Abstract
Background
Many genetic risk variants are now well established in multiple sclerosis (MS), but the impact on clinical phenotypes is unclear.
Objective
To investigate the impact of established MS genetic risk variants on MS phenotypes, in well-characterized MS cohorts.
Methods
Norwegian MS patients (n = 639) and healthy controls (n = 530) were successfully genotyped for 61 established MS-associated single nucleotide polymorphisms (SNPs). Data including and excluding Major Histocompatibility Complex (MHC) markers were summed to a MS Genetic Burden (MSGB) score. Study replication was performed in a cohort of white American MS patients (n = 1997) and controls (n = 708).
Results
The total human leukocyte antigen (HLA) and the non-HLA MSGB scores were significantly higher in MS patients than in controls, in both cohorts (P << 10 −22). MS patients, with and without cerebrospinal fluid (CSF) oligoclonal bands (OCBs), had a higher MSGB score than the controls; the OCB-positive patients had a slightly higher MSGB than the OCB-negative patients. An early age at symptom onset (AAO) also correlated with a higher MSGB score, in both cohorts.
Conclusion
The MSGB score was associated with specific clinical MS characteristics, such as OCBs and AAO. This study underlines the need for well-characterized, large cohorts of MS patients, and the usefulness of summarizing multiple genetic risk factors of modest effect size in genotype-phenotype analyses.
Keywords: Age of onset, cerebrospinal fluid, genetic association, genetic risk, genotype, multiple sclerosis, oligoclonal bands, Multiple Sclerosis Genetic Burden
Introduction
Multiple sclerosis (MS) is an inflammatory, demyelinating disease of the central nervous system (CNS),1 caused most likely by the interaction between genetic and environmental factors.2 Recent advances in single nucleotide polymorphism (SNP) array methodologies and improved analytic capabilities, combined with a cooperative global effort to clarify the genetic underpinnings of MS, have succeeded in elucidating over 50 common DNA variants that are unequivocally associated with this complex disorder.2–5 The majority of risk markers are located nearby or within genes that are known to have roles in the immune system, arguing for a primary immune, and likely autoimmune, etiology for MS.
Another principle of the genetic landscape of MS is that, with the sole exception of the MHC region, each variant confers only a very small contribution to MS risk (odds ratio (OR) < 1.2). Given the small effect sizes, even in aggregate, the identified genetic MS risk variants confer only a fraction of the total inherited risk, as estimated from family studies including twin studies, and at an individual level they are insensitive for risk assessments; however, at a population level, genetic risk factors could represent important new tools to better understand the clinical biology of MS, including heterogeneity. For example, we recently identified genetic networks that are associated with different patterns of MS lesion distribution in the CNS.6
The MS Genetic Burden (MSGB) score is a useful statistic that sums the aggregate genetic risk of MS, based on validated association signals derived from genetic studies of MS.7 Beyond the association with susceptibility, this score provides a simple metric to explore the influence, if any, of genetic burden on disease expression.
A registry for MS was created in the high-risk population of Oslo, Norway, with a population of more than 500,000 inhabitants, that supports population-based investigations of this well-characterized and longitudinally monitored cohort of MS patients,8 facilitating standardization of data collection in this region. For example: systematic lumbar punctures are obtained, enabling population-wide assessments of cerebrospinal fluid (CSF) parameters, like oligoclonal bands (OCBs). The current study’s aim was to investigate the associations of the MSGB score with clinical and paraclinical data in this highly characterized Oslo MS cohort. To replicate our findings, we used an independent well-characterized dataset from the University of California San Francisco (UCSF), US, representing a large sample of white American MS patients and controls recruited at that site.
Materials and methods
The Oslo MS registry established in 1992 contains clinical data from 1648 patients, all collected at the Oslo University Hospital, Ullevål, Oslo, Norway. The Oslo MS clinic diagnoses and prospectively follows closely all MS cases within the Oslo population. Approximately one-half of these patients have donated blood for genetic studies. In this study, 639 Norwegian MS patients and 530 healthy controls obtained from the Norwegian Bone Marrow Registry were successfully genotyped (Table 1(a)) for 61 well-established MS-associated SNPs, using TaqMan® OpenArray® genotyping technology (Life Technologies, Carlsbad, CA, US) (Supplementary Table 1). For the MSGB calculation we used 61 successfully genotyped SNPs, 3 SNPs from the intended 64 SNPs showed call rates below 90% (rs2028597, rs2150702 and rs10411936) and were excluded before MSGB calculation. From the HLA region, we included rs3129889 tagging HLA-DRB1*15:01 and rs2523393, tagging HLA-B*44.9 The samples had not been genotyped for this SNP panel in previous studies. Classical HLA typing data from the HLA-A, -B, -C and -DRB1 loci were available for a subgroup of the included study participants (Table 1(b)). The Oslo MS samples are described in further detail, in earlier studies.10
Table I.
(a) Clinical data of our study’s included MS and control cohorts. (b) HLA data of our study’s included MS and control cohorts.
| 1(a) | |||||
|---|---|---|---|---|---|
|
| |||||
| Clinical data of the Oslo MS-control cohort |
MS patients from the Oslo MS registry |
Oslo MS patients genotyped |
Oslo controls genotyped |
UCSF MS patients genotyped |
UCSF controls genotyped |
| N = 1648 | N = 639 | N = 530 | N = 1997 | N = 708 | |
| Female, % | 68.5 | 73.0 | 68.6 | 73.0 | 52.7 |
| RRMS, % | 79.5a | 82.6 | 90.4 | ||
| PPMS, % | 20.5a | 17.4 | – | 8.5 | |
| AAO, mean (SE) | 32.4 (0.27)b | 32.4 (0.42) | – | 31.6 (0.21) | |
| Age at EDSS exam, mean (SE) | 49.1 (0.42)c | 50.2 (0.55) | – | 43.6 (0.24) | |
| OCB positive, % | 84.8d | 88.7d | 69.3d | ||
| EDSS, mean (SE) | 4.5 (0.09)c | 4.6 (0.12) | – | 3.49 (0.05) | |
| MSSS, mean (SE) | 4.76 (0.11)e | 4.80 (0.14) | – | 4.75 (0.06) | |
|
| |||||
| 1(b) | |||||
|
| |||||
| Associated HLA and DRB1 alleles in Oslo MS cohort subsetf |
Allele | Allele frequency in MS (%) |
Allele frequency in Control (%) |
p-value | |
|
| |||||
| 2n = 1156 | 2n = 698 | ||||
| HLA-A | *02 | 326 (28.2) | 236 (33.8) | 0.0092 | |
| *03 | 228 (19.7) | 108 (15.5) | 0.0251 | ||
| 2n = 1140 | 2n = 699 | ||||
| HLA-B | *07 | 285 (25) | 102 (14.6) | 0.0001 | |
| *44 | 58 (5.1) | 92 (13.2) | 0.0001 | ||
| 2n = 1211 | 2n = 625 | ||||
| HLA-C | *07 | 467 (39.3) | 178 (28.5) | 0.0001 | |
| *03 | 207 (17.4) | 133 (21.3) | 0.0428 | ||
| *06 | 63 (5.3) | 52 (8.3) | 0.0146 | ||
| 2n = 1183 | 2n = 700 | ||||
| HLA-DRB1 | *04 | 181 (15.3) | 156 (22.3) | 0.6119g | |
| *15 | 460 (38.8) | 103 (14.8) | 1*10−26 | ||
| *01 | 103 (8.7) | 88 (12.6) | 0.7536g | ||
| *13 | 100 (8.4) | 86 (12.3) | 0.7506g | ||
| *03 | 128 (10.8) | 83 (11.9) | 0.0824g | ||
| *07 | 68 (5.7) | 79 (11.3) | 0.0277g | ||
From 1485 patients.
From 1504 patients.
From 927 patients.
Based on 1293 patients in the Oslo MS registry, 568 genotyped patients from the Oslo MS registry and 760 patients from UCSF. OCB measured by isoelectric focusing and agarose gel electrophoresis.
From 735 patients.
Available classical HLA typing data for the included MS cases and controls, from the Oslo cohort.
HLA-DRB1*15 was excluded from the analysis.
AOO: Age at onset; EDSS: Expanded Disability Status Scale; HLA: human leukocyte antigen; MS: multiple sclerosis; MSSS: MS severity score; OCB: oligoclonal bands; PPMS: primary progressive MS; RRMS: relapsing–remitting MS; SE: standard error; UCSF: University of CA - San Francisco.
By using the OpenArray® Genotyping Analysis Software, we visually inspected genotype plots and we were able to exclude samples in overlapping or questionable clusters from the analyses. We excluded samples from individuals with an average call rate below 90%. The resulting sample call rate was 98.1% in the Oslo MS cases and 98.6% in the Oslo controls.
By using identical criteria for quality control as in the Norwegian dataset, the replication sample collected at UCSF consisted of 1997 MS cases and 708 healthy controls (Table 1(a)). These were genotyped for the same SNP panel, using the same technology mentioned above. The UCSF replication sample is an extension of the dataset that was described in detail previously.7 The sample call rates for the UCSF replication dataset were 97.9% in MS patients and 98.7% in controls.
We performed the clinical and paraclinical examinations in both cohorts according to established international guidelines and practices. All MS patients were diagnosed according to the updated McDonald diagnostic criteria.11 Gender, disease course, age at symptom onset (AAO), Expanded Disability Status Scale (EDSS),12 multiple sclerosis severity score (MSSS),13 OCB in CSF and information about MS in the family was recorded. OCB positivity was determined either by isoelectric focusing or agarose gel electrophoresis, both in the Oslo14 and UCSF cohorts. The IgG index was not available for all patients, so was therefore not included in the analysis.
We calculated the MSGB score per sample using a log-additive model, as described by Gourraud et al.,7 and it was repeated after excluding the SNPs of the MHC region (6p21.3), in order to evaluate the contribution of non-HLA MS risk variants. In the rare event that an individual’s genotype for a SNP was missing, we used the risk allele frequency in the healthy control population for that SNP, to estimate the complete MSGB score for that individual. We analyzed the phenotype-MSGB correlations using Chi-square statistics, t-test and linear logistic regression, including MSGB scores as a predictor and gender as a covariate. We performed analyses using IBM SPSS 20.0 and R statistical software. The P-values provided were not corrected.
Results
Clinical and demographic characteristics are shown, in Table 1(a), of the total of 1648 Oslo MS cases, the 639 Oslo MS cases and 530 Oslo controls that were successfully genotyped, as well as the UCSF replication set of 1997 MS cases and 708 controls. There were no significant differences between all Oslo MS cases and the genotyped cases from the Oslo cohort regarding sex, AAO, disease course nor disability. As expected, classical HLA data available for a large subset of the Oslo MS cases and controls that were included in the MSGB analysis confirmed that there was a strong association of MS to HLA-DRB1*15:01. The strongest associations for HLA class I alleles were to HLA-A*02, HLA-B*7, HLA-B*44 and HLA-C*07 (Table 1(b)).
The clinical characteristics of the white American replication cohort are shown for comparison in Table 1(a). The relative proportion of relapsing–remitting MS patients was higher in the UCSF cohort; also, they were on average somewhat younger at examination and had slightly lower EDSS and MSSS scores. This may be explained by the differences in recruitment to the two MS clinics: candidates recruited at UCSF are often referred for evaluation of FDA-approved immune modulatory therapies, whereas MS patients in the Oslo clinic represent the whole MS population in Oslo.
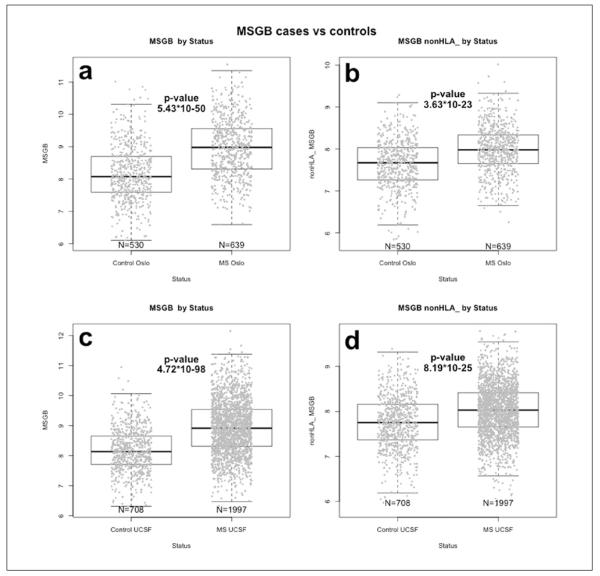
The total MSGB score in the Oslo MS cases, ranged from 6.59–11.54, whereas the non-HLA MSGB ranged from 6.25–10.02. Oslo MS patients had a significantly higher total MSGB score as compared to controls, respectively: mean MSGB (standard error (SE)) 8.97 (0.03) versus 8.18 (0.04), P = 5.44*10−50 and non-HLA MSGB score (mean (SE) 7.98 (0.02) versus 7.64 (0.03), P = 3.63*10−23, as shown in Figure 1(a) and Figure 1(b). This finding was replicated in the UCSF cohort (MSGB score mean (SE) 8.94 (0.02) versus 8.19 (0.03), P = 4.72*10−98; non-HLA MSGB score mean (SE) 8.02 (0.01) versus 7.76 (0.02), P = 8.19*10−25 (Figure 1(c) and Figure 1(d)). In the Oslo cohort, there were no statistical differences in the total MSGB scores between MS cases reporting any family history of MS (n = 124) and those reporting no family history (n = 515) (P = 0.629), in contrast to what was observed in the UCSF family studies earlier7 and in the present UCSF dataset (n = 433 reported family history of MS, versus n = 1143 whom reported no familial history of MS; P = 1.35*10−3). The Oslo MS subgroup with a family history of MS was too small to have sufficient power for analyses stratified for first-degree or second-degree relatives with MS in the family.
Figure 1.
Comparison of MSGB according to disease status.
(a) Total MSGB boxplots for patients and controls in the Oslo cohort.
(b) Total MSGB excluding HLA contribution (MSGB non-HLA) boxplots for patients and controls in the Oslo cohort.
(c) Total MSGB boxplots for patients and controls collected at UCSF.
(d) Total MSGB excluding HLA contribution (non-HLA MSGB) boxplots for patients and controls collected at UCSF.
HLA: Human leukocyte antigen; MS: multiple sclerosis; MSGB: MS genetic burden; UCSF: University of CA, San Francisco.
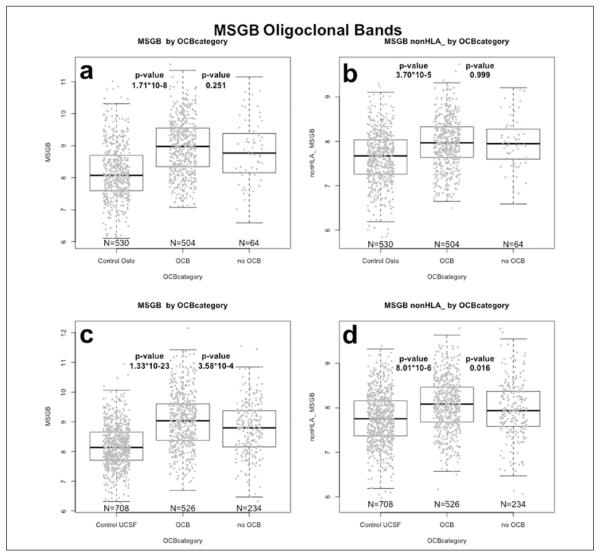
The OCB-positive MS patients from Oslo (n = 504) had a higher MSGB score than OCB-negative patients (n = 64), when including HLA in the score estimation (MSGB mean (SE) 8.96 (0.04) versus 8.83 (0.12), although this did not reach significance in the Oslo sample (P = 0.251). No difference was observed for the non-HLA MSGB for the OCB positives, versus OCB-negative Oslo MS patients (MSGB mean (SE) 7.96 (0.02) versus 7.96 (0.07), P = 0.999); however, there was a very significant difference between OCB-positive MS patients and controls, for both the total MSGB score and the non-HLA MSGB score (P = 1.71 *10−8 and P = 3.70 *10−5 (Figure 2(a) and 2(b)). In the UCSF samples, the difference in MSGB between OCB-positive (n = 526) and OCB-negative MS patients (n = 234) was significant, both when including and excluding the HLA region markers (MSGB mean (SE) 9.04 (0.04) versus 8.78 (0.06), P = 3.58*10−4; non-HLA MSGB mean (SE) 8.07 (0.03) versus 7.96 (0.04), P = 0.016) (Figure 2(c) through Figure 2(d)).
Figure 2.
Comparison of MSGB, according to OCB positivity.
(a) Total MSGB boxplots for OCB-negative and OCB-positive MS patients, and controls, in Oslo cohort.
(b) Total MSGB excluding HLA contribution (non-HLA MSGB) boxplots for OCB-negative and OCB-positive MS patients, and controls, in Oslo cohort.
(c) Total MSGB boxplots for OCB-negative and OCB-positive MS patients, and controls, in UCSF cohort.
(d) Total MSGB excluding HLA contribution (non-HLA MSGB) boxplots for OCB-negative and OCB-positive MS patients in the UCSF cohort.
HLA: Human leukocyte antigen; MS: multiple sclerosis; MSGB: MS genetic burden; OCB: oligoclonal band; UCSF: University of CA, San Francisco.
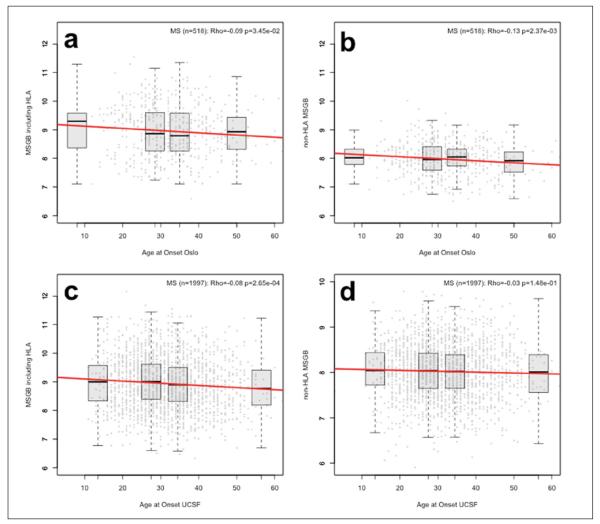
MSGB score (P = 0.035) in the Oslo MS patients, and this was more pronounced for the non-HLA MSGB score (P = 0.003) (Figure 3(a) through 3(b)). An earlier age at onset was also observed for the UCSF patients with a higher total MSGB score (P = 2.65*10−4); however, this association was observed only when including the HLA in the score calculation (Figure 3(c) and Figure 3(d)). No differences were observed between the disease courses of RRMS nor PPMS, nor female versus male gender (data not shown). Also, the total MSGB and non-HLA MSGB scores were not associated with the MS severity score (MSSS)13 in the Oslo samples (Supplementary Figure 1(a) and Supplementary Figure 1(b)), and the MSSS did not differ between patients with and without OCBs in CSF (data not shown).
Figure 3.
Association between MSGB and AAO, illustrated as scatterplots, showing MSGB versus AAO in each cohort. A linear regression line is drawn in red. We provide Rho estimates and p-values from the Spearman’s non-parametric correlation test. Boxplots for each quartile are displayed and located at the mean value of quartile group on the x-axis.
(a) Oslo cohort scatterplot of MSGB versus AAO.
(b) Oslo cohort scatterplot of non-HLA MSGB versus AAO. (c) UCSF sample scatterplot of MSGB versus AAO.
(d) UCSF sample scatterplot of non-HLA MSGB versus AAO.
AAO: Age at onset of MS; HLA: human leukocyte antigen; MS: multiple sclerosis; MSGB: MS genetic burden; UCSF: University of CA, San Francisco.
Discussion
This study reports that the total MSGB and non-HLA MSGB scores were significantly higher in MS cases from the population-based Oslo MS registry, compared to healthy controls, as well as from a large white American sample collected at UCSF. Indeed, it is remarkable how similar the results from the two datasets were, despite the independent collections and the genetic architecture of the populations sampled. This observation strongly confirms the validity of the MSGB score in diverse MS cohorts.
The observation that the total MSGB score is higher in both OCB-positive and -negative MS patients, compared to controls, may indicate that these two MS phenotypes share the same degree of genetic risk factors. In the UCSF cohort, which was better powered than the Oslo cohort, we also confirmed a higher MSGB score in OCB-positive versus OCB-negative MS patients. This concurs with our recent Scandinavian study, which reported a stronger association with HLA-DRB1*15:01 in OCB-positive than OCB-negative MS patients.14 As the total MSGB score represented an aggregated risk score that was strongly influenced by the effect of HLA-DRB1*15:01, it is possible that the MSGB-OCB relationship could be due to an effect of HLA-DRB1*1501, independent of the other 60 loci comprising the MSGB estimation in our analysis. However, in the larger UCSF sample, the difference between OCB-positive and OCB-negative MS patients remained significant, even when excluding the HLA markers in the MSGB estimation. This novel finding indicates that non-HLA genes also have impact on the OCB-positive phenotype. OCBs in CSF are present in the majority of MS patients.15,16 Of note, the proportion of OCB-negative MS patients was larger in the white American than the Norwegian cohort, which may be due to population-specific differences.17,18 We cannot rule out some differences in laboratory methods between the centers, although both clinics evaluated OCB in CSF with sensitive, modern techniques. Earlier, inconsistent reports on the clinical differences between MS patients with and without OCB in the CSF were published.19–24 In our study, we did not identify differences in MSGB scores in the patients with phenotypes like gender, disease course and severity, but we did identify a lower AAO leave “occurred” away with higher MSGB scores.
Interestingly, the higher MSGB scores observed in patients with lower AAO were identified in both populations. A possible interpretation is that the accumulation of genetic risk factors is lowering the threshold for the expression of the disease thereby lowering AAO. A lower age at onset in MS patients carrying the MS-associated HLA-DRB1*15:01 allele was previously reported.3,25–28 In agreement with earlier observations in the UCSF cohort,7 the MSGB score was not associated with disease course (RRMS versus PPMS) in the Oslo dataset. In addition, we could not find an association between MSGB and disability trajectories, as measured by MSSS.
The MSGB score is helpful for summarizing the per-patient genetic burden, although this score cannot be used for disease risk prediction in the clinic for an individual patient. Our study illustrates the usefulness of this score in the assessment of differences between groups of patients, by being able to show differences in genetic impact on phenotypes. Genotyping costs are reduced by estimating the summarized genetic risk only by genotyping the established risk loci; however, aggregate scores for disease risk loci are vulnerable for missing SNP genotypes, especially in the absence of proxy markers (as in a genome-wide screening), that may be used to impute genotypes. We applied a conservative approach after strict quality control, by replacing the few missing genotypes in both patients and controls with the risk allele frequencies in the healthy control population, for that specific locus, when estimating the MSGB scores. The MSGB score is not affected much by this replacement, except reducing the power to detect deviations on an individual level, but it saves power to detect the difference between the groups, by keeping as many individuals as possible in the study.
By conducting discovery and replication studies in two independent MS centers, recruitment biases or potential differences in the manner in which clinical measurements are obtained can be overcome, and results are thus likely to be widely applicable across different practice settings. Evaluations of AAO were assessed in the same manner at the time of the initial visit, in both MS centers. OCB status didn’t prove amenable to standardization across different laboratories; and thus, it was particularly encouraging that similar associations were identified in our two study populations. Analyses of other phenotypic parameters, such as metrics derived from magnetic resonance imaging (MRI) of brain and spinal cord, will likely require prospective studies using standardized MRI protocols across the recruiting clinics. Establishment of well-defined protocols applicable both in clinical practice and in research across different clinics and continents are needed for well-powered genotype-phenotype analyses, but the current data indicate that at least for some parameters, large sample sizes coupled with the use of discovery and replication cohorts can overcome the variability inherent in comparison of data from different cohorts.
In conclusion, analysis of a population-based MS cohort from Oslo, Norway and a replication study in a MS cohort from San Francisco revealed that aggregated MS genetic burden scores, calculated from the current list of MS-associated genetic variants, is associated with OCB positivity as well as AAO. The MSGB summary scoring methods open a new metric for assessment of differences that may prove useful in exploring how genetic risk contributes to clinical phenotypes, environmental risk and underlying immuno-pathogenic heterogeneity in MS.
Supplementary Material
Acknowledgement
We acknowledge the UCSF Neurology Laboratory at University of CA, US, for providing genotyping facilities; and acknowledge the Norwegian Bone Marrow Registry and Benedicte A Lie for access to the Norwegian controls included in the study.
Funding This work was supported by the Research Council of Norway (HFH grant numbers 189639 and 196776). Genotyping of the Oslo cohort was partly supported by Novartis Norway (unrestricted grant). Data collection and genotyping in the US was supported by the US National Institute of Health (grant number RO1-NS26799). Postdoctoral fellow NI was supported by the Japan Society for Promotion of Science. PAG was supported by the Nancy Davis Foundation for Multiple Sclerosis (Junior Faculty Award).
Footnotes
Conflict of interest The authors declare that there is no conflict of interest.
Notes HFH initiated the project and participated in the study design, contributed to the establishment and funding of the Oslo dataset, performed the experiments, contributed to the statistical analysis, and drafted and edited the manuscript. NI participated in experiments, statistical analysis and edited the manuscript. PBH contributed to establishment of the Oslo dataset, clinical analyses, drafting and editing of the manuscript. SDB contributed to the statistical analysis, drafting and editing of the manuscript. SJC assisted in sample preparation, genotyping and management. MWG and ILM contributed to establishment of the Oslo dataset, sample preparation and management. EGC contributed to establishment, funding and clinical analyses of the Oslo dataset, and editing of the manuscript. SLH contributed to the design, as well as phenotypic characterization, of the UCSF study participants and critically revised the manuscript. JRO contributed to the initiation and study design of the project, supervised the study and edited the manuscript. PAG contributed to the initiation and study design of the project, supervised the experiments and statistical analyses, and edited the manuscript.
References
- 1.Compston A, Coles A. Multiple sclerosis. Lancet. 2008;372:1502–1517. doi: 10.1016/S0140-6736(08)61620-7. [DOI] [PubMed] [Google Scholar]
- 2.Gourraud PA, Harbo HF, Hauser SL, et al. The genetics of multiple sclerosis: An up-to-date review. Immunol Rev. 2012;248:87–103. doi: 10.1111/j.1600-065X.2012.01134.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sawcer S, Hellenthal G, Pirinen M, et al. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature. 2011;476:214–219. doi: 10.1038/nature10251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Jager PL, Jia X, Wang J, et al. Meta-analysis of genome scans and replication identify CD6, IRF8 and TNFRSF1A as new multiple sclerosis susceptibility loci. Nat Genet. 2009;41:776–782. doi: 10.1038/ng.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Patsopoulos NA, Esposito F, Reischl J, et al. Genome-wide meta-analysis identifies novel multiple sclerosis susceptibility loci. Ann Neurol. 2011;70:897–912. doi: 10.1002/ana.22609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gourraud PA, Sdika M, Khankhanian P, et al. A genome-wide association study of brain lesion distribution in multiple sclerosis. Brain. 2013;136:1012–1024. doi: 10.1093/brain/aws363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gourraud PA, McElroy JP, Caillier SJ, et al. Aggregation of multiple sclerosis genetic risk variants in multiple and single case families. Ann Neurol. 2011;69:65–74. doi: 10.1002/ana.22323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smestad C, Sandvik L, Holmoy T, et al. Marked differences in prevalence of multiple sclerosis between ethnic groups in Oslo, Norway. J Neurol. 2008;255:49–55. doi: 10.1007/s00415-007-0659-8. [DOI] [PubMed] [Google Scholar]
- 9.Cree BA, Rioux JD, McCauley JL, et al. A Major Histocompatibility Class I locus contributes to multiple sclerosis susceptibility independently from HLA-DRB1*15:01. PLoS One. 2010;5:e11296. doi: 10.1371/journal.pone.0011296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lorentzen AR, Karlsen TH, Olsson M, et al. Killer immunoglobulin-like receptor ligand HLA-Bw4 protects against multiple sclerosis. Ann Neurol. 2009;65:658–666. doi: 10.1002/ana.21695. [DOI] [PubMed] [Google Scholar]
- 11.Polman CH, Reingold SC, Banwell B, et al. Diagnostic criteria for multiple sclerosis: 2010 Revisions to the McDonald criteria. Ann Neurol. 2011;69:292–302. doi: 10.1002/ana.22366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kurtzke JF. Rating neurologic impairment in multiple sclerosis: An Expanded Disability Status Scale (EDSS) Neurology. 1983;33:1444–1452. doi: 10.1212/wnl.33.11.1444. [DOI] [PubMed] [Google Scholar]
- 13.Roxburgh RH, Seaman SR, Masterman T, et al. Multiple Sclerosis Severity Score: Using disability and disease duration to rate disease severity. Neurology. 2005;64:1144–1151. doi: 10.1212/01.WNL.0000156155.19270.F8. [DOI] [PubMed] [Google Scholar]
- 14.Mero IL, Gustavsen MW, Saether HS, et al. Oligoclonal band status in Scandinavian multiple sclerosis patients is associated with specific genetic risk alleles. PLoS One. 2013;8:e58352. doi: 10.1371/journal.pone.0058352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andersson M, Alvarez-Cermeno J, Bernardi G, et al. Cerebrospinal fluid in the diagnosis of multiple sclerosis: A consensus report. J Neurol Neurosurg Psychiatry. 1994;57:897–902. doi: 10.1136/jnnp.57.8.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Link H, Huang YM. Oligoclonal bands in multiple sclerosis cerebrospinal fluid: An update on methodology and clinical usefulness. J Neuroimmunol. 2006;180:17–28. doi: 10.1016/j.jneuroim.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 17.Lechner-Scott J, Spencer B, De MT, et al. The frequency of CSF oligoclonal banding in multiple sclerosis increases with latitude. Mult Scler. 2012;18:974–982. doi: 10.1177/1352458511431729. [DOI] [PubMed] [Google Scholar]
- 18.Rinker JR, Trinkaus K, Naismith RT, et al. Higher IgG index found in African Americans versus Caucasians with multiple sclerosis. Neurology. 2007;69:68–72. doi: 10.1212/01.wnl.0000265057.79843.d9. [DOI] [PubMed] [Google Scholar]
- 19.Balnyte R, Rastenyte D, Uloziene I, et al. The significance of HLA DRB1*1501 and oligoclonal bands in multiple sclerosis: Clinical features and disability. Medicina (Kaunas) 2011;47:368–373. [PubMed] [Google Scholar]
- 20.Idiman E, Ozakbas S, Dogan Y, et al. The significance of oligoclonal bands in multiple sclerosis: Relevance of demographic and clinical features, and immunogenetic backgrounds. J Neuroimmunol. 2009;212:121–124. doi: 10.1016/j.jneuroim.2009.04.014. [DOI] [PubMed] [Google Scholar]
- 21.Imrell K, Landtblom AM, Hillert J, et al. Multiple sclerosis with and without CSF bands: Clinically indistinguishable but immunogenetically distinct. Neurology. 2006;67:1062–1064. doi: 10.1212/01.wnl.0000237343.93389.35. [DOI] [PubMed] [Google Scholar]
- 22.Imrell K, Greiner E, Hillert J, et al. HLA-DRB115 and cerebrospinal fluid-specific oligoclonal immunoglobulin G bands lower age at attainment of important disease milestones in multiple sclerosis. J Neuroimmunol. 2009;210:128–130. doi: 10.1016/j.jneuroim.2009.03.005. [DOI] [PubMed] [Google Scholar]
- 23.Joseph FG, Hirst CL, Pickersgill TP, et al. CSF oligoclonal band status informs prognosis in multiple sclerosis: A case control study of 100 patients. J Neurol Neurosurg Psychiatry. 2009;80:292–296. doi: 10.1136/jnnp.2008.150896. [DOI] [PubMed] [Google Scholar]
- 24.Siritho S, Freedman MS. The prognostic significance of cerebrospinal fluid in multiple sclerosis. J Neurol Sci. 2009;279:21–25. doi: 10.1016/j.jns.2008.12.029. [DOI] [PubMed] [Google Scholar]
- 25.Hensiek AE, Seaman SR, Barcellos LF, et al. Familial effects on the clinical course of multiple sclerosis. Neurology. 2007;68:376–383. doi: 10.1212/01.wnl.0000252822.53506.46. [DOI] [PubMed] [Google Scholar]
- 26.Masterman T, Ligers A, Olsson T, et al. HLA-DR15 is associated with lower age at onset in multiple sclerosis. Ann Neurol. 2000;48:211–219. [PubMed] [Google Scholar]
- 27.Smestad C, Brynedal B, Jonasdottir G, et al. The impact of HLA-A and -DRB1 on age at onset, disease course and severity in Scandinavian multiple sclerosis patients. Eur J Neurol. 2007;14:835–840. doi: 10.1111/j.1468-1331.2007.01825.x. [DOI] [PubMed] [Google Scholar]
- 28.Celius EG, Harbo HF, Egeland T, et al. Sex and age at diagnosis are correlated with the HLA-DR2, DQ6 haplotype in multiple sclerosis. J Neurol Sci. 2000;178:132–135. doi: 10.1016/s0022-510x(00)00389-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.