Abstract
Mild cognitive impairment (MCI) represents a cognitive state intermediate between normal aging and early Alzheimer Disease (AD). To investigate if the molecular signature of MCI parallels the clinical picture, we use microarrays to extensively profile gene expression in 4 cortical brain regions (entorhinal cortex, hippocampus, superior frontal gyrus, post-central gyrus) using post-mortem tissue from cognitively normal aged controls, MCI, and AD cases. Our data reveal that gene expression patterns in MCI are not an extension of aging, and for the most part, are not intermediate between aged controls and AD. Functional enrichment analysis of significant genes revealed prominent upregulation in MCI brains of genes associated with anabolic and biosynthetic pathways (notably transcription, protein biosynthesis, protein trafficking and turnover) as well as mitochondrial energy generation. In addition, many synaptic genes showed altered expression in MCI, predominantly upregulation, including genes for central components of the vesicle fusion machinery at the synapse, synaptic vesicle trafficking, neurotransmitter receptors, and synaptic structure and stabilization. These data suggest that there is a rebalancing of synaptic transmission in the MCI brain. To investigate if synaptic gene expression levels in MCI were related to cognitive function, Pearson’s correlation coefficient between MMSE and region-specific mRNA expression were computed for MCI cases. A number of synaptic genes showed strong significant correlations (r>0.8, p<0.01) most notably in the EC, with fewer in the HC, and very few in neocortical regions. The synaptic genes with highly significant correlations were predominantly related to synaptic transmission and plasticity, and myelin composition. Unexpectedly, we found that gene expression changes that facilitate synaptic excitability and plasticity were overwhelmingly associated with poorer MMSE, and conversely that gene expression changes that inhibit plasticity were positively associated with MMSE. These data suggest that there is excessive excitability and apparent plasticity in limbic brain regions in MCI, that is associated with impaired synaptic and cognitive function. Such changes would be predicted to contribute to increased excitability, in turn leading to greater metabolic demand and ultimately progressive degeneration and AD, if not controlled.
Keywords: Microarray, Synapse, Electron transport, Mitochondria, SNARE, Neurotransmitter receptor, Hippocampus, Entorhinal cortex, Superior frontal gyrus, Somatosensory cortex
INTRODUCTION
Mild cognitive impairment (MCI) is characterized as a state of cognitive function intermediate between normal aging and the criteria for dementia, most commonly Alzheimer’s disease (AD), and is thought to represent a transitional condition between aging and dementia (Morris, 2012,Petersen, 2011). While much is known about the clinical and neuropathological profiles of MCI (Markesbery, 2010,Mufson, et al., 2012,Storandt, et al., 2006), no studies have extensively characterized the molecular state of the MCI brain at the gene expression level. It is possible that the gene expression signature of MCI parallels the clinical picture, with gene expression levels intermediate between the profiles characterizing non-demented aged and AD brains. Alternatively, MCI gene expression patterns may represent a unique molecular state that does not follow a continuum of change between aged and AD profiles. Thus, this study uses microarrays and bioinformatics analyses to address three main questions: (1) are gene expression patterns in MCI intermediate between aged and AD molecular profiles, similar to the clinical picture? (2) What functional gene categories are primarily affected in MCI? (3) What is the response of genes regulating core cellular processes of neuronal function in MCI, specifically synaptic signaling, energy production and protein homeostasis?
Microarray studies suggest that the integrity of these core processes (synaptic signaling, energy production and protein homeostasis) declines in the brain with aging (particularly after the 6th-7th decades of life) (Berchtold, et al., 2008,Lu, et al., 2004) and further degrades in AD (Berchtold, et al., 2013,Blalock, et al., 2004,Ginsberg, et al., 2012,Liang, et al., 2008,J.A. Miller, et al., 2008,Tan, et al., 2010), contributing to the spiraling decline in cognitive function, plasticity, and brain health (Blalock, et al., 2004,Gibson, et al., 2010,Ginsberg, et al., 2010,Liang, et al., 2008,Lynn, et al., 2010,Mattson, et al., 2008,Muller, et al., 2010,Nixon and Yang, 2012,Selkoe, 2002). While these classes of genes are well established to show decreased expression in moderate-severe AD, there is conflicting evidence on the direction of expression changes in the early stages of AD, and uncertainty regarding regional differences in response patterns. For example, incipient AD has been associated with downregulation in the hippocampus of these gene classes (Blalock, et al., 2004), while another report indicates increased expression in the prefrontal cortex in very early (presymptomatic) stages of AD (Bossers, et al., 2010).
In this study, we use microarrays to comprehensively profile gene expression patterns in MCI, aged control, and AD cases. Microarray technology in combination with bioinformatics analytical software constitutes a powerful strategy for gaining insight into the pathways, processes, and functional endpoints that impact brain health and that underlie cognitive decline with neurodegeneration. Thus, to obtain a global perspective on brain changes in MCI and how these changes relate to normal aged and AD brains, we investigated gene expression patterns in 4 brain regions, 3 of which are vulnerable to accumulation of pathology in AD (neocortical superior frontal gyrus (SFG), hippocampus (HC), entorhinal cortex (EC)), and a 4th region (neocortical post-central gyrus (PCG)) that is relatively spared.
MATERIALS AND METHODS
Tissue was obtained from 6 well-established National Institute on Aging Alzheimer’s Disease brain banks located at Sun Health Research Institute, UC Irvine, University of Rochester, Johns Hopkins University, University of Pennsylvania, and the University of Southern California. Unfixed frozen tissue from 4 brain regions (EC, HC, SFG, PCG) from MCI cases (n=16), AD cases (n=25) and age-matched non-demented controls (n=24), was processed, as described previously (Berchtold, et al., 2013,Berchtold, et al., 2008,Cribbs, et al., 2012) (Table 1, Supplemental Table 1). Of the MCI cases, 12 were run on microarrays, and an additional 4 samples were obtained to expand PCR validation. Average MMSE across groups was as follows: aged controls (29 ± 1.56), MCI (26.5 ± 2.47), AD (11.9 ± 9.02). Average post-mortem intervals for the aged control, MCI, and AD groups were as follows, respectively: 3.8 ± 1.7 hrs., 5.6 ± 4.9 hrs., 4.4 ± 2.9 hrs. Tangle pathology of aged controls and MCI cases was distributed across Braak 0-IV, with no Braak V-VI. AD cases were predominantly Braak V-VI (n=15), with some Braak stage III-IV (n=10), and 1 case at Braak stage II. RNA was extracted from approximately 50-100 mg tissue, and RNA integrity number (RIN) was assessed using the Agilent Bioanalyzer. Detailed case information and RIN number for tissue samples run on microarrays can be found in Supplemental Table 1. 179 samples were processed individually on Affymetrix (HgU133 plus 2.0) microarrays (Table 1). Microarray data were analyzed with GeneSpring 7.3 software (Agilent Technologies) and GC-Robust Multi-array average (GC-RMA) summarized expression values. Hierarchical cluster analysis (Pearson correlation and average linkage) was used to visualize the relationship between aged, MCI, and AD cases.
TABLE 1.
Clinical, demographic and neuropathology characteristics by diagnosis category of cases included in the analysis. n/a indicates data not available
| Clinical Diagnosis | ||||
|---|---|---|---|---|
| Aged controls (n=24) |
MCI (n=16) |
AD (n=27) |
||
| Age (years) at death : |
mean ± stdev
(range) |
85.2 ± 6.8 (74-95) |
87.7 ± 4.4 (74-98) |
84.9 ± 6.2 (74-95) |
| Gender distribution: |
females
males |
15 9 |
8 8 |
15 12 |
| MMSE: | mean ± stdev (range) | 29 ± 1.56 (25-30) |
26.5 ± 2.47 (22-30) |
11.9 ± 9.02 (0-27) |
| ApoE4 allele: | number (%) | 2 (8%) | 4 (25%) | 17 (63%) |
| Post-mortem interval (hours): |
mean ± stdev
(range) |
3.8 ± 1.7 (2-6.25) |
5.8 ± 5 (2-14) |
4.4 ± 2.9 (1.5-10) |
| Distribution of Braak scores (n): |
0-II
III-IV V-VI n/a |
15 9 0 0 |
3 11 0 2 |
1 10 15 1 |
| microarrrays used/group (n): | Total | 57 | 44 | 78 |
| microarrays used/region (n): |
EC
HC PCG SFG EC |
9 16 15 17 |
10 8 11 15 |
15 18 24 21 |
|
RNA integrity (RIN)
mean ± stdev: |
EC
HC PCG SFG |
8.67 ± 0.49 8.14 ± 0.75 8.63 ± 0.61 8.66 ± 0.84 |
8.21 ± 0.76 8.94 ± 0.36 8.63 ± 0.84 8.48 ± 0.96 |
8.21 ± 0.77 7.81 ± 0.68 8.44 ± 0.70 8.53 ± 0.76 |
Differential gene expression analysis (p<0.01) was followed by data-mining using bioinformatics software to identify Gene Ontology functional categories that were significantly over-represented. Comprehensive lists were generated for in-depth analysis of genes related to protein metabolism (2,075 probesets related to protein biosynthesis, folding, trafficking, turnover), mitochondrial bioenergetics (142 probesets coding for mitochondrial electron transport chain subunits and translocases of the inner and outer mitochondrial membranes), and synaptic function (561 probesets). The relationship between gene expression and MMSE was evaluated in MCI cases with Pearson correlations and a statistical threshold of p<0.05 using Excel. Quantitative real time PCR (RT-PCR) was used to validate microarray results for a subset of synaptic and mitochondrial electron transport genes in the SFG, across aged controls, MCI, and AD groups (Supplemental Figure 1). Microarray data is available at the Gene Expression Omnibus (GEO) website (http://www.ncbi.nlm.nih.gov/geo), accession number GSE11882. For detailed methods, see the online Supplemental material.
RESULTS
MCI cases cluster separately from aged and AD cases in all brain regions
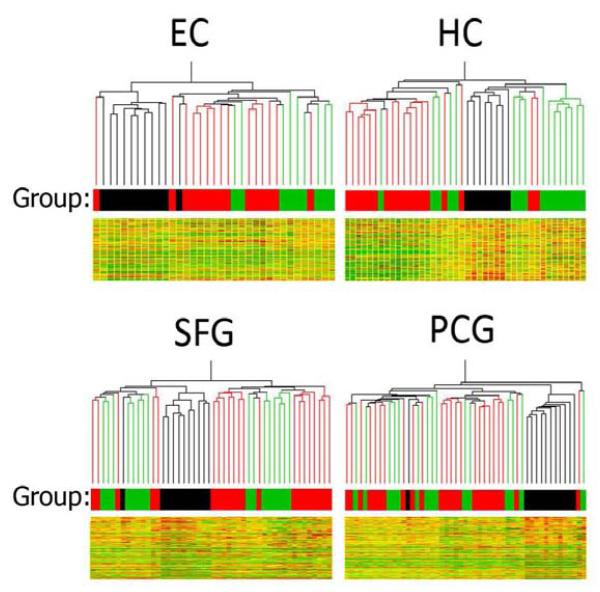
1 way ANOVA (p<.01) across the 3 groups (aged control, MCI, AD) was first applied to the 33,021 reliably detectable probeset list independently for each region to generate a region-specific significant gene set. Next, the relationship between aged, MCI, and AD cases in each region was visualized using hierarchical cluster analysis by region of the region-specific significant gene sets. In each brain region, MCI cases were closely clustered, constituting a unique branch of the hierarchical tree that was distinctly separate from both aged and AD samples (Figure 1). Overall, these data demonstrate in multiple brain regions that gene expression patterns in MCI are relatively similar across MCI cases – showing surprising uniformity across 11 of the 12 MCI cases - and are unique from the expression profiles of age-matched non-demented controls and AD cases.
Figure 1.
Hierarchical cluster analysis of region-specific expression was used to visualize the relationship between MCI (black), aged control (green), and AD cases (red) in the entorhinal cortex (EC), hippocampus (HC), superior frontal gyrus (SFG) and post-central gyrus (PCG). In each brain region, MCI cases were closely clustered, constituting a unique branch of the hierarchical tree that was distinctly separate from both aged and AD samples. Interestingly, one MCI case (#102, see supplemental table 1 for case details) was consistently an exception in the SFG, PCG and EC where it tended to cluster most closely with AD cases in the EC and PCG, while clustering with aged controls in the SFG. Other than case #102, no single MCI case was repeatedly an outlier across multiple brain regions, although there was one additional MCI outlier in the EC (case #41), which clustered with a moderate AD case (case #05).
MCI is more differentiated from AD than from age-matched controls
To identify the extent to which genes are differentially expressed in MCI, and to examine region-specific patterns of change, gene expression was compared in MCI relative to aged non-demented control or AD cases in each of the 4 brain regions (t-tests, p<0.01). In all brain regions examined, gene expression patterns in MCI bear more similarity to age-matched controls than to AD.
In MCI relative to age-matched controls, there were 2,600-5,800 significant probesets depending on brain region (Supplemental Table 3) with the greatest differences in the neocortical regions (SFG>PCG) and more modest differences in limbic regions (EC>HC). Numbers of upregulated vs. downregulated genes were relatively balanced in all brain regions. Far more extensive differences in gene expression emerged in the comparison of MCI with AD, with the greatest number of differentially expressed genes apparent in the HC, followed by the PCG, SFG and EC. The majority of genes were upregulated in MCI relative to AD.
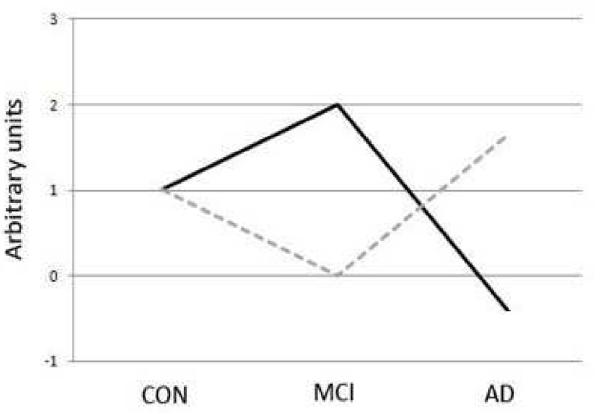
While it may be predicted that gene expression patterns in MCI would be intermediate between the profiles of non-demented aged and AD brains, we found that genes differentially expressed in MCI followed the general pattern of expression change of a pyramid or inverted pyramid across aged controls-MCI-AD (Figure 2). These data reveal that gene expression patterns in MCI represent a unique molecular state that does not follow a continuum of change between aged and AD profiles.
Figure 2.
Schematization of the general gene expression patterns across aged controls, MCI, and AD. The microarray data demonstrate that gene expression patterns in MCI do not follow a continuum of change between aged and AD profiles. The solid line represents genes that are upregulated in MCI vs. age-matched controls followed by downregulation in AD, while the dashed line represents the opposite pattern.
GO classification reveals upregulation of anabolic/metabolic and energy generation pathways in MCI relative to age-matched controls and AD
We next used GO classification to identify enriched functional gene classes among the genes significantly upregulated or downregulated in MCI (Supplemental tables 4A-D). Overall, MCI was associated with widespread upregulation of genes associated with biosynthetic and energy production, specifically genes related to protein biosynthesis, turnover and trafficking, mitochondrial energy generation, and, to a lesser degree, synaptic signaling and structure. Changes in these gene classes are described in detail below.
Protein-related function (biosynthesis, trafficking/localization, turnover) is upregulated in MCI
A comprehensive list of probesets related to protein metabolism (2075 probesets) was next analyzed for in depth analysis of gene expression responses in MCI and AD, in each brain region, using p<0.01. GO categories related to protein function consisted of genes that could be broadly categorized into protein biosynthesis, protein trafficking and localization, and protein turnover. The analysis revealed that many aspects of protein biosynthesis and intracellular trafficking are upregulated in the MCI brain relative to age-matched controls and AD, with protein trafficking and localization most extensively affected.
Specific genes related to protein biosynthesis that were significantly different in MCI vs. aged-controls or AD included upregulation of numerous translation initiation factors, ribosomal proteins, and tRNA synthetases (Supplemental Table 5). Few significant genes related to biosynthesis were downregulated in MCI relative to aged controls and AD, and were restricted to a subset of genes involved in translation. Similarly, many genes related to protein trafficking and localization were significantly different in MCI vs. aged-controls or AD, predominantly with upregulation of genes important for endocytosis, vesicle formation, endosomal sorting/signaling and exocytosis (Supplemental Table 6). For example, clathrin, several adaptor related protein complexes involved in vesicle formation, sorting nexins, vacuolar protein sorting molecules, multiple RABs and exocyst/Sec genes were upregulated in MCI. Similarly, in the category related to protein turnover (Supplemental Table 7), several genes are significantly different in MCI vs. aged-controls or AD, mainly with upregulation of autophagy related proteins, proteosome subunits, and numerous ubiquitin-related enzymes.
Mitochondrial electron transport chain genes are upregulated in MCI
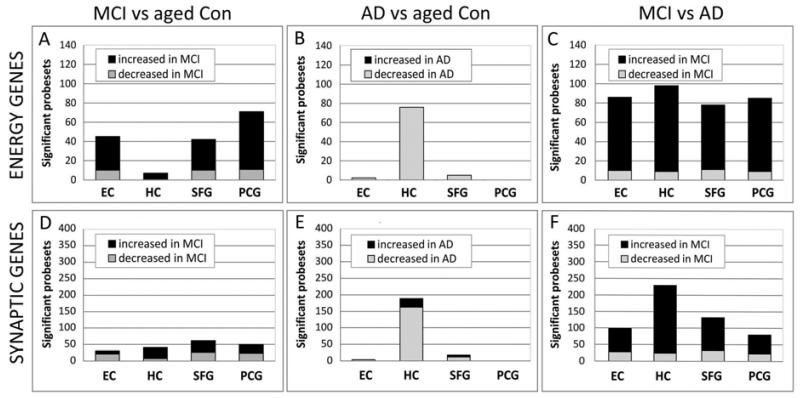
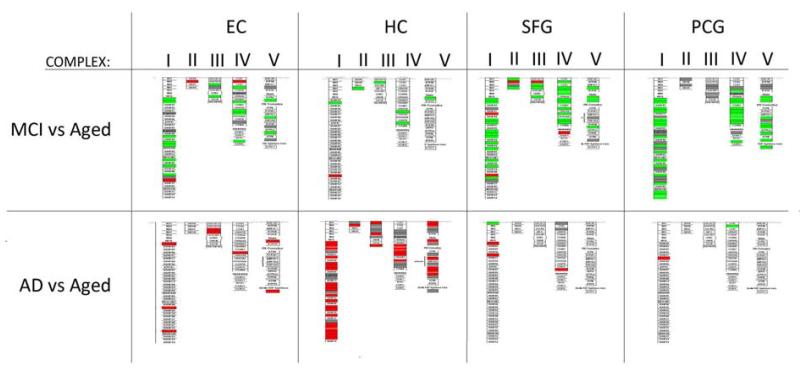
GO enrichment analysis also revealed that many genes related to mitochondrial function are upregulated in the MCI brain, particularly in the EC and cortical regions (Supplemental Table 8). We next undertook a comprehensive analysis of expression changes in each brain region of genes related to mitochondrial function, based on 142 probesets for electron transport chain subunits and translocases of the inner and outer mitochondrial membranes (p<0.01). Our analysis revealed that MCI was associated with upregulated expression of oxidative phosphorylation genes in each of the 4 brain regions assessed, relative to age-matched controls (Figure 3A). In contrast, this class of genes was notably downregulated in AD (Figure 3B), and the extent to which genes are differentially expressed in MCI vs. AD is shown in Figure 3C. The extent to which different mitochondrial complexes are affected is illustrated in Figure 4. Validation of expression differences between controls, MCI, and AD was undertaken with qPCR for a subset of genes related to mitochondrial function, with RT-PCR data consistent with the microarray findings (Supplemental Figure 1). Overall, these data indicate that the MCI brain undergoes dramatic upregulation of energy metabolism genes, in contrast to the transcriptional downregulation associated with AD.
Figure 3.
Many genes for mitochondrial electron transport and synaptic genes show altered expression in MCI, with predominant upregulation in MCI relative to aged controls (A) and relative to AD (C). In contrast, gene expression changes in AD are predominantly downregulated relative to age-matched controls (C). Electron-transport genes (A-C), Synaptic genes (D-F). The lists of mitochondrial bioenergetic genes (142 probesets) and synaptic genes (561 probesets) were each analyzed with a statistical threshold of p≤ 0.01.
Figure 4.
Mapping genes to the electron transport chain GenMAPP pathway demonstrates the extent of expression change in mitochondrial complexes I-V genes in MCI and AD relative to age-matched controls. In MCI, genes are predominantly upregulated (green) across complex I-V, with the greatest extent of response observed in the neocortical regions (superior frontal gyrus, SFG; post-central gyrus, PCG). In contrast to the response in MCI, genes in mitochondrial complexes I-V subunits are extensively downregulated (red) in the AD brain, most notably in the hippocampus (HC). Each box represents a gene. Complex I: NADH-ubiquinone oxidoreductase, Complex II: Succinate-ubiquinone oxidoreductase, Complex III: ubiquinol-cytochrome C reductase, complex IV: cytochrome C oxidase, complex V: ATP synthase. Each box represents a gene. Increased relative expression is indicated in green and decreased expression in red.
Synaptic genes are upregulated in MCI
We next examined synaptic gene responses in each brain region in MCI, using a comprehensive list of 340 synapse-related genes (p<0.01). Consistent with the pattern observed for protein biosynthesis and energy metabolism, the majority of synaptic genes that were significantly different in MCI relative to age-matched controls were upregulated in MCI, particularly in the HC, SFG, and PCG (Figure 3D). In contrast, synaptic genes were notably downregulated in AD (Figure 3E). The percentage of synapse-related genes showing significantly different expression in MCI was even more pronounced when expression levels were compared to AD (Figure 3F) with differentially expressed genes predominantly upregulated in MCI. Multiple aspects of synaptic function were represented, including the synaptic vesicle trafficking and neurotransmitter release machinery, neurotransmitter receptors, and synaptic structure and stabilization, as discussed in detail below. Assessment of gene expression for a subset of synaptic genes was undertaken using with RT-PCR, with qRT-PCR data consistent with the microarray findings (Supplemental Figure 1).
Synaptic vesicle trafficking and release
Many genes related to synaptic vesicle trafficking and release showed significantly altered expression in MCI, with the majority upregulated (Supplemental Table 9). For example, these included upregulation of several members of the soluble NSF attachment protein receptor (SNARE) family, a family of genes that is intimately involved in the membrane fusion machinery controlling neurotransmitter release into the synapse. Few genes related to synaptic vesicle trafficking and release showed decreased expression in MCI. Nearly all of the genes showing significantly altered expression in MCI vs. age-matched controls were similarly significantly altered when comparing MCI vs. AD. The increased expression of genes related to the presynaptic vesicle trafficking and release machinery suggests the possibility that there is increased neurotransmitter release in various regions of the MCI brain.
Neurotransmitter receptor-related
Similarly, many genes related to neurotransmitter receptors and neurotransmitter receptor trafficking showed significantly altered expression in MCI, predominantly upregulated (Supplemental Table 10). Upregulated neurotransmitter receptor genes included several glutamate receptor subtypes, multiple GABAA-associated genes and serotonin receptors, genes involved in glutamate receptor trafficking, and glutamate recycling. Downregulated neurotransmitter receptor genes included several cholinergic receptor subtypes (muscarinic, nicotinic), glutamate receptors, GABA-A subtypes, and the dopamine D2 receptor. Glutamate and GABA neurotransmitter systems were most extensively altered in MCI, with specific receptor subtypes that serve different functions being differentially up or downregulated. For example, while the glutamate receptor subtype NMDA2a was upregulated the NMDA2b and 2c subtypes were downregulated in MCI. Overall, these data suggest that there is a rebalancing of synaptic transmission and a reset of the excitatory/inhibitory balance that modulates synaptic plasticity show altered expression in MCI.
Synaptic structure and stabilization
Similar to neurotransmitter receptor and synaptic vesicle trafficking genes, most significant genes involved in synaptic structure and stabilization showed increased expression in MCI vs. aged matched controls (Supplemental Table 11). Examples of upregulated genes include several neuroligins, neurexins, and integrins, along with NCAM, EPH receptor B2, cofilin, spectrin, and protocadherins. Interestingly, different subunits in these same gene families were downregulated, including integrins, protocadherins, and EPH receptors, suggesting that the subunit availability of many gene families is altered in MCI. Overall, the increased expression of genes involved in multiple aspects of synaptic function suggests the potential for enhanced synaptic transmission and plasticity in the MCI brain.
Cognitive function correlates with synaptic gene expression in MCI, especially in the EC
Finally, to investigate if synaptic gene changes in MCI were related to cognitive function, Pearson’s correlation coefficient between MMSE and region-specific mRNA expression were computed for all synaptic genes. A number of synaptic genes showed strong significant correlations (r>0.8, p<0.01) between mRNA level and MMSE scores. Notably, the majority of the significant correlations with MMSE were with gene expression in the EC, with fewer significant correlations with HC mRNA expression, and very few with neocortical gene expression (Supplemental Table 3). The set of synaptic genes with highly significant correlations were predominantly related to synaptic transmission and plasticity, actin dynamics, adhesion molecules, and myelin composition, with both positive and negative correlations often observed within the same general functional category.
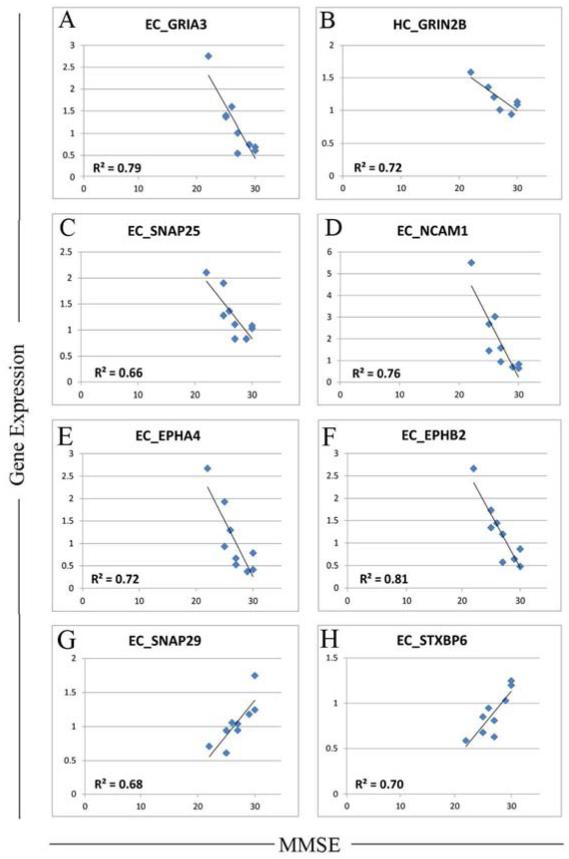
Next, to determine if any concepts emerged from the correlation data, we catalogued the functional output effect of the genes showing significant correlations to cognition, taking into consideration the direction of the correlation. While at first glance the presence of both positive and negative correlations between gene expression and MMSE seemed conflicting, comprehensive analysis of gene function revealed several consistent functional themes, with a major theme centering around synaptic excitability and plasticity. Specifically, of genes showing significant correlations with MMSE, gene changes that facilitate synaptic excitability/plasticity were overwhelmingly associated with poorer MMSE. For example, strong negative correlations between gene expression and MMSE were found for glutamate receptors that enhance plasticity (GluR3 (r = −0.89, EC) (Figure 5A)), NR2b (r = −0.93, HC) (Figure 5B)), as well as SNAP-25 (r = −0.81, EC) (Figure 5C) -- an essential SNARE component involved in synaptic vesicle exocytosis, and neural cell adhesion molecule (NCAM1, r= −0.87, EC) (Figure 5D), an adhesion molecule that facilitates AMPA receptor delivery to the synapse (Dityatev, et al., 2000,Doherty, et al., 1995). In addition, strong negative correlations between mRNA and MMSE were found for two ephrin receptors that enhance plasticity: ephrin A4 (EPHA4: r= −0.85, EC) (Figure 5E) which is expressed on spines and shapes spine morphology/excitability (Tremblay, et al., 2007,Yamaguchi and Pasquale, 2004), and Ephrin B2 (EFNB2) which facilitates NMDA signaling (Nolt, et al., 2011). Notably, EFNB2 showed a strong negative correlation with MMSE in both the EC (r= −0.9)( Figure 5F) and HC (r= −0.88). Conversely, gene changes that inhibit plasticity were positively associated with MMSE. These include SNAP29 (r= + 0.82, EC) (Figure 5G) which inhibits SNARE disassembly and post-fusion SNARE recycling, and syntaxin binding protein 6/amysin (STXBP6) (Figure 5H) which negatively regulates vesicle exocytosis. STXBP6 showed a strong positive correlation with MMSE in both the EC (r= +0.84) and the HC (r= + 0.88). Taken together, these data suggest that there is excessive excitability and apparent plasticity in limbic regions of the brain in MCI, which is associated with poorer cognitive function.
Figure 5.
Correlations between gene expression and MMSE in MCI. Genes that facilitate synaptic excitability/plasticity were overwhelmingly associated with poorer MMSE, with strong negative correlations found for glutamate receptors that enhance plasticity including A) GRIA3/GluR3, B) NR2b/GRIN2B, along with CC) SNAP-25, an essential SNARE component involved in synaptic vesicle exocytosis, and D) neural cell adhesion molecule, an adhesion molecule that facilitates AMPA receptor delivery to the synapse, E) ephrin A4 (EPHA4) which is expressed on spines and shapes spine morphology/excitability, and F) Ephrin B2 (EFNB2) which facilitates NMDA signaling. Gene changes that inhibit plasticity were positively associated with MMSE, including G) SNAP29 which inhibits SNARE disassembly and post-fusion SNARE recycling, and H) syntaxin binding protein 6/amysin (STXBP6) which negatively regulates vesicle exocytosis. Statistical threshold for correlations was set at p<0.05.
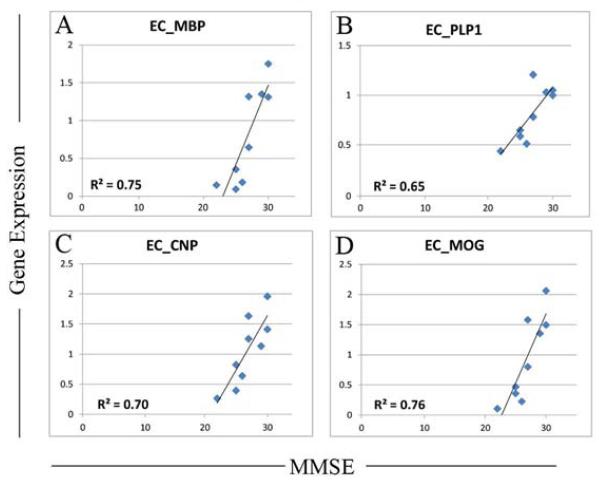
A second theme that emerged from the correlation data was that EC expression of several abundant myelin-related genes showed a strong positive relationship with MMSE. These include myelin basic protein (MBP; r=+.87) (Figure 6A), proteolipid protein 1 (PLP1: r= +0.80, EC) (Figure 6B) and multiple probesets for 2′3′-cyclic nucleotide 3′ phosphodiesterase (CNP; r=0.84, r=0.92) (Figure 6C), an abundant CNS myelin protein involved in the very rapid growth of myelin membrane during early membrane biogenesis (Sprinkle, 1989)), and multiple probesets for myelin oligodendrocyte glycoprotein (MOG) (r=+0.91, r= +0.89, r=+0.87, r= +0.82) (Figure 6D), one of a family of myelin associated inhibitors (MAIs). Interestingly, recent literature reveals that MAIs potently limit activity and experience-dependent plasticity in the intact, adult CNS (Akbik, et al., 2012,Cafferty and Strittmatter, 2006). This is interesting because the strong positive correlations found in the EC between MOG expression and MMSE indicate that factors that inhibit activity-dependent plasticity in the EC are associated with better cognitive function in MCI, supporting our theme that poorer cognitive function in MCI are related to excessive excitability and plasticity in the EC.
Figure 6.
Correlation analysis reveals strong positive relationship in MCI between MMSE and expression of several abundant myelin-related genes. These include A) myelin basic protein (MBP), B) proteolipid protein 1 (PLP1), multiple probesets for 2′3′-cyclic nucleotide 3′ phosphodiesterase (CNP), an abundant CNS myelin protein involved in the very rapid growth of myelin membrane during early membrane biogenesis, and multiple probesets for myelin oligodendrocyte glycoprotein (MOG) one of a family of myelin associated inhibitors (MAIs), that potently limit activity and experience-dependent plasticity in the intact, adult CNS. Statistical threshold for correlations was set at p<0.05.
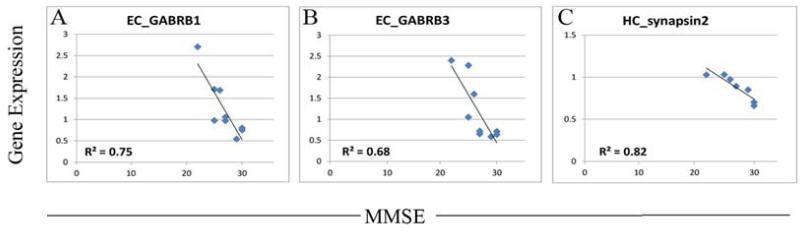
A third theme emerging from the correlation data reflects altered GABA function in the limbic region in MCI. Specifically, several genes involved in GABA signaling showed strong relationships with MMSE, such as synapsin II (SYN2: r= −0.90, HC) (Figure 7A) which promotes GABA asynchronous release, and several GABA receptor subtypes including (GABARR1; r= +0.94, HC), GABA receptor beta 1 (GABARB1: r= −0.90, EC) (Figure 7B) and GABA receptor beta 3 (GABARB3: r= −0.83, EC) (Figure 7C). In the EC, these relationships suggest that decreased GABAergic signaling is beneficial to cognitive function, again suggestive of altered excitability and plasticity in MCI. However, to evaluate if the relationship of GABAergic gene expression with MMSE indeed reflects a change in excitability, it will first be necessary to determine which types of neurons express these receptor subtypes (e.g. GABA or Glutamatergic), and ultimately assess the functional effects of specific receptor manipulations on limbic excitability, plasticity, and cognition, using animal models.
Figure 7.
Correlations analysis reveals altered GABA function in the limbic region in MCI. Expression of several genes involved in GABA signaling showed strong relationships with MMSE in MCI, including several GABA receptor subtypes such as (A) GABA receptor beta 1 (GABARB1) and (B) GABA receptor beta 3 (GABARB3) as well as synapsin II (SYN2), which promotes GABA asynchronous release. The strong correlations between MMSE and expression of specific components regulating GABA signaling suggest that there is altered excitability and plasticity in limbic regions in MCI. Statistical threshold for correlations was set at p<0.05.
Finally, there were a number of additional genes with strong correlations to MMSE in MCI cases. These included modulatory neurotransmitter receptors (dopaminergic D5 receptor (DRD5: r= −0.87, HC) (Supplemental Figure 2A), several cell adhesion molecules (contactin-associated proteins, neurexin 1, integrin alpha 2, cell adhesion molecule 1 (CADM1: r = −0.94, HC) (Supplemental Figure 2B), and genes regulating actin and cytoskeleton dynamics (gelsolin, cofilin 2 (CFL2: r= +0.88, EC) (Supplemental Figure 2C), (S100B: r= −0.81, EC) (Supplemental Figure 2D), in addition to bridging integrator 1/amphiphysin 2 (BIN1: r= +0.89, EC) (Supplemental Figure 2E), recently been identified as the most important risk locus for late onset Alzheimer’s disease after apolipoprotein E (Bertram, et al., 2007,Tan, et al., 2013).
DISCUSSION
Because MCI represents a clinical transition state between aging and dementia, we investigated the hypothesis that gene expression patterns in the MCI brain might show a profile intermediate between the molecular profiles of normal aged controls and AD. This analysis revealed that the gene expression profiles of MCI cases clustered closely together in each brain region, forming a unique branch of the hierarchical tree that was distinctly separate from both aged and AD samples. That is, expression patterns in MCI are unique from those seen in age-matched controls and AD but are relatively similar across MCI cases, showing surprising uniformity across 11 of the 12 MCI cases.
Microarray analysis revealed extensive gene expression differences in all brain regions in MCI relative to both age-matched controls and AD. Relative to age matched controls the greatest differences in MCI were observed in the neocortical regions (SFG>PCG) with more modest differences in limbic regions (EC>HC). Many of the same genes that differentiated MCI from non-demented controls also differentiated MCI from AD, thus following the expression pattern across aged controls-MCI-AD schematized in Figure 2. For example ~75% of genes upregulated in MCI vs. age-matched controls also showed significant upregulation in MCI when compared to AD. Expression patterns of genes related to mitochondrial bioenergetics, protein homeostasis, and synaptic function were predominantly upregulated in MCI, in stark contrast to the extensive downregulation that occurs in aging, particularly after the 6th-7th decades of life (Berchtold, et al., 2013,Lu, et al., 2004). Taken together, these results demonstrate that gene expression patterns in MCI are not an extension of aging, and for the most part, are not intermediate between aged controls and AD.
Functional enrichment analysis revealed that genes upregulated in MCI were prominently enriched in categories related to anabolic/metabolic function and mitochondrial energy generation pathways, while downregulated genes did not tend to show notable GO category enrichment. Upregulated categories were predominantly related to transcription, protein biosynthesis, protein trafficking and localization, and protein turnover, revealing that many aspects of protein homeostasis are upregulated in the MCI brain. These data demonstrate that the MCI brain undergoes a generalized effort to bolster protein synthesis and intracellular trafficking. In parallel, many genes related to mitochondrial energy generation were upregulated. The upregulation of genes for energy generation is consistent with the increased anabolic/metabolic effort, as increased energy production would be necessary to support an increased demand for protein synthesis and trafficking throughout the cell. The largest responses were observed in the EC and neocortical regions (with 75-85% of significant genes upregulated), with fewer changes in these gene categories apparent in the HC, relative to age-matched controls.
In addition to widespread responses of genes related to protein homeostasis and energy production, many genes important in synaptic function showed altered expression in MCI, predominantly upregulation in the HC and neocortical regions. These included several essential members of the SNARE complex (involved in membrane fusion events controlling neurotransmitter release) as well as many classes of neurotransmitter receptors that modulate synaptic plasticity, set the excitatory-inhibitory balance and support synaptic structure and stabilization. Taken together, increased expression of genes involved in multiple aspects of synaptic function suggests the potential for enhanced synaptic transmission and plasticity in the MCI brain. While these endpoints have traditionally been considered to be beneficial for cognitive function, we propose that these changes likely lead to dysfunctional plasticity and networks. For example, the extensive synaptic gene upregulation likely results in a higher baseline expression that impairs gene-mediated plasticity and encoding, leading to an inability to register new information.
To examine potential relationships between gene expression and cognitive function in MCI, we next analyzed Pearson correlations between mRNA levels and MMSE in MCI cases. Expression of a number of synaptic genes showed strong correlations with MMSE, with EC gene expression showing the most extensive relationship to cognitive function. The extent to which gene expression correlated with MMSE was less evident in the HC, and was absent in the neocortical regions. Comprehensive analysis of the functional effect of the significant genes, taking into consideration the direction of the correlations, revealed several consistent functional themes, with a major theme centering on synaptic excitability and plasticity. For example, of genes showing significant correlations with MMSE, gene changes that facilitate synaptic excitability and plasticity were overwhelmingly associated with poorer MMSE. Conversely, gene changes that inhibit plasticity were positively associated with MMSE. These data suggest that there is excessive excitability and apparent plasticity in limbic brain regions in MCI, which lead to impaired synaptic and cognitive function.
In parallel, we also found that several abundant myelin-related genes showed strong positive relationships with MMSE in MCI cases, suggesting that myelogenic events that promote myelin biogenesis and stability help preserve cognitive function in MCI. Interestingly, one of the myelin genes, myelin oligodendrocyte glycoprotein (MOG), belongs to a family of myelin associated inhibitors that modulate cytoskeletal rearrangements that stabilize synaptic ultrastructure (Lee, et al., 2008,Zagrebelsky, et al., 2010), and potently limit activity and experience-dependent plasticity in the intact, adult CNS (Akbik, et al., 2012,Cafferty and Strittmatter, 2006). The strong positive correlations found in the EC between MOG gene expression and MMSE provides further support for our developing hypothesis that factors that inhibit activity-dependent plasticity in MCI, at least in the EC, are associated with better cognitive function. These data contribute to the accumulating evidence of white matter changes in MCI, supporting the emerging idea that a myelin-related dysfunction may contribute to MCI and onset of AD, particularly in tracts that connect gray matter structures associated with memory function (Carmeli, et al., 2013,Gold, et al., 2012a,Gold, et al., 2012b).
Importantly, of the 4 regions assessed, the correlation of cognition with gene expression was especially strong in the EC. As a critical gateway for cortical information to and from the HC, the EC plays a key role in normal cognitive function, particularly memory formation and retrieval (Scharfman and Chao, 2013). EC dysfunction is thought to play a prominent and early role in cognitive decline in AD, as accumulation of pathology and neuronal degeneration in this brain region is one of the earliest changes on the pathway to AD. An emerging theme in the literature suggests that neuronal dysfunction and degeneration in the EC drives HC dysfunction and degeneration in AD, in part through loss of neurotrophic support along with other mechanisms (Scharfman and Chao, 2013). Our finding that in MCI, cognition correlated strongly with EC synaptic gene expression is consistent with a critical role of EC synaptic function in cognition, and supports the idea that EC synaptic dysfunction occurs early in the course of cognitive decline in MCI.
Overall, our microarray analysis revealed in multiple brain regions that MCI is associated with widespread upregulation of genes associated with biosynthetic and energy production, specifically genes related to protein biosynthesis, turnover and trafficking, mitochondrial energy generation, and, to a lesser degree, synaptic signaling and structure. In addition, the correlation data suggest that there is excessive excitability and apparent plasticity in limbic brain regions in MCI, particularly in the EC. These data are consistent with numerous imaging studies suggesting that very early stages of cognitive decline are associated with increased synaptic activity and plasticity. For example, functional magnetic resonance imaging (fMRI) studies have identified a state of hyperexcitability in the hippocampus and medial temporal lobe network in early to mild MCI (Bakker, et al., 2012,Dickerson, et al., 2005,Hamalainen, et al., 2007,Putcha, et al., 2011,Vannini, et al., 2007,Yassa, et al., 2010), in mildly impaired, amyloid positive older adults (Sperling, et al., 2009), low-performing clinically normal controls (S.L. Miller, et al., 2008a) and even in cognitively normal controls at genetic risk for AD (Bassett, et al., 2006,Yassa, et al., 2008). Further, a longitudinal fMRI study where MCI subjects were followed over approximately 6 years revealed that greater HC activation in MCI predicted a greater degree and rate of subsequent cognitive decline (S.L. Miller, et al., 2008b). This hyperactivation in MCI/early stage AD has been suggested to reflect a failure of deactivation (Bejanin, et al., 2012,Celone, et al., 2006), which may be determined in part by the alterations that we observed in MCI in activity of synaptic genes that set the excitatory-inhibitory balance.
Extensive discussion has focused on whether the limbic hyperactivation in MCI/incipient AD is beneficial or represents a dysfunctional state. Our data suggest that excessive excitability and apparent plasticity in limbic brain regions in MCI is associated with poorer cognitive function, consistent with the emerging idea that greater limbic activation in MCI is a dysfunctional condition. Building on the literature, a recent study in amnestic MCI patients demonstrated that cognition is improved when hippocampal hyperactivity is reduced with the antiepileptic agent leviracetam, (Bakker, et al., 2012). It is likely that the hyperexcitable “noisy” networks in the MCI increase the risk for excitotoxicity, oxidative damage, and impending metabolic burnout (Cohen, et al., 2009,Sperling, et al., 2010). Increased metabolism, altered excitatory/inhibitory balance, and augmented synaptic machinery in the MCI brain may all contribute to increased excitability, with increased excitability in turn leading to greater metabolic demand, and ultimately progressive degeneration and AD, if not controlled.
Our microarray data are also consistent with the growing evidence that there is a non-linear trajectory of change in the brain in the transition from the cognitively normal condition to MCI and AD, and even across the continuum of impairment within the spectrum of MCI. For example, a recent fMRI study revealed that when MCI patients are stratified by degree of impairment, only the less-impaired MCI patients showed hyperactivation in the HC relative to controls, whereas more impaired MCI subjects demonstrated significant hypoactivation (Celone, et al., 2006). Similarly, PET assessment of fluorodeoxyglucose (FDG) metabolism of the basal forebrain in MCI revealed an inverted U-relationship with MMSE, in that neuronal activity initially increased, then decreased with further cognitive decline (Kim, et al., 2012). These studies suggest that hyperexcitability is present in multiple brain regions in MCI and is associated with very early stages of cognitive impairment, and that hypoactivation ensues as cognitive impairment progresses to AD. Supporting the imaging data, anatomical and molecular analysis also reveals a non-linear trajectory of change in the brain with the transition from the cognitively normal condition to MCI and AD. For example, the human literature has reported elevated glutamatergic presynaptic bouton density in the midfrontal cortex in MCI (Bell, et al., 2007), increases in drebrin (a synaptic protein that regulates spine morphogenesis and receptor density on spines) in the superior frontal cortex in incipient AD (Counts, et al., 2006), and a biphasic synaptic protein response across Braak staging in the isocortex, with levels of synaptophysin, syntaxin, SNAP25, alpha synuclein, and MAP2 increasing in Braak stages III-IV, and subsequently declining in later Braak stages (Mukaetova-Ladinska, et al., 2000). Similarly, analysis of the medial prefrontal cortex in incipient AD revealed upregulation of genes associated with synaptic function, energy metabolism and protein homeostasis in the very early “pre-symptomatic” stage of AD, in association with increasing Braak pathology (Bossers, et al., 2010). Further, in synaptoneurosomes derived from prefrontal cortex of incipient AD, increased gene and protein expression was found for multiple neuroplasticity genes, notably those involved in vesicle mediated transport, synaptic transmission, and intracellular transport (Williams, et al., 2009), similar to our findings in the 4 cortical regions assessed. Importantly, many of the same genes reported to be upregulated in synaptosomes derived from prefrontal cortex (of incipient AD) were similarly upregulated in the prefrontal cortex (SFG) in our MCI cases, including glutamate receptors (GRIA2/GluR2), GABA receptors (GABBR2, GABRA1), neurexins (NRXN1, NRXN2), RABS (RAB14, RAB15), GAP43, HTR2a, and synaptotagmin 11, among others (Williams, et al., 2009). In parallel, molecular analysis of transgenic mouse models of AD have revealed increased density of presynaptic boutons during early stages of the amyloid pathology (Bell, et al., 2003,Hu, et al., 2003), and aberrant excitatory neuronal activity and compensatory remodeling of inhibitory hippocampal circuits (Palop, et al., 2007). This literature strongly supports the concept that the brain undergoes a non-linear trajectory of change in the transition from the cognitively normal condition to cognitively impaired, with the inflection point of the change likely most pronounced in the early stages of MCI.
Some controversy currently exists in the literature regarding the stage of cognitive decline at which pronounced molecular changes occur, and how response patterns compare across brain regions. For example, downregulation of genes related to synaptic function, energy metabolism and protein homeostasis has been reported in the HC in incipient AD (Blalock, et al., 2004), while analysis of the cingulate cortex and amygdala identified upregulated protein synthesis, but downregulated energy and synaptic vesicle function (Loring, et al., 2001). Integration of these multiple varying microarray findings into a fully consistent concept is complicated by a number of variables in the published studies. These include that the analyses across human studies target differing brain regions (e.g., multiple cortical regions, hippocampus, entorhinal cortex, cingulate cortex, amygdala, among others), target regional subfields within a brain structure (e.g. hippocampal CA1 containing both grey/white matter (Colangelo, et al., 2002), CA1 grey matter vs CA1 white matter (Blalock, et al., 2011)), or investigate changes at the single cell (laser-captured) level (pyramidal cells of the CA1 (Counts, et al., 2014,Ginsberg, et al., 2012,Ginsberg, et al., 2010,Ginsberg, et al., 2000)). In addition, the variability in findings across studies likely reflects the heterogeneity of clinical MCI as well as the current difficulty in clinically defining the transition between late MCI and early AD, which remains an evolving frontier. Ultimately, continuing studies are needed to elucidate and differentiate the varying underlying mechanisms across the spectrum of MCI and incipient AD.
Finally, there are a number of additional variables to consider that are not addressed in this analysis, which are known to affect AD risk and disease progression and that likely contribute to gene expression changes in MCI and AD. For example, aging, MCI, and AD are accompanied by region-specific synaptic loss and remodeling along with neuronal loss in later stages of AD (Morrison and Baxter, 2012), which may factor into the gene expression changes in MCI and AD. Other factors include genotype, such as ApoE4 status (the strongest currently-known genetic risk factor for late-onset AD (Corder, et al., 1993,Genin, et al., 2011), pathological features such as beta amyloid accumulation and polymerization state, and tau pathology, among others. Recent evidence demonstrates that ApoE status affects even normal brain function (Filippini, et al., 2011), and that the ApoE4 genotype accelerates onset of age-related and disease-related changes (Liu, et al., 2013), including augmented pathology accumulation in AD, and even the capacity to generate glutamate from glutamine (Dumanis, et al., 2013). Moreover, ApoE4 carriers show a greater extent of immune/inflammation-related gene responses in the brain in aging and AD (Cribbs, et al., 2012). It is possible that the presence of ApoE4 may similarly exacerbate/accelerate the disease-related changes in energy, synaptic, and protein-related genes that we observe here for MCI and AD. Other genetic factors likely also come into play, including polymorphisms in AD risk factor genes recently identified in genome-wide association studies (2013) and genetic differences due to ethnicities (Li, et al., 2010). While the effects of these variables on gene expression profiles could not be addressed using the current dataset, they are important considerations for future studies exploring gene expression changes in MCI and AD, as well as in normal aging.
Despite the above-mentioned caveats, our study reveals that that the MCI brain undergoes vast molecular reprogramming, with gene expression signatures undergoing a non-linear progression in the transitions to MCI and AD. Taken together with the current literature, our data reveal that the MCI brain undergoes vast molecular reprogramming, and lend support to the emerging concept that, during the very early stages of cognitive decline, mechanisms are engaged in the brain that increase synaptic activity and plasticity, particularly in the pre-symptomatic and prodromal stages of AD, and that these mechanisms are likely not beneficial to cognitive function (Bossers, et al., 2010,Williams, et al., 2009). These data and the literature suggest that therapeutic approaches aimed at reducing excitability and plasticity, and potentially increasing synaptic stabilization, may be particularly beneficial for improving cognition in MCI, an approach adopted in recent promising clinical trials (Bakker, et al., 2012).
Supplementary Material
ACKNOWLEDGEMENTS
Funding for this research was provided by the N.I.H. to CWC (AG034667, AG16573, AG00538). The authors Nicole C. Berchtold and Carl W. Cotman had full access to all of the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author’s contributions:
NCB: data analysis, interpretation, wrote manuscript
MNS: case selection, clinical data, provided tissue
TGB: case selection, clinical data, provided tissue
RCK: case selection, clinical data, provided tissue
DHC: manuscript assistance
CWC: designed study, obtained funding, data interpretation, wrote manuscript
Author contributions: design and conduct of the study (AG034667, AG16573, AG00538), collection, management analysis and interpretation of the data (AG034667, AG16573, AG00538), preparation, review, approval of the manuscript (AG034667, AG16573, AG00538).
Conflict of interest:
The authors declare no conflict of interest.
REFERENCES
- Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet. 2013;45(12):1452–8. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbik F, Cafferty WB, Strittmatter SM. Myelin associated inhibitors: a link between injury-induced and experience-dependent plasticity. Experimental neurology. 2012;235(1):43–52. doi: 10.1016/j.expneurol.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker A, Krauss GL, Albert MS, Speck CL, Jones LR, Stark CE, Yassa MA, Bassett SS, Shelton AL, Gallagher M. Reduction of hippocampal hyperactivity improves cognition in amnestic mild cognitive impairment. Neuron. 2012;74(3):467–74. doi: 10.1016/j.neuron.2012.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett SS, Yousem DM, Cristinzio C, Kusevic I, Yassa MA, Caffo BS, Zeger SL. Familial risk for Alzheimer’s disease alters fMRI activation patterns. Brain. 2006;129(Pt 5):1229–39. doi: 10.1093/brain/awl089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejanin A, Viard A, Chetelat G, Clarys D, Bernard F, Pelerin A, de La Sayette V, Eustache F, Desgranges B. When Higher Activations Reflect Lower Deactivations: A PET Study in Alzheimer’s Disease during Encoding and Retrieval in Episodic Memory. Front Hum Neurosci. 2012;6:107. doi: 10.3389/fnhum.2012.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell KF, Bennett DA, Cuello AC. Paradoxical upregulation of glutamatergic presynaptic boutons during mild cognitive impairment. J Neurosci. 2007;27(40):10810–7. doi: 10.1523/JNEUROSCI.3269-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell KF, de Kort GJ, Steggerda S, Shigemoto R, Ribeiro-da-Silva A, Cuello AC. Structural involvement of the glutamatergic presynaptic boutons in a transgenic mouse model expressing early onset amyloid pathology. Neuroscience letters. 2003;353(2):143–7. doi: 10.1016/j.neulet.2003.09.027. [DOI] [PubMed] [Google Scholar]
- Berchtold NC, Coleman PD, Cribbs DH, Rogers J, Gillen DL, Cotman CW. Synaptic genes are extensively downregulated across multiple brain regions in normal human aging and Alzheimer’s disease. Neurobiology of aging. 2013;34(6):1653–61. doi: 10.1016/j.neurobiolaging.2012.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berchtold NC, Cribbs DH, Coleman PD, Rogers J, Head E, Kim R, Beach T, Miller C, Troncoso J, Trojanowski JQ, Zielke HR, Cotman CW. Gene expression changes in the course of normal brain aging are sexually dimorphic. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(40):15605–10. doi: 10.1073/pnas.0806883105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram L, McQueen MB, Mullin K, Blacker D, Tanzi RE. Systematic meta-analyses of Alzheimer disease genetic association studies: the AlzGene database. Nat Genet. 2007;39(1):17–23. doi: 10.1038/ng1934. [DOI] [PubMed] [Google Scholar]
- Blalock EM, Buechel HM, Popovic J, Geddes JW, Landfield PW. Microarray analyses of laser-captured hippocampus reveal distinct gray and white matter signatures associated with incipient Alzheimer’s disease. J Chem Neuroanat. 2011;42(2):118–26. doi: 10.1016/j.jchemneu.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blalock EM, Geddes JW, Chen KC, Porter NM, Markesbery WR, Landfield PW. Incipient Alzheimer’s disease: microarray correlation analyses reveal major transcriptional and tumor suppressor responses. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(7):2173–8. doi: 10.1073/pnas.0308512100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossers K, Wirz KT, Meerhoff GF, Essing AH, van Dongen JW, Houba P, Kruse CG, Verhaagen J, Swaab DF. Concerted changes in transcripts in the prefrontal cortex precede neuropathology in Alzheimer’s disease. Brain. 2010;133(Pt 12):3699–723. doi: 10.1093/brain/awq258. [DOI] [PubMed] [Google Scholar]
- Cafferty WB, Strittmatter SM. The Nogo-Nogo receptor pathway limits a spectrum of adult CNS axonal growth. J Neurosci. 2006;26(47):12242–50. doi: 10.1523/JNEUROSCI.3827-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmeli C, Donati A, Antille V, Viceic D, Ghika J, von Gunten A, Clarke S, Meuli R, Frackowiak RS, Knyazeva MG. Demyelination in mild cognitive impairment suggests progression path to Alzheimer’s disease. PLoS ONE. 2013;8(8):e72759. doi: 10.1371/journal.pone.0072759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celone KA, Calhoun VD, Dickerson BC, Atri A, Chua EF, Miller SL, DePeau K, Rentz DM, Selkoe DJ, Blacker D, Albert MS, Sperling RA. Alterations in memory networks in mild cognitive impairment and Alzheimer’s disease: an independent component analysis. J Neurosci. 2006;26(40):10222–31. doi: 10.1523/JNEUROSCI.2250-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen AD, Price JC, Weissfeld LA, James J, Rosario BL, Bi W, Nebes RD, Saxton JA, Snitz BE, Aizenstein HA, Wolk DA, Dekosky ST, Mathis CA, Klunk WE. Basal cerebral metabolism may modulate the cognitive effects of Abeta in mild cognitive impairment: an example of brain reserve. J Neurosci. 2009;29(47):14770–8. doi: 10.1523/JNEUROSCI.3669-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colangelo V, Schurr J, Ball MJ, Pelaez RP, Bazan NG, Lukiw WJ. Gene expression profiling of 12633 genes in Alzheimer hippocampal CA1: transcription and neurotrophic factor down-regulation and up-regulation of apoptotic and pro-inflammatory signaling. J Neurosci Res. 2002;70(3):462–73. doi: 10.1002/jnr.10351. [DOI] [PubMed] [Google Scholar]
- Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, Roses AD, Haines JL, Pericak-Vance MA. Science. 5123. Vol. 261. New York, NY: 1993. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families; pp. 921–3. [DOI] [PubMed] [Google Scholar]
- Counts SE, Alldred MJ, Che S, Ginsberg SD, Mufson EJ. Synaptic gene dysregulation within hippocampal CA1 pyramidal neurons in mild cognitive impairment. Neuropharmacology. 2014;79:172–9. doi: 10.1016/j.neuropharm.2013.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Counts SE, Nadeem M, Lad SP, Wuu J, Mufson EJ. Differential expression of synaptic proteins in the frontal and temporal cortex of elderly subjects with mild cognitive impairment. J Neuropathol Exp Neurol. 2006;65(6):592–601. doi: 10.1097/00005072-200606000-00007. [DOI] [PubMed] [Google Scholar]
- Cribbs DH, Berchtold NC, Perreau V, Coleman PD, Rogers J, Tenner AJ, Cotman CW. Extensive innate immune gene activation accompanies brain aging, increasing vulnerability to cognitive decline and neurodegeneration: a microarray study. Journal of neuroinflammation. 2012;9:179. doi: 10.1186/1742-2094-9-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickerson BC, Salat DH, Greve DN, Chua EF, Rand-Giovannetti E, Rentz DM, Bertram L, Mullin K, Tanzi RE, Blacker D, Albert MS, Sperling RA. Increased hippocampal activation in mild cognitive impairment compared to normal aging and AD. Neurology. 2005;65(3):404–11. doi: 10.1212/01.wnl.0000171450.97464.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dityatev A, Dityateva G, Schachner M. Synaptic strength as a function of post-versus presynaptic expression of the neural cell adhesion molecule NCAM. Neuron. 2000;26(1):207–17. doi: 10.1016/s0896-6273(00)81151-4. [DOI] [PubMed] [Google Scholar]
- Doherty P, Fazeli MS, Walsh FS. The neural cell adhesion molecule and synaptic plasticity. J Neurobiol. 1995;26(3):437–46. doi: 10.1002/neu.480260315. [DOI] [PubMed] [Google Scholar]
- Dumanis SB, DiBattista AM, Miessau M, Moussa CE, Rebeck GW. APOE genotype affects the pre-synaptic compartment of glutamatergic nerve terminals. Journal of neurochemistry. 2013;124(1):4–14. doi: 10.1111/j.1471-4159.2012.07908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippini N, Ebmeier KP, MacIntosh BJ, Trachtenberg AJ, Frisoni GB, Wilcock GK, Beckmann CF, Smith SM, Matthews PM, Mackay CE. Differential effects of the APOE genotype on brain function across the lifespan. NeuroImage. 2011;54(1):602–10. doi: 10.1016/j.neuroimage.2010.08.009. [DOI] [PubMed] [Google Scholar]
- Genin E, Hannequin D, Wallon D, Sleegers K, Hiltunen M, Combarros O, Bullido MJ, Engelborghs S, De Deyn P, Berr C, Pasquier F, Dubois B, Tognoni G, Fievet N, Brouwers N, Bettens K, Arosio B, Coto E, Del Zompo M, Mateo I, Epelbaum J, Frank-Garcia A, Helisalmi S, Porcellini E, Pilotto A, Forti P, Ferri R, Scarpini E, Siciliano G, Solfrizzi V, Sorbi S, Spalletta G, Valdivieso F, Vepsalainen S, Alvarez V, Bosco P, Mancuso M, Panza F, Nacmias B, Bossu P, Hanon O, Piccardi P, Annoni G, Seripa D, Galimberti D, Licastro F, Soininen H, Dartigues JF, Kamboh MI, Van Broeckhoven C, Lambert JC, Amouyel P, Campion D. APOE and Alzheimer disease: a major gene with semi-dominant inheritance. Mol Psychiatry. 2011;16(9):903–7. doi: 10.1038/mp.2011.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson GE, Starkov A, Blass JP, Ratan RR, Beal MF. Cause and consequence: mitochondrial dysfunction initiates and propagates neuronal dysfunction, neuronal death and behavioral abnormalities in age-associated neurodegenerative diseases. Biochim Biophys Acta. 2010;1802(1):122–34. doi: 10.1016/j.bbadis.2009.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsberg SD, Alldred MJ, Che S. Gene expression levels assessed by CA1 pyramidal neuron and regional hippocampal dissections in Alzheimer’s disease. Neurobiology of disease. 2012;45(1):99–107. doi: 10.1016/j.nbd.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsberg SD, Alldred MJ, Counts SE, Cataldo AM, Neve RL, Jiang Y, Wuu J, Chao MV, Mufson EJ, Nixon RA, Che S. Microarray analysis of hippocampal CA1 neurons implicates early endosomal dysfunction during Alzheimer’s disease progression. Biol Psychiatry. 2010;68(10):885–93. doi: 10.1016/j.biopsych.2010.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsberg SD, Hemby SE, Lee VM, Eberwine JH, Trojanowski JQ. Expression profile of transcripts in Alzheimer’s disease tangle-bearing CA1 neurons. Annals of neurology. 2000;48(1):77–87. [PubMed] [Google Scholar]
- Gold BT, Jiang Y, Powell DK, Smith CD. Multimodal imaging evidence for axonal and myelin deterioration in amnestic mild cognitive impairment. J Alzheimers Dis. 2012a;31(Suppl 3):S19–31. doi: 10.3233/JAD-2012-112165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold BT, Johnson NF, Powell DK, Smith CD. White matter integrity and vulnerability to Alzheimer’s disease: preliminary findings and future directions. Biochim Biophys Acta. 2012b;1822(3):416–22. doi: 10.1016/j.bbadis.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamalainen A, Pihlajamaki M, Tanila H, Hanninen T, Niskanen E, Tervo S, Karjalainen PA, Vanninen RL, Soininen H. Increased fMRI responses during encoding in mild cognitive impairment. Neurobiology of aging. 2007;28(12):1889–903. doi: 10.1016/j.neurobiolaging.2006.08.008. [DOI] [PubMed] [Google Scholar]
- Hu L, Wong TP, Cote SL, Bell KF, Cuello AC. The impact of Abeta-plaques on cortical cholinergic and non-cholinergic presynaptic boutons in alzheimer’s disease-like transgenic mice. Neuroscience. 2003;121(2):421–32. doi: 10.1016/s0306-4522(03)00394-4. [DOI] [PubMed] [Google Scholar]
- Kim MJ, Lee KM, Son YD, Jeon HA, Kim YB, Cho ZH. Increased basal forebrain metabolism in mild cognitive impairment: an evidence for brain reserve in incipient dementia. J Alzheimers Dis. 2012;32(4):927–38. doi: 10.3233/JAD-2012-120133. [DOI] [PubMed] [Google Scholar]
- Lee H, Raiker SJ, Venkatesh K, Geary R, Robak LA, Zhang Y, Yeh HH, Shrager P, Giger RJ. Synaptic function for the Nogo-66 receptor NgR1: regulation of dendritic spine morphology and activity-dependent synaptic strength. J Neurosci. 2008;28(11):2753–65. doi: 10.1523/JNEUROSCI.5586-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Liu Y, Kim T, Min R, Zhang Z. Gene expression variability within and between human populations and implications toward disease susceptibility. PLoS Comput Biol. 2010;6(8) doi: 10.1371/journal.pcbi.1000910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang WS, Reiman EM, Valla J, Dunckley T, Beach TG, Grover A, Niedzielko TL, Schneider LE, Mastroeni D, Caselli R, Kukull W, Morris JC, Hulette CM, Schmechel D, Rogers J, Stephan DA. Alzheimer’s disease is associated with reduced expression of energy metabolism genes in posterior cingulate neurons. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(11):4441–6. doi: 10.1073/pnas.0709259105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CC, Kanekiyo T, Xu H, Bu G. Apolipoprotein E and Alzheimer disease: risk, mechanisms and therapy. Nat Rev Neurol. 2013;9(2):106–18. doi: 10.1038/nrneurol.2012.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loring JF, Wen X, Lee JM, Seilhamer J, Somogyi R. A gene expression profile of Alzheimer’s disease. DNA Cell Biol. 2001;20(11):683–95. doi: 10.1089/10445490152717541. [DOI] [PubMed] [Google Scholar]
- Lu T, Pan Y, Kao SY, Li C, Kohane I, Chan J, Yankner BA. Gene regulation and DNA damage in the ageing human brain. Nature. 2004:883–91. doi: 10.1038/nature02661. [DOI] [PubMed] [Google Scholar]
- Lynn BC, Wang J, Markesbery WR, Lovell MA. Quantitative changes in the mitochondrial proteome from subjects with mild cognitive impairment, early stage, and late stage Alzheimer’s disease. J Alzheimers Dis. 2010;19(1):325–39. doi: 10.3233/JAD-2010-1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markesbery WR. Neuropathologic alterations in mild cognitive impairment: a review. J Alzheimers Dis. 2010;19(1):221–8. doi: 10.3233/JAD-2010-1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattson MP, Gleichmann M, Cheng A. Mitochondria in neuroplasticity and neurological disorders. Neuron. 2008;60(5):748–66. doi: 10.1016/j.neuron.2008.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JA, Oldham MC, Geschwind DH. A systems level analysis of transcriptional changes in Alzheimer’s disease and normal aging. J Neurosci. 2008;28(6):1410–20. doi: 10.1523/JNEUROSCI.4098-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SL, Celone K, DePeau K, Diamond E, Dickerson BC, Rentz D, Pihlajamaki M, Sperling RA. Age-related memory impairment associated with loss of parietal deactivation but preserved hippocampal activation. Proceedings of the National Academy of Sciences of the United States of America. 2008a;105(6):2181–6. doi: 10.1073/pnas.0706818105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SL, Fenstermacher E, Bates J, Blacker D, Sperling RA, Dickerson BC. Hippocampal activation in adults with mild cognitive impairment predicts subsequent cognitive decline. Journal of neurology, neurosurgery, and psychiatry. 2008b;79(6):630–5. doi: 10.1136/jnnp.2007.124149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JC. Revised criteria for mild cognitive impairment may compromise the diagnosis of Alzheimer disease dementia. Archives of neurology. 2012;69(6):700–8. doi: 10.1001/archneurol.2011.3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison JH, Baxter MG. The ageing cortical synapse: hallmarks and implications for cognitive decline. Nature reviews. 2012;13(4):240–50. doi: 10.1038/nrn3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mufson EJ, Binder L, Counts SE, DeKosky ST, de Toledo-Morrell L, Ginsberg SD, Ikonomovic MD, Perez SE, Scheff SW. Mild cognitive impairment: pathology and mechanisms. Acta neuropathologica. 2012;123(1):13–30. doi: 10.1007/s00401-011-0884-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukaetova-Ladinska EB, Garcia-Siera F, Hurt J, Gertz HJ, Xuereb JH, Hills R, Brayne C, Huppert FA, Paykel ES, McGee M, Jakes R, Honer WG, Harrington CR, Wischik CM. Staging of cytoskeletal and beta-amyloid changes in human isocortex reveals biphasic synaptic protein response during progression of Alzheimer’s disease. Am J Pathol. 2000;157(2):623–36. doi: 10.1016/s0002-9440(10)64573-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller WE, Eckert A, Kurz C, Eckert GP, Leuner K. Mitochondrial dysfunction: common final pathway in brain aging and Alzheimer’s disease--therapeutic aspects. Mol Neurobiol. 2010;41(2-3):159–71. doi: 10.1007/s12035-010-8141-5. [DOI] [PubMed] [Google Scholar]
- Nixon RA, Yang DS. Autophagy and neuronal cell death in neurological disorders. Cold Spring Harb Perspect Biol. 2012;4(10) doi: 10.1101/cshperspect.a008839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolt MJ, Lin Y, Hruska M, Murphy J, Sheffler-Colins SI, Kayser MS, Passer J, Bennett MV, Zukin RS, Dalva MB. EphB controls NMDA receptor function and synaptic targeting in a subunit-specific manner. J Neurosci. 2011;31(14):5353–64. doi: 10.1523/JNEUROSCI.0282-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palop JJ, Chin J, Roberson ED, Wang J, Thwin MT, Bien-Ly N, Yoo J, Ho KO, Yu GQ, Kreitzer A, Finkbeiner S, Noebels JL, Mucke L. Aberrant excitatory neuronal activity and compensatory remodeling of inhibitory hippocampal circuits in mouse models of Alzheimer’s disease. Neuron. 2007;55(5):697–711. doi: 10.1016/j.neuron.2007.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen RC. Clinical practice. Mild cognitive impairment. The New England journal of medicine. 2011;364(23):2227–34. doi: 10.1056/NEJMcp0910237. [DOI] [PubMed] [Google Scholar]
- Putcha D, Brickhouse M, O’Keefe K, Sullivan C, Rentz D, Marshall G, Dickerson B, Sperling R. Hippocampal hyperactivation associated with cortical thinning in Alzheimer’s disease signature regions in non-demented elderly adults. J Neurosci. 2011;31(48):17680–8. doi: 10.1523/JNEUROSCI.4740-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharfman HE, Chao MV. The entorhinal cortex and neurotrophin signaling in Alzheimer’s disease and other disorders. Cogn Neurosci. 2013;4(3-4):123–35. doi: 10.1080/17588928.2013.826184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selkoe DJ. Science. 5594. Vol. 298. New York, NY: 2002. Alzheimer’s disease is a synaptic failure; pp. 789–91. [DOI] [PubMed] [Google Scholar]
- Sperling RA, Dickerson BC, Pihlajamaki M, Vannini P, LaViolette PS, Vitolo OV, Hedden T, Becker JA, Rentz DM, Selkoe DJ, Johnson KA. Functional alterations in memory networks in early Alzheimer’s disease. Neuromolecular Med. 2010;12(1):27–43. doi: 10.1007/s12017-009-8109-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperling RA, Laviolette PS, O’Keefe K, O’Brien J, Rentz DM, Pihlajamaki M, Marshall G, Hyman BT, Selkoe DJ, Hedden T, Buckner RL, Becker JA, Johnson KA. Amyloid deposition is associated with impaired default network function in older persons without dementia. Neuron. 2009;63(2):178–88. doi: 10.1016/j.neuron.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprinkle TJ. 2′,3′-cyclic nucleotide 3′-phosphodiesterase, an oligodendrocyte-Schwann cell and myelin-associated enzyme of the nervous system. Crit Rev Neurobiol. 1989;4(3):235–301. [PubMed] [Google Scholar]
- Storandt M, Grant EA, Miller JP, Morris JC. Longitudinal course and neuropathologic outcomes in original vs revised MCI and in pre-MCI. Neurology. 2006;67(3):467–73. doi: 10.1212/01.wnl.0000228231.26111.6e. [DOI] [PubMed] [Google Scholar]
- Tan MG, Chua WT, Esiri MM, Smith AD, Vinters HV, Lai MK. Genome wide profiling of altered gene expression in the neocortex of Alzheimer’s disease. J Neurosci Res. 2010;88(6):1157–69. doi: 10.1002/jnr.22290. [DOI] [PubMed] [Google Scholar]
- Tan MS, Yu JT, Tan L. Bridging integrator 1 (BIN1): form, function, and Alzheimer’s disease. Trends Mol Med. 2013;19(10):594–603. doi: 10.1016/j.molmed.2013.06.004. [DOI] [PubMed] [Google Scholar]
- Tremblay ME, Riad M, Bouvier D, Murai KK, Pasquale EB, Descarries L, Doucet G. Localization of EphA4 in axon terminals and dendritic spines of adult rat hippocampus. The Journal of comparative neurology. 2007;501(5):691–702. doi: 10.1002/cne.21263. [DOI] [PubMed] [Google Scholar]
- Vannini P, Almkvist O, Dierks T, Lehmann C, Wahlund LO. Reduced neuronal efficacy in progressive mild cognitive impairment: a prospective fMRI study on visuospatial processing. Psychiatry research. 2007;156(1):43–57. doi: 10.1016/j.pscychresns.2007.02.003. [DOI] [PubMed] [Google Scholar]
- Williams C, Mehrian Shai R, Wu Y, Hsu YH, Sitzer T, Spann B, McCleary C, Mo Y, Miller CA. Transcriptome analysis of synaptoneurosomes identifies neuroplasticity genes overexpressed in incipient Alzheimer’s disease. PLoS ONE. 2009;4(3):e4936. doi: 10.1371/journal.pone.0004936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Pasquale EB. Eph receptors in the adult brain. Current opinion in neurobiology. 2004;14(3):288–96. doi: 10.1016/j.conb.2004.04.003. [DOI] [PubMed] [Google Scholar]
- Yassa MA, Stark SM, Bakker A, Albert MS, Gallagher M, Stark CE. High-resolution structural and functional MRI of hippocampal CA3 and dentate gyrus in patients with amnestic Mild Cognitive Impairment. NeuroImage. 2010;51(3):1242–52. doi: 10.1016/j.neuroimage.2010.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yassa MA, Verduzco G, Cristinzio C, Bassett SS. Altered fMRI activation during mental rotation in those at genetic risk for Alzheimer disease. Neurology. 2008;70(20):1898–904. doi: 10.1212/01.wnl.0000312288.45119.d1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagrebelsky M, Schweigreiter R, Bandtlow CE, Schwab ME, Korte M. Nogo-A stabilizes the architecture of hippocampal neurons. J Neurosci. 2010;30(40):13220–34. doi: 10.1523/JNEUROSCI.1044-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.