Abstract
Rac1-GTPases serve as intermediary cellular switches which conduct transient and constitutive signals from upstream cues, including those from Ras oncoproteins. While the sirtuin1 (SIRT1) deacetylase is overexpressed in several human cancers and has recently been linked to cancer cell motility as a context-dependent regulator of multiple pathways, its role in Rac1 activation has not been reported. Likewise, SIRT2 has been demonstrated to be upregulated in some cancers; however, studies have also reported its role in tumor suppression. Here, we demonstrate that SIRT1 and SIRT2 positively regulate the levels of Rac1-GTP and the activity of T-cell lymphoma invasion and metastasis 1 (TIAM1), a Rac guanine nucleotide exchange factor (GEF). Transient inhibition of SIRT1 and SIRT2 resulted in increased acetylation of TIAM1 whereas chronic SIRT2 knockdown resulted in enhanced acetylation of TIAM1. SIRT1 regulates Dishevelled (DVL) protein levels in cancer cells and DVL along with TIAM1 are known to augment Rac activation; however, SIRT1 or 2 have not been previously linked with TIAM1. We found that diminished sirtuin activity led to the disruption of the DVL1-TIAM1 interaction. We hence propose a model for Rac activation where SIRT1/2 positively modulate the DVL/TIAM1/Rac axis and promote sustained pathway activation.
Keywords: SIRT1, SIRT2, TIAM1, Rac1-GTPase, Dishevelled, acetylation
Introduction
Small GTPases are signaling proteins that cycle between active GTP-bound and an inactive GDP-bound form in normal cells modulated by diverse upstream signals. Rac1 is a member of the Ras superfamily and Rho subfamily of GTPases, and its GTP-bound form activates multiple pathways downstream which can regulate cell motility, polarity, vesicular trafficking or proliferation (1;2). GEFs or GTPase activating proteins (GAPs) facilitate the addition of GTP or its hydrolysis back to GDP respectively, thus aiding in activation or inactivation of small GTPases (3;4). TIAM1, the GEF that activates Rac1 (5–8), is overexpressed in multiple malignant cancers and is associated with cell proliferation and increased metastasis (9–11). How TIAM1 activity is regulated is unclear but limited information suggests that phosphorylation and membrane localization are involved (12;13). While cell type and substrate specificity appears to be important in TIAM1-dependent cell migration or cell adhesion, the upstream regulators directing these processes are poorly understood (14–17). Reports have also shown nuclear localization of TIAM1 and its role in transcriptional regulation (18).
Histone deacetylases (HDACs) are enzymes that remove the post-translationally added acetyl group on the ε-amino group of lysine residues of histones and non-histone proteins in both the nucleus and cytoplasm, thereby affecting multiple cellular processes (19). HDACs have been shown to be overexpressed in multiple types of cancers suggesting that perturbed acetylation status of proteins may contribute to tumorigenesis (20;21). Mammalian sirtuins (SIRT1-7) are NAD+-dependent class III HDACs. The most well studied sirtuin family member, SIRT1, has been shown to alter cellular metabolism and responses to stress and thereby influence programs such as transcription, apoptosis, DNA damage repair and senescence (22;23). SIRT1 is overexpressed in both cancer cell lines and human cancers, and mediates cell survival in several tumor types such as the breast, colon, prostate, liver and also some types of leukemia (24–27). The other sirtuin family member, SIRT2, also targets cytosolic and nuclear proteins for deacetylation thus affecting their function (28;29). SIRT2 has been implicated in tumor suppression and the maintenance of genomic stability (30). However, recent studies have also shown tumor-promoting effects of SIRT2 where it was identified as a KRAS deacetylase (along with HDAC6) to positively regulate its activity or to stabilize Myc oncoprotein levels in cancer cells (31;32). It was also reported to be overexpressed in hepatocellular carcinoma and to promote epithelial to mesenchymal transition of hepatocellular carcinoma cells (33).
SIRT1 and SIRT2 have also been reported to influence cell migration via regulation of cytoskeletal protein dynamics in collaboration with HDAC6 (a class II HDAC). While SIRT1 is known to deacetylate and stabilize cortactin (34), SIRT2 colocalizes with and deacetylates tubulin (28). Consistent with this observation, we reported decreased basal or induced migration of cancer cells upon inhibition of the deacetylase activity of SIRT1 and SIRT2 using a small molecule inhibitor, cambinol (35;36). Additionally, a recent report shows the overexpression of SIRT1 in pancreatic tumors and demonstrates that knockdown of SIRT1 expression upregulates E-cadherin expression which influences cell migration (37). SIRT1 has also been demonstrated to positively regulate epithelial to mesenchymal transition and metastatic growth in prostate cancer (38), localize to the nucleus and cytosol in cancer cells (39), but the mechanistic basis remains unclear. Likewise, SIRT2 expression has been positively correlated with cortactin expression and tumor progression in prostate cancer (40).
The Wnt signaling pathway orchestrates complex processes such as early embryonic development, cell movement during gastrulation and cell polarity, and is pathway frequently activated during tumorigenesis. Dishevelled (DVL) proteins are versatile regulators of Wnt signaling. For example, DVL2 has been shown to recruit β-arrestin2 to mediate Wnt-stimulated endocytosis of Frizzled receptors (41). The non-canonical Wnt pathway regulates cell motility via Wnt/DVL-mediated activation of Rho proteins, and DVL has been implicated in this process (42;43). We have shown previously that SIRT1 regulates DVL and β-catenin protein expression and thus regulates the expression of Wnt target genes in cancer cells (35).
Here, we report for the first time that SIRT1 and SIRT2 regulate Rac1 activation in cancer cells. Additionally, we report the identification of TIAM1 as an acetylated protein and demonstrate that SIRT1 and 2 regulate its acetylation, GEF activity and binding to DVL. We also demonstrate that multiple small molecule inhibitors of sirtuins significantly inhibit TIAM1 interaction with DVL1 but not with other DVL proteins. Together, these data elucidate a new signaling axis critical for cancer cell migration.
Results
Inhibition or knockdown of SIRT1/2 decreases active Rac1 levels in cancer cells
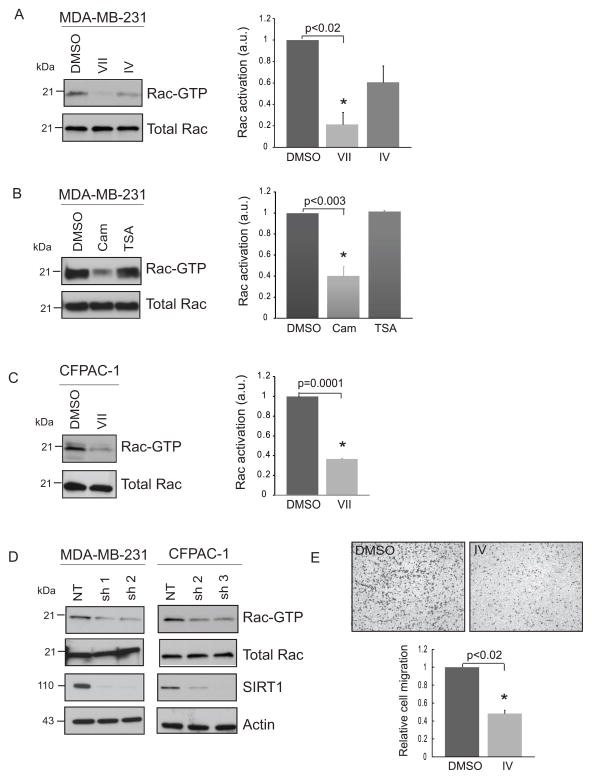
Rac1-GTPases regulate not only cell motility but also a number of genomic and cytoplasmic events (2). In recent years SIRT1 has been linked with enhancement of the migratory capability of cancer cells (34;37). As part of a screen to identify key mediators of this property, we identified the Rac1-GTPase. To further investigate this unexpected link between the sirtuins and Rac1, we first performed active Rac1-GTPase pull down assays to assess the levels of Rac1-GTP utilizing the p21-binding domain of p21-activated kinase, PAK1. To inhibit sirtuin activity three commercially available SIRT1/2 or SIRT1 inhibitors, namely, SIRT1/2 inhibitors VII (abbreviated as VII from here on) and cambinol (Cam) (36) or SIRT1 inhibitor IV (IV), an S-35 analog of EX-527 (44) were utilized. First, MDA-MB-231 breast cancer cells treated with inhibitors VII or IV showed a decrease in Rac1-GTP levels (Fig. 1A). While inhibitor IV showed a trend towards decreased levels, we observed a statistically significant decrease for VII compared to DMSO (vehicle control) treated cells. Similarly, when treated with cambinol, a significant decrease in Rac1-GTP levels was observed whereas inhibition of class I/II HDACs with trichostatin A (TSA) did not affect Rac1 activity (Fig. 1B). The same lysates were probed for acetyl-tubulin to verify the activity of TSA which showed an increase in acetylated tubulin level as expected (Fig. S1A). To extend this analysis to another cell line, CFPAC-1 pancreatic cancer cells also showed a significant decrease in active Rac1 upon treatment with inhibitor VII (Fig. 1C). To determine the specific contribution of SIRT1 or SIRT2, we stably knocked down SIRT1 in MDA-MB-231 and CFPAC-1 and SIRT2 in MDA-MB-231 cells. Consistent with pharmacologic inhibition, a decrease in Rac1-GTP levels was observed upon SIRT1 knockdown in two independent stable knockdown clones (Fig. 1D). Active Rac1 levels also decreased in MDA-MB-231 cells harboring stable SIRT2 knockdown (Fig. S1B). We have previously shown that cambinol decreases migration of cancer cells both basally and under Wnt-stimulated conditions (35). Since SIRT1 inhibitor IV, (S-35) had not been previously applied in this context, we performed a transwell migration assay with the MDA-MB-231 cells treated with vehicle or IV and observed a significant decrease in migration of these cells across the transwell membrane (Fig. 1E). A similar decrease in migration of 231 cells was observed upon SIRT1 knockdown (Fig. S1C). A few reports have shown the effect of inhibition of SIRT1 by small molecule inhibitor, sirtinol (45), or SIRT1 RNAi on Ras/ERK pathway (46). Here, we report for the first time that activation of Rac1-GTPase can be controlled in a SIRT1-dependent manner in cancer cells.
Figure 1. Pharmacological inhibition or knockdown of SIRT1 decreases active Rac1 levels in cancer cells.
(A) Decrease in active Rac1 (Rac1-GTP) levels upon inhibition of SIRT1 and SIRT2 deacetylase activity by inhibitor VII or IV compared to DMSO, vehicle control. MDA-MB-231 cells were treated with DMSO or 50 μM VII or 20 μM inhibitor IV for 1 h. Active Rac1-GTP pull down was performed on fresh lysates followed by western blotting for active and total Rac1 levels. Inhibitors VII and IV resulted in a 79% and 39% decrease in Rac-GTP levels compared to DMSO control. (B) Decrease in active Rac1 levels upon treatment of MDA-MB-231 cells with cambinol compared to DMSO control. Cells were treated with DMSO or 100 μM cambinol or 1 μM TSA for 1 h. Treatment with TSA showed no change whereas cambinol decreased active Rac levels by 60%. (C) CFPAC-1 cells were treated with DMSO or 50 μM inhibitor VII for 1 h. Inhibitor VII led to a 64% decrease in active Rac1 levels compared to DMSO control. (D) Decrease in active Rac1 levels in MDA-MB-231 and CFPAC-1 cells upon stable SIRT1 knockdown. 231 or CFPAC-1 cells were infected with lentivirus carrying non-targeting shRNA (NT) or SIRT1-targeting shRNA and maintained stably under puromycin selection. (E) SIRT1 inhibitor IV, (S-35) decreases migration of MDA-MB-231 cells in a transwell migration assay. 231 cells were plated in the top chamber at 50,000 cells per transwell insert in duplicate in serum-free DMEM medium containing either DMSO or 20 μM inhibitor IV. Migration towards 10% serum containing medium in the lower chamber occurred for 14 hours. NIH ImageJ was used for densitometric and cell migration analyses. Rac activation is the normalized ratio of Rac-GTP to total Rac and is represented in arbitrary units on the y-axis. Relative cell migration is the mean of normalized migrated cell counts for DMSO or inhibitor IV treatments. Values are mean ± SEM, n = 3.
SIRT1 co-immunoprecipitates with TIAM1 in cancer cells
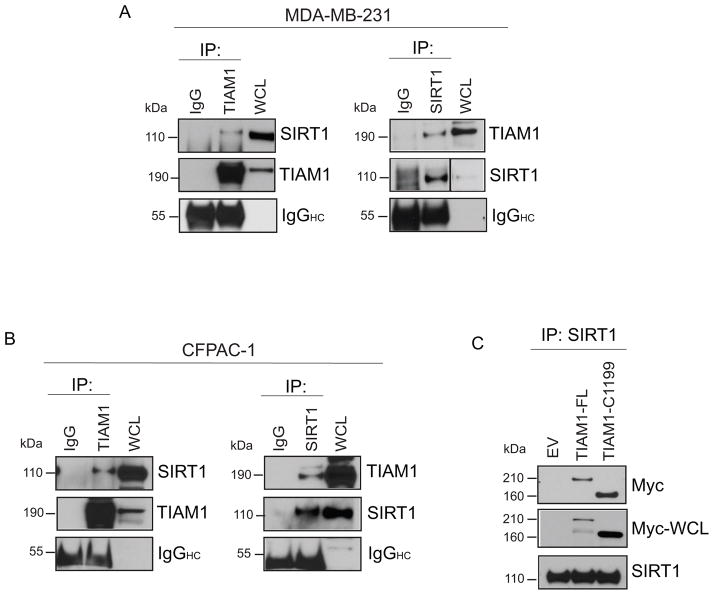
Multiple GEFs that can activate Rac1-GTPase under the influence of a specific upstream signal or cell or tissue type are also overexpressed in cancer (4;8;47). As TIAM1 is generally considered a Rac-specific GEF, we first wanted to test whether SIRT1 was physically interacting with TIAM1. Upon immunoprecipitation of endogenous TIAM1 from MDA-MB-231 cells we observed a co-precipitation of SIRT1, indicating that SIRT1 was directly or indirectly bound to this Rac-GEF (Fig. 2A, left panel). Similar results were observed upon reciprocal immunoprecipitation with a SIRT1 or SIRT2 antibody (Figs. 2A, right panel and S2A). Extending this observation to CFPAC-1 and human embryonic kidney (HEK) 293 cells yielded a similar binding pattern between endogenous SIRT1 and TIAM1 (Figs. 2B and S2B). To further verify the association between SIRT1 and TIAM1, we transiently transfected a full-length Myc-tagged TIAM1 or N-terminally truncated Myc-tagged C1199-TIAM1 (stable and more potently expressed form of TIAM1, (6)) in HEK293 cells and were able to immunoprecipitate endogenous SIRT1 and pull down Myc-TIAM1 in both cases (Fig. 2C). These data demonstrate that SIRT1-TIAM1 binding occurs basally in HEK293 and cancer cells and the constitutively active TIAM1 mutant lacking amino acids 1-392 of TIAM1 retains its ability to bind endogenous SIRT1.
Figure 2. SIRT1 co-immunoprecipitates with the Rac-GEF, TIAM1.
(A) SIRT1 and TIAM1 co-immunoprecipitate in MDA-MB-231 Untreated MDA-MB-231 cells were used to extract whole cell lysate for immunoprecipitation of endogenous TIAM1 (left panel) or SIRT1 (right panel) with specific antibodies. Western blotting was performed for SIRT1 and TIAM1 respectively from immunoprecipitated samples along with whole cell lysates (WCL). Species-matched IgG was used as a negative control. The membranes were stripped and re-probed for input TIAM1 or SIRT1. IgG heavy chain (IgGHC) was blotted for as a control for equal antibody addition to samples. (B) SIRT1 and TIAM1 were immunoprecipitated from CFPAC-1 cells as in (A) followed by western blotting for TIAM1 and SIRT1 respectively. (C) Exogenously expressed Myc-tagged TIAM1 (transiently transfected for 24 h) co-immunoprecipitates with SIRT1 in HEK293 cells. EV (Empty vector, pcDNA3); TIAM1-FL (Myc-tagged full-length TIAM1); TIAM1-C1199 (Myc-tagged truncated TIAM1-C1199).
TIAM1 is acetylated and SIRT1 and SIRT2 deacetylate TIAM1 in cancer cells
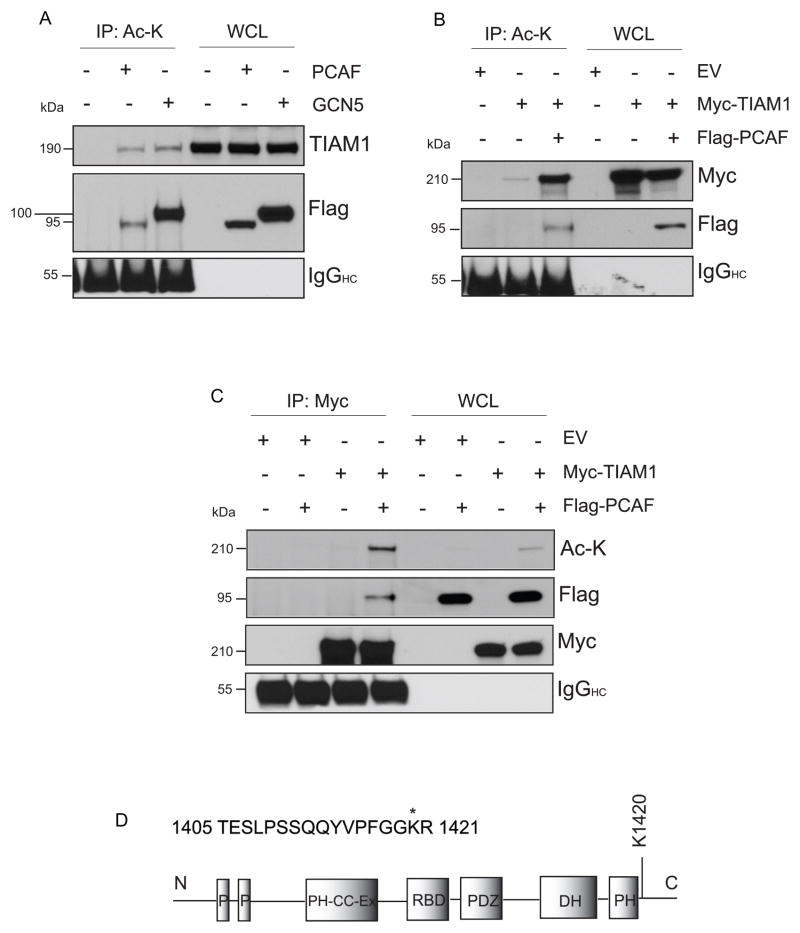
The outcome of the co-immunoprecipitation assay suggested that SIRT1 or SIRT2 binding to TIAM1 could be a direct interaction. So, we hypothesized that TIAM1 is an acetylated protein that may be deacetylated by these sirtuins. To detect acetylation of TIAM1, we performed transient overexpression of two lysine acetyltransferases, p300/CBP-associated factor (PCAF) and general control non-repressible 5 (GCN5), in HEK293 cells. PCAF and GCN5 are structurally and functionally related members of the lysine acetyltransferase 2 (KAT2) family of acetyltransferases (48). An increase in acetylation of endogenous TIAM1 was observed upon overexpression of the acetyltransferases (Fig. 3A). Co-transfection of Myc-tagged full-length TIAM1 with PCAF showed an increased acetyl-Myc-TIAM1 expression, compared to Myc-TIAM1-only transfected cells, in immunoprecipitates with either acetyl-lysine (Fig. 3B) or Myc (Fig. 3C) antibodies. In order to determine the specific lysine residues that are acetylated, Myc-TIAM1 (full-length or C1199 mutant) were overexpressed in HEK293 cells along with the acetyltransferase GCN5. Immunoprecipitation was performed for Myc and 1D SDS PAGE gel band corresponding to Myc-TIAM1 was excised for detection of acetylation by mass spectrometry using matrix-assisted laser desorption/ionization-time-of-flight/time-of-flight (MALDI-TOF/TOF). Of the 48 matching peptides from full-length protein and 46 from C1199-TIAM1, 10 peptides showed the presence of potentially acetylated lysines (supplementary table 1). Three of these peptides showed immonium ion peaks at mass/charge (m/z) ratio of ~126, when subjected to tandem mass spectrometry (MS/MS analysis), demonstrating the presence of acetylated lysines at amino acids 1068, 1226 and 1420 (Fig. S3 A; manually verified and detailed in supplementary file 1). However, only K1420 was detected with a high ion score (Fig. 3 D; supplementary file 1).
Figure 3. TIAM1 is an acetylated protein.
(A) PACF and GCN5 increase acetylation of endogenous TIAM1. HEK293 cells overexpressing Flag-tagged PCAF or GCN5 show enhanced acetyl-TIAM1. Cells were transiently transfected with indicated plasmids for 24 hours followed by immunoprecipitation with acetyl-lysine antibody. Blot with anti-Flag M2 antibody shows the presence of Flag-tagged plasmid expression in transfected lanes. IgG heavy chain (IgGHC) was blotted for as a control for equal antibody loading. (B) Overexpression of PCAF increases acetyl-Myc-TIAM1 levels. HEK293 cells were transiently transfected with either empty vector or full-length Myc-TIAM1 or co-transfected with full-length Myc-TIAM1 and Flag-PCAF for 24 hours. Immunoprecipitation with acetyl-lysine antibody was performed as in (A) followed by western blot for Myc or Flag. (C) HEK293 cells were transfected with empty vector or Myc-TIAM1-full-length with or without co-transfection with PCAF. Immunoprecipitation was performed with a Myc-specific antibody and blotted for acetyl-lysine or Flag. Acetyl-lysine blot was stripped and re-probed for Myc demonstrating equal immunoprecipitation in Myc-TIAM1-only or Myc-TIAM1+PCAF lanes. (D) Schematic showing the acetylated peptide with an ion score of 16 and a percent confidence interval of 99 for peptide sequence ID from MS/MS data analysis of Myc-TIAM1 full-length. Acetylated lysine is marked with “*”. Approximate representation of the acetylated lysine on the TIAM1 domain map is shown. N, N-terminal; P, PEST sequence; PH-CC-Ex, Pleckstrin-homology, coiled-coil and extra; RBD, Ras-binding domain; PDZ, PSD-95/Dlg A/ZO-1; DH, Dbl-homology; PH, Pleckstrin-homology; C, C-terminal
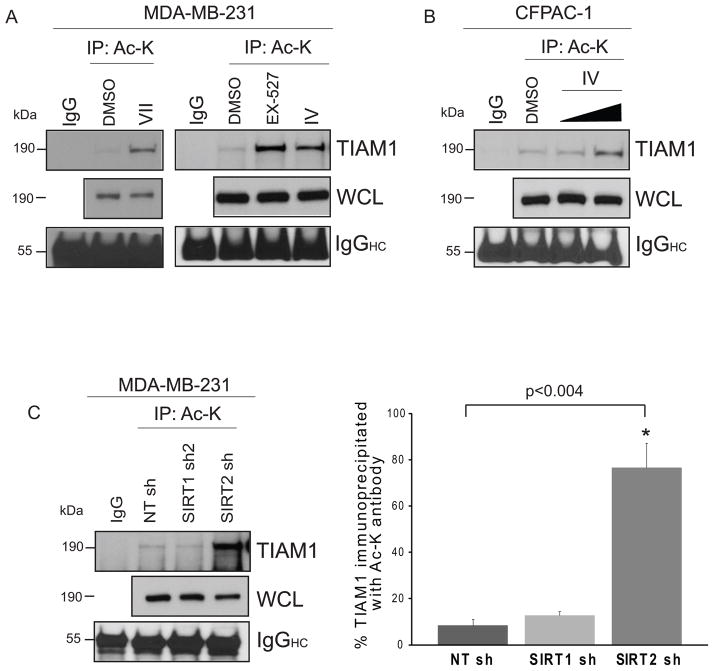
To determine the effect of inhibition of sirtuin activity on endogenous TIAM1 acetylation MDA-MB-231 or CFPAC-1 cells were treated with the SIRT1 and/or SIRT2 described earlier in addition to EX-527, a SIRT1-specific inhibitor. MDA-MB-231 cells treated with EX-527, IV or VII showed a strong acetylation signal for TIAM1 upon immunoprecipitation of acetyl lysine followed by TIAM1-specific blotting (Fig. 4A). A dose-dependent increase in TIAM1 acetylation was observed in inhibitor IV-treated CFPAC-1 cells (Fig. 4B). HEK293 cells also showed a moderate increase in acetylation of TIAM1 upon treatment with VII, cambinol or nicotinamide (NAM), a broad sirtuin family inhibitor (Fig. S3B). To compare acute sirtuin inhibition pharmacologically with a sustained inhibition via SIRT1 or SIRT2 knockdown, we performed the acetyl-lysine immunoprecipitations using MDA-MB-231 cells stably expressing SIRT1 or 2 shRNA. To our surprise, SIRT1 knockdown did not result in an increase in TIAM1 acetylation while SIRT2 knockdown did so significantly (Fig. 4C). This suggests that the magnitude of acetylation may depend on whether the sirtuins are inhibited acutely or chronically or that SIRT2 may play a dominant role in acetylation induction under conditions of chronic inhibition. Thus, we show for the first time that under basal conditions TIAM1 is predominantly in a deacetylated state while bound to SIRT1 or 2 but shows a significant increase in acetylation upon inhibition of sirtuin activity or expression in vivo. Only one report has demonstrated TIAM1 acetylation in a high-throughput MS/MS screen of HCT116 colon carcinoma cells (PhosphoSitePlus Database, Cell Signaling Technology, (49)). This report involved immunoprecipitation of multiple acetylated proteins in HCT116 cells with an acetyl-lysine antibody followed by mass spectrometry. Here, we use a different approach for MS/MS analysis where we immunoprecipitate TIAM1 specifically using a Myc tag. We further identify at least two potential acetyltransferases and deacetylases associated with this modification and provide data linking TIAM1 acetylation with a physiological role.
Figure 4. Inhibition or knockdown of sirtuins increase TIAM1 acetylation in vivo.
(A) Acetylation of TIAM1 is enhanced upon inhibition of SIRT1/2 activity compared to DMSO control in MDA-MB-231 cells. Cells were treated with DMSO or 50 μM inhibitor VII or 10 μM EX-527 or inhibitor IV, (S-35) for 45 minutes. Equal μgs of protein was loaded for each immunoprecipitation set up using acetyl-lysine (Ac-K) antibody as per protocol. IP and whole cells lysate samples were blotted for TIAM1. Species-matched IgG was used as a negative control. IgG heavy chain (IgGHC) was blotted for as a control for equal antibody loading for immunoprecipitation. (B) CFPAC-1 cells were treated with 10 or 20 μM inhibitor IV for 45 minutes and immunoprecipitation was performed as in (A). Acetylation of TIAM1 was detected by western blotting. (C) SIRT2 knockdown increases TIAM1 acetylation by ~68% compared to NT shRNA samples. MDA-MB-231 cells stably expressing non-targeting (NT) or SIRT1 or SIRT2 shRNA were used for immunoprecipitation of Ac-K and blotted for TIAM1 as in (A). Percent TIAM1 immunoprecipitated with Ac-K antibody was calculated by dividing densitometric signal values for immunoprecipitated samples by the vales obtained from whole cell extracts. NIH ImageJ was used for densitometry. Values are mean ± SEM, n=3.
Inhibition of sirtuin activity affects TIAM1 GEF activity but not its localization in the cells
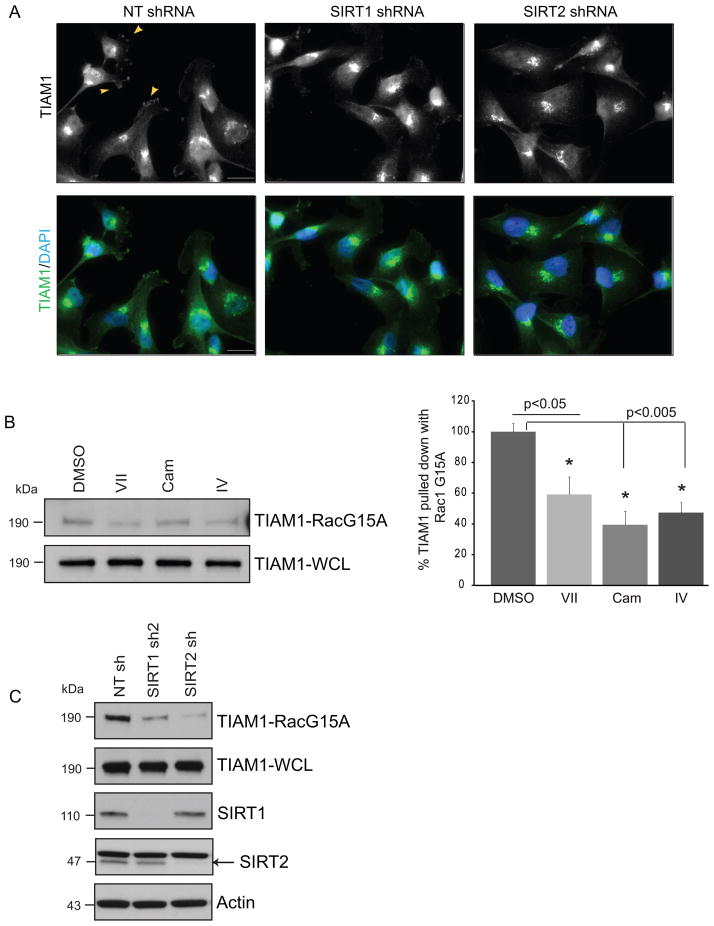
TIAM1 is well known to be localized in the cytosol and at cell membranes facilitating the recruitment and activation of Rac at the membrane, maintenance of adherens junctions and regulation of the polarity and random migration of cells (12;15;50). In cancer cells TIAM1 can be localized to the cell junctions (51), juxtanuclear compartments such as Golgi (52) or leading edges of migrating cells (53) thereby establishing front-rear polarity of cells. To determine the effect of sirtuin inhibition on TIAM1 localization, we treated MDA-MB-231 cells with vehicle, inhibitors VII, Cam or IV (Fig. S4) or utilized stable SIRT1 or SIRT2 knockdown cells (Fig. 5 A). In agreement with Adams et al., we observed a TIAM1 localization to be juxtanuclear with a punctuate appearance (Figs. 5A and S4). These cells lack cell-cell junctions and E-cadherin expression, however, regions of the cells with lamellipodial protrusions readily stained for TIAM1. While cells expressing non-targeting shRNA showed presence of TIAM1 on membrane ruffles (Fig. 5A, arrowheads in top left image), this pattern was diminished or lost upon SIRT1 or 2 knockdown (Fig. 5A, middle and right panels). Thus, sirtuin inhibition or knockdown did not alter the gross TIAM1 staining pattern in MDA-MB-231 cells but had a moderate effect on localization at the membrane ruffles, suggesting that acetylation may not alter subcellular localization extensively.
Figure 5. Inhibition or knockdown of sirtuins alter TIAM1 GEF activity.
(A) MDA-MB-231 stably expressing NT, SIRT1 or SIRT2 shRNA cells show endogenous TIAM1 localization in representative images. Arrowheads in the top left box point at TIAM1 staining on the membrane ruffles in NTshRNA expressing cells; not observed in SIRT2 and moderately observed in SIRT1 knockdown cells. Upper panel represents green channel only. Lower panel shows merged images for TIAM1 and nuclear (DAPI) staining. Scale bar, 20 μm. (B) TIAM1 binding to nucleotide-free Rac1 G15A decreases upon sirtuin inhibition. MDA-231 cells were treated with DMSO control or inhibitors VII (50 μM), cambinol (100 μM) or IV (20 μM) for 45 minutes. %TIAM1 bound to Rac1 G15A was calculated as a ratio of G15A-bound TIAM1 to TIAM1 in whole cell extracts. Inhibitors VII, Cam and IV show 41, 60 and 52% decrease compared to DMSO respectively. NIH ImageJ was used for densitometry. Values are mean ± SEM, n = 3. (C) Stable SIRT1 and SIRT2 knockdown decrease TIAM1 GEF activity compared to non-targeting control. MDA-MB-231 cell stably expressing NT, SIRT1 or SIRT2 shRNA were used in TIAM1 GEF activity assay with Rac1 G15A as in (B).
During the GDP to GTP exchange the GTPase exists in a nucleotide-free form which favors the binding of a GEF which facilitates the exchange process (54). Utilizing a nucleotide-free form of Rac1, Rac1 G15A, conjugated to agarose beads, we performed experiments to pull-down active TIAM1 which shows a high affinity towards the nucleotide-free form of Rac (55). In MDA-MB-231 cells treated with inhibitors VII, IV or cambinol, active TIAM1 pulled down with Rac1 G15A decreased (Fig. 5B) as compared to vehicle control. This suggested that inhibition of sirtuin activity with the same doses and duration that induced TIAM1 acetylation and decreased Rac1 activation also decreased the GEF activity of TIAM1. These observations were confirmed with SIRT1 or -2 knockdowns. The decrease in GEF activity upon SIRT2 knockdown was greater than that observed upon knocking down SIRT1 expression (Fig. 5C), raising the possibility that the enhancement of TIAM1 acetylation upon SIRT2 knockdown had a more pronounced effect on decreasing TIAM1 GEF activity.
TIAM1 co-immunoprecipitates with DVL proteins in cancer cells
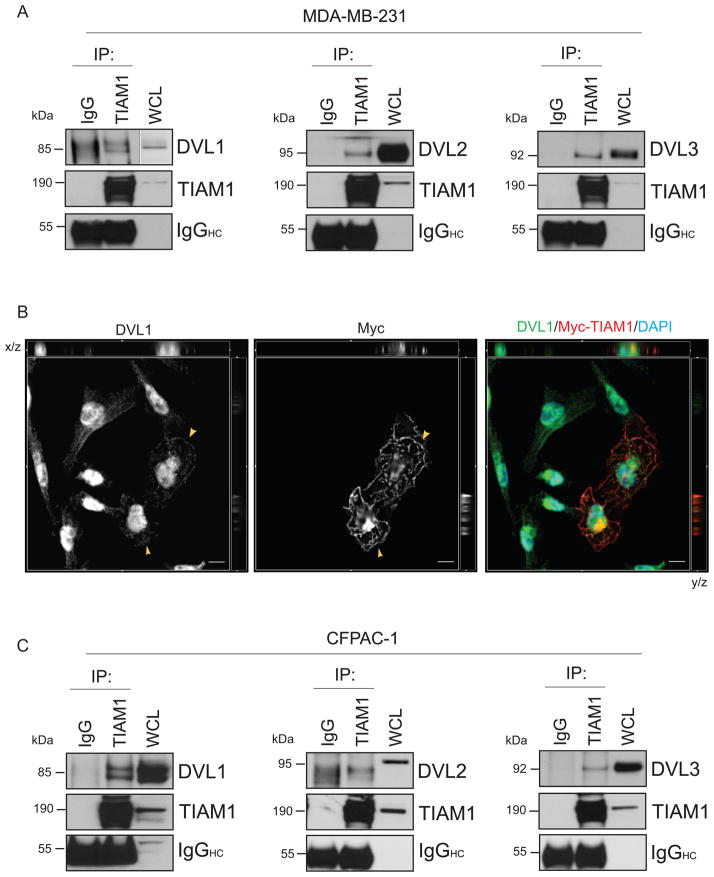
We have previously shown that SIRT1 positively regulates the Wnt signaling pathway in cancer cells by positively regulating Wnt signaling proteins such as DVL or β-catenin. Stimulation of the non-canonical Wnt pathway leads to the activation of small GTPases via the DVL proteins. Since SIRT1 stabilizes DVL protein levels, we hypothesized that SIRT1 may regulate Rac activation in a DVL-dependent manner. However, the changes in Rac activation, TIAM1 acetylation or GEF activity upon sirtuin inhibition were observed over a brief period of 30 minutes to one hour which preceded changes in DVL protein levels. In order to further elucidate the role of sirtuin activity during this brief period, we first performed an immunoprecipitation of TIAM1 in MDA-MB-231 and CFPAC-1 cells to determine whether it binds DVL in these cells. DVL was shown to facilitate Rho activation via Daam1 which recruited Rho-GEF (43) but the identification of a protein that could co-activate Rac1 downstream of DVL was lacking. We observed that endogenous TIAM1 successfully co-immunoprecipitates with all the three DVL family proteins in MDA-MB-231 and CFPAC-1 cancer cells (Figs. 6 A and C). We also noted co-staining of Myc-TIAM1-C1199 with endogenous DVL1 on the membrane ruffles in MDA-MB-231 cells (Figs. 6 B and S5). The morphology of TIAM1 overexpressing cells displaying extensive membrane ruffling was in corroboration with previously published reports (Fig. 6 B, middle panel). Endogenous DVL1 was localized in and around the nucleus and showed diffused staining in the cytosol. It followed the staining pattern of Myc-C1199-TIAM1 and co-localizes with Myc in the two TIAM1 overexpressing cells in the view (Fig. 6 B, arrowheads in left image showing DVL1 localization and merged image to the right; Fig. S5). Exogenous co-expression of both TIAM1 and DVL1 resulted largely in apoptotic cells where DVL1 was observed in large punctuate structures (data not shown). While we independently made these observations regarding TIAM1 and DVL binding in cancer cells, a study published by Cajánek et al. (56) found similar results in dopaminergic neuronal cells.
Figure 6. DVL 1, 2 and 3 bind to TIAM1 in cancer cells.
(A) TIAM1 co-immunoprecipitates with DVL1, 2 or 3 in MDA-MB-231 cells. The membranes were stripped and re-probed to blot for TIAM1 input. Species-matched IgG was used as a negative control. IgG heavy chain (IgGHC) was blotted for as a control for equal antibody loading for immunoprecipitation. (B) DVL1 and TIAM1 co-localize in MDA-MB-231 cells expressing My-tagged TIAM1. Cells were transfected with Myc-TIAM1-C1199 for 24 h and processed for immunostaining for endogenous DVL1 (left image) or Myc tag (two cells showing successful transfection in the middle image). Right box shows merged channels for DVL1 (green), Myc-TIAM1 (red) and DAPI (blue) in the representative image. Co-staining can be observed in the x-y focal plane and orthogonal view of the displayed section of a z-stack. Cut-lines are hidden from view. Arrowheads in left and middle images show DVL1 and Myc-TIAM1 localization respectively on the membrane ruffles in Myc-TIAM1 expressing cells. Scale bar, 10 μm. (C) TIAM1 co-immunoprecipitates with DVL1, 2 or 3 in CFPAC-1 cells. Immunoprecipitation for endogenous TIAM1 was followed by blotting with DVL1, 2 or 3 specific antibodies as in (A).
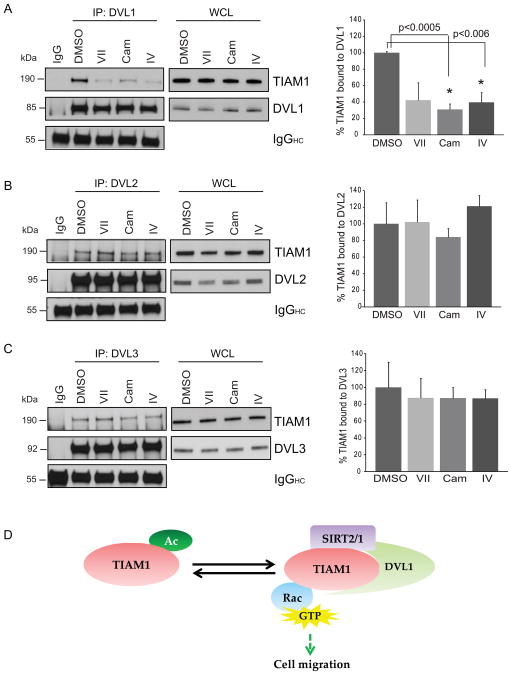
To define the role of sirtuin activity in TIAM1/DVL-mediated Rac activation, we treated 231 cells with inhibitors VII, cambinol or IV and immunoprecipitated DVL1, 2 or 3 from whole cell extracts. While all the three DVL proteins pulled down TIAM1 in DMSO control treated cells, only DVL1-TIAM1 complex was significantly dissociated upon sirtuin inhibition (Fig. 7 A). DVL2-TIAM1 and DVL3-TIAM1 binding remained intact (Figs. 7 B and C).
Figure 7. Inhibition of sirtuin activity disrupts DVL1-TIAM1 but not DVL2/3-TIAM1 binding in MDA-MB-231 cells.
(A–C) Cells were treated with inhibitors VII (50 μM), Cam (100 μM), IV (20 μM) or DMSO control for 45 minutes followed by immunoprecipitation with DVL1, 2 or 3-specific antibodies. Percent TIAM1 bound to DVL was calculated first by taking a ratio of immunoprecipitated TIAM1 or DVL to its own whole cell extract followed by calculating percent TIAM1/DVL. DVL1 showed a loss binding with TIAM1 with all three inhibitors at 58, 69 and 60% each compared to control. Treatment with inhibitor VII showed near-significance with p-value = 0.0535. DVL2 and DVL3 binding with TIAM1 are not affected significantly with inhibitor treatments. NIH ImageJ was used for densitometry and each treatment was compared to DMSO for statistical analysis. Values are mean ± SEM, n = 3. (D) Proposed model for reversible TIAM1 acetylation/deacetylation, the role of SIRT2 and SIRT1 in the process and resulting DVL/TIAM1/Rac1 pathway activation.
These findings lead us to believe that DVL1 plays a unique role in sirtuin-regulated Rac1 activation and is sensitive to the inhibition of deacetylase activity. We propose that SIRT2 and SIRT1 act on the TIAM1/DVL1/Rac axis in cancer cells resulting in stabilization of TIAM1-DVL1 complex and chronic pathway activation (Fig. 7 D). Loss of deacetylase activity results in dissociation of TIAM1-DVL1 complex, increased TIAM1 acetylation, decreased GEF activity, and subsequent loss of Rac1 activation.
Discussion
In this study we describe for the first time an important link between the sirtuins 1 and 2, TIAM1 and DVL1. We have demonstrated that TIAM1 is acetylated and binds to SIRT1 and SIRT2 endogenously in immortalized and cancer cells under basal conditions. SIRT2 serves as a TIAM1 deacetylase and appears to maintain it in a chronically deacetylated form. SIRT1/2 inhibition, but not, class I/II HDAC inhibition also led to a significant reduction in active Rac, suggesting that specificity in the deacetylase regulates Rac activity. There is a significant decrease in active Rac levels upon SIRT1 or SIRT2 knockdown and a decrease in GEF activity of TIAM1 suggesting a role for acetylation/deacetylation of TIAM1 in regulating Rac activation. Gross TIAM1 localization was not affected with inhibition of deacetylase activity, but its binding with DVL1 was disrupted. Future mutagenesis analyses will further delineate the mechanistic basis for this. We also demonstrate the biological use of a novel SIRT1/2 inhibitor in cancer cells.
TIAM1 can activate Rac by phosphatidyl-inositol-3-kinase-dependent or independent mechanisms (7;15). Here we describe a novel post-translational modification of TIAM1 which could regulate Rac activation in a manner not previously reported. Upregulation of SIRT1 and SIRT2 in malignant tumors could result in increased formation of the TIAM1-SIRT1/2-DVL1 complex. Increased deacetylation of TIAM1 could result in increased active Rac levels, which could augment the metastatic potential of neoplastic cells. The findings reported here suggest that alterations in HDAC/HAT activity could have important implications for TIAM1 function and Rac activation. Emerging reports on protein acetylation associated with critical cellular processes underline the potential of this post-translational modification to be targeted for therapeutic purposes.
While a decrease in active Rac levels and TIAM1 GEF activity were observed upon both SIRT1 and SIRT2 knockdown, increase in TIAM1 acetylation was observed only upon SIRT2 knockdown. A simple explanation is that SIRT2 could be the primary TIAM1 deacetylase positively regulating its activity, DVL complex formation and Rac activation. While both SIRT1 and SIRT2 bind to TIAM1 in vivo, it is possible they target distinct lysine residues resulting in varying degrees of deacetylation. Additionally, acute vs chronic inhibition each sirtuin may create different acetylation portraits. Under either scenario, this may differentially influence downstream effects such as GEF activity and Rac activation. Since interaction of DVL with Rac and TIAM1 can regulate Rac activity, the disruption of DVL1-TIAM1 interaction upon sirtuin inhibition has important implications. In cancer cells DVL1-TIAM1 mediated Rac activation could add an extra layer of activity which pushes the pathway into a “hyperactivated” state and hence increase metastatic potential of cancer cells. This could be a more prominent event in cancers overexpressing DVL1 protein. In our experiments inhibition of sirtuin activity did not abolish Rac activation or TIAM1 GEF activity completely. This could be an effect of a residual deacetylase activity from sirtuins and class I/II deacetylases. It could also be an effect of the persistent binding of DVL2 and DVL3 to TIAM1. Hence, SIRT1 and/or SIRT2 may be selectively acting on the DVL1/TIAM1/Rac axis short-term. Since knockdown or sirtuin inhibition leads to loss of all three DVL proteins to differing extents (35), and here we have seen that Rac activation is also affected upon shRNA mediated SIRT1 and SIRT2 knockdown, it remains to be determined how the DVL-TIAM1 binding states are affected upon chronic inhibition or knockdown.
Historically, the mechanism of action of histone deacetylase inhibitors has been associated with altering chromatin structure and gene expression patterns. There is increasing evidence for the presence of SIRT1 in the cytosol of cancer cells, which was originally reported to be an exclusively nuclear protein (39), while SIRT2 is already known to be a cytosolic protein (57). The presence of these and other deacetylases in cytosolic compartments positions them to mediate post-translational modifications on cytosolic proteins. While conventional GTPase-targeting therapy has been unsuccessful in the clinic (58), targeting GTPases that promote tumor properties via inhibition of such regulatory proteins like sirtuins could open new avenues for therapeutic development in cancers. Currently, FDA-approved class I/II HDAC inhibitors, such as vorinostat and romidepsin, are being used in clinic for treating multiple cancer types (59;60). However, there are no sirtuin inhibitors approved for clinically applications so far. Our study and others have shown the significance of SIRT1 and SIRT2 in regulating events in cancer cells either independent of class I/II HDACs or in collaboration with them. This provides further rationale for developing sirtuin inhibitors that may show promise in clinical applications.
Materials and methods
Cell culture
MDA-MB-231 breast cancer and CFPAC-1 pancreatic cancer cells purchased from ATCC were grown in DMEM (Genesee) or Iscove’s Modified Dulbecco’s Medium (ATCC) respectively, containing 10% FBS and penicillin/streptomycin. HEK293 cells obtained from ATCC were grown in EMEM containing 10% FBS and penicillin/streptomycin under normal culture conditions.
Antibodies and inhibitors
RNAi and plasmids
Lentiviruses encoding non-targeting (SHC002V), human SIRT1-targeting shRNA (sh1-TRCN0000018979, sh2-TRCN0000018981, sh3-TRCN0000018983,) or human SIRT2-targeting shRNA (sh-TRCN0000010435) were purchased from Sigma. MDA-MB-231 or CFPAC-1 cells were infected with lentivirus in serum-free media containing polybrene (Sigma). A no-infection control was used for observing cell death during the selection process and the cells were stably maintained under puromycin (Sigma) selection. Plasmids were transiently transfected in HEK293 or MDA-MB-231 cells using Lipofectamine 2000 reagent from Invitrogen. pcDNA3-Myc-TIAM1 full-length and C1199 plasmids were a kind gift from Dr. Channing Der, University of North Carolina, School of Medicine, Chapel Hill, NC, USA. pCMVSport6-Flag-mPCAF and Flag-mGCN5 plasmids were a kind gift from Dr. Sharon Dent, University of Texas M D Anderson Cancer Center, Houston, TX, USA.
Active Rac1-GTPase pull-down assay
Active Rac1 pull-down assay was performed on treated CFPAC-1 or MDA-MB-231 cells or cells expressing stable NT, SIRT1 or SIRT2 shRNA using active Rac1-GTPase pull down assay kit (Thermo Scientific). Details in supplementary information.
Co-immunoprecipitations and Western blotting
CFPAC-1, MDA-MB-231 or HEK293 cells were used for immunoprecipitating endogenous SIRT1, SIRT2, TIAM1, DVL1/2/3 or Myc-TIAM1. Briefly, untreated or treated cells were washed in ice-cold PBS and lysed in Mammalian Protein Extraction Reagent (Thermo Scientific) in the presence of protease inhibitor cocktail (Calbiochem) and deacetylase inhibitors followed by immunoprecipitation of desired protein using the specific antibody along with species matched control IgG. Immunoprecipitation was followed by western blotting for protein of interest expected to be pulled down with the target. Details in supplementary information.
Acetyl-lysine IP
CFPAC-1, MDA-MB-231 or HEK293 cells treated with DMSO or sirtuin inhibitors or transfected with indicated plasmids, washed and harvested in IP lysis buffer (25mM Tris-Cl pH 8.0, 150 mM NaCl, 1mM EDTA, 1% NP-40, 5% glycerol) containing protease and deacetylase inhibitors. Protein was quantified to load equal μgs per IP. Samples were incubated overnight with acetyl-lysine antibody. Antigen-antibody complex was incubated with Dynabeads protein A for 2.5 hours following which samples were washed in IP lysis buffer, eluted in 2x Laemmli sample buffer and used for blotting. Same protocol (except the treatment) was followed for IPs utilizing stable knockdown cell lines.
Mass spectrometry
HEK293 cells were co-transfected with Myc-TIAM1 (full-length or C1199) and GCN5, harvested in IP lysis buffer as mentioned above and the lysates incubated with anti-c-Myc affinity gel for 2 hours at 4°C with gentle rocking. Affinity gel was washed with lysis buffer and samples eluted in 2x Laemmli sample buffer. SDS-PAGE was followed by staining with GelCode Blue Stain Reagent (Thermo Scientific). See Supplementary information.
Transwell migration assay
24-well transwell 8 micron inserts were purchased from BD. 50,000 MDA-MB-231 cells were plated in top chamber in serum-free DMEM containing 0.2% BSA and either DMSO or inhibitor IV, each condition in duplicate. Lower chamber had DMEM with 10% FBS. Cells were allowed to migrate for 14 hours. Same protocol (except for treatment) was followed for migration using stable shRNA expressing cells. Details in supplementary information.
Active Rac GEF assay for TIAM1
MDA-MB-231 cells were grown in complete medium which was changed to serum-free medium containing 0.2% BSA four hours prior to treatment with inhibitors or DMSO control. Cells were washed in ice cold PBS and harvested in lysis buffer (20mM HEPES pH 7.5, 150mM NaCl, 1% Triton-X100 and 5mM MgCl2) containing protease and deacetylase inhibitors and protein was isolated by centrifugation at 14,000 ×g for 15 minutes. Equal amount of protein per sample was used for incubation with Rac G15A-conjugated agarose beads (Cell Biolabs, Inc.) for 1 hour and 15 minutes at 4°C with gentle rotation. Beads were washed three times in the lysis buffer and protein eluted in Laemmli buffer was used for western blotting for TIAM1 along with whole cell lysates. Same protocol (except for treatment) was followed for GEF assay using stable shRNA expressing cells.
Immunofluorescence
For TIAM1 staining in shRNA expressing 231 cells or Myc-TIAM1/DVL1 co-staining cells were grown (and transfected in case of Myc/DVL1 staining) in chamber slides (BD Falcon). Detailed protocol in supplementary information. Images were captured as z-stacks for all channels on Zeiss AxioObserver.Z1 with AxioCam MRm CCD camera. For comparison of TIAM1 in NT, SIRT1 or SIRT2 shRNA expressing cells, images were captured at same exposure. A 40x/1.3NA PlanApo oil immersion objective was used. The acquisition and image processing software for was AxioVision v4.8.2. DVL1/Myc-TIAM1 co-staining images were subject to 3D deconvolution using AxioVision v4.8.2. Brightness and contrast were adjusted in Adobe Photoshop.
Quantification and statistical analysis
Image J (NIH) was used for densitometric analysis of the western blots. One sample t-test for a hypothetical mean or two-tailed Student’s t-test, whichever applicable, was performed for statistical analysis.
Supplementary Material
Acknowledgments
We thank Dr. Channing Der at University of North Carolina and Dr. Sharon Dent at University of Texas M. D. Anderson Cancer Center for providing the plasmids. The work described here is funded by a Feist-Weiller Cancer Center Idea Award to K.P. M.S. is supported by the Carroll-Feist Predoctoral Fellowship Award.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
Reference List
- 1.Heasman SJ, Ridley AJ. Mammalian Rho GTPases: new insights into their functions from in vivo studies. Nat Rev Mol Cell Biol. 2008 Sep;9(9):690–701. doi: 10.1038/nrm2476. [DOI] [PubMed] [Google Scholar]
- 2.Bosco EE, Mulloy JC, Zheng Y. Rac1 GTPase: a “Rac” of all trades. Cell Mol Life Sci. 2009 Feb;66(3):370–4. doi: 10.1007/s00018-008-8552-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rossman KL, Der CJ, Sondek J. GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nat Rev Mol Cell Biol. 2005 Feb;6(2):167–80. doi: 10.1038/nrm1587. [DOI] [PubMed] [Google Scholar]
- 4.Vigil D, Cherfils J, Rossman KL, Der CJ. Ras superfamily GEFs and GAPs: validated and tractable targets for cancer therapy? Nat Rev Cancer. 2010 Dec;10(12):842–57. doi: 10.1038/nrc2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Habets GG, Scholtes EH, Zuydgeest D, van der Kammen RA, Stam JC, Berns A, et al. Identification of an invasion-inducing gene, Tiam-1, that encodes a protein with homology to GDP-GTP exchangers for Rho-like proteins. Cell. 1994 May 20;77(4):537–49. doi: 10.1016/0092-8674(94)90216-x. [DOI] [PubMed] [Google Scholar]
- 6.Michiels F, Habets GG, Stam JC, van der Kammen RA, Collard JG. A role for Rac in Tiam1-induced membrane ruffling and invasion. Nature. 1995 May 25;375(6529):338–40. doi: 10.1038/375338a0. [DOI] [PubMed] [Google Scholar]
- 7.Lambert JM, Lambert QT, Reuther GW, Malliri A, Siderovski DP, Sondek J, et al. Tiam1 mediates Ras activation of Rac by a PI(3)K-independent mechanism. Nat Cell Biol. 2002 Aug;4(8):621–5. doi: 10.1038/ncb833. [DOI] [PubMed] [Google Scholar]
- 8.Cook DR, Rossman KL, Der CJ. Rho guanine nucleotide exchange factors: regulators of Rho GTPase activity in development and disease. Oncogene. 2013 Sep 16; doi: 10.1038/onc.2013.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Minard ME, Herynk MH, Collard JG, Gallick GE. The guanine nucleotide exchange factor Tiam1 increases colon carcinoma growth at metastatic sites in an orthotopic nude mouse model. Oncogene. 2005 Apr 7;24(15):2568–73. doi: 10.1038/sj.onc.1208503. [DOI] [PubMed] [Google Scholar]
- 10.Ding Y, Chen B, Wang S, Zhao L, Chen J, Ding Y, et al. Overexpression of Tiam1 in hepatocellular carcinomas predicts poor prognosis of HCC patients. Int J Cancer. 2009 Feb 1;124(3):653–8. doi: 10.1002/ijc.23954. [DOI] [PubMed] [Google Scholar]
- 11.Liu H, Shi G, Liu X, Wu H, Fan Q, Wang X. Overexpression of Tiam1 predicts poor prognosis in patients with esophageal squamous cell carcinoma. Oncol Rep. 2011 Mar;25(3):841–8. doi: 10.3892/or.2010.1122. [DOI] [PubMed] [Google Scholar]
- 12.Michiels F, Stam JC, Hordijk PL, van der Kammen RA, Ruuls-Van SL, Feltkamp CA, et al. Regulated membrane localization of Tiam1, mediated by the NH2-terminal pleckstrin homology domain, is required for Rac-dependent membrane ruffling and C-Jun NH2-terminal kinase activation. J Cell Biol. 1997 Apr 21;137(2):387–98. doi: 10.1083/jcb.137.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fleming IN, Elliott CM, Collard JG, Exton JH. Lysophosphatidic acid induces threonine phosphorylation of Tiam1 in Swiss 3T3 fibroblasts via activation of protein kinase C. J Biol Chem. 1997 Dec 26;272(52):33105–10. doi: 10.1074/jbc.272.52.33105. [DOI] [PubMed] [Google Scholar]
- 14.Hordijk PL, ten Klooster JP, van der Kammen RA, Michiels F, Oomen LC, Collard JG. Inhibition of invasion of epithelial cells by Tiam1-Rac signaling. Science. 1997 Nov 21;278(5342):1464–6. doi: 10.1126/science.278.5342.1464. [DOI] [PubMed] [Google Scholar]
- 15.Sander EE, van DS, ten Klooster JP, Reid T, van der Kammen RA, Michiels F, et al. Matrix-dependent Tiam1/Rac signaling in epithelial cells promotes either cell-cell adhesion or cell migration and is regulated by phosphatidylinositol 3-kinase. J Cell Biol. 1998 Nov 30;143(5):1385–98. doi: 10.1083/jcb.143.5.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Malliri A, van der Kammen RA, Clark K, van d V, Michiels F, Collard JG. Mice deficient in the Rac activator Tiam1 are resistant to Ras-induced skin tumours. Nature. 2002 Jun 20;417(6891):867–71. doi: 10.1038/nature00848. [DOI] [PubMed] [Google Scholar]
- 17.Woodcock SA, Rooney C, Liontos M, Connolly Y, Zoumpourlis V, Whetton AD, et al. SRC-induced disassembly of adherens junctions requires localized phosphorylation and degradation of the rac activator tiam1. Mol Cell. 2009 Mar 13;33(5):639–53. doi: 10.1016/j.molcel.2009.02.012. [DOI] [PubMed] [Google Scholar]
- 18.Buongiorno P, Pethe VV, Charames GS, Esufali S, Bapat B. Rac1 GTPase and the Rac1 exchange factor Tiam1 associate with Wnt-responsive promoters to enhance beta-catenin/TCF-dependent transcription in colorectal cancer cells. Mol Cancer. 2008;7:73. doi: 10.1186/1476-4598-7-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat Rev Genet. 2009 Jan;10(1):32–42. doi: 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krusche CA, Wulfing P, Kersting C, Vloet A, Bocker W, Kiesel L, et al. Histone deacetylase-1 and -3 protein expression in human breast cancer: a tissue microarray analysis. Breast Cancer Res Treat. 2005 Mar;90(1):15–23. doi: 10.1007/s10549-004-1668-2. [DOI] [PubMed] [Google Scholar]
- 21.Weichert W, Roske A, Niesporek S, Noske A, Buckendahl AC, Dietel M, et al. Class I histone deacetylase expression has independent prognostic impact in human colorectal cancer: specific role of class I histone deacetylases in vitro and in vivo. Clin Cancer Res. 2008 Mar 15;14(6):1669–77. doi: 10.1158/1078-0432.CCR-07-0990. [DOI] [PubMed] [Google Scholar]
- 22.Brooks CL, Gu W. How does SIRT1 affect metabolism, senescence and cancer? Nat Rev Cancer. 2009 Feb;9(2):123–8. doi: 10.1038/nrc2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu T, Liu PY, Marshall GM. The critical role of the class III histone deacetylase SIRT1 in cancer. Cancer Res. 2009 Mar 1;69(5):1702–5. doi: 10.1158/0008-5472.CAN-08-3365. [DOI] [PubMed] [Google Scholar]
- 24.Huffman DM, Grizzle WE, Bamman MM, Kim JS, Eltoum IA, Elgavish A, et al. SIRT1 is significantly elevated in mouse and human prostate cancer. Cancer Res. 2007 Jul 15;67(14):6612–8. doi: 10.1158/0008-5472.CAN-07-0085. [DOI] [PubMed] [Google Scholar]
- 25.Lee H, Kim KR, Noh SJ, Park HS, Kwon KS, Park BH, et al. Expression of DBC1 and SIRT1 is associated with poor prognosis for breast carcinoma. Hum Pathol. 2011 Feb;42(2):204–13. doi: 10.1016/j.humpath.2010.05.023. [DOI] [PubMed] [Google Scholar]
- 26.Chen J, Zhang B, Wong N, Lo AW, To KF, Chan AW, et al. Sirtuin 1 is upregulated in a subset of hepatocellular carcinomas where it is essential for telomere maintenance and tumor cell growth. Cancer Res. 2011 Jun 15;71(12):4138–49. doi: 10.1158/0008-5472.CAN-10-4274. [DOI] [PubMed] [Google Scholar]
- 27.Holloway KR, Barbieri A, Malyarchuk S, Saxena M, Nedeljkovic-Kurepa A, Cameron MM, et al. SIRT1 positively regulates breast cancer associated human aromatase (CYP19A1) expression. Mol Endocrinol. 2013 Mar;27(3):480–90. doi: 10.1210/me.2012-1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.North BJ, Marshall BL, Borra MT, Denu JM, Verdin E. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol Cell. 2003 Feb;11(2):437–44. doi: 10.1016/s1097-2765(03)00038-8. [DOI] [PubMed] [Google Scholar]
- 29.Vaquero A, Scher MB, Lee DH, Sutton A, Cheng HL, Alt FW, et al. SirT2 is a histone deacetylase with preference for histone H4 Lys 16 during mitosis. Genes Dev. 2006 May 15;20(10):1256–61. doi: 10.1101/gad.1412706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim HS, Vassilopoulos A, Wang RH, Lahusen T, Xiao Z, Xu X, et al. SIRT2 maintains genome integrity and suppresses tumorigenesis through regulating APC/C activity. Cancer Cell. 2011 Oct 18;20(4):487–99. doi: 10.1016/j.ccr.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang MH, Laurent G, Bause AS, Spang R, German N, Haigis MC, et al. HDAC6 and SIRT2 Regulate the Acetylation State and Oncogenic Activity of Mutant K-RAS. Mol Cancer Res. 2013 Sep;11(9):1072–7. doi: 10.1158/1541-7786.MCR-13-0040-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu PY, Xu N, Malyukova A, Scarlett CJ, Sun YT, Zhang XD, et al. The histone deacetylase SIRT2 stabilizes Myc oncoproteins. Cell Death Differ. 2013 Mar;20(3):503–14. doi: 10.1038/cdd.2012.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen J, Chan AW, To KF, Chen W, Zhang Z, Ren J, et al. SIRT2 overexpression in hepatocellular carcinoma mediates epithelial to mesenchymal transition by protein kinase B/glycogen synthase kinase-3beta/beta-catenin signaling. Hepatology. 2013 Jun;57(6):2287–98. doi: 10.1002/hep.26278. [DOI] [PubMed] [Google Scholar]
- 34.Zhang Y, Zhang M, Dong H, Yong S, Li X, Olashaw N, et al. Deacetylation of cortactin by SIRT1 promotes cell migration. Oncogene. 2009 Jan 22;28(3):445–60. doi: 10.1038/onc.2008.388. [DOI] [PubMed] [Google Scholar]
- 35.Holloway KR, Calhoun TN, Saxena M, Metoyer CF, Kandler EF, Rivera CA, et al. SIRT1 regulates Dishevelled proteins and promotes transient and constitutive Wnt signaling. Proc Natl Acad Sci U S A. 2010 May 18;107(20):9216–21. doi: 10.1073/pnas.0911325107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heltweg B, Gatbonton T, Schuler AD, Posakony J, Li H, Goehle S, et al. Antitumor activity of a small-molecule inhibitor of human silent information regulator 2 enzymes. Cancer Res. 2006 Apr 15;66(8):4368–77. doi: 10.1158/0008-5472.CAN-05-3617. [DOI] [PubMed] [Google Scholar]
- 37.Zhao G, Cui J, Zhang JG, Qin Q, Chen Q, Yin T, et al. SIRT1 RNAi knockdown induces apoptosis and senescence, inhibits invasion and enhances chemosensitivity in pancreatic cancer cells. Gene Ther. 2011 Sep;18(9):920–8. doi: 10.1038/gt.2011.81. [DOI] [PubMed] [Google Scholar]
- 38.Byles V, Zhu L, Lovaas JD, Chmilewski LK, Wang J, Faller DV, et al. SIRT1 induces EMT by cooperating with EMT transcription factors and enhances prostate cancer cell migration and metastasis. Oncogene. 2012 Oct 25;31(43):4619–29. doi: 10.1038/onc.2011.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Byles V, Chmilewski LK, Wang J, Zhu L, Forman LW, Faller DV, et al. Aberrant cytoplasm localization and protein stability of SIRT1 is regulated by PI3K/IGF-1R signaling in human cancer cells. Int J Biol Sci. 2010;6(6):599–612. doi: 10.7150/ijbs.6.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hou H, Chen W, Zhao L, Zuo Q, Zhang G, Zhang X, et al. Cortactin is associated with tumour progression and poor prognosis in prostate cancer and SIRT2 other than HADC6 may work as facilitator in situ. J Clin Pathol. 2012 Dec;65(12):1088–96. doi: 10.1136/jclinpath-2012-200940. [DOI] [PubMed] [Google Scholar]
- 41.Chen W, ten BD, Brown J, Ahn S, Hu LA, Miller WE, et al. Dishevelled 2 recruits beta-arrestin 2 to mediate Wnt5A-stimulated endocytosis of Frizzled 4. Science. 2003 Sep 5;301(5638):1391–4. doi: 10.1126/science.1082808. [DOI] [PubMed] [Google Scholar]
- 42.Eaton S, Wepf R, Simons K. Roles for Rac1 and Cdc42 in planar polarization and hair outgrowth in the wing of Drosophila. J Cell Biol. 1996 Dec;135(5):1277–89. doi: 10.1083/jcb.135.5.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Habas R, Kato Y, He X. Wnt/Frizzled activation of Rho regulates vertebrate gastrulation and requires a novel Formin homology protein Daam1. Cell. 2001 Dec 28;107(7):843–54. doi: 10.1016/s0092-8674(01)00614-6. [DOI] [PubMed] [Google Scholar]
- 44.Napper AD, Hixon J, McDonagh T, Keavey K, Pons JF, Barker J, et al. Discovery of indoles as potent and selective inhibitors of the deacetylase SIRT1. J Med Chem. 2005 Dec 15;48(25):8045–54. doi: 10.1021/jm050522v. [DOI] [PubMed] [Google Scholar]
- 45.Ota H, Tokunaga E, Chang K, Hikasa M, Iijima K, Eto M, et al. Sirt1 inhibitor, Sirtinol, induces senescence-like growth arrest with attenuated Ras-MAPK signaling in human cancer cells. Oncogene. 2006 Jan 12;25(2):176–85. doi: 10.1038/sj.onc.1209049. [DOI] [PubMed] [Google Scholar]
- 46.Li Y, Xu W, McBurney MW, Longo VD. SirT1 inhibition reduces IGF-I/IRS-2/Ras/ERK1/2 signaling and protects neurons. Cell Metab. 2008 Jul;8(1):38–48. doi: 10.1016/j.cmet.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barrio-Real L, Kazanietz MG. Rho GEFs and cancer: linking gene expression and metastatic dissemination. Sci Signal. 2012 Oct 2;5(244):e43. doi: 10.1126/scisignal.2003543. [DOI] [PubMed] [Google Scholar]
- 48.Allis CD, Berger SL, Cote J, Dent S, Jenuwien T, Kouzarides T, et al. New nomenclature for chromatin-modifying enzymes. Cell. 2007 Nov 16;131(4):633–6. doi: 10.1016/j.cell.2007.10.039. [DOI] [PubMed] [Google Scholar]
- 49.Hornbeck PV, Kornhauser JM, Tkachev S, Zhang B, Skrzypek E, Murray B, et al. PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 2012 Jan;40(Database issue):D261–D270. doi: 10.1093/nar/gkr1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pegtel DM, Ellenbroek SI, Mertens AE, van der Kammen RA, de RJ, Collard JG. The Par-Tiam1 complex controls persistent migration by stabilizing microtubule-dependent front-rear polarity. Curr Biol. 2007 Oct 9;17(19):1623–34. doi: 10.1016/j.cub.2007.08.035. [DOI] [PubMed] [Google Scholar]
- 51.Adam L, Vadlamudi RK, McCrea P, Kumar R. Tiam1 overexpression potentiates heregulin-induced lymphoid enhancer factor-1/beta -catenin nuclear signaling in breast cancer cells by modulating the intercellular stability. J Biol Chem. 2001 Jul 27;276(30):28443–50. doi: 10.1074/jbc.M009769200. [DOI] [PubMed] [Google Scholar]
- 52.Adams HC, III, Chen R, Liu Z, Whitehead IP. Regulation of breast cancer cell motility by T-cell lymphoma invasion and metastasis-inducing protein. Breast Cancer Res. 2010;12(5):R69. doi: 10.1186/bcr2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang S, Watanabe T, Matsuzawa K, Katsumi A, Kakeno M, Matsui T, et al. Tiam1 interaction with the PAR complex promotes talin-mediated Rac1 activation during polarized cell migration. J Cell Biol. 2012 Oct 15;199(2):331–45. doi: 10.1083/jcb.201202041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Worthylake DK, Rossman KL, Sondek J. Crystal structure of Rac1 in complex with the guanine nucleotide exchange region of Tiam1. Nature. 2000 Dec 7;408(6813):682–8. doi: 10.1038/35047014. [DOI] [PubMed] [Google Scholar]
- 55.Arthur WT, Ellerbroek SM, Der CJ, Burridge K, Wennerberg K. XPLN, a guanine nucleotide exchange factor for RhoA and RhoB, but not RhoC. J Biol Chem. 2002 Nov 8;277(45):42964–72. doi: 10.1074/jbc.M207401200. [DOI] [PubMed] [Google Scholar]
- 56.Cajanek L, Ganji RS, Henriques-Oliveira C, Theofilopoulos S, Konik P, Bryja V, et al. Tiam1 regulates the Wnt/Dvl/Rac1 signaling pathway and the differentiation of midbrain dopaminergic neurons. Mol Cell Biol. 2013 Jan;33(1):59–70. doi: 10.1128/MCB.00745-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.North BJ, Verdin E. Interphase nucleo-cytoplasmic shuttling and localization of SIRT2 during mitosis. PLoS One. 2007;2(8):e784. doi: 10.1371/journal.pone.0000784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gysin S, Salt M, Young A, McCormick F. Therapeutic strategies for targeting ras proteins. Genes Cancer. 2011 Mar;2(3):359–72. doi: 10.1177/1947601911412376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Drummond DC, Noble CO, Kirpotin DB, Guo Z, Scott GK, Benz CC. Clinical development of histone deacetylase inhibitors as anticancer agents. Annu Rev Pharmacol Toxicol. 2005;45:495–528. doi: 10.1146/annurev.pharmtox.45.120403.095825. [DOI] [PubMed] [Google Scholar]
- 60.Wagner JM, Hackanson B, Lubbert M, Jung M. Histone deacetylase (HDAC) inhibitors in recent clinical trials for cancer therapy. Clin Epigenetics. 2010 Dec;1(3–4):117–36. doi: 10.1007/s13148-010-0012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.