Abstract
The Lhx2 transcription factor plays essential roles in morphogenesis and patterning of ectodermal derivatives as well as in controlling stem cell activity. Here, we show that during murine skin morphogenesis, Lhx2 is expressed in the hair follicle (HF) buds, whereas in postnatal telogen HFs Lhx2+ cells reside in the stem cell-enriched epithelial compartments (bulge, secondary hair germ) and co-express selected stem cell markers (Sox9, Tcf4 and Lgr5). Remarkably, Lhx2+ cells represent the vast majority of cells in the bulge and secondary hair germ that proliferate in response to skin injury. This is functionally important, as wound re-epithelization is significantly retarded in heterozygous Lhx2 knockout (+/–) mice, whereas anagen onset in the HFs located closely to the wound is accelerated compared with wild-type mice. Cell proliferation in the bulge and the number of Sox9+ and Tcf4+ cells in the HFs closely adjacent to the wound in Lhx2+/– mice are decreased in comparison with wild-type controls, whereas expression of Lgr5 and cell proliferation in the secondary hair germ are increased. Furthermore, acceleration of wound-induced anagen development in Lhx2+/– mice is inhibited by administration of Lgr5 siRNA. Finally, Chip-on-chip/ChIP-qPCR and reporter assay analyses identified Sox9, Tcf4 and Lgr5 as direct Lhx2 targets in keratinocytes. These data strongly suggest that Lhx2 positively regulates Sox9 and Tcf4 in the bulge cells, and promotes wound re-epithelization, whereas it simultaneously negatively regulates Lgr5 in the secondary hair germ and inhibits HF cycling. Thus, Lhx2 operates as an important regulator of epithelial stem cell activity in the skin response to injury.
Keywords: Skin, Stem cells, Wound healing
INTRODUCTION
Skin development is governed by interactions between the epithelium and mesenchyme, which result in formation of the epidermis and a number of skin appendages, including hair follicles (HFs) (Driskell et al., 2011; Fuchs, 2007; Schmidt-Ullrich and Paus, 2005). During postnatal life, epidermis and HFs self-renew and regenerate with an involvement of stem cells that are capable of differentiating into distinct epithelial cell lineages (Ambler and Maatta, 2009; Blanpain and Fuchs, 2009; Cotsarelis, 2006; Watt and Jensen, 2009). Epithelial stem cells also contribute to epidermal regeneration after injury, a complex process that includes tightly regulated recruitment of undifferentiated progenitor cells to the wound epithelium (Gurtner et al., 2008; Lau et al., 2009; Schafer and Werner, 2007).
The HF is an important source of epithelial stem cells, which are capable of generating daughter cells that re-build epithelial hair bulb during the hair cycle or migrate to the wound epithelium and promote the regenerative process (Cotsarelis et al., 1990; Ghazizadeh and Taichman, 2001; Hsu et al., 2011; Ito et al., 2005; Ito et al., 2007; Kasper et al., 2011; Langton et al., 2008; Levy et al., 2005; Levy et al., 2007; Snippert et al., 2010; Taylor et al., 2000). HFs contain several populations of epithelial stem cells, each characterized by distinct, yet partially overlapping, expression patterns of surface receptors (Cd34, Cd71, Cd200, Lgr5, Lgr6, Lrig1, Sca-1), cytoskeletal proteins (keratins 15 and 19), signalling/transcriptional regulators (Lhx2, Sox9, Tcf3/4, Nfatc1, Gli-1) and epigenetic markers (Blanpain et al., 2004; Brownell et al., 2011; Frye et al., 2007; Garza et al., 2011; Horsley et al., 2008; Jaks et al., 2008; Jensen et al., 2009; Jensen et al., 2008; Morris et al., 2004; Nguyen et al., 2009; Nowak et al., 2008; Rhee et al., 2006; Snippert et al., 2010; Trempus et al., 2007; Tumbar et al., 2003).
In normal skin, distinct populations of epithelial stem cells residing in the bulge, secondary hair germ or infundibulum of the HFs differentially contribute to the renewal of the epidermis versus the regeneration of the HF during the hair cycle (Ito et al., 2005; Jaks et al., 2008; Jensen et al., 2009; Li and Clevers, 2010). However, wounding perturbs homeostasis, resulting in activation of those populations of epithelial stem cells that normally supply progenies only into the HF during hair cycling (Ito et al., 2005; Kasper et al., 2011; Levy et al., 2007). Although HFs are not absolutely required for skin regeneration after injury, re-epithelization in hair-bearing skin occurs faster and more efficiently when compared with non-hair-bearing areas (Ansell et al., 2011; Ito and Cotsarelis, 2008; Langton et al., 2008; Romagosa et al., 2008).
The Lim-homeodomain transcription factor Lhx2 is an important regulator controlling the switch between stem cell maintenance and activation in the HFs (Rhee et al., 2006; Tiede and Paus, 2006; Tornqvist et al., 2010). Lhx2 is expressed in the bulge and hair germ of mouse telogen (resting) HFs, and Lhx2 deficiency in mice leads to incapability in maintaining a quiescent state in the bulge stem cells (Rhee et al., 2006). In human HFs, Lhx2 shows more broad expression patterns and, in addition to the bulge cells, is seen in the isthmus, the infundibulum and the lower outer root sheath (Kloepper et al., 2008). In other organs (eye, pituitary gland, limb, brain, hematopoietic system), Lhx2 operates as a central link in the genetic networks that coordinate multiple signalling pathways controlling organ development and cell fate determination, as well as stem cell maintenance, differentiation and self-renewal (Chou et al., 2009; Dahl et al., 2008; Hirota and Mombaerts, 2004; Porter et al., 1997; Tetreault et al., 2009; Yun et al., 2009).
Despite the advantages in identification of distinct stem cell populations in the skin achieved during past few years (Blanpain and Fuchs, 2009; Watt and Jensen, 2009), regulatory mechanisms involved in controlling their activity and expression of stem cell markers (membrane receptors, cytoskeleton components, transcription factors) during their transition towards the epidermal or HF cell lineages in normal and injured skin are still unclear. In addition, the mechanisms underlying differential involvement of these stem cell populations in the control of distinct types of the regenerative processes in the epithelial tissues (epidermal regeneration after injury, physiological or wound-induced hair follicle cycling) remain to be elucidated.
Here, we show that Lhx2 plays an important role as a switchboard regulator of the activity of distinct epithelial stem cell populations in the HF during wound healing. In particular, we demonstrate that Lhx2 promotes wound re-epithelization via positive regulation of Sox9, and Tcf4, while simultaneously inhibiting HF cycling via negative regulation of Lgr5. Thus, Lhx2 serves as a key factor integrating signalling and transcriptional networks that modulate activity of the HF stem cells during epidermal regeneration after injury.
MATERIALS AND METHODS
Animal experiments
All animal experiments were performed under the Home Office Project License (University of Bradford, UK). Lhx2+/– mice (Porter et al., 1997) were crossbred to obtain homozygous Lhx2–/– and wild-type embryos, which were collected on embryonic day 16.5 (E16.5), as described before (Botchkarev et al., 1999a). A full-thickness 3 mm wound was introduced by punch biopsy onto back skin of 8-week-old Lhx2+/– and wild-type mice at the telogen stage of the hair cycle (8-10 female mice of each strain per time point, one wound per mouse). Lgr5 siRNA or control siRNA (Dharmacon, Chicago, IL) were injected into the wounds of Lhx2+/– mice at a concentration of 20 μM as described previously (Mardaryev et al., 2010). siRNA treatment was performed on days 1-6 after wound infliction, and skin samples were collected on day 7 of the experiment. In each experiment, at least 4-5 mice per time point were used for analyses in both experimental and control groups. Samples of the skin closely (within 5 mm) and distantly located to the wound edge were collected at days 0, 1, 3 and 5 after wounding and were snap-frozen in liquid nitrogen.
Human skin biopsies and wound healing organ culture assay
Biopsies of human skin with chronic wounds after diagnostic skin surgery and scalp skin of patients undergoing elective cosmetic surgery were obtained after informed consent and Ethics Committee Approval according to the Helsinki Ethical Guidelines for medical research involving human subjects. For skin organ culture, 2 mm punches were cut into full-thickness skin samples from patients. Then, 4 mm punches were set in the skin surrounding each 2 mm hole to obtain ‘punch within a punch’ skin samples (Moll et al., 1998). Samples were frozen immediately for analysis (day 0) or transferred to six-well plates containing Williams E culture medium supplemented with insulin and hydrocortisone (Philpott et al., 1990; Lu et al., 2007). Each well contained two skin punches in 3 ml medium. After 48 hours the incubation medium was changed, and skin punches from each experimental condition were frozen at day 3. All experiments were performed in triplicates.
Microarray and qRT-PCR analyses
Total RNA was isolated from snap-frozen tissue samples using miRNeasy kit (Qiagen, Hilden, Germany). For microarray analysis, 0.5 μg of total RNA was processed for one-round amplification using RiboAmp RNA Amplification Kit (Molecular Devices, Sunnyvale, CA). mRNA microarray analysis was performed by Mogene (St Louis, MO) using 41K Whole Mouse Genome 60-mer oligo-microarray (manufactured by Agilent Technologies). For qRT-PCR, 1 μg of total RNA was converted into cDNA using Reverse Transcription System (Promega, Madison, WI). Gene expression was performed on a MyiQ single-colour real-time PCR detection system (Bio-Rad, Hercules, CA) using PerfeCta SYBR Green FastMix for iQ (Quanta BioSciences, Gaithersburg, MD), as described previously (Sharov et al., 2003). PCR primers were designed using the Beacon Designer software (Premier Biosoft International, Palo Alto, CA; supplementary material Table S2). Differences between samples were calculated using the Genex database software (Bio-Rad) based on the Ct (ΔΔCt) equitation method and normalized to the house-keeping gene Gapdh. Data from triplicates were pooled, mean±s.e.m. was calculated, and statistical analysis was performed using unpaired Student’s t-test. Microarray data have been deposited in the Gene Expression Omnibus database (Accession Numbers GSE32511 and GSE32514)
Chip-on-chip and Chip-qPCR
Mouse embryonic, neonatal and adult epidermal keratinocytes were isolated from E16.5 embryos, 2- to 3-day-old and 8-week-old FVB mice, respectively, as described previously (Sharov et al., 2006) with modifications. Single cell suspensions were prepared after overnight digestion in 0.25% trypsin (Invitrogen) and epidermal separation. Cells were dual cross-linked with 2 mM disuccinimidyl glutarate (DSG) for 45 minutes and then in 1% paraformaldehyde for 15 minutes at room temperature as described previously (Nowak et al., 2005). Fixed cells were lysed in 10 ml of Lysis Buffer 1 [50 mM HEPES (pH 7.5), 140 mM NaCl, 1 mM EDTA, 0.1% IGEPAL 630 (Sigma-Aldrich)], containing 0.05% Triton X100, 2.5 % glycerol and supplemented with 1× protease inhibitor cocktail (Roche, Rotkreuz, Switzerland) for 10 minutes on ice, followed by incubation in Buffer 2 [0.1 M Tris HCl (pH 8) and 200 mM NaCl with protease inhibitors] for 15 minutes at room temperature. Chromatin was sonicated at 30% of amplitude for 10 minutes on a Branson Sonifier 450 (Branson Ultrasonics, Danbury, CT) using a cup horn. The samples were centrifuged (2×14,000 g for 5 minutes each), and soluble chromatin was transferred to a fresh tube. Crosslinked DNA after sonication was precipitated with 5 μg of anti-Lhx2 antibody (C-20, sc-19344, Santa Cruz, Santa Cruz, CA) or non-immune goat IgG (Vector Laboratories, Burlingame, CA) overnight at 4°C. Chromatin/antibody complex was pulled down with Dynal Protein G magnetic beads (Invitrogen, Carlsbad, CA) and washed in the low- and high-salt buffers. After de-crosslinking (65°C for 4 hours) and Proteinase K treatment, chromatin was purified by phenol-chloroform extraction and ethanol precipitation. Precipitated DNA after one round of amplification (WGA2, Sigma) was processed with NimbleGen MM8 Deluxe Promoter HX1 array (Mogene, St Louis, MO), or was analyzed by qPCR using primers generated for predicted Lhx2 binding sites (supplementary material Table S2). ChiIP-qPCR data were pooled, mean±s.e.m. was calculated, and statistical analysis was performed using Student’s t-test.
Reporter assay
Lhx2 expressing plasmid (pLhx2) was generated by subcloning the mouse Lhx2 cDNA from an I.M.A.G.E. clone (6413339, Gene Service Ltd, UK) into the pCMV-SPORT6 vector (Invitrogen) at SalI and NotI sites. pSox9-luc construct (also known as pGL6.8-luc) bearing –6.8 kb promoter region of the mouse Sox9 gene was kindly provided by Dr P. Koopman (Kanai and Koopman, 1999). To generate a Tcf4 reporter construct, the Tcf4 promoter region (–3598/+462) was PCR amplified from mouse genomic DNA with Advantage HD DNA polymerase (Clontech) using the following primers (BglII and HindIII restriction sites are underlined): 5′-CCCGGGCTCGAGATCTTGACCACACCCCCACCACTT-3′ (F) and 5′-CCGGAATGCCAAGCTTGCATTTTTCACCCACCAGCAGC-3′ (R). The purified fragment was then cloned into pGl3-basic vector (Promega) at BglII and HindIII sites using In-Fusion HD Cloning System (Clontech, Mountain View, CA). For reporter assay, HaCaT cells were seeded into 96-well plates (104 cells per well) 1 day before transfection. Cells were co-transfected with pLhx2 or pCDN3 plasmids (120 ng/well) along with one of the reporter constructs (pSox9-Luc or pTcf4-Luc; 80 ng/well) using Lipofectamine 2000 (0.5 μl/well) (Invitrogen). After 48 hours, reporter activity was detected using the Dual-Glo Luciferase Assay System (Promega) on Infinite 200 PRO microplate reader (Tecan, Männedorf, Switzerland). Two independent assays were performed in triplicates and results were normalized to the pnull-Renilla construct activity. Data were pooled, mean±s.e.m. was calculated, and statistical analysis was performed using Student’s t-test.
Western blotting
Proteins were extracted from snap-frozen skin samples or cultured cells with lysis buffer as described previously (Sharov et al., 2005; Sharov et al., 2006). Protein (5 μg) was processed for western blot analysis as described previously, followed by incubation with primary antibodies against Lhx2 [goat, 1:100, Santa Cruz (sc19344)] or Lgr5 [rabbit, 1:100, Abcam (ab75732)] overnight at 4°C. Horseradish peroxidase-tagged IgG antibody was used as secondary antibody (1:5000; Thermo Scientific, Rockford, IL). Antibody binding was visualized using an enhanced chemiluminescence system (SuperSignal West Pico Kit, Thermo Scientific) followed by autoradiography with X-ray film (CL-Xposure Film, Thermo Scientific). Densitometric analysis was performed using Total Lab v1.10 software (Biogenetic, USA).
Histology, histomorphometry and immunohistochemistry
For histological analyses, 8 μm sections from the central parts of the wounds were stained with Hematoxylin-Eosin and photographed using an Eclipse 50i microscope equipped with a DS-C1 digital camera and ACT-2U image analysis software (Nikon, Tokyo, Japan). The length and area of the hyperproliferative wound epithelium were determined using the ImageJ software (http://rsbweb.nih.gov/ij/) as described previously (Kumin et al., 2007). Histomorphometry of the distinct hair cycle stages was performed on skin cryosections stained by alkaline phosphatase as described previously (Botchkarev et al., 1999b; Botchkareva et al., 2000).
For immunohistochemical analyses, cryosections were incubated with primary antisera against Lhx2, Sox9, Tcf4, Lgr5, Ki-67, Lef1, Krt16, Krt17, CD4, F4/80 and Ly-6G [CD4, rat, 1:100, Dako; F4/80, rat, 1:100, Dianova (T-2006); Ki-67, rabbit, 1:100, Abcam (ab15580); Krt14, rabbit, 1:200, Sigma (c-8791); Krt16, rabbit, 1:100, Abcam (ab76416); Krt17, rabbit, 1:5000, Abcam (ab53707); Lef1, rabbit, 1:100, Cell Signaling (C12A5); Lhx2, goat, 1:100, Santa Cruz (sc-19344); Ly-6G, rat, 1:150, BD Farmingen (551459); Sox9, rabbit, 1:100, Santa Cruz (sc20095); Tcf4, rabbit, 1:100, Cell Signaling (c48H11)] overnight at 4°C, followed by application of corresponding Alexa-488 and Cy-555-coupled secondary antibodies (Invitrogen; 1:200) for 60 minutes at 37°C, as described previously (Botchkareva et al., 2000). Image acquisition was performed on a Zeiss confocal microscope or a Nikon fluorescent microscope. Statistical analysis of the histomorphometric parameters and the number of Ki67+ cells was performed using an unpaired Student’s t-test or Mann-Whitney’s test after the data from triplicate experiments were pooled and mean±s.e.m. was calculated.
In vitro migration assay
Primary keratinocytes from Lhx2+/– and Lhx2+/+ mice were isolated as mentioned above and plated on collagen-coated dishes. Cells were grown at 33°C with 5% CO2 until 100% confluency. Using a P10 tip, a scratch was made in the middle of the plate. The distance between leading edges of the migrating keratinocytes was measured using the ImageJ software, as described previously (Chmielowiec et al., 2007). Data from triplicates were pooled, mean±s.d. was calculated, and statistical analysis was performed using unpaired Student’s t-test.
RESULTS
Lhx2 ablation results in altered expression of stem cells markers in the skin
Lhx2 is one of the hair bud-specific markers, whereas in postnatal telogen HFs Lhx2 is expressed in the bulge and secondary hair germ, the compartments enriched by epithelial stem cells (Rhee et al., 2006). To gain mechanistic insight into the role of Lhx2 in the regulation of the activity of epithelial stem cells, samples of total RNA isolated from E16.5 Lhx2–/– and wild-type embryos were processed for microarray analysis (Sharov et al., 2006). In addition, the formaldehyde-cross-linked chromatin fragments from primary epidermal keratinocytes were immunoprecipitated with anti-Lhx2 antibody, and ChiP-on-chip analysis was performed. Microarray and ChiP-on-chip data were merged, and 621 genes, the expression of which was significantly (two-fold and higher) changed in Lhx2–/– mice and showed association of Lhx2 with the corresponding promoter regions, were selected as the genuine Lhx2 targets in keratinocytes (Fig. 1A).
Fig. 1.
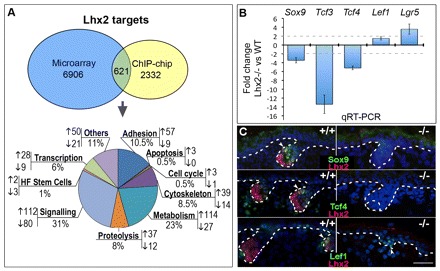
Altered expression of stem cell markers in the Lhx2-null skin. E16.5 Lhx2–/– and wild-type embryos were processed for RNA extraction and subsequent microarray studies and qRT-PCR analyses, as well as for immunohistochemistry. Primary keratinocytes isolated from mouse back skin were processed for ChIP-on-chip analysis with an antibody against the Lhx2 or purified goat IgG. (A) The ontology of the genes whose expression was altered in the E16.5 Lhx2–/– mice versus wild-type controls and which showed association of Lhx2 with the corresponding promoter regions in ChIP-on-chip assay. Full list of the genes is shown in Table S1 (supplementary material). (B) qRT-PCR analysis of the stem cell markers and regulators of stem cell activity in the Lhx2–/– versus wild-type skin. Data are mean±s.e.m. (C) Hair follicle buds of Lhx2–/– mice show lack of the Sox9- and Tcf4-positive cells, and presence of Lef1, which are all visible in wild-type mice (arrowheads). Borders between the epithelial and mesenchymal skin compartments are indicated by a broken line. Scale bar: 50 μm.
Interestingly, among the Lhx2 targets involved in the control of cell adhesion/extracellular matrix remodelling, metabolism, cytoskeleton, signalling and transcription, several genes were implicated in the control of stem cell activity (Fig. 1B; supplementary material Table S1). These included Sox9, Tcf3 and Tcf4, which were downregulated in the Lhx2–/– mice, as well as Lgr5, which was upregulated (Fig. 1B, supplementary material Table S1). Alterations in the expression of these genes in Lhx2–/– versus wild-type mice were confirmed by qRT-PCR and immunohistochemistry (Fig. 1B,C). In particular, Sox9 and Tcf4 were not seen in the HF placodes in the embryonic Lhx2–/– skin, thus serving as an additional control for the microarray and Chip-on-chip data (Fig. 1C). Interestingly, expression of Lef1, which is closely related to Tcf3 and Tcf4 (Nguyen et al., 2009; Nguyen et al., 2006), was not affected by the deletion of Lhx2 (Fig. 1C). This is consistent with data demonstrating differential expression patterns and functional roles for Lef1 versus Tcf3/4 in the developing and postnatal skin (DasGupta and Fuchs, 1999; Merrill et al., 2001; Nguyen et al., 2009).
Lhx2 expression is increased in the skin during wound healing
Regeneration of skin after injury includes not only re-epithelization of the epidermis, but also growth activation in the HFs closely adjacent to the wound (Ansell et al., 2011; Ito and Cotsarelis, 2008; Langton et al., 2008; Romagosa et al., 2008). To determine a potential role of Lhx2 in wound repair, we first studied its expression in mouse skin at distinct time-points after wounding, as well as in human skin biopsies obtained from individuals with chronic wounds and in punch-wounded human skin cultured under serum-free conditions (Moll et al., 1998; Lu et al., 2007). In the skin of 8- to 9-week-old mice, levels of Lhx2 transcripts increased on days 3 and 5 after application of full-thickness 3 mm wounds (Fig. 2A). Lhx2+ cells showed a marked increase in their proliferation on days 3 and 5 after wounding (Fig. 2B,C). In both bulge and secondary hair germ of the HFs adjacent to the wound more than 80% of proliferating cells were Lhx2+ (Fig. 2C). However, similar to unwounded skin (Rhee et al., 2006), Lhx2+ cells were seen only in the bulge and secondary hair germ of mouse HFs adjacent to the wound, whereas lack of Lhx2 expression was seen in the infundibulum, interfollicular epidermis and wound epithelium (Fig. 2D).
Fig. 2.
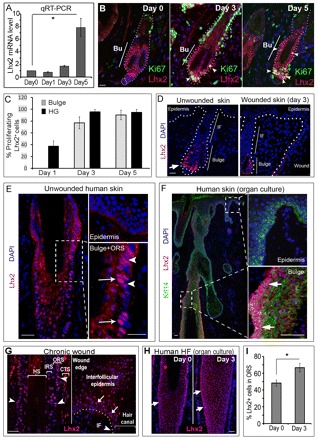
Lhx2 expression increases in hair follicles during wound healing. qRT-PCR for Lhx2 was performed with RNA from mouse skin wounds at different days after wounding. Cryosections of the wounded human and mouse skin were immunolabelled with anti-Lhx2 antiserum alone or together with anti-Ki-67 or CK14 antibody. In microscopic images, borders between the epidermis or HF and dermis are depicted by the broken line. (A) Increase in Lhx2 transcripts in mouse skin 5 days after wounding (mean±s.e.m.; *P<0.05, Student’s t-test); (B,C)Lhx2+ cells proliferate in response to skin wounding in mice. Double immunodetection shows localization of the proliferating Lhx2+ cells in the hair germ and bulge of HFs adjacent to the wound (B, arrowheads). The majority (>80%) of proliferating cells in the hair germ and bulge are Lhx2 positive (C). Bu, bulge. Data are mean±s.e.m. (D) During wound healing, Lhx2+ cells are seen only in the hair follicle bulge and secondary hair germ (arrow) and are absent in the wound epithelium. (E) In normal unwounded human skin, Lhx2 shows nuclear (arrows) and cytoplasmic (arrowheads) expression only in the HF bulge and outer root sheath (bottom image, arrows), and is absent in the epidermis (upper image). (F) In organ cultures of human skin, Lhx2+ cells that also expressed keratin 14 are seen in the bulge/outer root sheath (lower inset, arrows). Lack of Lhx2 is seen in the epidermis (upper inset, arrow). (G) In the biopsies of chronic skin wounds, Lhx2 is expressed in the wound edge epithelium and interfollicular epidermis (arrows), as well as in the follicular infundibulum (arrowheads). (H,I) Increase in the number of Lhx2+ cells in the HF outer root sheath in organ cultured human skin after wounding (mean±s.e.m., *P<0.05, Student’s t-test). Arrows indicate Lhx2+ cells in the outer root sheath. CTS, connective tissue sheath; HS, hair shaft; IF, infundibulum; IRS, inner root sheath; ORS, outer root sheath. Scale bars: 50 μm; 25 μm in insets.
In normal human skin, Lhx2 was expressed in the HF bulge and outer root sheath, whereas in the skin biopsies from chronic wounds, Lhx2+ cells were seen in the wound edge epithelium and upper region of the infundibulum of the HFs located closely to the wound edge (Fig. 2E,G). In organ-cultured full-thickness fragments of normal human skin, the Lhx2+ cells (expressing also keratin 14) were located in the bulge and outer root sheath of the HFs and their number was significantly (P<0.05) increased on day 3 after injury compared with unwounded skin (Fig. 2F,H,I).
Lhx2 deficiency results in impaired wound repair and accelerated entry of the hair follicles into anagen
To assess the role of Lhx2 in the control of wound re-epithelization after injury, Lhx2 heterozygous knockout (+/–) mice were used as a model (Porter et al., 1997). Lhx2 knockout (–/–) mice die between E15.5 and E16.5 from abnormalities in the development of the liver, brain and hematopoietic system (Porter et al., 1997). Lhx2+/– mice were viable, fertile and showed ∼50% decrease of Lhx2 protein and mRNA levels in the skin compared with wild-type mice (Fig. 3A,B).
Fig. 3.
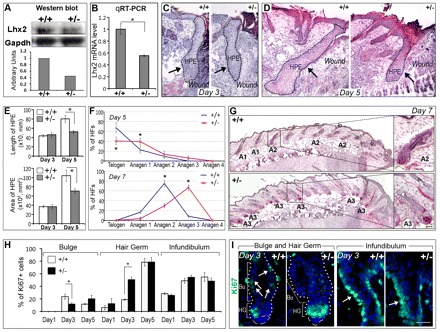
Lhx2+/– mice show impaired wound re-epithelization and accelerated anagen development associated with alterations of cell proliferation in the hair follicles. Full-thickness wounds were introduced to back skin of 8-week-old wild-type (+/+) or Lhx2 heterozygous (+/–) mice, and skin was collected at days 1-7 after wounding. Skin samples (n=8-10 from each strain) from the central part of the wounds were processed for Hematoxylin and Eosin, alkaline phosphatase or Ki67 staining, and morphometric analyses. (A,B) Decrease in levels of Lhx2 protein (A) and transcripts (B) in the skin of 8-week-old Lhx2+/– mice versus age-matched wild-type controls, as determined by western blot analysis (A) or qRT-PCR (B). Data are mean±s.e.m. (C,D) Impaired development of the wound epithelium in Lhx2+/– skin on day 5 post wounding (D) compared with day 3 (C) and with corresponding wild-type controls. Broken line and arrows show the areas of hyper-proliferative epithelium (HPE). (E) Significantly reduced area and length of the hyper-proliferative epithelium (HPE) in the Lhx2+/– skin at day 5 after wounding versus the controls (mean±s.e.m., *P<0.05). (F,G) Acceleration of anagen development in the HFs located closely to the wounds in Lhx2+/– mice versus wild-type controls on days 5 and 7 after wound infliction (mean±s.e.m., *P<0.05). HFs at distinct stages of the hair cycle on day 7 of experiment are shown [G; T, telogen or resting stage; A1, anagen 1, a stage characterized by onset of keratinocyte proliferation resulted in the enlargement of secondary hair germ; A2, anagen 2, a stage characterized by partial incorporation of the dermal papilla into enlarged secondary hair germ; A3, a stage characterized by full incorporation of the dermal papilla into the forming hair bulb (see Muller-Rover et al., 2001)]. Images of the skin on day 5 of the experiment are shown in Fig. S1A (supplementary material). (H,I) Analysis of cell proliferation in distinct HF compartments (days 1-5 after wounding) shows inverse changes in the number of Ki67+ cells in the bulge versus secondary hair germ of the HFs in Lhx2+/– mice compared with wild-type controls on day 3 after wound infliction (mean±s.e.m., *P<0.05, Student’s t-test). Number of Ki67+ cells in the infundibulum (I, arrows) does not change between the Lhx2+/– mice and wild-type controls. Data on cell proliferation in the interfollicular epidermis and wound epithelium in Lhx2+/– and wild-type mice at distinct time-points after wounding are shown in Fig. S1B (supplementary material). Bu, bulge; HG, secondary hair germ. Scale bars: 50 μm; 100 μm in G (insets).
Histological and histomorphometric analysis of the skin sections at distinct time-points after wound infliction showed progressive alterations in the development of the hyperproliferative wound epithelium in Lhx2+/– mice compared with wild-type mice (Fig. 3C-E). Despite the fact that on day 3 after wounding the length and area of the hyperproliferative wound epithelium used as established parameters of the regenerative process (Kumin et al., 2007) were quite similar between wild-type and Lhx2+/– mice, both parameters were significantly reduced (P<0.05) in Lhx2+/– mice on day 5 after wounding compared to wild-type controls (Fig. 3C-E). Interestingly, on days 5-7 after wounding, HFs in Lhx2+/– mice located closely (within 5 mm) to the wound showed significantly more advanced anagen development compared with the HFs in wild-type mice (Fig. 3F,G; supplementary material Fig. S1A), whereas no differences in hair cycle progression between Lhx2+/– and wild-type mice were seen in the HFs distantly located to the wounds (data not shown).
Analysis of cell proliferation in the distinct epithelial skin compartments (wound epithelium, interfollicular epidermis, HF infundibulum, bulge and secondary germ) on days 1-5 after wounding revealed significant differences between Lhx2+/– and WT mice only in the bulge and secondary hair germ of the HFs (Fig. 3H,I; supplementary material Fig. S1B). Interestingly, cell proliferation in the bulge of the HFs in Lhx2+/– mice was significantly lower (P<0.05) on day 3 after wounding compared with wild-type mice, whereas the number of Ki67+ cells in the secondary hair germ was significantly higher (P<0.001) compared with wild-type mice at this time point (Fig. 3H,I). However, other parameters of the wound-healing process, such as expression of wound-associated keratins (Krt16, Krt17), presence of immune cells (macrophages, neutrophils, CD4+ T cells) in the dermis/granulation tissue, as well as keratinocyte migration (as determined by scratch assay with cultured keratinocytes) were not changed in the Lhx2+/– versus wild-type mice (supplementary material Fig. S1C-H).
Thus, the impaired re-epithelization in Lhx2+/– mice was accompanied by inverse changes in cell proliferation in the bulge versus secondary hair germ and by more rapid development of anagen in the HFs adjacent to the wound (Fig. 3C-I). These data suggest that Lhx2 differentially regulates the activity of those populations of the HF stem cells that respond to skin injury versus those that regulate HF cycling by supplying their progenies either into the wound epithelium or into the growing HF, respectively (Ito et al., 2005).
Lhx2 stimulates the expression of Sox9, Tcf4 and inhibits Lgr5 expression in hair follicle stem cells during wound healing
Because, in mice, Lhx2 was expressed exclusively in the bulge and secondary hair germ and was not seen in the follicular infundibulum, inter-follicular epidermis or wound epithelium (Fig. 2B,D), we hypothesized that Lhx2 contributes to wound re-epithelization rather indirectly and most likely via regulation of Sox9 and Tcf4 in hair follicle stem cells. Genetic ablation of Sox9 or Tcf3/4 resulted in marked retardation of the wound-healing process (Nguyen et al., 2009; Nowak et al., 2008). In addition, Sox9 and Tcf3 proteins are ectopically expressed in the epidermis of K14-Lhx2 transgenic mice, suggesting an involvement of Lhx2 in the control of their expression in keratinocytes (Rhee et al., 2006).
In early anagen HFs of unwounded adult mouse skin, Sox9+ and Tcf4+ cells were only seen in the bulge, but were absent in the follicular infundibulum (supplementary material Fig. S2A). In wounded skin, double immunolabelling of Lhx2 with Sox9 or Tcf4 revealed their colocalization within the bulge area (Fig. 4A). However, in wounded skin Sox9+ and Tcf4+ cells were also seen in the follicular infundibulum (Fig. 4B), and these cells did not express Lhx2 (data not shown). To assess whether Lhx2 deficiency may affect the expression of Sox9 and Tcf4, the number Sox9+ and Tcf4+ cells was compared in the HFs closely adjacent to the wounds between the Lhx2+/– and wild-type mice. The percentage of Sox9+ and Tcf4+ cells was markedly decreased in the infundibulum but not in the bulge of the HFs adjacent to the wound in Lhx2+/– mice versus wild-type controls (Fig. 4B,C; supplementary material Fig. S2B). Furthermore, qRT-PCR data showed significant decrease of the Sox9 and Tcf4 transcript levels in wounded skin of Lhx2+/– mice compared with wild-type controls, whereas no such changes were seen in unwounded skin (supplementary material Fig. S2C). These data suggest that Lhx2 is indeed involved in regulating the expression of Sox9 and Tcf4 in the bulge progenitor cells during wound healing.
Fig. 4.
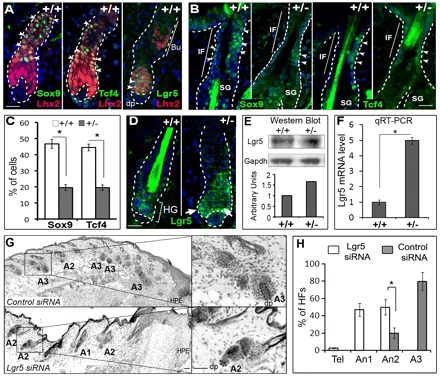
Interrelations between Lhx2 and Sox9, Tcf4 and Lgr5 in hair follicle progenitor cells during wound healing. Skin of Lhx2+/– and wild-type mice at days 5-7 after wounding was processed for immunohistochemical detection of Lhx2 and/or Sox9, Tcf4, Lgr5 (A-D), as well as processed for detection of the Lgr5 proteins and transcripts by western blot and qRT-PCR (E,F). Lhx2+/– mice were treated with Lgr5 or control siRNA on days 1-6 after wound infliction, and skin was processed for histoenzymatic detection of alkaline phosphatase and histomorphometry of hair cycle stages (G,H). (A) Co-expression of Lhx2 and/or Sox9, Tcf4 and Lgr5 in HFs of wild-type mice (arrowheads). Borders between the epidermis or HF and dermis are depicted by the broken line. (B,C) Immunodetection of the Sox9+ and Tcf4+ cells in the HFs of wild-type and Lhx2+/– mice 5 days after wounding. The percentage of Sox9+ and Tcf4+ cells (B, arrowheads) is reduced in the infundibulum of Lhx2+/– mice versus wild-type controls (C, mean±s.e.m., *P<0.05, Student’s t-test). (D) Expression of Lgr5 increased in the secondary hair germ of the HFs in Lhx2+/– mice (arrows) compared with wild-type controls on day 5 after wounding. (E,F) Increase in expression of Lgr5 protein (E) and transcripts (F) in the skin of Lhx2+/– mice versus age-matched wild-type controls on day 5 after wounding, as determined by western blot analysis (E) or qRT-PCR (F) (mean±s.e.m., *P<0.05). (G,H) Inhibition of wound-induced anagen development and significant increase in the number of anagen 2 HFs and decrease of anagen 3 HFs on day 7 of experiment in Lhx2+/– mice after treatment with Lgr5 siRNA compared with control siRNA (mean±s.e.m., *P<0.05, Student’s t-test). High magnifications of the HFs at distinct stages of the hair cycle are shown in the insets (G; T, telogen; A1-A3, anagen 1-3; dermal papilla of the HFs is depicted by a broken line). Data on the validations of the Lgr5 siRNA activity are shown in Fig. S2 (supplementary material). Bu, bulge; DP, dermal papilla; HF, secondary hair germ; IF, infundibulum; SG, sebaceous gland. Scale bars: 50 μm; 100 μm in G (insets).
Lgr5 also showed colocalization with Lhx2 in the secondary hair germ (Fig. 4A). Furthermore, expression of Lgr5 in the secondary hair germ, as well as Lgr5 mRNA and protein levels were increased in total skin of Lhx2+/– mice on day 5 after wounding compared with wild-type controls, but not in the unwounded skin (Fig. 4D-F; supplementary material Fig. S2C). Importantly, acceleration of anagen development in Lhx2+/– mice versus wild-type mice on day 7 after wounding was significantly (P<0.05) inhibited by intracutaneous administration of Lgr5 siRNA (Fig. 4G,H). Lgr5 siRNA treatment also inhibited wound-associated anagen development in wild-type mice (data not shown). The efficient Lgr5 knockdown was verified by the detection of significantly reduced levels of Lgr5 transcripts in the skin, as well as of the levels of Lgr5 protein and mRNA in primary keratinocytes after Lgr5 siRNA treatment (supplementary material Fig. S3A,B). These data suggest that Lhx2 inhibits wound-induced anagen development, at least in part, via negative regulation of Lgr5 expression in the HF progenitor cells residing in the secondary hair germ.
Sox9, Tcf4 and Lgr5 are direct Lhx2 targets in keratinocytes
To further validate whether Sox9, Tcf4 and Lgr5 are indeed direct targets of Lhx2, MatInspector analysis (Cartharius et al., 2005) for putative Lhx2-binding sites was performed and revealed several AT-rich Lim-homeodomain DNA-binding sites within the promoter regions of the Sox9, Tcf4 and Lgr5 genes. To confirm the ChIP-on-chip data, we carried out ChIP-qPCR on chromatin samples isolated from keratinocytes of E16.5 embryos, newborn or adult mice (Fig. 5A,B; supplementary material Fig. S4). ChIP-qPCR analyses of newborn or adult keratinocytes showed Lhx2 association with chromatin at distinct sites in the genomic regions of the Sox9, Tcf4 and Lgr5 promoters. In all three promoters tested, several sites among the predicted binding regions showed distinct (from twofold to fourfold) degrees of enrichment of binding to anti-Lhx2 antibody versus controls (Fig. 5A,B). Lhx2 binding to the Sox9 and Tcf4 promoters was also seen in keratinocytes isolated from E16.5 embryos (supplementary material Fig. S4). However, in contrast to keratinocytes from newborn or adult mice, lack of Lhx2 binding to the Lgr5 promoter was seen in E16.5 keratinocytes (supplementary material Fig. S4). This is consistent with low Lgr5 expression levels in embryonic skin (Jaks et al., 2008) and suggests that Lhx2 controls expression of Lgr5 only in postnatal skin. However, there was no difference in precipitation with the anti-Lhx2 antibody of the control sites on each promoter compared with control IgG, thus serving as internal negative control for this assay (Fig. 5B).
Fig. 5.
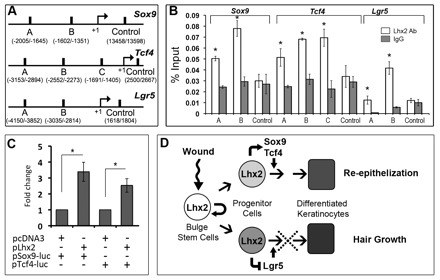
Lhx2 binds to the promoter regions of the Sox9, Tcf4 and Lgr5 genes in keratinocytes. Mouse keratinocytes isolated from adult telogen skin were processed for ChIP-qPCR analysis with an antibody against Lhx2 or purified goat IgG. HaCaT keratinocytes were processed for detection of the Sox9 and Tcf4 promoter activities. (A) Sox9, Tcf4 and Lgr5 promoter regions with putative Lim-homeodomain DNA-binding sites or control sites predicted by bioinformatic analysis. (B) ChIP-qPCR analysis of the distinct regions in the Sox9, Tcf4 and Lgr5 promoters, showing enrichment with Lhx2 antibody compared with IgG controls (mean±s.e.m., n=3; *P<0.05, Student’s t-test). (C) HaCaT keratinocytes were transfected with the reporter plasmids containing mouse Sox9 or Tcf4 promoter fragments, as well as with Lhx2-containing vector or with pCDN3 control vector. Increase in the pSox9-Luc or pTcf4-Luc activities versus controls (mean±s.e.m., n=3; *P<0.05, Student’s t-test). (D) The involvement of Lhx2 in the control of the hair follicle response to skin injury.
To assess whether Lhx2 controls the activities of the Sox9 and Tcf4 promoters, HaCaT keratinocytes were transfected with the corresponding reporter plasmids containing mouse Sox9 or Tcf4 promoters, respectively. Cells were co-transfected with an Lhx2 expressing vector, which resulted in a 2.5- to 3.5-fold increase in pSox9-Luc or pTcf4-Luc activities when compared with control (Fig. 5C). These data suggested that Lhx2 positively regulates the activity of the both promoters. They are consistent with the results of microarray, ChIP-on-chip and immunohistochemical analyses (Figs 1, 4) and demonstrate that Lhx2 indeed controls the expression of these markers in distinct populations of HF progenitor cells during epidermal regeneration after injury.
DISCUSSION
Skin repair after injury is a complex process, which involves recruitment of distinct populations of undifferentiated progenitor cells from adjacent HFs into regenerating epidermis (Blanpain and Fuchs, 2009; Cotsarelis, 2006; Gurtner et al., 2008; Lau et al., 2009). The data presented in this manuscript unravel a previously unrecognized role for Lhx2 in the promotion of epidermal regeneration after wounding and demonstrate that Lhx2 operates as one of the key regulators controlling the differential response of the distinct populations of the HF stem cells to skin injury.
We show that Lhx2+ cells residing in the bulge and secondary hair germ of the HF represent the vast majority of cells that proliferate after skin injury in mice and expand in number in the HF outer root sheath in human skin (Fig. 2). This suggests a high degree of conservation of the molecular control of the wound healing response between humans and mice. Although the precise choreography of the molecular events by which Lhx2 impacts on epidermal regeneration in human skin remain to be defined, we demonstrate here that in mice Lhx2 causes opposite effects on cell proliferation in the bulge and secondary hair germ and differentially regulates the activity of the distinct populations of the progenitor cells residing in these follicular compartments.
Because, in mice, Lhx2 is not expressed in the wound epithelium or interfollicular epidermis (Fig. 2), we hypothesize that it contributes to the wound-healing process rather indirectly and, at least in part, via positive regulation of Sox9 and Tcf4 in the bulge stem cells. Sox9 and Tcf4 transcription factors play essential roles in the control of HF morphogenesis and in the maintenance of the HF stem cells during postnatal life (Nguyen et al., 2009; Nowak et al., 2008), and Lhx2 co-localizes with these markers in the HF bulge (Fig. 4). Furthermore, lineage tracing experiments showed that bulge-derived Sox9+ and Tcf4+ progenitor cells contribute to wound re-epithelization, and genetic ablation of Sox9 or Tcf3/4 resulted in marked retardation of the wound-healing process (Nguyen et al., 2009; Nowak et al., 2008). ChIP-on-chip/Chip-qPCR and reporter assay data presented here provide evidence that Lhx2 serves as a direct upstream regulator of Sox9 and Tcf4 and, most likely, promotes expansion of the Sox9+ and Tcf4+ progenitor cells towards the regenerating epidermis during the response of the skin to injury (Figs 4, 5).
In addition to the marked retardation of wound re-epithelization, Lhx2+/– mice show significant acceleration of anagen development in the HFs adjacent to the wounds (Fig. 3; supplementary material Fig. S1). We demonstrate here that Lhx2 negatively regulates the expression of Lgr5 (Figs 4, 5), a marker of the cycling population of the HF stem cells predominantly residing in secondary hair germ and lower bulge, which is capable of generating all HF cell lineages during the hair cycle (Jaks et al., 2008). It has recently been shown that Lgr5-lacZ-expressing cells could also be seen in the bulge and interfollicular epidermis at late stages of the healing process, suggesting their contribution to re-epithelization (Kasper et al., 2011). We show that Lhx2 and Lgr5 are co-expressed in the progenitor cells residing in secondary hair germ, that levels of Lgr5 transcripts and protein are increased in the skin of Lhx2+/– mice, and that acceleration of the wound-induced anagen development in Lhx2+/– mice is inhibited by administration of Lgr5 siRNA (Fig. 4). Together with the ChIP-on-chip and Chip-qPCR data (Fig. 5), these data strongly suggest that, during wound healing, Lhx2 inhibits HF cycling, most likely via negative regulation of Lgr5 in the progenitor cell population located in the secondary hair germ.
It has previously been shown that Lhx2 operates as a transcriptional activator or repressor during development depending on the site, timing and levels of expression, as well as on the availability of distinct co-factors that form the Lhx2 transcription complexes (Chou et al., 2009; Hobert and Westphal, 2000; Xu et al., 2007; Yun et al., 2009). Our data on the wound-induced acceleration of anagen development in Lhx2+/– mice are consistent with data showing accelerated entry of the HFs into anagen phase during the normal hair cycle in the Lhx2-null skin grafted onto nude mice (Rhee et al., 2006). However, these data are quite opposite to the results obtained with CreER:Lhx2fl/fl mice, in which the Lhx2 gene is inactivated in adult skin after topical tamoxifen application leading to retardation of anagen development (Tornqvist et al., 2010). It remains to be determined whether timing and/or degree of Lhx2 inactivation, as well as availability of its co-factors in the skin may contribute to the differences in hair growth phenotype between two genetic models. These data also suggest that the role of Lhx2 in the control of hair-specific genetic programs in the follicular stem cells is more complex than previously anticipated. Elucidating its precise role in different developmental processes will require further investigations using mice with tissue-specific knockout or overexpression of Lhx2.
Interestingly, Lhx2 protein was not seen in the isthmus and infundibulum of the HFs during wound healing, the areas containing two other stem cell populations expressing Lgr6 and Lrig1, respectively (Jensen et al., 2009; Snippert et al., 2010). Thus, Lhx2 involvement in the control of Lgr6+ and Lrig1+ stem cell populations is rather unlikely, supporting a concept that several independent mechanisms control the recruitment of the distinct HF stem cell populations to the regenerating epidermis in wounded skin (Ito et al., 2005; Jaks et al., 2008; Jensen et al., 2009; Levy et al., 2005; Levy et al., 2007; Snippert et al., 2010).
Taken together, our data demonstrating that Lhx2 positively regulates Sox9 and Tcf4 and negatively regulates Lgr5 in the bulge and secondary hair germ progenitor cells, suggest that Lhx2 operates as a central switchboard controlling the differential response of the distinct HF stem cell populations to wounding. Indeed, by stimulating the expression of Sox9 and Tcf4, Lhx2 may promote generation of the HF-derived progenitor cells that contribute to regeneration of the epidermis after injury. Instead, by inhibiting Lgr5 expression, Lhx2 suppresses stem cell activation for construction of new anagen HF, and thereby HF entry into anagen (Fig. 5D).
Through its dual activity, which includes stimulation of epidermal regeneration on the one hand and inhibition of hair cycling on the other hand, Lhx2 serves as one of the key regulators in the molecular network that control the response of the HFs to skin injury. Although detailed mechanisms underlying its involvement in reciprocal control of the activity of distinct HF stem cell populations remain to be further defined, these data suggest that Lhx2 may serve as a novel target for promoting the production and recruitment of the progenitor cells into regenerating epithelia in conditions associated with an impaired stem cell activity, including ageing and chronic inflammation.
Supplementary Material
Acknowledgments
We thank Dr H. Westphal and Dr P. Koopman for providing Lhx2+/– mice and Sox9-Luc plasmid, respectively.
Footnotes
Funding
This study was supported by the grants from the Biotechnology and Biological Sciences Research Council [BB/E023010/1 to V.A.B.] and from Deutsche Forschungsgemeinschaf [Pa 345/12-2 to R.P.].
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.070284/-/DC1
References
- Ambler C. A., Maatta A. (2009). Epidermal stem cells: location, potential and contribution to cancer. J. Pathol. 217, 206–216 [DOI] [PubMed] [Google Scholar]
- Ansell D. M., Kloepper J. E., Thomason H. A., Paus R., Hardman M. J. (2011). Exploring the ‘hair growth-wound healing connection’: anagen phase promotes wound re-epithelization. J. Invest. Dermatol. 131, 512–528 [DOI] [PubMed] [Google Scholar]
- Blanpain C., Fuchs E. (2009). Epidermal homeostasis: a balancing act of stem cells in the skin. Nat. Rev. Mol. Cell Biol. 10, 207–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanpain C., Lowry W. E., Geoghegan A., Polak L., Fuchs E. (2004). Self-renewal, multipotency, and the existence of two cell populations within an epithelial stem cell niche. Cell 118, 635–648 [DOI] [PubMed] [Google Scholar]
- Botchkarev V. A., Botchkareva N. V., Roth W., Nakamura M., Chen L.-H., Herzog W., Lindner G., McMahon J. A., Peters C., Lauster R., et al. (1999a). Noggin is a mesenchymally-derived stimulator of hair follicle induction. Nature Cell Biol. 1, 158–164 [DOI] [PubMed] [Google Scholar]
- Botchkarev V. A., Peters E. M., Botchkareva N. V., Maurer M., Paus R. (1999b). Hair cycle-dependent changes in adrenergic skin innervation, and hair growth modulation by adrenergic drugs. J. Invest. Dermatol. 113, 878–887 [DOI] [PubMed] [Google Scholar]
- Botchkareva N. V., Botchkarev V. A., Welker P., Airaksinen M., Roth W., Suvanto P., Muller-Rover S., Hadshiew I. M., Peters C., Paus R. (2000). New roles for glial cell line-derived neurotrophic factor and neurturin: involvement in hair cycle control. Am. J. Pathol. 156, 1041–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownell I., Guevara E., Bai C. B., Loomis C. A., Joyner A. L. (2011). Nerve-derived sonic hedgehog defines a niche for hair follicle stem cells capable of becoming epidermal stem cells. Cell Stem Cell 8, 552–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartharius K., Frech K., Grote K., Klocke B., Haltmeier M., Klingenhoff A., Frisch M., Bayerlein M., Werner T. (2005). MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics 21, 2933–2942 [DOI] [PubMed] [Google Scholar]
- Chmielowiec J., Borowiak M., Morkel M., Stradal T., Munz B., Werner S., Wehland J., Birchmeier C., Birchmeier W. (2007). c-Met is essential for wound healing in the skin. J. Cell Biol. 177, 151–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou S. J., Perez-Garcia C. G., Kroll T. T., O’Leary D. D. (2009). Lhx2 specifies regional fate in Emx1 lineage of telencephalic progenitors generating cerebral cortex. Nat. Neurosci. 12, 1381–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotsarelis G. (2006). Epithelial stem cells: a folliculocentric view. J. Invest. Dermatol. 126, 1459–1468 [DOI] [PubMed] [Google Scholar]
- Cotsarelis G., Sun T. T., Lavker R. M. (1990). Label-retaining cells reside in the bulge area of pilosebaceous unit: implications for follicular stem cells, hair cycle, and skin carcinogenesis. Cell 61, 1329–1337 [DOI] [PubMed] [Google Scholar]
- Dahl L., Richter K., Hagglund A. C., Carlsson L. (2008). Lhx2 expression promotes self-renewal of a distinct multipotential hematopoietic progenitor cell in embryonic stem cell-derived embryoid bodies. PLoS One 3, e2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DasGupta R., Fuchs E. (1999). Multiple roles for activated LEF/TCF transcription complexes during hair follicle development and differentiation. Development 126, 4557–4568 [DOI] [PubMed] [Google Scholar]
- Driskell R. R., Clavel C., Rendl M., Watt F. M. (2011). Hair follicle dermal papilla cells at a glance. J. Cell Sci. 124, 1179–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye M., Fisher A. G., Watt F. M. (2007). Epidermal stem cells are defined by global histone modifications that are altered by Myc-induced differentiation. PLoS One 2, e763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs E. (2007). Scratching the surface of skin development. Nature. 445, 834–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garza L. A., Yang C. C., Zhao T., Blatt H. B., Lee M., He H., Stanton D. C., Carrasco L., Spiegel J. H., Tobias J. W., et al. (2011). Bald scalp in men with androgenetic alopecia retains hair follicle stem cells but lacks CD200-rich and CD34-positive hair follicle progenitor cells. J. Clin. Invest. 121, 613–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghazizadeh S., Taichman L. B. (2001). Multiple classes of stem cells in cutaneous epithelium: a lineage analysis of adult mouse skin. EMBO J 20, 1215–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurtner G. C., Werner S., Barrandon Y., Longaker M. T. (2008). Wound repair and regeneration. Nature 453, 314–321 [DOI] [PubMed] [Google Scholar]
- Hirota J., Mombaerts P. (2004). The LIM-homeodomain protein Lhx2 is required for complete development of mouse olfactory sensory neurons. Proc. Natl. Acad. Sci. USA 101, 8751–8755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert O., Westphal H. (2000). Functions of LIM-homeobox genes. Trends Genet. 16, 75–83 [DOI] [PubMed] [Google Scholar]
- Horsley V., Aliprantis A. O., Polak L., Glimcher L. H., Fuchs E. (2008). NFATc1 balances quiescence and proliferation of skin stem cells. Cell 132, 299–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu Y. C., Pasolli H. A., Fuchs E. (2011). Dynamics between stem cells, niche, and progeny in the hair follicle. Cell 144, 92–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M., Cotsarelis G. (2008). Is the hair follicle necessary for normal wound healing? J. Invest. Dermatol 128, 1059–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M., Liu Y., Yang Z., Nguyen J., Liang F., Morris R. J., Cotsarelis G. (2005). Stem cells in the hair follicle bulge contribute to wound repair but not to homeostasis of the epidermis. Nat. Med. 11, 1351–1354 [DOI] [PubMed] [Google Scholar]
- Ito M., Yang Z., Andl T., Cui C., Kim N., Millar S. E., Cotsarelis G. (2007). Wnt-dependent de novo hair follicle regeneration in adult mouse skin after wounding. Nature 447, 316–320 [DOI] [PubMed] [Google Scholar]
- Jaks V., Barker N., Kasper M., van Es J. H., Snippert H. J., Clevers H., Toftgard R. (2008). Lgr5 marks cycling, yet long-lived, hair follicle stem cells. Nat. Genet. 40, 1291–1299 [DOI] [PubMed] [Google Scholar]
- Jensen K. B., Collins C. A., Nascimento E., Tan D. W., Frye M., Itami S., Watt F. M. (2009). Lrig1 expression defines a distinct multipotent stem cell population in mammalian epidermis. Cell Stem Cell 4, 427–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen U. B., Yan X., Triel C., Woo S. H., Christensen R., Owens D. M. (2008). A distinct population of clonogenic and multipotent murine follicular keratinocytes residing in the upper isthmus. J. Cell Sci. 121, 609–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai Y., Koopman P. (1999). Structural and functional characterization of the mouse Sox9 promoter: implications for campomelic dysplasia. Hum. Mol. Genet. 8, 691–696 [DOI] [PubMed] [Google Scholar]
- Kasper M., Jaks V., Are A., Bergström A., Schwäger A., Barker N., Toftgård R. (2011). Wounding enhances epidermal tumorigenesis by recruiting hair follicle keratinocytes. Proc. Natl. Acad. Sci. USA 108, 4099–4104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloepper J. E., Tiede S., Brinckmann J., Reinhardt D. P., Meyer W., Faessler R., Paus R. (2008). Immunophenotyping of the human bulge region: the quest to define useful in situ markers for human epithelial hair follicle stem cells and their niche. Exp. Dermatol. 17, 592–609 [DOI] [PubMed] [Google Scholar]
- Kumin A., Schafer M., Epp N., Bugnon P., Born-Berclaz C., Oxenius A., Klippel A., Bloch W., Werner S. (2007). Peroxiredoxin 6 is required for blood vessel integrity in wounded skin. J. Cell Biol. 179, 747–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langton A. K., Herrick S. E., Headon D. J. (2008). An extended epidermal response heals cutaneous wounds in the absence of a hair follicle stem cell contribution. J. Invest. Dermatol. 128, 1311–1318 [DOI] [PubMed] [Google Scholar]
- Lau K., Paus R., Tiede S., Day P., Bayat A. (2009). Exploring the role of stem cells in cutaneous wound healing. Exp. Dermatol. 18, 921–933 [DOI] [PubMed] [Google Scholar]
- Levy V., Lindon C., Harfe B. D., Morgan B. A. (2005). Distinct stem cell populations regenerate the follicle and interfollicular epidermis. Dev. Cell 9, 855–861 [DOI] [PubMed] [Google Scholar]
- Levy V., Lindon C., Zheng Y., Harfe B. D., Morgan B. A. (2007). Epidermal stem cells arise from the hair follicle after wounding. FASEB J. 21, 1358–1366 [DOI] [PubMed] [Google Scholar]
- Li L., Clevers H. (2010). Coexistence of quiescent and active adult stem cells in mammals. Science 327, 542–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z., Hasse S., Bodo E., Rose C., Funk W., Paus R. (2007). Towards the development of a simplified long-term organ culture method for human scalp skin and its appendages under serum-free conditions. Exp. Dermatol. 16, 37–49 [DOI] [PubMed] [Google Scholar]
- Mardaryev A. N., Ahmed M. I., Vlahov N. V., Fessing M. Y., Gill J. H., Sharov A. A., Botchkareva N. V. (2010). Micro-RNA-31 controls hair cycle-associated changes in gene expression programs of the skin and hair follicle. FASEB J. 24, 3869–3881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrill B. J., Gat U., DasGupta R., Fuchs E. (2001). Tcf3 and Lef1 regulate lineage differentiation of multipotent stem cells in skin. Genes Dev. 15, 1688–1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll I., Houdek P., Schmidt H., Moll R. (1998). Characterization of epidermal wound healing in a human skin organ culture model: acceleration by transplanted keratinocytes. J. Invest. Dermatol. 111, 251–258 [DOI] [PubMed] [Google Scholar]
- Morris R. J., Liu Y., Marles L., Yang Z., Trempus C., Li S., Lin J. S., Sawicki J. A., Cotsarelis G. (2004). Capturing and profiling adult hair follicle stem cells. Nat. Biotechnol. 22, 411–417 [DOI] [PubMed] [Google Scholar]
- Muller-Rover S., Handjiski B., van der Veen C., Eichmuller S., Foitzik K., McKay I. A., Stenn K. S., Paus R. (2001). A comprehensive guide for the accurate classification of murine hair follicles in distinct hair cycle stages. J. Invest. Dermatol. 117, 3–15 [DOI] [PubMed] [Google Scholar]
- Nguyen H., Rendl M., Fuchs E. (2006). Tcf3 governs stem cell features and represses cell fate determination in skin. Cell 127, 171–183 [DOI] [PubMed] [Google Scholar]
- Nguyen H., Merrill B. J., Polak L., Nikolova M., Rendl M., Shaver T. M., Pasolli H. A., Fuchs E. (2009). Tcf3 and Tcf4 are essential for long-term homeostasis of skin epithelia. Nat. Genet. 41, 1068–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak D. E., Tian B., Brasier A. R. (2005). Two-step cross-linking method for identification of NF-kappaB gene network by chromatin immunoprecipitation. Biotechniques 39, 715–725 [DOI] [PubMed] [Google Scholar]
- Nowak J. A., Polak L., Pasolli H. A., Fuchs E. (2008). Hair follicle stem cells are specified and function in early skin morphogenesis. Cell Stem Cell 3, 33–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philpott M. P., Green M. R., Kealey T. (1990). Human hair growth in vitro. J. Cell Sci. 97, 463–471 [DOI] [PubMed] [Google Scholar]
- Porter F. D., Drago J., Xu Y., Cheema S. S., Wassif C., Huang S. P., Lee E., Grinberg A., Massalas J. S., Bodine D., et al. (1997). Lhx2, a LIM homeobox gene, is required for eye, forebrain, and definitive erythrocyte development. Development 124, 2935–2944 [DOI] [PubMed] [Google Scholar]
- Rhee H., Polak L., Fuchs E. (2006). Lhx2 maintains stem cell character in hair follicles. Science 312, 1946–1949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romagosa Y., Hu S., Kirsner R. S. (2008). Wound healing without hair. J. Invest. Dermatol. 128, 1058 [DOI] [PubMed] [Google Scholar]
- Schafer M., Werner S. (2007). Transcriptional control of wound repair. Annu. Rev. Cell Dev. Biol. 23, 69–92 [DOI] [PubMed] [Google Scholar]
- Schmidt-Ullrich R., Paus R. (2005). Molecular principles of hair follicle induction and morphogenesis. BioEssays 27, 247–261 [DOI] [PubMed] [Google Scholar]
- Sharov A. A., Weiner L., Sharova T. Y., Siebenhaar F., Atoyan R., McNamara C. A., Funa K., Gilchrest B. A., Brissette J. L., Botchkarev V. A. (2003). Noggin overexpression inhibits eyelid opening by altering epidermal apoptosis and differentiation. EMBO J. 22, 2992–3003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharov A. A., Fessing M., Atoyan R., Sharova T. Y., Haskell-Luevano C., Weiner L., Funa K., Brissette J. L., Gilchrest B. A., Botchkarev V. A. (2005). Bone morphogenetic protein (BMP) signaling controls hair pigmentation by means of cross-talk with the melanocortin receptor-1 pathway. Proc. Natl. Acad. Sci. USA 102, 93–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharov A. A., Sharova T. Y., Mardaryev A. N., Tommasi di Vignano A., Atoyan R., Weiner L., Yang S., Brissette J. L., Dotto G. P., Botchkarev V. A. (2006). BMP signaling regulates size of the hair follicles and modulates the expression of cell cycle-associated genes.. Proc. Natl. Acad. Sci. USA 103, 18166–18171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snippert H. J., Haegebarth A., Kasper M., Jaks V., van Es J. H., Barker N., van de Wetering M., van den Born M., Begthel H., Vries R. G., et al. (2010). Lgr6 marks stem cells in the hair follicle that generate all cell lineages of the skin. Science 327, 1385–1389 [DOI] [PubMed] [Google Scholar]
- Taylor G., Lehrer M. S., Jensen P. J., Sun T. T., Lavker R. M. (2000). Involvement of follicular stem cells in forming not only the follicle but also the epidermis. Cell 102, 451–461 [DOI] [PubMed] [Google Scholar]
- Tetreault N., Champagne M. P., Bernier G. (2009). The LIM homeobox transcription factor Lhx2 is required to specify the retina field and synergistically cooperates with Pax6 for Six6 trans-activation. Dev. Biol. 327, 541–550 [DOI] [PubMed] [Google Scholar]
- Tiede S., Paus R. (2006). Lhx2-decisive role in epithelial stem cell maintenance, or just the ‘tip of the iceberg’? BioEssays 28, 1157–1160 [DOI] [PubMed] [Google Scholar]
- Tornqvist G., Sandberg A., Hagglund A. C., Carlsson L. (2010). Cyclic expression of lhx2 regulates hair formation. PLoS Genet. 6, e1000904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trempus C. S., Morris R. J., Ehinger M., Elmore A., Bortner C. D., Ito M., Cotsarelis G., Nijhof J. G., Peckham J., Flagler N., et al. (2007). CD34 expression by hair follicle stem cells is required for skin tumor development in mice. Cancer Res. 67, 4173–4181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumbar T., Guasch G., Greco V., Blanpain C., Lowry W. E., Rendl M., Fuchs E. (2003). Defining the epithelial stem cell niche in skin. Science 303, 359–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt F. M., Jensen K. B. (2009). Epidermal stem cell diversity and quiescence. EMBO Mol. Med. 1, 260–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Mannik J., Kudryavtseva E., Lin K. K., Flanagan L. A., Spencer J., Soto A., Wang N., Lu Z., Yu Z., et al. (2007). Co-factors of LIM domains (Clims/Ldb/Nli) regulate corneal homeostasis and maintenance of hair follicle stem cells. Dev. Biol. 312, 484–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun S., Saijoh Y., Hirokawa K. E., Kopinke D., Murtaugh L. C., Monuki E. S., Levine E. M. (2009). Lhx2 links the intrinsic and extrinsic factors that control optic cup formation. Development 136, 3895–3906 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


