Background: The RNA polymerase II (Pol II) subunit Rpb4 was reported to function in mRNA degradation.
Results: Rpb4 deletion causes defects in mRNA synthesis and compensatory changes in mRNA degradation. Covalent attachment of Rpb4 to the Pol II core largely restores mRNA metabolism.
Conclusion: Rpb4 is an integral part of Pol II and functions mainly in Pol II transcription.
Significance: Rpb4 functions mainly in transcription.
Keywords: mRNA Decay, RNA Metabolism, RNA Polymerase II, RNA Turnover, Transcription, mRNA Degradation
Abstract
RNA polymerase II (Pol II) is the central enzyme that carries out eukaryotic mRNA transcription and consists of a 10-subunit catalytic core and a subcomplex of subunits Rpb4 and Rpb7 (Rpb4/7). Rpb4/7 has been proposed to dissociate from Pol II, enter the cytoplasm, and function there in mRNA translation and degradation. Here we provide evidence that Rpb4 mainly functions in nuclear mRNA synthesis by Pol II, as well as evidence arguing against an important cytoplasmic role in mRNA degradation. We used metabolic RNA labeling and comparative Dynamic Transcriptome Analysis to show that Rpb4 deletion in Saccharomyces cerevisiae causes a drastic defect in mRNA synthesis that is compensated by down-regulation of mRNA degradation, resulting in mRNA level buffering. Deletion of Rpb4 can be rescued by covalent fusion of Rpb4 to the Pol II core subunit Rpb2, which largely restores mRNA synthesis and degradation defects caused by Rpb4 deletion. Thus, Rpb4 is a bona fide Pol II core subunit that functions mainly in mRNA synthesis.
Introduction
The level of an mRNA in the cell is determined by its synthesis rate (SR)3 and its degradation rate (DR). Although in eukaryotes gene transcription and mRNA degradation have been intensively studied, the regulation and interplay between these two spatially separated processes remain poorly understood. We and others have shown recently that mRNA synthesis and degradation are coupled and that this coupling serves to buffer cellular mRNA levels (1–6). It has been shown that a single point mutation near the active site of RNA polymerase (Pol) II (7) leads to a reduction of SRs but unexpectedly also DRs (1). Vice versa, impaired DRs also lead to decreased SRs, resulting in a buffering of mRNA levels (1, 2, 6).
All eukaryotic mRNAs are synthesized by the 12-subunit Pol II enzyme that consists of a 10-subunit catalytic core and a 2-subunit subcomplex made of subunit Rpb4 and Rpb7 (Rpb4/7). Only two Pol II subunits are dispensable for viability in the yeast Saccharomyces cerevisiae, Rpb4 and Rpb9. Although Rpb9 has been shown to be involved in transcription fidelity (8, 9), Rpb4 has been shown to be dispensable for optimum growth at 18–23 °C and has been proposed to protect Pol II from inactivation at high temperatures (10–13). The Rpb4/7 heterodimer can dissociate from core Pol II in S. cerevisiae (14). Rpb4 and Rpb7 have been reported to be in excess over core Pol II subunits within cells (10), but a recent study using quantitative proteomics could not confirm this (15). Although Rpb4 has been proposed to associate poorly with core Pol II at 27 °C during logarithmic growth (13), it could be detected on all genes at 30 °C using chromatin immunoprecipitation (16).
It has been reported that Rpb4 and Rpb7 shuttle between the nucleus and the cytoplasm and that Rpb4 and Rpb7 accumulate in the cytoplasm during heat shock (4, 17). Rpb4/7 has been reported to shuttle in a transcription-dependent manner, and it has been reported to first associate with Pol II in order for it to exert its post-transcriptional functions in the cytoplasm (4, 18–20). Furthermore, Rpb4 has been reported to function in the degradation of specific mRNAs (21), whereas an effect of Rpb7 on mRNA degradation was reported to be general (22), but shuttling of Rpb4 and Rpb7 has been reported to depend on each other (19). Additionally, Rpb4 has been proposed to function directly in translation initiation (18). These studies led to the conclusion that Rpb4/7 influence transcription in the nucleus and mRNA degradation in the cytoplasm and were crucial for deriving the model of “mRNA imprinting” (20, 23, 24).
Here we test the direct functions of Rpb4 in mRNA synthesis and degradation. Determining whether a protein is directly involved in both processes is not trivial because perturbation of one process leads to compensatory effects of the other. Indeed, such mutual compensation between SRs and DRs is a general phenomenon in cells that has been observed for multiple mutants (1, 2, 6). To test for a direct function of Rpb4 in mRNA degradation, a mutant has to be used in which the putative functions in transcription and degradation are uncoupled. We created such a mutant by covalently linking Rpb4 to the Pol II core subunit Rpb2. We show that this Rpb2-Rpb4 fusion mutant rescues all growth and transcription defects of the Rpb4 deletion strain Δrpb4 at 30 and at 37 °C. Furthermore, we could not detect strong differences in mRNA degradation between the Rpb2-Rpb4 mutant and wild-type yeast. These results demonstrate that Rpb4 is a bona fide Pol II subunit with functions in transcription and do argue against the proposed direct function of Rpb4 in mRNA degradation.
EXPERIMENTAL PROCEDURES
Yeast and Plasmids
Δrpb4a strain was obtained from the knock-out library from Euroscarf and was compared against an isogenic wild type (BY4741). The freshly prepared Δrpb4 was created in a different background for comparability with the Rpb2-Rpb4 fusion strain. To this end a strain carrying a genomic deletion of Rpb2 was created in our laboratory. Plasmids pRS316-RPB2, pRS315-RPB2, and pRS315-RPB2-RPB4 were generated by cloning the respective ORF plus sequences 500 bp upstream and 250 bp downstream into pRS315 (ATCC) and pRS316 (ATCC) using XhoI/SacI restriction sites. For pRS315-RPB2-RPB4 overlap, PCRs were used to introduce RPB4 in-frame between RPB2 and the RPB2 3′-UTR (25). We added three glycines as a linker and removed the start methionine in the Rpb4 sequence. The heterozygous RPB2/rpb2Δ S. cerevisiae yeast strain (BY4743, rpb2::KanMX6/RPB2) was generated and transformed with pRS316-RPB2. Diploids were sporulated and tetrads were dissected on YPD plates. The shuffle strain carrying pRS316-RPB2 was transformed with either pRS315-RPB2 or pRS315-RPB2-RPB4 and streaked on 5-fluoroorotic acid plates and then on synthetic complete-Leu plates. The resulting strain carrying pRS315-RPB2 was used as corresponding wild type for the fusion strain carrying pRS315-RPB2-RPB4. Genotypes of the strains are MATa; his3Δ1; leu2Δ0; met15Δ0; LYS2; ura3Δ0 rpb2::KanMX6 pRS315-Rpb2 (wild type); MATa; his3Δ1; leu2Δ0; met15Δ0; LYS2; ura3Δ0 rpb2::KanMX6 rpb4::clonNAT pRS315-Rpb2-Rpb4 (Rpb2-Rpb4 fusion); and MATa; his3Δ1; leu2Δ0; met15Δ0; LYS2; ura3Δ0 rpb2::KanMX6 rpb4::clonNAT pRS315-Rpb2 (Δrpb4 knock-out). Genomic deletion of Rpb4 in the fusion strain as well as in the corresponding wild type was carried out via homologous recombination using a clonNAT cassette amplified from a pYM-natNT2-3HA vector (26). Directly after generation and strain validation via PCR, yeast strains were stored in 30% glycerol stocks at −80 °C. Experiments were performed from fresh plates from −80 °C stocks, and yeast strains were generally either grown in rich medium (YPD) at optimal growth temperature (30 °C) or under heat shock (37 °C).
Western Blotting
Overnight cultures of yeast cells were diluted to an A600 of 0.1 and grown until ∼A600 of 0.8. A total of 5 optical densities were sedimented and resuspended in 100 μl of lysis buffer (10 mm MOPS, pH 6.8, 10% SDS, 48% urea, 10 mm EDTA), and 100 μl of glass beads were added. Samples were vortexed for 3 min, and 100 μl of 2× loading buffer (75 mm MOPS, pH 6.8, 4% SDS, 48% urea, 3% DTT, 0.2% bromphenol blue) were added and heated to 55 °C for 10 min. Samples were separated on precast gels (NuPAGE 4–12% Bis-Tris, Invitrogen) and analyzed by immunoblotting with antibodies against Rpb4 (sc-101604, Santa Cruz Biotechnologies), Rpb1 (8WG16, Abcam), Rpb3 (1Y26, NeoClone), and tubulin (sc-69971, Santa Cruz Biotechnologies).
Immunostaining
Overnight cultures of yeast cells were diluted to A600 ∼0.1 and grown until A600 of 0.5–1. Cells were sedimented and resuspended in 4% paraformaldehyde in PBS and cross-linked for 10 min. Cross-linking was quenched with 125 mm glycine for 10 min, and cells were sedimented and resuspended in KIP/sorbitol buffer (0.1 m KH2PO4, 1 m sorbitol). Cell walls of cross-linked cells were digested with Zymolyase for 30–45 min at 37 °C. Spheroplasts were washed with PBS twice and resuspended in PBS. Different amounts of cells were spotted on poly-l-lysine (Sigma, P8920)-coated cover glasses. Cells were permeabilized with ice-cold methanol for 6 min, washed with PBS three times, washed one time with PBS + 0.1% Triton X-100, and incubated for 20 min with blocking solution (PBS, 0.1% Tween, 2% bovine serum albumin). 100 μl of a 1:1000 dilution of Rpb4 antibody (sc-101604, Santa-Cruz Biotechnologies) in PBS were used to stain cells for 1 h. Cells were washed six times with PBS before staining with Alexa Fluor® 488 goat anti-mouse antibody (Invitrogen, A31619) for 1 h. Cells were washed five times with PBS and stained with 1 μg/ml DAPI for 5 min. Cells were washed five times with PBS before imaging. Images were taken on a Leica DM2500 microscope with Leica LAS AF software.
Comparative Dynamic Transcriptome Analysis (cDTA)
cDTA was carried out as described (1). Briefly, S. cerevisiae cells were grown to an A600 of 0.8 and labeled with 4-thiouracil for 6 min. Schizosaccharomyces pombe cultures for normalization were grown to A600 of 0.8 in YES (5 g/liter yeast extract; 30 g/liter glucose; supplements: 225 mg/liter adenine, histidine, leucine, uracil, and lysine hydrochloride) medium and labeled with 4-thiouridine. Heat-shock samples were grown at 30 °C overnight, and an amount equivalent to an A600 of 0.1 was transferred to 37 °C and grown until it reached an A600 of 0.8. Equal numbers of S. pombe cells from one batch were added to equal cell numbers of S. cerevisiae samples for global intensity normalization (1). Cell lysis was performed on the S. pombe/S. cerevisiae cell mixture for 8 × 40 s with 1 min on ice in between on a MP-Biosciences FastPrep-24 machine. Total RNA was extracted with the RiboPure yeast kit (Ambion/Applied Biotechnologies), following the manufacturer's protocol. Labeled RNA was extracted from 100 μg of total RNA via biotinylation, and total and labeled RNA were analyzed with Affymetrix gene expression arrays as described (1).
Data Analysis
Analysis of data from cDTA was carried out as described (1) using R/Bioconductor and the DTA package (27). Because we used a mixture of S. cerevisiae and S. pombe cells, we used a custom probe annotation environment (cdf) to exclude cross-hybridization probes from our analyses. Labeling bias correction was done as described in Miller et al. (28), and proportional rescaling of microarray intensities to the internal S. pombe standard was done as described in Sun et al. (1). mRNA synthesis rates and decay rates were obtained as described in Miller et al. (28). Microarray data were deposited in ArrayExpress under accession number E-MTAB-2539.
RESULTS
Rpb4 Deletion Causes a Severe Growth Defect
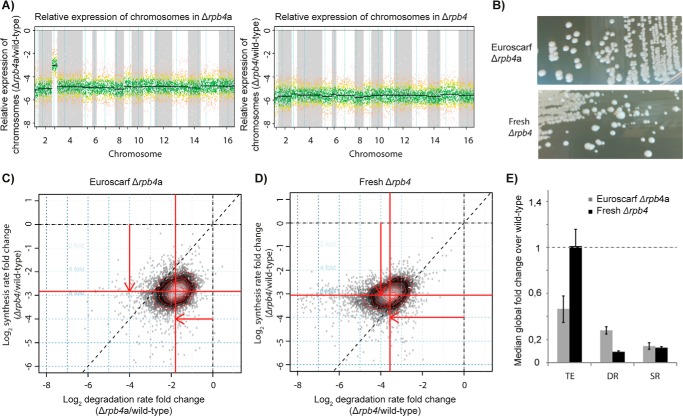
To investigate the general function of Rpb4 in mRNA metabolism under optimum growth conditions at 30 °C, we used a knock-out strain from Euroscarf (Δrpb4a) and performed microarray gene expression analysis. The obtained data revealed that the strain was aneuploid and carried two copies of chromosome 3 (Fig. 1A). Therefore we prepared a fresh knock-out strain of Rpb4 (Δrpb4). We found that the fresh Δrpb4 strain was not aneuploid and was even more growth-defective than the aneuploid Δrpb4a library strain. At optimum growth temperatures, the Δrpb4 strain had a doubling time of 5.3 h (1.6 h for wild type) and the Δrpb4a strain had a doubling time of 3 h (1.5 h for wild type). Our freshly prepared Δrpb4 strain also showed strong variations in colony growth on YPD agar plates, indicating selection of secondary mutations, which was not observed for the Euroscarf Δrpb4a strain (Fig. 1B). These results supported the importance of Rpb4 for cellular mRNA metabolism.
FIGURE 1.
Rpb4 deletion causes severe defects in mRNA metabolism. A, labeled RNA expression -fold changes in Δrpb4a and the Δrpb4 of genes plotted against their position in chromosomes. Black lines represent the median expression of all genes of the respective chromosome. B, colony growth on YPD plates for the freshly prepared Δrpb4 and the Euroscarf Δrpb4a strain. Cells were plated from cryo-cultures and incubated for 4 days at 30 °C. C, -fold changes in DRs (log folds, x axis) and SRs (log folds, y axis) for the Euroscarf Δrpb4a mutant against the isogenic wild type. Each point corresponds to one mRNA, and the density of points is encoded by their brightness. Red contour lines define regions of equal density. The center of the distribution is shifted from the origin, indicating a global shift in median SRs and DRs (SR shift is reflected by red line relative to dashed x axis line; DR shift is reflected by red line relative to dashed y axis line). D, DR and SR -fold changes for the freshly prepared Δrpb4 mutant as in C. E, median -fold changes of total RNA levels (TE), mRNA DRs, and mRNA SRs for the Δrpb4 (black bars) and the Δrpb4a (gray bars) mutants over wild type determined from cDTA. The dashed gray line reflects the respective wild-type value, which is normalized to one. Error bars indicate S.D.
Rpb4 Deletion Leads to Defects in mRNA Synthesis and Degradation
To investigate to which extent Rpb4 is required for RNA synthesis, we used cDTA (1). cDTA is based on non-perturbing metabolic labeling of newly synthesized RNAs and makes use of fission yeast cells (S. pombe) as an internal standard. cDTA can measure SRs, determine DRs, and detect global -fold changes in rates and mRNA levels (1, 2). This is particularly important when working with factors that can potentially affect the expression of all genes (29). Application of cDTA to our freshly prepared Δrpb4 strain revealed a strong global defect in both SRs and DRs under optimal growth conditions at 30 °C (Fig. 1C). Although we observed a median 8-fold decrease in SRs as compared with the wild type, the level of total RNA in the Δrpb4 strain was essentially unchanged, due to an 11-fold decrease in DRs (Fig. 1E). mRNA synthesis rates were decreased at least 2-fold for 99.6% of genes, consistent with a global function of Rpb4 in Pol II transcription.
When we repeated cDTA with the commercially available knock-out strain of Rpb4 (Δrpb4a), a similarly strong global defect in SRs was observed (7-fold). However, the defect in mRNA degradation was less severe, with a median 3.6-fold decrease in DRs (Fig. 1D), which resulted in 54% lower total RNA levels in the Δrpb4a strain (Fig. 1E). SRs of 99.4% of genes were decreased more than 2-fold in the Δrpb4a strain. The differences in total mRNA levels were comparable with a previous study, where a global decrease of 46% of mRNAs had been observed at 24 °C upon deletion of Rpb4 (12). SRs in the Δrpb4 and the Δrpb4a mutants were at 12–14% of wild-type values, indicating that the increase in growth rates of the Δrpb4a strain was due to secondary mutations.
The simplest explanation for our results is that Rpb4 is a bona fide Pol II subunit. The previously defined general cellular mechanism for buffering of mRNA levels (1, 2, 28) is able to compensate for the transcription defect by a lowering of DRs, i.e. stabilizing the transcripts. This explanation is in line with our previous data and with published work (1, 2, 10–12), and does not require the assumption that Rpb4 has a major, active role in mRNA degradation.
Rpb2-Rpb4 Fusion Rescues Δrpb4 Growth Phenotypes
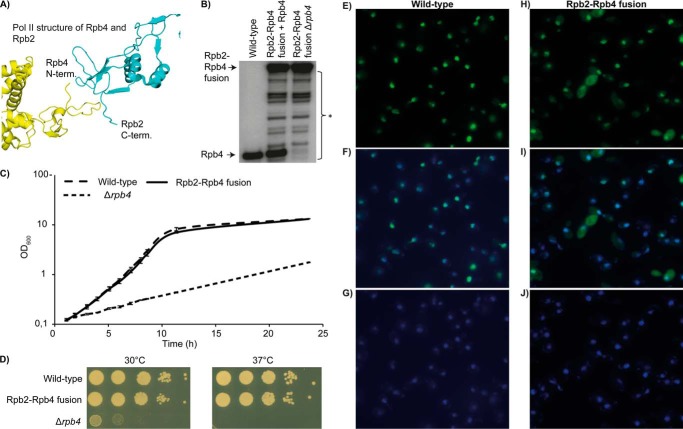
The above results could alternatively be interpreted as evidence for the previously proposed active function of Rpb4 in mRNA degradation in the cytoplasm (4, 18, 21, 22, 30) because cause and consequence in mRNA synthesis-degradation compensation are not readily distinguished. To address whether the effect of Rpb4 deletion on mRNA degradation is direct, via an active role of Rpb4 in the cytoplasm, or indirect, via a general mRNA buffering mechanism, we covalently attached Rpb4 to the Pol II core enzyme by fusing it to the core subunit Rpb2. We prepared a Rpb2-Rpb4 fusion construct because the C terminus of Rpb2 is in close proximity to the N terminus of Rpb4 (Fig. 2A) in the crystal structure of Pol II (31).
FIGURE 2.
Covalent fusion of Rpb2 and Rpb4 compensates growth defects of Rpb4 deletion. A, the C terminus of Rpb2 (cyan) lies in close proximity to the N terminus of Rpb4 (yellow) and suggested the possibility of direct fusion of the two proteins with a linker containing three glycine residues. B, Western blot against Rpb4 for the wild type, the Rpb2-Rpb4 fusion mutant with free Rpb4, and the fusion mutant after genomic deletion of Rpb4. An asterisk marks low amounts of various N-terminal degradation products of the fusion protein that do not lead to significant amounts of free Rpb4. C, growth curves of wild-type, fusion mutant, and Δrpb4 strains in culture at 30 °C. Doubling times were determined from the slope during logarithmic growth. OD, optical density. D, cultures of wild-type, Rpb2–4 fusion, and Δrpb4 cells were grown to the logarithmic phase at 30 °C. For each strain, 1 A600 was spotted on plates in 10-fold serial dilutions on YPD plates. Plates were incubated either at 30 or 37 °C for 2–3 days. E, immunostaining of Rpb4 (green signal) in wild-type cells. F, colocalization of the nucleus stained with DAPI (blue signal) and Rpb4 (green signal) in wild-type cells. G, nuclei stained with DAPI in wild-type cells. H–J, immunostaining as in E–G, for the Rpb2-Rpb4 fusion mutant reveals a major nuclear localization.
We generated three yeast strains: one carrying a genomic deletion of RPB2 with RPB2 expressed from a plasmid under the native promoter (wild type), one carrying a genomic deletion of RPB2 with RPB2 expressed from a plasmid under the native promoter and with a genomic deletion of RPB4 (Δrpb4), and one carrying a genomic deletion of RPB2 and RPB4 and a plasmid expressing the Rpb2-Rpb4 fusion construct under the native RPB2 promoter (Rpb2-Rpb4 fusion strain). Thus, in the fusion strain, the Rpb2-Rpb4 fusion protein is the only source of Rpb2 and Rpb4. Expression of the fusion protein was verified (Fig. 2B).
We tested whether the Rpb2-Rpb4 fusion protein rescued the growth defects of the Δrpb4 strain. Growth of the Δrpb4 strain at 30 °C was very slow, but the fusion protein completely rescued the growth defect (Fig. 2, C and D). The doubling time (1.7 h) was essentially the same as for the isogenic wild type (1.6 h) but very different from the Δrpb4 strain (5.6 h) (Fig. 2C). Additionally, the Rpb2–4 fusion protein completely rescued growth at 37 °C, whereas the Δrpb4 strain was inviable at 37 °C (Fig. 2D). We also investigated the localization of Rpb4 and the Rpb2-Rpb4 fusion protein. Both proteins were localized mainly in the nucleus (Fig. 2, E–J). However, cytoplasmic signals of the Rpb2-Rpb4 fusion protein appeared to be slightly stronger than those of Rpb4 in the wild-type strain. Taken together, our data indicate that Rpb4 mainly functions in mRNA synthesis.
Rpb2-Rpb4 Fusion Rescues Transcription and mRNA Degradation Defects of Δrpb4
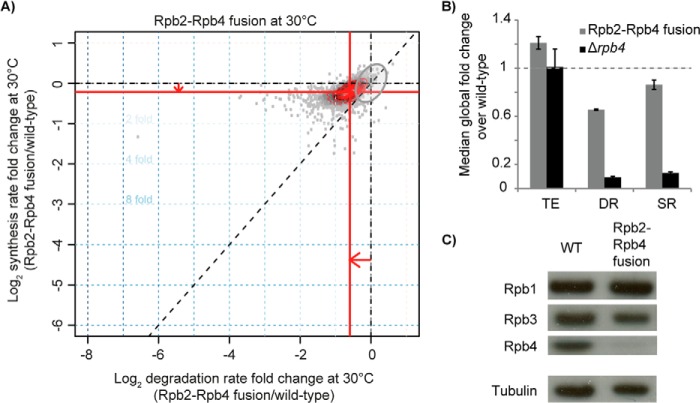
Further analysis showed that the Rpb2-Rpb4 fusion protein could mostly rescue the transcription defects caused by Rpb4 deletion. With the use of cDTA, we observed that median SRs were restored to 86% of wild-type levels (Fig. 3,A and B). Only 0.8% of genes showed SRs that were changed more than 2-fold in the fusion mutant, and these were not enriched in specific Gene Ontology (GO) terms (adjusted p value 0.05). We also tested whether the protein levels of other Pol II subunits were changed in the fusion strain and found similar levels of Rpb1 and Rpb3 in all strains (Fig. 3C). Thus, the Rpb2-Rpb4 fusion protein functioned almost as well as the two separate proteins Rpb2 and Rpb4 in mRNA synthesis in vivo. The mutant also showed only slightly compromised mRNA degradation, and the median DR was restored to 66% of wild-type levels (Fig. 3, A and B). DRs of only 13.7% of transcripts were changed more than 2-fold, but again these genes were not enriched in certain GO terms. These data indicate that Rpb4 fusion to the core Pol II enzyme largely restores mRNA synthesis and thereby also mRNA degradation, although a minor defect remained.
FIGURE 3.
Rpb2-Rpb4 fusion rescues SR and DR defects of Rpb4 deletion. A, -fold changes in DRs and SRs for the Rpb2-Rpb4 fusion mutant as in Fig. 1C. The gray ellipse reflects the 95% confidence region of the median SR and DR of the wild type (1). B, median -fold changes of total RNA expression (TE), mRNA DRs, and mRNA SRs for the Rpb2-Rpb4 fusion mutant (gray bars). For comparison the values for Δrpb4 were plotted again (black bars). The dashed gray line reflects the respective wild-type value, which is normalized to one. Error bars indicate S.D. C, Western blot of Rpb1, Rpb3, Rpb4, and tubulin (loading control) in wild-type and Rpb2-Rpb4 fusion mutant cells.
Rpb2-Rpb4 Fusion Enables Normal Heat-shock Response
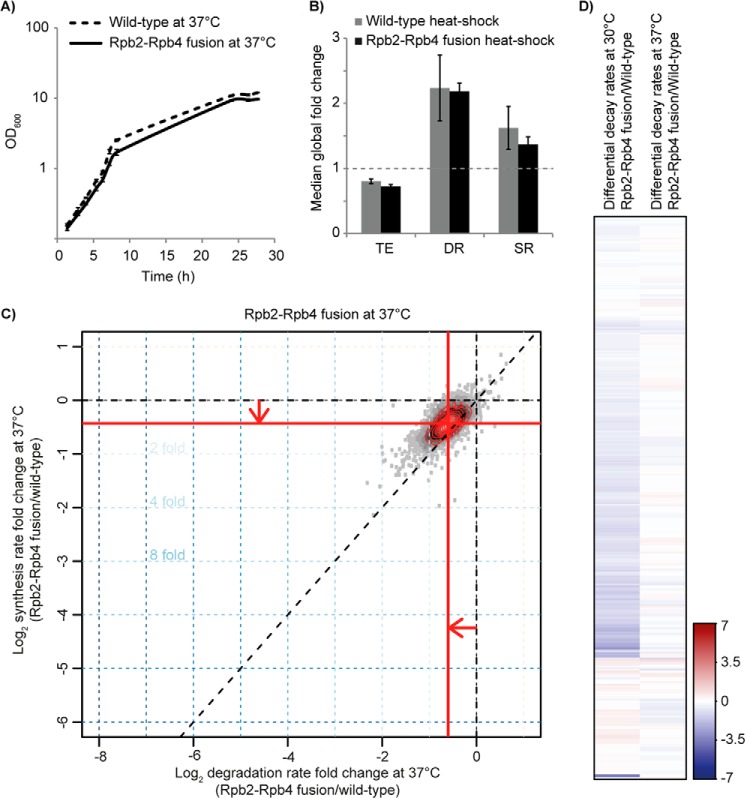
The above data showed that the strong effects of Rpb4 deletion on mRNA degradation stem to the largest extent from mRNA synthesis-degradation compensation and not from a direct role of Rpb4 in degradation. Otherwise a much stronger effect on mRNA degradation should have been observed in the Rpb2-Rpb4 fusion strain, which does not express any free Rpb4. However, Rpb4 has been reported to shuttle between the nucleus and the cytoplasm and to accumulate in the cytoplasm upon heat shock caused by incubation at 37 °C (17, 19). If free cytosolic Rpb4 would be important for regulating mRNA degradation during heat shock, the Rpb2-Rpb4 fusion strain should grow poorly or not at all after heat shock because the Δrpb4 mutant is inviable at 37 °C (Fig. 2D). However, we observed the contrary. We found that the Rpb2-Rpb4 fusion mutant grows essentially like wild type at 37 °C, with a similar doubling time (2.1 h, as compared with 1.9 h for wild type) (Fig. 4A). This indicated that free cytosolic Rpb4 is not required for growth under heat-shock conditions.
FIGURE 4.
Rpb2-Rpb4 fusion mutant cells are fully capable of adapting to 37 °C. A, growth curves of wild-type and Rpb2-Rpb4 fusion cells at 37 °C in culture. Doubling times were calculated from the slope during logarithmic growth. OD, optical density. B, median -fold changes of total RNA expression (TE), mRNA DRs, and mRNA SRs between the Rpb2-Rpb4 fusion mutant at 30 and 37 °C (gray bars) and between the wild type at 30 and 37 °C (black bars). The dashed gray line reflects the respective wild-type value that is normalized to one. Error bars indicate S.D. C, -fold changes in DRs and SRs for the Rpb2-Rpb4 fusion at 37 °C as compared with the wild type at 37 °C. D, heat map comparing DRs between Rpb2-Rpb4 fusion and wild type at 30 and 37 °C. Transcripts colored in blue show lower DRs, and transcripts colored in red show higher DRs. Differences are shown as variation from the median global -fold change after median centering.
We next analyzed changes in mRNA metabolism upon heat shock. We carried out cDTA after incubation for 6 h at 37 °C for wild type and the Rpb2-Rpb4 fusion mutant. We could not observe differences in median total RNA levels, SRs, or DRs between the heat-shock response in the two strains (Fig. 4B), indicating that the mutant is fully capable of adapting to 37 °C. When we compared the heat-shock profile of the Rpb2-Rpb4 mutant with the heat-shock profile of the wild type, we found essentially what we had observed at 30 °C (Fig. 4C). As observed before for wild-type cells (1), mRNA metabolism was slightly elevated at 37 °C as compared with 30 °C. Despite generally higher rates at 37 °C, the fusion mutant still showed slightly decreased median SRs (76%) and DRs (68%) as compared with the wild type at 37 °C. Also, selected transcripts showed even more decreased DRs at 30 °C, but this was not observed at 37 °C (Fig. 4D). These results show that the fusion mutant can adopt its mRNA metabolism after heat shock at 37 °C and that this does not depend on free Rpb4, further arguing against a direct role of Rpb4 in mRNA degradation.
DISCUSSION
Here we addressed the question of whether the Pol II subunit Rpb4 functions only in nuclear mRNA synthesis during transcription or whether it additionally functions in cytoplasmic mRNA degradation. With the use of cDTA, we showed that Rpb4 is generally required for mRNA synthesis at all genes in S. cerevisiae at a growth temperature of 30 °C. We observed a severe reduction in median SRs that was 8-fold lower in the Δrpb4 mutant than in wild-type cells. Thus, at 30 °C Δrpb4 mutants can still transcribe DNA but at low rates. However, the transcription capacity of the Δrpb4 mutant is strongly impaired, and this could explain why perturbations that require transcriptional adaption such as heat shock can lead to cell death.
Previous studies could not detect the requirement of Rpb4 for normal mRNA synthesis at all genes under optimum growth conditions as they monitored only mRNA levels that are not strongly changed (12, 32) because of mRNA level buffering. Our results are in line with previous findings that Rpb4 occupancy over genes determined by chromatin immunoprecipitation is essentially the same as for the core Pol II subunit Rpb3, indicating that the complete 12-subunit Pol II is the genome-associated form of the enzyme (16, 33). Our results also support previous findings that Rpb4 is required to prevent Pol II inactivation at higher temperatures (10–12).
Our cDTA data further revealed that the severe defects in SRs caused by Rpb4 deletion are compensated by a strong decrease in DRs, leading to a buffering of mRNA levels that is a general phenomenon (1, 2). To further address this, we constructed a yeast strain in which Rpb4 cannot dissociate from the Pol II core enzyme because it is covalently attached to a core subunit. In this Rpb2-Rpb4 fusion strain, the median SR was only slightly lower than in wild type, indicating that the fusion protein could largely rescue the defect caused by Rpb4 deletion. In addition, the median DR was decreased, again compensating for the mild loss in transcription activity caused by the fusion.
The slightly higher effect on mRNA degradation as compared with the effect on mRNA synthesis may indicate a minor, additional role of Rpb4 in mRNA degradation as suggested (4, 17, 21). It remains possible that at 30 °C, transient dissociation of Rpb4 (15) favors correct degradation of some mRNAs as can be seen from decreased DRs of some mRNAs at 30 °C but not at 37 °C (Fig. 4D). However, if this occurs, it is not a global phenomenon. Also, the observed minor effects in the fusion mutant are too small as compared with the very large effects of Rpb4 deletion to take them as evidence for a general role of Rpb4/7 in mRNA degradation. The Rpb2-Rpb4 fusion mRNA is expressed from a plasmid, which may result in higher protein levels of the fusion protein as compared with Rpb4 in wild-type cells. However, excess fusion protein is not incorporated into Pol II and cannot function in coupling of transcription and mRNA degradation as suggested (4, 18–20). Additionally, if Rpb4 had a function in mRNA degradation independent of its function through Pol II, we would expect higher degradation rates due to higher levels in the cytoplasm (Fig. 2, E–J), but this is not at all observed. Indeed, the observed mild effects on mRNA degradation in the Rpb2-Rpb4 fusion mutant may be explained in part by the general mRNA buffering mechanism and in part by impaired RNA 3′-end processing that depends on Rpb4 (34) and likely enhances mRNA degradation during RNA surveillance. This is supported by the fact that there is no difference in mRNA metabolism during heat shock in wild type and Rpb2-Rpb4 fusion mutant. Thus, Rpb4 functions predominantly in nuclear mRNA synthesis by Pol II.
Acknowledgment
We thank Björn Schwalb for bioinformatics advice and critically reading the manuscript.
This article was selected as a Paper of the Week.
- SR
- synthesis rate
- DR
- degradation rate
- cDTA
- comparative Dynamic Transcriptome Analysis
- Pol
- RNA polymerase
- YPD
- yeast extract peptone dextrose
- Bis-Tris
- 2-(bis(2-hydroxyethyl)amino)-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1. Sun M., Schwalb B., Schulz D., Pirkl N., Etzold S., Larivière L., Maier K. C., Seizl M., Tresch A., Cramer P. (2012) Comparative dynamic transcriptome analysis (cDTA) reveals mutual feedback between mRNA synthesis and degradation. Genome Res. 22, 1350–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sun M., Schwalb B., Pirkl N., Maier K. C., Schenk A., Failmezger H., Tresch A., Cramer P. (2013) Global analysis of eukaryotic mRNA degradation reveals Xrn1-dependent buffering of transcript levels. Mol. Cell 52, 52–62 [DOI] [PubMed] [Google Scholar]
- 3. Shalem O., Dahan O., Levo M., Martinez M. R., Furman I., Segal E., Pilpel Y. (2008) Transient transcriptional responses to stress are generated by opposing effects of mRNA production and degradation. Mol. Syst. Biol. 4, 223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Goler-Baron V., Selitrennik M., Barkai O., Haimovich G., Lotan R., Choder M. (2008) Transcription in the nucleus and mRNA decay in the cytoplasm are coupled processes. Genes Dev. 22, 2022–2027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bregman A., Avraham-Kelbert M., Barkai O., Duek L., Guterman A., Choder M. (2011) Promoter elements regulate cytoplasmic mRNA decay. Cell 147, 1473–1483 [DOI] [PubMed] [Google Scholar]
- 6. Haimovich G., Medina D. A., Causse S. Z., Garber M., Millán-Zambrano G., Barkai O., Chávez S., Pérez-Ortín J. E., Darzacq X., Choder M. (2013) Gene expression is circular: factors for mRNA degradation also foster mRNA synthesis. Cell 153, 1000–1011 [DOI] [PubMed] [Google Scholar]
- 7. Malagon F., Kireeva M. L., Shafer B. K., Lubkowska L., Kashlev M., Strathern J. N. (2006) Mutations in the Saccharomyces cerevisiae RPB1 gene conferring hypersensitivity to 6-azauracil. Genetics 172, 2201–2209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ruan W., Lehmann E., Thomm M., Kostrewa D., Cramer P. (2011) Evolution of two modes of intrinsic RNA polymerase transcript cleavage. J. Biol. Chem. 286, 18701–18707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Walmacq C., Kireeva M. L., Irvin J., Nedialkov Y., Lubkowska L., Malagon F., Strathern J. N., Kashlev M. (2009) Rpb9 subunit controls transcription fidelity by delaying NTP sequestration in RNA polymerase II. J. Biol. Chem. 284, 19601–19612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rosenheck S., Choder M. (1998) Rpb4, a subunit of RNA polymerase II, enables the enzyme to transcribe at temperature extremes in vitro. J. Bacteriol. 180, 6187–6192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Maillet I., Buhler J. M., Sentenac A., Labarre J. (1999) Rpb4p is necessary for RNA polymerase II activity at high temperature. J. Biol. Chem. 274, 22586–22590 [DOI] [PubMed] [Google Scholar]
- 12. Miyao T., Barnett J. D., Woychik N. A. (2001) Deletion of the RNA polymerase subunit RPB4 acts as a global, not stress-specific, shut-off switch for RNA polymerase II transcription at high temperatures. J. Biol. Chem. 276, 46408–46413 [DOI] [PubMed] [Google Scholar]
- 13. Choder M., Young R. A. (1993) A portion of RNA polymerase II molecules has a component essential for stress responses and stress survival. Mol. Cell. Biol. 13, 6984–6991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Edwards A. M., Kane C. M., Young R. A., Kornberg R. D. (1991) Two dissociable subunits of yeast RNA polymerase II stimulate the initiation of transcription at a promoter in vitro. J. Biol. Chem. 266, 71–75 [PubMed] [Google Scholar]
- 15. Mosley A. L., Hunter G. O., Sardiu M. E., Smolle M., Workman J. L., Florens L., Washburn M. P. (2013) Quantitative proteomics demonstrates that the RNA polymerase II subunits Rpb4 and Rpb7 dissociate during transcriptional elongation. Mol. Cell. Proteomics 12, 1530–1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jasiak A. J., Hartmann H., Karakasili E., Kalocsay M., Flatley A., Kremmer E., Strässer K., Martin D. E., Söding J., Cramer P. (2008) Genome-associated RNA polymerase II includes the dissociable Rpb4/7 subcomplex. J. Biol. Chem. 283, 26423–26427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Farago M., Nahari T., Hammel C., Cole C. N., Choder M. (2003) Rpb4p, a subunit of RNA polymerase II, mediates mRNA export during stress. Mol. Biol. Cell 14, 2744–2755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Harel-Sharvit L., Eldad N., Haimovich G., Barkai O., Duek L., Choder M. (2010) RNA polymerase II subunits link transcription and mRNA decay to translation. Cell 143, 552–563 [DOI] [PubMed] [Google Scholar]
- 19. Selitrennik M., Duek L., Lotan R., Choder M. (2006) Nucleocytoplasmic shuttling of the Rpb4p and Rpb7p subunits of Saccharomyces cerevisiae RNA polymerase II by two pathways. Eukaryot. Cell 5, 2092–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Haimovich G., Choder M., Singer R. H., Trcek T. (2013) The fate of the messenger is pre-determined: a new model for regulation of gene expression. Biochim. Biophys. Acta 1829, 643–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lotan R., Bar-On V. G., Harel-Sharvit L., Duek L., Melamed D., Choder M. (2005) The RNA polymerase II subunit Rpb4p mediates decay of a specific class of mRNAs. Genes Dev. 19, 3004–3016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lotan R., Goler-Baron V., Duek L., Haimovich G., Choder M. (2007) The Rpb7p subunit of yeast RNA polymerase II plays roles in the two major cytoplasmic mRNA decay mechanisms. J. Cell Biol. 178, 1133–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Choder M. (2011) mRNA imprinting: additional level in the regulation of gene expression. Cell. Logist. 1, 37–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dahan N., Choder M. (2013) The eukaryotic transcriptional machinery regulates mRNA translation and decay in the cytoplasm. Biochim. Biophys. Acta 1829, 169–173 [DOI] [PubMed] [Google Scholar]
- 25. Higuchi R., Krummel B., Saiki R. K. (1988) A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 16, 7351–7367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961 [DOI] [PubMed] [Google Scholar]
- 27. Schwalb B., Schulz D., Sun M., Zacher B., Dümcke S., Martin D. E., Cramer P., Tresch A. (2012) Measurement of genome-wide RNA synthesis and decay rates with Dynamic Transcriptome Analysis (DTA). Bioinformatics 28, 884–885 [DOI] [PubMed] [Google Scholar]
- 28. Miller C., Schwalb B., Maier K., Schulz D., Dümcke S., Zacher B., Mayer A., Sydow J., Marcinowski L., Dölken L., Martin D. E., Tresch A., Cramer P. (2011) Dynamic transcriptome analysis measures rates of mRNA synthesis and decay in yeast. Mol. Syst. Biol. 7, 458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lovén J., Orlando D. A., Sigova A. A., Lin C. Y., Rahl P. B., Burge C. B., Levens D. L., Lee T. I., Young R. A. (2012) Revisiting global gene expression analysis. Cell 151, 476–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Duan R., Rhie B. H., Ryu H. Y., Ahn S. H. (2013) The RNA polymerase II Rpb4/7 subcomplex regulates cellular lifespan through an mRNA decay process. Biochem. Biophys. Res. Commun. 441, 266–270 [PubMed] [Google Scholar]
- 31. Armache K. J., Mitterweger S., Meinhart A., Cramer P. (2005) Structures of complete RNA polymerase II and its subcomplex, Rpb4/7. J. Biol. Chem. 280, 7131–7134 [DOI] [PubMed] [Google Scholar]
- 32. Pillai B., Verma J., Abraham A., Francis P., Kumar Y., Tatu U., Brahmachari S. K., Sadhale P. P. (2003) Whole genome expression profiles of yeast RNA polymerase II core subunit, Rpb4, in stress and nonstress conditions. J. Biol. Chem. 278, 3339–3346 [DOI] [PubMed] [Google Scholar]
- 33. Verma-Gaur J., Rao S. N., Taya T., Sadhale P. (2008) Genomewide recruitment analysis of Rpb4, a subunit of polymerase II in Saccharomyces cerevisiae, reveals its involvement in transcription elongation. Eukaryot. Cell 7, 1009–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Runner V. M., Podolny V., Buratowski S. (2008) The Rpb4 subunit of RNA polymerase II contributes to cotranscriptional recruitment of 3′ processing factors. Mol. Cell. Biol. 28, 1883–1891 [DOI] [PMC free article] [PubMed] [Google Scholar]