Abstract
The tumor suppressor gene RASSF1A is inactivated through point mutation or promoter hypermethylation in many human cancers. In this study, we performed a Sleeping Beauty transposon-mediated insertional mutagenesis screen in Rassf1a null mice to identify candidate genes that collaborate with loss of Rassf1a in tumorigenesis. We identified 10 genes, including the transcription factor Runx2, a transcriptional partner of Yes-associated protein (YAP1) that displays tumor suppressive activity through competing with the oncogenic TEA domain family of transcription factors (TEAD) for YAP1 association. While loss of RASSF1A promoted the formation of oncogenic YAP1-TEAD complexes, the combined loss of both RASSF1A and RUNX2 further increased YAP1-TEAD levels, demonstrating that loss of RASSF1A, together with RUNX2, is consistent with the multi-step model of tumorigenesis. Clinically, RUNX2 expression was frequently down-regulated in various cancers, and reduced RUNX2 expression was associated with poor survival in patients with diffuse large B-cell or atypical Burkitt’s/Burkitt’s-like lymphomas. Interestingly, decreased expression levels of RASSF1 and RUNX2 were observed in both precursor T-cell acute lymphoblastic leukemia and colorectal cancer, further supporting the hypothesis that dual regulation of YAP1-TEAD promotes oncogenic activity. Together, our findings provide evidence that loss of RASSF1A expression switches YAP1 from a tumor suppressor to an oncogene through regulating its association with transcription factors, thereby suggesting a novel mechanism for RASSF1A-mediated tumor suppression.
Keywords: RASSF1A, RUNX2, YAP, tumor suppressor, transposon, hippo
INTRODUCTION
The RASSF1A tumor suppressor exhibits epigenetic or genetic inactivation in the majority of human tumors and plays a role in a variety of key biological processes that restrain the development of cancer, including apoptosis, cell cycle regulation, mitosis, and microtubule dynamics (reviewed in 1, 2). Although the precise mechanisms by which RASSF1A functions as a tumor suppressor are still under investigation, the most likely hypothesis is that it serves as a scaffold to modulate, localize, and perhaps integrate multiple tumor suppressor pathways. The components of these tumor suppressor pathways remain largely unknown; however the hippo pathway is a key downstream pathway that also restricts tumorigenesis.
The first four components of the hippo pathway were discovered in genetic screens for tumor suppressor genes in Drosophila, and include the NDR family protein kinase Warts (Wts), the WW domain-containing protein Salvador (Sav), the Ste20-like protein kinase Hippo (Hpo) and the adaptor protein Mob-as-tumor-suppressor (Mats) (reviewed in 3). Loss-of-function mutant clones for any of these four genes lead to a strong tissue overgrowth phenotype characterized by increased proliferation and diminished cell death. Biochemically, these four tumor suppressors form a kinase cascade in which the Hpo-Sav kinase complex phosphorylates and activates the Wts-Mats kinase complex (4, 5), to restrict proliferation via inactivation of the transcriptional complex formed by Yorkie (Yki) and Scalloped (Sd) (6, 7).
The hippo pathway is conserved in mammalian systems (reviewed in 3) with MST and LATS kinases (orthologs of Hippo and Warts respectively) functioning as tumor suppressors that phosphorylate the mammalian homolog of Yorkie, yes-associated protein YAP1 (8). The regulation of the Yki-Sd (YAP1-TEAD) complex by the hippo pathway is similarly conserved, being responsible for restricting YAP-induced overgrowth, epithelial–mesenchymal transition (EMT), and oncogenic transformation (7-9). However, in mammals YAP1 displays a more pleiotropic role serving as a coactivator of multiple transcription factors such as p73 (affecting tumor suppression) (10), ErbB4 (11) and RUNX2 (inducing differentiation) (12).
RASSF1A is an upstream component of the MST/LATS pathway, as it binds MST kinases and promotes active YAP1/p73 transcriptional complexes (13, 14). Therefore, hippo pathway activation leads to opposing effects on the tumor suppressive YAP1/p73 and oncogenic YAP1/TEAD transcription factor complexes. Loss of RASSF1A in tumors leads to a failure in formation of YAP1/p73 complexes and concomitantly, Rassf1a homozygous null mice die faster than wildtype controls due to an increased incidence of tumor formation (15). Herein we describe how deregulation of oncogenic YAP1-TEAD complexes can contribute to the tumor suppressor functions of RASSF1A. Moreover, the decrease in tumor latency following exposure to mutagens (15) suggests that additional genes collaborate with loss of Rassf1a in tumorigenesis. Thus to identify these cooperating genetic events, we performed a Sleeping Beauty transposon-mediated insertional mutagenesis screen in Rassf1a null mice. This analysis allowed us to identify 10 genes potentially associated with tumor formation in the context of loss of Rassf1a. We selected the YAP1 transcriptional partner, Runx2, for follow-up analysis and show that loss of RUNX2 further enhances YAP1-TEAD complex levels initiated by loss of RASSF1A. Thus we provide evidence for RASSF1A-dependent switching of YAP1 between transcription factor complexes that regulate proliferation (TEAD), differentiation (RUNX2) and tumor suppression (p73), providing new insights into RASSF1A-mediated tumor suppression.
MATERIALS & METHODS
Mice and genotyping
Generation of the Rassf1a null mice (Rassf1aBrdm2) (15), mice carrying the SB transposon array (T2/Onc) (16), and mice carrying the SB transposase (Rosa26SB11) (17) have been described previously. All mice were on a mixed 129/Sv-C57BL/6J background. Mice were housed in accordance with Home Office regulations (UK), and fed a diet of mouse pellets and water ad libitum. PCR genotyping for the Rassf1a (15), T2/Onc (16), and Rosa26SB11 (17) alleles, as well ‘excision’ of the transposon from the donor array (16) was performed as described previously.
Tumor watch analysis
Heterozygous Rassf1a (Rassf1a+/−) mice were bred with mice carrying either the Sleeping Beauty transposon (T2/OncTg/Tg) or transposase (Rosa26 SB11/SB11). The resulting offspring (Rassf1a+/−, T2/Onc+/Tg, Rosa26+/SB11) were intercrossed to generate offspring of each genotype (Rassf1a+/+, T2/Onc+/Tg, Rosa26+/SB11 and Rassf1a−/−, T2/Onc+/Tg, Rosa26+/SB11), which were subsequently placed on tumor watch from birth. All mice on tumor watch were examined twice daily for signs of disease, at which time they were sacrificed and a full necropsy performed.
Histology and immunohistochemistry
Tissues were fixed in 10% neutral-buffered formalin (NBF) at room temperature overnight. Samples were then transferred to 50% ethanol, embedded in paraffin, sectioned and stained with hematoxylin and eosin (H&E). Immunophenotyping was performed on formalin-fixed, paraffin-embedded tissue sections that had undergone antigen retrieval (microwaving in citrate buffer pH 6 for 20 min) using antibodies for CD3 (clone SP7; Abcam, Cambridge, UK), CD45R/B220 (clone RA3-6B2, R&D systems) and MPO (DAKO, Ely, UK). Immunohistochemical signal was detected by secondary biotinylated goat anti-rabbit antibody (Vector Laboratories, Burlingame, CA), followed by Vectorstain Elite ABC kit (Vector Laboratories) according to the manufacturer’s instructions.
Isolation and statistical analysis of transposon insertion sites
Isolation of the transposon insertion sites from tumors of both cohorts was performed using splinkerette PCR to produce barcoded PCR products that were pooled and sequenced as described previously (18). The pooled PCRs were sequenced on the 454 GS-FLX platform (Roche) over four separate lanes, with one lane per restriction enzyme and a maximum of 48 tumors per lane. Processing of 454 reads, identification of insertion sites, and the Gaussian Kernel Convolution statistical methods used to identify common insertion sites (CIS) have been described previously (18, 19). The P-value for each CIS was calculated using an adjusted-by-chromosome cut-off of P<0.05. CIS on mouse chromosome 1 were not reported as this is the ‘donor chromosome’ where the transposon array is located and as such there is a significantly higher than background level of transposon insertion events due to local hoping which complicates CIS analysis (16). Genotype-specific CIS analysis was performed by i) calling CISs on a per-chromosome basis with a cut-off of P<0.1 and by comparing the CIS calls between groups to identify a discovery set of insertions, and ii) by pooling genotypes together, calling CISs on a per-chromosome basis and then deconvoluting the CIS peaks using the Fishers Exact test to identify genotypes enriched at each CIS peak. Only CISs that survived both calling methods were listed as ‘Rassf1a−/−-specific CIS’.
Reagents and cells
U2OS cells (ATCC® HTB-96™) and HCT116 cells were grown in DMEM containing 10% fetal calf serum (Gibco). Transient transfection used Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. U2OS Tet-On cells (Clontech) that inducibly express FLAG-RASSF1A upon doxycycline induction were established following puromycin selection as described in the manufacturer’s protocol (20). Authentication of the cell lines was provided with their purchase from ATCC and Clontech, and the cell lines were cultured from the original stocks and maintained for no longer than 2 months.
siRNA
50 ng/mL siRNA duplexes either non-targeting or targeted against RUNX2, RASSF1A, TEAD or p73 were transfected with Lipofectamine 2000. In order to avoid unspecific effects, several different siRNAs were used for the knockdown of each protein. For detailed information, see Supplementary Table S1.
Cell assays. Colony forming
U2OS cells were transfected with siRNAs and 24 h later trypsinized and replated in 6 cm dishes at a density of 350 cells/dish. Plates were stained with crystal violet (0.5% w/v crystal violet, 50% v/v methanol and 10% v/v ethanol) 11 days later and colonies were counted.
Viability
U2OS cells were transfected with siRNA and 24 h later trypsinized and replated in 6-well dishes at a density of 7.5 × 103/well. Cell viability was determined using the resazurin assay. The cells were incubated with media containing 10ug/ml resazurin (Sigma) at 37°C in a humidified 5% CO2 in-air atmosphere for 2 h. Resazurin reduction was then measured fluorometrically using a plate reader (Wallace Perkin Elmer) at excitation 530nm and emission wavelength 590nm.
Proliferation
Growth curves were measured by plating 5 × 103 cells onto specialized conductance plates and growth was determined as a steady reduction in individual well conductivity in real time on an xcelligence system (Roche).
Immunoprecipitation and immunoblotting
Whole cell lysate preparation, immunoprecipitation and western blotting were performed as previously described (13, 14). Nuclei were isolated as previously described (21) prior to incubation in lysis buffer (150 mM NaCl, 20 mM HEPES pH 7.5, 0.5 mM EDTA, 1% NP-40). Antibodies were used for RUNX2 (M-70; Santa Cruz Biotechnology), YAP1 (H125; Santa Cruz Biotechnology), RASSF1A (3F3; Santa Cruz Biotechnology), RASSF1 (Epitomics), TEAD (TEF-1; BD Biosciences), p73 (Epitomics), FLAG (Stratagene), GAPDH (Epitomics), Hsp70 (W27; Santa Cruz Biotechnology) and Lamin B1 (Abcam).
Bioinformatic meta-analysis of RUNX2 and RASSF1 expression
Microarray expression data from six independent data sets were downloaded from the Oncomine repository (http://www.oncomine.org/) to examine the relative mRNA expression levels of RUNX2 between normal and cancer samples in a variety of tissue types. The distributions of log2 median-centered signal intensities were plotted using boxplots and differential gene expression was computed using the Welch two sample t-test, which is appropriate for subsets of unequal variances. Only tumor sets showing the same differential mode of expression in at least three independent datasets were included in this analysis. To correlate gene expression of RUNX2 with patient survival, a Univariate Cox proportional hazard regression model (22) was applied to a lymphoma dataset of n=272 samples and the Likelihood ratio test, Wald test, and Score (logrank) test were all used to compute the P value (P <5.3×10−7 for all three tests). To visualize the result obtained from the survival analysis, the samples were ranked according to RUNX2 gene expression and Kaplan-Meier survival curves were plotted for lymphomas with the lowest (<25th percentile) versus highest (>25th percentile) RUNX2 expression giving a P value of 1.87×10−6 (logrank test). The expression microarray data of bone marrow from precursor T-cell lymphoblastic leukemias (Array ID: E-MEXP-313, n=27) and the human colo-rectal data series (Array ID: GSE5206, n=105) were downloaded from ArrayExpress and Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) respectively, and the data were normalized using the RMA (Robust Multi-Array) method. Pearson’s correlation coefficient analysis was performed to correlate RUNX2 and RASSF1 expression and the P value was computed using an asymptotic confidence interval based on Fisher’s Z transform. The samples were clustered using the Euclidean distance metric and the complete linkage algorithm. Full details of the specific microarray data used are supplied in Supplementary Table S2).
RESULTS
Tumor watch analysis
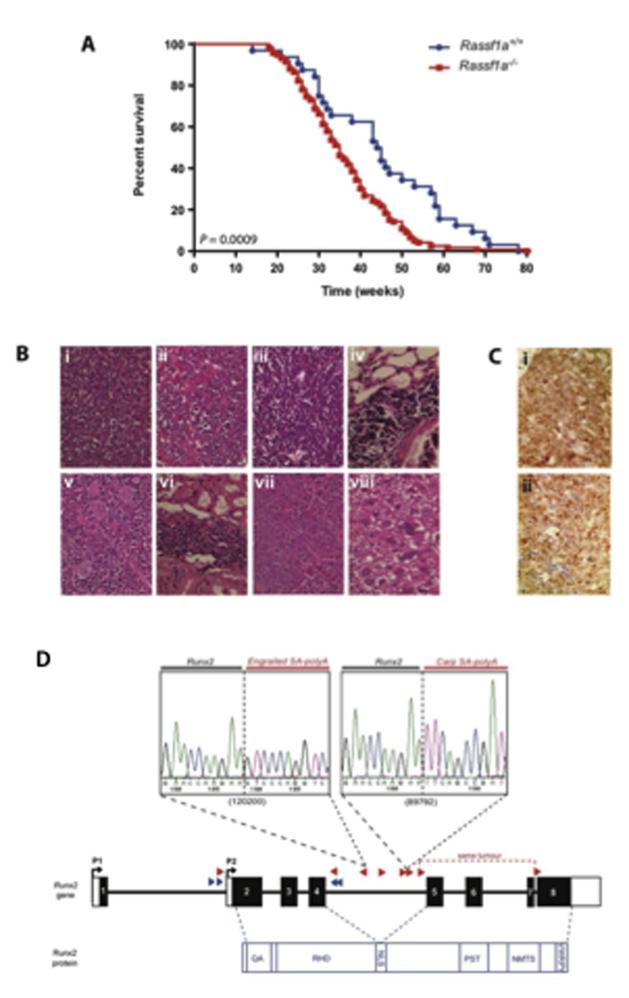
Mice homozygous or wildtype for the Rassf1aBrdm2 allele (hereafter referred to as Rassf1a−/− or Rassf1a+/+ mice, respectively) with Sleeping Beauty (SB) transposition occurring (i.e., on a T2/Onc+/Tg, Rosa26+/SB11 background) were aged until they became moribund. Rassf1a−/− SB mice developed tumors significantly faster than their wildtype SB littermates (average lifespan of 35 and 44 weeks for Rassf1a−/− and Rassf1a+/+ mice, respectively; Figure 1a). As previously reported for the T2Onc transposon that carries the murine stem cell virus (MSCV) promoter that is preferentially active in the hematopoietic system (16), all mice in both cohorts developed leukemia/lymphoma (see Figure 1b), although a small proportion of mice did develop an additional tumor, typically an hepatocellular carcinoma (Figure 1b).
Figure 1. Loss of Rassf1a promotes tumorigenesis.
A. Kaplan-Meier curves showing the tumor latency for Rassf1a+/+ and Rassf1a−/− mice on a ‘jumping’ background (i.e., T2Onc+/Tg; Rosa+/SB) are significantly different using the Log-rank (Mantel-Cox) Test: P<0.0009. B. Photomicrographs of formalin-fixed, hematoxylin and eosin-stained sections of (i) a thymic lymphoma (ii) that metastasized to the liver; (iii) a splenic lymphoma (iv) that metastasized to the lung; a leukemia that infiltrated the (v) spleen and (vi) kidney; and a mouse that developed two independent tumors, specifically (vii) splenic lymphoma and (viii) hepatocellular carcinoma. C. Photomicrographs of an immunohistochemically-stained (i) lung section infiltrated by a lymphoma of T-cell origin (CD3-positive), and (ii) a spleen section infiltrated by myeloid leukemia (MPO-positive). All sections shown are representative and images are at x400 magnification. D. Seven of the Rassf1a−/− SB mice that developed leukemia/lymphoma carried transposons which had inserted into the Runx2 gene (indicated by the red triangles; direction of triangle indicates orientation of the transposon). The blue triangles are Rassf1a+/+ SB mice that developed leukemia/lymphoma and carried Runx2 insertions, although these insertions did not constitute a statistically significant common insertion site (CIS). Sequencing of the insertion-genome junction from splenic cDNA of two of these mice showed the splicing of Runx2 directly onto the splice acceptor (SA)-polyA from the transposon. The protein structure of RUNX2 is shown in blue and the key domains are labeled, including a glutamine/alanine (QA) rich tract, a Runt domain (RHD), a nuclear localization signal (NLS), a proline/serine/threonine (PST) rich tract, a nuclear matrix targeting signal (NMTS) and the C-terminal VWRPY domain for TLE/Groucho co-repressor interactions.
Immunohistochemical analysis of a selection of the leukemias/lymphomas showed the predominant disease subtypes were poorly differentiated lymphomas, not staining positively for either T-cell (CD3) or B-cell (CD45R/B220) antigens (29/69, 42%) and CD3-positive T-cell lymphomas (27/69, 39%), with only a small amount of MPO-positive high-grade myeloid leukemias (13/69, 19%; Figure 1c).
Statistical analysis of transposon insertion sites in leukemias/lymphomas
To identify tumor-associated genotype-enriched somatically mutated genes, i.e., those genes mutated by transposon insertion found specifically in tumors on a Rassf1a−/− background, we identified common insertion sites (CISs) in leukemias/lymphomas from 126 wildtype SB mice (25 Rassf1a+/+ SB mice and 101 wildtype mice from other SB studies carried out in our facility at the same time and on the same mixed C57-129 genetic background) and 111 Rassf1a−/− SB mice using Gaussian Kernel Convolution (GKC) (19) statistical analysis in two ways. Firstly, we treated the insertions from wildtype and Rassf1a−/− tumors as two independent groups and identified 209 and 165 CISs, respectively, which we then analysed to determine which were common to both groups and which were found specifically on a Rassf1a−/− background. Using a chromosome-adjusted P-value cut-off of P<0.1 ensured that an extra degree of stringency was implemented when detecting shared CISs. This meant that the CISs found in the Rassf1a−/− group were compared with the CIS in the wildtype group that were marginally P>0.05 (up to a significance of 0.1) and would otherwise have been missed. We then pooled the insertions from both groups (using a P value generated by the Fisher Exact Test), and identified CIS found to be ‘enriched’ only in the Rassf1a−/− tumors. The Rassf1a−/− CIS calls generated by both methods were compared, and resulted in the identification of 10 CISs that were present in both lists (Table 1).
Table 1. Gaussian kernel convolution (GKC) analysis of Rassf1a−/−-specific common insertion sites (CISs).
| CIS identification | Predicted affected gene | Other genes in CIS | Genomic location of CIS (chr:start-end) | Insertions (tumors) | P value |
|---|---|---|---|---|---|
|
| |||||
| CIS12:100543835_75k | Foxn3 | 3300002A11Rik | 12: 100477940-100609729 | 20 (16) | 0.02848 |
| CIS14:67677451_15k | Ppp2r2a | Bnip3l | 14: 67657900-67692812 | 11 (11) | 0.00026 |
| CIS5:34769677_15k | Fam193a | Rnf4, Sh3bp2, Tnip2 | 5: 34753537-34781415 | 9 (9) | 0.00261 |
| CIS12:93020818_15k | Ston2 | Sel1l | 12: 93004760-93032497 | 9 (9) | 0.01352 |
| CIS3:104653212_15k | Capza1 | Fam19a3, St7l, Mov10, Ppm1j, Rhoc, Wnt2b | 3: 104639966-104662043 | 8 (8) | 0.01081 |
| CIS17:44818488_30k | Runx2 | Supt3h | 17: 44801068-44830102 | 8 (7) | 0.03697 |
| CIS4:97738853_15k | Nfia | - | 4: 97727123-97749117 | 7 (7) | 0.00078 |
| CISX:39556669_15k | Stag2 | Xiap | X: 39546415-39563993 | 7 (7) | 0.01552 |
| CIS16:4187397_15k | Crebbp | Adcy9, Trap1 | 16: 4175786-4197556 | 7 (7) | 0.02029 |
| CIS5:34056588_15k | Fgfr3 | Fam53a, Letm1, Slbp, Tacc3, Tmem129 | 5: 34047785-34062457 | 7 (7) | 0.02839 |
Insertions in the Runx2 gene
Given that RASSF1A is a component of the hippo signaling pathway (13) and the transcription factor RUNX2 is activated by the Hippo pathway member, YAP1 (12), we focused our attention on ‘CIS17:44818488_30k’, which was predicted to affect the Runx2 gene. There were 8 insertions from a total of 7 tumors that contributed to ‘CIS17:44818488_30k’ (Figure 1d). Interestingly, 6/7 of the tumors had insertions in intron 4 of the gene and the transposons were in the reverse orientation, thus predicted to be truncating mutations. In agreement with this, analysis of cDNA from these tumors showed splicing of Runx2 directly onto the splice acceptor/polyA of the transposon (Figure 1d). This resulted in the premature truncation of the gene after exon 4 such that if a protein were translated, it would only contain the Runt domain, and not the activation domain (AD) or repression domain (RD), and thus be unable to carry out the functions of a full-length Runx2 protein. Importantly, the YAP1/RUNX2 protein interaction occurs between the ‘WW domain’ in YAP1 (23) and the ‘PY motif’ located within a 10 amino acid sequence (HTYLPPPYPG) in the C-terminal region of RUNX2 (12); Figure 1d. Thus premature truncation of the transcript before the PY motif would prevent Yap1/Runx2 complex formation.
Although these tumors contained only heterozygous loss of Runx2 (i.e., insertions were only in one Runx2 allele), Runx2 is haploinsufficient, as human cleidocranial dysplasia (CCD) is an autosomal dominant disease that results from heterozygous inactivation of RUNX2 (24), and heterozygous Runx2 mice recapitulate the CCD phenotype (25-26).
Loss of RUNX2 in the absence of RASSF1A enhances YAP-TEAD complex levels
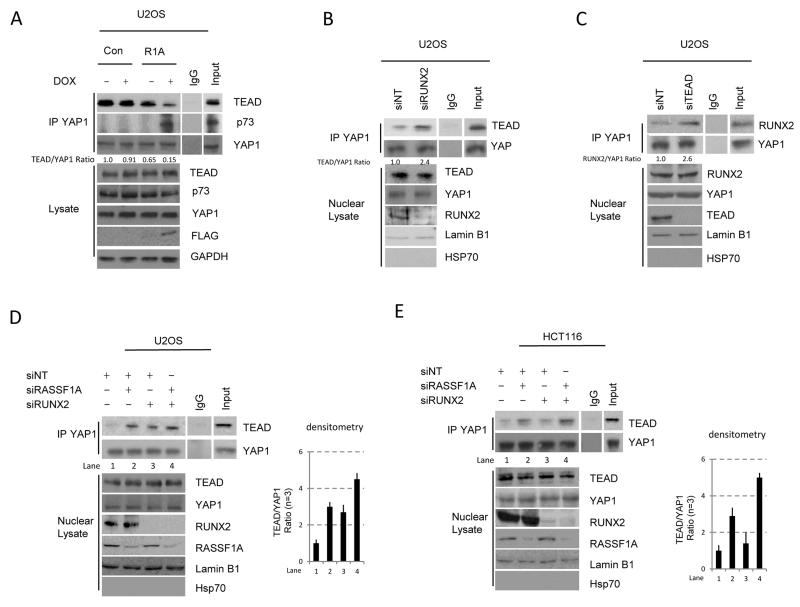
We first reasoned that the RASSF1A-mediated modulation of YAP1 that promotes the formation of YAP1-p73 complexes may occur at the expense of YAP1-TEAD complexes, thus serving as a comprehensive switch away from proliferation to active tumor suppression (20). As RUNX2 is also a transcriptional partner of YAP1, the enhanced tumorigenesis indicated by the Rassf1a−/− SB mice may be due to an additional layer of competition for YAP1 association. In tumors that have lost RASSF1A, YAP1 fails to associate with p73 and where this occurs in conjunction with RUNX2 loss, YAP1-TEAD complexes may be more likely and exacerbate oncogenic proliferation. We were unable to test this hypothesis in the leukemia/lymphoma samples containing transposon insertions in Runx2 due to the frequently oligoclonal or polyclonal nature of tumors induced by insertional mutagens (27). Thus, we utilized the RUNX2 expressing human osteosarcoma cell line U2OS, that has low levels of RASSF1A and in which the association of YAP1 with TEAD was readily observed (Figure 2a). Doxycycline-inducible expression of RASSF1A restricted the ability of YAP1 to associate with TEAD and promoted association with p73 (Figure 2a).
Figure 2. RASSF1A and RUNX2 limit the formation of YAP1-TEAD complexes.
A. Human osteosarcoma U2OS tet-on inducible cells expressing control vector (CON) or FLAG-RASSF1A (R1A) were treated with doxycycline (0.25 ug/uL) for 24 h. Endogenous YAP1 immunoprecipitates and whole cell lysates were western blotted with indicated antibodies (on the right). B. U2OS cells were transfected with either non-targeting siRNA (siNT) or siRNA targeted against RUNX2. YAP1 immunoprecipitates from nuclear lysates were western blotted with the indicated antibodies (lamin B1 being used as a nuclear control). C. U2OS cells transfected with either siNT or siRNA targeted against TEAD. YAP1 immunoprecipitates from nuclear lysates were western blotted with the indicated antibodies (lamin B1 being used as a nuclear control). D. U2OS cells were transfected with either siNT or siRNA against RUNX2 as above, in the presence or absence of siRNA targeting RASSF1A, and endogenous YAP1 immunoprecipitates were blotted with the indicated antibodies. E. HCT116 cells were transfected with either siNT or siRNA targeted against RUNX2 or RASSF1A, and endogenous YAP1 immunoprecipitates were blotted with the indicated antibodies. Densitometry for D & E was performed on a Licor Odessey imager and p<0.01 (students t-Test) for the control versus dual siRNA. All western blots (A-E) are representative of at least 3 independent experiments.
To test whether RUNX2 was similarly able to compete with TEAD for YAP1 association in U2OS cells we focused on complexes within the nuclear compartment, due to alternative functions for YAP1 at cell junctions. We found that reduction of RUNX2 expression increased TEAD association with YAP1, indicating competition between transcription factors (Figure 2b). To confirm the competition between TEAD and RUNX2, we down-regulated TEAD by siRNA and observed an increase in YAP1 association with RUNX2 (Figure 2c). Thus, low levels of either RASSF1A or RUNX2 independently elevated levels of the oncogenic YAP1-TEAD complex. We next targeted both RASSF1A together with RUNX2 and observed that knock down of RASSF1A further enhanced YAP1-TEAD complex formation compared to loss of RUNX2 expression alone, in U2OS and the colo-rectal cell line HCT116 (Figures 2d, 2e and Supplementary Figure S1).
TEAD dependent proliferation and clonogenicity requires loss of RASSF1A-p73
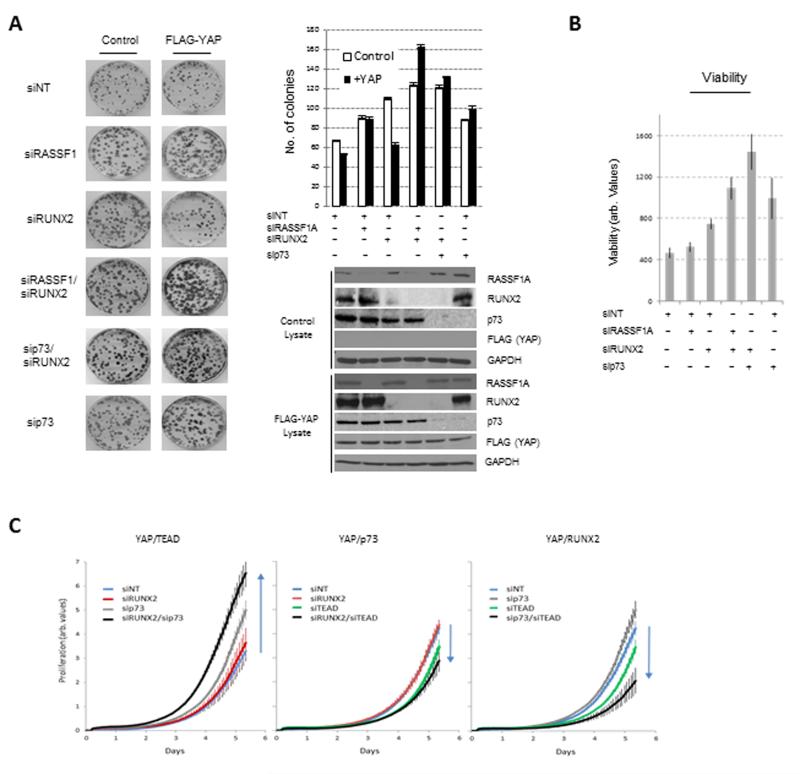
To determine whether the regulation of YAP1-TEAD observed above played a role in tumorigenic potential of cells, we performed colony formation assays in U2OS cells. siRNA-mediated reduction of both RASSF1A and RUNX2 expression increased the clonogenic capacity of tumor cells to a greater extent than knock-down of either RASSF1A or RUNX2 alone (Figure 3a). In support of a model where RASSF1A restricts proliferation by promoting YAP1-p73, targeting of either p73 or RASSF1A with RUNX2 resulted in equivalent enhanced clonogenicity and increased viability compared to controls (Figures 3a and 3b). The clonogenic potential of both controls and dual RASSF1A/RUNX2 were decreased by concomitant reduction of TEAD by siRNA, confirming that the additional proliferation was due to TEAD oncogenic activity (Supplementary Figure S2a). We reasoned that exogenous over-expression of YAP1 may resolve competition and permit all three complexes. However, YAP1 expression only increased clonogenicity in the absence of RASSF1A or p73 together with RUNX2 (Figure 3a). Interestingly, while the absence of RUNX2 favored YAP1-TEAD complexes and promoted colony formation, over-expression of YAP1 suppressed the growth advantage, perhaps through simultaneous enhancement of YAP1-p73 (Figure 3a).
Figure 3. RASSF1 and RUNX2 restrict YAP1-TEAD oncogenic behavior.
A. U2OS cells were transfected with control vector or plasmid expressing FLAG-YAP1 and indicated siRNAs before colonies were allowed to grow for 11 d, fixed and visualized with crystal violet. Whole cell lysates were probed with indicated antibodies. Error bars indicate SEM of n=3. Significance was determined by student t-Test RASSF1A vs RUNX2/RASSF1A p<0.001; Runx2 vs RUNX2/RASSF1A p=0.023; p73 vs RUNX2/p73 p<0.001. B. U2OS cells were transfected with indicated siRNAs, as shown in A, and cell viability was determined using the resazurin assay after 48 hrs. Error bars indicate SEM of at least n=4. Significance was determined by student t-Test RASSF1A vs RUNX2/RASSF1A p=0.008; Runx2 vs RUNX2/RASSF1A p=0.0101; p73 vs RUNX2/p73 p=0.0185. C. U2OS cells were transfected with indicated siRNAs to promote the different YAP transcriptional partner, as shown in A, and cell proliferation was monitored by xcelligenceTM in real time. Error bars indicate SEM of n=3. Arrows indicate increase or decrease in cell growth, significance determined at end point as below p=0.05 in set of dual knockdown compared to controls.
To definitively address the transcription factor competition we utilized combinations of siRNA that should promote YAP1-TEAD, YAP1-p73 or YAP1-RUNX2 and monitored U2OS cell growth curves in real time. As observed above, reduction of RUNX2 and p73 levels (favoring YAP1-TEAD complexes) increased proliferation, while dual reduction of RUNX2 and TEAD (favoring YAP1-p73) suppressed cell growth (Figure 3c). The combination of p73 and TEAD siRNA (favoring YAP1-RUNX2) surprisingly increased viability compared to TEAD loss alone, but significantly reduced cellular proliferation (Figure 3c) and clonogenicity which is consistent with a potential switch to a RUNX2 differentiation program (Supplementary Figures S2b and S2a).
Loss of RUNX2 and RASSF1 in human tumors
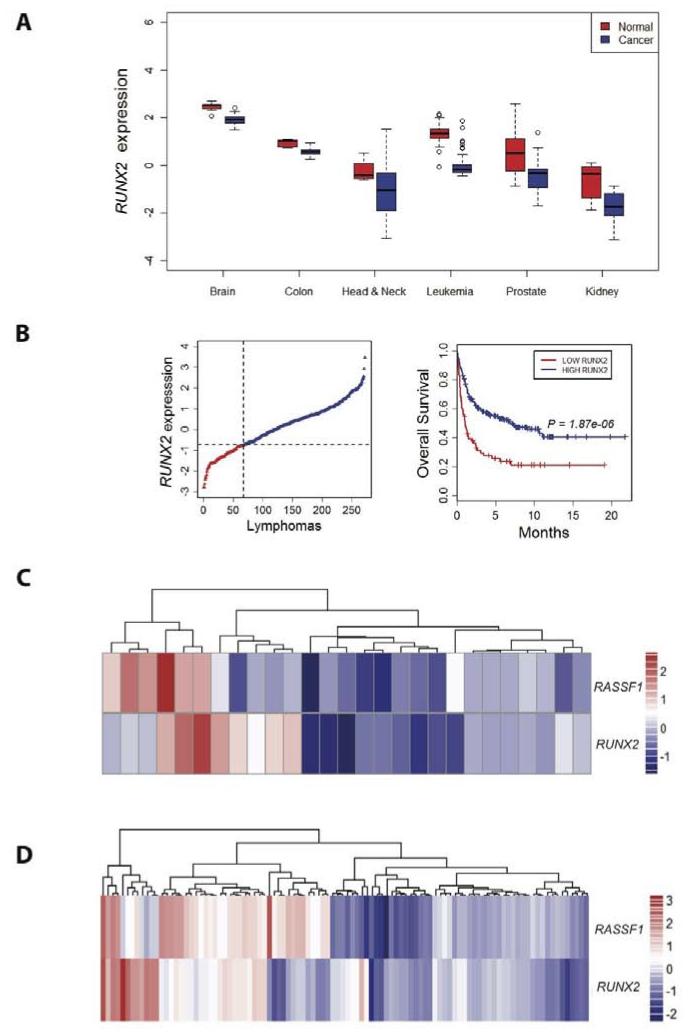
Loss of RASSF1A expression is found in almost all types of human cancer (reviewed in 2). Investigation of RUNX2 expression in microarray analysis performed across different tumor types revealed that RUNX2 mRNA levels were significantly lower in many tumor types compared to their corresponding normal tissues, particularly in tumors of the brain, colon, head and neck, prostate, kidney and leukemias (Figure 4a), and loss of RUNX2 expression showed a strong association with poorer survival in patients with diffuse large B-cell or atypical Burkitt’s/Burkitt’s-like lymphomas (Figure 4b). Importantly, a strong concordance in expression levels of RUNX2 and RASSF1 was observed in precursor T-cell lymphoblastic leukemias (Spearman’s rank correlation coefficient = 0.56, P value = 0.002; Figure 4c). RASSF1A is frequently methylated in tumors of the gastrointestinal tract (2) and we have shown that loss of Rassf1a co-operates with loss of Apc to accelerate intestinal tumorigenesis (28). In agreement with the promotion of YAP1-TEAD in HCT116 cells (Figure 2e), we found that loss of RUNX2 expression strongly correlated with loss of RASSF1 expression in human colo-rectal cancers (Pearson’s correlation coefficient = 0.379, P value = 9.86×10−5; Figure 4d).
Figure 4. Analysis of RUNX2 and RASSF1 expression across different tumor types.
A. Box plots indicating that RUNX2 expression is significantly lower in many tumor types compared with their corresponding normal tissues (only the tumor types that showed significantly lower RUNX2 expression in cancer vs. normal in at least three independent microarray datasets are included (from left to right: RUNX2 expression in normal brain (white matter, n= 7) vs. brain tumor (astrocytoma, glioblastoma, oligodendroglioma, n=35), normal colon (n=5) vs. colo-rectal cancer (n=100), normal uvula (n=4) vs. head and neck squamous cell carcinoma (n=34), peripheral blood mononuclear cell (n=58) vs. B-Cell Acute Lymphoblastic Leukemia (n=114), prostate gland (n=21) vs. prostate carcinoma (n=30), normal kidney (n=10) vs. clear cell renal cell carcinoma (n=10). B. (left) Ranked RUNX2 expression in a large dataset of 272 lymphomas, and (right) Kaplan-Meier survival curves comparing disease-free survival between lymphomas with the lowest (<25th percentile) versus highest (>25th percentile) RUNX2 expression (Log rank test, P = 1.87×10-6). C. Unsupervised hierarchical clustering and heatmap of RUNX2 and RASSF1 relative transcript levels in precursor T-cell lymphoblastic leukemias (n=27), of which 18 show concordant changes for these two transcripts. D. Unsupervised hierarchical clustering and heatmap of RUNX2 and RASSF1 relative transcript levels indicating a strong positive correlation (predominantly concordant down-regulated expression) between RASSF1 and RUNX2 expression in colo-rectal adenocarcinomas (n=105) (44). The color bar indicates normalized expression levels with red signifying the magnitude of up-regulation and blue of down-regulation. In all microarray studies, the probe(s) that detected RASSF1 were found in the part of the gene common to all RASSF1 transcripts.
DISCUSSION
Since its discovery in 2000 (29), hypermethylation of the RASSF1A promoter, and ensuing transcriptional silencing of RASSF1A, has been frequently observed in almost all tumor types (reviewed in 2). We have previously shown that Rassf1a null mice die faster than their wildtype littermates due to increased incidence of tumorigenesis, predominantly lymphoma/leukemias (15), and that loss of Rassf1a co-operates with loss of Apc to accelerate intestinal tumorigenesis (28). In this study, we used Sleeping Beauty (SB) insertional mutagenesis to identify candidate genes that are associated with tumorigenesis in the context of loss of Rassf1a. We have shown that Rassf1a−/− SB mice develop tumors significantly faster than their wildtype SB littermates, specifically poorly differentiated lymphomas or CD3-positive T-cell lymphomas, with a small amount of MPO-positive high-grade myeloid leukemias.
Isolating the transposon insertion sites from these SB-induced tumors allowed the discovery of a set of 10 genes enriched in the Rassf1a−/− SB tumors (Table 1). Similar to RASSF1A, several of these genes have roles in regulating the cell cycle, mitosis and mitotic progression, with reported loss of expression being found in tumors, including Foxn3 (30-31), Ppp2r2a (32-33), Stag2 (34-35) and Runx2 (36). In addition, some of these genes are known to interact with the Ras signaling pathway, and mutations in these genes are associated with tumorigenesis, including Crebbp (37-38) and Fgfr3 (39-40). Given that RASSF1A is a component of the hippo signaling pathway (13) that can activate RUNX2 (via YAP1) (12), we focused our attention on the Runx2 gene.
The three members of the Runx family of mammalian transcription factors, RUNX1-3, are related to Runt, the Drosophila pair rule gene (41) and share a highly conserved DNA-binding domain and a common DNA-binding cofactor. However, the RUNX2 gene (also known as PEPBP2A, AML3, CCD1, CBFA1 and OSF2) is a unique member of the family in that it produces the largest protein product which possesses two domains distinct from its homologues: a short stretch of glutamine-alanine (QA) repeats at the N-terminus and a C-terminal proline/serine/threonine (PST) rich tract, both regions of which are necessary for full transactivation activity (42) (Figure 1d). Members of the RUNX family regulate multiple cell fate decisions and have been implicated in a wide range of cancers where there is unequivocal evidence that members of this family can act as oncogenes or as tumor suppressors according to context (43). Specifically, Runx2-deficient (Runx2−/−) mouse embryonic fibroblasts (MEFs) are prone to spontaneous immortalization and display an early growth advantage that is resistant to stress-induced growth arrest (44). Thus RUNX2 can function as a tumor suppressor gene and loss of its expression is an important step in oncogenic transformation. Based on the location and orientation of these transposons in the Runx2 gene, they were predicted to result in the premature truncation of the transcript, therefore could be described as a loss-of-function allele. Interestingly, insertions in Runx2 were only significantly associated with tumourigenesis in the Rassf1a−/− mice and not wild type counterparts, indicating that loss of RUNX2 may not be sufficient for tumorigenesis.
In hippo pathway signaling, RASSF1A ensures that YAP1 associates with the proapoptotic p73 (13), whereas loss of hippo pathway signaling allows YAP1 to associate with TEAD, leading to oncogenic proliferation (8). We provided the first evidence that RASSF1A restricts the ability of YAP1 to associate with TEAD as part of its tumor suppressor activity (Figure 2). YAP1 (also known as YAP65) is a key regulator of organ size and has been implicated as an oncogene due to amplification in human cancers (8). In agreement with others, we found that over-expression of YAP1 enhances oncogenic behavior. However, high YAP1 levels, in the absence of RUNX2, promoted a p73 dependent suppression of clonogenicity (Figure 3a), therefore indicating that YAP1 amplification and RUNX2 loss are unlikely to be sufficient to promote tumorigenesis and require deregulation of either RASSF1A or p73. Importantly, clinical correlations of outcome and YAP1 levels may benefit from further stratification of tumors displaying reduced RASSF1A or p73 (45).
Hypermethylation of the RASSF1A promoter is a frequent occurrence in a wide variety of tumors, and together with the fact that point mutations have also been found in up to 15% of primary tumors, this makes RASSF1A one of the most frequently inactivated proteins in human cancer (reviewed in 2). We found that RUNX2 mRNA levels were significantly lower in many different tumor types compared to their corresponding normal tissues and loss of RUNX2 expression showed a strong association with poorer survival in some cancer types (Figure 4b). Importantly, a strong concordance in expression levels of RUNX2 and RASSF1 was observed in human precursor T-cell lymphoblastic leukemias (Figure 4c) and loss of RUNX2 expression is strongly correlated with loss of RASSF1 expression in human colo-rectal cancers (Figure 4d).
In summary, we have provided evidence that loss of RASSF1A expression switches YAP1 from a tumor suppressor to an oncogene through regulating its association with the transcription factors p73 and TEAD. Furthermore, the terminal differentiation factor RUNX2 also competes with TEAD for YAP1 association, independently of RASSF1A and p73, and combined knock down exacerbates YAP1-TEAD levels (Figure 5). We found that the resulting enhancement in proliferative signals results in elevated tumor indices in both genetic systems and human disease. Thus loss of RASSF1A, together with RUNX2, is concomitant with the multi-step model of tumorigenesis.
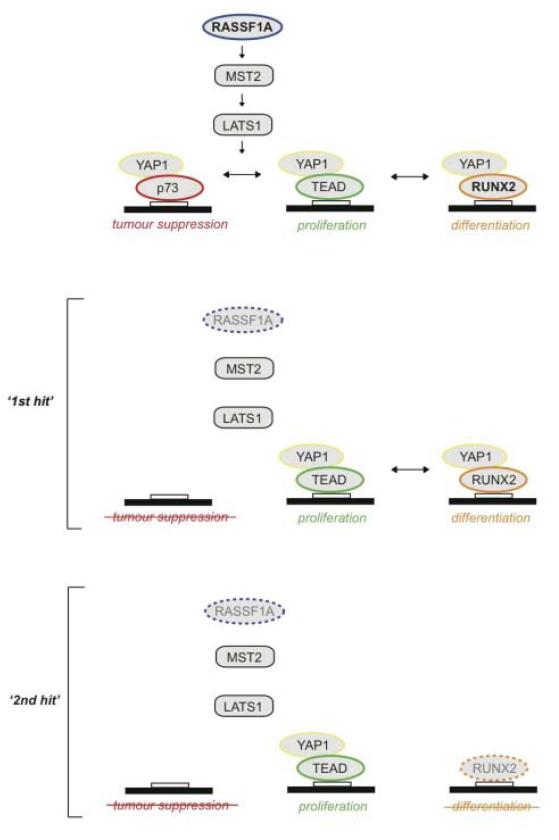
Figure 5. Proposed model of YAP complex formation.
YAP exists in a dynamic equilibrium with a number of transcriptional partners, p73, TEAD and RUNX2, modulated by upstream signals. Reduction of RASSF1A, either genetically or through promoter methylation, leaves cells susceptible to changes in other tumor suppressor genes that can exacerbate tumorigenesis (Table 1). By ablating RUNX2 we observed an upregulation of the YAP1-TEAD transcriptional complex (Figure 2d), which is further enhanced in the context of RASSF1A loss.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Mahrokh Nohadani for performing the tissue processing and immunohistochemistry, and the staff of Team 83 at the Wellcome Trust Sanger Institute for looking after the mice.
Financial support: A Kay Kendall Leukemia Fund Intermediate Fellowship KKL309 (L. van der Weyden), a Cancer Research UK grant C20510/A6997 and Wellcome Trust grant 082356 (D.J. Adams), a Cancer Research UK grant C20510/A6997 (MJ. Arends), a Cancer Research UK grant A12932 (EON), and a Medical Research Council UK studentship (A. Papaspyropoulos). G. Poulogiannis is a Pfizer Fellow of the Life Sciences Research Foundation.
Footnotes
Disclosure of potential conflicts of interest: The authors declare they have no potential conflicts of interest.
REFERENCES
- 1.van der Weyden L, Adams DJ. The Ras-association domain family (RASSF) members and their role in human tumourigenesis. Biochim Biophys Acta. 2007;1776:58–85. doi: 10.1016/j.bbcan.2007.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Donninger H, Vos MD, Clark GJ. The RASSF1A tumor suppressor. J Cell Sci. 2007;120:3163–72. doi: 10.1242/jcs.010389. [DOI] [PubMed] [Google Scholar]
- 3.Pan D. The Hippo Signaling Pathway in Development and Cancer. Developmental Cell. 2010;19:491–505. doi: 10.1016/j.devcel.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu S, Huang J, Dong J, Pan D. Hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell. 2003;114:445–56. doi: 10.1016/s0092-8674(03)00549-x. [DOI] [PubMed] [Google Scholar]
- 5.Wei X, Shimizu T, Lai ZC. Mob as tumor suppressor is activated by Hippo kinase for growth inhibition in Drosophila. EMBO J. 2007;26:1772–81. doi: 10.1038/sj.emboj.7601630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang J, Wu S, Barrera J, Matthews K, Pan D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell. 2005;122:421–34. doi: 10.1016/j.cell.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 7.Bandura JL, Edgar BA. Yorkie and Scalloped: partners in growth activation. Dev Cell. 2008;14:315–6. doi: 10.1016/j.devcel.2008.02.010. [DOI] [PubMed] [Google Scholar]
- 8.Zhao B, Ye X, Yu J, Li L, Li W, Li S, et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008;22:1962–71. doi: 10.1101/gad.1664408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vassilev A, Kaneko KJ, Shu H, Zhao Y, DePamphilis ML. TEAD/TEF transcription factors utilize the activation domain of YAP65, a Src/Yes-associated protein localized in the cytoplasm. Genes Dev. 2001;15:1229–41. doi: 10.1101/gad.888601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Strano S, Munarriz E, Rossi M, Castagnoli L, Shaul Y, Sacchi A, et al. Physical interaction with Yes-associated protein enhances p73 transcriptional activity. J Biol Chem. 2001;276:15164–73. doi: 10.1074/jbc.M010484200. [DOI] [PubMed] [Google Scholar]
- 11.Komuro A, Nagai M, Navin NE, Sudol M. WW domain-containing protein YAP associates with ErbB-4 and acts as a co-transcriptional activator for the carboxyl-terminal fragment of ErbB-4 that translocates to the nucleus. J Biol Chem. 2003;278:33334–41. doi: 10.1074/jbc.M305597200. [DOI] [PubMed] [Google Scholar]
- 12.Yagi R, Chen LF, Shigesada K, Murakami Y, Ito Y. A WW domain-containing yes-associated protein (YAP) is a novel transcriptional co-activator. EMBO J. 1999;18:2551–62. doi: 10.1093/emboj/18.9.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matallanas D, Romano D, Yee K, Meissl K, Kucerova L, Piazzolla D, et al. RASSF1A elicits apoptosis through an MST2 pathway directing proapoptotic transcription by the p73 tumor suppressor protein. Mol Cell. 2007;27:962–75. doi: 10.1016/j.molcel.2007.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hamilton G, Yee KS, Scrace S, O’Neill E. ATM regulates a RASSF1A-dependent DNA damage response. Curr Biol. 2009;19:2020–5. doi: 10.1016/j.cub.2009.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van der Weyden L, Tachibana KK, Gonzalez MA, Adams DJ, Ng BL, Petty R, et al. The RASSF1A isoform of RASSF1 promotes microtubule stability and suppresses tumorigenesis. Mol Cell Biol. 2005;25:8356–67. doi: 10.1128/MCB.25.18.8356-8367.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Collier LS, Carlson CM, Ravimohan S, Dupuy AJ, Largaespada DA. Cancer gene discovery in solid tumours using transposon-based somatic mutagenesis in the mouse. Nature. 2005;436:272–6. doi: 10.1038/nature03681. [DOI] [PubMed] [Google Scholar]
- 17.Dupuy AJ, Akagi K, Largaespada DA, Copeland NG, Jenkins NA. Mammalian mutagenesis using a highly mobile somatic Sleeping Beauty transposon system. Nature. 2005;436:221–6. doi: 10.1038/nature03691. [DOI] [PubMed] [Google Scholar]
- 18.March HN, Rust AG, Wright NA, ten Hoeve J, de Ridder J, Eldridge M, et al. Insertional mutagenesis reveals multiple networks of co-operating genes driving intestinal tumorigenesis. Nature Genetics. 2011;43:1202–9. doi: 10.1038/ng.990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Ridder J, Uren A, Kool J, Reinders M, Wessels L. Detecting statistically significant common insertion sites in retroviral insertional mutagenesis screens. PLoS Comput Biol. 2006;2:e166. doi: 10.1371/journal.pcbi.0020166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yee KS, O’Neill E. YAP1--friend and foe. Cell Cycle. 2010;9:1447–8. doi: 10.4161/cc.9.8.11480. [DOI] [PubMed] [Google Scholar]
- 21.Schreiber E, Matthias P, Muller MM, Schaffner W. Rapid detection of octamer binding proteins with ‘mini-extracts’, prepared from a small number of cells. Nucleic Acids Res. 1989;17:6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andersen P, Gill R. Cox’s regression model for counting processes, a large sample study. Annals of Statistics. 1982;10:1100–20. [Google Scholar]
- 23.Hofmann K, Bucher P. The rsp5-domain is shared by proteins of diverse functions. FEBS Lett. 1995;358:153–7. doi: 10.1016/0014-5793(94)01415-w. [DOI] [PubMed] [Google Scholar]
- 24.Mundlos S, Otto F, Mundlos C, Mulliken JB, Aylsworth AS, Albright S, et al. Mutations involving the transcription factor CBFA1 cause cleidocranial dysplasia. Cell. 1997;89:773–9. doi: 10.1016/s0092-8674(00)80260-3. [DOI] [PubMed] [Google Scholar]
- 25.Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, et al. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89:755–64. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- 26.Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KC, Rosewell IR, et al. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. 1997;89:765–71. doi: 10.1016/s0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- 27.Kool J, Berns A. High-throughput insertional mutagenesis screens in mice to identify oncogenic networks. Nat Rev Cancer. 2009;9:389–99. doi: 10.1038/nrc2647. [DOI] [PubMed] [Google Scholar]
- 28.van der Weyden L, Arends MJ, Dovey OM, Harrison HL, Lefebvre G, Conte N, et al. Loss of Rassf1a cooperates with Apc(Min) to accelerate intestinal tumorigenesis. Oncogene. 2008;27:4503–8. doi: 10.1038/onc.2008.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dammann R, Li C, Yoon JH, Chin PL, Bates S, Pfeifer GP. Epigenetic inactivation of a RAS association domain family protein from the lung tumor suppressor locus 3p21.3. Nat Genet. 2000;25:315–9. doi: 10.1038/77083. [DOI] [PubMed] [Google Scholar]
- 30.Busygina V, Kottemann MC, Scott KL, Plon SE, Bale AE. Multiple endocrine neoplasia type 1 interacts with forkhead transcription factor CHES1 in DNA damage response. Cancer Res. 2006;66:8397–403. doi: 10.1158/0008-5472.CAN-06-0061. [DOI] [PubMed] [Google Scholar]
- 31.Chen YJ, Liao CT, Chen PJ, Lee LY, Li YC, Chen IH, et al. Downregulation of Ches1 and other novel genes in oral cancer cells chronically exposed to areca nut extract. Head Neck. 2011;33:257–66. doi: 10.1002/hed.21442. [DOI] [PubMed] [Google Scholar]
- 32.Clarke PR, Hoffmann I, Draetta G, Karsenti E. Dephosphorylation of cdc25-C by a type-2A protein phosphatase: specific regulation during the cell cycle in Xenopus egg extracts. Mol Biol Cell. 1993;4:397–411. doi: 10.1091/mbc.4.4.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu X, Sempere LF, Ouyang H, Memoli VA, Andrew AS, Luo Y, et al. MicroRNA-31 functions as an oncogenic microRNA in mouse and human lung cancer cells by repressing specific tumor suppressors. J Clin Invest. 2010;120:1298–309. doi: 10.1172/JCI39566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hauf S, Roitinger E, Koch B, Dittrich CM, Mechtler K, Peters JM. Dissociation of cohesin from chromosome arms and loss of arm cohesion during early mitosis depends on phosphorylation of SA2. PLoS Biol. 2005;3:e69. doi: 10.1371/journal.pbio.0030069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rocquain J, Gelsi-Boyer V, Adélaïde J, Murati A, Carbuccia N, Vey N, et al. Alteration of cohesin genes in myeloid diseases. Am J Hematol. 2010;85:717–9. doi: 10.1002/ajh.21798. [DOI] [PubMed] [Google Scholar]
- 36.Dickey-Sims C, Robertson AJ, Rupp DE, McCarthy JJ, Coffman JA. Runx-dependent expression of PKC is critical for cell survival in the sea urchin embryo. BMC Biol. 2005;3:18–29. doi: 10.1186/1741-7007-3-18. 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Foulds CE, Nelson ML, Blaszczak AG, Graves BJ. Ras/mitogen-activated protein kinase signaling activates Ets-1 and Ets-2 by CBP/p300 recruitment. Mol Cell Biol. 2004;24:10954–64. doi: 10.1128/MCB.24.24.10954-10964.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mullighan CG, Zhang J, Kasper LH, Lerach S, Payne-Turner D, Phillips LA, et al. CREBBP mutations in relapsed acute lymphoblastic leukemia. Nature. 2011;471:235c9. doi: 10.1038/nature09727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kompier LC, Lurkin I, van der Aa MN, van Rhijn BW, van der Kwast TH, Zwarthoff EC. FGFR3, HRAS, KRAS, NRAS and PIK3CA mutations in bladder cancer and their potential as biomarkers for surveillance and therapy. PLoS One. 2010;5:e13821. doi: 10.1371/journal.pone.0013821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Al-Ahmadie HA, Iyer G, Janakiraman M, Lin O, Heguy A, Tickoo SK, et al. Somatic mutation of fibroblast growth factor receptor-3 (FGFR3) defines a distinct morphological subtype of high-grade urothelial carcinoma. J Pathol. 2011;224:270–9. doi: 10.1002/path.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gergen JP, Butler BA. Isolation of the Drosophila segmentation gene runt and analysis of its expression during embryogenesis. Genes Dev. 1988;2:1179–93. doi: 10.1101/gad.2.9.1179. [DOI] [PubMed] [Google Scholar]
- 42.Thirunavukkarasu K, Mahajan M, McLarren KW, Stifani S, Karsenty G. Two domains unique to osteoblast-specific transcription factor Osf2/Cbfa1 contribute to its transactivation function and its inability to heterodimerize with Cbfbeta. Mol Cell Biol. 1998;18:4197–208. doi: 10.1128/mcb.18.7.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blyth K, Cameron ER, Neil JC. The Runx gene family: gain or loss of function in cancer. Nat Rev Cancer. 2005;5:376–87. doi: 10.1038/nrc1607. [DOI] [PubMed] [Google Scholar]
- 44.Kilbey A, Blyth K, Wotton S, Terry A, Jenkins A, Bell M, et al. Runx2 disruption promotes immortalization and confers resistance to oncogene-induced senescence in primary murine fibroblasts. Cancer Res. 2007;67:11263–71. doi: 10.1158/0008-5472.CAN-07-3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang X, George J, Deb S, Degoutin LJ, Takano EA, Fox SB, et al. The Hippo pathway transcriptional co-activator, YAP, is an ovarian cancer oncogene. Oncogene. 2011;30:2810–22. doi: 10.1038/onc.2011.8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.