIEs, and apparent values for Al/Ah that can be much larger than unity (Figure 1b, extensive tunneling regime) (27).
In many studies of reactions of small molecules, TS models based on a rigid 1D potential surface, with or without a tunneling correction, can rationalize temperature-dependent KIEs (27). However, these models fail to explain the hallmark of enzyme-catalyzed C–H bond cleavages, that temperature-independent KIEs exist under conditions in which there is a significant energy of activation for the isotopically sensitive step (see below). A major shortcoming of the Bell tunneling correction and similar TS-based models is the exclusive focus on the hydrogen-transfer reaction coordinate, to the neglect of the contribution of motions from the heavy-atom environment. A successful treatment of H-tunneling can be accommodated only within a multidimensional treatment that includes the role of environmental/protein dynamics.
Full tunneling models
To accommodate both temperature-dependent and temperature-independent KIEs for reactions near room temperature that are characterized by measurable and often large enthalpic barriers, separating the temperature dependence of the KIE (ΔEa) from that of the reaction rate (Ea) is very useful. Phenomenological models constructed by both experimentalists and theoreticians capable of explaining such observations are often called Marcus-like models. These are an extension of the Marcus theory of electron tunneling (28), in which the tunneling particle is a hydrogen atom rather than an electron. This model, which has been described under different names [e.g., Marcus-like models (29, 30), environmentally coupled tunneling (31, 32), vibrationally enhanced tunneling (12)], proposes a mechanism whereby heavy atom reorganization leads to a tunneling-ready state (TRS). Within the TRS, the potential energy surfaces of the reactants and products become transiently degenerate, thereby permitting donor-acceptor wave function overlap.
Several groups have independently developed the functional form of the rate constant (k) for H-transfer reactions (33–38):
| 4 |
The terms in front of the integral sign resemble the standard Marcus theory (28) for the rate of reaching a TRS (Figure 2a) and contain the electronic coupling (V), the reorganization energy (λ), the reaction-driving force (ΔG°), and the absolute temperature (T). The constants, ℏ and kB, are the Planck constant over 2π and the Boltzmann constant, respectively. The term C(T) designates the fraction of reactive enzyme complexes that can proceed to product and is discussed later in the context of protein conformational landscapes. Although these terms are temperature dependent, they are largely insensitive to the mass of the transferring particle.
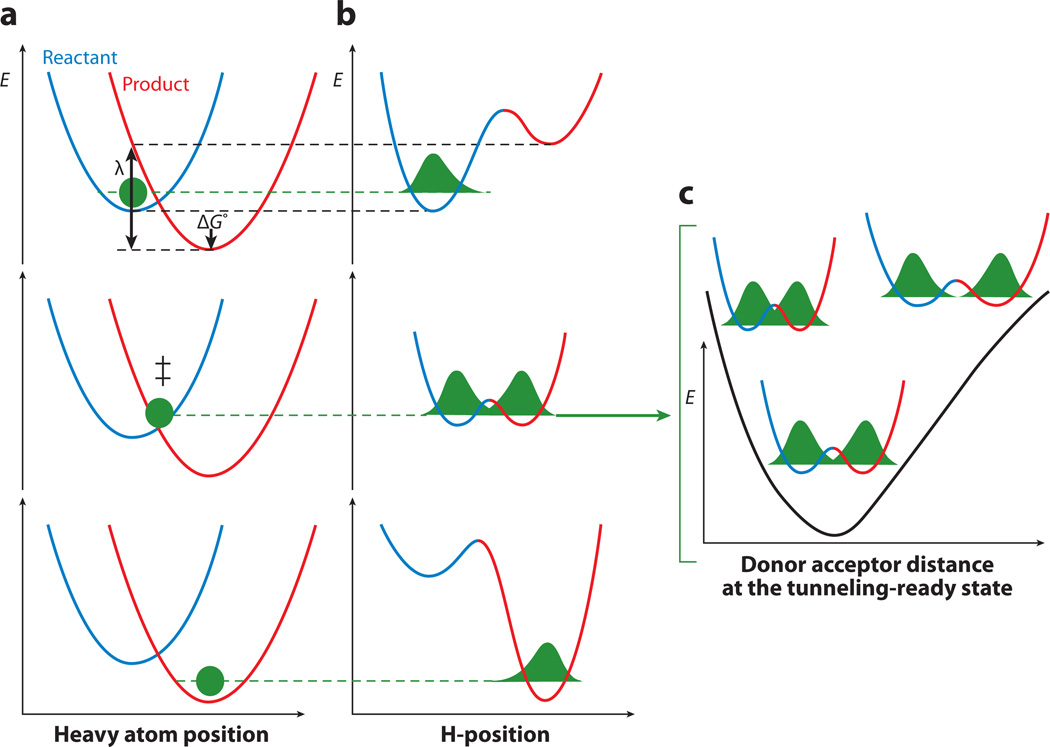
Figure 2.
Marcus-like models of H-tunneling. Three slices of the potential energy surface along components of the collective reaction coordinate, showing the effect of heavy-atom motions on the zero-point energy in the reactant (blue) and product (red ) potential well. Panel a presents the heavy atoms’ coordinate, and panel b the H-atom position. In the top graphs, the hydrogen is localized in the reactant well. Heavy-atom reorganization brings the system to the tunneling-ready state (TRS; middle graphs in panels a and b), in which the zero-point energy levels in the reactant and product wells are degenerate and the hydrogen can tunnel between the wells as a function of the donor-acceptor distance (DAD), represented within E(rx) in Equation 4. The rate of reaching the TRS depends on the reorganization energy (λ) and driving force (ΔG°). Further heavy-atom reorganization breaks the transient degeneracy and traps the hydrogen in the product state (bottom graphs). The tunneling of the particle is essentially instantaneous in the context of these and other heavy-atom motions. Panel c presents the effective potential surface along the DAD coordinate at the TRS and shows the effect of DAD sampling on the wave function overlap at the TRS. Adapted from Reference 151.
By contrast, within the integral sign, there is:
| 5 |
where mH, ωH, and r represent the mass, frequency, and distance transferred, respectively, for the tunneling particle, and 0,0 refers to tunneling from ground-state vibrational modes. The second exponential term inside the integral sign contains E(rx), the energetic barrier for the fluctuation of the donor-acceptor distance (DAD); this depends on the frequency (ωx) and the collective mass (mx) of the heavy atoms controlling distance sampling. As written, the origin of the isotope effect resides entirely within this integral sign, which computes the probability of tunneling over all possible DADs once the system has reached the TRS (Figure 2b,c).
Marcus-like models can rationalize a wide range of kinetic data observed in enzymatic and nonenzymatic reactions. Because the thermal activation to reach the TRS is unconnected to the tunneling probability, this model can account for temperature-dependent rates with temperature-independent KIEs, behavior that is inconsistent with simple tunneling corrections to TS theory (27). The second exponential term within the integral sign is mass and temperature dependent, a consequence of the fact that heavier transferred particles may require further adjustment of the DAD [i.e., larger E(rx) values] to achieve optimal wave function overlap. Importantly, when significant DAD sampling at the TRS contributes to the reaction coordinate, the KIE becomes temperature dependent. The great flexibility of Equation 4 has allowed experimentalists to interpret their results in concise, physically relevant terms, and quantitative fitting to data for enzymatic systems has been quite successful, particularly for nonadiabatic reactions (38–41). In related studies, Kiefer & Hynes (42, 43) showed that the free-energy barrier is substantially affected by the DAD vibration, which together with solvent polarization defines the reaction coordinate. Several QM/MM (quantum mechanics/molecular mechanics) simulations have also used similarly designated coordinates and report outcomes in good agreement with these phenomenological Marcus models (e.g., 44–48).
Mulholland and colleagues (49) recently discussed the possible impact of multiple reacting conformers with different DAD values on the temperature dependence of the KIE. Mulholland’s study focused specifically on temperature-independent KIEs and, although able to reproduce experimental parameters, required extremely high and unrealistic reaction-coordinate frequencies; additionally, the fitting was not transparent with regard to other parameters. Quantitative extension of Equation 4 to adiabatic H-transfer reactions remains a challenge, especially in cases in which the experimental KIEs are small and show a very large temperature dependence. Roston et al. (50) recently adapted the concept of multiple protein conformers to explain reactions with these properties. Their model focuses on the DAD and its distribution, with transfer occurring by a combination of wave function overlap above the barrier at very short distances and tunneling below the barrier at longer distances; for temperature-independent or weakly temperature-dependent KIEs, data can be modeled by a single population of reactants, in keeping with the physical picture illustrated in Figure 2. This study points out the possible contribution of an ensemble of reacting states that varies depending on the enzyme under study.
Impact of pressure on tunneling
The examination of hydrostatic pressure on the KIE of enzymatic reactions has also been suggested as a method to follow enzyme dynamics (51, 52). The isotope effect on the activation volume can be expressed in the context of Equation 4, with dominant effects expected for the H-transfer distance (r) and the force constant for DAD sampling [contained within E(rx)] (52):
| 6 |
In contrast to tunneling reactions, semiclassical models of the KIE that depend on differences in zero-point energy lack DAD dependence (25). Northrop (53) initiated a systematic study of high-pressure effects in enzymatic H-transfer using steady-state turnover rates, and then Scrutton and coworkers (54) extended the work to single-turnover kinetics. If we rule out protein denaturation at greatly elevated pressures (1–2 kbar), the impact of pressure on H-transfer efficiency can be highly system dependent and capable of yielding opposite pressure effects on the KIE, attributed to the opposing impacts of pressure on the H-transfer distance versus the force constant of its fluctuations (cf. 35, 55). As discussed in the recent literature, the pressure effects described in Equation 6 may be propagated across a protein structure in an anisotropic manner that is distinct from the directional changes required for a measurable perturbation to the reaction coordinate (56). Finally, there is an anticipated impact of elevated pressure on the ensemble of protein conformational substates that influence H-tunneling efficiency. Until the manifold impacts of high pressure on proteins are better understood, the temperature dependence of the KIE presents the more robust and predictive probe for understanding the physical parameters that control H-tunneling.
PROTEIN DYNAMICS LINKED TO HYDROGEN TUNNELING EFFICIENCY
General Features
The role of dynamics in enzyme-catalyzed reactions appears to lead to significant controversy among scientists. To discuss this topic, we first wish to carefully define statistical vs. nonstatistical motions and their timescales. We suggest that some of the apparent controversy indeed represents different understandings of enzymatic reactions, whereas others stem from different definitions of the term dynamics.
Statistical versus nonstatistical effects
The Marcus-like model described above is predicated on heavy-atom motions that are in thermal equilibrium with their environment on a timescale equal to or faster than that of the experimental rate constant for the chemical event (C–H→X transfer). In many previous publications, the use of the word dynamics simply meant motions, without distinguishing these equilibrated motions from ballistic or nonstatistical dynamics, which are directly coupled to the bond cleavage event (31, 32, 57–60). This confusion led to controversy over whether protein dynamics are expected to contribute at all to enzyme-catalyzed reactions (11, 61). However, Marcus-like models neither require nor assume nonequilibrium dynamics.
The issue of nonstatistical dynamics continues to generate controversy. Mulholland and coworkers (49) recently suggested that the absence of experimental data for such effects indicates that, by Occam’s razor, they are unimportant to enzyme function. By counterpoint, Hay & Scrutton (13) pointed out that nonstatistical dynamics may be critical, even if no experimental data have yet been reported. Computations by Miller and coworkers (62) on dihydrofolate reductase (DHFR) indicate highly localized nonstatistical heavy-atom dynamics that are restricted to the atoms of the H-donor and -acceptor. Schwartz and coworkers (63–65) are among the few that invoke an extended network of nonstatistical dynamics, specifically for lactate dehydrogenase and purine nucleoside phosphorylase. A recent experimental test of the latter involved the preparation of protein labeled with 13C, 15N, and 2H followed by comparison of its catalytic behavior with that of the native protein (66). The small impact of the heavy protein on the chemical step, to the exclusion of other steps such as product release, has been interpreted as direct evidence for nonstatistical dynamics during the bond cleavage events. However, extensive isotopic labeling alters vibrational frequencies throughout the entire protein (Equation 1), leading to perturbation of the conformational landscape as the possible origin of the observed effects (see Conformational Landscapes, below).
Timescales of motions
The timescale of protein motions has received considerable attention in the literature, with researchers often dividing these into separate regimes that involve large-scale conformational motions (millisecond to microsecond), local changes in protein configuration (nanosecond to picosecond), and barrier crossings (picosecond to femtosecond). Representative spectroscopic methods for quantifying these motions in the context of H-tunneling are given in the sidebar, Representative Spectroscopic Methods for Detecting Protein Motions and Their Link to Hydrogen Tunneling. Importantly, for multidimensional H-tunneling models, QM wave function overlap is assumed to be an instantaneous event, with the slower heavy-atom motions controlling the barrier crossing. This feature was initially counterintuitive and contrary to earlier treatises on KIEs, in which researchers concluded that for KIEs to be measurable, all steps (including protein conformation changes) would need to be rapid in relation to the H-transfer (17, 67). The separation of timescales for protein motions and hydrogenic wave function overlap that results from Equation 4 alters the underlying timescales, such that hydrogen transfer by wave function overlap is always the fastest step.
Local and Global Dynamics
The presence within proteins of a hierarchy of motions that range in time from milliseconds to femtoseconds has implications regarding the degree of locality of such motions. Although some investigators prefer to treat all protein motions in the aggregate, the properties of KIEs in C–H activating enzymes provide a window into the spatial resolution of motions that contribue to catalysis. As described, the impact of isotopic labelling at the substrate can distinguish local, donor-acceptor distance sampling modes from more global, isotope-independent conformational sampling.
Donor-acceptor distance sampling
At this time, the temperature dependence of the KIE offers the most effective tool to estimate time constants for fluctuations in DADs that affect the tunneling probability. As outlined above (see Tunneling, above), the temperature dependence of the KIE can be quite variable for a full tunneling model and depends on the average DAD and its fluctuations. In all instances, achieving the TRS requires a shorter DAD than van der Waals distances, and this can occur through either ground-state effects or the DAD sampling process represented in Equation 4. Unexpectedly, the KIE is largely temperature independent for many native enzyme systems (e.g., 38, 41, 77–85). The physical process that achieves the requisite short (compressed) DAD is most generally ascribed to a protein conformational sampling process that transiently creates substates in which donor and acceptor are close enough to permit effective wave function overlap (discussed below).
There are also many examples in which enzymes show temperature-dependent KIEs. In some instances, this occurs with native enzymes (e.g., 86, 87), but most often, with enzymes that have either undergone site-specific mutagenesis or have been studied under nonoptimal conditions (e.g., 38, 77, 88–96). The two situations—temperature-independent and temperature-dependent—segue from one to the other as the frequency for DAD decreases into a range accessible at or near room temperature (approximately 200 cm−1). Although the magnitude of the DAD frequency must be estimated from experimental data and is thus expected to be model dependent, virtually all investigators have converged on DAD frequencies between 300 and 50 cm−1 corresponding to time constants for the DAD sampling of nanoseconds to picoseconds) (12, 38, 48).
Direct fitting of equations analogous to Equation 4 is possible for nonadiabatic (weak electronic coupling between reactants) H-transfer reactions, either in isolation or in combination with methods that take into account the protein structure and dynamics [QM/MM, MD (molecular dynamics)] (38, 41, 48). Additionally, Scrutton and coworkers (35, 97) have explored the applicability of Equation 4 to adiabatic H-transfer reactions catalyzed by aromatic amine dehydrogenase and morphinone reductase.
Marcus (98, 99) has also formulated theoretical equations for the free-energy barrier of H-transfer in enzymatic reactions, describing models with different extents of coupling between the chemical coordinate and the protein reorganization coordinate. Moderate coupling produces significant contributions from the donor-acceptor atoms and from the interactions of protein charges and dipoles with one another and the reacting atoms. Marcus noted high sensitivity of the H-transfer to the DAD and took it into account in the reorganization energy expression.
Using QM/MM simulations, Cui & Karplus (100) studied environmental effects on the proton transfer TS for triosephosphate isomerase and found that the tunneling coefficients fluctuate considerably with DAD fluctuations associated with reorganization of the environment. They compared these findings for H-transfer with electron-transfer reactions according to Marcus theory. Carrying out calculations with fixed DADs, they found that the reaction is coupled to vibrational motions in the active site. The donor-acceptor stretch seems to modulate the effective barrier for the proton transfer as well as the magnitude of tunneling, which becomes more important at large DADs. This study of a charged particle (proton activation) is illustrative of models that allow for a progression from extensive tunneling to over-the-barrier H-transfer.
Given the extensive experimental studies of DHFR from Escherichia coli, many investigators have pursued computational approaches to model the chemical (hydride transfer) step. The work done in this field is so extensive that we mention only studies that directly address the question of DAD and its designated potential surface. Warshel and coworkers (101, 102) compared the contributions of solute and solvent molecules, including an enzymatic environment, presenting a 2D energy surface for a given reaction as a function of the solute and solvent coordinates. In particular, Liu & Warshel (46) examined DHFR and its mutants, in which experimental studies suggest that mutations remote from the active site affect the dynamics of the DAD (91, 103, 104). The free-energy surface was described as a function of the DAD. The authors found that the remote mutations change the reaction coordinate and suggested that the main reduction in catalysis is associated with the change of the solvent contribution to the reorganization along the reaction coordinate, as well as with the effect of increasing the DAD. Miller and coworkers (62) have also studied the H-transfer in DHFR using quantized ring polymer MD and characterized nonstatistical and statistical dynamics correlations. The DAD strongly correlates with the dynamics of the hydride transfer step. In this study, the DAD was both statistically and nonstatistically correlated with the H-transfer step, but the impact of the nonstatistical coupling was limited to the few atoms covalently bound to the donor and acceptor and dissipated within ~150 fs. A recent QM/MM study of DHFR that examined the role of dynamics on the chemical step suggested that nonstatistical dynamics co-occur with the chemical step but are not coupled to it (105). Studies of DHFR by Major and coworkers (106) demonstrate that addition of nuclear quantum corrections to the classical free-energy hypersurface brings about a significant change in the exact DAD determined at the TS.
Conformational landscapes
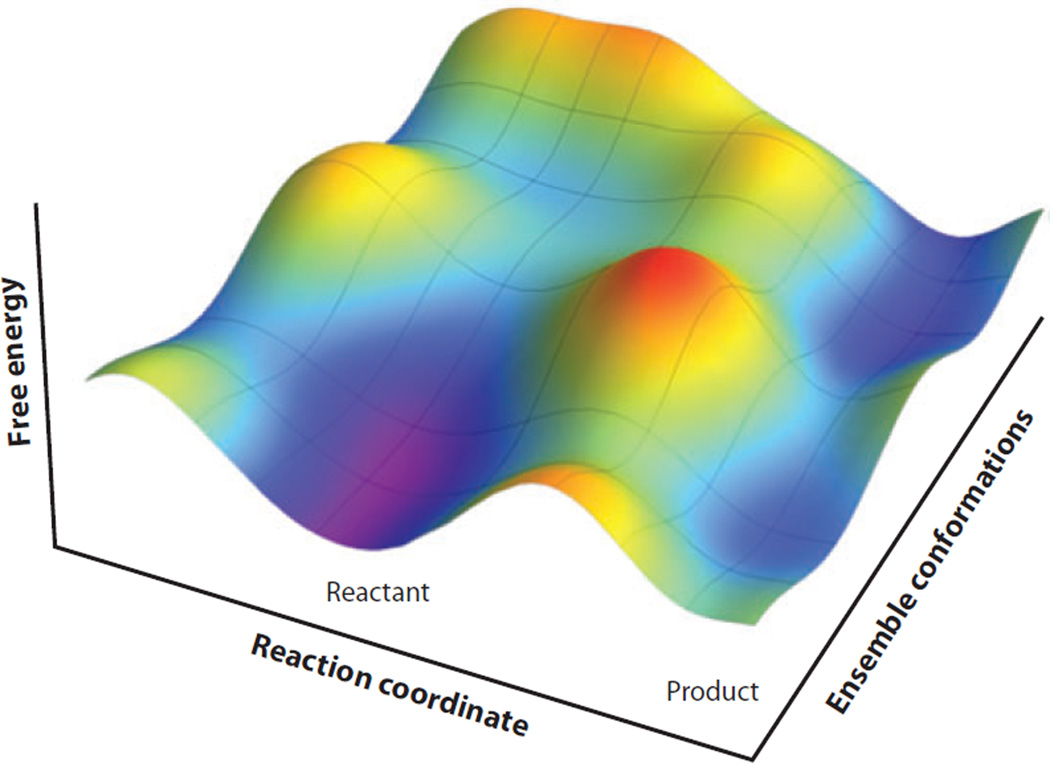
The heavy reliance on static 3D structures to infer mechanism and origins of catalysis in enzymology has given way to the recognition of the importance of protein conformational landscapes in ligand binding (4, 107), allostery (108–110), catalytic rate enhancement (111–114), and possibly substrate specificity. That mutant forms of enzymes with very different rate properties are frequently characterized by virtually identical 3D structures is now an expectation, rather than a surprise. The expected dependence of catalysis on conformational ensembles is represented in Figure 3 in a manner analogous to that described in Reference 112.
Figure 3.
Schematic representation of a free-energy landscape for an enzyme reaction. Protein motions occur along both axes. The reaction coordinate axis corresponds to the local environmental reorganization that facilitates the chemical reaction. In contrast, the ensemble conformations axis represents the sampling of multiple substates that occur at all stages along the reaction coordinate. The reactant and product valleys represent the ground states before and after the chemical conversion, and the dividing surfaces between them (maxima along the reaction coordinate) represent the ensemble of TRSs, or more generally, of the TS. This figure was produced by Dr. Russell Larsen and is printed here with permission.
In these previous treatises, all the motions that control catalysis are generally lumped together when describing the ground-state conformers that proceed to product with high rate constants. In the context of the Marcus-like formalisms that have been developed for H-tunneling, distinguishing the specific motions that control tunneling (reorganization) from those that create the appropriate ground-state structures that can proceed to product with high rates (preorganization) is conceptually very useful (95, 96). The reason for this separation arose initially from the unique information that can be obtained from the temperature dependence of KIEs, which is contained explicitly within the integral sign of Equation 4. The electrostatic reorganization at the active site, which achieves transient donor-acceptor degeneracy, is included within the first exponential term in Equation 4. This follows from the definition of the Marcus barrier for electron tunneling that is dependent on environmental reorganization, λ, and reaction driving force, ΔG°. The exponential barriers that contribute to Equation 4 can be thought of as local changes in active-site configuration (bond distances, angles, and charge distribution) that occur on rapid timescales (e.g., nanosecond to picosecond), whereas the remainder of the changes in protein conformation (e.g., millisecond to nanosecond) arise from the preexponential term, C(T), of Equation 4. Once again, following the formalism for electron tunneling, C(T) can be thought of as the Marcus work term that converts a bimolecular to a unimolecular process (28, 98) and that for H-tunneling corresponds to the conformational sampling that brings about the optimal alignment of all protein side chains in relation to the H-donor and -acceptor. This conceptual separation of the components of the conformational landscape also has interesting consequences for understanding the physical processes that underlie the enthalpic versus entropic barriers in enzymatic catalysis (see next section).
INTERRELATIONSHIP BETWEEN DONOR-ACCEPTOR DISTANCE SAMPLING AND CONFORMATIONAL LANDSCAPES
Experimentally, the simplest distinction between these DAD and conformational samplings is their sensitivity to isotopic labeling at the C–H bond that is cleaved during reaction. The contribution of DAD sampling to enzyme systems that are optimized for H-tunneling, as determined from the temperature dependence of the KIE, is generally quite weak (31, 32, 115), with the temperature-independent KIEs easily fitted to Equation 4 within a single population of protein (50) displaying a high frequency for DAD sampling (95, 96). Such apparent active-site compaction/compression could arise, in principle, from ground-state interactions within the dominant E-S complex. However, extensive X-ray structural data for enzymes catalyzing C–H abstraction routinely show distances between a carbon donor and acceptor (analog) of >3.2 Å, requiring tunneling over distances >1.0 Å. By contrast, theoretical modeling almost invariably leads to shorter predicted tunneling distances that are approximately 0.7 Å. The achievement of a TRS thus implicates a time-dependent sampling process that is inherently linked to a protein’s overall dynamical features.
Thermophilic Alcohol Dehydrogenase
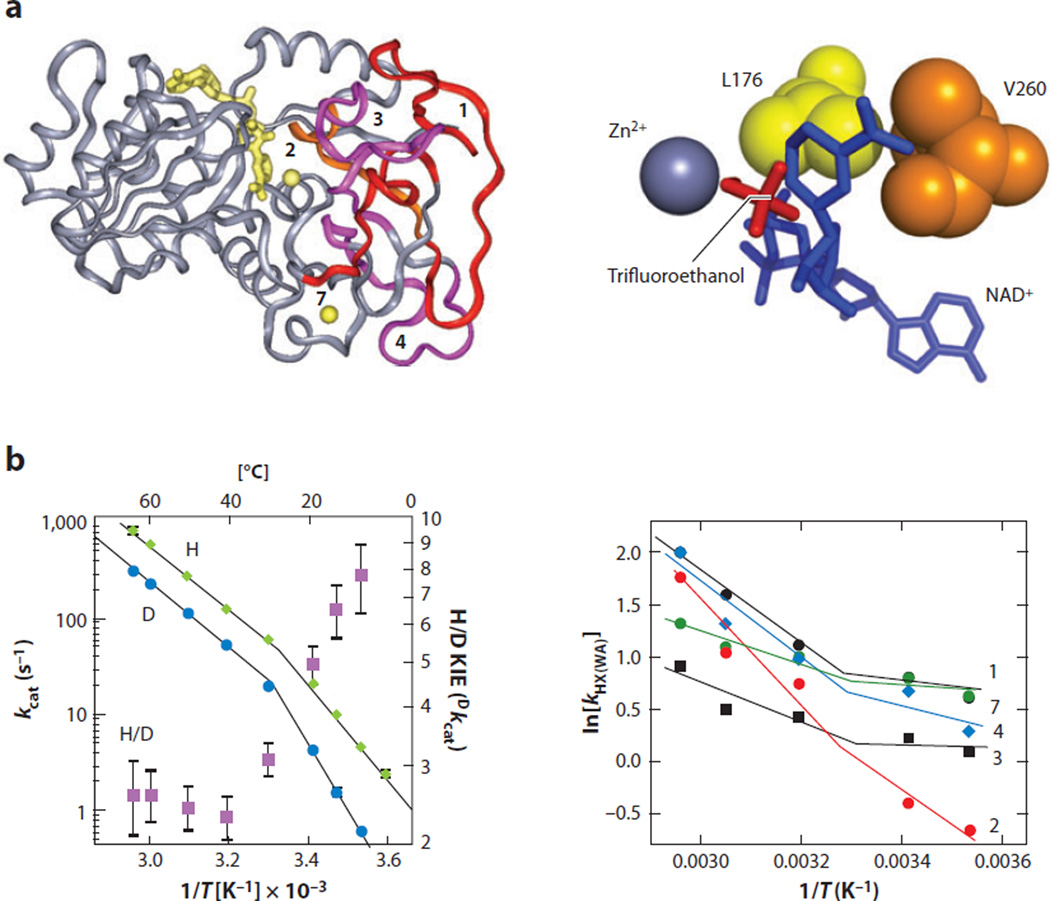
Thermophilic enzymes offer attractive systems in which to pursue dynamical features’ link to tunneling efficiency, given their enhanced stability at high temperatures and propensity to rigidify at reduced temperatures (116). The thermophilic alcohol dehydrogenase (ht-ADH) from Bacillus stearothermophilus has emerged as a paradigmatic system for linking protein conformational substates to tunneling efficiency. The ht-ADH has been characterized by a range of methods including X-ray crystallography, kinetics of enzyme turnover, KIEs and their temperature dependencies, and H/D exchange (77, 117–121). X-ray studies show a functional tetramer with each subunit composed of separate cofactor- and substrate-binding domains that converge at an active-site zinc ion (Figure 4a, left) (120).
Figure 4.
Identification of the regions of thermophilic alcohol dehydrogenase (ht-ADH) that are more flexible above 30°C. Hydrogen/deuterium (H/D) exchange coupled to mass spectrometric analyses of protein-derived peptide shows five peptides that undergo a temperature-dependent transition between 10°C and 65°C, analogous to the temperature dependence of the kcat and KIE. (a, left) The five peptides are mapped onto the ht-ADH structure and colored red/orange/magenta over a gray background. The active-site Zn2+ is shown as a yellow ball near peptide two, and the cofactor NAD+ is modeled into the active site (also in yellow). Reprinted with permission from Proc. Natl. Acad. Sci. USA. Copyright © 2004, National Academy of Sciences USA (119). (a, right) The active-site hydrophobic side chains (Leu176 and Val260) that sit behind the nicotinamide ring of cofactor are illustrated. Reprinted with permission from Biochemistry. Copyright © 2012, American Chemical Society (123). (b) The 30°C transition in behavior occurs in kcat, Dkcat, and kHX(WA); the third is the weighted average rate constant for the multiexponential exchange of deuterium from solvent into the peptide backbone. The temperature dependence of kcat and Dkcat is on the left, and the temperature dependence of kHX(WA) is on the right. Panel b (left) reproduced with permission from Nature. Copyright © 1999, Nature Publishing Group (77). Panel b (right) reproduced with permission from Proc. Natl. Acad. Sci. USA. Copyright © 2004, National Academy of Sciences USA (119).
An early, exhaustive kinetic study of ht-ADH demonstrated a rate-limiting C–H bond cleavage step (benzyl alcohol as substrate) across a wide temperature range, with a cooperative break in behavior at 30°C. This break is accompanied by an increase of the enthalpy of activation from approximately 14.6 kcal mol−1 to approximately 23.6 kcal mol−1 as the temperature is reduced (77). Significantly, the temperature dependence of the KIE also undergoes a transition from temperature independent in the high-temperature regime to temperature dependent below 30°C, indicating that under the physiologically relevant high-temperature conditions, DAD distance sampling contributes little to the tunneling rate constant.
Local protein dynamics were also interrogated using the technique of H/D exchange, followed by limited proteolysis and mass spectrometric analysis of the resulting peptides (110). The pattern of exchange under conditions in which the native protein is in rapid conformational equilibrium between open and closed states, relative to the chemical exchange rate of deuterium into the amides of the protein backbone, highlighted five peptides that, above 30°C, undergo a transition to enhanced protein flexibility that correlates with the changes in rate and KIEs (Figure 4b, left). The peptides that undergo this increase in flexibility are distributed throughout the substrate-binding domain and, in several instances, are quite remote from the active site, which is colored orange, magenta, and red in Figure 4a (left). These data provided one of the first examples of a link between distal conformational flexibility and active-site compactness. A picture has emerged in which a dynamically interconverting conformational landscape is necessary for a protein to transiently achieve the conformational ensemble that affords the TRS (95, 96).
The structural origin of the 30°C transition in ht-ADH is also becoming apparent. At a distance 20 Å from the substrate pocket and located across the surface of the substratebinding domain, an intersubunit contact has been identified in ht-ADH at position 25. This involves π-stacking of a pair of tyrosines in ht-ADH that is converted to an alanine in the less stable/more flexible psychrophilic ortholog (ps-ADH) (118). The kinetic consequences of interchanging tyrosine and alanine in ht-ADH and ps-ADH indicate that a distal subunit interaction controls the ability of these enzymes to generate the compact TRS (153).
Insight into the nature of the rigidification of ht-ADH below 30°C (linked to temperature-dependent KIEs for the native enzyme) comes from two recent studies of mutated variants that reduce the bulk of hydrophobic side chains (Leu176 and Val260; Figure 4a, right) residing directly behind the nicotinamide ring of the bound cofactor (yellow in Figure 4a, left). First, the kinetic impact of the mutants at low temperature is startling, yielding values for the Arrhenius prefactor that exceed the expected semiclassical limit of 1013 s−1 by as much as ten orders of magnitude (122). The resulting elevated values for Ah almost exactly parallel the increases in Ea below 30°C, a consequence of consistently similar rate constants immediately above and below the 30°C break. The trapping of enzyme into energy wells that are either inactive or have greatly reduced activity can explain this behavior. For catalysis to proceed efficiently, the enzyme must exit from such traps into a region of conformational space that allows sampling among substates that are amenable to tunneling (Figure 5). The behavior of mutants at positions 176 and 260 is significantly more extreme than for wild-type (WT) enzyme, and in the case of Val260Ala, the Ah is 1024 s−1.
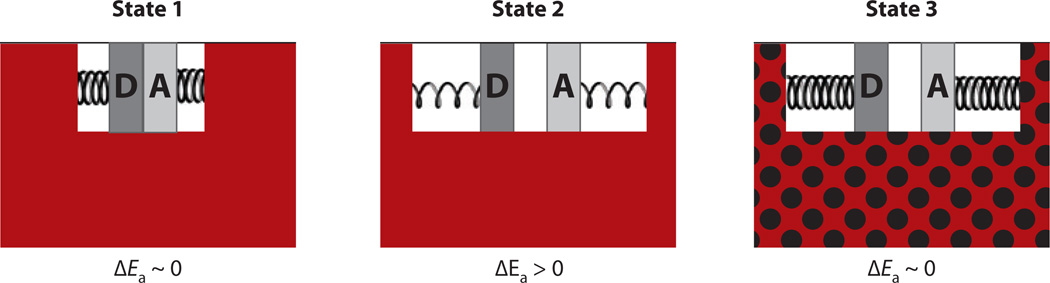
Figure 5.
Illustration of a perturbed landscape for conformational sampling that explains the greatly elevated values for Ah in ht-ADH below 30°C. The trapping of the mutant protein at low temperatures into noncatalytic or low catalytic conformational states (asterisks) is expected to affect both ΔH‡ and Ah in the manner observed. In the equations, ΔH‡ int and ln Ah(int) represent the expected parameters in the absence of the energetics required to exit from the low catalytic conformational states (asterisk) (cf. 122).
Extending the studies of cofactor site mutants to include the size and temperature dependence of the KIE led to another set of unexpected observations in which the increased temperature dependence of the KIE seen for native ht-ADH below 30°C is eliminated for almost all of the mutants (123). Instead, the mutants show an increase in the temperature dependence of the KIE above 30°C, in which enzyme flexibility allows DAD sampling to compensate for the loss of precise alignment between theH-donor and -acceptor. This is similar to a behavior seen earlier in other, unrelated systems (cf. 40, 92). The value of the KIE for the mutants in ht-ADH at approximately 30°C is similar to the KIE for the WT enzyme at 5°C, i.e., both WT and mutants reach a similar KIE but in different temperature regimes. The combination of low temperature and active-site mutation is proposed to lead to conformational substates that are exceedingly rigid (123). Although the DAD is increased in the mutants, no reduction in distance is possible at low temperature via DAD sampling because the DAD fluctuations rigidify and the rate falls off, whereas the KIE remains constant (Figure 6). Recently, Roston et al. (50) proposed that the highly temperature-dependent KIEs for WT ht-ADH below 30°C and mutant enzyme forms of ht-ADH above 30°C may also arise from the contribution of several protein conformers with different DADs and KIEs. A quantitative model that combines the impact of side-chain bulk on DAD sampling with temperature-dependent changes in the distribution of conformational substates is not yet available.
Figure 6.
A model to illustrate the impact of active-site mutations in ht-ADH above and below the transition temperature at 30°C, ΔEa = Ea(h) − Ea(l), according to Equation 3. State 1 is the ideal state and represents the wild-type (WT) protein above 30°C. The protein is optimally flexible (red region), and this generates active-site compression accompanied by a close approach of the H-donor and -acceptor. State 2 represents the situation following active-site mutation above 30°C. The white region, representing the active site, is artificially enlarged to allow a depiction of the resulting increased distance between the H-donor and -acceptor that is accompanied by a decrease in the force constant for DAD sampling. State 3 is the situation that results from the combination of active-site mutation and low temperature (black dots on red region). Once again, the region representing the active site (white) is enlarged to allow depiction of the increase in distance between the H-donor and -acceptor. Under these conditions, the increased rigidity of the protein below 30°C is proposed to prevent any recovery via DAD sampling. Adapted from Reference 123.
Dihydrofolate Reductase
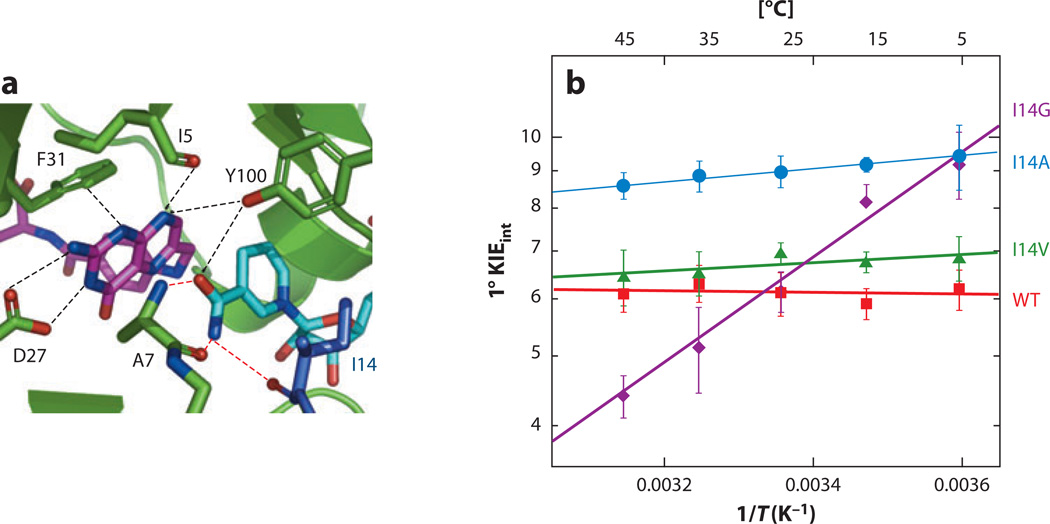
A second paradigmatic system for examining the link of DAD sampling to a conformational landscape is DHFR. In the E. coli DHFR, a series of active-site mutants was constructed, focusing on Ile14 (Figure 7), which was gradually reduced to Val, Ala, and Gly. Examination of the H-transfer rates and intrinsic KIEs and their temperature dependence, together with MD simulations, allowed the effect of mutations on different protein dynamics to be studied (92). As expected, the smaller the side chain, the longer the DAD and the broader its distribution, leading to a gradual increase in the temperature dependence of intrinsic KIEs (Figure 7). For the most extreme mutant (I14G), MD simulation revealed larger scale motions, showing new conformations of the donor and acceptor that were far from the TRS (e.g., >3.5 Å and at an angle far from linear C–H–C). In a recent treatment, the highly temperature-dependent KIEs for I14G were found to be fit using two active-site conformations with very different DADs and KIEs (50).
Figure 7.
(a) Active site of WT DHFR (dihydrofolate reductase) from Escherichia coli [Protein Data Bank (PDB) ID 1RX2] emphasizing the role of Ile14 (metallic blue) as a support of the nicotinamide ring of NADP+. The nicotinamide ring is highlighted in light blue and the folate in magenta. (b) Arrhenius plot of intrinsic H/T KIEs (on a log scale) for WT DHFR (red ), I14V DHFR mutant ( green), I14A DHFR (blue), and I14G DHFR ( purple). The lines represent the nonlinear regression to an exponential equation. Reproduced with permission from J. Am. Chem. Soc. Copyright © 2012, the American Chemical Society (92).
A moderately thermophilic DHFR (ht-DHFR) from B. stearothermophilus has enabled studies into the role of conformational barriers and enthalpic traps during catalysis. The ht-DHFR is highly homologous to the well-studied E. coli DHFR (124) but permits the extension of kinetic and physical characterizations to elevated temperatures (125, 126). The H/D exchange methodology described above for ht-ADH was applied to ht-DHFR, with the observation of much more rapid deuterium exchange into the small ht-DHFR monomer (18.7 kDa). This property refocused attention on the extent of H/D exchange within the plateau regions of H/D exchange versus time, providing a measure of the changes in the extent of protein that is protected from exchange as a function of temperature (125). Among the 11 peptides examined, covering 91% of the total protein sequence, almost all showed a very small enthalpic barrier of 1 to 2 kcal mol−1, in the range of room temperature. The regions of native protein represented by the peptides with a low enthalpic barrier for exchange include the M20 and F/G loops, whose motions had previously been directly linked to the H-transfer step. A model has been put forth in which the energetic barriers for the interconversion of protein conformational substates may be largely entropic, reflective of the probability of arriving at the states that are optimal for tunneling (i.e., TRS). Wand and coworkers (127) have also investigated via NMR the contribution of side-chain dynamics to the role of entropy in protein ligand binding and catalysis. The maintenance of small enthalpic barriers for substate interconversion ensures a smooth energetic landscape that facilitates continuous sampling of a manifold of active-site configurations. This is in contrast to the impairment of catalysis that occurs with WT ht-ADH, and especially its mutants below 30°C, in which the protein increasingly resides within low-enthalpy traps (Figure 5) (122).
TOWARD A COMPREHENSIVE MODEL FOR ENZYME CATALYSIS
Generalizing Beyond C–H Activation
Although several light-activated, enzyme-catalyzed rearrangements are documented (e.g., 128), most enzymes, including those catalyzing H-tunneling, occur via thermally activated processes. The strong dependence of thermally activated H-tunneling on the DAD, emphasized throughout this review, can be viewed in the context of an empirical proportionality factor (β) for the dependence of the tunneling rate on transfer distance [estimated as 23 Å−1 for H-transfer (129) versus only approximately 1 Å−1 for electron transfer (130, 131)]. This link of short DADs to H-tunneling efficiency raises the question of the importance of barrier width in other classes of enzyme reactions.
Studies by Schowen and coworkers (132) addressed this question for the catechol O-methyl transferase (COMT), in which S-adenosylmethionine (AdoMet) transfers a methyl group to a ring oxygen on an acceptor catechol. The measured secondary KIE was smaller (inverse) for the enzymatic reaction in relation to the comparable reaction in solution, providing one of the earliest demonstrations of a likely role for compaction in an enzymatic process. Quite recently, NMR and X-ray studies of the SET domain lysine methyl transferases have led to the proposal of close H-bonding interactions between amino acid side chains and the carbon of the transferred methyl group as a means of restricting the trajectory of the methyl group from AdoMet toward its acceptor (133). Site-specific mutagenesis was extended to COMT to test the role of a conserved active-site side chain (tyrosine) that resides behind the sulfur donor of AdoMet in creating a compact active site. One of us found a remarkable linear correlation, in which the second-order rate constant for the reaction of dopamine with the enzyme-bound AdoMet decreased more than three orders of magnitude as the secondary KIE became less inverse (more like the solution reaction) (134). These results with COMT indicate a trend similar to that seen in H-tunneling reactions, i.e., a dependence of active-site compaction on strategically placed active-site side chains. Whether the observed effects have their origin in the dominant ground-state enzyme–AdoMet complex, versus a dynamically achieved activated complex, is under investigation.
Interactive experimental and computational work has also examined dynamics in purine nucleoside phosphorylase, which catalyzes phosphate addition to a nucleotide at the anomeric carbon of the ribose ring. The findings are in accordance with significant compaction of the active site leading the system toward its TS (65). The picture that has emerged from these studies may resemble the conformational sampling and active-site reorganization of the reactants toward the TRS that are characteristic of H-transfer reactions.
Model Reactions for Hydrogen Tunneling
The direct comparison of the properties of H-transfer for model reactions with those for an analogous enzyme reaction is fairly rare. In one well-executed (and -publicized) example, the size of the KIE and its temperature dependence were shown to be very similar for the B-12-dependent methylmalonyl-CoA mutase and ethanolamine ammonia lyase and for a model thereof (135, 136). In both the model and enzyme reactions, the magnitude of the KIE was quite elevated above the semiclassical limit and showed a strong temperature dependence that produced inverse ratios for Al/Ah (cf. middle bar of Figure 1b). In light of the common observation of temperature-independent KIEs for native enzyme systems, the data for these B-12-dependent reactions suggest an optimization of the thermal homolysis of the cobalt-carbon bond of the cofactor and not the C–H bond cleavage step. In the ensuing years, the data have made it increasingly clear that tunneling may contribute significantly to the room temperature reaction of small molecules in condensed phase as well as enzymatic reactions. However, the phenomenon of weak or temperature-independent KIEs that become temperature dependent upon either local or distal structural perturbation has not been reported for nonenzymatic reactions. The rare examples in which temperature-independent KIEs are also seen for small molecule H-transfer generally represent intramolecular processes in which the structure of the reactant constrains the DAD (137–141).
In a second comparative study, Pollock and coworkers (142) analyzed the interrelationship of secondary KIEs using multiply labeled substrates (see Table 1) for a model reaction of ketosteroid isomerase as well as the enzyme itself. Whereas semiclassical deviations were prominent in the enzyme reaction (especially for C–D activation at the primary position), they were absent from the model system. This behavior has since been attributed to a very close approach between the H-donor and -acceptor at the enzyme active site that becomes exaggerated when deuterium is transferred, owing to the much smaller wavelength for the heavier isotope (31, 32, 58, 143). The different properties of the model versus enzyme reaction are fully compatible with an increased compaction of reactants at the TRS in the enzymatic reaction. The concept of dynamically enhanced active-site compaction is, perhaps, the most important feature to emerge from the observed properties of enzymatic reactions that have been optimized for H-tunneling, distinguishing these reactions from those of small molecules.
BIOCHEMICAL RELEVANCE
Understanding the origin of enzyme catalysis in the broader biological context remains important. Kinetic studies of enzymes frequently show that substrate-binding and product-release steps control turnover (67), indicating that during evolution, the barriers for chemical catalysis have been reduced to the point at which they are no longer rate limiting. Evolutionary pressure will continue to be effective even in such highly evolved systems to ensure that the chemical step does not slow to a point at which it begins to limit or abolish turnover. This explains the strict retention of residues whose loss would dramatically eliminate chemical catalysis under conditions in which the rate-determining step(s) is independent of a role for such residues. In many instances, chemical rate acceleration with enzymes may be expected to exceed estimates based on steady-state or even presteady-state kinetics, which can be governed by nonchemical processes.
A major goal for many decades has been the de novo design of proteins on the basis of the principles of enzyme catalysis. Catalytic antibodies (3) and biomimetic systems (144–146), as well as computational efforts (e.g., 147), have not yet been able to approach the rate accelerations observed in most biological catalysts. This points toward the importance of the size and specific folds of proteins in effecting catalysis. As this review has emphasized, motions within a protein, both proximal and distal to the active site, are increasingly expected to be crucial to their chemical catalysis. The linking of protein motions to the evolution of new enzyme activities, as well as to the origin of substrate specificity, is also emerging as a major theme in biology.
Finally, we address the repeated question of how the concepts and data discussed here relate to the design of inhibitors and, in particular, drug development. The use of high-throughput computer docking frequently ignores dynamics when searching for possible drug candidates. This may eliminate the most promising leads early in the selection process. There is also the question of how the textbook concept of enhanced TS binding relates to drug design. The ideas presented here support the view that catalysis arises from a subset of the total protein conformations that achieves, simultaneously, all of the specific interactions that allow for rapid bond cleavage in the aggregate. By contrast, tight binding of a TS analog targets an energy minimum that mimics the saddle point of the TS. A suggested reconciliation of these differences attributes tight binding of TS analogs to a clamping down of the inherent motions of an enzyme upon inhibitor complex formation (148). Integrating the growing knowledge of the essential role of protein dynamics in catalytic rate enhancement may promote a resurgence of rational drug design.
CLARIFICATION OF CONCEPTS REGARDING PROTEIN DYNAMICS.
The role of protein motions, often referred to as protein dynamics, is distinct from the dynamics invoked by physical chemists in their description of the activated complex and its probability of proceeding to product. The latter is defined by the transmission coefficient in TS theory. Calculations support the view that the transmission coefficient will be similar for enzyme versus solution reactions (cf. 11).
Protein motions can be either statistically distributed or directly coupled to the bond cleavage event. This review is focused on stochastic protein sampling processes, though recent controversy has centered on the possibility of direct coupling of long-range networks of protein motions with the reaction coordinate (12, 13).
Most enzymes turn over their substrates in milliseconds, and slower motions are unlikely to be a part of the barriers discussed here. Although technical hurdles have, thus far, prevented the study of single-enzyme molecule catalysis on timescales at or faster than milliseconds, single-molecule methodologies do validate the presence of multiple protein substates that interconvert on the timescale of seconds and are characterized by variable catalytic parameters (14–16).
REPRESENTATIVE SPECTROSCOPIC METHODS FOR DETECTING PROTEIN MOTIONS AND THEIR LINK TO HYDROGEN TUNNELING.
Millisecond to nanosecond motions: Nuclear magnetic resonance (NMR) studies of loop closures and other conformational changes in DHFR have indicated that millisecond dynamics are relevant to the kinetic turnover cascade (68). In another example, NMR studies of proline isomerase suggested that the protein conformational changes drive the bond isomerization (7). However, the movements in DHFR at this slower timescale are now attributed to an accommodation of protein to different enzyme-ligand complexes along the reaction coordinate (57).
Nanosecond to picosecond motions: Fluorescence lifetime and Stokes shift measurements, using either an intrinsic or extrinsic fluorescent probe, are established techniques for detecting local nanosecond and picosecond motions within a protein (69). Such fluorescence measurements, when coupled to protein perturbation (for example, by site-specific mutagenesis), offer the possibility of correlating changes in active-site dynamics to changes in H-tunneling parameters.
Picosecond to femtosecond motions: 2D-IR (infrared) measurements are nonlinear vibrational spectroscopy relaxation experiments that can probe the direct environment of an IR-chromophore at these rapid timescales. For formate dehydrogenase, increased rigidity of a TS analog was observed upon formation of the ternary complex. As the intrinsic KIEs of formate dehydrogenase are temperature independent, the spectroscopic and kinetic observations suggest a common phenomenon of active-site compaction (70, 71).
Techniques that are associated with the rigidity/flexibility of the protein but that cannot be directly assigned to a dynamics timescale: These include X-ray diffraction data of protein crystals analyzed either for anisotropic B-factors (e.g., 72) or via new computational tools for electron density sampling, model refinement, and molecular packing analysis using room temperature data (8) and hydrogen-deuterium (H/D) exchange methods (73–76). These alternate methods are proving very useful in allowing correlations between spatially resolved regions of protein rigidity/flexibility and active-site function.
SUMMARY POINTS.
The persistent deviation of empirical observations for enzymatic C–H activation from semiclassical predictions leads to the incorporation of QM nuclear tunneling into theoretical models.
The pervasiveness of tunneling properties in enzymatic C–H activation that cannot be explained by a 1D tunneling correction leads to a multidimensional physical picture that resembles Marcus models for electron transfer.
The availability of an analytical rate expression for nonadiabatic H-transfer, which can be used to represent physical parameters that control tunneling and to quantitatively reproduce or even be fitted to experimental data, is discussed.
The importance of proximal, active-site side-chain packing in achieving an optimal approach of the H-donor to the H-acceptor is apparent in numerous systems.
The importance of a stochastic protein conformational sampling process is presented in the context of achieving the optimal TRS.
The link of protein conformational substates to the properties of active-site tunneling is demonstrated in two paradigmatic enzyme systems, ht-ADH and DHFR.
The role for active-site compaction in enzymatic methyl transfer reactions, which is modulated by active-site side chains, may represent an extension of the concepts introduced for H-transfer reactions to larger group transfer reactions.
The above features are likely to be relevant to future designs of highly active/selective catalysts and, possibly, to new strategies for drug design.
FUTURE ISSUES.
More efforts are needed for the development of an accessible analytical rate expression for H-tunneling that can accommodate, within a cohesive physical picture, the processes underlying both nonadiabatic (e.g., hydrogen atom transfer) and adiabatic (e.g., hydride and proton transfer) reactions.
A possible role for nonstatistical dynamics in promoting dynamics of atoms remote from the H-donor and -acceptor is yet to be deciphered theoretically and experimentally.
The structural networks within proteins that act, via dynamical sampling, to establish the TRS in enzyme active sites will need to be identified in a system-dependent manner.
More work is required before the above information can be used to develop a set of rules for predicting the regions of proteins that will be most influential in linking protein flexibility/motions to active-site compactness.
Researchers need to continue the exploration of spectroscopic methods that allow specific time constants to be linked to H-transfer, in particular in the picosecond to femtosecond range. Time-dependent fluorescence is a promising tool, as is nonlinear vibrational spectroscopy, which probes the dynamics of the reacting ligands.
A focus on the principles that arise from these studies may enable the sought-after goal of de novo design of enzyme catalysts that reproduce the activity of native enzymes.
ACKNOWLEDGMENTS
Support came from the following agencies: National Institutes of Health (GM025765 to J.P.K. and GM65368 to A.K.), National Science Foundation (MCB0446395 to J.P.K. and CHE-1149023 to A.K.), and the United States-Israel Binational Science Foundation (2007256 to A.K.).
Glossary
- TS
transition state
- Tunneling
a phenomenon in which a particle crosses a barrier between reactants and products owing to its wave-like nature
- Isotope effect
the difference in a kinetic or thermodynamic property that results solely from differences in the mass of the reacting molecules
- KIE
kinetic isotope effect
- Reorganization
the motions of the heavy atoms that bring the donor and acceptor from the reactant’s ground state to the tunneling-ready state(s)
- Tunneling-ready state (TRS)
the ensemble of reactants that achieve the energetics and distances compatible with tunneling between the H-donor and -acceptor
- DAD
donor-acceptor distance
- QM/MM
quantum mechanics/molecular mechanics
- Nonstatistical dynamics
motions not in thermal equilibrium with their environment; along the reaction coordinate, these must be at the timescale of the bond cleavage event(s)
- DHFR
dihydrofolate reductase
- MD
molecular dynamics
- Statistical dynamics
motions in thermal equilibrium with their environment; along the reaction coordinate, these must be faster than the net reaction
- Preorganization
all components of the reaction that lead to the ground-state ensemble of catalytically relevant conformers
- Active-site compaction
the compact arrangement of active-site residues and reactants that affords the tunneling-ready state
- Tunneling distance (r in Equation 4)
the distance traversed by hydrogen as it moves from its reactant well to product well; Figure 1a
- ht-ADH
high-temperature (thermophilic) alcohol dehydrogenase
- H/D
hydrogen/deuterium
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Pauling L. Chemical achievement and hope for the future. Am. Sci. 1948;36:51–58. [PubMed] [Google Scholar]
- 2.Wolfenden R. Degrees of difficulty of water-consuming reactions in the absence of enzymes. Chem. Rev. 2006;106:3379–3396. doi: 10.1021/cr050311y. [DOI] [PubMed] [Google Scholar]
- 3.Hilvert D. Critical analysis of antibody catalysis. Annu. Rev. Biochem. 2000;69:751–793. doi: 10.1146/annurev.biochem.69.1.751. [DOI] [PubMed] [Google Scholar]
- 4.Boehr DD, Dyson HJ, Wright PE. Conformational relaxation following hydride transfer plays a limiting role in dihydrofolate reductase catalysis. Biochemistry. 2008;47:9227–9233. doi: 10.1021/bi801102e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hammes-Schiffer S, Benkovic SJ. Relating protein motion to catalysis. Annu. Rev. Biochem. 2006;75:519–541. doi: 10.1146/annurev.biochem.75.103004.142800. [DOI] [PubMed] [Google Scholar]
- 6.Williams JC, McDermott AE. Dynamics of the flexible loop of triosephosphate isomerase: The loop motion is not ligand gated. Biochemistry. 1995;34:8309–8319. doi: 10.1021/bi00026a012. [DOI] [PubMed] [Google Scholar]
- 7.Henzler-Wildman K, Kern D. Dynamic personalities of proteins. Nature. 2007;450:964–972. doi: 10.1038/nature06522. [DOI] [PubMed] [Google Scholar]
- 8.Fraser JS, Clarkson MW, Degnan SC, Erion R, Kern D, Alber T. Hidden alternative structures of proline isomerase essential for catalysis. Nature. 2009;462:669–673. doi: 10.1038/nature08615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koshland DE., Jr Application of a theory of enzyme specificity to protein synthesis. Proc. Natl. Acad. Sci. USA. 1958;44:98–104. doi: 10.1073/pnas.44.2.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koshland DE., Jr The key-lock theory and the induced fit theory. Angew. Chem. Int. Ed. 1994;33:2375–2378. [Google Scholar]
- 11.Adamczyk AJ, Cao J, Kamerlin SC, Warshel A. Catalysis by dihydrofolate reductase and other enzymes arises from electrostatic preorganization, not conformational motions. Proc. Natl. Acad. Sci. USA. 2011;108:14115–14120. doi: 10.1073/pnas.1111252108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schwartz SD. Vibrationally enhanced tunneling and kinetic isotope effects in enzymatic reactions. See Ref. 152. 2006:475–498. [Google Scholar]
- 13.Hay S, Scrutton NS. Good vibrations in enzyme-catalysed reactions. Nat. Chem. 2012;4:161–168. doi: 10.1038/nchem.1223. [DOI] [PubMed] [Google Scholar]
- 14.Lu HP. Biochemistry. Enzymes in coherent motion. Science. 2012;335:300–301. doi: 10.1126/science.1217170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu HP, Xun L, Xie XS. Single-molecule enzymatic dynamics. Science. 1998;282:1877–1882. doi: 10.1126/science.282.5395.1877. [DOI] [PubMed] [Google Scholar]
- 16.Tan YW, Yang H. Seeing the forest for the trees: fluorescence studies of single enzymes in the context of ensemble experiments. Phys. Chem. Chem. Phys. 2011;13:1709–1721. doi: 10.1039/c0cp02412k. [DOI] [PubMed] [Google Scholar]
- 17.Klinman JP. Kinetic isotope effects in enzymology. Adv. Enzymol. Relat. Areas Mol. Biol. 1978;46:415–494. doi: 10.1002/9780470122914.ch7. [DOI] [PubMed] [Google Scholar]
- 18.Cleland WW. The use of isotope effects to determine transition-state structure for enzymic reactions. Methods Enzymol. 1982;87:625–641. doi: 10.1016/s0076-6879(82)87033-x. [DOI] [PubMed] [Google Scholar]
- 19.Schowen RL. Hydrogen bonds, transition-state stabilization, and enzyme catalysis. See Ref. 152. 2006:765–792. [Google Scholar]
- 20.Bigeleisen J. Theoretical basis of isotope effects from an autobiographical perspective. See Ref. 152. 2006:1–40. [Google Scholar]
- 21.Blanchard JS, Cleland WW. Kinetic and chemical mechanisms of yeast formate dehydrogenase. Biochemistry. 1980;19:3543–3550. doi: 10.1021/bi00556a020. [DOI] [PubMed] [Google Scholar]
- 22.Hermes JD, Cleland WW. Evidence from multiple isotope effect determinations for coupled hydrogen motion and tunneling in the reaction catalyzed by glucose-6-phosphate dehydrogenase. J. Am. Chem. Soc. 1984;106:7263–7264. [Google Scholar]
- 23.Cha Y, Murray CJ, Klinman JP. Hydrogen tunneling in enzyme reactions. Science. 1989;243:1325–1330. doi: 10.1126/science.2646716. [DOI] [PubMed] [Google Scholar]
- 24.Melander L, Saunders WH. Reaction Rates of Isotopic Molecules. Malabar, FL: R.E. Krieger; 1987. [Google Scholar]
- 25.Isaacs NS. The effect of pressure on kinetic isotope effects. In: Buncel E, Lee CC, editors. Isotope Effects in Organic Chemistry. London: Elsevier; 1984. pp. 67–105. [Google Scholar]
- 26.Kohen A, Klinman JP. Enzyme catalysis: beyond classical paradigms. Acc. Chem. Res. 1998;31:397–404. [Google Scholar]
- 27.Bell RP. The Tunnel Effect in Chemistry. New York: Chapman & Hall; 1980. [Google Scholar]
- 28.Marcus RA, Sutin N. Electron transfers in chemistry and biology. Biochim. Biophys. Acta Bioenerg. 1985;811:265–322. [Google Scholar]
- 29.Dutta S, Li YL, Rock W, Houtman JC, Kohen A, Cheatum CM. 3-Picolyl azide adenine dinucleotide as a probe of femtosecond to picosecond enzyme dynamics. J. Phys. Chem. B. 2012;116:542–548. doi: 10.1021/jp208677u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sen A, Kohen A. Enzymatic tunneling and kinetic isotope effects: chemistry at the crossroads. J. Phys. Org. Chem. 2010;23:613–619. [Google Scholar]
- 31.Nagel ZD, Klinman JP. Update 1 of: Tunneling and dynamics in enzymatic hydride transfer. Chem. Rev. 2010;110:PR41–PR67. doi: 10.1021/cr1001035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nagel ZD, Klinman JP. Tunneling and dynamics in enzymatic hydride transfer. Chem. Rev. 2006;106:3095–3118. doi: 10.1021/cr050301x. [DOI] [PubMed] [Google Scholar]
- 33.Kuznetsov AM, Ulstrup J. Proton and hydrogen atom tunneling in hydrolytic and redox enzyme catalysis. Can. J. Chem. 1999;77:1085–1096. [Google Scholar]
- 34.Knapp MJ, Klinman JP. Environmentally coupled hydrogen tunneling. Linking catalysis to dynamics. Eur. J. Biochem. 2002;269:3113–3121. doi: 10.1046/j.1432-1033.2002.03022.x. [DOI] [PubMed] [Google Scholar]
- 35.Pudney CR, Johannissen LO, Sutcliffe MJ, Hay S, Scrutton NS. Direct analysis of donor-acceptor distance and relationship to isotope effects and the force constant for barrier compression in enzymatic H-tunneling reactions. J. Am. Chem. Soc. 2010;132:11329–11335. doi: 10.1021/ja1048048. [DOI] [PubMed] [Google Scholar]
- 36.Hammes-Schiffer S. Kinetic isotope effects for proton-coupled electron transfer reactions. See Ref. 152. 2006:499–520. doi: 10.1021/jp809122y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Borgis D, Hynes JT. Proton transfer reactions. In: Cooper A, Houben J, Chien L, editors. The Enzyme Catalysis Process. New York: Plenum; 1989. pp. 293–303. [Google Scholar]
- 38.Knapp MJ, Rickert K, Klinman JP. Temperature-dependent isotope effects in soybean lipoxygenase-1: correlating hydrogen tunneling with protein dynamics. J. Am. Chem. Soc. 2002;124:3865–3874. doi: 10.1021/ja012205t. [DOI] [PubMed] [Google Scholar]
- 39.Meyer MP, Klinman JP. Modeling temperature dependent kinetic isotope effects for hydrogen transfer in a series of soybean lipoxygenase mutants: the effect of anharmonicity upon transfer distance. Chem. Phys. 2005;319:283–296. doi: 10.1016/j.chemphys.2005.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meyer MP, Tomchick DR, Klinman JP. Enzyme structure and dynamics affect hydrogen tunneling: the impact of a remote side chain (I553) in soybean lipoxygenase-1. Proc.Natl. Acad. Sci. USA. 2008;105:1146–1151. doi: 10.1073/pnas.0710643105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Francisco WA, Knapp MJ, Blackburn NJ, Klinman JP. Hydrogen tunneling in peptidylglycine α-hydroxylating monooxygenase. J. Am. Chem. Soc. 2002;124:8194–8195. doi: 10.1021/ja025758s. [DOI] [PubMed] [Google Scholar]
- 42.Kiefer PM, Hynes JT. Kinetic isotope effects for nonadiabatic proton transfer reactions in a polar environment. 1. Interpretation of tunneling kinetic isotopic effects. J. Phys. Chem. A. 2004;108:11793–11808. [Google Scholar]
- 43.Kiefer PM, Hynes JT. Kinetic isotope effects for adiabatic proton transfer reactions in a polar environment. J. Phys. Chem. A. 2003;107:9022–9039. [Google Scholar]
- 44.Olsson MH, Siegbahn PE, Warshel A. Simulations of the large kinetic isotope effect and the temperature dependence of the hydrogen atom transfer in lipoxygenase. J. Am. Chem. Soc. 2004;126:2820–2828. doi: 10.1021/ja037233l. [DOI] [PubMed] [Google Scholar]
- 45.Liu H, Warshel A. The catalytic effect of dihydrofolate reductase and its mutants is determined by reorganization energies. Biochemistry. 2007;46:6011–6025. doi: 10.1021/bi700201w. [DOI] [PubMed] [Google Scholar]
- 46.Liu H, Warshel A. Origin of the temperature dependence of isotope effects in enzymatic reactions: the case of dihydrofolate reductase. J. Phys. Chem. B. 2007;111:7852–7861. doi: 10.1021/jp070938f. [DOI] [PubMed] [Google Scholar]
- 47.Edwards SJ, Soudackov AV, Hammes-Schiffer S. Impact of distal mutation on hydrogen transfer interface and substrate conformation in soybean lipoxygenase. J. Phys. Chem. B. 2010;114:6653–6660. doi: 10.1021/jp100133p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hatcher E, Soudackov AV, Hammes-Schiffer S. Proton-coupled electron transfer in soybean lipoxygenase: dynamical behavior and temperature dependence of kinetic isotope effects. J. Am. Chem. Soc. 2007;129:187–196. doi: 10.1021/ja0667211. [DOI] [PubMed] [Google Scholar]
- 49.Glowacki DR, Harvey JN, Mulholland AJ. Taking Ockham’s razor to enzyme dynamics and catalysis. Nat. Chem. 2012;4:169–176. doi: 10.1038/nchem.1244. [DOI] [PubMed] [Google Scholar]
- 50.Roston D, Cheatum CM, Kohen A. Hydrogen donor-acceptor fluctuations from kinetic isotope effects: a phenomenological model. Biochemistry. 2012;51:6860–6870. doi: 10.1021/bi300613e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Northrop DB. Effects of high hydrostatic pressure on isotope effects. See Ref. 152. 2006:837–846. [Google Scholar]
- 52.Hay S, Scrutton NS. Incorporation of hydrostatic pressure into models of hydrogen tunneling highlights a role for pressure-modulated promoting vibrations. Biochemistry. 2008;47:9880–9887. doi: 10.1021/bi8005972. [DOI] [PubMed] [Google Scholar]
- 53.Northrop DB. Effects of high pressure on isotope effects and hydrogen tunneling. J. Am. Chem. Soc. 1999;121:3521–3524. [Google Scholar]
- 54.Hay S, Sutcliffe MJ, Scrutton NS. Promoting motions in enzyme catalysis probed by pressure studies of kinetic isotope effects. Proc. Natl. Acad. Sci. USA. 2007;104:507–512. doi: 10.1073/pnas.0608408104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Johannissen LO, Scrutton NS, Sutcliffe MJ. How does pressure affect barrier compression and isotope effects in an enzymatic hydrogen tunneling reaction? Angew. Chem. Int. Ed. 2011;50:2129–2132. doi: 10.1002/anie.201006668. [DOI] [PubMed] [Google Scholar]
- 56.Hay S, Johannissen LO, Hothi P, Sutcliffe MJ, Scrutton NS. Pressure effects on enzyme-catalyzed quantum tunneling events arise from protein-specific structural and dynamic changes. J. Am. Chem. Soc. 2012;134:9749–9754. doi: 10.1021/ja3024115. [DOI] [PubMed] [Google Scholar]
- 57.Bhabha G, Lee J, Ekiert DC, Gam J, Wilson IA, et al. A dynamic knockout reveals that conformational fluctuations influence the chemical step of enzyme catalysis. Science. 2011;332:234–238. doi: 10.1126/science.1198542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Klinman JP. Linking protein structure and dynamics to catalysis: the role of hydrogen tunnelling. Philos. TransRSoc. Lond. B. 2006;361:1323–1331. doi: 10.1098/rstb.2006.1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kohen A, Klinman JP. Hydrogen tunneling in biology. Chem. Biol. 1999;6:R191–R198. doi: 10.1016/s1074-5521(99)80058-1. [DOI] [PubMed] [Google Scholar]
- 60.Boehr DD, McElheny D, Dyson HJ, Wright PE. The dynamic energy landscape of dihydrofolate reductase catalysis. Science. 2006;313:1638–1642. doi: 10.1126/science.1130258. [DOI] [PubMed] [Google Scholar]
- 61.Pisliakov AV, Cao J, Kamerlin SC, Warshel A. Enzyme millisecond conformational dynamics do not catalyze the chemical step. Proc. Natl. Acad. Sci. USA. 2009;106:17359–17364. doi: 10.1073/pnas.0909150106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Boekelheide N, Salomon-Ferrer R, Miller TF., 3rd Dynamics and dissipation in enzyme catalysis. Proc. Natl. Acad. Sci. USA. 2011;108:16159–16163. doi: 10.1073/pnas.1106397108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Antoniou D, Schwartz SD. Protein dynamics and enzymatic chemical barrier passage. J. Phys. Chem. B. 2011;115:15147–15158. doi: 10.1021/jp207876k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Davarifar A, Antoniou D, Schwartz SD. The promoting vibration in human heart lactate dehydrogenase is a preferred vibrational channel. J. Phys. Chem. B. 2011;115:15439–15444. doi: 10.1021/jp210347h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Saen-Oon S, Quaytman-Machleder S, Schramm VL, Schwartz SD. Atomic detail of chemical transformation at the transition state of an enzymatic reaction. Proc. Natl. Acad. Sci. USA. 2008;105:16543–16548. doi: 10.1073/pnas.0808413105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Silva RG, Murkin AS, Schramm VL. Femtosecond dynamics coupled to chemical barrier crossing in a Born-Oppenheimer enzyme. Proc. Natl. Acad. Sci. USA. 2011;108:18661–18665. doi: 10.1073/pnas.1114900108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cleland WW. What limits the rate of an enzyme-catalyzed reaction? Acc. Chem. Res. 1975;8:145–151. [Google Scholar]
- 68.Boehr DD, McElheny D, Dyson HJ, Wright PE. Millisecond timescale fluctuations in dihydrofolate reductase are exquisitely sensitive to the bound ligands. Proc. Natl. Acad. Sci. USA. 2010;107:1373–1378. doi: 10.1073/pnas.0914163107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Maroncelli M, Fleming GR. Comparison of time-resolved fluorescence Stokes shift measurements to a molecular theory of solvation dynamics. J. Chem. Phys. 1988;89:875–881. [Google Scholar]
- 70.Bandaria JN, Dutta S, Nydegger MW, Rock W, Kohen A, Cheatum CM. Characterizing the dynamics of functionally relevant complexes of formate dehydrogenase. Proc. Natl. Acad. Sci. USA. 2010;107:17974–17979. doi: 10.1073/pnas.0912190107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bandaria JN, Cheatum CM, Kohen A. Examination of enzymatic H-tunneling through kinetics and dynamics. J. Am. Chem. Soc. 2009;131:10151–10155. doi: 10.1021/ja902120t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Newby Z, Lee TT, Morse RJ, Liu Y, Liu L, et al. The role of protein dynamics in thymidylate synthase catalysis: variants of conserved 2′-deoxyuridine 5′-monophosphate (dUMP)-binding Tyr-261. Biochemistry. 2006;45:7415–7428. doi: 10.1021/bi060152s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bai Y, Milne JS, Mayne L, Englander SW. Primary structure effects on peptide group hydrogen exchange. Proteins. 1993;17:75–86. doi: 10.1002/prot.340170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hoofnagle AN, Resing KA, Ahn NG. Protein analysis by hydrogen exchange mass spectrometry. Annu. Rev. Biophys. Biomol. Struct. 2003;32:1–25. doi: 10.1146/annurev.biophys.32.110601.142417. [DOI] [PubMed] [Google Scholar]
- 75.Englander JJ, Del Mar C, Li W, Englander SW, Kim JS, et al. Protein structure change studied by hydrogen-deuterium exchange, functional labeling, and mass spectrometry. Proc. Natl. Acad. Sci. USA. 2003;100:7057–7062. doi: 10.1073/pnas.1232301100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kan ZY, Mayne L, Chetty PS, Englander SW. ExMS: data analysis for HX-MS experiments. J. Am. Soc. Mass Spectrom. 2011;22:1906–1915. doi: 10.1007/s13361-011-0236-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kohen A, Cannio R, Bartolucci S, Klinman JP. Enzyme dynamics and hydrogen tunnelling in a thermophilic alcohol dehydrogenase. Nature. 1999;399:496–499. doi: 10.1038/20981. [DOI] [PubMed] [Google Scholar]
- 78.Pudney CR, Hay S, Levy C, Pang J, Sutcliffe MJ, et al. Evidence to support the hypothesis that promoting vibrations enhance the rate of an enzyme catalyzed H-tunneling reaction. J. Am. Chem. Soc. 2009;131:17072–17073. doi: 10.1021/ja908469m. [DOI] [PubMed] [Google Scholar]
- 79.Basran J, Sutcliffe MJ, Scrutton NS. Enzymatic H-transfer requires vibration-driven extreme tunneling. Biochemistry. 1999;38:3218–3222. doi: 10.1021/bi982719d. [DOI] [PubMed] [Google Scholar]
- 80.Harris RJ, Meskys R, Sutcliffe MJ, Scrutton NS. Kinetic studies of the mechanism of carbonhydrogen bond breakage by the heterotetrameric sarcosine oxidase of Arthrobacter sp. 1-IN. Biochemistry. 2000;39:1189–1198. doi: 10.1021/bi991941v. [DOI] [PubMed] [Google Scholar]
- 81.Abad JL, Camps F, Fabrias G. Is hydrogen tunneling involved in acylCoA desaturase reactions? The case of a Δ9 desaturase that transforms (E)-11-tetradecenoic acid into (Z,E)-9,11-tetradecadienoic acid. Angew. Chem. Int. Ed. 2000;39:3279–3281. doi: 10.1002/1521-3773(20000915)39:18<3279::aid-anie3279>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 82.Sikorski RS, Wang L, Markham KA, Rajagopalan PT, Benkovic SJ, Kohen A. Tunneling and coupled motion in the Escherichia coli dihydrofolate reductase catalysis. J. Am. Chem. Soc. 2004;126:4778–4779. doi: 10.1021/ja031683w. [DOI] [PubMed] [Google Scholar]
- 83.Fan F, Gadda G. An internal equilibrium preorganizes the enzyme-substrate complex for hydride tunneling in choline oxidase. Biochemistry. 2007;46:6402–6408. doi: 10.1021/bi700255v. [DOI] [PubMed] [Google Scholar]
- 84.Gupta A, Mukherjee A, Matsui K, Roth JP. Evidence for protein radical-mediated nuclear tunneling in fatty acid α-oxygenase. J. Am. Chem. Soc. 2008;130:11274–11275. doi: 10.1021/ja8042273. [DOI] [PubMed] [Google Scholar]
- 85.Danish HH, Doncheva IS, Roth JP. Hydrogen tunneling steps in cyclooxygenase-2 catalysis. J. Am. Chem. Soc. 2011;133:15846–15869. doi: 10.1021/ja2059523. [DOI] [PubMed] [Google Scholar]
- 86.Grant KL, Klinman JP. Evidence that both protium and deuterium undergo significant tunneling in the reaction catalyzed by bovine serum amine oxidase. Biochemistry. 1989;28:6597–6605. doi: 10.1021/bi00442a010. [DOI] [PubMed] [Google Scholar]
- 87.Jonsson T, Edmondson DE, Klinman JP. Hydrogen tunneling in the flavoenzyme monoamine oxidase B. Biochemistry. 1994;33:14871–14878. doi: 10.1021/bi00253a026. [DOI] [PubMed] [Google Scholar]
- 88.Murakawa T, Okajima T, Kuroda S, Nakamoto T, Taki M, et al. Quantum mechanical hydrogen tunneling in bacterial copper amine oxidase reaction. Biochem. Biophys. Res. Commun. 2006;342:414–423. doi: 10.1016/j.bbrc.2006.01.150. [DOI] [PubMed] [Google Scholar]
- 89.Maglia G, Allemann RK. Evidence for environmentally coupled hydrogen tunneling during dihydrofolate reductase catalysis. J. Am. Chem. Soc. 2003;125:13372–13373. doi: 10.1021/ja035692g. [DOI] [PubMed] [Google Scholar]
- 90.Quaye O, Gadda G. Effect of a conservative mutation of an active site residue involved in substrate binding on the hydride tunneling reaction catalyzed by choline oxidase. Arch. Biochem. Biophys. 2009;489:10–14. doi: 10.1016/j.abb.2009.07.021. [DOI] [PubMed] [Google Scholar]
- 91.Wang L, Goodey NM, Benkovic SJ, Kohen A. Coordinated effects of distal mutations on environmentally coupled tunneling in dihydrofolate reductase. Proc. Natl. Acad. Sci. USA. 2006;103:15753–15758. doi: 10.1073/pnas.0606976103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Stojkovic V, Perissinotti LL, Willmer D, Benkovic SJ, Kohen A. Effects of the donor-acceptor distance and dynamics on hydride tunneling in the dihydrofolate reductase catalyzed reaction. J. Am. Chem. Soc. 2012;134:1738–1745. doi: 10.1021/ja209425w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Seymour SL, Klinman JP. Comparison of rates and kinetic isotope effects using PEG-modified variants and glycoforms of glucose oxidase: the relationship of modification of the protein envelope to C–H activation and tunneling. Biochemistry. 2002;41:8747–8758. doi: 10.1021/bi020054g. [DOI] [PubMed] [Google Scholar]
- 94.Tsai S, Klinman JP. Probes of hydrogen tunneling with horse liver alcohol dehydrogenase at subzero temperatures. Biochemistry. 2001;40:2303–2311. doi: 10.1021/bi002075l. [DOI] [PubMed] [Google Scholar]
- 95.Klinman JP. An integrated model for enzyme catalysis emerges from studies of hydrogen tunneling. Chem. Phys. Lett. 2009;471:179–193. doi: 10.1016/j.cplett.2009.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nagel ZD, Klinman JP. A 21st century revisionist’s view at a turning point in enzymology. Nat. Chem. Biol. 2009;5:543–550. doi: 10.1038/nchembio.204. [DOI] [PubMed] [Google Scholar]
- 97.Johannissen LO, Hay S, Scrutton NS, Sutcliffe MJ. Proton tunneling in aromatic amine dehydrogenase is driven by a short-range sub-picosecond promoting vibration: consistency of simulation and theory with experiment. J. Phys. Chem. B. 2007;111:2631–2638. doi: 10.1021/jp066276w. [DOI] [PubMed] [Google Scholar]
- 98.Marcus RA. Enzymatic catalysis and transfers in solutionITheory and computations, a unified view. J. Chem. Phys. 2006;125:194504. doi: 10.1063/1.2372496. [DOI] [PubMed] [Google Scholar]
- 99.Marcus RA. H and other transfers in enzymes and in solution: theory and computations, a unified view. 2. Applications to experiment and computations. J. Phys. Chem. B. 2007;111:6643–6654. doi: 10.1021/jp071589s. [DOI] [PubMed] [Google Scholar]
- 100.Cui Q, Karplus M. Quantum mechanics/molecular mechanics studies of triosephosphate isomerase-catalyzed reactions: effect of geometry and tunneling on proton-transfer rate constants. J. Am. Chem. Soc. 2002;124:3093–3124. doi: 10.1021/ja0118439. [DOI] [PubMed] [Google Scholar]
- 101.Warshel A, Olsson MHM, Villà-Freixa J. Computer simulations of isotope effects in enzyme catalysis. See Ref. 152. 2006:621–644. [Google Scholar]
- 102.Warshel A, Sharma PK, Kato M, Xiang Y, Liu H, Olsson MH. Electrostatic basis for enzyme catalysis. Chem. Rev. 2006;106:3210–3235. doi: 10.1021/cr0503106. [DOI] [PubMed] [Google Scholar]
- 103.Wang L, Goodey NM, Benkovic SJ, Kohen A. The role of enzyme dynamics and tunnelling in catalysing hydride transfer: studies of distal mutants of dihydrofolate reductase. Philos. TransRSoc. Lond. B. 2006;361:1307–1315. doi: 10.1098/rstb.2006.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wang L, Tharp S, Selzer T, Benkovic SJ, Kohen A. Effects of a distal mutation on active site chemistry. Biochemistry. 2006;45:1383–1392. doi: 10.1021/bi0518242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Dametto M, Antoniou D, Schwartz SD. Barrier crossing in dihydrofolate reductase does not involve a rate-promoting vibration. Mol. Phys. 2012;110:531–536. doi: 10.1080/00268976.2012.655337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Engel H, Doron D, Kohen A, Major DT. Momentum distribution as a fingerprint of quantum delocalization in enzymatic reactions: open-chain path-integral simulations of model systems and the hydride transfer in dihydrofolate reductase. J. Chem. Theory Comput. 2012;8:1223–1234. doi: 10.1021/ct200874q. [DOI] [PubMed] [Google Scholar]
- 107.Schrank TP, Bolen DW, Hilser VJ. Rational modulation of conformational fluctuations in adenylate kinase reveals a local unfolding mechanism for allostery and functional adaptation in proteins. Proc. Natl. Acad. Sci. USA. 2009;106:16984–16989. doi: 10.1073/pnas.0906510106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Motlagh HN, Hilser VJ. Agonism/antagonism switching in allosteric ensembles. Proc. Natl. Acad. Sci. USA. 2012;109:4134–4139. doi: 10.1073/pnas.1120519109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Adamczyk AJ, Warshel A. Converting structural information into an allosteric-energy-based picture for elongation factor Tu activation by the ribosome. Proc. Natl. Acad. Sci. USA. 2011;108:9827–9832. doi: 10.1073/pnas.1105714108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sours KM, Kwok SC, Rachidi T, Lee T, Ring A, et al. Hydrogen-exchange mass spectrometry reveals activation-induced changes in the conformational mobility of p38α MAP kinase. J. Mol. Biol. 2008;379:1075–1093. doi: 10.1016/j.jmb.2008.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Arora K, Brooks CL., 3rd Functionally important conformations of the Met20 loop in dihydrofolate reductase are populated by rapid thermal fluctuations. J. Am. Chem. Soc. 2009;131:5642–5647. doi: 10.1021/ja9000135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Benkovic SJ, Hammes GG, Hammes-Schiffer S. Free-energy landscape of enzyme catalysis. Biochemistry. 2008;47:3317–3321. doi: 10.1021/bi800049z. [DOI] [PubMed] [Google Scholar]
- 113.Swint-Kruse L, Fisher HF. Enzymatic reaction sequences as coupled multiple traces on a multidimensional landscape. Trends Biochem. Sci. 2008;33:104–112. doi: 10.1016/j.tibs.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 114.Roca M, Messer B, Hilvert D, Warshel A. On the relationship between folding and chemical landscapes in enzyme catalysis. Proc. Natl. Acad. Sci. USA. 2008;105:13877–13882. doi: 10.1073/pnas.0803405105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sen A, Kohen A. Quantum effects in enzyme kinetics. In: Allemann RK, Scrutton N, editors. Quantum Tunneling in Enzyme Catalyzed Reactions. London: R. Soc. Chem.; 2009. pp. 161–178. [Google Scholar]
- 116.Somero GN. Proteins and temperature. Annu. Rev. Physiol. 1995;57:43–68. doi: 10.1146/annurev.ph.57.030195.000355. [DOI] [PubMed] [Google Scholar]
- 117.Kohen A, Klinman JP. Protein flexibilty correlates with degree of hydrogen tunneling in thermophilic and mesophilic alcohol dehydrogenases. J. Am. Chem. Soc. 2000;122:10738–10739. [Google Scholar]
- 118.Liang ZX, Tsigos I, Lee T, Bouriotis V, Resing KA, et al. Evidence for increased local flexibility in psychrophilic alcohol dehydrogenase relative to its thermophilic homologue. Biochemistry. 2004;43:14676–14683. doi: 10.1021/bi049004x. [DOI] [PubMed] [Google Scholar]
- 119.Liang ZX, Lee T, Resing KA, Ahn NG, Klinman JP. Thermal-activated protein mobility and its correlation with catalysis in thermophilic alcohol dehydrogenase. Proc. Natl. Acad. Sci. USA. 2004;101:9556–9561. doi: 10.1073/pnas.0403337101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ceccarelli C, Liang ZX, Strickler M, Prehna G, Goldstein BM, et al. Crystal structure and amide H/D exchange of binary complexes of alcohol dehydrogenase from Bacillus stearothermophilus: insight into thermostability and cofactor binding. Biochemistry. 2004;43:5266–5277. doi: 10.1021/bi049736p. [DOI] [PubMed] [Google Scholar]
- 121.Liang ZX, Tsigos I, Bouriotis V, Klinman JP. Impact of protein flexibility on hydride-transfer parameters in thermophilic and psychrophilic alcohol dehydrogenases. J. Am. Chem. Soc. 2004;126:9500–9501. doi: 10.1021/ja047087z. [DOI] [PubMed] [Google Scholar]
- 122.Nagel ZD, Dong M, Bahnson BJ, Klinman JP. Impaired protein conformational landscapes as revealed in anomalous Arrhenius prefactors. Proc. Natl. Acad. Sci. USA. 2011;108:10520–10525. doi: 10.1073/pnas.1104989108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Nagel ZD, Meadows CW, Dong M, Bahnson BJ, Klinman JP. Active site hydrophobic residues impact hydrogen tunneling differently in a thermophilic alcohol dehydrogenase at optimal versus nonoptimal temperatures. Biochemistry. 2012;51:4147–4156. doi: 10.1021/bi3001352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kim HS, Damo SM, Lee SY, Wemmer D, Klinman JP. Structure and hydride transfer mechanism of a moderate thermophilic dihydrofolate reductase from Bacillus stearothermophilus and comparison to its mesophilic and hyperthermophilic homologues. Biochemistry. 2005;44:11428–11439. doi: 10.1021/bi050630j. [DOI] [PubMed] [Google Scholar]
- 125.Oyeyemi OA, Sours KM, Lee T, Resing KA, Ahn NG, Klinman JP. Temperature dependence of protein motions in a thermophilic dihydrofolate reductase and its relationship to catalytic efficiency. Proc. Natl. Acad. Sci. USA. 2010;107:10074–10079. doi: 10.1073/pnas.1003678107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Oyeyemi OA, Sours KM, Lee T, Kohen A, Resing KA, et al. Comparative hydrogen-deuterium exchange for a mesophilic versus thermophilic dihydrofolate reductase at 25°C: identification of a single active site region with enhanced flexibility in the mesophilic protein. Biochemistry. 2011;50:8251–8260. doi: 10.1021/bi200640s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Marlow MS, Dogan J, Frederick KK, Valentine KG, Wand AJ. The role of conformational entropy in molecular recognition by calmodulin. Nat. Chem. Biol. 2010;6:352–358. doi: 10.1038/nchembio.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Heyes DJ, Sakuma M, de Visser SP, Scrutton NS. Nuclear quantum tunneling in the light-activated enzyme protochlorophyllide oxidoreductase. J. Biol. Chem. 2009;284:3762–3767. doi: 10.1074/jbc.M808548200. [DOI] [PubMed] [Google Scholar]
- 129.Hammes-Schiffer S, Soudackov AV. Proton-coupled electron transfer in solution, proteins, and electrochemistry. J. Phys. Chem. B. 2008;112:14108–14123. doi: 10.1021/jp805876e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Moser CC, Keske JM, Warncke K, Farid RS, Dutton PL. Nature of biological electron transfer. Nature. 1992;355:796–802. doi: 10.1038/355796a0. [DOI] [PubMed] [Google Scholar]
- 131.Gray HB, Winkler JR. Electron transfer in proteins. Annu. Rev. Biochem. 1996;65:537–561. doi: 10.1146/annurev.bi.65.070196.002541. [DOI] [PubMed] [Google Scholar]
- 132.Hegazi MF, Borchardt RT, Schowen RL. α-Deuterium and carbon-13 isotope effects for methyl transfer catalyzed by catechol O-methyltransferase. SN2-like transition state. J. Am.Chem. Soc. 1979;101:4359–4365. [Google Scholar]
- 133.Horowitz S, Yesselman JD, Al-Hashimi HM, Trievel RC. Direct evidence for methyl group coordination by carbon-oxygen hydrogen bonds in the lysine methyltransferase SET7/9. J. Biol. Chem. 2011;286:18658–18663. doi: 10.1074/jbc.M111.232876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zhang J, Klinman JP. Enzymatic methyl transfer: role of an active site residue in generating active site compaction that correlates with catalytic efficiency. J. Am. Chem. Soc. 2011;133:17134–17137. doi: 10.1021/ja207467d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Doll KM, Finke RG. A compelling experimental test of the hypothesis that enzymes have evolved to enhance quantum mechanical tunneling in hydrogen transfer reactions: the β-neopentylcobalamin system combined with prior adocobalamin data. Inorg. Chem. 2003;42:4849–4856. doi: 10.1021/ic0300722. [DOI] [PubMed] [Google Scholar]
- 136.Doll KM, Bender BR, Finke RG. The first experimental test of the hypothesis that enzymes have evolved to enhance hydrogen tunneling. J. Am. Chem. Soc. 2003;125:10877–10884. doi: 10.1021/ja030120h. [DOI] [PubMed] [Google Scholar]
- 137.Kwart H, Briechbiel MW, Acheson RM, Ward DC. Observations on the geometry of hydrogen transfer in [1,5] sigmatropic rearrangements. J. Am. Chem. Soc. 1982;104:4671–4672. [Google Scholar]
- 138.Kwart H. Temperature dependence of the primary kinetic hydrogen isotope effect as amechanistic criterion. Acc. Chem. Res. 1982;15:401–408. [Google Scholar]
- 139.Roth WR, König J. Hydrogen displacements. IV. Kinetic isotope effect of the 1.5 hydrogen displacement in cis-pentadiene-(1.3) Justus Liebigs Ann. Chem. 1966;699:24–32. doi: 10.1002/jlac.19666990103. (In German) [DOI] [PubMed] [Google Scholar]
- 140.Braun J, Schwesinger R, Williams PG, Morimoto H, Wemmer DE, Limbach HH. KineticH/D/T isotope and solid state effects on the tautomerism of the conjugate porphyrin monoanion. J. Am. Chem. Soc. 1966;118:11101–11110. [Google Scholar]
- 141.Limbach HH, Miguel Lopez J, Kohen A. Arrhenius curves of hydrogen transfers: tunnel effects, isotope effects and effects of pre-equilibria. Philos. TransRSoc. Lond. B. 2006;361:1399–1415. doi: 10.1098/rstb.2006.1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wilde TC, Blotny G, Pollack RM. Experimental evidence for enzyme-enhanced coupled motion/quantum mechanical hydrogen tunneling by ketosteroid isomerase. J. Am. Chem. Soc. 2008;130:6577–6585. doi: 10.1021/ja0732330. [DOI] [PubMed] [Google Scholar]
- 143.Roston D, Kohen A. Elusive transition state of alcohol dehydrogenase unveiled. Proc. Natl. Acad. Sci. USA. 2010;107:9572–9577. doi: 10.1073/pnas.1000931107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Mahammed A, Gross Z. The importance of developing metal complexes with pronounced catalaselike activity. Catal. Sci. Technol. 2011;1:535–540. [Google Scholar]
- 145.Xuereb DJ, Raja R. Design strategies for engineering selectivity in bio-inspired heterogeneous catalysts. Catal. Sci. Technol. 2011;1:517–534. [Google Scholar]
- 146.Bhandari R, Coppage R, Knecht MR. Mimicking nature’s strategies for the design of nanocatalysts. Catal. Sci. Technol. 2012;2:256–266. [Google Scholar]
- 147.Jiang L, Althoff EA, Clemente FR, Doyle L, Rothlisberger D, et al. De novo computational design of retro-aldol enzymes. Science. 2008;319:1387–1391. doi: 10.1126/science.1152692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Schramm VL. Enzymatic transition states and transition state analogues. Curr. Opin. Struct. Biol. 2005;15:604–613. doi: 10.1016/j.sbi.2005.10.017. [DOI] [PubMed] [Google Scholar]
- 149.Glickman MH, Klinman JP. Extremely large isotope effects in the soybean lipoxygenase-linoleic acid reaction. J. Am. Chem. Soc. 1994;116:793–794. [Google Scholar]
- 150.McCusker KP, Klinman JP. An active-site phenylalanine directs substrate binding and C-Hcleavage in the α-ketoglutarate-dependent dioxygenase TauD. J. Am. Chem. Soc. 2010;132:5114–5120. doi: 10.1021/ja909416z. [DOI] [PubMed] [Google Scholar]
- 151.Cheatum CM, Kohen A. Relationship of femtosecond-picosecond dynamics to enzyme catalyzed H-transfer. In: Hammes-Schiffer S, Klinman JP, editors. Topics in Current Chemistry: Dynamics in Enzyme Catalysis. Heidelberg, Ger.: Springer; 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Kohen A, Limbach HH. Isotope Effects in Chemistry and Biology. Boca Raton, FL: CRC; 2006. [Google Scholar]
- 153.Nagel ZD, Cun S, Klinman JP. Identification of a long-range protein network that modulates active site dynamics in extremophilic alcohol dehydrogenases. J. Biol. Chem. 2013 doi: 10.1074/jbc.M113.453951. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]