Abstract
Correct centriole/basal body positioning is required for numerous biological processes, yet how the cell establishes this positioning is poorly understood. Analysis of centriolar/basal body duplication provides a key to understanding basal body positioning and function. Chlamydomonas basal bodies contain structural features that enable specific triplet microtubules to be specified. Electron tomography of cultures enriched in mitotic cells allowed us to follow basal body duplication and identify a specific triplet at which duplication occurs. Probasal bodies elongate in prophase, assemble transitional fibers (TF) and are segregated with a mature basal body near the poles of the mitotic spindle. A ring of nine-singlet microtubules is initiated at metaphase, orthogonal to triplet eight. At telophase/cytokinesis, triplet microtubule blades assemble first at the distal end, rather than at the proximal cartwheel. The cartwheel undergoes significant changes in length during duplication, which provides further support for its scaffolding role. The uni1-1 mutant contains short basal bodies with reduced or absent TF and defective transition zones, suggesting that the UNI1 gene product is important for coordinated probasal body elongation and maturation. We suggest that this site-specific basal body duplication ensures the correct positioning of the basal body to generate landmarks for intracellular patterning in the next generation.
Keywords: Chlamydomonas, basal bodies, uni1, tomography
Introduction
Correct centriole/basal body positioning is required for a number of key biological processes including coordinated ciliary beating, orienting mitotic spindle poles, cell migration and establishment of cell geometry [Bornens, 2012]. The direction of ciliary beating is dictated by the orientation of the basal bodies that nucleate them [Tamm et al., 1975; Boisvieux-Ulrich et al., 1985]. Defects in correct positioning of these organelles have profound effects. For example, defects in cilia-driven flow cause a range of disease symptoms that include hydrocephalus, situs inversus, and middle ear and respiratory infections [reviewed in Wallingford, 2010; Ostrowski et al., 2011]. Understanding what controls centriole and basal body positioning is therefore important.
The correct positioning of the basal body complex in Chlamydomonas reinhardtii is particularly important in establishing the stereotypical cell geometry found in this algal cell [reviewed in Dutcher, 2007]. Organelles such as the eyespot, mating structures, contractile vacuole, and cleavage furrow are associated with specific MT bundles of the basal body complex and contribute to the cellular asymmetry that is inherited by daughter cells [Holmes and Dutcher, 1989; Feldman et al., 2007; Mittelmeier et al., 2011].
In Chlamydomonas, the interphase cell contains two mature basal bodies and two probasal bodies, separated by two sets of rootlet MT [Ringo, 1967; Johnson and Porter, 1968; Rosenbaum et al., 1969; Cavalier-Smith, 1974; O’Toole et al., 2003; Geimer and Melkonian, 2004]. A two membered rootlet MT bundle separates a mother basal body and its probasal body. The two mature basal bodies are connected at their proximal ends by two proximal striated fibers (psf) and at the midregion by a distal striated fiber. The distal-most region of the basal body contains transitional fibers (TF) that radiate out from the triplet blades and anchor the organelle to the plasma membrane [Weiss et al., 1977]. The transition zone is the region of conversion from triplet to doublet MTs and the assembly of a remarkable nine-pointed stellate array of fibers. Beyond the transition zone, the 9 + 2 MT arrangement of the flagellum proper is then assembled.
Centrioles and basal bodies share a common cylindrical structure built from nine blades of triplet MTs. Although centrioles and basal bodies are cylindrical they are not rotationally symmetric and have a variety of internal and external ultrastructural features that are only found at specific angular positions around their circumference that allow the rotational orientation to be specified [Hoops and Witman, 1983; Beisson and Jerka-Dziadosz, 1999; Silflow et al., 2001; Geimer and Melkonian, 2004]. Likewise, the centriole/basal body exhibits asymmetries along their length; the proximal base contains a prominent cartwheel structure and the distal region can assemble appendages. Recently, an elegant, high resolution cryotomography and subvolume averaging study has revealed that there are multiple longitudinal variations along the basal body that suggest a multistep assembly mechanism [Li et al., 2012]. In addition, the centrioles in an animal centrosome or flagellated protists are nonequivalent; they differ in age with one, older centriole and a newly formed centriole. In animal cells, the older centriole can be distinguished by the presence of subdistal and distal fibers and is competent to assemble the primary cilium [reviewed in Kobayashi and Dynlacht, 2011].
The uni mutants in Chlamydomonas illustrate the nonequivalence of the basal body pair. As the name implies, cells assemble one flagellum on the basal body that is on the side of the cell away from the eyespot (designated trans) [Huang et al., 1982; Holmes and Dutcher, 1989; Dutcher and Trabuco, 1998; Piasecki and Silflow, 2009]. This basal body is the older of the two and has undergone maturation during the cell cycle. The uni3 mutant cells assemble 0, 1, or 2 flagella and like in uni1 cells, the single flagellum is trans to the eyespot. The UNI3 gene encodes delta-tubulin and cells with mutations in this gene contain basal bodies with mostly doublet, rather than triplet MTs. The uni2 mutant also assembles 0, 1, or 2 flagella. The UNI2 protein, which is the homolog of Cep120, is present in different phosphorylation states during and after mitosis and localizes to the region of triplet to doublet transition [Piasecki and Silflow, 2009]. Cep120 is found on daughter centrioles in mammalian cells [Mahjoub et al., 2010]. To date, the UNI1 gene product is not known. Images from thin sections of uni1 and uni2 mutants show a number of structural defects, including elongated transition zone structures, triplet MTs in the transition zone proper, reduced TF and basal bodies that are not docked at the plasma membrane [Huang et al., 1982; Piasecki and Silflow, 2009]. These results suggest that UNI1 and UNI2 are important for the conversion of the daughter basal body into a mature basal body that is then competent to build a proper transition zone and subsequent flagellum.
In this paper, we describe a comprehensive overview of the structural events that occur during duplication of the basal body complex and how the basal body complex contributes to the establishment of cell geometry. We followed the process of basal body duplication using cultures of cells grown in a light/dark cycle to maximize the number of cells entering mitosis. Electron tomography and modeling of basal body complex structures were then performed to study its 3D spatial organization during duplication. We confirm and extend observations from numerous landmark studies in Chlamydomonas and other green algae. We found that daughter basal bodies arise through extension of probasal body triplets and assemble TF during prophase. Basal bodies from uni1-1 mutants are defective in new daughter assembly. New basal body duplication occurs at a specific site orthogonal to triplet #8 of the mother basal body. This coordinated duplication and assembly of the basal body complex ensures the transmission of intracellular patterning to the next generation.
Results
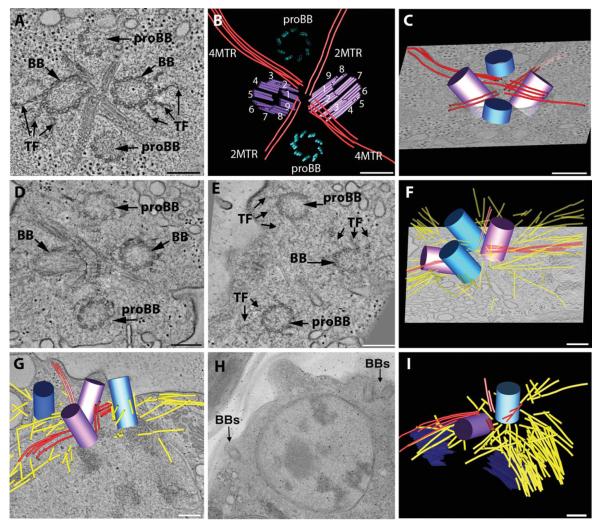
Interphase cells contain basal body complexes with a highly stereotypical organization; two mature basal bodies (which are called mother and daughter basal bodies), and two probasal bodies are arranged in a cross-like arrangement that are separated by four rootlet MT bundles (Figs. 1A–1C; see Movie S1 in Supporting Information) [Ringo, 1967; Johnson and Porter, 1968; Cavalier-Smith, 1974; O’Toole et al., 2003]. Unlike mammalian cells the probasal bodies are present throughout interphase; they contain nine triplet MT blades, a proximal cartwheel and have an average length of 86 ± 12 nm (−sd; n = 13) (Figs. 1A–1C; proBB, blue cylinder). The triplet MT blades of the mature basal bodies have an established numbering relative to the rootlet MTs; this asymmetry of the basal bodies is hypothesized to contribute to the structural asymmetry of the flagellar waveform (Fig. 1B (purple) [Hoops and Witman,1983]. Interestingly, we found that the basal body complex maintains the rootlet MT organization throughout the duplication cycle, and this enabled the identification of specific triplet MT blades at which duplication intermediates assemble.
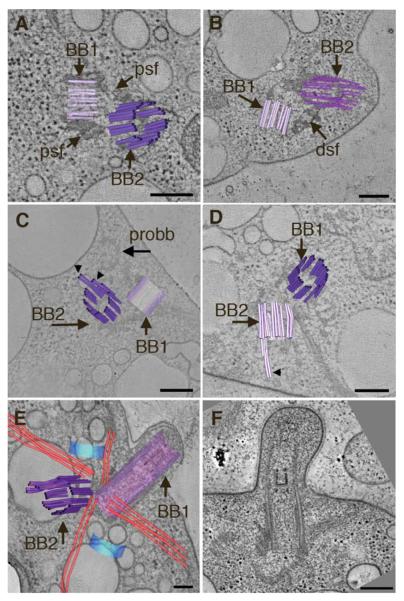
Fig. 1. Early events in basal body duplication in Chlamydomonas.
(A, B, C) Interphase cells contain a basal body complex formed from two mature (BB) and two immature basal bodies (proBB) (purple and blue, respectively) separated by four bundles of rootlet MTs in a cross-like arrangement (red lines). A movie of the complete tomographic volume is shown in Supporting Information movie S1. (D, E, F) Cell cultures were synchronized using cycles of light and dark and examined by electron tomography. Soon after entering the dark cycle, the probasal bodies elongate (blue cylinders), the rootlet MTs remain intact (red lines) and there is an increase in cytoplasmic MTs (yellow lines). The elongated probasal bodies (proBB) assemble TF similar to the mature basal bodies (BB). A movie of the complete tomographic volume is shown in Supporting Information movie S2. (G, H, I) At late prophase, the basal body complex separates and two basal bodies migrate to opposite sides of the nucleus (BBs) and there is a noticeable increase in cytoplasmic MTs (yellow lines). Bar = 200 nm.
Daughter Basal Bodies Assemble Transitional Fibers in Prophase
To understand more about the duplication process, wild-type cells were grown in a 14/10 light/dark cycle to enrich cultures for mitotic cells in order to examine the early steps in basal body duplication. When shifted to the dark, the cells enter prophase and the two probasal bodies (Figs. 1D–1F; proBB) elongate to a mean length of 351 ± 28 nm (±sd; n = 10) and orient to a more vertical, parallel position relative to the mother basal bodies (see Movie S2 in Supporting Information; purple cylinders; [Gould, 1975; Gaffal, 1988]. The two mature basal bodies are connected by a distal striated fiber (Fig. 1D, dsf) [Gaffal, 1988]. TF assemble on the distal triplet blades of the elongated probasal bodies, anchoring them beneath the plasma membrane (Fig. 1E; TF) [Gaffal, 1988]. At this point in the cell cycle, we refer to them as daughter basal bodies. The rootlet MT bundles maintain the cross-like arrangement (Fig. 1F; red lines; Movie S2) typical of interphase, and many additional cytoplasmic MTs assemble (Fig. 1F; yellow lines). In late prophase, tomographic analysis shows that the distal striated fiber disassembles, the flagella are released proximal to the flagellar transition zone [Gaffal, 1988; Parker et al., 2010], and the basal bodies separate into two complexes that migrate to opposite sides of the nucleus (Figs. 1G–1H; BBs). Rootlet MT bundles assemble at the two complexes to generate the cross-shaped array and there is a noticeable further increase in the number of cytoplasmic MTs near an opening in the nuclear envelope (Fig. 1I; yellow, dark blue). Thus, each pole of the mitotic spindle contains one older basal body (Fig. 1I, purple cylinder) and the newly elongated daughter basal (Fig. 1I, light blue cylinder), whose duplication was initiated in the previous generation [Dutcher, 2007].
Chlamydomonas Exhibits Site-Specific Basal Body Duplication
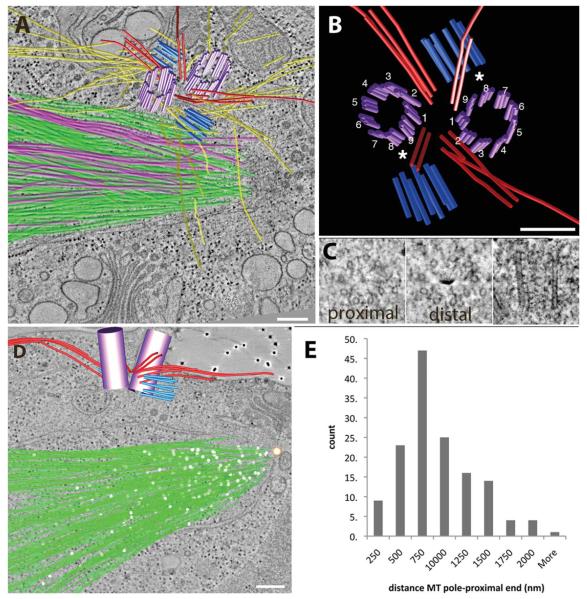
Microtubule cylinders of new probasal bodies assemble during mitosis, which was observed previously using thin sections (Fig. 2; see Movie S3 in Supporting Information) [Johnson and Porter, 1968; Cavalier-Smith, 1974; Gaffal, 1988]. At metaphase, the new probasal bodies form from a ring of nine singlet MTs and are separated from the mother and daughter basal bodies by four bundles of rootlet MTs that tended to bend at steep angles toward the center of the complex (Fig. 2A, red lines) [Gaffal, 1988]. Because the rootlet MT organization was established at this stage, we confirmed that the daughter basal body assembled orthogonal to triplet #8 of the mother basal body (Fig. 2B; see Movie S3 in Supporting Information). Cross sections of the new probasal bodies from the proximal and distal ends and profiles in longitudinal view show that the new probasal bodies are formed as extended singlet MTs (Fig. 2C). However, we were not able to identify a structure connecting the new probasal body to triplet #8. Interestingly, the singlet MTs forming the probasal body cylinder are much longer than the triplet MTs in interphase, with a mean length of 180 ± 36 nm (±sd; n = 13).
Fig. 2. The basal body complex remains associated with the plasma membrane at metaphase.
(A) Kinetochore and non-kinetochore MTs were arranged much like a bouquet (pink and green lines, respectively). (B, C) New probasal bodies (light blue) are formed from a ring of singlet MTs at triplet eight of the mother BB (*; purple). (D, E) Pole-proximal MT ends (white spheres) are distributed along the length of the spindle with a mean distance of 750 nm from the spindle pole. A movie of the complete tomographic volume is shown in Supporting Information movie S3. Bar = 200 nm.
Mitotic Spindle Poles Differ From Mammalian Centrosomes
Both the mature and daughter basal bodies remain associated with the plasma membrane throughout mitosis by means of TF attachment at the cell surface (see Movie S3 in Supporting Information for complete volume). Numerous kinetochore and non-kinetochore spindle MTs are arranged like a bouquet, with their pole-proximal ends focused towards a large polar fenestra in the nuclear envelope (Fig. 2A; pink and green lines, respectively; see Movie S3 in Supporting Information) [Johnson and Porter, 1968]. In addition, the 3D models show that the spindle MTs are separated from the basal body complex by the nuclear envelope. Reconstruction of complete half spindles contain ~300 MTs per spindle pole (Fig. 2D; green lines; see Movie S3 in Supporting Information for the complete volume). Pole-proximal MT ends have a distinctive closed, capped morphology, and these ends are distributed throughout the length of the spindle (Fig. 2D; white spheres). This closed structure is likely to represent the gamma-tubulin complex [Moritz et al., 2000; Kollman et al., 2010]. A distribution of the 3D distances of pole-proximal ends relative to a reference point marking the spindle pole at the nuclear envelope fenestra (Fig. 2D white and tan sphere, respectively) shows that pole-proximal ends have a broad distribution throughout the spindle, with the majority present at a distance of 750 nm from the spindle pole (Fig. 2E). Thus, the MT arrangement of the Chlamydomonas spindle pole is in marked contrast to the radial MT arrays found in mammalian centrosomes. However, it is interesting to note that the basal bodies in Chlamydomonas become the focus of an increased number of cytoplasmic microtubules in mitosis compared to interphase (Figs. 1C, 1F, and 1I). Thus, they show properties expected of centrosome maturation in which the centrioles gain the ability to promote the assembly of cytoplasmic (astral) microtubules [Dobbelaere et al., 2008].
Triplet MTs Assemble at the Distal Region of the Probasal Body
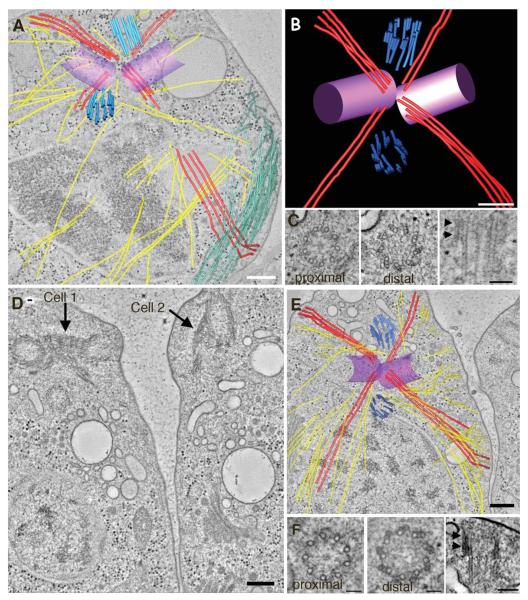
Cells grown under these conditions go through rapid stages of cell division resulting in four to eight daughter cells that are surrounded by a mother cell wall. If a cell has not reached a critical volume, it will divide again without going through an intervening interphase [Umen and Goodenough, 2001]. Tomographic reconstructions of basal body complexes in cells from telophase (Figs. 3A–3C) and those undergoing cytokinesis (Figs. 3D–3F) show that the rootlet MT arrangement is maintained, and that the four-MT rootlet bundle is oriented to define the position of the cleavage furrow (Fig. 3A; red lines). In Fig. 3A, numerous MTs are also detected in the cleavage furrow proper and this MT array is known as the phycoplast (Fig. 3A; green lines) [Johnson and Porter, 1968; Ehler et al., 1995; Ehler and Dutcher, 1998]. Remarkably, doublet and triplet MTs assemble at the distal end of new probasal bodies rather than the proximal end as reported in Paramecium [Dippell, 1968] (Figs. 3B and 3C; arrowheads). A movie showing serial tomographic slices from the proximal to the distal end of a new probasal body illustrates the assembly of triplet blades at the distal end (see Movie S4 in Supporting Information). This orientation is more similar to that reported in the related green alga, Polytoma papillatum [Wolf, 1992] and images of proximal A-tubule extensions have also been reported in S. similus, and P. parva [Beech et al., 1991; Lechtreck and Grunow, 1999]. In addition, B and C tubules of human centrioles have also been reported to assemble along singlet A tubules in a bidirectional manner [Guichard et al., 2010]. At this stage, the new probasal bodies are angled in a position similar to that of interphase probasal bodies (Fig. 3B). However, the mean length of the probasal body is 237 ± 47 nm (±sd; n = 14), which is much longer than the probasal bodies in interphase. Interestingly, the portion containing triplet blades has a mean length of 83 ± 14 nm (±sd; n = 13), which is similar to the interphase probasal body length, although we observed one probasal body had triplets assembled to a length of 234 nm. A similar arrangement of probasal bodies is observed at cytokinesis (Figs. 3D–3F; see Movie S5 in Supporting Information). The cells shown in Fig. 3D are in the final division, due to the orientation of the basal body complexes at the anterior of both cells [Holmes and Dutcher, 1989]. Thus, at telophase and cytokinesis the distal end of the developing probasal body containing triplet MT blades that are placed in the same orientation and at the same distance with respect to the mature basal body as is observed for the interphase probasal body.
Fig. 3. Daughter basal bodies assemble triplet MTs in late mitosis/cytokinesis.
(A, B, C) In telophase, the rootlet MT organization is maintained (red lines) and the four-MT rootlet bundle defines the position of the cleavage furrow. Numerous cytoplasmic microtubules are present (yellow lines) as well as MTs of the phycoplast (green lines). (B,C) Doublet and triplet MTs assemble at the distal end of new daughter basal bodies (E; light blue). A movie showing triplet MTs at the distal end of the new daughter basal body is shown in Supporting Information movie S5. (D, E, F) At cytokinesis, the probasal bodies reside in a position similar to that in interphase and triplet blades are present at the distal end (F; arrowheads). Bar = 200 nm A, B, D, E and 100 nm in C, F.
The Cartwheel is a Dynamic Structure
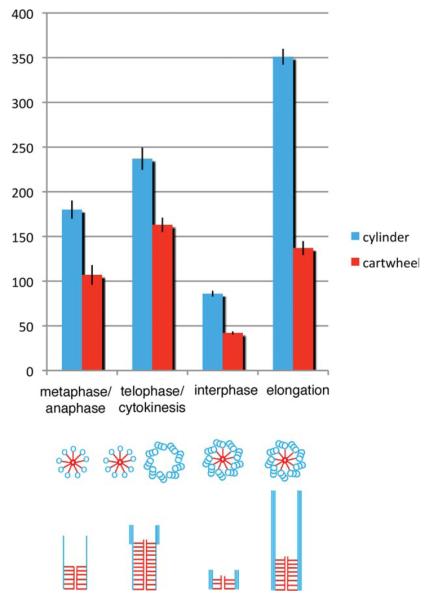
Measurements from the tomographic reconstructions show that cartwheel undergoes significant changes during the cell cycle (Fig. 4; See representative longitudinal images in Supporting Information Fig. 1). New probasal bodies that assemble during metaphase have longer cartwheel structures 107 ± 36 nm (±sd; n = 11) and by telephase/cytokinesis, the cartwheel has increased to 163 ± 31 nm (±sd; n = 15). Singlet MTs are present in the metaphase probasal body and the elongated cartwheel may act to stabilize this forming cylinder. Likewise, the cartwheel in probasal bodies in telophase/cytokinesis extends to the region where distal triplet MT blades assemble (arrowhead, Supporting Information Fig. 1). Elongation of the cartwheel is concurrent with the elongation of triplet MTs of the daughter basal body prior to mitosis. Similar to the triplet MTs, the cartwheel in the developing daughter basal body during elongation is also significantly longer than that found in interphase (137 ± 23 nm (±sd; n = 11) vs. 42 ± 5nm (±sd; n = 10), respectively). Taken together, these results suggest a scaffolding role of the cartwheel during probasal body assembly and maturation.
Fig. 4. The cartwheel is a dynamic structure throughout the basal body duplication cycle.
New probasal bodies are detected at metaphase and contain singlet MTs and elongated cartwheels. Triplet MTs are added at the distal end of the MT cylinder at telophase/cytokinesis. The interphase probasal body MT cylinder and cartwheel elongate at prophase to form the daughter basal body. Displayed are mean ± SEM. Representative longitudinal images at each stage are shown in Supporting Information Fig. 1.
UNI1 is Required for Proper Probasal Body Elongation and Transitional Fiber Assembly
Thin section analysis of eleven basal body complexes and 10 tomographic volumes from the uni1-1 basal bodies confirmed a number of structural defects reported previously by thin section EM, including elongated transition zone structures and defects in triplet to doublet transition (see Figs. 2A–2D Supporting Information and Fig. 5); [Huang et al., 1982; Piasecki and Silflow, 2009]. However, a careful examination of the tomographic volumes revealed that six of 10 cells had at least one basal body whose triplet blades that did not extend past the region where the distal striated fiber attaches (mean length 195 + 20 nm ± sd; n = 6); these basal bodies contain reduced or absent TF. Shown in Figs. 5A–5D are examples of uni1-1 cells containing at least one short basal body (<250 nm) that is connected by a dsf to the older basal body. In Fig. 5A, the uni1-1 basal bodies are connected by two psf, but both basal bodies terminate prematurely with lengths of only ~ 164 and 296 nm, respectively. The serial tomographic slices of the cell shown in Fig. 5A (see Movie S6 Supporting Information) shows that BB2 ends just immediately after the distal striated fiber and only two blades assemble partial TF. In some cells, one basal body terminates at the level of the dsf (Fig. 5B; BB1, dsf) and the other basal body has extended triplets beyond this region to the transition zone (Fig. 5B). The complete volume of this basal body complex (see Movie S7 Supporting Information) shows that TF assemble onto six triplet blades, but this basal body does not build a complete transition zone and two blades end prematurely and are replaced with electron-dense material. Basal bodies with an extension of a subset of triplet blades were also detected (Figs. 5C–5D; arrows). Figure 5E shows a uni1-1 cell that has assembled two basal bodies with altered transition zones. The basal body in longitudinal view (Fig. 5E, BB1; see Movie S8 in Supporting Information) contains multiple, stacked transition zone structures. The triplets in the other basal body (Fig. 5E, BB2; see Movie S8 in Supporting Information) extend beyond the region of the distal striated fiber but do not assemble prominent TF. Six doublet blades end at the transition zone and are replaced with a ring of electron-dense material. Cells with short, bulbous flagella were also detected (Fig. 5F, Fig. S2), which is a common phenotype observed in mutants with defects in intraflagellar transport. Taken together, these observations support the hypothesis that UNI1 is important for the initial elongation of probasal bodies and their subsequent maturation that includes the assembly of TF and a proper transition zone.
Fig. 5. Basal body defects in uni1-1 cells.
(A, B) Basal bodies are connected by two psf but some basal bodies do not extend beyond the dsf and are not docked at the plasma membrane. Movies showing the complete tomographic volumes of the cells in Figs. 5A and 5B are shown in Supporting Information movies S6 and S7, respectively. (C, D) Examples of cells with basal bodies that contain only a subset of MT blades that extend beyond the distal striated fiber (BB2, arrowheads). The assembly of the transition zone is incomplete. Bar = 100 nm. (E) uni1-1 cell that builds altered transition zones in both basal bodies. BB1 has stacked transition zone material and BB2 contains a subset of four-MT blades with triplet and doublet MTs that form and altered transition zone. A movie showing the complete tomographic volume of this cell is shown in Supporting Information movie S8. (F) uni1-1 cell that has built a short, bulbous flagellum. Bar = 200 nm.
Discussion
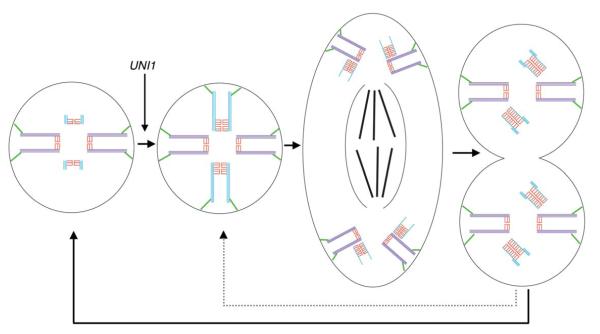
Figure 6 summarizes the steps of basal body duplication in Chlamydomonas. New probasal bodies are initiated during mitosis orthogonal to triplet MT eight of the mother, with the assembly of a ring of nine singlet MTs. The initial singlet MT cylinder elongates to a length sufficient to position its distal end at the correct distance from the mature basal body for the future interphase probasal body. Doublet and triplet blades then assemble at the distal end to generate a length that is similar to the short probasal bodies found in interphase, similar to that reported for P. papillatum [Wolf, 1992]. Our observations support the idea of a simple geometric mechanism to ensure the placement of the new probasal bodies during interphase at the correct position relative to the mature basal bodies. However, if the cell remains large and surpasses a critical size that ensures another cell division, the triplet blades extend fully to the proximal base and the cell enters mitosis again. Our tomograms did not reveal how the probasal bodies found in interphase become shortened; it appears to be a step that occurs very rapidly. We hypothesize that the singlet MTs disassemble from the proximal end, leaving the distal triplet MT blades in a position that is characteristic of the position found in interphase.
Fig. 6. Summary diagram of the basal body duplication cycle in Chlamydomonas.
The probasal body MTs (light blue) and cartwheel (red) elongate and assemble TF (green) at prophase in a UNI1 dependent manner. New daughter BBs are first detected at metaphase and contain singlet MTs and an elongated cartwheel. Doublet MTs, then triplet MTs are assembled at the distal end of the dBB at late mitosis and are oriented in the correct position for interphase (solid line). If the cell has not reached a critical size, it will quickly undergo another mitosis without an intervening interphase (dotted line).
The cartwheel is a dynamic structure that lengthens and shortens throughout the duplication cycle (Fig. 5a) [Beech et al., 1991; Geimer and Melkonian, 2004]. Chlamydomonas contains a presumptive pre-cartwheel protein, CRC70 [Shiratsuchi et al., 2011], and at least the BLD10/Cep135 and BLD12/Sas6 in the cartwheel proper [Hiraki et al., 2007; Nakazawa et al., 2007]. Strains defective in these proteins show severe basal body assembly defects as they do in other organisms [Reviewed in Nigg, 2007].
Cells must have a mechanism to trim both the singlet MTs and the cartwheel to the correct size. Dynamic assembly patterns of specific basal body components have also been reported in Tetrahymena [Pearson et al., 2009a,b]. In wild-type basal bodies, the proximal end is capped with an amorphous ring of material [Ringo, 1967; O’Toole et al., 2003]. The assembly of this amorphous material may serve to limit the assembly of the cartwheel. In mammalian cells, the Sas-6 protein of the cartwheel is missing from centrioles in G1 [Strnad et al., 2007]. This dynamic change in cartwheel length during new basal body assembly supports the hypothesis that the cartwheel serves as a scaffold during new basal body assembly, as reported in other organisms [Strnad and Gönczy, 2008].
A hallmark of centriole/basal body maturation is the assembly of subdistal and distal appendages. In animal cells, these structures are present on the older centriole and appendage proteins such as ODF2/cenexin, Cep164 and ninein are important for cilogenesis [Ishikawa 2005, Graser, 2007]. The TFs in Chlamydomonas are thought to be the structural equivalent of distal appendages and assemble onto newly elongated daughter basal bodies during late prophase. The UNI1 protein is important in the maturation of basal bodies competent to assem a proper transition zone and flagellum [Huang et al., 1982; Piasecki and Silflow, 2009]. Lengthening the cell cycle (Holmes and Dutcher, 1992) does not increase the proportion of biflagellated cells; this suggests that maturation of the daughter basal body requires progression through the cell cycle as has been observed for maturation in mammalian centrioles. Maturation in mammalian cells takes about 1.5 cell cycles and involves the recruitment of distal appendages [Vorobjev and Chentsov, 1982]. Quantitative proteomics has identified five candidate distal appendage proteins in mammalian cells [Tanos et al., 2013]. The Chlamydomonas genome has homologs of four of these genes (CCDC41, SCLT1, FBF1, and CEP164). However, the role of these genes in Chlamydomonas is unknown.
A second line of evidence that maturation requires progress through the cell cycle comes from analysis of dikaryons. When Chlamydomonas cells mate, the flagella adhere to each other, leading within ~10 min to cell fusion and formation of a single cell with four flagella and two nuclei, which is known as a dikaryon. If a mutant parent is mated to a wild-type parent, rescue of a mutant phenotype can be determined. Most flagellar motility phenotypes can be rescued [Luck et al., 1977]. The uni1 mutant, although recessive in stable diploid strains, is not rescued by wild-type cytoplasm. Because gametes and dikaryons are in G1 of the cell cycle, the timing may be wrong.
Although previously published ultrastructural reports using serial thin sections showed a strong defect in the region of the transition zone structures and defects in triplet to doublet transition, analysis of the tomographic volumes revealed additional defects. In the tomographic volumes, where both basal bodies could be examined, some basal bodies (presumably the daughters) were defective in triplet elongation and assembly of TF. Profiles of uni1, uni2 and uni1; uni2 double mutant basal bodies containing reduced TF were observed by others [Piasecki and Silflow, 2009]. This delay in elongation of the distal region of the basal body and subsequent assembly of TF could lead to defects in proper anchoring and positioning of the basal body and its subsequent assembly of a flagellum. Future work to identify the UNI1 protein, its localization and role in flagellar assembly is needed.
This is the first study to identify a specific triplet at which basal body duplication occurs. Our results were made possible by the unique arrangement of the basal body complex and rootlet MTs in Chlamydomonas that allow the numbering of triplet blades to be defined. This result explains how the cell can establish the stereotypical geometry of the basal body complex found in this algal cell. In Chlamydomonas the rootlet MTs are important for positioning of organelles like the eyespot, contractile vacuole, mating structures, and cleavage furrow. Defects in the assembly of basal bodies and their proper placement in the cell can have profound effects on cell division, ciliary, and flagellar function. In cells with a primary cilia, super resolution microscopy has suggested that Plk4, which is needed for building new centrioles, is localized to a single triplet microtubule [Sonnen et al., 2012]. However, it is not known if the Plk4 spot occurs on a specific microtubule triplet blade. In multiciliated epithelia cells, orientation of the basal bodies and the cilia is important for coordination between cilia into metachronal waves. Similarly, the intrinsic asymmetry geometry of the basal bodies and their daughters may be modulated by signals from the planar polarity gene, Dishevelled, that controls the docking and orientation of the basal bodies [Park et al., 2008; Mitchell et al., 2009; Vladar et al., 2012]. This site-specific duplication of basal bodies contributes to establishing and maintaining the intracellular infrastructure of the cell and the subsequent function of cilia and flagella.
Experimental Procedures
Cell culture
The wild-type C. reinhardtii cells 137c mt+ (CC-125) were grown in minimal medium in a 14:10 hour light: dark cycle as previously described [Holmes and Dutcher, 1989] The uni1-1 strain (CC-1901) was obtained from the Chlamydomonas Center and backcrossed twice to ensure a single mutation affecting the flagellar assembly defect.
Preparation Cells for Electron Microscopy
Aliquots were high pressure frozen within the first 1.5 hour of entering the dark cycle, and the frozen cells were freeze-substituted and embedded in epon/araldite using methods described in detail in [O’Toole et al., 2007, 2010]. Serial thick (250–300 nm) sections were collected onto formvar-coated copper slot grids. The grids were post-stained with Reynold’s lead citrate and aqueous uranyl acetate. Colloidal gold particles (15 nm) were affixed to both sides of the grid to serve as fiducial markers for tilt series alignment.
Electron Tomography and Image Analysis
Tomography was performed using a Tecnai F30 microscope operating at 300 kV as essentially described in O’Toole et al. [2007]. The complete basal body complex was reconstructed from 2 to 3 adjacent serial, thick sections. Mitotic stages were assessed by the chromosome arrangement in the tomogram overviews (see Supporting Information Fig. 3). Microtubules of the basal bodies, rootlet bundles, mitotic spindle, and other cytoplasmic MTs were modeled in the tomographic volume using the IMOD software package [Kremer et al., 1995; Mastronarde, 1997]. Basal body dimensions and cartwheel lengths were measured using the imodinfo program. Thirty, serial, tomographic datasets were collected that spanned the cell cycle stages of interphase (9), elongation/prophase (6), metaphase/anaphase (7), and telophase/cytokinesis (10). Serial thin section analysis of eleven cells was performed to document defects in uni1-1 basal bodies. A total of ten dual-axis tomograms were reconstructed to examine the 3-D fine structure of the uni1-1 basal body defects. Measurements of basal body cylinder and cartwheel length were restricted to those datasets where the entire basal body was present in the volume of the reconstruction.
Supplementary Material
Acknowledgments
The authors thank Thomas H. Giddings, Jr. of the MCDB EM Suite for the preparation of the uni1-1 strain. This work was supported by grants 8P41GM103431 from National Institute of General Medical Sciences to Andreas Hoenger and GM032843 from the National Institute of General Medical Sciences to S.K.D.
Abbreviations used
- BB
basal body
- dsf
distal striated fiber
- MT
microtubule
- proBB
probasal body
- psf
proximal striated fibers
- TF
transitional fibers
Footnotes
Additional Supporting Information may be found in the online version of this article.
References
- Beech PL, Heimann K, Melkonian M. Development of the flagellar apparatus during the cell cycle in unicellular algae. Protoplasma. 1991;164:23–37. [Google Scholar]
- Beisson J, Jerka-Dziadosz M. Polarities of the centriolar structure: morphogenetic consequences. Biol Cell. 1999;91:367–378. [PubMed] [Google Scholar]
- Boisvieux-Ulrich E, Laine MC, Sandoz D. The orientation of ciliary basal bodies in quail oviduct is related to the ciliary beating cycle commencement. Biol Cell. 1985;55:147–150. doi: 10.1111/j.1768-322x.1985.tb00417.x. [DOI] [PubMed] [Google Scholar]
- Bornens M. The centrosome in cells and organisms. Science. 2012;335:422–426. doi: 10.1126/science.1209037. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. Basal body and flagellar development during the vegetative cell cycle and the sexual cycle of Chlamydomonas reinhardii. J Cell Sci. 1974;16:529–556. doi: 10.1242/jcs.16.3.529. [DOI] [PubMed] [Google Scholar]
- Dippell RV. The development of basal bodies in paramecium. Proc Natl Acad Sci USA. 1968;61:461–468. doi: 10.1073/pnas.61.2.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbelaere J, Josué F, Suijkerbuijk S, Baum B, Tapon N, Raff J. A genome-wide RNAi screen to dissect centriole duplication and centrosome maturation in Drosophila. PLoS Biol. 2008;6:e224. doi: 10.1371/journal.pbio.0060224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutcher SK. Basal bodies: their roles in generating asymmetry. Harvey Lectures. 2007;102:7–50. doi: 10.1002/9780470593042.ch2. [DOI] [PubMed] [Google Scholar]
- Dutcher SK, Trabuco EC. The UNI3 gene is required for assembly of basal bodies of Chlamydomonas and encodes delta-tubulin, a new member of the tubulin superfamily. Mol Biol Cell. 1998;9:1293–1308. doi: 10.1091/mbc.9.6.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehler LL, Dutcher SK. Pharmacological and genetic evidence for a role of rootlet and phycoplast microtubules in the positioning and assembly of cleavage furrows in Chlamydomonas reinhardtii. Cell Motil Cytoskeleton. 1998;40:193–207. doi: 10.1002/(SICI)1097-0169(1998)40:2<193::AID-CM8>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Feldman JL, Geimer S, Marshall WF. The mother centriole plays an instructive role in defining cell geometry. PLoS Biol. 2007;5:e149. doi: 10.1371/journal.pbio.0050149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaffal KP. The basal body-root complex of Chlamydomonas reinhardtii during mitosis. Protoplasma. 1988;143:118–129. [Google Scholar]
- Geimer S, Melkonian M. The ultrastructure of the Chlamydomonas reinhardtii basal apparatus: identification of an early marker of radial asymmetry inherent in the basal body. J Cell Sci. 2004;117:2663–2674. doi: 10.1242/jcs.01120. [DOI] [PubMed] [Google Scholar]
- Gould RR. The basal bodies of Chlamydomonas reinhardtii. Formation from probasal bodies, isolation, and partial characterization. J Cell Biol. 1975;65:65–74. doi: 10.1083/jcb.65.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guichard P, Chr etien D, Marco S, Tassin AM. Procentriole assembly revealed by cryo-electron tomography. EMBO J. 2010;29:1565–1572. doi: 10.1038/emboj.2010.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraki M, Nakazawa Y, Kamiya R, Hirono M. Bld10p constitutes the cartwheel-spoke tip and stabilizes the 9-fold symmetry of the centriole. Curr Biol. 2007;17:1778–1783. doi: 10.1016/j.cub.2007.09.021. [DOI] [PubMed] [Google Scholar]
- Holmes JA, Dutcher SK. Cellular asymmetry in Chlamydomonas reinhardtii. J Cell Sci. 1989;94:273–285. doi: 10.1242/jcs.94.2.273. [DOI] [PubMed] [Google Scholar]
- Hoops HJ, Witman GB. Outer doublet heterogeneity reveals structural polarity related to beat direction in Chlamydomonas flag-ella. J Cell Biol. 1983;97:902–908. doi: 10.1083/jcb.97.3.902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Ramanis Z, Dutcher SK, Luck DJ. Uniflagellar mutants of Chlamydomonas: evidence for the role of basal bodies in transmission of positional information. Cell. 1982;29:745–753. doi: 10.1016/0092-8674(82)90436-6. [DOI] [PubMed] [Google Scholar]
- Johnson UG, Porter KR. Fine structure of cell division in Chlamydomonas reinhardi. Basal bodies and microtubules. J Cell Biol. 1968;38:403–425. doi: 10.1083/jcb.38.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Dynlacht BD. Regulating the transition from centriole to basal body. J Cell Biol. 2011;193:435–444. doi: 10.1083/jcb.201101005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollman JM, Polka JK, Zelter A, Davis TN, Agard DA. Microtubule nucleating gamma-TuSC assembles structures with 13-fold microtubule-like symmetry. Nature. 2010;466:879–882. doi: 10.1038/nature09207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1995;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- Lechtreck KF, Grunow A. Evidence for a direct role of nas-cent basal bodies during spindle pole initiation in the green alga Spermatozopsis similis. Protist. 1999;150:163–181. doi: 10.1016/S1434-4610(99)70019-2. [DOI] [PubMed] [Google Scholar]
- Li S, Fernandez J-J, Marshall WF, Agard DA. Three-dimensional structure of basal body triplet revealed by electron cryo-tomography. The EMBO J. 2012;31:552–562. doi: 10.1038/emboj.2011.460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luck D, Piperno G, Ramanis Z, Huang B. Flagellar mutants of Chlamydomonas: studies of radial spoke-defective strains by dikaryon and revertant analysis. Proc Natl Acad Sci of the USA. 1977;74:3456–3460. doi: 10.1073/pnas.74.8.3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahjoub MR, Xie Z, Stearns T. Cep120 is asymmetrically localized to the daughter centriole and is essential for centriole assembly. J Cell Biol. 2010;191:331–346. doi: 10.1083/jcb.201003009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall WF, Kintner C. Cilia orientation and the fluid mechanics of development. Curr Opin Cell Biol. 2008;20:48–52. doi: 10.1016/j.ceb.2007.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde DN. Dual-axis tomography: an approach with alignment methods that preserve resolution. J Struct Biol. 1997;120:343–352. doi: 10.1006/jsbi.1997.3919. [DOI] [PubMed] [Google Scholar]
- Mitchell B, Stubbs JL, Huisman F, Taborek P, Yu C, Kintner C. The PCP pathway instructs the planar orientation of ciliated cells in the Xenopus larval skin. Curr Biol. 2009;19:924–929. doi: 10.1016/j.cub.2009.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittelmeier TM, Boyd JS, Lamb MR, Dieckmann CL. Asymmetric properties of the Chlamydomonas reinhardtii cytoskeleton direct rhodopsin photoreceptor localization. J Cell Biol. 2011;193:741–753. doi: 10.1083/jcb.201009131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz M, Braunfeld MB, Guénebaut V, Heuser J, Agard DA. Structure of the gamma-tubulin ring complex: a template for microtubule nucleation. Nat Cell Biol. 2000;2:365–370. doi: 10.1038/35014058. [DOI] [PubMed] [Google Scholar]
- Nakazawa Y, Hiraki M, Kamiya R, Hirono M. SAS-6 is a cartwheel protein that establishes the 9-fold symmetry of the centriole. Current Biol. 2007;17:2169–2174. doi: 10.1016/j.cub.2007.11.046. [DOI] [PubMed] [Google Scholar]
- Nigg EA. Centrosome duplication: of rules and licenses. Trends Cell Biol. 2007;17:215–221. doi: 10.1016/j.tcb.2007.03.003. [DOI] [PubMed] [Google Scholar]
- O’Toole ET. Chlamydomonas cryopreparation methods for the 3-D analysis of cellular organelles. Methods Cell Biol. 2010;96:71–91. doi: 10.1016/S0091-679X(10)96004-4. [DOI] [PubMed] [Google Scholar]
- O’Toole ET, Giddings TH, McIntosh JR, Dutcher SK. Three-dimensional organization of basal bodies from wild-type and delta-tubulin deletion strains of Chlamydomonas reinhardtii. Mol Biol Cell. 2003;14:2999–3012. doi: 10.1091/mbc.E02-11-0755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Toole ET, Giddings TH, Dutcher SK. Understanding microtubule organizing centers by comparing mutant and wild-type structures with electron tomography. Methods Cell Biol. 2007;79:125–143. doi: 10.1016/S0091-679X(06)79005-7. [DOI] [PubMed] [Google Scholar]
- Ostrowski LE, Dutcher SK, Lo CW. Cilia and models for studying structure and function. Proc Am Thorac Soc. 2011;8:423–429. doi: 10.1513/pats.201103-027SD. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park TJ, Mitchell BJ, Abitua PB, Kintner C, Wallingford JB. Dishevelled controls apical docking and planar polarization of basal bodies in ciliated epithelial cells. Nat Genet. 2008;40:871–879. doi: 10.1038/ng.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker JDK, Hilton LK, Diener DR, Rasi MQ, Mahjoub MR, Rosenbaum JL, Quarmby LM. Centrioles are freed from cilia by severing prior to mitosis. Cytoskeleton (Hoboken, N.J.) 2010;67:425–430. doi: 10.1002/cm.20454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson CG, Giddings TH, Winey M. Basal body components exhibit differential protein dynamics during nascent basal body assembly. Mol Biol Cell. 2009a;20:904–914. doi: 10.1091/mbc.E08-08-0835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson CG, Osborn DPS, Giddings TH, Beales PL, Winey M. Basal body stability and ciliogenesis requires the conserved component Poc1. J Cell Biol. 2009b;187:905–920. doi: 10.1083/jcb.200908019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piasecki BP, Silflow CD. The UNI1 and UNI2 genes function in the transition of triplet to doublet microtubules between the centriole and cilium in Chlamydomonas. Mol Biol Cell. 2009;20:368–378. doi: 10.1091/mbc.E08-09-0900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringo DL. Flagellar motion and fine structure of the flagellar apparatus in Chlamydomonas. J Cell Biol. 1967;33:543–571. doi: 10.1083/jcb.33.3.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum JL, Moulder JE, Ringo DL. Flagellar elongation and shortening in Chlamydomonas. The use of cycloheximide and colchicine to study the synthesis and assembly of flagellar proteins. J Cell Biol. 1969;41:600–619. doi: 10.1083/jcb.41.2.600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiratsuchi G, Kamiya R, Hirono M. Scaffolding function of the Chlamydomonas procentriole protein CRC70, a member of the conserved Cep70 family. J Cell Sci. 2011;124:2964–2975. doi: 10.1242/jcs.084715. [DOI] [PubMed] [Google Scholar]
- Silflow CD, LaVoie M, Tam LW, Tousey S, Sanders M, Wu W, Borodovsky M, Lefebvre PA. The Vfl1 Protein in Chlamydomonas localizes in a rotationally asymmetric pattern at the distal ends of the basal bodies. J Cell Biol. 2001;153:63–74. doi: 10.1083/jcb.153.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnen KF, Schermelleh L, Leonhardt H, Nigg EA. 3D-structured illumination microscopy provides novel insight into architecture of human centrosomes. Biol Open. 2012;1:965–976. doi: 10.1242/bio.20122337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strnad P, Gönczy P. Mechanisms of procentriole formation. Trends Cell Biol. 2008;18:389–396. doi: 10.1016/j.tcb.2008.06.004. [DOI] [PubMed] [Google Scholar]
- Strnad P, Leidel S, Vinogradova T, Euteneuer U, Khodjakov A, Gönczy P. Regulated HsSAS-6 levels ensure formation of a single procentriole per centriole during the centrosome duplication cycle. Dev Cell. 2007;13:203–213. doi: 10.1016/j.devcel.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamm SL, Sonneborn TM, Dippell RV. The role of cortical orientation in the control of the direction of ciliary beat in Paramecium. J Cell Biol. 1975;64:98–112. doi: 10.1083/jcb.64.1.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umen JG, Goodenough UW. Control of cell division by a retinoblastoma protein homolog in Chlamydomonas. Genes Dev. 2001;15:1652–1661. doi: 10.1101/gad.892101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vladar EK, Bayly RD, Sangoram AM, Scott MP, Axelrod JD. Microtubules enable the planar cell polarity of airway cilia. Curr Biol. 2012;22:2203–2212. doi: 10.1016/j.cub.2012.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallingford JB. Planar cell polarity signaling, cilia and polarized ciliary beating. Curr Opin Cell Biol. 2010;22:597–604. doi: 10.1016/j.ceb.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss RL, Goodenough DA, Goodenough UW. Membrane particle arrays associated with the basal body and with contractile vacuole secretion in Chlamydomonas. J Cell Biol. 1977;72:133–43. doi: 10.1083/jcb.72.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf K. Assembly and fate of basal bodies in the colourless phytoflagellate Polytoma papillatum. Biol Cell. 1992;76:193–200. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.