Abstract
Epigenetic deregulation is intimately associated with the development of human diseases. Intensive studies are currently underway to clarify the mechanism for the sake of achieving ideal diagnostic and therapeutic goals. It has been demonstrated that enzymes with histone-modifying activities can also target non-histone proteins, with the underlying mechanism remaining obscure. In this review, we focus on a novel histone mimicry strategy that may be wildly adapted during the non-histone substrate recognition process. Its potential clinical implications are also discussed.
Keywords: epigenetics, histone mimicry
Epigenetics and human diseases: an overview
Coined as early as 1942, the term “epigenetics” is undoubtedly showing its popularity nowadays (Waddington, 2011). While the field of genetics focuses on the study of inherited genes, epigenetics deals with the issue beyond the genetic code: the epigenetic code, which has been introduced to describe the determinants of gene features other than the changes in DNA sequence (Wang et al., 2004; Turner, 2007; Jenuwein and Allis, 2001). More specifically, within this immense field we are looking into processes such as DNA and RNA methylation, protein modifications and chromatin structure remodeling, and digging out the underlying mechanism of how they cooperatively take control of development. With the advancing knowledge in epigenetics, we have been able to better appreciate the diversity and delicacy of gene expression regulation, which makes every single creature in one species distinct and unique. Among the various areas of epigenetics that embraces DNA, RNA and proteins, histone modification is perhaps the most dynamic. As the chief protein components of chromatin intimately wrapped by DNA, histones are subject to a wide variety of post-translational modifications such as phosphorylation, acetylation, ubiquitination and methylation, as well as their counterparts, i.e. dephosphorylation, deacetylation, deubiquitination and demethylation. Carried out by the corresponding enzymes on specific amino acid residues, these modifications function to affect histone-DNA affinity and chromatin structure, and provide docking sites for nuclear proteins with further chromatin structure modulation or transcription regulation activities, in such a way that they present and orchestrate epigenetic information to manipulate gene expression (Kouzarides, 2007).
Not surprisingly, epigenetic deregulation has been linked to many human diseases, including but not limited to diabetes (Villeneuve and Natarajan, 2010), asthma (Adcock et al., 2005), neuronal disorder (Urdinguio et al., 2009) and cancer (Jones and Baylin, 2007; Esteller, 2008). The very first case was documented in 1983 when researchers found an aberrant level of DNA methylation in colorectal cancer patient tissues (Feinberg and Vogelstein, 1983). Since then, a growing number of studies have demonstrated the contribution of abnormal DNA methylation to human diseases, a typical example being promoter hypermethylation and therefore silencing of tumor suppressor p16 frequently found in multiple human carcinomas (Shima et al., 2011). Besides DNA methylation, the far more diversified histone modifications play crucial roles in epigenetics and are often misrepresented in human diseases. The undesirable and disease-causing epigenetic patterns are reminiscent of anomalous chromatin regulator activities (Rodenhiser and Mann, 2006). In a recent example, Jiao and colleaguesidentified common genetic mutations in pancreatic neuroendocrine tumors, including mutations in MEN1 that encodes the transcription factor Menin with the H3K4me3 histone methyltransferase mixed-lineage leukemia (MLL) complex recruiting activity; and mutations in ATRX and DAXX that encode proteins to form a complex harboring chromatin-remodeling adenosine triphosphatase activity (Elsässer and Allis, 2011; Jiao et al., 2011). Attributed to the exome sequencing technology that reveals all protein-coding regions in the human genome, many chromatin-regulating candidates remain to be discovered.
The nature of diversity and specificity of enzymes with histone-modifying activities makes them ideal yet challenging therapeutic targets. While earlier research focused on their function on histones, as the initial name “histone-modifying” indicated, recent studies have confirmed their roles on nonhistone proteins, rendering their targets to be more universal (Morgunkova and Barlev, 2006; Lee and Stallcup, 2009; Egorova et al., 2010; Singh et al., 2010; Peng and Seto, 2011). This adds another layer to the still elusive mechanism governing the process of epigenetic regulation. What is the determinant, or driving force, of these enzymes to target certain groups of proteins? What is the biologic consequence of histones and nonhistone proteins “sharing” specific enzymes? Going further beyond the level of molecular biology, does this “sharing” give us any medical implications? In this review we put our focus on the recently emerged “histone mimicry” concept, which ideally targets these questions.
Beginning with the “histone fold” idea
As early as two decades ago, Arents and colleagues identified a common motif shared by four core histones when they performed crystallographic analysis to determine the structure of the histone octamer (Arents et al., 1991). They found that individual polypeptides in the central portion of histone chains are folded in a similar manner and named this helix-loop-helix motif the histone fold (Arents et al., 1991). Driven by previous findings that a couple of proteins such as TAFII40 and TAFII60 from Drosophila and macro-H2A from rat also contain histone-like components, Arents and colleagues reasoned that histone fold might also be prevalent in certain groups of nonhistone proteins. They performed computer-based motif searches and sequence analysis, and indeed confirmed the presence of the histone fold in a number of nonhistone proteins, most of which are involved in protein-protein or protein-DNA interactions (Baxevanis et al., 1995). These proteins, which include transcription factors and enzymes, were defined to form a distinct histone fold superfamily (Arents and Moudrianakis, 1995). Further research by Arents and colleagues demonstrated that the histone fold plays a role in DNA compaction and protein dimerization (Arents and Moudrianakis, 1995).
The histone fold superfamily represents the earliest example of histones and nonhistone proteins sharing a common motif. Recent studies, as reviewed below, have identified additional cases. It should be noted, however, that when a certain group of proteins share a common histone-like structure or sequence, they might appear to have significantly different biologic functions. In addition, it remains to be determined whether these nonhistone proteins are also subject to post-translational modifications by specific histone-modifying enzymes. How the modifications of the nonhistone proteins coordinate with those of histones to regulate the transcription process under certain biologic circumstances is worth further study. Several excellent reviews have covered the findings of histone-modifying enzymes targeting non-histone proteins, although the underlying mechanism is still unclear. In the following paragraphs we aim to encompass recent discoveries featuring a novel “histone-mimicking” mechanism used by nonhistone proteins to share with histones for enzyme recognition. Its biologic significance, as well as potential therapeutic implications, will also be discussed.
The introduction of the “histone mimicry” concept
While it has been over a century since histones were discovered, only recently did we begin to appreciate their roles in transcription regulation (Kayne et al., 1988). Noticeably, the histone code hypothesis was introduced one decade ago suggesting genetic information be regulated in part by histone modifications (Jenuwein and Allis, 2001). These modifications, as the hypothesis states, serve to recruit nuclear proteins with specialized recognition domains to regulate gene expression. As the expansion of the list of nonhistone proteins that are under post-translational modifications in a similar pattern to that of histones, a critical question surfaced: does the histone code hypothesis also apply to nonhistone proteins?
Recently, Sampath and colleagues addressed this question by providing a typical example in the name of G9a, which belongs to the Suv(3-9) family of SET domain histone methyltransferases and possesses histone H3 lysine 9 methylation abilities (Sampath et al., 2007). Through sequence alignment, they identified a short conserved sequence within the N-terminal of G9a as well as its family member GLP (G9a-like protein). Interestingly, they found that the sequence highly resembled the histone H3K9 methylation site. They continued to confirm that G9a was subject to auto-methylation at lysine 165 within the conserved sequence; and the auto-methylation was required for G9a to form corepressor complex with HP1γ, the euchromatic isoform of HP1 which was previously found to recognize methylated lysines at the histone H3 tail through its prototypical chromodomain. Nuclear magnetic resonance(NMR) spectroscopy analysis further defined that HP1γ binds methylated G9a with similar manner to that of HP1γ-methyl-H3K9.
Evidence presented by Sampath and colleagues offered several implications. First, there is a complicated interplay between histone and nonhistone methylation systems and one of their linkages would be the histone-like modification cassettes: the histone mimics. Second, the histone code hypothesis might go beyond histone and apply more universally to nonhistone proteins. Third, it might be possible to predict methylation targets based on their structure or sequence similarity to histone methylation sites. Indeed, the initial studies performed by Sampath and colleagues, as well as by others, have identified a number of proteins harboring H3K9-mimicking cassettes and are potential targets of lysine methylation. Many of these proteins are associated with chromatin regulation, including mAM (Sampath et al., 2007), which is a murine ATFa-associated factor and stimulates the activity of histone H3K9 methyltransferase ESET; as well as NSD1 and NSD3 (Henkels and Khorasanizadeh, 2007), which possess histone H3K36 methyltransferase activities. Last but certainly not least, the identification and structural analysis of histone methylation mimics may aid in the design of more potent and specific methyltransferase inhibitors, which are discussed in the later portion of this review. In sum, the introduction of the histone mimicry concept helps to improve our appreciation of epigenetic regulation network and diversify the epigenetic research methodologies.
Taking it further: the Snail-LSD1 story and beyond
The Snail transcription factor is well known as the master regulator of epithelial-mesenchymal transition (EMT) (Batlle et al., 2000; Cano et al., 2000; Savagner, 2001; Nieto, 2002; Zhou et al., 2004), a signature process during embryonic development as well as metastatic transformation (Thiery, 2002; Shook and Keller, 2003; Yang and Weinberg, 2008; Thiery et al., 2009). Loss of E-cadherin expression is a hallmark of EMT (Nieto, 2002; Thiery, 2002). Typically, Snail binds to E-cadherin promoter to repress its transcription. It should be noted that as a transcription factor that “read” DNAs, Snail itself does not have chromatin “editing” activities. To exert its function of transcriptional repression, it needs to cooperate with molecules harboring chromatin modulation activities, such as histone-modifying enzymes.
Using protein affinity-coupled mass spectrometry analysis, we recently identified lysine specific demethylase 1 (LSD1) as a partner of Snail (Lin et al., 2010). LSD1 is the first identified histone demethylase that specifically removes methylations of histone H3 lysine 4 (H3K4), a transcriptional activation mark (Shi et al., 2004). Overexpression of LSD1 has been correlated with an adverse clinical outcome for patients with ER-negative breast cancer (Lim et al., 2010). We speculated that Snail and LSD1 form a repressor complex to regulate E-cadherin expression. Interestingly, we found Snail interacts with LSD1 through its highly conserved N-terminal SNAG (Snail/Gfi-1) domain, the sequence of which strongly resembles that of histone H3 tail. Computer modeling analysis further revealed that the SNAG domain of Snail adopts a conformation that is superimposed by the histone tail and binds to the enzymatic cleft of LSD1. Noticeably, Arg3, Arg8 and Lys9 of the SNAG domain participated in critical contacts with LSD1, similar to the case of their counterparts Arg2, Arg8 and Lys9 of histone H3 (Lin et al., 2010). Based on our studies, we proposed a model in which Snail uses its histone H3-resembling SNAG domain as a molecular “hook” (or pseudo-substrate) to recruit LSD1 to the target gene promoters, where LSD1 demethylates histone H3K4 and suppresses transcription (Lin et al., 2010). Baron and colleagues performed a more detailed structural analysis of SNAG-LSD1 binding and found that the positively charged groups and hydroxyl side chains shared by N-terminal tails of Snail and H3 enable them to fit into the catalytic cavity of LSD1 in a similar conformation (Baron et al., 2011).
The histone H3-mimicry by SNAG may represent a common mechanism for some transcription factors to recruit chromatin-modifying enzymes and cofactors. Since the other members of the Snail/Scratch family also contain the SNAG domain, it is very likely that they interact with LSD1 in the similar fashion. In addition, it is possible that Snail is modified by LSD1 on its SNAG domain just like the histone H3 tail. To make an even more adventurous hypothesis, LSD1 cooperates with a potential lysine methyltransferase to define the methylation status of the SNAG domain of Snail: under specific biologic circumstances, one might become tightly bound to and modify SNAG while the other goes off, or vice versa. It would be tantalizing to characterize this dynamic “switch,” as well as specify the other potential cofactors involved in this regulatory process. Chan and colleagues recently reported an unbiased proteomic screen for histone H3 peptide-bound proteins (Chan et al., 2009). Through a sensitive pull-down approach in combination with LC-MS/ MS analysis, they identified 86 nuclear proteins that associated with histone H3 peptides harboring different lysine modifications (Chan et al., 2009). Given the resemblance between the histone H3 tail and the SNAG domain of Snail, we consider it reasonable to take a similar strategy to identify interacting proteins of SNAG with or without lysine methylations. The identification of the candidate proteins, followed by extensive structural analysis of the potential complexes, could provide valuable resources for the characterization of the Snail-involved epigenetic regulation network.
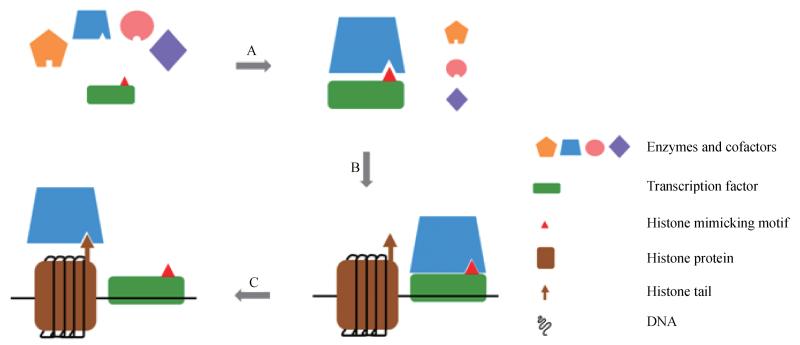
Shaking things up on an even broader scale, other families of transcription factors might harbor some yet-to-know histone-mimicking motif, through which they compete with histones for enzyme recognition. If that was the case, thosemotifs might first appear as inhibitors of the chromatin-modifying enzymes, preventing them from access to their bona fide targets: the histones. This possibility is somewhat confirmed through the SNAG-LSD1 structure analysis by Baron and colleagues (Baron et al., 2011). A fascinating question then remains: after binding and bringing enzymes to chromatin, how will the histone-mimicking motif get released to make room for histones? In the specific case of Snail, while we hypothesized that it is overabundant amount of histone H3 at the chromatin region that outcompetes the binding of SNAG, further experiments are required to address this issue (Lin et al., 2010). Overall, the Snail-LSD1 story advises us to think of histone mimicry in a more extensive and sophisticated setting (Fig. 1).
Figure 1.
A model of transcription regulation using “histone mimicry”. A) Transcription factor(s) interacts with a specific histone-modifying enzyme using its histone-mimicking motif. B) The transcription factor binds its target gene using the DNA-binding domain, thus recruiting the enzyme to the chromatin region. C) The overabundant amount of histone proteins at the chromatin region outcompetes the histone-mimicking motif for enzyme-binding, allowing the enzyme to modify histone and regulate target gene transcription.
Implications on drug development
As mentioned above, epigenetic deregulation is almost always associated with human diseases. Accordingly, epigenetic therapy, which aims to correct these defects by targeting specific epigenetic regulators, has emerged as a promising area of pharmacology given the fact that unlike genetic alterations, epigenetic aberrations are potentially reversible (Egger et al., 2004; Yoo and Jones, 2006; Kelly et al., 2010a). In 2004, Azacitidine, with the trade name Vidaza, became the first FDA-approved epigenetic drug to treat bone-marrow cancer and blood disorder (Issa et al., 2005). It is a cytidine analog and functions as a potent inhibitor of DNA methyltransferases. In addition, innovative drugs targeting enzymes involved in histone modifications are under development as well. Vorinostat (Zolinza, formerly known as SAHA), for example, is a histone deacetylase inhibitor that was approved by the FDA in 2006 to treat the rare cancer cutaneous T cell lymphoma (CTCL) (Marks and Breslow, 2007). Since then, research has endeavored to investigate more DNA methyltransferase inhibitors and histone deacetylase inhibitors, many of which are currently under clinical or preclinical tests.
Also potentiality exists that several histone methyltransferase inhibitors will enter clinical trials in the near future (Spannhoff et al., 2009). Recently, Chang and colleagues reported that adding a lysine mimic to an established G9a inhibitor BIX-01294 could enhance its potency in vitro and reduce cell toxicity in vivo (Chang et al., 2010). It remains to be evaluated whether a certain inhibitor is substrate-specific, in this very case, prefers to protect histones rather than G9a itself from methylation. Furthermore, Nicodeme and colleagues reported that I-BET, a synthetic compound mimicking acetylated histones, can bind the bromodomain and extra terminal domain (BET) proteins with high affinity, thus disrupting BET protein-mediated assembly of chromatin complexes responsible for the expression of key inflammatory genes (Nicodeme et al., 2010). In vivo studies further showed that I-BET treatment conferred protection effect and suppressed inflammation in mice (Nicodeme et al., 2010). While further efforts are still needed for the development of ideal small molecule inhibitors in specific pathological circumstances, histone mimicry-based compounds do show great promise. Clarification and systemical exploration of histone mimicry will cement the molecular basis for drug development and eventually accelerate the bench-to-bedside translation process. It would be definitely exciting, for example, to develop a SNAG-mimicking compound that inhibits Snail recruitment of its accomplice LSD1, and therefore denies pathogenic EMT.
Staying tuned: histone-mimicking goes global?
The abovementioned studies are highlighted by unprecedented findings of histone mimicry adapted by enzymes and transcription factors. We speculate the histone-mimicking strategy may be applied more broadly by the epigenetic system. A recent study on chromosome segregation dynamics during mitosis provides us with another example. In this particular study, Kelly and colleagues demonstrated that Thr3 phosphorylation on histone H3, a hallmark of mitosis, is recognized by the Survivin subunit of the chromosomal passenger complex (CPC) (Kelly et al., 2010b), which is an essential modulator of chromosome segregation (Ruchaud et al., 2007). Based on their results, this interaction mediates the recruitment of the CPC to chromosomes. Jeyaprakash and colleagues further demonstrated that a phospho-mimic N-terminal sequence, which is also found in several mitotic proteins including human Shugoshin 1, is responsible for Survivin binding (Jeyaprakash et al., 2011). The identification of the epitopes present in both the histone H3 and several mitotic proteins indicated their mutually exclusive interactions with the CPC (Jeyaprakash et al., 2011). Together, these studies offer us a clue: histone mimicry also applies to mitotic-associated proteins and makes a contribution to the fine-tuning of the cell division process.
The good news is that the histone mimicry concept is becoming more recognized. It gives us a fresh glimpse of the amazing post-translational modification network and stretches our brains to think beyond the chromatin-based epigenetic landscape. Even in the world of microorganisms, histone mimicry may be applied by some pathogens to compete with the host chromatin for histone readers or editors (Tarakhovsky, 2010). Given the presence of histone-mimicking proteins based on genome analysis, it remains to be seen, for example, whether a certain virus uses this strategy to hijack the host epigenetic program. Considering the far more complicated genomes of mammalians, especially we human beings, histone mimicry may lurk in much wilder territories than expected. As an up-and-comer in the flourishing field of epigenetic pharmaceutics, histone-mimicking compounds might work together with other therapeutic approaches to ideally target specific human diseases. With the deluge of modern molecular technology, more fascinating aspects about histone mimicry are sure to be unearthed.
Compliance with ethics guidelines
The authors Yiwei Lin and Binhua P. Zhou declare that they have no conflict of interest. This review article does not contain any unpublished data of human or animal studies.
Acknowledgements
We thank Dr. Nathan L. Vanderford for critical reading and editing of this manuscript. This work was supported by grants from NIH (RO1CA125454), Susan G Komen Foundation (KG081310), Mary Kay Ash Foundation (to B.P. Zhou) and pre-doctoral fellowship (BC101068) from DoD Breast Cancer Research Program (to Y. Lin).
References
- Adcock IM, Ito K, Barnes PJ. Histone deacetylation: an important mechanism in inflammatory lung diseases. COPD. 2005;2(4):445–455. doi: 10.1080/15412550500346683. [DOI] [PubMed] [Google Scholar]
- Arents G, Burlingame RW, Wang BC, Love WE, Moudrianakis EN. The nucleosomal core histone octamer at 3.1 A resolution: a tripartite protein assembly and a left-handed superhelix. Proc Natl Acad Sci USA. 1991;88(22):10148–10152. doi: 10.1073/pnas.88.22.10148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arents G, Moudrianakis EN. The histone fold: a ubiquitous architectural motif utilized in DNA compaction and protein dimerization. Proc Natl Acad Sci USA. 1995;92(24):11170–11174. doi: 10.1073/pnas.92.24.11170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron R, Binda C, Tortorici M, McCammon JA, Mattevi A. Molecular mimicry and ligand recognition in binding and catalysis by the histone demethylase LSD1-CoREST complex. Structure. 2011;19(2):212–220. doi: 10.1016/j.str.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batlle E, Sancho E, Francí C, Domínguez D, Monfar M, Baulida J, García De Herreros A. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat Cell Biol. 2000;2(2):84–89. doi: 10.1038/35000034. [DOI] [PubMed] [Google Scholar]
- Baxevanis AD, Arents G, Moudrianakis EN, Landsman D. A variety of DNA-binding and multimeric proteins contain the histone fold motif. Nucleic Acids Res. 1995;23(14):2685–2691. doi: 10.1093/nar/23.14.2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cano A, Pérez-Moreno MA, Rodrigo I, Locascio A, Blanco MJ, del Barrio MG, Portillo F, Nieto MA. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat Cell Biol. 2000;2(2):76–83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- Chan DW, Wang Y, Wu M, Wong J, Qin J, Zhao Y. Unbiased proteomic screen for binding proteins to modified lysines on histone H3. Proteomics. 2009;9(9):2343–2354. doi: 10.1002/pmic.200800600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y, Ganesh T, Horton JR, Spannhoff A, Liu J, Sun A, Zhang X, Bedford MT, Shinkai Y, Snyder JP, Cheng X. Adding a lysine mimic in the design of potent inhibitors of histone lysine methyltransferases. J Mol Biol. 2010;400(1):1–7. doi: 10.1016/j.jmb.2010.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429(6990):457–463. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- Egorova KS, Olenkina OM, Olenina LV. Lysine methylation of nonhistone proteins is a way to regulate their stability and function. Biochemistry (Mosc) 2010;75(5):535–548. doi: 10.1134/s0006297910050019. [DOI] [PubMed] [Google Scholar]
- Elsässer SJ, Allis CD, Lewis PW. Cancer. New epigenetic drivers of cancers. Science. 2011;331(6021):1145–1146. doi: 10.1126/science.1203280. [DOI] [PubMed] [Google Scholar]
- Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358(11):1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- Feinberg AP, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983;301(5895):89–92. doi: 10.1038/301089a0. [DOI] [PubMed] [Google Scholar]
- Henkels CH, Khorasanizadeh S. Implications of a histone code mimic in epigenetic signaling. Mol Cell. 2007;27(4):521–522. doi: 10.1016/j.molcel.2007.08.002. [DOI] [PubMed] [Google Scholar]
- Issa JP, Kantarjian HM, Kirkpatrick P. Azacitidine. Nat RevDrug Discov. 2005;4(4):275–276. doi: 10.1038/nrd1698. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293(5532):1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Jeyaprakash AA, Basquin C, Jayachandran U, Conti E. Structural basis for the recognition of phosphorylated histone h3 by the survivin subunit of the chromosomal passenger complex. Structure. 2011;19(11):1625–1634. doi: 10.1016/j.str.2011.09.002. [DOI] [PubMed] [Google Scholar]
- Jiao Y, Shi C, Edil BH, de Wilde RF, Klimstra DS, Maitra A, Schulick RD, Tang LH, Wolfgang CL, Choti MA, Velculescu VE, Diaz LA, Jr, Vogelstein B, Kinzler KW, Hruban RH, Papadopoulos N. DAXX/ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science. 2011;331(6021):1199–1203. doi: 10.1126/science.1200609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128(4):683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayne PS, Kim UJ, Han M, Mullen JR, Yoshizaki F, Grunstein M. Extremely conserved histone H4 N terminus is dispensable for growth but essential for repressing the silent mating loci in yeast. Cell. 1988;55(1):27–39. doi: 10.1016/0092-8674(88)90006-2. [DOI] [PubMed] [Google Scholar]
- Kelly AE, Ghenoiu C, Xue JZ, Zierhut C, Kimura H, Funabiki H. Survivin reads phosphorylated histone H3 threonine 3 to activate the mitotic kinase Aurora B. Science. 2010b;330(6001):235–239. doi: 10.1126/science.1189505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly TK, De Carvalho DD, Jones PA. Epigenetic modifications as therapeutic targets. Nat Biotechnol. 2010a;28(10):1069–1078. doi: 10.1038/nbt.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128(4):693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Lee YH, Stallcup MR. Minireview: protein arginine methylation of nonhistone proteins in transcriptional regulation. Mol Endocrinol. 2009;23(4):425–433. doi: 10.1210/me.2008-0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim S, Janzer A, Becker A, Zimmer A, Schüle R, Buettner R, Kirfel J. Lysine-specific demethylase 1 (LSD1) is highly expressed in ER-negative breast cancers and a biomarker predicting aggressive biology. Carcinogenesis. 2010;31(3):512–520. doi: 10.1093/carcin/bgp324. [DOI] [PubMed] [Google Scholar]
- Lin Y, Wu Y, Li J, Dong C, Ye X, Chi YI, Evers BM, Zhou BP. The SNAG domain of Snail1 functions as a molecular hook for recruiting lysine-specific demethylase 1. EMBO J. 2010;29(11):1803–1816. doi: 10.1038/emboj.2010.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks PA, Breslow R. Dimethyl sulfoxide to vorinostat: development of this histone deacetylase inhibitor as an anticancer drug. Nat Biotechnol. 2007;25(1):84–90. doi: 10.1038/nbt1272. [DOI] [PubMed] [Google Scholar]
- Morgunkova A, Barlev NA. Lysine methylation goes global. Cell Cycle. 2006;5(12):1308–1312. doi: 10.4161/cc.5.12.2820. [DOI] [PubMed] [Google Scholar]
- Nicodeme E, Jeffrey KL, Schaefer U, Beinke S, Dewell S, Chung CW, Chandwani R, Marazzi I, Wilson P, Coste H, White J, Kirilovsky J, Rice CM, Lora JM, Prinjha RK, Lee K, Tarakhovsky A. Suppression of inflammation by a synthetic histone mimic. Nature. 2010;468(7327):1119–1123. doi: 10.1038/nature09589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto MA. The snail superfamily of zinc-finger transcription factors. Nat Rev Mol Cell Biol. 2002;3(3):155–166. doi: 10.1038/nrm757. [DOI] [PubMed] [Google Scholar]
- Peng L, Seto E. Deacetylation of nonhistone proteins by HDACs and the implications in cancer. Handb Exp Pharmacol. 2011;206:39–56. doi: 10.1007/978-3-642-21631-2_3. [DOI] [PubMed] [Google Scholar]
- Rodenhiser D, Mann M. Epigenetics and human disease: translating basic biology into clinical applications. CMAJ. 2006;174(3):341–348. doi: 10.1503/cmaj.050774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruchaud S, Carmena M, Earnshaw WC. Chromosomal passengers: conducting cell division. Nat Rev Mol Cell Biol. 2007;8(10):798–812. doi: 10.1038/nrm2257. [DOI] [PubMed] [Google Scholar]
- Sampath SC, Marazzi I, Yap KL, Sampath SC, Krutchinsky AN, Mecklenbräuker I, Viale A, Rudensky E, Zhou MM, Chait BT, Tarakhovsky A. Methylation of a histone mimic within the histone methyltransferase G9a regulates protein complex assembly. Mol Cell. 2007;27(4):596–608. doi: 10.1016/j.molcel.2007.06.026. [DOI] [PubMed] [Google Scholar]
- Savagner P. Leaving the neighborhood: molecular mechanisms involved during epithelial-mesenchymal transition. Bioessays. 2001;23(10):912–923. doi: 10.1002/bies.1132. [DOI] [PubMed] [Google Scholar]
- Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119(7):941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Shima K, Nosho K, Baba Y, Cantor M, Meyerhardt JA, Giovannucci EL, Fuchs CS, Ogino S. Prognostic significance of CDKN2A (p16) promoter methylation and loss of expression in 902 colorectal cancers: Cohort study and literature review. Int J Cancer. 2011;128(5):1080–1094. doi: 10.1002/ijc.25432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shook D, Keller R. Mechanisms, mechanics and function of epithelial-mesenchymal transitions in early development. Mech Dev. 2003;120(11):1351–1383. doi: 10.1016/j.mod.2003.06.005. [DOI] [PubMed] [Google Scholar]
- Singh BN, Zhang G, Hwa YL, Li J, Dowdy SC, Jiang SW. Nonhistone protein acetylation as cancer therapy targets. Expert Rev Anticancer Ther. 2010;10(6):935–954. doi: 10.1586/era.10.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spannhoff A, Hauser AT, Heinke R, Sippl W, Jung M. The emerging therapeutic potential of histone methyltransferase and demethylase inhibitors. ChemMedChem. 2009;4(10):1568–1582. doi: 10.1002/cmdc.200900301. [DOI] [PubMed] [Google Scholar]
- Tarakhovsky A. Tools and landscapes of epigenetics. Nat Immunol. 2010;11(7):565–568. doi: 10.1038/ni0710-565. [DOI] [PubMed] [Google Scholar]
- Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2(6):442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139(5):871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Turner BM. Defining an epigenetic code. Nat Cell Biol. 2007;9(1):2–6. doi: 10.1038/ncb0107-2. [DOI] [PubMed] [Google Scholar]
- Urdinguio RG, Sanchez-Mut JV, Esteller M. Epigenetic mechanisms in neurological diseases: genes, syndromes, and therapies. Lancet Neurol. 2009;8(11):1056–1072. doi: 10.1016/S1474-4422(09)70262-5. [DOI] [PubMed] [Google Scholar]
- Villeneuve LM, Natarajan R. The role of epigenetics in the pathology of diabetic complications. Am J Physiol Renal Physiol. 2010;299(1):F14–F25. doi: 10.1152/ajprenal.00200.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddington CH. The epigenotype. Int J Epidemiol. 2011 doi: 10.1093/ije/dyr184. online available December 20, 2011. [DOI] [PubMed] [Google Scholar]
- Wang Y, Fischle W, Cheung W, Jacobs S, Khorasanizadeh S. Beyond the double helix: writing and reading the histone code. Novartis Found Symp. 2004;259:3–17. [PubMed] [Google Scholar]
- Yang J, Weinberg RA. Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev Cell. 2008;14(6):818–829. doi: 10.1016/j.devcel.2008.05.009. [DOI] [PubMed] [Google Scholar]
- Yoo CB, Jones PA. Epigenetic therapy of cancer: past, present and future. Nat Rev Drug Discov. 2006;5(1):37–50. doi: 10.1038/nrd1930. [DOI] [PubMed] [Google Scholar]
- Zhou BP, Deng J, Xia W, Xu J, Li YM, Gunduz M, Hung MC. Dual regulation of Snail by GSK-3beta-mediated phosphorylation in control of epithelial-mesenchymal transition. Nat Cell Biol. 2004;6(10):931–940. doi: 10.1038/ncb1173. [DOI] [PubMed] [Google Scholar]