Background: T-cell immunoglobulin and ITIM domain (TIGIT) was recently defined as an inhibitory receptor that is expressed on NK cells and T cells.
Results: TIGIT/poliovirus receptor (PVR) ligation signaling mediates suppression of IFN-γ production through NF-κB pathway via β-arrestin 2-mediated negative signaling.
Conclusion: TIGIT/PVR signaling suppresses IFN-γ production of NK cells.
Significance: TIGIT/PVR signaling acts as a potent negative mediator to down-regulate NK cell response for immune homeostasis.
Keywords: Immunology, Innate Immunity, Interferon, Natural Killer Cells (NK Cells), Signaling
Abstract
Natural killer (NK) cell activation is well orchestrated by a wide array of NK cell receptor repertoire. T-cell immunoglobulin and ITIM domain (TIGIT) receptor was recently defined as an inhibitory receptor that is expressed on NK cells and T cells. TIGIT receptor/poliovirus receptor (PVR) ligand engagement signaling inhibits cytotoxicity mediated by NK and CD8+ T cells. However, it is unclear how TIGIT/PVR signaling regulates cytokine secretion in NK cells. Here we show that TIGIT/PVR engagement suppresses interferon-γ (IFN-γ) production of NK cells. TIGIT transgenic NK cells generate less IFN-γ undergoing TIGIT/PVR ligation. Moreover, TIGIT knock-out NK cells produce much more IFN-γ. TIGIT/PVR ligation signaling mediates suppression of IFN-γ production via the NF-κB pathway. We identified a novel adaptor β-arrestin 2 that associates with phosphorylated TIGIT for further recruitment of SHIP1 (SH2-containing inositol phosphatase 1) through the ITT-like motif. Importantly, SHIP1, but not other phosphatases, impairs the TNF receptor-associated factor 6 (TRAF6) autoubiquitination to abolish NF-κB activation, leading to suppression of IFN-γ production in NK cells.
Introduction
Natural killer (NK)3 cells are originally defined as an effector cell of innate immunity, acting as the first defense line against transformed and virus-infected cells (1). NK cells also play an important role in orchestrating adaptive immunity (2, 3). Cytokine secretion and cytotoxicity are the two major effector functions of NK cells whose activation is finely regulated by the activating and inhibitory receptors on NK cell surface (4). Unlike T or B cells, NK cells do not utilize a single receptor to dictate NK cell activation. Ligation of activating NK cell receptors, such as NKG2D, 2B4, NKp30, NKp44, and NKp46, acts in synergy to initiate NK cell activation (5–7). On the other hand, NK cells also possess a large array of inhibitory receptors to protect host self cells by recognizing their major histocompatibility complex class I molecules or other corresponding ligands (8).
TIGIT was recently characterized as an inhibitory receptor, which is expressed mainly on NK, Treg, CD8+ T, and CD4+ T cells (9–11). TIGIT harbors an immunoglobulin tail tyrosine (ITT)-like phosphorylation motif and an ITIM (immunoreceptor tyrosine-based inhibition motif) in its cytoplasmic tail (12). The poliovirus receptor (PVR or CD155) was identified as the physical ligand of TIGIT with high affinity, and PVRL2 (Nectin2 or CD112) also binds to TIGIT with a weaker binding capacity (9, 11). TIGIT/PVR engagement suppresses T cell activation through IL-10 secretion mediated by dendritic cells (9). Furthermore, TIGIT also exerts an intrinsic inhibitory function to T cell activation (13). TIGIT/PVR signaling can also down-regulate both human and mouse NK cell cytotoxicity (10, 14). We have defined that the ITT-like motif of TIGIT recruits Grb2 and SHIP1 to initiate negative signaling, leading to suppression of granule polarization and cytotoxicity of NK cells (15). However, whether and how TIGIT/PVR signaling regulates cytokine secretion of NK cells remains elusive.
β-Arrestin 2 is ubiquitously expressed as an adaptor protein that is recruited to activated G protein-coupled receptors (16). It has been defined that β-arrestin 2 also modulates other G protein-independent receptor signaling cascades (17). More recently, β-arrestin 2 is involved in modulation of immune responses. It has been shown that β-arrestin 2 interacts with an inhibitory receptor KIR2DL1 to recruit tyrosine phosphatases SHP1 and SHP2, leading to inhibitory signaling of NK cells (18). Here we show that TIGIT/PVR engagement inhibits IFN-γ production of NK cells. TIGIT associates with β-arrestin 2 through the ITT-like motif to mediate SHIP1 recruitment for suppression of IFN-γ production.
EXPERIMENTAL PROCEDURES
Cell Lines and Reagents
YTS, Yac1, and 721.221 cells were cultured in RPMI 1640 supplemented with 10% (v/v) heat-inactivated FBS (Invitrogen), 50 μm β-mercaptoethanol, 100 units/ml of penicillin, and 100 μg/ml of streptomycin. YTS-TIGIT cells, its mutants, and 721.221-PVR cells were maintained with additional 0.5 μg/ml of puromycin (Sigma) as described (15). HEK293T cells were grown in DMEM supplemented with 10% (v/v) heat-inactivated FBS (Invitrogen), 50 μm β-mercaptoethanol, 100 units/ml of penicillin, and 100 μg/ml of streptomycin. NF-κB inhibitor Bay11-7085 was purchased from Sigma. Protein A/G-agarose was purchased from Santa Cruz Biotechnology. GST-Sepharose was from GE Healthcare. The SuperReal premix plus quantitative PCR (qPCR) buffer was from TIANGEN Biotech. Mouse anti-c-Myc, mouse anti-GST(B14)(sc-138), mouse anti-Ub, and mouse anti-TRAF6 antibodies were from Santa Cruz Biotechnology. Monoclonal mouse anti-FLAG M2 antibody, monoclonal mouse anti-FLAG M2 Affinity Gel (A2220), and mouse anti-β-actin were from Sigma. Rabbit polyclonal antibodies against phosphorylated IκBα, phosphorylated p38, p38, p65, β-arrestin 2, phosphorylated p65, and SHIP1 (C4099) were all from Cell Signaling Technology; mouse anti-GFP antibody was from Sanjian (Tianjing, China).
Generation of FLAG-TIGIT Transgenic Mice
Eggs from hormonally superovulated female C57BL/6 mice were microinjected with purified linear CD2-FLAG-TIGIT expression vector. Injected fertilized eggs were transplanted into the oviduct of pseudo-pregnant F1 hybrids of C57BL/6 mice. Then, founder mice were identified by PCR analysis of genomic DNA from the tails of transgenic mouse offspring. Founder mice were crossed with WT C57BL/6 mice to generate transgenic and littermate Ctrl mice used for all experiments. Mouse experiments were approved by the Institutional Animal Care and Use Committee at the Institute of Biophysics, Chinese Academy of Sciences.
Generation of TIGIT−/− Mice
The target vector of TIGIT conditional knock-out mice (TIGITflox/flox) was constructed to flank exon 2 of the mouse TIGIT gene with two loxP sites. The vector was transfected into embryonic stem (ES) cells of 129 mice. After neomycin selection, the ES clones flanking loxP sites were microinjected into blastocysts of C57BL/6 mice. TIGIT+/flox mice were obtained after several rounds of selection. TIGITflox/flox mice were generated via intercrossing of TIGIT+/flox mice. TIGIT−/− mice were produced by crossing TIGITflox/flox mice with NKP46-Cre transgenic mice.
Isolation of Mouse Primary NK Cells
NK cells were purified from spleens of TIGIT transgenic mice, TIGIT-deficient mice, and WT mice by APC-NK1.1 staining followed by anti-APC beads (MACS). Purified NK cells were cultured in media containing 200 units/ml of IL-2.
Yeast Two-hybrid Screening
Yeast two-hybrid screening was performed using MatchmakerTM Gold Yeast Two-Hybrid system (Clontech/Takara) as described (19). Briefly, TIGIT was subcloned into pGBKT7 vector (BD-TIGIT). Yeast AH109 cells were transfected with BD-TIGIT and plasmids containing a human spleen cDNA library (Clontech/Takara) and then plated on SD medium lacking adenine, histidine, tryptophan, and leucine. Selected clones were isolated and identified by DNA sequencing. AD-p53 and BD-Large T antigen were co-transfected as a positive control.
Plasmid Construction
TIGIT plasmids were generated as described previously (15). Human full-length β-arrestin 2 was a gift from Dr. Gang Pei (Institute of Biochemistry and Cell Biology, Shanghai) and was subcloned into pcDNA4.0/myc-HisB vector and pEGFP-C1 vector (Invitrogen). Human full-length TRAF6 was cloned from cDNA of YTS cells and subcloned into pRK vector. GFP-SHIP1 was a gift from Dr. Gerald Krystal (BC Cancer Agency).
RT-PCR and ELISA
For comparing IFN-γ mRNA expression, YTS cells were cultured with 721.221 cells at an effector/target ratio of 1:1 for 6 h, total RNAs were extracted with an RNA Kit (LC Biosystems) according to the manufacturer's instructions. For detecting TIGIT mRNA expression, mouse primary NK cells are isolated from Tg mice, TIGIT conditional knock-out mice, or WT mice, total RNAs were extracted with an RNA Kit (LC Biosystems) according to the manufacturer's instructions. Then 2 μg of total RNA per aliquot was used for synthesizing cDNA using the Moloney MLV reverse transcriptase (Promega). qPCR analysis of IFN-γ mRNA was performed with Corbett 6200 qPCR System using human IFN-γ primer: forward, 5′-CAGGTCATTCAGATGTAGCG-3′; reverse, 5′-TTCATGTATTGCTTTGCGTT-3′ and mouse TIGIT primer: forward, 5′-CCACAGCAGGCACGATAGATA-3′; reverse, 5′-CATGCCACCCCAGGTCAAC-3′. Quantitation was normalized to an endogenous GAPDH control. For an ELISA, YTS cells were cultured with 721.221 cells at an effector/target ratio of 1:1 for 24 h, culture supernatants were analyzed by a human-specific ELISA kit (Ebioscience). Mouse primary NK cells and Yac1 cells were co-cultured at an effector/target ratio of 10:1 for 24 h or mouse primary NK cells were stimulated with IL-2 (200 units/ml) plus IL-12 (10 ng/ml) for 24 h. Concentrations of IFN-γ or TNF-α in culture supernatants were detected by a mouse-specific ELISA kit (Ebioscience).
Flow Cytometry
Effector cells and target cells were cultured as the indicated E/T ratios for 6 h. Then cells were fixed for 10 min and permeabilized for 10 min, then stained with APC-CD56 antibody, APC-NK1.1 antibody, human phycoerythrin-IFN-γ antibody, or mouse phycoerythrin-IFN-γ antibody. For flow cytometry analysis of TIGIT expression, mouse primary NK cells from Tg mice, or TIGIT conditional KO (cKO) mice were stained with phycoerythrin-mouse TIGIT antibody.
Laser Scanning Confocal Microscopy
YTS-TIGIT cells were incubated with 721.221 or 721.221-PVR cells for 30 min. Treated cells were loaded on polylysine-pretreated cover glass. Cells were permeabilized with 1% Triton X-100 for p65 nuclear translocation after 30 min fixation in 4% paraformaldehyde. Cells were stained with anti-p65 or anti-FLAG antibody, then stained with fluorescence-labeled second antibody for microscopy as described (20).
RNA Interference
siRNA duplexes against human β-arrestin 2 or SHIP1 were synthesized by GenePharma (Shanghai, China). siRNAs for β-arrestin 2 and SHIP are as follows: β-arrestin 2, 5′-GGACCGCAAAGUGUUUGUG-3′; 5′-CCAACCUCAUUGAAUUUGA-3′ SHIP, 5′-GCCCAUAUCACCCAAGAAGUU-3′; 5′-GCCACAUCUGUACUGACAACG-3′. Scrambled siRNAs for each specific siRNA were as a negative control. siRNAs were transfected into YTS cells with Program O-017 from the manufacturer's introductions of the NucleofectorTM Kit R (Amaxa Biosystems).
Immunoprecipitation Assay
HEK293T cells were transfected with the indicated vectors for 24 h. Cells were harvested and lysed in a lysis buffer (20 mm Tris-HCl (pH 7.4), 150 mm NaCl, 1 mm Na2EDTA (pH 8.0), 0.5% (v/v) Nonidet P-40, 1 mm Na3VO4, 10% (v/v) glycerol and protease inhibitors) for 30 min on ice. Supernatants were achieved by centrifugation (14,000 × g, 10 min, 4 °C), and incubated with the indicated antibodies for 4 h at 4 °C followed by immunoprecipitation with 30 μl of protein A/G-agarose. The precipitates were washed four times with lysis buffer and tested by immunoblotting. For pervanadate treatment, pervanadate (0.1 mm sodium orthovanadate and 10 mm H2O2) was added into YTS cells and HEK293T cells, then cells were incubated at 37 °C for 10 min. Cells were lysed in lysis buffer for immunoblotting.
GST Pulldown Assay
GST-TIGIT fusion proteins were as described previously (15). YTS cell lysates were incubated with equal amounts of the indicated fusion proteins at 4 °C for 4 h, followed by incubation with GST-Sepharose beads for 4 h. Beads were isolated by centrifugation, boiled in SDS loading buffer, and analyzed by immunoblotting.
NF-κB Activation Reporter Assay
HEK293T cells were transfected with the indicated plasmids combined with pNF-κΒ-TA-Luc for NF-κB reporter plus pRL-TK for 24 h. For β-arrestin 2 knockdown assay, HEK293T cells were transfected with siRNAs for 30 h prior to transfection of the reporter detection system. Cells were lysed and NF-κB reporter activity was assayed with the Dual Luciferase Reporter Assay System (Promega).
Statistical Analysis
Student's t test was used as statistical analysis by using Sigma Plot as described (20).
RESULTS
TIGIT/PVR Engagement Inhibits IFN-γ Production of NK Cells
We previously demonstrated that TIGIT/PVR signaling suppresses granule polarization and NK cell cytotoxicity (15). Cytokine secretion is another role of NK cells in defense against viruses or transformed tumor cells. However, whether TIGIT exerts a regulatory role in IFN-γ secretion of NK cells is unclear. We used NK cell line YTS cells as an effector killer and the 721.221 cells as a target, which executes a restricted killing mainly through the interaction between the 2B4 receptor on YTS cells and its ligand CD48 on the target cells. Notably, CD226 and CD96 are expressed on YTS cells, but they are unable to bind to PVR for an unknown reason (10). Actually, YTS cells did not express TIGIT, and 721.221 cells did not harbor PVR. YTS cells stably expressing FLAG-tagged TIGIT (named YTS-TIGIT) and 721.221 cells with PVR expression (named 721.221-PVR) were established (15). The cytoplasmic segment of TIGIT has two tyrosines, Tyr-225 in the ITT-like motif and Tyr-231 in the ITIM motif. These two motifs have distinct binding features to mediate restricted signaling. Y225A mutation impairs the ITT-like motif, whereas the Y231A mutation disrupts the ITIM motif.
We established four stable cell lines, YTS-TIGIT, Y225A, Y231A, and Y225A,Y231A cells with overexpression of TIGIT as effector cells as previously described (15). The surface expression levels on their transfectant cell lines for TIGIT and mutant proteins were analyzed by flow cytometry. They showed similar expression levels as our previous description (15). 721.221-PVR cells expressing PVR were used as target cells. We incubated YTS or YTS-TIGIT cells with 721.221 or 721.221-PVR cells followed by analysis of IFN-γ expression. We found that TIGIT/PVR engagement dramatically reduced IFN-γ expression in its mRNA and protein levels (Fig. 1, A and B). YTS and YTS-TIGIT cells were incubated with 721.221 or 721.221-PVR cells followed by flow cytometry. We gated CD56+ cells and further analyzed IFN-γ+CD56+ cells to detect rates of IFN-γ secreted cells. IFN-γ expression levels appeared to be similar in YTS cells treated with 721.221 or 721.221-PVR cells (Fig. 1C). However, IFN-γ expression was dramatically declined in YTS-TIGIT cells incubated with 721.221-PVR cells compared with those of 721.221 cells. These results indicate that TIGIT/PVR signaling also inhibits IFN-γ secretion of NK cells.
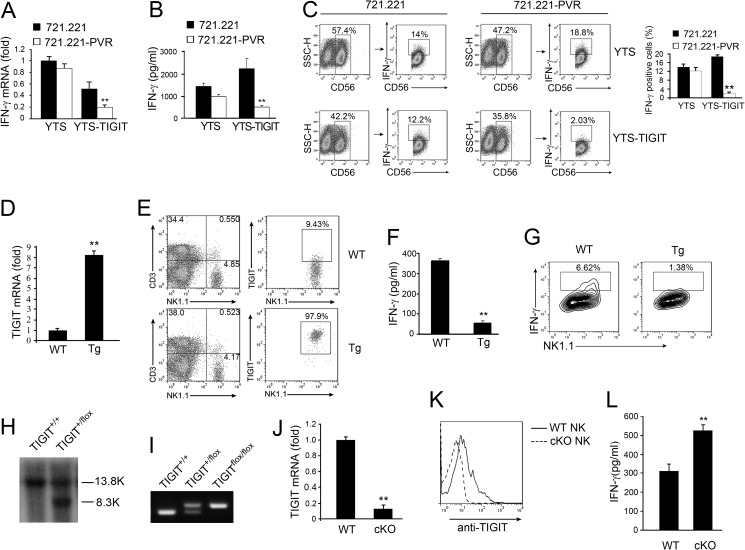
FIGURE 1.
TIGIT/PVR signaling inhibits IFN-γ production. A and B, YTS or YTS-TIGIT cells were incubated with 721.221 or 721.221-PVR cells. A, for 6 h, followed by quantitative RT-PCR analysis of IFN-γ mRNA. B, after 24 h incubation, IFN-γ in supernatants of YTS or YTS-TIGIT cells was analyzed by ELISA. **, p < 0.01. C, YTS or YTS-TIGIT cells were incubated with 721.221 or 721.221-PVR cells for 6 h in the presence of brefeldin A. Brefeldin A was used to block IFN-γ secretion. YTS or YTS-TIGIT cells were stained with anti-CD56 and anti-IFN-γ antibodies. CD56+ cells were first gated and then further gated with IFN-γ+CD56+ cells (left panel). IFN-γ+ cells were calculated as shown as mean ± S.D. (right panel). **, p < 0.01. D, quantitative RT-PCR analysis of TIGIT mRNA expression in NK cells isolated from WT mice and TIGIT transgenic mice (Tg). **, p < 0.01. E, flow cytometry analysis of TIGIT expression in NK cells from WT mice and TIGIT Tg mice. Primary NK cells were stained with anti-NK1.1 and anti-TIGIT antibodies. F, IFN-γ in supernatants of NK cells from WT mice and TIGIT Tg mice cultured with Yac1 cells for 24 h (n = 6 mice per group). **, p < 0.01. G, IFN-γ intracellular staining of NK cells from WT mice and TIGIT Tg mice cultured with Yac1 cells in the presence of brefeldin A for 6 h (n = 6 mice per group). H and I, the floxed TIGIT gene by gene targeting was analyzed by southern blotting and PCR. J, quantitative RT-PCR analysis of TIGIT mRNA expression in NK cells isolated from WT mice and TIGIT cKO mice. TIGIT cKO mice were generated as described under “Experimental Procedures.” **, p < 0.01. K, flow cytometry analysis of TIGIT expression in NK cells isolated from WT mice and TIGIT cKO mice. L, IFN-γ in supernatants of NK cells from WT mice or TIGIT cKO cultured with Yac1 cells for 24 h (n = 6 mice per group). **, p < 0.01. Data are representative of at least three independent experiments.
To verify the inhibitory function of TIGIT in vivo, we generated TIGIT transgenic (Tg) mice with a CD2 promoter-driven construct, by which TIGIT was restrictively expressed in NK and T cells. TIGIT mRNA was highly overexpressed in primary NK cells of TIGIT Tg mice compared with wide type (WT) mice through qPCR analysis (Fig. 1D). Higher expression of TIGIT in Tg NK cells was confirmed by flow cytometry (Fig. 1E). Yac1 cells, a murine cell line, express PVR on their cell surface. We isolated NK cells from TIGIT Tg or WT mice and then incubation with Yac1 cells for 24 h and detection by IFN-γ expression. Consistently, NK cells from TIGIT Tg mice produced much less IFN-γ than those of WT mice (Fig. 1F). Similar results were obtained by IFN-γ intracellular staining (Fig. 1G).
To further confirm the physiological role of TIGIT signaling, we also generated TIGIT conditional knock-out mice (TIGITflox/flox). Exon 2 of the mTIGIT gene was deleted by flanking with loxP sites (data not shown). TIGITflox/flox mice were screened via genotyping (Fig. 1, H and I). We crossed TIGITflox/flox mice with NKp46-Cre transgenic mice to obtain TIGIT cKO mice in NK cells. TIGIT mRNA was not expressed in TIGIT−/− NK cells through qPCR analysis (Fig. 1J). Of note, TIGIT was not expressed in TIGIT−/− NK cells via flow cytometry (Fig. 1K). Then we incubated TIGIT−/− NK cells with Yac1 cells for 24 h and detected IFN-γ expression. Consequently, TIGIT−/− NK cells produced much more IFN-γ than those of WT mice (Fig. 1L). Additionally, TIGIT−/− NK cells produced much more TNF-α compared with those of WT mice (data not shown). However, TIGIT−/− NK cells did not affect IL-12/IL-2-induced IFN-γ production (data not shown). Collectively, TIGIT/PVR signaling significantly suppresses IFN-γ secretion in NK cells.
TIGIT/PVR Signaling Suppresses NF-κB Activation
NF-κB is an important transcription factor that controls the production of many inflammatory cytokines in various immune cells (21). The NF-κB pathway is critical for IFN-γ production through engagement of multiple immunoreceptor tyrosine-based activation motif-coupled cell receptors in NK cells (22). To examine whether TIGIT/PVR signaling impairs IFN-γ production is dependent on the NF-κB pathway, we incubated YTS-TIGIT cells with 721.221 or 721.221-PVR cells treated with the NF-κB inhibitor Bay11-7085. We observed that YTS-TIGIT cells treated with the inhibitor produced much less IFN-γ (Fig. 2A). Additionally, incubation of YTS-TIGIT cells with 721.221-PVR cells significantly suppressed phosphorylation of IκBα (Fig. 2B). Because YTS cell activating receptors could initiate NF-κB activation signaling with incubation of 721.221 cells, IκB phosphorylation was dynamically activated without TIGIT/PVR ligation. Consequently, TIGIT/PVR engagement impaired p65 nucleus translocation (Fig. 2C). However, incubation of YTS-TIGIT cells with 721.221-PVR cells failed to change the level of phosphorylated p38 kinase (Fig. 2D), suggesting TIGIT/PVR signaling is independent of MAPK activation. Thus, TIGIT/PVR engagement impairs NF-κB activation in NK cells.
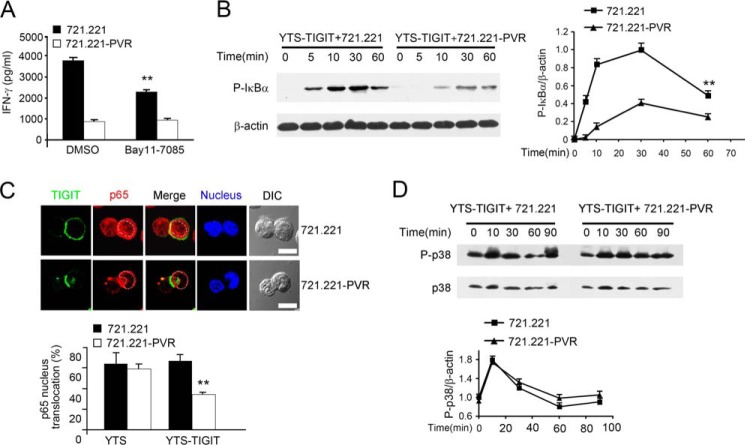
FIGURE 2.
TIGIT/PVR engagement suppresses NF-κB signaling. A, inhibition of IFN-γ production by the NF-κB inhibitor in YTS cells. IFN-γ in supernatants of YTS-TIGIT cells, pretreated with DMSO or 10 μm Bay11-7085 (NF-κB inhibitor) for 2 h followed by incubation with 721.221 or 721.221-PVR cells for 24 h. **, p < 0.01. B, immunoblotting analysis of phosphorylated IκBα (left panel). β-Actin served as a loading control. Graphical representation of densitometric analysis (right panel). **, p < 0.01. C, TIGIT/PVR engagement decreases p65 nucleus translocation in YTS-TIGIT cells. YTS-TIGIT cells were incubated with 721.221 or 721.221-PVR cells at 37 °C for 30 min. Treated cells were stained with anti-FLAG (green) and anti-p65 (red) antibody, and nuclei were stained with Hochest dye (blue) (upper panel). Scale bar, 10 μm. p65 nucleus translocation was counted at 50 conjugates (lower panel). **, p < 0.01. D, immunoblotting analysis of phosphorylated p38 (upper panel). β-Actin served as a loading control. Graphical representation of densitometric analysis (lower panel). Data are representative of at least three separate experiments.
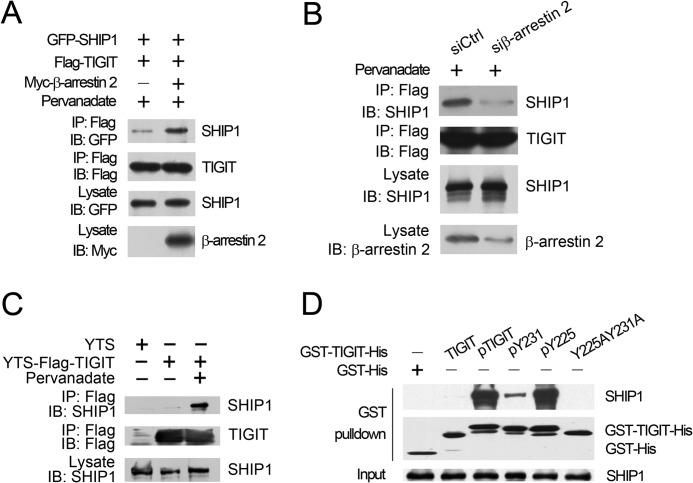
TIGIT Associates with β-Arrestin 2
To explore the underlying mechanism by which TIGIT inhibits NF-κB pathway, we used the cytoplasmic segment of TIGIT as a bait and screened a human spleen cDNA library to identify TIGIT interactors using a yeast two-hybrid system. By screening 2 × 107 transformants, we isolated five clones encoding β-arrestin 2. Moreover, β-arrestin 2 was identified to interact with TIGIT (Fig. 3A). Their association was also verified in HEK293T cells transfected with FLAG-tagged TIGIT and Myc-tagged β-arrestin 2 by a co-immunoprecipitation assay (Fig. 3B). Importantly, with treatment of pervanadate, a pan inhibitor of phosphatases, TIGIT was able to precipitate much more β-arrestin 2 (Fig. 3B). Similar results were obtained in TIGIT-overexpressed YTS cells (Fig. 3C). Moreover, the interaction of TIGIT with β-arrestin 2 was detectable in IL-2-activated primary NK cells (data not shown). These data suggest that phosphorylation of the TIGIT cytoplasmic domain may mediate its binding capacity with β-arrestin 2.
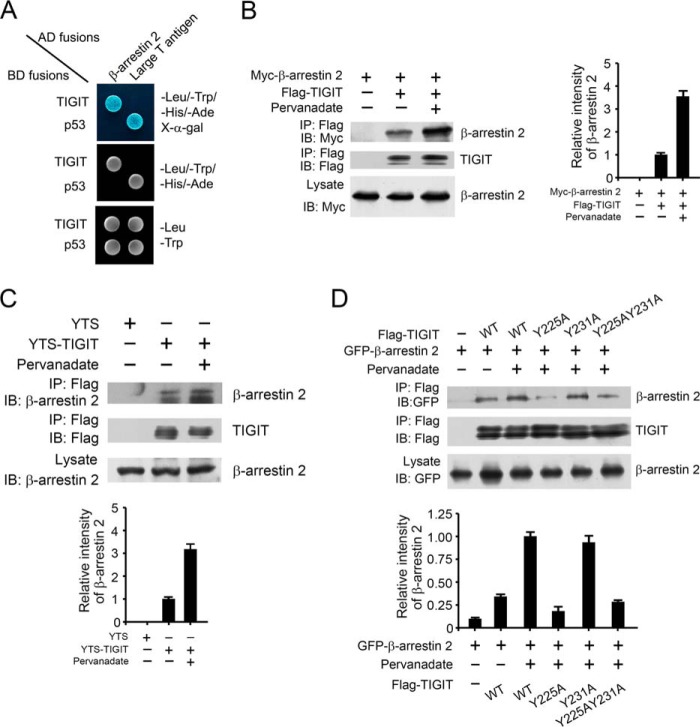
FIGURE 3.
TIGIT associates with β-arrestin 2. A, TIGIT associates with β-arrestin 2 by a yeast two-hybrid assay. The yeast strain AH109 was co-transfected with the Gal4 DNA-binding domain (BD) fused TIGIT and Gal4 activating domain (AD)-fused β-arrestin 2. The interaction of BD-p53 and AD-large T antigen was used as a positive control. B, HEK293T cells were transfected with Myc-tagged β-arrestin 2 and FLAG-tagged TIGIT for 24 h. With or without pervanadate treatment, cell lysates were immunoprecipitated (IP) with anti-FLAG antibody, followed by immunoblotting (IB) with antibody against Myc or FLAG (left panel). Graphical representation of densitometric analysis (right panel). C, phosphorylated TIGIT associates with more β-arrestin 2 in YTS cells. YTS-TIGIT or YTS cell lysates with or without pervanadate treatment were immunoprecipitated by anti-FLAG antibody and immunoblotted with anti-β-arrestin 2 antibody (upper panel). Graphical representation of densitometric analysis is shown in the lower panel. D, HEK293T cells were transfected with GFP-β-arrestin 2 and FLAG-TIGIT or FLAG-TIGIT mutants (Y225A, Y231A, or Y225A,Y231A) for 24 h (upper panel). Graphical representation of densitometric analysis is shown in the lower panel. Data represent at least three independent experiments.
TIGIT has an ITT-like motif nearby the ITIM motif in the cytoplasmic segment (12, 15). In our previous study, we have shown that in HEK293T cells with pervanadate treatment, Y225A mutation almost abolishes phosphorylation, however, the Y231A mutation can be phosphorylated as wild type does. Notably, we found that the Y225A mutation dramatically impaired the association of TIGIT with β-arrestin 2 (Fig. 3D), whereas the Y231A mutation had no such activity. Consequently, the two tyrosine mutations (Y225A,Y231A) indeed disrupted the interaction of TIGIT with β-arrestin 2, which was reminiscent of the Y225A mutation (Fig. 3D). Taken together, the ITT-like motif of TIGIT is also required for the interaction of TIGIT with β-arrestin 2.
β-Arrestin 2 Is Involved in TIGIT/PVR-mediated Inhibition of the NF-κB Pathway
To determine whether β-arrestin 2 participates in TIGIT-mediated inhibitory signaling, we silenced β-arrestin 2 in YTS-TIGIT cells by RNA interference (RNAi) (Fig. 4A). Two siRNA duplexes against β-arrestin 2 were transfected into YTS-TIGIT cells and obtained similar knockdown efficiency (data not shown). Interestingly, β-arrestin 2 knockdown enhanced phosphorylation of IκBα when TIGIT/PVR ligated (Fig. 4B). Consequently, β-arrestin 2 knockdown significantly augmented p65 phosphorylation with TIGIT/PVR engagement (Fig. 4C). The phosphorylated forms of P-p65 and P-IκBα indeed declined with prolongation, suggesting phosphorylation is a dynamic regulation in the process of NK cell activation. To further validate these observations, we detected NF-κB activation by using a luciferase reporter assay system. TRAF6 was used to activate the NF-κB pathway in HEK293T cells as reported (23). Expectedly, β-arrestin 2 silencing dramatically increased NF-κB activation (Fig. 4D). By contrast, β-arrestin 2 overexpression apparently declined NF-κB activation (Fig. 4E). In summary, β-arrestin 2 is an important adaptor molecule to mediate the TIGIT/PVR-mediated suppression of NF-κB pathway.
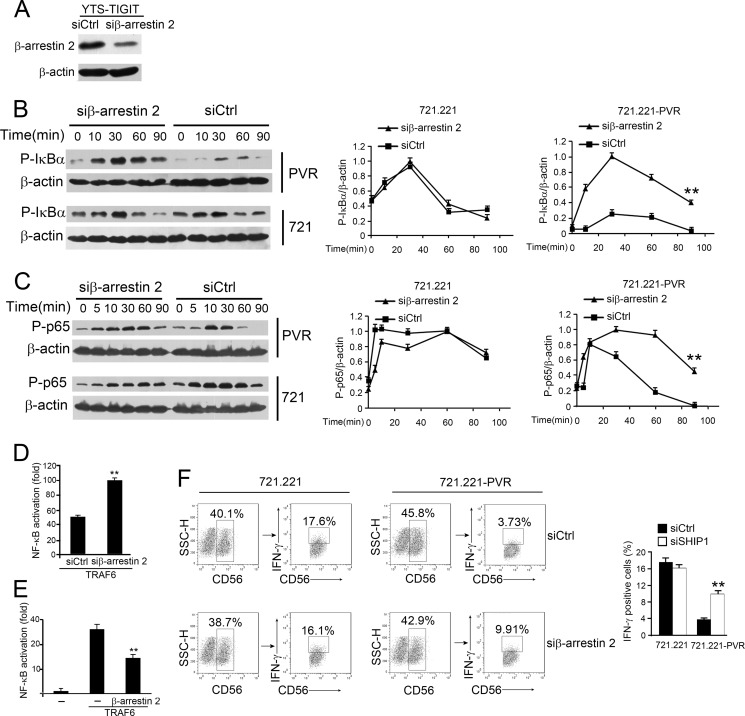
FIGURE 4.
β-Arrestin 2 is involved in TIGIT-mediated inhibition of NF-κB pathway. A, β-arrestin 2 was silenced in YTS-TIGIT cells. YTS-TIGIT cells were transfected with control siRNA (siCtrl) or β-arrestin 2-specific siRNA (siβ-arrestin 2). β-Actin was used as a loading control. Two siRNA duplexes against β-arrestin 2 were transfected into YTS-TIGIT cells and obtained similar knockdown efficiency (data not shown). B and C, immunoblotting analysis of phosphorylated IκBα and p65 in lysates of YTS-TIGIT cells transfected with control or β-arrestin 2-specific siRNA and cultured with 721.221 or 721.221-PVR cells (left panel). Treatment with 1% paraformaldehyde in 721.221 or 721.221-PVR cells blocks IκBα and p65 phosphorylation in target cells. β-Actin served as a loading control. Graphical representation of densitometric analysis (right panel). **, p < 0.01. D, β-arrestin 2 knockdown enhances NF-κB activation. β-Arrestin 2-silenced HEK293T cells were transfected with FLAG-TRAF6 plasmid plus pNF-κB-TA-Luc with pRL-TK for 24 h, followed by luciferase assays. **, p < 0.01. E, HEK293T cells were transfected with FLAG-TRAF6 and β-arrestin 2 plasmids plus pNF-κB-TA-Luc with pRL-TK for 24 h. **, p < 0.01. F, IFN-γ intracellular staining in YTS-TIGIT cells transfected with control (siCtrl) or siβ-arrestin 2 and cultured with 721.221 or 721.221-PVR cells for 6 h in the presence of brefeldin A. YTS or YTS-TIGIT cells were stained with anti-CD56 and anti-IFN-γ antibodies. CD56+ cells were first gated and further gated with IFN-γ+CD56+ cells (left panel). IFN-γ+ cells were calculated as shown as mean ± S.D. (right panel). **, p < 0.01. Data are representative of at least three independent experiments.
β-Arrestin 2-silenced or siCtrl-treated YTS-TIGIT cells were incubated with 721.221 or 721.221-PVR cells followed by flow cytometry. We gated CD56+ cells and further analyzed IFN-γ+CD56+ cells to assay rates of IFN-γ-secreted cells. IFN-γ expression levels in β-arrestin 2-silenced cells were restored with TIGIT/PVR engagement (Fig. 4F, right panel). By contrast, IFN-γ expression levels in β-arrestin 2-silenced cells were unchangeable without TIGIT/PVR engagement (Fig. 4F, left panel). These data suggest that β-arrestin 2 is involved in suppression of IFN-γ production of NK cells.
β-Arrestin 2 Facilitates the Recruitment of SHIP1 to TIGIT
NK cell inhibitory receptors usually initiate inhibitory signaling through recruiting either the inositol phosphatases (SHIP1 and SHIP2) or Src homology (SH) 2 domain-containing protein-tyrosine phosphatases (SHP1 and SHP2) by the ITIM domains in their cytoplasmic tails (18, 24, 25). We recently demonstrated that TIGIT associates with Grb2 to recruit SHIP1, leading to inhibition of NK cell cytotoxicity (15). Intriguingly, we found that β-arrestin 2 overexpression dramatically enhanced the interaction of TIGIT with SHIP1 (Fig. 5A), but was not detectable for other phosphatases such as SHIP2, SHP1, and SHP2 (data not shown). In contrast, β-arrestin 2 knockdown significantly impaired the association of TIGIT with SHIP1 (Fig. 5B).
FIGURE 5.
β-Arrestin 2 facilitates the recruitment of SHIP1 to TIGIT. A, β-arrestin 2 augments the association of TIGIT with SHIP1. HEK293T cells were transfected with the indicated vectors for 24 h. With pervanadate treatment, cell lysates were immunoprecipitated (IP) with anti-FLAG antibody followed by immunoblotting (IB). B, β-arrestin 2 silencing reduces SHIP1 recruitment. YTS-TIGIT cells were transfected with siRNA duplexes either for β-arrestin 2 (siβ-arrestin 2) or scrambled siRNA control (siCtrl) for 48 h. C, phosphorylated TIGIT interacts with SHIP1. YTS-TIGIT or YTS cell lysates with or without pervanadate treatment were immunoprecipitated by anti-FLAG antibody and immunoblotted with anti-SHIP1 antibody. D, Tyr-225 mutation fails to recruit SHIP1. Precipitation of endogenous SHIP1 in YTS cells with the indicated TIGIT fusion proteins is shown. SHIP1 in YTS lysates was probed as a loading control. Data were repeated for at least four independent experiments and obtained similar results.
Actually, β-arrestin 2-mediated recruitment of SHIP1 required phosphorylated TIGIT (Fig. 5C), whereas TIGIT failed to precipitate SHIP1 in YTS cells without pervanadate treatment (Fig. 5C). We previously defined that Fyn and Lck can phosphorylate TIGIT (15). We generated phosphorylated TIGIT (pTIGIT) and its tyrosine-phosphorylated variants (pY231 and pY225) by coexpression of GST-TIGIT-His and its variants with tyrosine kinase Fyn as described (15). Importantly, through an in vitro pulldown assay, pTIGIT indeed pulled down SHIP1 (Fig. 5D), whereas non-phosphorylated TIGIT or the Y225A,Y231A mutation abrogated the association of TIGIT with SHIP1. Additionally, phosphorylated Tyr-225 (pY225) was still associated with SHIP1 (Fig. 5D), whereas phosphorylated Tyr-231 (pY231) abolished this interaction. Together, the recruitment of SHIP1 to TIGIT mediated by β-arrestin 2 depends on Tyr-225 phosphorylation of TIGIT.
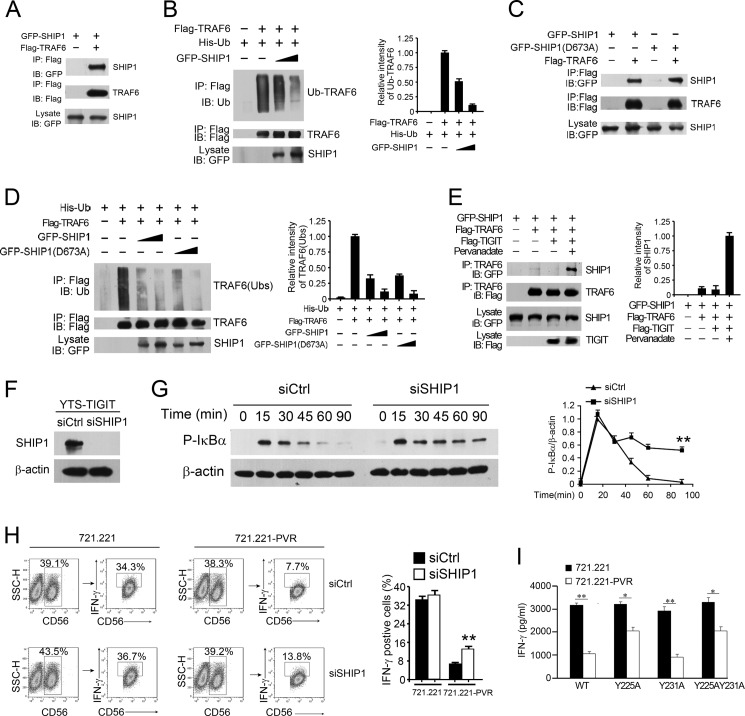
SHIP1 Impedes TRAF6 Ubiquitination to Impair NF-κB Activation Leading to Suppression of IFN-γ Production
TRAF6 has been defined to be a key mediator in NF-κB signaling (26). TRAF6 autoubiquitination mediates IκB kinase activation that is necessary for the NF-κB activation (27, 28). To determine whether SHIP1 impairs the autoubiquitination of TRAF6 in TIGIT signaling, we first performed a co-immunoprecipitation assay. Of note, SHIP1 was associated with TRAF6 in co-transfected HEK293T cells (Fig. 6A). Moreover, SHIP1 overexpression suppressed TRAF6 autoubiquitination in a dose-dependent manner (Fig. 6B). Given that SHIP1 catalyzes the product of PI3K, phosphatidylinositol 3,4,5-triphosphate (PI(3,4,5)P3) to PI(3,4)P2 as an inositol phosphatase (25, 29), we next wanted to explore whether SHIP1-mediated inhibition of TRAF6 autoubiquitination is dependent on its lipid phosphatase activity. We constructed a phosphatase-inactive SHIP1 mutant (SHIP1(D673A)) as reported (30). We found that the SHIP1(D673A) mutant did not impair its interaction with TRAF6 as WT-SHIP1 did (Fig. 6C). Furthermore, the SHIP1(D673A) mutant still inhibited TRAF6 autoubiquitination compared with WT-SHIP1 (Fig. 6D). These observations indicate that suppression of TRAF6 autoubiquitination is independent of SHIP1 lipid phosphatase activity.
FIGURE 6.
TIGIT augments the binding of TRAF6 to SHIP1 to impair TRAF6 ubiquitination. A, SHIP1 associates with TRAF6. HEK293T cells were transfected with GFP-SHIP1 and FLAG-TRAF6. Cell lysates were immunoprecipitated with anti-FLAG antibody, followed by immunoblotting. B, SHIP1 inhibits autoubiquitination of TRAF6. HEK293T cells were transfected with His-Ub and FLAG-TRAF6 vectors plus increasing doses of GFP-SHIP1 vectors for 24 h (left panel). Graphical representation of densitometric analysis is shown in the right panel. C, HEK293T cells were transfected with GFP-SHIP1 or GFP-SHIP1(D673A) and FLAG-TRAF6 vectors for 24 h. D, HEK293T cells were transfected with His-Ub and FLAG-TRAF6 plus increasing doses of GFP-SHIP1 or GFP-SHIP1(D673A) vectors for 24 h (left panel). Graphical representation of densitometric analysis is shown in the right panel. E, TIGIT enhances the binding of TRAF6 to SHIP1 (left panel). HEK293T cells were transfected with the indicated vectors with or without pervanadate treatment. Graphical representation of densitometric analysis is shown in the right panel. F, YTS-TIGIT cells were transfected with control siRNA (siCtrl) or SHIP1-specific siRNA (siSHIP1) for 48 h. β-Actin served as a loading control. Two siRNA duplexes against SHIP1 were transfected into YTS-TIGIT cells and obtained similar knockdown efficiency (data not shown). G, immunoblotting analysis of phosphorylated IκBα in lysates of YTS-TIGIT cells transfected with control (siCtrl) or SHIP1-specific siRNA and cultured with 721.221-PVR cells (left panel). Graphical representation of densitometric analysis is shown in the right panel. **, p < 0.01. H, IFN-γ intracellular staining in YTS-TIGIT cells transfected with control (siCtrl) or SHIP1-specific siRNA and cultured with 721.221 or 721.221-PVR cells for 6 h in the presence of brefeldin A (left panel). YTS or YTS-TIGIT cells were stained with anti-CD56 and anti-IFN-γ antibodies. CD56+ cells were first gated and the further gated with IFN-γ+CD56+ cells (left panel). IFN-γ+ cells were calculated as shown, mean ± S.D. (right panel). **, p < 0.01. I, IFN-γ was detected in supernatants of YTS-TIGIT or YTS-TIGIT mutant (Y225A, Y231A, or Y225A,Y231A) cells cultured with 721.221 or 721.221-PVR cells for 24 h. *, p < 0.05; **, p < 0.01. Data are representative of at least three separate experiments.
Importantly, overexpression of TIGIT in HEK293T cells augmented the association of SHIP1 with TRAF6 with pervanadate treatment (Fig. 6E), but this association was almost abrogated without pervanadate treatment. Similar results were observed in TIGIT overexpressed YTS cells (data not shown). These data indicate that phosphorylated TIGIT recruits SHIP1 to suppress the autoubiquitination of TRAF6.
To further confirm whether SHIP1 participates in TIGIT-mediated inhibition of IFN-γ production in NK cells, we silenced SHIP1 in YTS-TIGIT cells by RNA interference (Fig. 6F). Two siRNA duplexes against SHIP1 were used and obtained similar depletion efficiency (data not shown). Notably, SHIP1 knockdown remarkably increased phosphorylation of IκBα undergoing TIGIT/PVR engagement (Fig. 6G). Consequently, SHIP1 knockdown restored IFN-γ production with TIGIT/PVR engagement (Fig. 6H). Without TIGIT/PVR engagement, SHIP1 knockdown had no such activity. More importantly, the Y225A mutation displayed less inhibitory activity to IFN-γ production of YTS cells once TIGIT/PVR engaged (Fig. 6I), whereas the Y231A mutation had inhibitory activity as WT-TIGIT did. Furthermore, the Y225A,Y231A mutation had less inhibitory activity as the Y225A mutation. Together, these results confirmed that SHIP1 is required for TIGIT/PVR-mediated suppression of IFN-γ production.
DISCUSSION
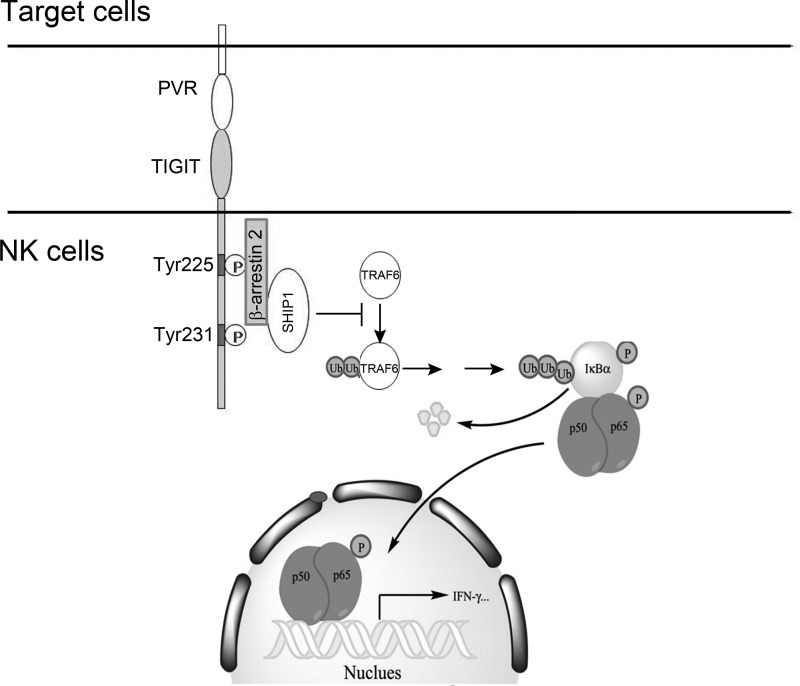
NK cell activation is well orchestrated by a wide array of NK cell receptor repertoire, including activating and inhibitory receptors. TIGIT was recently defined as an inhibitory receptor that is expressed on NK cells and T cells (9–11). TIGIT directly down-regulates cytotoxicity mediated by NK and CD8+ T cells (10, 13, 14, 31). Our recent study showed that TIGIT/PVR signaling inhibits NK cell cytotoxicity through recruitment of Grb2 and SHIP1 by the ITT-like motif of TIGIT. However, it is unclear how TIGIT/PVR signaling modulates cytokine secretion of NK cells. Herein we show that TIGIT/PVR engagement inhibits IFN-γ production of YTS cells. Moreover, TIGIT Tg NK cells generate less IFN-γ undergoing TIGIT/PVR ligation. Although TIGIT cKO in NK cells significantly promotes IFN-γ production. TIGIT/PVR signaling mediates suppression of IFN-γ production via the NF-κB pathway. We identified a new adaptor β-arrestin 2 that associates with phosphorylated TIGIT and mediates recruitment of inositol phosphatase SHIP1 through the ITT-like motif (Fig. 7). Finally, SHIP1 impairs TRAF6 autoubiquitination to abolish NF-κB activation, leading to inhibition of IFN-γ production in NK cells.
FIGURE 7.
Work model for TIGIT/PVR-mediated suppression of IFN-γ production in NK cells. With TIGIT/PVR engagement, cytoplasmic TIGIT was phosphorylated at Tyr-225 and Tyr-231 residues. Phosphorylated Tyr-225 recruits adaptor protein β-arrestin 2, which further mediates recruitment of SHIP1. Finally, SHIP1 impairs TRAF6 autoubiquitination to inhibit NF-κB activation, leading to suppression of IFN-γ production in NK cells.
TIGIT contains an ITT-like phosphorylation motif and a classical ITIM motif in the cytoplasmic tail (12). Inhibitory ITIM-containing receptors usually undergo tyrosine phosphorylation of cytoplasmic tails in the first step for negative signaling transduction (32). Then, they recruit either SH2 domain-containing tyrosine phosphatases (such as SHP1 and SHP2) or inositol phosphatases (such as SHIP1 and SHIP2) to induce inhibitory signaling (25). We recently showed that the ITT-like phosphorylation motif of TIGIT plays a predominant role in TIGIT/PVR-mediated negative signaling (15). Phosphorylated TIGIT binds to adaptor molecule Grb2 and mediates recruitment of SHIP1, which initiates negative signaling to suppress cytotoxicity of NK cells. Here we show that phosphorylated TIGIT binds to another adaptor molecule, β-arrestin 2, and recruits SHIP1 to inhibit NF-κB activation, leading to down-regulation of cytokine production. Our observations suggest that TIGIT may utilize different adaptors to mediate separate cytotoxicity and cytokine secretion of NK cells, respectively.
The two β-arrestin isoforms, β-arrestin 1 and β-arrestin 2, are ubiquitously expressed as adaptor molecules to mediate multifunctional signaling (17, 33). β-Arrestins have been characterized to interact with many signaling molecules, including Src kinases, Akt, and MAPK kinases (17, 33). Importantly, β-arrestins are implicated in the modulation of immune responses. β-Arrestin 2 directly interacts with IκBα to prevent its phosphorylation and degradation (34). More recently, it has also been shown that inhibitory receptor KIR2DL1 directly associates with β-arrestin 2 and mediates recruitment of SHP1 and SHP2, leading to an inhibitory signaling of NK cell cytotoxicity (18). However, we demonstrate that phosphorylated TIGIT interacts with β-arrestin 2 to recruit SHIP1, but not other phosphatases such as SHIP2, SHP1, or SHP2, resulting in suppression of cytokine secretion of NK cells. Thus our findings reveal a novel role for β-arrestin 2 in ITIM receptor signaling.
SHIP1 is an SH2-domain containing inositol phosphatase that degrades PI(3,4,5)P3 to PI(3,4)P2. PI(3,4,5)P3, a product of PI3K, is accumulated to the plasma membrane that provides recruitment regions for signaling mediators (25). SHIP1 is mainly expressed in hematopoietic cells (35). It has been defined as a potent negative regulator of immune cells (25, 36). SHIP1 deficiency impairs development of T and B cells (37, 38). SHIP1-deleted mice have severe immune deficiency due to aberrations in the expression of NK cell receptors or downstream signaling (29). A recent study showed that SHIP1 is required for the transition from immature to mature NK cells (39). Generally, hydrolysis of PI(3,4,5)P3 impairs the recruitment of PH domain-containing kinases to the plasma membrane, resulting in inhibition of PI3K signaling. For NK cell cytotoxicity, TIGIT associates with Grb2 and mediates recruitment of SHIP1, which causes inhibition of PI3K downstream effectors, such as ERK, MEK, and p38 MAPK (15). However, for NK cell production of IFN-γ, TIGIT associates with β-arrestin 2 and mediates recruitment of SHIP1, which impairs NF-κB activation, but not affect p38 MAPK signaling. Additionally, SHIP1-mediated inhibition of TRAF6 autoubiquitination is lipid phosphatase independent. SHIP1 silencing restores IFN-γ production, but it does not completely abolish the inhibition of TIGIT signaling. This may be because there are some tyrosine-independent mechanisms in inhibition of IFN-γ production by TIGIT.
TRAF6 functions as a core mediator in NF-κB signaling activation (26, 40). TRAF6 knock-out in mice blocks IL-1β and lipopolysaccharide-mediated NF-κB activation (41). TRAF6 overexpression in cells activates NF-κB signaling (42). TRAF6 is a RING domain-containing E3 ligase that catalyzes itself on lysine 63-linked ubiquitin chain as an autocatalytic Lys-63 ubiquitin ligase (27, 28, 43). TRAF6 autoubiquitination initiates IκB kinase activation (28, 40). Recently, a large amount of negative regulators were reported to suppress autoubiquitination of TRAF6 that impairs NF-κB signaling (23, 44, 45). Another recent study showed that the bacterial ITIM-containing protein Tir mediates recruitment of SHP1 to the adaptor TRAF6 and represses the autoubiquitination of TRAF6 (46). In this study, we show that TIGIT associates with β-arrestin 2 and only recruits SHIP1 but not other phosphatases, leading to suppression of autoubiquitination of TRAF6 via a lipid phosphatase-independent manner.
NK cell effector functions are finely tuned and timely regulated by many activating receptors and inhibitory receptors (4). Excessive NK cell activation can cause severe immunopathology of the infected tissues or autoimmune diseases (8, 47). At an early stage during viral infection, NK cell-produced cytokines play a pivotal role in antiviral immunity and augment further priming of virus-specific CD8+ T cell responses (8, 48). Once viruses are eradicated, NK cell activation needs to be shut down immediately. Thus, the inhibitory receptors are necessary to down-regulate NK cell function, which protects the host from excessive virus-induced immune injury. Bi et al. (49) just reported that blockade of TIGIT in vivo or Tigit deficiency enhances NK cell activation and aggravates poly(I:C)/D-GalN-induced liver injury. Moreover, TIGIT-deficient mice are more susceptible to autoimmune diseases (13, 50). Here we show that TIGIT Tg mice display declined secretion of IFN-γ production in NK cells. By contrast, TIGIT cKO mice in NK cells promote IFN-γ generation. Altogether, TIGIT/PVR signaling acts as a potent negative mediator to down-regulate NK cell responses for immune homeostasis.
Acknowledgments
We thank Dr. Gang Pei for providing the β-arrestin 2 plasmid. We thank Dr. Vivier for providing NKp46-Cre mice. We thank Dr. Gerald Krystal for providing the GFP-SHIP1 plasmid.
This work was supported by National Natural Science Foundation of China Grants 81330047, 31170837, 30830030, and 30972676, 973 Program of the MOST of China Grant 2010CB911902, and Strategic Priority Research Programs of the Chinese Academy of Sciences Grant XDA01010407.
- NK
- natural killer
- TIGIT
- T-cell immunoglobulin and ITIM domain
- PVR
- poliovirus receptor
- ITT
- immunoglobulin tail tyrosine
- qPCR
- quantitative PCR
- Tg
- transgenic
- SH
- Src homology
- cKO
- conditional KO
- PI(3,4,5)P3
- phosphatidylinositol 3,4,5-triphosphate
- PI(3,4)P2
- phosphatidylinositol 3,4-bisphosphate.
REFERENCES
- 1. Vivier E., Tomasello E., Baratin M., Walzer T., Ugolini S. (2008) Functions of natural killer cells. Nat. Immunol. 9, 503–510 [DOI] [PubMed] [Google Scholar]
- 2. Sun J. C., Beilke J. N., Lanier L. L. (2009) Adaptive immune features of natural killer cells. Nature 457, 557–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fan Z., Yu P., Wang Y., Wang Y., Fu M. L., Liu W., Sun Y., Fu Y. X. (2006) NK-cell activation by LIGHT triggers tumor-specific CD8+ T-cell immunity to reject established tumors. Blood 107, 1342–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bryceson Y. T., March M. E., Ljunggren H. G., Long E. O. (2006) Activation, coactivation, and costimulation of resting human natural killer cells. Immunol. Rev. 214, 73–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Raulet D. H., Gasser S., Gowen B. G., Deng W., Jung H. (2013) Regulation of ligands for the NKG2D activating receptor. Annu. Rev. Immunol. 31, 413–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Narni-Mancinelli E., Jaeger B. N., Bernat C., Fenis A., Kung S., De Gassart A., Mahmood S., Gut M., Heath S. C., Estellé J., Bertosio E., Vely F., Gastinel L. N., Beutler B., Malissen B., Malissen M., Gut I. G., Vivier E., Ugolini S. (2012) Tuning of natural killer cell reactivity by NKp46 and Helios calibrates T cell responses. Science 335, 344–348 [DOI] [PubMed] [Google Scholar]
- 7. Hoffmann S. C., Cohnen A., Ludwig T., Watzl C. (2011) 2B4 engagement mediates rapid LFA-1 and actin-dependent NK cell adhesion to tumor cells as measured by single cell force spectroscopy. J. Immunol. 186, 2757–2764 [DOI] [PubMed] [Google Scholar]
- 8. Long E. O., Kim H. S., Liu D., Peterson M. E., Rajagopalan S. (2013) Controlling natural killer cell responses: integration of signals for activation and inhibition. Annu. Rev. Immunol. 31, 227–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yu X., Harden K., Gonzalez L. C., Francesco M., Chiang E., Irving B., Tom I., Ivelja S., Refino C. J., Clark H., Eaton D., Grogan J. L. (2009) The surface protein TIGIT suppresses T cell activation by promoting the generation of mature immunoregulatory dendritic cells. Nat. Immunol. 10, 48–57 [DOI] [PubMed] [Google Scholar]
- 10. Stanietsky N., Simic H., Arapovic J., Toporik A., Levy O., Novik A., Levine Z., Beiman M., Dassa L., Achdout H., Stern-Ginossar N., Tsukerman P., Jonjic S., Mandelboim O. (2009) The interaction of TIGIT with PVR and PVRL2 inhibits human NK cell cytotoxicity. Proc. Natl. Acad. Sci. U.S.A. 106, 17858–17863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Boles K. S., Vermi W., Facchetti F., Fuchs A., Wilson T. J., Diacovo T. G., Cella M., Colonna M. (2009) A novel molecular interaction for the adhesion of follicular CD4 T cells to follicular DC. Eur. J. Immunol. 39, 695–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Engels N., Wienands J. (2011) The signaling tool box for tyrosine-based costimulation of lymphocytes. Curr. Opin. Immunol. 23, 324–329 [DOI] [PubMed] [Google Scholar]
- 13. Joller N., Hafler J. P., Brynedal B., Kassam N., Spoerl S., Levin S. D., Sharpe A. H., Kuchroo V. K. (2011) Cutting edge: TIGIT has T cell-intrinsic inhibitory functions. J. Immunol. 186, 1338–1342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Stanietsky N., Rovis T. L., Glasner A., Seidel E., Tsukerman P., Yamin R., Enk J., Jonjic S., Mandelboim O. (2013) Mouse TIGIT inhibits NK-cell cytotoxicity upon interaction with PVR. Eur. J. Immunol. 43, 2138–2150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liu S., Zhang H., Li M., Hu D., Li C., Ge B., Jin B., Fan Z. (2013) Recruitment of Grb2 and SHIP1 by the ITT-like motif of TIGIT suppresses granule polarization and cytotoxicity of NK cells. Cell Death Differ. 20, 456–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Benovic J. L., Kühn H., Weyand I., Codina J., Caron M. G., Lefkowitz R. J. (1987) Functional desensitization of the isolated β-adrenergic receptor by the β-adrenergic receptor kinase: potential role of an analog of the retinal protein arrestin (48-kDa protein). Proc. Natl. Acad. Sci. U.S.A. 84, 8879–8882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. DeWire S. M., Ahn S., Lefkowitz R. J., Shenoy S. K. (2007) β-Arrestins and cell signaling. Annu. Rev. Physiol. 69, 483–510 [DOI] [PubMed] [Google Scholar]
- 18. Yu M. C., Su L. L., Zou L., Liu Y., Wu N., Kong L., Zhuang Z. H., Sun L., Liu H. P., Hu J. H., Li D., Strominger J. L., Zang J. W., Pei G., Ge B. X. (2008) An essential function for β-arrestin 2 in the inhibitory signaling of natural killer cells. Nat. Immunol. 9, 898–907 [DOI] [PubMed] [Google Scholar]
- 19. Xia P., Wang S., Du Y., Zhao Z., Shi L., Sun L., Huang G., Ye B., Li C., Dai Z., Hou N., Cheng X., Sun Q., Li L., Yang X., Fan Z. (2013) WASH inhibits autophagy through suppression of Beclin 1 ubiquitination. EMBO J. 32, 2685–2696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang S., Xia P., Ye B., Huang G., Liu J., Fan Z. (2013) Transient activation of autophagy via Sox2-mediated suppression of mTOR is an important early step in reprogramming to pluripotency. Cell Stem Cell 13, 617–625 [DOI] [PubMed] [Google Scholar]
- 21. Hayden M. S., Ghosh S. (2004) Signaling to NF-κB. Genes Dev. 18, 2195–2224 [DOI] [PubMed] [Google Scholar]
- 22. Gross O., Grupp C., Steinberg C., Zimmermann S., Strasser D., Hannesschläger N., Reindl W., Jonsson H., Huo H., Littman D. R., Peschel C., Yokoyama W. M., Krug A., Ruland J. (2008) Multiple ITAM-coupled NK-cell receptors engage the Bcl10/Malt1 complex via Carma1 for NF-κB and MAPK activation to selectively control cytokine production. Blood 112, 2421–2428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang X., Zhang J., Zhang L., van Dam H., ten Dijke P. (2013) UBE2O negatively regulates TRAF6-mediated NF-κB activation by inhibiting TRAF6 polyubiquitination. Cell Res. 23, 366–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Long E. O. (2008) Negative signaling by inhibitory receptors: the NK cell paradigm. Immunol. Rev. 224, 70–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kerr W. G., Colucci F. (2011) Inositol phospholipid signaling and the biology of natural killer cells. J. Innate Immun. 3, 249–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sun L., Deng L., Ea C. K., Xia Z. P., Chen Z. J. (2004) The TRAF6 ubiquitin ligase and TAK1 kinase mediate IKK activation by BCL10 and MALT1 in T lymphocytes. Mol. Cell 14, 289–301 [DOI] [PubMed] [Google Scholar]
- 27. Chen Z. J. (2005) Ubiquitin signalling in the NF-κB pathway. Nat. Cell Biol. 7, 758–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang C., Deng L., Hong M., Akkaraju G. R., Inoue J., Chen Z. J. (2001) TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature 412, 346–351 [DOI] [PubMed] [Google Scholar]
- 29. Wang J. W., Howson J. M., Ghansah T., Desponts C., Ninos J. M., May S. L., Nguyen K. H., Toyama-Sorimachi N., Kerr W. G. (2002) Influence of SHIP on the NK repertoire and allogeneic bone marrow transplantation. Science 295, 2094–2097 [DOI] [PubMed] [Google Scholar]
- 30. Trotta R., Parihar R., Yu J., Becknell B., Allard J., 2nd, Wen J., Ding W., Mao H., Tridandapani S., Carson W. E., Caligiuri M. A. (2005) Differential expression of SHIP1 in CD56bright and CD56dim NK cells provides a molecular basis for distinct functional responses to monokine costimulation. Blood 105, 3011–3018 [DOI] [PubMed] [Google Scholar]
- 31. Lozano E., Dominguez-Villar M., Kuchroo V., Hafler D. A. (2012) The TIGIT/CD226 axis regulates human T cell function. J. Immunol. 188, 3869–3875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lanier L. L. (2008) Up on the tightrope: natural killer cell activation and inhibition. Nat. Immunol. 9, 495–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lefkowitz R. J., Shenoy S. K. (2005) Transduction of receptor signals by β-arrestins. Science 308, 512–517 [DOI] [PubMed] [Google Scholar]
- 34. Gao H., Sun Y., Wu Y., Luan B., Wang Y., Qu B., Pei G. (2004) Identification of β-arrestin2 as a G protein-coupled receptor-stimulated regulator of NF-κB pathways. Mol. Cell 14, 303–317 [DOI] [PubMed] [Google Scholar]
- 35. Hazen A. L., Smith M. J., Desponts C., Winter O., Moser K., Kerr W. G. (2009) SHIP is required for a functional hematopoietic stem cell niche. Blood 113, 2924–2933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Veillette A., Latour S., Davidson D. (2002) Negative regulation of immunoreceptor signaling. Annu. Rev. Immunol. 20, 669–707 [DOI] [PubMed] [Google Scholar]
- 37. Karlsson M. C., Guinamard R., Bolland S., Sankala M., Steinman R. M., Ravetch J. V. (2003) Macrophages control the retention and trafficking of B lymphocytes in the splenic marginal zone. J. Exp. Med. 198, 333–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Collazo M. M., Wood D., Paraiso K. H., Lund E., Engelman R. W., Le C. T., Stauch D., Kotsch K., Kerr W. G. (2009) SHIP limits immunoregulatory capacity in the T-cell compartment. Blood 113, 2934–2944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Banh C., Miah S. M., Kerr W. G., Brossay L. (2012) Mouse natural killer cell development and maturation are differentially regulated by SHIP-1. Blood 120, 4583–4590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chen Z. J. (2012) Ubiquitination in signaling to and activation of IKK. Immunol. Rev. 246, 95–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lomaga M. A., Yeh W. C., Sarosi I., Duncan G. S., Furlonger C., Ho A., Morony S., Capparelli C., Van G., Kaufman S., van der Heiden A., Itie A., Wakeham A., Khoo W., Sasaki T., Cao Z., Penninger J. M., Paige C. J., Lacey D. L., Dunstan C. R., Boyle W. J., Goeddel D. V., Mak T. W. (1999) TRAF6 deficiency results in osteopetrosis and defective interleukin-1, CD40, and LPS signaling. Genes Dev. 13, 1015–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cao Z., Xiong J., Takeuchi M., Kurama T., Goeddel D. V. (1996) TRAF6 is a signal transducer for interleukin-1. Nature 383, 443–446 [DOI] [PubMed] [Google Scholar]
- 43. Deng L., Wang C., Spencer E., Yang L., Braun A., You J., Slaughter C., Pickart C., Chen Z. J. (2000) Activation of the IκB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell 103, 351–361 [DOI] [PubMed] [Google Scholar]
- 44. Sanada T., Kim M., Mimuro H., Suzuki M., Ogawa M., Oyama A., Ashida H., Kobayashi T., Koyama T., Nagai S., Shibata Y., Gohda J., Inoue J., Mizushima T., Sasakawa C. (2012) The Shigella flexneri effector OspI deamidates UBC13 to dampen the inflammatory response. Nature 483, 623–626 [DOI] [PubMed] [Google Scholar]
- 45. Schneider M., Zimmermann A. G., Roberts R. A., Zhang L., Swanson K. V., Wen H., Davis B. K., Allen I. C., Holl E. K., Ye Z., Rahman A. H., Conti B. J., Eitas T. K., Koller B. H., Ting J. P. (2012) The innate immune sensor NLRC3 attenuates Toll-like receptor signaling via modification of the signaling adaptor TRAF6 and transcription factor NF-κB. Nat. Immunol. 13, 823–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yan D., Wang X., Luo L., Cao X., Ge B. (2012) Inhibition of TLR signaling by a bacterial protein containing immunoreceptor tyrosine-based inhibitory motifs. Nat. Immunol. 13, 1063–1071 [DOI] [PubMed] [Google Scholar]
- 47. Zhang Z., Zhang S., Zou Z., Shi J., Zhao J., Fan R., Qin E., Li B., Li Z., Xu X., Fu J., Zhang J., Gao B., Tian Z., Wang F. S. (2011) Hypercytolytic activity of hepatic natural killer cells correlates with liver injury in chronic hepatitis B patients. Hepatology 53, 73–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Orange J. S., Wang B., Terhorst C., Biron C. A. (1995) Requirement for natural killer cell-produced interferon γ in defense against murine cytomegalovirus infection and enhancement of this defense pathway by interleukin 12 administration. J. Exp. Med. 182, 1045–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bi J., Zhang Q., Liang D., Xiong L., Wei H., Sun R., Tian Z. (2014) TIGIT regulates NK cell activation in murine acute viral hepatitis. Hepatology 59, 1715–1725 [DOI] [PubMed] [Google Scholar]
- 50. Levin S. D., Taft D. W., Brandt C. S., Bucher C., Howard E. D., Chadwick E. M., Johnston J., Hammond A., Bontadelli K., Ardourel D., Hebb L., Wolf A., Bukowski T. R., Rixon M. W., Kuijper J. L., Ostrander C. D., West J. W., Bilsborough J., Fox B., Gao Z., Xu W., Ramsdell F., Blazar B. R., Lewis K. E. (2011) Vstm3 is a member of the CD28 family and an important modulator of T-cell function. Eur. J. Immunol. 41, 902–915 [DOI] [PMC free article] [PubMed] [Google Scholar]