Abstract
Aberrant NF-κB activation is frequently observed in human cancers. Genome characterization efforts have identified genetic alterations in multiple components of the NF-κB pathway, some of which have been shown to be essential for cancer initiation and tumor maintenance. Here using patient tumors and cancer cell lines, we identify the NF-κB regulator, TRAF2 as an oncogene that is recurrently amplified and rearranged in 15% of human epithelial cancers. Suppression of TRAF2 in cancer cells harboring TRAF2 copy number gain inhibits proliferation, NF-κB activation, anchorage-independent growth and tumorigenesis. Cancer cells that are dependent on TRAF2 also require NF-κB for survival. The phosphorylation of TRAF2 at serine 11 is essential for the survival of cancer cells harboring TRAF2 amplification. Together these observations identify TRAF2 as a frequently amplified oncogene.
Keywords: TRAF2, NF-κB, cancer, 9q34 amplification
Introduction
Nuclear Factor kappa B (NF-κB) transcription factors play pivotal roles in immunity, inflammation, cell differentiation, proliferation and survival. In addition to its roles in immunity, constitutive NF-κB activity is frequently detected in both hematopoietic and epithelial cancers (1). In tumor cells, activation of NF-κB occurs in response to inflammatory stimuli within a tumor microenvironment, and cancers associated with chronic inflammation are dependent on NF-κB (2–7).
Cancer genome characterization efforts have identified alterations in many components of the NF-κB pathway. Translocations and mutations of NF-κB regulators have been identified in several different cancer types. In particular, CARD11 is both mutated and amplified in diffuse large B cell lymphomas, and CYLD and A20 are tumor suppressor genes deleted in familial cylindromatosis and marginal zone B cell lymphomas, respectively (8–12). Other NF-κB components such as CD40, NIK, NFKB1, NFKB2 are amplified in multiple myeloma (8, 13–15). In solid tumors, amplification, somatic mutations, chromosomal translocations of IKBKA, IKBKE and IKBKB are observed in breast and prostate cancers respectively (16–18). Moreover, NF-κB activity is essential in KRas-driven lung and pancreatic cancer progression that occur in a p53-deficient background (19–22). Similarly, TRAF6 is an amplified oncogene present in non-small cell lung cancers with activated RAS (23), and loss of the tumor suppressor DAB2IP contributes to prostate cancer progression in part through activating NF-κB signaling (24). These observations implicate aberrant NF-κB signaling in the initiation or progression of many types of human cancers.
TRAF2 is an adaptor molecule that assembles active NF-κB signaling scaffolds. After TNF receptor engagement, TRAF2 forms multimeric complexes with several intracellular proteins including CIAP1, RIPK, TANK, and TAK1, initiating a kinase cascade that activates NF-κB and JNK (25, 26). One key function of TRAF2 is to facilitate Lys63 ubiquitination of components in these scaffolds (27). TRAF2-mediated Lys63 ubiquitination is essential for the recruitment of the canonical IKK complex, the central mediator of NF-κB activation.
Several studies suggest that TRAF2 plays an important role in cancer. In Ras-transformed cells, TRAF2 promotes resistance to stress-induced apoptosis (28). Similarly, TRAF2 also facilitates resistance to MAPK pathway inhibitors in BRAF V600E mutant melanoma (29). We recently identified TRAF2 as a substrate of the IKKε breast oncogene (30). IKKε phosphorylates TRAF2 at Ser11 to activate NF-κB and promote malignant transformation. Here we report that TRAF2 is amplified in a substantial fraction of human epithelial cancers where it functions independently of IKKε to induce tumorigenicity.
Results
TRAF2 is amplified in a substantial fraction of human epithelial cancers
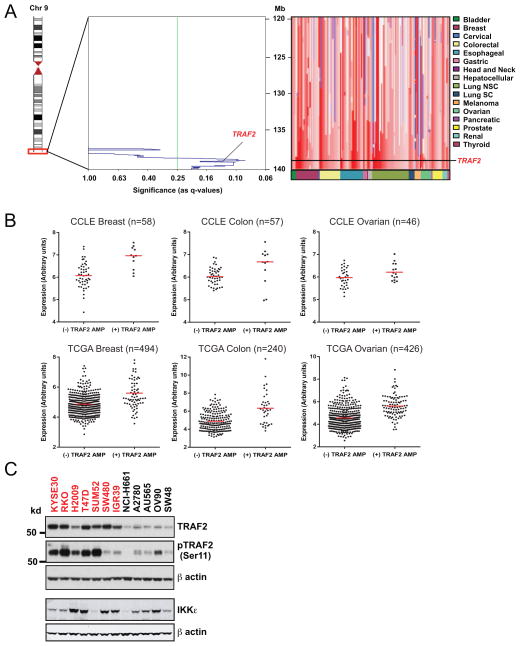
In prior work, we identified TRAF2 and the tumor suppressor CYLD as key effectors in IKKε driven tumorigenesis in breast cancer (30, 31). We found that expression of TRAF2 could replace IKKε to confer anchorage independent growth in NIH3T3 cells and immortalized human embryonic kidney cells (HA1EM) in a manner that is dependent on TRAF2 Ser11 phosphorylation, an activity that promotes NF-κB activation (Supplementary Figure 1A). To determine whether genetic alterations involving TRAF2 occur in human cancers, we analyzed genome-wide somatic copy number alterations in 3131 cancer samples including 2520 carcinomas and 611 cancer cell lines (32). We identified a focal region of recurrent amplification (9q34) that encompasses the TRAF2 locus. We found increased copy number of TRAF2 in 15.1% of epithelial cancers and 13.1% of all human cancers across multiple tissue types including breast, lung, colorectal, gastric, melanoma, ovarian, and esophageal cancers (Figure 1A). In contrast to broad regions of amplification that include more than half of the chromosome arm, 9q34 is significantly amplified (q = 0.11) across all lineages and TRAF2 lies within a peak region containing genes most likely to be the targets of these amplifications (32). To validate this finding, we performed FISH on a panel of cancer cell lines using a TRAF2-specific fosmid probe and confirmed increased TRAF2 copy number in six cancer cell lines classified by GISTIC as harboring a 9q34 amplification (RKO, KYSE30, KYSE510, MDA-MB-453, H2009, SUM52) in comparison to two copy neutral cell lines (A2780, AU565) (Table 1). We further observed that TRAF2 is rearranged to alternative chromosomes in the six cancer cell lines that harbor TRAF2 amplification, and one additional cell line (MCF7) without TRAF2 amplification (Supplementary Figure 2A). These observations suggest that 9q34 amplification and rearrangement drives TRAF2 dysregulation in a subset of human cancers.
Figure 1. TRAF2 is amplified in human cancers.
(A) Copy number profiles at the 9q34 locus. Left panel, Significance of TRAF2 amplifications across 3131 cancer samples was determined by GISTIC (genomic identification of significant targets in cancer) and shown as q-values (false discovery rate corrected significance of amplification frequency). Right panel, copy number profiles for 50 cancer samples harboring TRAF2 amplifications. Genomic location and the TRAF2 locus are indicated on the vertical axis and lineages on the horizontal axis are denoted by color. Copy number gain and loss are indicated as red and blue signals respectively. (B) Scatterplots of TRAF2 mRNA expression in TRAF2 amplified or non-amplified primary breast, ovarian, and colon tumors in TCGA (33–35) and cell lines in CCLE (43). These data are log2-transformed signal intensities with the median of each sample set denoted by a red line. (C) Immunoblot of TRAF2 and Ser11 phosphorylated TRAF2, and IKKε in 9q34 amplified (red) and copy neutral (black) cell lines. β-actin is displayed as a loading control.
Table 1.
FISH analysis of TRAF2 in cancer cell lines
| Cell line | TRAF2 Copy Number | 9q34 Amplification | 9q34 Rearrangement |
|---|---|---|---|
| KYSE30 | 4.1 | + | + |
| RKO | 3.8 | + | + |
| MDA-MB-453 | 3.7 | + | + |
| H2009 | 2.7 | + | + |
| SUM52 | 2.6 | + | + |
| MCF7 | 2.5 | − | + |
| KYSE510 | 2.3 | + | + |
| A2780 | 2.0 | − | − |
| AU565 | 1.8 | − | − |
To determine whether 9q34 amplification influences TRAF2 expression, we analyzed 9q34 copy number and transcript levels in both the Cancer Cell Line Encyclopedia (CCLE) and The Cancer Genome Atlas (TCGA) datasets (33–35). We determined that median TRAF2 expression levels were increased in breast, ovarian and colon samples with 9q34 amplification in comparison to samples without 9q34 amplification in both the TCGA and CCLE data sets (Figure 1B). In contrast, samples that harbor 9q34 amplification did not exhibit increased IKKε expression in comparison to non-amplified samples, suggesting that TRAF2 amplification is unrelated to IKKε expression or dependency (Supplementary Figure 2B). Comparative marker selection analysis using the CCLE collection of cell lines with matched copy number and expression data likewise revealed that TRAF2 transcript levels scored as the 5th most highly correlated with 9q34 amplification among 18,988 transcripts (Supplementary Table S1). We then profiled TRAF2 expression in a panel of cancer cell lines and found that TRAF2 protein levels were elevated in all cell lines harboring 9q34 amplification in comparison to cell lines lacking this amplification (Figure 1C). TRAF2-amplified cell lines also exhibited higher levels of TRAF2 Ser11 phosphorylation, suggesting that 9q34 gain facilitates both TRAF2 expression and activation (30, 36). IKKε expression levels and dependency, however, did not correlate with TRAF2-amplification in these cell lines, suggesting that IKKε-independent mechanisms also facilitate TRAF2 amplification, overexpression and activation (Figure 1C, Supplementary Figure 1B). Combined, these findings indicate that TRAF2 is frequently altered and overexpressed in cancers harboring 9q34 amplification and implicate a role for TRAF2 as a driver oncogene in 9q34 amplified cancers independently of IKKε expression or amplification.
9q34 amplification confers TRAF2 dependency
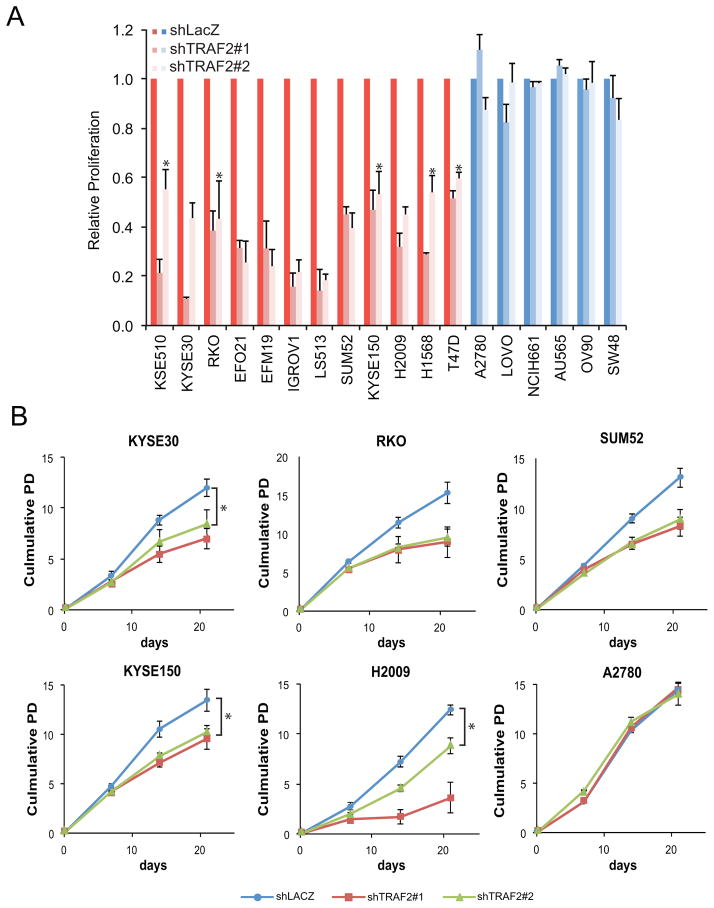
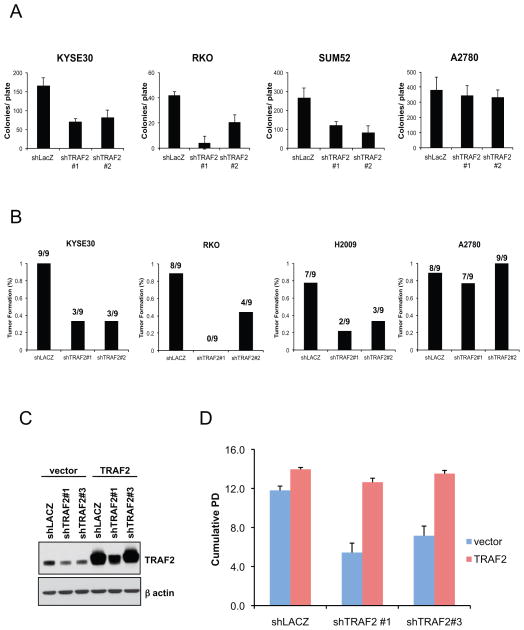
We next determined whether cell lines that harbor increased copy number of TRAF2 depend on TRAF2 for proliferation and/or survival. We suppressed TRAF2 expression in a panel of eighteen cancer cell lines derived from various lineages that either harbor or lack TRAF2 copy number gain and measured proliferation, apoptosis, and anchorage-independent growth (Supplementary Figure 3). Twelve cell lines (KYSE510, KYSE30, RKO, EFO21, EFM19, IGROV1, LS513, SUM52, KYSE150, H2009, H1568, and T47D) with increased TRAF2 copy number exhibited decreased proliferative potential in a 7 day proliferation assay when we suppressed TRAF2 with two distinct TRAF2-specific shRNA (Figure 2A, Supplementary Figure 3). In contrast, suppression of TRAF2 failed to affect the proliferation of cell lines (A2780, LOVO, NCI-H661, AU565, OV90, and SW48) that lacked TRAF2 amplification (Figure 2A). In long term proliferation assays, we found that suppression of TRAF2 in cells harboring increased TRAF2 copy number (KYSE30, RKO, SUM52, KYSE150, H2009, led to a mean 41.6% decrease in the doubling time of such cells (Figure 2B). In contrast, we failed to detect evidence of apoptosis after TRAF2 suppression (Supplementary Figure 4). Depletion of TRAF2 in three cell lines that exhibited delayed proliferative capacity (KYSE30, RKO, SUM52) also inhibited anchorage-independent growth (Figure 3A). We further found that TRAF2 suppression in three 9q34 amplified cell lines, KYSE30, RKO and H2009 cells inhibited tumorigenesis in immunodeficient mice (Figure 3B).
Figure 2. Cancer cells with TRAF2 amplification depend on TRAF2 for proliferation.
(A) Proliferation of cells harboring (red) and lacking (blue) TRAF2 copy number gain after transduction with two distinct TRAF2-specific shRNA (shTRAF2#1 and shTRAF#2). Proliferation was normalized to cells expressing control shRNA (shLacZ). Results reported as mean ± SD of three experiments. * p<= 0.02 as calculated by a standard t test (B) Long-term proliferative capacity of cell lines following TRAF2 suppression. Population doubling (PD) of cells harboring (KYSE30, RKO, SUM52, KYSE50, and H2009) and lacking (A2780) TRAF2 copy number gain after transduction with control shRNA (shLACZ), shTRAF2#1, or shTRAF2#2. Cells were assayed for 21d. * p<= 0.04 as calculated by a standard t test.
Figure 3. TRAF2 is essential for tumorigenicity in cancer cells with TRAF2 amplification.
(A) Anchorage-independent growth of cell lines harboring (KYSE30, RKO, SUM52) and lacking (A2780) TRAF2 copy number gain. Cells were transduced with control shRNA (shLACZ) or two distinct TRAF2-specific shRNA (shTRAF2#1 and shTRAF2#2). Colony formation was measured after 21 d. p values were calculated by a standard t-test. (B) Tumorigenesis of cancer cells following TRAF2 suppression. Indicated cells transduced with a control shRNA (shLACZ), shTRAF2 #1 or shTRAF2#2 were subcutaneously introduced into immunodeficient mice. Tumor formation was assessed after 21 d. (C) TRAF2 expression in MCF7 cells transduced to coexpress either control (shLACZ), shTRAF2#1 or a UTR-specific shTRAF2 (shTRAF2 #3) with TRAF2 or control (vector). (D) Long term proliferative capacity of MCF7 cells after TRAF2 suppression and overexpression. Cumulative population doubling (PD) of MCF7 cells from (C) 21d after transduction.
To confirm that the proliferative defect induced by our TRAF2-directed shRNAs was due to specific TRAF2 suppression, we transduced TRAF2-dependent cells with both an shRNA targeting the 3′UTR of TRAF2 (shTRAF2 #3) and a TRAF2 cDNA lacking the 3′ UTR. In both RKO and MCF7 cells, forced expression of TRAF2 restored TRAF2 protein levels and rescued the proliferative defect (Figure 3C, 3D and 4F). Combined, these findings demonstrate that epithelial cancers with 9q34 amplification depend on aberrant TRAF2 expression.
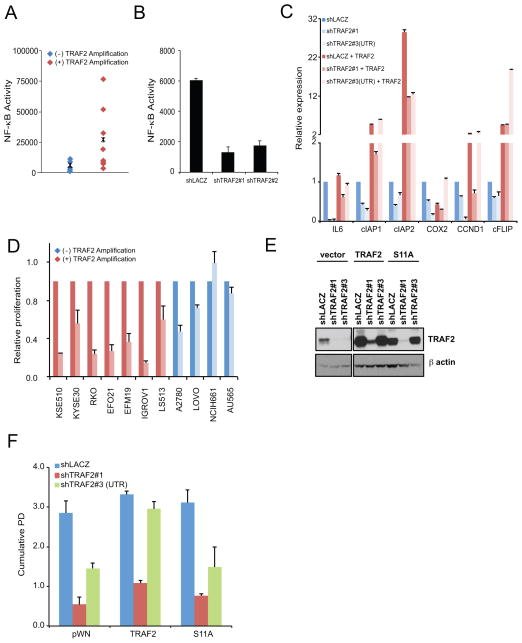
Figure 4. TRAF2 activates NF-κB in cancer cells with TRAF2 amplification.
(A) NF-κB activity in cancer cell lines with (RKO, KYSE30, H2009, SUM52, T47D, LS513, EFO21, EFM19) or without (A2780, NCIH661, OV90, SW48, BT474) TRAF2 amplification. Cell lines were transduced with a stable NF-κB luciferase reporter and raw light united (RLU) activity was measured and normalized to baseline viability activity (B) NF-κB activity in RKO cells after TRAF2 suppression. Stable NF-κB reporter RKO cells were transduced as indicated with TRAF2-specific shRNA and luciferase raw light unit (RLU) activity was measured and normalized to activity observed with control vector. Results reported as mean ± SD of three experiments. (C) NF-κB target gene expression in RKO cells following expressing TRAF2 specific shRNA (TRAF2#1 or shTRAF2 #3(UTR)) specific shTRAF2 (shTRAF2#3) with TRAF2 or control (vector). Relative expression was quantified by RT-PCR using ΔΔCT and normalized to levels observed with control (V5 and shLACZ) cells. (D) Proliferative capacity of cell lines after NF-κB inhibition. Cells line with (red) and without (blue) TRAF2 amplification were transduced with the NF-κB super-repressor and proliferation was normalized to cells expressing control vector (Dark colors). Results reported as mean ± SD of three experiments. (E) TRAF2 expression in RKO cells coexpressing either control (shLACZ), shTRAF2#1 or a UTR-specific shTRAF2 (shTRAF2 #3) with TRAF2, TRAF2 S11A, or control (vector). (F) Long term proliferative capacity of RKO cells after TRAF2 suppression and forced expression of TRAF2 or TRAF2 S11A. Cumulative population doubling (PD) of RKO cells from (E) 21d after transduction.
TRAF2 dependency is conferred through NF-κB activation
TRAF2 promotes NF-κB activation through its ability to facilitate recruitment and activation of the canonical IKK complex (37). To assess the consequences of TRAF2 amplifications on NF-κB activity, we stably expressed a NF-κB reporter in cell lines that do and do not harbor 9q34 amplification. In comparison to copy-neutral cell lines, the mean NF-κB activity was higher in the TRAF2 amplified cells by 5.3 fold (Figure 4A). Suppression of TRAF2 in RKO and cells also resulted in decreased NF-κB activity as measured by the stable NF-κB reporter (Figure 4B). We further evaluated the expression of several NF-κB target genes including IL6, CIAP1, CIAP2, CCND1, and cFLIP, and observed that the expression of these genes were decreased in RKO cells. Under conditions where TRAF2 overexpression rescues TRAF2 dependency, we found that TRAF2 expression restored the expression of these NF-κB target genes (Figure 4C).
To determine whether TRAF2 dependent cell lines are also dependent on canonical NF-κB activation, we introduced a dominant interfering allele of IκBα (IκBα super repressor) in cells with and without 9q34 copy number gain and assessed cell proliferation. We found that TRAF2 dependent cell lines were particularly sensitive to inhibition of NF-κB activity, as proliferation was inhibited by up to 7-fold in comparison to control cells (Figure 4). We note that we also found cell lines that lacked the 9q34 amplification, which depended on NF-κB, suggesting that there are other TRAF2-independent mechanisms that activate NF-κB signaling.
In prior work, we found that TRAF2 phosphorylation at Serine 11 promotes Lys63 ubiquitination, recruitment of the IKK complex and downstream NF-κB activation necessary for tumorigenesis (30). Since we observed elevated TRAF2 Ser11 phosphorylation in 9q34 amplified cells, we assessed whether overexpression of a TRAF2 S11A mutant is sufficient to rescue the proliferative defect induced by TRAF2 suppression. We overexpressed both wildtype TRAF2 and the TRAF2 S11A mutant in RKO cells prior to TRAF2 depletion with shRNAs (Figure 4E). We found that in contrast to wildtype TRAF2, TRAF2 S11A failed to rescue the proliferative defect after TRAF2 depletion (Figure 4F). Collectively, these observations provide evidence that TRAF2 is an oncogene that promotes NF-κB activation that is essential in human cancers that harbor 9q34 amplification.
Discussion
Here we identified TRAF2 as a bona fide oncogene that is essential for the proliferation and transformation of several types of epithelial cancer cell lines. By analyzing more than 3000 primary tumor samples, we found recurrent amplifications at 9q34 involving TRAF2 that are present in 15% of human epithelial cancers across multiple lineages. Although other candidate oncogenic driver genes may be present at 9q34, we demonstrated that human cancer cell lines harboring TRAF2 amplifications are dependent on TRAF2 expression as suppression of TRAF2 in these cells inhibited proliferative capacity, anchorage independent growth, and tumorigenesis. Moreover, although we previously showed that TRAF2 expression is required for IKKε to transform mammary epithelial cells (30), TRAF2 promotes transformation in NIH3T3 cells and immortalized HA1EM cells independently of IKKε. Taken together, these observations support the notion that TRAF2 is an oncogene.
Our observations provide evidence that TRAF2 can contribute to cell transformation in two ways. As a substrate of IKKε in cancer cells that harbor amplification or overexpression of IKKε, TRAF2 is required to mediate the activation of NF-κB signaling induced by IKKε amplification (30). Here we demonstrate that in other tumors where 9q34 is amplified, TRAF2 dysregulation occurs as a consequence of increased copy number of TRAF2. We observed increased TRAF2 Ser11 phosphorylation in cells that harbor TRAF2 amplifications. Although such cells do not always exhibit increased IKKε expression, the basal levels of IKKε may suffice to induce this phosphorylation. Alternatively, other kinases may also induce TRAF2 phosphorylation in cells that harbor TRAF2 amplifications (30, 36, 38). Thus, TRAF2 induces cell transformation through both IKKε-dependent and IKKε-independent mechanisms.
Genetic alterations of the NF-κB pathway have recently emerged in different cancer types (39). In addition to TRAF2 amplifications identified here, TRAF2 mutations are detected in diffuse large cell lymphoma, multiple myeloma and marginal zone lymphomas (8, 11, 14). More recently, focal of amplifications of TRAF6, a structural and functional counterpart of TRAF2, were identified in non-small cell lung cancer (23). Like TRAF2, TRAF6 overexpression induces malignant transformation and TRAF6 depletion inhibits lung cancer cell proliferation and tumorigenesis. We also identified lung cancer cell lines that exhibit TRAF2 dependency, suggesting that TRAF2 may share a similar role as TRAF6 in driving the pathogenesis of non-small cell lung cancers.
TRAF2 regulation and activation involves a dynamic interplay of multiple postranslational events. Phosphorylation of TRAF2 occurs at multiple residues including Thr117, Ser55 and Ser11 and is mediated by PKC and IKKε kinases respectively (30, 36, 38). Both PKC and IKKε-induced phosphorylation of TRAF2 results in Lys63-ubiquitination of TRAF2 required for recruitment and activation of the IKK complex (30, 36, 38). We demonstrated that TRAF2-dependent cancer cells have increased levels of TRAF2 Ser11 phosphorylation, suggesting that IKKε is important for TRAF2 transformation. Coincidentally, TRAF2 promotes Lys63 ubiquitination of IKKε, and both TRAF2 Ser11 phosphorylation and IKKε Lys63-ubiquitination are required for IKKε oncogenesis in breast cancer (40). This feed-forward regulation suggests a co-dependency between TRAF2 and IKKε in cancer cells with either TRAF2 or IKKε amplification.
Since TRAF2 plays a key role in the assembly of protein complexes necessary for canonical NF-κB activation, TRAF2 amplification and overexpression would naturally lead to constitutive NF-κB activation. Upon activation, TRAF2 itself undergoes Lys63 ubiquitination and also recruits CIAP1, RIPK, TANK, and TAK1 (25, 26). This activity is a prerequisite for the phosphorylation and activation of the IKK complex, which is essential for the removal of IκBα, the central inhibitor of NF-κB transcription factors. TRAF2-amplified cells are dependent on both TRAF2 and NF-κB activation for proliferation. Thus, dysregulation of TRAF2 by increased copy number facilitates a continuous proliferative signal through the release of NF-κB.
We demonstrate that TRAF2 is essential for the proliferation of many epithelial cancers that harbor TRAF2 amplification. TRAF2-dependent cancer cells also appear to be dependent on NF-κB activity, suggesting that TRAF2 amplification and overexpression may underscore constitutive NF-κB activity in carcinomas. Identification of TRAF2 dependency may therefore provide a targeted therapy in NF-κB activated or 9q34 amplified cancers.
Materials and Methods
Antibodies and plasmids
The antibodies used include: HSP90, Lamin A/C, p50, p52/p100, TRAF2 (Cell Signaling Technologies, Danvers, MA, USA), p65 (Abcam, Cambridge, MA, USA), and β actin (Sigma-Aldrich, St. Louis, MO, USA). The phospho-TRAF2 (Ser11) was a gift from Dr. Hasem Habelhah.
pWZL-TRAF2, pLEX-V5-TRAF2, pLKO-shLACZ, pLKO-shTRAF2#1 and pLKO-shTRAF2#2 lentiviral constructs are previously described(30). Additional shRNA constructs were obtained from the RNAi Consortium (Broad Institute, Cambridge, MA, USA) and include pLKO-shTRAF2#3 (TRCN0000004572). The plasmid for the NF-κB super-repressor (pWZL-IκBαMUT) was previously described (18).
Cell Culture, Transfection, Subcellular Fractionation
HEK293T, MCF7, MDA-MB-453 and RKO cells were obtained from ATCC (Manassas, VA, USA) and were grown in DMEM containing 10% FBS. A2780, EFM19, EFO21, H1568, H2009, IGROV1, KYSE30, KYSE510, LOVO, LS513, NCIH661, OV90, SW48, and T47D were obtained from ATCC (Manassas, VA, USA) and maintained in RPMI1640 containing 10% FBS. KYSE150 were maintained in RPMI1640:HamsF-12 (1:1) with 2% FBS. SUM52 cells were maintained in HamsF-12:MEGM (1:1) with 10% FBS. Transfection experiments were performed using Fugene (Roche, Indianapolis, IN, USA). Subcellular fractionation experiments were performed as previously described (19).
FISH analysis
Fluorescent in situ hybridization (FISH) analysis is previously described (41) and was performed using a TRAF2 specific fosmid probes (PR11-769N4) and a Chromosome 9 centromeric reference probe on metaphase spreads of indicated cell lines.
NF-κB reporter assays and quantitative RT-PCR
For NF-κB luciferase reporter assays, cell lines were transduced with a lentiviral construct containing a 10xNF_B response element cloned into the PstI and NheI sites of the previously described 7TFP reporter. (42) NF-κB activity was measured using the Dual-Glo Luciferase assay (Promega, Madison, WI, USA) 4 days post-transduction. Luciferase values were normalized to CTG values to yield relative NF-κB activity. Quantitative RT-PCR for NF-κB target genes was conducted as described (18).
Tumorigenicity assay
2 × 106 cells were subcutaneously implanted into immunodeficient mice (Balb/c Nude, Charles River Laboratories, Wilmington, MA USA) anesthetized with isofluorane. Three mice were used per group, and three implantation sites were made per mouse. Tumors were measured at 21 d after implantation.
Viability and proliferation measurements
Relative proliferation was measured 7d post-infection using Cell-Titer Glo (Promega, Madison, WI, USA) in triplicate. Apoptosis was assessed with Annexin V/ PI staining (BD, Franklin Lakes, NJ, USA) and flow cytometric analysis 6d post–infection according to the manufacturer’s protocol. Additional proliferation assays were performed my measuring duplicate population doubling (PD) using a Vi-Cell counter every 7d for 21 d. PD were defined as [log2(cells counted/cells plated)].
Supplementary Material
Acknowledgments
We thank member of the Hahn lab and the Cichowski lab for thoughtful discussion, reagents, and technical assistance. We also thank Hasem Habelhah for providing the pTRAF2 (Ser11) antibody. We further thank Shumei Wang, Anita Hawkins, Chengzeng Zhang and Cynthia Morton at the DFCI Cytogenetics Core for TRAF2 FISH analysis and discussion. This work was supported in part by R01 CA130988 (W.C.H.), a Ruth L. Kirschstein National Research Service Award F32 CA128265 (R.R.S.) and The Aid for Cancer Research (R.R.S.).
Footnotes
Conflict of interest
RB and WCH are consultants for Novartis.
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc)
References
- 1.Baud V, Karin M. Is NF-kappaB a good target for cancer therapy? Hopes and pitfalls. Nat Rev Drug Discov. 2009;8(1):33–40. doi: 10.1038/nrd2781. Epub 2009/01/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Greten FR, Eckmann L, Greten TF, Park JM, Li ZW, Egan LJ, et al. IKKbeta links inflammation and tumorigenesis in a mouse model of colitis-associated cancer. Cell. 2004;118(3):285–96. doi: 10.1016/j.cell.2004.07.013. Epub 2004/08/06. [DOI] [PubMed] [Google Scholar]
- 3.Karin M. Nuclear factor-kappaB in cancer development and progression. Nature. 2006;441(7092):431–6. doi: 10.1038/nature04870. Epub 2006/05/26. [DOI] [PubMed] [Google Scholar]
- 4.Karin M, Greten FR. NF-kappaB: linking inflammation and immunity to cancer development and progression. Nat Rev Immunol. 2005;5(10):749–59. doi: 10.1038/nri1703. Epub 2005/09/22. [DOI] [PubMed] [Google Scholar]
- 5.Pikarsky E, Porat RM, Stein I, Abramovitch R, Amit S, Kasem S, et al. NF-kappaB functions as a tumour promoter in inflammation-associated cancer. Nature. 2004;431(7007):461–6. doi: 10.1038/nature02924. Epub 2004/08/27. [DOI] [PubMed] [Google Scholar]
- 6.Aggarwal BB, Sung B. NF-kappaB in cancer: a matter of life and death. Cancer Discov. 2011;1(6):469–71. doi: 10.1158/2159-8290.CD-11-0260. Epub 2012/05/16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwitalla S, Fingerle AA, Cammareri P, Nebelsiek T, Goktuna SI, Ziegler PK, et al. Intestinal tumorigenesis initiated by dedifferentiation and acquisition of stem-cell-like properties. Cell. 2013;152(1–2):25–38. doi: 10.1016/j.cell.2012.12.012. Epub 2013/01/01. [DOI] [PubMed] [Google Scholar]
- 8.Compagno M, Lim WK, Grunn A, Nandula SV, Brahmachary M, Shen Q, et al. Mutations of multiple genes cause deregulation of NF-kappaB in diffuse large B-cell lymphoma. Nature. 2009;459(7247):717–21. doi: 10.1038/nature07968. Epub 2009/05/05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kovalenko A, Chable-Bessia C, Cantarella G, Israel A, Wallach D, Courtois G. The tumour suppressor CYLD negatively regulates NF-kappaB signalling by deubiquitination. Nature. 2003;424(6950):801–5. doi: 10.1038/nature01802. Epub 2003/08/15. [DOI] [PubMed] [Google Scholar]
- 10.Lenz G, Davis RE, Ngo VN, Lam L, George TC, Wright GW, et al. Oncogenic CARD11 mutations in human diffuse large B cell lymphoma. Science. 2008;319(5870):1676–9. doi: 10.1126/science.1153629. Epub 2008/03/08. [DOI] [PubMed] [Google Scholar]
- 11.Novak U, Rinaldi A, Kwee I, Nandula SV, Rancoita PM, Compagno M, et al. The NF-{kappa}B negative regulator TNFAIP3 (A20) is inactivated by somatic mutations and genomic deletions in marginal zone lymphomas. Blood. 2009;113(20):4918–21. doi: 10.1182/blood-2008-08-174110. Epub 2009/03/05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brummelkamp TR, Nijman SM, Dirac AM, Bernards R. Loss of the cylindromatosis tumour suppressor inhibits apoptosis by activating NF-kappaB. Nature. 2003;424(6950):797–801. doi: 10.1038/nature01811. Epub 2003/08/15. [DOI] [PubMed] [Google Scholar]
- 13.Annunziata CM, Davis RE, Demchenko Y, Bellamy W, Gabrea A, Zhan F, et al. Frequent engagement of the classical and alternative NF-kappaB pathways by diverse genetic abnormalities in multiple myeloma. Cancer cell. 2007;12(2):115–30. doi: 10.1016/j.ccr.2007.07.004. Epub 2007/08/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keats JJ, Fonseca R, Chesi M, Schop R, Baker A, Chng WJ, et al. Promiscuous mutations activate the noncanonical NF-kappaB pathway in multiple myeloma. Cancer cell. 2007;12(2):131–44. doi: 10.1016/j.ccr.2007.07.003. Epub 2007/08/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kan Z, Jaiswal BS, Stinson J, Janakiraman V, Bhatt D, Stern HM, et al. Diverse somatic mutation patterns and pathway alterations in human cancers. Nature. 2010;466(7308):869–73. doi: 10.1038/nature09208. Epub 2010/07/30. [DOI] [PubMed] [Google Scholar]
- 16.Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458(7239):719–24. doi: 10.1038/nature07943. Epub 2009/04/11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pflueger D, Terry S, Sboner A, Habegger L, Esgueva R, Lin PC, et al. Discovery of nonETS gene fusions in human prostate cancer using next-generation RNA sequencing. Genome research. 2011;21(1):56–67. doi: 10.1101/gr.110684.110. Epub 2010/11/03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boehm JS, Zhao JJ, Yao J, Kim SY, Firestein R, Dunn IF, et al. Integrative genomic approaches identify IKBKE as a breast cancer oncogene. Cell. 2007;129(6):1065–79. doi: 10.1016/j.cell.2007.03.052. Epub 2007/06/19. [DOI] [PubMed] [Google Scholar]
- 19.Meylan E, Dooley AL, Feldser DM, Shen L, Turk E, Ouyang C, et al. Requirement for NF-kappaB signalling in a mouse model of lung adenocarcinoma. Nature. 2009;462(7269):104–7. doi: 10.1038/nature08462. Epub 2009/10/23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Basseres DS, Ebbs A, Levantini E, Baldwin AS. Requirement of the NF-kappaB subunit p65/RelA for K-Ras-induced lung tumorigenesis. Cancer Res. 2010;70(9):3537–46. doi: 10.1158/0008-5472.CAN-09-4290. Epub 2010/04/22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barbie DA, Tamayo P, Boehm JS, Kim SY, Moody SE, Dunn IF, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462(7269):108–12. doi: 10.1038/nature08460. Epub 2009/10/23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ling J, Kang Y, Zhao R, Xia Q, Lee DF, Chang Z, et al. KrasG12D-induced IKK2/beta/NF-kappaB activation by IL-1alpha and p62 feedforward loops is required for development of pancreatic ductal adenocarcinoma. Cancer cell. 2012;21(1):105–20. doi: 10.1016/j.ccr.2011.12.006. Epub 2012/01/24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Starczynowski DT, Lockwood WW, Delehouzee S, Chari R, Wegrzyn J, Fuller M, et al. TRAF6 is an amplified oncogene bridging the RAS and NF-kappaB pathways in human lung cancer. J Clin Invest. 2011;121(10):4095–105. doi: 10.1172/JCI58818. Epub 2011/09/14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Min J, Zaslavsky A, Fedele G, McLaughlin SK, Reczek EE, De Raedt T, et al. An oncogene-tumor suppressor cascade drives metastatic prostate cancer by coordinately activating Ras and nuclear factor-kappaB. Nature medicine. 2010;16(3):286–94. doi: 10.1038/nm.2100. Epub 2010/02/16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Au PY, Yeh WC. Physiological roles and mechanisms of signaling by TRAF2 and TRAF5. Adv Exp Med Biol. 2007;597:32–47. doi: 10.1007/978-0-387-70630-6_3. Epub 2007/07/18. [DOI] [PubMed] [Google Scholar]
- 26.Pomerantz JL, Baltimore D. NF-kappaB activation by a signaling complex containing TRAF2, TANK and TBK1, a novel IKK-related kinase. EMBO J. 1999;18(23):6694–704. doi: 10.1093/emboj/18.23.6694. Epub 1999/12/03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen ZJ. Ubiquitin signalling in the NF-kappaB pathway. Nat Cell Biol. 2005;7(8):758–65. doi: 10.1038/ncb0805-758. Epub 2005/08/02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thomas GS, Zhang L, Blackwell K, Habelhah H. Phosphorylation of TRAF2 within its RING domain inhibits stress-induced cell death by promoting IKK and suppressing JNK activation. Cancer Res. 2009;69(8):3665–72. doi: 10.1158/0008-5472.CAN-08-4867. Epub 2009/04/02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wood KC, Konieczkowski DJ, Johannessen CM, Boehm JS, Tamayo P, Botvinnik OB, et al. MicroSCALE screening reveals genetic modifiers of therapeutic response in melanoma. Science signaling. 2012;5(224):rs4. doi: 10.1126/scisignal.2002612. Epub 2012/05/17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bivona TG, Hieronymus H, Parker J, Chang K, Taron M, Rosell R, et al. FAS and NF-kappaB signalling modulate dependence of lung cancers on mutant EGFR. Nature. 2011;471(7339):523–6. doi: 10.1038/nature09870. Epub 2011/03/25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hutti JE, Shen RR, Abbott DW, Zhou AY, Sprott KM, Asara JM, et al. Phosphorylation of the tumor suppressor CYLD by the breast cancer oncogene IKKepsilon promotes cell transformation. Mol Cell. 2009;34(4):461–72. doi: 10.1016/j.molcel.2009.04.031. Epub 2009/06/02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463(7283):899–905. doi: 10.1038/nature08822. Epub 2010/02/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. Epub 2012/09/25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474(7353):609–15. doi: 10.1038/nature10166. Epub 2011/07/02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–7. doi: 10.1038/nature11252. Epub 2012/07/20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blackwell K, Zhang L, Thomas GS, Sun S, Nakano H, Habelhah H. TRAF2 phosphorylation modulates tumor necrosis factor alpha-induced gene expression and cell resistance to apoptosis. Mol Cell Biol. 2009;29(2):303–14. doi: 10.1128/MCB.00699-08. Epub 2008/11/05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karin M, Gallagher E. TNFR signaling: ubiquitin-conjugated TRAFfic signals control stop-and-go for MAPK signaling complexes. Immunol Rev. 2009;228(1):225–40. doi: 10.1111/j.1600-065X.2008.00755.x. Epub 2009/03/18. [DOI] [PubMed] [Google Scholar]
- 38.Li S, Wang L, Dorf ME. PKC phosphorylation of TRAF2 mediates IKKalpha/beta recruitment and K63-linked polyubiquitination. Mol Cell. 2009;33(1):30–42. doi: 10.1016/j.molcel.2008.11.023. Epub 2009/01/20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Staudt LM. Oncogenic activation of NF-kappaB. Cold Spring Harb Perspect Biol. 2010;2(6):a000109. doi: 10.1101/cshperspect.a000109. Epub 2010/06/03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou AY, Shen RR, Kim E, Lock YJ, Xu M, Chen ZJ, et al. IKKepsilon-mediated tumorigenesis requires K63-linked polyubiquitination by a cIAP1/cIAP2/TRAF2 E3 ubiquitin ligase complex. Cell reports. 2013;3(3):724–33. doi: 10.1016/j.celrep.2013.01.031. Epub 2013/03/05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bassing CH, Suh H, Ferguson DO, Chua KF, Manis J, Eckersdorff M, et al. Histone H2AX: a dosage-dependent suppressor of oncogenic translocations and tumors. Cell. 2003;114(3):359–70. doi: 10.1016/s0092-8674(03)00566-x. Epub 2003/08/14. [DOI] [PubMed] [Google Scholar]
- 42.Fuerer C, Nusse R. Lentiviral vectors to probe and manipulate the Wnt signaling pathway. PloS one. 2010;5(2):e9370. doi: 10.1371/journal.pone.0009370. Epub 2010/02/27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483(7391):603–7. doi: 10.1038/nature11003. Epub 2012/03/31. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.