Abstract
Signal Transducers and Activator of Transcription-3 (STAT3) are latent transcription factors that are regulated by post-translational modifications (PTMs) in response to cellular activation by the IL-6 superfamily of cytokines to regulate cell cycle progression and/or apoptosis. Here we observe that STAT3 is inducibly mono-ubiquitinated and investigate its consequences. Using domain mapping and highly specific selected reaction monitoring- mass spectrometric assays, we identify lysine (K) 97 in its NH2-terminal domain as the major mono-ubiquitin conjugation site. We constructed a mono-ubiquitinated mimic consisting of a deubiquitinase-resistant monomeric ubiquitin fused to the NH2 terminus of STAT3 (ubiquitinated-STAT3 FP). In complex assays of ectopically expressed ubi-STAT3-FP, we observed enhanced complex formation with bromodomain -containing protein 4 (BRD4), a component of the activated positive transcriptional elongation factor (P-TEFb) complex. Chromatin immunoprecipitation experiments in STAT3+/− and STAT3−/− MEFs showed BRD4 recruitment to STAT3-dependent suppressor of cytokine signaling-3 gene (SOCS3). The effect of a selective small molecule inhibitor of BRD4, JQ1, to inhibit SOCS3 expression demonstrated the functional role of BRD4 for STAT3-dependent transcription. Additionally, ectopic ubiquitinated-STAT3 FP expression upregulated BCL2, BCL2L1, APEX1, SOD2, CCND1 and MYC expression indicating the role of ubiquitinated STAT3 in anti-apoptosis and cellular proliferation. Finally we observed that ubiquitinated-STAT3 FP suppressed TNFα-induced apoptotic cell death, indicating the functional importance of mono-ubiquitinated STAT3 in antiapoptotic gene expression. We conclude that STAT3 mono-ubiquitination is a key trigger in BRD4-dependent antiapoptotic and pro-proliferative gene expression programs. Thus, inhibiting the STAT3 mono-ubiquitination - BRD4 pathway may be a novel therapeutic target for the treatment of STAT3-dependent proliferative diseases.
Keywords: Mono-ubiquitination, STAT3, BRD4, Proliferation, Apoptosis
1. Introduction
STAT3 is a transcription factor that participates in critical cellular processes involving cell-cycle progression and the anti-apoptotic program [1]. STAT3 is activated by tyrosine and serine phosphorylation in response to IL-6 type of cytokines and growth factors, leading to the formation of dimers that rapidly translocate into the nucleus and activate the promoters of target genes [2,3]. Aberrant constitutive STAT3 activation is frequently found in various type of cancers [4,5] and in autoimmune and inflammatory disorders, where it plays a deleterious role through prolonged activation of BCL2L1, BCL2, MYC and CCND1, target genes [6]. STAT3 has been also classified as an oncogene because of its predominant role in mediating cellular transformation by v-src and in promoting tumor progression in nude mice [7,8].
The regulation of STAT3 transcriptional activity mainly depends on posttranslational modifications (PTMs); these PTMS trigger STAT3 dimerization, nuclear translocation, complex formation with nuclear coactivators and/or its degradation. Although phosphorylation is the hallmark of STAT3 transcriptional activity [9–11] (tyrosine 705 and serine 727 phosphorylation are required for STAT3 dimerization and maximal transactivation, respectively), several other PTMs are also known to tightly regulate STAT3 function. For instance, the recently reported lysine (K) methylation at residue 140 (K140) is known to negatively regulate STAT3 activity [12] whereas lysine acetylation at its NH2 terminal (K49 and 87) and –COOH terminal (K685) is associated with positive regulation and DNA binding [13,14]. PTM by polyubiquitination also plays a key role in promoting STAT3 degradation by the 26S proteasome, a process for maintaining cellular homeostasis by clearance of activated STAT3, after execution of its transcriptional role [15]. Polyubiquitination and proteosomal degradation of STAT3 by the nuclear ubiquitin E3 ligase PDLIM2, has been reported in human granulomatous inflammation; this process inhibits T helper 17 cell development [16]. However, it remains unclear whether STAT3 could be modified directly by ubiquitination without leading to its degradation.
Ubiquitins are highly conserved small protein modifiers (MW~8 kDa) that are covalently attached to ε–amino group of K residues of target proteins. Depending on the ligase complex, K residues within the ubiquitin molecule form different types of branched chain chemistry [17]. Covalent attachment of polyubiquitin chains through K48 linkages mediate the well-characterized role of ubiquitin in targeting protein substrates for degradation by the 26S proteasome, whereas mono-ubiquitination and K63-linked polyubiquitination chemistries function as signal-dependent devices for inducing networks of protein-protein interactions. Mono-ubiquitination or multi-mono-ubiquitination on K residues of non-histone substrate can cause changes in a protein’s response to cellular trafficking, chromatin remodeling, DNA repair and transcriptional activation/repression [18–23]. Ubiquitin adducts target proteins can be disassembled by deubiquitin enzymes (DUB) and therefore is considered a dynamic, reversible, covalent modification [24].
In this paper, we report that STAT3 transcriptional activity is regulated by mono-ubiquitination. We have identified the K97 residue in the NH2-terminal domain of STAT3 as a site of ubiquitination using protein domain mapping and high resolution mass spectrometry. Expression of an ubiquitin-STAT3 fusion protein (ubiquitinated-STAT3 FP) mimic of the NH2 terminal mono-ubiquitinated protein complexes with bromodomain containing 4 (BRD4), a member of the bromodomain and extra-terminal bromodomain protein family. BRD4 is a structural scaffold with kinase activity that influences transcription by recruiting the positive transcription elongation factor complex-b (P-TEFb) to the transcription pre-initiation complex of inducible genes [25][26–28], directly phosphorylating serine 2 (Ser2) sites of the carboxy-terminal domain (CTD) of RNA polymerase II (Pol II). In our previous work, we showed that IL-6 promotes a nuclear complex of STAT3 and CDK9, a major component of active P-TEFb complex in derepression and activation of RNA Pol II on IL-6 inducible genes [29]. Here, we extend this work by providing evidence that mono-ubiquitinated STAT3 recruits BRD4 to regulate cellular proliferation and apoptosis. We show that small molecule inhibition of BRD4 downregulates STAT3-dependent MYC expression, a proliferation maker. Finally, we observed ubi-STAT3 FP protects against TNF-α induced apoptosis. These data suggest that mono-ubiquitinated STAT3 mediates BRD4 recruitment to promote a pro-proliferative and anti-apoptotic cellular state.
2. Materials and Methods
2.1. Cell Culture, Reagents, Transfection and Western Blot
Human embryonic kidney 293 and HepG2 cells were obtained from ATCC (Manassas, VA) and cultured as previously described [30]. STAT3−/− and STAT3+/− MEFs were obtained from Dr. Stephanie Watowitch at the University of Texas M.D. Anderson Cancer Center. Recombinant human IL-6 and sIL-6Rα were obtained from Pepro Tech Inc. (RockyHill, NJ). JQ1was purchased from Cayman Chemicals (Ann Arbor, MI). Sources of primary antibody (Ab) included Santa Cruz Biotechnology (Dallas TX) for anti-STAT3 Ab (C20) and anti-HA Ab; anti-V5 Ab was from Invitrogen (Grand Island, NY); anti β-actin Ab, anti-Flag M2Ab, Flag agarose beads and 3X Flag peptides were from Sigma-Aldrich (St. Louis, MO); anti- mouse monoclonal anti-HA Ab was from Millipore Billerica, MA). The STAT3 deletion plasmids [amino acids (aa) 1–770, 130–770, 1–688, 1–30] were constructed by PCR using wild type STAT3 as a template and gene-specific primers [13]. Transient transfections in exponentially growing cells were performed using Lipofectamine PLUS reagent (Life Technologies, Inc.) following the manufacturer’s protocol. For IP and Western blot analysis, whole cell lysates (WCLs) were prepared by lysing cells in modified RIPA buffer [31]. Proteins were fractionated by 10% SDS-PAGE and were transferred to polyvinylidine difluoride membranes (Millipore, Bedford, Mass) and blocked with 5% milk for 1 h followed by primary antibody treatment for overnight at 4°C. Membranes were washed in TBST (0.1%) tween and incubated with secondary antibody for 1 h. Signals were detected by the Odyssey Infrared Imaging system (LICOR Biosciences, Lincoln, NE).
2.2. Ni-NTA Affinity Chromatography
Cells transfected with 6X His-V5-tagged STAT3 expression plasmids were lysed in ice-cold ‘NTA buffer’ (where mentioned) containing 20 mM Tris (pH 8.0), 500 mM NaCl, 0.05% Tween 20, 10mM imidazole, 2μM MG132 (Sigma), Proteinase Inhibitor Cocktail (EDTA free; Roche, Cambridge, MA, USA), and phenylmethylsulfonyl fluoride. After centrifugation, the supernatant was incubated with Ni-NTA magnetic beads (Qiagen, Germantown, MD, USA) for 20 min at 4°C. The beads were washed with the NTA buffer containing 10 and 20mM imidazole and the bound proteins were eluted with the NTA buffer with 200 mM imidazole.
2.3. Flag-tagged protein Extraction with 3X Flag peptide
Flag-tagged STAT3 (1-130) in pECFP or Flag-tagged STAT3 K49/87R (1-130) proteins were IPed with anti-Flag M2 agarose beads (Sigma) for overnight at 4°C. Beads were washed three times with cold PBS and for elution, 100 μl of 3X Flag peptide (200 μg/ml, final concentration), diluted in PBS, were added to each IP sample and incubated for 2 h at 4°C with shaking. Proteins were eluted after centrifuging at 8000 × g for 30 sec at 4°C.
2.4. Identification of ubiquitination sites of STAT3
The ubiquitinated STAT3 was fractionated by 1D SDS-PAGE. The top band was excised and digested with trypsin as described before [32]. The tryptic digest was analyzed by LC-MS/MS on a TSQ Vantage triple quadrupole mass spectrometer equipped with nanospray source (Thermo Scientific, San Jose, CA). The online chromatography were performed using an Eksigent NanoLC-2D HPLC system (AB SCIEX, Dublin, CA). An aliquot of 10 μL of each of tryptic digests were injected on a C18 reverse-phase nano-HPLC column (PicoFrit™, 75 μm × 10 cm; tip ID 15 μm) at a flow rate of 500 nL/min with a 20-min 98% phase A (0.1 % formic acid), followed by a 15-min linear gradient from 2–30% mobile phase B (0.1 % formic acid-90 % acetonitrile) in phase A. The proteins were identified by searching the mass spectrometry data against Swissprot Human protein database with Proteome Discoverer 1.3 (Thermo Scientific). The false positive ratio cutoff of protein identification was 1%.
The identification of STAT3 ubiquitination sites was performed with selected reaction monitoring mass spectrometry (SRM-MS). Specific SRM-MS assays were developed for ubiquitinated-Lys48-STAT3, ubiquitinated-Lys87-STAT3 and ubiquitinated-Lys97-STAT3 modifications. Because the tryptic peptide for ubiquitinated-Lys48-STAT3 has 39 amino acid residues which does not yield a suitable signature peptide, we used Glu-C digestion instead. This strategy produced SQDWAYAASK[Ubiquitin]E, the signature peptide. The mass spectrometry parameter for each SRM assay is listed in Table 1.
Table 1. Mass spectrometry parameters for the SRM assays of ubiquitination sites of STAT3 (1-130).
| Signature peptides | Ubiquitination site | Q1, m/z | Q3, m/z | CE (V) | Precursor, Z | Product, Z | ion type |
|---|---|---|---|---|---|---|---|
|
| |||||||
| IK[Ubiquitin]QFLQSR | Lys87 | 378.5524 | 503.2931 | 20 | 3 | 1 | y4 |
| 378.5524 | 650.3615 | 20 | 3 | 1 | y5 | ||
| 378.5524 | 778.42 | 20 | 3 | 1 | y6 | ||
| 378.5524 | 1020.558 | 20 | 3 | 1 | y7 | ||
| 378.5524 | 1133.642 | 20 | 3 | 1 | y8 | ||
| 567.3249 | 650.3615 | 23 | 2 | 1 | y5 | ||
| 567.3249 | 778.42 | 23 | 2 | 1 | y6 | ||
| 567.3249 | 1020.558 | 23 | 2 | 1 | y7 | ||
| 567.3249 | 1133.642 | 23 | 2 | 1 | y8 | ||
|
| |||||||
| SQDWAYAASK[Ubiquitin]E | Lys48 | 457.2055 | 619.304 | 23 | 3 | 1 | y5 |
| 457.2055 | 782.3673 | 23 | 3 | 1 | y6 | ||
| 457.2055 | 853.4044 | 23 | 3 | 1 | y7 | ||
| 457.2055 | 1039.484 | 23 | 3 | 1 | y8 | ||
| 685.3046 | 782.3673 | 26 | 2 | 1 | y6 | ||
| 685.3046 | 853.4044 | 26 | 2 | 1 | y7 | ||
| 685.3046 | 1039.484 | 26 | 2 | 1 | y8 | ||
| 685.3046 | 1154.511 | 26 | 2 | 1 | y9 | ||
|
| |||||||
| YLEK[Ubiquitin]PM[Oxid]EIAR | Lys97 | 460.5711 | 488.2822 | 24 | 3 | 1 | y4 |
| 460.5711 | 635.3176 | 24 | 3 | 1 | y5 | ||
| 460.5711 | 732.3704 | 24 | 3 | 1 | y6 | ||
| 460.5711 | 974.5082 | 24 | 3 | 1 | y7 | ||
| 460.5711 | 1103.551 | 24 | 3 | 1 | y8 | ||
| 460.5711 | 1216.635 | 24 | 3 | 1 | y9 | ||
| 690.353 | 732.3704 | 27 | 2 | 1 | y6 | ||
| 690.353 | 974.5082 | 27 | 2 | 1 | y7 | ||
| 690.353 | 1103.551 | 27 | 2 | 1 | y8 | ||
| 690.353 | 1216.635 | 27 | 2 | 1 | y9 | ||
|
| |||||||
| YLEK[Ubiquitin]PMEIAR | Lys97 | 455.2395 | 488.2822 | 23 | 3 | 1 | y4 |
| 455.2395 | 619.3226 | 23 | 3 | 1 | y5 | ||
| 455.2395 | 716.3754 | 23 | 3 | 1 | y6 | ||
| 455.2395 | 958.5133 | 23 | 3 | 1 | y7 | ||
| 455.2395 | 1087.556 | 23 | 3 | 1 | y8 | ||
| 455.2395 | 1200.64 | 23 | 3 | 1 | y9 | ||
| 682.3555 | 716.3754 | 26 | 2 | 1 | y6 | ||
| 682.3555 | 958.5133 | 26 | 2 | 1 | y7 | ||
| 682.3555 | 1087.556 | 26 | 2 | 1 | y8 | ||
| 682.3555 | 1200.64 | 26 | 2 | 1 | y9 | ||
Abbreviations: Q, quadrupole; m/z, mass to charge ratio; CE, collision energy; Z, charge state.
The ubiquitinated STAT3 isoform, representing the upper STAT3 band on an SDS-PAGE was digested with trypsin or Glu-C, respectively. Each digestion was analyzed by LC-SRM-MS on a TSQ Vantage triple quadrupole mass spectrometer equipped with nanospray source (Thermo Scientific, San Jose, CA). The online chromatography were performed using an Eksigent NanoLC-2D HPLC system (AB SCIEX, Dublin, CA). An aliquot of 10 μL of each of tryptic digests were injected on a C18 reverse-phase nano-HPLC column (PicoFrit™, 75 μm × 10 cm; tip ID 15 μm) at a flow rate of 500 nL/min with a 20-min 98% phase A, followed by a 15-min linear gradient from 2–30% mobile phase B in mobile phase A. The TSQ Vantage was operated in high-resolution SRM mode with Q1 and Q3 set to 0.2 and 0.7-Da Full Width Half Maximum (FWHM). All acquisition methods used the following parameters: 1800 V ion spray voltage, a 275 °C ion transferring tube temperature, a collision-activated dissociation pressure at 1.5 mTorr, and the S-lens voltage used the values in S-lens table generated during MS calibration.
2.5. Stable isotope labeled (SID)-selected Reaction Monitoring (SRM)-mass spectrometry (MS) analysis of STAT3 complex
The SRM assays for total STAT3, P300, CDK9, and BRD4 proteins were developed using a workflow described in previous publications [33,34]. The signature peptides and SRM parameters are listed in Table 2. The stable isotope standard (SIS) peptides were chemically synthesized incorporating isotopically labeled [13C615N4] Arginine or [13C615N2] Lysine to a 99% isotopic enrichment (Thermo Scientific).
Table 2. Mass spectrometry parameters for SRM assays of STAT3 and. Masses listed are for the native forms of the peptides.
| Gene Name | Swissprot No | Sequence | Q1 m/z | Q3 m/z | Ion type | CE (V) |
|---|---|---|---|---|---|---|
| STAT3 | P40763 | TLTDEELADWK | 660.82 | 761.382 | y6 | 26 |
| 660.82 | 890.425 | y7 | 26 | |||
| 660.82 | 1005.452 | y8 | 26 | |||
| 660.82 | 1106.5 | y9 | 26 | |||
|
| ||||||
| BRD4 | O60885 | DAQEFGADVR | 554.257 | 664.341 | y6 | 22 |
| 554.257 | 793.383 | y7 | 22 | |||
| 554.257 | 921.442 | y8 | 22 | |||
Abbreviations: CE, collision energy; Q, quadropole,
The proteins were immunoprecipitated with anti-STAT3 antibody were captured by protein A magnetic beads (Dynal Inc). The proteins on the beads were digested with trypsin as described previously [35]. Briefly, the beads were washed with PBS for three times and then resuspended in 30 μL of 50 mM ammonium hydrogen carbonate (pH 7.8) and 20 μL of 0.1 μg/ μL of trypsin was added. The samples were mixed and trypsinized by gentle vortexing overnight at 37 °C. After digestion, the supernatant was collected. The beads were washed with 50 μL of 50% acetonitrile (ACN) three times and the supernatant was pooled, and dried. The tryptic digests were then reconstituted in 30 μL of 5% formic acid-0.01% TFA. An aliquot of 10 μL of diluted SIS peptides were added to each tryptic digest. These samples were desalted with ZipTip C18. The peptides were eluted with 80% ACN and dried. The dried peptides were reconstituted in 30 μL of 5% formic acid-0.01% TFA and were directly used for LC-SRM-MS analysis.
LC-SRM-MS analysis was performed with a TSQ Vantage triple quadrupole mass spectrometer equipped with nanospray source (Thermo Scientific, San Jose, CA) as described above. All SRM data were manually inspected to ensure peak detection and accurate integration. The chromatographic retention time and the relative product ion intensities of the analyte peptides were compared to those of the SIS peptides. The variation of the retention time between the analyte peptides and their SIS counterparts were within 0.05 min, and no significant difference in the relative product ion intensities of the analyte peptides and SIS peptides were observed. The peak area in the extract ion chromatography of the native and SIS version of each signature peptide were integrated using Xcalibur® 2.1. The default values for noise percentage and base-line subtraction window were used. The ratio between the peak area of native and SIS version of each peptide were calculated.
2.6. Generation of ubiquitinated-STAT3 fusion protein
Plasmid DNA for the ‘ubiquitin STAT3 fusion protein (FP)’ was generated by inserting the ubiquitin (ubiG76A [36], where the COOH terminal Gly 76 is replaced with Ala, a gift from Dr. T Izumi, University of Kentucky at Lexington) gene in the NH2 terminus of STAT3 in pCDNA 3.1 Flag-monomeric Cherry fluorescent protein (mcherry) expression vector backbone.
2.7. RT-PCR Analysis
Total cellular RNA was extracted by Tri Reagent (Sigma). One μg of RNA was used for reverse transcription using iScript cDNA Synthesis Kit (Bio Rad). Three μl of cDNA products was amplified in 20 μl reaction system containing 10 μl iQ SYBR Green Super Mix (Bio Rad) and 400 nM primer mix. All reactions were processed in MyiQ Single-Color Real-Time PCR Detection System (Bio Rad) and results were analyzed by IQ5 program (Bio Rad). To normalize template input, GAPDH (endogenous control) transcript level was measured for each sample. Data are expressed as fold change after normalizing to GAPDH. The human primers used for RT-PCR are as follows: SOCS3, SP: 5′CAAGGACGGAGACTTCGATT-3′ and AS: 5′ GACTGGGTCTTGACGCTGA-3′; BCL2L1, SP: 5′GTTGAAGCGTTCCTGGCCCTTT-3′ and AS: 5′CAGAATGGACTGAATCGGAGAT-3′; MYC, SP: 5′ATTCTCTGCTCTCCTCGA-3′ and AS: 5′TCTTGGCAGCAGGATAGT-3′; CCND1, SP: 5′GTGCTGCGAAGTGGAAACC-3′ and AS: 5′ATCCAGGTGGCGACGATCT-3′; Primers for APEX1, SOD2 and BCL2 were ordered from IDT DNA integrated technology.
2.8. Two-step chromatin immunoprecipitation (XChIP)
XChIP was performed using sequential 2 mM disuccinimidyl glutarate (Pierce) protein-protein cross-linking followed by formaldehyde protein-DNA cross-linking as described [29]. Equal amounts of sheared chromatin were immunoprecipitated overnight at 4 °C with 4 μg indicated Ab in ChIP dilution buffer. Immunoprecipitates were collected with 40 μL protein-A magnetic beads (Dynal Inc), washed and eluted in 250 μl elution buffer for 15 min at room temperature. Samples were de-cross-linked in 0.2 M NaCl at 65 °C for 2 h. The precipitated DNA was phenol/chloroform extracted, precipitated by 100% ethanol and dried. Quantitative genomic PCR was performed using the mouse SOCS3 promoter specific primer pairs [29]. Data are corrected for variations in input and expressed as fold change relative to IgG controls.
2.9. Electroporation in STAT3−/− MEF
STAT3−/− MEF after trypsinization were resuspended in 100 μl of Nucleofactor solution (Amexa, Lonza) along with mcherry-ubi-STAT3 FP or mcherry-STAT3 expression plasmid or mcherry empty vector (EV) in an electroporator cuvettes. The cell suspension was electroporated using a Nucleofactor I device (Amexa, Lonza). The Nucleofactor program A-23 was used for MEF cells according to the manufacturer’s instruction.
2.10. Apoptosis Assay and Flow Cytometry
STAT3−/− MEFs were electroporated with mcherry-STAT3, mcherry ubi-STAT3 FP or EV expression plasmids. 40 h after transfection, cells were treated with TNFα (15 ng/ml) and cyclohexamide (30 μg/ml) for 0, 3 and 5 h. Cells were harvested with accumax (Sigma) for single cell suspension and 1×106 cells first were stained with Live/Dead Fixable Aqua Dead Cell Stain Kit, 405 nm excitation, (from invitrogen), followed by FITC conjugated Annexin V staining (from eBioscience) for detecting early apoptotic cell death, following manufacturer’ protocol. Samples were analyzed by FACS Canto (BD) and data was analyzed with FACSDiva software (BD Bioscience).
2.11. Statistics
All experiments were carried out in triplicates (n=3) and the Q-RT-PCR data are expressed as mean ± SD. Statistical analysis was performed using the two-tailed student t-test, with significance set at p<0.05.
3. Results
3.1. STAT3 is mono-ubiquitinated in cellulo
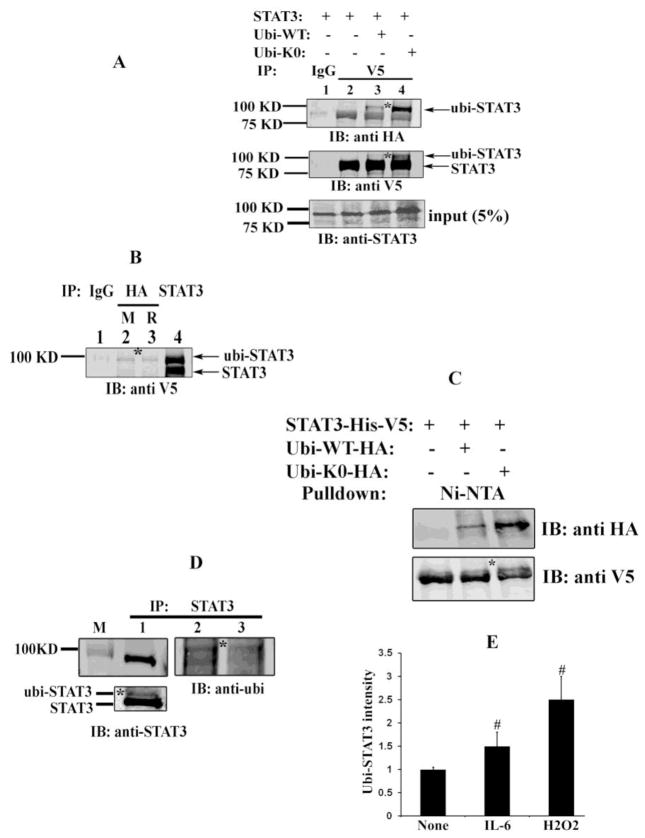
Although previous studies have implicated STAT3 poly-ubiquitination and degradation by the proteasome pathway [16,37], mono-ubiquitination or K63 linkages of STAT3 have not been reported. To examine the physiological significance of ubiquitination of STAT3 in detail, HEK293 cells were transfected with V5-tagged STAT3 alone (Fig. 1A, lane1 and 2) or V5-tagged STAT3 and HA- tagged ubiquitin (ubi-WT) (lane 3), and IL-6 stimulated whole cell lysates (WCL) were immunoprecipitated (IPed) with anti-V5 Ab (Fig. 1A, lane 2 and 3) or mouse IgG (lane 1) as a negative control. IPs were then resolved by SDS-PAGE, followed by immunoblotting with anti-HA Ab. In comparison with IgG IPs, STAT3 in the V5 IP was weakly recognized by the anti-HA Ab (Fig. 1A, top panel, lane 2). By contrast, in cells transfected with HA-tagged ubiquitin (ubi), we observed a prominent 100 kDa band (Fig. 1A, top panel, lane 3). We noted that the relative molecular mass of the ubi-dependent band corresponded with the molecular weight of full length STAT3 plus 8 kDa (91+ ~10 =100 kDa) suggesting that STAT3 can be mono-ubiquitinated. A western blot of the input was performed using anti-STAT3 Ab, confirming equivalent input of STAT3 in the immunoprecipitates. We note that the 100 kDa monoubiquitinated form was not detectable in the Western blot of the starting material (input), indicating that this form represents a low abundance modification of the total STAT3 pool.
Fig. 1.
STAT3 mono-ubiquitination in cellulo. (A) HEK 293 cells were transfected either with V5- tagged STAT3 alone (lane 1 and 2) or along with HA- tagged WT ubi (lane 3) or HA- tagged K0 ubi (lane 4). 40 h after transfection, cells were treated with 1.5 μM MG132 for 4 h prior to 15 min stimulation with IL-6 (8ng/ml) and sIL-6R (6 μg/ml). Cells were harvested with modified RIPA and WCL were IP’ed either with IgG (lane 1) or with anti-V5 Ab (lane 2, 3, 4) and immunoblotted with anti-HA Ab (top) or with anti-V5 Ab (middle). The bottom panel shows input samples (5% WCL without IP) subjected to western immunoblot analysis wih anti-STAT3 Ab.
(B) HEK 293 cells were transfected with V5-STAT3 and HA-ubi-K0 and WCL were IP’ed either with IgG (lane 1) or with anti-HA Ab from mouse (M) (lane 2) or anti-HA Ab from rabbit (R) (lane 3) or with anti-STAT3 Ab (lane 4) and Western immunoblot was performed with anti-V5 Ab.
(C) Cells were transfected with either V5-His-STAT3 alone (lane 1) or with V5-His-STAT3 and HA-ubi-WT (lane 2) or V5-His-STAT3 and HA-ubi-K0 (lane 3). WCL were subjected to Ni-affinity NTA pull down assay. The eluted fraction were resolved in SDS-PAGE and immunoblotted with anti-HA Ab (top) and anti-V5 Ab (bottom).
(D) 293 cells were transfected either with ubi-K0 (lane 1and 2) or ubi-WT (lane 3) expression plasmid and WCL were IPed with anti-STAT3 Ab and immunoblotted with either anti-STAT3 Ab (left) or with anti-ubi Ab (right). The bottom left panel is lane 1 after longer exposure, showing ubiquitin modified STAT3.
(E) Cells were transfected with V5-His-STAT3 and HA-ubi-K0 and prior to harvest, cells were stimulated either with IL-6 or with H2O2 or left untreated. WCL were pull down by Ni-NTA assay and eluted proteins were resolved and immunoblotted with anti-HA Ab. Shown is the average relative intensity of ubi-STAT3 expression (pulled down in Ni-NTA assay (n=3)) in the Western Blot. #, represents p<0.05, students t test.
* represents monomeric ubiquitin conjugated STAT3.
To further confirm the observation that STAT3 is mono-ubiquitinated, we used a lysine (K) free HA-Ubiquitin construct in which all seven K residues were mutated to arginine (ubi-K0), and is therefore unable to form K48 or K63 linked polymers. After transfection of ubi-K0, followed by IP-Western immunoblot as above, a similar pattern with a strong ~100 kDa band was observed in lane 4 (top panel), supporting the evidence that STAT3 is mono-ubiquitinated. The middle panel is a Western blot with V5 Ab that shows equal amount of STAT3 in the immunocomplex, along with a high MW ~100 kDa band particularly in lane 4, confirming the presence of modified, mono-ubiquitinated STAT3. Conversely, a co-IP assay of the extract containing ectopically expressed V5-STAT3 and HA-ubi-K0 with anti-HA Ab followed by immunoblot with anti-V5 Ab confirms that STAT3 is present in HA immune complex (Fig. 1B). The migration of this band matches exactly with the high molecular weight modified band of V5-STAT3 in the STAT3 immune complex (Fig. 1B, lane 4).
To further validate the finding that STAT3 is mono-ubiquitinated, WCL of V5-His-tagged STAT3 (Fig. 1C, lane1) or V5-His-tagged STAT3 and HA-ubi-WT (Fig. 1C, lane 2) /or HA-ubi-K0 (Fig. 1C, lane 3) were subjected to Ni-NTA affinity pull down assay and Western blotted with either anti-HA Ab (top) or anti-V5 Ab (bottom). As shown in Fig. 1C, (top panel) the histidine hexamer (6X His) tagged STAT3 proteins that were bound with HA-ubiquitin were eluted and only immunoreacted with HA Ab (Fig. 1C, lane 2 and 3), confirming that STAT3 is conjugated with ubiquitin monomer in cellulo.
To detect if endogenous STAT3 is mono-ubiquitinated, cells were transfected with either ubi-K0 or ubi-WT (because the sub-population of mono-ubiquitinated proteins is extremely low and difficult to detect) and endogenous STAT3 ubiquitination was determined. For this purpose endogenous STAT3 was enriched by IP with pan-STAT3 Ab from the WCL prior to immunoblotting with anti-ubi Ab (Fig. 1D, lane 2 and lane 3, right) or with anti-STAT3 Ab (lane 1, left). In Fig. 1D left bottom panel, when WCL from ubi-K0 transfected cells were IPed and immunoblotted with anti-STAT3 Ab, we again detected presence of an additional slower migrating species around ~100 kDa (longer exposure) confirming presence of endogenous mono-ubiquitinated STAT3 species.
Next, to determine if STAT3 mono-ubiquitination is stimulus-dependent, cells were co-transfected with STAT3 and ubi-K0 and stimulated with either IL-6 and sIL-6R or H2O2 or left untreated prior to harvest and pull down assay by Ni-NTA. Proteins were fractionated by SDS-PAGE and ubiquitin conjugated STAT3 was measured by anti-HA immunoblot and band intensity were plotted. As seen in Fig. 1E, increased STAT3 mono-ubiquitination is observed when cells were exposed to a known inducer of mono-and /or polyubiquitination hydrogen peroxide and also with IL-6 stimulation.
3.2. Mapping of the STAT3 domain for mono-ubiquitination
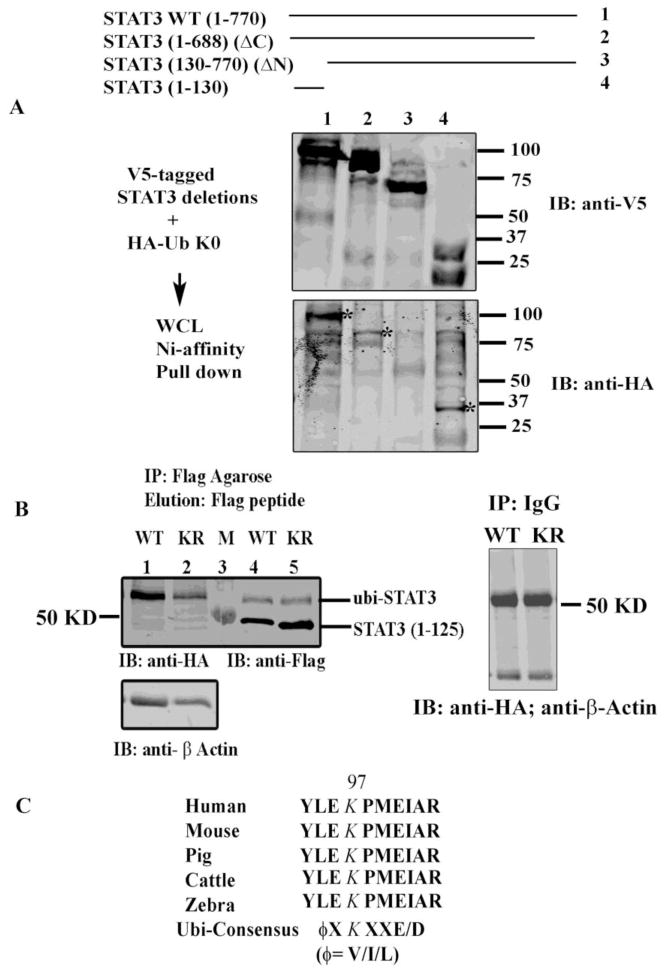
Ubiquitination is a covalent adduct on the ε-amino group of K residue of the target protein. To determine the specific K residues of STAT3, we examined its deletion mutants in the ubiquitin assay. For this purpose we transfected HEK293 cells with a series of 6XHis-V5-tagged STAT3 deletion mutants (aa 1-770, aa 1-688, aa 130-770 and aa 1-130) and HA-tagged ubi-K0. Cells were harvested, and proteins were purified from WCL on Ni-NTA magnetic beads after stringent washing and elution with 200 mM imidazole. Proteins were resolved in SDS-PAGE prior to immunoblotting with anti-V5 Ab (Fig 2A, top) or anti-HA Ab (Fig 2A, bottom). In Fig 2A (bottom), the WT STAT3 (1-770), and STAT3 deletion mutants STAT3 (1-688) and STAT3 (1-130) were covalently modified by ubiquitin conjugation, however, STAT3 deletion mutant lacking aa 1-130 of the NH2 terminal domain did not (lane 3) indicating that the acceptor site for mono-ubiquitination resides in the NH2 terminal regulatory domain of STAT3. As a negative control when cells were transfected with each 6X His V5-tagged STAT3 construct alone (without HA-ubi plasmid) and processed as described above and immunoblotted with anti-HA Ab, we did not observe any immune-reactive bands although each of the deletion mutants was sufficiently expressed with expected mobility on anti-V5 immunoblots (data not shown).
Fig 2.
STAT3 is ubiquitinated on its NH2 terminal domain. (A) HEK293 cells were transfected with the indicated V5-His-tagged STAT3 WT and deletion constructs and HA-ubi-K0. WCL were subjected to Ni-NTA affinity assay and the proteins were resolved and immunoblotted with either anti-V5 Ab (top) or anti-HA Ab (bottom). * represent monomeric ubiquitin conjugated STAT3. (B) HEK 293 cells were transfected with either Flag-tagged STAT3 WT (1-125) or with Flag-tagged K49/87R (1-125) as indicated. WCL were IPed with Flag-agarose beads and proteins were eluted with 3X Flag peptide prior to resolved in SDS-PAGE. Western immunoblot analysis was performed either with anti-HA Ab (left) or with anti-Flag Ab (right). Bottom left panel is the immunoblots of lane 1 and 2 with β-Actin Ab. Right panel shows WCL IP’ed with IgG as a negative control and immunoblotted with anti-HA and β-Actin Ab. M represents Marker lane. (C) ClustalW sequence analysis of STAT3 (1-125) showed conservation of K residue 97 among different mammalian species and contains an ubiquitin/sumoylation consensus sites.
The STAT3 NH2 terminal domain (aa 1-130) contains the three K residues at amino acids (aa) 48, 87 and 97 representing the primary NH2-terminal acetyl-acceptor sites identified previously by our group was at K residue 49 and 87 [13]. We therefore tested if K residue at 97 could be the possible ubiquitin acceptor site in STAT3 NH2 terminal domain. For this purpose we transfected 293 cells with a Flag-tagged STAT3 (1-125) expression vector or with a Flag-tagged STAT3K49/87R (1-125) mutant with K 49 and 87 residues mutated to arginine (R), along with HA-ubi-K0. WCL were IPed with Flag agarose beads, and competitively eluted with 3X-Flag peptide. Eluted proteins were then separated in SDS-PAGE and immunoblotted with either anti-HA Ab (Fig. 2B, left) or anti-Flag Ab (Fig. 2B, right). Both WT STAT3 (1-125) and STAT3K49/87R (1-125) mutant with intact K97 were modified by ubiquitination (left panel) indicating that K97 could be the primary acceptor site for STAT3 mono-ubiquitination. As a negative control when WCL were IP’ed with mouse IgG, we did not observe any immunoreactive band when blotted with anti-HA Ab (right panel). However, when we generated a STAT3 K97 to Arginine (R) point mutants both in full length STAT3 and in STAT3 (1-125) and performed ubiquitination assay, surprisingly STAT3 mono-ubiquitination still occurred on this mutation, suggesting that mono-ubiquitin conjugation takes place in alternative lysine residues following mutation of the primary site, as reported by many other groups [38].
Moreover, interestingly, in Fig 2C, a ClustalW analysis revealed that K 97 in STAT3 NH2 terminus is evolutionary conserved in various species, implying a potentially important function for this lysine and it is also located in a sumolation/ubiquitination consensus sequence ϕKXe [39, where the acceptor K residue is flanked by NH2-terminal hydrophobic aa (ϕ is hydrophobic residue I, L or V) and a COOH-terminal acidic aa (e stands for E or D).
3.3. Identification of the lysine residues of STAT3 mono-ubiquitination
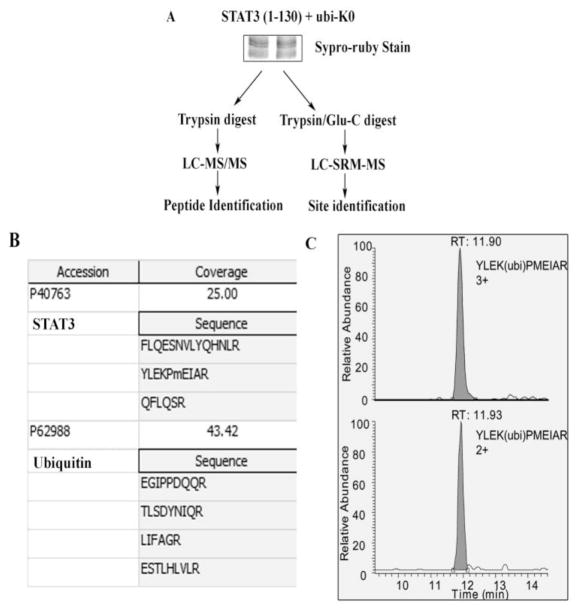
Next, we sought to confirm K97 mono-ubiquitination directly by MS analysis. V5- tagged STAT3 (1-130) and HA-ubi K0 were transfected in HEK293 cells. Immune complexes of WCL IPed with anti-V5 Ab were separated by SDS-PAGE and stained with supro ruby (Fig 3A). The upper band was excised trypsin digested and analyzed by LC-MS/MS. Both STAT3 and ubiquitin were identified from this gel band, demonstrating that STAT3 was ubiquitinated. The peptides identified with LC-MS/MS are tabulated in Fig 3B. In order to identify which lysine residues on STAT3 (1-130) was the target of mono-ubiquitination, we developed specific SRM assays for each of the three putative K ubiquitination sites contained within STAT3 (1-130). The mass spectrometry parameters for the assays are listed on Table 1. For this purpose, the upper gel band was digested with trypsin and Glu-C, respectively and the tryptic digest and Glu-C digest of the upper band were analyzed by LC-SRM-MS. Out of all the targeted ubiquitinated-STAT3 signature peptides, only the signature peptide of ubiquitinated-Lys97-STAT3, YLEK(ubi)PMEIAR, showed detectable signal (Fig 3C). These highly specific assays indicated that the primary mono-ubiquitination site occurred on K97, not K48 nor K87.
Fig. 3.
Mapping of NH2 terminal STAT3 residue K97 as the ubiquitin acceptor by mass spectrometry. (A) HEK293 cells were transfected with STAT3 (1-130) and ubi-K0 and resolved in SDS-PAGE. Gel was stained with supro-ruby and upper band was excised and either trypin digested and subjected to LC-MS/MS for identification of peptide in (B) or the band was digested with trypsin and Glu-C and subjected to LC-SRM-MS for K residue identification in (C). (B), shows peptide sequence of STAT3 and ubiquitin as identified in LC-MS/MS. (C) The SRM analysis of ubiquinitated STAT3 peptide YLEK(ubi)PMEIAR is shown. The upper panel is the extract ion chromatogram of triply charged YLEK(ubi)PMEIAR, and the bottom panel is extract ion chromatogram of doubly charged YLEK(ubi)PMEIAR.
3.4. Enhanced transcriptional activity of ubiquitinated-STAT3 fusion protein
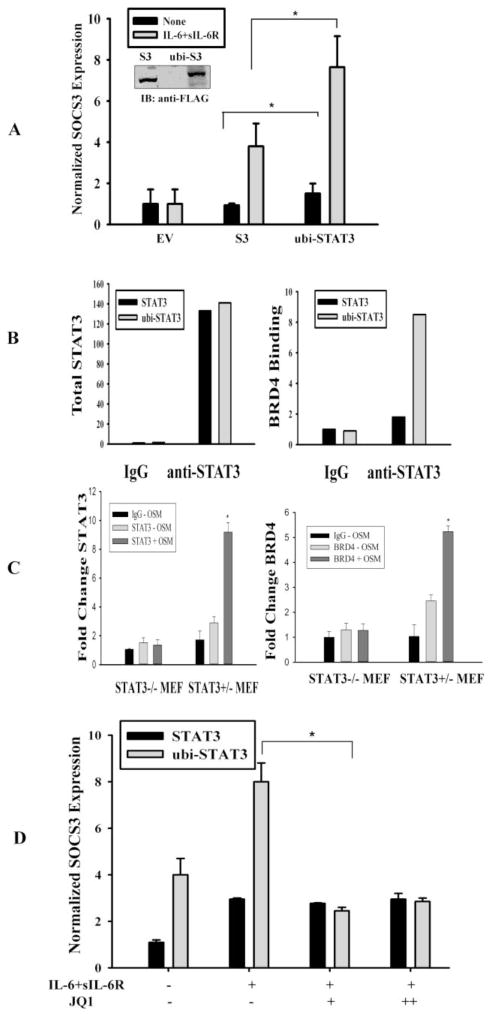
To determine the functional consequence of STAT3 mono-ubiquitination, we made an in-frame fusion protein of ubiquitin monomer and STAT3 (ubi-STAT3 FP) that mimics endogenously generated mono-ubiquitinated protein. To avoid cleavage of ubiquitin by cellular DUBs, we used a C-terminal glycine 76 to alanine mutant of ubiquitin (UbiG76A) [36] to fused with the 5′ end of STAT3 gene, representing a constitutively ‘ubiquitinated’ STAT3 mimic, as shown by its expression in Fig 4A inset. To determine the effect of transcriptional activity of ubi-STAT3 FP, we transfected 293 cells either with Flag tagged STAT3 expression vector or Flag tagged ubi-STAT3 FP and total RNA was isolated after IL-6 and sIL-6R stimulation, for quantitative real time-PCR (Q-RT-PCR) analysis for suppressor of cytokine signaling-3 (SOCS3) gene, a highly inducible STAT3-dependent gene [31,40]. We observed that the expression of human SOCS3 gene is induced both in basal and in response to IL-6 stimulation when ubi-STAT3 FP is transfected in cell in contrast to that seen in STAT3 expression alone (Fig. 4A). As a negative control, empty vector (EV) expression showed no effect on hSOCS3 expression (Fig. 4A). We therefore concluded that ubiquitination enhanced the transcriptional activity of STAT3.
Fig. 4.
ono-ubiquitinated STAT3 has increased transcriptional activity and is BRD4-dependent. (A) HEK 293 cells were transfected either with empty vector (EV) or with Flag-mcherry-STAT3 expression vector (S3) or with Flag-mcherry-ubiquitin-STAT3 fusion protein (ubi-STAT3). 40 h after transfection cells were either treated with IL-6 and sIL-6R for 20 mins or left untreated and total RNA was subjected to Q-RT-PCR for human SOCS3 expression. Data are expressed as mean±SD, * indicates P< 0.05. The inset figure shows expression of Flag tagged STAT3 and ubi-STAT3 FP. (B) HEK 293 cells were transfected with Flag-tagged STAT3 or Flag- tagged ubi-STAT3 FP and WCL were IP’ed either with IgG or anti-STAT3 Ab and subjected to SID-SRM analysis (described experimental procedures) for STAT3 and BRD4 abundance. The results of SID-SRM assay were normalized with total input concentrations of the WCL. The left panel figure shows the total STAT3 detected in each sample and right panel shows relative ratio of BRD4 associated with STAT3 in each samples. (C) STAT3 and BRD4 binding to SOCS3 promoter by XChIP assay. STAT3−/− and STAT3+/− MEF cells were treated with or without the STAT3 agonist, OSM (20 ng/ml) for 30 min. Double cross-linked chromatin was IPed with either IgG, anti-STAT3 (STAT3) or anti-BRD4 antibody (BRD4) and bound chromatin was amplified by Q-gPCR using a mSOCS3 promoter-specific primer pair. Fold change is expressed relative to IgG. *p< 0.01. (D) HEK 293 cells were transfected with either STAT3 or ubi-STAT3 FP. 24 h after transfection cells were treated with JQ1 at a concentration of 10 μM and 50 μM for overnight and total RNA was isolated prior to IL-6 and sIL-6R treatment. Human SOCS3 mRNA expression was measured by Q-RT-PCR. Data are expressed as mean ±SD, * indicates p< 0.05.
3.5. Interaction of ubi-STAT3 FP with BRD4
To understand the mechanism of enhanced transcriptional activity of mono-ubiquitinated STAT3, we sought to find its interacting partners by nondenaturing co-IP. In our previous work, we found that STAT3 forms a nuclear complex with cyclin- dependent kinase 9 (CDK9), a major component of P-TEFb, involved in derepression and activation of RNA Pol II [29]. Within the PTEFb complex, bromodomain containing protein 4 (BRD4) is a protein unique to the activated state of the P-TEFb complex. In this complex, BRD4 stabilizes P-TEFb interaction with chromatin by binding acetylated histones and is involved in Ser2 phosphorylation of the RNA PolII CTD. We therefore transfected cells either with ubi-STAT3 FP or with STAT3 expression plasmid followed by measurement of BRD4 association using immunoprecipitation-selected reaction monitoring-based mass spectrometry (IP-SRM). IP-SRM was selected as a technique because of its accurate quantification capabilities and its absolute specificity for the target protein. Here, we observed that the abundance of STAT3-associated BRD4 was significantly increased in ubi-STAT3 FP WCL immnoprecipitate in contrast to that seen in unmodified STAT3 lysates (Fig 4B, right), although the amount of total STAT3 in both immunoprecipitates are the same (Fig 4B, left). This observation indicated that a mono-ubiquitinated STAT3 promotes complex formation with an active component of the PTEFb complex, BRD4, and therefore suggested a novel role for BRD4 in STAT3-mediated transcription.
These above data indicated that BRD4 may functionally important in STAT3-dependent transcription. To examine whether BRD4 was recruited to STAT3-dependent genes, a two-step chromatin immunoprecipitation (XChIP) assay was performed in STAT3−/− and STAT3−/+ cells. Because we previously showed that MEFs do not express high amounts of IL-6R, cells were stimulated in the absence or presence of a potent gp130-STAT3 activator, oncostatin-M (OSM) [31]. Chromatin was crosslinked and the recruitment of STAT3 and BRD4 to mSOCS3 promoter was determined by XChIP. We observed that although ~2.5-fold increase of STAT3 binding could be detected on mSOC3 in unstimulated STAT3+/− MEFs, OSM induced a 9-fold fold increase of STAT3 binding (Fig. 4C). Similarly, 2.5-fold increase in BRD4 was observed in untreated STAT3−/+ MEFs, and OSM induced a 5.5-fold increase BRD4 binding in STAT3−/+ MEFs. The absence of BRD4 binding to mSOCS3 promoter in STAT3−/− cells indicates that STAT3 is required for targeting BRD4 to STAT3 dependent target genes.
To further investigate the functional role BRD4 in STAT3 transcription, we measured the effect of a selective small-molecule bromodomain inhibitor (JQ1). We observed an increase in SOCS3 expression in presence of ubi-STAT3 FP both in basal condition and with IL-6 stimulation (Fig. 4D), reproducing our observation in Fig 4A; however, in the presence of JQ1, SOCS3 mRNA expression was significantly abrogated in cells transfected mainly with ubi-STAT3 FP. This result indicates that IL-6 inducible SOCS3 expression is JQ1 sensitive and therefore further confirms the functional role of BRD4 in STAT3 mediated gene transcription.
3.6. Expression of anti-apoptotic and cellular proliferation marker genes by ubi-STAT3 FP
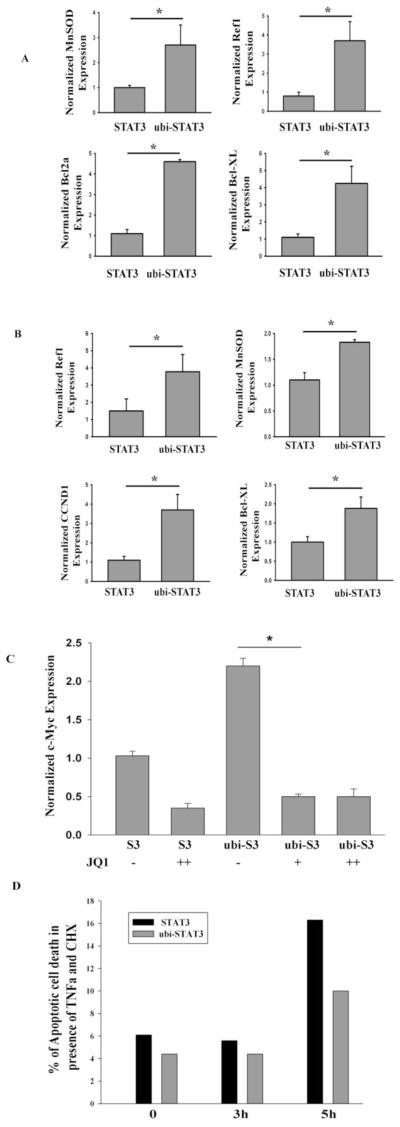
Because STAT3 activates cellular proliferation and anti-apoptosis programs through regulating the expression of its target genes, we next asked whether expression of mono-ubiquitinated STAT3 would lead to an increased expression of proliferation and anti-apoptotic marker gene expression. Expression of ubi-STAT3 FP in HepG2 cells showed an increased mRNA expression of CCND1, BCL2L1, APEX1, SOD2 and BCL2 genes relative to that seen in unmodified STAT3 expression (Fig. 5A). A similar finding was also observed in STAT3−/− MEFs (Fig. 5B). To further confirm that increased gene expression by ubi-STAT3 FP is mediated through BRD4, we tested the effect of JQ1 on expression of the master cellular proliferation regulator MYC. Expression of ubi-STAT3 FP elevated MYC expression compared to that produced by STAT3 alone and treatment with JQ1 inhibited MYC expression approx ~3 fold in ubi-STAT3 transfected cells (Fig. 5C), consistent with other studies [41]. These results further supported our earlier findings that BRD4 is a functionally important as a regulatory factor for ubi-STAT3 mediated transcription of MYC expression.
Fig. 5.
Mono-ubiquitinated STAT3 induces expression of proliferation and anti-apoptotic marker gene expression. (A) HepG2 cells were transfected with either STAT3 or ubi-STAT3 FP. 40 h after transfection total RNA was isolated and Q-RT-PCR was performed for SOCS3, SOD2, APEX1, BCL2, BCL2L1expression. Data are expressed as mean ± SD, * indicates P< 0.05. (B) STAT3−/− MEF were transfected either with STAT3 or ubi-STAT3 FP and mRNA expression of Ref1, BCL2L1, CCND1, and SOD2 were measured by Q-RT-PCR. (C) HEK 293 cells were transfected with either STAT3 (S3) or ubi-STAT3 FP (ubi-S3). 24 h after transfection cells were treated with JQ1 (10 μM and 50 μM for overnight) and total RNA was isolated for mRNA expression of MYC. Data are expressed as mean±SD, * indicates P< 0.05. (D) STAT3−/− MEF were electroporated with either STAT3 or ubi-STAT3 FP. 40 h after transfection cells were treated with TNFα (15 ng/ml) and cyclohexamide (30μg/ml) for 0, 3 and 5 h. Cells were gated for mcherry positive cells and early apoptosis was measured by FITC conjugated Annexin V staining. Shown is the percentage of apoptotic cell death as measured by flow cytometry (FACS Canto, BD).
Cellular treatment with TNFα and cyclohexamide (CHX) induces ROS/H2O2 - mediated apoptosis [42]. Next, to determine if anti-apoptotic gene expression induced by mono-ubiquitinated STAT3 is sufficient to protect against TNFα/CHX-mediated apoptosis, STAT3−/− MEF cells were electroporated with either mcherry-tagged STAT3 or mcherry-tagged-ubi-STAT3 FP. After allowing for the tagged STAT3 expression, the cells were treated with TNFα and CHX for various times and early apoptosis was measured in cherry-positive cell population by annexin V staining in flow cytometry. We observed that the percent of apoptotic cell death in STAT3 transfected cells were ~3.5 fold higher than that in ubi-STAT3 FP transfected cells (Fig. 5D). As controls, cells transfected with mcherry alone showed similar levels of apoptotic cell death as that in STAT3 transfected cells (data not shown). This result indicated that expression of critical level of anti-apoptotic protein expression in presence of ubi-STAT3 FP protects against TNFα induced cytotoxicity.
4. Discussion
STAT3 is a cytokine inducible transcription factor regulated by site-specific PTMs to regulate acute-phase response, cell cycle progression and anti-apoptosis. Mono-ubiquitination is a molecular event that controls important cellular pathways and regulates complex nuclear functions of histones and non-histone proteins. In this study, we show for the first time, that STAT3 is mono-ubiquitinated in its NH2 terminal domain using specific mass spectrometry analysis, domain mapping and cell-based ubiquitination assays. Although polyubiquitination of STAT3 has been recently implicated in neonatal brain development and in T helper 17 cell development [16,43], the presence and effect of site-specific mono-ubiquitination of STAT3 protein is unknown. Here, we detected that the NH2 terminus of endogenous STAT3 is mono-ubiquitinated in cellulo and is under stimulus-dependent control. Analysis of the mono-ubiquitinated STAT3 mimic shows that this modification is associated with enhanced complex formation with activated P-TEFb; within the P-TEFb complex, BRD4 mediates STAT3-dependent cell cycle and anti-apoptotic gene expression programs.
Previously, the transcription factor Forkhead box O has been shown to become mono-ubiquitinated in response to increased cellular oxidative stress [21] with hydrogen peroxide. Similarly, we have observed here an increased STAT3 mono-ubiquitination upon treatment of cells with IL-6 and sIL-6R that are also known to induce oxidative stress in various cell types [44,45] and at the same time induce tyrosine phosphorylation, nuclear import, DNA binding and transcriptional complex assembly. More work will be required to understand how intracellular ROS controls mono-ubiquitination of these oxidative stress-regulated transcription factors.
Our mass spectrometry and western-based ubiquitination assays both show that K97 is a major mono-ubiquitination acceptor site in the STAT3 NH2 terminus. We had previously shown that among the three K residues (at 49, 87 and 97) in the NH2 terminus of STAT3, K 49 and K87 are the two major sites for P300 mediated acetylation [13]. To investigate if acetylation and mono-ubiquitination in STAT3 are two independent phenomena and occur in different K residues, we used STAT3 WT and STAT3K49/87R mutant in in vitro ubiquitination assay. From our observation that STAT3K49/87R is still mono-ubiquitinated, we concluded that ubiquitin conjugation occurs at K residue 97, and is independent of prior NH2 terminal acetylation (Fig. 2B). This conclusion was further directly confirmed by LC-MS/MS and LC-SRM-MS that K 97 residue is indeed the major mono-ubiquitination acceptor site in the STAT3 NH2 terminus. Interestingly, a ClustalW analysis further showed that K97 is found within an evolutionary conserved sumoylation/ubiquitination consensus site (ϕKXe) found in SRC3 [46], SHIP2 [39] and other mammalian proteins regulated by small ubiquitin modifiers.
Construction and ectopic expression of a monomeric ubiquitin-conjugated fusion protein has been used in determining the functions of various ubiquitinated proteins and transcription factors [22]. To address the functional consequences STAT3 monoubiquitination, we tested whether this PTM could modulate the transcriptional activity of STAT3. Our observation that the ectopic expression of ubi-STAT3 FP increases both basal and IL-6 mediated mRNA expression of STAT3 target gene, SOCS3 (Fig 4A), indicate that STAT3 monoubiquitination and its target gene expression is IL-6 inducible. We also determined whether increased STAT3 transcriptional activity correlates with increased nuclear import in endogenous fluorescent protein mcherry-conjugated ubi-STAT3 FP transfected HepG2 cells. We indeed observed an increase in monomeric cherry fluorescence of ubi-STAT3 FP in the nucleus of HepG2 cells upon IL-6 stimulation by confocal microscopy (data not shown). Further detailed study is required to confirm its role in nuclear import.
One striking finding of our present study is that we have observed for the first time by IP-SID-SRM-MS that mono-ubiquitinated STAT3 induces complex formation with BRD4. BRD4 is a chromatin adapter protein that binds to the acetylated histone H3 and H4 and can also bridge and enhance the binding of a sequence-specific transcription factor to chromatin and promote the stability via direct protein-protein interaction [47]. We previously showed that IL-6 strongly induces a nuclear complex of STAT3 with CDK9 in regulation of acute phase response gene and showed a direct evidence that IL-6 induces CDK9 recruitment to the γ-Fibrinogen promoter along with enhanced RNA Pol II and phospho-Ser2CTD Pol II loading on the coding region [29]. In this study we show that BRD4 is associated with mono-ubiquitinated STAT3 and is recruited to STAT3-dependent target genes. The strong transcriptional dependence of STAT3-dependent target genes on BRD4 binding suggests that BRD4 may also play a role with CDK9 on RNA Pol II CTD phosphorylation to induce transcriptional elongation. Alternatively, it could be possible that binding of BRD4 to mono-ubiquitinated STAT3 may induce a conformational changes in the NH2 terminus of STAT3 that may facilitates binding of the P300/CBP bromodomain to the NH2 terminal acetylated K49 and K87 leading to stable enhancer complex formation [48]. It will be of interest in future studies to determine if K97 mono-ubiquitination facilitates K49/87 acetylation. Furthermore, as Brd4 is also known to function as a nucleosome-binding factor that plays an active role in recruiting sequence-specific transcription factors to their chromatin target sites, it could be possible that BRD4 binding to mono-ubiquitinated STAT3 may also facilitate binding of STAT3 to the hyper-acetylated histone at the promoter of its target gene and thereby modulates gene activity.
Our findings on the recruitment of BRD4 to STAT3 dependent target genes and the inhibitory effect of JQ1 supports the conclusion that BRD4 is an essential component for STAT3-mediated transcription. It is interesting to note that JQ1 was also shown to downregulate MYC oncogene transcription in an experimental model of multiple myeloma [41]. Here it was shown that JQ1 downregulates the expression of MYC by dislocating BRD4 from the MYC promoter. Coincidently, we also recently showed that MYC expression is augmented in human post-burn hypertrophic scar fibroblast (HSF) leading to exaggerated proliferation of the fibroblast. Treatment of HSFs with STAT3 siRNA significantly decreased MYC expression in HSF but not in normal fibroblasts, indicating an important role of STAT3 in MYC expression and fibroblast proliferation [40]. In this study, when ubi-STAT3 FP was overexpressed, we observed an increase in MYC expression compared to unmodified STAT3 which indicated recruitment of BRD4 by mono-ubiquitinated STAT3. Our observation that treatment of these cells with JQ1 at much lower concentration attenuated MYC expression in ubi-STAT3 FP transfected cells further indicated an essential role of BRD4 in STAT3 mediated gene expression. Thus, BRD4 inhibitors may serve as a novel therapeutic tool to treat different STAT3-driven inflammatory and/or oncogenic diseases.
Furthermore, it is known that constitutive STAT3 expression plays an important role in tumorigenesis and proliferative diseases through the upregulation of genes involved in anti-apoptosis and proliferation. STAT3 activation can support cell survival through up-regulating the expression of the CCND1, BCL2L1, APEX1, SOD2, BCL2 and others. Our observation that mono-ubiquitinated STAT3 upregulated these anti-apoptotic and proliferation marker genes further indicates an essential role of mono-ubiquitination in cell survival. In this context, we also noted that when STAT3−/− MEFs were induced to undergo apoptosis by treatment with TNFα/CHX, cells that expressed ubi-STAT3 FP, were protected. We interpret these data to mean that mono-ubiquitinated STAT3 upregulates anti-apoptotic gene program, and confers protection against apoptotic cell death.
Taken together, our results suggest that PTM of STAT3 by NH2 terminal mono-ubiquitination plays an important regulatory mechanism for cellular proliferation and apoptosis. Mono-ubiquitination of STAT3 augments its transcriptional activity by recruiting bromodomain protein BRD4, a component of active PTEFb complex in RNA Pol II mediated transcriptional elongation. Therefore, identification of BRD4 as a novel regulator for STAT3 mono-ubiquitination not only provides new insights into STAT3 function but also provides potential strategies for the prevention and treatment of STAT3 driven proliferative diseases by targeting BRD4.
Highlights.
STAT3 undergoes PTM by monoubiquitination at NH2 terminal domain at K 97.
Monoubiquitinated STAT3 recruits BRD4 and regulates STAT3 mediated transcription.
BRD4 inhibitors may serve as a novel therapeutic approach to inhibit activated STAT3.
Acknowledgments
This work is supported in part by grants from NCATS UL1TR000071, NHLBI Proteomics Center for Airway Inflammation NIH-NHLBI-HHSN268201000037C, and NIEHS P30 ES006676.
Abbreviations
- BRD4
Bromodomain-containing protein 4
- CDK9
cyclin dependent kinase 9
- CHX
Cyclohexamide
- EV
empty vector
- FP
fusion protein
- Lys/K
Lysine
- mcherry
monomeric cherry fluorescent protein
- MEF
mouse embryonic fibroblast
- MS
mass spectrometry
- P-TEFb
positive elongation factor b
- PTM
post-translational modification
- SIS
stable isotope standard
- SRM
selected reaction monitoring
- S3
STAT3
- TNFα
tumor necrotic factor alpha
- ubi
ubiquitinated
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
Authors Contributions
S. Ray wrote the paper and designed and performed experiments; Y. Zhao and E.C. Edeh performed mass spectrometric analysis, C. Lee performed Ni-NTA affinity chromatography; M. Jamaluddin performed ChIP assay and A.R. Brasier supervised the research design, data interpretation and editing. All authors have read and commented on the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bromberg JF. Bioessays. 2001;23:161–169. doi: 10.1002/1521-1878(200102)23:2<161::AID-BIES1023>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 2.Darnell JE, Jr, Kerr IM, Stark GR. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 3.Darnell JE., Jr Science. 1997;277:1630–1635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- 4.Chiarle R, Simmons WJ, Cai H, Dhall G, Zamo A, Raz R, Karras JG, Levy DE, Inghirami G. Nat Med. 2005;11:623–629. doi: 10.1038/nm1249. [DOI] [PubMed] [Google Scholar]
- 5.Levy DE, Darnell JE., Jr Nat Rev Mol Cell Biol. 2002;3:651–662. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- 6.Jones SA, Richards PJ, Scheller J, Rose-John S. J Interferon Cytokine Res. 2005;25:241–253. doi: 10.1089/jir.2005.25.241. [DOI] [PubMed] [Google Scholar]
- 7.Bromberg JF, Horvath CM, Besser D, Lathem WW, Darnell JE., Jr Mol Cell Biol. 1998;18:2553–2558. doi: 10.1128/mcb.18.5.2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bromberg JF, Wrzeszczynska MH, Devgan G, Zhao Y, Pestell RG, Albanese C, Darnell JE., Jr Cell. 1999;98:295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- 9.Zhong Z, Wen Z, Darnell JE., Jr Science. 1994;264:95–98. doi: 10.1126/science.8140422. [DOI] [PubMed] [Google Scholar]
- 10.Darnell JE., Jr Recent Prog Horm Res. 1996;51:391–403. [PubMed] [Google Scholar]
- 11.Horvath CM, Wen Z, Darnell JE., Jr Genes Dev. 1995;9:984–994. doi: 10.1101/gad.9.8.984. [DOI] [PubMed] [Google Scholar]
- 12.Yang J, Huang J, Dasgupta M, Sears N, Miyagi M, Wang B, Chance MR, Chen X, Du Y, Wang Y, An L, Wang Q, Lu T, Zhang X, Wang Z, Stark GR. Proc Natl Acad Sci U S A. 2010;107:21499–21504. doi: 10.1073/pnas.1016147107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ray S, Boldogh I, Brasier AR. Gastroenterology. 2005;129:1616–1632. doi: 10.1053/j.gastro.2005.07.055. [DOI] [PubMed] [Google Scholar]
- 14.Yuan ZL, Guan YJ, Chatterjee D, Chin YE. Science. 2005;307:269–273. doi: 10.1126/science.1105166. [DOI] [PubMed] [Google Scholar]
- 15.Perry E, Tsruya R, Levitsky P, Pomp O, Taller M, Weisberg S, Parris W, Kulkarni S, Malovani H, Pawson T, Shpungin S, Nir U. Oncogene. 2004;23:8908–8919. doi: 10.1038/sj.onc.1208149. [DOI] [PubMed] [Google Scholar]
- 16.Tanaka T, Yamamoto Y, Muromoto R, Ikeda O, Sekine Y, Grusby MJ, Kaisho T, Matsuda T. Sci Signal. 2011;4:ra85. doi: 10.1126/scisignal.2001637. [DOI] [PubMed] [Google Scholar]
- 17.Hoeller D, Hecker CM, Wagner S, Rogov V, Dotsch V, Dikic I. Mol Cell. 2007;26:891–898. doi: 10.1016/j.molcel.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 18.Tanaka Y, Tanaka N, Saeki Y, Tanaka K, Murakami M, Hirano T, Ishii N, Sugamura K. Mol Cell Biol. 2008;28:4805–4818. doi: 10.1128/MCB.01784-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karpiuk O, Najafova Z, Kramer F, Hennion M, Galonska C, Konig A, Snaidero N, Vogel T, Shchebet A, Begus-Nahrmann Y, Kassem M, Simons M, Shcherbata H, Beissbarth T, Johnsen SA. Mol Cell. 2012;46:705–713. doi: 10.1016/j.molcel.2012.05.022. [DOI] [PubMed] [Google Scholar]
- 20.Bienko M, Green CM, Sabbioneda S, Crosetto N, Matic I, Hibbert RG, Begovic T, Niimi A, Mann M, Lehmann AR, Dikic I. Mol Cell. 2010;37:396–407. doi: 10.1016/j.molcel.2009.12.039. [DOI] [PubMed] [Google Scholar]
- 21.van der Horst A, de Vries-Smits AM, Brenkman AB, van Triest MH, van den Broek, Colland F, Maurice MM, Burgering BM. Nat Cell Biol. 2006;8:1064–1073. doi: 10.1038/ncb1469. [DOI] [PubMed] [Google Scholar]
- 22.Li M, Brooks CL, Wu-Baer F, Chen D, Baer R, Gu W. Science. 2003;302:1972–1975. doi: 10.1126/science.1091362. [DOI] [PubMed] [Google Scholar]
- 23.Johnsen SA. PLoS Genet. 2012;8:e1002860. doi: 10.1371/journal.pgen.1002860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoeller D, Crosetto N, Blagoev B, Raiborg C, Tikkanen R, Wagner S, Kowanetz K, Breitling R, Mann M, Stenmark H, Dikic I. Nat Cell Biol. 2006;8:163–169. doi: 10.1038/ncb1354. [DOI] [PubMed] [Google Scholar]
- 25.Dey A, Nishiyama A, Karpova T, McNally J, Ozato K. Mol Biol Cell. 2009;20:4899–4909. doi: 10.1091/mbc.E09-05-0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patel MC, Debrosse M, Smith M, Dey A, Huynh W, Sarai N, Heightman TD, Tamura T, Ozato K. Mol Cell Biol. 2013;33:2497–2507. doi: 10.1128/MCB.01180-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Devaiah BN, Lewis BA, Cherman N, Hewitt MC, Albrecht BK, Robey PG, Ozato K, Sims RJ, Singer DS. Proc Natl Acad Sci U S A. 2012;109:6927–6932. doi: 10.1073/pnas.1120422109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu SY, Chiang CM. J Biol Chem. 2007;282:13141–13145. doi: 10.1074/jbc.R700001200. [DOI] [PubMed] [Google Scholar]
- 29.Hou T, Ray S, Brasier AR. J Biol Chem. 2007;282:37091–37102. doi: 10.1074/jbc.M706458200. [DOI] [PubMed] [Google Scholar]
- 30.Sherman CT, Brasier AR. Mol Endocrinol. 2001;15:441–457. doi: 10.1210/mend.15.3.0609. [DOI] [PubMed] [Google Scholar]
- 31.Ray S, Lee C, Hou T, Bhakat KK, Brasier AR. Mol Endocrinol. 2010;24:391–401. doi: 10.1210/me.2009-0319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao Y, Zhang YY, Kho Y, Zhao Y. Anal Chem. 2004;76:1817–1823. doi: 10.1021/ac0354037. [DOI] [PubMed] [Google Scholar]
- 33.Zhao Y, Tian B, Edeh CB, Brasier AR. Mol Cell Proteomics. 2013;12:1513–1529. doi: 10.1074/mcp.M112.023465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao Y, Brasier AR. METHODS. 2013;61:313–322. doi: 10.1016/j.ymeth.2013.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao Y, Widen SG, Jamaluddin M, Tian B, Wood TG, Edeh CB, Brasier AR. Mol Cell Proteomics. 2011;10:M111. doi: 10.1074/mcp.M111.008771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Busso CS, Iwakuma T, Izumi T. Oncogene. 2009;28:1616–1625. doi: 10.1038/onc.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Safhi MM, Rutherford C, Ledent C, Sands WA, Palmer TM. Mol Pharmacol. 2010;77:968–978. doi: 10.1124/mol.109.062455. [DOI] [PubMed] [Google Scholar]
- 38.Li Y, Sun X, Elferich J, Shinde U, David LL, Dai M. J Biol Chem. 2014;289:5097–5108. doi: 10.1074/jbc.M113.533109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.De SJ, Guillabert A, Imbault V, Degraef C, Erneux C, Communi D, Pirson I. J Biol Chem. 2009;284:36062–36076. doi: 10.1074/jbc.M109.064923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ray S, Ju X, Sun H, Finnerty CC, Herndon DN, Brasier AR. J Invest Dermatol. 2013;133:1212–1220. doi: 10.1038/jid.2012.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, Kastritis E, Gilpatrick T, Paranal RM, Qi J, Chesi M, Schinzel AC, McKeown MR, Heffernan TP, Vakoc CR, Bergsagel PL, Ghobrial IM, Richardson PG, Young RA, Hahn WC, Anderson KC, Kung AL, Bradner JE, Mitsiades CS. Cell. 2011;146:904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jin S, Ray RM, Johnson LR. Am J Physiol Gastrointest Liver Physiol. 2008;294:G928–G937. doi: 10.1152/ajpgi.00219.2007. [DOI] [PubMed] [Google Scholar]
- 43.Murase S. J Biol Chem. 2013;288:20151–20161. doi: 10.1074/jbc.M113.470583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wassmann S, Stumpf M, Strehlow K, Schmid A, Schieffer B, Bohm M, Nickenig G. Circ Res. 2004;94:534–541. doi: 10.1161/01.RES.0000115557.25127.8D. [DOI] [PubMed] [Google Scholar]
- 45.Nieto N. Hepatology. 2006;44:1487–1501. doi: 10.1002/hep.21427. [DOI] [PubMed] [Google Scholar]
- 46.Wu RC, Feng Q, Lonard DM, O’Malley BW. Cell. 2007;129:1125–1140. doi: 10.1016/j.cell.2007.04.039. [DOI] [PubMed] [Google Scholar]
- 47.Lee AY, Chiang CM. J Biol Chem. 2009;284:2778–2786. doi: 10.1074/jbc.M805835200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hou T, Ray S, Lee C, Brasier AR. J Biol Chem. 2008;283:30725–30734. doi: 10.1074/jbc.M805941200. [DOI] [PMC free article] [PubMed] [Google Scholar]