Abstract
Mesenchymal chondrosarcomas are fast-growing tumors that account for 2–10% of primary chondrosarcomas. Cytogenetic information is restricted to 12 cases that did not show a specific aberration pattern. Recently, two fusion genes were described in mesenchymal chondrosarcomas: a recurrent HEY1-NCOA2 found in tumors that had not been cytogenetically characterized and an IRF2BP2-CDX1 found in a tumor carrying a t(1;5)(q42;q32) translocation as the sole chromosomal abnormality. Here, we present the cytogenetic and molecular genetic analysis of a mesenchymal chondrosarcoma in which the patient had two histologically indistinguishable tumor lesions, one in the neck and one in the thigh. An abnormal clone with the G-banding karyotype 46,XX,add(6)(q23),add(8)(p23),del(10)(p11),+12,−15[6] was found in the neck tumor whereas a normal karyotype, 46,XX, was found in the tumor of the thigh. RT-PCR and Sanger sequencing showed that exon 4 of HEY1 was fused to exon 13 of NCOA2 in the sample from the thigh lesion; we did not have spare material to perform a similar analysis of the neck tumor. Examining the published karyotypes we observed numerical or structural aberrations of chromosome 8 in the majority of the karyotyped mesenchymal chondrosarcomas. Chromosome 8 was also structurally affected in the present study. The pathogenetic mechanisms behind this nonrandom involvement are unknown, but the presence on 8q of two genes, HEY1 and NCOA2, now known to be involved in mesenchymal chondrosarcoma tumorigenesis is, of course, suggestive.
Keywords: mesenchymal chondrosarcoma, cytogenetics, fusion gene, HEY1-NCOA2
Introduction
Mesenchymal chondrosarcomas are rare tumors that account for 2–10% of primary chondrosarcomas (1). Their typical histological appearance includes a biphasic pattern with areas of round, primitive mesenchymal cells and interspersed islands of well differentiated hyaline cartilage (2). They are two to three times more common in bone than in soft tissue and are mostly found in the head and neck area, particularly the orbit, the cranial and spinal dura mater, and the lower extremities, especially the thigh (2). However, rare cases of mesenchymal chondrosarcoma have been described in virtually every anatomic site. Unlike other types of chondrosarcoma, mesenchymal chondrosarcomas grow fast and often give rise to local recurrences and metastases. The majority of the cases are diagnosed in the second and third decade of life and the prognosis is highly variable with published 10-year overall survival rates ranging from 21% to 67% (1). Moreover, some patients live for long periods with metastatic disease, whereas others die shortly after diagnosis (1). Adequate surgery is the treatment of choice for localized disease (3). The role of chemotherapy and radiotherapy remains poorly defined (4,5).
To date, only 12 mesenchymal chondrosarcomas have been karyotyped (6–15) and no consistent aberration pattern has been established. Recently, however, two fusion genes were reported in mesenchymal chondrosarcomas. Wang et al used a genome-wide exon-resolution expression screen to identify a fusion between the hairy/enhancer-of-split related with YRPW motif 1 (HEY1; on 8q21.13) gene and the nuclear receptor coactivator 2 (NCOA2; on 8q13.3) gene; no karyotypic data were available on the tumors thus examined (16). Nyquist et al used karyotyping followed by RNA-Seq to identify an IRF2BP2-CDX1 fusion gene in a case of mesenchymal chondrosarcoma carrying a solitary t(1;5)(q42;q32) chromosomal translocation (12). Here, we present a mesenchymal chondrosarcoma which proved to have an informative karyotype and which, by RT-PCR, was found to carry a HEY1-NCOA2 fusion gene.
Materials and methods
Ethics statement
The study was approved by the regional ethics committee (Regional komité for medisinsk forskningsetikk Sør-Øst, Norge, http://helseforskning.etikkom.no) and written informed consent was obtained from the patient.
Case report - pathology
The patient was a 26-year-old woman who had noticed a tumor in the right thigh three months prior to diagnosis. Radiologic evaluation revealed a 9.0×8.0×5.4 cm large tumor in the large adductor muscle. A soft tissue lesion, presumed to be a metastasis, was detected in the neck, and another lesion of unknown origin was found in the left iliac bone. The patient received four cycles of chemotherapy; two cycles of vincristine, doxorubicin and cyclophosphamide, one cycle of vincristine, ifosfamide and actinomycin D, and one cycle of etoposide and ifosfamide. Radiologic evaluation after chemotherapy revealed no significant change in size of any of the tumors. A wide resection of the primary tumor (thigh) was performed (Fig. 1A). Radiotherapy and subsequent surgery of the metastasis in the neck is planned.
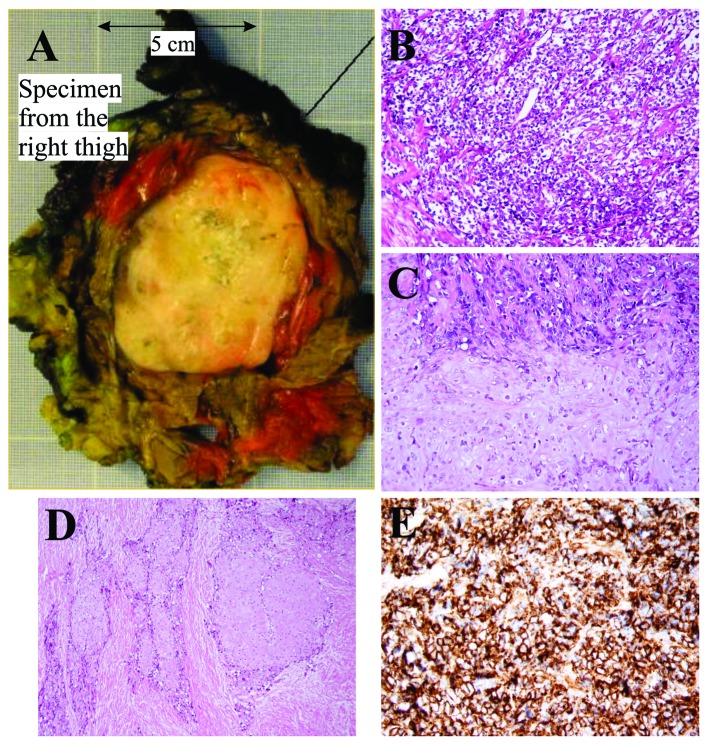
Figure 1.
Pathological features of the mesenchymal chondrosarcoma from the thigh. (A) Macroscopic image of the tumor from the right thigh. (B) Areas with small round cells. (C and D) Biphasic growth pattern showing chondroid areas with transition to small round cells. (E) Positive immunohistochemical reaction of CD99.
Microscopic examination of the specimen from the thigh showed a biphasic tumor with cellular areas with high-grade, malignant-looking, small undifferentiated round cells and some more pleomorphic cells (Fig. 1B) alternating with cartilage of hyaline type consistent with a low-grade malignant chondrosarcoma (Fig. 1C). The transition between the two components was mostly abrupt. There was bone formation close to the chondroid areas. A desmoplastic stroma was seen both around the small round cells and the chondroid areas (Fig. 1D). Immunohistochemical analysis showed a strong positive reaction to the antibody CD99 in the small round cells (Fig. 1E). Microscopic examination of the lesions in the neck and iliac bone showed tumor tissue consistent with mesenchymal chondrosarcoma.
Karyotyping
Tumor samples from both the neck and thigh were removed by core needle biopsy. The samples were mechanically and enzymatically disaggregated and then short-term cultured as described elsewhere (17). The cultures were harvested and the chromosomes G-banded using Wright stain. The subsequent cytogenetic analysis and karyotype description followed the recommendations of the ISCN (18).
Reverse transcription-polymerase chain reaction (RT-PCR)
Total RNA was extracted using TRIzol reagent from the core needle biopsy of the tumor of the thigh. No material was available from the tumor of the neck. Then, 1 μg of total RNA was reverse-transcribed in a 20 μl reaction volume using iScript Advanced cDNA Synthesis Kit for RT-qPCR according to the manufacturer’s instructions (Bio-Rad). The cDNA was diluted to 50 μl and 2 μl (corresponding to 40 ng of total RNA) were used as template in subsequent PCR assays. As a positive control, a mesenchymal chondrosarcoma known to carry the HEY1-NCOA2 fusion was used (12). The 25 μl PCR-volume contained 12.5 μl of Premix Taq (Takara Bio Europe/SAS, Saint-Germain-en-Laye, France), 2 μl of diluted cDNA, and 0.2 μM of each of the forward HEY1-F1 (CGAGGTGGAGAAGGAGAGTG) and reverse NCOA2-E13-R3 (AGTTGGGCTTTGCAATGTGA) primers. Both samples were tested for expression of the HEY1 gene to assess the quality of RNA and cDNA synthesis. The PCR components were the same as above except that the primer combination HEY1-F1 and HEY1-551R (CTCCGATAGTCCATAGCAAGG) was used. The PCRs were run on a C-1000 Thermal Cycler (Bio-Rad) using the following cycling conditions: an initial denaturation at 94°C for 30 sec followed by 35 cycles of 7 sec at 98°C, 30 sec at 55°C and 2 min at 68°C, and a final extension for 5 min at 68°C.
Four microliters of the PCR products were stained with GelRed (Biotium, Hayward, CA, USA), analyzed by electrophoresis through 1.0% agarose gel, and photographed. The amplified fragment was purified using the Qiagen gel extraction kit (Qiagen). Direct (Sanger) sequencing was performed using the light run sequencing service of GATC Biotech (http://www.gatc-biotech.com/en/sanger-services/lightrun-sequencing.html). The BLAST software (http://www.ncbi.nlm.nih.gov/BLAST/) was used for computer analysis of sequence data.
Results
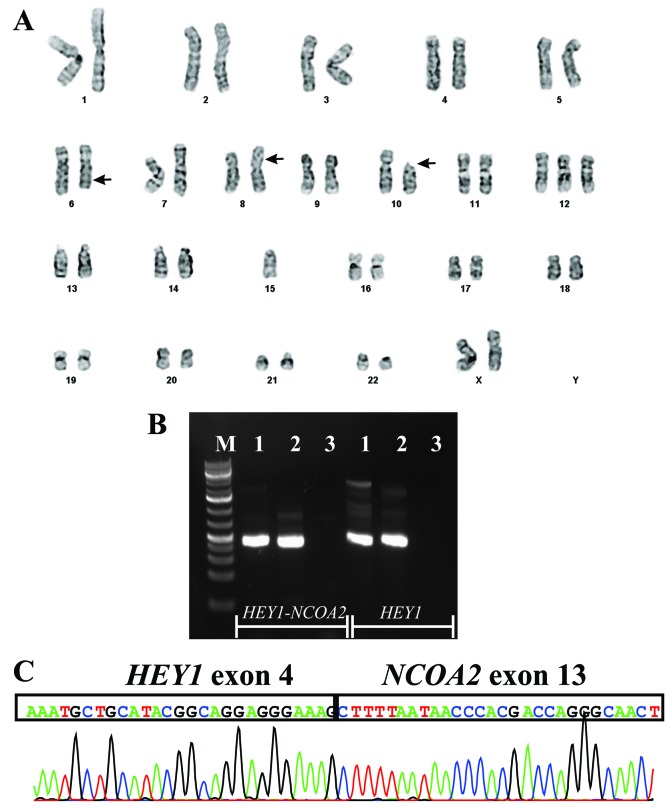
The G-banding analysis of cells cultured from the tumor of the neck yielded the karyotype 46,XX,add(6)(q23),add(8)(p23),del(10)(p11),+12, −15[6]/46,XX[5] (Fig. 2A), whereas a normal karyotype, 46,XX, was found in the tumor of the thigh.
Figure 2.
Cytogenetic and PCR analyses of the mesenchymal chondrosarcoma. (A) Karyotype of the tumor of the neck; breakpoint positions are indicated by arrows. (B) RT-PCR of RNA extracted from the tumor of the thigh. cDNA fragment amplifications of HEY1-NCOA2 using primers HEY1-F1 and NCOA2-E13-R3 and normal HEY1 with primers HEY1-F1 and HEY1-551R. M, 1 Kb Plus DNA ladder (GeneRuler, Fermentas). (C) Partial sequence chromatogram showing that exon 4 of HEY1 is fused to exon 13 of NCOA2.
RT-PCR with the HEY1-F1/NCOA2-E13-R3 primer combination amplified a single cDNA fragment in both the tumor of the thigh and the positive control (Fig. 2B). Sequencing of the amplified fragment showed that exon 4 of HEY1(nt 531 in sequence with accession number NM_012258 version 3) was fused to exon 13 of NCOA2 (nt 2768 in sequence with accession number NM_006540 version 2) (Fig. 2C). Normal HEY1cDNA fragments were amplified in both cases (Fig. 2B).
Discussion
Despite being a recognized entity for more than 50 years, mesenchymal chondrosarcoma continues to present substantial diagnostic, prognostic and management challenges, due, in large part, to its rarity (1). The cytogenetic information is restricted to 12 cases with variable karyotypes and no consistent aberration pattern has been established. Dobin et al reported an intrathoracal mesenchymal chondrosarcoma with a near-tetraploid karyotype which included structural chromosomal abnormalities such as add(7)(p13), add(22)(q13), markers and double minutes (7). Gatter et al reported trisomy 8 as the sole cytogenetic abnormality (9), while three other reports described different chromosomal translocations, t(4;19)(q35;q13), t(6;10)(p21;q22), and t(1;5)(q42;q32), as the sole cytogenetic abnormality (6,12,13). Nevertheless, examining the karyotypes as part of this study we observed that involvement of chromosome 8 was found in as many as 7 out of the 12 karyotyped mesenchymal chondrosarcomas with aberrations: +8 was found in three cases (7,9,14), −8 was found in two cases (8,10), whereas structural aberrations of 8q were found in three cases (in one case together with −8) (8,11). Chromosome 8 was also structurally affected in the present study. The pathogenetic mechanisms behind this nonrandom involvement are unknown, but the presence on 8q of two genes, HEY1 and NCOA2, now known to be involved in mesenchymal chondrosarcoma tumorigenesis is, of course, suggestive.
Recently, Wang et al identified a HEY1-NCOA2 fusion in mesenchymal chondrosarcomas. Using a combination of FISH and RT-PCR methodologies they found that 10 out of 15 examined mesenchymal chondrosarcomas carried the HEY1-NCOA2 (16). The findings were verified in two other studies: Nyquist et al (12) showed by RT-PCR the presence of HEY1-NCOA2 in 3 of 4 mesenchymal chondrosarcomas, and Nakayama et al (19) found HEY1-NCOA2 in 8 of 10 mesenchymal chondrosarcomas using FISH on formalin-fixed and paraffin-embedded samples. The absence of HEY1-NCOA2 fusion in some cases has been explained as being due to methodological inadequacy (16,19) but the possibility of other disease-specific fusion gene(s), and thus pathogenetic heterogeneity in this diagnostic entity, should not be ruled out. As demonstrated by Nyquist et al, the chromosomal translocation t(1;5)(q42;q32) resulted in fusion of IRF2BP2 (located on 1q42) with CDX1 (on 5q32) to generate an IRF2BP2-CDX1 fusion gene in the mesenchymal chondrosarcoma they studied (12). In spite of this, the HEY1-NCOA2 fusion does seem to be common as well as specific for mesenchymal chondrosarcomas since it was not found in conventional and dedifferentiated chondrosarcomas (16). Both genes are located on the long arm of chromosome 8, HEY1 in 8q21.13 and NCOA2 in 8q13.3, therefore the fusion may result from an interstitial deletion (16) or a t(8;8)(q13;q21) chromosomal translocation.
The involvement of the NCOA2 gene in neoplasia was first reported by Carapeti et al who showed that in acute myeloid leukemia the cytogenetic aberration inv(8)(p11q13) resulted in a KAT6A-NCOA2, also known as MOZ-TIF2, fusion gene (20,21). Since then, the gene has also been implicated in various other malignancies. Strehl et al identified a novel recurrent t(8;12)(q13;p13) resulting in a fusion between the transcriptional repressor ETV6 (TEL) and NCOA2 in six cases of childhood leukemia expressing both T-lymphoid and myeloid antigens (22). A PAX3-NCOA2 gene was found as a rare variant fusion in alveolar rhabdomyosarcoma; it was brought about by a t(2;8)(q35;q13) translocation (23). The AHRR-NCOA2 and GTF2I-NCOA2 fusion genes were described in soft tissue angiofibroma through translocations t(5;8)(p15;q13) and t(7;8)(q11;q13), respectively, emphasizing the role of NCOA2 in soft tissue angiofibroma development (24,25). Recently, SRF-NCOA2 and TEAD1-NCOA2 fusions were reported in rhabdomyosarcomas (26). In all the above mentioned fusions, NCOA2 is the 3′-partner gene and all fusion proteins contain the two C-terminal activation domains AD1/CID (activation domain 1/CREB binding protein interacting domain) and AD2 (20–26). The transforming activities of KAT6A-NCOA2 and PAX3-NCOA2 have been demonstrated experimentally (23,27). In addition, KAT6A-NCOA2 was shown to induce acute myeloid leukemia in transgenic fish (28). Deguchi et al (27) showed that the KAT6A-NCOA2 interaction with CREBBP through AD1/CID is essential for transformation. Similarly, Sumegi et al (23) showed that while deletion of the AD2 portion of PAX3-NCOA2 fusion protein reduced transforming activity, deletion of the AD1/CID domain fully abrogated the transforming activity of the chimeric protein. Thus, the presence of the AD1/CID and AD2 domains of NCOA2 seems to be essential for the transformation capacity of the various cancer fusion genes.
HEY1 encodes a nuclear protein belonging to the hairy and enhancer of split-related (HESR) family of basic helix-loop-helix (bHLH)-type transcriptional repressors (29). Expression of this gene is induced by the Notch and c-Jun signal transduction pathways (30). HEY1 protein binds to specific DNA sequences in the promoter regions of target genes as a dimer, recruiting co-repressors to repress the target genes of Notch signaling. The HEY1-NCOA2 fusion replaces the C-terminal portion of HEY1 by the NCOA2 AD1/CID and AD2 domains, while retaining the HEY1 bHLH DNA-binding/dimerization domain. Therefore, the HEY1-NCOA2 fusion protein, instead of recruiting co-repressors, may recruit co-activators through its NCOA2 part to some Notch/HEY1 target genes (16). Additional experiments are required to confirm or falsify the validity of this hypothesis.
Acknowledgements
The authors thank Saeedeh Shahmohammadi for the technical help. This work was supported by grants from the Norwegian Cancer Society and The South-East Norway Regional Health Authority.
References
- 1.Shakked RJ, Geller DS, Gorlick R, Dorfman HD. Mesenchymal chondrosarcoma: clinicopathologic study of 20 cases. Arch Pathol Lab Med. 2012;136:61–75. doi: 10.5858/arpa.2010-0362-OA. [DOI] [PubMed] [Google Scholar]
- 2.Weiss SW, Goldblum JR, editors. Enzinger and Weiss’s Soft Tissue Tumors. Mosby; St Louis, MO: 2001. [Google Scholar]
- 3.Riedel RF, Larrier N, Dodd L, Kirsch D, Martinez S, Brigman BE. The clinical management of chondrosarcoma. Curr Treat Options Oncol. 2009;10:94–106. doi: 10.1007/s11864-009-0088-2. [DOI] [PubMed] [Google Scholar]
- 4.Cesari M, Bertoni F, Bacchini P, Mercuri M, Palmerini E, Ferrari S. Mesenchymal chondrosarcoma. An analysis of patients treated at a single institution. Tumori. 2007;93:423–427. doi: 10.1177/030089160709300503. [DOI] [PubMed] [Google Scholar]
- 5.Dantonello TM, Int-Veen C, Leuschner I, et al. Mesenchymal chondrosarcoma of soft tissues and bone in children, adolescents, and young adults: experiences of the CWS and COSS study groups. Cancer. 2008;112:2424–2431. doi: 10.1002/cncr.23457. [DOI] [PubMed] [Google Scholar]
- 6.Crosswell H, Buchino JJ, Sweetman R, Reisner A. Intracranial mesenchymal chondrosarcoma in an infant. Med Pediatr Oncol. 2000;34:370–374. doi: 10.1002/(sici)1096-911x(200005)34:5<370::aid-mpo14>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 7.Dobin SM, Donner LR, Speights VO., Jr Mesenchymal chondrosarcoma. A cytogenetic, immunohistochemical and ultrastructural study. Cancer Genet Cytogenet. 1995;83:56–60. doi: 10.1016/0165-4608(95)00031-3. [DOI] [PubMed] [Google Scholar]
- 8.Fletcher CD, Dal Cin P, de Wever I, et al. Correlation between clinicopathological features and karyotype in spindle cell sarcomas. A report of 130 cases from the CHAMP study group. Am J Pathol. 1999;154:1841–1847. doi: 10.1016/S0002-9440(10)65441-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gatter KM, Olson S, Lawce H, Rader AE. Trisomy 8 as the sole cytogenetic abnormality in a case of extraskeletal mesenchymal chondrosarcoma. Cancer Genet Cytogenet. 2005;159:151–154. doi: 10.1016/j.cancergencyto.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 10.Mandahl N, Gustafson P, Mertens F, et al. Cytogenetic aberrations and their prognostic impact in chondrosarcoma. Genes Chromosomes Cancer. 2002;33:188–200. doi: 10.1002/gcc.10012. [DOI] [PubMed] [Google Scholar]
- 11.Naumann S, Krallman PA, Unni KK, Fidler ME, Neff JR, Bridge JA. Translocation der(13;21)(q10;q10) in skeletal and extraskeletal mesenchymal chondrosarcoma. Mod Pathol. 2002;15:572–576. doi: 10.1038/modpathol.3880565. [DOI] [PubMed] [Google Scholar]
- 12.Nyquist KB, Panagopoulos I, Thorsen J, et al. Whole-transcriptome sequencing identifies novel IRF2BP2-CDX1 fusion gene brought about by translocation t(1;5)(q42;q32) in mesenchymal chondrosarcoma. PLoS One. 2012;7:e49705. doi: 10.1371/journal.pone.0049705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Richkind KE, Romansky SG, Finklestein JZ. t(4;19)(q35;q13.1): a recurrent change in primitive mesenchymal tumors? Cancer Genet Cytogenet. 1996;87:71–74. doi: 10.1016/0165-4608(95)00240-5. [DOI] [PubMed] [Google Scholar]
- 14.Sainati L, Scapinello A, Montaldi A, et al. A mesenchymal chondrosarcoma of a child with the reciprocal translocation (11;22)(q24;q12) Cancer Genet Cytogenet. 1993;71:144–147. doi: 10.1016/0165-4608(93)90020-m. [DOI] [PubMed] [Google Scholar]
- 15.Szymanska J, Tarkkanen M, Wiklund T, et al. Cytogenetic study of extraskeletal mesenchymal chondrosarcoma. A case report. Cancer Genet Cytogenet. 1996;86:170–173. doi: 10.1016/0165-4608(95)00200-6. [DOI] [PubMed] [Google Scholar]
- 16.Wang L, Motoi T, Khanin R, et al. Identification of a novel, recurrent HEY1-NCOA2 fusion in mesenchymal chondrosarcoma based on a genome-wide screen of exon-level expression data. Genes Chromosomes Cancer. 2012;51:127–139. doi: 10.1002/gcc.20937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mandahl N. Methods in solid tumour cytogenetics. In: Rooney DE, editor. Human Cytogenetics: Malignancy and Acquired Abnormalities. Oxford University Press; New York: 2001. pp. 165–203. [Google Scholar]
- 18.Schaffer LG, Slovak ML, Campbell LJ, editors. ISCN 2009: An International System for Human Cytogenetic Nomenclature. Karger; Basel: 2009. [Google Scholar]
- 19.Nakayama R, Miura Y, Ogino J, et al. Detection of HEY1-NCOA2 fusion by fluorescence in-situ hybridization in formalin-fixed paraffin-embedded tissues as a possible diagnostic tool for mesenchymal chondrosarcoma. Pathol Int. 2012;62:823–826. doi: 10.1111/pin.12022. [DOI] [PubMed] [Google Scholar]
- 20.Carapeti M, Aguiar RC, Goldman JM, Cross NC. A novel fusion between MOZ and the nuclear receptor coactivator TIF2 in acute myeloid leukemia. Blood. 1998;91:3127–3133. [PubMed] [Google Scholar]
- 21.Carapeti M, Aguiar RC, Watmore AE, Goldman JM, Cross NC. Consistent fusion of MOZ and TIF2 in AML with inv(8)(p11q13) Cancer Genet Cytogenet. 1999;113:70–72. doi: 10.1016/s0165-4608(99)00007-2. [DOI] [PubMed] [Google Scholar]
- 22.Strehl S, Nebral K, Konig M, et al. ETV6-NCOA2: a novel fusion gene in acute leukemia associated with coexpression of T-lymphoid and myeloid markers and frequent NOTCH1 mutations. Clin Cancer Res. 2008;14:977–983. doi: 10.1158/1078-0432.CCR-07-4022. [DOI] [PubMed] [Google Scholar]
- 23.Sumegi J, Streblow R, Frayer RW, et al. Recurrent t(2;2) and t(2;8) translocations in rhabdomyosarcoma without the canonical PAX-FOXO1 fuse PAX3 to members of the nuclear receptor transcriptional coactivator family. Genes Chromosomes Cancer. 2010;49:224–236. doi: 10.1002/gcc.20731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arbajian E, Magnusson L, Mertens F, Domanski HA, Vult von Steyern F, Nord KH. A novel GTF2I/NCOA2 fusion gene emphasizes the role of NCOA2 in soft tissue angiofibroma development. Genes Chromosomes Cancer. 2013;52:330–331. doi: 10.1002/gcc.22033. [DOI] [PubMed] [Google Scholar]
- 25.Jin Y, Möller E, Nord KH, et al. Fusion of the AHRR and NCOA2 genes through a recurrent translocation t(5;8)(p15;q13) in soft tissue angiofibroma results in upregulation of aryl hydrocarbon receptor target genes. Genes Chromosomes Cancer. 2012;51:510–520. doi: 10.1002/gcc.21939. [DOI] [PubMed] [Google Scholar]
- 26.Mosquera JM, Sboner A, Zhang L, et al. Recurrent NCOA2 gene rearrangements in congenital/infantile spindle cell rhabdomyosarcoma. Genes Chromosomes Cancer. 2013;52:538–550. doi: 10.1002/gcc.22050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deguchi K, Ayton PM, Carapeti M, et al. MOZ-TIF2-induced acute myeloid leukemia requires the MOZ nucleosome binding motif and TIF2-mediated recruitment of CBP. Cancer Cell. 2003;3:259–271. doi: 10.1016/s1535-6108(03)00051-5. [DOI] [PubMed] [Google Scholar]
- 28.Zhuravleva J, Paggetti J, Martin L, et al. MOZ/TIF2-induced acute myeloid leukaemia in transgenic fish. Br J Haematol. 2008;143:378–382. doi: 10.1111/j.1365-2141.2008.07362.x. [DOI] [PubMed] [Google Scholar]
- 29.Steidl C, Leimeister C, Klamt B, et al. Characterization of the human and mouse HEY1, HEY2, and HEYL genes: cloning, mapping, and mutation screening of a new bHLH gene family. Genomics. 2000;66:195–203. doi: 10.1006/geno.2000.6200. [DOI] [PubMed] [Google Scholar]
- 30.Maier MM, Gessler M. Comparative analysis of the human and mouse Hey1 promoter: Hey genes are new Notch target genes. Biochem Biophys Res Commun. 2000;275:652–660. doi: 10.1006/bbrc.2000.3354. [DOI] [PubMed] [Google Scholar]