Background: The AP-2 adaptor is a central protein-protein interaction hub during clathrin-mediated endocytosis.
Results: Introducing a specific phosphotyrosine mimetic on the large AP-2 β2 subunit selectively regulates protein interactions with the adaptor complex.
Conclusion: A directed post-translational modification can locally rewire the AP-2 interaction network.
Significance: This provides a plausible mechanism for regional cargo uncoupling at cell surface clathrin coats.
Keywords: Endocytosis, G Protein-coupled Receptor (GPCR), Intracellular Trafficking, Lipoprotein, Phosphoproteomics, Phosphotyrosine Signaling, Clathrin
Abstract
The AP-2 clathrin adaptor complex oversees endocytic cargo selection in two parallel but independent manners. First, by physically engaging peptide-based endocytic sorting signals, a subset of clathrin-dependent transmembrane cargo is directly collected into assembling buds. Synchronously, by interacting with an assortment of clathrin-associated sorting proteins (CLASPs) that independently select different integral membrane cargo for inclusion within the incipient bud, AP-2 handles additional cargo capture indirectly. The distal platform subdomain of the AP-2 β2 subunit appendage is a privileged CLASP-binding surface that recognizes a cognate, short α-helical interaction motif. This signal, found in the CLASPs β-arrestin and the autosomal recessive hypercholesterolemia (ARH) protein, docks into an elongated groove on the β2 appendage platform. Tyr-888 is a critical constituent of this spatially confined β2 appendage contact interface and is phosphorylated in numerous high-throughput proteomic studies. We find that a phosphomimetic Y888E substitution does not interfere with incorporation of expressed β2-YFP subunit into AP-2 or alter AP-2 deposition at surface clathrin-coated structures. The Y888E mutation does not affect interactions involving the sandwich subdomain of the β2 appendage, indicating that the mutated appendage is folded and operational. However, the Y888E, but not Y888F, switch selectively uncouples interactions with ARH and β-arrestin. Phyogenetic conservation of Tyr-888 suggests that this residue can reversibly control occupancy of the β2 platform-binding site and, hence, cargo sorting.
Introduction
Post-translational phosphorylation of Ser, Thr, and Tyr residues is a very prevalent modification across eukaryotic phyla, occurring in at least 30% of expressed proteins from the unicellular Saccharomyces cerevisiae to complex metazoans like primates (1–4). There are estimates of 500,000 potential phosphorylation acceptor residues (5) and >100,000 phosphosites/cell (4); a data set of 68,379 phosphopeptides has been compiled from growth-arrested HeLa cell populations (6). Appropriately, it is widely accepted that phosphorylation regulates the activity of endocytosis. Perhaps the best established mechanism is when extracellular ligand binding elicits receptor phosphorylation upon cytosol-oriented residues, which then act either as physical internalization signals or initiate the display or generation of a controlled uptake signal (7, 8). Internalization of G protein-coupled receptors (9) and the epidermal growth factor receptor (10) are emblematic of this mode of post-translationally activated clathrin-mediated endocytosis. The reciprocal is also known, where site-specific phosphorylation can cloak or incapacitate a constitutively internalization signal. For instance, in the ionotropic γ-aminobutyric acid A (GABAA) receptor, phosphorylation of flanking Tyr side chains within a YXYXXØ-type sorting signal (where Ø represents a bulky hydrophobic residue) on the γ2 subunit prevents efficient clathrin-mediated endocytosis (11). Analogously, the prompt internalization directed by a YXXØ sorting signal in the co-receptor CTLA4 expressed on activated T lymphocytes is negatively regulated by Tyr phosphorylation (12).
In addition to trafficking transmembrane cargo, numerous soluble core components of the endocytic coat are also phosphorylated. Vertebrate clathrin coat proteins are dubbed “dephosphins” (13) because many of these factors are swiftly dephosphorylated in concert by calcineurin upon depolarization of the nerve terminal (14). This established rapid change in overall phosphostatus is not, however, accompanied by a clear mechanistic understanding of the precise regulatory consequences of phosphorylation-dephosphorylation cycles. It is known that there are global phosphorylation differences in the heterotetrameric clathrin adaptor AP-2 when exchanging between cytosolic and membrane-associated pools and that hyperphosphorylation impedes AP-2 interactions with clathrin triskelia (15). Also, phosphorylation of Thr-156 on the AP-2 μ2 subunit promotes recognition of YXXØ-type endocytic sorting signals (16–18). The PhosphoSitePlus discovery mode database (19) currently identifies Thr(P)-156 in at least 35 independent mass spectrometry studies. Yet the crystal structure of the AP-2 heterotetramer core in the open, cargo-binding conformation fails to reveal precisely why this phosphorylation event biases cargo capture (20). More recently, phosphoproteomics has categorized numerous additional phosphorylated side chains in the AP-2 α, β2, μ2, and σ2 subunits from many different organisms, cell types, and tissues (19).
Proteins can be multiply, sequentially, and redundantly phosphorylated (19, 21), often making categorical dissection of the precise physiological role of individual modifications challenging. In fact, the biological role of the vast majority of the phosphorylated residues catalogued in high-throughput mass spectrometry-based screens is presently elusive and unsettled, and only about 2% or less of mapped phosphosites are Tyr(P), the bulk being Ser(P) (4, 5, 22). The low relative frequency of Tyr(P) makes delineation of phosphoacceptor function more tractable. Also germane is that binding interfaces are predisposed to evolutionarily conserved phosphorylation (23), and the preponderance of phosphorylation sites map to intrinsically disordered regions of proteins (22, 24). Nevertheless, overall, Tyr(P) tends to be enriched on folded domains and at structured contact interfaces (24). Here, we examine the physiological consequences of the most prevalent Tyr phosphosite modification, Tyr(P)-888, upon the independently folded appendage of the AP-2 adaptor β2 subunit (the product of the AP2B1 gene).
EXPERIMENTAL PROCEDURES
All the experiments described here were carried out at least three times independently, with comparable results obtained. Selected but representative data are shown for each set of experiments.
Cell Culture
Spontaneously immortalized mouse embryonic fibroblasts (β2+/+ and β2−/− iMEFs)2 (25), derived from OVE427 AP-2 β2 subunit homozygous wild-type or null embryos (26), were maintained in DMEM supplemented with 10% fetal calf serum (FCS), 2 mm l-glutamine, and 1× antibiotic/antimycotic mixture (Sigma-Aldrich). The human embryonic kidney (HEK) 293 cells expressing Cre recombinase (Cre 8 cells) used for adenovirus generation (27, 28) were cultured in DMEM supplemented with 10% defined FCS and 1× penicillin/streptomycin (Invitrogen). Transfection with Lipofectamine 2000 (Invitrogen) and the appropriate amount of DNA was as directed by the manufacturer.
Antibodies and Recombinant DNA Constructs
We used the following primary antibodies: an AP-2 α subunit monoclonal antibody (mAb) AP.6 (29) and anti-clathrin heavy chain mAb TD.1 (30) generously provided by Frances Brodsky (University of California, San Francisco, CA), the affinity-purified rabbit anti-Eps15 polyclonal antibody (shared by Ernst Ungewickell, Medizinische Hochschule Hannover, Germany) (31), and an anti-GFP polyclonal antibody, B5 (a gift from Phyllis Hanson, Washington University School of Medicine, St. Louis, MO) (32). Preparation and use of the anti-epsin 1 polyclonal antibody (33) and the affinity-purified anti-AP-1/2 β1/β2 subunit antibody GD/1 (34), the anti-AP-1 γ subunit antibody AE/1 (34), and the anti-autosomal recessive hypercholesterolemia (ARH) antibody (35) is described elsewhere. Commercial primary antibodies were the anti-AP-2 α subunit mAb C-8 from Santa Cruz Biotechnology, Inc. (Dallas, TX), an anti-β-arrestin polyclonal antibody from Sigma-Aldrich, the anti-tubulin mAb E7 from the Developmental Studies Hybridoma Bank (University of Iowa, Iowa City, IA), and the anti-DsRed monoclonal antibody obtained from Clontech (Mountain View, CA). Alexa Fluor 568-conjugated goat anti-mouse IgG secondary antibody was purchased from Molecular Probes. The horseradish peroxidase-conjugated donkey anti-mouse and anti-rabbit secondary antibodies were procured from GE Healthcare.
The Rattus norvegicus AP-2 β2-YFP fusion plasmid (provided by Alexander Sorkin, University of Pittsburgh, Pittsburgh, PA) has been described (25, 36). QuikChange site-directed mutagenesis was used to engineer the β2 platform subdomain (Y888E or Y888F) mutants using this AP-2 β2-YFP plasmid as template. A full-length cDNA encoding R. norvegicus epsin 1 that was utilized to amplify residues 229–407 for cloning into pGEX-4T-1 (GST-DPW) was a gift from Pietro de Camilli (Yale University School of Medicine, New Haven, CT). The tandem dimer Tomato red fluorescent protein (tdRFP)-tagged Bos taurus β-arrestin 1 (I386A/V387A) (tdRFP-β-arrestin 1 (IV → AA) double mutant was generated as we have described previously (25). The wild-type β2-YFP or Y888E or Y888F mutants were subcloned into pAdLox using the Cold Fusion Cloning Kit (SBI Systems Biosciences, Mountain View, CA) following the manufacturer's instructions. The wild-type GST-tagged AP-2 β2 (R. norvegicus residues 714–951) appendage (GST-β2) and the sandwich subdomain site mutant β2 (Y815A) appendage in pGEX-4T-1 vector were constructed as outlined previously (37–39). The wild-type β2 appendage in pGEX-4T-1 was used as the template to engineer the GST-β2 (Y888E) or GST-β2 (Y888F) platform mutants using QuikChange site-directed mutagenesis. The GST-β-arrestin 1 CT (residues 331–418) (37) and GST-ARH C1 (residues 180–308) (38) are described elsewhere, and the required mutations were similarly generated using the QuikChange procedure. All constructs were verified by automated dideoxynucleotide sequencing. Complete details of primers and sequences are available upon request.
Protein Preparation
GST and various GST fusion proteins were expressed in Escherichia coli strain BL21 with a standard protocol (33, 37–39). In brief, protein expression was induced in 2× TY medium by adding isopropyl-1-thio-β-d-galactopyranoside (0.2 mm final concentration) to log phase bacterial cultures (A600 ∼0.7) at room temperature. Following the addition of isopropyl-1-thio-β-d-galactopyranoside, cultures were allowed to grow for an additional 16 h with constant shaking. Bacteria were harvested by centrifugation at 15,000 × g for 15 min at 4 °C and stored at −80 °C. Bacterial pellets were resuspended in 50 mm Tris-HCl, pH 7.5, 10 mm β-mercaptoethanol, 300 mm NaCl, and 0.2% (w/v) Triton X-100 before sonication on ice. Cellular debris was removed by centrifugation at 23,700 × g for 30 min at 4 °C, and GST fusion proteins were recovered on glutathione-Sepharose. After extensive washing with phosphate-buffered saline (PBS), GST fusion proteins were eluted with 25 mm Tris-HCl, pH 8.0, 5 mm DTT, 200 mm NaCl, and 10 mm glutathione on ice, dialyzed into PBS supplemented with 1 mm DTT, and stored at −80 °C until further use.
Cell and Tissue Extract Preparation
Rat brain cytosol was prepared from frozen rat brains (PelFreez) essentially as described (33, 37–39). In brief, the cytosol was prepared by sequential differential centrifugation of homogenized rat brains in a buffer containing 25 mm HEPES-KOH, pH 7.2, 2 mm EDTA, 2 mm EGTA, 250 mm sucrose, supplemented with 1 mm phenylmethylsulfonyl fluoride and Complete protease inhibitor (Roche Applied Science). The supernatant fraction obtained after centrifugation at 105,000 × g is designated the cytosol and was stored in small aliquots at −80 °C until further use. Prior to the binding assays, rat brain cytosol aliquots were thawed and adjusted to 25 mm HEPES-KOH, pH 7.2, 125 mm potassium acetate, 5 mm magnesium acetate, 2 mm EDTA, 2 mm EGTA, and 1 mm DTT (1× assay buffer) by dilution of a 10× stock and then centrifuged at 245,000 × g (TLA-100.4 rotor) at 4 °C for 20 min to remove any remaining insoluble debris.
For fibroblast lysates, control β2−/− iMEFs, β2−/− iMEFs transfected with the tdRFP-β-arrestin 1 (IV → AA) mutant or β2−/− iMEFs infected with adenovirus encoding either β2-YFP wild type or Y888E or Y888F mutant at multiplicities of infection (MOI) of 500 for 24 h, were trypsinized, washed in PBS, and lysed on ice in 25 mm Hepes-KOH, pH 7.2, 250 mm sucrose, 1% Triton X-100, 2 mm EDTA, 2 mm EGTA, and Complete miniprotease inhibitor mixture (Roche Applied Science) for 30 min, followed by brief sonication on ice. Cell debris was removed by centrifugation at 18,000 × g for 20 min at 4 °C, and the supernatants were stored at −80 °C until further use. Prior to use in binding assays, thawed lysates were adjusted to 1× assay buffer by diluting a 10× stock and then centrifuged at 18,000 × g at 4 °C for 20 min to remove any remaining insoluble debris. For experiments other than the binding assays, control β2−/− iMEFs or those infected with adenovirus encoding β2WT-YFP, β2Y888E-YFP, or β2Y888F-YFP at various MOIs for 24 h or at an MOI of 500 for 24–72 h were trypsinized, washed in PBS, and lysed in SDS-PAGE sample buffer using sonication and then heated at 95 °C for 5 min.
Binding Assays
GST fusion protein pull-down assays were performed as described previously (33, 37–39). In brief, GST or GST fusion proteins (100–200 μg) were immobilized on ∼25 μl of packed glutathione-Sepharose beads at 4 °C for 1 h. Beads were then collected by centrifugation (10,000 × g, 1 min), washed, and resuspended in 50 μl of assay buffer. Aliquots of clarified rat brain cytosol (250 μl) or β2−/− iMEF lysate (200 μl) was added, and the tubes were incubated at 4 °C for 2 h with continuous mixing. The glutathione-Sepharose beads were recovered by centrifugation (10,000 × g, 1 min), and then an aliquot of 40 μl of each supernatant was retained and adjusted to 100 μl with SDS sample buffer (designated the supernatant). The pellets were washed thoroughly (four times with ∼1.5 ml of ice-cold PBS/wash), and then each pellet was resuspended in SDS sample buffer up to ∼80 μl (designated the pellet fraction). Unless otherwise mentioned, 10-μl aliquots, corresponding to ∼2% (β2−/− iMEF lysate) or ∼1.5% (rat brain cytosol) of each supernatant and ∼12.5% of each pellet fraction, were analyzed using SDS-PAGE and immunoblotting.
Electrophoresis and Immunoblotting
Samples were resolved on freshly prepared 11% polyacrylamide gels formulated from a stock solution of acrylamide/bisacrylamide (30:0.4 ratio). After SDS-PAGE, proteins were either stained with Coomassie Blue or transferred to nitrocellulose membranes. Blots were blocked overnight or for 2 h in 5% skim milk in 10 mm Tris-HCl, pH 7.8, 150 mm NaCl, 0.1% Tween 20 (TBST), and then portions of the blots were incubated with primary antibodies as indicated in the figure legends. After incubation with horseradish peroxidase (HRP)-conjugated anti-mouse and/or anti-rabbit IgG, immunoreactive protein bands were visualized with enhanced chemiluminescence. For the portions of the blots probed with anti-β-arrestin antibody, the blots were stripped using 62 mm Tris-HCl, pH 6.8, 100 mm β-mercaptoethanol, 2% SDS, washed with TBST, blocked, and reprobed with anti-DsRed monoclonal antibody. Densitometric quantitation was carried out using ImageJ (National Institutes of Health) (40).
Recombinant Adenovirus Preparation and Infection
The recombinant YFP-tagged wild-type, Y888E mutant, or Y888F mutant AP-2 β2 subunit adenoviruses were generated (27) and purified as described (28, 41). Viral titer was estimated by measuring A260 of the final preparation and was typically 7–9 × 1012 particles/ml. Adenoviral infection was performed as described previously (28). In brief, AP-2 β2−/− iMEFs were counted, and an equal number of cells were seeded onto tissue culture dishes in the presence or absence of glass coverslips and allowed to attach and grow for 24 h. After 24 h, cells were washed with PBS supplemented with 1 mm MgCl2 (PBS-M) and incubated in fresh PBS-M for an additional 5 min at 37 °C. Following incubation, PBS-M was removed, and adenovirus was added to cells (at various MOIs) in PBS-M and allowed to adsorb for several h before the addition of culture medium.
Immunofluorescence and Confocal Microscopy
AP-2 β2−/− iMEFs seeded on round glass coverslips and infected with adenovirus encoding β2WT-YFP at a MOI of 500 for 24, 48, or 72 h were fixed with 2% paraformaldehyde, permeabilized, and immunolabeled with anti-AP-2 α subunit mAb AP.6 and Alexa568-labeled goat anti-rabbit secondary antibody. AP-2 β2−/− iMEFs infected with adenovirus encoding β2-YFP wild type, Y888E, or Y888F at a MOI of 500 for 24 h and transfected with tdRFP-β-arrestin 1 (IV → AA) mutant were fixed with 2% paraformaldehyde and washed with PBS. The cells were then mounted with Cytoseal. Cell surface transferrin binding experiments were performed by switching AP-2 β2−/− iMEFs infected with adenovirus encoding the various β2-YFP proteins at a MOI of 500 for 24 h to prewarmed DMEM supplemented with 25 mm Hepes, pH 7.2, and 0.5% BSA (starvation medium) and incubated at 37 °C for 1 h. Following incubation, the cells were switched to ice-cold starvation medium for 1 h at 4 °C and incubated for an additional 1 h at 4 °C in ice-cold starvation medium containing 25 μg/ml Alexa 568-labeled transferrin (Molecular Probes) and then washed with PBS, fixed, and mounted with Cytoseal.
Confocal images were acquired using Olympus Fluoview 1000 microscope with a PlanApo N (×60/1.42 numerical aperture) oil objective. Sequential scanning was applied for acquiring individual emission channels when multiple fluorophores were involved. Data images were acquired using FV10-ASW software and exported in TIFF format. The TIFF files were then imported into Adobe Photoshop CS for making minor adjustments to brightness and/or contrast. The final figures were imported and assembled in Adobe Illustrator software.
Through-the-objective-type total internal reflection fluorescence microscopy (TIR-FM) was conducted as described (25, 42), utilizing a ×100 CFI Apochromat series 1.49 numerical aperture TIR-FM objective mounted on a Nikon Eclipse Ti inverted microscope equipped with a Perfect Focus System device, Agilent 4 color launch, and Elements AR software. Images were collected using an Andor Zyla 5.5 camera (Andor Technology USA, South Windsor, CT), and all colors were collected with a single quad pass (405, 488, 561, 633 nm) cube to ensure registration. Emission images were sorted with a high speed (20 ms) FLI (Finger Lakes Instrumentation, Lima, NY) filter wheel.
RESULTS
All heterotetrameric adaptor complexes (AP-1 to AP-5) have a common architecture, being composed of two different large chains (designated α–ζ subunits), a medium chain (termed a μ subunit), and a small chain (the σ subunit) (43, 44). Despite the clear overall structural homology and shared quaternary organization of the complexes, in general, these subunits display limited (<50%) primary sequence identity (43, 44). The exceptions are the ubiquitously expressed large β1 and β2 subunits of AP-1 and AP-2, respectively, which are ∼85% identical (45–47). Indeed, endogenous β1 incorporates promiscuously into AP-2 and β2 into AP-1 in vivo (25, 26, 48–50), whereas the Drosophila melanogaster (51), Caenorhabditis elegans (43, 52), and Dictyostelium discoideum (53) genomes each encode only a single β chain that is incorporated into both AP-1 and AP-2 heterotetramers. Compared with the early embryonic lethality of AP-2 μ2 subunit gene disruption (54), mice nullizygous for the co-assembled β2 subunit have a surprisingly mild developmental phenotype (26). This is because the standard β1 subunit reconstitutes functional AP-2, albeit at lower levels than normal (25, 26). β2−/− mice are born but die perinatally because a cleft palate prevents proper nursing. The newborns are otherwise remarkably normal in anatomy and physiology (26).
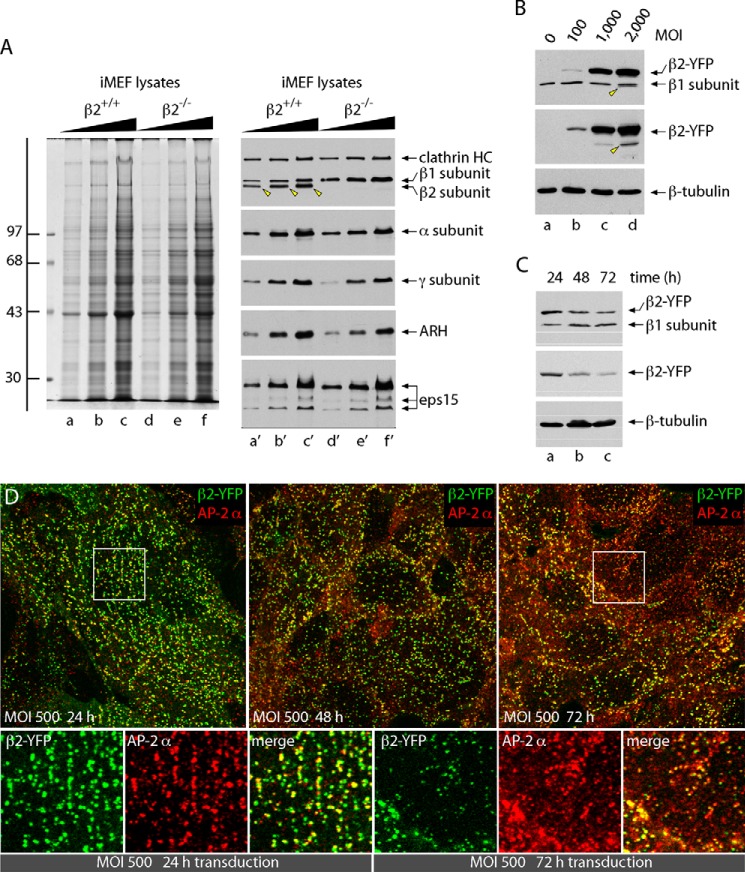
We have utilized iMEFs derived from homozygous β2−/−animals (25, 26) for cell-based AP-2 reconstitution experiments. As expected, the gross protein compositions of wild-type and null iMEF whole-cell lysates are very similar (Fig. 1A, lanes a–f), while immunoblots of littermate-derived β2+/+ iMEFs demonstrate that in these cells, the β1 and β2 chains are present in roughly similar amounts at steady state (Fig. 1A, lanes a′–c′). In many cultured cells and tissues, expression levels from the AP1B1 and AP2B1 genes are similar (55), although in neuronal tissue, the AP2B1 product predominates considerably (56, 57). By contrast, the β2-nullizygous iMEFs lack completely the faster migrating β2 subunit (arrowheads) and evidently compensate by expressing about 2-fold-elevated levels of β1 (Fig. 1A, lanes d′–f′), yet the clathrin heavy chain protein levels and the abundance of adaptors (AP-2 α subunit and AP-1 γ subunit) and several clathrin-associated sorting proteins (CLASPs; ARH, and Eps15) are analogous in the two lines of iMEFs (Fig. 1A, lanes a′–f′).
FIGURE 1.
Adenovirus-mediated re-expression of the AP-2 β2 subunit in β2−/− iMEFs. A, serial dilutions of whole-cell lysates from either β2+/+ (lanes a–c and a′–c′) or β2−/− iMEFs (lanes d–f and d′–f′) were resolved by SDS-PAGE. Duplicate gels were either stained with Coomassie Blue (left) or transferred to nitrocellulose (right) and probed with antibodies directed against the clathrin heavy chain (HC) mAb TD.1 and the anti-β1/β2 subunit antibody GD/1 or with an anti-AP-2 α subunit mAb C-8 or affinity-purified anti-AP-1 γ subunit antibody AE/1 or affinity-purified anti-ARH or anti-Eps15 antibodies. The position of the molecular mass standards (in kDa) is shown on the left, and only the relevant portion of each blot is presented. Homozygous gene disruption of the Ap2b1 locus results in loss of the ∼100-kDa β2 subunit product (identified by arrowheads in the β2+/+ lysates) and an approximate 2-fold up-regulation of the β1 subunit in compensation. B, control β2−/− MEF cells (MOI = 0, lane a) or iMEFs infected for 24 h with an adenovirus encoding a YFP-tagged full-length wild-type AP-2 β2 subunit (β2-YFP) at the indicated viral MOIs (lanes b–d) were lysed and resolved by SDS-PAGE, and replicate immunoblots were probed with anti-β1/β2 subunit antibody GD/1 (top), polyclonal anti-GFP antibodies (middle), or mouse anti-tubulin mAb E7 (bottom). The appearance of a minor β2-YFP degradation product, migrating just below the native β1 subunit (arrowhead), is indicated. C, β2−/− iMEF cells infected with β2-YFP at a MOI of 500 for 24 (lane a), 48 (lane b), or 72 h (lane c) were lysed and resolved by SDS-PAGE, and duplicate immunoblots were probed as in A. Note that the expression level of β2WT-YFP decreases as a function of infection duration with a concomitant increase in the level of the endogenous cellular β1 subunit. D, β2−/− cells infected with β2-YFP (green) at a MOI of 500 for 24, 48, or 72 h were fixed, permeabilized, and immunolabeled with anti-AP-2 α subunit mAb AP.6 (red). Representative confocal sections of the adherent ventral plasma membrane region are shown. Enlarged, color-separated views of selected regions from the 24- and 72-h transduction panels (boxed) are shown below.
The reconstitution approach entails infecting the β2−/− iMEFs with adenovirus encoding a full-length β2 subunit polypeptide tagged at the C terminus with YFP; numerous studies attest to the proper folding and operation of this β2-YFP subunit (25, 36, 55, 58). Preliminary titration experiments show dose-dependent expression of the wild-type β2-YFP in the β2−/− iMEFs (Fig. 1B, lanes b–d compared with lane a). On immunoblots, anti-β1/β2 antibodies reveal that adding adenovirus at a MOI of 500 results in production of the β2-YFP chain in excess of the endogenous β1 subunit at 24 h postinfection (Fig. 1C, lane a) and that the level of the virus-encoded protein drops substantially by 72 h postinfection (Fig. 1C, lane c). This reconstitution procedure therefore approximates the normal β1/β2 expression ratios (Fig. 1A, lanes a′–c′) 1 day following infection. Also, the β2-YFP fluorescent signal is located primarily in heterogeneously sized puncta distributed irregularly over the ventral surface of infected iMEFs at 24 h postinfection (Fig. 1D), with a high degree of coincidence with the endogenous AP-2 α subunit. By 72 h postinfection, the intensity of the β2-YFP drops, and a diffuse, featureless pool of α subunit immunoreactivity is increasingly evident, suggesting that the ectopic β2 chain is becoming limiting.
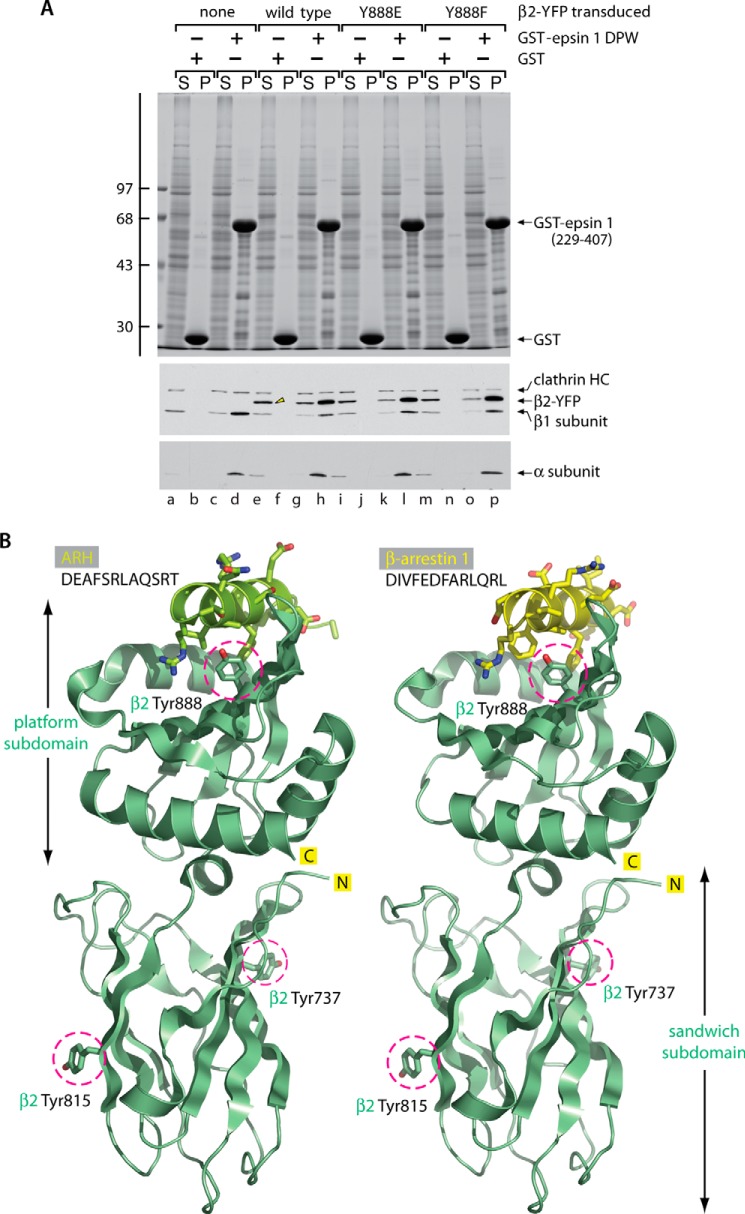
The proper incorporation of the virally transduced β2-YFP into heterotetrameric AP-2 complexes is verified biochemically by pull-down assays using cell lysates from uninfected or infected iMEFs (Fig. 2A). In uninfected β2−/− iMEF lysates, only the ∼105-kDa β1 subunit is present (25), and this chain is recovered in the pellet fraction, along with the AP-2 α subunit (Fig. 2A, lane d), which binds physically to the tandem Asp-Pro-Trp (DPW) repeats positioned within a GST fusion of the central region of the endocytic protein epsin 1 (33, 59). Because this tract from epsin 1 also contains a clathrin box sequence, a fraction of the cytosolic clathrin heavy chain associates independently with the immobilized GST fusion protein (33, 59, 60). Neither AP-1/2 subunits nor soluble clathrin trimers interact appreciably with GST alone (Fig. 2A, lane b). Similar analysis of the lysates from virus-infected iMEFs shows the incorporation of the ectopic ∼140-kDa β2-YFP chain into GST-epsin 1-bound AP-2 at the expense of the native β1 subunit (Fig. 2A, lane h, arrowhead). However, the expressed larger β2-YFP chain remains entirely in the supernatant, along with the endogenous α and β1 subunits, when incubated with the immobilized GST control (Fig. 2A, lanes e and f, arrowhead).
FIGURE 2.
Stable incorporation of adenovirus-encoded wild-type or mutant β2-YFP subunits into AP-2 complexes in β2−/− iMEFs. A, aliquots of ∼200 μg of either GST (lanes a, b, e, f, i, j, m, and n) or GST-epsin 1 DPW (residues 229–407; lanes c, d, g, h, k, l, o, and p) immobilized on glutathione-Sepharose were incubated with precleared lysates prepared from either uninfected β2−/− iMEFs (lanes a–d) or cells transduced for 24 h at a MOI of 500 with adenovirus encoding β2-YFP (lanes e–h), β2-YFP (Y888E) (lanes i–l), or β2-YFP (Y888F) (lanes m–p). After incubation at 4 °C for 2 h, the Sepharose beads were centrifuged and washed. Aliquots corresponding to ∼2% of each supernatant (S) and 12.5% of each pellet (P) fraction were resolved by SDS-PAGE and either stained with Coomassie Blue or transferred to nitrocellulose. Replicate blots were probed with a mixture of anti-clathrin HC mAb TD.1 and the anti-β1/β2 subunit antibody GD/1 or with an anti-AP-2 α subunit mAb. The position of the molecular mass standards (in kDa) is shown on the left, and only the relevant portion of each blot is presented. Note that when β2-YFP is expressed, this chain reduces the recovery of the endogenous β1 subunit in the pellet fractions containing AP-2 (lanes h, l, and p compared with lane d). B, ribbon representation of the structure of the β2 appendage domain (pale green) complexed with either the ARH (Protein Data Bank code 2G30; chartreuse) or β-arrestin 1 (Protein Data Bank code 2IV8; yellow) (D/E)nX1–2FXX(F/L)XXXR peptide. Selected Tyr residues on the β2 appendage (in a stick representation) are indicated (magenta circles). The primary sequence of each bound peptide is indicated, and the side chains on the bound α-helical peptides are shown in a stick representation with oxygen colored red and nitrogen blue.
Post-translational Modification at a Secondary Cargo Selection Site on the β2 Appendage
The bilobal α and β2 subunit appendages of AP-2 both operate as protein-protein interaction hubs, each with two spatially separate binding sites and numerous shared binding partners (7, 61–63). However, despite this structural homology, we have proposed that the distal platform subdomain of the AP-2 β2 appendage is a privileged docking site (25, 37, 38). This interaction surface is used primarily by the CLASPs ARH and β-arrestins to accomplish prompt cargo packaging in the face of ongoing uptake of different cargo (Fig. 2B) (37, 62, 64). Dissimilar to the extended interaction motifs that engage the three other binding surfaces on the α and β2 appendages, the (D/E)nX1–2FXX(F/L)XXXR motif found in β-arrestins and ARH contacts the AP-2 β2 platform subdomain structured as a three-turn α helix, packing the protruding anchor aromatic/aliphatic anchor side chains located on one face of the helix into the complementary β2 surface groove (Fig. 2B) (37, 62). Main chain hydrogen bonding of the α-helical scaffold reinforces the hydrophobic nature of the protein-protein interaction surface, analogous to a transmembrane α helix. This altered binding form accounts for β2-selective interactions. A vital residue on the cognate AP-2 β2 platform surface is Tyr-888, which contributes structurally to the hydrophobic acceptor pockets for both the proximal Phe and the trailing Phe/Leu motif side chains (Fig. 2B). A Y888A (65) or a Y888V (37, 62, 66) substitution eliminates ARH and β-arrestin binding, and Tyr-888 is virtually phylogenetically invariant from unicellular invertebrates to mammals. Intriguingly, natively phosphorylated Tyr-888 (Tyr(P)-888) has been identified at least 50 times in discovery mode high-throughput mass spectrometry investigations (see the PhosphoSitePlus Web site) (19, 67, 68). This is thus a natural post-translational modification and, because of its location, suggests that Tyr-888 could be a key phosphoregulatory side chain that could determine occupancy of the β2 platform (Fig. 3A), a hypothesis we explored using, in part, the β2-null iMEF reconstitution system.
FIGURE 3.
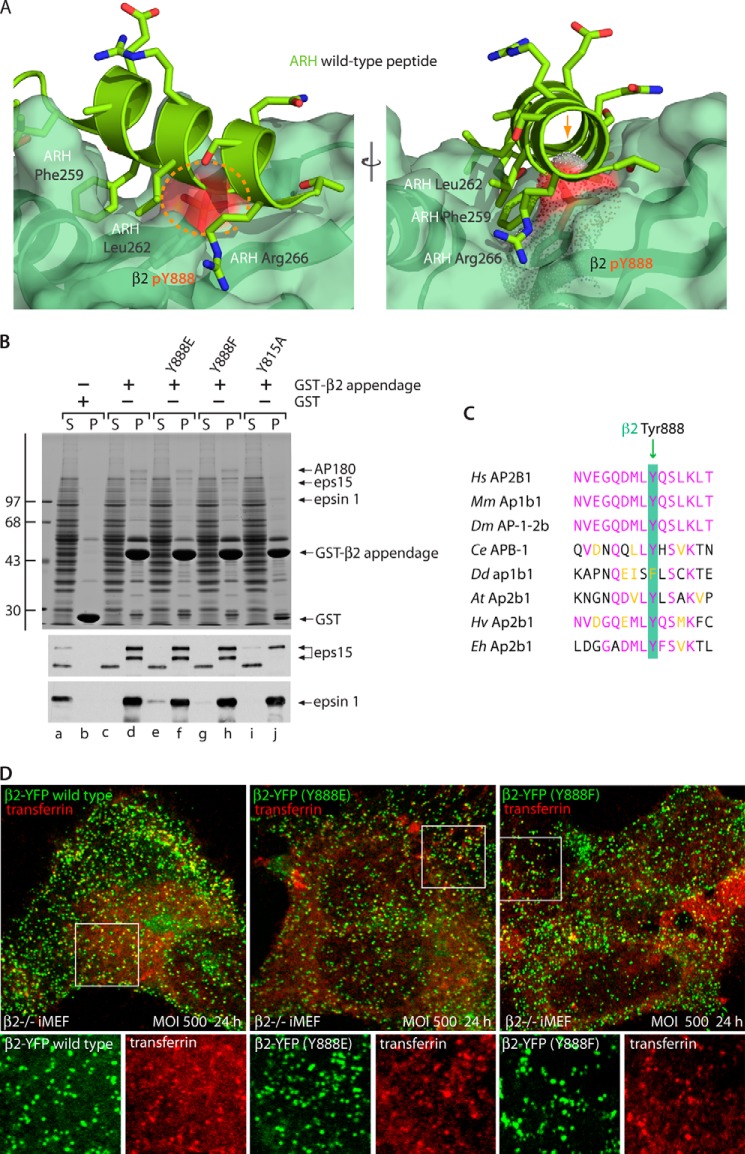
Tyr-888 mutations in the β2 appendage. A, steric clash at the β2-platform contact site with a Tyr(P)-888 modification. A combined ribbon and space-filling surface representation of the β2 appendage platform subdomain (pale green) bound to the ARH DEAFSRLAQSRT interaction peptide (Protein Data Bank code 2G30; chartreuse) (37) is shown with side chains in a stick representation. Element coloring is as follows: red, oxygen; blue, nitrogen; orange, phosphorus. A Tyr(P) modification is modeled on Tyr-888 (orange broken line on the left or the molecular surface of Tyr(P)-888 demarcated with spheres on the right). Obvious steric discordance with the ARH main chain α helix is indicated (orange arrow). B, aliquots of ∼100 μg of GST (lanes a and b); the wild-type GST-β2 appendage (lanes c and d); or mutant GST-β2 (Y888E) (lanes e and f), GST-β2 (Y888F) (lanes g and h), or GST-β2 (Y815A) (lanes i and j) mutant appendage immobilized on glutathione-Sepharose were incubated with precleared rat brain cytosol. After incubation at 4 °C for 2 h, the Sepharose beads were recovered by centrifugation and washed. Aliquots corresponding to ∼1.5% of each supernatant (S) and 12.5% of each pellet (P) were resolved by SDS-PAGE and either stained with Coomassie Blue or transferred to nitrocellulose. Replicate blots were probed with polyclonal anti-Eps15 or anti-epsin antibodies. The position of the molecular mass standards (in kDa) is shown at the left, and only the relevant portion of each blot is presented. C, phylogenetic alignment of the primary amino acid sequence surrounding Tyr-888 in selected β1/2 chains. The genus/species abbreviations and accession numbers are as follows. Hs, Homo sapiens (NP_001118); Mm, Mus musculus (NP_001229972); Dm, D. melangaster (NP_523415); Ce, C. elegans (NP_001022937; Dd, D. discoideum (XP_641846); At, Arabidopsis thaliana (NP_194077); Hv, Hydra vulgaris (XP_002159430; Eh, Emiliania huxleyi (a unicellular coccolithophore plankton) (XP_005767414). Identical residues are colored magenta, and conservatively substituted residues are yellow. Note that only D. discoideum has a Phe at the position equivalent to Tyr-888, and Phe is also present at this position in another slime mold, Polysphondylium pallidum (EFA78033). D, β2−/− iMEF cells infected with adenovirus encoding β2-YFP, β2-YFP (Y888E), or β2-YFP (Y888F) at a MOI of 500 for 24 h, as indicated, were serum-starved for 1 h and then surface-labeled with Alexa568-transferrin at 4 °C for 1 h before fixation. Representative confocal optical sections of the ventral plasma membrane are shown. Enlarged views of selected regions (boxed) are shown. Note the substantial colocalization of fluorescent transferrin with β2-YFP-, β2-YFP (Y888E)-, or β2-YFP (Y888F)-positive structures.
First, in pull-down assays utilizing the AP-2 β2 appendage domain (residues 714–951) fused to GST, the general consequence of selected mutation of Tyr-888 within the platform site illustrates that either a conservative Y888F or a phosphomimetic Y888E change is well tolerated (Fig. 3B, lanes e–h compared with lanes c and d). These results rule out gross misfolding or serious structural perturbations in these Tyr-888 mutants. The binding of the endocytic partner Eps15 to the mutants is very similar to the wild-type appendage (Fig. 3B, lanes f and h compared with lane d) and distinct from the inhibitory consequence of a Y815A mutation (Fig. 3B, lane j) within the proximal sandwich site on the adjacent β2 appendage subdomain (see Fig. 2B) (37, 62). By contrast, engagement of the platform site partner epsin 1 (37) is diminished weakly by the Y888E but not by the Y888F or Y815A mutations (Fig. 3B, lane f); some epsin 1 is still present in the supernatant fraction (Fig. 3B, lane e). The fact that the Y888F substitution has no appreciable effect on β2-platform binding indicates that the Tyr hydroxyl group is not essential for establishing or maintaining these interactions. Indeed, the crystal structures of either the ARH (37) or β-arrestin (62) (D/E)nX1–2FXX(F/L)XXXR peptide complexed with the β2 appendage reveal that Tyr-888 engages in aromatic stacking interactions without hydrogen bond donation through the hydroxyl group. Our interpretation of these findings is that the strict selection/retention of Tyr at this position over millions of years of evolution (with the exception of slime molds, where a Phe is instead present; Fig. 3C) is consistent with a primary (reversible) regulatory role for the hydroxyl group.
In addition, the full-length wild-type, Y888F-, and Y888E-encoding cDNAs transduced transiently into β2−/− nullizygous iMEFs are equally expressed and equivalently incorporated into heterotetrameric AP-2 complexes at ∼25 h postinfection (Fig. 2A). Again, in each case, co-assembly of the ectopic β2-YFP chain reduces the amount of endogenous β1 subunit incorporated into the AP-2 population (Fig. 2A, lanes h, i, and p compared with lane d). After infection, the β2-YFP wild type represents on average 85.9% of the total affinity-purified AP-2-associated β chain and 80.0 and 78.3% in the β2-YFP (Y888E) and β2-YFP (Y888F) mutants, respectively. Also, on the immunoblots, there is no evidence of extensive or major degradation products within the cell extracts from cells expressing the mutant forms of the β2-YFP protein. The three forms of β2-YFP each display random punctate distributions (69) on the ventral surface of infected iMEFs (Fig. 3D). These structures colocalize well with ligand-labeled surface transferrin receptors irrespective of whether β2 residue 888 is Tyr, Phe, or Glu, identifying the spots as plasma membrane clathrin-coated structures. We conclude that all three β2-YFP polypeptides are similarly incorporated into functional AP-2 heterotetramers in β2−/− iMEFs.
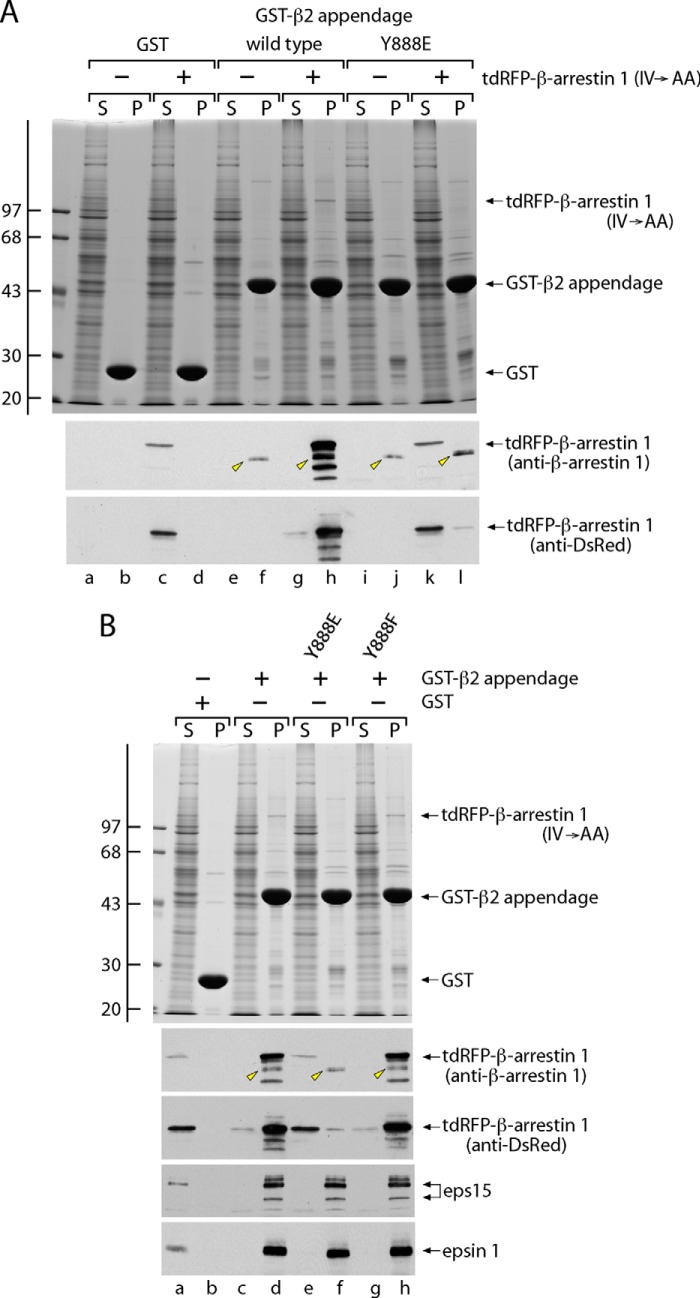
To investigate a selective effect of modification Tyr-888 on β2 appendage platform binding partners, we used a constitutively active form of β-arrestin 1, the so-called IV → AA mutant (25, 64). In the closed basal state, a tract of the C-terminal β2 platform-binding (D/E)nX1–2FXX(F/L)XXXR sequence of the β-arrestins is bound intramolecularly to the globular core of the protein (37, 70–72). By perturbing this intermolecular contact, the vicinal di-Ala substitution of Ile-386 and Val-387 shifts the equilibrium of β-arrestin 1 to the open, endocytically competent form (25, 64, 73) (see Fig. 7), which now binds robustly in pull-down assays. Compared with lysates from untransfected cells (Fig. 4A, lane f), a ∼110 kDa band is visible on Coomassie Blue-stained gels in the pellet fractions from incubations of the GST-β2 appendage with lysates of ectopic tdRFP-β-arrestin 1 (IV → AA)-producing cells (Fig. 4A, lane h). The identity of this band is substantiated with antibodies against both β-arrestin 1 and the DsRed RFP, and the distinctive stained band is not seen in parallel pellet fractions using immobilized control GST (Fig. 4A, lane d) and remains fully in the supernatant fraction (Fig. 4A, lane c). Nor is this immunoreactivity detected in lysates not transfected with the mutant β-arrestin 1 (Fig. 4A, lanes a, b, e, f, i, and j). In the same experiment, the GST-β2 appendage containing a Y888E substitution behaves quite differently; a dramatic decrease in tdRFP-β-arrestin 1 binding is seen, and no equivalent stained band is evident in the pellet (Fig. 4, A (lane l) and B (lane f)). The majority of the RFP-tagged β-arrestin is recovered in the supernatant fraction (Fig. 4, A (lane k) and B (lane e)). However, with the Y888F switch, the particular Coomassie-stained 110-kDa β-arrestin band is present in the pellet, and the GST fusion protein affinity-isolates immunoreactive tdRFP-β-arrestin 1 (Fig. 4B, lane h). Across phyla, only slime molds display a Phe at the Tyr-888 equivalent position (Fig. 3B), but these results restate that for platform binding, Tyr and Phe are functionally interchangeable at this location. Additional immunoblots confirm that interactions of Eps15 and epsin 1 with the wild-type, Y888E, and Y888F GST-β2 proteins are similar (Fig. 4B, lanes d, f, and h). Our interpretation of these results is that the Y888E mutant is selectively defective in interactions with (D/E)nX1–2FXX(F/L)XXXR-containing binding partners. The disparity between β-arrestin and epsin 1 in engaging the β2 appendage Y888E mutation stems from the fact that although epsin 1 contains a (D/E)nX1–2FXX(F/L)XXXR sequence, the KD for the β2 platform surface is at least 10-fold weaker than ARH or β-arrestins (37); therefore, epsin 1 utilizes a molecularly distinct manner to productively engage the β2 appendage.
FIGURE 7.
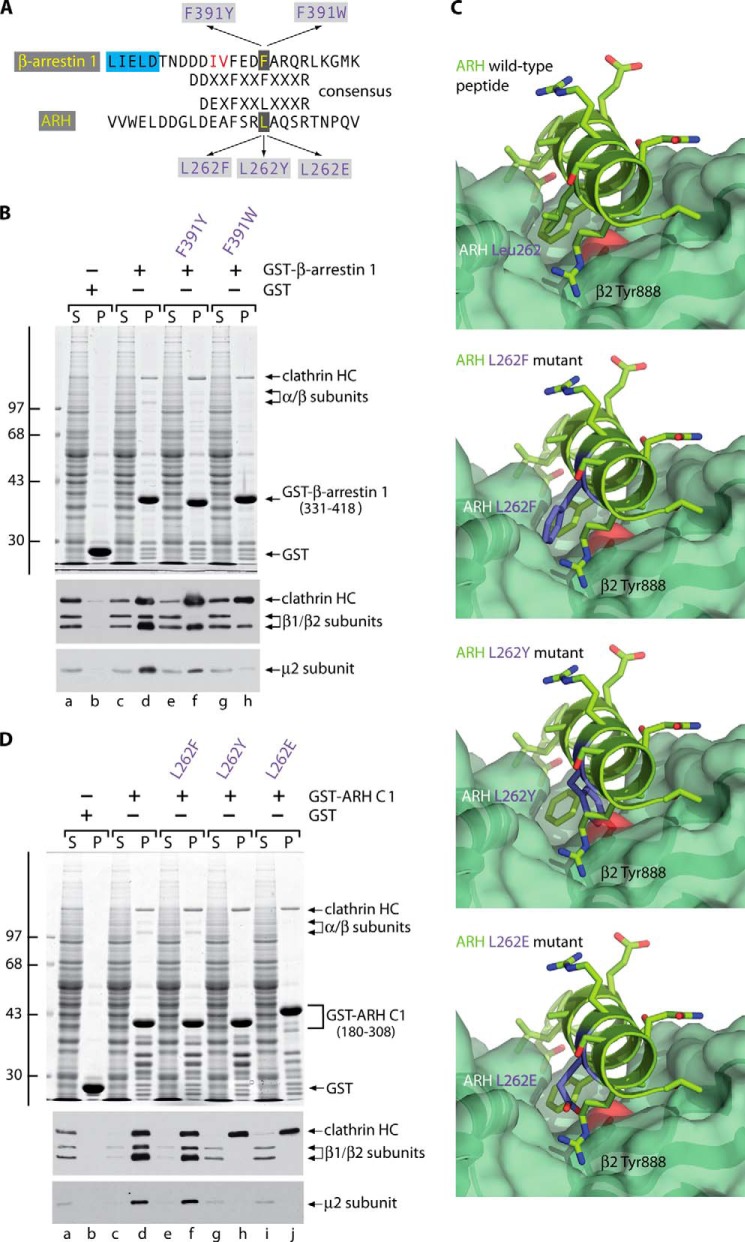
Complementary mutagenesis of the β2-platform binding partner interface. A, primary sequence alignment of the β-arrestin 1 and ARH (D/E)nX1–2FXX(F/L)XXXR-containing regions. The location and selected substitutions for residues mutated are indicated, as are LIELD clathrin box 1 (blue highlight) and the IV residues (red) mutated to Ala in the IV → AA mutation of β-arrestin 1. In ARH, the functionally analogous LLDLE clathrin box motif is positioned 35 residues proximal to the β2 platform binding sequence shown. B, aliquots of ∼100 μg of GST (lanes a and b), the wild-type GST-β-arrestin 1 CT (residues 331–418) (lanes c and d), or the GST-β-arrestin 1 CT (F391Y) (lanes e and f) or GST-β-arrestin 1 CT (F391W) (lanes g and h) single point mutants each immobilized on glutathione-Sepharose were incubated with precleared rat brain cytosol. After incubation at 4 °C for 2 h, the Sepharose beads were recovered by centrifugation and washed. Aliquots corresponding to ∼1.5% of each supernatant (S; lanes a, c, e, and g) and ∼12.5% of each pellet (P; lanes b, d, f, and h) were resolved by SDS-PAGE and either stained with Coomassie Blue (top) or transferred to nitrocellulose. Replicate blots were probed with a mixture of anti-clathrin HC mAb TD.1 and affinity-purified anti-β1/β2 antibody GD/1 or anti-AP-2 μ2 subunit antibodies. The position of the molecular mass standards (in kDa) is shown at the left, and only the relevant portion of each blot is presented. C, combined ribbon and space-filling surface representation of the β2 appendage platform subdomain bound to the ARH DEAFSRLAQSRT interaction peptide (Protein Data Bank code 2G30) (37) shown in a stick representation with oxygen colored red and nitrogen blue. Compared with the native structure, models of allowed rotomers of the L262F, L262Y, and L262E substitutions (shown in a purple stick representation) are illustrated. D, aliquots of ∼100 μg of GST (lanes a and b); wild-type GST-ARH C1 (residues 180–308) (lanes c and d); or GST-ARH C1 (L262F) (lanes e and f), GST-ARH C1 (L262Y) (lanes g and h), or GST-ARH C1 (L262E) (lanes i and j) single point mutants each immobilized on glutathione-Sepharose were incubated with precleared rat brain cytosol. After incubation at 4 °C for 2 h, the Sepharose beads were recovered by centrifugation and washed. Aliquots corresponding to ∼1.5% of each supernatant (S) and ∼12.5% of each pellet (P) were resolved by SDS-PAGE and analyzed as in B with antibodies against the clathrin HC, β1/β2, and μ2 adaptor subunits.
FIGURE 4.
Selective disruption of protein-protein interactions on the β2 appendage platform subdomain. A, aliquots of ∼100 μg of GST (lanes a–d), the wild-type GST-β2 appendage (lanes e–h), or the GST-β2 (Y888E) mutant appendage (lanes i–l) immobilized on glutathione-Sepharose were incubated with precleared lysates prepared from either control β2−/− iMEFs or from iMEFs ectopically expressing tdRFP-β-arrestin 1 (IV → AA). Following incubation with the lysates at 4 °C for 2 h, the Sepharose beads were recovered by centrifugation and washed. Aliquots corresponding to ∼1.5% of each supernatant (S; lanes a, c, e, g, i, and k) and 12.5% of each pellet (P; lanes b, d, f, h, j, and l) fraction were resolved by SDS-PAGE and either stained with Coomassie Blue (top) or transferred to nitrocellulose. Replicate blots were probed with anti-β-arrestin polyclonal antibody or an anti-DsRed mAb. The position of the molecular mass standards (in kDa) on the Coomassie Blue-stained gel is shown at the left, and only the relevant portion of each blot is presented. A nonspecific band on immunoblots probed with the anti-β-arrestin antibody is indicated (arrowheads) and is not detected with anti-DsRed antibody; the apparent mass of this band (∼95 kDa) rules out the possibility that it represents the endogenous cytosolic β-arrestin (∼50 kDa), which also binds the β2 appendage very poorly (37). B, aliquots of ∼100 μg of GST (lanes a and b), the wild-type GST-β2 appendage (lanes c and d), or GST-β2 (Y888E) (lanes e and f) or GST-β2 (Y888F) (lanes g and h) mutant appendages immobilized on glutathione-Sepharose were incubated with precleared lysates obtained from β2−/− iMEFs transfected with the tdRFP-β-arrestin 1 (IV → AA) mutant. Gels and blots were prepared as in A and immunoblotted with antibodies directed against β-arrestin 1, DsRed, Eps15, or epsin 1. Note that the tdRFP-β-arrestin 1 (IV → AA) mutant protein remains almost completely in the supernatant fraction after incubation with the Y888E β2 appendage mutant.
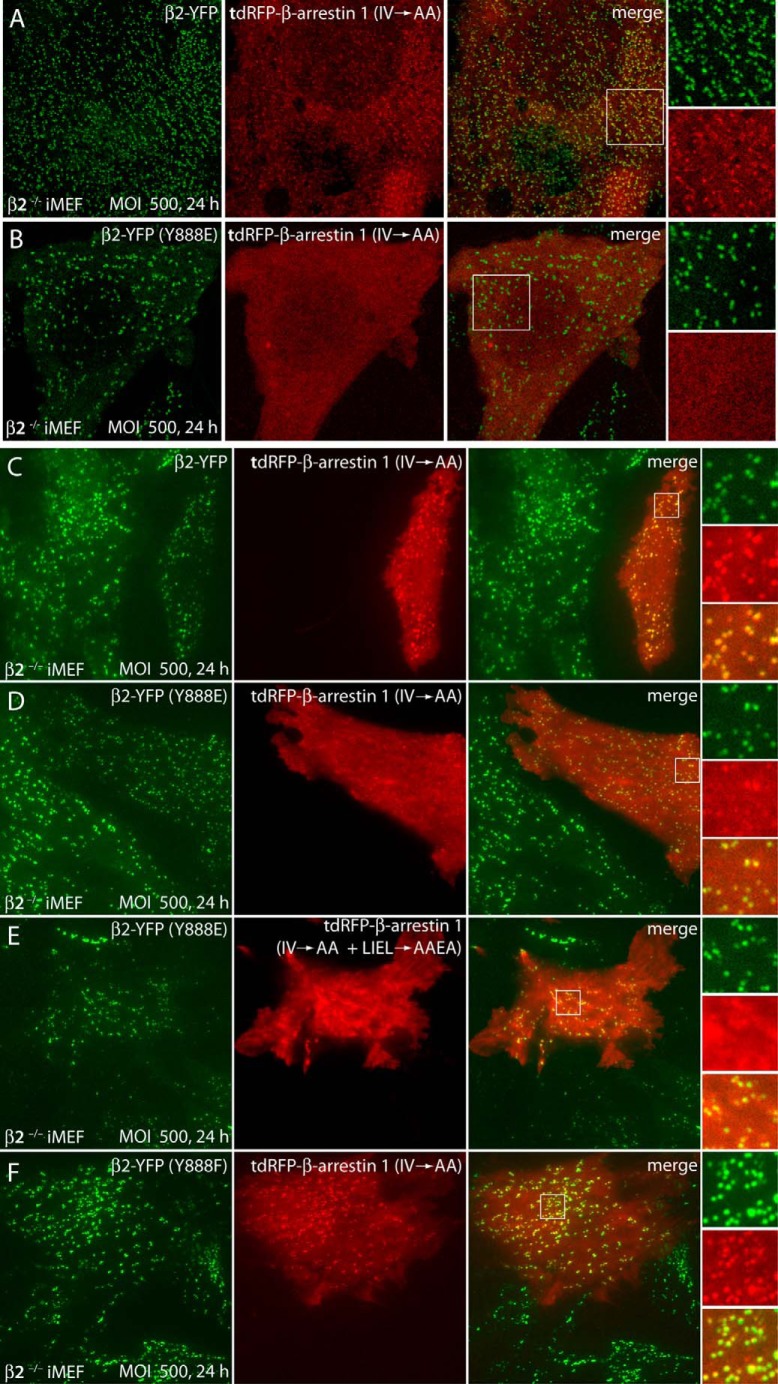
Last, infecting the β2−/− iMEFs with adenovirus encoding a β2-YFP subunit reconstitutes cellular AP-2 and the recruitment of RFP-tagged β-arrestin 1 to surface clathrin puncta in the absence of any acute G protein-coupled receptor activation. The fluorescently-tagged β-arrestin 1 (IV → AA) reporter is distributed together with the random array of β2-YFP-containing clathrin puncta (Fig. 5A). Parallel infections with a β2-YFP (Y888E) mutant, while still being incorporated into AP-2 and targeted to bud sites, result in strongly diminished RFP-β-arrestin 1 deposition at AP-2-positive coated structures and hence loss of close proximity of the clustered signals (Fig. 5B).
FIGURE 5.
Deposition of artificially active β-arrestin 1 at wild-type and mutant β1-containing clathrin-coated structures in transduced iMEFs. A and B, β2−/− iMEFs were infected with adenovirus encoding either β2-YFP (A) or β2-YFP (Y888E) (B) at a MOI of 500. Following 6 h of infection, cells were transfected with the tdRFP-β-arrestin 1 (IV → AA) mutant. After an additional 18-h incubation, the cells were fixed. Representative single confocal optical sections of the ventral plasma membrane region are shown. Color-separated enlarged views of selected regions (boxed) are shown on the right. Note that the ectopic tdRFP-β-arrestin 1 (IV → AA) shows a combined diffuse and punctate localization pattern in cells infected with adenovirus encoding wild-type β2-YFP, whereas the CLASP is essentially diffuse in cells infected with adenovirus encoding a phosphorylation-mimicking β2-YFP (Y888E) mutant. C–F, β2−/− iMEFs transduced and transfected as in A and B were analyzed by live cell TIR-FM without fixation. A single representative image frame from iMEFs transduced with wild-type β2-YFP (C), β2-YFP (Y888E) (D and E), or β2-YFP (Y888F) (F) and subsequently transfected with either tdRFP-β-arrestin 1 (IV → AA) (C, D, and F) or the tdRFP-β-arrestin 1 (IV → AA + LIEL → AAEA) compound mutant (E) is presented, with enlargements of selected regions (boxed) shown on the right.
The deposition and concentration of mutated β-arrestin 1 at β2-YFP puncta are better visualized with TIR-FM (25) (Figs. 5D and 6). Again, the extent of tdRFP-β-arrestin 1 (IV → AA) colocalizing with the Y888E mutant β2-YFP is clearly reduced, but not completely abolished, relative to the transduced wild-type β2 chain (Figs. 5C and 6). Following a 24-h transduction with β2-YFP-encoding adenovirus, the cellular AP-2 pool in the β2−/− iMEFs still contains some endogenous β1 subunit (Fig. 2). The reason for incomplete loss of tdRFP-β-arrestin 1 (IV → AA) clustering at β2-YFP-positive patches, therefore, may be due to imperfect reconstitution of AP-2 with the Y888E mutant, because native AP-2 has a biological half-life of ∼35 h (49), or due to the capability of the exposed clathrin box motif to still engage assembled clathrin in the absence of β2 platform binding, or both. Parallel TIR-FM experiments with a tdRFP-β-arrestin 1 (IV → AA + LIEL → AAEA) compound mutant show that disrupting the LIELD clathrin box motif diminishes further the residual coincidence between the tagged β-arrestin 1 and the β2-YFP (Y888E) surface puncta (Figs. 5E and 6). However, corroborating the biochemical data, AP-2 reconstituted with the Y888F mutant operates analogously to the wild-type β2-YFP (Fig. 5F), with tdRFP-β-arrestin 1 (IV → AA) clustered at β2-YFP-positive structures at the cell surface. This assessment is supported by quantitative line scan analysis of the TIR-FM data (Fig. 6). A β-arrestin 1 variant (IV → AA), which displays constitutive physical interaction with AP-2 through the wild-type β2 platform contact site, thus interacts only poorly with plasma membrane-tethered AP-2 when a single structural modification, the phosphomimicking Y888E, is present in the β2 chain.
FIGURE 6.
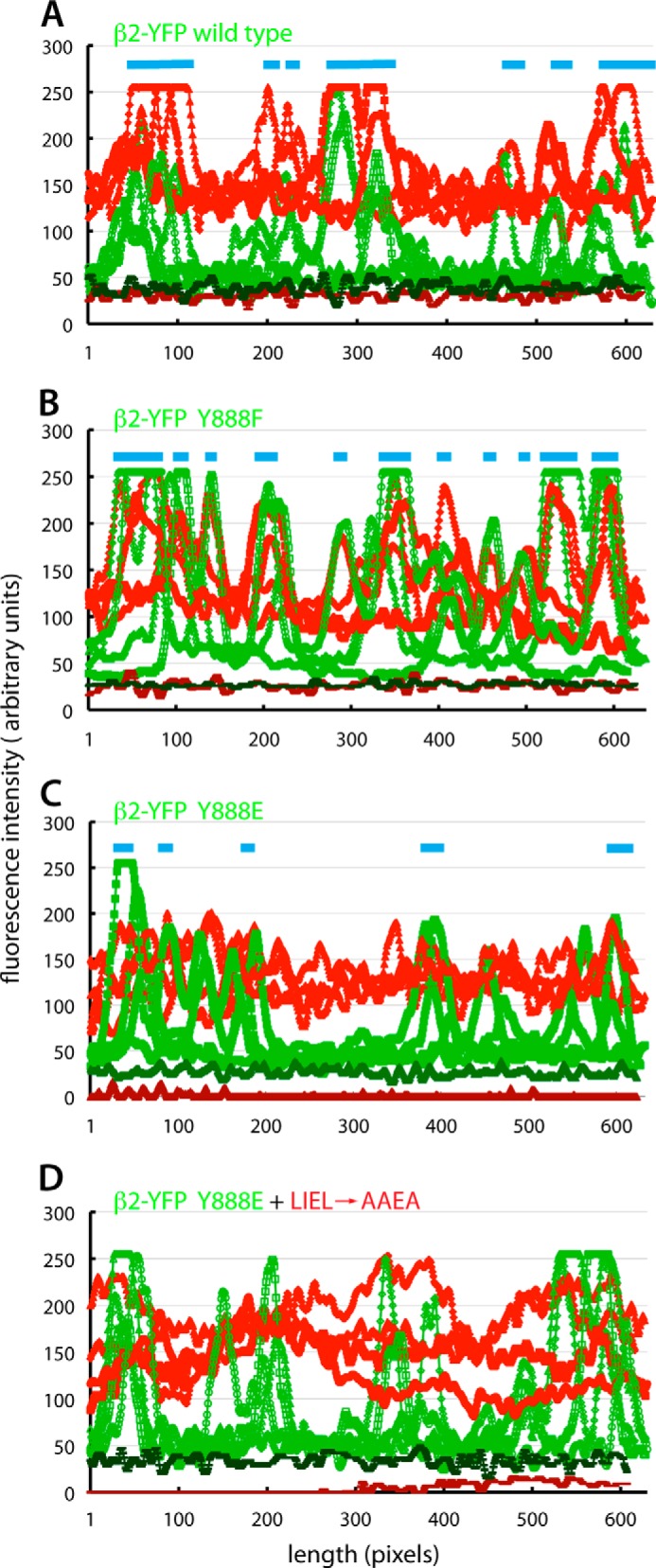
Quantitative line scan analysis of β-arrestin 1 (IV → AA) translocation into clathrin coats. A–D, single time point images from the live cell TIR-FM experimental data shown in Fig. 5 were analyzed for AP-2 (β2-YFP; green) and tdRFP-β-arrestin 1 (IV → AA) (red) fluorescence intensity along 620–645-pixel-long linear segments. Four randomly selected lines from the β2-YFP wild type (A), β2-YFP (Y888F) (B), or β2-YFP (Y888E) (C and D) virally transduced β2−/− iMEFs subsequently transfected with either the constitutively active tdRFP-β-arrestin 1 (IV → AA) reporter (A–C) or the tdRFP-β-arrestin 1 (IV → AA + LIEL → AAEA) compound mutant reporter (D) are superimposed by plotting together. The background fluorescence intensities for the red and green channels (colored brick red and forest green, respectively) are shown for each image analyzed. Note that the amplitudes of β2-YFP intensity are similar in the four reconstituted iMEF populations, supporting the conclusion that all three products of the YFP-tagged β2 transgene are incorporated into functional AP-2 that is deposited at maturing clathrin assemblies at the cell surface. However, unlike the wild-type and Y888F β2-YFP (light blue bars), the Y888E mutant does not have strongly colocalized peaks of red tdRFP-β-arrestin 1 fluorescence, and, in general, the maximal peak amplitude above cytosolic baseline fluorescence for this mutant is considerably smaller than that for wild-type or Y888F β2-YFP-expressing cells.
Perturbation of the β2·(D/E)nX1–2FXX(F/L)XXXR Motif Interaction Interface
Although strongly suggestive, a legitimate concern with the studies described above is that although Ser/Thr → Asp/Glu substitutions are well accepted functional phosphomimetics (21, 74, 75), the Y888E mutation loses the aromatic character of the side chain. Active phosphomimetic Tyr → Glu mutations are reported in the literature (76–87), but this is clearly a suboptimal substitution in certain instances (82, 88–91). To further demonstrate that Tyr(P)-888 modification interferes with motif accommodation, we also tested whether complex formation can be disrupted by artificially altering the reciprocal binding partner. The rationale is to probe whether the physical bulk of an additional aromatic ring or the hydration shell of an attached hydroxyl or carboxyl group impedes the close apposition of interfacial hydrophobic surfaces (Fig. 7, A and C).
Using immobilized GST-β-arrestin 1 CT (residues 331–418) in pull-down assays with brain cytosol, both F391Y and F391W substitutions at the F/L anchor site interfere with the binding of soluble AP-2 without similarly affecting clathrin association (Fig. 7B, lanes f and h compared with lane d), which is dictated by the separate adjacent sequence (92, 93). Neither clathrin nor AP-2 interacts with GST alone (Fig. 7B, lane b), and these soluble coat components remain completely in the supernatant (Fig. 7B, lane a).
Analogous but stronger inhibitory effects are seen with L262Y or L262E mutations in GST-ARH C1 (residues 180–308) (Fig. 7D, lanes h and j), whereas a L262F change does not perturb AP-2 binding substantially, as expected (Fig. 7D, lane f). Cytosolic AP-2 remains fully within the supernatant fraction in pull-down assays with GST-ARH C1 (L262Y) or GST-ARH C1 (L262E) (Fig. 7D, lanes g and i), indicating that the lack of AP-2 recovery in the pellets is not due to the substitution altering the off rate and causing the adaptor to be lost during washing. The different substitutions to ARH Leu-262 have comparably minor effects on clathrin binding through the independent interaction motif (35, 94). Although the C-terminal regions of both the β-arrestins and ARH each contain a single (D/E)nX1–2FXX(F/L)XXXR interaction motif and a clathrin-binding motif, we have previously documented that ARH engages AP-2 and clathrin more avidly in pull-down assays (38) (Fig. 7, B and D). This may be related to the relative spacing between the two interaction motifs, which is much shorter in β-arrestins (Fig. 7A), or perhaps due to inhibitory residues in β-arrestin 1 (72). Nevertheless, the impact of the β-arrestin 1 F391Y and ARH L262Y mutations is significant; introducing only a para-hydroxyl group has an unmistakable inhibitory effect on productive AP-2 binding. Most important, the highly disruptive ARH L262E substitution does not replace the aliphatic character of the natural leucine at this position, unlike in the case of the β2 Y888E switch. We conclude that engineered bulky or polar alterations to the β2 platform interaction interface can directly obstruct proper CLASP engagement. The electrostatic perturbation of a new dianionic charge, in addition to the physical bulk of the added phosphate group on Tyr-888 (Fig. 3A), will function similarly in vivo.
DISCUSSION
At a rudimentary level, reversible Tyr phosphorylation refashions the local binding features of protein interaction partners. Although this often operates in a positive manner, like the engagement of Tyr(P) motifs by Src homology 2, phosphotyrosine binding (and structurally related FERM), and C2 domains (95–98), obstruction of protein-protein contacts is also known to accompany Tyr(P) modification (11, 12, 99, 100). Parsing the phosphoproteome (19), we noticed that a functionally critical, solvent-exposed Tyr side chain upon the β2 subunit platform interface is repeatedly catalogued in the database. The vast majority of Tyr(P)-888 is reported from leukocyte cell extracts. Tantalizingly, the LDL receptor, which in leukocytes depends on ARH (and hence on the AP-2 β2 appendage) for packaging into clathrin coats, is not concentrated at surface AP-2-positive clathrin puncta in lymphocytes (101), quite dissimilar to fibroblasts (102) and other cultured cells (55, 103). Our structure-guided argument is that the positioning, charge, and large ionic hydration shell of the affixed phosphate will change the chemical properties of the β1/2 platform interaction surface, interfering sterically with successful docking and engagement of the (D/E)nX1–2FXX(F/L)XXXR interaction motif (Fig. 3A). Our results attest that indeed there is a spatially confined environment upon the β2 platform for productive α-helical peptide encounters. It seems a remote possibility that the Tyr(P)-888 modification serves as a signal for the binding of a Tyr(P)-selective modular fold to AP-2. First, the phosphotyrosine binding domain typically recognizes Tyr(P) in the context of a conserved sequence tract ((F/Y)XNPXpY) and a structurally defined orientation (a tight β-turn) (104), which is unrelated to the flanking β2 primary sequence and impossible for Tyr-888 to assume in the context of a residue projecting off of a laterally hydrogen-bonded antiparallel β-strand within the β2 appendage platform subdomain (66). Second, similar structural considerations indicate that Tyr(P)-888 is too buried and inaccessible and lacks flexible adjacent residues to engage an Src homology 2 domain in the accepted fashion (97). But even were this to occur, the functional consequence would be to compete with the engagement of ARH or β-arrestins through an overlapping site upon the β2 platform, so the Tyr(P) modification would still be molecularly translated into an inhibitory input on regular β2 platform interactions.
Besides operating at the cell surface for receptor internalization, both the β-arrestins and ARH have been localized to endosomal compartments under various conditions (105–112). This is dissimilar to many other AP-2 appendage-binding CLASPs and accessory proteins, which cycle strictly between plasma membrane assemblages and a reserve cytosolic pool. In this regard, it is significant that the high degree of similarity between the AP-1 β1 and AP-2 β2 chains is also seen in common post-translational modifications. The positionally equivalent side chain to Tyr-888 in the β1 chain is Tyr-897, and Tyr(P)-897 is also represented in the phosphoproteome database (113). Actually, Tyr(P)-897 is currently the most frequently detected phosphorylation event on the β1 subunit (50 reports), like the Tyr(P)-888 in β2, and is present in numerous other cell types besides leukocytes. A phosphopeptide encompassing the Tyr-888 analogous residue in the D. melanogaster β-adaptin (Tyr-871) has also been detected in Kc167 cells (114). Thus, because Tyr-888 is structurally and functionally conserved in the AP-1 β1 appendage platform subdomain (66), phosphorylation probably regulates engagement of this surface as well. This reversible modification may underlie the variable assembly of ARH and β-arrestins on AP-1-positive endosomal structures (112).
It is instructive to compare the representation of Tyr(P)-888 with the frequency of other phosphosites currently mapped within the β2 chain (19). In the basal AP-2 heterotetramer conformation, the first occurring Tyr at the β2 subunit N terminus (Tyr-6) along with adjacent Phe-7 occlude the binding site for the (D/E)XXXL(L/I/M)-type dileucine sorting signal on the α·σ2 hemicomplex (115). Phosphorylation of Tyr-6 is linked to dileucine signal recognition (116) and may dislodge the N-terminal region of β2 from the α·σ2 molecular surface (115) when AP-2 attaches to the inner leaflet of the plasma membrane to permit transmembrane cargo capture. However, Tyr(P)-6 is only reported once in low throughput (116) and currently only once in high throughput experiments and is recorded a single time in the β1 chain (19). On the C-terminal β2 appendage, Tyr-737 within the sandwich subdomain is connected to the regulation of β-arrestin interactions through phosphorylation by the non-receptor Tyr kinase Src (58, 117, 118). Curiously, Tyr(P)-737 is remote from the platform site utilized by β-arrestins (and ARH) (see Fig. 2B) and has been observed nine times by high throughput detection, compared with 53 instances for Tyr(P)-888.
The widespread occurrence of phosphoregulation through eukaryotic evolution is argued to be due, in part, to the fact that the fundamental chemical properties of phosphoamino acids are unlike any natural side chains within proteins (119). Even so, there is evidence that phosphosites can evolve to replace acidic residues by Ser/Thr/Tyr substitution to impose conditional regulation (120). In the Raf-1 Ser/Thr kinase, residues Tyr340-Tyr341 within the catalytic domain require phosphorylation for kinase activation (76, 77). These residues correspond to vicinal Asp448-Asp449 in the enzymatically more active B-Raf (76), so Asp/Glu can functionally substitute for Tyr(P) under at least some conditions, and in invertebrate Raf kinases, this vicinal pair is composed of Asp and/or Glu residues (120). Furthermore, there is a computational argument that a Glu-for-Tyr(P) substitution has occurred in the CMGC kinase group (120). Taken together, it appears that Asp/Glu can mimic Tyr(P) in certain biological contexts. Still, because of our lingering reservation with the Y888E substitution on the molecular surface of the β2 platform subdomain, we mutated the core F/L position side chain in ARH and β-arrestin 1 (D/E)nX1–2FXX(F/L)XXXR motifs with similar inhibitory outcomes.
The identity of the protein kinase(s) that reversibly phosphorylates Tyr-888 on the β2 appendage and Tyr-897 on the β1 chain remains to be established. Because both AP-1 and AP-2 are modified at this position, it is possible that a non-receptor Tyr kinase catalyzes this post-translational modification as opposed to a receptor Tyr kinase activated at the cell surface. An interesting aspect of the recognition of this particular substrate is that the acceptor side chain is not located within a flexible loop or a largely unstructured polypeptide region housing an identifiable phosphorylation motif (121, 122). Instead, Tyr-888 projects out from strand β11 of the five-stranded antiparallel β-sheet that forms the base of the β2 platform subdomain (37, 62, 66) (Fig. 2B) and displays a computed 6.87% of the surface area as accessible (similar to the 4.95% surface accessibility for Tyr-737) (123). This suggests that conformational recognition as opposed to a linear peptide-based identification motif depending on specificity-determining flanking residues is necessary for the Tyr(P)-888 modification of this solvent-exposed side chain. The relevant protein Tyr phosphatase that reverses this modification is also not yet known.
Finally, an attractive emerging concept is the existence of compositionally distinct clathrin-coated structures that can relay distinct populations of cargo from the surface into the cell interior in parallel (124–130). On one level, this idea seems generally improbable because the majority of inchoate clathrin-coated structures at the surface show a similar sequence of pioneer protein arrival (131), and at steady state, clathrin assembly zones are populated with mostly the same adaptors, CLASPs, and accessory factors (55, 131–134). Our results illustrate that specialized or cargo-biased clathrin-coated vesicles are not necessarily characterized by different sets of co-assembled adaptors and CLASPs but could possibly arise through different local, switchable binding capabilities of these cargo-sorting components at individual clathrin internalization zones. The ultimate effectiveness and precision of this type of toggle mechanism will depend upon the rate and extent of post-translational modification and the half-life of the covalent adduct and also upon the degree of functional redundancy in sorting. In several cell types, Tyr(P)-888 will be more effective at uncoupling G protein-coupled receptor internalization than LDL receptor family uptake because alternate CLASPs like Dab2 (55, 103) and possibly Ced-6 (135) can also recognize the (F/Y)XNPX(Y/F) sorting signal decoded by ARH, and these proteins do not engage the AP-2 β2 subunit platform surface.
Acknowledgments
We are extremely grateful to Eric Everett for generously supplying the β2+/+ and β2−/− MEFs. John (Nick) Johnson, Stuart Kornfeld, and David Owen each provided thoughtful comments on the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 GM106963-17 (to L. M. T.).
- iMEF
- immortalized mouse embryonic fibroblast
- ARH
- autosomal recessive hypercholesterolemia
- CLASP
- clathrin-associated sorting protein
- HC
- heavy chain
- MOI
- multiplicity of infection
- tdRFP
- Tomato tandem dimer red fluorescent protein
- TIR-FM
- total internal reflection fluorescence microscopy
- CT
- C terminus.
REFERENCES
- 1. Kreegipuu A., Blom N., Brunak S. (1999) PhosphoBase, a database of phosphorylation sites: release 2.0. Nucleic Acid Res. 27, 237–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sadowski I., Breitkreutz B. J., Stark C., Su T. C., Dahabieh M., Raithatha S., Bernhard W., Oughtred R., Dolinski K., Barreto K., Tyers M. (2013) The PhosphoGRID Saccharomyces cerevisiae protein phosphorylation site database: version 2.0 update. Database 2013, bat026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Shiromizu T., Adachi J., Watanabe S., Murakami T., Kuga T., Muraoka S., Tomonaga T. (2013) Identification of missing proteins in the neXtProt database and unregistered phosphopeptides in the PhosphoSitePlus database as part of the Chromosome-centric Human Proteome Project. J. Proteome Res. 12, 2414–2421 [DOI] [PubMed] [Google Scholar]
- 4. Zhou H., Di Palma S., Preisinger C., Peng M., Polat A. N., Heck A. J., Mohammed S. (2013) Toward a comprehensive characterization of a human cancer cell phosphoproteome. J. Proteome Res. 12, 260–271 [DOI] [PubMed] [Google Scholar]
- 5. Lemeer S., Heck A. J. (2009) The phosphoproteomics data explosion. Curr. Opin. Chem. Biol. 13, 414–420 [DOI] [PubMed] [Google Scholar]
- 6. Dephoure N., Zhou C., Villén J., Beausoleil S. A., Bakalarski C. E., Elledge S. J., Gygi S. P. (2008) A quantitative atlas of mitotic phosphorylation. Proc. Natl. Acad. Sci. U.S.A. 105, 10762–10767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Traub L. M. (2009) Tickets to ride: selecting cargo for clathrin-regulated internalization. Nat. Rev. Mol. Cell Biol. 10, 583–596 [DOI] [PubMed] [Google Scholar]
- 8. Bonifacino J. S., Traub L. M. (2003) Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu. Rev. Biochem. 72, 395–447 [DOI] [PubMed] [Google Scholar]
- 9. Sibley D. R., Strasser R. H., Benovic J. L., Daniel K., Lefkowitz R. J. (1986) Phosphorylation/dephosphorylation of the β-adrenergic receptor regulates its functional coupling to adenylate cyclase and subcellular distribution. Proc. Natl. Acad. Sci. U.S.A. 83, 9408–9412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hunter T., Cooper J. A. (1981) Epidermal growth factor induces rapid tyrosine phosphorylation of proteins in A431 human tumor cells. Cell 24, 741–752 [DOI] [PubMed] [Google Scholar]
- 11. Kittler J. T., Chen G., Kukhtina V., Vahedi-Faridi A., Gu Z., Tretter V., Smith K. R., McAinsh K., Arancibia-Carcamo I. L., Saenger W., Haucke V., Yan Z., Moss S. J. (2008) Regulation of synaptic inhibition by phospho-dependent binding of the AP2 complex to a YECL motif in the GABAA receptor γ2 subunit. Proc. Natl. Acad. Sci. U.S.A. 105, 3616–3621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shiratori T., Miyatake S., Ohno H., Nakaseko C., Isono K., Bonifacino J. S., Saito T. (1997) Tyrosine phosphorylation controls internalization of CTLA-4 by regulating its interaction with clathrin-associated adaptor complex AP-2. Immunity 6, 583–589 [DOI] [PubMed] [Google Scholar]
- 13. Clayton E. L., Evans G. J., Cousin M. A. (2007) Activity-dependent control of bulk endocytosis by protein dephosphorylation in central nerve terminals. J. Physiol. 585, 687–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Slepnev V. I., Ochoa G. C., Butler M. H., Grabs D., De Camilli P. (1998) Role of phosphorylation in regulation of the assembly of endocytic coat complexes. Science 281, 821–824 [DOI] [PubMed] [Google Scholar]
- 15. Wilde A., Brodsky F. M. (1996) In vivo phosphorylation of adaptors regulates their interaction with clathrin. J. Cell Biol. 135, 635–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Olusanya O., Andrews P. D., Swedlow J. R., Smythe E. (2001) Phosphorylation of threonine 156 of the μ2 subunit of the AP2 complex is essential for endocytosis in vitro and in vivo. Curr. Biol. 11, 896–900 [DOI] [PubMed] [Google Scholar]
- 17. Ricotta D., Conner S. D., Schmid S. L., von Figura K., Honing S. (2002) Phosphorylation of the AP2 μ subunit by AAK1 mediates high affinity binding to membrane protein sorting signals. J. Cell Biol. 156, 791–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Höning S., Ricotta D., Krauss M., Späte K., Spolaore B., Motley A., Robinson M., Robinson C., Haucke V., Owen D. J. (2005) Phosphatidylinositol-(4,5)-bisphosphate regulates sorting signal recognition by the clathrin-associated adaptor complex AP2. Mol. Cell 18, 519–531 [DOI] [PubMed] [Google Scholar]
- 19. Hornbeck P. V., Kornhauser J. M., Tkachev S., Zhang B., Skrzypek E., Murray B., Latham V., Sullivan M. (2012) PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 40, D261–D270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jackson L. P., Kelly B. T., McCoy A. J., Gaffry T., James L. C., Collins B. M., Höning S., Evans P. R., Owen D. J. (2010) A large-scale conformational change couples membrane recruitment to cargo binding in the AP2 clathrin adaptor complex. Cell 141, 1220–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hao N., Budnik B. A., Gunawardena J., O'Shea E. K. (2013) Tunable signal processing through modular control of transcription factor translocation. Science 339, 460–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Collins M. O., Yu L., Campuzano I., Grant S. G., Choudhary J. S. (2008) Phosphoproteomic analysis of the mouse brain cytosol reveals a predominance of protein phosphorylation in regions of intrinsic sequence disorder. Mol. Cell Proteomics 7, 1331–1348 [DOI] [PubMed] [Google Scholar]
- 23. Nishi H., Hashimoto K., Panchenko A. R. (2011) Phosphorylation in protein-protein binding: effect on stability and function. Structure 19, 1807–1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nishi H., Fong J. H., Chang C., Teichmann S. A., Panchenko A. R. (2013) Regulation of protein-protein binding by coupling between phosphorylation and intrinsic disorder: analysis of human protein complexes. Mol. Biosyst. 9, 1620–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Keyel P. A., Thieman J. R., Roth R., Erkan E., Everett E. T., Watkins S. C., Heuser J. E., Traub L. M. (2008) The AP-2 adaptor β2 appendage scaffolds alternate cargo endocytosis. Mol. Biol. Cell 19, 5309–5326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li W., Puertollano R., Bonifacino J. S., Overbeek P. A., Everett E. T. (2010) Disruption of the murine Ap2b1 gene causes nonsyndromic cleft palate. Cleft Palate Craniofac. J. 47, 566–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hardy S., Kitamura M., Harris-Stansil T., Dai Y., Phipps M. L. (1997) Construction of adenovirus vectors through Cre-lox recombination. J. Virol. 71, 1842–1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Henkel J. R., Apodaca G., Altschuler Y., Hardy S., Weisz O. A. (1998) Selective perturbation of apical membrane traffic by expression of influenza M2, an acid-activated ion channel, in polarized Madin-Darby canine kidney cells. Mol. Biol. Cell 9, 2477–2490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chin D. J., Straubinger R. M., Acton S., Näthke I., Brodsky F. M. (1989) 100-kDa polypeptides in peripheral clathrin-coated vesicles are required for receptor-mediated endocytosis. Proc. Natl. Acad. Sci. U.S.A. 86, 9289–9293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Näthke I. S., Heuser J., Lupas A., Stock J., Turck C. W., Brodsky F. M. (1992) Folding and trimerization of clathrin subunits at the triskelion hub. Cell 68, 899–910 [DOI] [PubMed] [Google Scholar]
- 31. Hinrichsen L., Harborth J., Andrees L., Weber K., Ungewickell E. J. (2003) Effect of clathrin heavy chain- and α-adaptin specific small interfering RNAs on endocytic accessory proteins and receptor trafficking in HeLa cells. J. Biol. Chem. 278, 45160–45170 [DOI] [PubMed] [Google Scholar]
- 32. Dalal S., Rosser M. F., Cyr D. M., Hanson P. I. (2004) Distinct roles for the AAA ATPases NSF and p97 in the secretory pathway. Mol. Biol. Cell 15, 637–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Drake M. T., Downs M. A., Traub L. M. (2000) Epsin binds to clathrin by associating directly with the clathrin-terminal domain: evidence for cooperative binding through two discrete sites. J. Biol. Chem. 275, 6479–6489 [DOI] [PubMed] [Google Scholar]
- 34. Traub L. M., Kornfeld S., Ungewickell E. (1995) Different domains of the AP-1 adaptor complex are required for Golgi membrane binding and clathrin recruitment. J. Biol. Chem. 270, 4933–4942 [DOI] [PubMed] [Google Scholar]
- 35. Mishra S. K., Watkins S. C., Traub L. M. (2002) The autosomal recessive hypercholesterolemia (ARH) protein interfaces directly with the clathrin-coat machinery. Proc. Natl. Acad. Sci. U.S.A. 99, 16099–16104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sorkina T., Huang F., Beguinot L., Sorkin A. (2002) Effect of tyrosine kinase inhibitors on clathrin-coated pit recruitment and internalization of epidermal growth factor receptor. J. Biol. Chem. 277, 27433–27441 [DOI] [PubMed] [Google Scholar]
- 37. Edeling M. A., Mishra S. K., Keyel P. A., Steinhauser A. L., Collins B. M., Roth R., Heuser J. E., Owen D. J., Traub L. M. (2006) Molecular switches involving the AP-2 β2 appendage regulate endocytic cargo selection and clathrin coat assembly. Dev. Cell 10, 329–342 [DOI] [PubMed] [Google Scholar]
- 38. Mishra S. K., Keyel P. A., Edeling M. A., Dupin A. L., Owen D. J., Traub L. M. (2005) Functional dissection of an AP-2 β2 appendage-binding sequence within the autosomal recessive hypercholesterolemia (ARH) protein. J. Biol. Chem. 280, 19270–19280 [DOI] [PubMed] [Google Scholar]
- 39. Thieman J. R., Mishra S. K., Ling K., Doray B., Anderson R. A., Traub L. M. (2009) Clathrin regulates the association of PIPKIγ661 with the AP-2 adaptor β2 appendage. J. Biol. Chem. 284, 13924–13939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Abramoff M. D., Magelhaes P. J., Ram S. J. (2004) Image processing with ImageJ. Biophotinics Int. 11, 36–42 [Google Scholar]
- 41. Green M., Pina M. (1963) Biochemical studies on adenovirus multiplication. IV. Isolation, purification, and chemical analysis of adenovirus. Virology 20, 199–207 [DOI] [PubMed] [Google Scholar]
- 42. Keyel P. A., Watkins S. C., Traub L. M. (2004) Endocytic adaptor molecules reveal an endosomal population of clathrin by total internal reflection fluorescence microscopy. J. Biol. Chem. 279, 13190–13204 [DOI] [PubMed] [Google Scholar]
- 43. Boehm M., Bonifacino J. S. (2001) Adaptins: the final recount. Mol. Biol. Cell 12, 2907–2920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hirst J., Barlow L. D., Francisco G. C., Sahlender D. A., Seaman M. N., Dacks J. B., Robinson M. S. (2011) The fifth adaptor protein complex. PLoS Biol. 9, e1001170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kirchhausen T., Nathanson K. L., Matsui W., Vaisberg A., Chow E. P., Burne C., Keen J. H., Davis A. E. (1989) Structural and functional division into two domains of the large (100- to 115-kDa) chains of the clathrin-associated protein complex AP-2. Proc. Natl. Acad. Sci. U.S.A. 86, 2612–2616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ponnambalam S., Robinson M. S., Jackson A. P., Peiperl L., Parham P. (1990) Conservation and diversity in families of coated vesicle adaptins. J. Biol. Chem. 265, 4814–4820 [PubMed] [Google Scholar]
- 47. Guilbaud C., Peyrard M., Fransson I., Clifton S. W., Roe B. A., Carter N. P., Dumanski J. P. (1997) Characterization of the mouse beta-prime adaptin gene: cDNA sequence, genomic structure, and chromosomal localization. Mamm. Genome 8, 651–656 [DOI] [PubMed] [Google Scholar]
- 48. Page L. J., Robinson M. S. (1995) Targeting signals and subunit interactions in coated vesicle adaptor complexes. J. Cell Biol. 131, 619–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sorkin A., McKinsey T., Shih W., Kirchhausen T., Carpenter G. (1995) Stoichiometric interaction of the epidermal growth factor receptor with the clathrin-associated protein complex AP-2. J. Biol. Chem. 270, 619–625 [DOI] [PubMed] [Google Scholar]
- 50. Traub L. M., Bannykh S. I., Rodel J. E., Aridor M., Balch W. E., Kornfeld S. (1996) AP-2-containing clathrin coats assemble on mature lysosomes. J. Cell Biol. 135, 1801–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Camidge D. R., Pearse B. M. (1994) Cloning of the Drosophila β-adaptin and its localization on expression in mammalian cells. J. Cell Sci. 107, 709–718 [PubMed] [Google Scholar]
- 52. Gu M., Liu Q., Watanabe S., Sun L., Hollopeter G., Grant B. D., Jorgensen E. M. (2013) AP2 hemicomplexes contribute independently to synaptic vesicle endocytosis. Elife 2, e00190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sosa R. T., Weber M. M., Wen Y., O'Halloran T. J. (2012) A single β adaptin contributes to AP1 and AP2 complexes and clathrin function in Dictyostelium. Traffic 13, 305–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Mitsunari T., Nakatsu F., Shioda N., Love P. E., Grinberg A., Bonifacino J. S., Ohno H. (2005) Clathrin adaptor AP-2 is essential for early embryonal development. Mol. Cell. Biol. 25, 9318–9323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Keyel P. A., Mishra S. K., Roth R., Heuser J. E., Watkins S. C., Traub L. M. (2006) A single common portal for clathrin-mediated endocytosis of distinct cargo governed by cargo-selective adaptors. Mol. Biol. Cell 17, 4300–4317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Robinson M. S. (1987) 100-kDa coated vesicle proteins: molecular heterogeneity and intracellular distribution studied with monoclonal antibodies. J. Cell Biol. 104, 887–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ahle S., Mann A., Eichelsbacher U., Ungewickell E. (1988) Structural relationships between clathrin assembly proteins from the Golgi and the plasma membrane. EMBO J. 7, 919–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Fessart D., Simaan M., Zimmerman B., Comeau J., Hamdan F. F., Wiseman P. W., Bouvier M., Laporte S. A. (2007) Src-dependent phosphorylation of β2-adaptin dissociates the β-arrestin-AP-2 complex. J. Cell Sci. 120, 1723–1732 [DOI] [PubMed] [Google Scholar]
- 59. Rosenthal J. A., Chen H., Slepnev V. I., Pellegrini L., Salcini A. E., Di Fiore P. P., De Camilli P. (1999) The epsins define a family of proteins that interact with components of the clathrin coat and contain a new protein module. J. Biol. Chem. 274, 33959–33965 [DOI] [PubMed] [Google Scholar]
- 60. Dannhauser P. N., Ungewickell E. J. (2012) Reconstitution of clathrin-coated bud and vesicle formation with minimal components. Nat. Cell Biol. 14, 634–639 [DOI] [PubMed] [Google Scholar]
- 61. Edeling M. A., Smith C., Owen D. (2006) Life of a clathrin coat: insights from clathrin and AP structures. Nat. Rev. Mol. Cell Biol. 7, 32–44 [DOI] [PubMed] [Google Scholar]
- 62. Schmid E. M., Ford M. G., Burtey A., Praefcke G. J., Peak-Chew S. Y., Mills I. G., Benmerah A., McMahon H. T. (2006) Role of the AP2 β-appendage hub in recruiting partners for clathrin coated vesicle assembly. PLoS Biol. 4, e262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. McMahon H. T., Boucrot E. (2011) Molecular mechanism and physiological functions of clathrin-mediated endocytosis. Nat. Rev. Mol. Cell Biol. 12, 517–533 [DOI] [PubMed] [Google Scholar]
- 64. Burtey A., Schmid E. M., Ford M. G., Rappoport J. Z., Scott M. G., Marullo S., Simon S. M., McMahon H. T., Benmerah A. (2007) The conserved isoleucine-valine-phenylalanine motif couples activation state and endocytic functions of β-arrestins. Traffic 8, 914–931 [DOI] [PubMed] [Google Scholar]
- 65. Laporte S. A., Miller W. E., Kim K. M., Caron M. G. (2002) β-Arrestin/AP-2 interaction in G protein-coupled receptor internalization. Identification of a β-arrestin binding site in β2-adaptin. J. Biol. Chem. 277, 9247–9254 [DOI] [PubMed] [Google Scholar]
- 66. Owen D. J., Vallis Y., Pearse B. M., McMahon H. T., Evans P. R. (2000) The structure and function of the β2-adaptin appendage domain. EMBO J. 19, 4216–4227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Rikova K., Guo A., Zeng Q., Possemato A., Yu J., Haack H., Nardone J., Lee K., Reeves C., Li Y., Hu Y., Tan Z., Stokes M., Sullivan L., Mitchell J., Wetzel R., Macneill J., Ren J. M., Yuan J., Bakalarski C. E., Villen J., Kornhauser J. M., Smith B., Li D., Zhou X., Gygi S. P., Gu T. L., Polakiewicz R. D., Rush J., Comb M. J. (2007) Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell 131, 1190–1203 [DOI] [PubMed] [Google Scholar]
- 68. Villén J., Beausoleil S. A., Gerber S. A., Gygi S. P. (2007) Large-scale phosphorylation analysis of mouse liver. Proc. Natl. Acad. Sci. U.S.A. 104, 1488–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Lagache T., Lang G., Sauvonnet N., Olivo-Marin J. C. (2013) Analysis of the spatial organization of molecules with robust statistics. PLoS One 8, e80914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Han M., Gurevich V. V., Vishnivetskiy S. A., Sigler P. B., Schubert C. (2001) Crystal structure of β-arrestin at 1.9 Å: possible mechanism of receptor binding and membrane Translocation. Structure 9, 869–880 [DOI] [PubMed] [Google Scholar]
- 71. Kang D. S., Kern R. C., Puthenveedu M. A., von Zastrow M., Williams J. C., Benovic J. L. (2009) Structure of an arrestin2/clathrin complex reveals a novel clathrin binding domain that modulates receptor trafficking. J. Biol. Chem. 284, 29860–29872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kern R. C., Kang D. S., Benovic J. L. (2009) Arrestin2/clathrin interaction is regulated by key N- and C-terminal regions in arrestin2. Biochemistry 48, 7190–7200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Shukla A. K., Manglik A., Kruse A. C., Xiao K., Reis R. I., Tseng W. C., Staus D. P., Hilger D., Uysal S., Huang L. Y., Paduch M., Tripathi-Shukla P., Koide A., Koide S., Weis W. I., Kossiakoff A. A., Kobilka B. K., Lefkowitz R. J. (2013) Structure of active β-arrestin-1 bound to a G-protein-coupled receptor phosphopeptide. Nature 497, 137–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Thorsness P. E., Koshland D. E., Jr. (1987) Inactivation of isocitrate dehydrogenase by phosphorylation is mediated by the negative charge of the phosphate. J. Biol. Chem. 262, 10422–10425 [PubMed] [Google Scholar]
- 75. Hazra S., Szewczak A., Ort S., Konrad M., Lavie A. (2011) Post-translational phosphorylation of serine 74 of human deoxycytidine kinase favors the enzyme adopting the open conformation making it competent for nucleoside binding and release. Biochemistry 50, 2870–2880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Bosch E., Cherwinski H., Peterson D., McMahon M. (1997) Mutations of critical amino acids affect the biological and biochemical properties of oncogenic A-Raf and Raf-1. Oncogene 15, 1021–1033 [DOI] [PubMed] [Google Scholar]
- 77. Chong H., Lee J., Guan K. L. (2001) Positive and negative regulation of Raf kinase activity and function by phosphorylation. EMBO J. 20, 3716–3727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Coló G. P., Hernández-Varas P., Lock J., Bartolomé R. A., Arellano-Sánchez N., Strömblad S., Teixidó J. (2012) Focal adhesion disassembly is regulated by a RIAM to MEK-1 pathway. J. Cell Sci. 125, 5338–5352 [DOI] [PubMed] [Google Scholar]
- 79. Kruchten A. E., Krueger E. W., Wang Y., McNiven M. A. (2008) Distinct phospho-forms of cortactin differentially regulate actin polymerization and focal adhesions. Am. J. Physiol. Cell Physiol. 295, C1113–C1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Funakoshi-Tago M., Tago K., Kasahara T., Parganas E., Ihle J. N. (2008) Negative regulation of Jak2 by its auto-phosphorylation at tyrosine 913 via the Epo signaling pathway. Cell. Signal. 20, 1995–2001 [DOI] [PubMed] [Google Scholar]
- 81. Rescher U., Ludwig C., Konietzko V., Kharitonenkov A., Gerke V. (2008) Tyrosine phosphorylation of annexin A2 regulates Rho-mediated actin rearrangement and cell adhesion. J. Cell Sci. 121, 2177–2185 [DOI] [PubMed] [Google Scholar]
- 82. Zisch A. H., Pazzagli C., Freeman A. L., Schneller M., Hadman M., Smith J. W., Ruoslahti E., Pasquale E. B. (2000) Replacing two conserved tyrosines of the EphB2 receptor with glutamic acid prevents binding of SH2 domains without abrogating kinase activity and biological responses. Oncogene 19, 177–187 [DOI] [PubMed] [Google Scholar]
- 83. Aran V., Bryant N. J., Gould G. W. (2011) Tyrosine phosphorylation of Munc18c on residue 521 abrogates binding to Syntaxin 4. BMC Biochem. 12, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Huang Y., Day R. N., Gunst S. J. (2014) Vinculin phosphorylation at Tyr1065 regulates vinculin conformation and tension development in airway smooth muscle tissues. J. Biol. Chem. 289, 3677–3688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Lie P. P., Mruk D. D., Mok K. W., Su L., Lee W. M., Cheng C. Y. (2012) Focal adhesion kinase-Tyr407 and -Tyr397 exhibit antagonistic effects on blood-testis barrier dynamics in the rat. Proc. Natl. Acad. Sci. U.S.A. 109, 12562–12567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Chowdhury D., Marco S., Brooks I. M., Zandueta A., Rao Y., Haucke V., Wesseling J. F., Tavalin S. J., Pérez-Otaño I. (2013) Tyrosine phosphorylation regulates the endocytosis and surface expression of GluN3A-containing NMDA receptors. J. Neurosci. 33, 4151–4164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Uezu A., Okada H., Murakoshi H., del Vescovo C. D., Yasuda R., Diviani D., Soderling S. H. (2012) Modified SH2 domain to phototrap and identify phosphotyrosine proteins from subcellular sites within cells. Proc. Natl. Acad. Sci. U.S.A. 109, E2929–E2938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Xie J., Supekova L., Schultz P. G. (2007) A genetically encoded metabolically stable analogue of phosphotyrosine in Escherichia coli. ACS Chem. Biol. 2, 474–478 [DOI] [PubMed] [Google Scholar]
- 89. Muratore K. E., Cole P. A. (2007) A lock on phosphotyrosine signaling. ACS Chem. Biol. 2, 454–456 [DOI] [PubMed] [Google Scholar]
- 90. Honda M., Okuno Y., Yoo J., Ha T., Spies M. (2011) Tyrosine phosphorylation enhances RAD52-mediated annealing by modulating its DNA binding. EMBO J. 30, 3368–3382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Dephoure N., Gould K. L., Gygi S. P., Kellogg D. R. (2013) Mapping and analysis of phosphorylation sites: a quick guide for cell biologists. Mol. Biol. Cell 24, 535–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Krupnick J. G., Goodman O. B., Jr., Keen J. H., Benovic J. L. (1997) Arrestin/clathrin interaction. Localization of the clathrin binding domain of nonvisual arrestins to the carboxy terminus. J. Biol. Chem. 272, 15011–15016 [DOI] [PubMed] [Google Scholar]
- 93. Laporte S. A., Oakley R. H., Zhang J., Holt J. A., Ferguson S. S., Caron M. G., Barak L. S. (1999) The β2-adrenergic receptor/βarrestin complex recruits the clathrin adaptor AP-2 during endocytosis. Proc. Natl. Acad. Sci. U.S.A. 96, 3712–3717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. He G., Gupta S., Yi M., Michaely P., Hobbs H. H., Cohen J. C. (2002) ARH is a modular adaptor protein that interacts with the LDL receptor, clathrin, and AP-2. J. Biol. Chem. 277, 44044–44049 [DOI] [PubMed] [Google Scholar]
- 95. Schlessinger J., Lemmon M. A. (2003) SH2 and PTB domains in tyrosine kinase signaling. Sci. STKE 2003, RE12. [DOI] [PubMed] [Google Scholar]
- 96. Sondermann H., Kuriyan J. (2005) C2 can do it, too. Cell 121, 158–160 [DOI] [PubMed] [Google Scholar]
- 97. Wagner M. J., Stacey M. M., Liu B. A., Pawson T. (2013) Molecular mechanisms of SH2- and PTB-domain-containing proteins in receptor tyrosine kinase signaling. Cold Spring Harb. Perspect. Biol. 5, a008987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Ghai R., Bugarcic A., Liu H., Norwood S. J., Skeldal S., Coulson E. J., Li S. S., Teasdale R. D., Collins B. M. (2013) Structural basis for endosomal trafficking of diverse transmembrane cargos by PX-FERM proteins. Proc. Natl. Acad. Sci. U.S.A. 110, E643–E652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Boll W., Ohno H., Songyang Z., Rapoport I., Cantley L. C., Bonifacino J. S., Kirchhausen T. (1996) Sequence requirements for the recognition of tyrosine-based endocytic signals by clathrin AP-2 complexes. EMBO J. 15, 5789–5795 [PMC free article] [PubMed] [Google Scholar]
- 100. Betts G. N., van der Geer P., Komives E. A. (2008) Structural and functional consequences of tyrosine phosphorylation in the LRP1 cytoplasmic domain. J. Biol. Chem. 283, 15656–15664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Eden E. R., Patel D. D., Sun X. M., Burden J. J., Themis M., Edwards M., Lee P., Neuwirth C., Naoumova R. P., Soutar A. K. (2002) Restoration of LDL-receptor function in cells from patients with autosomal recessive hypercholesterolemia by retroviral expression of ARH1. J. Clin. Invest. 110, 1695–1702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Anderson R. G., Goldstein J. L., Brown M. S. (1976) Localization of low density lipoprotein receptors on plasma membrane of normal human fibroblasts and their absence in cells from a familial hypercholesterolemia homozygote. Proc. Natl. Acad. Sci. U.S.A. 73, 2434–2438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Maurer M. E., Cooper J. A. (2006) The adaptor protein Dab2 sorts LDL receptors into coated pits independently of AP-2 and ARH. J. Cell Sci. 119, 4235–4246 [DOI] [PubMed] [Google Scholar]
- 104. Uhlik M. T., Temple B., Bencharit S., Kimple A. J., Siderovski D. P., Johnson G. L. (2005) Structural and evolutionary division of phosphotyrosine binding (PTB) domains. J. Mol. Biol. 345, 1–20 [DOI] [PubMed] [Google Scholar]
- 105. Nagai M., Meerloo T., Takeda T., Farquhar M. G. (2003) The adaptor arotein ARH escorts megalin to and through endosomes. Mol. Biol. Cell 14, 4984–4996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Shah M., Baterina O. Y., Jr., Taupin V., Farquhar M. G. (2013) ARH directs megalin to the endocytic recycling compartment to regulate its proteolysis and gene expression. J. Cell Biol. 202, 113–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Mahabaleshwar H., Tarbashevich K., Nowak M., Brand M., Raz E. (2012) β-Arrestin control of late endosomal sorting facilitates decoy receptor function and chemokine gradient formation. Development 139, 2897–2902 [DOI] [PubMed] [Google Scholar]
- 108. Zhang J., Barak L. S., Anborgh P. H., Laporte S. A., Caron M. G., Ferguson S. S. (1999) Cellular trafficking of G protein-coupled receptor/β-arrestin endocytic complexes. J. Biol. Chem. 274, 10999–11006 [DOI] [PubMed] [Google Scholar]
- 109. Déry O., Thoma M. S., Wong H., Grady E. F., Bunnett N. W. (1999) Trafficking of proteinase-activated receptor-2 and β-arrestin-1 tagged with green fluorescent protein: β-arrestin-dependent endocytosis of a proteinase receptor. J. Biol. Chem. 274, 18524–18535 [DOI] [PubMed] [Google Scholar]
- 110. Groarke D. A., Wilson S., Krasel C., Milligan G. (1999) Visualization of agonist-induced association and trafficking of green fluorescent protein-tagged forms of both β-arrestin-1 and the thyrotropin-releasing hormone receptor-1. J. Biol. Chem. 274, 23263–23269 [DOI] [PubMed] [Google Scholar]
- 111. Mundell S. J., Benovic J. L. (2000) Selective regulation of endogenous G protein-coupled receptors by arrestins in HEK293 cells. J. Biol. Chem. 275, 12900–12908 [DOI] [PubMed] [Google Scholar]
- 112. Kang R. S., Fölsch H. (2011) ARH cooperates with AP-1B in the exocytosis of LDLR in polarized epithelial cells. J. Cell Biol. 193, 51–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Hornbeck P. V., Chabra I., Kornhauser J. M., Skrzypek E., Zhang B. (2004) PhosphoSite: a bioinformatics resource dedicated to physiological protein phosphorylation. Proteomics 4, 1551–1561 [DOI] [PubMed] [Google Scholar]
- 114. Bodenmiller B., Malmstrom J., Gerrits B., Campbell D., Lam H., Schmidt A., Rinner O., Mueller L. N., Shannon P. T., Pedrioli P. G., Panse C., Lee H. K., Schlapbach R., Aebersold R. (2007) PhosphoPep: a phosphoproteome resource for systems biology research in Drosophila Kc167 cells. Mol. Syst. Biol. 3, 139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Kelly B. T., McCoy A. J., Späte K., Miller S. E., Evans P. R., Höning S., Owen D. J. (2008) A structural explanation for the binding of endocytic dileucine motifs by the AP2 complex. Nature 456, 976–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Huang F., Jiang X., Sorkin A. (2003) Tyrosine phosphorylation of the β2 subunit of clathrin adaptor complex AP-2 reveals the role of a di-leucine motif in the epidermal growth factor receptor trafficking. J. Biol. Chem. 278, 43411–43417 [DOI] [PubMed] [Google Scholar]
- 117. Fessart D., Simaan M., Laporte S. A. (2005) c-Src regulates clathrin adapter protein 2 interaction with β-arrestin and the angiotensin II type 1 receptor during clathrin-mediated internalization. Mol. Endocrinol. 19, 491–503 [DOI] [PubMed] [Google Scholar]
- 118. Zimmerman B., Simaan M., Lee M. H., Luttrell L. M., Laporte S. A. (2009) c-Src-mediated phosphorylation of AP-2 reveals a general mechanism for receptors internalizing through the clathrin pathway. Cell. Signal. 21, 103–110 [DOI] [PubMed] [Google Scholar]
- 119. Hunter T. (2012) Why nature chose phosphate to modify proteins. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367, 2513–2516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Pearlman S. M., Serber Z., Ferrell J. E., Jr. (2011) A mechanism for the evolution of phosphorylation sites. Cell 147, 934–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Johnson L. N., Lowe E. D., Noble M. E., Owen D. J. (1998) The eleventh Datta lecture: the structural basis for substrate recognition and control by protein kinases. FEBS Lett. 430, 1–11 [DOI] [PubMed] [Google Scholar]
- 122. Amanchy R., Periaswamy B., Mathivanan S., Reddy R., Tattikota S. G., Pandey A. (2007) A curated compendium of phosphorylation motifs. Nat. Biotechnol. 25, 285–286 [DOI] [PubMed] [Google Scholar]
- 123. Lu C. T., Huang K. Y., Su M. G., Lee T. Y., Bretaña N. A., Chang W. C., Chen Y. J., Huang H. D. (2013) DbPTM 3.0: an informative resource for investigating substrate site specificity and functional association of protein post-translational modifications. Nucleic Acids Res. 41, D295–D305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Di Fiore P. P., De Camilli P. (2001) Endocytosis and signaling: an inseparable partnership. Cell 106, 1–4 [DOI] [PubMed] [Google Scholar]
- 125. Mundell S. J., Luo J., Benovic J. L., Conley P. B., Poole A. W. (2006) Distinct clathrin-coated pits sort different G protein-coupled receptor cargo. Traffic 7, 1420–1431 [DOI] [PubMed] [Google Scholar]
- 126. Cao T. T., Mays R. W., von Zastrow M. (1998) Regulated endocytosis of G-protein-coupled receptors by a biochemically and functionally distinct subpopulation of clathrin-coated pits. J. Biol. Chem. 273, 24592–24602 [DOI] [PubMed] [Google Scholar]
- 127. Leonard D., Hayakawa A., Lawe D., Lambright D., Bellve K. D., Standley C., Lifshitz L. M., Fogarty K. E., Corvera S. (2008) Sorting of EGF and transferrin at the plasma membrane and by cargo-specific signaling to EEA1-enriched endosomes. J. Cell Sci. 121, 3445–3458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Lakadamyali M., Rust M. J., Zhuang X. (2006) Ligands for clathrin-mediated endocytosis are differentially sorted into distinct populations of early endosomes. Cell 124, 997–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Puthenveedu M. A., von Zastrow M. (2006) Cargo regulates clathrin-coated pit dynamics. Cell 127, 113–124 [DOI] [PubMed] [Google Scholar]
- 130. Grossier J. P., Xouri G., Goud B., Schauer K. (2014) Cell adhesion defines the topology of endocytosis and signaling. EMBO J. 33, 35–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Taylor M. J., Perrais D., Merrifield C. J. (2011) A high precision survery of the molecular dynamics of mammalian clathrin mediated endocytosis. PLoS Biol. 9, e1000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Hawryluk M. J., Keyel P. A., Mishra S. K., Watkins S. C., Heuser J. E., Traub L. M. (2006) Epsin 1 is a polyubiquitin-selective clathrin-associated sorting protein. Traffic 7, 262–281 [DOI] [PubMed] [Google Scholar]
- 133. Chetrit D., Ziv N., Ehrlich M. (2009) Dab2 regulates clathrin assembly and cell spreading. Biochem. J. 418, 701–715 [DOI] [PubMed] [Google Scholar]
- 134. Sochacki K. A., Larson B. T., Sengupta D. C., Daniels M. P., Shtengel G., Hess H. F., Taraska J. W. (2012) Imaging the post-fusion release and capture of a vesicle membrane protein. Nat. Commun. 3, 1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Jha A., Watkins S. C., Traub L. M. (2012) The apoptotic engulfment protein Ced-6 participates in clathrin-mediated yolk uptake in Drosophila egg chambers. Mol. Biol. Cell 23, 1742–1764 [DOI] [PMC free article] [PubMed] [Google Scholar]