Abstract
Botulinum neurotoxins (BoNTs) inhibit neurotransmitter release by hydrolysing SNARE proteins. The most important serotype BoNT/A employs the synaptic vesicle glycoprotein 2 (SV2) isoforms A-C as neuronal receptors. Here, we identified their binding site by blocking SV2 interaction using monoclonal antibodies with characterised epitopes within the cell binding domain HC. The site is located on the backside of the conserved ganglioside binding pocket at the interface of the HCC and HCN subdomains. The dimension of the binding pocket was characterised in detail by site directed mutagenesis allowing the development of potent inhibitors as well as modifying receptor binding properties.
Keywords: botulinum neurotoxin A, protein receptor binding site, SV2, monoclonal antibody, neutralisation
Introduction
The fatal disease botulism, a flaccid paralysis of the muscles, is caused by blockade of the acetylcholine release at the neuromuscular junction due to intoxication with botulinum neurotoxins (BoNTs) even at minute quantities. On the other hand, of the seven major serotypes A-G the serotype BoNT/A constitutes a highly effective, widespread medicine to treat diseases like cervical dystonia, blepharospasm, hyperhidrosis and overactive bladder syndrome. All BoNTs are composed of four functional domains which play individual roles in their intoxication mechanism. The N-terminal 50 kDa light chain (LC) proteolyses specifically a member of the soluble N-ethlymaleimide sensitive attachment protein receptors family of proteins which are part of the vesicular fusion machinery [1,2]. The C-terminal 100 kDa heavy chain (HC) consists of three domains. Juxtaposed to LC is the 50 kDa α-helical translocation domain HN, and the remaining 50 kDa HC-fragment comprises the HCN domain which might trigger the translocation step [3] and the HCC domain which harbours the binding sites for gangliosides and in case of BoNT/B, DC, and G for the synaptic vesicle (SV) protein receptor synaptotagmin [4].
BoNTs are taken up by neurons via a double receptor mechanism [5–7]. First, neuron specific abundant polysialo gangliosides like GT1b, GD1a and GD1b enrich BoNTs on the plasma membrane to facilitate contact with the intravesicular parts of the transiently occurring SV protein receptors [8]. E.g., BoNT/B, DC and G bind the membrane neighbouring 15–17 amino acids of the intraluminal domain of the synaptic vesicle type I membrane proteins synaptotagmin (Syt)-I and Syt-II in the distal tip of their HC-fragment (Syt site) in the direct neighbourhood of the conserved ganglioside binding pocket in different arrangements [6,9–16].
Later, the large intravesicular domain 4 (LD4) of the synaptic vesicle glycoprotein 2 (SV2) isoforms A, B, and C was discovered as receptor domains for BoNT/A [17,18], the unglycosylated, isolated LD4 of SV2C exhibiting the highest affinity to BoNT/A HC. However, since all three SV2 isoforms are expressed in α-motoneurons [17], it is still not clear which SV2 isoform is most relevant for the physiological uptake of BoNT/A.
In contrast to BoNT/A, functional studies revealed that BoNT/E only interacts with SV2A and SV2B, but not with SV2C [19] while the biological activity of BoNT/D primarily requires the presence of SV2B and to a lesser extent of SV2A and C [20]. However, a direct protein-protein interaction between SV2 and BoNT/D remains to be shown like for BoNT/F which co-precipitated SV2A-C from rat brain synaptic vesicle lysates [21,22]. Altogether, BoNT/A, D, E and F utilise seemingly diverse SV2-binding mechanisms.
Only recently, the first SV2 binding pocket could be mapped in BoNT/E to an extended surface area located on the back of the conserved ganglioside binding site comprising the HCN and HCC interface, part of the corresponding Syt site in BoNT/B and the region in between (ODA1 site), the latter two also constituting a partial epitope of the monoclonal antibody (mAb) 4E13 which neutralises BoNT/E activity [23]. The SV2A binding site in BoNT/E seems to represent the prototypic region for all other BoNTs that exploit SV2 as a cell surface receptor. In accordance, a co-crystal structure allocated the binding site of human SV2C in BoNT/A to the HCN and HCC interface adjacent to the SV2A binding site in BoNT/E [24].
Here, we identified the SV2 binding site at the interface of HCC and HCN domains of BoNT/A employing neutralising mAbs with defined epitopes used as therapeutics [25]. Site-directed mutagenesis characterised the SV2 binding pocket in detail and exhibited the single mutation G1292R decreasing the biological activity of BoNT/A ~300-fold. Employment of ganglioside deficient mice and BoNT/A mutants comprising deactivated receptor binding sites proved that the double receptor model also accounts for BoNT/A.
Materials and Methods
Anti-BoNT/A mAb CR2 and scFvs RAZ1, S25 and CR2 were generated as described previously [25–29]. Figure 1B was prepared using the program Discovery Studio Visualizer 2.5 (Accelrys Software Inc.) based on 2NYY.pdb [28].
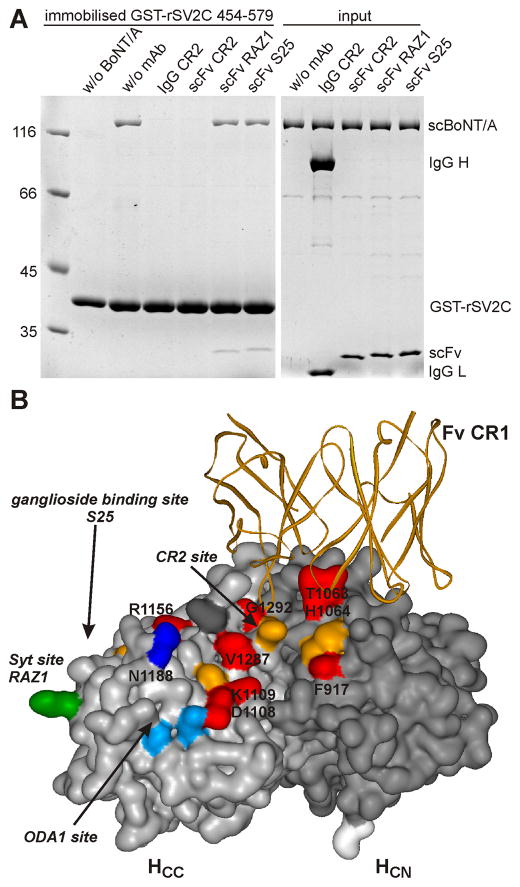
Fig. 1.
A Binding of full-length BoNT/A to GST-rSV2C 454–579 is blocked by monoclonal antibody IgG CR2 and its corresponding scFv. H6tBoNTA (50 pmol) serving as prey was preincubated with mAb (400 pmol) for 30 min at RT (“input”) in 200 μl 100 mM Tris–HCl, pH 8.0, 150 mM NaCl, 0.5% Triton X-100 and subsequently incubated for 2 h at 4°C with the bait, immobilised GST-rSV2C 454–579 (75 pmol). Bound H6tBoNTA was detected by 12.5% SDS-PAGE analysis and Coomassie blue staining. B Complex crystal structure of BoNT/A HC (HCC, light gray surface; HCN, dark grey surface) and the variable part of CR1 Fab (orange ribbon; modified from 2NYY.pdb). Residues in the analogous Syt site/RAZ1 epitope are depicted in green, those of the ODA1 site in light blue or dark blue (effect on SV2C binding). All positions analysed in the CR2 site are coloured in orange, those with clear effect on SV2C binding in red.
Plasmid constructions and production of proteins
Rat GST-rSV2C 454–579 encoded by pGEXrSV2C 454–579 and rat GST-rSV2A 468–594 encoded by pGEXrSV2A 468–594 were produced as described previously [18]. The plasmid pH6tBoNTA1 encoding BoNT/A1 fused to an N-terminal His6tag was generated by modifying pBoNTAS [30]. The plasmids pHCAS, pH6F3HCES and pH6F3HCGS were described previously [15,23,30], pH6F3HCES serving as parental vector for pH6F3HCAS and pH6F3HCFS which were generated by ligation of DNA sequences encoding HCA (aa 871–1296) from pS3HCAH6 and HCF (aa 866–1277) from pS3HCFH6 [22]. All pH6tBoNTA and pH6F3HCAS mutants were prepared using the GeneTailor method (Invitrogen GmbH, Karlsruhe, Germany) employing suitable primers and pH6tBoNTA and pH6F3HCAS, respectively, as template DNA. Nucleotide sequences of all mutants were verified by DNA sequencing.
Wild-type and mutated recombinant H6F3HCXS (= N-terminal 6x His tag, 3x Flag tag, HC fragment of serotype A-G, C-terminal Streptag), HCAS and full-length neurotoxins, the latter under biosafety level 2 containment (project number GAA A/Z 40654/3/123), were produced utilizing the E. coli strain M15pREP4 (Qiagen, Germany) during 16 h of induction at 22°C in the presence of 0.2 mM IPTG, and were purified on Co2+-Talon matrix (Takara Bio Europe S.A.S., France). Full-length neurotoxins were eluted using 50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 250 mM imidazole, subjected to gel filtration (Superdex-200 16/60 column, GE Healthcare, Germany) in 100 mM Tris-HCl, pH 8.0, 150 mM NaCl, frozen in liquid nitrogen and kept at −70°C. H6F3HcXS proteins were eluted using 100 mM Tris-HCl pH 8.0, 150 mM NaCl, 100 mM Imidazole and further purified on StrepTactin-sepharose beads (IBA GmbH, Germany). Proteins were eluted by 10 mM desthiobiotin in 100 mM Tris-HCl pH 8.0, 150 mM NaCl, frozen in liquid nitrogen and kept at −70°C. For CD analysis desired volume of protein was dialysed against 1x PBS pH 7.4
Protein concentrations were determined subsequent to SDS-PAGE and Coomassie blue staining by using a LAS-3000 imaging system (Fuji Photo Film), the AIDA 3.51 program, and various known concentrations of BSA as reference.
GST-pull down assays
For competition experiments HCAS (100 pmol) or H6tBoNTA (50 pmol) were preincubated with mAb (400 pmol) for 30 min at RT in 187.5 μl in 100 mM Tris– HCl, pH 8.0, 150 mM NaCl.
Glutathion-S-transferase (GST, 150/75 pmol), GST-rSV2A 468–594 (150 pmol) and GST-rSV2C 454–579 (75 pmol) immobilised to 10 μl of glutathione-sepharose-4B matrix (GE healthcare, Germany) were incubated with HCAS (100 pmol), H6tBoNTA wild-type (with or without preincubated mAbs) or mutants (50 pmol each) in a total volume of 200 μl 100 mM Tris–HCl, pH 8.0, 150 mM NaCl, supplemented with 0.5% Triton X-100 (Tris/NaCl/Triton) buffer for 2 h at 4 °C. Beads were collected by centrifugation and washed two times each with 200 μl Tris/NaCl/Triton buffer. Washed pellet fractions were denatured in SDS sample buffer for 20 min at 37°C and analysed by SDS-PAGE. Proteins were detected by Coomassie blue staining and subsequently quantified by densitometry.
Mouse phrenic nerve hemidiaphragm (MPN) assay
The MPN assay was performed as described previously employing 20–30 g NMRI mice (Janvier SA, France) [31] or complex polysialo ganglioside deficient C57BL/6 mice lacking the genes B4galnt1 and/or St8sia1 [22]. The phrenic nerve was continuously stimulated at 5–25 mA with a frequency of 1 Hz and with a 0.1 ms pulse duration. Isometric contractions were transformed using a force transducer and recorded with VitroDat Online software (FMI GmbH, Germany). The time required to decrease the amplitude to 50 % of the starting value (paralytic half-time) was determined. To allow comparison of the altered neurotoxicity of mutants with H6tBoNTA wild-type, a power function (y(H6tBoNTA; 10, 30, 80 pM) = 139.6×−0.1957, R2 = 0.9991) was fitted to a concentration-response-curve consisting of three concentrations determined minimum in triplicates. Resulting paralytic half-times of the H6tBoNTA mutants were converted to concentrations of the wild-type employing the above power functions and finally expressed as relative neurotoxicity (Fig. 3 and 5)
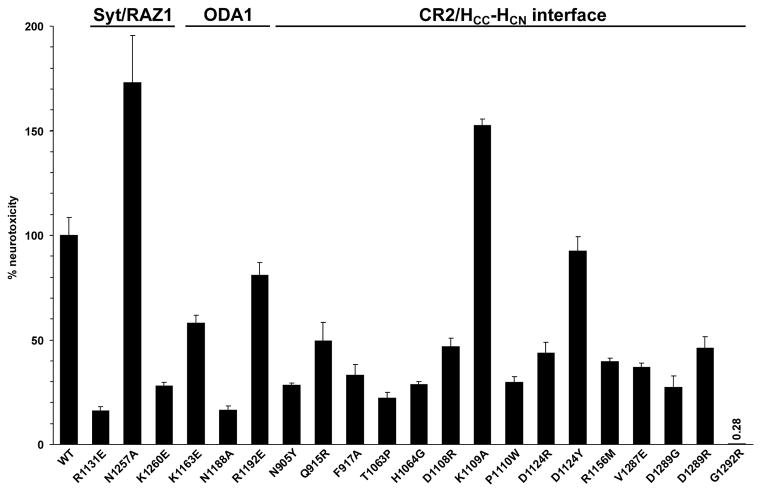
Fig. 3.
The impact of mutations on the neurotoxicity of BoNT/A was analysed employing the MPN preparations. The paralytic halftimes were determined and converted to the corresponding concentrations of wild-type BoNT/A using a dose-response curve. The resulting toxicities were finally expressed relative to wild-type BoNT/A (n= 3–5, ±SD).
Fig. 5.
The neurotoxicity of wild-type and BoNT/A mutants either with inactivated GBS (W1266L), inactivated SV2 binding site (R1156M, G1292R) or both was determined employing wild-type and ganglioside-deficient tissue for the MPN assay (n = 7–16, ±SD).
Co-immunoprecipitation
All centrifugation and incubation steps were carried out at 4°C, except where otherwise stated. The preparation of rat brain synaptosomes was performed as previously described [31]. Synaptosomes were solubilised in lysis buffer (20 mM Tris-HCl, pH 7.4, 80 mM NaCl, 0.5% Triton X-100) for one hour, subsequently centrifuged at 21,000 × g for 10 min and the supernatant was transferred into a fresh tube. Co-immunoprecipitation of native SV2A and B was done in lysis buffer supplemented with 0.1% BSA in a total volume of 200 μl employing 10 μl Protein G agarose beads (Amersham Biosciences), 6 μg of anti-Flag M2 monoclonal antibody (Sigma-Aldrich), 12 μg of the indicated BoNT HC-fragment and 50 μg of synaptosomal proteins in the presence of 125 μg mixed bovine brain gangliosides for 3 h. Thereafter, beads were collected by centrifugation at 2,000 × g and washed three times using lysis buffer. Washed pellet fractions were incubated in Laemmli sample buffer for 20 min at 37°C and analyzed by SDS-PAGE and Western blotting (rabbit polyclonal anti-SV2A & anti-SV2B, Synaptic Systems, Germany; goat anti-rabbit, Pierce; StrepMAP Classic-HRP, IBA GmbH, Germany).
Results and Discussion
The HC-fragment of BoNT/A displays the highest affinity to the luminal domain 4 (residues 454–579) of the isoform SV2C independent of its glycosylation status followed by SV2A and SV2B [17,18]. This direct protein-protein interaction is clearly demonstrated in a GST pull down assay employing full-length recombinant H6tBoNTA and rat GST-rSV2C 454–579 (Fig. 1A). The weaker binding of HCA to recombinant rat GST-rSV2A 468–594 is also shown (Fig. S1) whereas binding to non-glycosylated rSV2B is hardly detectable. Well characterised neutralising monoclonal antibodies directed against different epitopes within the HC-fragment of BoNT/A [25–29] were screened for interference with the SV2C interaction. To avoid putative cross-linking of two BoNT/A molecules by an intact IgG which could impair SV2 binding, we initially pre-incubated H6tBoNTA with the single-chain variable fragments (scFv) S25, RAZ1 and CR2 in 8-fold excess. scFv S25 is directed against residues 1254–56 forming a part of the rim of the single ganglioside binding site (GBS) [30]; RAZ1 is an affinity matured mAb based on 3D12 directed against residues 1129–31 comprising the region analogous to the Syt binding site in BoNT/B and G [6,14]. CR2 and the closely related CR1 are engineered mAbs derived from C25 which binds predominantly to residues 918–20, F953 and 1062–64 at the interface of the HCC and HCN domain [28,29] (Fig. 1B). Whereas scFv S25 did not and RAZ1 only marginally altered the interaction of GST-rSV2C with H6tBoNT/A, the scFv CR2 eliminated the precipitation of H6tBoNTA by GST-rSV2C (Fig. 1A). Analogously, the 4-fold excess of IgG CR2 terminated binding of H6tBoNTA to GST-rSV2C, which explains its potency in ex vivo MPN assay as well as in in vivo mice toxin neutralisation assays [26]. Hence, a main interaction site of SV2C with BoNT/A resides in the epitope region of CR2. Analogously, CR2 was able to inhibit the precipitation of HCA by GST-rSV2A LD4 indicating that both SV2 isoforms interact with the same surface area of BoNT/A (Fig. S1). Previously, the CR2 epitope was mapped in detail by two crystal structure complexes consisting of BoNT/A and Fabs of CR1 or AR2, respectively, closely related mAbs recognising identical epitopes [28] (Fig. 1B). Based on this structural data site directed mutagenesis was performed in the CR2 site, the Syt/RAZ1 site as well as in the region connecting both surface areas called ODA1 site according to Mahrhold et al. [23].
The full-length recombinant H6tBoNTA mutants were screened in pull down assays employing rat GST-rSV2C 454–579. Charge reversal by R1131E in the RAZ1 site and K1163E in the ODA1 site decreased precipitation of H6tBoNTA, even more pronounced for mutation N1188A in the ODA1 site (Fig. 2). Furthermore, 15 mutations in the CR2 site were generated. Here, the H6tBoNTA single mutants N905Y, T1063P, H1064G, D1108R, R1156M, V1287E and G1292R displayed a 50% or greater decrease of binding to GST-rSV2C which is in good agreement with binding assay data obtained for the CR2-related humanised C25 [29] and the crystallised complexes BoNT/A-CR1 or AR2 which identified i.a. T1063 and H1064 as key residues in their epitopes [28]. With the exception of D1124R also the remaining seven mutants showed at least moderately reduced affinity to GST-rSV2C (Fig. 2).
Fig. 2.
Binding of H6tBoNTA single mutants (50 pmol) for 2 h at 4°C to immobilised GST-rSV2C 454–579 (75 pmol) in 200 μl 100 mM Tris–HCl, pH 8.0, 150 mM NaCl, 0.5% Triton X-100. Bound H6tBoNTA mutants were detected by 10% SDS-PAGE analysis and Coomassie blue staining (wt n=15, mutants n= 3–5, ±SD), quantified by densitometry and compared versus wild-type binding.
To verify whether the reduced affinity of the BoNT/A mutants to recombinant rSV2C LD4 also translates into reduced biological activity all 21 mutants were tested in the MPN assay. Correspondingly, mutants H6tBoNTA R1131E and K1260E in the RAZ1 site and N1188A within the ODA1 site showed diminished potency (Fig. 3). Also mutations N905Y, F917A, T1063P, H1064G, D1108R, P1110W, D1124R, R1156M, V1287E and D1289G/R in the CR2 site caused a decrease of more than 50% in neurotoxicity. The single mutant G1292R even displayed a 300-fold reduction in potency. Thus the MPN data largely confirms the previous in vitro binding data obtained with isolated rat GST-rSV2C 454–579, also because the LD4 of mouse and rat SV2C differ only in three conservative amino acid exchanges. Noteworthy, the isoforms SV2A and B are also expressed in motoneurons [17], although neither their differential expression levels nor their affinity to BoNT/A have been quantified. The amino acid sequence identity between the LD4 of rSV2A, rSV2B and rSV2C is only 48% and 44%, respectively. Hence, differences between the GST-rSV2C pull down and the MPN assay as observed for mutants P1110W, D1124R and D1289G/R could be explained by subtle differences in the mode of interaction of the three SV2 isoforms. In conclusion, the main SV2C binding site is allocated in the CR2 epitope site at the interface of HCC and HCN domains on the backside of the GBS extending towards the Syt/RAZ1 binding site. Our finding is in good agreement with a recently published co-crystal structure of human SV2C residues 456–579 bound to the BoNT/A HC-fragment where one β-strand of the SV2C quadrilateral β-helix interacts with the convex β-sheet (1140–56) at the BoNT/A HCC-HCN interface [24]. This β-sheet is adjacent to the CR2 epitope in HCN-HCC. Here, T1145/46 and R1156 dominate the few proposed side chain-side chain interactions. Analogously, our mutant R1156M showed the lowest affinity to GST-rSV2C. Secondly, the strong effects observed for mutation of G1292 is in accordance with data of Benoit et al. showing impaired hSV2C interaction upon mutation of the neighbouring R1294 to alanine [24]. Furthermore, comparison with the SV2A-binding site in BoNT/E shows that mutation of Y879, E1246 and K1084 in BoNT/E, like the corresponding residues N905, V1287 and D1108 in BoNT/A for SV2C, interferes with SV2A-binding of BoNT/E. This illustrates that the SV2 binding site of BoNT/A covers the homologous region of BoNT/E [23].
To ensure that the single mutations on the HC-fragment surface did not impair its structural integrity and thereby caused the loss in SV2C binding, the mutations displaying the most severe effects (R1156M, V1287E and G1292R) were transferred into HC-fragments fused to an N-terminal 3xFLAG tag. The thermal stability of the HC-fragment mutants was virtually identical to the wild-type (51±1°C; Fig. S2) indicating that the secondary structure was not altered due to the single amino acid exchanges.
To further deepen the conclusion obtained for the interaction site of SV2 in BoNT/A, co-immunoprecipitation experiments were conducted employing 3xFLAG tagged BoNT/A HC-fragment mutants (H6F3HCAS; Fig. 4). Since the tissue from motoneurons as physiological target cannot be accessed in sufficient quantities, rat brain synaptosomes were generated and solubilised in Triton X-100. However, whereas SV2A is ubiquitously expressed in virtually all synapses and SV2B was also found in a wide variety of synapses throughout the brain, SV2C was observed only in few brain areas like the pallidum, the substantia nigra, the midbrain, the brainstem and the olfactory bulb, but was undetectable in the cerebral cortex and the hippocampus, and found at low levels in the cerebellar cortex [32]. Hence, SV2C constitutes only a minor fraction in synaptosomal lysates and HCA wild-type co-precipitated only minor amounts. In contrast, SV2A and SV2B were satisfactorily precipitated by HCA wild-type (Fig. 4). HCA mutants R1156M and V1287E showed similar affinity to SV2A and SV2B as wild-type HCA indicating subtle differences between the mode of interaction of SV2A/B and of SV2C with BoNT/A. R1156 is suggested to interact with F563 of human SV2C by cation-π-stacking interaction [24]. However, mouse and rat SV2C comprise L563 which does not allow this cation-π-stacking, but possibly R1156 establishes a salt bridge to the neighbouring E543 in rat SV2C. However, the corresponding residues Y557 in rSV2A and Y500 in SV2B would form a weaker H-bond to R1156. Consequently, mutant R1156M does only moderately interfere with SV2A and B binding. The lack of effect for mutant V1287E cannot be deduced from the HCA-hSV2C complex structure. In contrast, mutant HCA G1292R did neither co-precipitate SV2A nor SV2B which confirms the conclusion that also SV2A and SV2B interact at the similar region on the HCA surface. Hereby, the strong effect of mutation G1292R on SV2A-C interaction also explains the drastic decrease of biological activity of H6tBoNTA G1292R in the MPN assay compared to all other mutants in the CR2 site.
Fig. 4.
Co-immunoprecipitation of SV2A and B by selected HCA mutants was performed by incubating 12 μg of 3xFlag-tagged wild-type and mutant H6F3HCAS, H6F3HCES, H6F3HCFS and H6F3HCGS with 6 μg anti-Flag-tag M2 antibody and Triton X-100 solubilised rat brain synaptosomal membranes. Bound SV2A and B were detected by western blotting, amount of precipitated H6F3HCXS was verified by HRP conjugated StrepMAB classic. MWM denotes molecular weight marker.
The single BoNT/A mutant G1292R with eliminated SV2 protein receptor recognition, the mutant W1266L lacking ganglioside binding [30] as well as ganglioside deficient mice [33] provided powerful tools to prove the dual receptor hypothesis for BoNT/A analogously to BoNT/B and G [6]. Like previously shown [22] the sensitivity of tissue expressing GM3 only was decreased by ~600-fold for wild-type H6tBoNTA (Fig. 5) which corresponds to the decrease in sensitivity of wild-type mice hemidiaphragm tissue towards H6tBoNTA W1266L bearing a deactivated GBS [30](Fig. 5). Whereas the neurotoxicity of the mutant W1266L at GM3-only tissue remained roughly the same, the H6tBoNTA G1292R with inactivated SV2 binding site (= 300-fold reduced neurotoxicity at wild-type tissue) showed further reduced neurotoxicity at ganglioside-deficient tissue indicating the importance of both receptor interactions (Fig. 5). Finally, the double mutant comprising W1266L and G1292R turning both receptor sites inactive displayed a 250,000-fold reduced biological activity in the MPN assay (Fig. 5). In comparison, the individual mutations each decreased the potency by ~200- and 300-fold demonstrating the synergy of both binding sites for the intoxication mechanism.
In conclusion, exchange of two critical amino acids in the GBS and the newly identified SV2 binding site generates a virtually atoxic BoNT/A which adds another piece of evidence for the dual receptor mechanism by which botulinum neurotoxins specifically enter nerve cells [4,5]. Furthermore, the knowledge of both receptor binding sites now allows the design of bivalent inhibitor molecules blocking the uptake of BoNT/A in case of its deliberate or accidental release. Also the retargeting of BoNT/A to e.g. non-neuronal cells to treat diseases caused by overactive secretion mechanisms which has so far been achieved by replacing the whole HC-fragment [34] could now be accomplished with minimal alterations in the SV2 binding site. And in combination with three critical substitutions in the LC interfering with the catalytic mechanism [35–37] such a quintuple mutant represents an ideal atoxic antigen to generate vaccines against physiologically folded BoNT/A.
Supplementary Material
Binding of BoNT/A HC fragment to GST-rSV2A 468–594 is blocked by monoclonal antibody IgG CR2 and its corresponding scFv. HCAS (100 pmol) serving as prey was preincubated with mAb (400 pmol) for 30 min at RT (“input”) in 200 μl of 20 mM Tris, pH 7.4, 80 mM NaCl, 0.5% Triton X-100 and subsequently incubated for 2 h at 4°C with the bait, immobilised GST-rSV2A 468–594 (150 pmol). Precipitated HCAS was detected by 12.5% SDS-PAGE analysis and Coomassie blue staining.
Thermal denaturation experiments of wild-type and mutated H6F3HCAS yielded steep sigmoidal curves. The melting temperature (Tm), defined by the inflection point, of wild-type HCA was 51°C. Tm values of the mutants were virtually indistinguishable from the wild-type value taking into account deviations of ±2°C of Tm for separate measurements of the same sample. In conclusion, the results do not indicate structural disturbances in any of these mutants.
CD data was collected with a Jasco J-810 spectropolarimeter in a 1 mm path length cuvette with a concentration of 11–13 μM degassed HCA in PBS pH 7.4. Initial spectra were recorded at 22°C from 195–250 nm with 100 nm/min, response of 1 s, standard sensitivity, bandwidth of 1 nm and five accumulations. Subsequent temperature-induced denaturation was performed by monitoring the CD signal at 210 nm with an average temperature increase of 0.5°C/min from 30°C to 70°C.
Structured summary of protein interactions.
HcF physically interacts with SV2C and SV2A by anti tag coimmunoprecipitation (View interaction) rSV2A binds to HcA by pull down (View interaction) rSV2C binds to BoNT/A by pull down (1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11) HcE physically interacts with SV2A and SV2C by anti tag coimmunoprecipitation (View interaction) HcA physically interacts with SV2A and SV2C by anti tag coimmunoprecipitation (View interaction)
Highlights.
Neutralising mechanism of potent therapeutic monoclonal BoNT/A antibody is exhibited.
Single mutation G1292R eliminates interaction of BoNT/A with all three SV2 isoforms.
Proof for neuronal uptake of BoNT/A by a double receptor mechanism.
Quintuple inactive BoNT/A mutant represents ideal antigen for vaccination.
Acknowledgments
We thank Weihua Wen and Catharina Melzer for technical assistance. This work was supported in part by grants from the Bundesministerium für Bildung und Forschung (FK031A212A) to A.R., from the Deutsche Forschungsgemeinschaft (BI 660/3-1) to T.B and National Institute of Allergy and Infectious Diseases (NIAID) cooperative agreement U54 AI065359 to J.D.M.
Abbreviations
- BoNT
botulinum neurotoxin
- GBS
ganglioside binding site
- GST
glutathion-S-transferase
- HC
heavy chain
- HC
carboxyl-terminal half of HC
- HCA
HCE, HCF, HCG, HCX, HC fragment of BoNT serotype A, E, F, G or A-G, respectively
- HN
amino-terminal half of HC
- H6F3HCXS
N-terminal 6x His tag, 3x Flag tag, HC fragment of serotype A-G, C-terminal Streptag
- LC
light chain
- mAb
monoclonal antibody
- MPN assay
mice phrenic nerve hemidiaphragm assay
- scFv
single-chain variable fragments
- rSV2A
B, C, rat synaptic vesicle glycoprotein 2 isoform A, B or C
- Syt
synaptotagmin
- CD
circular dichroism
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Montal M. Botulinum neurotoxin: a marvel of protein design. Annu Rev Biochem. 2010;79:591–617. doi: 10.1146/annurev.biochem.051908.125345. [DOI] [PubMed] [Google Scholar]
- 2.Binz T. Clostridial neurotoxin light chains: devices for SNARE cleavage mediated blockade of neurotransmission. Curr Top Microbiol Immunol. 2013;364:139–57. doi: 10.1007/978-3-642-33570-9_7. [DOI] [PubMed] [Google Scholar]
- 3.Fischer A, Mushrush DJ, Lacy DB, Montal M. Botulinum neurotoxin devoid of receptor binding domain translocates active protease. PLoS Pathog. 2008;4:e1000245. doi: 10.1371/journal.ppat.1000245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rummel A. Double receptor anchorage of botulinum neurotoxins accounts for their exquisite neurospecificity. Curr Top Microbiol Immunol. 2013;364:61–90. doi: 10.1007/978-3-642-33570-9_4. [DOI] [PubMed] [Google Scholar]
- 5.Montecucco C. How do tetanus and botulinum toxins bind to neuronal membranes? Trends Biochemical Science. 1986;11:314–317. [Google Scholar]
- 6.Rummel A, et al. Identification of the protein receptor binding site of botulinum neurotoxins B and G proves the double-receptor concept. Proc Natl Acad Sci U S A. 2007;104:359–64. doi: 10.1073/pnas.0609713104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berntsson RP, Peng L, Dong M, Stenmark P. Structure of dual receptor binding to botulinum neurotoxin B. Nat Commun. 2013;4:2058. doi: 10.1038/ncomms3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Binz T, Rummel A. Cell entry strategy of clostridial neurotoxins. J Neurochem. 2009;109:1584–95. doi: 10.1111/j.1471-4159.2009.06093.x. [DOI] [PubMed] [Google Scholar]
- 9.Dong M, Richards DA, Goodnough MC, Tepp WH, Johnson EA, Chapman ER. Synaptotagmins I and II mediate entry of botulinum neurotoxin B into cells. J Cell Biol. 2003;162:1293–303. doi: 10.1083/jcb.200305098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nishiki T, Kamata Y, Nemoto Y, Omori A, Ito T, Takahashi M, Kozaki S. Identification of protein receptor for Clostridium botulinum type B neurotoxin in rat brain synaptosomes. J Biol Chem. 1994;269:10498–503. [PubMed] [Google Scholar]
- 11.Rummel A, Karnath T, Henke T, Bigalke H, Binz T. Synaptotagmins I and II act as nerve cell receptors for botulinum neurotoxin G. J Biol Chem. 2004;279:30865–70. doi: 10.1074/jbc.M403945200. [DOI] [PubMed] [Google Scholar]
- 12.Peng L, Berntsson RP, Tepp WH, Pitkin RM, Johnson EA, Stenmark P, Dong M. Botulinum neurotoxin D-C uses synaptotagmin I and II as receptors, and human synaptotagmin II is not an effective receptor for type B, D-C and G toxins. J Cell Sci. 2012;125:3233–42. doi: 10.1242/jcs.103564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chai Q, Arndt JW, Dong M, Tepp WH, Johnson EA, Chapman ER, Stevens RC. Structural basis of cell surface receptor recognition by botulinum neurotoxin B. Nature. 2006;444:1096–100. doi: 10.1038/nature05411. [DOI] [PubMed] [Google Scholar]
- 14.Jin R, Rummel A, Binz T, Brunger AT. Botulinum neurotoxin B recognizes its protein receptor with high affinity and specificity. Nature. 2006;444:1092–5. doi: 10.1038/nature05387. [DOI] [PubMed] [Google Scholar]
- 15.Willjes G, Mahrhold S, Strotmeier J, Eichner T, Rummel A, Binz T. Botulinum Neurotoxin G Binds Synaptotagmin-II in a Mode Similar to That of Serotype B: Tyrosine 1186 and Lysine 1191 Cause Its Lower Affinity. Biochemistry. 2013;52:3930–8. doi: 10.1021/bi4003502. [DOI] [PubMed] [Google Scholar]
- 16.Berntsson RP, Peng L, Svensson LM, Dong M, Stenmark P. Crystal structures of botulinum neurotoxin DC in complex with its protein receptors synaptotagmin I and II. Structure. 2013;21:1602–11. doi: 10.1016/j.str.2013.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dong M, Yeh F, Tepp WH, Dean C, Johnson EA, Janz R, Chapman ER. SV2 is the protein receptor for botulinum neurotoxin A. Science. 2006;312:592–6. doi: 10.1126/science.1123654. [DOI] [PubMed] [Google Scholar]
- 18.Mahrhold S, Rummel A, Bigalke H, Davletov B, Binz T. The synaptic vesicle protein 2C mediates the uptake of botulinum neurotoxin A into phrenic nerves. FEBS Lett. 2006;580:2011–4. doi: 10.1016/j.febslet.2006.02.074. [DOI] [PubMed] [Google Scholar]
- 19.Dong M, Liu H, Tepp WH, Johnson EA, Janz R, Chapman ER. Glycosylated SV2A and SV2B mediate the entry of botulinum neurotoxin E into neurons. Mol Biol Cell. 2008;19:5226–37. doi: 10.1091/mbc.E08-07-0765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peng L, Tepp WH, Johnson EA, Dong M. Botulinum Neurotoxin D Uses Synaptic Vesicle Protein SV2 and Gangliosides as Receptors. PLoS Pathog. 2011;7:e1002008. doi: 10.1371/journal.ppat.1002008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fu Z, Chen C, Barbieri JT, Kim JJ, Baldwin MR. Glycosylated SV2 and gangliosides as dual receptors for botulinum neurotoxin serotype F. Biochemistry. 2009;48:5631–41. doi: 10.1021/bi9002138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rummel A, et al. Botulinum neurotoxins C, E and F bind gangliosides via a conserved binding site prior to stimulation-dependent uptake with botulinum neurotoxin F utilising the three isoforms of SV2 as second receptor. J Neurochem. 2009;110:1942–1954. doi: 10.1111/j.1471-4159.2009.06298.x. [DOI] [PubMed] [Google Scholar]
- 23.Mahrhold S, Strotmeier J, Garcia-Rodriguez C, Lou J, Marks JD, Rummel A, Binz T. Identification of the SV2 protein receptor-binding site of botulinum neurotoxin type E. Biochem J. 2013;453:37–47. doi: 10.1042/BJ20130391. [DOI] [PubMed] [Google Scholar]
- 24.Benoit RM, et al. Structural basis for recognition of synaptic vesicle protein 2C by botulinum neurotoxin A. Nature. 2014;505:108–11. doi: 10.1038/nature12732. [DOI] [PubMed] [Google Scholar]
- 25.Mullaney BP, Pallavicini MG, Marks JD. Epitope mapping of neutralizing botulinum neurotoxin A antibodies by phage display. Infect Immun. 2001;69:6511–4. doi: 10.1128/IAI.69.10.6511-6514.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nowakowski A, et al. Potent neutralization of botulinum neurotoxin by recombinant oligoclonal antibody. Proc Natl Acad Sci U S A. 2002;99:11346–50. doi: 10.1073/pnas.172229899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Razai A, et al. Molecular evolution of antibody affinity for sensitive detection of botulinum neurotoxin type A. J Mol Biol. 2005;351:158–69. doi: 10.1016/j.jmb.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 28.Garcia-Rodriguez C, et al. Molecular evolution of antibody cross-reactivity for two subtypes of type A botulinum neurotoxin. Nat Biotechnol. 2007;25:107–16. doi: 10.1038/nbt1269. [DOI] [PubMed] [Google Scholar]
- 29.Levy R, Forsyth CM, LaPorte SL, Geren IN, Smith LA, Marks JD. Fine and domain-level epitope mapping of botulinum neurotoxin type A neutralizing antibodies by yeast surface display. J Mol Biol. 2007;365:196–210. doi: 10.1016/j.jmb.2006.09.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rummel A, Mahrhold S, Bigalke H, Binz T. The HCC-domain of botulinum neurotoxins A and B exhibits a singular ganglioside binding site displaying serotype specific carbohydrate interaction. Mol Microbiol. 2004;51:631–43. doi: 10.1046/j.1365-2958.2003.03872.x. [DOI] [PubMed] [Google Scholar]
- 31.Rummel A, Bade S, Alves J, Bigalke H, Binz T. Two carbohydrate binding sites in the HCC-domain of tetanus neurotoxin are required for toxicity. J Mol Biol. 2003;326:835–47. doi: 10.1016/s0022-2836(02)01403-1. [DOI] [PubMed] [Google Scholar]
- 32.Janz R, Sudhof TC. SV2C is a synaptic vesicle protein with an unusually restricted localization: anatomy of a synaptic vesicle protein family. Neuroscience. 1999;94:1279–90. doi: 10.1016/s0306-4522(99)00370-x. [DOI] [PubMed] [Google Scholar]
- 33.Kawai H, et al. Mice expressing only monosialoganglioside GM3 exhibit lethal audiogenic seizures. J Biol Chem. 2001;276:6885–8. doi: 10.1074/jbc.C000847200. [DOI] [PubMed] [Google Scholar]
- 34.Chaddock J. Transforming the domain structure of botulinum neurotoxins into novel therapeutics. Curr Top Microbiol Immunol. 2013;364:287–306. doi: 10.1007/978-3-642-33570-9_13. [DOI] [PubMed] [Google Scholar]
- 35.Binz T, Bade S, Rummel A, Kollewe A, Alves J. Arg(362) and Tyr(365) of the botulinum neurotoxin type a light chain are involved in transition state stabilization. Biochemistry. 2002;41:1717–23. doi: 10.1021/bi0157969. [DOI] [PubMed] [Google Scholar]
- 36.Gu S, et al. Botulinum neurotoxin is shielded by NTNHA in an interlocked complex. Science. 2012;335:977–81. doi: 10.1126/science.1214270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li L, Binz T, Niemann H, Singh BR. Probing the mechanistic role of glutamate residue in the zinc-binding motif of type A botulinum neurotoxin light chain. Biochemistry. 2000;39:2399–405. doi: 10.1021/bi992321x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Binding of BoNT/A HC fragment to GST-rSV2A 468–594 is blocked by monoclonal antibody IgG CR2 and its corresponding scFv. HCAS (100 pmol) serving as prey was preincubated with mAb (400 pmol) for 30 min at RT (“input”) in 200 μl of 20 mM Tris, pH 7.4, 80 mM NaCl, 0.5% Triton X-100 and subsequently incubated for 2 h at 4°C with the bait, immobilised GST-rSV2A 468–594 (150 pmol). Precipitated HCAS was detected by 12.5% SDS-PAGE analysis and Coomassie blue staining.
Thermal denaturation experiments of wild-type and mutated H6F3HCAS yielded steep sigmoidal curves. The melting temperature (Tm), defined by the inflection point, of wild-type HCA was 51°C. Tm values of the mutants were virtually indistinguishable from the wild-type value taking into account deviations of ±2°C of Tm for separate measurements of the same sample. In conclusion, the results do not indicate structural disturbances in any of these mutants.
CD data was collected with a Jasco J-810 spectropolarimeter in a 1 mm path length cuvette with a concentration of 11–13 μM degassed HCA in PBS pH 7.4. Initial spectra were recorded at 22°C from 195–250 nm with 100 nm/min, response of 1 s, standard sensitivity, bandwidth of 1 nm and five accumulations. Subsequent temperature-induced denaturation was performed by monitoring the CD signal at 210 nm with an average temperature increase of 0.5°C/min from 30°C to 70°C.