Summary
Despite the great potential of stem cells for basic research and clinical applications, obstacles – such as their scarce availability and difficulty in controlling their fate – need to be addressed to fully realize their potential. Recent achievements of cellular reprogramming have enabled the generation of induced pluripotent stem cells (iPSCs) or other lineage-committed cells from more accessible and abundant somatic cell types by defined genetic factors. However, serious concerns remain about the efficiency and safety of current genetic approaches to cell reprogramming and traditional culture systems that are used for stem cell maintenance. As a complementary approach, small molecules that target specific signaling pathways, epigenetic processes and other cellular processes offer powerful tools for manipulating cell fate to a desired outcome. A growing number of small molecules have been identified to maintain the self-renewal potential of stem cells, to induce lineage differentiation and to facilitate reprogramming by increasing the efficiency of reprogramming or by replacing genetic reprogramming factors. Furthermore, mechanistic investigations of the effects of these chemicals also provide new biological insights. Here, we examine recent achievements in the maintenance of stem cells, including pluripotent and lineage-specific stem cells, and in the control of cell fate conversions, including iPSC reprogramming, conversion of primed to naïve pluripotency, and transdifferentiation, with an emphasis on manipulation with small molecules.
Key words: Stem cells, Small molecules, Self-renewal, Reprogramming
Introduction
Stem cells, which are characterized by the ability to self-renew and the potential to differentiate into diverse cell types (Box 1), have essential roles in embryonic development and tissue homeostasis. Owing to various promising applications in basic research, disease modeling, drug screening and regenerative medicine, stem cells have attracted enormous interest in the last three decades.
Box 1. Introduction to stem cell biology.
Stem cells are unspecialized cells that are characterized by their capacity for self-renewal and differentiation. They can give rise either to cells that bear characteristics identical to themselves and, thus, maintain self-renewal, or to more specialized cells with more-limited developmental potential, thereby resulting in differentiation. Noticeably, stem cells exist not only in embryos but also in adults throughout their whole life. Stem cells that are derived from distinct developmental stages may display different developmental potential.
Totipotent stem cells have the potential to generate an entire functional organism, including not only the embryo but also the extra-embryonic tissues. In mammals, the fertilized eggs and early embryonic cells, such as blastomeres, are totipotent.
Pluripotent stem cells (PSCs) can give rise to all the cell types of the entire embryo, including ectoderm, mesoderm and endoderm, as well as germ cells, but not the extraembryonic tissues, such as placenta. To date, several kinds of pluripotent stem cells (PSCs) have been reported, including embryonic stem cells (ESCs) derived from the inner cell mass of preimplantation embryos (Evans and Kaufman, 1981; Martin, 1981; Thomson et al., 1998), epiblast stem cells (EpiSCs) derived from the epiblast layer of the implanted embryos (Brons et al., 2007; Tesar et al., 2007), and induced pluripotent stem cells (iPSCs) generated from somatic cells by reprogramming (Takahashi et al., 2007; Takahashi and Yamanaka, 2006; Yu et al., 2007). Their pluripotency can be evaluated by a series of assays, such as the formation of teratomas or chimeras, germline contribution or tetraploid complementation.
Multipotent stem cells have the ability to develop into different cell types within the same cell lineage and are, therefore, also referred to as lineage-specific stem cells or progenitors. These cells have essential roles in maintaining tissue homeostasis under both physiological and pathological conditions. For example, hematopoietic stem cells in bone marrow can give rise to all types of blood cell and replenish peripheral blood.
Induced pluripotent stem cells (iPSCs) are artificially generated from somatic cells by ectopic expression of certain pluripotency-related factors. They closely resemble natural PSCs in many features, such as cellular biological properties, pluripotency and epigenetic signatures.
Ever since embryonic stem cells (ESCs) were first derived from mouse embryos (Evans and Kaufman, 1981; Martin, 1981), advances in stem cell biology and stem cell engineering have resulted in a number of methods to maintain stem cell self-renewal and direct lineage-specific differentiation, and to induce the reprogramming of somatic cells either to a pluripotent state or into another somatic cell type (Fig. 1). Despite this substantial progress, a number of diverse challenges remain. For example, defined culture conditions, which ideally are compatible with clinical applications, remain highly desired to maintain the long-term pluripotency of human ESCs (hESCs). Additionally, although lineage-specific stem cells that reside in many adult tissues have considerable self-renewal capacity under physiological or pathological conditions, it is still technically challenging to expand most types of lineage-specific stem cell ex vivo. The advances in cell reprogramming have stimulated an increased interest in generating patient-specific cell types from easily accessible and healthy cell types. However, reprogramming remains largely an inefficient and non-specific process, with efficiencies of transduced cells becoming fully reprogrammed induced pluripotent stem cells (iPSCs) lower than 0.01% (Hasegawa et al., 2010; Takahashi et al., 2007; Yu et al., 2007). Moreover, safety concerns – as well as other practical issues (e.g. large-scale cell production) – still represent major challenges for clinical translation. An improved understanding of the complex regulation of stem cells through cell-intrinsic and -extrinsic signals is essential to rationally devise appropriate conditions for controlling stem cell fate, state and function.
Fig. 1.
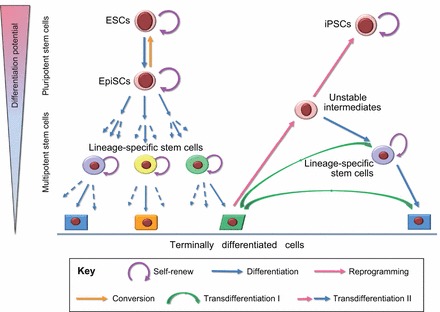
Chemical manipulation of stem cell fate. Small molecules can control stem cell fate, including stem cell self-renewal (purple curved arrows) and induction of lineage-specific differentiation (blue arrows). In iPSC reprogramming (pink arrows), small molecules can replace certain transcription factors, enhance the efficiency of reprogramming and accelerate the reprogramming processes. Small molecules can also facilitate the conversion of primed pluripotent stem cells into naïve stem cells (orange arrow). Transdifferentiation of one somatic cell type into another, bypassing pluripotency, can be mediated either by lineage-specific factors (transdifferentiation I; green arrows) or the curtailed reprogramming and subsequent lineage-specific differentiation (transdifferentiation II; pink and subsequent blue arrows). Small molecules known to be involved in reprogramming and differentiation can potentially also be employed for transdifferentiation II. ESCs, embryonic stem cells; EpiSCs, epiblast stem cells; iPSCs induced pluripotent stem cells.
Small molecules provide an attractive approach to addressing these challenges, as they offer a number of compelling advantages. First, the biological effects of small molecules are typically rapid, reversible and dose-dependent, allowing precise control over specific outcomes by fine-tuning their concentrations and combinations. Second, the structural diversity that can be provided by synthetic chemistry allows the functional optimization of small molecules. Third, compared with genetic interventions, the relative ease of the handling and administration of small molecules make them more practical for in vitro and in vivo applications, and for further therapeutic development. However, small molecules have their own disadvantages. A specific small molecule may have more than one target. Moreover, unexpected toxicity or other side effects in vivo may interfere with the clinical application of small molecules. However, the potential of small molecules to advance the field of stem cell research should not be underestimated.
In fact, phenotypic screening of chemical libraries, i.e. using expression of markers or cellular functions as readouts of biological effects, not only represents a powerful strategy for identifying the conditions that maintain, differentiate or reprogram cells, but also provides a chemical tool to dissect the underlying molecular mechanisms of these phenomena (Boitano et al., 2010; Chen et al., 2006; Desbordes et al., 2008; Zhu et al., 2010). Owing to the explosion of interest in applying chemical approaches to stem cell biology and regenerative medicine (Ding and Schultz, 2004; Xu et al., 2008), many compounds that regulate cell fate and function have been identified and characterized in recent years (summarized in Table 1; Fig. 2). For more general discussions of stem cell differentiation, readers are encouraged to consult comprehensive reviews (Efe and Ding, 2011; Lyssiotis et al., 2011). In this Commentary, we will focus on recent advances in the area of stem cell maintenance and reprogramming, and place a special emphasis on chemical strategies.
Table 1. Known compounds that modulate stem cell fate and reprogramming.
| Compound name | Identity | Function | References |
| Epigenetic-related compounds | |||
| Valproic acid (VPA) | HDAC inhibitor | Promotes MEF reprogramming efficiency, and enables Oct4- and Sox2-mediated reprogramming of human fibroblasts; | (Huangfu et al., 2008a; Huangfu et al., 2008b) |
| facilitates proteins mediated reprogramming of MEFs | (Zhou et al., 2009) | ||
| Suberoylanilide hydroxamc acid (SAHA) | HDAC inhibitor | Promotes MEF reprogramming efficiency | (Huangfu et al., 2008a) |
| Trichostatin A (TSA) | HDAC inhibitor | Promotes MEF reprogramming efficiency | (Huangfu et al., 2008a) |
| Sodium butyrate (NaB) | HDAC inhibitor | Enhances reprogramming efficiency of human adult or fetal fibroblasts; | (Mali et al., 2010) |
| facilitates Oct4-only mediated reprogramming when combined with A-83-01/PD0325901/PS48 | (Zhu et al., 2010) | ||
| BIX-01294 | G9a HMT inhibitor | Enables NPC reprogramming mediated by Oct4 and Klf4, or substitutes for Oct4 in NPC reprogramming; | (Shi et al., 2008b) |
| promotes MEF reprogramming mediated by Oct4 and Klf4 | (Shi et al., 2008a) | ||
| RG108 | DNMT inhibitor | Promotes MEF reprogramming mediated by Oct4 when combined with BIX-01294 | (Shi et al., 2008a) |
| 5-azazcytidine (5-aza) | DNMT inhibitor | Increases MEF reprogramming efficiency | (Huangfu et al., 2008a; Mikkelsen et al., 2008) |
| Parnate | LSD1 inhibitor | Enables reprogramming of human keratinocytes mediated by Oct4 and Klf4; | (Li et al., 2009b) |
| facilitates the conversion of mEpiSCs to naïve pluripotent state | (Zhou et al., 2010) | ||
| Signaling-pathway- or kinase-related compounds | |||
| PD0325901 | MEK inhibitor | Blocks differentiation pathway of ESCs and supports self-renewal; | (Ying et al., 2008; Tsutsui et al.) |
| supports ESC derivation from refractory strains or species; | (Nichols et al., 2009;Buehr et al., 2008; Li et al., 2008) | ||
| facilitates conversion of mEpiSCs and hESCs to naïve pluripotent state; | (Hanna et al., 2010; Zhou et al., 2010) | ||
| facilitates generation and maintenance of mESC-like rat or human iPSCs; | (Li et al., 2009a) | ||
| facilitates rapid and efficient generation of fully reprogrammed hiPSCs; | (Lin et al., 2009) | ||
| enables Oct4-mediated reprogramming when combined with A-83-01/NaB/PS48 | (Zhu et al., 2010) | ||
| CHIR99021 | GSK3 inhibitor | Supports ESCs self-renewal; facilitates ESCs derivation from refractory stains or species | (Ying et al., 2008; Tsutsui et al. 2011) |
| captures and maintains lineage-specific stem cells, like pNSCs; facilitates the conversion of mEpiSCs and hESCs to naïve pluripotent state; | (Nichols et al., 2009; Buehr et al., 2008; Li et al., 2008) | ||
| enables Oct4- and Klf4-mediated reprogramming of MEFs or human primary keratinocytes with Parnate; | (Li et al., 2009a; Hanna et al., 2010; Zhou et al., 2010; Li et al., 2009b) | ||
| facilitates generation and maintenance of mESC-like rat or human iPSCs; | (Li et al., 2009a) | ||
| facilitates the neural conversion of human fibroblasts mediated by Ascl1 and Ngn2 | (Ladewig et al., 2012) | ||
| 6-bromoindirubin-3′-oxime (BIO) | GSK3 inhibitor | Promotes self-renewal of ESCs and Isl+ cardiovascular progenitors | (Sato et al., 2004; Qyang et al., 2007) |
| Kenpaullone | GSK3 and CDK inhibitor | Replaces Klf4 in MEF reprogramming | (Lyssiotis et al., 2009) |
| PD173074 | FGF receptor inhibitor | Supports mESC self-renewal; | (Buehr et al., 2008) |
| facilitates the conversion of mEpiSCs to naïve pluripotent state | (Zhou et al., 2010) | ||
| SU5402 | FGF receptor inhibitor | Supports mESC self-renewal | (Buehr et al., 2008) |
| A-83-01 | ALK4, ALK5, ALK7 inhibitor | Facilitates the conversion of mEpiSCs to naïve pluripotent state; | (Zhou et al., 2010) |
| enables generation and long-term maintenance of mESC-like human iPSCs; | (Li et al., 2009a) | ||
| enables Oct4-mediated reprogramming when combined with PD0325901/NaB/PS48 | (Zhu et al., 2010) | ||
| SB431542 | ALK4, ALK5, ALK7 inhibitor | Captures and maintains pNSCs when combined with CHIR99021; | (Li et al., 2011) |
| facilitates rapid and efficient generation of fully reprogrammed human iPSCs; | (Lin et al., 2009) | ||
| Facilitates the neural conversion of human fibroblasts mediated by Ascl1 and Ngn2 | (Ladewig et al., 2012) | ||
| E-616452 | ALK4, ALK5 and ALK7 inhibitor | Replaces Sox2 in MEF reprogramming. | (Ichida et al., 2009; Maherali and Hochedlinger, 2009) |
| LDN193189 | ALK2, ALK3 and ALK6 inhibitor | Facilitates the neural conversion of human fibroblasts mediated by Ascl1 and Ngn2 | (Ladewig et al., 2012) |
| Compound E | γ-secretase inhibitor | Accelerates the generation of pNSCs | (Li et al., 2011) |
| JAK Inhibitor I | JAK inhibitor | Inhibits the generation of iPSCs in iPSC-TF-based transdifferentiaion | (Efe et al., 2011; Kim et al., 2011a) |
| Pluripotin (SC1) | RasGAP and ERK inhibitor | Maintains mESC self-renewal | (Chen et al., 2006) |
| Y-27632 | ROCK inhibitor | Improves survival of hESCs upon dissociation | (Chen et al., 2010; Ohgushi et al., 2010; Xu et al., 2010) |
| Thiazovivin (Tzv) | ROCK inhibitor | Improves survival of hESCs upon dissociation; | (Xu et al., 2010) |
| facilitates rapid and efficient generation of fully reprogrammed hiPSCs | (Lin et al., 2009) | ||
| StemRegenin1 | AhR antagonist | Enables ex vivo expansion of CD34+ HSCs ex vivo | (Boitano et al., 2010) |
| PS48 | PDK1 activator | Enables OCT4-mediated reprogramming with A-83-01, NaB and PD0325901 | (Zhu et al., 2010) |
| BayK8644 | L-type Ca2+ channel agonist | Promotes MEF reprogramming mediated by Oct4 and Klf4 when combined with BIX-01294 | (Shi et al., 2008a) |
| Forskolin | PKA agonist | Induces Klf4 and Klf2 expression to facilitate hESCs conversion into a naïve pluripotent state | (Hanna et al., 2010) |
Notice that, in this table, reprogramming generally refers to iPSC-TF-mediated reprogramming; in most cases the iPSC-TFs are Oct4, Sox2, Klf4 and Myc.
HDAC, histone deacetylase; HMT, histone methyltransferase; DNMT, DNA methyltransferase; LSD1, lysine-specific demethylase 1; MEK, MAPK kinase; GSK3, glycogen synthase kinase 3; FGF, fibroblast growth factor; activin A receptor-like kinase (ALK) ALK2, ACVR1; ALK3, BMPR1A; ALK4, ACVR1B; ALK5, TGFBBR1; ALK6, BMPR1B; ALK7, ACVR1C; JAK, Janus kinase; RasGAP, Ras GTPase-activating protein; ERKs, extracellular signal-regulated kinases; PDK1, phosphoinositide-dependent kinase 1; ROCK, Rho-associated coiled-coil-containing protein kinase; AhR, aryl hydrocarbon receptor; PKA, protein kinase A; MEF, mouse embryonic fibroblasts; ESC, embryonic stem cell; mESC, mouse ESC; hESC, human ESC; mEpiSC, mouse epiblast stem cell; iPSC, induced pluripotent stem cell; NPC, neural progenitor cell; pNSC, primitive neural stem cell; HSC, hematopoietic stem cell.
Fig. 2.
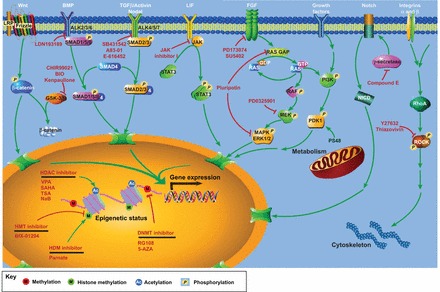
Mech anisms underlying the chemical manipulation of stem cell fate and reprogramming. Small molecules (red) and the signaling pathways they act on are given in above their respective receptors (white). Red blunt-headed arrows indicate inhibition, and green arrows indicate activation. Epigenetic modulators can modify chromatin structure and make it more permissive to changes in the epigenome and cell fate during reprogramming. Small molecules that target signaling pathways can regulate the expression of target genes and, eventually, influence cell fate. Other chemical manipulations of, for example, metabolism or the cytoskeleton, can also influence cell fate as described in the main text. See Table 1 for abbreviations.
The role of small molecules in stem cell maintenance
Here, we discuss strategies and new developments, particularly chemical approaches that have been employed to maintain the self-renewal of ESCs or lineage-specific stem cells.
Embryonic stem cell culture systems
Conventionally, ESCs are cultured in the presence of feeder cells – typically human or mouse fibroblasts that have been growth inactivated through chemicals or γ-irradiation – serum products, e.g. fetal bovine serum (FBS), or knockout serum replacement (KSR), and growth factors. Until now, several crucial signaling pathways as well as related growth factors have been indentified that participate in the maintenance of ESC pluripotency (Fig. 2). For mouse ESCs (mESCs), these include leukemia inhibitory factor (LIF)-signal transducer and activator for transcription 3 (STAT3) (Niwa et al., 1998), as well as bone morphogenetic protein (BMP) (Chambers and Smith, 2004; Ying et al., 2003). Human ESCs rely on fibroblast growth factor 2 (FGF2, also known as basic fibroblast growth factor) and Activin or NODAL signaling (James et al., 2005; Vallier et al., 2005). In addition, Wnt signaling was reported to contribute to the maintenance of both mESCs and hESCs (Sato et al., 2004).
However, the presence of undefined culture components raises a number of possible issues. First, feeder cells and other animal products including serum or serum replacements might entail the risk of xenogeneic or pathogenic contaminations. Second, the batch-to-batch variability of feeders and serum might make it difficult to achieve culture consistency between different laboratories or even different experiments. Third, with regards to mechanistic investigation, these uncharacterized components might impede the elucidation of the exact molecular circuitry underlying pluripotency by exerting unrecognized roles (e.g. signaling crosstalk) within the pluripotency network. Although more-defined culture systems have been developed in recent years (Valamehr et al., 2011; Vallier et al., 2005; Ying et al., 2003), some animal products, such as bovine serum albumin (BSA) and cell-derived matrix components, continue to be used even under these more defined conditions. Therefore, a fully defined, scalable and reproducible culture system for ESCs is necessary for both basic research and clinical applications.
Small molecules and the self-renewal of embryonic stem cells
A cell-based high-throughput screening (HTS), using an mESC line that harbors green fluorescent protein (GFP) under the control of the Oct4 (also known as Pou5f1) promoter as a molecular reporter, led to the identification of a synthetic small molecule Pluripotin (also known as SC1) that can maintain long-term self-renewal of mESCs under feeder-, serum- and LIF-free conditions (Chen et al., 2006). Pluripotin was found to exert its function by simultaneously inhibiting two endogenous differentiation-inducing proteins, Ras GTPase-activating protein (RasGAP) and extracellular-signal-regulated kinase 1 (ERK1, officially known as MAPK3). This was the first study to show that ESCs possess the intrinsic ability to sustain pluripotency in the absence of activation of pluripotency-associated pathways by exogenous growth factors (including the LIF–Stat3, Bmp–Smad and Wnt–β-catenin pathways) that, previously, have been regarded as essential for ESC self-renewal (Ying et al., 2003; Chambers and Smith, 2004; Niwa et al., 1998; (Sato et al., 2004). Furthermore, the identification of Pluripotin revealed a fundamental strategy for maintaining stem cell self-renewal through the inhibition of endogenous differentiation mechanisms, and explained how combining the activation of differentiation-inducing pathways (e.g. BMP signaling) with the modulation of other pathways [e.g. inhibition of the MAPK kinase (MEK)–ERK pathway] can sustain self-renewal, i.e. by effectively balancing out the differentiation activity of stem cells.
This new concept was further substantiated by a more recent study, in which a combination of the MEK inhibitor PD0325901 and the glycogen synthase kinase-3 (GSK3) inhibitor CHIR99021 (which activates Wnt signaling) was used to support the maintenance of mESCs without the need for feeder cells or exogenous cytokines (Buehr et al., 2008). In addition to the conceptual advance in these studies, these small molecules also allow derivation of ESCs from other species – such as rats (Buehr et al., 2008; Li et al., 2008) – as well as from some mouse strains, including the nonobese diabetic (NOD), severe combined immunodeficiency (SCID) and NOD-SCID beige mice, which have specific biological significance but previously had been technically challenging (Hanna et al., 2009; Yang et al., 2009).
Until recently, the vulnerability of hESCs to cell death upon single-cell dissociation was a substantial hurdle to their practical applications (e.g. in large-scale cell passaging, genetic manipulation and selection). This obstacle was overcome with the identification of Rho-associated coiled-coil-containing protein kinase (ROCK) inhibitors, such as Y-27632 and thiazovivin (Tzv), which substantially enhance the survival of dissociated single hESCs (Chen et al., 2010; Ohgushi et al., 2010; Watanabe et al., 2007; Xu et al., 2010). These studies revealed that integrin-mediated cell–ECM interactions and E-cadherin-mediated cell–cell interactions form a positive feedback loop by inhibiting Rho–ROCK signaling, which promotes hESC survival. When cells become detached and dissociate into single cells, this positive feedback loop is interrupted, and culminates in the hyperactivation of ROCK signaling and the irreversible disruption of cell–cell and cell–ECM adhesion. Eventually, this disruption leads to cell death but can be overcome when using a ROCK inhibitor. Recently, a cocktail of PD0325901, CHIR99021 and Y27632 combined with bFGF was reported to maintain the pluripotency and karyotype integrity of hESCs following long-term single-cell passaging on a fibronectin-coated surface (Tsutsui et al., 2011). Although the conditions still contain KSR, the chemical manipulation overcomes some technical hurdles to the expansion of hESCs.
Small molecules and the expansion of lineage-specific stem cells
In contrast to ESCs, the long-term maintenance of most lineage-specific adult stem cells (Box 1) remains challenging. As they have more restricted differentiation potentials than pluripotent stem cells (PSCs), lineage-specific adult stem cells are less likely to form teratomas (Box1), a crucial safety concern in the clinical application of PSCs. Besides, adult stem cells can be isolated from patients for autologous therapy, which avoids immunocompatibility issues. Therefore, lineage-specific stem cells may prove to be more suitable for therapeutic applications than PSCs. However, their scarcity and technical challenges to their ex vivo expansion significantly impair their therapeutic potential.
Chemical approaches offer substantial promise for sustaining lineage-specific stem (or progenitor) cells collected from differentiated tissues. In a cell-based chemical screening, the compound StemRegenin1 (SR1), in combination with conventional hematopoietic stem cell (HSC) growth cytokines, was found to promote the ex-vivo expansion of CD34+ HSCs that have the potential to differentiate into all blood cell types from human cord blood. Moreover, SR1 substantially increases the number of cells that retain the capacity of multilineage long-term engraftment in NOD-SCID mice, which is the functional standard to assess the quality of HSCs (Boitano et al., 2010). Mechanistic investigation revealed that SR1 exerts its function by antagonizing the aryl hydrocarbon rceptor (AhR), thus implying a role for AhR signaling in hematopoiesis. In another study, the potent GSK3 inhibitor 6-bromoindirubin-3′-oxime (BIO) has been found to promote the expansion of murine and human Isl+ cardiovascular progenitors, which are regarded as the stem cells of the second heart field (Qyang et al., 2007). It was demonstrated that BIO exerts this function by activating canonical Wnt signaling, which implies that Wnt–β-catenin signaling has an evolutionary conserved role in the self-renewal of cardiovascular progenitors. An alternative strategy for obtaining lineage-specific stem cells is to induce their differentiation from PSCs, which can avoid the technical challenges of isolating resident stem cells from most tissues and organs. Recently, we have established conditions under which defined small molecules allow the rapid differentiation and capture of primitive neural stem cells (pNSCs) from hESCs (Li et al., 2011). Here, combining human LIF, CHIR99021 and SB431542 (an inhibitor of the TGF-β type I receptors ALK4, ALK5 and ALK7) efficiently induces monolayer-cultured hESCs into homogenous pNSCs. Addition of γ-secretase inhibitor XXI (also called compound E), further accelerates differentiation and leads to complete neural induction within one week (>97% cells are SOX2 positive but OCT4 negative), which might be due to the suppression of Notch signaling (Li et al., 2011). Importantly, in the presence of LIF, CHIR99021 and SB431542, these pNSCs can stably self-renew over long-term serial passages, while still maintaining their spatial plasticity and high neurogenic differentiation propensity. This study thus created a new paradigm in which hESCs can be differentiated into homogenous lineage-committed progenitors that can be stabilized and expanded under chemically defined conditions. Such a strategy might make it possible to overcome some of the limitations that are inherent in the step-wise uninterrupted differentiation schemes, which result in the accumulation of undesired cells in each differentiation step, eventually leading to increased heterogeneity and a substantially lower yield of target cells.
Owing to their potency and flexibility in manipulating protein functions, small molecules have become increasingly popular in maintaining ESC self-renewal, inducing their lineage differentiation and expanding lineage-specific stem cells. Unbiased chemical screening could thus be an especially powerful approach to interrogate the so-far unknown mechanisms that govern stem cell self-renewal.
The role of small molecules in reprogramming
Reprogramming of somatic cells towards pluripotency
The reversal of differentiation and the generation of PSCs from somatic cells have fascinated researchers for years. Early studies of somatic cell nuclear transfer (SCNT) revealed that a somatic nucleus can be fully reprogrammed into a totipotent state by factors from an enucleated egg, proceed to the generation of an entire organism or be used to derive ESCs (Agarwal, 2006; Campbell et al., 1996). Despite many advances in SCNT, the process remains technically challenging and, in the human system, there are ethical concerns to use SCNT to generate hESCs. Cell fusion between somatic cells and ESCs to form heterokaryons can also reprogram somatic nuclei into the pluripotent state, although this typically results in the presence of extra sets of chromosomes (Cowan et al., 2005; Ying et al., 2002). Another limitation of SCNT and cell fusion is that, to mediate reprogramming, they use largely undefined cellular contents, which makes it difficult to investigate the underlying mechanisms.
In 2006, Yamanaka and colleagues demonstrated that virus-mediated overexpression of Oct4, Sox2, Klf4 and Myc (collectively termed OSKM) can convert mouse fibroblasts into induced PSCs (iPSCs), which closely resemble mESCs in terms of global gene expression, epigenetic state and developmental potential (Takahashi and Yamanaka, 2006). Soon after this discovery, human fibroblasts were reprogrammed into iPSCs by overexpressing OSKM factors (Takahashi et al., 2007), or OCT4, SOX2, NANOG and LIN28 (OSNL) (Yu et al., 2007). Compared with SCNT and cell fusion approaches, the generation of iPSCs relies on defined factors, is a much simpler process and is unaffected by ethical controversy. However, there have been concerns over the use of integrating retroviruses to deliver the iPSC factors, which could potentially compromise the quality of or even cause tumorigenicity in the resultant iPSCs.
Recent advances in iPSC technology have largely resolved the concerns over genome modification through exogenous sequences when new methods were introduced to deliver the reprogramming factors that included the use of episomal plasmids (Yu et al., 2009) or excisable expression systems (Soldner et al., 2009), recombinant cell-penetrating reprogramming proteins (Kim et al., 2009; Zhou et al., 2009) and reprogramming mRNAs (Warren et al., 2010; Yakubov et al., 2010) or microRNAs (Anokye-Danso et al., 2011; Miyoshi et al., 2011). For details regarding the technical achievements in the reprogramming field, readers are encouraged to examine more comprehensive reviews on this subject (González et al., 2011; Patel and Yang, 2010). Despite these technical advancements, a challenging and more fundamental issue is how to change the current iPSC reprogramming procedure from a slow, inefficient and non-deterministic process that involves stochastic events, to one that is highly directed, specific and efficient. Another important question to be answered is how to achieve reprogramming by using only defined small molecules – an approach that is fundamentally different from the exogenous transcription-factor-based reprogramming that is employed in SCNT, cell fusion and current iPSC methods. These advances would also address other unresolved safety concerns with regards to the generation and use of iPSCs, such as the potential effects on epigenetic memory and other subtle genetic and epigenetic changes that might occur during reprogramming (Kim et al., 2010; Ohi et al., 2011).
Using both phenotypic screening and hypothesis-driven approaches, a growing number of compounds have been identified that can functionally replace reprogramming transcription factors, enhance efficiency of iPSC generation and accelerate the reprogramming process (Table 1).
Given that reprogramming is accompanied by remodeling of the epigenome, modulations of the epigenetic processes may facilitate such conversion of cell fate by making cells more permissive to these epigenenomic changes. Therefore, it is no surprise to find that compounds that modulate epigenetic enzymes, such as histone deacetylase (HDAC), histone methyltransferase (HMT), histone demethylase (HDM) and DNA methyltransferase (DNMT), can improve the efficiency of reprogramming, or even replace the need to use certain transcription factors. In a chemical screening, BIX-01294 (an inhibitor of the HMT EHMT2; also known as G9a) was found to substantially enhance Oct4–Klf4-mediated reprogramming of neural progenitor cells (NPCs) into iPSCs to a level that is comparable with that when using OSKM factors, as well as to enable reprogramming mediated by only Klf4–Sox2–Myc without the need for Oct4 (Shi et al., 2008b), albeit with much reduced efficiency. In another study, BIX-01294 was shown to enable Oct4-Klf4-mediated reprogramming of mouse embryonic fibroblasts (MEFs), which could be further enhanced by BayK8644 (an agonist of L-type Ca2+ channels) or RG108 (an inhibitor of DNMT) (Shi et al., 2008a). HDAC inhibitors [e.g. suberoylanilide hydroxamic acid (SAHA), Trichostatin A (TSA) and valproic acid (VPA)] have also been shown to improve reprogramming efficiency (Huangfu et al., 2008a; Mikkelsen et al., 2008). In particular, VPA has been used in the reprogramming of human fibroblasts with Oct4 and Sox2 (Huangfu et al., 2008b), in the reprogramming of MEFs with recombinant cell-penetrating reprogramming proteins (Zhou et al., 2009), and in mir-302/367-mediated mouse fibroblasts reprogramming (Anokye-Danso et al., 2011). Other compounds that affect epigenetic processes as well as their effects on reprogramming are summarized in Table 1.
As in the maintenance of pluripotency, some signaling pathways and their chemical modulators also help to re-establish pluripotency during reprogramming. The Wnt–β-catenin signaling pathway has been reported to enhance reprogramming through alleviating the inhibitory effect of T-cell factor-3 (TCF3) on pluripotency (Niwa, 2011). Consistently, CHIR99021, a GSK3 inhibitor that activates Wnt signaling, enables the reprogramming of MEFs into iPSCs by leading to overexpression of Oct4 and Klf4 only, and also facilitates the Oct4–Klf4-mediated reprogramming of human primary keratinocytes when combined with Parnate, an inhibitor of lysine-specific demethylase1 (LSD1) (Li et al., 2009b). Kenpaullone, which inhibits GSK3 and several other kinases, was identified from a HTS to be able to substitute for Klf4 in OSM-mediated reprogramming of MEFs (Lyssiotis et al., 2009).
TGF-β signaling is crucial to epithelial–mesenchymal transition (EMT), an important hallmark of embryonic development (Xie et al., 2004b). TGF-β induces EMT through canonical Smad signaling, non-canonical Ras–MEK–ERK MAP kinase signaling and Rho signaling (Xu et al., 2009). The reverse process, mesenchymal–epithelial transition (MET), is a crucial early event in reprogramming to pluripotency. It can thus be anticipated that small molecules that block TGF-β signaling or its downstream effectors facilitate MET and enhance reprogramming. Consistent with this idea, inhibitors of TGF-β receptors, indeed, enhance reprogramming and can replace Sox2 in the reprogramming of MEFs (Ichida et al., 2009; Maherali and Hochedlinger, 2009). In another hypothesis-driven study, small molecules that are known to promote MET or inhibit EMT – including SB431542, PD0325901 and Tzv, which inhibit TGF-β receptors, MEK and Rho-ROCK, respectively – were tested in human fibroblasts and found to not only substantially enhance reprogramming efficiency, but also accelerate the speed of reprogramming, partially through derepression of the epithelial phenotype (Lin et al., 2009).
Recently, we have reported a chemical cocktail that enabled the generation of human iPSCs from several human primary somatic cell types when only Oct4 was exogenously expressed (Zhu et al., 2010). This cocktail contains sodium butyrate (NaB, an inhibitor of HDAC), A-83-01 (an inhibitor of TGF-β type I receptors ALK4, ALK5 and ALK7) and PD0325901, as well as PS48 (an activator of 3′-phosphoinositide-dependent kinase-1, PDK1) (Zhu et al., 2010). Detailed mechanistic studies have revealed that PS48 acts at the early phase of reprogramming in order to facilitate a metabolic switch from mitochondrial oxidation (typically utilized by adult somatic cells) to glycolysis (almost exclusively used by PSCs) (Zhu et al., 2010). Other compounds that promote glycolytic metabolism have also been shown to enhance reprogramming. They include fructose 2,6-bisphosphate (an activator of phosphofructokinase 1, a key rate-limiting enzyme of glycolysis) and N-oxaloylglycine and quercetin, both activators of hypoxia-inducible factor-1 (Zhu et al., 2010). This study represents another significant step towards the ultimate goal of chemical reprogramming and also reveals metabolic modulation as another fundamental mechanism underlying somatic cell reprogramming.
Conversion from the primed to naïve pluripotent stem cells
In addition to mESCs, which originate from the inner cell mass (ICM) of preimplantation mouse embryos, a different kind of PSCs, mouse epiblast stem cells (mEpiSCs) can be derived from late postimplantation epiblasts of mouse embryos (Brons et al., 2007; Tesar et al., 2007). mESCs and mEpiSCs exhibit distinct properties in terms of gene expression, epigenetic profile, cell behavior and their response to different signals (Table 2) (Brons et al., 2007; Guo et al., 2009; Tesar et al., 2007). Importantly, when mEpiSCs are injected into a preimplantation blastocyst, they contribute poorly to chimerism and are unable to transmit through the germline in contrast to the robust chimera formation and germline transmission seen with mESCs (Brons et al., 2007). These different characteristics indicate that, consistent with their origins, mEpiSCs represent a later or ‘primed’ state that is reminiscent of post-implantation epiblasts, whereas mESCs represent the ‘naïve’ state that corresponds to preimplantation ICM (Nichols and Smith, 2009). This observation raises the interesting question of whether it is feasible to convert mEpiSCs back into the mESC-like naïve pluripotent state. Indeed, overexpression of Klf4, Nanog or nuclear receptor subfamily 5 group A (Nr5a) has been shown to facilitate the reversion of mEpiSCs into mESC-like cells (Guo and Smith, 2010; Guo et al., 2009; Silva et al., 2009). Concurrent with these genetic approaches, we identified small-molecule conditions that also induce this conversion, but in a more specific and efficient manner. The combination of the inhibitors Parnate, A-83-01, PD0325901, PD173074 and CHIR99021 (Table 1), which antagonize LSD1, ALK5, MEK, FGF receptor and GSK3, respectively, can fully convert mEpiSCs back to the mESC-like naïve state and restore their chimera competence (Zhou et al., 2010). Another group also demonstrated that the treatment of PD0325901, CHIR99021 and LIF allow the conversion from mEpiSCs to a mESC-like status that notably allowed for germline transmission (Greber et al., 2010).
Table 2. Comparison of mouse ESCs, EpiSCs and human ESCs.
| Mouse ESCs | Mouse EpiSCs | Human ESCs | Putative human naïve state PSCs | |
| Derivation | ||||
| Origin | ICM | Epiblast | ICM | Human ESCs or fibroblasts |
| Developmental stage | Preimplantation | Post-implantation | Preimplantation | N/A |
| Epigenetic character | ||||
| XX status in female cells | XaXa | XaXi | XaXi with sporadic reactivation (XaXa) in some lines | XaXa |
| Differentiation potential | ||||
| Three germ layers | Yes | Yes | Yes | Yes |
| Germline cells | Yes | Yes | Yes | ND |
| Teratoma formation | Yes | Yes | Yes | Yes |
| Chimera formation | Yes | No | N/A | N/A |
| Growth properties | ||||
| Colony morphology | Domed, multilayer | Flat, monolayer | Flat, monolayer | Domed, multilayer |
| Tolerate single-cell dissociation | Yes | No | No | Yes |
| Signal response | ||||
| Self-renewal | LIF, BMP4, inhibition of MEK | bFGF, activin | bFGF, activin | LIF, MEKi |
| Differentiation | TGF-β/activin, bFGF, ERK1/2 | BMP4 | BMP4 | TGF-β |
| Gene expression profile | ||||
| Pluripotency-related genes (e.g. Oct4, Nanog, Sox2) | + | + | + | + |
| ICM associated genes (e.g. Stella, Tbx3, Gbx2) | High | Low or undetectable | Low or undetectable | Increase |
| Genetic manipulation | ||||
| Gene targeting | Easy | ND | Difficult | Easy |
N/A, not applicable; ND, not determined; ESC, embryonic stem cell; EpiSC, epiblast stem cell; PSC, pluripotent stem cell; ICM, for inner cell mass; LIF, leukemia inhibitory factor; BMP4, bone morphogenetic protein 4; bFGF, basic fibroblast growth factor; TGF-β, transforming growth factor β; ERKs, extracellular signal-regulated kinases (MAPKs); Xa, active X chromosome; Xi, inactive X chromosome.
Interestingly, even though the preparation of hESCs typically starts with preimplantation embryos, in many ways these cells more closely resemble mEpiSCs than mESCs (Table 2), implying that hESCs represent a primed pluripotent state and not the naïve state (Tesar et al., 2007). Technically, hESCs are more resistant to gene targeting mediated by homogenous recombination, which is routinely used in mESCs. This might be owing to the intolerance of hESCs to single-cell dissociation, slow proliferation and a less-open chromatin structure (Li and Ding, 2011). It has been speculated that naïve hPSCs – if they could be generated – might have a number of advantages over primed hPSCs in various applications (Li and Ding, 2011). For example, a more robust growth and survival of naïve state hPSCs would facilitate practical cell expansion and genetic manipulation. In addition, the knowledge gained from studies in mESCs might be more readily translatable to naïve state hPSCs. From both scientific and technical viewpoints, it is of great interest to generate naïve hESCs either from conventional hESCs or directly from somatic cells by reprogramming. We first demonstrated that mESC-like human PSCs can be generated and stably expanded from human fibroblasts in an approach that included genetic reprogramming (i.e. the overexpression of OCT4, SOX2, NANOG and LIN28) in a culture medium containing LIF, PD0325901, A-83-01 and CHIR99021 (Li et al., 2009a). Another study combined the transient induction of Oct4, Klf4 and Klf2 with a cocktail containing of PD0325901, CHIR99021, LIF and forskolin (an agonist of protein kinase A), which was able to convert hESCs into a naïve pluripotent state and also facilitated the derivation of naïve hiPSCs that shared many features with miPSCs (Hanna et al., 2010). Later, naïve hPSCs were also generated directly from fibroblasts by constitutively overexpressing OSKM factors and NANOG in the presence of LIF (yielding the so-called hLR5 cells) (Buecker et al., 2010), or by transient coexpression of retinoic acid receptor γ (RARG), liver receptor homolog 1 (Nr5a2) and OSKM factors with subsequent culturing in a medium containing PD0325901, CHIR99021 and LIF (Wang et al., 2011). These converted naïve hESCs closely resemble mESCs in certain aspects, such as morphology, signaling dependence and gene expression. More importantly, as preliminarily identified in hLR5 cells, these naïve hESCs allow for efficient gene targeting, thus making the genetic manipulation of hESCs more feasible (Buecker et al., 2010). However, it remains a substantial challenge to derive hPSCs at the naïve pluripotent state and maintain them long-term without the expression of any exogenous reprogramming factors.
Lineage-specific reprogramming – transdifferentiation
Unlike the reprogramming of iPSCs, transdifferentiation refers to the direct reprogramming of one somatic cell type into another one without passing through a pluripotent state. Transdifferentiation may have several advantages over PSC-based strategies in generating somatic cells, as it is potentially faster and more efficient and can result in a higher yield of target cells. Additionally, transdifferentiation might be safer for cell-based therapeutic applications as it eliminates the risk of tumorigenesis inherent to PSCs. To date, two distinct transdifferentiation strategies have been developed, conventional transdifferentiation mediated by lineage-specific factors and iPSC-transcription factor (TF)-based transdifferentiation, which relies on curtailed or hijacked reprogramming followed by lineage-specific culture conditions to generate the desired cell type (Fig. 1).
Long before the development of iPSC technology, various transdifferentiation procedures have been attempted. An early example demonstrated the conversion of fibroblasts into skeletal muscle cells by ectopic expression of MyoD, a master regulatory transcription factor involved in myogenesis (Davis et al., 1987). However, this conversion is considered incomplete, as the acquired phenotype relies on a sustained overexpression of MyoD. Other groups also reported the conversion of somatic cells to other closely related cell types (Izumikawa et al., 2005; Xie et al., 2004a; Zhou et al., 2008). Recently, several studies have demonstrated that transdifferentiation between distantly related cell types can be achieved by ectopic expression of multiple lineage-specific factors. For example, mouse fibroblasts can be converted into functional neurons by overexpressing Brn2, Ascl1 and Myt1L (Vierbuchen et al., 2010) and into cardiomyocytes by overexpression of Gata4, Mef2c and Tbx5 (Ieda et al., 2010; Qian et al., 2012), and into hepatocyte-like cells by overexpressing Gata4, Hnf1a and Foxa3, while at the same time inactivating p19Arf (Huang et al., 2011), or by overexpressing Hnf4a and Foxa1, Foxa2 or Foxa3 (Sekiya and Suzuki, 2011). In these studies, the transdifferentiated cells closely resembled native cell types in terms of morphology, gene expression and functions in vitro or even in vivo. Of note, studies from the Wernig laboratory verified that not only fibroblasts (Vierbuchen et al., 2010), but also terminally differentiated hepatocytes can be converted into functional neurons by using the same set of neuronal master factors (Marro et al., 2011). As shown in this work, converted cells can acquire the transcriptional neuronal program and robustly silence the transcriptional network of their cell type of origin. In addition, several groups have independently reported that human fibroblasts can be converted into functional neurons by overexpressing different combinations of master factors, such as miR-124, MYT1L and BRN2 (Ambasudhan et al., 2011), Brn2, Ascl1, Myt1L and NeuroD1 (Pang et al., 2011), or miR-9/9*, miR-124 and NEUROD2 (Yoo et al., 2011). From these studies it is apparent that several combinations or classes of master factor exist that can perform a similar, if not identical, transdifferentiation. In addition, some specific neural cell types, such as spinal motor neurons (Son et al., 2011) and dopaminergic neurons (Caiazzo et al., 2011; Kim et al., 2011b; Pfisterer et al., 2011) could also be generated from both mouse and human fibroblasts with the forced expression of cell-subtype-specific transcription factors. More recently, it was reported that mouse fibroblasts can also be converted into neural stem cells by overexpression of Brn2, Sox2 and FoxG1 (Lujan et al., 2012), or of Brn4, Sox2, Myc, Klf4 and Tcf3 (Han et al., 2012). Because these induced neural stem cells (iNSCs) retain the ability to proliferate and the potential to differentiate into neurons, astrocytes and oligodendrocytes, they might be more desirable than terminally differentiated cells for therapeutic cell replacement, where proliferation and multilineage potential are needed to repopulate the damaged tissue.
Collectively, in conventional transdifferentiation approaches, overexpression of lineage-specific master factors can drive somatic cell conversion within or across germ layer boundaries. This approach – although unlikely to generate iPSCs – has a reduced risk of teratoma formation. However, to generate any desired cell type, the required master factors need to be defined for each type of target cell, and the underlying molecular and epigenetic mechanisms of each possible transdifferentiation need to be fully elucidated, which – undoubtedly – represents an enormous research effort.
The second approach, iPSC-TF-based transdifferentiation, was developed on the basis of curtailed reprogramming and subsequent lineage differentiation. iPSC reprogramming is a non-deterministic process that involves step-wise stochastic events, and only a very small percentage of induced cells eventually become iPSCs. We, therefore, hypothesized that it is possible to guide cells that have initially been epigentically ‘activated’ through temporally restricted iPSC-TF expression, towards lineage-specific cell types, by switching to different sets of signaling inputs without going through the pluripotent state. Driven by this hypothesis, we transiently expressed the OSKM factors that are able to induce the iPSC state, but for a period that is insufficient to generate iPSCs (Efe et al., 2011; Kim et al., 2011a). This stage was followed by an exposure of the cells to signals that guide them to the desired lineage, while simultaneously inhibiting the establishment of pluripotency (Fig. 1). This procedure not only allows the generation of terminally differentiated somatic cells but also provides a time window for capturing lineage-specific stem or progenitor cells (Efe et al., 2011; Kim et al., 2011a). Following the transient overexpression of the OSKM factors, we induced cardiac differentiation by treating cells with BMP4 and obtained spontaneously contracting cardiomyocytes from MEFs as early as day 11 (Efe et al., 2011). Cardiac mesoderm precursor cells were also observed during the transdifferentiation. In another study, we captured neural progenitors from fibroblasts when cells were treated with FGFs and epidermal growth factor (EGF) following transient overexpression of OSKM factors (Kim et al., 2011a). Using a similar concept, another group demonstrated that the curtailed reprogramming with transient Oct4 expression during the initial phase of reprogramming (within 5 days) converts fibroblasts into expandable and functional neural stem cells (NSCs) (Thier et al., 2012). In another study, multilineage blood progenitors were generated from human dermal fibroblasts through the ectopic expression of OCT4 and specific cytokine treatment, without going through a pluripotent state (Szabo et al., 2010).
Compared with conventional transdifferentiation, iPSC-TF-based transdifferentiation provides a general platform with several practical advantages. First, it can be applied to induce reprogramming towards several lineage-specific cell types by using a single combination of transcription factors and well-established lineage induction conditions. Moreover, not only terminally differentiated cells but also their lineage-committed progenitors can be generated in one process. Second, because this method relies on ‘curtailed reprogramming’, advances in reprogramming – such as small-molecule treatment and non-integrating factor delivery – can easily be implemented to improve iPSC-TF-based transdifferentiation. This approach might also represent an ideal platform to transfer the chemical manipulations in iPSC reprogramming and lineage differentiation directly into lineage-specific transdifferentiation.
Despite these advances of genetically based transdifferentiation, it has the inherent problems of transgene expression, such as the genomic intrgration of viral DNA fragments, as well as issues of low efficiency or unclear epigenetic status that need to be addressed before its clinical applications can be realized. For this, small molecules might provide a promising alternative for increasing the efficiency or replacing some, if not all, components needed for transdifferentiation. Given the substantial resetting of epigenetic and gene expression patterns during transdifferentiation, it is reasonable to suspect that chemicals that affect epigenetic or signaling pathways have roles in this process. In fact, long before the aforementioned genetically based transdifferentiations were described, it had already been reported that 5-azazcytidine (5-aza) induces the conversion of a mesenchymal cell line into muscles, adipocytes and chondroblasts (Taylor and Jones, 1979) by suppressing DNA methylation (Jones and Taylor, 1980). In the iPSC-TF-based transdifferentiation approach described above, the JAK inhibitor, which antagonizes the LIF–STAT3 pathway, an essential pathway for mESC maintenance, is used to suppress the establishment of pluripotency, while at the same time facilitating the generation of epigenetically plastic intermediate cells (Efe et al., 2011; Kim et al., 2011a). More recently, it was reported that inhibition of GSK-3β and Smad by CHIR99021, SB431542 and Noggin (a protein antagonist of BMP signaling) substantially enhances the efficiency of Ascl- and Ngn2-driven conversion of human fibroblasts into neural cells (Ladewig et al., 2012). It was demonstrated that the concentration of Noggin can be reduced significantly when used in combination with LDN193189 (a chemical inhibitor of BMP type I receptors ALK2, ALK3 and ALK6). As these small molecules act not only in reprogramming but also in neural differentiation (Chambers et al., 2009), their exact role during this conversion needs to be further elucidated. Although to date there are only a few reports of the use of small molecules in the nascent field of transdifferentiation, we expect to see more chemical applications, especially those that have effects on iPSC reprogramming and lineage differentiation, can be directly applied to the transdifferentiation field. Additionally, in HTS or hypothesis-driven studies, some small molecules have been shown to induce certain conversions into specific cell types (Chen et al., 2004; Yau et al., 2011) and these approaches might, therefore, provide additional insight into the exact molecular mechanisms that underlie transdifferentiation.
Conclusions
Although still very young, the field of stem cell research has already offered enormous unprecedented opportunities for basic research, disease modeling, drug screening and regenerative medicine. Especially the recent achievements in reprogramming and transdifferentiation have made it possible to generate highly desired cell types from more accessible cell populations, and bringing us closer to cell-based autotherapy. Complementary to conventional strategies that include the overexpression of reprogramming factors, chemical manipulation not only represents a powerful tool for controlling stem cell fate, or facilitating reprogramming or transdifferentiation, but also provides the means for dissecting the underlying mechanisms. There is no doubt that chemical approaches and the ongoing discovery of new small molecules will continue to have essential roles in the study and control of stem cell fate, state and function towards the ultimate development of safe regenerative-medicine-based treatments for various injuries and diseases.
Supplementary Material
Acknowledgments
S.D. is supported by funding from National Institute of Child Health and Human Development, National Heart, Lung, and Blood Institute, National Eye Institute, and National Institute of Mental Health/National Institute of Health [grant numbers HD064610, HL107436, EY021374], California Institute for Regenerative Medicine, Prostate Cancer Foundation, and the Gladstone Institute. We thank Gary Howard, and Anna Lisa Lucido for editing of this manuscript, and Saiyong Zhu, Jianghwan Kim, Tianhua Ma, Peng Liu, Baoming Nie and other members of the Ding lab for helpful discussions. The authors apologize to all scientists whose research could not be discussed and/or cited in this Commentary to space limitations. Deposited in PMC for release after 12 month.
References
- Agarwal S. (2006). Cellular Reprogramming. Methods in Enzymology, Vol. 420 (ed. Irina K, Robert L.), pp. 265–283 New York, NY: Academic Press; [DOI] [PubMed] [Google Scholar]
- Ambasudhan R., Talantova M., Coleman R., Yuan X., Zhu S., Lipton S. A., Ding S. (2011). Direct reprogramming of adult human fibroblasts to functional neurons under defined conditions. Cell Stem Cell 9, 113–118 10.1016/j.stem.2011.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anokye–Danso F., Trivedi C. M., Juhr D., Gupta M., Cui Z., Tian Y., Zhang Y., Yang W., Gruber P. J., Epstein J. A.et al. (2011). Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell 8, 376–388 10.1016/j.stem.2011.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boitano A. E., Wang J., Romeo R., Bouchez L. C., Parker A. E., Sutton S. E., Walker J. R., Flaveny C. A., Perdew G. H., Denison M. S.et al. (2010). Aryl hydrocarbon receptor antagonists promote the expansion of human hematopoietic stem cells. Science 329, 1345–1348 10.1126/science.1191536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brons I. G., Smithers L. E., Trotter M. W., Rugg–Gunn P., Sun B., Chuva de Sousa Lopes S. M., Howlett S. K., Clarkson A., Ahrlund–Richter L., Pedersen R. A.et al. (2007). Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature 448, 191–195 10.1038/nature05950 [DOI] [PubMed] [Google Scholar]
- Buecker C., Chen H. H., Polo J. M., Daheron L., Bu L., Barakat T. S., Okwieka P., Porter A., Gribnau J., Hochedlinger K.et al. (2010). A murine ESC-like state facilitates transgenesis and homologous recombination in human pluripotent stem cells. Cell Stem Cell 6, 535–546 10.1016/j.stem.2010.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buehr M., Meek S., Blair K., Yang J., Ure J., Silva J., McLay R., Hall J., Ying Q. L., Smith A. (2008). Capture of authentic embryonic stem cells from rat blastocysts. Cell 135, 1287–1298 10.1016/j.cell.2008.12.007 [DOI] [PubMed] [Google Scholar]
- Caiazzo M., Dell'Anno M. T., Dvoretskova E., Lazarevic D., Taverna S., Leo D., Sotnikova T. D., Menegon A., Roncaglia P., Colciago G.et al. (2011). Direct generation of functional dopaminergic neurons from mouse and human fibroblasts. Nature 476, 224–227 10.1038/nature10284 [DOI] [PubMed] [Google Scholar]
- Campbell K. H. S., McWhir J., Ritchie W. A., Wilmut I. (1996). Sheep cloned by nuclear transfer from a cultured cell line. Nature 380, 64–66 10.1038/380064a0 [DOI] [PubMed] [Google Scholar]
- Chambers I., Smith A. (2004). Self-renewal of teratocarcinoma and embryonic stem cells. Oncogene 23, 7150–7160 10.1038/sj.onc.1207930 [DOI] [PubMed] [Google Scholar]
- Chambers S. M., Fasano C. A., Papapetrou E. P., Tomishima M., Sadelain M., Studer L. (2009). Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 27, 275–280 10.1038/nbt.1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Zhang Q., Wu X., Schultz P. G., Ding S. (2004). Dedifferentiation of lineage-committed cells by a small molecule. J. Am. Chem. Soc. 126, 410–411 10.1021/ja037390k [DOI] [PubMed] [Google Scholar]
- Chen S., Do J. T., Zhang Q., Yao S., Yan F., Peters E. C., Schöler H. R., Schultz P. G., Ding S. (2006). Self-renewal of embryonic stem cells by a small molecule. Proc. Natl. Acad. Sci. USA 103, 17266–17271 10.1073/pnas.0608156103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G., Hou Z., Gulbranson D. R., Thomson J. A. (2010). Actin-myosin contractility is responsible for the reduced viability of dissociated human embryonic stem cells. Cell Stem Cell 7, 240–248 10.1016/j.stem.2010.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan C. A., Atienza J., Melton D. A., Eggan K. (2005). Nuclear reprogramming of somatic cells after fusion with human embryonic stem cells. Science 309, 1369–1373 10.1126/science.1116447 [DOI] [PubMed] [Google Scholar]
- Davis R. L., Weintraub H., Lassar A. B. (1987). Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell 51, 987–1000 [DOI] [PubMed] [Google Scholar]
- Desbordes S. C., Placantonakis D. G., Ciro A., Socci N. D., Lee G., Djaballah H., Studer L. (2008). High-throughput screening assay for the identification of compounds regulating self-renewal and differentiation in human embryonic stem cells. Cell Stem Cell 2, 602–612 10.1016/j.stem.2008.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding S., Schultz P. G. (2004). A role for chemistry in stem cell biology. Nat. Biotechnol. 22, 833–840 10.1038/nbt987 [DOI] [PubMed] [Google Scholar]
- Efe J. A., Ding S. (2011). The evolving biology of small molecules: controlling cell fate and identity. Philos. Trans. R. Soc. Lond. B Biol. Sci. 366, 2208–2221 10.1098/rstb.2011.0006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efe J. A., Hilcove S., Kim J., Zhou H., Ouyang K., Wang G., Chen J., Ding S. (2011). Conversion of mouse fibroblasts into cardiomyocytes using a direct reprogramming strategy. Nat. Cell Biol. 13, 215–222 10.1038/ncb2164 [DOI] [PubMed] [Google Scholar]
- Evans M. J., Kaufman M. H. (1981). Establishment in culture of pluripotential cells from mouse embryos. Nature 292, 154–156 10.1038/292154a0 [DOI] [PubMed] [Google Scholar]
- González F., Boué S., Izpisúa Belmonte J. C. (2011). Methods for making induced pluripotent stem cells: reprogramming à la carte. Nat. Rev. Genet. 12, 231–242 10.1038/nrg2937 [DOI] [PubMed] [Google Scholar]
- Greber B., Wu G., Bernemann C., Joo J. Y., Han D. W., Ko K., Tapia N., Sabour D., Sterneckert J., Tesar P.et al. (2010). Conserved and divergent roles of FGF signaling in mouse epiblast stem cells and human embryonic stem cells. Cell Stem Cell 6, 215–226 10.1016/j.stem.2010.01.003 [DOI] [PubMed] [Google Scholar]
- Guo G., Smith A. (2010). A genome-wide screen in EpiSCs identifies Nr5a nuclear receptors as potent inducers of ground state pluripotency. Development 137, 3185–3192 10.1242/dev.052753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G., Yang J., Nichols J., Hall J. S., Eyres I., Mansfield W., Smith A. (2009). Klf4 reverts developmentally programmed restriction of ground state pluripotency. Development 136, 1063–1069 10.1242/dev.030957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han D. W., Tapia N., Hermann A., Hemmer K., Höing S., Araúzo–Bravo M. J., Zaehres H., Wu G., Frank S., Moritz S.et al. (2012). Direct reprogramming of fibroblasts into neural stem cells by defined factors. Cell Stem Cell 10, 465–472 10.1016/j.stem.2012.02.021 [DOI] [PubMed] [Google Scholar]
- Hanna J., Markoulaki S., Mitalipova M., Cheng A. W., Cassady J. P., Staerk J., Carey B. W., Lengner C. J., Foreman R., Love J.et al. (2009). Metastable pluripotent states in NOD-mouse-derived ESCs. Cell Stem Cell 4, 513–524 10.1016/j.stem.2009.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J., Cheng A. W., Saha K., Kim J., Lengner C. J., Soldner F., Cassady J. P., Muffat J., Carey B. W., Jaenisch R. (2010). Human embryonic stem cells with biological and epigenetic characteristics similar to those of mouse ESCs. Proc. Natl. Acad. Sci. USA 107, 9222–9227 10.1073/pnas.1004584107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa K., Zhang P., Wei Z., Pomeroy J. E., Lu W., Pera M. F. (2010). Comparison of reprogramming efficiency between transduction of reprogramming factors, cell-cell fusion, and cytoplast fusion. Stem Cells 28, 1338–1348 10.1002/stem.466 [DOI] [PubMed] [Google Scholar]
- Huang P., He Z., Ji S., Sun H., Xiang D., Liu C., Hu Y., Wang X., Hui L. (2011). Induction of functional hepatocyte-like cells from mouse fibroblasts by defined factors. Nature 475, 386–389 10.1038/nature10116 [DOI] [PubMed] [Google Scholar]
- Huangfu D., Maehr R., Guo W., Eijkelenboom A., Snitow M., Chen A. E., Melton D. A. (2008a). Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat. Biotechnol. 26, 795–797 10.1038/nbt1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu D., Osafune K., Maehr R., Guo W., Eijkelenboom A., Chen S., Muhlestein W., Melton D. A. (2008b). Induction of pluripotent stem cells from primary human fibroblasts with only Oct4 and Sox2. Nat. Biotechnol. 26, 1269–1275 10.1038/nbt.1502 [DOI] [PubMed] [Google Scholar]
- Ichida J. K., Blanchard J., Lam K., Son E. Y., Chung J. E., Egli D., Loh K. M., Carter A. C., Di Giorgio F. P., Koszka K.et al. (2009). A small-molecule inhibitor of tgf-Beta signaling replaces sox2 in reprogramming by inducing nanog. Cell Stem Cell 5, 491–503 10.1016/j.stem.2009.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ieda M., Fu J. D., Delgado–Olguin P., Vedantham V., Hayashi Y., Bruneau B. G., Srivastava D. (2010). Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell 142, 375–386 10.1016/j.cell.2010.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumikawa M., Minoda R., Kawamoto K., Abrashkin K. A., Swiderski D. L., Dolan D. F., Brough D. E., Raphael Y. (2005). Auditory hair cell replacement and hearing improvement by Atoh1 gene therapy in deaf mammals. Nat. Med. 11, 271–276 10.1038/nm1193 [DOI] [PubMed] [Google Scholar]
- James D., Levine A. J., Besser D., Hemmati–Brivanlou A. (2005). TGFbeta/activin/nodal signaling is necessary for the maintenance of pluripotency in human embryonic stem cells. Development 132, 1273–1282 10.1242/dev.01706 [DOI] [PubMed] [Google Scholar]
- Jones P. A., Taylor S. M. (1980). Cellular differentiation, cytidine analogs and DNA methylation. Cell 20, 85–93 10.1016/0092-8674(80)90237-8 [DOI] [PubMed] [Google Scholar]
- Kim D., Kim C. H., Moon J. I., Chung Y. G., Chang M. Y., Han B. S., Ko S., Yang E., Cha K. Y., Lanza R.et al. (2009). Generation of human induced pluripotent stem cells by direct delivery of reprogramming proteins. Cell Stem Cell 4, 472–476 10.1016/j.stem.2009.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K., Doi A., Wen B., Ng K., Zhao R., Cahan P., Kim J., Aryee M. J., Ji H., Ehrlich L. I. R.et al. (2010). Epigenetic memory in induced pluripotent stem cells. Nature 467, 285–290 10.1038/nature09342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Efe J. A., Zhu S., Talantova M., Yuan X., Wang S., Lipton S. A., Zhang K., Ding S. (2011a). Direct reprogramming of mouse fibroblasts to neural progenitors. Proc. Natl. Acad. Sci. USA 108, 7838–7843 10.1073/pnas.1103113108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Su S. C., Wang H., Cheng A. W., Cassady J. P., Lodato M. A., Lengner C. J., Chung C. Y., Dawlaty M. M., Tsai L. H.et al. (2011b). Functional integration of dopaminergic neurons directly converted from mouse fibroblasts. Cell Stem Cell 9, 413–419 10.1016/j.stem.2011.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladewig J., Mertens J., Kesavan J., Doerr J., Poppe D., Glaue F., Herms S., Wernet P., Kögler G., Müller F. J.et al. (2012). Small molecules enable highly efficient neuronal conversion of human fibroblasts. Nat. Methods 9, 575–578 10.1038/nmeth.1972 [DOI] [PubMed] [Google Scholar]
- Li W., Ding S. (2011). Human pluripotent stem cells: Decoding the naïve state. Science translational medicine 3, 76ps10–76ps10 [DOI] [PubMed] [Google Scholar]
- Li P., Tong C., Mehrian–Shai R., Jia L., Wu N., Yan Y., Maxson R. E., Schulze E. N., Song H., Hsieh C. L.et al. (2008). Germline competent embryonic stem cells derived from rat blastocysts. Cell 135, 1299–1310 10.1016/j.cell.2008.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Wei W., Zhu S., Zhu J., Shi Y., Lin T., Hao E., Hayek A., Deng H., Ding S. (2009a). Generation of rat and human induced pluripotent stem cells by combining genetic reprogramming and chemical inhibitors. Cell Stem Cell 4, 16–19 10.1016/j.stem.2008.11.014 [DOI] [PubMed] [Google Scholar]
- Li W., Zhou H., Abujarour R., Zhu S., Young Joo J., Lin T., Hao E., Schöler H. R., Hayek A., Ding S. (2009b). Generation of human-induced pluripotent stem cells in the absence of exogenous Sox2. Stem Cells 27, 2992–3000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Sun W., Zhang Y., Wei W., Ambasudhan R., Xia P., Talantova M., Lin T., Kim J., Wang X.et al. (2011). Rapid induction and long-term self-renewal of primitive neural precursors from human embryonic stem cells by small molecule inhibitors. Proc. Natl. Acad. Sci. USA 108, 8299–8304 10.1073/pnas.1014041108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T., Ambasudhan R., Yuan X., Li W., Hilcove S., Abujarour R., Lin X., Hahm H. S., Hao E., Hayek A.et al. (2009). A chemical platform for improved induction of human iPSCs. Nat. Methods 6, 805–808 10.1038/nmeth.1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lujan E., Chanda S., Ahlenius H., Südhof T. C., Wernig M. (2012). Direct conversion of mouse fibroblasts to self-renewing, tripotent neural precursor cells. Proc. Natl. Acad. Sci. USA 109, 2527–2532 10.1073/pnas.1121003109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyssiotis C. A., Foreman R. K., Staerk J., Garcia M., Mathur D., Markoulaki S., Hanna J., Lairson L. L., Charette B. D., Bouchez L. C.et al. (2009). Reprogramming of murine fibroblasts to induced pluripotent stem cells with chemical complementation of Klf4. Proc. Natl. Acad. Sci. USA 106, 8912–8917 10.1073/pnas.0903860106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyssiotis C. A., Lairson L. L., Boitano A. E., Wurdak H., Zhu S., Schultz P. G. (2011). Chemical control of stem cell fate and developmental potential. Angew. Chem. Int. Ed. Engl. 50, 200–242 10.1002/anie.201004284 [DOI] [PubMed] [Google Scholar]
- Maherali N., Hochedlinger K. (2009). Tgfbeta signal inhibition cooperates in the induction of iPSCs and replaces Sox2 and cMyc. Curr. Biol. 19, 1718–1723 10.1016/j.cub.2009.08.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P., Chou B. K., Yen J., Ye Z., Zou J., Dowey S., Brodsky R. A., Ohm J. E., Yu W., Baylin S. B.et al. (2010). Butyrate greatly enhances derivation of human induced pluripotent stem cells by promoting epigenetic remodeling and the expression of pluripotency-associated genes. Stem Cells 28, 713–720 10.1002/stem.402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marro S., Pang Z. P., Yang N., Tsai M. C., Qu K., Chang H. Y., Südhof T. C., Wernig M. (2011). Direct lineage conversion of terminally differentiated hepatocytes to functional neurons. Cell Stem Cell 9, 374–382 10.1016/j.stem.2011.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G. R. (1981). Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc. Natl. Acad. Sci. USA 78, 7634–7638 10.1073/pnas.78.12.7634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen T. S., Hanna J., Zhang X., Ku M., Wernig M., Schorderet P., Bernstein B. E., Jaenisch R., Lander E. S., Meissner A. (2008). Dissecting direct reprogramming through integrative genomic analysis. Nature 454, 49–55 10.1038/nature07056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi N., Ishii H., Nagano H., Haraguchi N., Dewi D. L., Kano Y., Nishikawa S., Tanemura M., Mimori K., Tanaka F.et al. (2011). Reprogramming of mouse and human cells to pluripotency using mature microRNAs. Cell Stem Cell 8, 633–638 10.1016/j.stem.2011.05.001 [DOI] [PubMed] [Google Scholar]
- Nichols J., Smith A. (2009). Naive and primed pluripotent states. Cell Stem Cell 4, 487–492 10.1016/j.stem.2009.05.015 [DOI] [PubMed] [Google Scholar]
- Niwa H. (2011). Wnt: what's needed to maintain pluripotency? Nat. Cell Biol. 13, 1024–1026 10.1038/ncb2333 [DOI] [PubMed] [Google Scholar]
- Niwa H., Burdon T., Chambers I., Smith A. (1998). Self-renewal of pluripotent embryonic stem cells is mediated via activation of STAT3. Genes Dev. 12, 2048–2060 10.1101/gad.12.13.2048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohgushi M., Matsumura M., Eiraku M., Murakami K., Aramaki T., Nishiyama A., Muguruma K., Nakano T., Suga H., Ueno M.et al. (2010). Molecular pathway and cell state responsible for dissociation-induced apoptosis in human pluripotent stem cells. Cell Stem Cell 7, 225–239 10.1016/j.stem.2010.06.018 [DOI] [PubMed] [Google Scholar]
- Ohi Y., Qin H., Hong C., Blouin L., Polo J. M., Guo T., Qi Z., Downey S. L., Manos P. D., Rossi D. J.et al. (2011). Incomplete DNA methylation underlies a transcriptional memory of somatic cells in human iPS cells. Nat. Cell Biol. 13, 541–549 10.1038/ncb2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang Z. P., Yang N., Vierbuchen T., Ostermeier A., Fuentes D. R., Yang T. Q., Citri A., Sebastiano V., Marro S., Südhof T. C.et al. (2011). Induction of human neuronal cells by defined transcription factors. Nature 476, 220–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel M., Yang S. (2010). Advances in reprogramming somatic cells to induced pluripotent stem cells. Stem Cell Rev. 6, 367–380 10.1007/s12015-010-9123-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfisterer U., Kirkeby A., Torper O., Wood J., Nelander J., Dufour A., Björklund A., Lindvall O., Jakobsson J., Parmar M. (2011). Direct conversion of human fibroblasts to dopaminergic neurons. Proc. Natl. Acad. Sci. USA 108, 10343–10348 10.1073/pnas.1105135108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian L., Huang Y., Spencer C. I., Foley A., Vedantham V., Liu L., Conway S. J., Fu J. D., Srivastava D. (2012). In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature 485, 593–598 10.1038/nature11044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qyang Y., Martin–Puig S., Chiravuri M., Chen S., Xu H., Bu L., Jiang X., Lin L., Granger A., Moretti A.et al. (2007). The renewal and differentiation of Isl1+ cardiovascular progenitors are controlled by a Wnt/beta-catenin pathway. Cell Stem Cell 1, 165–179 10.1016/j.stem.2007.05.018 [DOI] [PubMed] [Google Scholar]
- Sato N., Meijer L., Skaltsounis L., Greengard P., Brivanlou A. H. (2004). Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nat. Med. 10, 55–63 10.1038/nm979 [DOI] [PubMed] [Google Scholar]
- Sekiya S., Suzuki A. (2011). Direct conversion of mouse fibroblasts to hepatocyte-like cells by defined factors. Nature 475, 390–393 10.1038/nature10263 [DOI] [PubMed] [Google Scholar]
- Shi Y., Desponts C., Do J. T., Hahm H. S., Schöler H. R., Ding S. (2008a). Induction of pluripotent stem cells from mouse embryonic fibroblasts by Oct4 and Klf4 with small-molecule compounds. Cell Stem Cell 3, 568–574 10.1016/j.stem.2008.10.004 [DOI] [PubMed] [Google Scholar]
- Shi Y., Do J. T., Desponts C., Hahm H. S., Schöler H. R., Ding S. (2008b). A combined chemical and genetic approach for the generation of induced pluripotent stem cells. Cell Stem Cell 2, 525–528 10.1016/j.stem.2008.05.011 [DOI] [PubMed] [Google Scholar]
- Silva J., Nichols J., Theunissen T. W., Guo G., van Oosten A. L., Barrandon O., Wray J., Yamanaka S., Chambers I., Smith A. (2009). Nanog is the gateway to the pluripotent ground state. Cell 138, 722–737 10.1016/j.cell.2009.07.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldner F., Hockemeyer D., Beard C., Gao Q., Bell G. W., Cook E. G., Hargus G., Blak A., Cooper O., Mitalipova M.et al. (2009). Parkinson's disease patient-derived induced pluripotent stem cells free of viral reprogramming factors. Cell 136, 964–977 10.1016/j.cell.2009.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son E. Y., Ichida J. K., Wainger B. J., Toma J. S., Rafuse V. F., Woolf C. J., Eggan K. (2011). Conversion of mouse and human fibroblasts into functional spinal motor neurons. Cell Stem Cell 9, 205–218 10.1016/j.stem.2011.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo E., Rampalli S., Risueño R. M., Schnerch A., Mitchell R., Fiebig–Comyn A., Levadoux–Martin M., Bhatia M. (2010). Direct conversion of human fibroblasts to multilineage blood progenitors. Nature 468, 521–526 10.1038/nature09591 [DOI] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S. (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 10.1016/j.cell.2006.07.024 [DOI] [PubMed] [Google Scholar]
- Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. (2007). Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861–872 10.1016/j.cell.2007.11.019 [DOI] [PubMed] [Google Scholar]
- Taylor S. M., Jones P. A. (1979). Multiple new phenotypes induced in 10T1/2 and 3T3 cells treated with 5-azacytidine. Cell 17, 771–779 10.1016/0092-8674(79)90317-9 [DOI] [PubMed] [Google Scholar]
- Tesar P. J., Chenoweth J. G., Brook F. A., Davies T. J., Evans E. P., Mack D. L., Gardner R. L., McKay R. D. (2007). New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature 448, 196–199 10.1038/nature05972 [DOI] [PubMed] [Google Scholar]
- Thier M., Wörsdörfer P., Lakes Y. B., Gorris R., Herms S., Opitz T., Seiferling D., Quandel T., Hoffmann P., Nöthen M. M.et al. (2012). Direct conversion of fibroblasts into stably expandable neural stem cells. Cell Stem Cell 10, 473–479 10.1016/j.stem.2012.03.003 [DOI] [PubMed] [Google Scholar]
- Thomson J. A., Itskovitz–Eldor J., Shapiro S. S., Waknitz M. A., Swiergiel J. J., Marshall V. S., Jones J. M. (1998). Embryonic stem cell lines derived from human blastocysts. Science 282, 1145–1147 10.1126/science.282.5391.1145 [DOI] [PubMed] [Google Scholar]
- Tsutsui H., Valamehr B., Hindoyan A., Qiao R., Ding X., Guo S., Witte O. N., Liu X., Ho C. M., Wu H. (2011). An optimized small molecule inhibitor cocktail supports long-term maintenance of human embryonic stem cells. Nat. Commun 2, 167 10.1038/ncomms1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valamehr B., Tsutsui H., Ho C. M., Wu H. (2011). Developing defined culture systems for human pluripotent stem cells. Regen. Med. 6, 623–634 10.2217/rme.11.54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallier L., Alexander M., Pedersen R. A. (2005). Activin/Nodal and FGF pathways cooperate to maintain pluripotency of human embryonic stem cells. J. Cell Sci. 118, 4495–4509 10.1242/jcs.02553 [DOI] [PubMed] [Google Scholar]
- Vierbuchen T., Ostermeier A., Pang Z. P., Kokubu Y., Südhof T. C., Wernig M. (2010). Direct conversion of fibroblasts to functional neurons by defined factors. Nature 463, 1035–1041 10.1038/nature08797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Yang J., Liu H., Lu D., Chen X., Zenonos Z., Campos L. S., Rad R., Guo G., Zhang S.et al. (2011). Rapid and efficient reprogramming of somatic cells to induced pluripotent stem cells by retinoic acid receptor gamma and liver receptor homolog 1. Proc. Natl. Acad. Sci. USA 108, 18283–18288 10.1073/pnas.1100893108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren L., Manos P. D., Ahfeldt T., Loh Y. H., Li H., Lau F., Ebina W., Mandal P. K., Smith Z. D., Meissner A.et al. (2010). Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell 7, 618–630 10.1016/j.stem.2010.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K., Ueno M., Kamiya D., Nishiyama A., Matsumura M., Wataya T., Takahashi J. B., Nishikawa S., Nishikawa S., Muguruma K.et al. (2007). A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nat. Biotechnol. 25, 681–686 10.1038/nbt1310 [DOI] [PubMed] [Google Scholar]
- Xie H., Ye M., Feng R., Graf T. (2004a). Stepwise reprogramming of B cells into macrophages. Cell 117, 663–676 10.1016/S0092-8674(04)00419-2 [DOI] [PubMed] [Google Scholar]
- Xie L., Law B. K., Chytil A. M., Brown K. A., Aakre M. E., Moses H. L. (2004b). Activation of the Erk pathway is required for TGF-β1-induced EMT in vitro. Neoplasia 6, 603–610 10.1593/neo.04241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Shi Y., Ding S. (2008). A chemical approach to stem-cell biology and regenerative medicine. Nature 453, 338–344 10.1038/nature07042 [DOI] [PubMed] [Google Scholar]
- Xu J., Lamouille S., Derynck R. (2009). TGF-beta-induced epithelial to mesenchymal transition. Cell Res. 19, 156–172 10.1038/cr.2009.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Zhu X., Hahm H. S., Wei W., Hao E., Hayek A., Ding S. (2010). Revealing a core signaling regulatory mechanism for pluripotent stem cell survival and self-renewal by small molecules. Proc. Natl. Acad. Sci. USA 107, 8129–8134 10.1073/pnas.1002024107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakubov E., Rechavi G., Rozenblatt S., Givol D. (2010). Reprogramming of human fibroblasts to pluripotent stem cells using mRNA of four transcription factors. Biochem. Biophys. Res. Commun. 394, 189–193 10.1016/j.bbrc.2010.02.150 [DOI] [PubMed] [Google Scholar]
- Yang W., Wei W., Shi C., Zhu J., Ying W., Shen Y., Ye X., Fang L., Duo S., Che J.et al. (2009). Pluripotin combined with leukemia inhibitory factor greatly promotes the derivation of embryonic stem cell lines from refractory strains. Stem Cells 27, 383–389 10.1634/stemcells.2008-0974 [DOI] [PubMed] [Google Scholar]
- Yau W. W., Tang M. K., Chen E., Yaoyao, Wong I. W., Lee H. S., Lee K. K. (2011). Cardiogenol C can induce Mouse Hair Bulge Progenitor Cells to Transdifferentiate into Cardiomyocyte-like Cells. Proteome Sci. 9, 3 10.1186/1477-5956-9-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying Q. L., Nichols J., Evans E. P., Smith A. G. (2002). Changing potency by spontaneous fusion. Nature 416, 545–548 10.1038/nature729 [DOI] [PubMed] [Google Scholar]
- Ying Q. L., Nichols J., Chambers I., Smith A. (2003). BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell 115, 281–292 10.1016/S0092-8674(03)00847-X [DOI] [PubMed] [Google Scholar]
- Ying Q. L., Wray J., Nichols J., Batlle–Morera L., Doble B., Woodgett J., Cohen P., Smith A. (2008). The ground state of embryonic stem cell self-renewal. Nature 453, 519–523 10.1038/nature06968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo A. S., Sun A. X., Li L., Shcheglovitov A., Portmann T., Li Y., Lee–Messer C., Dolmetsch R. E., Tsien R. W., Crabtree G. R. (2011). MicroRNA-mediated conversion of human fibroblasts to neurons. Nature 476, 228–231 10.1038/nature10323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Vodyanik M. A., Smuga–Otto K., Antosiewicz–Bourget J., Frane J. L., Tian S., Nie J., Jonsdottir G. A., Ruotti V., Stewart R.et al. (2007). Induced pluripotent stem cell lines derived from human somatic cells. Science 318, 1917–1920 10.1126/science.1151526 [DOI] [PubMed] [Google Scholar]
- Yu J., Hu K., Smuga–Otto K., Tian S., Stewart R., Slukvin I. I., Thomson J. A. (2009). Human induced pluripotent stem cells free of vector and transgene sequences. Science 324, 797–801 10.1126/science.1172482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q., Brown J., Kanarek A., Rajagopal J., Melton D. A. (2008). In vivo reprogramming of adult pancreatic exocrine cells to beta-cells. Nature 455, 627–632 10.1038/nature07314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Wu S., Joo J. Y., Zhu S., Han D. W., Lin T., Trauger S., Bien G., Yao S., Zhu Y.et al. (2009). Generation of induced pluripotent stem cells using recombinant proteins. Cell Stem Cell 4, 381–384 10.1016/j.stem.2009.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Li W., Zhu S., Joo J. Y., Do J. T., Xiong W., Kim J. B., Zhang K., Schöler H. R., Ding S. (2010). Conversion of mouse epiblast stem cells to an earlier pluripotency state by small molecules. J. Biol. Chem. 285, 29676–29680 10.1074/jbc.C110.150599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S., Li W., Zhou H., Wei W., Ambasudhan R., Lin T., Kim J., Zhang K., Ding S. (2010). Reprogramming of human primary somatic cells by OCT4 and chemical compounds. Cell Stem Cell 7, 651–655 10.1016/j.stem.2010.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


