Abstract
We develop and test a new pipeline in CASP10 to predict protein structures based on an interplay of I-TASSER and QUARK for both free-modeling (FM) and template-based modeling (TBM) targets. The most noteworthy observation is that sorting through the threading template pool using the QUARK-based ab initio models as probes allows the detection of distant-homology templates which might be ignored by the traditional sequence profile-based threading alignment algorithms. Further template assembly refinement by I-TASSER resulted in successful folding of two medium-sized FM targets with >150 residues. For TBM, the multiple threading alignments from LOMETS are, for the first time, incorporated into the ab initio QUARK simulations, which were further refined by I-TASSER assembly refinement. Compared with the traditional threading assembly refinement procedures, the inclusion of the threading-constrained ab initio folding models can consistently improve the quality of the full-length models as assessed by the GDT-HA and hydrogen-bonding scores. Despite the success, significant challenges still exist in domain boundary prediction and consistent folding of medium-size proteins (especially beta-proteins) for nonhomologous targets. Further developments of sensitive fold-recognition and ab initio folding methods are critical for solving these problems.
Keywords: protein structure prediction, CASP10, threading, ab initio folding, I-TASSER, QUARK
INTRODUCTION
After nearly four decades of effort and progress,1–7 computational protein structure prediction has evolved into a problem of strict hierarchy in modeling strategy and accuracy. For proteins homologous to solved structures, high-resolution models can be built by comparative modeling, which copies and refines structure frameworks from homologous templates.5 For proteins without homology templates, one has to construct the structural models from scratch, which generally has low-resolution, with success reported only on small proteins below 100 residues.6,8–11
We have recently developed two methods for template-based and template-free protein structure predictions. In I-TASSER,7,9,12 we construct structural models by reassembling the continuous segments excised from homologous templates generated by multiple threading programs.13 One of the major advantages of I-TASSER is that the best structural templates can be consistently identified and driven closer to the native state by consensus spatial restraints from multiple threading alignments. In QUARK,10,14 structural models are assembled from small continuous fragments (1–20 residues) excised from unrelated proteins. An essential difference between QUARK and I-TASSER is that the QUARK-based fragment assembly simulations starts from random conformation without relying on global threading templates, which enables it to construct new protein folds from scratch. The strategy, like other ab initio modeling approaches,6,8,9 only works for proteins of short length.
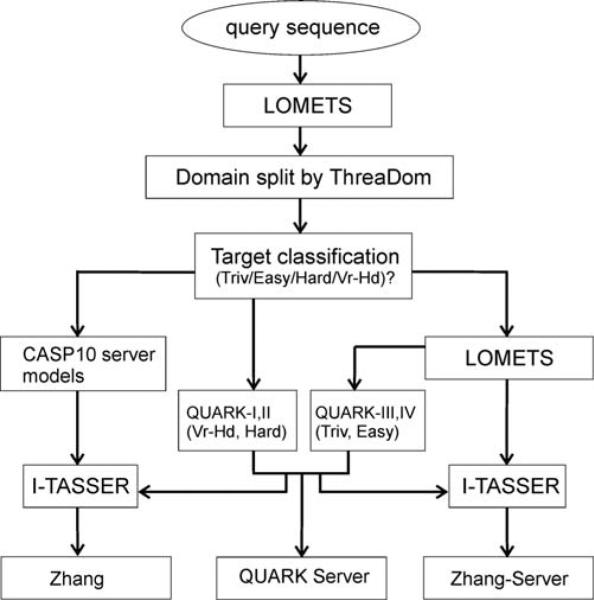
In this CASP experiment, we develop and test a new strategy to combine I-TASSER (template-based modeling) and QUARK (ab initio modeling) for protein structure construction; one goal is to fold distant-homology proteins, especially those with size beyond the traditional ab initio modeling regime. The focus of this report is mainly on the results generated by the automated pipeline of “Zhang-Server,” although three pipelines (i.e. “QUARK” and “Zhang-Server” in the automated Server Section, and “Zhang” in Human Section) have been tested in CASP10 experiments (see: http://predictioncenter.org/casp10/groups_analysis.cgi), where models in “QUARK” were generated by the QUARK-based ab initio folding programs and those in “Zhang-Server” and “Zhang” by an interplay of I-TASSER and QUARK programs (Fig. 1).
Figure 1.
The flowchart of the interplay of I-TASSER and QUARK methods for automated model generation for “Zhang,” “Zhang-Server,” and “QUARK” in CASP10. In general, models in QUARK Server are generated by QUARK, with the LOMETS restraints incorporated for the Trivial (Triv) and Easy targets. Models in Zhang-Server and Zhang Human use both threading and QUARK models as starting conformations. The only difference between Zhang-Server and Zhang Human is that Zhang-Server uses the in-house templates from LOMETS while Zhang Human uses templates from the CASP10 server models. “Vr-Hd” denotes “Very-Hard” targets.
METHODS
The methods of I-TASSER7,9 and QUARK10,14 have been published elsewhere, with the online servers and the standalone I-TASSER package freely available at http://zhanglab.ccmb.med.umich.edu/I-TASSER and http://zhan glab.ccmb.med.umich.edu/QUARK, respectively. Here, we briefly outline the pipelines of the algorithms and then discuss in some detail the most recent developments which are relevant to the structural modeling conducted in the CASP10 experiment.
I-TASSER outline
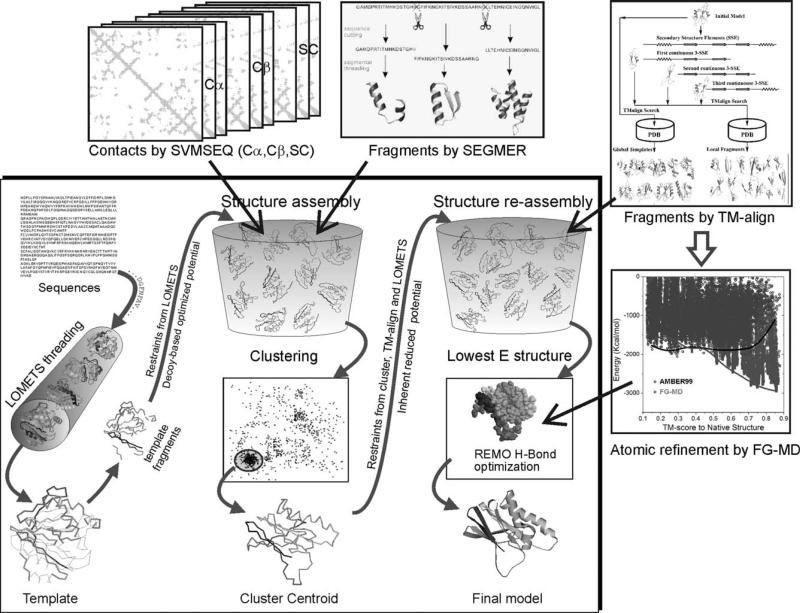
I-TASSER is a threading template-based, iterative fragment assembly approach to protein structure prediction whereby a flowchart is shown in the middle square of Figure 2. For a query sequence, I-TASSER first identifies structural templates from the PDB using LOMETS.13 Continuous fragments are then excised from the templates in the threading aligned regions, which are used to reassemble full-length models by replica-exchange Monte Carlo simulations. The structure trajectories are clustered by SPICKER15 to identify the low free-energy states. Starting from the SPICKER clusters, a second round fragment reassembly simulation is conducted to further refine the structural models. The final models from the low energy conformations are further refined by the atomic-level simulations.16,17
Figure 2.
The new I-TASSER pipeline used in CASP10 which combines recent developments from SVMSEQ,21 SEGMER,23 and FG-MD17 for enhanced spatial restraint predictions and atomic-level structural refinements.
The I-TASSER force field for simulation contains both knowledge- and physics-based terms, including generic Cα and side-chain contact potential calculated from structures in the PDB library, orientation-specific backbone hydrogen-bonding, de novo contact prediction from machine learning, segment-based Cα correlation, and spatial restraints from threading alignments. These terms were systematically optimized on large-scale sequence and structure decoy sets by maximizing the correlation between TM-score and the total energy.18
Recent developments in I-TASSER based structural assembly
I-TASSER was previously tested in large-scale benchmark and blind tests,9,12,19,20 which demonstrates significant efficiency in refining threading template structures; the quantitative data analyses showed that in around 81% of cases the RMSD of the I-TASSER models to the native is lower than that of the best initial templates, and in 46% of cases the RMSD reduction is more than 1 Å in the same threading aligned regions. Despite the ability of multiple template reassembly and template refinement, I-TASSER has limits in constructing correct structural models for distant-homology proteins. The limits mainly stem from the lack of long-range interaction information as the weakly aligned threading templates often contain only the local structural information of short-range interactions which are less useful for global fold construction. Meanwhile, the coarse-grained combination of multiple templates often results in unphysical local structural models, where efficient atomic structural refinement algorithms to refine the coarse-grained models and yet maintain the physical realism are required. To address these issues, most of our recent developments have been focused on the generation and combination of medium-to-long range residue interactions in the I-TASSER simulations, as well as the high-resolution atomic structure refinement (see the up-right boxes in Fig. 2).
Multiple de novo contact prediction by SVMSEQ
SVMSEQ21 was designed to predict residue-residue contacts based on a support vector machine, which was trained on the sequence profile, secondary structure, solvation and in-between residue segment features. In a recent study,22 SVMSEQ was extended to train on nine sets of contacts, between three atom types (Cα, Cβ, and side-chain center of mass) and with three different distance cutoffs (7, 8, and 9 Å), which are used coherently to constrain different subunits of the I-TASSER conformations.
Fragment identification by SEGMER
Considering the fact that distant-homology templates are often difficult to identify from global sequence threading, we developed a new program, SEGMER,23 to detect the structural motifs of super-secondary structures, where medium-to-long range interaction information could be reliably obtained. The target sequence is first split into segments of two to four (consecutive and nonconsecutive) secondary structure elements; each of the segments is then threaded through the PDB to identify structural motifs by MUSTER.24 The spatial restraints, including Cα distance and side-chain contacts, are derived from the high-scoring segment motifs which are finally incorporated into the global template-based restraints from LOMETS13 to guide the I-TASSER simulations.
Fragment-guided MD simulation for atomic structure refinement
To refine the I-TASSER models in atomic-level, the structures are split into segments of two to four consecutive secondary structural elements, which are then used as probes to detect the segment analogs by TM-align25 from the PDB. It has been shown that the distance maps extracted from the TM-align structure segments have a higher accuracy than the maps from the entire probe structures, although the TM-score of the overall structure of the analogous templates is still lower.17 These distance maps are used for two purposes. First, they are used as restraints to guide the second round of I-TASSER simulations. Second, they were found to be able to reshape the energy funnel of the physics-based force field, especially for the models which have correct folds (TM-score >0.5). Therefore, we use the fragment distance maps to guide the molecular dynamics simulations by FG-MD for full-atomic structure refinements after the I-TASSER simulations17 (Fig. 2).
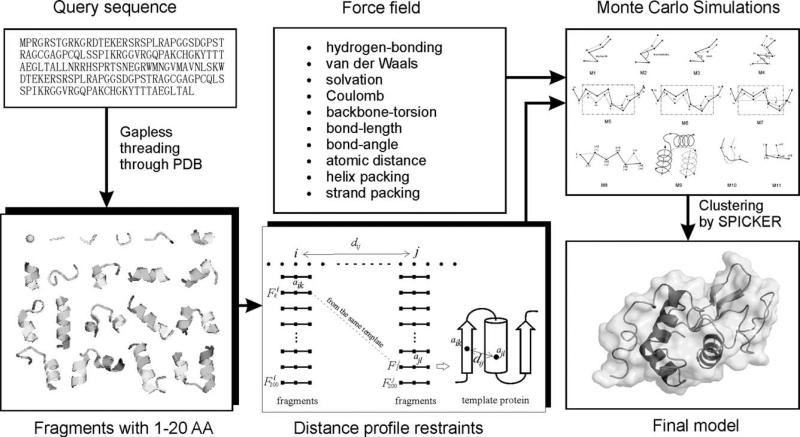
QUARK for ab initio and template-based structure prediction
The flowchart of QUARK is shown in Figure 3, which starts from the collection of continuously distributed fragments (1–20 residues) by gapless threading from non-redundant PDB structure libraries. A distance profile containing long-range interactions is then obtained from the fragments in a two-step procedure:14 (1) for each pair of residues (i and j) a histogram of distances dij is calculated from the pairs of the top 200 fragments at ith and jth positions if they come from the same PDB structure; (2) the histograms will be converted into the distance profiles if there is a histogram peak in the middle range of the distances. Finally, the fragments are assembled into full-length models by replica-exchange Monte Carlo simulations which are under the guidance of the distance profile and a composite physics- and knowledge-based potentials, including hydrogen-bonding, van der Waals, solvation, Coulomb, backbone-torsion, radius of gyration, and chiral-specific packing of regular secondary structure elements.10
Figure 3.
The flowchart of QUARK for ab initio structural assembly.10,14
Two PDB libraries were used for the QUARK fragment collections. First, a small library containing 6,023 high-resolution structures was culled from the PDB which have a pair-wise sequence identity cutoff 25%. This library is used for the Very-Hard protein targets >100 residues (see below for definition of target category). For other proteins, a larger library is used which contains all the PDB structures with a pair-wise sequence identity cutoff 70%. This library is the same as that used by LOMETS and currently contains 47,742 entries (http://zhanglab.ccmb.med.umich.edu/library). The selection of the template library is based on benchmark results, where the preference for the smaller library by the Very-Hard targets is probably because using the smaller library can somewhat avoid the bias towards artificial homology templates since the Very-Hard targets are not supposed to have homologous templates.
Interplay of I-TASSER and QUARK
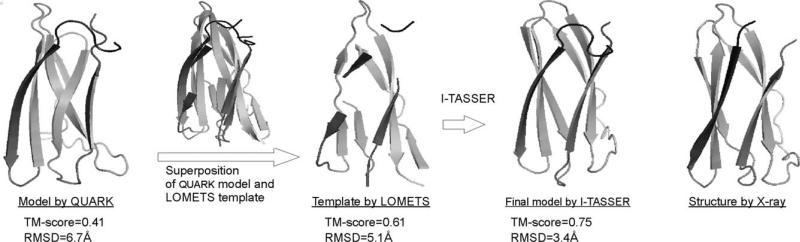
The interplay of I-TASSER and QUARK includes twofolds. First, we use the QUARK models as a probe to sort the LOMETS templates by the TM-score between the QUARK model and the threading templates, that is, the templates, which have the highest TM-score to the QUARK models, are used as the initial models and as a source of constraints for the I-TASSER model simulations. Since the QUARK models are built from ab initio folding simulations, any reasonable match of the real protein structures to the QUARK models (e.g. a TM-score >0.35)26 can be considered significant and may indicate a correct template hit. Figure 4 shows an example of testing on a CASP9 FM target (T0612: 37–128), where a low resolution QUARK model fishes out a good template structure from the low-rank LOMETS alignments which has a higher TM-score to the native and helps I-TASSER to build a much-improved full-length model. In addition to the QUARK-based template sorting, we have run another pipeline which merges the QUARK models into the pool of default LOMETS templates. The inclusion of the QUARK models as templates can improve the accuracy of the spatial restraints for the I-TASSER simulation.
Figure 4.
An illustration of the QUARK-assisted template identification from the CASP9 target T0612 (S38-S128). The original QUARK model has the correct shape but with incorrect beta-strand arrangements (TM-score = 0.41). The superposition of the QUARK model with the top 200 templates by LOMETS picked up the template 1xf1A which has a TM-score = 0.61 to the native, where the TM-score between the QUARK model and the template is only 0.46. This template is the best template but ranked low (41st) in LOMETS due to low alignment score. After the template was refined by I-TASSER, it resulted in the final model with a TM-score = 0.75.
Second, although QUARK was originally developed for ab initio protein structure prediction without using global template structures, in the new developments we found that the threading-based alignments, even for those with weak scores, can be exploited to assist the QUARK structural assembly simulations for both distant-and close-homology targets (Xu and Zhang, in preparation). As part of the second way in which QUARK and I-TASSER interplay, we implemented four different versions of the QUARK program, depending on how the templates and restraints from I-TASSER are used:
QUARK-I: the default simulation without using threading templates;
QUARK-II: similar to QUARK-1 but with the initial conformation starting from the top 200 threading templates by LOMETS;13
QUARK-III: similar to QUARK-II but with the distance profile restraints collected from the top 200 threading alignments;
QUARK-IV: similar to QUARK-III but with the full-set of spatial restraints (Cα distance map and side-chain contact restraints same as used in I-TASSER) exploited in QUARK simulations.
Target categorization and modeling method assignments
Different structural modeling methods have different accuracies and are suitable for different targets. It is therefore essential to apply appropriate methods on the correct targets. In CASP10, we categorized the protein targets into four groups (“Trivial,” “Easy,” “Hard,” and “Very-Hard”) based on the significance score and the consensus of the LOMETS threading alignments. Considering the first template from M threading programs in LOMETS, nine quality scores are calculated:
| (1) |
where Zi is the highest Z-score of the alignment by the ith threading program and Z0i is the Z-score cutoff decided for the ith program in the way that the average TM-score of all templates with Zi > Z0i equals approximately to 0.65. TM ’j (j 5 1, 2, 3, . . ., N = M × (M – 1)/2) represents jth pair-wise TM-score among the M templates which have been sorted in a decreasing order. Za, TM, and ZTM thus represent the average significance and the consensus scores of the threading alignments, respectively.
Two sets of cutoffs for the nine quality scores are defined in Table I. If there are more than eight quality scores defined in Eq. (1) >1.8 × cut2, the target is defined as a Trivial target; otherwise, if there are more than eight quality scores >cut2, the target is an Easy target; otherwise, if there are more than eight quality scores <cut1, the target is a Very-Hard target; all others are Hard targets. The defined categories are highly correlated with the actual TM-score of the templates to the native. In a benchmark containing 200 evenly-distributed domains, the average TM-scores of the best in top five templates are 0.773, 0.655, 0.417, and 0.274 for Trivial, Easy, Hard, and Very-Hard targets, respectively, with a standard deviation of TM-score <0.08 in each of the categories. If we defined the categories based only on Z-score as what we did in LOMETS,13 the average TM-score in these categories are 0.701, 0.555, 0.387, and 0.314, respectively, with the average standard deviation of 0.137. These data show that the consideration of template consensus increases the specificity of the target category definitions.
Table I.
Quality Score Cutoffs used for Target Categorization
| Quality score (s) | Cut1 | N(s < cut1) | Cut2 | N(s > cut2) |
|---|---|---|---|---|
| (Za) | 0.55 | 85 | 0.9 | 83 |
| TM1 | 0.3 | 69 | 0.53 | 96 |
| TM2 | 0.26 | 77 | 0.43 | 96 |
| TM3 | 0.225 | 76 | 0.38 | 95 |
| TM4 | 0.195 | 73 | 0.34 | 92 |
| zaTM1 | 0.135 | 61 | 0.34 | 95 |
| zaTM2 | 0.125 | 75 | 0.28 | 95 |
| zaTM3 | 0.1 | 63 | 0.25 | 93 |
| zaTM4 | 0.09 | 64 | 0.21 | 93 |
N is the number of the targets below or above the cutoffs when tested on 200 nonhomologous benchmark proteins.
Different procedures were used to generate model predictions for protein targets in different categories. In the QUARK server, the program QUARK-I was implemented for the Very-Hard targets; QUARK-I and II were implemented for the Hard targets; and QUARK-III and IV were implemented for the Easy and Trivial targets. In Zhang and Zhang-Server, for the Very-Hard and Hard targets, the models generated by QUARK-I and II simulations were used to sort the LOMETS templates, where the top templates which are structurally closest to the QUARK ab initio models were used by I-TASSER for further structure assembly; for the Easy and Trivial targets, the default I-TASSER simulations were implemented to generate the structural decoys but the QUARK-III and -IV models were added and treated as a new set of threading templates in addition to the LOMETS templates (see Figure 1).
Meta-MQAP model selections
To select models generated from different pipelines, we implemented a set of seven MQAP programs, including the I-TASSER C-score,27 structural consensus measured by pair-wise TM-score,28 and five statistical potentials (RW,29 RWplus,29 Dfire,30 Dope31, and verify3D32). Finally, a meta-MQAP consensus score was calculated as the sum of the rank of the seven MQAP scores. The models with the lowest consensus scores are selected for submission.
Domain prediction and structure assembly of multiple-domain proteins
For a given target sequence, we used ThreaDom33 to predict the boundary locations of protein domains based on the domain conservation score which is designed to combine information from template domain structure and the terminal and internal gaps/insertions in the LOMETS alignments. If the target was judged as multiple-domain by ThreaDom, the I-TASSER simulations would be run for both the whole chain and the separate domains. The final full-length models were generated by docking the domain models using the whole-chain model as a reference template, where the reference template was selected from the whole-chain I-TASSER models that have the highest TM-score to the individual domain models. Once the full-chain template is selected, the docking is conducted through a quick Metropolis Monte Carlo simulation, where the simulation energy is defined as the RMSD of the domain models to the whole-chain model template plus the reciprocal of the number of interdomain steric clashes.
RESULTS AND DISCUSSION
There are 144 domains from 110 protein entries which were eventually assessed in the Server Section, and 88 domains in the Human Section. Following the assessor's assignments, 110 out of the 144 domains are Template-Base Modeling (TBM) targets which have a length range in [24, 498] and an average length of 181 residues; the remaining 34 targets are Free Modeling (FM) targets (including the CASP Roll targets) which have lengths in the range [33, 383] and an average length of 137 residues. Because more targets were tested in the Server Section, and the methods used in our server and human predictions are essentially identical, our report will mainly focus on the server predictions, in particular the failed cases by the current modeling methods. The data analysis on template refinements, accuracy of spatial restraints and human-versus-server comparison, which have been discussed in detail in previous CASP experiments12,19,20 and are largely unchanged in CASP10, will be ignored or summarized briefly.
Threading template identification and QUARK-based template sorting
As expected, the threading templates identified by LOMETS have a much higher quality for the TBM targets than that for the FM targets. Eighty-seven out of the 110 TBM targets (~80%) have the best in top five templates with a TM-score >0.5, while this is true only in 1 out of the 34 FM targets (R0003). That FM target, R0003, is a specially designed Knottin 2.5D protein consisting of 33 residues with a sequence identity of 70% to the EETI-II template protein that is 3.1 Å away from the target with a TM-score 5 0.530 (this target has a sequence identity of 88% to another CASP10 target T0711, both of which were included in CASP10 probably for testing the ability of predicting big structure variances on a small number of residue mutations). Overall, the average TM-scores for the best in top five templates are 0.657 and 0.256 for the TBM and FM targets, respectively. The average TM-score for all the 144 targets is 0.618.
Due to the limits of current threading methods, the templates by LOMETS are far from the best possible templates in the PDB. When we use the target structure as the probe, the structure alignment program TM-align25 can identify the top template with an average TM-score 0.761 and 0.611 for the TBM and FM targets, respectively, which are 16% and 139% higher than that by LOMETS. The TM-align templates have at least a TM-score >0.43 for all the targets. These data demonstrate significant room for further improvement of the current fold-recognition algorithms, as well as the possibility to increase the TM-score by re-ranking the template alignments.
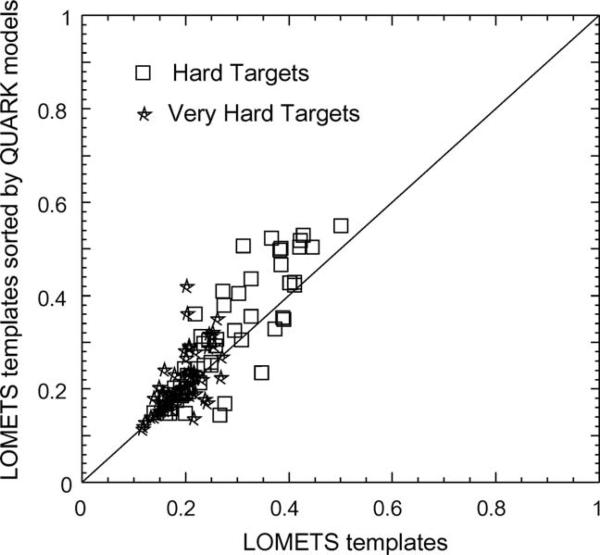
As described in Methods, for the Hard and Very-Hard targets we re-ordered the LOMETS templates based on the maximum TM-score to the top five QUARK models. In Figure 5, we plot the average TM-score of the top five LOMETS templates in the original threading order versus that of the templates sorted by the QUARK models. The data are shown for the 120 Hard and Very-Hard targets which have a length <250 residues, the maximum length of the targets for which QUARK simulations were conducted. After sorting by the QUARK models, the average TM-score becomes improved for 81 targets, where the TM-score decreased for only 38 targets. If considering the best in top 20 templates, the average TM-score increased in 90 cases and decreased in 25 cases. In 46 cases, the top threading templates after sorting have a TM-score higher than the probe QUARK models despite the short length of alignment in the templates, which shows the potential to recognize higher quality templates even when the template structures are only partially matched to the ab initio models.
Figure 5.
Average TM-score of the top five templates by LOMETS versus that of the templates after sorting by the TM-score to the QUARK models. The data are shown for the 120 Hard and Very-Hard targets with size below 250 residues. The domains were defined by ThreaDom with the target types assigned by Eq. (1).
Here, we note that the domains defined by ThreaDom based on the target sequence may be different from the domain assigned by the assessors based on the target structure. The data in Figure 5 are thus presented based on the ThreaDom domains, which include the Hard and Very-Hard targets defined by Eq. (1).
Template-based modeling
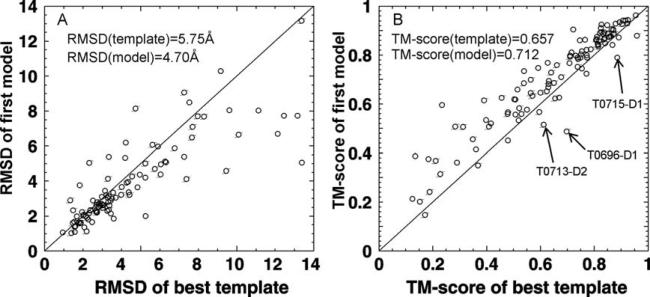
One of the major challenges in template-based structure modeling is the refinement of the threading templates relative to the native structures. In Figure 6, we plot the RMSD and TM-score comparisons of the first full-length models submitted by Zhang-Server versus the best template structures identified by LOMETS. Here, only the 110 TBM domains are counted. If considering the RMSD in the same aligned regions by threading, there are 87 cases that the final models are driven closer to the native than that of the initial templates, where in 23 cases the models have a higher RMSD to the native than the initial templates. The average RMSDs in the same threading aligned region are 4.70 and 5.75 Å for the final model and threading templates, respectively. These improvements are consistent with data observed in previous CASP experiments12,19,20 and benchmark tests,9,34 which are mainly attributed to the optimized knowledge-based potential of I-TASSER assembly and the fact that multiple templates have been used by I-TASSER and the consensus restraints from multiple templates have a higher accuracy than that from the individual templates.
Figure 6.
The quality of the first models by Zhang-Server versus that of the best templates identified by LOMETS for the 110 TBM targets. (A) RMSD to the native calculated in the same threading aligned regions; (B) TM-score. The arrows label the notable targets for which I-TASSER modeling makes the final models significantly worse than the initial templates.
The inclusion of full-length QUARK models as starting conformations also contributes to model quality improvement. Compared with the traditional I-TASSER pipeline starting from LOMETS templates, the I-TASSER pipeline with LOMETS and QUARK models generated the first models with the average TM-score increased by 1.1%, GDT-HA by 2.5%, and hydrogen-bond (HB) score by 2.7%, respectively. The inclusion of the QUARK models contribute apparently more onto the improvement of local structures as the latter two scores (GDT-HA and HB-score) are more sensitive to the quality of the local structures of the predicted models.
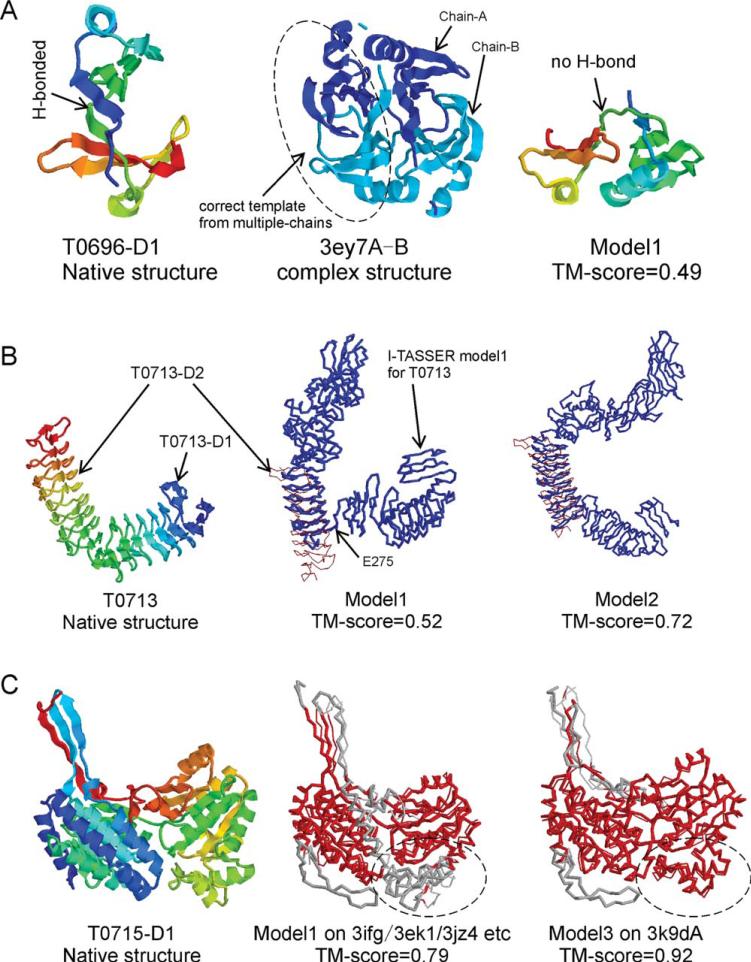
Nevertheless, there are several cases in which the ITASSER reassembly made the models significantly worse than the best templates [labeled in Fig. 6(B)]. These cases highlight the typical issues of the current pipelines in template-based structure modeling. First, T0696-D1 is a beta protein consisting of 111 residues and containing eight anti-parallel beta-strands, where the first strand is paired with the fifth strand by hydrogen-bonding [Fig. 7(A)]. The dominant template hit from LOMETS, 3ey7A, is one chain of the 3ey7A-B dimer complex, which is a domain swap of the target protein. As a result, the first model predicted by I-TASSER has the correct beta-hairpin topology but with the orientation of N-terminal domain tilted relative to the C-terminal domain where the hydrogen-bonds between the first and fifth strands are broken, which results in a TM-score = 0.49, much lower than the best template. This example of the failed cases highlights the requirement of including domain-swapped structures from multi-chain complexes into the threading library. In our post-CASP test, we constructed an artificial template by connecting the two chains in 3ey7, where an alignment to the template generated a better model of TM-score = 0.741 with the domain orientation correctly built.
Figure 7.
Three typical examples where I-TASSER made the best threading templates worse. (A) The best templates for T0696-D1 is from a domain-swap of a multi-chain complex which was missed by the single-chain based threading programs. (B) Incorrect domain split results in inappropriate first model for T0713-D2. (C) Incorrect model selection for T0715-D1 where the best model is from the minority of threading templates. The red color in the superposition refers to the regions with a pair-wise distance below 5 Å.
T0713-D2 is a typical example of failure due to incorrect domain parsing. The target T0713 includes 739 residues, while X-ray crystallography only solved the two domains (T0713-D1 (A33-N207) and T0713-D2 (Y208-F406)) with 374 residues. As shown in Figure 7(B) (middle panel), the first I-TASSER model split the domain boundary at E275 following the ThreaDom prediction which was based on the template alignments from 3sb4A. Therefore, the orientation of the region in (Y208-E275) is tilted away from the main-body of T0713-D2, resulting in a TM-score = 0.52. In the second model, I-TASSER takes another domain parsing based on 2id5B, which has the entire domain modeled correctly with a much improved TM-score = 0.72.
T0715-D1 is a two domain protein but the assessor assigned it as one target unit in the assessment because full-length homologous templates of the target exist in the PDB [Fig. 7(C)]. Most of the LOMETS programs assigned the dehydrogenase proteins with PDB IDs 3ifgA, 3ek1A, 3jz4A, 1o04A, 2wmeA as top templates. The templates in this group have a high pair-wise sequence identity (>60%). The first I-TASSER model from the largest cluster is very close (TM-score >0.9) to these templates, due to the consensus. However, the TM-score of the first model to the target structure is only 0.79, with the major error being due to the big loop region in (E292–F345) as highlighted in Figure 7(C). The best template is 3k9dA which is also a dehydrogenase from listeria monocyto-genes EGD-e but has the loop region tilted compared with other dehydrogenases. This template was hit only by MUSTER24 and the structure conformations similar to it are the minority in the I-TASSER simulations. The SPICKER cluster program15 therefore ranks them as the third cluster, which has a TM-score = 0.92 to the native. This target highlights a significant problem of the consensus-based structure modeling approaches, where the best templates can be ignored when they are hit by a minority set of threading programs.
Free modeling
As demonstrated in previous CASP experiments,8,12,20,35 the traditional fragment assembly approaches have the ability to construct correct folds from scratch. However, the success of this approach is limited to small proteins below 100 residues and most of the successful cases are alpha proteins. The major reason is that the small alpha proteins have, in general, much smaller conformational space compared with the big proteins and those with complicated beta-strand topologies.
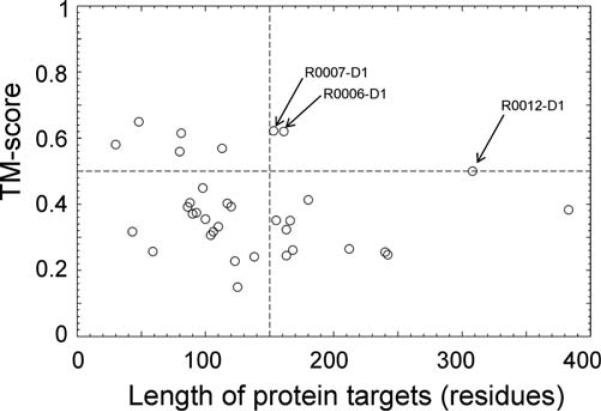
In Figure 8, we present a summary of the I-TASSER and QUARK based modeling results on the 34 FM targets versus the length of the protein targets. Interestingly, there are three successfully modeled targets (R0006-D1, R0007-D1, and R0012-D1) which are longer than 150 residues and for which the best I-TASSER models have the correct fold, that is, with a TM-score >0.5.26 A closer check on R0012-D1 (308 residues) shows that the success of this target is attributed to the template from 3cl6A which was correctly recognized by several hidden Markov model based threading approaches. The TM-score of the template is 0.48 and the final model after I-TASSER refinement is 0.50. This target was involved in the CASP ROLL experiment probably due to the other two domains (R0012-D2 and R0012-D3) for which no significant templates were identified by LOMETS.
Figure 8.
TM-score of the best models by Zhang-Server in the FM category versus the length of the protein targets. Proteins with TM-score >0.5 and length >150 residues are labeled.
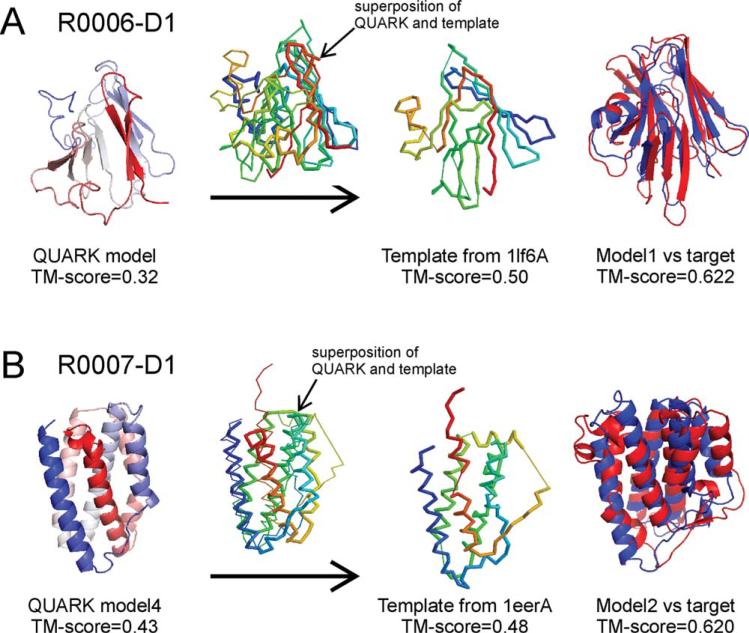
R0006-D1 is a challenging target with a beta-barrel topology consisting 12 beta-strands from 169 residues. None of the QUARK models have the topology correctly assembled. However, the first model from the QUARK simulations has the beta-hairpin in the right-hand side approximately constructed, which results in an overall TM-score = 0.32 [Fig. 9(A)]. When superimposed with the structures in the LOMETS template pool, this model fishes out a template (PDB ID: 1lf6A) which has a TM-score = 0.50 but ranked relatively low (52nd) in LOMETS. The best match between the QUARK model and the template is in the beta-hairpin regions, which demonstrates that only partially modeled structures from the ab initio modeling could be sufficient to pick up correct templates from the PDB library. After the I-TASSER refinement, the first submitted model has a TM-score 5 0.622 for this target [Fig. 9(A) right panel].
Figure 9.
Successful modeling of two FM targets by Zhang-Server. (A) R0006-D1 is a beta-barrel protein of 169 residues encoded in the genome of bacteroides thetaiotaomicron, VPI-5482. The ab initio folding algorithm QUARK generates five models with the highest TM-score = 0.32; based on the QUARK models, the structural superposition fishes out a template of TM-score = 0.5, which results in the final submitted model with a TM-score = 0.622 after the I-TASSER refinements. (B) R0007-D1 is an alpha protein with 161 residues from the human interleukin-34 protein. The best structure generated by QUARK has a TM-score of 0.43; based on the QUARK models the structural superposition picks up a template of TM-score = 0.48 from the LOMETS template pool which results in a final model of TM-score = 0.620 after the I-TASSER refinements.
R0007-D1 is a medium-size alpha-protein of 161 residues with a six-helix bundle topology. The best QUARK model has the global topology approximately constructed; but the spatial order of the two loops between helix-1, -2 and helix-5, -6 is swapped compared with the target structure. The QUARK model has a TM-score = 0.43 to the native [Fig. 9(B) left panel]. The TM-score sort based on the QUARK models ranks the best template 1eerA on the top of all LOMETS templates, which has a TM-score = 0.48 to the native. Interestingly, the loop swapping error in the QUARK model is now fixed in the template, that is, the two loops have the same order to the target structure, showing the possibility to exploit the natural template structures to amend the local structural errors of ab initio folding. Finally, after the I-TASSER refinement, the second submitted full-length model has a TM-score = 0.620 to the native structure [Fig. 9(B) right panel].
The two successfully modeled examples shown in Figure 9 are all from the CASP10-ROLL experiment and considered as difficult targets for structural modeling with the predictions generated before the normal CASP season. For the official CASP10 experiment, however, we found that no target in the FM category has the I-TASSER models with a TM-score >0.5. One reason is probably due to the fact that a cutoff on the maximum GDT-score of the final models was used for the definition of the FM targets.36 In fact, there was no target in the FM category of the official CASP10 experiment which has a GDT-score >0.45 by any groups according to the assessor's report.37
Nevertheless, there are four domains in the I-TASSER predictions which have reasonable folds with a TM-score >0.4. The models of these examples (from T0666-D1, T0735-D2, T0737-D1, and T0756-D2) are showed in Figure S1 in the Supporting Information. Most of these domains are a-proteins with length ranging from 86 to 188 residues. The QUARK ab initio folding simulations generated models with TM-score = 0.302, 0.316, 0.290, and 0.361, respectively, for T0666-D1, T0735-D2, T0737-D1, and T0756-D2. The TM-scores for the best in top five templates after the QUARK-model based template sorting are slightly increased to 0.361, 0.307, 0.324, and 0.375, which result in the final full-length models with TM-score = 0.413, 0.404, 0.402, and 0.401, respectively, after the I-TASSER reassembly refinements (see Supporting Information Fig. S1).
CONCLUSIONS
We have tested two methods of the I-TASSER and QUARK algorithms and the combinations for protein structure prediction in the CASP10 experiment. The most notable new observation is that the interplay of ab initio modeling and template-based modeling methods can help improve the accuracy of protein structures in both categories of FM and TBM targets. First, the structural models by ab initio models can help pick up correct folds by structural alignments from a list of low-rank threading templates for the distant-homology proteins. In CASP10, this strategy helped successfully fold two FM proteins (R0006-D1 and R0007-D1) with size >150 residues, a length range that has never been reached for the FM targets in previous CASP experiments. Second, the spatial restraints from threading alignments can be used to guide the structural assembly simulation of ab initio folding methods. In our experiment, different levels of restraint information are collected from the LOMETS alignments, which are used to guide the QUARK simulations for TBM targets. The QUARK models are then used as input templates for the I-TASSER refinements. The structural accuracy of the final models, especially the local structure quality as assessed by GDT-HA and HB-scores, outperforms that of the models generated by the original ITASSER pipeline starting directly from the LOMETS threading alignments.
Another relatively new observation in our tests is that the fragment structures found their usefulness in different steps of the I-TASSER structure predictions. First, the fragments recognized by the segmental threading SEGMER23 were used to improve the medium-to-long range distance restraints for the regions that the global alignments have often missed. Second, the fragment structures identified by TM-align from the PDB using segments from the I-TASSER models as probes can be used to improve the energy funnel shape of the physics-based force field and therefore improve the ability of structural refinement of molecular dynamics simulations.17 In the CASP10 data, both the GDT-HA score and the hydrogen-bonding network of the final models were improved by the use of fragment-guided FG-MD simulations, as compared with our models in previous CASPs12,19,20 which used only reduced modeling for full-length structure constructions.16
There are several other advancements of the I-TASSER pipelines which have been discussed in detail in previous CASP reports12,19,20 but not in this report although they have critical importance to the success of our structure modeling in CASP10. These include (1) the consistent template refinements driven by the consensus of multiple template restraints, (2) the successful ab initio folding of small proteins by QUARK driven by the fragment-based distance profiles, and (3) the usefulness of the sequence-based contact predictions for both TBM and FM modeling. As a new application to the I-TASSER pipeline, we also found that the meta-MQAP approach, combining both consensus- and statistics-based scores, can improve the overall model selection from SPICKER, as demonstrated by the improved total TM-score of all modeling targets (data not shown).
Despite the success, there are still significant challenges in the current pipelines. For the TBM targets, one of the major errors comes from the uncertainty of the domain split. The current domain prediction by ThreaDom is based on threading alignments, which often has difficulty for distantly-homologous targets. This issue also affects the modeling of the FM targets if the sequence of the FM domains cannot be correctly isolated out. The second challenge for the TBM targets is the correct selection of templates when the consensus hits do not correspond to the best template alignments.
For the FM targets, the folding of beta-proteins with long-range beta-strand contacts remains a significant challenge since these proteins have generally a much more complicated topology than the alpha-proteins or the beta-protein with short-range contacts. The current ab initio folding methods with limited simulation time can rarely reach the conformation of such complicated topology when starting from scratch. One temporary solution to the issue might be to start the ab initio folding simulations from an enumeration of all typical beta-protein topologies in the PDB considering that the structure space of the PDB library is approaching to being complete.38 Second, although the interplay of QUARK with I-TASSER has the potential to fold medium-size FM proteins, the QUARK program on its own has difficulty to consistently assemble correct structure for proteins >100–120 residues from scratch. This significantly limits the potential of the hybrid methodology for reliable construction of the medium-to-large size protein structures. Thus, the development of more reliable methods for distant-homology template recognition and for medium-size ab initio folding remains the major bottleneck to overcome for the current structure prediction pipelines.
Supplementary Material
ACKNOWLEDGMENTS
The author thanks Drs. D Xu, J Yang, A Roy, and R Yan for assistance in CASP10.
Grant sponsor: NSF Career Award; Grant number: DBI 0746198; Grant sponsor: National Institute of General Medical Sciences; Grant number: R01GM083107.
REFERENCES
- 1.Levitt M, Warshel A. Computer-simulation of protein folding. Nature. 1975;253:694–698. doi: 10.1038/253694a0. [DOI] [PubMed] [Google Scholar]
- 2.Lewis PN, Momany FA, Scheraga HA. Folding of polypeptide chains in proteins - proposed mechanism for folding. Proc Natl Acad Sci USA. 1971;68:2293–2297. doi: 10.1073/pnas.68.9.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mccammon JA, Gelin BR, Karplus M. Dynamics of folded proteins. Nature. 1977;267:585–590. doi: 10.1038/267585a0. [DOI] [PubMed] [Google Scholar]
- 4.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 6.Simons KT, Kooperberg C, Huang E, Baker D. Assembly of protein tertiary structures from fragments with similar local sequences using simulated annealing and Bayesian scoring functions. J Mol Biol. 1997;268:209–225. doi: 10.1006/jmbi.1997.0959. [DOI] [PubMed] [Google Scholar]
- 7.Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liwo A, Lee J, Ripoll DR, Pillardy J, Scheraga HA. Protein structure prediction by global optimization of a potential energy function. Proc Natl Acad Sci USA. 1999;96:5482–5485. doi: 10.1073/pnas.96.10.5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu S, Skolnick J, Zhang Y. Ab initio modeling of small proteins by iterative TASSER simulations. BMC Biol. 2007;5:17. doi: 10.1186/1741-7007-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu D, Zhang Y. Ab initio protein structure assembly using continuous structure fragments and optimized knowledge-based force field. Proteins. 2012;80:1715–1735. doi: 10.1002/prot.24065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klepeis JL, Wei Y, Hecht MH, Floudas CA. Ab initio prediction of the three-dimensional structure of a de novo designed protein: a double-blind case study. Proteins. 2005;58:560–570. doi: 10.1002/prot.20338. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Y. Template-based modeling and free modeling by I-TASSER in CASP7. Proteins. 2007;69:108–117. doi: 10.1002/prot.21702. [DOI] [PubMed] [Google Scholar]
- 13.Wu ST, Zhang Y. LOMETS: a local meta-threading-server for protein structure prediction. Nucleic Acids Res. 2007;35:3375–3382. doi: 10.1093/nar/gkm251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu D, Zhang Y. Towards optimal fragment generations for ab initio protein structure assembly. Proteins. 2013;81:229–239. doi: 10.1002/prot.24179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Y, Skolnick J. SPICKER: a clustering approach to identify near-native protein folds. J Comput Chem. 2004;25:865–871. doi: 10.1002/jcc.20011. [DOI] [PubMed] [Google Scholar]
- 16.Li Y, Zhang Y. REMO: a new protocol to refine full atomic protein models from C-alpha traces by optimizing hydrogen-bonding networks. Proteins. 2009;76:665–676. doi: 10.1002/prot.22380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang J, Liang Y, Zhang Y. Atomic-level protein structure refinement using fragment-guided molecular dynamics conformation sampling. Structure. 2011;19:1784–1795. doi: 10.1016/j.str.2011.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Y, Kolinski A, Skolnick J. TOUCHSTONE II: a new approach to ab initio protein structure prediction. Biophys J. 2003;85:1145–1164. doi: 10.1016/S0006-3495(03)74551-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y. I-TASSER: fully automated protein structure prediction in CASP8. Proteins. 2009;77:100–113. doi: 10.1002/prot.22588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu D, Zhang J, Roy A, Zhang Y. Automated protein structure modeling in CASP9 by I-TASSER pipeline combined with QUARK-based ab initio folding and FG-MD-based structure refinement. Proteins. 2011;79:147–160. doi: 10.1002/prot.23111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu S, Zhang Y. A comprehensive assessment of sequence-based and template-based methods for protein contact prediction. Bioinformatics. 2008;24:924–931. doi: 10.1093/bioinformatics/btn069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu S, Szilagyi A, Zhang Y. Improving protein structure prediction using multiple sequence-based contact predictions. Structure. 2011;19:1182–1191. doi: 10.1016/j.str.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu S, Zhang Y. Recognizing protein substructure similarity using segmental threading. Structure. 2010;18:858–867. doi: 10.1016/j.str.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu S, Zhang Y. MUSTER: improving protein sequence profile-profile alignments by using multiple sources of structure information. Proteins. 2008;72:547–556. doi: 10.1002/prot.21945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y, Skolnick J. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 2005;33:2302–2309. doi: 10.1093/nar/gki524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu J, Zhang Y. How significant is a protein structure similarity with TM-score = 0.5? Bioinformatics. 2010;26:889–895. doi: 10.1093/bioinformatics/btq066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Y, Skolnick J. Scoring function for automated assessment of protein structure template quality. Proteins. 2004;57:702–710. doi: 10.1002/prot.20264. [DOI] [PubMed] [Google Scholar]
- 29.Zhang J, Zhang Y. A novel side-chain orientation dependent potential derived from random-walk reference state for protein fold selection and structure prediction. PLoS One. 2010;5:e15386. doi: 10.1371/journal.pone.0015386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou H, Zhou Y. Distance-scaled, finite ideal-gas reference state improves structure-derived potentials of mean force for structure selection and stability prediction. Protein Sci. 2002;11:2714–2726. doi: 10.1110/ps.0217002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shen MY, Sali A. Statistical potential for assessment and prediction of protein structures. Protein Sci. 2006;15:2507–2524. doi: 10.1110/ps.062416606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eisenberg D, Luthy R, Bowie JU. VERIFY3D: assessment of protein models with three-dimensional profiles. Methods Enzymol. 1997;277:396–404. doi: 10.1016/s0076-6879(97)77022-8. [DOI] [PubMed] [Google Scholar]
- 33.Xue Z, Xu D, Wang Y, Zhang Y. ThreaDom: extracting protein domain boundary information from multiple threading alignments. Bioinformatics. doi: 10.1093/bioinformatics/btt209. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, Skolnick J. Automated structure prediction of weakly homologous proteins on a genomic scale. Proc Natl Acad Sci USA. 2004;101:7594–7599. doi: 10.1073/pnas.0305695101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bradley P, Malmstrom L, Qian B, Schonbrun J, Chivian D, Kim DE, Meiler J, Misura KM, Baker D. Free modeling with Rosetta in CASP6. Proteins. 2005;61:128–134. doi: 10.1002/prot.20729. [DOI] [PubMed] [Google Scholar]
- 36.Lee BK. 10th Community wide experiment on the critical assessment of techniques for protein structure prediction. Gaeta; Italy: 2012. Domain definition and classification. [Google Scholar]
- 37.Lee BK. 10th Community wide experiment on the critical assessment of techniques for protein structure prediction. Gaeta; Italy: 2012. Template free modeling assessment in CASP10. [Google Scholar]
- 38.Zhang Y, Skolnick J. The protein structure prediction problem could be solved using the current PDB library. Proc Natl Acad Sci USA. 2005;102:1029–1034. doi: 10.1073/pnas.0407152101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.