Background: Kv4 and its auxiliary subunit KChIP form an octameric complex (4:4) in crystal structure.
Results: Biophysical properties of Kv4 gradually changed depending on the amount of expressed KChIP.
Conclusion: The stoichiometry of the Kv4·KChIP complex is not fixed but variable.
Significance: Results suggest that excitable cells, such as cardiac cells, can be finely tuned via the relative expression levels of Kv4 and KChIP.
Keywords: Electrophysiology, Fluorescence, Imaging, Ion Channel, Potassium Channel, Protein Complex, K+ Channel-interacting Protein (KChIP), Single Molecule Imaging, Stoichiometry
Abstract
Kv4 is a voltage-gated K+ channel, which underlies somatodendritic subthreshold A-type current (ISA) and cardiac transient outward K+ (Ito) current. Various ion channel properties of Kv4 are known to be modulated by its auxiliary subunits, such as K+ channel-interacting protein (KChIP) or dipeptidyl peptidase-like protein. KChIP is a cytoplasmic protein and increases the current amplitude, decelerates the inactivation, and accelerates the recovery from inactivation of Kv4. Crystal structure analysis demonstrated that Kv4 and KChIP form an octameric complex with four Kv4 subunits and four KChIP subunits. However, it remains unknown whether the Kv4·KChIP complex can have a different stoichiometry other than 4:4. In this study, we expressed Kv4.2 and KChIP4 with various ratios in Xenopus oocytes and observed that the biophysical properties of Kv4.2 gradually changed with the increase in co-expressed KChIP4. The tandem repeat constructs of Kv4.2 and KChIP4 revealed that the 4:4 (Kv4.2/KChIP4) channel shows faster recovery than the 4:2 channel, suggesting that the biophysical properties of Kv4.2 change, depending on the number of bound KChIP4s. Subunit counting by single-molecule imaging revealed that the bound number of KChIP4 in each Kv4.2·KChIP4 complex was dependent on the expression level of KChIP4. Taken together, we conclude that the stoichiometry of Kv4·KChIP complex is variable, and the biophysical properties of Kv4 change depending on the number of bound KChIP subunits.
Introduction
Membrane proteins, including ion channels, often form a heteromultimeric complex with auxiliary subunits in physiological conditions. The stoichiometry of the protein complex, therefore, needs to be appropriately regulated. Kv4 is a member of the voltage-gated K+ channel family. In hippocampal pyramidal neurons, Kv4 produces the somatodendritic subthreshold A-type current (ISA) and attenuates the back-propagation of action potentials (1–5). In ventricular cardiac myocytes, Kv4 produces transient outward K+ current (Ito), which shapes the first phase of repolarization (6, 7). Kv4 is also well known to form an ion channel complex with its auxiliary subunits. Although Kv4 generates a current as a homotetramer and exhibits fast inactivation and slow recovery from inactivation, auxiliary subunits, such as K+ channel-interacting protein (KChIP)2 or dipeptidyl peptidase-like protein (DPP), change the properties of Kv4 (8–12). Kv4, KChIP, and DPP even form a ternary complex (13).
KChIP is a member of the neuronal calcium sensor protein family (14–16), and four subtypes have been identified (KChIP1–4) (12, 16, 17). KChIP is known to bind to the N terminus of Kv4 and to modulate multiple Kv4 properties (18–21). First, KChIP promotes the surface expression of Kv4 and increases the current amplitude of Kv4 (22, 23). Second, KChIP binding slows the inactivation of Kv4 by restricting the movement of the N terminus, which may occlude the ion channel pore (24). Third, KChIP accelerates the recovery from inactivation of Kv4 (2, 25, 26). It has been suggested that the C terminus of KChIP plays an important role in acceleration of the recovery from inactivation of Kv4 (25, 27). However, the detailed mechanism of the acceleration of the recovery from inactivation remains unknown. The Kv4 channel is composed of four α-subunits. This means that one Kv4 channel has four identical binding sites for KChIP. A previous biochemical study suggested that the purified Kv4.2·KChIP2 complex consists of four subunits for each (28). Crystal structure analysis showed that Kv4.3 and KChIP1 form a 4:4 channel complex (29, 30). However, it still remains uncertain whether the Kv4·KChIP complex is expressed with a stoichiometry of 4:4 exclusively or whether complexes with different stoichiometries also exist. It is also unknown whether the different stoichiometries, if they exist, affect the biophysical properties of the Kv4 channel.
In this study, we examined how the expression level of KChIP4 affects the properties of Kv4.2. We first co-expressed Kv4.2 and KChIP4 in different complementary RNA (cRNA) ratios and evaluated the biophysical properties with two-electrode voltage clamp. We observed that the biophysical properties of Kv4.2 changed gradually with an increase in KChIP4 expression, suggesting the existence of Kv4.2·KChIP4 complexes with variable stoichiometries. We subsequently confirmed that the stoichiometry of the Kv4.2·KChIP4 complex affects the properties of Kv4.2 by using tandem repeat constructs in which the stoichiometry is strictly controlled. We finally applied the subunit-counting method by single-molecule imaging (31–35). Although KChIP4 is a cytoplasmic protein, we could successfully apply the method to determine the number of KChIP4 subunits included in a Kv4.2·KChIP4 complex and confirmed that the stoichiometry changes depending on the expression level of KChIP4.
EXPERIMENTAL PROCEDURES
Molecular Biology
Human Kv4.2 (NM_012281.2) and rat KChIP4 (NM_181365.2) were subcloned into the pGEMHE expression vector. For the KChIP4-Kv4.2 tandem construct, a flexible GGS linker (GGSGGSGGSGGSGG) and a NotI site were introduced into the N terminus of Kv4.2 by PCR using KOD Plus version 2 (Toyobo) (“NotI-linker-Kv4.2”). A NotI site was also introduced into the C terminus of KChIP4 by PCR (“KChIP4-NotI”). Two fragments were fused at the NotI site and inserted into the pGEMHE expression vector. For the KChIP4-Kv4.2-Kv4.2 tandem construct, we first made a Kv4.2-Kv4.2 construct by PCR, in which two Kv4.2 sequences were linked with a flexible GGS linker. A fragment was produced from this construct by cleavage at the unique BamHI site in Kv4.2 and inserted into the BamHI site of the KChIP4-Kv4.2 tandem construct. For the monomeric enhanced green fluorescent protein (mEGFP)-tagged constructs, mEGFP was fused to the C terminus of Kv4.2 or KChIP4 with a flexible GGS linker (Kv4.2-mEGFP and KChIP4-mEGFP). For Kv4.2-KChIP4-mEGFP, we first made a Kv4.2-KChIP4 construct. The fragment obtained from KChIP4-mEGFP by cleavage at the unique EcoT22I site in KChIP4 and the unique HindIII site in the vector was inserted between the EcoT22I and HindIII sites in the Kv4.2-KChIP4 construct. All constructs were confirmed by DNA sequencing. cRNA was then prepared from the linearized plasmid cDNA using the T7 mMessage mMachine kit (Ambion).
Preparation of Xenopus laevis Oocytes
Oocytes were collected from X. laevis anesthetized in water containing 0.15% Tricaine and treated with collagenase (2 mg/ml; type 1; Sigma-Aldrich) for 6–7 h to remove the follicular cell layer. Oocytes of similar size at stage V or VI were injected with 50 nl of cRNA solution. The injected oocytes were incubated for 1–3 days at 17 °C in frog Ringer solution containing 88 mm NaCl, 1 mm KCl, 2.4 mm NaHCO3, 0.3 mm Ca(NO3)2, 0.41 mm CaCl2, and 0.82 mm MgSO4, pH 7.6, with 0.1% penicillin-streptomycin solution (Sigma-Aldrich). All experiments were approved by the Animal Care Committee of the National Institute for Physiological Sciences and performed following the institutional guidelines.
Two-electrode Voltage Clamp
K+ currents were recorded under two-electrode voltage clamp 1–3 days after cRNA injection, using an OC-725C amplifier (Warner Instruments). All experiments were performed at room temperature. The microelectrodes were drawn from borosilicate glass capillaries (World Precision Instruments) to a resistance of 0.2–0.5 megaohms and filled with 3 m potassium acetate and 10 mm KCl (pH 7.2). ND-96 was used for the bath solution containing 96 mm NaCl, 2 mm KCl, 1.8 mm CaCl2, 1 mm MgCl2, and 5 mm HEPES, pH 7.2. Holding potential was at −80 mV. Stimulation, data acquisition, and data analysis were performed using a Digidata 1440 system and pCLAMP version 10.3 software (Molecular Devices). Data from the amplifier were digitized at 10 kHz and filtered at 1 kHz.
Western Blotting
One day after cRNA injection, oocytes were sonicated in PBS containing 137 mm NaCl, 2.7 mm KCl, 10 mm Na2HPO4, 1.76 mm KH2PO4 with protein inhibitor (Complete, Mini, EDTA-free, Roche Applied Science). The homogenates were centrifuged at 3000 rpm and 4 °C for 10 min to remove yolk, and the supernatants were retained as protein extracts. These were treated at 100 °C for 5 min in SDS-DTT solution containing 50 mm Tris-HCl (pH 6.8), 100 mm DTT, 10% glycerol, 0.1% bromphenol blue, 2% SDS. SDS-PAGE was performed using 2–15% gradient gels in SDS-PAGE buffer containing 25 mm Tris-base, 192 mm glycine, 0.1% SDS. Protein concentration for each sample was measured by a Bradford protein assay (36). The amount of each loaded protein was adjusted to 10 μg. The proteins were transferred to polyvinylidene fluoride (PVDF) membranes in Western blot buffer containing 25 mm Tris-base, 192 mm glycine. The membranes with bound protein were treated with 5% skim milk in TBST solution containing 20 mm Tris-HCl (pH 8.0), 150 mm NaCl, 0.05% Tween 20. Membranes were then incubated with primary antibody (1:250) at 37 °C for 1 h. The primary antibody against KChIP4 used in this study was anti-KCNIP4 (HPA022862, Sigma-Aldrich). Bound primary antibodies were detected with HRP-labeled donkey anti-rabbit IgG (1:1000) (GE Healthcare). Immunoreactivity was visualized using an enhanced chemiluminescence (ECL) detection kit (GE Healthcare) and was detected using a LAS-3000 image analyzer (Fujifilm, Tokyo, Japan).
Single Molecule Subunit Counting
Oocytes of similar size at stage V or VI were injected with 50 nl of cRNA solution. At 12–20 h after cRNA injection, the oocytes were enzymatically treated with 1 unit/ml neuraminidase and 1 mg/ml hyaluronidase for 15 min at 17 °C to remove the extracellular matrix. Oocytes were osmotically shrunk with 2× frog Ringer solution and devitellinized manually by forceps. Oocytes were placed on a high refractive index coverslip (n = 1.78). The coverslip matched the reflective index of the objective lens (Olympus; ×100, numerical aperture 1.65) mounted on an inverted microscope (Olympus; IX71). ND-96 was used as the extracellular solution. mEGFP was excited with a Cyan 488-nm solid state laser (Spectra-Physics), and the fluorescent spots were captured by an iXon3 EMCCD camera (Andor). To obtain total internal reflection fluorescence (TIRF) conditions, the orientation of the laser was adjusted using a manual mirror. Movies of 400 frames were acquired at 20 Hz for 20 s. The sampling area was 25.6 × 25.6 μm2. The fluorescence intensity of each spot was measured off-line. Only non-overlapping and stable (not moving during the 20-s recording) spots were used for analysis and data collection (31).
Data Analysis
We analyzed the ionic currents using pCLAMP version 10.3 (Molecular Devices) and Igor Pro software (WaveMetrics, Inc.). The analyses of inactivation, recovery from inactivation, and steady-state inactivation were all performed without leak subtraction. For the analysis of inactivation, the currents from each potential were fitted with either a single or a double exponential function. The time course of recovery from inactivation was fitted with a single exponential function.
For steady-state inactivation analysis, the current amplitudes after 5-s depolarization at each command potential were plotted and fitted with a two-state Boltzmann equation,
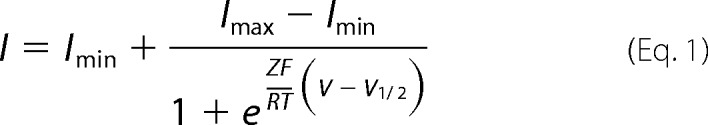 |
where Imax and Imin are the maximum and minimum current amplitudes during the test pulse, V½ is the half-inactivation voltage, Z is the effective charge, F is Faraday's constant, R is the gas constant, and T is the absolute temperature. For normalization, I was divided by Imax.
In subunit counting, the distributions of observed bleaching steps were fitted with a binomial distribution P(X = a),
where A is the total number of fluorescent spots (including “0 step” spot), n is the number of bound subunits, a is the observed number of bleaching steps, and p is the probability of GFP being fluorescent.
Statistical Analyses
All data are presented as the mean ± S.E., and n represents the number of samples. Statistical differences were evaluated by Student's unpaired t test (for two-group comparisons) or Dunnett's test (for multiple comparisons). Values of p < 0.05 were considered significant (***, p < 0.001).
RESULTS
KChIP4 Enhances the Expression and Modifies the Biophysical Properties of Kv4.2 in a Dose-dependent Manner
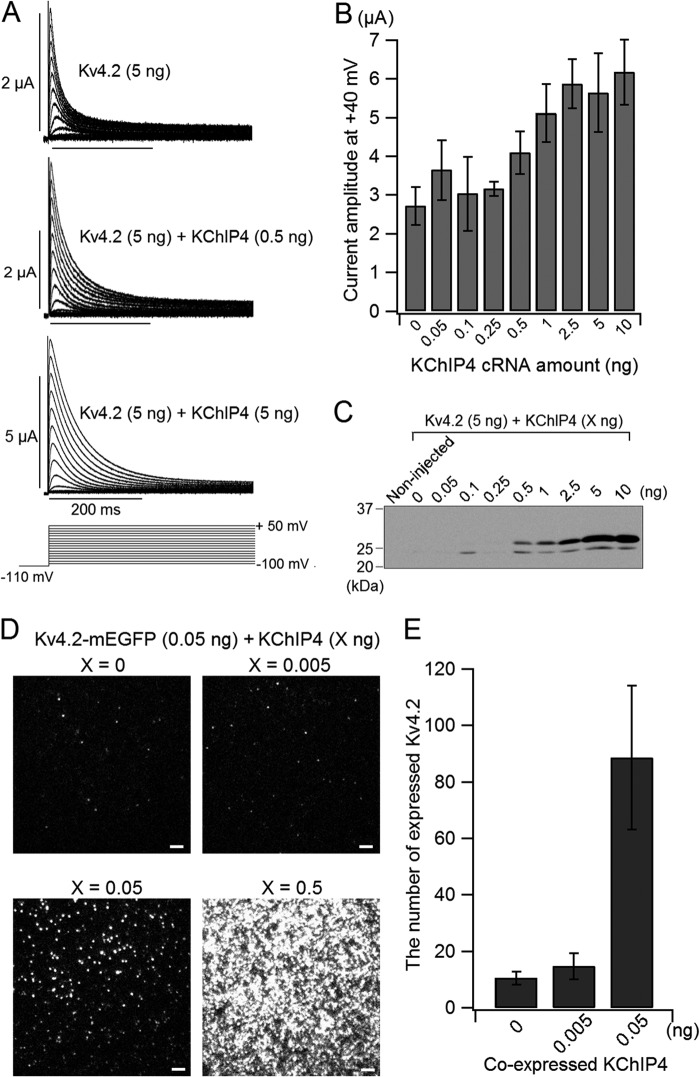
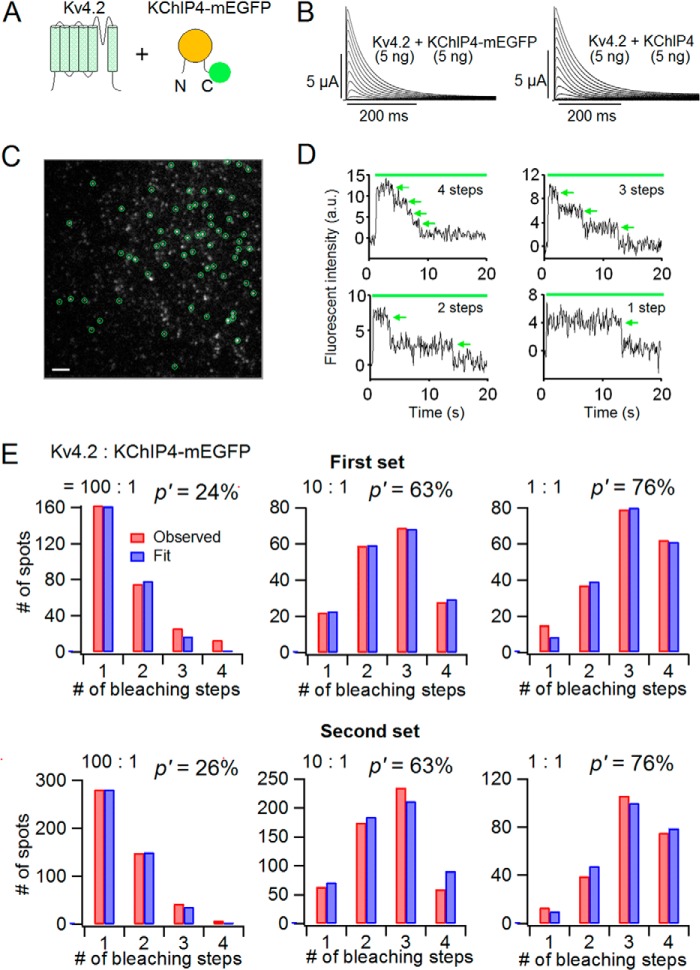
KChIP is known to change some properties of Kv4, such as peak current amplitude, inactivation kinetics, steady state inactivation, and recovery from inactivation (2, 13, 17, 26). First, we examined how the amount of co-expressed KChIP4 affects Kv4.2 expression. We injected different amounts of KChIP4 cRNA (0–10 ng) with 5 ng of Kv4.2 cRNA in X. laevis oocytes and analyzed the currents with two-electrode voltage clamp. The current amplitude was augmented with an increase in the amount of co-expressed KChIP4 (Fig. 1, A and B). The current amplitude of Kv4.2 was almost doubled, with KChIP4 over 2.5 ng (Fig. 1B). To make sure that the expression of KChIP4 in oocytes varied in proportion to the injected KChIP4 cRNA, Western blotting of KChIP4 prepared from oocytes injected with different amounts of KChIP4 cRNA along with 5 ng of Kv4.2 cRNA was performed. 10 μg of sample protein was loaded in each lane. We observed two bands with different sizes in the lanes of samples from oocytes injected with more than 0.5 ng of KChIP4 cRNA. The larger bands gradually intensified in accordance with the amount of injected KChIP4 (Fig. 1C). To examine the effect of KChIP4 on the expression of Kv4.2, we expressed Kv4.2-mEGFP (0.05 ng) with different amounts of KChIP4 cRNA (0, 0.005, 0.05, and 0.5 ng) in oocytes and analyzed the expression of Kv4.2-mEGFP by counting the fluorescent spots on the cell surface under TIRF microscopy. (Note that the cRNA of Kv4.2 was very low (0.05 ng) for single molecule imaging, compared with the cRNA for electrophysiology (5 ng)). The number of Kv4.2-mEGFP spots showed a steep increase from 0.005 to 0.05 ng of cRNA of co-expressed KChIP4 (Fig. 1, D and E), which is well below the detection limit of Western blotting. Another large increment of expression was observed between 0.05 and 0.5 ng, although there were too many spots to count (Fig. 1D).
FIGURE 1.
The expression of Kv4.2 gradually changes with increasing co-expression of KChIP4. A, representative current traces of Kv4.2 (5 ng), Kv4.2 (5 ng) with KChIP4 (0.5 ng), and Kv4.2 (5 ng) with KChIP4 (5 ng). The holding potential was at −80 mV. After a 500-ms hyperpolarization at −110 mV to remove the inactivation, the currents were elicited by depolarizing voltage steps for 1 s between −100 and +50 mV in 10-mV increments. Only the current traces of the first 400 ms are shown. B, comparison of the peak current amplitudes of Kv4.2 at +40 mV with various amounts of KChIP4 (n = 6–12). C, dependence of KChIP4 protein expression on the amount of cRNA injected. The different amounts of KChIP4 cRNA (0–10 ng) were injected along with 5 ng of Kv4.2 cRNA into oocytes. Non-injected, oocytes without any injection. D, comparison of the images of Kv4.2-mEGFP expressed on the oocyte surface with various amounts of co-expressed KChIP4 under TIRF microscopy. Each fluorescent spot corresponds to a single tetrameric Kv4.2 channel. Scale bar, 2 μm. E, the numbers of fluorescent spots from Kv4.2-mEGFP are plotted for different amounts of KChIP4 injected (n = 10 for each). Error bars, S.E.
We next examined the biophysical properties of Kv4.2 with different amounts of KChIP4. The normalized I–V relationships were not changed by the amount of KChIP4 (Fig. 2A). This means that KChIP4 does not influence the voltage dependence of activation of Kv4.2. The steady-state inactivation curve shifted to depolarized potentials by increasing KChIP4. The amount of change in V½ was approximately +10 mV between Kv4.2 alone (−66.7 ± 2.2 mV; n = 3) and Kv4.2 (5 ng) with KChIP4 (10 ng) (−56.0 ± 1.3 mV; n = 5) (Fig. 2B). We also observed that the inactivation of Kv4.2 was decelerated with an increase in co-expressed KChIP4 (Fig. 1A). The inactivation kinetics at each potential from Kv4.2 (5 ng) with low KChIP4 expression (0, 0.05, 0.1, 0.25, 0.5, and 1 ng) were best fitted with a double exponential function, whereas the inactivation kinetics from Kv4.2 (5 ng) with high KChIP4 expression (2.5, 5, and 10 ng) were fitted with a single exponential function (Fig. 2C). When the currents were fitted with a double exponential function with low KChIP4 expression, the faster component was dominant at all membrane potentials studied (Fig. 2D). The faster inactivation time constant was increased with an increase in the amount of KChIP4 up to 2.5 ng, where the inactivation kinetics can be fitted with a single exponential function.
FIGURE 2.
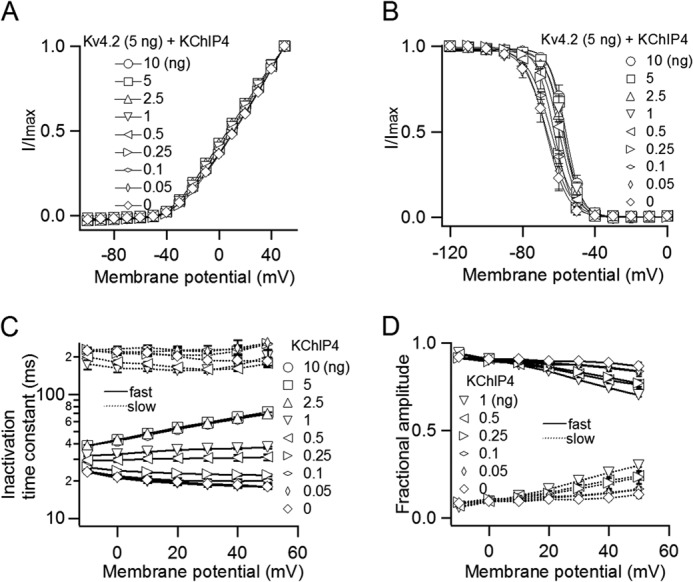
The biophysical properties of Kv4.2 gradually change with increasing co-expression of KChIP4. A, I–V relationships with various amounts of KChIP4. The peak current amplitudes at each potential are normalized to the peak current amplitude at +50 mV (n = 3–13). B, the steady-state inactivation curves of Kv4.2 with various amounts of KChIP4. A 5-s prepulse from −120 to 0 mV in 10-mV increments was applied before a test pulse at +40 mV (n = 3–5). C, voltage dependence of fast and slow inactivation time constants for Kv4.2 with various amounts of KChIP4 (n = 3–13). Slow time constants only are shown for 0–1 ng. D, fractional contributions of the fast and slow inactivation components for 0–1 ng. Error bars, S.E.
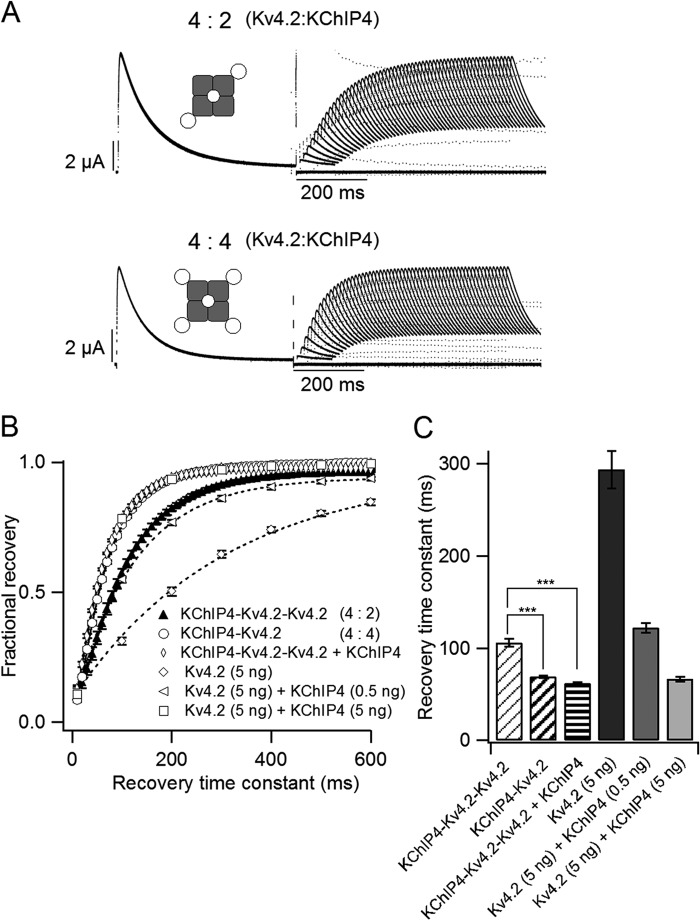
The Recovery from Inactivation of Kv4.2 Is Gradually Modulated with an Increase in the Amount of Co-expressed KChIP4
We also investigated whether the expression level of KChIP4 affects the recovery from inactivation of Kv4.2. The same range of KChIP4 cRNA as used in Fig. 1 (0–10 ng) was co-injected with Kv4.2 cRNA (5 ng), and the kinetics of the recovery from inactivation were tested using a two-pulse stimulation protocol. A +40-mV depolarizing prepulse was applied for 500 ms to make the channel completely inactivated, and a +40-mV test pulse was subsequently applied for 100 ms after an interval from 10 to 800 ms (Fig. 3A, inset). As reported in previous studies (15), KChIP4 accelerated the recovery from inactivation of Kv4.2. The time course of the recovery for each group is shown in Fig. 3B. The recovery from inactivation of Kv4.2 was gradually accelerated with an increase in KChIP4 cRNA, up to 2.5 ng of KChIP4 cRNA. Each time course of recovery from inactivation was fitted with a single exponential function. The recovery time constants were gradually shortened from 358 ± 72 ms (n = 3) without KChIP4 to 50 ± 3 ms (n = 8) with 2.5 ng of KChIP4 (Fig. 3C). This gradual change implies that each Kv4.2 channel may have fewer than four KChIP4s when the expression of KChIP4 is low and that the recovery process from inactivation is increasingly facilitated as more KChIP4 subunits are bound.
FIGURE 3.
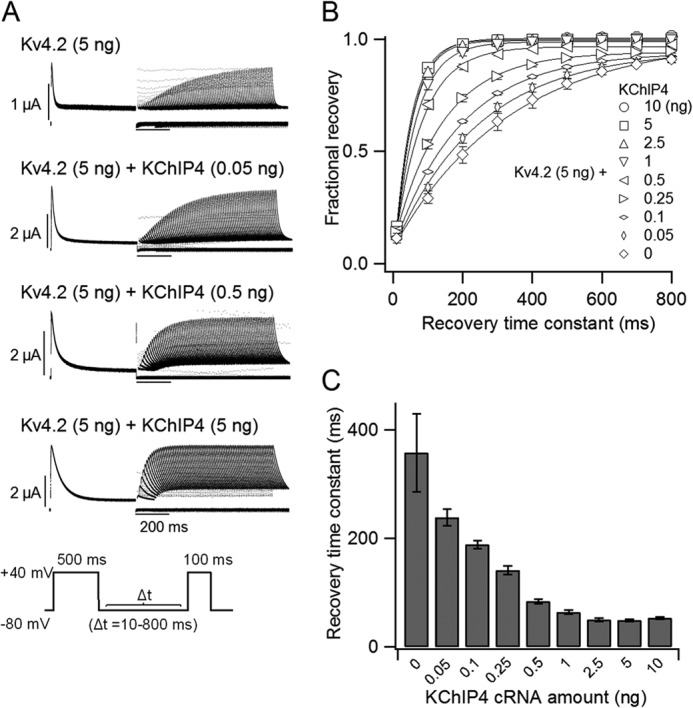
The recovery from inactivation of Kv4.2 is gradually accelerated with increasing expression of KChIP4. A, representative current traces of the recovery from inactivation of Kv4.2 (5 ng) with various amounts of KChIP4 mRNA (0–5 ng). The currents were elicited by a two-pulse protocol, as shown in the inset. B, the kinetics of the recovery from inactivation of Kv4.2 at −80 mV with various amounts of KChIP4. The current amplitudes were normalized to the average of the prepulse current amplitudes at +40 mV (n = 3–13). C, time constants of the recovery from inactivation of Kv4.2 at −80 mV with various amounts of KChIP4. The time constants were obtained from B by fitting with a single exponential function. Error bars, S.E.
Acceleration of Recovery from Inactivation of Kv4.2 by Increasing the Number of Bound KChIP4 Subunits, as Shown by Tandem Constructs
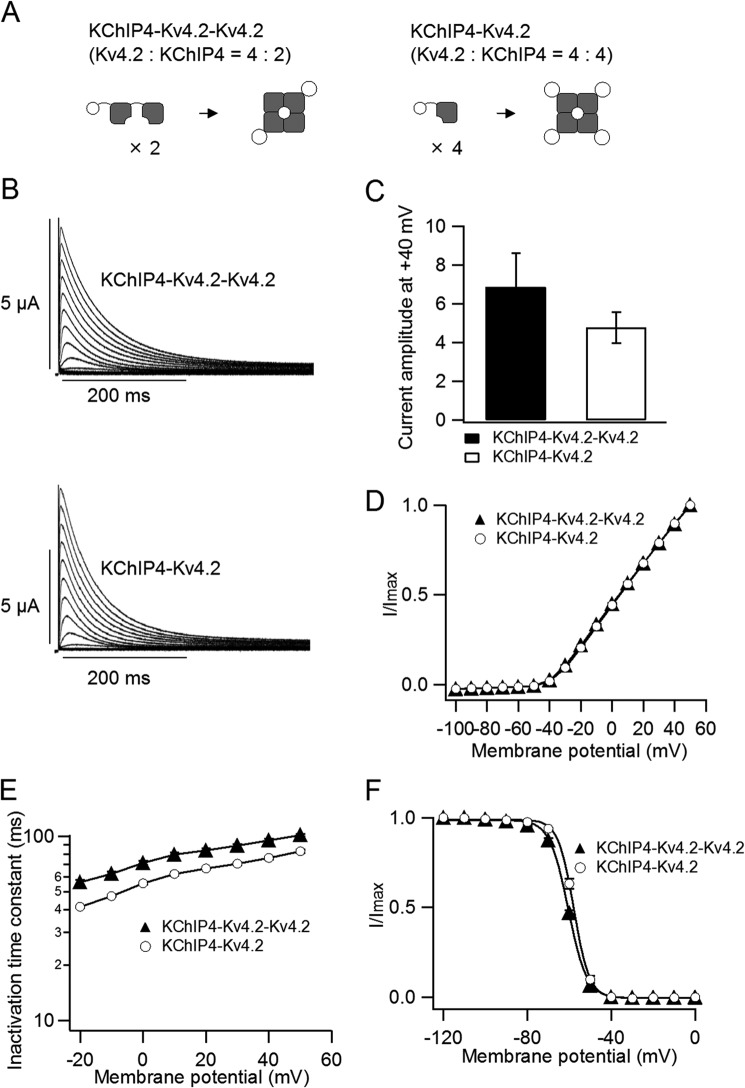
We hypothesized that the number of bound KChIP4 subunits affects the properties of Kv4.2. To tightly control the stoichiometry of the Kv4.2·KChIP4 complex, we made two tandem repeat constructs. The KChIP4-Kv4.2-Kv4.2 tandem construct was expected to form a 4:2 (Kv4.2/KChIP4) channel, and the KChIP4-Kv4.2 tandem construct was expected to form a 4:4 channel (Fig. 4A). Their electrophysiological properties were evaluated by two-electrode voltage clamp. The peak current amplitudes and the normalized I–V relationships were not significantly different between the 4:2 and the 4:4 channels (Fig. 4, B–D). The 4:2 channel showed slightly slower inactivation than the 4:4 channel (Fig. 4E). The V½ of the steady-state inactivation was slightly but significantly (p < 0.01) different between the 4:2 (−60.6 ± 0.3 mV, n = 3) and the 4:4 channels (−57.9 ± 0.5 mV, n = 4) (Fig. 4F). On the other hand, the recovery from inactivation was significantly faster in the 4:4 channel than in the 4:2 channel (Fig. 5A). The 4:4 channel showed kinetics similar to that of Kv4.2 (5 ng) with KChIP4 (5 ng), whereas the 4:2 channel showed kinetics similar to that of Kv4.2 (5 ng) with KChIP4 (0.5 ng) (Fig. 5, B and C). This indicates that KChIP4 binding is not saturated in the case of Kv4.2 (5 ng) with KChIP4 (0.5 ng). Because the 4:2 channel should have two open slots for KChIP4, extra KChIP4 is expected to make the recovery of the 4:2 channel faster. To examine this possibility, we expressed the KChIP4-Kv4.2-Kv4.2 tandem construct with additional KChIP4 (5 ng). As expected, the recovery kinetics of the 4:2 channel with KChIP4 were similar to that of the 4:4 channel (Fig. 5B). Each time course of recovery was fitted with a single exponential function. The recovery time constants of the 4:4 channel (68.6 ± 1.5 ms, n = 4) and the 4:2 channel with KChIP4 (61.8 ± 1.0 ms, n = 4) were significantly (p < 0.001) smaller than that of the 4:2 channel (105.8 ± 4.2 ms, n = 4) (Fig. 5C). This observation further confirmed that the recovery from inactivation of Kv4.2 changes, depending on the number of bound KChIP4 subunits.
FIGURE 4.
Biophysical properties of the tandem repeat constructs. A, schematic illustrations of the tandem repeat constructs. The KChIP4-Kv4.2-Kv4.2 construct forms a 4:2 (Kv4.2/KChIP4) channel (left), and the KChIP4-Kv4.2 construct forms a 4:4 channel (right). B, representative current traces from the tandem repeat constructs, KChIP4-Kv4.2-Kv4.2 (4:2 channel; top) and KChIP4-Kv4.2 (4:4 channel; bottom). The currents were elicited by step pulses from −100 to +50 mV in 10-mV increments. C, the peak current amplitudes at +40 mV for the 4:2 channel (left) and the 4:4 channel (right) (n = 5). D, comparison of I–V relationships for the 4:2 channel (filled triangles) and the 4:4 channel (open circles). The current amplitude at each potential is normalized to the current amplitude at +50 mV. E, comparison of the inactivation time constants from the 4:2 channel (n = 3) and the 4:4 channel (n = 4). The time constants were deduced by fitting a single exponential function to the current traces. F, steady-state inactivation curves for the 4:2 channel (filled triangles) and the 4:4 channel (open circles). The mean values of V½ for the 4:2 and the 4:4 channels are −60.6 ± 0.3 (n = 3) and −57.9 ± 0.5 (n = 4), respectively. Error bars, S.E.
FIGURE 5.
The 4:4 (Kv4.2/KChIP4) channel shows faster recovery than the 4:2 channel. A, representative current traces showing the recovery from inactivation of the 4:2 channel (top) and the 4:4 channel (bottom). The currents were evoked by a two-pulse protocol, as shown in Fig. 3A (inset). B, comparison of the recovery kinetics of the tandem constructs with those of Kv4.2/KChIP4 co-expression. The 4:2 channel shows kinetics similar to that of Kv4.2 (5 ng) with KChIP4 (0.5 ng). The 4:4 channel shows kinetics similar to that of Kv4.2 (5 ng) with KChIP4 (5 ng). The recovery kinetics of the 4:2 channel with additional KChIP4 was comparable with that of the 4:4 channel (n = 4). C, the time constants of single exponential fits of the recovery taken from B. ***, statistically significant difference (p < 0.001 by Dunnett's test). Error bars, S.E.
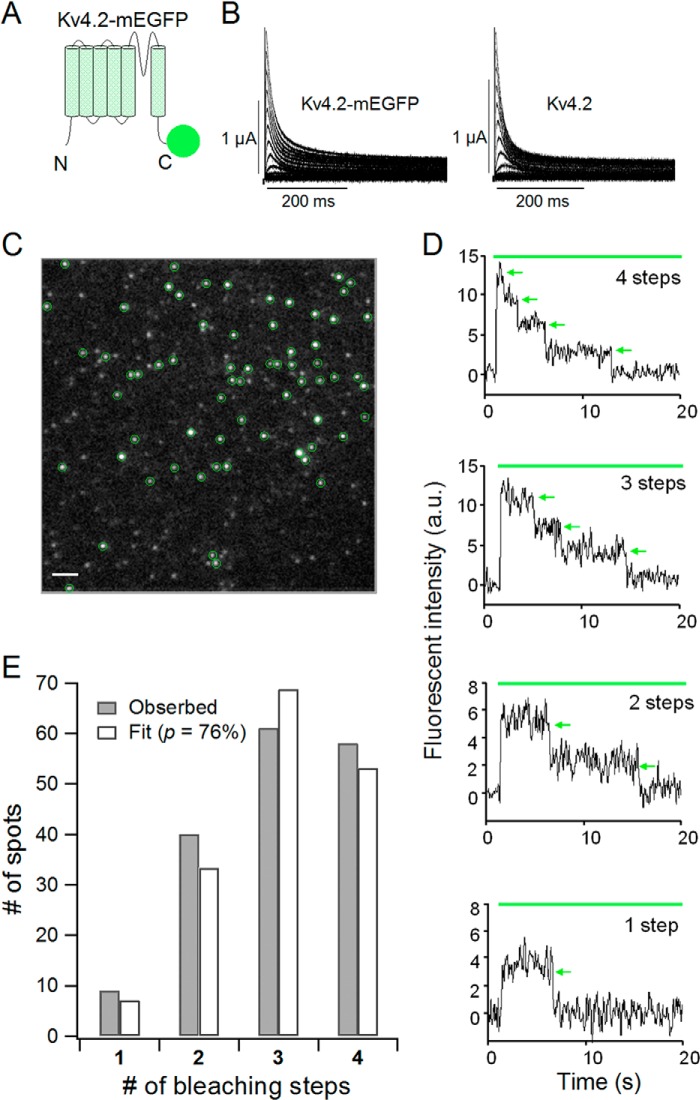
Kv4.2 Is a Tetramer
Before examining the assembly of the Kv4.2 α-subunit and KChIP4 as an ion channel complex, we first attempted to confirm the tetrameric stoichiometry of Kv4.2 in the absence of KChIP4 to examine the validity of our subunit-counting method by single-molecule imaging (31). This method enables us to detect a bleaching event from a single GFP molecule and count the number of GFP-tagged subunits. mEGFP was attached to the C terminus of Kv4.2 with an 18-amino acid linker (Kv4.2-mEGFP) (Fig. 6A). We confirmed that Kv4.2-mEGFP (5 ng) showed fast inactivating current similar to that of wild-type Kv4.2 (5 ng) under two-electrode voltage clamp (Fig. 6B). Kv4.2-mEGFP (0.5 ng) was expressed on the plasma membrane of X. laevis oocytes, and we identified discrete fluorescent spots under TIRF microscopy (Fig. 6C). The density of ion channels was kept low enough (less than 1 spot/μm2) to minimize the probability of two spots overlapping within the same diffraction-limited spot. Most of the Kv4.2-mEGFP channels were observed as immobile spots. We counted bleaching steps from each single fluorescent spot (Fig. 6D). Because mEGFP is not always fluorescent due to misfolding or other reasons, there were not always four bleaching steps from a single fluorescent spot, even for the tetrameric Kv4 channel. The distributions of the bleaching steps were fitted by a binomial distribution formula with a probability (p) of mEGFP being fluorescent as a free parameter (see “Experimental Procedures”). We counted bleaching steps from a total of 168 spots for Kv4.2-mEGFP, and the numbers of spots for each bleaching step were plotted as a histogram (Fig. 6E, gray bars). The observed distribution was best fitted by a binomial distribution with p = 76% (Fig. 6E, white bars). This means that Kv4.2 forms a tetramer, and the florescent probability of mEGFP tagged to Kv4.2 is 76%. We performed another independent set of experiments by counting bleaching steps of 188 spots from another batch and obtained p = 76%. This value is similar to those from previous experiments for the CNG channel, the NMDA receptor, and the TRPP2 channel (79–83%) (31, 33).
FIGURE 6.
Kv4.2 forms a tetramer. A, a schematic illustration of the Kv4.2-mEGFP construct. mEGFP was connected to the C terminus of Kv4.2 with a flexible GGS linker. B, currents from the Kv4.2-mEGFP construct (left) and WT Kv4.2 (right). The currents were elicited by step pulses from −100 to +50 mV in 10-mV increments. C, a single image from a TIRF movie of a Xenopus oocyte expressing Kv4.2-mEGFP. Green circles, countable spots for the bleaching step analysis. Scale bar, 2 μm. D, representative traces of 4, 3, 2, and 1 bleaching step(s) from Kv4.2-mEGFP. Green bar, illumination with 488 nm to excite mEGFP. Green arrows, bleaching events. a.u., arbitrary units. E, distributions of the number of spots with observed bleaching steps (gray) were fitted by a binomial distribution with p = 76% (white). p is the probability that the mEGFP attached to Kv4.2 is fluorescent.
The Stoichiometry of the Kv4.2·KChIP4 Complex Changes, Depending on the Expression Level of KChIP4
To directly count the number of KChIP4 subunits bound to a Kv4.2 tetramer, we next made a KChIP4-mEGFP construct, in which mEGFP was attached to the C terminus of KChIP4 with a flexible 18-amino acid linker (Fig. 7A). The stoichiometry of the Kv4.2·KChIP4 complex with different expression levels of Kv4.2 and KChIP4 was estimated using the subunit-counting method. We first confirmed that KChIP4-mEGFP (5 ng) decelerated the inactivation of Kv4.2 (5 ng) as wild-type KChIP4 (WT KChIP4) (5 ng) did under two-electrode voltage clamp (Fig. 7B). To investigate a possible expression-dependent stoichiometry change of the Kv4.2·KChIP4-mEGFP complex, three different ratios of injected cRNA (Kv4.2/KChIP4-mEGFP = 100:1 (0.5 ng:0.005 ng), 10:1 (0.1 ng:0.01 ng), and 1:1 (0.05 ng:0.05 ng)) were examined. Under TIRF illumination, we detected clear fluorescent spots of Kv4.2·KChIP4-mEGFP complex and counted bleaching steps from each spot, with four steps being the largest number (Fig. 7, C and D, and supplemental Movie 1). In oocytes expressing only KChIP4-mEGFP, on the other hand, we only observed mobile and fuzzy spots, which were probably moving in the cytosol adjacent to the membrane (supplemental Movie 2). Therefore, fluorescent spots circled in green in Fig. 7C should be KChIP4 with Kv4.2 on the plasma membrane. In oocytes with a 100:1 ratio of Kv4.2 to KChIP4-mEGFP, the fractions of three and four bleaching steps were minor, and those of one and two bleaching steps were mainly observed. On the other hand, when the ratio of Kv4.2 to KChIP4-mEGFP were 10:1 and 1:1, the spots with two, three, and four bleaching steps were considerably increased, and those with one bleaching step were minor (Fig. 7E, red bars). The observed distributions of the bleaching steps were fitted by a binomial distribution with an apparent fluorescence probability (p′), which should be the product of (fluorescence probability of mEGFP) × (association probability of KChIP4-mEGFP with Kv4.2) (Fig. 7E, blue bars). The value of p′ from each cRNA ratio was 24% (100:1), 63% (10:1), and 76% (1:1), respectively (Fig. 7E, top row). p′ values from another set of independent experiments were similar to those of the first set (100:1, 26%; 10:1, 63%; and 1:1, 76%) (Fig. 7E, bottom row). To deduce the association probability of KChIP4 with Kv4.2, we made a Kv4.2-KChIP4-mEGFP tandem construct, in which KChIP4-mEGFP was fused to the C terminus of Kv4.2 with a 16-amino acid linker (Fig. 8A). Because it was expected that this construct formed only the 4:4 channel (Kv4.2-KChIP4-mEGFP), we assumed that the association probability of KChIP4 with Kv4.2 in this tandem construct would be 100%. The tandem constructs were successfully expressed on the plasma membrane, and the fluorescent spots were observed under TIRF illumination (Fig. 8B). The observed distribution was best fitted by a binomial distribution with 77% of fluorescence probability of mEGFP attached to KChIP4 (Fig. 8C). By dividing p′ values by the fluorescence probability of the Kv4.2-KChIP4-mEGFP tandem construct (77%), association probabilities of KChIP4 to Kv4.2 with different cRNA ratios could be estimated. With 100:1, 10:1, and 1:1 ratios, calculated association probabilities from the first data set were 32, 83, and 99%, and the probabilities from the second data set were 35, 83, and 99%, respectively (Fig. 8D). We concluded that the stoichiometry of the Kv4.2·KChIP4 complex is variable, with no specific preference and changes depending on the expression level of KChIP4.
FIGURE 7.
The stoichiometry of the Kv4.2·KChIP4 complex changes, depending on the co-expression level of KChIP4. A, a schematic illustration of Kv4.2 α-subunit and KChIP4-mEGFP. B, currents from WT Kv4.2 with KChIP4-mEGFP (left) and WT Kv4.2 with WT KChIP4 (right). The currents were elicited by step pulses from −100 to +50 mV in 10-mV increments. C, a single image from a TIRF movie of a Xenopus oocyte expressing Kv4.2 and KChIP4-mEGFP with a cRNA ratio of 10:1. Green circles, countable spots for the bleaching step analysis. Scale bar, 2 μm. D, representative traces of KChIP4-mEGFP with four, three, two, and one bleaching step(s). The green bar indicates illumination with a 488-nm laser. Green arrows, bleaching steps. E, distributions of the number of bleaching steps from oocytes expressing Kv4.2 and KChIP4-mEGFP in the ratios indicated (red bars). Two independent sets of the experiments were performed from two different batches (top row, first set; bottom row, second set). The observed distributions were fitted to a binomial distribution with p′ of apparent fluorescence probability given by (fluorescence probability of mEGFP) × (association probability of KChIP4-mEGFP to Kv4.2) (blue bars). The values of p′ for each cRNA ratio are as follows: first set, 100:1 (24%), 10:1 (63%), 1:1 (76%); second set, 100:1 (26%), 10:1 (63%), 1:1 (76%).
FIGURE 8.
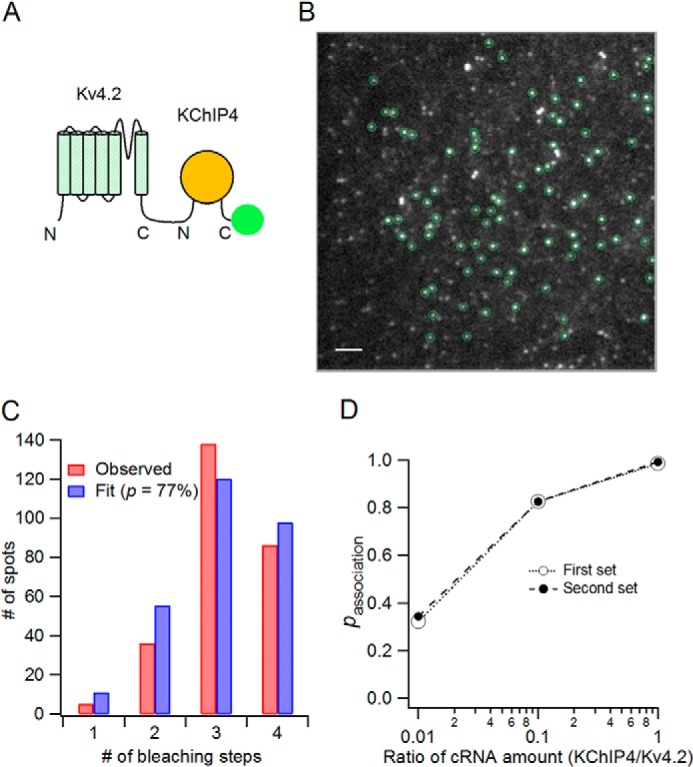
Association probability of KChIP4 with Kv4.2 shows gradual augmentation with increasing KChIP4 expression. A, schematic illustration of the Kv4.2-KChIP4-mEGFP construct. KChIP4 (yellow circle) is bound to the C terminus of Kv4.2, and mEGFP (green circle) is tagged to the C terminus of KChIP4. B, TIRF image of a Xenopus oocyte expressing Kv4.2-KChIP4-mEGFP. Green circles, spots suitable for counting bleaching steps. Scale bar, 2 μm. C, distribution of the number of bleaching steps (red bars) and fit to a binomial distribution (blue bars). Fluorescence probability of mEGFP here (p = 77%) was also used for the calculation of association probability in Fig. 9D. D, association probability of KChIP4-mEGFP with Kv4.2 for each cRNA ratio. Association probability was deduced by dividing p′ (apparent fluorescence probability in Fig. 7E) by the fluorescence probability of mEGFP attached to KChIP4 (77%; C).
Co-expression of DPP10 Does Not Markedly Affect the Stoichiometry of the Kv4.2·KChIP4 Complex
DPP is another auxiliary subunit for Kv4 channels and forms ternary complexes with Kv4 and KChIP. It is known that DPP10 is a membrane protein and increases Kv4.2 current amplitude and accelerates the inactivation and the recovery from inactivation of Kv4.2 (10, 13, 37). We first confirmed that co-expression of DPP10 accelerates the inactivation of Kv4.2·KChIP4 (Fig. 9, A and B). To examine whether DPP10 affects the stoichiometry of the Kv4.2·KChIP4 complex, we expressed DPP10 with Kv4.2 and KChIP4-mEGFP and counted the number of KChIP4 subunits in the complex by single-molecule imaging. The expression level of each subunit was controlled by the amount of injected cRNA (Kv4.2/KChIP4/DPP10 = 100:1:100 (0.5 ng:0.005 ng:0.5 ng) and 10:1:10 (0.1 ng:0.01 ng:0.1 ng)). The observed distributions of bleaching steps for each cRNA ratio (Fig. 9C, red bars) were fitted by a binomial distribution to obtain p′ as an apparent fluorescence probability (Fig. 9C, blue bars). The values of p′ were 27% (100:1:100) and 69% (10:1:10), respectively (Fig. 9C). Calculated association probabilities, based on the fluorescence probability of mEGFP (77%; Fig. 8C) were 36% (100:1:100) and 90% (10:1:10), respectively (Fig. 9D). Association probabilities were comparable with those in the absence of DPP10 (Fig. 8D, Kv4.2/KChIP4 = 100:1 (32–35%) and 10:1 (83%)), showing that DPP10 does not markedly influence the stoichiometry of the Kv4.2·KChIP4 complex.
FIGURE 9.
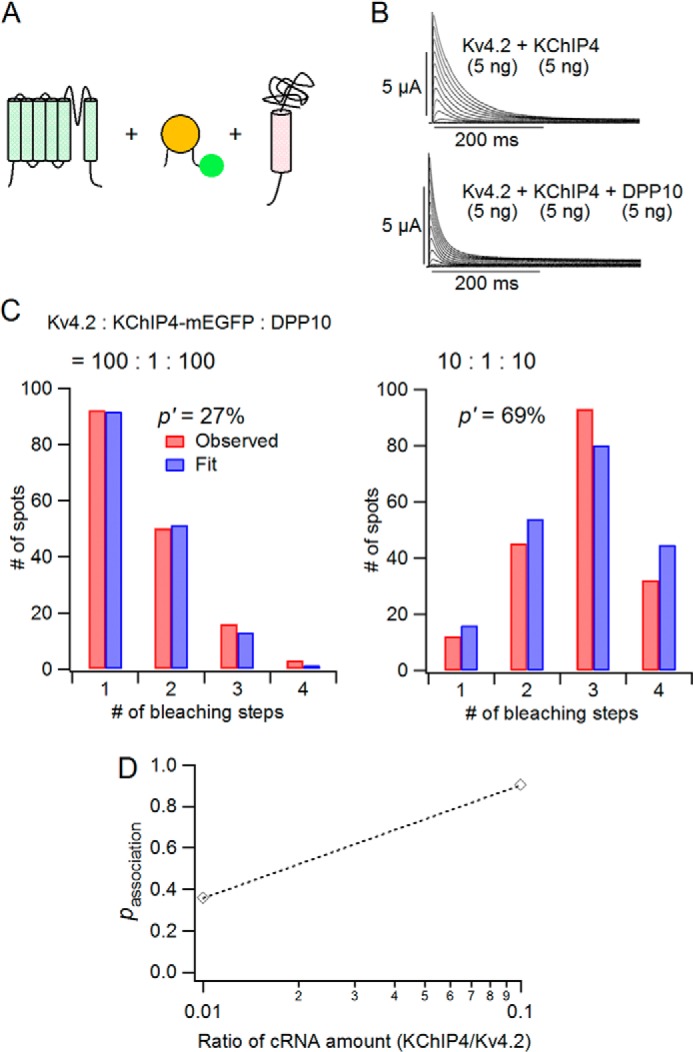
DPP10 does not markedly affect the stoichiometry of the Kv4.2·KChIP4 complex. A, schematic illustrations of Kv4.2 α-subunit, KChIP4-mEGFP, and DPP10. B, currents from Kv4.2 and KChIP4 in the absence (top) and presence of DPP10 (bottom). C, distributions of number of bleaching steps from oocytes expressing Kv4.2, KChIP4-mEGFP, and DPP10 for the cRNA ratios indicated (red bars). The observed distributions were fitted to a binomial distribution with a p′ of apparent fluorescence probability (blue bars). The p′ values were 27% (Kv4.2/KChIP4-mEGFP/DPP10 = 100:1:100) and 69% (10:1:10). D, association probability of KChIP4-mEGFP with Kv4.2 in the presence of DPP10.
DISCUSSION
Stoichiometry-dependent Changes of the Biophysical Properties of Kv4.2·KChIP4
We expressed Kv4.2 with various amounts of KChIP4 and evaluated the electrophysiological properties under two-electrode voltage clamp. As described in previous studies, KChIP4 (not KChIP4a containing a K channel inactivation suppressor domain (38–41)) increased the current amplitude of Kv4.2, decelerated the inactivation, and accelerated the recovery from inactivation of Kv4.2, as did KChIP1, KChIP2, and KChIP3 (2, 13, 25). Our main focus was to examine whether or not the stoichiometry of the Kv4.2·KChIP4 complex affects the biophysical properties of Kv4.2. We observed that these biophysical properties of Kv4.2 gradually changed with an increase in the expression level of KChIP4, as shown in Figs. 1–3. By applying the subunit counting experiment, we revealed that the stoichiometry of the Kv4.2·KChIP4 complex actually changed, depending on the amount of co-expressed KChIP4 (Fig. 7). These findings suggest that the biophysical properties of Kv4.2 changes, depending on the number of bound KChIP4 subunits. Interestingly, the recovery from inactivation of Kv4.2 (5 ng) was highly sensitive to the expression of KChIP4 because it was accelerated even with as little as 0.05 ng of KChIP4 (Fig. 3C). On the other hand, the current amplitude and the inactivation kinetics of Kv4.2 (5 ng) required 0.25–0.5 ng of KChIP4 to be affected (Fig. 1, A and B). These observations suggest that the number of KChIP4 subunits required for speeding up the recovery from inactivation of Kv4.2 may be smaller than the number of KChIP4 subunits required for the increase in current amplitude and for the deceleration of inactivation.
Electrophysiological experiments using KChIP4-Kv4.2 tandem constructs also support the idea of variable stoichiometry and function (Fig. 5). The 4:2 (Kv4.2/KChIP4) channel showed slower recovery from inactivation than the 4:4 channel did, as we expected. The time constant of the recovery from inactivation of the 4:2 channel was similar to that of Kv4.2 (5 ng) with KChIP4 (0.5 ng). This means that approximately two KChIP4 subunits bind to each Kv4.2 channel on average with Kv4.2 (5 ng) and KChIP4 (0.5 ng).
Unexpectedly, however, the 4:2 channel showed slower inactivation than the 4:4 channel. These results do not go well with the inactivation change observed in the WT Kv4.2 with different co-expression levels of KChIP4 (Figs. 2C and 4E). According to a previous study, an N terminus truncation mutant of Kv4.2 (Kv4.2Δ40) shows slower inactivation than WT Kv4.2, and the inactivation of Kv4.2Δ40 is accelerated by an additional Kv4.2 N terminus peptide; therefore, it was concluded that the inactivation of Kv4.2 is produced by occluding the channel pore with the N terminus (24). Because binding of KChIP to the N terminus slows down the inactivation kinetics of Kv4.2, free N terminus without KChIP binding may be required for the fast inactivation. In the 4:2 channel-forming tandem repeat construct we used, the N terminus of the second Kv4.2 was bound to the C terminus of the first Kv4.2; therefore, movement of the N terminus region of the second Kv4.2 is restricted even more severely than the ligation with KChIP4 and not available for occluding the pore. We speculate that this is why the 4:2 channel shows slower inactivation than expected.
We also observed that the current amplitude of Kv4.2 was gradually augmented when the co-expression of KChIP4 increased. There are some possible mechanisms for the increase in the Kv4.2 current amplitude by KChIP4. According to previous studies, the single channel conductance of Kv4 (∼5 picosiemens) was not affected by the co-expression of KChIP proteins (38, 42). It was reported that KChIP binding to the N terminus of Kv4.2 promotes the trafficking of Kv4.2 to the plasma membrane from the endoplasmic reticulum or Golgi (22, 23, 39, 43). Consistent with these findings, we also observed that the surface expression of Kv4.2 increased in accordance with the increase in co-expressed KChIP4 (Fig. 1, D and E). Therefore, the increase of Kv4.2 current amplitude induced by KChIP is mostly due to the increase in the number of Kv4.2 on the plasma membrane rather than an increase in single channel conductance.
Does the Stoichiometry of the Kv4.2·KChIP4 Complex Change on the Cell Membrane?
As mentioned above, KChIP is known to promote the translocation of Kv4.2 from the endoplasmic reticulum or Golgi to the cell surface (22, 23, 39, 43). Thus, KChIP binds to Kv4.2 in the endoplasmic reticulum and is required for the transport of the complex to the cell surface. In addition to this function, crystal structure analysis has revealed that Kv4 and KChIP form a 4:4 channel complex (28–30, 44). Taking these results together, Kv4 and KChIP have been thought to form a 4:4 channel in the endoplasmic reticulum or Golgi and then be expressed on the membrane without changing the stoichiometry. On the other hand, here we have found that the stoichiometry of the Kv4.2·KChIP4 complex is variable: Kv4.2 with fewer than four KChIP4 is functionally expressed if the expression level of KChIP4 is low (Figs. 7 and 10). We also observed freely moving KChIP4 in the cytosol (supplemental Movie 2), consistent with the diffuse distribution throughout the cell of KChIP1, -2, and -3 alone (22). These observations raise the possibility that free KChIPs in the cytosol bind to the open slots in the Kv4.2 N termini of the 4:0, 4:1, 4:2, and 4:3 channel complexes and that bound KChIPs are released from the N terminus of Kv4.2. In other words, the stoichiometry of the Kv4.2·KChIP4 complex might dynamically change on the cell membrane, depending on the expression level of free KChIP4 in the cytosol.
FIGURE 10.
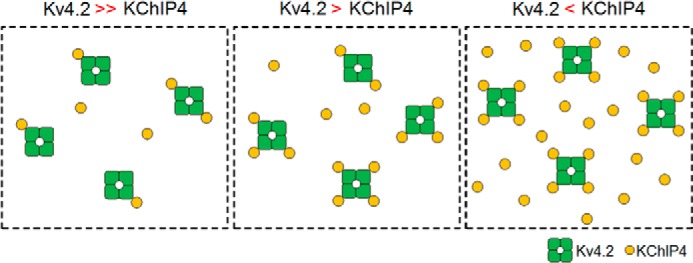
A schematic drawing of the stoichiometry change of the Kv4.2·KChIP4 complex depending on the expression level of KChIP4. The number of KChIP4 subunits bound to Kv4.2 gradually increases with increasing KChIP4 expression. Green square with a hole in the center, Kv4.2 tetramer. Yellow circles, KChIP4 subunits.
Interestingly, in cardiac myocytes, the expression levels of Kv4.2 and KChIP2 oscillate in accordance with the endogenous circadian rhythm, and the ratio of the expression levels of Kv4.2 and KChIP2 changes during a single day (45, 46). The repolarizing duration of the ventricular action potential is affected by the expression level of KChIP2; therefore, a circadian rhythmic change of KChIP2 expression could change cardiac excitability during the day and increase the risk of ventricular arrhythmia and sudden cardiac death at certain times of day (46). By changing the expression level of KChIP in the cytoplasm, the stoichiometry and function of the Kv4.2·KChIP2 complex could be controlled in a circadian rhythm-dependent manner in cardiac myocytes. The variability of stoichiometry with expression level and the hypothetical dynamic nature of stoichiometry on the cell surface may occur under physiological conditions.
Supplementary Material
Acknowledgments
We thank M. Ulbrich (University of Frieburg) for providing the analysis software for the subunit counting and the vectors for constructing the mEGFP fusion proteins. We also thank B. Keceli, I. Yamamoto, and T. Yamamoto (National Institute for Physiological Sciences) for technical guidance on Western blotting experiments. We thank A. Collins (Queen's University, Belfast) for comments and editing of the English.
This study was supported by Japan Society for the Promotion of Science Grant KAKENHI (to K. N. and Y. K.).

This article contains supplemental Movies 1 and 2.
- KChIP
- K+ channel-interacting protein
- DPP
- dipeptidyl peptidase-like protein
- cRNA
- complementary RNA
- mEGFP
- monomeric enhanced green fluorescent protein
- TIRF
- total internal reflection fluorescence.
REFERENCES
- 1. Serôdio P., Kentros C., Rudy B. (1994) Identification of molecular components of A-type channels activating at subthreshold potentials. J. Neurophysiol. 72, 1516–1529 [DOI] [PubMed] [Google Scholar]
- 2. An W. F., Bowlby M. R., Betty M., Cao J., Ling H. P., Mendoza G., Hinson J. W., Mattsson K. I., Strassle B. W., Trimmer J. S., Rhodes K. J. (2000) Modulation of A-type potassium channels by a family of calcium sensors. Nature 403, 553–556 [DOI] [PubMed] [Google Scholar]
- 3. Rhodes K. J., Carroll K. I., Sung M. A., Doliveira L. C., Monaghan M. M., Burke S. L., Strassle B. W., Buchwalder L., Menegola M., Cao J., An W. F., Trimmer J. S. (2004) KChIPs and Kv4 α subunits as integral components of A-type potassium channels in mammalian brain. J. Neurosci. 24, 7903–7915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen X., Yuan L. L., Zhao C., Birnbaum S. G., Frick A., Jung W. E., Schwarz T. L., Sweatt J. D., Johnston D. (2006) Deletion of Kv4.2 gene eliminates dendritic A-type K+ current and enhances induction of long-term potentiation in hippocampal CA1 pyramidal neurons. J. Neurosci. 26, 12143–12151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Norris A. J., Foeger N. C., Nerbonne J. M. (2010) Interdependent roles for accessory KChIP2, KChIP3, and KChIP4 subunits in the generation of Kv4-encoded IA channels in cortical pyramidal neurons. J. Neurosci. 30, 13644–13655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dixon J. E., Shi W., Wang H. S., McDonald C., Yu H., Wymore R. S., Cohen I. S., McKinnon D. (1996) Role of the Kv4.3 K+ channel in ventricular muscle. A molecular correlate for the transient outward current. Circ. Res. 79, 659–668 [DOI] [PubMed] [Google Scholar]
- 7. Nerbonne J. M., Kass R. S. (2005) Molecular physiology of cardiac repolarization. Physiol. Rev. 85, 1205–1253 [DOI] [PubMed] [Google Scholar]
- 8. Hatano N., Ohya S., Imaizumi Y. (2002) Functional interaction between KChIP1 and GFP-fused Kv4.3L co-expressed in HEK293 cells. Pflugers Arch. 444, 80–88 [DOI] [PubMed] [Google Scholar]
- 9. Ren X., Hayashi Y., Yoshimura N., Takimoto K. (2005) Transmembrane interaction mediates complex formation between peptidase homologues and Kv4 channels. Mol. Cell. Neurosci. 29, 320–332 [DOI] [PubMed] [Google Scholar]
- 10. Zagha E., Ozaita A., Chang S. Y., Nadal M. S., Lin U., Saganich M. J., McCormack T., Akinsanya K. O., Qi S. Y., Rudy B. (2005) DPP10 modulates Kv4-mediated A-type potassium channels. J. Biol. Chem. 280, 18853–18861 [DOI] [PubMed] [Google Scholar]
- 11. Seikel E., Trimmer J. S. (2009) Convergent modulation of Kv4.2 channel α subunits by structurally distinct DPPX and KChIP auxiliary subunits. Biochemistry 48, 5721–5730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pongs O., Schwarz J. R. (2010) Ancillary subunits associated with voltage-dependent K+ channels. Physiol. Rev. 90, 755–796 [DOI] [PubMed] [Google Scholar]
- 13. Jerng H. H., Kunjilwar K., Pfaffinger P. J. (2005) Multiprotein assembly of Kv4.2, KChIP3 and DPP10 produces ternary channel complexes with ISA-like properties. J. Physiol. 568, 767–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nakamura T. Y., Pountney D. J., Ozaita A., Nandi S., Ueda S., Rudy B., Coetzee W. A. (2001) A role for frequenin, a Ca2+-binding protein, as a regulator of Kv4 K+-currents. Proc. Natl. Acad. Sci. U.S.A. 98, 12808–12813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Morohashi Y., Hatano N., Ohya S., Takikawa R., Watabiki T., Takasugi N., Imaizumi Y., Tomita T., Iwatsubo T. (2002) Molecular cloning and characterization of CALP/KChIP4, a novel EF-hand protein interacting with presenilin 2 and voltage-gated potassium channel subunit Kv4. J. Biol. Chem. 277, 14965–14975 [DOI] [PubMed] [Google Scholar]
- 16. Burgoyne R. D. (2007) Neuronal calcium sensor proteins: generating diversity in neuronal Ca2+ signalling. Nat. Rev. Neurosci. 8, 182–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jerng H. H., Pfaffinger P. J., Covarrubias M. (2004) Molecular physiology and modulation of somatodendritic A-type potassium channels. Mol. Cell. Neurosci. 27, 343–369 [DOI] [PubMed] [Google Scholar]
- 18. Ren X., Shand S. H., Takimoto K. (2003) Effective association of Kv channel-interacting proteins with Kv4 channel is mediated with their unique core peptide. J. Biol. Chem. 278, 43564–43570 [DOI] [PubMed] [Google Scholar]
- 19. Scannevin R. H., Wang K., Jow F., Megules J., Kopsco D. C., Edris W., Carroll K. C., Lü Q., Xu W., Xu Z., Katz A. H., Olland S., Lin L., Taylor M., Stahl M., Malakian K., Somers W., Mosyak L., Bowlby M. R., Chanda P., Rhodes K. J. (2004) Two N-terminal domains of Kv4 K+ channels regulate binding to and modulation by KChIP1. Neuron 41, 587–598 [DOI] [PubMed] [Google Scholar]
- 20. Zhou W., Qian Y., Kunjilwar K., Pfaffinger P. J., Choe S. (2004) Structural insights into the functional interaction of KChIP1 with Shal-type K+ channels. Neuron 41, 573–586 [DOI] [PubMed] [Google Scholar]
- 21. Wang K. (2008) Modulation by clamping: Kv4 and KChIP interactions. Neurochem. Res. 33, 1964–1969 [DOI] [PubMed] [Google Scholar]
- 22. Shibata R., Misonou H., Campomanes C. R., Anderson A. E., Schrader L. A., Doliveira L. C., Carroll K. I., Sweatt J. D., Rhodes K. J., Trimmer J. S. (2003) A fundamental role for KChIPs in determining the molecular properties and trafficking of Kv4.2 potassium channels. J. Biol. Chem. 278, 36445–36454 [DOI] [PubMed] [Google Scholar]
- 23. Hasdemir B., Fitzgerald D. J., Prior I. A., Tepikin A. V., Burgoyne R. D. (2005) Traffic of Kv4 K+ channels mediated by KChIP1 is via a novel post-ER vesicular pathway. J. Cell Biol. 171, 459–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gebauer M., Isbrandt D., Sauter K., Callsen B., Nolting A., Pongs O., Bähring R. (2004) N-type inactivation features of Kv4.2 channel gating. Biophys. J. 86, 210–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Patel S. P., Campbell D. L., Strauss H. C. (2002) Elucidating KChIP effects on Kv4.3 inactivation and recovery kinetics with a minimal KChIP2 isoform. J. Physiol. 545, 5–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Patel S. P., Parai R., Parai R., Campbell D. L. (2004) Regulation of Kv4.3 voltage-dependent gating kinetics by KChIP2 isoforms. J. Physiol. 557, 19–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hovind L. J., Campbell D. L. (2011) The “structurally minimal” isoform KChIP2d modulates recovery of K(v)4.3 N-terminal deletion mutant Δ2–39. Channels 5, 225–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kim L. A., Furst J., Butler M. H., Xu S., Grigorieff N., Goldstein S. A. (2004) Ito channels are octomeric complexes with four subunits of each Kv4.2 and K+ channel-interacting protein 2. J. Biol. Chem. 279, 5549–5554 [DOI] [PubMed] [Google Scholar]
- 29. Pioletti M., Findeisen F., Hura G. L., Minor D. L., Jr. (2006) Three-dimensional structure of the KChIP1-Kv4.3 T1 complex reveals a cross-shaped octamer. Nat. Struct. Mol. Biol. 13, 987–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang H., Yan Y., Liu Q., Huang Y., Shen Y., Chen L., Chen Y., Yang Q., Hao Q., Wang K., Chai J. (2007) Structural basis for modulation of Kv4 K+ channels by auxiliary KChIP subunits. Nat. Neurosci. 10, 32–39 [DOI] [PubMed] [Google Scholar]
- 31. Ulbrich M. H., Isacoff E. Y. (2007) Subunit counting in membrane-bound proteins. Nat. Methods 4, 319–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tombola F., Ulbrich M. H., Isacoff E. Y. (2008) The voltage-gated proton channel Hv1 has two pores, each controlled by one voltage sensor. Neuron 58, 546–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yu Y., Ulbrich M. H., Li M. H., Buraei Z., Chen X. Z., Ong A. C., Tong L., Isacoff E. Y., Yang J. (2009) Structural and molecular basis of the assembly of the TRPP2/PKD1 complex. Proc. Natl. Acad. Sci. U.S.A. 106, 11558–11563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nakajo K., Ulbrich M. H., Kubo Y., Isacoff E. Y. (2010) Stoichiometry of the KCNQ1-KCNE1 ion channel complex. Proc. Natl. Acad. Sci. U.S.A. 107, 18862–18867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hastie P., Ulbrich M. H., Wang H. L., Arant R. J., Lau A. G., Zhang Z., Isacoff E. Y., Chen L. (2013) AMPA receptor/TARP stoichiometry visualized by single-molecule subunit counting. Proc. Natl. Acad. Sci. U.S.A. 110, 5163–5168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bradford M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 37. Nadal M. S., Ozaita A., Amarillo Y., Vega-Saenz de Miera E., Ma Y., Mo W., Goldberg E. M., Misumi Y., Ikehara Y., Neubert T. A., Rudy B. (2003) The CD26-related dipeptidyl aminopeptidase-like protein DPPX is a critical component of neuronal A-type K+ channels. Neuron 37, 449–461 [DOI] [PubMed] [Google Scholar]
- 38. Holmqvist M. H., Cao J., Hernandez-Pineda R., Jacobson M. D., Carroll K. I., Sung M. A., Betty M., Ge P., Gilbride K. J., Brown M. E., Jurman M. E., Lawson D., Silos-Santiago I., Xie Y., Covarrubias M., Rhodes K. J., Distefano P. S., An W. F. (2002) Elimination of fast inactivation in Kv4 A-type potassium channels by an auxiliary subunit domain. Proc. Natl. Acad. Sci. U.S.A. 99, 1035–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jerng H. H., Pfaffinger P. J. (2008) Multiple Kv channel-interacting proteins contain an N-terminal transmembrane domain that regulates Kv4 channel trafficking and gating. J. Biol. Chem. 283, 36046–36059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liang P., Wang H., Chen H., Cui Y., Gu L., Chai J., Wang K. (2009) Structural insights into KChIP4a modulation of Kv4.3 inactivation. J. Biol. Chem. 284, 4960–4967 [DOI] [PubMed] [Google Scholar]
- 41. Tang Y. Q., Liang P., Zhou J., Lu Y., Lei L., Bian X., Wang K. (2013) Auxiliary KChIP4a suppresses A-type K+ current through endoplasmic reticulum (ER) retention and promoting closed-state inactivation of Kv4 channels. J. Biol. Chem. 288, 14727–14741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Beck E. J., Bowlby M., An W. F., Rhodes K. J., Covarrubias M. (2002) Remodelling inactivation gating of Kv4 channels by KChIP1, a small-molecular-weight calcium-binding protein. J. Physiol. 538, 691–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Foeger N. C., Marionneau C., Nerbonne J. M. (2010) Co-assembly of Kv4 α subunits with K+ channel-interacting protein 2 stabilizes protein expression and promotes surface retention of channel complexes. J. Biol. Chem. 285, 33413–33422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kim L. A., Furst J., Gutierrez D., Butler M. H., Xu S., Goldstein S. A., Grigorieff N. (2004) Three-dimensional structure of Ito: Kv4.2-KChIP2 ion channels by electron microscopy at 21 Å resolution. Neuron 41, 513–519 [DOI] [PubMed] [Google Scholar]
- 45. Yamashita T., Sekiguchi A., Iwasaki Y. K., Sagara K., Iinuma H., Hatano S., Fu L. T., Watanabe H. (2003) Circadian variation of cardiac K+ channel gene expression. Circulation 107, 1917–1922 [DOI] [PubMed] [Google Scholar]
- 46. Jeyaraj D., Haldar S. M., Wan X., McCauley M. D., Ripperger J. A., Hu K., Lu Y., Eapen B. L., Sharma N., Ficker E., Cutler M. J., Gulick J., Sanbe A., Robbins J., Demolombe S., Kondratov R. V., Shea S. A., Albrecht U., Wehrens X. H., Rosenbaum D. S., Jain M. K. (2012) Circadian rhythms govern cardiac repolarization and arrhythmogenesis. Nature 483, 96–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.