Summary
Understanding the molecular basis of how reproductive isolation evolves between individuals from the same species offers valuable insight into patterns of genetic differentiation as well as the onset of speciation [1, 2]. The yeast Saccharomyces cerevisiae constitutes an ideal model partly due to its vast ecological range, high level of genetic diversity [3–6] and laboratory amendable sexual reproduction. Between S. cerevisiae and its sibling species in the Saccharomyces sensu stricto complex, reproductive isolation acts post-zygotically and could be attributed to chromosomal rearrangements [7], cyto-nuclear incompatibility [8, 9] and anti-recombination [10, 11]; although the implication of these mechanisms at the incipient stage of speciation remains unclear due to further divergence in the nascent species. Recently, several studies assessed the onset of intraspecific reproductive isolation in S. cerevisiae by evaluating the effect of the mismatch repair system [12–14] or by fostering incipient speciation using the same initial genetic background [15–18]. Nevertheless, the overall genetic diversity within this species was largely overlooked and no systematic evaluation has been performed. Here, we carried out the first species-wide survey for post-zygotic reproductive isolation in S. cerevisiae. We crossed 60 natural isolates sampled from diverse niches with the reference strain S288c, and identified 16 cases of reproductive isolation with reduced offspring viabilities ranging from 44% to 86%. Using different mapping strategies, we identified reciprocal translocations in a large fraction of all isolates surveyed, indicating that large-scale chromosomal rearrangements might play a major role to the onset of reproductive isolation in this species.
Keywords: intraspecific diversity, reproductive isolation, translocations, yeast
Results and discussion
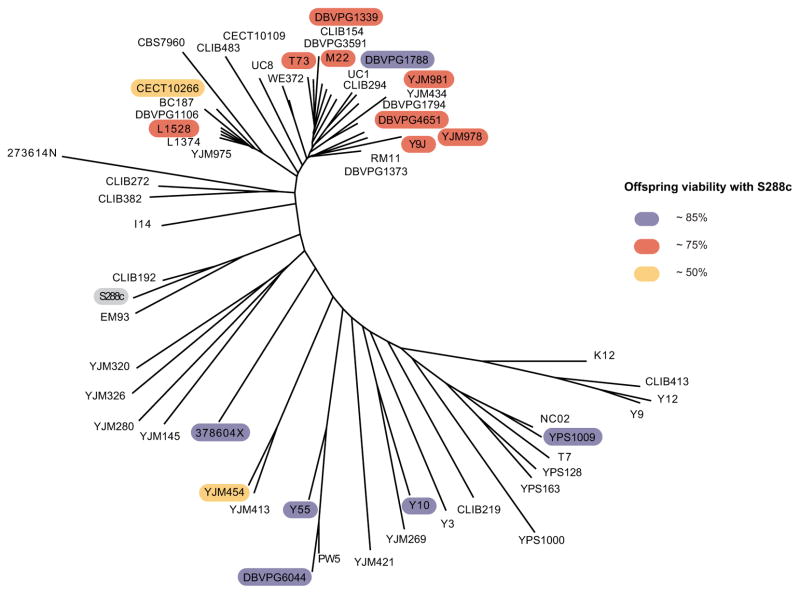
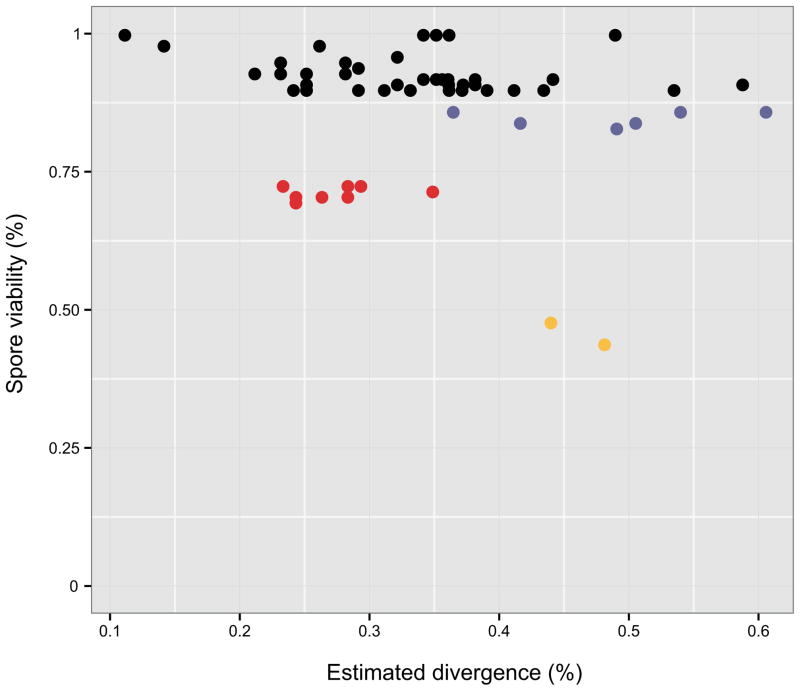
To obtain a global view of the landscape of intraspecific reproductive isolation in S. cerevisiae, we selected 60 natural isolates from diverse ecological and geographical niches (Table S1). Estimated genetic divergence within these strains ranges from 0.11% to 0.60%, which is a relatively comprehensive representation of the genetic diversity currently observed in this species (Figure 1). We crossed all isolates with the reference strain S288c and estimated the offspring viability for each cross. A relatively large fraction of crosses (16 out of 60) qualified as cases of reproductive isolation, with reduced offspring viabilities ranging from 44% to 86% (Table S1). No apparent correlation was observed between the estimated genetic divergence of the parental pairs and the resulting offspring viability (Figure 2), indicating that general DNA sequence differences were not sufficient to explain the observed reproductive isolation.
Figure 1. Neighbor-joining tree of 60 studied S. cerevisiae isolates.
A majority-rule consensus tree of the surveyed strains was built based on the 101,343 segregating sites identified in [3]. Branch lengths are proportional to the number of segregating sites that differentiate each pair of strains. Isolates that are incompatible were color coded according to the offspring viability resulting from the cross with the reference S288c. See also Table S1 and Figure S1 for detailed strain origins and phenotype segregations for the incompatible crosses.
Figure 2. Sequence divergence does not correlate with the observed offspring viability.
The estimated sequence divergence between each pair of parental strains (horizontal axis) was plotted against the observed offspring viability (vertical axis). All strains were crossed with the reference strain S288c and the offspring viability was estimated by dissecting 20 tetrads. Crosses with offspring viabilities <90% were color coded. Blue: crosses with offspring viability of ~85%. Red: crosses with offspring viability of ~75%. Yellow: crosses with offspring viability of ~50%.
To understand the molecular basis and complexity underlying the identified cases, additional tetrads were dissected for all 16 incompatible crosses (Table S2) and the segregation of the lethal phenotype was analyzed (Figure S1). 6 cases showed mild reduction of offspring viability (78% to 87%, mean=82%; 65 tetrads analyzed on average) (Figure 2; Table S2), which resulted in a Poisson distribution with decreasing number of full tetrads (4 viable spores, Figure S1). This segregation pattern suggests these cases were probably caused by a mutator [13, 14] or anti-recombination [12] effect of the mismatch repair system, as previously observed. The remaining 10 cases with a higher degree of progeny loss (44% to 74%) were further analyzed.
Bulk segregant analysis revealed a unique reciprocal translocation responsible for cases of reduced offspring viability of ~75%
According to the segregation, 8 crosses (between S288c and DBVPG1339, DBVPG4651, M22, T73, Y9J, L-1528, YJM978 and YJM981) showed predominantly 3 types of tetrads with 4, 3 or 2 viable spores (Figure 1, Figure S1). The ratio between these tetrad types was roughly 1:2:1, resulting in reduced spore viability of ~75% (66% to 74%, mean=71%; 228 tetrads analyzed on average) (Table S2). In addition, pairwise crosses among all 8 strains showed offspring viabilities higher than 90% (data not shown), indicating these cases represented a unique genetic origin. To map the genomic regions involved, we focused on one cross (between DBVPG1339 and S288c) and performed bulk segregant analysis by sequencing a pool of 50 independent segregants from tetrads with only two viable spores, where the lethal genotype combination was absent (Figure S2-A). Following this selection, genomic regions involved were expected to have allele frequencies skewed from 0.5 whereas the rest of the genome should have equal proportions of alleles from each parent.
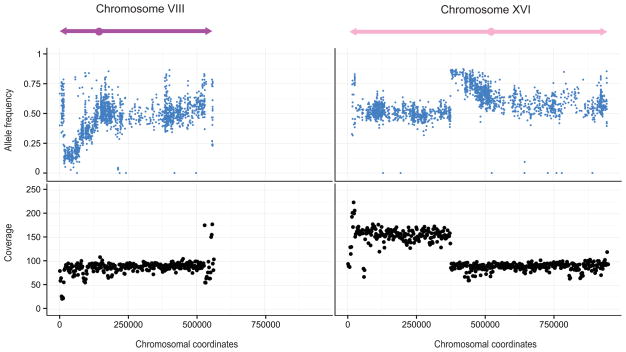
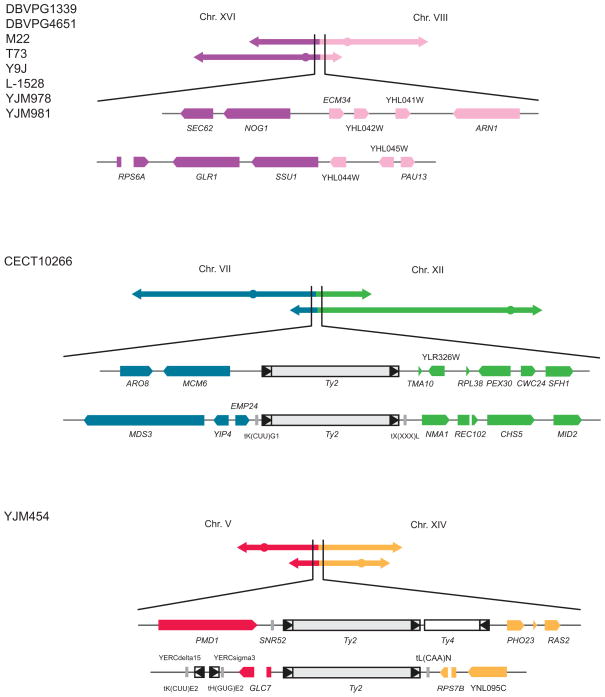
Two regions with significantly skewed allele frequencies were mapped (Figure 3, Figure S3). The first one was located at the left-arm region of chromosome VIII and the second near the centromeric region of chromosome XVI (Figure 3). Additionally, the end of chromosome VIII (~15 kb) showed a low coverage (~30X) whereas the left-arm of chromosome XVI (~370 kb) showed a coverage that was nearly 200X, indicating that two copies of the left-arm of chromosome XVI might be present (Figure 3). This unbalanced inheritance suggests the presence of a reciprocal translocation between chromosome VIII and XVI. In fact, when crossing strains bearing the translocation with the reference strain S288c, offspring would inherit either a balanced or unbalanced set of chromosomes (Figure S2-B) [19]. As the region involved on chromosome VIII was near the telomere and does not contain any essential genes, only unbalanced spores with two copies of the left-arm of chromosome XVI were viable, as was evident by the abnormal coverage. PCR results demonstrated in all 8 strains that the translocation occurred between the promoter region of ECM34 (YHL043W) on chromosome VIII and the promoter region of SSU1 (YPL092C) on chromosome XVI (Figure 4). Analysis of the junctions revealed no significant homology, suggesting that the translocation originated via a Non-Homologous End-Joining (NHEJ) event.
Figure 3. Bulk segregant analysis mapped two regions with skewed allele frequencies and abnormal coverage.
Plot was obtained using bulk segregant data from cross between DBVPG1339 and S288c. The horizontal axis represents the coordinates of chromosome VIII and XVI. The upper vertical axis corresponds to the allele frequencies of S288c: values close to 1 imply that only alleles of S288c are present and vice versa. The lower vertical axis represents the sequencing coverage in a 2 kb window. The theoretical coverage was expected to be 100X if a single copy was present. Two regions showed significant allele frequency variations: the left-arm region of chromosome VIII (position 15000 to 71000) and the centromeric region of chromosome XVI (position 374000 to 453000). See also Figure S3 for the complete mapping results.
Figure 4. Identified translocations responsible for the observed reproductive isolation.
Schematics of translocations identified in this study. Chromosome pairs involved are color-coded. Chromosome and gene sizes are scaled according to SGD annotations. See also Figure S4 for the complete mapping results for the backcrossed segregants.
Successive backcross strategy identified multiple reciprocal translocations responsible for the reduced offspring viability of ~50%
The remaining 2 crosses (CECT10266 and YJM454 with S288c) showed a reduced spore viability of 50% (44% to 48%, mean=46%; 100 tetrads analyzed on average) (Table S2), where 3 major types of tetrads were observed, each contained 4, 2 or 0 viable spores with a ratio of 1:2:1 (Figure S1). Based on the segregation pattern, we reasoned that the most plausible explanation was the presence of a reciprocal translocation involving two large chromosomal regions, each of which contains at least one essential gene [19]. In this context, any meiotic recombination will lead to mis-segregation of essential genes and consequently only the progeny that inherited a balanced set of chromosomes would be viable (Figure S2-B). Moreover, the cross between CECT10266 and YJM454 demonstrated a further reduction of offspring viability (~25%, data not shown), indicating that these two strains probably underwent different events leading to the observed reproductive isolation.
Since in these cases, all viable F1 segregants would have an equal probability of inheriting either balanced parental genome, no allele frequency variation would be observed by pooling the F1 segregants. We then developed a strategy based on successive backcrossing and next-generation sequencing to map the regions involved. F1 segregants that maintained the phenotype of 50% offspring viability were successively backcrossed to S288c for 5 generations, in order to obtain a single segregant enriched for the S288c genome but still retaining the original translocation. Each 5th generation backcross segregant, namely CS-B5 (from the cross between CECT10266 and S288c) and YS-B5 (from the cross between YJM454 and S288c), was subjected to whole genome sequencing. Due to limited recombination around the junctions, the genomes of these backcrossed segregants would be otherwise allelic to S288c except for regions involved in the translocation.
Identification of a reciprocal translocation between chromosome VII and XII in CECT10266
Genome sequencing of the segregant CS-B5 (from the cross between CECT10266 and S288c) revealed two regions that are polymorphic to S288c. The first region was approximately located on the left-arm of chromosome VII and the second on the right-arm of chromosome XII (Figure S4-A). Breakpoints of the putative translocation were identified using PCR. The first breakpoint was located between MCM6 (YGL200C) and EMP24 (YGL201C) on chromosome VII and the second between YLR326W and NMA1 (YLR328W) on chromosome XII. Considering the relative position of the centromeres on these two chromosomes, the translocation likely occurred between the left-arm of chromosome VII and the right-arm of chromosome XII (Figure 4), leading to two new chimeric chromosomes with functional centromeres. The junctions of this putative translocation were confirmed using PCR. Sequencing of the amplified fragments revealed a full-length Ty2 transposon at both junctions (Figure 4), suggesting that the translocation likely originated from Homologous Recombination (HR) between Ty elements.
Identification of a reciprocal translocation between chromosome V and XIV in YJM454
Similarly, we also mapped two regions in the genome of YS-B5 (from the cross between YJM454 and S288c). The first one was found on the right-arm of chromosome V and the second on the left-arm of chromosome XIV (Figure S4-B). Based on the same principle, we identified two breakpoints: the first located between PMD1 (YER132C) and GLC7 (YER133W) on chromosome V and the second between PHO23 (YNL097C) and RPS7B (YNL096C) on chromosome XIV. In this case, the right-arm of chromosome V was likely exchanged with the left-arm of chromosome XIV to ensure centromeric functions of the chimeric chromosomes (Figure 4). Indeed, PCRs confirmed the presence of both junctions involved in this putative translocation (Figure 4). Sequence analysis of the junctions revealed a full-length Ty2 transposon at both junctions, and an additional 3 kb fragment containing a partial Ty4 element at the junction uniting the right-arms of chromosome V and XIV (Figure 4). The presence of multiple Ty elements suggests that the breakpoints might overlap with potential Ty insertion hotspots. This translocation was probably also mediated by Homologous Recombination (HR) through Ty elements.
Relative importance of chromosomal rearrangements in yeast speciation
The process of speciation is often quantitative, as the strength of reproductive isolation varies continuously at different levels of divergence [2]. The yeast Saccharomyces cerevisiae and its close relatives in the Saccharomyces sensu strico complex offer a unique opportunity to explore the possible mechanisms leading to the onset of intrinsic reproductive isolation at both “short” (within species) and “long” (between species) evolutionary scales.
Including S. cerevisiae, six species are currently circumscribed in this group [20], all of which readily cross with each other to form viable hybrids [21]. Yet, interspecific hybrids showed strong post-zygotic reproductive isolation, producing only ~1% of viable offspring [21, 22]. Many species in this group differ by chromosomal rearrangements [7, 22, 23], however, as this only partially explains the substantial loss of hybrid progeny due to the extant high interspecific divergence, the relative role of translocations in the onset of reproductive isolation and speciation in these species was largely debated [7, 24, 25].
In this study, we found that chromosomal rearrangements, especially reciprocal translocations, play a substantial role in the onset of reproductive isolation in S. cerevisiae. The fact that this type of mechanism exists at different temporal levels of genetic divergence, both within and between species, suggests that reciprocal translocations might have a larger impact to the onset of speciation in yeast than previously thought.
Adaptation through chromosomal rearrangements is common in S. cerevisiae
Chromosomal rearrangements including polyploidies, aneuploidies, segmental duplications and translocations, are frequently observed in wild and domesticated strains of S. cerevisiae [26–29] and such rearrangements could readily be associated with adaptation to environmental stress. For example, the translocation between chromosome VIII and XVI observed in this study was previously identified in several wine strains, conferring to an advantageous sulfite resistant phenotype, as this compound was commonly used in wine making [30, 31]. Interestingly, among the 8 strains identified here, only 4 were associated with wine (T73, Y9J, L-1528, M22 and DBVPG1339) whereas the others were from various niches including clinical sources (YJM978 and YJM981), and white truffle (DBVPG4651) (Figure 1; Table S1), suggesting this translocation was dispersed and might have been selectively maintained across different populations.
In fact, adaptive chromosomal rearrangements were frequently observed on different spatiotemporal scales, both in nature [32] and in short-term laboratory evolution [33–35]. These observations, in agreement with our data, suggest that chromosomal rearrangements might offer a mechanism of rapid response to stress and become fixed in the population, despite the potential loss of offspring.
Do Dobzhansky-Müller incompatibilities exist in yeast?
In theory, the Dobzhansky-Müller model of genetic incompatibility offers the inherent link between divergent adaptation and reproductive isolation. If two populations evolved to adapt to different environments, mutations accumulated independently in each specialized group may cause negative interactions which reduce hybrid fitness or viability [36]. To date, few pairs of “Dobzhansky-Muller genes” have been identified in plants, insects and animals, both among and within species [1, 37–41]. Curiously, between different yeast species, genetic incompatibilities appear to be scarce and hardly any examples have been described [42–44]. Moreover, by screening a large collection of ecologically diverse strains of S. cerevisiae, we found no classic Dobzhansky-Müller gene pairs, which would generally affect 50% to 25% of the offspring depending on the dominance or recessivity of the genes involved. The only cases found which could implicate genic interactions are from the 85% spore viability class. Yet, the segregation pattern of theses cases (Figure S1) strongly suggests a “mutator” or “anti-recombinogenic” phenotype [12–14] and not a classic two-gene interaction model. Overall, these observations suggest that the classic Dobzhansky-Müller genetic incompatibility scenario is probably rare and might have a modest effect in the onset of post-zygotic reproductive isolation in this species.
The lack of awareness concerning such incompatibilities in yeast might be due to the incomplete penetrance of antagonic genetic interactions on permissive rich media. Future research should explore the possibility of incompatibilities related to different conditions such as temperature, media composition or exposure to various chemical compounds in order to obtain a more complete picture of the molecular mechanisms involved in the onset of intraspecific reproductive isolation in S. cerevisiae.
Experimental procedures
Strains
A collection of 60 strains isolated from diverse ecological (tree exudate, wine, different fermentations and clinical) and geographical (Europe, Asia, Africa and America) origins was used in this study (Table S1). Laboratory strains isogenic to S288c, FY4 (MATa) and FY5 (MATα) were also used.
Media and culture conditions
Yeast cells were grown on YPD media (1% yeast extract, 2% peptone and 2% glucose) using liquid culture or solid plates. Crosses were carried out on YPD plates by mixing freshly grown cells with the opposite mating type. Sporulation was induced on potassium acetate plates (1% potassium acetate, 2% agar). All procedures were done at 30°C.
Test of spore viability
All strains screened in this study are MATα and were systematically crossed to FY4 (MATa). Diploids obtained from different crosses were sporulated and the spore viability for each cross was scored after tetrad dissection. Tetrad asci were gently digested by zymolyase (MT ImmunO™ 20T) and then dissected using micromanipulator Singer MSM-400. Spores were aligned on YPD plate and cultured for 48 hours. Viable spores will form colonies and the spore viability corresponds to the ratio between the number of viable spores and the total number of spores dissected. The first screening was done by analyzing 20 tetrads for each cross. Additional tetrads were dissected for incompatible crosses as listed in Table S2.
Bulk segregant analysis strategy
For cases with 75% spore viability, the segregation of the lethal phenotype resulted in predominantly 3 types of tetrads: tetrads with 4 viable spores or parental ditypes (PD), 3 viable spores or tetratypes (TT) and 2 viable spores or non-parental ditypes (NPD) (Figure S2). To map the genomic regions involved, we used bulk segregant analysis strategy by pooling a set of viable spores from NPD tetrads. The cross between DBVPG1339 and FY4 was selected for the mapping. In total, 300 tetrads were dissected and 50 independent spores from NPD tetrads were separately cultured then pooled by equal O.D. readings at 600 nm. Regions involved were mapped by analyzing the allele frequency variation along the genome.
Successive backcrossing strategy
For both cases with 50% spore viability, only segregants which inherited either parental genotypes were viable, resulting in a segregation of predominantly 3 types of tetrads: parental ditypes (PD) with four viable spores, tetratypes (TT) with 2 viable spores, and non-parental ditypes (NPD) with 0 viable spores (Figure S2). To map the genomic regions involved, we used a successive backcrossing strategy. For each cross i.e. the cross between CECT10266 and S288c and the cross between YJM454 and S288c, one F1 parental ditype tetrad (PD, 4 viable spores) was selected, and all four spores were backcrossed to S288c with opposite mating types (FY4 or FY5). Spore viabilities were analyzed, and a segregant which has retained the 50% spore viability segregation was selected for a subsequent backcross to S288c. Five generations of backcrosses were performed and one 5th generation-backcrossed segregant (B5) was obtained for each cross, namely CS-B5 for the segregant derived from the cross between CECT10266 and S288c and YS-B5 for the segregant derived from the cross between YJM454 and S288c. Using this strategy, the majority of the genome was enriched for S288c alleles except for regions involved in low spore viability.
DNA extraction, sequencing, and SNP calling
Genomic DNA was extracted using the Qiagen Genomic-tip kit. Sequencing of the samples was performed using Illumina Hiseq 2000 technology. We used paired-end libraries, 101 bp/read, and 100X coverage for bulk segregants and 50X coverage for backcrossed segregants. Quality controlled reads were aligned to the S288c genome using BWA with “-n 5 -o 2” options. SNP calling was done using SAMtools [45]. The allele frequency of S288c was scored at each polymorphic position. Coverage along the genome was calculated by averaging the number of reads aligned at each genomic position within a 2 kb window.
Neighbor joining tree
A majority-rule consensus tree of the surveyed strains was built based on the 101,343 segregating sites identified by Schacherer et al. 2009. For strains that were not represented in the original tree [3], the publicly available sequences [46] were recovered and aligned against the S288c reference sequence with BWA (-bwasw option), except for the CECT10266 strain, for which we computed our own reads mapping (see DNA extraction, sequencing, and SNP calling section). Polymorphic positions were called with SAMtools and used to complete the segregating sites matrix. We constructed a neighbour-joining tree of the strains studied from these SNP data using the software package Splitstree [47], with branch lengths proportional to the number of segregating sites that differentiate each node.
Supplementary Material
Highlights.
First systematic survey of post-zygotic reproductive isolation in S. cerevisiae
Progeny loss was not correlated with sequence divergence at the intraspecific scale
Reproductive isolation in S. cerevisiae was mainly related to genome rearrangements
Pervasiveness of such rearrangements suggest an essential role to yeast speciation
Acknowledgments
We sincerely thank Bernard Dujon for helpful discussions and Kelle Freel for critical reading of the manuscript. We are most grateful to the GeneCore sequencing team (EMBL, Heidelberg, Germany). This work was supported by an Agence nationale de la recherche grant (2011-JSV6-004-01) and a National Institutes of Health (NIH) grant R01 GM101091-01 (J.S.). J.H. was supported by a grant from the French “Ministère de l’Enseignement Supérieur et de la Recherche”.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Maheshwari S, Barbash DA. The genetics of hybrid incompatibilities. Annual review of genetics. 2011;45:331–355. doi: 10.1146/annurev-genet-110410-132514. [DOI] [PubMed] [Google Scholar]
- 2.Nosil P. Ecological speciation. Oxford: Oxford University Press; 2012. [Google Scholar]
- 3.Schacherer J, Shapiro JA, Ruderfer DM, Kruglyak L. Comprehensive polymorphism survey elucidates population structure of Saccharomyces cerevisiae. Nature. 2009;458:342–345. doi: 10.1038/nature07670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liti G, Carter DM, Moses AM, Warringer J, Parts L, James SA, Davey RP, Roberts IN, Burt A, Koufopanou V, et al. Population genomics of domestic and wild yeasts. Nature. 2009;458:337–341. doi: 10.1038/nature07743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang QM, Liu WQ, Liti G, Wang SA, Bai FY. Surprisingly diverged populations of Saccharomyces cerevisiae in natural environments remote from human activity. Molecular ecology. 2012;21:5404–5417. doi: 10.1111/j.1365-294X.2012.05732.x. [DOI] [PubMed] [Google Scholar]
- 6.Cromie GA, Hyma KE, Ludlow CL, Garmendia-Torres C, Gilbert TL, May P, Huang AA, Dudley AM, Fay JC. Genomic sequence diversity and population structure of Saccharomyces cerevisiae assessed by RAD-seq. G3 (Bethesda) 2013;3:2163–2171. doi: 10.1534/g3.113.007492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fischer G, James SA, Roberts IN, Oliver SG, Louis EJ. Chromosomal evolution in Saccharomyces. Nature. 2000;405:451–454. doi: 10.1038/35013058. [DOI] [PubMed] [Google Scholar]
- 8.Chou JY, Hung YS, Lin KH, Lee HY, Leu JY. Multiple molecular mechanisms cause reproductive isolation between three yeast species. PLoS biology. 2010;8:e1000432. doi: 10.1371/journal.pbio.1000432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee HY, Chou JY, Cheong L, Chang NH, Yang SY, Leu JY. Incompatibility of nuclear and mitochondrial genomes causes hybrid sterility between two yeast species. Cell. 2008;135:1065–1073. doi: 10.1016/j.cell.2008.10.047. [DOI] [PubMed] [Google Scholar]
- 10.Chambers SR, Hunter N, Louis EJ, Borts RH. The mismatch repair system reduces meiotic homeologous recombination and stimulates recombination-dependent chromosome loss. Molecular and cellular biology. 1996;16:6110–6120. doi: 10.1128/mcb.16.11.6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hunter N, Chambers SR, Louis EJ, Borts RH. The mismatch repair system contributes to meiotic sterility in an interspecific yeast hybrid. The EMBO journal. 1996;15:1726–1733. [PMC free article] [PubMed] [Google Scholar]
- 12.Greig D, Travisano M, Louis EJ, Borts RH. A role for the mismatch repair system during incipient speciation in Saccharomyces. Journal of evolutionary biology. 2003;16:429–437. doi: 10.1046/j.1420-9101.2003.00546.x. [DOI] [PubMed] [Google Scholar]
- 13.Heck JA, Argueso JL, Gemici Z, Reeves RG, Bernard A, Aquadro CF, Alani E. Negative epistasis between natural variants of the Saccharomyces cerevisiae MLH1 and PMS1 genes results in a defect in mismatch repair. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:3256–3261. doi: 10.1073/pnas.0510998103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Demogines A, Wong A, Aquadro C, Alani E. Incompatibilities involving yeast mismatch repair genes: a role for genetic modifiers and implications for disease penetrance and variation in genomic mutation rates. PLoS genetics. 2008;4:e1000103. doi: 10.1371/journal.pgen.1000103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dettman JR, Sirjusingh C, Kohn LM, Anderson JB. Incipient speciation by divergent adaptation and antagonistic epistasis in yeast. Nature. 2007;447:585–588. doi: 10.1038/nature05856. [DOI] [PubMed] [Google Scholar]
- 16.Anderson JB, Funt J, Thompson DA, Prabhu S, Socha A, Sirjusingh C, Dettman JR, Parreiras L, Guttman DS, Regev A, et al. Determinants of divergent adaptation and Dobzhansky-Muller interaction in experimental yeast populations. Current biology : CB. 2010;20:1383–1388. doi: 10.1016/j.cub.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maclean CJ, Greig D. Reciprocal gene loss following experimental whole-genome duplication causes reproductive isolation in yeast. Evolution; international journal of organic evolution. 2011;65:932–945. doi: 10.1111/j.1558-5646.2010.01171.x. [DOI] [PubMed] [Google Scholar]
- 18.Kroll E, Coyle S, Dunn B, Koniges G, Aragon A, Edwards J, Rosenzweig F. Starvation-associated genome restructuring can lead to reproductive isolation in yeast. PloS one. 2013;8:e66414. doi: 10.1371/journal.pone.0066414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loidl J, Jin QW, Jantsch M. Meiotic pairing and segregation of translocation quadrivalents in yeast. Chromosoma. 1998;107:247–254. doi: 10.1007/s004120050304. [DOI] [PubMed] [Google Scholar]
- 20.Scannell DR, Zill OA, Rokas A, Payen C, Dunham MJ, Eisen MB, Rine J, Johnston M, Hittinger CT. The Awesome Power of Yeast Evolutionary Genetics: New Genome Sequences and Strain Resources for the Saccharomyces sensu stricto Genus. G3 (Bethesda) 2011;1:11–25. doi: 10.1534/g3.111.000273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Naumov GI, Naumova ES, Masneuf I, Aigle M, Kondratieva VI, Dubourdieu D. Natural polyploidization of some cultured yeast Saccharomyces sensu stricto: auto- and allotetraploidy. Systematic and applied microbiology. 2000;23:442–449. doi: 10.1016/S0723-2020(00)80076-4. [DOI] [PubMed] [Google Scholar]
- 22.Liti G, Barton DB, Louis EJ. Sequence diversity, reproductive isolation and species concepts in Saccharomyces. Genetics. 2006;174:839–850. doi: 10.1534/genetics.106.062166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ryu SL, Murooka Y, Kaneko Y. Genomic reorganization between two sibling yeast species, Saccharomyces bayanus and Saccharomyces cerevisiae. Yeast. 1996;12:757–764. doi: 10.1002/(sici)1097-0061(19960630)12:8<757::aid-yea970>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 24.Delneri D, Colson I, Grammenoudi S, Roberts IN, Louis EJ, Oliver SG. Engineering evolution to study speciation in yeasts. Nature. 2003;422:68–72. doi: 10.1038/nature01418. [DOI] [PubMed] [Google Scholar]
- 25.Greig D. Reproductive isolation in Saccharomyces. Heredity. 2009;102:39–44. doi: 10.1038/hdy.2008.73. [DOI] [PubMed] [Google Scholar]
- 26.Albertin W, Marullo P, Aigle M, Bourgais A, Bely M, Dillmann C, DEV D, Sicard D. Evidence for autotetraploidy associated with reproductive isolation in Saccharomyces cerevisiae: towards a new domesticated species. Journal of evolutionary biology. 2009;22:2157–2170. doi: 10.1111/j.1420-9101.2009.01828.x. [DOI] [PubMed] [Google Scholar]
- 27.Rachidi N, Barre P, Blondin B. Multiple Ty-mediated chromosomal translocations lead to karyotype changes in a wine strain of Saccharomyces cerevisiae. Molecular & general genetics : MGG. 1999;261:841–850. doi: 10.1007/s004380050028. [DOI] [PubMed] [Google Scholar]
- 28.Bidenne C, Blondin B, Dequin S, Vezinhet F. Analysis of the chromosomal DNA polymorphism of wine strains of Saccharomyces cerevisiae. Current genetics. 1992;22:1–7. doi: 10.1007/BF00351734. [DOI] [PubMed] [Google Scholar]
- 29.Casaregola S, Nguyen HV, Lepingle A, Brignon P, Gendre F, Gaillardin C. A family of laboratory strains of Saccharomyces cerevisiae carry rearrangements involving chromosomes I and III. Yeast. 1998;14:551–564. doi: 10.1002/(SICI)1097-0061(19980430)14:6<551::AID-YEA260>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 30.Goto-Yamamoto N, Kitano K, Shiki K, Yoshida Y, Suzuki T, Iwata T, Yamane Y, Hara S. SSU1-R, a sulfite resistance gene of wine yeast, is an allele of SSU1 with a different upstream sequence. J Ferment Bioeng. 1998;86:427–433. [Google Scholar]
- 31.Perez-Ortin JE, Querol A, Puig S, Barrio E. Molecular characterization of a chromosomal rearrangement involved in the adaptive evolution of yeast strains. Genome research. 2002;12:1533–1539. doi: 10.1101/gr.436602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang SL, Lai HY, Tung SY, Leu JY. Dynamic large-scale chromosomal rearrangements fuel rapid adaptation in yeast populations. PLoS genetics. 2013;9:e1003232. doi: 10.1371/journal.pgen.1003232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dunham MJ, Badrane H, Ferea T, Adams J, Brown PO, Rosenzweig F, Botstein D. Characteristic genome rearrangements in experimental evolution of Saccharomyces cerevisiae. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:16144–16149. doi: 10.1073/pnas.242624799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rancati G, Pavelka N, Fleharty B, Noll A, Trimble R, Walton K, Perera A, Staehling-Hampton K, Seidel CW, Li R. Aneuploidy underlies rapid adaptive evolution of yeast cells deprived of a conserved cytokinesis motor. Cell. 2008;135:879–893. doi: 10.1016/j.cell.2008.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yona AH, Manor YS, Herbst RH, Romano GH, Mitchell A, Kupiec M, Pilpel Y, Dahan O. Chromosomal duplication is a transient evolutionary solution to stress. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:21010–21015. doi: 10.1073/pnas.1211150109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dobzhansky T. Genetics and the origin of species. New York: Columbia Univ. Press; 1937. [Google Scholar]
- 37.Coyne JA, Orr HA. Speciation. Sunderland, Mass: Sinauer Associates; 2004. [Google Scholar]
- 38.Seidel HS, Ailion M, Li J, van Oudenaarden A, Rockman MV, Kruglyak L. A novel sperm-delivered toxin causes late-stage embryo lethality and transmission ratio distortion in C. elegans. PLoS biology. 2011;9:e1001115. doi: 10.1371/journal.pbio.1001115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seidel HS, Rockman MV, Kruglyak L. Widespread genetic incompatibility in C. elegans maintained by balancing selection. Science. 2008;319:589–594. doi: 10.1126/science.1151107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bikard D, Patel D, Le Mette C, Giorgi V, Camilleri C, Bennett MJ, Loudet O. Divergent evolution of duplicate genes leads to genetic incompatibilities within A. thaliana. Science. 2009;323:623–626. doi: 10.1126/science.1165917. [DOI] [PubMed] [Google Scholar]
- 41.Presgraves DC. The molecular evolutionary basis of species formation. Nature reviews Genetics. 2010;11:175–180. doi: 10.1038/nrg2718. [DOI] [PubMed] [Google Scholar]
- 42.Greig D. A screen for recessive speciation genes expressed in the gametes of F1 hybrid yeast. PLoS genetics. 2007;3:e21. doi: 10.1371/journal.pgen.0030021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kao KC, Schwartz K, Sherlock G. A genome-wide analysis reveals no nuclear dobzhansky-muller pairs of determinants of speciation between S. cerevisiae and S. paradoxus, but suggests more complex incompatibilities. PLoS genetics. 2010;6:e1001038. doi: 10.1371/journal.pgen.1001038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li C, Wang Z, Zhang J. Toward genome-wide identification of bateson-dobzhansky-muller incompatibilities in yeast: a simulation study. Genome biology and evolution. 2013;5:1261–1272. doi: 10.1093/gbe/evt091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Skelly DA, Merrihew GE, Riffle M, Connelly CF, Kerr EO, Johansson M, Jaschob D, Graczyk B, Shulman NJ, Wakefield J, et al. Integrative phenomics reveals insight into the structure of phenotypic diversity in budding yeast. Genome research. 2013 doi: 10.1101/gr.155762.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Molecular biology and evolution. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.