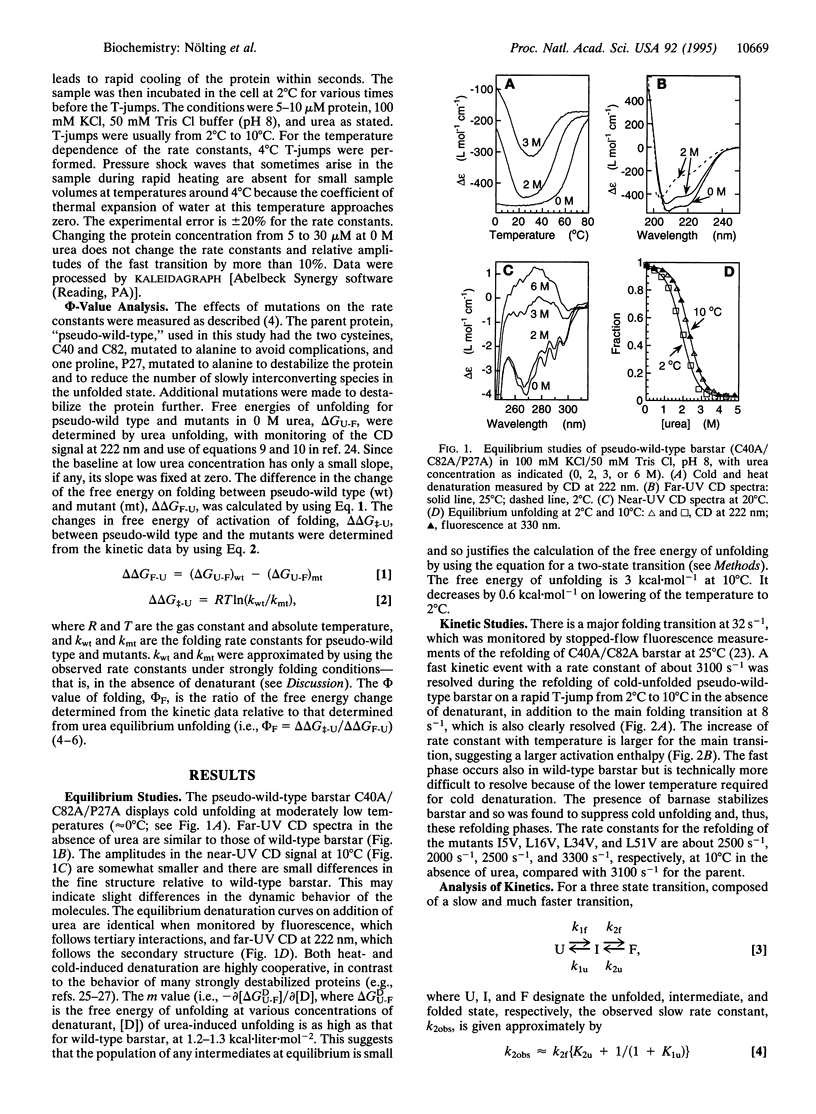
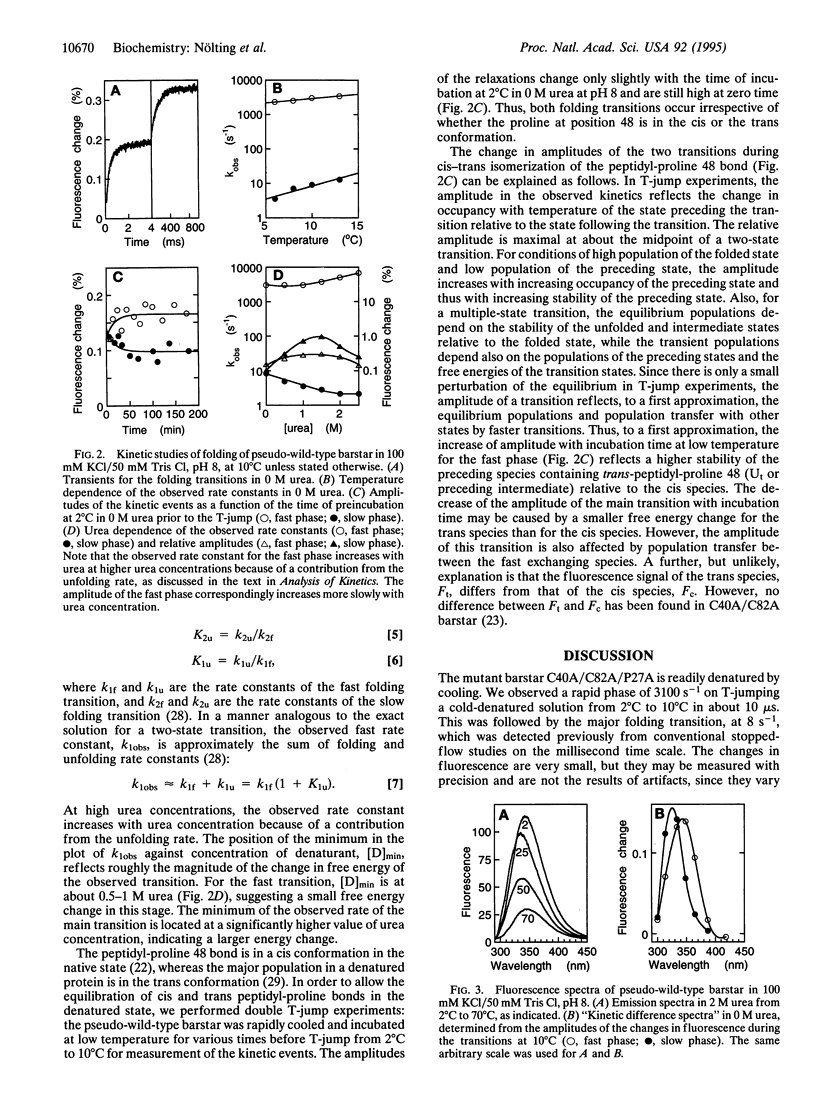
Abstract
The pathway of protein folding is now being analyzed at the resolution of individual residues by kinetic measurements on suitably engineered mutants. The kinetic methods generally employed for studying folding are typically limited to the time range of > or = 1 ms because the folding of denatured proteins is usually initiated by mixing them with buffers that favor folding, and the dead time of rapid mixing experiments is about a millisecond. We now show that the study of protein folding may be extended to the microsecond time region by using temperature-jump measurements on the cold-unfolded state of a suitable protein. We are able to detect early events in the folding of mutants of barstar, the polypeptide inhibitor of barnase. A preliminary characterization of the fast phase from spectroscopic and phi-value analysis indicates that it is a transition between two relatively solvent-exposed states with little consolidation of structure.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Agashe V. R., Udgaonkar J. B. Thermodynamics of denaturation of barstar: evidence for cold denaturation and evaluation of the interaction with guanidine hydrochloride. Biochemistry. 1995 Mar 14;34(10):3286–3299. doi: 10.1021/bi00010a019. [DOI] [PubMed] [Google Scholar]
- Clarke J., Fersht A. R. Engineered disulfide bonds as probes of the folding pathway of barnase: increasing the stability of proteins against the rate of denaturation. Biochemistry. 1993 Apr 27;32(16):4322–4329. doi: 10.1021/bi00067a022. [DOI] [PubMed] [Google Scholar]
- Fersht A. R. Characterizing transition states in protein folding: an essential step in the puzzle. Curr Opin Struct Biol. 1995 Feb;5(1):79–84. doi: 10.1016/0959-440x(95)80012-p. [DOI] [PubMed] [Google Scholar]
- Fersht A. R., Matouschek A., Serrano L. The folding of an enzyme. I. Theory of protein engineering analysis of stability and pathway of protein folding. J Mol Biol. 1992 Apr 5;224(3):771–782. doi: 10.1016/0022-2836(92)90561-w. [DOI] [PubMed] [Google Scholar]
- Fersht A. R. The sixth Datta Lecture. Protein folding and stability: the pathway of folding of barnase. FEBS Lett. 1993 Jun 28;325(1-2):5–16. doi: 10.1016/0014-5793(93)81405-o. [DOI] [PubMed] [Google Scholar]
- Fisher M. T., Sligar S. G. Temperature jump relaxation kinetics of the P-450cam spin equilibrium. Biochemistry. 1987 Jul 28;26(15):4797–4803. doi: 10.1021/bi00389a029. [DOI] [PubMed] [Google Scholar]
- French T. C., Hammes G. G. Relaxation spectra of ribonuclease. II. Isomerization of ribonuclease at neutral pH values. J Am Chem Soc. 1965 Nov 5;87(21):4669–4673. doi: 10.1021/ja00949a002. [DOI] [PubMed] [Google Scholar]
- Grathwohl C., Wüthrich K. The X-Pro peptide bond as an nmr probe for conformational studies of flexible linear peptides. Biopolymers. 1976 Oct;15(10):2025–2041. doi: 10.1002/bip.1976.360151012. [DOI] [PubMed] [Google Scholar]
- Griko Y. V., Privalov P. L. Calorimetric study of the heat and cold denaturation of beta-lactoglobulin. Biochemistry. 1992 Sep 22;31(37):8810–8815. doi: 10.1021/bi00152a017. [DOI] [PubMed] [Google Scholar]
- Hartley R. W. Barnase and barstar. Expression of its cloned inhibitor permits expression of a cloned ribonuclease. J Mol Biol. 1988 Aug 20;202(4):913–915. doi: 10.1016/0022-2836(88)90568-2. [DOI] [PubMed] [Google Scholar]
- Huang G. S., Oas T. G. Submillisecond folding of monomeric lambda repressor. Proc Natl Acad Sci U S A. 1995 Jul 18;92(15):6878–6882. doi: 10.1073/pnas.92.15.6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones C. M., Henry E. R., Hu Y., Chan C. K., Luck S. D., Bhuyan A., Roder H., Hofrichter J., Eaton W. A. Fast events in protein folding initiated by nanosecond laser photolysis. Proc Natl Acad Sci U S A. 1993 Dec 15;90(24):11860–11864. doi: 10.1073/pnas.90.24.11860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubienski M. J., Bycroft M., Freund S. M., Fersht A. R. Three-dimensional solution structure and 13C assignments of barstar using nuclear magnetic resonance spectroscopy. Biochemistry. 1994 Aug 2;33(30):8866–8877. [PubMed] [Google Scholar]
- Matouschek A., Kellis J. T., Jr, Serrano L., Bycroft M., Fersht A. R. Transient folding intermediates characterized by protein engineering. Nature. 1990 Aug 2;346(6283):440–445. doi: 10.1038/346440a0. [DOI] [PubMed] [Google Scholar]
- Matouschek A., Kellis J. T., Jr, Serrano L., Fersht A. R. Mapping the transition state and pathway of protein folding by protein engineering. Nature. 1989 Jul 13;340(6229):122–126. doi: 10.1038/340122a0. [DOI] [PubMed] [Google Scholar]
- Nath U., Udgaonkar J. B. Perturbation of a tertiary hydrogen bond in barstar by mutagenesis of the sole His residue to Gln leads to accumulation of at least one equilibrium folding intermediate. Biochemistry. 1995 Feb 7;34(5):1702–1713. doi: 10.1021/bi00005a027. [DOI] [PubMed] [Google Scholar]
- Otzen D. E., Itzhaki L. S., elMasry N. F., Jackson S. E., Fersht A. R. Structure of the transition state for the folding/unfolding of the barley chymotrypsin inhibitor 2 and its implications for mechanisms of protein folding. Proc Natl Acad Sci U S A. 1994 Oct 25;91(22):10422–10425. doi: 10.1073/pnas.91.22.10422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeil W., Bychkova V. E., Ptitsyn O. B. Physical nature of the phase transition in globular proteins. Calorimetric study of human alpha-lactalbumin. FEBS Lett. 1986 Mar 31;198(2):287–291. doi: 10.1016/0014-5793(86)80422-7. [DOI] [PubMed] [Google Scholar]
- Pfeil W., Nölting B. O., Jung C. Apocytochrome P450cam is a native protein with some intermediate-like properties. Biochemistry. 1993 Aug 31;32(34):8856–8862. doi: 10.1021/bi00085a017. [DOI] [PubMed] [Google Scholar]
- Privalov P. L. Cold denaturation of proteins. Crit Rev Biochem Mol Biol. 1990;25(4):281–305. doi: 10.3109/10409239009090612. [DOI] [PubMed] [Google Scholar]
- Pryse K. M., Bruckman T. G., Maxfield B. W., Elson E. L. Kinetics and mechanism of the folding of cytochrome c. Biochemistry. 1992 Jun 9;31(22):5127–5136. doi: 10.1021/bi00137a006. [DOI] [PubMed] [Google Scholar]
- Radford S. E., Dobson C. M., Evans P. A. The folding of hen lysozyme involves partially structured intermediates and multiple pathways. Nature. 1992 Jul 23;358(6384):302–307. doi: 10.1038/358302a0. [DOI] [PubMed] [Google Scholar]
- Roder H., Elöve G. A., Englander S. W. Structural characterization of folding intermediates in cytochrome c by H-exchange labelling and proton NMR. Nature. 1988 Oct 20;335(6192):700–704. doi: 10.1038/335700a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber G., Fersht A. R. The refolding of cis- and trans-peptidylprolyl isomers of barstar. Biochemistry. 1993 Oct 19;32(41):11195–11203. doi: 10.1021/bi00092a032. [DOI] [PubMed] [Google Scholar]
- Sligar S. G. Coupling of spin, substrate, and redox equilibria in cytochrome P450. Biochemistry. 1976 Nov 30;15(24):5399–5406. doi: 10.1021/bi00669a029. [DOI] [PubMed] [Google Scholar]
- Tanford C. Protein denaturation. C. Theoretical models for the mechanism of denaturation. Adv Protein Chem. 1970;24:1–95. [PubMed] [Google Scholar]
- Udgaonkar J. B., Baldwin R. L. NMR evidence for an early framework intermediate on the folding pathway of ribonuclease A. Nature. 1988 Oct 20;335(6192):694–699. doi: 10.1038/335694a0. [DOI] [PubMed] [Google Scholar]



