Abstract
Despite a substantial evidence base, implementation of pharmacogenetics into routine patient care has been slow due to a number of non-trivial practical barriers. We implemented a Personalized Anti-platelet Pharmacogenetics Program (PAP3) for cardiac catheterization patients at the University of Maryland Medical Center and the Baltimore Veterans Administration Medical Center Patients are offered CYP2C19 genetic testing, which is performed in our Clinical Laboratory Improvement Amendment (CLIA)-certified Translational Genomics Laboratory. Results are returned within five hours along with clinical decision support that includes interpretation of results and prescribing recommendations for anti-platelet therapy based on the Clinical Pharmacogenetics Implementation Consortium guidelines. Now with a working template for PAP3, implementation of other drug-gene pairs is in process. Lessons learned as described in this article may prove useful to other medical centers as they implement pharmacogenetics into patient care, a critical step in the pathway to personalized and genomic medicine.
Keywords: Pharmacogenomics, individualized medicine, personalized medicine, translational research, implementation science, CYP2C19, clopidogrel, anti-platelet pharmacogenetics
INTRODUCTION
Pharmacogenetics, namely the use of a patient’s genetic information to treat disease more effectively and minimize adverse drug effects, is an area within the broader scope of genomic medicine that holds great promise for personalizing healthcare practice [McCarthy et al., 2013]. As the number and availability of evidence-based pharmacogenetic tests increase and the cost of testing decreases, we anticipate a surge in consumer and clinician demand. However, despite a substantial evidence base for a number of drug-gene pairs, the barriers to implementing pharmacogenetics in clinical settings are many, and adoption has been slow [Manolio et al., 2013]. In this article, we describe the University of Maryland Personalized Anti-platelet Pharmacogenetics Program (PAP3). We provide practical information that may be useful to other institutions and for dissemination of evidence-based pharmacogenetics and personalized medicine practices.
BARRIERS TO IMPLEMENTING PHARMACOGENETICS
Barriers to routine use of pharmacogenetics in a clinical setting that we encountered include: logistics of performing timely genotyping in a Clinical Laboratory Improvement Amendments (CLIA)-certified laboratory; electronic health records (EHRs) do not support formats needed to record genetic results; lack of prospective genotype-directed randomized clinical trials validating the advantage of using pharmacogenetic based dosing over standard of care treatment algorithms; inexperience and lack of knowledge of many clinicians in interpreting and acting on pharmacogenetic information; paucity of clear recommendations for pharmacogenetic testing by professional associations; lack of a robust infrastructure to provide decision support for genomic medicine; cost and reimbursement issues; and ethical and medico-legal concerns. These barriers need to be addressed for pharmacogenetics to become a routine part of health care.
The University of Maryland is one of eight implementation sites of the Translational Pharmacogenomics Program (TPP) funded by the National Institutes of Health Pharmacogenomics Research Network [Shuldiner et al., 2013]. The goal of the TPP is to implement into patient care drug-gene pairs published in the Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines [Relling and Klein, 2011], and to identify barriers to implementation and develop and disseminate real-world solutions. Below, we describe in some detail the University of Maryland’s PAP3 initiative.
THE UNIVERSITY OF MARYLAND PERSONALIZED ANTI-PLATELET PHARMACOGENETICS PROGRAM (PAP3)
Background
Clopidogrel is the most commonly prescribed anti-platelet agent for prevention of myocardial infarction in patients who undergo percutaneous coronary intervention (PCI) procedures. Despite its widespread use, there is great inter-individual variation in response as determined by residual on-treatment platelet aggregation and recurrent cardiac events [Gurbel et al., 2003]. Early studies published in 2006 – 2007 indicated that clopidogrel is a pro-drug requiring activation by cytochrome P450 2C19 (CYP2C19) and that healthy subjects and coronary artery disease patients with common loss-of-function mutations in the CYP2C19 gene had decreased blood levels of active metabolite and increased residual on-treatment platelet reactivity [Brandt et al., 2007; Hulot et al., 2006]. Subsequent larger studies [Mega et al., 2010a; Simon et al., 2009] as well as a genome-wide association study [Shuldiner et al., 2009] and meta analysis [Mega et al., 2010b] subsequently replicated these findings and linked the most common loss of function variant, CYP2C19*2, to an increased incidence of cardiovascular events, especially stent thrombosis. CYP2C19 loss of function heterozygotes (intermediate metabolizers) and homozygotes (poor metabolizers) constitute approximately 22 - 25% and 3 - 4%, respectively, of the general U.S. population [Fisch et al., 2013; Scott et al., 2013b]. Most studies indicate a gene-dose dependent decrease in clopidogrel responsiveness, with poor metabolizers at greatest risk for recurrent cardiovascular events and intermediate metabolizers at intermediate risk relative to clopidogrel-treated patients without any loss of function alleles (extensive metabolizers). Interestingly, the association between CYP2C19 loss of function alleles and poorer outcomes in clopidogrel-treated patients appears to be most marked in patients in whom coronary stents have been placed, and less so (if at all) for other indications for clopidogrel, e.g., atrial fibrillation, peripheral vascular disease, stable angina, stroke [Johnson et al., 2012]. Furthermore, the role of concurrent use of CYP2C19 inhibitors, e.g., proton pump inhibitors, on clopidogrel efficacy remains controversial [Depta and Bhatt, 2012; Scott et al., 2013a].
The newer anti-platelet medications, prasugrel and ticagrelor, are unaffected by CYP2C19 metabolizer status but are associated with higher bleeding rates than clopidogrel and have other contraindications, suggesting that these medications not be used in all patients, but rather be reserved for CYP2C19 loss of function carriers. Indeed, in March 2010, FDA added a boxed warning to the clopidogrel label to alert health-care professionals and patients to the potential decreased efficacy of the drug in CYP2C19 poor metabolizers, the availability of CYP2C19 genetic tests, and consideration of alternate therapy in CYP2C19 poor metabolizers (FDA Drug Safety Communication 3/12/2010).
Despite the aggregate of pharmacokinetic, pharmacodynamic, and clinical outcomes evidence, the uptake of CYP2C19 genetic testing by clinicians is relatively low. Perhaps the most significant barrier to widespread adoption is the absence of prospective randomized clinical trials of genotype-guided anti-platelet algorithms to provide a more definitive evidence base [Perry and Shuldiner, 2013]. As adverse event rates such as stent thrombosis have dropped over the last decade, a properly powered definitive trial would require several thousand participants to demonstrate such a benefit. Recently, smaller prospective clinical trials have begun to provide further support for CYP2C19 genetic testing. The non-randomized prospective GIANT Trial studied 1,445 PCI patients of whom 319 (22%) were CYP2C19 intermediate or poor metabolizers (IM/PMs). Eighty-five percent of IMs/PMs received alternative anti-platelet therapy and experienced lower cardiovascular event rates than IMs/PMs who did not receive alternative anti-platelet therapy (3.3% versus 15.6%) [Chevalier, 2013]. In another recent study by Xie and colleagues, 600 Chinese PCI patients were randomized and then received personalized anti-platelet therapy, which included CYP2C19 genotyping and alternative anti-platelet therapy in CYP2C19 IMs/PMs versus conventional treatment. Twenty-seven patients (9.3%) assigned to the conventional treatment group experienced a cardiovascular event compared to 8 patients (2.7%) in the personalized anti-platelet group (p<0.01) [Xie et al., 2013].
In the absence of a definitive prospective randomized clinical trial, the current PCI practice guidelines, which were published by major professional organizations (ACCF/AHA/SCAI) prior to the results of the GIANT and Xie studies, do not support the routine clinical use of genetic testing to screen patients undergoing PCI [Levine et al., 2011]. Similarly, the FDA’s boxed warning on the clopidogrel label provides latitude, falling short of mandating CYP2C19 genetic testing, stating: “Tests are available to identify a patient’s CYP2C19 genotype; these tests can be used as an aid in determining therapeutic strategy. Consider alternative treatment or treatment strategies in patients identified as CYP2C19 poor metabolizers.” Thus the option to perform genetic testing is up to the individual clinician, an option that for various reasons, including medical-legal considerations, is rarely taken at the current time. However, growth in the evidence base for clinical benefit, such as seen in the GIANT and Xie [Chevalier, 2013; Xie et al., 2013] and expanding awareness of cost savings to the patient and health care systems [Reese et al., 2012] are expected to gradually shift routine care to a more personalized anti-platelet therapy approach.
Overview of Personalized Anti-Platelet Pharmacogenetics Program PAP3 Implementation Process
The objective of the PAP3 is to implement a process whereby: i) health care providers order CYP2C19 genetic testing in patients undergoing cardiac catheterization; ii) CLIA-certified CYP2C19 genetic testing is performed with high precision and a rapid turnaround time in the University of Maryland Translational Genomics Laboratory (TGL); iii) results are returned to prescribers in standard report formats that include patient genotype, metabolizer phenotype, and suggested treatment algorithm; iv) in-service education programs regarding pharmacogenetic testing and individualized therapy are offered to promote adoption of genotype-guided anti-platelet therapy by clinicians; and v) effectiveness of the implementation is measured, e.g., results turn-around time, changes in provider test ordering and prescribing practices, efficacy of education programs, provider satisfaction, and clinical outcomes and cost effectiveness.
Recognizing that the success of the implementation depends on the contributions of diverse functional groups, a PAP3 advisory and working group was convened. Membership included leaders in cardiology (physicians, cardiac catheterization laboratory nurse), biomedical informatics (hospital Chief Information Officer), the TGL (directors, genetic counselor), pathology, finance, and other strategic representatives from the University of Maryland Medical Center (UMMC) and the Baltimore Veterans Administration Medical Center (BVAMC). The group met on a monthly basis to identify potential issues and discuss approaches to managing necessary clinical processes, workflows, laboratory and information system requirements, and financial implications.
At an early juncture the PAP3 advisory and working group decided to initiate CYP2C19 testing in cardiac catheterization patients under the auspices of an IRB-approved research protocol to enhance our ability to obtain implementation metrics and outcome data for analysis and sharing to establish best practices for clinical implementation. The last step in the implementation process would be the transition of the optimized research protocol to routine patient care. We decided to implement CYP2C19 genetic testing in the cardiac catheterization laboratories at two clinical sites, UMMC and BVAMC, anticipating that distinct processes and workflows would need to be established at each. The UMMC is an 800 bed academic medical center, serving Baltimore and the state of Maryland, southern Delaware, and portions of southern Pennsylvania. The UMMC Cardiac Catheterization Lab (CCL) performs approximately 900 left heart catheterizations per year. Adjacent to UMMC is the BVAMC, a 137 bed acute medical and surgical care facility for the VA Maryland Health Care System. The BVAMC performs approximately 250 left heart catheterizations per year. Patient care is provided by the same UMSOM faculty interventional cardiology physicians in both facilities.
Instrumental to the implementation process was an acknowledgement of the need for rapid turn-around time from sample collection to return of results to the patient’s medical record in order for clinicians to use the results to direct drug selection prior to patient discharge. Several parallel efforts were initiated to bring this clinical service to a functional state, including close attention to patient enrollment, sample acquisition/handling, TGL genotype testing, result delivery to the medical record, and education and engagement of care providers. Individualized workflows for each of these components were woven together to comprise the full project (Figure 1).
Figure 1.
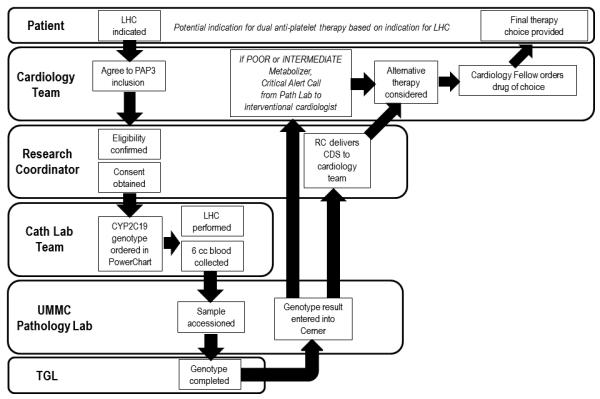
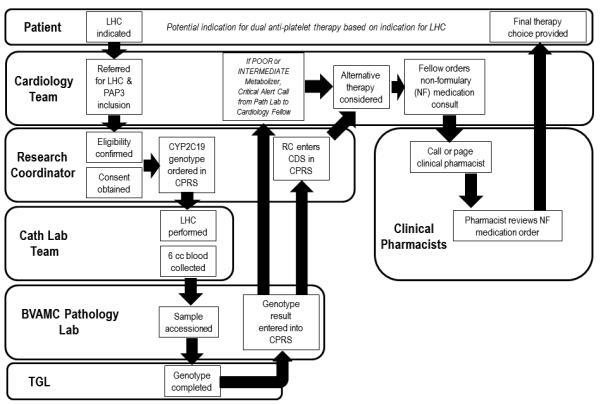
Workflow for CYP2C19 genetic testing at the University of Maryland Cardiac Catheterization Laboratory (Panel A) and the Baltimore Veterans Administration Medical Center (Panel B).
Implementation of Personalized Anti-Platelet Pharmacogenetics Program at UMMC (Figure 1A)
Eligible individuals are male and female patients, 18 years of age and older, who undergo left heart catheterization. These patients are at a high likelihood of requiring anti-platelet therapy. An advantage of this approach is that elective patients undergoing cardiac catheterization are funneled through the Catheterization Preparation and Recovery Unit (CPRU) and thus recruitment efforts can be focused in one location. For hospitalized patients undergoing cardiac catheterization on an urgent basis, close communication with the inpatient cardiology and cardiology consultation services have been essential to patient enrollment from these units. In the CPRU, research coordinators review patient charts, approach and consent patients for participation, verify eligibility, and collect baseline demographic and clinical information from patients before they are taken to the CCL for their procedure. Together with the UMMC laboratory information technology team, we implemented computerized provider order entry for CYP2C19 test ordering. Once ordered, a blood sample is collected, in most cases at the time of sheath insertion during the catheterization procedure. Samples are accessioned in the hospital Laboratory Information System (Cerner PathNet) and transported (“sent out”) to the TGL in the UMSOM for processing and analysis.
In the TGL, genotype analysis is performed using the Verigene® System and the 2C19/CLO+ Nucleic Acid Assay (Nanosphere, Northbrook, IL), which measures the three common variants, *2, *3 and *17. Other loss of function variants (*4 - *10) are very rare and not measured in this assay in its current release. CLIA-compliant reports are prepared, reviewed and signed by the laboratory director or designee. These reports specify which variants were measured and provide limitations with regard to potentially missing rare known or unknown variants that are not measured. Results are entered into Cerner Pathnet, so results are available in the EHR for clinicians to use at the point of care. The protocol procedure specifies calling clinicians by telephone when there are “actionable” genotypes (i.e., predicted clopidogrel poor responders) so patient therapy may be changed to alternative anti-platelet agent at the earliest possible time. With a turn-around time of approximately five hours from sample receipt in laboratory to result, genotype results are available before the next day’s maintenance dose in the majority of patients. However, CYP2C19 testing is currently not available 24/7 and tests performed the morning following blood draw may not be completed prior to patient discharge from the hospital. In these cases, telephone follow-up with the referring physician is necessary.
Treating physicians are provided anti-platelet therapy treatment recommendations based on current published CYP2C19 CPIC guidelines in which both IMs and PMs are recommended alternative anti-platelet therapy [Scott et al., 2013b]; however, final selection of an anti-platelet agent is determined by the physician’s clinical judgment. Clinical decision support (CDS) delivered through the EHR is vital for scaling implementation of pharmacogenetic (and ultimately pharmacogenomic) applications in diverse health care setting. Our institution, like others, is adding new features and functions to the EHR systems. Currently, however, the infrastructure for building computational algorithms and CDS within our EHR is limited. For the time being, genotype-based anti-platelet therapy prescribing information is hand delivered, faxed or emailed to the prescribing clinician once testing is completed. Additionally, the recommendations are posted in common areas in the CPRU, and Cardiac and Progressive Care units in the UMMC. Although not sustainable for more complex genomic data, we have found these manual methods to be reasonably effective, highlighting that implementing pharmacogenetic applications can be accomplished even in the absence of a fully functioning integrated EHR. Since UMMC is a regional referral center, genotype results with interpretation are also mailed to each patient and referring primary physicians following discharge (see Supplemental Information).
Implementation of PAP3 at BVAMC (Figure 1B)
Similar to the UMMC effort, blood samples are sent to the TGL. In contrast to the UMMC workflow, at the BVAMC the clinical pharmacist plays an important role to assist in ordering non-formulary alternative anti-platelet agents, when indicated. A second distinction is that results are linked with CDS in the Computerized Patient Record System (CPRS), the VA EHR that is available to all authorized VA providers. Another difference is that notification letters are uploaded into CPRS which are then electronically cosigned by the primary care physician. Since all VAMCs use the same CPRS, CDS developed as part of PAP3 should be readily exportable nationally. In addition, all veterans receive the results of their tests in the mail.
Progress to date
From March 2013 to December 9, 2013 at UMMC and BVAMC, 203 patients (170 at UMMC and 33 at BVAMC) have been approached to participate and 166 (81.8%) consented and enrolled under this protocol. Of those enrolled, 53 (31.9%) were IM/PMs; 27 of these had a PCI of which 17 (63%) were prescribed an alternative anti-platelet therapy. The most common reasons for not prescribing alternative therapy to clopidogrel in IMs/PMs were: physician preference and patient factors; discharged before or right after CYP2C19 results were available; already on alternate treatment; and lack of insurance.
Tracking Implementation Metrics
Case report forms are used to capture data from enrolled participants including the times of test order entry, sample collection, sample accessioning, and recording of test result to the EHR Examples of measurable outcomes at this stage of the project are time between test order, sample delivery in TGL and report delivery to prescribers, genotype distribution, and the proportion of tested patients with actionable genotypes. Additional evaluations include how CDS was delivered, to whom it was communicated, and whether there was successful contact with the primary decision maker. Finally, we track the number of new or revised prescriptions written based upon genotype result, the time between test result and new or revised drug order, and physician adherence to CPIC guidelines.
Education and Provider Support
To maximize involvement and utilization, providers must have a reasonable understanding of both the scientific and clinical knowledge which forms the basis for genotype-directed personalized anti-platelet therapy. Communicating the benefits of genotype directed anti-platelet therapy to providers is accomplished by presenting the cumulative evidence from pharmacokinetic, pharmacodynamic, and retrospective and small prospective clinical outcomes studies and has been time consuming. In addition to a rigorous review of the evidence base, a significant portion of the education process for health care providers is developing an understanding and comfort using personalized genetic information within their medical decision making process. Educational efforts targeting these objectives include formal presentations at conferences such as Cardiovascular Medicine and Internal Medicine Grand Rounds as well as multiple informal interactions such as in-service sessions for providers and health care delivery teams. These education sessions and interactions must be repeated over time, given provider turnover and reluctance to deviate from established practices and adoption of new approaches. Our PAP3 implementation team includes cardiologists, genetic counselors, nurses, and pharmacists. In our experience, education sessions appear to be most effective when educators of similar training background to the audience deliver the presentations, e.g., interventional cardiologists to other cardiologists; nurses to other nurses. Periodically, we provide updates to providers regarding the PAP3 project itself including the number of subjects recruited, turnaround time, number of actionable genotypes, and the number of prescription changes. It has been our experience that when education is approached in this manner, and providers are engaged in the project, they are excited to participate in the program.
FUTURE DIRECTIONS AND CONCLUDING REMARKS
Herein, we describe our initial experience implementing a pharmacogenetics application for patient care. Despite significant challenges navigating a complex and fragmented health care system, we have realized successes in implementing a personalized anti-platelet pharmacogenetics program. We found that establishing an infrastructure, garnering and maintaining institutional support at high levels, and engaging and educating stakeholders to establish a culture of early adopters were critical ingredients to our success.
As we move forward, we will continue to track implementation metrics and to utilize this information iteratively to optimize workflow. A critical final step to any implementation research project is to remove the research component altogether and establish a new standard of care. We anticipate implementing additional drug-gene pairs based on published CPIC guidelines (e.g., warfarin and CYP2C9/VKORC1 [Johnson et al., 2011], thiopurines and TPMT [Relling et al., 2013], statins and SLCO1B1 [Wilke et al., 2012], codeine and CYP2D6 [Crews et al., 2012], and seamless incorporation of the entire process into the EHR. As others have done successfully [Bell et al., 2013] we plan to establish a pharmacogenomics consult service which will be instrumental to providing providers with up to date recommendations regarding the application of pharmacogenomic knowledge to patient care. This service will consist of a multidisciplinary team of providers, including physicians, genetic counselors and pharmacists. As reported by others [Farrugia and Weinshilboum, 2013; Johnson et al., 2013; Pulley et al., 2012] we anticipate initiating preemptive pharmacogenomic testing in patients at high risk for exposure to medications with known pharmacogenomic impact.
Ultimately, to demonstrate clinical utility and cost effectiveness of pharmacogenomics in patient care, large multicenter pragmatic clinical trials in diverse health care settings will be necessary. Establishing the infrastructure and workflow such as that described is a first step toward enabling these trials.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grants U01HL105198 and S10OD012357, and institutional funding from the University of Maryland School of Medicine and the University of Maryland Medical Center. We thank, Amie Branham, Danielle Sewell, Meghan Farrell, Lauren Doyle, Christopher Gallagher, Sandford Stass, and the staff of the University of Maryland Medical Center Catheterization Preparation and Recovery Unit and Cardiac Catheterization Laboratory for their support and extraordinary efforts on behalf of the Personalized Anti-platelet Pharmacogenetics Program and other Program for Personalized and Genomic Medicine initiatives.
Biography
Alan R. Shuldiner, MD is the John L. Whitehurst Professor at the University of Maryland School of Medicine, Associate Dean of Personalized Medicine, Director of the Program for Personalized and Genomic Medicine, and Head of the Division of Endocrinology, Diabetes and Nutrition. He leads a multidisciplinary clinical and translational research program that includes protocols conducted at the Amish Research Clinic in Lancaster, PA, the University of Maryland, and Baltimore Veterans Administration Medical Center.
Kathleen Palmer, RN is the Clinical Research Manager for the Program for Personalized and Genomic Medicine at the University of Maryland School of Medicine.
Ruth Pakyz, BS is Program Manager for the Program for Personalized and Genomic Medicine at the University of Maryland School of Medicine.
Tameka Alestock, MS is a Clinical Research Coordinator for the University of Maryland School of Medicine Program for Personalized and Genomic Medicine and the Division of Cardiovascular Medicine.
Kristin Maloney, MS, MGC is a certified Genetic Counselor at the University of Maryland Center for Medical Genetics and Personalized Medicine and is a member of the University of Maryland School of Medicine Program for Personalized and Genomic Medicine. Her current position includes both research and clinical responsibilities.
Courtney O’Neill, MSN, RN is a Cardiac Catheterization Nurse and Clinical Research Coordinator at the Baltimore Veterans Administration Medical Center. She has 8 years of critical care and research experience. She collaborates with Drs. Vesley and Robinson to develop research protocols at that facility and with the University of Maryland research team.
Shaun Bhatty, MD is a Cardiology Fellow at the University of Maryland Medical Center.
Jamie Schub, MS is Research Study Coordinator for the University of Maryland School of Medicine Program for Personalized and Genomic Medicine.
Casey Lynnette Overby, PhD is an Assistant Professor in the Program for Personalized and Genomic Medicine and a member of the Center for Health-related Informatics and Bio-imaging at the University of Maryland School of Medicine. Her research seeks to develop new approaches for interpreting genome information. She also seeks to understand processes and factors associated with the successful integration of these approaches into healthcare settings.
Richard B. Horenstein, MD, JD is an Assistant Professor at the University of Maryland School of Medicine and a member of the Program for Personalized and Genomic Medicine. He is a physician scientist with over 12 years of experience performing clinical and basic studies including a genotype related clopidogrel dose escalation study in collaboration with NIH and FDA scientists. He also served as site-PI of the NHLBI sponsored COAG Trial.
Toni I. Pollin, PhD is an Associate Professor at the University of Maryland School of Medicine and a member of the Program for Personalized and Genomic Medicine. She is a human geneticist and genetic counselor with 15 years of research experience in the area of the genetics of common complex diseases such as diabetes and its cardiovascular complications, including study leadership, management and statistical analysis and interpretation.
Mark Kelemen MD, MBA is Clinical Associate Professor of Medicine in the Division of Cardiovascular Medicine and Senior Vice President and Chief Medical Informatics Officer of the University of Maryland Medical System. He leads the implementation of advanced clinical decision support and electronic health records for the health system.
Amber Beitelshees, PharmD, MPH is an Assistant Professor at the University of Maryland School of Medicine and a member of the Program for Personalized and Genomic Medicine. Her expertise resides in clinical pharmacology, pharmacogenomics, public health, and translational research.
Shawn W. Robinson, MD is Cardiology Section Chief for the VA Maryland Health Care System. He is board certified in both general cardiovascular diseases and in advanced heart failure and transplant cardiology. He has over 15 years of experience serving as co-investigator on several acute and chronic heart failure clinical trials. His research effort in the area of pharmacogenomics investigates how various genetic polymorphisms influence myocardial function and treatment.
Miriam G. Blitzer, PhD, is Professor at the University of Maryland School of Medicine. She is Head of the Division of Human Genetics in the Department of Pediatrics. She is a board-certified in clinical biochemical genetics. She co-directs the medical school curriculum that focuses on medical genetics.
Patrick McArdle, PhD is an Assistant Professor at the University of Maryland School of Medicine and a member of the Program for Personalized and Genomic Medicine. He is an epidemiologist with extensive experience in human subjects research with a focus on genetic epidemiology, particularly the analysis and interpretation of genome-wide genetic data and complex phenotypic data.
Lawrence A. Brown, MD is a Staff Pathologist at the Baltimore VA Medical Center, and Assistant Professor of Pathology at the University of Maryland School of Medicine.
Linda Jo Bone Jeng, MD, PhD is an Associate Professor in Medicine, Pathology and Pediatrics at the University of Maryland School of Medicine and a member of the Program for Personalized and Genomic Medicine. She is Director of Genomic Medicine in the Translational Genomics Laboratory. She is board-certified in clinical genetics and clinical molecular genetics.
Richard Y. Zhao, PhD is a Professor of Pathology and Microbiology-Immunology at the University of Maryland School of Medicine and a member of the Program for Personalized and Genomic Medicine. He is the Director of the Translational Genomics Laboratory where the CYP2C19 clinical tests are performed. He has nearly 20 years of experience in molecular diagnostics and molecular pathology.
Nicholas P. Ambulos, Jr., Ph.D. is Executive Director of the University of Maryland School of Medicine Research Cores and Technical Supervisor of the Translational Genomics Laboratory. He is also a member of the Program for Personalized and Genomic Medicine.
Mark R. Vesely, MD is an Assistant Professor of Medicine at the University of Maryland School of Medicine and a member of the Program for Personalized and Genomic Medicine. He is Director of the Interventional Cardiology Fellowship Training Program and the Baltimore VA Medical Center Cardiac Catheterization Lab. His major clinical focus is interventional cardiology where he provides invasive diagnostic and interventional therapeutic care for the entire range of clinical cardiac pathology, most prominently ischemic heart disease and congestive heart failure with an expanding focus on structural and valvular heart disease. He has been involved in clinical research for over 10 years. He has held the position of principal investigator on numerous studies and trials as well as being a sub-investigator on many others.
Footnotes
CONFLICT OF INTEREST DISCLOSURE Dr. Shuldiner receives support from NIH for anti-platelet pharmacogenomics research, and is a consultant to United States Diagnostic Standards, Inc. and Merck, Inc. Other authors have no conflicts of interest to declare.
REFERENCES
- Bell GC, Crews KR, Wilkinson MR, Haidar CE, Hicks JK, Baker DK, Kornegay NM, Yang W, Cross SJ, Howard SC, Freimuth RR, Evans WE, Broeckel U, Relling MV, Hoffman JM. Development and use of active clinical decision support for preemptive pharmacogenomics. J Am Med Inform Assoc. 2013 doi: 10.1136/amiajnl-2013-001993. [epub ahead of print DOI:10.1136/amiajnl-2013-001993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt JT, Close SL, Iturria SJ, Payne CD, Farid NA, Ernest CS, 2nd, Lachno DR, Salazar D, Winters KJ. Common polymorphisms of CYP2C19 and CYP2C9 affect the pharmacokinetic and pharmacodynamic response to clopidogrel but not prasugrel. J Thromb Haemost. 2007;5:2429–2436. doi: 10.1111/j.1538-7836.2007.02775.x. [DOI] [PubMed] [Google Scholar]
- Chevalier BR. Transcatheter Cardiovascular Therapeutics. San Francisco, CA: 2013. GIANT: A Prospective Registry Study of CYP2C19 Genetic Profiling for Thienopyridine Management After Primary PCI in Acute Myocardial Infarction. [Google Scholar]
- Crews KR, Gaedigk A, Dunnenberger HM, Klein TE, Shen DD, Callaghan JT, Kharasch ED, Skaar TC. Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines for codeine therapy in the context of cytochrome P450 2D6 (CYP2D6) genotype. Clin Pharmacol Ther. 2012;91:321–326. doi: 10.1038/clpt.2011.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depta JP, Bhatt DL. Antiplatelet therapy and proton pump inhibition: cause for concern? Curr Opin Cardiol. 2012;27:642–650. doi: 10.1097/HCO.0b013e32835830b6. [DOI] [PubMed] [Google Scholar]
- Farrugia G, Weinshilboum RM. Challenges in implementing genomic medicine: the Mayo Clinic Center for Individualized Medicine. Clin Pharmacol Ther. 2013;94:204–206. doi: 10.1038/clpt.2013.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisch AS, Perry CG, Stephens SH, Horenstein RB, Shuldiner AR. Pharmacogenomics of anti-platelet and anti-coagulation therapy. Curr Cardiol Rep. 2013;15:381. doi: 10.1007/s11886-013-0381-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurbel PA, Bliden KP, Hiatt BL, O’Connor CM. Clopidogrel for coronary stenting: response variability, drug resistance, and the effect of pretreatment platelet reactivity. Circulation. 2003;107:2908–2913. doi: 10.1161/01.CIR.0000072771.11429.83. [DOI] [PubMed] [Google Scholar]
- Hulot JS, Bura A, Villard E, Azizi M, Remones V, Goyenvalle C, Aiach M, Lechat P, Gaussem P. Cytochrorne P4502C19 loss-of-function polymorphism is a major determinant of clopidogrel responsiveness in healthy subjects. Blood. 2006;108:2244–2247. doi: 10.1182/blood-2006-04-013052. [DOI] [PubMed] [Google Scholar]
- Johnson JA, Elsey AR, Clare-Salzler MJ, Nessl D, Conlon M, Nelson DR. Institutional profile: University of Florida and Shands Hospital Personalized Medicine Program: clinical implementation of pharmacogenetics. Pharmacogenomics. 2013;14:723–726. doi: 10.2217/pgs.13.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JA, Gong L, Whirl-Carrillo M, Gage BF, Scott SA, Stein CM, Anderson JL, Kimmel SE, Lee MT, Pirmohamed M, Wadelius M, Klein TE, Altman RB. Clinical Pharmacogenetics Implementation Consortium Guidelines for CYP2C9 and VKORC1 genotypes and warfarin dosing. Clin Pharmacol Ther. 2011;90:625–629. doi: 10.1038/clpt.2011.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JA, Roden DM, Lesko LJ, Ashley E, Klein TE, Shuldiner AR. Clopidogrel: a case for indication-specific pharmacogenetics. Clin Pharmacol Ther. 2012;91:774–776. doi: 10.1038/clpt.2012.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine GN, Bates ER, Blankenship JC, Bailey SR, Bittl JA, Cercek B, Chambers CE, Ellis SG, Guyton RA, Hollenberg SM, Khot UN, Lange RA, Mauri L, Mehran R, Moussa ID, Mukherjee D, Nallamothu BK, Ting HH. 2011 ACCF/AHA/SCAI Guideline for Percutaneous Coronary Intervention: executive summary: a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines and the Society for Cardiovascular Angiography and Interventions. Circulation. 2011;124:2574–2609. doi: 10.1161/CIR.0b013e31823a5596. [DOI] [PubMed] [Google Scholar]
- Manolio TA, Chisholm RL, Ozenberger B, Roden DM, Williams MS, Wilson R, Bick D, Bottinger EP, Brilliant MH, Eng C, Frazer KA, Korf B, Ledbetter DH, Lupski JR, Marsh C, Mrazek D, Murray MF, O’Donnell PH, Rader DJ, Relling MV, Shuldiner AR, Valle D, Weinshilboum R, Green ED, Ginsburg GS. Implementing genomic medicine in the clinic: the future is here. Genet Med. 2013;15:258–267. doi: 10.1038/gim.2012.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy JJ, McLeod HL, Ginsburg GS. Genomic medicine: a decade of successes, challenges, and opportunities. Sci Transl Med. 2013;5:189sr184. doi: 10.1126/scitranslmed.3005785. [DOI] [PubMed] [Google Scholar]
- Mega JL, Simon T, Collet JP, Anderson JL, Antman EM, Bliden K, Cannon CP, Danchin N, Giusti B, Gurbel P, Horne BD, Hulot JS, Kastrati A, Montalescot G, Neumann FJ, Shen L, Sibbing D, Steg PG, Trenk D, Wiviott SD, Sabatine MS. Reduced-Function CYP2C19 Genotype and Risk of Adverse Clinical Outcomes Among Patients Treated With Clopidogrel Predominantly for PCI A Meta-analysis. Jama-J Am Med Assoc. 2010a;304:1821–1830. doi: 10.1001/jama.2010.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mega JL, Simon T, Collet JP, Anderson JL, Antman EM, Bliden K, Cannon CP, Danchin N, Giusti B, Gurbel P, Horne BD, Hulot JS, Kastrati A, Montalescot G, Neumann FJ, Shen L, Sibbing D, Steg PG, Trenk D, Wiviott SD, Sabatine MS. Reduced-function CYP2C19 genotype and risk of adverse clinical outcomes among patients treated with clopidogrel predominantly for PCI: a meta-analysis. JAMA. 2010b;304:1821–1830. doi: 10.1001/jama.2010.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry CG, Shuldiner AR. Pharmacogenomics of anti-platelet therapy: how much evidence is enough for clinical implementation? J Hum Genet. 2013;58:339–345. doi: 10.1038/jhg.2013.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pihoker C, Gilliam LK, Ellard S, Dabelea D, Davis C, Dolan LM, Greenbaum CJ, Imperatore G, Lawrence JM, Marcovina SM, Mayer-Davis E, Rodriguez BL, Steck AK, Williams DE, Hattersley AT. Prevalence, Characteristics and Clinical Diagnosis of Maturity Onset Diabetes of the Young Due to Mutations in HNF1A, HNF4A, and Glucokinase: Results From the SEARCH for Diabetes in Youth. J Clin Endocrinol Metab. 2013;98:4055–4062. doi: 10.1210/jc.2013-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollin TI, Damcott CM, Shen H, Ott SH, Shelton J, Horenstein RB, Post W, McLenithan JC, Bielak LF, Peyser PA, Mitchell BD, Miller M, O’Connell JR, Shuldiner AR. A null mutation in human APOC3 confers a favorable plasma lipid profile and apparent cardioprotection. Science. 2008;322:1702–1705. doi: 10.1126/science.1161524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulley JM, Denny JC, Peterson JF, Bernard GR, Vnencak-Jones CL, Ramirez AH, Delaney JT, Bowton E, Brothers K, Johnson K, Crawford DC, Schildcrout J, Masys DR, Dilks HH, Wilke RA, Clayton EW, Shultz E, Laposata M, McPherson J, Jirjis JN, Roden DM. Operational implementation of prospective genotyping for personalized medicine: the design of the Vanderbilt PREDICT project. Clin Pharmacol Ther. 2012;92:87–95. doi: 10.1038/clpt.2011.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampersaud E, Damcott CM, Fu M, Shen H, McArdle P, Shi X, Shelton J, Yin J, Chang YP, Ott SH, Zhang L, Zhao Y, Mitchell BD, O’Connell J, Shuldiner AR. Identification of novel candidate genes for type 2 diabetes from a genome-wide association scan in the Old Order Amish: evidence for replication from diabetes-related quantitative traits and from independent populations. Diabetes. 2007;56:3053–3062. doi: 10.2337/db07-0457. [DOI] [PubMed] [Google Scholar]
- Reese ES, Daniel Mullins C, Beitelshees AL, Onukwugha E. Cost-Effectiveness of Cytochrome P450 2C19 Genotype Screening for Selection of Antiplatelet Therapy with Clopidogrel or Prasugrel. Pharmacotherapy. 2012;32:323–332. doi: 10.1002/PHAR.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relling MV, Gardner EE, Sandborn WJ, Schmiegelow K, Pui CH, Yee SW, Stein CM, Carrillo M, Evans WE, Hicks JK, Schwab M, Klein TE. Clinical pharmacogenetics implementation consortium guidelines for thiopurine methyltransferase genotype and thiopurine dosing: 2013 update. Clin Pharmacol Ther. 2013;93:324–325. doi: 10.1038/clpt.2013.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relling MV, Klein TE. CPIC: Clinical Pharmacogenetics Implementation Consortium of the Pharmacogenomics Research Network. Clin Pharmacol Ther. 2011;89:464–467. doi: 10.1038/clpt.2010.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott SA, Owusu Obeng A, Hulot JS. Antiplatelet drug interactions with proton pump inhibitors. Expert Opin Drug Metab Toxicol. 2013a doi: 10.1517/17425255.2014.856883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott SA, Sangkuhl K, Stein CM, Hulot JS, Mega JL, Roden DM, Klein TE, Sabatine MS, Johnson JA, Shuldiner AR. Clinical Pharmacogenetics Implementation Consortium Guidelines for CYP2C19 Genotype and Clopidogrel Therapy: 2013 Update. Clin Pharmacol Ther. 2013b;94:317–323. doi: 10.1038/clpt.2013.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuldiner AR, O’Connell JR, Bliden KP, Gandhi A, Ryan K, Horenstein RB, Damcott CM, Pakyz R, Tantry US, Gibson Q, Pollin TI, Post W, Parsa A, Mitchell BD, Faraday N, Herzog W, Gurbel PA. Association of cytochrome P450 2C19 genotype with the antiplatelet effect and clinical efficacy of clopidogrel therapy. JAMA. 2009;302:849–857. doi: 10.1001/jama.2009.1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuldiner AR, Relling MV, Peterson JF, Hicks JK, Freimuth RR, Sadee W, Pereira NL, Roden DM, Johnson JA, Klein TE, Vesely M, Robinson SW, Ambulos N, Jr., Stass SA, Kelemen MD, Brown LA, Pollin TI, Beitelshees AL, Zhao RY, Pakyz RE, Palmer K, Alestock T, O’Neill C, Maloney K, Branham A, Sewell D, Crews K, Hoffman J, Cross S, Haidar C, Baker D, Bell G, Greeson F, Gaur A, Reiss U, Huettel A, Cheng C, Gajjar A, Pappo A, Howard S, Hudson M, Pui CH, Jeha S, Evans WE, Broeckel U, Altman RB, Gong L, Whirl-Carrillo M, Manickam K, Sweet KM, Embi PJ, Roden D, Peterson J, Denny J, Schildcrout J, Bowton E, Pulley J, Beller M, Mitchell J, Danciu I, Price L, Weinshilboum R, Wang L, Nelson D, Clare-Salzler M, Elsey A, Burkley B, Langaee T, Liu F, Nessl D, Dong HJ, Lesko L, Chute CG. The Pharmacogenomics Research Network Translational Pharmacogenetics Program: overcoming challenges of real-world implementation. Clin Pharmacol Ther. 2013;94:207–210. doi: 10.1038/clpt.2013.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon T, Verstuyft C, Mary-Krause M, Quteineh L, Drouet E, Meneveau N, Steg PG, Ferrieres J, Danchin N, Becquemont L. Genetic determinants of response to clopidogrel and cardiovascular events. N Engl J Med. 2009;360:363–375. doi: 10.1056/NEJMoa0808227. [DOI] [PubMed] [Google Scholar]
- Wilke RA, Ramsey LB, Johnson SG, Maxwell WD, McLeod HL, Voora D, Krauss RM, Roden DM, Feng Q, Cooper-Dehoff RM, Gong L, Klein TE, Wadelius M, Niemi M. The clinical pharmacogenomics implementation consortium: CPIC guideline for SLCO1B1 and simvastatin-induced myopathy. Clin Pharmacol Ther. 2012;92:112–117. doi: 10.1038/clpt.2012.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Ma YT, Yang YN, Li XM, Zheng YY, Ma X, Fu ZY, Ba B, Li Y, Yu ZX, Chen Y, Chen BD, Liu F, Huang Y, Liu C, Baituola G. Personalized antiplatelet therapy according to CYP2C19 genotype after percutaneous coronary intervention: a randomized control trial. Int J Cardiol. 2013;168:3736–3740. doi: 10.1016/j.ijcard.2013.06.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


