Background: To generate active Nodal suited for directed stem cell differentiation, we purified it as a heterodimer with Gdf1.
Results: Heterodimeric Nodal·Gdf1 copurified with their cleaved prodomains, potentiated receptor activation and endoderm differentiation, and enabled paracrine signaling in serum-free conditions.
Conclusion: Cell-non-autonomous high Nodal signaling thresholds depend on low molecular weight heterodimers.
Significance: Nodal·Gdf1 is a new reagent to differentiate endoderm.
Keywords: Embryonic Stem Cell, Protein Purification, Serum, Signaling, Transforming Growth Factor β (TGFβ), Growth and Differentiation Factor 1 (GDF1), Nodal, Prodomains, Proprotein Convertase, Stem Cell Differentiation
Abstract
The TGFβ family member Nodal is central to control pluripotent stem cell fate, but its use as a stem cell differentiation factor is limited by low specific activity. During development, Nodal depends on growth and differentiation factor (Gdf)-1 and on the shared co-receptor Cryptic to specify visceral left-right axis asymmetry. We therefore asked whether the functionality of Nodal can be augmented by Gdf1. Because Nodal and Gdf1 coimmunoprecipitate each other, they were predicted to form heterodimers, possibly to facilitate diffusion or to increase the affinity for signaling receptors. Here, we report that Gdf1 suppresses an unexpected dependence of Nodal on serum proteins and that it is critically required for non-autonomous signaling in cells expressing Cryptic. Nodal, Gdf1, and their cleaved propeptides copurified as a heterodimeric low molecular weight complex that stimulated Activin receptor (Acvr) signaling far more potently than Nodal alone. Although heterodimerization with Gdf1 did not increase binding of Nodal to Fc fusions of co-receptors or Acvr extracellular domains, it was essential for soluble Acvr2 to inhibit Nodal signaling. This implies that Gdf1 potentiates Nodal activity by stabilizing a low molecular weight fraction that is susceptible to neutralization by soluble Acvr2. Finally, in differentiating human ES cells, endodermal markers were more efficiently induced by Nodal·Gdf1 than by Nodal, suggesting that Nodal·Gdf1 is an attractive new reagent to direct stem cell differentiation.
Introduction
Pluripotent stem cells hold considerable promise for regenerative medicine as their differentiation might be directed toward specific cell fates in vitro by signaling pathways that guide normal embryonic development in vivo. Endodermal progenitors that give rise to therapeutically relevant cell types such as hepatocytes or insulin-producing pancreatic β cells can be induced by Activin A. However, during normal embryogenesis, endoderm is specified by Nodal signals. In mammalian embryos, Nodal signaling maintains pluripotent epiblast progenitors from the inner cell mass and governs their allocation to mesoderm and endoderm lineages during gastrulation (for a review, see Ref. 1). Despite the central role of Nodal in specifying endoderm during development, current protocols for directed in vitro differentiation of endoderm derivatives rely on the related ligand Activin A as a surrogate, presumably because Activin is more active than commercially available recombinant Nodal (2–8). However, a recent study showed that Nodal induces endoderm progenitors that are more competent to mature into functional insulin-producing cells if it is added to cultured mouse ES cells at a 20-fold higher concentration than Activin A (9). As this concentration likely exceeds physiological levels, it is important to determine how the specific activity of recombinant Nodal might be improved.
Nodal assembles heteromeric complexes of the type I and II Activin receptors (Acvr1b2 and Acvr2a or Acvr2b) that stimulate the phosphorylation and nuclear translocation of Smad2 and Smad3 transcription factors (10). Nodal is secreted as a precursor that must be cleaved by the proprotein convertases Furin or Pace4 to remove an N-terminal prodomain. Although Nodal processing is essential to potentiate Smad signaling in cultured cells, a cleavage mutant Nodal precursor retains significant activity during gastrulation (11–13). To activate Smad2/3, Nodal must also bind one of the co-receptors, Cripto or Cryptic from the EGF-CFC family of glycosylphosphatidylinositol-anchored proteoglycans (14). Analysis in cultured cells suggested a dual role for Cripto in concentrating the Nodal precursor in lipid rafts during processing and in stabilizing a complex of mature ligand with signaling receptors at the limiting membrane of signaling endosomes (13, 15, 16). Compound mutant mouse embryos lacking both Cripto and Cryptic phenocopy Nodal−/− null mutants (17). In addition, analysis of Cripto−/− and Cryptic−/− single mutants revealed unique functions in different tissues: Cripto expression in the epiblast enables Nodal autoinduction and thereby promotes germ layer formation (14, 18). By contrast, Cryptic is primarily required after gastrulation to mediate paracrine Nodal signaling from the ventral node to the left lateral plate mesoderm during left-right axis formation (19, 20). Whether these differences simply reflect distinct expression patterns is unclear.
Growth and differentiation factor (Gdf) 1 and Gdf3 barely stimulate Smad2/3 signaling except if they are coexpressed in cis together with Nodal (21, 22). This lack of intrinsic activity was initially attributed to inefficient precursor processing because substitution of the Gdf1 or Gdf3 prodomains by heterologous BMP prodomains facilitates the accumulation of mature ligands (21, 23, 24). Furthermore, Gdf1 stimulates the Smad3 reporter CAGA-luc independently of Nodal when overexpressed together with the proprotein convertase Furin (21). However, this activity was very limited. Also in conditioned medium of Xenopus oocytes where mature Gdf1 can accumulate at high levels independently of Nodal, coexpression of Nodal was still essential for signaling (22). This implies that Gdf1 and Nodal synergize by somehow promoting the bioavailability of mature ligand even after precursor cleavage. Because Gdf1 and Nodal also coimmunoprecipitated each other, the authors predicted that synergistic signaling may involve heterodimers (22). During development, Gdf1 primarily enables long range Nodal signaling from the midline to left lateral plate mesoderm for left-right patterning. In addition, analysis of Gdf1−/−; Gdf3−/− double mutants revealed that Gdf1 cooperates with Nodal during gastrulation to induce prechordal plate and overlying forebrain structures in a manner that appears to be redundant with the function of Gdf3 (21, 25). However, the actual composition of Nodal·Gdf1 complexes and a mechanism to explain their increased activity are elusive.
To address this, we analyzed the effect of Gdf1 coexpression on Nodal protein level, signal strength, and protein-protein interactions. We found that although the Gdf1 precursor must be cleaved to synergize with Nodal, proteolytic processing of Nodal is not essential. Furthermore, we report that soluble Nodal activity depends on high molecular weight serous proteins and that coexpression of Gdf1 bypasses this requirement and enables Nodal to accumulate in a low molecular weight (LMW) complex with the Nodal and Gdf1 prodomains. This LMW complex of Nodal·Gdf1 can be purified and remains active in serum-free conditions, making it attractive for cell fate-directing experiments as shown here by increased differentiation of human ES cells (hESCs) into endoderm progenitors.
EXPERIMENTAL PROCEDURES
Expression Vectors and Lentivirus Production
Full-length Nodal precursor tagged with a FLAG epitope immediately after the RQRR cleavage motif was generated by overlap extension PCR using primers that replaced the wild-type sequence AGGCAACGCCGACATCAT by the sequence CAACGTCGCGATTACAAAGACGATGACGATAAAGGACCCGGACAGGTGCAGCTGCAGCATCAT. Expression vectors for Nodal prodomain (Npro) or constitutively mature mutant form (Nmat) and cleavage-resistant Nodal precursor (Nr) have been described (26, 27). Although Nmat is FLAG-tagged at the N terminus of the mature domain, Npro and Nr carry a FLAG epitope in the prodomain after Met-45. All are in the pCS2+ backbone except Nmat, which is in pEF IRES-Puro (28).
To generate Gdf1 expression vectors, an EcoRI-AgeI fragment of a mouse expressed sequence tag clone in plasmid pME18S-FL3 (Genome Systems Inc., St. Louis, MO) was cloned into pCS2+. Linkers composed of the oligonucleotides 5′-CTAGACATCATCACCATCACCACC-3′ and 5′-CTAGGGTGGTGATGGTGATGATGT-3′ encoding a His6 tag or of the oligonucleotides 5′-ATATCCATATGATGTACCAGATTACG-3′ and 5′-CCGGCGTAATCTGGTACATCATATGGATAT-3′ encoding an HA epitope were inserted between engineered NruI and AvrII sites at the N terminus of the mature domain. In a second step, inverse PCR using the primers 5′-CAGTATGGCCCCTCACAGCGGTGAA-3′ and 5′-AGCCACGACATCATCACCATCACCA-3′ followed by self-ligation added the first 4 amino acids of mature Gdf1 proximal to the His6 epitope to improve the efficiency of precursor cleavage. The expression vector pEF-Gmat for constitutively mature Gdf1 was derived using overlap extension PCR to fuse HA-tagged mature Gdf1 (primers 5′-CATCATCACCATCACCACCCTAGGGTA-3′ and 5′-TGTCACGAATTCTCAACGGCAGCCACACTC-3′) and the secretory signal sequence of Nmat (primers 5′-ATAGAAACTGGGCTTGTCGAGACAG-3′ and 5′-GTGGTGATGGTGATGATGATCGCCCCGCACAGC-3′). The fused amplicon was digested with NheI and EcoRI and cloned into pEF IRES-Puro. Gdf1 prodomain was amplified from CS-Gdf1 using the primers 5′-CTGCCTGGGGACGTCGGAGC-3′ and 5′-AATCATACTAGTTCAGCGTCGCAATCGCGGCAAG-3′, cut with EcoRI and SpeI, and inserted in pCS-Gdf1 to yield CS-Gpro. To create CS-Gr, the cleavage site of CS-Gdf1 was mutated by inverse PCR using the primers 5′-GGCCAATCCCGGCAAGGGACACAGG-3′ and 5′-GGCCACACGGAGCCCAGGGTAGA-3′ followed by self-ligation. To derive lentiviral vectors pLenti-Gdf1(HA)-puro and pLenti-Gdf1(His6)-puro, the inserts CS-Gdf1(HA) or Gdf1(His6) were subcloned into pLenti hEF1α-MCS//SV40-BsdR (pCF520) using BamHI and SpeI.
To generate the Smad3 reporter pLenti-CAGA-lux-neo, a CAGA-luciferase cassette was isolated from CAGA-lux (a gift from Olov Anderson) using XmaI and XbaI and ligated into AgeI-SpeI sites of pLenti hEF1α-MCS//SV40-NeoR (pCF521). The Cripto coding sequence of pCS-Cripto was cloned into BamHI-HpaI of pCF520, giving rise to pLenti-Cripto-bsd. To derive pLenti-Cryptic-bsd, Cripto was replaced by Cryptic using BamHI and XbaI. Finally, pLenti-Rluc-puro was generated by cloning Renilla luciferase as an NheI-XbaI fragment into the XbaI site pLenti hEF1α-MCS//SV40-PuroR (pCF519). pCF519, pCF520, and pCF521 have been described earlier (29).
To generate receptor-Fc fusion constructs, a human ACVR1B cDNA fragment was amplified using primers 5′-TTTAAGCTTCCACCATGGCGGAGTCGGCCGGA-3′ and 5′-TTTGTCGACTTCCACCGGGCCCCACATGG-3′ and cloned into the pCR3-MCS-Presci-Fc vector (a gift from Pascal Schneider, Lausanne, Switzerland) using HindIII and SalI. The cDNA encoding mouse Acvr2b extracellular domain was amplified using primers 5′-TTTAAGCTTCCACCATGACGGCGCCCTGGGCGG-3′ and 5′-TTTGTCGACGGTGGGGGCTGTCGGGGGT-3′ and cloned similarly. Cripto was amplified from pIg-CriptoFc7 (a gift from Gabriella Minchiotti, Naples, Italy) using primers 5′-CACTATAGGGAGACCCAAGCTTCGAAGATGG-3′ and 5′-AATTAAGTCGACAGTGGTCGTCACAGACGG-3′ and cloned similarly. Cryptic cDNA (a gift from Gabriella Minchiotti) was amplified with 5′-CATTACACCGGTACCATGAGAGCGAACTCACCA-3′ and 5′-GAGTCAGTCGACGCTGCATTCTCTTGATCC-3′, cut with Age I (T4-blunted) and SalI, and inserted into pCR3-MCS-Presci-Fc cut with HindIII (T4-blunted) and SalI. The receptor-Fc sequences were excised from these plasmids using HindIII (T4-blunted) and XbaI (or Acc65I (T4-blunted) and XbaI in the case of Cryptic) and cloned into pEF IRES-Puro (28) cut with EcoRI (T4-blunted) and XbaI to yield the final Fc fusion expression plasmids. Acvr2 was amplified from mouse E8.5 cDNA using primers 5′-CGACGCAAGCTTACCATGGGAGCTGCTGCAAAGT-3′ and 5′-AATGTTGTCGACGGGTGGCTTCGGTGTAACAGGAT-3′, cut with HindIII and SalI, and inserted into pCR3-MCS-Presci-Fc. The fusion sequence was excised with NcoI and XbaI and transferred to the pEF IRES-Puro backbone of ACVR1B vector. The cDNA of Tgfbr2 extracellular domain was amplified from mouse E17.5 cDNA using primers 5′-AATCATGCTAGCACCATGGGTCGGGGGCTG-3′ and 5′-AATCTAGTCGACGCTGGTGGTGTATTCTTCCG-3′, cut with NheI and SalI, and inserted into pEF Acvr2a IRES-Puro cut with SalI (partially) and NheI. Bmpr2 was amplified from mouse E17.5 cDNA using primers 5′-AATCTTGCTAGCACCATGACTTCCTCGCTGCAT-3′ and 5′-ATAGATGTCGACTCGATTAAATGAATGAGGTGG-3′ and cloned like Tgfbr2.
Lentiviruses were produced by transient transfection of HEK293T with a 20:13:7 mixture of lentivirus:packaging:envelope vectors. Packaging and envelope vectors were pCMVΔR8.74 and pMD2.VSVg, respectively (30). In brief, 5 × 106 cells were seeded into a 10-cm dish and transfected on the following day with 20 μg of total DNA. The medium was replaced after 16 h and conditioned twice for 24 h. Conditioned media were pooled, filtered through a 0.45-μm polyethersulfone filter, and either used as such or centrifuged at 50,000 × g for 140 min. After centrifugation, the viral pellet was resuspended in 400 μl of PBS containing 0.1% bovine serum albumin (BSA).
Cell Culture and Establishment of Stable Cell Lines
HEK293T and HepG2 cells were cultured in a humidified incubator at 37 °C under 5% CO2 in DMEM (Sigma) containing 10% fetal bovine serum (FBS) (Invitrogen) and 1% gentamicin (Invitrogen). HEK293T cells stably expressing Nodal or its mutant derivatives from pEF IRES-Puro vectors were selected in 1 μg/ml puromycin (Sigma) starting 1 day after transfection. To generate Gdf1 stable cells, HEK293T or Nodal-expressing cells were infected with serial dilutions of pLenti-Gdf1(HA)-puro or pLenti-Gdf1(His6)-puro lentiviruses. Selection in 10 μg/ml blasticidin S (Invitrogen) was started 2 days later. Resistant cell populations selected after a multiplicity of infection of 1 were pooled. Western blotting and coimmunoprecipitation experiments were done with Gdf1(HA). All activity, pulldown, and purification experiments were performed with Gdf1(His6). To create Smad3 reporter cells, HepG2 liver hepatocellular carcinoma cells were first infected with serial dilutions of pLenti-CAGA-lux-neo lentivirus and selected with 1 mg/ml G418 (Sigma). Cells infected at the lowest multiplicity of infection were chosen and superinfected with either pLenti-Cripto-bsd or pLenti-Cryptic-bsd lentiviruses and selected with 10 μg/ml blasticidin S. Superinfection of these cell lines with pLenti-Rluc-puro and selection in 1 μg/ml puromycin gave rise to the reporter cell lines HepG2-Cripto·CAGA·Rluc (H-CrCR) and HepG2-CAGA·Rluc·Cryptic (H-Cryptic), respectively.
Production of Conditioned Medium
Cells stably expressing Nodal and/or Gdf1 proteins were seeded in cell culture dishes containing complete medium. On the following day, medium was either left unchanged (standard conditioning in complete medium) or replaced by serum-free medium (Opti-MEM I from Invitrogen). Medium was conditioned for 3 or 4 days. After conditioning, media were collected, cleared by centrifugation at 4,000 × g for 5 min, and filtered through a 0.45-μm filter (Minisart-N GF AC, Sartorius Stedim Biotech). Serum-free conditioned media were not filtered to avoid protein loss. For transient transfection (see Fig. 2C), HEK293T cells were seeded in 24-well plates at a density of 105 cells/well and transfected 8 h later with a 1:1 mixture of JetPEI and Nodal- and Gdf1-expressing vectors (1 μg total) (Polyplus). Conditioned media were collected after 3 days, spun at 4000 × g for 5 min, and added onto reporter cells.
FIGURE 2.
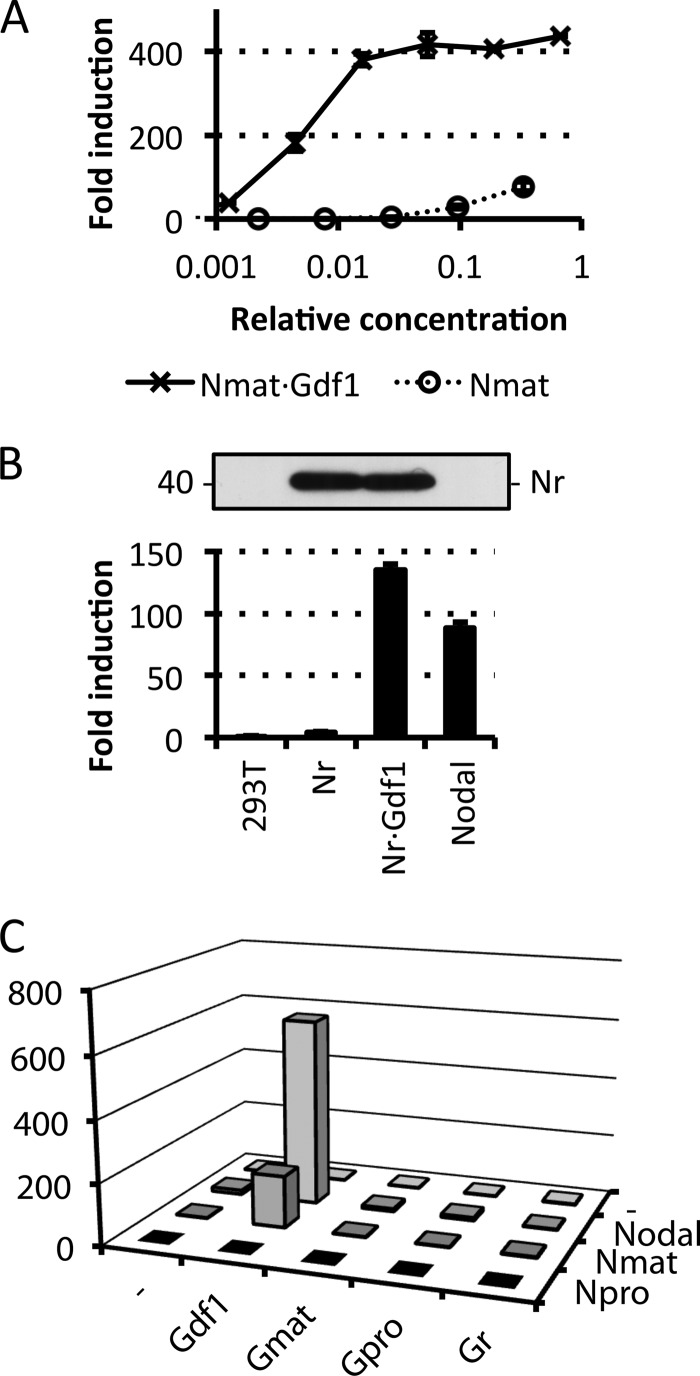
Nodal and Gdf1 can synergize independently of the Nodal prodomain and precursor cleavage motif. A, induction of CAGA-luc by Nmat, which lacks the prodomain, or Nmat·Gdf1(HA) in complete conditioned media. B, induction of CAGA-luc by complete media conditioned with Nr or Nr·Gdf1(His6). 600 ng/ml rNodal was analyzed for comparison. Top, full-length Nodal precursor detected with the M2 anti-FLAG antibody. C, induction of CAGA-luc by complete media conditioned by transient transfection of combinations of Nodal (N) and Gdf1(HA). mat, mature domain only; pro, prodomain only; r, resistant to proprotein convertase cleavage. In A, error bars represent S.D.
Luciferase Reporter Assay
Firefly and Renilla activities were measured as described (31). Briefly, reporter cells in 100 μl of medium were seeded in 96-well plates at a density of 5 × 104/well. One day later, 200 μl of conditioned medium or medium containing recombinant factors was added. After overnight incubation, medium was removed, cells were lysed in 100 μl of potassium phosphate buffer with Triton X-100 for 20 min, and 5 μl was transferred to white 96-well plates (Nunc). Firefly and Renilla luciferase luminescence was revealed using a Centro LB960 luminometer (Berthold) by adding 50 μl of P/R Luc A and B reagents, respectively. Firefly luciferase values were normalized to Renilla luciferase values. Error bars represent S.D. of triplicate values of representative experiments. To measure the activity of serum-free samples or in samples where serum had been heated or replaced by other components, reporter cells were first washed thrice and then overlaid with 100 μl of serum-free medium prior to sample addition. To determine the half-minimal effective concentration (EC50), experimental data points were fitted to a sigmoidal dose-response curve by the least square method. Calculated EC50 values were 4, 105, and 10 ng/ml for Nodal·Gdf1, rNodal, and recombinant Activin, respectively. These values were obtained from nine, six, and two biological replicates with an S.E. of 8, 11, and 11%, respectively.
Western Blotting and Antibodies
Western blot analysis was conducted using standard techniques. Upon addition of Laemmli buffer, samples were boiled at 95 °C for 5 min prior to loading. For detection of purified products after gel filtration, samples were concentrated 20-fold by acetone precipitation and resuspension in 2× Laemmli buffer. The antibodies used were mouse M1 anti-FLAG, rabbit anti-HA (Sigma), mouse HIS-1 (Sigma), and rabbit anti-Alix (Covalab). A custom polyclonal rabbit antiserum against Nodal (CAN3) was raised against the peptide sequence KQYNAYRCEGECPNPV (Eurogentec, Belgium). Secondary antibodies (sheep anti-mouse, donkey anti-rabbit, and sheep anti-human) were from GE Healthcare. Antibodies were used at 1:1000 in PBS with Tween 20 containing 5% milk (Sigma) except anti-Alix, which was used at 1:500, and CAN3, which was used at 1:200 in PBS with Tween 20 containing 5% milk and 1% BSA. 1 mm CaCl2 was added during all steps of M1 anti-FLAG immunoblotting.
Determination of Protein Concentration
To determine Nodal protein concentrations, serial dilutions of crude conditioned media or purified fractions were compared side by side with known concentrations of recombinant Nodal protein (R&D Systems) by Western blotting using CAN3 anti-Nodal antibody. All estimates are thus solely based on the amount of Nodal detected without taking into account Gdf1. To quantify Fc fusion proteins, 1-ml aliquots of conditioned media were first incubated with Protein A Dynabeads for 30 min at 4 °C. Beads were washed with PBSN and added to sample buffer. Protein concentration in the eluates was compared side by side with known concentrations of recombinant leukemia inhibitory factor-Fc (a gift from David Hacker, Ecole Polytechnique Fédérale de Lausanne protein purification core facility). This purification was necessary because BSA in crude conditioned medium co-migrated with receptor fusions and thus confounded their accurate quantification in Western blots. Total precipitation of the Fc fusion proteins was verified by probing the depleted conditioned medium. Estimation of total protein content by Bradford (Bio-Rad) was done following the microassay procedure for microtiter plate protocol using BSA as a standard.
Analysis of Protein Oligomerization, Exosome Partitioning, and Effects of Serum
To study protein aggregation, conditioned media were centrifuged at 16,000 × g for 1 h at 4 °C. Pellet and supernatant were resuspended in sample buffer and analyzed for Nodal protein content using the M1 anti-FLAG antibody. To monitor the reversible interconversion of Nodal·Gdf1 high molecular weight (HMW) forms into LMW forms, conditioned media were cleared by centrifugation at 4000 × g for 5 min, filtered through 0.45-μm filters (Minisart-N GF AC, Sartorius Stedim Biotech), and subjected to ultrafiltration using Amicon 100-kDa cut off filters (Millipore). Retentates were brought back to their original volume with serum-free DMEM prior to the next round of filtration.
Heat inactivation of complete serum (FBS) involved heating at 95 °C for 0, 15, or 30 min and equilibration to 37 °C prior to addition of rNodal (250 ng/ml). For size fractionation, complete medium was filtered successively through 100-, 50-, and 3-kDa-cutoff Amicon ultrafiltration columns (Millipore). Retentates were brought back to their original volume with serum-free DMEM, and rNodal (250 ng/ml) was added to each fraction. For serum replacement, rNodal (500 ng/ml) was added to medium previously supplemented with 5 g/liter BSA, 3.4 g/liter yeast extract), or 3.8 g/liter FBS. For serum titration, rNodal and Nodal·Gdf1 were added to medium containing serial dilutions of FBS. In all these experiments, the solutions were first incubated for 6 h at 37 °C before assessing Nodal activity on reporter cells in serum-free medium.
Exosome isolation was performed by differential centrifugation following a modification of the protocol of Théry et al. (32). Briefly, conditioned media were cleared by centrifugation at 4000 × g for 5 min and then at 10,000 × g for 30 min. Exosomes were pelleted by ultracentrifugation at 100,000 × g for 2 h followed by resuspension in sample buffer.
Immunoprecipitation Experiments
To immunoprecipitate Nodal·Gdf1 complex, 20 μl of anti-HA-agarose beads (HA-7, Sigma) precleared for 30 min in 1 ml of complete medium at 4 °C were added to Nodal·Gdf1 conditioned medium, incubated overnight, washed thrice with complete medium, and resuspended in 2× sample buffer. To immunoprecipitate serous proteins that associated with Nodal, 100 μl of M1 anti-FLAG-agarose beads (Sigma) were blocked for 2 h at 4 °C with 1 ml of complete medium, then split in two, washed thrice with DMEM with or without the indicated concentration of FBS, and resuspended in 200 μl of DMEM (±FBS). The beads were subsequently incubated overnight at 4 °C with 1 ml of conditioned medium, washed four times in TBS containing 1 mm Ca2+, and resuspended in 2× sample buffer.
To pull down soluble Fc fusion receptors, citrate buffers at pH 5, 5.4, 5.8, 6.4, 6.6, and 7.0 were prepared from 0.2 m Na2HPO4 and 0.1 m citrate solutions. Nonidet P-40 was added to 0.01% (v/v) in each solution or in PBS to make PBSN. 25 μl of Protein A Dynabeads (Invitrogen) was washed in 1 ml of PBSN. After incubation for 30 min at 4 °C with 1 ml of conditioned medium containing Fc fusion protein, the beads were washed thrice with 1 ml of the appropriate citrate buffer. 450-μl aliquots of Fc-bound beads were distributed to tubes containing 450 μl of citrate buffer of the indicated pH and incubated overnight at 4 °C with 100 μl of Nodal or Nodal·Gdf1 conditioned media, each containing 7.2 ng of Nodal protein. After washing with citrate buffer of the corresponding pH, beads were resuspended in 2× sample buffer. For the type II receptor pulldowns shown in Fig. 8C, 100 μl of Protein A beads was washed in 2 ml of PBSN, added to 2 ml of receptor-Fc conditioned medium, incubated at 4 °C for 30 min, and washed thrice with PBSN. 240 μl of Fc-bound bead slurry was added to tubes containing 1 ml of Nodal or Nodal·Gdf1 conditioned media (72 ng of Nodal). Beads were incubated overnight at 4 °C, washed in PBSN, and resuspended in 2× sample buffer.
FIGURE 8.
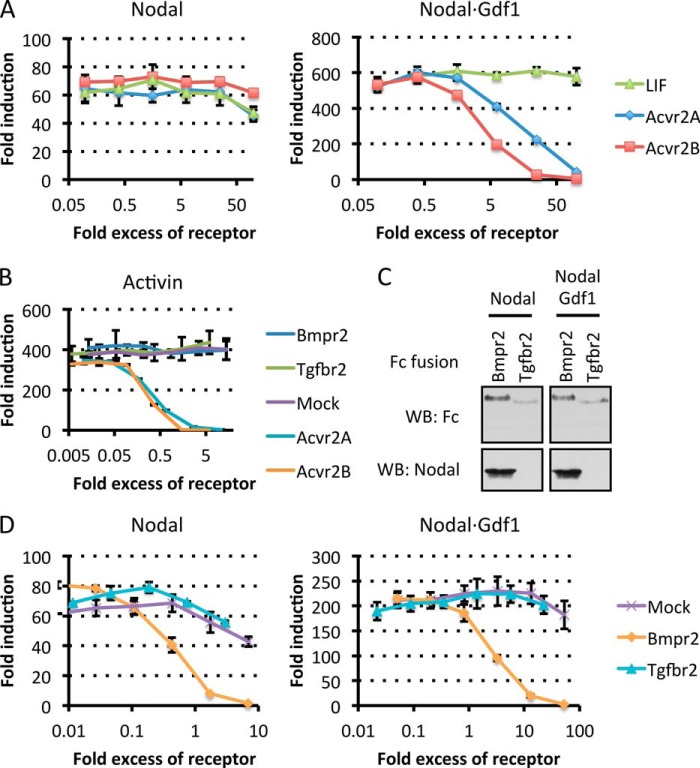
Differential inhibition of Nodal and Nodal·Gdf1 by soluble type II receptors. A, induction of CAGA-luc by Nodal or Nodal·Gdf1(HA) conditioned medium preincubated with increasing concentrations of the indicated type II receptor extracellular domain fused to Fc. B, induction of CAGA-luc by Activin conditioned medium that was preincubated with increasing concentrations of the indicated Fc-receptor fusion protein. C, Western blot (WB) analysis of Fc-receptor fusion constructs and associated Nodal proteins following pulldown of the fusion proteins. D, induction of CAGA-luc by Nodal or Nodal·Gdf1(HA) conditioned medium that was preincubated with increasing concentrations of the indicated type II receptor extracellular domain fused to Fc. In A, B, and D, error bars represent S.D. LIF, leukemia inhibitory factor.
To assess dominant negative activity of soluble extracellular domains of type II Activin or BMP receptors, Nodal or Nodal·Gdf1 conditioned media were mixed with serial dilutions of conditioned media of cells expressing the indicated Fc fusion proteins and rotated for 2 h at 37 °C prior to incubation with H-CrCR Smad3 reporter cells in 96-well plates. Concentrations of the proteins were estimated as described above and converted to molarities using the predicted molecular weights.
Nodal·Gdf1 Purification
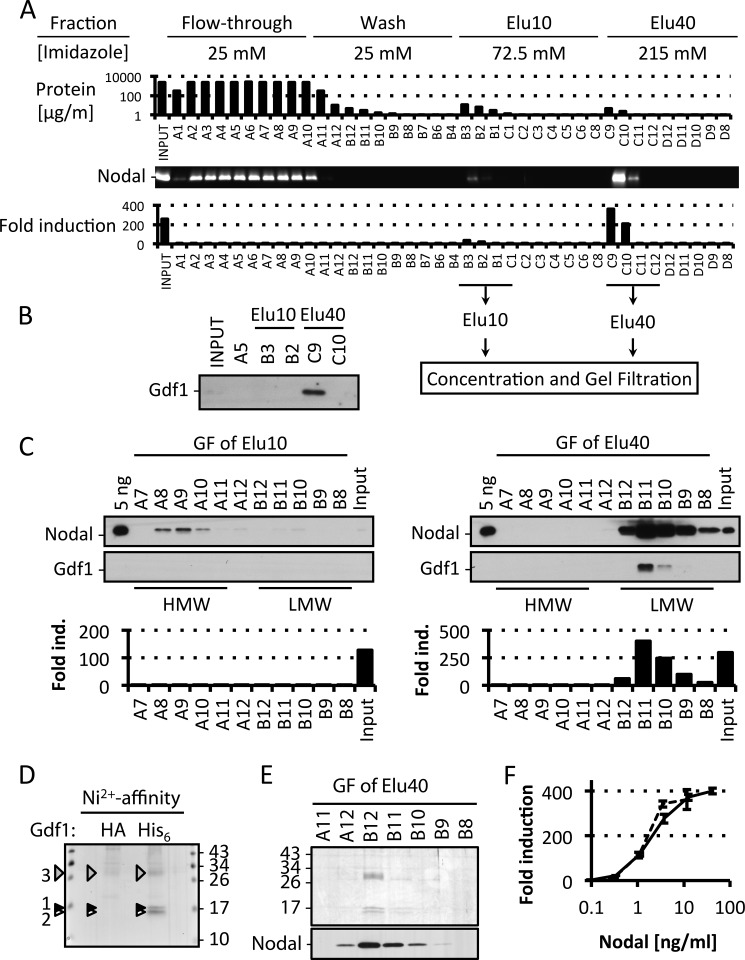
Nodal·Gdf1 protein complexes were purified in two steps by affinity purification and gel filtration using an ÄKTA FPLC system (GE Healthcare). For affinity purification, 50 ml of Nodal·Gdf1 medium was conditioned for 4 days and cleared by centrifugation. 1 ml of sodium phosphate buffer, pH 7.4; 5 ml of 5 m NaCl; and 400 μl of 3 m imidazole were added to 43.6 ml of conditioned medium (25 mm imidazole final). The solution was loaded on a HisTrap FF Nickel column (GE Healthcare) previously equilibrated with 5 volumes of 20 mm sodium phosphate, pH 7.4 containing 0.5 m NaCl and 25 mm imidazole (equilibration buffer). After extensive washing with 10 volumes of equilibration buffer, bound proteins were eluted with 10 volumes of 20 mm sodium phosphate, pH 7.4, 0.5 m NaCl, and 72.5 mm imidazole (Elu10 fractions) followed by 10 volumes of 20 mm sodium phosphate, pH 7.4, 0.5 m NaCl, and 215 mm imidazole (Elu40 fractions). Eluates were collected in 5-ml fractions and analyzed for protein content and activity. Flow rate was 5 ml/min for 5-ml columns and 1 ml/min for 1-ml columns. For gel filtration, pools of the first four Elu10 and Elu40 fractions were each concentrated using 3-kDa-cutoff Amicon ultrafiltration devices (Millipore) and diluted 1:1 with PBS containing 0.5 m NaCl and 1% CHAPS prior to size separation on a Superdex 200 10/300 GL column (GE Healthcare) in PBS with 0.5 m NaCl and 0.5% CHAPS at 0.5 ml/min. Eluates were collected in 5-ml fractions and analyzed for protein content and activity. Two peaks scored positive in anti-Nodal Western blots; one corresponded to the void volume of the column (HMW fraction) and one corresponded to a size of ∼60 kDa (LMW fraction). Because gel filtration did not increase specific activity of the complex, this step was omitted, and Elu40 fractions were pooled, concentrated, and buffer-exchanged into PBS using Zeba spin columns (Thermo Scientific), and BSA was added to 0.1% final concentration. Activity assays using gel-filtrated material are presented in Fig. 5, C and F.
FIGURE 5.
Purification of Nodal·Gdf1 reveals active LMW complexes containing both prodomains. A, representative Nodal·Gdf1(His6) affinity purification profile from 45 ml of conditioned medium. Each 5-ml fraction was analyzed for total protein content by Bradford assay, Nodal protein content was analyzed by immunoblotting, and Nodal activity was analyzed in CAGA-luc reporter assays. B, immunoblot analysis of Gdf1 in relevant fractions. C, typical Nodal·Gdf1 gel filtration (GF) profile from pooled and concentrated Elu10 and Elu40 fractions from the affinity purification step. Each 5-ml fraction was analyzed by immunoblotting for Nodal and Gdf1 protein and by luciferase reporter assays for Nodal activity. D, silver staining of material eluted from a nickel affinity column using either HA- or His6-tagged Gdf1 in Nodal·Gdf1 conditioned medium. LC-MS/MS analysis identified mature Nodal and Gdf1 (black and white arrowheads, respectively) as well as the prodomains of Nodal and Gdf1 (gray arrowhead). E, selected fractions from the gel filtration of an Elu40 pool. Top, silver staining. Bottom, immunoblotting against Nodal. F, induction of CAGA-luc by affinity-purified (dashed line) or gel-filtered (solid line) Nodal·Gdf1. In F, error bars represent S.D. ind., induction.
Silver Staining and Mass Spectrometry
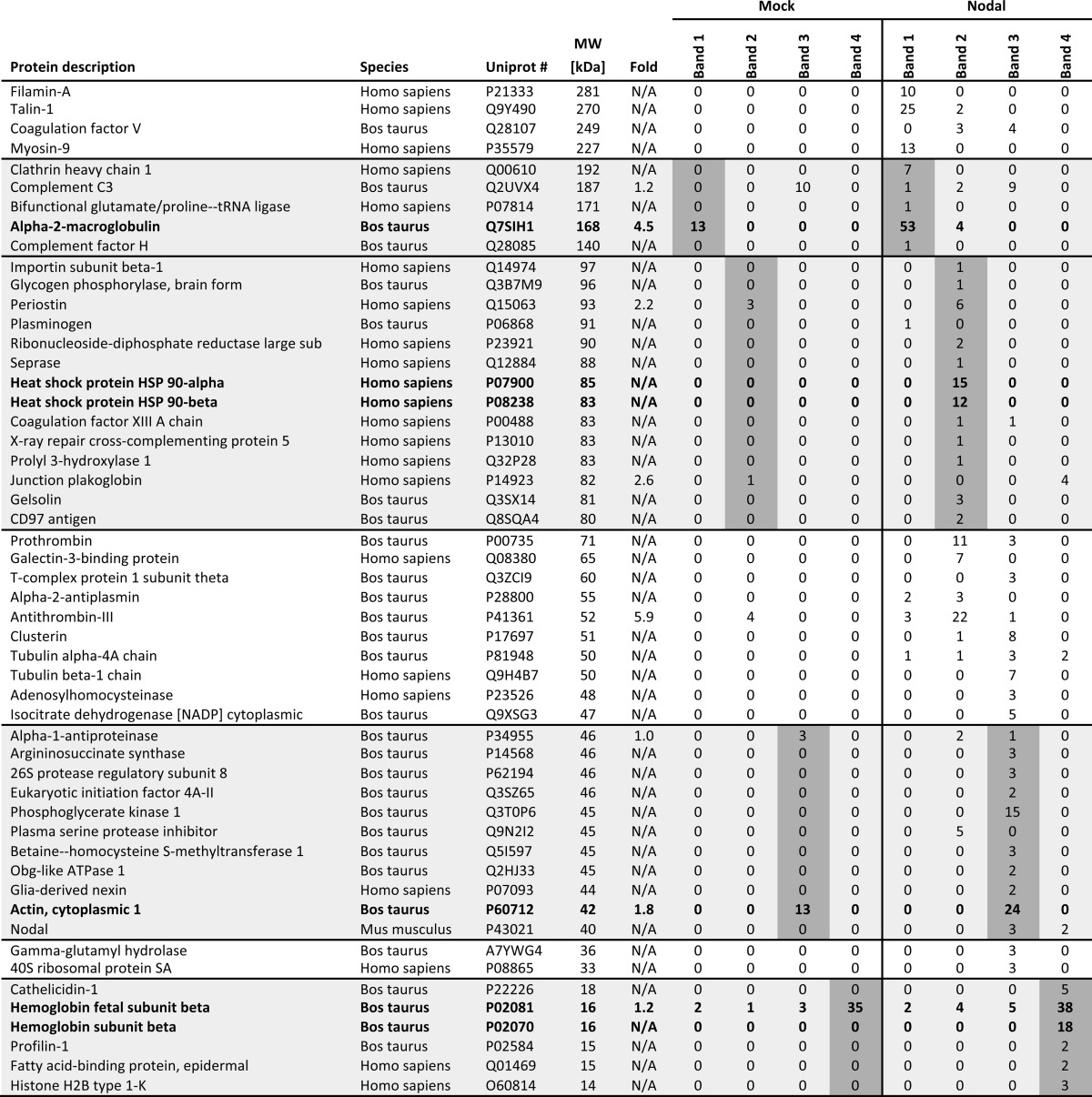
Polyacrylamide gels were fixed in 30% EtOH and 10% acetic acid for 30 min, rinsed 2 × 10 min in 20% EtOH and 2 × 10 min in water, and sensitized for 1 min in 0.02% Na2S2O3. After washing 2 × 1 min in water, gels were then impregnated for 30 min in 0.1% silver nitrate, dipped for 10 s in water, and developed in 3% (w/v) K2CO3 containing 0.025% (v/v) freshly added formalin. After development, gels were transferred for 30 min to stop solution containing 4% (w/v) Tris and 2% (v/v) acetic acid and finally washed in water. Bands were submitted to Ecole Polytechnique Fédérale de Lausanne proteomics core facility for LC-MS/MS analysis. Proteins were identified based on a 1% false discovery rate for both peptides and proteins and a minimum of two unique peptides using the software Scaffold 4. Hits corresponding to irrelevant species were manually removed from the data set, and numbers represent normalized spectral count values related to the amount of proteins (number of spectra recorded for a given protein). Only hits enriched more than 1-fold and with a spectral count higher than 3 are displayed except when they fell within the size range of the cut bands (shaded areas in Table 1).
TABLE 1.
Proteins identified after Nodal pulldown
Nodal protein was pulled down from mock or Nodal conditioned media in the presence of serum. Fold, -fold change between mock and Nodal conditioned medium (N/A indicates the protein was absent from mock). In bold are proteins specifically mentioned in the text. Other proteins were specifically enriched in the Nodal sample (phosphoglycerate kinase, periostin, and clathrin heavy chain). Shaded areas include proteins with a size corresponding to that of the isolated gel band. sub, subunit.
hES Cell Differentiation Assays
H1 (WA01) cell line was differentiated toward definitive endoderm (DE) in monolayer format according to an established protocol (2) using 100 ng/ml Activin A (R&D Systems), 100 ng/ml rNodal (R&D Systems), or purified Nodal·Gdf1 in combination with 25 ng/ml Wnt3a (R&D Systems) for 1 day and 100 ng/ml Activin A (R&D Systems), 100 ng/ml rNodal (R&D Systems), or purified Nodal·Gdf1 in combination with 5 ng/ml basic FGF for 2 days. Cells were dissociated and stained with anti-CXCR4-allophycocyanin (BD Biosciences) and anti-CD326/EpCam (eBioscience) at day 3 of differentiation. Total RNA was extracted at day 3 of differentiation using an RNAqueous-Micro kit (Ambion) and treated with RNase-free DNase (Ambion). 0.5–1 μg of RNA was reverse transcribed into cDNA using random hexamers and oligo(dT) with Superscript III reverse transcriptase (Invitrogen). Quantitative PCR was performed on a MasterCycler EP RealPlex (Eppendorf) using a QuantiFast SYBR Green PCR kit (Qiagen) as described previously (33). SOX17, FOXA2, SOX3, and OCT4 expression levels were normalized to the housekeeping gene TATA box-binding protein. The oligonucleotide sequences are available on request.
Indirect Immunofluorescence Staining
Monolayer culture immunostaining was performed as described previously (2). The following primary and secondary antibodies were used: mouse anti-OCT3/4 (BD Transduction Laboratories; 1:500), rabbit anti-SOX17 (Beta Cell Biology Consortium; 1:500), donkey anti-mouse Alexa Fluor 647 (Invitrogen; 1:800), and donkey anti-rabbit Alexa Fluor 488 (Jackson ImmunoResearch Laboratories; 1:800). The stained cells were visualized using a digital inverted fluorescence microscope (Advanced Microscopy Group EVOS).
RNAi of Type II Receptors in Reporter Cells
The sequences of previously validated siRNAs against ACVR2B and BMPR2 were as described (34, 35). On day 0, H-CrCR reporter cells were seeded in a 24-well plate (2.5 × 104/well). On day 1, cells were transfected with 40 nm siRNA against ACVR2B (5′-GGUGUACUUCUGCUGCUGUuu-3′) or BMPR2 (5′-GGAUGAGCGUCCAGUUGCUuu-3′) (Microsynth) or control scrambled siRNA (Silencer negative control siRNA, Ambion) using Lipofectamine 2000 (Invitrogen) following the manufacturers' instructions. The transfection was repeated on day 2. On day 3, Nodal or Nodal·Gdf1 conditioned media were added to the wells. Luciferase activity was measured on day 4.
RESULTS
Gdf1 Accumulates in HEK293T-conditioned Medium When Nodal Is Coexpressed, and the Two Proteins Synergize in a Cripto-dependent Manner
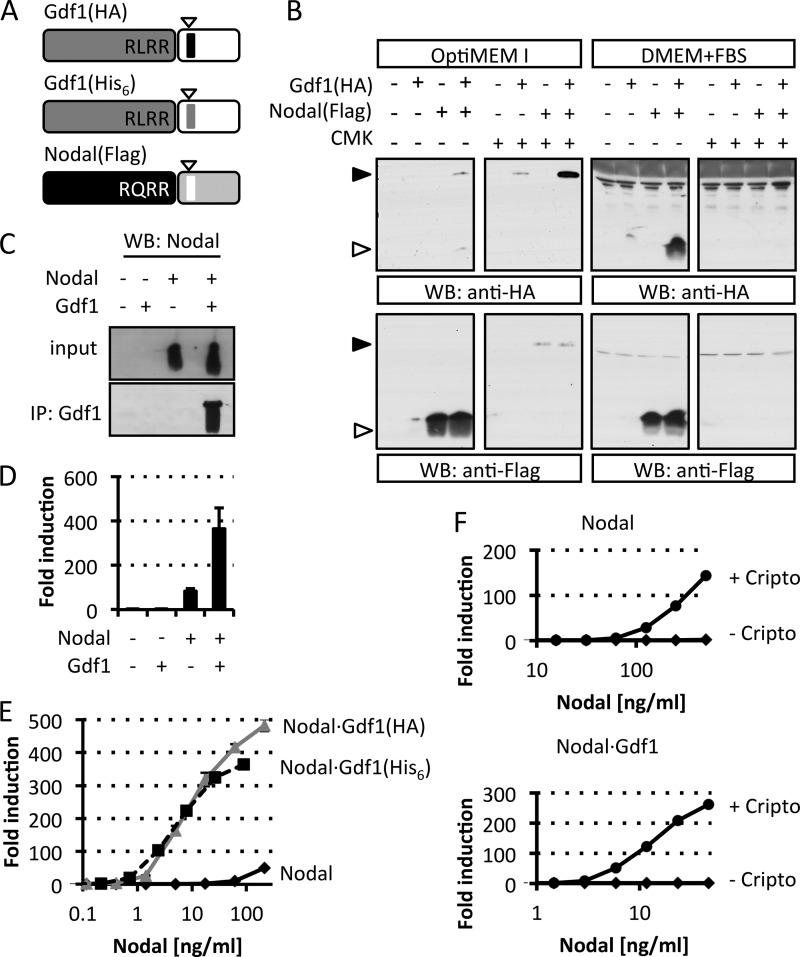
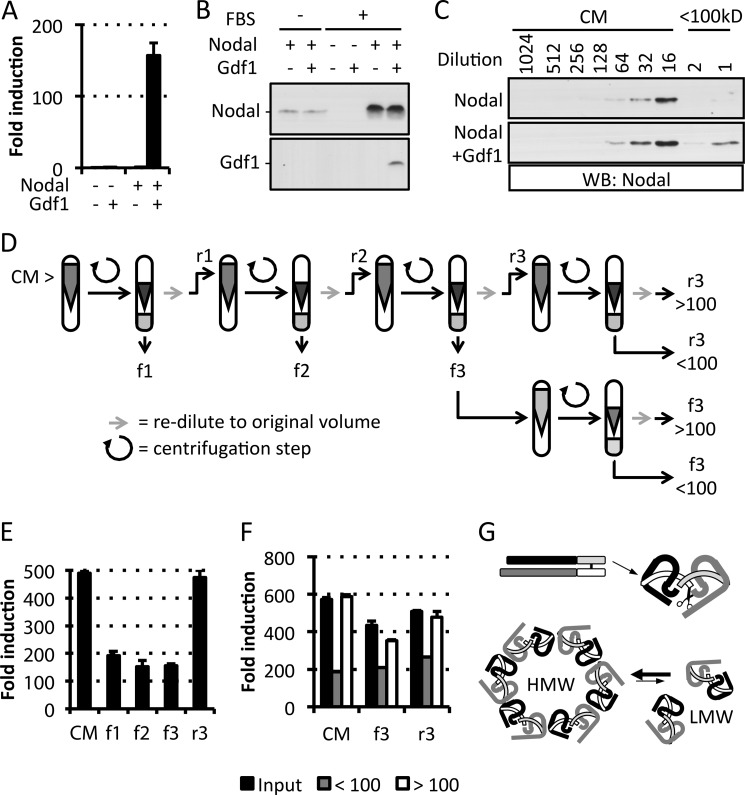
In Xenopus oocytes and COS1 cells, coexpression with Nodal increases the electrophoretic mobility but not the accumulation of mature mouse Gdf1 (22). To monitor secretion and precursor processing, we tagged Nodal and Gdf1 with FLAG, HA, or His6 epitopes after their unique PC cleavage motifs and stably transduced them into HEK293T cells (Fig. 1A). Western blot analysis revealed similar levels of mature Nodal in conditioned medium irrespective of the presence or absence of Gdf1. By contrast, mature Gdf1(HA) and Gdf1(His6) only accumulated if Nodal was coexpressed (Fig. 1B and data not shown). Nodal also increased the levels of Gdf1 precursor especially if cells were treated with the PC inhibitor decanoyl-RVKR-chloromethyl ketone. In complete medium supplemented with serum, a specific signal corresponding to Gdf1 precursor was not detected, but instead the levels of mature Gdf1 accumulating with Nodal further increased. Moreover, coimmunoprecipitation analysis of complete conditioned medium indicated that Nodal and Gdf1 form a complex as described previously (22) (Fig. 1C). Experiments performed in the absence of serum showed that this interaction was serum-independent (data not shown). To monitor signaling activities, we generated the HepG2 reporter cell line H-CrCR, which stably expresses the Nodal co-receptor Cripto and the Smad3 luciferase reporter CAGA-luc as lentiviral transgenes together with Renilla luciferase for signal normalization. Conditioned medium of HEK293T cells expressing Gdf1 alone failed to induce CAGA-luc expression in H-CrCR reporter cells. However, when coexpressed with Nodal, Gdf1 increased the basal activity of Nodal in a synergistic manner (Fig. 1D). To quantify this synergism, we compared serial dilutions of complete conditioned media containing defined amounts of Nodal alone or together with co-transfected Gdf1. The concentration of Nodal required to induce a half-maximal response (EC50) was too high to be estimated but clearly exceeded 100 ng/ml. Coexpression of Gdf1 lowered the EC50 of Nodal to 5 ng/ml, indicating a dramatic increase in specific activity, irrespective of whether Gdf1 was tagged with HA or with His6 (Fig. 1E). Furthermore, incubation with CAGA-luc reporter cells lacking Cripto confirmed that Nodal·Gdf1 activity strictly relies on the presence of an EGF-CFC co-receptor (Fig. 1F).
FIGURE 1.
Dependence of extracellular Gdf1 on Nodal. A, schematic depiction of HA-, His6-, or FLAG-tagged Gdf1 and Nodal precursors. The epitopes (arrowheads) were inserted 4 residues after the precursor cleavage sites RLRR and RQRR. N-terminal prodomains are shaded dark gray or black. B, Western blot (WB) analysis of media conditioned by HEK293T cell lines that were stably transduced with Gdf1, Nodal, or both. Where specified, cells were treated with 10 μm decanoyl-RRKR-chloromethyl ketone (CMK) during medium conditioning. Opti-MEM I and DMEM samples were analyzed separately on different blots with distinct exposure times in this experiment. C, Western blot analysis of Nodal following immunoprecipitation (IP) of Gdf1(HA) from complete conditioned media (DMEM+FBS). D, induction of the Smad3 luciferase reporter CAGA-luc in stable H-CrCR reporter cells by serum-containing conditioned DMEM. Gdf1 is HA-tagged. E, induction of CAGA-luc by serum-containing Nodal (black, solid line), Nodal·Gdf1(HA) (gray, solid line), or Nodal·Gdf1(His6) (black, dashed line) conditioned media. HA-tagged Gdf1 and His6-tagged Gdf1 similarly potentiate Nodal activity. HA reacted more strongly than His6 in Western blots, but His6 was later preferred for affinity purification. F, induction of CAGA-luc by recombinant Nodal or Nodal·Gdf1(His6) in naïve (diamonds) or Cripto-overexpressing (circles) reporter cells. Note that concentrations in E and F only refer to total amounts of Nodal because tagged Gdf1 was not quantified. In D, E, and F, error bars represent S.D.
Nodal, but Not Gdf1, Can Synergize Independently of Its Prodomain and Precursor Cleavage Motif
In TGFβ, the prodomain acts as a chaperone that usually remains associated with mature ligand even after proteolytic maturation to mask receptor-binding epitopes (36, 37). To evaluate how prodomains and precursor processing influence the synergism of Nodal with Gdf1, we tested Nr (27), Npro, or Nmat fused directly to the secretory signal sequence (26). Surprisingly, both Nr and Nmat robustly signaled with Gdf1, whereas Npro did not (Fig. 2, A–C). By contrast, neither the mutated PC-resistant Gdf1 precursor (Gr), Gdf1 prodomain (Gpro), nor Gdf1 constitutively mature form (Gmat) stimulated Nodal activity (Fig. 2C). These results suggest that the prodomain of Gdf1 is required but has to be cleaved from the mature domain for the synergy to occur. In sharp contrast, the Nodal precursor can synergize with Gdf1 independently of a functional PC cleavage motif.
Nodal Signaling Requires Heat-labile, High Molecular Weight Serum Components
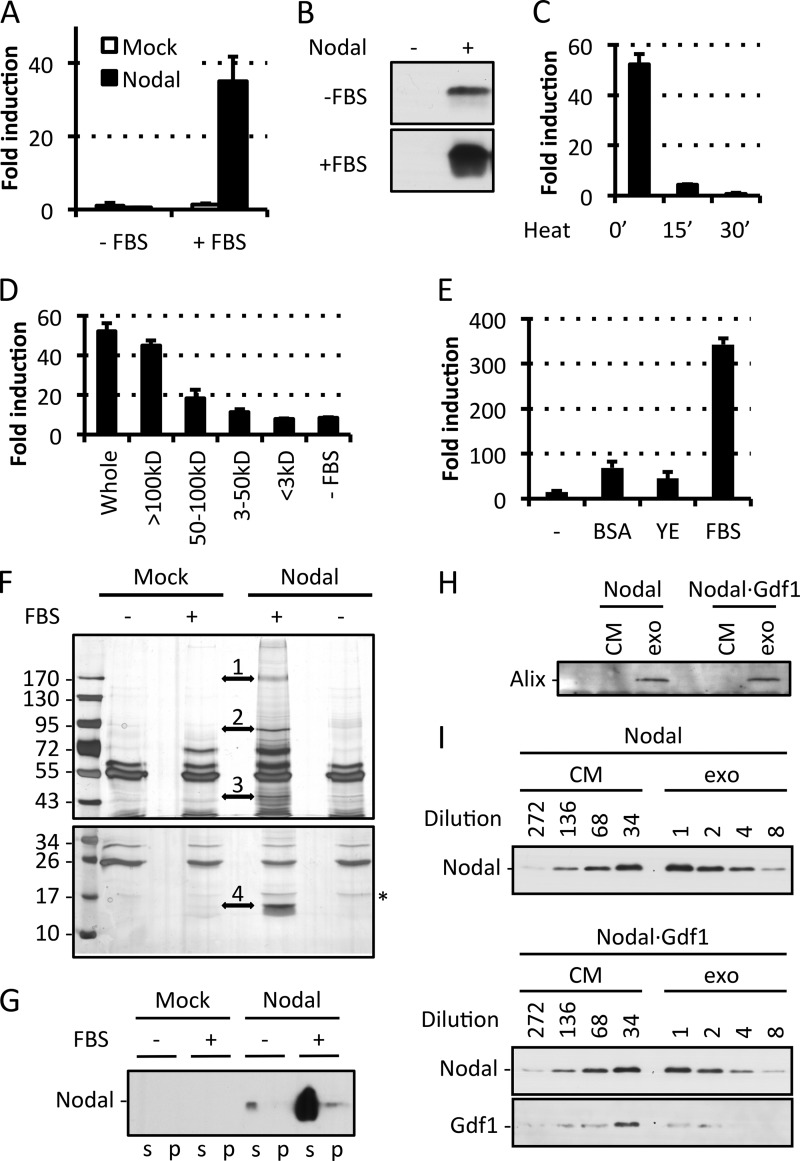
In Xenopus embryos that are cultured in modified Barth's solution without serum, coexpression of Gdf1 is essential for secreted Nodal to signal (22). By contrast, in our conditioned medium, which contains 10% FBS, secreted Nodal activity only partially depended on Gdf1 (Fig. 1D). We found that serum was essential to detect cell-non-autonomous Nodal activity (Fig. 3A) and that it boosted the levels of secreted Nodal protein in conditioned DMEM (Fig. 3B). To address how serum promotes Nodal signaling, complete medium was heated at 95 °C for 15 or 30 min prior to addition of recombinant Nodal. Alternatively, conditioned medium was subjected to ultrafiltration. Transient heating or ultrafiltration of the conditioned medium by a size cutoff below 100 kDa abolished the stimulatory effect of FBS (Fig. 3, C and D). Neither BSA nor yeast extract mimicked the effect of FBS (Fig. 3E), suggesting that Nodal activity depends on specific serum proteins or protein complexes. We therefore analyzed Nodal immunoprecipitates from conditioned medium by mass spectrometry. Several serum proteins were specifically enriched in Nodal immunoprecipitates, including α2-macroglobulin, Hsp90, and actin (Fig. 3F and Table 1). An additional prominent protein band migrating with a molecular weight of 15 kDa (region 4) contained hemoglobin. Taken together, these results suggest that Nodal alone requires serum for paracrine signaling and that this supporting activity is likely conveyed by a complex with one or several other proteins.
FIGURE 3.
Paracrine Nodal activity requires serum proteins. A, induction of CAGA-luc by control (Mock) or Nodal conditioned medium (DMEM) previously supplemented with or without FBS. B, immunoblot analysis of Nodal in the conditioned media analyzed in A. C, induction of CAGA-luc by 250 ng/ml rNodal in complete medium previously heated at 95 °C for the indicated duration. ′, minutes. D, induction of CAGA-luc by 250 ng/ml rNodal in complete medium previously separated by ultrafiltration membranes with the indicated size cutoffs. E, induction of CAGA-luc by 500 ng/ml rNodal in medium previously supplemented with 5 g/liter BSA, 3.4 g/liter yeast extract (YE), or 3.8 g/liter FBS. F, silver staining of associated proteins after immunoprecipitation of Nodal (asterisk) from control (Mock) or Nodal conditioned medium previously supplemented with or without FBS. Four bands (1–4) were cut from the gel on each side of the arrows and subjected to mass spectrometry (see Table 1). G, immunoblot analysis of Nodal in the supernatant (s) or pellet (p) after centrifugation of Nodal conditioned medium at 16,000 × g. H, immunoblot analysis of the exosomal marker Alix in whole conditioned medium (CM) or the exosomal fraction (exo). I, immunoblot analysis of Nodal and Gdf1(HA) in whole conditioned medium (CM) or the exosomal fraction (exo). In A, C, D, and E, error bars represent S.D.
Coexpression of Gdf1 Induces a Low Molecular Weight Form of Nodal and Suppresses Its Dependence on Serum
Previous analysis of the unrelated factor Wnt3a suggested a role for serum in preventing protein aggregation (29). To assess whether Nodal similarly aggregates, conditioned media were subjected to high speed centrifugation. Under these conditions, the large majority of Nodal with or without serum remained in the supernatant (Fig. 3G), indicating that serum is not essential for Nodal solubility. Because immunoprecipitation of Nodal from HEK293T-conditioned medium enriched Hsp90 and actin (Fig. 3F) and because these proteins associate with exosomes (38), we asked whether Nodal has an affinity for exosomes that is modulated by Gdf1. To address this, conditioned media were submitted to differential centrifugation and probed for the exosomal marker Alix and for Nodal protein. We found that the fraction that was enriched for Alix contained less than 5% of the total Nodal protein and that this amount was unchanged upon Gdf1 coexpression (Fig. 3, H and I). Less than 2% of Gdf1 partitioned to the Alix-positive fraction, indicating that Gdf1 is unlikely to recruit Nodal to exosomes under the conditions examined (Fig. 3I). We therefore asked whether Gdf1 instead might be required to alleviate the dependence of secreted Nodal activity on serum. In keeping with this idea, coexpression of Nodal with Gdf1 enabled robust signaling in serum-free conditions (Fig. 4A). Similar to what we observed in the presence of serum (Fig. 1B), Western blot analysis showed that Nodal was also essential in these serum-free samples to detect Gdf1 (Fig. 4B). Taken together, these results suggest that the synergism with Gdf1 diminishes a newly found dependence of Nodal on factors in the serum by a mechanism that probably involves neither exosomes nor an overt change in protein solubility.
FIGURE 4.
Coexpression of Gdf1 suppresses serum requirement and induces an LMW form of Nodal. A, induction of CAGA-luc by serum-free conditioned media using Gdf1(HA). B, immunoblot analysis of Nodal and Gdf1(HA) in the corresponding conditioned media. C, immunoblot analysis of Nodal in crude conditioned medium (CM) or conditioned medium filtered through a 100 kDa-cutoff filter (<100kD). Gdf1(His6) was used, and samples were diluted for equivalent signal intensity. D, schematic description of the ultrafiltration experiments described in E and F. E, induction of CAGA-luc by Nodal·Gdf1(HA) conditioned medium (CM), the corresponding LMW fractions obtained by three successive filtrations through 100 kDa cutoff filters (f1, f2, and f3), and the third retentate (r3) corresponding to HMW species. F, induction of CAGA-luc by the filtrate (<100) and retentate (>100) of the third fractions described above (Input). G, schematic description of the heterodimer of Nodal and Gdf1 (top) as well as the relation between LMW and HMW species. Prodomains and covalently linked mature regions and their color coding are as in Fig. 1A. Cleavage of precursor dimers is indicated by a scissor. Note that the actual number of heterodimers and their precise arrangement in non-covalent high molecular weight complexes are unknown. In A, E, and F, error bars represent S.D. WB, Western blot.
To further address how Gdf1 might increase Nodal activity, conditioned media were analyzed by ultrafiltration. We found that although Nodal alone was unable to pass through filters with a cutoff of 100 kDa, coexpression of Gdf1 enabled recovery of a small but significant amount (∼2%) in the flow-through (Fig. 4C). To determine whether this LMW form of Nodal·Gdf1 corresponds to a static pool of protein or whether it exists in a dynamic equilibrium with HMW complexes, we subjected conditioned medium to successive rounds of ultrafiltration and redilution (Fig. 4D). To our surprise, the relative activities in the retentate and flow-through fractions remained constant even after three rounds of ultrafiltration (Fig. 4E). Furthermore, when the final flow-through (flow-through fraction 3) and rediluted retentate (retentate fraction 3) were ultrafiltrated further, they each partitioned again into HMW and LMW fractions (Fig. 4F). We conclude that Gdf1 enriches Nodal in an LMW complex that appears to be in a dynamic equilibrium with a HMW form (Fig. 4G).
To directly assess whether Gdf1 potentiates the activity of an LMW form of Nodal, we isolated the complex from conditioned medium using a two-step purification protocol. First, a nickel column was used to bind a His6 tag in mature Gdf1. Increasing concentrations of imidazole eluted bound proteins in two peaks termed Elu10 and Elu40 (a typical profile of this purification step is shown in Fig. 5A). Although Elu40 was enriched for both Nodal and Gdf1, Elu10 only contained Nodal (Fig. 5, A and B). In a second step, the first four fractions of each peak were pooled and concentrated for gel filtration analysis. The Elu10 pool yielded inactive HMW Nodal without any detectable Gdf1 (Fig. 5C, left). In contrast, the Elu40 pool contained LMW Nodal·Gdf1 associated with high signaling activity (Fig. 5C, right). Silver staining of affinity-purified material (Fig. 5D) or of gel-filtrated fractions (Fig. 5E) revealed two highly enriched proteins in a 1:1 ratio that migrated at 14.9 and 13.6 kDa together with another prominent band at 28.8 kDa. Mass spectrometry analysis by LC-MS/MS revealed that the two lower bands corresponded to mature Nodal and Gdf1 (Fig. 5D, black and white arrowheads, respectively), whereas the upper band comprised the prodomains of Nodal and Gdf1 (Fig. 5D, gray arrowhead, and Table 2). Affinity-purified and gel-filtrated Nodal·Gdf1 displayed similar specific activities (Fig. 5F). Overall, these results suggest that mature Nodal and Gdf1 form heterodimers that remain bound to propeptides in a biologically active complex even after proteolytic maturation.
TABLE 2.
Identification of Nodal and Gdf1 from purified material
Listed are normalized spectral counts of Nodal and Gdf1 identified after affinity purification of Nodal·Gdf1(His6) and Nodal·Gdf1(HA) using a nickel column. Fold, -fold change between Gdf1(HA) and Gdf1(His6). All other proteins did not show a -fold change higher than 1 or more than 3 spectral counts. Mature and pro-Nodal are found in bands 1 and 3, respectively. Mature and pro-Gdf1 are found in bands 2 and 3, respectively.
| Protein description | Species | UniProt no. | Molecular weight | -Fold | Nodal·Gdf1(His6) |
Nodal·Gdf1(HA) |
||||
|---|---|---|---|---|---|---|---|---|---|---|
| Band 1 | Band 2 | Band 3 | Band 1 | Band 2 | Band 3 | |||||
| kDa | ||||||||||
| Embryonic growth/differentiation factor 1 | M. musculus | P20863 | 39 | 7.6 | 40 | 83 | 12 | 5 | 13 | 0 |
| Nodal | M. musculus | P43021 | 40 | 10 | 31 | 9 | 49 | 5 | 2 | 2 |
Purified Nodal·Gdf1 Complex Has High Specific Activity in Serum-free Conditions and Stimulates Differentiation of hESCs into Endoderm Progenitors
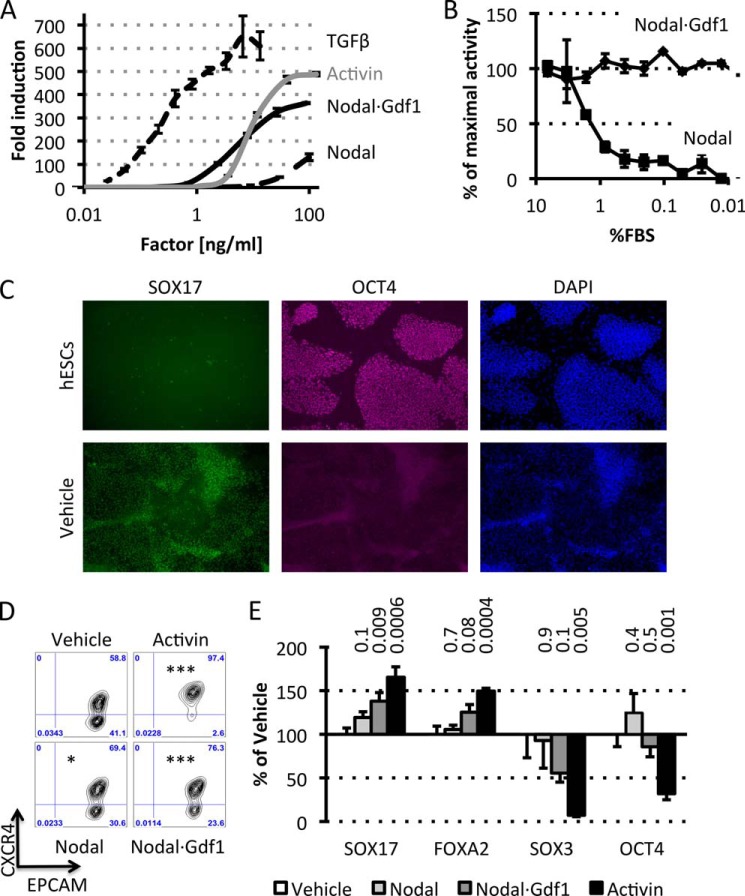
To functionally characterize purified Nodal·Gdf1, we compared its specific activity with that of commercially available recombinant proteins. In luciferase reporter assays, the EC50 values of TGFβ1, Nodal·Gdf1, Activin A, and Nodal were 0.3, 4, 10, and >100 ng/ml, respectively (Fig. 6A). Furthermore, although recombinant Nodal lost most of its activity at concentrations of FBS below 2%, purified Nodal·Gdf1 was able to signal without FBS (Fig. 6B). Because Nodal·Gdf1 had a higher specific activity than Nodal and was independent of serum, we compared its potential with that of commercially available Nodal and Activin A to differentiate pluripotent human ES cells into DE (2). First, treatment of hESCs with only Wnt3a and basic FGF already resulted in the down-regulation of the pluripotency marker OCT4 and significant up-regulation of DE markers such as SOX17 (Fig. 6C). Next, we probed the effect of Nodal·Gdf1, Nodal, and Activin A. Enrichment of an hESC-derived definitive endoderm population (CXCR4+EpCamhi) was more efficient with Nodal·Gdf1 compared with Nodal but below that obtained with Activin A (Fig. 6D). Analysis of DE markers showed a significant increase of SOX17 mRNA with Nodal·Gdf1 (Fig. 6E). FOXA2 showed a similar trend but did not reach significance. In both cases, Nodal did not lead to any significant increase in DE marker expression. SOX3, an early ectoderm marker, decreased upon Nodal·Gdf1 treatment, whereas OCT4 (pluripotency marker) was not further decreased by either Nodal or Nodal·Gdf1 treatment. These results indicate that Nodal·Gdf1 is superior to Nodal to induce endoderm progenitor differentiation in vitro.
FIGURE 6.
Purified Nodal·Gdf1 complex has high specific activity in serum-free conditions and stimulates differentiation of hESCs into endoderm progenitors. A, induction of CAGA-luc by recombinant TGFβ (dotted line), Activin (gray line), Nodal (dashed line), and Nodal·Gdf1 (black line). B, induction of CAGA-luc by recombinant Nodal (50 ng/ml) or affinity-purified Nodal·Gdf1 (12.5 ng/ml) previously incubated in medium containing decreasing FBS concentrations. C, immunohistochemistry analysis of SOX17 and OCT4 in undifferentiated hESCs and cells differentiated with Wnt3a and basic FGF (Vehicle). D, CXCR4 and EpCam FACS profile of hESCs treated with 100 ng/ml Activin A, Nodal, Nodal·Gdf1, or vehicle. Asterisks indicate p values relative to the vehicle control. *, p ≤ 0.05; ***, p ≤ 0.001. E, RT-PCR analysis of selected markers following treatment of hESCs with the same factors. p values (above the graph) were calculated relative to the vehicle control. In A and B, error bars represent S.D. In E, error bars represent S.E.
Cryptic Shows Increased Selectivity for Nodal Heterodimers
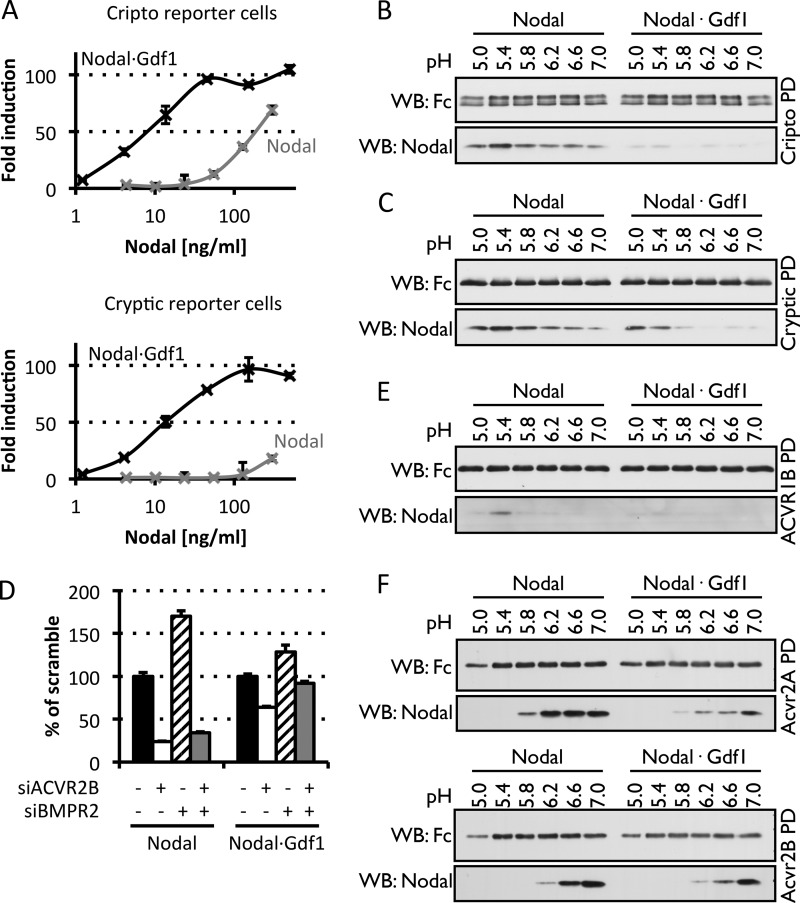
Although Nodal can signal independently of Gdf1 when co-transfected together with Cripto or Cryptic in the same cells (39, 40), signaling via Cryptic during left-right patterning seems to strictly depend on Gdf1 (22). To compare Nodal·Gdf1 signaling with each co-receptor side by side, we generated the HepG2 reporter cell line H-Cryptic, which expresses CAGA-luc and Renilla luciferase together with Cryptic instead of Cripto. H-CrCR and H-Cryptic cells induced similar levels of CAGA-luc when treated with Nodal·Gdf1. In sharp contrast, Nodal signaling without Gdf1 was clearly attenuated in H-Cryptic cells compared with H-CrCR cells (Fig. 7A). To investigate why Cryptic barely potentiates secreted Nodal activity when Gdf1 is absent, we monitored binding of soluble Fc fusions of each co-receptor to Nodal and Nodal·Gdf1 in pulldown assays. Within a pH range that mimics conditions at the plasma membrane or in endosomes, Fc-Cripto readily pulled down Nodal with optimal binding around pH 5.4 (Fig. 7B). Fc-Cripto also pulled down Nodal·Gdf1 although to a lesser extent. By contrast, Fc-Cryptic at acidic pH strongly interacted with both Nodal and Nodal·Gdf1 (Fig. 7C). These results indicate that Nodal·Gdf1 binds soluble Cryptic with a higher affinity than soluble Cripto. However, they do not support the idea that secreted Nodal depends on Gdf1 simply to enhance binding to co-receptors.
FIGURE 7.
Gdf1 is needed for efficient signaling via Cripto or Cryptic but does not enhance binding of Nodal to soluble receptor Fc fusion proteins. A, induction of CAGA-luc by recombinant Nodal (gray) or purified Nodal·Gdf1 (black) in reporter cells expressing Cripto (top) or Cryptic (bottom). Western blot (WB) analysis of Fc-receptor fusion constructs and associated Nodal proteins following pulldown (PD) of Cripto-Fc (B) or Cryptic-Fc (C) fusion proteins using Gdf1(HA) is shown. D, induction of CAGA-luc by recombinant Nodal or Nodal·Gdf1(His6) following treatment of the reporter cells with siRNAs against ACVR2B or BMPR2. Western blot analysis of Fc-receptor fusion constructs and associated Nodal proteins following pulldown of ACVR1B-Fc (E) or Acvr2a-Fc and Acvr2b-Fc (F) fusion proteins using Gdf1(HA) is shown. In A and D, error bars represent S.D.
Heterodimerization of Nodal with Gdf1 Allows Inhibition by Soluble Activin Receptors
Genetic evidence suggests that Nodal signaling in utero is mediated by complexes of ACVR1B (ALK4) with ACVR2A or ACVR2B (for a review, see Ref. 10). To directly assess the effects of different type II receptors on Nodal versus Nodal·Gdf1 signaling, we transfected H-CrCR reporter cells with established siRNAs (34, 35). Depletion of 70% of ACVR2B mRNA in H-CrCR cells reduced the induction of CAGA-luc by recombinant Nodal to less than 30% (Fig. 7D). RNAi of ACVR2B also reduced Nodal·Gdf1 signaling, albeit to a lesser extent. Among possible alternative type II receptors, we also considered Bmpr2 because a subset of Nodal target genes fails to be induced both in Bmpr2−/− mouse embryos and in double mutants lacking Gdf1 and Gdf3 (21, 41, 42). We found that depletion of BMPR2 in H-CrCR cells neither inhibited the activity of Nodal alone nor that of Nodal·Gdf1 but rather increased it (Fig. 7D). To directly assess binding to signaling receptors, we monitored pull down of Nodal and Nodal·Gdf1 by soluble Fc fusions of type I or type II receptor extracellular domains. ACVR1B-Fc only bound Nodal at an acidic pH around pH 5.4 and only in trace amounts that were further reduced down to the detection limit if Nodal was coexpressed with Gdf1 (Fig. 7E). By comparison, Nodal bound more efficiently to Fc fusions of Acvr2a or Acvr2b through an interaction that was optimal at neutral pH but that again decreased in the presence of Gdf1 rather than being increased (Fig. 7F). As an alternative readout of ligand binding, we also evaluated the potential of soluble receptor Fc fusions to inhibit signaling in H-CrCR reporter cells. Preincubation with excess soluble Cripto, Cryptic, or ACVR1B had no specific effect on Nodal or Nodal·Gdf1 signaling compared with mock treatment (data not shown). Also, an Fc fusion of leukemia inhibitory factor used as a negative control had no effect (Fig. 8A). By contrast, soluble Acvr2a and Acvr2b dose-dependently blocked Activin A and Nodal·Gdf1 (Fig. 8, A and B). Unexpectedly, however, neither of the Acvr2 receptor fusions significantly inhibited Gdf1-independent Nodal signaling even when added in large molar excess (Fig. 8A). The only Fc fusion that bound and inhibited both Nodal·Gdf1 and Nodal consisted of Bmpr2 (Fig. 8, C and D). This inhibition was specific because Bmpr2-Fc did not significantly affect the activity of Activin A (Fig. 8B). Thus, although Nodal alone is sufficient to bind receptors and co-receptors, a biologically active conformation susceptible to inhibition by soluble Acvr2 depends on Gdf1.
DISCUSSION
Current efforts in regenerative medicine seek to derive functional tissue transplants from pluripotent stem cells by in vitro differentiation. Here, we have produced a heterodimer of Nodal with Gdf1 and reported its superiority to Nodal alone in directing the derivation of endoderm progenitors from hESCs. A purified complex of Nodal and Gdf1 contained mature heterodimers together with the corresponding prodomains. When added to a novel Smad3 luciferase reporter cell line expressing the co-receptor Cripto, processed heterodimeric Nodal·Gdf1 displayed a more than 25-fold higher specific activity than Nodal alone. Gdf1 also signaled synergistically with a cleavage-resistant mutant Nodal precursor, albeit less efficiently. Furthermore, although recombinant Nodal without Gdf1 required heat-sensitive, high molecular weight components from serum to remain active, heterodimerization with Gdf1 suppressed this serum dependence. Not only in the absence but also in the presence of serum, Gdf1 further increased Nodal activity. Surprisingly, however, Gdf1 did not stimulate binding of Nodal to extracellular domains of either type I or type II activin receptors or to EGF-CFC co-receptors in cell-free assays. We therefore propose that heterodimerization with Gdf1 potentiates Nodal signaling indirectly by stabilizing a free LMW form of the mature protein in a complex with its propeptides.
Nodal·Gdf1 Forms an LMW Complex with Prodomains
To study the synergism between Nodal and Gdf1, we analyzed crude conditioned media of stably transduced HEK293T cells, and we purified active complexes by affinity and size exclusion chromatography. In conditioned medium lacking Gdf1, Nodal was unable to pass an ultrafiltration membrane with a size cutoff below 100 kDa. By contrast, ultrafiltration of Nodal·Gdf1 conditioned medium yielded ∼2% of Nodal in the flow-through. This difference in mobility was unexpected because mature Nodal and Nodal·Gdf1 dimers have similar predicted molecular weights of 24 and 25 kDa, respectively. The predicted sizes of their precursor dimers are also comparable (78 and 79 kDa, respectively). A likely explanation is that Nodal forms HMW complexes and only passes the ultrafiltration membrane if an LMW form is stabilized by Gdf1. When the HMW form in the retentate of ultrafiltrated conditioned medium was rediluted, it yielded each time a constant amount of Nodal·Gdf1 activity in the LMW flow-through in multiple successive rounds of ultrafiltration. Conversely, if the flow-through was subjected to another round of ultrafiltration, the LMW fraction passing the 100-kDa cutoff a second time was not increased. Instead, most of its activity returned to HMW complexes found in the retentate. We conclude that heterodimerization with Gdf1 shifts the equilibrium between HMW and LMW forms of Nodal toward an LMW form.
Affinity purification of Nodal·Gdf1 utilized a His6 tag after the precursor cleavage site of Gdf1. Bound proteins were eluted in two fractions termed Elu10 and Elu40. Elu10 only contained inactive Nodal that eluted at a low imidazole concentration without detectable Gdf1 and that associated with the void volume in a subsequent size exclusion purification, indicating that it consists of nonspecifically bound HMW complexes. By contrast, Nodal·Gdf1 in Elu40 showed high specific activity and eluted in the subsequent size exclusion chromatography as a 60-kDa LMW complex. Size exclusion chromatography did not further increase the specific activity, and affinity-purified Nodal·Gdf1 was stable at 4 °C or frozen for at least 1 year with no noticeable loss of activity. We have not further analyzed the composition of Nodal HMW complexes. In latent TGFβ, Cys-33 of the propeptide can mediate the formation of a large complex with latent TGFβ-binding protein-1 that promotes secretion and matrix binding (43, 44), but an analogous cysteine is not found in the propeptides of either Nodal or Gdf1 (37). The shift in the size of Nodal complexes by Gdf1 concurs with our interpretation that Gdf1 stabilizes an LMW form. Analysis by LC-MS/MS and SDS-PAGE indicates that purified Nodal·Gdf1 contains equimolar amounts of mature Nodal and Gdf1. The cleaved prodomains copurified with mature ligand, suggesting that they remain associated after precursor processing. Association of mature Nodal·Gdf1 with prodomains can explain the size of the LMW form observed by gel filtration (60 kDa).
Effects of Precursor Cleavage and Prodomains on Nodal·Gdf1 Activity
Prodomains may affect mature ligands in multiple ways. Here, we found that Gmat carrying only a heterologous signal sequence instead of the prodomain was inactive both with and without Nodal. The Gdf1 prodomain thus may be needed to secrete active heterodimers, stabilize active heterodimers, or both. In keeping with a stabilizing function, prodomains influence the stability of mature Nodal, BMP4, and Drosophila Gbb apparently by modulating endocytosis (26, 27, 45, 46). In sharp contrast to what we observed with Gmat, deletion of the Nodal prodomain in the corresponding truncated construct Nmat only partially inhibited the synergism with Gdf1. Only the prodomain of Gdf1 thus is necessary and sufficient for active Nodal·Gdf1 heterodimers to accumulate.
It has been shown previously that a cleavage mutant Gdf1 precursor lacking its PC recognition motif fails to synergize with Nodal when expressed in Xenopus oocytes (22). We found that an analogous PC-resistant mutant Gdf1, Gr, also failed to stimulate Smad3 signaling in mammalian cells. Uncleaved Gdf1 prodomain thus likely masks essential receptor-binding epitopes of mature ligand as described for latent TGFβ, and precursor cleavage may be essential to unfasten this straitjacket (37). Even after cleavage, mature TGFβ can be inhibited by a homodimer of its prodomain that is held together by interchain cystine bonds in a latent complex (47). However, these disulfides are not conserved in Gdf1 (37). Precursor cleavage thus may suffice to unfasten the straitjacket of the prodomain and unmask receptor-binding epitopes of Gdf1. In sharp contrast to Gr, an analogous cleavage-resistant mutant Nodal precursor did synergize, albeit less efficiently than wild-type Nodal. This suggests that the Nodal prodomain does not abolish receptor binding even if it is covalently attached. Synergism with Gdf1 may contribute to the residual activity of a NodalNr knock-in allele encoding convertase-resistant mutant Nodal precursor (12).
Interaction of Nodal with Serum
Unexpectedly, Gdf1 suppressed a hitherto unknown dependence of Nodal on serum. In previous studies, Nodal was active independently of Gdf1 in conditioned Opti-MEM (40). Here, however, we found that decreasing the serum content from 10 to 1% in DMEM reduced Nodal activity in conditioned medium by more than 70% except if Nodal was coexpressed with Gdf1. This might explain why Gdf1 is also indispensable for paracrine signaling of Nodal in Xenopus embryo injection assays (22). How serum supports Nodal activity is unknown. High speed centrifugation of conditioned media did not precipitate Nodal·Gdf1, suggesting that both proteins were soluble independently of serum. The stabilizing effect of serum on Nodal activity could not be recapitulated by nonspecific proteins, and it was heat-sensitive. Mass spectrometry identified the plasma protein α2-macroglobulin in precipitates of both Nodal and Nodal·Gdf1 in pulldown experiments. When bound to α2-macroglobulin, the related ligand TGFβ is inhibited in a manner that is counteracted by heparin (48–50). Heparin also binds recombinant Nodal, and sulfated glycosaminoglycans are required for Nodal·Gdf1 signaling during left-right axis formation (20). It will be interesting, therefore, to assess in future studies whether α2-macroglobulin or other serum components promote Nodal signaling indirectly e.g. by facilitating its mobilization from higher order complexes.
Pulldown of Nodal or Nodal·Gdf1 also enriched Hsp90 and actin, two proteins associated with exosomes (38). However, irrespective of the presence or absence of Gdf1, less than 2% of Nodal protein in HEK293T-conditioned medium partitioned with exosome markers in high speed centrifugation. Exosomes thus are unlikely to account for HMW forms of Nodal or Nodal·Gdf1. Although these in vitro experiments do not rule out a role for exosomes in the secretion or transport of Nodal in vivo, they clearly suggest that Gdf1 does not depend on exosomes to potentiate Nodal signaling.
Gdf1 Is Essential for Efficient Paracrine Nodal Signaling via Cryptic
Even in complete medium containing serum, coexpression with Gdf1 still increased the specific activity of Nodal more than 100-fold (Fig. 1E). The synergism required the presence of Cripto as described previously (21). However, during left-right axis formation, Nodal·Gdf1 signaling depends on the related EGF-CFC co-receptor Cryptic (19). We found that although Cryptic stimulated Nodal·Gdf1 activity as efficiently as Cripto, it barely supported Nodal signaling. This was unexpected because Cryptic binds Nodal in cell-free assays and clearly stimulates its activity if coexpressed in the same cells (40). In contrast, our conditions selectively monitor cell-non-autonomous Nodal·Gdf1 activity and thus are more akin to paracrine signaling as it occurs from the node to the lateral plate mesoderm during left-right axis formation (20, 22). The striking bias of Cryptic to support cell-non-autonomous Nodal signaling only when Gdf1 is present might explain at least in part why loss of Gdf1 disrupts left-right axis formation.
Nodal Does Not Depend on Gdf1 to Bind Soluble Receptors in Pulldown Assays
To assess whether soluble secreted Nodal depends on Gdf1 to efficiently access EGF-CFC or signaling receptors or to stabilize binding in acidic compartments, we used soluble extracellular domains in pulldown assays. We found that both Nodal and Nodal·Gdf1 bound soluble EGF-CFC co-receptors within a broad pH range and that binding was optimal at pH 5.4. A similar acidic pH was necessary to detect binding to soluble ACVR1B. By contrast, the extracellular domains of Acvr2a and Acvr2b pulled down Nodal and Nodal·Gdf1 at a neutral pH optimum. These results are consistent both with a potential role for Acvr2 in Nodal uptake at the plasma membrane and with our earlier conclusion that Cripto stabilizes active signaling complexes in acidic endosomes (15, 16). Importantly, however, heterodimerization with Gdf1 did not enhance in vitro binding of Nodal to any of the receptors or co-receptors examined. These observations do not support a model whereby Gdf1 potentiates Nodal signaling simply by increasing its affinity for receptors.
As an alternative readout of receptor binding, we assessed the influence of soluble receptor fusion proteins on Nodal or Nodal·Gdf1 signaling in reporter cells. Although neither soluble ACVR1B nor EGF-CFC efficiently competed with endogenous receptors, preincubation with a 10-fold molar excess of Acvr2a or Acvr2b Fc fusion protein abrogated Nodal·Gdf1 activity, concurring with the dominant negative effect described in Xenopus (51). In sharp contrast, the same soluble Activin type II receptors failed to block Gdf1-independent Nodal signaling. Therefore, and because deletion of either Nodal or Bmpr2 in mice elicits partially overlapping phenotypes (41, 42), we considered whether Nodal and Nodal·Gdf1 assemble distinct heteromeric receptor complexes that differ in the content of BMP receptor subunits. We found that soluble Bmpr2, which does not inhibit Activin A (Ref. 52 and this study), pulled down Nodal as well as Nodal·Gdf1 and neutralized their activities. However, transfection of H-CrCR reporter cells with siRNA directed against BMPR2 did not inhibit Nodal or Nodal·Gdf1 signaling but rather stimulated it. In keeping with this result, deletion of Bmpr2 in early mouse embryos leads to increased pSmad2 activation by Acvr2b (42). These observations are compatible with a potential role for Bmpr2 as a decoy receptor of Acvr2 ligands as described in pulmonary smooth muscle cells (53). However, they do not explain why soluble Acvr2 selectively inhibits Nodal·Gdf1 but not Nodal.
In Drosophila, heterodimers of the related proteins Dpp and Scw synergize by assembling heteromeric complexes of two type I receptors, Tkv and Sax, with BMP type II receptor Punt, whereas the activity of Dpp homodimers is transduced by Punt with Tkv alone (54). Heterodimerization with Scw also increases the affinity of Dpp for a complex of short-of-gastrulation (Sog) and twisted gastrulation (Tsg) that facilitates transport in the extracellular matrix until Sog is cleaved by the metalloprotease Tolloid at the dorsal midline to liberate the Dpp·Scw heterodimer for signaling (54–57). Our results do not formally exclude a role for heteromeric receptor complexes in Nodal·Gdf1 signal transduction as we have not tested whether Acvr2b can be recruited together with Acvr2a or whether Acvr1b can synergize with the alternative Nodal/Activin type I receptor Alk7 (58). However, because neither Alk7 nor Acvr2a are essential in vivo, they are unlikely responsible for the synergism of Nodal and Gdf1 (59, 60). In our experimental setup, synergistic signaling is also not a consequence of facilitated transport through a tissue. Therefore, the heterodimerization with Gdf1 more likely increases the specific activity of Nodal by stimulating its release from poorly active HMW complexes or by stabilizing mature ligand in an LMW complex with propeptides.
Conclusion
In summary, our results suggest that Gdf1 can synergize with Nodal by several mechanisms: heterodimerization with Gdf1 bypasses the requirement of Nodal for extracellular proteins present in serum, it mobilizes an LMW form that shows greatly increased specific activity in both Cripto and Cryptic reporter cell lines, and it enables the use of Cryptic as a co-receptor in cells receiving Nodal·Gdf1 from separate cells. Gdf1 also renders Nodal susceptible to inhibition by the extracellular domains of truncated soluble Acvr2 receptors, indicating that soluble Acvr2 may selectively inhibit the LMW form of Nodal that depends on Gdf1.
Recombinant Nodal·Gdf1 emerges as a new tool for stem cell research. Current stem cell derivation and in vitro differentiation protocols rely on Activin as an alternative to non-physiological concentrations of commercially available recombinant Nodal. Our results show that commercial rNodal depends on serum to remain active, limiting its use in chemically defined media. By contrast, Nodal·Gdf1 activity was serum-independent, and it more potently induced definitive endoderm markers than Nodal in a serum-free protocol of human ES cell differentiation. However, Nodal·Gdf1 and Nodal alone failed to down-regulate OCT4 transcript expression, and endoderm progenitors did not form at the same frequency as with Activin A under the conditions examined. Possibly, Activin reduces OCT4 expression in hES cells by inhibiting the cell cycle (61). In keeping with this idea, gene expression profiling in Xenopus embryos suggests that cell proliferation is regulated by Activin but not by Nodal signaling (62). It will be interesting to determine in future studies whether endoderm progenitors obtained with Nodal·Gdf1 have a similar advantage as those induced by a high dose of Nodal to differentiate into functional mature cells and whether potential functional differences between Nodal·Gdf1 and Activin are linked to distinct effects on the cell cycle.
Acknowledgments
We thank David Hacker from the Ecole Polytechnique Fédérale de Lausanne (EPFL) Protein Expression Core Facility for the M1 antibody, leukemia inhibitory factor-Fc protein fusion standards, and helpful discussions; Marc Moniatte from the EPFL Proteomics Core Facility (PCF) for expert advice and discussions; Romain Hamelin and Diego Chiappe from the PCF for LC-MS/MS analyses; and Adrian Schmid (PCF) for support with the ÄKTA FPLC. We thank Aleksandra Deczkowska for the initial characterization of interactions between soluble Acvr2 receptor Fc fusion proteins and Nodal proteins and Stéphane Baflast for technical assistance.
Footnotes
- Acvr
- Activin receptor
- Gdf
- growth and differentiation factor
- BMP
- bone morphogenetic protein
- BMPR
- bone morphogenetic protein receptor
- luc
- luciferase
- hESC
- human ES cell
- Npro
- Nodal prodomain
- Nmat
- Nodal, constitutively mature mutant form
- Nr
- cleavage-resistant Nodal precursor
- H-CrCR
- HepG2-Cripto·CAGA·Rluc
- H-Cryptic
- HepG2-CAGA·Rluc·Cryptic
- EC50
- half-minimal effective concentration
- rNodal
- recombinant Nodal
- PBSN
- PBS containing Nonidet P-40
- DE
- definitive endoderm
- EpCam
- epithelial cell adhesion molecule
- PC
- proprotein convertase
- Gr
- mutated PC-resistant Gdf1 precursor
- Gpro
- Gdf1 prodomain
- Gmat
- Gdf1 constitutively mature form
- LMW
- low molecular weight
- HMW
- high molecular weight
- EGF-CFC
- epidermal growth factor-like, Cripto, FRL-1, Cryptic family.
REFERENCES
- 1. Arnold S. J., Robertson E. J. (2009) Making a commitment: cell lineage allocation and axis patterning in the early mouse embryo. Nat. Rev. Mol. Cell Biol. 10, 91–103 [DOI] [PubMed] [Google Scholar]
- 2. Nostro M. C., Sarangi F., Ogawa S., Holtzinger A., Corneo B., Li X., Micallef S. J., Park I. H., Basford C., Wheeler M. B., Daley G. Q., Elefanty A. G., Stanley E. G., Keller G. (2011) Stage-specific signaling through TGFβ family members and WNT regulates patterning and pancreatic specification of human pluripotent stem cells. Development 138, 861–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. D'Amour K. A., Agulnick A. D., Eliazer S., Kelly O. G., Kroon E., Baetge E. E. (2005) Efficient differentiation of human embryonic stem cells to definitive endoderm. Nat. Biotechnol. 23, 1534–1541 [DOI] [PubMed] [Google Scholar]
- 4. McLean A. B., D'Amour K. A., Jones K. L., Krishnamoorthy M., Kulik M. J., Reynolds D. M., Sheppard A. M., Liu H., Xu Y., Baetge E. E., Dalton S. (2007) Activin a efficiently specifies definitive endoderm from human embryonic stem cells only when phosphatidylinositol 3-kinase signaling is suppressed. Stem Cells 25, 29–38 [DOI] [PubMed] [Google Scholar]
- 5. Brons I. G., Smithers L. E., Trotter M. W., Rugg-Gunn P., Sun B., Chuva de Sousa Lopes S. M., Howlett S. K., Clarkson A., Ahrlund-Richter L., Pedersen R. A., Vallier L. (2007) Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature 448, 191–195 [DOI] [PubMed] [Google Scholar]
- 6. Vallier L., Touboul T., Brown S., Cho C., Bilican B., Alexander M., Cedervall J., Chandran S., Ahrlund-Richter L., Weber A., Pedersen R. A. (2009) Signaling pathways controlling pluripotency and early cell fate decisions of human induced pluripotent stem cells. Stem Cells 27, 2655–2666 [DOI] [PubMed] [Google Scholar]
- 7. Touboul T., Hannan N. R., Corbineau S., Martinez A., Martinet C., Branchereau S., Mainot S., Strick-Marchand H., Pedersen R., Di Santo J., Weber A., Vallier L. (2010) Generation of functional hepatocytes from human embryonic stem cells under chemically defined conditions that recapitulate liver development. Hepatology 51, 1754–1765 [DOI] [PubMed] [Google Scholar]
- 8. Tada S., Era T., Furusawa C., Sakurai H., Nishikawa S., Kinoshita M., Nakao K., Chiba T., Nishikawa S. (2005) Characterization of mesendoderm: a diverging point of the definitive endoderm and mesoderm in embryonic stem cell differentiation culture. Development 132, 4363–4374 [DOI] [PubMed] [Google Scholar]
- 9. Chen A. E., Borowiak M., Sherwood R. I., Kweudjeu A., Melton D. A. (2013) Functional evaluation of ES cell-derived endodermal populations reveals differences between Nodal and Activin A-guided differentiation. Development 140, 675–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schier A. F. (2003) Nodal signaling in vertebrate development. Annu. Rev. Cell Dev. Biol. 19, 589–621 [DOI] [PubMed] [Google Scholar]
- 11. Beck S., Le Good J. A., Guzman M., Ben Haim N., Roy K., Beermann F., Constam D. B. (2002) Extraembryonic proteases regulate Nodal signalling during gastrulation. Nat. Cell Biol. 4, 981–985 [DOI] [PubMed] [Google Scholar]
- 12. Ben-Haim N., Lu C., Guzman-Ayala M., Pescatore L., Mesnard D., Bischofberger M., Naef F., Robertson E. J., Constam D. B. (2006) The Nodal precursor acting via activin receptors induces mesoderm by maintaining a source of its convertases and BMP4. Dev. Cell 11, 313–323 [DOI] [PubMed] [Google Scholar]
- 13. Blanchet M.-H., Le Good J. A., Mesnard D., Oorschot V., Baflast S., Minchiotti G., Klumperman J., Constam D. B. (2008) Cripto recruits Furin and PACE4 and controls Nodal trafficking during proteolytic maturation. EMBO J. 27, 2580–2591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shen M. M. (2007) Nodal signaling: developmental roles and regulation. Development 134, 1023–1034 [DOI] [PubMed] [Google Scholar]
- 15. Blanchet M.-H., Le Good J. A., Oorschot V., Baflast S., Minchiotti G., Klumperman J., Constam D. B. (2008) Cripto localizes Nodal at the limiting membrane of early endosomes. Sci. Signal. 1, ra13. [DOI] [PubMed] [Google Scholar]
- 16. Constam D. B. (2009) Riding shotgun: a dual role for the epidermal growth factor-Cripto/FRL-1/Cryptic protein Cripto in Nodal trafficking. Traffic 10, 783–791 [DOI] [PubMed] [Google Scholar]
- 17. Chu J., Shen M. M. (2010) Functional redundancy of EGF-CFC genes in epiblast and extraembryonic patterning during early mouse embryogenesis. Dev. Biol. 342, 63–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. D'Andrea D., Liguori G. L., Le Good J. A., Lonardo E., Andersson O., Constam D. B., Persico M. G., Minchiotti G. (2008) Cripto promotes A-P axis specification independently of its stimulatory effect on Nodal autoinduction. J. Cell Biol. 180, 597–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yan Y. T., Gritsman K., Ding J., Burdine R. D., Corrales J. D., Price S. M., Talbot W. S., Schier A. F., Shen M. M. (1999) Conserved requirement for EGF-CFC genes in vertebrate left-right axis formation. Genes Dev. 13, 2527–2537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Oki S., Hashimoto R., Okui Y., Shen M. M., Mekada E., Otani H., Saijoh Y., Hamada H. (2007) Sulfated glycosaminoglycans are necessary for Nodal signal transmission from the node to the left lateral plate in the mouse embryo. Development 134, 3893–3904 [DOI] [PubMed] [Google Scholar]
- 21. Andersson O., Bertolino P., Ibáñez C. F. (2007) Distinct and cooperative roles of mammalian Vg1 homologs GDF1 and GDF3 during early embryonic development. Dev. Biol. 311, 500–511 [DOI] [PubMed] [Google Scholar]
- 22. Tanaka C., Sakuma R., Nakamura T., Hamada H., Saijoh Y. (2007) Long-range action of Nodal requires interaction with GDF1. Genes Dev. 21, 3272–3282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cheng S. K., Olale F., Bennett J. T., Brivanlou A. H., Schier A. F. (2003) EGF-CFC proteins are essential coreceptors for the TGF-β signals Vg1 and GDF1. Genes Dev. 17, 31–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wall N. A., Craig E. J., Labosky P. A., Kessler D. S. (2000) Mesendoderm induction and reversal of left-right pattern by mouse Gdf1, a Vg1-related gene. Dev. Biol. 227, 495–509 [DOI] [PubMed] [Google Scholar]
- 25. Andersson O., Reissmann E., Jörnvall H., Ibáñez C. F. (2006) Synergistic interaction between Gdf1 and Nodal during anterior axis development. Dev. Biol. 293, 370–381 [DOI] [PubMed] [Google Scholar]
- 26. Le Good J. A., Joubin K., Giraldez A. J., Ben-Haim N., Beck S., Chen Y., Schier A. F., Constam D. B. (2005) Nodal stability determines signaling range. Curr. Biol. 15, 31–36 [DOI] [PubMed] [Google Scholar]
- 27. Constam D. B., Robertson E. J. (1999) Regulation of bone morphogenetic protein activities by pro domains and proprotein convertases. J. Cell Biol. 144, 139–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hobbs S., Jitrapakdee S., Wallace J. C. (1998) Development of a bicistronic vector driven by the human polypeptide chain elongation factor 1α promoter for creation of stable mammalian cell lines that express very high levels of recombinant proteins. Biochem. Biophys. Res. Commun. 252, 368–372 [DOI] [PubMed] [Google Scholar]
- 29. Fuerer C., Habib S. J., Nusse R. (2010) A study on the interactions between heparan sulfate proteoglycans and Wnt proteins. Dev. Dyn. 239, 184–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dull T., Zufferey R., Kelly M., Mandel R. J., Nguyen M., Trono D., Naldini L. (1998) A third-generation lentivirus vector with a conditional packaging system. J. Virol. 72, 8463–8471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hampf M., Gossen M. (2006) A protocol for combined Photinus and Renilla luciferase quantification compatible with protein assays. Anal. Biochem. 356, 94–99 [DOI] [PubMed] [Google Scholar]
- 32. Théry C., Amigorena S., Raposo G., Clayton A. (2006) Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. Chapter 3, Unit 3.22 [DOI] [PubMed] [Google Scholar]
- 33. Nostro M. C., Cheng X., Keller G. M., Gadue P. (2008) Wnt, activin, and BMP signaling regulate distinct stages in the developmental pathway from embryonic stem cells to blood. Cell Stem Cell 2, 60–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Xia Y., Yu P. B., Sidis Y., Beppu H., Bloch K. D., Schneyer A. L., Lin H. Y. (2007) Repulsive guidance molecule RGMa alters utilization of bone morphogenetic protein (BMP) type II receptors by BMP2 and BMP4. J. Biol. Chem. 282, 18129–18140 [DOI] [PubMed] [Google Scholar]
- 35. Perron J. C., Dodd J. (2009) ActRIIA and BMPRII type II BMP receptor subunits selectively required for Smad4-independent BMP7-evoked chemotaxis. PLoS One 4, e8198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gray A. M., Mason A. J. (1990) Requirement for activin A and transforming growth factor-β 1 pro-regions in homodimer assembly. Science 247, 1328–1330 [DOI] [PubMed] [Google Scholar]
- 37. Shi M., Zhu J., Wang R., Chen X., Mi L., Walz T., Springer T. A. (2011) Latent TGF-β structure and activation. Nature 474, 343–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Théry C., Zitvogel L., Amigorena S. (2002) Exosomes: composition, biogenesis and function. Nat. Rev. Immunol. 2, 569–579 [DOI] [PubMed] [Google Scholar]
- 39. Chen C., Shen M. M. (2004) Two modes by which Lefty proteins inhibit nodal signaling. Curr. Biol. 14, 618–624 [DOI] [PubMed] [Google Scholar]
- 40. Yan Y. T., Liu J. J., Luo Y., E., C., Haltiwanger R. S., Abate-Shen C., Shen M. M. (2002) Dual roles of Cripto as a ligand and coreceptor in the nodal signaling pathway. Mol. Cell. Biol. 22, 4439–4449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Beppu H., Kawabata M., Hamamoto T., Chytil A., Minowa O., Noda T., Miyazono K. (2000) BMP type II receptor is required for gastrulation and early development of mouse embryos. Dev. Biol. 221, 249–258 [DOI] [PubMed] [Google Scholar]
- 42. Yamamoto M., Beppu H., Takaoka K., Meno C., Li E., Miyazono K., Hamada H. (2009) Antagonism between Smad1 and Smad2 signaling determines the site of distal visceral endoderm formation in the mouse embryo. J. Cell Biol. 184, 323–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Saharinen J., Taipale J., Keski-Oja J. (1996) Association of the small latent transforming growth factor-β with an eight cysteine repeat of its binding protein LTBP-1. EMBO J. 15, 245–253 [PMC free article] [PubMed] [Google Scholar]
- 44. Rifkin D. B. (2005) Latent transforming growth factor-β (TGF-β) binding proteins: orchestrators of TGF-β availability. J. Biol. Chem. 280, 7409–7412 [DOI] [PubMed] [Google Scholar]
- 45. Degnin C., Jean F., Thomas G., Christian J. L. (2004) Cleavages within the prodomain direct intracellular trafficking and degradation of mature BMP-4. Mol. Biol. Cell 15, 5012–5020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Akiyama T., Marqués G., Wharton K. A. (2012) A large bioactive BMP ligand with distinct signaling properties is produced by alternative proconvertase processing. Sci. Signal. 5, ra28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Brunner A. M., Marquardt H., Malacko A. R., Lioubin M. N., Purchio A. F. (1989) Site-directed mutagenesis of cysteine residues in the pro region of the transforming growth factor β1 precursor. Expression and characterization of mutant proteins. J. Biol. Chem. 264, 13660–13664 [PubMed] [Google Scholar]
- 48. O'Connor-McCourt M. D., Wakefield L. M. (1987) Latent transforming growth factor-β in serum. A specific complex with α2-macroglobulin. J. Biol. Chem. 262, 14090–14099 [PubMed] [Google Scholar]
- 49. Huang S. S., O'Grady P., Huang J. S. (1988) Human transforming growth factor β·α2-macroglobulin complex is a latent form of transforming growth factor β. J. Biol. Chem. 263, 1535–1541 [PubMed] [Google Scholar]
- 50. McCaffrey T. A., Falcone D. J., Borth W., Weksler B. B. (1994) α2-Macroglobulin/transforming growth factor-β1 interactions. Modulation by heparin-like molecules and effects on vascular smooth muscle cells. Ann. N.Y. Acad. Sci. 737, 368–382 [DOI] [PubMed] [Google Scholar]
- 51. Hemmati-Brivanlou A., Melton D. A. (1992) A truncated activin receptor inhibits mesoderm induction and formation of axial structures in Xenopus embryos. Nature 359, 609–614 [DOI] [PubMed] [Google Scholar]
- 52. Frisch A., Wright C. V. (1998) XBMPRII, a novel Xenopus type II receptor mediating BMP signaling in embryonic tissues. Development 125, 431–442 [DOI] [PubMed] [Google Scholar]
- 53. Yu P. B., Beppu H., Kawai N., Li E., Bloch K. D. (2005) Bone morphogenetic protein (BMP) type II receptor deletion reveals BMP ligand-specific gain of signaling in pulmonary artery smooth muscle cells. J. Biol. Chem. 280, 24443–24450 [DOI] [PubMed] [Google Scholar]
- 54. Shimmi O., Umulis D., Othmer H., O'Connor M. B. (2005) Facilitated transport of a Dpp/Scw heterodimer by Sog/Tsg leads to robust patterning of the Drosophila blastoderm embryo. Cell 120, 873–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Marqués G., Musacchio M., Shimell M. J., Wünnenberg-Stapleton K., Cho K. W., O'Connor M. B. (1997) Production of a DPP activity gradient in the early Drosophila embryo through the opposing action of the SOG and TLD proteins. Cell 91, 417–426 [DOI] [PubMed] [Google Scholar]
- 56. Srinivasan S., Rashka K. E., Bier E. (2002) Creation of a Sog morphogen gradient in the Drosophila embryo. Dev. Cell 2, 91–101 [DOI] [PubMed] [Google Scholar]
- 57. Sawala A., Sutcliffe C., Ashe H. L. (2012) Multistep molecular mechanism for bone morphogenetic protein extracellular transport in the Drosophila embryo. Proc. Natl. Acad. Sci. U.S.A. 109, 11222–11227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Reissmann E., Jörnvall H., Blokzijl A., Andersson O., Chang C., Minchiotti G., Persico M. G., Ibáñez C. F., Brivanlou A. H. (2001) The orphan receptor ALK7 and the Activin receptor ALK4 mediate signaling by Nodal proteins during vertebrate development. Genes Dev. 15, 2010–2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Jörnvall H., Reissmann E., Andersson O., Mehrkash M., Ibáñez C. F. (2004) ALK7, a receptor for nodal, is dispensable for embryogenesis and left-right patterning in the mouse. Mol. Cell. Biol. 24, 9383–9389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Matzuk M. M., Kumar T. R., Bradley A. (1995) Different phenotypes for mice deficient in either activins or activin receptor type II. Nature 374, 356–360 [DOI] [PubMed] [Google Scholar]
- 61. Pauklin S., Vallier L. (2013) The cell-cycle state of stem cells determines cell fate propensity. Cell 155, 135–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ramis J. M., Collart C., Smith J. C. (2007) Xnrs and activin regulate distinct genes during Xenopus development: activin regulates cell division. PLoS One 2, e213. [DOI] [PMC free article] [PubMed] [Google Scholar]