Abstract
The exterior surface of nanoparticles (NPs) dictates the behavior of these systems with the outside world. Understanding the interactions of NP surface functionality with biosystems enables the design and fabrication of effective platforms for therapeutics, diagnostics, and imaging agents. In this review, we highlight the role of chemistry in the engineering of nanomaterials, focusing on the fundamental role played by surface chemistry in controlling the interaction of NPs with proteins and cells.
Keywords: Nanoparticles, Surface functionality, Nano-bio interface, Stimuli responsive nanoparticles, Bionanotechnology
1. Introduction
Nanoparticles (NPs) provide platforms for crucial biomedical applications including delivery[1] and imaging.[2] Advances in these biomedical applications require a fundamental understanding of the complex interactions between NPs, biomolecules, and biosystems.[3] Using this insight, the tools of chemical synthesis can be used to create nanomaterials that interact efficiently and predictably with biosystems including proteins,[4] nucleic acids,[5] cells,[6] and tissues.[7]
In this review, we highlight selected systems that illustrate how the chemical structure of NP surface functionality can be used to tune the interactions of particle with proteins and regulate cell uptake and distribution in tissues.[8] We also show how the surface can contribute functional attributes to the NPs, including targeting modalities[9] and the ability to respond to stimuli.[10]
2. Interactions of proteins with surface-engineered nanoparticles
2.1. Control of protein structure and function using nanoparticle receptors
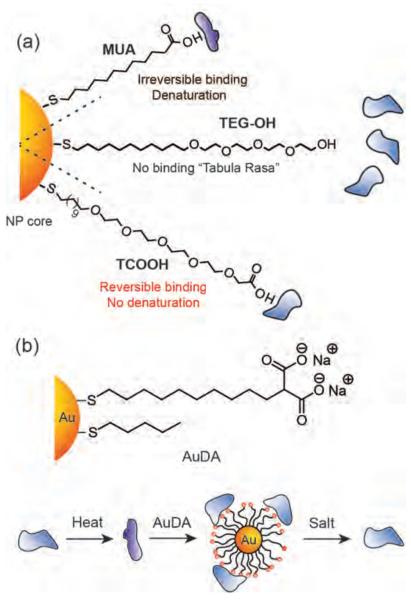
Selective binding of artificial receptors to proteins provides a means to modulate a variety of biomedically important cellular processes including protein-protein interactions, protein-nucleic acid interactions, and enzyme activity.[11] The nature of the interaction of NPs and proteins is an inherently biophysical question that builds upon the fundamentals of supramolecular chemistry.[12] In early studies, Rotello and coworkers demonstrated that 11-mercaptoundecanoic acid (MUA) functionalized gold NPs irreversibly inhibited the activity of chymotrypsin (ChT) with an apparent Ki of 10 nM (Figure 1a). [13] The mechanism of this interaction was attributed to a two-step process featuring a fast reversible association followed by a slower irreversible denaturation, and could be readily tuned by the ionic strength of the aqueous media[14] and addition of surfactants.[15]
Figure 1.
(a) Schematic representation of the effect of MUA, TEG-OH and TCOOH functionalized NPs on the structure and function of a cationic protein, chymotrypsin. (b) Schematic representation of the structure of AuDA, and AuDA-mediated refolding of thermally denatured proteins. Reproduced with permission from Ref [23]. Copyright 2008 Royal Society of Chemistry.
One inherent limitation of the use of simple ligands such as MUA is that protein structure and function were lost during the binding process. Preservation of the structure of NP surface-bound proteins is an essential prerequisite for many pragmatic applications including in vivo protein delivery and in vitro enzyme stabilization.[16] Structural integrity of proteins upon NP binding was achieved by introducing oligo(ethylene glycol) (OEG) tethers onto NP surfaces to provide TEG-OH and TCOOH (Figure 1a).[17] The incorporation of a short (at least four repeat units) [18] OEG segment on the exterior of MUA provided a non interacting `tabula rasa' of NPs that could be further functionalized with particular headgroups, allowing tailoring of the NP surface (Figure 1a). The stability of NP-ChT complexes was further investigated using gold NP featuring amino-acid side chains.[19] Binding studies using these NPs demonstrated that both electrostatic and hydrophobic interactions play a major role in the NP-ChT complex stability. Interestingly, the hydrophobic amino acid functionalized NPs did not affect the protein structure; however, hydrophilic amino acid functionalized NPs destabilized the native structure of ChT. This finding contradicts the general belief that hydrophobic surfaces lead to denaturation of proteins.[20] In a later study, detailed thermodynamic investigations of the interaction of protein and amino acid functionalized NPs demonstrated that the enthalpy and entropy changes for these interactions strongly mimic protein-protein interactions.[21]
Protein unfolding on NP surfaces can cause unwanted biological responses. For example, cryptic epitopes of proteins can be exposed that can further interact with cell membrane receptors, leading to aberrant inflammatory responses. Minchin et al. have shown that poly(acrylic acid)-coated gold NPs can selectively interact with fibrinogen in human plasma and expose an amino acid sequence of fibrinogen γ-chain, promoting the specific interaction with Mac-1 receptors on the cell surface. This interaction further leads to release of inflammatory cytokines from human monocyte cells via NF-κB signaling pathway.[22] However, appending appropriate functionality can improve the protein stability (vide supra), and even help in refolding the denatured proteins. Rotello et al. have demonstrated that 2-(10-mercaptodecyl) malonic acid functionalized gold NPs (AuDA) can refold thermally denatured cationic proteins, acting as artificial chaperones. The partially refolded proteins can be released from NP surfaces by changing the ionic strength of the solution (Figure 1b).[23]
2.2. NP surface chemistry regulates the affinity and identity of the `protein corona'
When surface-engineered NPs are exposed to complex protein biofluids (e.g. blood, plasma) a protein “corona” forms on the particle surface, masking its original chemical nature.[24] This non-specific adsorption of biomolecules on NP surfaces is important for delivery applications, increasing the clearance rate of NPs via the reticulo-endothelial system and decreasing the pharmacokinetic half-life of NPs in vivo. Blood plasma contains thousands of proteins that can bind to NPs with the interaction profile strongly depending on the NP surface chemistry. For example, positively charged polystyrene particles were shown to interact with proteins with isoelectric points less than 5.5 (e.g. serum albumin) while negatively charged particles preferentially bind to proteins with isoelectric points higher than 5.5 (e.g. IgG).[25] Muller et al. have further shown that negatively charged polymeric NPs of similar size and hydrophobicity adsorb higher amount of proteins on their surfaces with increasing surface charge density; however, no apparent difference was observed in the identity of the adsorbed proteins.[26]
The hydrophobicity of the NP surface is also an important parameter in NP-protein interactions.[27] For example, hydrophobic N-isopropylacrylamide-co-N-tert-butylacrylamide (NIPAM/BAM) copolymer NPs adsorb a greater amount of protein on their surfaces than their hydrophilic counterparts,[28] due to higher protein affinity and/or increased number of binding sites on hydrophobic NP surfaces.[29] NP surface hydrophobicity also regulates the nature of the protein corona formed on the surface. For example, hydrophobic NPs adsorb apolipoproteins on their surface; however, hydrophilic NPs tend to adsorb more abundant hydrophilic proteins in plasma including albumin and IgG.[30]
In addition to charge and hydrophobicity, interaction of proteins tends to depend on the specific functional groups on NP surfaces. For example, Shea et al. have synthesized three different cationic NIPAM polymeric NPs featuring guanidinium, primary amino, and quaternary ammonium functional groups (Figure 2a). The guanidinium functionalized polymer NPs showed significantly higher binding towards fibrinogen that is attributed to strong interactions with surface carboxylate groups on fibrinogen (Figure 2b).[31] Likewise, ~90 nm methylstyrene NPs showed significantly higher binding of plasma proteins compared to tert-butylstyrene NPs of similar charge and hydrophobicity.[27]
Figure 2.
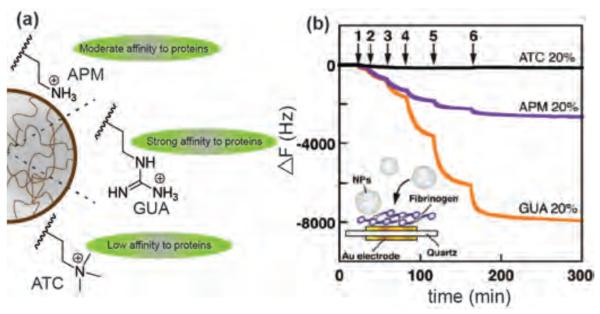
(a) Structure of guanidinium, primary amino, and quaternary ammonium functional groups in polymeric NPs. (b) Quartz crystal microbalance analysis of interactions between fibrinogen and positively charged NPs showing guanidinium functionalized polymeric NPs have the highest binding towards fibrinogen. Arrows indicate the time points of NP solution injections. Reproduced with permission from Ref [31]. Copyright 2012 American Chemical Society.
Even though numerous studies have documented the effect of surface chemistry on the formation of plasma protein coronas, a clear structure-function relationship still remains elusive. The inherent complexity of NP-protein interactions coupled with the rather disorganized array of particle sizes, shapes, and coverages studied makes it challenging to deduce general conclusions and providing impetus for parametric examination of this crucial process.
2.3. Engineering NP surface to prevent protein adsorption
Protein corona formation masks functionality and decreases circulatory lifetime of NPs. Grafting hydrophilic polymers on NP surfaces is a widely employed strategy to reduce non-specific protein adsorption. For example, polyethylene glycol (PEG), a charge-neutral highly hydrophilic polymer, is often incorporated on NP surfaces for in vitro and in vivo applications.[32] The advantage of NP surface modification with PEG is two-fold: 1) PEG delays the opsonization process, thereby reducing NP uptake in macrophage cells; 2) the improved circulation allows NPs to preferentially accumulate in tumor microenvironment exploiting enhanced permeability and retention effect.[33] Even though widely used for in vivo applications, PEG has several drawbacks. Chan et al. have shown that gold NPs with increasing surface PEG densities adsorb less serum proteins; however, even at high PEG grafting density the proteins were not fully removed.[34] Moreover, the different PEG densities influence the identity of the protein corona formed and subsequent macrophage uptake. Kiwada et al. have demonstrated siRNA encapsulated PEG-modified lipid nanocarriers can produce antibodies against them; therefore, subsequent doses of the same PEGylated NPs were rapidly cleared in vivo.[35] PEG can also create unwanted immune responses. Moghimi et al. have demonstrated activation of complement system by NPs functionalized with PEG,[36] harnessing adverse immune responses and facilitating clearance by macrophages.
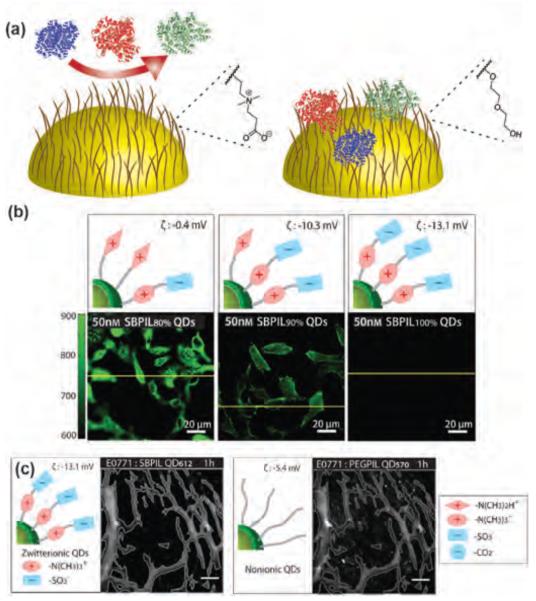
Recently, zwitterionic ligands including carboxybetaines and sulfobetaines have been introduced as `anti-fouling’ coatings on NP surfaces.[37] These functionalities provide much lower adsorption of proteins compared to PEG and very low non-specific cellular uptake, providing a potential alternative to PEG (Figure 3a).[38] Jiang et al. have demonstrated that poly (carboxybetaine)- functionalized gold NPs were highly stable in undiluted blood plasma and serum while PEG-coated NPs aggregated.[39] Likewise, Mattoussi and coworkers have synthesized zwitterionic quantum dots (QDs) that showed superior colloidal stability compared to simple lipoic acid functionalized QDs over a broad range of conditions including pH, salt, and undiluted serum.[40] Using zwitterionic mixed monolayer gold NPs, Rotello et al. have shown intracellular delivery of therapeutics while maintaining low uptake and minimal cytotoxicity from the NP itself (vide infra).[41] Mukherjee et al. have further demonstrated that zwitterionic gold NPs had higher blood circulation lifetime and enhanced tumor accumulation, while positively and negatively charged NPs were rapidly cleared.[42] However, the surface charge distribution of zwitterionic NPs can influence the uptake and biodistribution. Bawendi et al. have reported that zwitterionic QDs exposing positive charges in their outermost layer show non-specific adsorption in vitro and in vivo, whereas zwitterionic QDs exposing negative charges in their outermost layer are far less susceptible to interactions with proteins (Figure 3b–c).[43]
Figure 3.
(a) Illustration of protein adsorption on the surface of zwitterionic and PEGylated NPs. (b) Top: illustration of expected charge distribution on QDs. Bottom: non-specific binding of QDs with different charge distribution to HeLa cells. (c) Real-time intravital microscopy images of zwitterionic QDs and nonionic QDs in a murine breast tumor, demonstrating that extravasation of zwitterionic QDs from vessels is much less than extravasation of nonionic QDs. Scale bars: 100 mm. Reproduced with permission from Ref [43]. Copyright 2013 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim.
3. Interaction of cells with surface-engineered nanoparticles
3.1. Use of cell-penetrating peptides on NP surfaces
Penetration of the cell membrane and subsequent access of NPs to cytosol are two major hurdles for delivery of therapeutics. To overcome these challenges, NPs can be coated with peptide motifs that penetrate the cell membrane and can facilitate endosomal escape into the cytosol. Cell penetrating peptides (CPPs) including RGD,[44] allatostatin 1,[45] PLL,[46] and arginine-rich peptides[47] have been conjugated onto NP surfaces for efficient cellular delivery. Brust et al. have demonstrated cytosolic delivery of CPP conjugated gold NPs; addition of a nuclear localizing sequence (NLS) peptide resulted in localization to the nucleus.[48] In a similar study, gold NPs featuring NLS can penetrate to the nucleus after endosomal escape and induce DNA damage in cancer cells, demonstrating potential therapeutic strategy using surface-engineered NPs.[49] Gillies et al. have shown that superparamagnetic iron oxide NPs featuring dendritic guanidine moieties on their surface have similar cell-penetration efficiency with immunodeficiency virus-1 transactivator (HIV-TAT) peptide and showed better penetration efficiency than amine functionalized NPs.[50]
3.2. Effect of NP surface charge and hydrophobicity on cellular uptake
Engineering the NP surface provides a versatile alternative to CPPs for cellular delivery. The surface charge of the NP is a key parameter for NP-cell membrane interaction and subsequent intracellular internalization. In general, cationic NPs interact more strongly with the cell membrane due to the presence of negatively charged groups (e.g. sialic acid) onto cellular membranes, and hence show higher uptake efficiency compared to their anionic and neutral counterparts (Figure 4a).[51] However, efficient intracellular uptake of negatively charged NPs is known, presumably through pinocytosis or membrane diffusion.[52] Moreover, negatively charged QDs were shown to penetrate mouse skin, with higher degree of penetration observed upon exposure to UV light.[53] Likewise, DNA functionalized anionic gold NPs were shown to penetrate in epidermis layer of mouse skin with no apparent inflammation or toxicity.[54]
Figure 4.
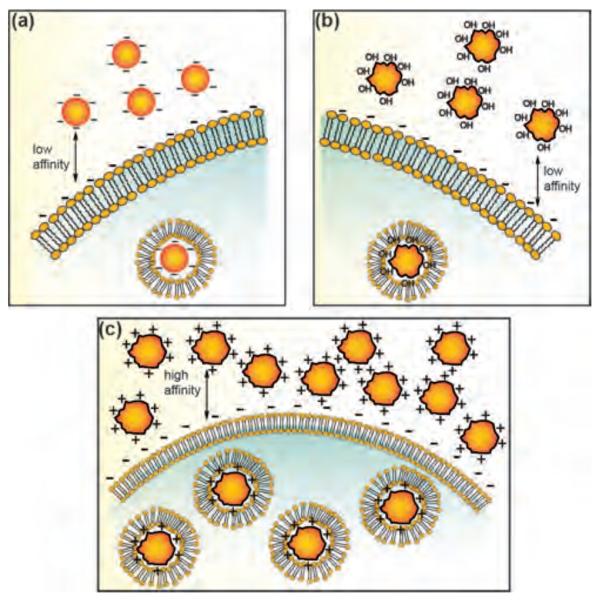
Schematic representation of the interaction between gold NPs bearing different surface charge and SK-BR-3 breast cancer cells. (a) Citrate-coated (negative), (b) PVA-coated (neutral), and (c) poly(allyamine hydrochloride)–coated (positive) NPs.
Two-dimensional cell culture models present a vastly different environment than tissues, complicating translation into in vivo systems. Rotello and Forbes et al. have investigated the role of NP surface charge in delivering covalently attached therapeutic drugs using a three-dimensional cell culture model. Significantly, cationic particles were better at delivering drugs on the proliferating peripheral cells due to their higher uptake; however, anionic particles had higher diffusion rates, delivering drug- and fluorophore-tagged NPs more rapidly to the center of the spheroid model (Figure 5).[55]
Figure 5.
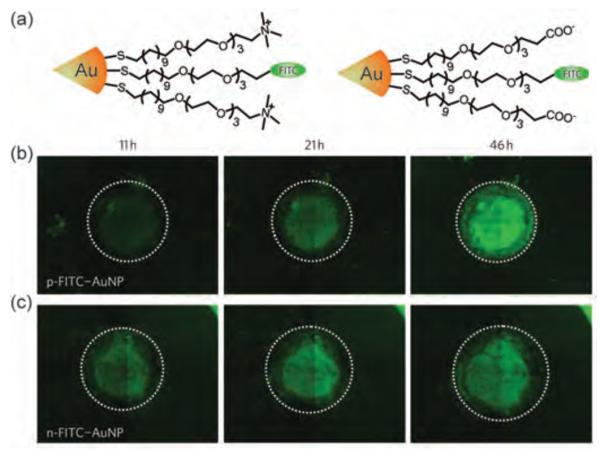
(a) Structure of mixed monolayer-protected cationic and anionic gold NPs loaded with thioalkylated fluorescein isothiocyanate (FITC). Green fluorescence images of tumor cylindroids treated with (b) cationic and (c) anionic particles. Reproduced with permission from Ref [55]. Copyright 2010 Nature Publishing Group.
In addition to surface charge, NP hydrophobicity plays an important role in the cellular uptake process. In one study, Mailänder et al. have showed that increasing surface hydrophobicity increases intracellular uptake of polymeric NPs in a variety of cell lines.[56] Rotello et al. have demonstrated a linear correlation of surface hydrophobicity and intracellular uptake of gold NPs (2 nm core), stemming from a stronger interaction of hydrophobic NPs with serum albumin.[57] However, in the absence of serum, no apparent trend between surface hydrophobicity and cellular uptake was observed, demonstrating the importance of serum proteins on the NP uptake process (vide supra).
4. Design of stimuli-responsive nanocarriers by tailoring surface functionality
In addition to regulating interactions with the environment, nanoparticle surfaces can be used to impart functionality to nanoparticles. “Smart” surface functionality provides a strategy for creating delivery systems that can release drugs at target site, minimizing potential off-target issues. Stimuli-responsive nanocarriers that respond to either a physiological (e.g. pH, enzyme) or external stimulus (e.g. magnetic field, light) can be used to enhance the therapeutic efficiency of delivery vehicles.[58]
4. 1. External Stimuli
4. 1. 1. Light-induced release of cargo
Light provides an excellent orthogonal stimulus for nanocarriers, providing spatio-temporal control of delivery. For example, Tamanoi and Zink et al. have reported the intracellular delivery and release of the anticancer drug camptothecin using nanoimpeller-controlled mesostructured silica nanoparticles that are functionalized with photo-switchable azobenzene moieties positioned in the pore interiors.[59] Lin et al. have presented a supramolecular assembly for visible light sensitive release of cargo from mesoporous silica nanoparticles (MS NPs) using Ru(bpy)2(PPh3) moieties coordinated to mercaptopropyl functional groups as gatekeepers.[60] Upon irradiation with visible light, the Ru–S coordination bond is cleaved and encapsulated molecules are released. Furthermore, Rotello et al. have shown the UV light-triggered release of fluorouracil from gold NP surface through a photoresponsive o-nitrobenzyl (ONB) linkage (Figure 6).[41]
Figure 6.
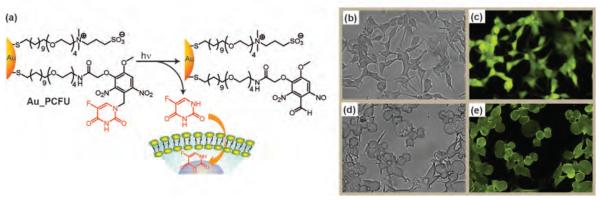
(a) Photochemical reaction of Au_PCFU and delivery of fluorouracil to cells. Bright-field and fluorescence-microscopy images of the cells exposed to UV before treated with Au_PCFU (b and c) and after treated with Au_PCFU (d and e). Reproduced with permission from Ref [41]. Copyright 2009 American Chemical Society.
UV light has a very short tissue penetration depth (~1 mm), making UV-activated systems unsuitable for deep tissue penetration. Branda et al. have addressed this issue by synthesizing NaYF4 upconverting NPs (UCNPs) doped with the lanthanides, Tm3+ and Yb3+ [NaYF4:TmYb] with a UV light activated caging group (3',5'-di(carboxymethoxy)benzoin) (Figure 7a).[61] These UCNPs were used as antennae for harvesting the NIR (980 nm) light and converting it into UV light (290 nm) (Figure 7a–b). This strategy was also employed in UCNPs containing hydrogels to release biomacromolecules on demand.[62]
Figure 7.
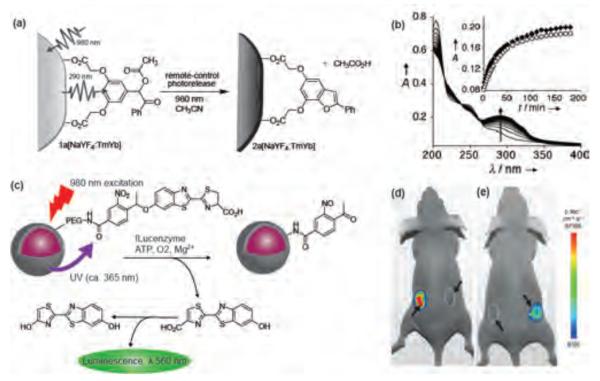
(a) Generation of UV light from NIR light using benzoin cage decorated UCNPs. (b) Changes in the UV/Vis absorption spectra of a solution of 1a[NaYF4:TmYb] when it is irradiated with 980 nm light. Reproduced with permission from Ref [61]. Copyright 2010 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim. (c) Schematic illustration of photocaged UCNPs synthesis and release of D-luciferin upon NIR irradiation and bioluminescence through the use of photocaged core-shell upconversion NPs. (d–e) Bioluminescent images of fLuc activity in living mice (d) Left: injection with D-luciferin and right: injection with photocaged nanoparticles without NIR light irradiation. (e) Left: injection with photocaged nanoparticles and irradiation with UV light and right: injection with photocaged nanoparticles and irradiation with NIR light. Reproduced with permission from Ref [63]. Copyright 2012 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim.
Recently, Xing et al. have demonstrated the use of UCNPs to uncage D-luciferin molecules caged with 1-(2-nitrophenyl)ethyl group in vitro and in vivo (Figure 7c).[63] The absorption band of the photocaged D-luciferin overlaps with the emission band of UCNPs in the UV region; thus disassociation of D-luciferin from the surface of the nanoparticle is triggered by excitation of UCNPs with NIR light. Released D-luciferin can recognize firefly luciferase (fLuc) reporter genes and generate a bioluminescence signal which is an indication of successful release of the cargo from the carrier (Figure 7d–e).[64] Their photocaged system has potential to selectively deliver payload in vivo with deep tissue penetration ability by NIR irradiation.
4.2. Endogenous stimulus for triggering the release of cargo
4.2.1 Non-covalent drug delivery through changes in environmental hydrophobicity
Endogenous release mechanisms use environmental changes in living systems to dictate their behavior. Non-covalent drug delivery systems using NPs can use either an encapsulation mechanism or a stabilizing pocket,[65] using electrostatic[66] and hydrophobic[67] interactions to reversibly bind the drug of interest.[68]
Burda et al. have demonstrated that encapsulating hydrophobic drug Pc4 inside the monolayer of PEG functionalized gold NPs increases the drug accumulation in the tumor. Significantly, little to no accumulation was observed when Pc4 was covalently attached to the gold NP surface, demonstrating that reversible association is required for therapeutic activity.[69] Rotello et al. have used hydrophobic pockets of gold NP monolayers to encapsulate highly hydrophobic dyes/therapeutics and release them upon interacting with cell membrane (Figure 8).[70] The zwitterionic surface of NPs further provided biocompatibility to the NP carriers. Hydrophobic payloads (tamoxifen and lapachone) were shown to be released into the cell by membrane-mediated diffusion without the uptake of the carrier NPs, demonstrating the desired biocompatibility of the carrier.
Figure 8.
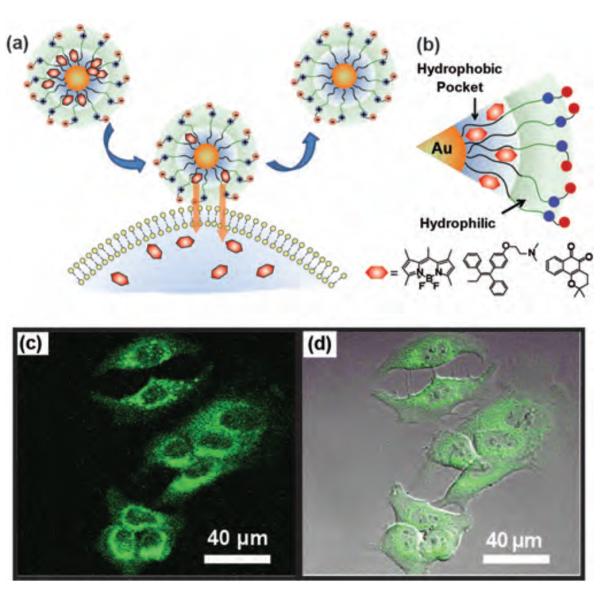
(a) Delivery of payload to cell due to change in hydrophobicity through monolayer-membrane interactions. (b) Schematic illustration of guest molecules entrapped in the hydrophobic pocket of zwitterionic NPs and structure of guest molecules: Bodipy, tamoxifen and lapachone. Confocal laser scanning microscopy images of MCF-7 cell treated with bodipy encapsulated zwitterionic NPs (c) green channel, (d) overlapped with bright field. Reproduced with permission from Ref [70]. Copyright 2009 American Chemical Society.
4.2.2. pH as an internal trigger to release the cargo
The average extracellular pH of solid tumors is 6 –7, lower than the pH of normal tissues and blood (7.4).[71] Surface functional groups that respond to acidic pH including acetal,[72] derivatized maleamate,[73] and hydrazone[74] have been used to attach the therapeutics to a variety of carriers for release at tumor sites. For example, Wang et al. have used carbamate linkage on a gold NP surface to release doxorubicin in response to the pH of acidic organelles after endocytosis.[75] They have further demonstrated that this delivery system can effectively inhibit the growth of multidrug-resistant MCF-7/ADR cancer cells, owing to the high endocytic uptake and subsequent pH responsive release in cells. Mirkin and Lippard et al. have combined the properties of DNA functionalized gold NPs and Pt(IV) prodrugs into a single agent for drug delivery.[76] They have engineered the surface of gold NPs to attach the prodrugs. They have demonstrated that the acidic environment in cancer cells facilitates reduction of the Pt(IV) and hence yields of the cytotoxic Pt(II) species (Figure 9).
Figure 9.
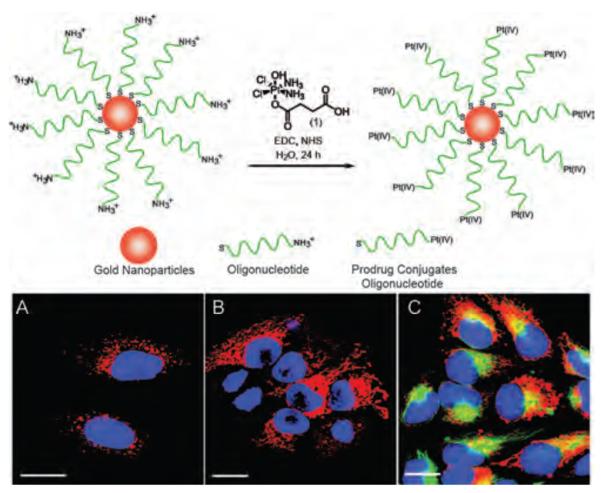
Top scheme-synthesis of Pt-DNA-Au NP. Bottom Live cell imaging of HeLa cells after incubation with platinum-tethered Cy5-DNA-Au NPs for (a) 6 h, (b) 12 h, and (c) colocalization of the particles with the cytoplasmic microtubules. Scale bars: 20 μm. Reproduced with permission from Ref [76]. Copyright 2009 American Chemical Society.
The acidic environment found in the endocytic compartments of cancer cells can also be used to release the drug molecules that are non-covalently attached to the nanoparticle surface. For example, Haam et al. have reported a pH-responsive drug delivering magnetic NPs utilizing non-covalent interactions (Figure 10).[77] They have used a nanoemulsion method to synthesize MnFe2O4 nanocrystals that are coated with α-pyrenyl- ω -carboxyl poly(ethylene glycol). Doxorubicin (DOX) molecules are loaded to the magnetic carrier via a strong π-π interaction between pyrene and DOX molecules. Upon intracellular uptake, protonation of DOX can decrease the π-π interaction, resulting in the release of DOX.[78] Receptor-mediated endocytosis was achieved by modifying surface of magnetic NPs with anti HER2/neu antibody, a tumor-targeting marker of the human HER2/ neu receptor of metastatic breast cancer.
Figure 10.
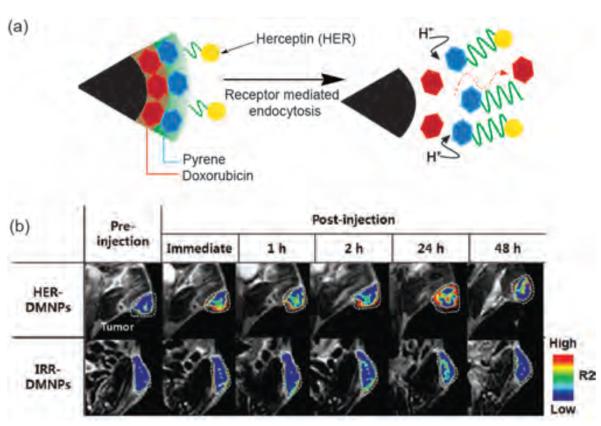
(a) Schematic illustration of anti HER2/neu antibody-modified pH-sensitive drug-releasing magnetic NPs. (b) Magnetic resonance images of tumor-bearing mice after the intravenous injection of human epidermal growth factor receptor (HER) and irrelevant antibody (IRR) functionalized pH sensitive magnetic NPs at various time intervals, demonstrating HER modified NPs accumulated in the tumor more than IRR modified NPs. Reproduced with permission from Ref [77]. Copyright 2011 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim.
In addition to cleaving the bond between drug and carrier, pH triggers can also be used to open molecular valves or lids, releasing encapsulated drugs. Zink et al. have loaded MS NPs with hydrophobic cargo and grafted pH- sensitive nanovalves to keep the cargo inside and release into the cells upon changes in pH (Figure 11).[79] In another study, Feng et al. have reported the controlled release of guest molecules from MS NPs that are capped by acid-labile acetal group linked gold NPs.[80] At neutral pH, no diffusion from pores is observed as pores are strongly blocked by gold NPs. Hovewer, at acidic pHs, the gold capping agent can be removed due to hydrolysis of the pH-responsive acetal group, releasing the trapped molecules. Feng et al. have also decorated MS NPs with poly(4-vinyl pyridine) (PVP) as a pH-sensitive capping nanoshell.[81] In their system, at low pH, the polymer becomes swollen and permeable to the encapsulated molecules after protonation.
Figure 11.
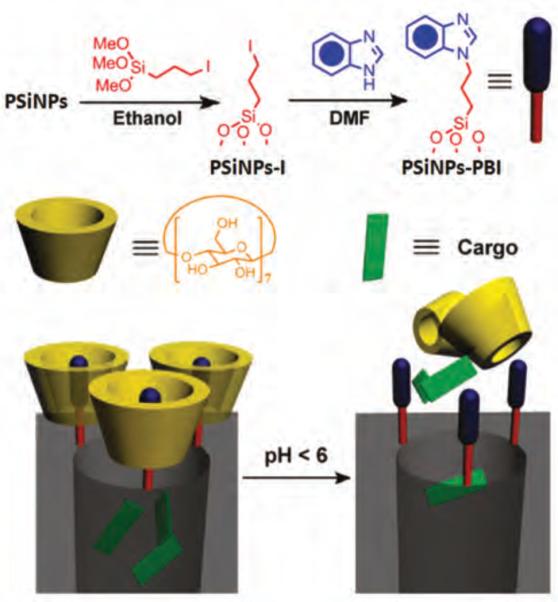
Top illustration-synthesis of pH-sensitive nanovalve. Bottom illustration-binding of the β-cyclodextrin (β-CD) cap to neutral benzimidazole stalk and releasing of β-CD and cargo from the carrier after protonation of the stalk. Reproduced with permission from Ref [79]. Copyright 2011 American Chemical Society.
4.2.3. Enzyme-responsive nanoparticles for delivery applications
Enzyme-responsive NPs can be used to deliver cargos via enzymatic action at targeted locations.[82] Tuning the surface chemistry of nanoparticles by introducing bioactive moieties or enzyme cleavable linkers is the most common way followed to fabricate enzyme-responsive delivery systems. Stoddart et al. have designed a delivery vehicle with an enzyme-responsive snap-top motif, activated by porcine liver esterase (PLE).[83] In their system, MS NPs loaded with luminescent cargo molecules (rhodamine B) were capped with the ester-linked adamantyl stopper. Hydrolysis of the adamantyl ester stopper by PLE resulted in the dethreading of the α-cyclodextrin and triggered the release of the rhodamine B from the pores. In another study, Akashi et al. have used hollow capsules prepared by MS NPs coated with chitosan and dextran sulfate for the delivery of the cargo.[84] Sustained release of the cargo was achieved through degradation of chitosan component by chitosanase.
Recently, Fukumura et al. have reported a multistage nanoparticle-based delivery system with a deep tumor-penetration feature.[85] In this work, enzyme-degradable type A gelatin was crosslinked with glutaraldehyde, and PEG-stabilized quantum dots (QDs, ~ 10 nm) were used to generate enzyme-degradable nanoparticles (~100 nm). Due to the enhanced permeability and retention (EPR) effect, the enzyme-responsive nanoparticles preferentially extravasate from leaky regions of tumor vasculature. However, after extravasation into the tumor tissue, these nanoparticles can be degraded by MMP-2 and smaller 10 nm QDs can be released from the surface, resulting a deeper penetration into the tumor parenchyma (Figure 12).
Figure 12.
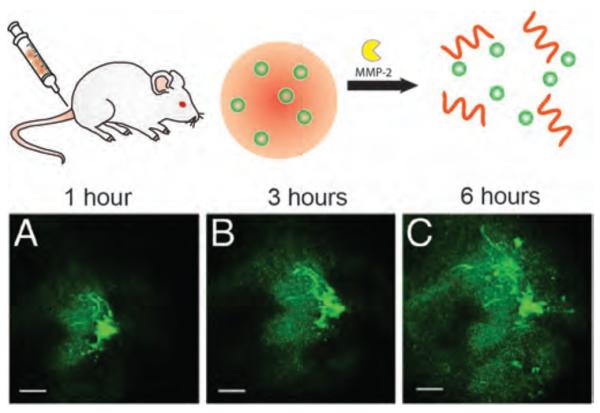
Top-schematic of 100-nm gelatin-QD changing size to 10 nm QD NPs in response to MMP-2. Bottom- in vivo images of gelatin-QD NPs (a) 1h, (b) 3 h, and (c) 6 h after intratumoral injection into the HT-1080 tumor. Reproduced with permission from Ref [85]. Copyright 2011 National Academy of Sciences.
5. Conclusions
Surface functionalization of NPs controls the interface between nanomaterials and biosystems. In this review, we have highlighted several examples that demonstrate the fundamental interaction of NP surface functional groups with proteins and cells, and showed how this understanding can be applied to delivery and imaging strategies.
This review focuses on innovative uses of surface functionalization to impart functional properties to NPs. In addition to these novel applications, surface functionalization dictates toxicity, immune response, and bioavailability. Integrated study of nanoparticle function and behavior in vitro and in vivo will be essential to establish the structure-activity correlation required to move these promising systems towards the clinical use.
Acknowledgements
This research was supported by the NIH (R01 GM077173 and R01 EB014277-01)
Biographies

Gulen Yesilbag Tonga received her B.Sc. and M.Sc. in Chemistry from Bogazici University, Turkey in 2008 and in 2010. Presently, she is a Ph.D. candidate at the Department of Chemistry, University of Massachusetts at Amherst, USA under the guidance of Professor Vincent M. Rotello. Her current research is focused on the use of supramolecular chemistry for delivery applications of nanoparticles.

Krishnendu Saha received his B.Sc. in Chemistry from Jadavpur University, India in 2006 and M.Sc. in Chemistry from Indian Institute of Technology-Madras, India in 2008. Currently, he is a Ph.D. candidate at the Department of Chemistry, University of Massachusetts at Amherst, USA under the guidance of Professor Vincent M. Rotello. His current research interest involves understanding the effect of nanoparticle surface functionality for cellular recognition and delivery applications.

Vincent Rotello is the Charles A. Goessmann Professor of Chemistry at the University of Massachusetts at Amherst, He is a Fellow of the American Association for the Advancement of Science (AAAS) and of the Royal Society of Chemistry (U.K) His research program focuses on using synthetic organic chemistry to engineer the interface between hard and soft materials, and spans the areas of devices, polymers, and nanotechnology/bionanotechnology, with over 380 papers published to date. He is actively involved in the development of new nanomanufacturing methods. In the area of bionanotechnology, his research includes programs in delivery, imaging, diagnostics and nanotoxicology.
References
- 1.a) Petros RA, DeSimone JM. Nat. Rev. Drug Discov. 2010;9:615. doi: 10.1038/nrd2591. [DOI] [PubMed] [Google Scholar]; b) Duncan B, Kim C, Rotello VM. J. Control. Release. 2010;148:122. doi: 10.1016/j.jconrel.2010.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.a) Popovic Z, Liu WH, Chauhan VP, Lee J, Wong C, Greytak AB, Insin N, Nocera DG, Fukumura D, Jain RK, Bawendi MG. Angew. Chem. Int. Ed. 2010;49:8649. doi: 10.1002/anie.201003142. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Erogbogbo F, Yong KT, Hu R, Law WC, Ding H, Chang CW, Prasad PN, Swihart MT. ACS Nano. 2010;9:5131. doi: 10.1021/nn101016f. [DOI] [PubMed] [Google Scholar]
- 3.a) Stark WJ. Angew. Chem. Int. Ed. 2011;50:1242. doi: 10.1002/anie.200906684. [DOI] [PubMed] [Google Scholar]; b) Moyano DF, Rotello VM. Langmuir. 2011;27:10376. doi: 10.1021/la2004535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.a) Saha K, Bajaj A, Duncan B, Rotello VM. Small. 2011;7:1903. doi: 10.1002/smll.201100478. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Mirshafiee V, Mahmoudi M, Lou K, Cheng JJ, Kraft ML. Chem. Commun. 2013;49:2557. doi: 10.1039/c3cc37307j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pellegrino T, Kudera S, Liedl T, Javier AM, Manna L, Parak WJ. Small. 2005;1:48. doi: 10.1002/smll.200400071. [DOI] [PubMed] [Google Scholar]
- 6.a) Saha K, Kim ST, Yan B, Miranda OR, Alfonso FS, Shlosman D, Rotello VM. Small. 2013;9:300. doi: 10.1002/smll.201201129. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Verma A, Stellacci F. Small. 2010;6:12. doi: 10.1002/smll.200901158. [DOI] [PubMed] [Google Scholar]; c) Gong YK, Winnik FM. Nanoscale. 2012;4:360. doi: 10.1039/c1nr11297j. [DOI] [PubMed] [Google Scholar]
- 7.a) Kim ST, Saha K, Kim C, Rotello VM. Acc. Chem. Res. 2013;46:681. doi: 10.1021/ar3000647. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Nel AE, Madler L, Velegol D, Xia T, Hoek EMV, Somasundaran P, Klaessig F, Castranova V, Thompson M. Nat. Mater. 2009;8:543. doi: 10.1038/nmat2442. [DOI] [PubMed] [Google Scholar]; c) Albanese A, Tang PS, Chan WCW. Annu. Rev. Biomed. Eng. 2012;14:1. doi: 10.1146/annurev-bioeng-071811-150124. [DOI] [PubMed] [Google Scholar]
- 8.a) Doane TL, Burda C. Chem. Soc. Rev. 2012;41:2885. doi: 10.1039/c2cs15260f. [DOI] [PubMed] [Google Scholar]; b) Fadeel B, Garcia-Bennett AE. Adv. Drug Deliv. Rev. 2010;62:362. doi: 10.1016/j.addr.2009.11.008. [DOI] [PubMed] [Google Scholar]
- 9.Xie J, Liu G, Eden HS, Ai H, Chen X. Acc. Chem. Res. 2011;44:883. doi: 10.1021/ar200044b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.a) Ganta S, Devalapally H, Shahiwala A, Amiji M. J. Control. Release. 2008;126:187. doi: 10.1016/j.jconrel.2007.12.017. [DOI] [PubMed] [Google Scholar]; b) Cayre OJ, Chagneux N, Biggs S. Soft Matter. 2011;7:2211. [Google Scholar]
- 11.Yin H, Hamilton AD. Angew. Chem. Int. Ed. 2005;44:4130. doi: 10.1002/anie.200461786. [DOI] [PubMed] [Google Scholar]
- 12.a) De M, Miranda OR, Rana S, Rotello VM. Chem. Commun. 2009;16:2157. doi: 10.1039/b900552h. [DOI] [PubMed] [Google Scholar]; b) Calzolai L, Franchini F, Gilliland D, Rossi F. Nano Lett. 2010;10:3101. doi: 10.1021/nl101746v. [DOI] [PubMed] [Google Scholar]
- 13.Fischer NO, McIntosh CM, Simard JM, Rotello VM. Proc. Natl. Acad. Sci. U.S.A. 2002;99:5018. doi: 10.1073/pnas.082644099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Verma A, Simard JM, Rotello VM. Langmuir. 2004;20:4178. doi: 10.1021/la036183v. [DOI] [PubMed] [Google Scholar]
- 15.Fischer NO, Verma A, Goodman CM, Simard JM, Rotello VM. J. Am. Chem. Soc. 2003;125:13387. doi: 10.1021/ja0352505. [DOI] [PubMed] [Google Scholar]
- 16.a) Bale SS, Kwon SJ, Shah DA, Banerjee A, Dordick JS, Kane RS. ACS Nano. 2010;4:1493. doi: 10.1021/nn901586e. [DOI] [PubMed] [Google Scholar]; b) Shah DA, Kwon SJ, Bale SS, Banerjee A, Dordick JS, Kane RS. Biomaterials. 2011;32:3210. doi: 10.1016/j.biomaterials.2010.11.077. [DOI] [PubMed] [Google Scholar]; c) Slowing II, Trewyn BG, Lin VSY. J. Am. Chem. Soc. 2007;129:8845. doi: 10.1021/ja0719780. [DOI] [PubMed] [Google Scholar]
- 17.Hong R, Fischer NO, Verma A, Goodman CM, Emrick T, Rotello VM. J. Am. Chem. Soc. 2004;126:739. doi: 10.1021/ja037470o. [DOI] [PubMed] [Google Scholar]
- 18.You CC, De M, Rotello VM. Org. Lett. 2005;7:5685. doi: 10.1021/ol052367k. [DOI] [PubMed] [Google Scholar]
- 19.You CC, De M, Han G, Rotello VM. J. Am. Chem. Soc. 2005;127:12873. doi: 10.1021/ja0512881. [DOI] [PubMed] [Google Scholar]
- 20.a) Dong HJ, Mukaiyama A, Tadokoro T, Koga YC, Takano K, Kanaya S. J. Mol. Biol. 2008;378:264. doi: 10.1016/j.jmb.2008.02.039. [DOI] [PubMed] [Google Scholar]; b) Sethuraman A, Belfort G. Biophys. J. 2005;88:1322. doi: 10.1529/biophysj.104.051797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De M, You CC, Srivastava S, Rotello VM. J. Am. Chem. Soc. 2007;129:10747. doi: 10.1021/ja071642q. [DOI] [PubMed] [Google Scholar]
- 22.Deng ZJ, Liang MT, Monteiro M, Toth I, Minchin RF. Nat. Nanotechnol. 2011;6:39. doi: 10.1038/nnano.2010.250. [DOI] [PubMed] [Google Scholar]
- 23.De M, Rotello VM. Chem. Commun. 2008;30:3504. doi: 10.1039/b805242e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.a) Walkey CD, Chan WCW. Chem. Soc. Rev. 2012;41:2780. doi: 10.1039/c1cs15233e. [DOI] [PubMed] [Google Scholar]; b) Monopoli MP, Aberg C, Salvati A, Dawson KA. Nat. Nanotechnol. 2012;7:779. doi: 10.1038/nnano.2012.207. [DOI] [PubMed] [Google Scholar]
- 25.Gessner A, Lieske A, Paulke BR, Muller RH. J. Biomed. Mater. Res. A. 2003;65A:319. doi: 10.1002/jbm.a.10371. [DOI] [PubMed] [Google Scholar]
- 26.Gessner A, Lieske A, Paulke BR, Muller RH. Eur. J. Pharm. Biopharm. 2002;54:165. doi: 10.1016/s0939-6411(02)00081-4. [DOI] [PubMed] [Google Scholar]
- 27.Gessner A, Waicz R, Lieske A, Paulke BR, Mader K, Muller RH. Int. J. Pharm. 2000;196:245. doi: 10.1016/s0378-5173(99)00432-9. [DOI] [PubMed] [Google Scholar]
- 28.Cedervall T, Lynch I, Lindman S, Berggard T, Thulin E, Nilsson H, Dawson KA, Linse S. Proc. Natl. Acad. Sci. U.S.A. 2007;104:2050. doi: 10.1073/pnas.0608582104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lindman S, Lynch I, Thulin E, Nilsson H, Dawson KA, Linse S. Nano Lett. 2007;7:914. doi: 10.1021/nl062743+. [DOI] [PubMed] [Google Scholar]
- 30.Cedervall T, Lynch I, Foy M, Berggad T, Donnelly SC, Cagney G, Linse S, Dawson KA. Angew. Chem. Int. Ed. 2007;46:5754. doi: 10.1002/anie.200700465. [DOI] [PubMed] [Google Scholar]
- 31.Yonamine Y, Yoshimatsu K, Lee SH, Hoshino Y, Okahata Y, Shea KJ. ACS Appl. Mater. Interfaces. 2013;5:374. doi: 10.1021/am302404q. [DOI] [PubMed] [Google Scholar]
- 32.Knop K, Hoogenboom R, Fischer D, Schubert US. Angew. Chem. Int. Ed. 2010;49:6288. doi: 10.1002/anie.200902672. [DOI] [PubMed] [Google Scholar]
- 33.Fang J, Nakamura H, Maeda H. Adv. Drug Deliv. Rev. 2011;63:136. doi: 10.1016/j.addr.2010.04.009. [DOI] [PubMed] [Google Scholar]
- 34.Walkey CD, Olsen JB, Guo HB, Emili A, Chan WCW. J. Am. Chem. Soc. 2012;134:2139. doi: 10.1021/ja2084338. [DOI] [PubMed] [Google Scholar]
- 35.Tagami T, Uehara Y, Moriyoshi N, Ishida T, Kiwada H. J. Control. Release. 2011;151:149. doi: 10.1016/j.jconrel.2010.12.013. [DOI] [PubMed] [Google Scholar]
- 36.a) Hamad I, Hunter AC, Rutt KJ, Liu Z, Dai H, Moghimi SM. Mol. Immunol. 2008;45:3797. doi: 10.1016/j.molimm.2008.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Andersen AJ, Robinson JT, Dai HJ, Hunter AC, Andresen TL, Moghimi SM. ACS Nano. 2013;7:1108. doi: 10.1021/nn3055175. [DOI] [PubMed] [Google Scholar]; c) Hamad I, Al-Hanbali O, Hunter AC, Rutt KJ, Andresen TL, Moghimi SM. ACS Nano. 2010;4:6629. doi: 10.1021/nn101990a. [DOI] [PubMed] [Google Scholar]
- 37.a) Cheng G, Zhang Z, Chen SF, Bryers JD, Jiang SY. Biomaterials. 2007;28:4192. doi: 10.1016/j.biomaterials.2007.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Zhang L, Xue H, Cao ZQ, Keefe A, Wang JN, Jiang SY. Biomaterials. 2011;32:4604. doi: 10.1016/j.biomaterials.2011.02.064. [DOI] [PubMed] [Google Scholar]
- 38.a) Breus VV, Heyes CD, Tron K, Nienhaus GU. ACS Nano. 2009;3:2573. doi: 10.1021/nn900600w. [DOI] [PubMed] [Google Scholar]; b) Muro E, Pons T, Lequeux N, Fragola A, Sanson N, Lenkei Z, Dubertret B. J. Am. Chem. Soc. 2010;132:4556. doi: 10.1021/ja1005493. [DOI] [PubMed] [Google Scholar]
- 39.Yang W, Zhang L, Wang SL, White AD, Jiang SY. Biomaterials. 2009;30:5617. doi: 10.1016/j.biomaterials.2009.06.036. [DOI] [PubMed] [Google Scholar]
- 40.Zhan NQ, Palui G, Grise H, Tang HL, Alabugin I, Mattoussi H. ACS Appl. Mater. Interfaces. 2013;5:2861. doi: 10.1021/am302788q. [DOI] [PubMed] [Google Scholar]
- 41.Agasti SS, Chompoosor A, You CC, Ghosh P, Kim CK, Rotello VM. J. Am. Chem. Soc. 2009;131:5728. doi: 10.1021/ja900591t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arvizo RR, Miranda OR, Moyano DF, Walden CA, Giri K, Bhattacharya R, Robertson JD, Rotello VM, Reid JM, Mukherjee P. Plos One. 2011;6:e24374. doi: 10.1371/journal.pone.0024374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Han H-S, Martin JD, Lee J, Harris DK, Fukumura D, Jain RK, Bawendi M. Angew. Chem. Int. Ed. 2013;52:1414. doi: 10.1002/anie.201208331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Green JJ, Chiu E, Leshchiner ES, Shi J, Langer R, Anderson DG. Nano Lett. 2007;7:874. doi: 10.1021/nl062395b. [DOI] [PubMed] [Google Scholar]
- 45.Biju V, Muraleedharan D, Nakayama K, Shinohara Y, Itoh T, Baba Y, Ishikawa M. Langmuir. 2007;23:10254. doi: 10.1021/la7012705. [DOI] [PubMed] [Google Scholar]
- 46.Mok H, Park JW, Park TG. Bioconjugate Chem. 2008;19:797. doi: 10.1021/bc700464m. [DOI] [PubMed] [Google Scholar]
- 47.Sun LL, Liu DJ, Wang ZX. Langmuir. 2008;24:10293. doi: 10.1021/la8015063. [DOI] [PubMed] [Google Scholar]
- 48.Nativo P, Prior IA, Brust M. ACS Nano. 2008;2:1639. doi: 10.1021/nn800330a. [DOI] [PubMed] [Google Scholar]
- 49.Kang B, Mackey MA, El-Sayed MA. J. Am. Chem. Soc. 2010;132:1517. doi: 10.1021/ja9102698. [DOI] [PubMed] [Google Scholar]
- 50.Martin AL, Bernas LM, Rutt BK, Foster PJ, Gillies ER. Bioconjugate Chem. 2008;19:2375. doi: 10.1021/bc800209u. [DOI] [PubMed] [Google Scholar]
- 51.a) Cho EC, Xie JW, Wurm PA, Xia YN. Nano Lett. 2009;9:1080. doi: 10.1021/nl803487r. [DOI] [PubMed] [Google Scholar]; b) Park J, Nam J, Won N, Jin H, Jung S, Jung S, Cho SH, Kim S. Adv. Funct. Mater. 2011;21:1558. [Google Scholar]
- 52.a) Shi XY, Thomas TP, Myc LA, Kotlyar A, Baker JR. Phys. Chem. Chem. Phys. 2007;9:5712. doi: 10.1039/b709147h. [DOI] [PubMed] [Google Scholar]; b) Wilhelm C, Billotey C, Roger J, Pons JN, Bacri JC, Gazeau F. Biomaterials. 2003;24:1001. doi: 10.1016/s0142-9612(02)00440-4. [DOI] [PubMed] [Google Scholar]; c) Villanueva A, Canete M, Roca AG, Calero M, Veintemillas-Verdaguer S, Serna CJ, Morales MD, Miranda R. Nanotechnology. 2009;20:115103. doi: 10.1088/0957-4484/20/11/115103. [DOI] [PubMed] [Google Scholar]; d) Patel PC, Giljohann DA, Daniel WL, Zheng D, Prigodich AE, Mirkin CA. Bioconjugate Chem. 2010;21:2250. doi: 10.1021/bc1002423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mortensen LJ, Oberdorster G, Pentland AP, DeLouise LA. Nano Lett. 2008;8:2779. doi: 10.1021/nl801323y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zheng D, Giljohann DA, Chen DL, Massich MD, Wang XQ, Iordanov H, Mirkin CA, Paller AS. Proc. Natl. Acad. Sci. U.S.A. 2012;109:11975. doi: 10.1073/pnas.1118425109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim B, Han G, Toley BJ, Kim CK, Rotello VM, Forbes NS. Nat. Nanotechnol. 2010;5:465. doi: 10.1038/nnano.2010.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lorenz S, Hauser CP, Autenrieth B, Weiss CK, Landfester K, Mailander V. Macromol. Biosci. 2010;10:1034. doi: 10.1002/mabi.201000099. [DOI] [PubMed] [Google Scholar]
- 57.Zhu ZJ, Posati T, Moyano DF, Tang R, Yan B, Vachet RW, Rotello VM. Small. 2012;8:2659. doi: 10.1002/smll.201200794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Timko BP, Dvir T, Kohane DS. Adv. Mater. 2010;22:4925. doi: 10.1002/adma.201002072. [DOI] [PubMed] [Google Scholar]
- 59.Lu J, Choi E, Tamanoi F, Zink JI. Small. 2008;4:421. doi: 10.1002/smll.200700903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Knežević NZ, Trewyn BG, Lin VS-Y. Chem. Commun. 2011;47:2817. doi: 10.1039/c0cc04424e. [DOI] [PubMed] [Google Scholar]
- 61.Carling CJ, Nourmohammadian F, Boyer JC, Branda NR. Angew. Chem. Int. Ed. 2010;49:3782. doi: 10.1002/anie.201000611. [DOI] [PubMed] [Google Scholar]
- 62.Yan B, Boyer JC, Habault D, Branda NR, Zhao Y. J. Am. Chem. Soc. 2012;134:16558. doi: 10.1021/ja308876j. [DOI] [PubMed] [Google Scholar]
- 63.Yang Y, Shao Q, Deng R, Wang C, Teng X, Cheng K, Cheng Z, Huang L, Liu Z, Liu X, Xing B. Angew. Chem. Int. Ed. 2012;51:3125. doi: 10.1002/anie.201107919. [DOI] [PubMed] [Google Scholar]
- 64.a) Zhou W, Valley MP, Shultz J, Hawkins EM, Bernad L, Good T, Good D, Riss TL, Klaubert DH, Wood KV. J. Am. Chem. Soc. 2006;128:3122. doi: 10.1021/ja058519o. [DOI] [PubMed] [Google Scholar]; b) Niu G, Xiong Z, Cheng Z, Cai W, Gambhir S, Chen X. Mol. Imaging Biol. 2007;9:126. doi: 10.1007/s11307-007-0079-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rosenholm JM, Peuhu E, Eriksson JE, Sahlgren C, Linden M. Nano Lett. 2009;9:3308. doi: 10.1021/nl901589y. [DOI] [PubMed] [Google Scholar]
- 66.Manju S, Sreenivasan K. Langmuir. 2011;27:14489. doi: 10.1021/la202470k. [DOI] [PubMed] [Google Scholar]
- 67.Lu J, Liong M, Zink JI, Tamanoi F. Small. 2007;3:1341. doi: 10.1002/smll.200700005. [DOI] [PubMed] [Google Scholar]
- 68.a) Muller-Dethlefs K, Hobza P. Chem. Rev. 2000;100:143. doi: 10.1021/cr9900331. [DOI] [PubMed] [Google Scholar]; b) Morgan TT, Muddana HS, Altinoglu EI, Rouse SM, Tabakovic A, Tabouillot T, Russin TJ, Shanmugavelandy SS, Butler PJ, Eklund PC, Yun JK, Kester M, Adair JH. Nano Lett. 2008;8:4108. doi: 10.1021/nl8019888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cheng Y, Samia AC, Li J, Kenney ME, Resnick A, Burda C. Langmuir. 2010;26:2248. doi: 10.1021/la902390d. [DOI] [PubMed] [Google Scholar]
- 70.Kim CK, Ghosh P, Pagliuca C, Zhu ZJ, Menichetti S, Rotello VM. J. Am. Chem. Soc. 2009;131:1360. doi: 10.1021/ja808137c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.a) Cardone RA, Casavola V, Reshkin SJ. Nat. Rev. Cancer. 2005;5:786. doi: 10.1038/nrc1713. [DOI] [PubMed] [Google Scholar]; b) Lee ES, Gao ZG, Bae YH. J. Control. Release. 2008;132:164. doi: 10.1016/j.jconrel.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Knorr V, Russ V, Allmendinger L, Ogris M, Wagner E. Bioconjugate Chem. 2008;19:1625. doi: 10.1021/bc8001858. [DOI] [PubMed] [Google Scholar]
- 73.Lee Y, Miyata K, Oba M, Ishii T, Fukushima S, Han M, Koyama H, Nishiyama N, Kataoka K. Angew. Chem. Int. Ed. 2008;47:5163. doi: 10.1002/anie.200800963. [DOI] [PubMed] [Google Scholar]
- 74.Yoo HS, Lee EA, Park TG. J. Control. Release. 2002;82:17. doi: 10.1016/s0168-3659(02)00088-3. [DOI] [PubMed] [Google Scholar]
- 75.Wang F, Wang Y-C, Dou S, Xiong M-H, Sun T-M, Wang J. ACS Nano. 2011;5:3679. doi: 10.1021/nn200007z. [DOI] [PubMed] [Google Scholar]
- 76.Dhar S, Daniel WL, Giljohann DA, Mirkin CA, Lippard SJ. J. Am. Chem. Soc. 2009;131:14652. doi: 10.1021/ja9071282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lim E-K, Huh Y-M, Yang J, Lee K, Suh J-S, Haam S. Adv. Mater. 2011;23:2436. doi: 10.1002/adma.201100351. [DOI] [PubMed] [Google Scholar]
- 78.Kim J, Lee JE, Lee SH, Yu JH, Lee JH, Park TG, Hyeon T. Adv. Mater. 2008;20:478. [Google Scholar]
- 79.Xue M, Zhong X, Shaposhnik Z, Qu YQ, Tamanoi F, Duan XF, Zink JI. J. Am. Chem. Soc. 2011;133:8798. doi: 10.1021/ja201252e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liu R, Zhang Y, Zhao X, Agarwal A, Mueller LJ, Feng P. J. Am. Chem. Soc. 2010;132:1500. doi: 10.1021/ja907838s. [DOI] [PubMed] [Google Scholar]
- 81.Liu R, Liao P, Liu J, Feng P. Langmuir. 2011;27:3095. doi: 10.1021/la104973j. [DOI] [PubMed] [Google Scholar]
- 82.de la Rica R, Aili D, Stevens MM. Adv. Drug Deliv. Rev. 2012;64:967. doi: 10.1016/j.addr.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 83.Patel K, Angelos S, Dichtel WR, Coskun A, Yang Y-W, Zink JI, Stoddart JF. J. Am. Chem. Soc. 2008;130:2382. doi: 10.1021/ja0772086. [DOI] [PubMed] [Google Scholar]
- 84.Itoh Y, Matsusaki M, Kida T, Akashi M. Biomacromolecules. 2006;7:2715. doi: 10.1021/bm060289y. [DOI] [PubMed] [Google Scholar]
- 85.Wong C, Stylianopoulos T, Cui J, Martin J, Chauhan VP, Jiang W, Popovic Z, Jain RK, Bawendi MG, Fukumura D. Proc. Natl. Acad. Sci. U. S. A. 2011;108:2426. doi: 10.1073/pnas.1018382108. [DOI] [PMC free article] [PubMed] [Google Scholar]