Summary
Objective
Coagulation phenotypes show strong intercorrelations, affect cardiovascular disease risk and are influenced by genetic variants. The objective of this study was to search for novel genetic variants influencing the following coagulation phenotypes: factor VII levels, fibrinogen levels, plasma viscosity and platelet count.
Methods and Results
We genotyped the British Women’s Heart and Health Study (n=3445) and the Whitehall II study (n=5059) using the Illumina HumanCVD BeadArray to investigate genetic associations and pleiotropy. In addition to previously reported associations (SH2B3, F7/F10, PROCR, GCKR, FGA/FGB/FGG, IL5), we identified novel associations at GRK5 (rs10128498, p=1.30×10−6), GCKR (rs1260326, p=1.63×10−6), ZNF259-APOA5 (rs651821, p=7.17×10−6) with plasma viscosity; and at CSF1 (rs333948, p=8.88×10−6) with platelet count. A pleiotropic effect was identified in GCKR which associated with factor VII (p=2.16×10−7) and plasma viscosity (p=1.63×10−6), and, to a lesser extent, ZNF259-APOA5 which associated with factor VII and fibrinogen (p<1.00×10−2) and additionally plasma viscosity (p<1.00×10−5). Triglyceride associated variants were overrepresented in Factor VII and plasma viscosity associations. Adjusting for triglyceride levels resulted in attenuation of associations at the GCKR and ZNF259-APOA5 loci.
Conclusions
In addition to confirming previously reported associations, we identified four SNPs associated with plasma viscosity and platelet count and found evidence of pleiotropic effects with SNPs in GCKR and ZNF259-APOA5. These triglyceride-associated, pleiotropic SNPs suggest a possible causal role for triglycerides in coagulation.
Keywords: Haemostasis, Thrombosis, HumanCVD, Clotting Factors, Genetic Association
Introduction
Blood coagulation is a process preventing blood loss from damaged vessels, but it can also be responsible for obstruction of blood vessels (thrombosis), potentially leading to venous thrombosis, or arterial thrombosis causing ischaemic heart disease, stroke or peripheral vascular disease (1). In the coagulation process, clots are formed from platelets (primary haemostasis) and fibrin (secondary haemostasis), with the latter being converted from fibrinogen as a consequence of a complex cascade of reactions between clotting factors. This coagulation cascade is triggered by the binding of circulating factor VII to tissue factor, when tissue damage exposes tissue factor to the blood stream (2). Amongst the numerous clotting factors and phenotypes within this coagulation cascade are factor VII levels, fibrinogen levels, platelet count and plasma viscosity.
Factor VII is a serine protease, and genes identified as associating with factor VII levels include the factor VII gene (F7), glucokinase (hexokinase 4) regulator (GCKR), and protein C receptor (PROCR) (3;4). Fibrinogen is a soluble glycoprotein circulating as a hexamer comprising two alpha, two beta and two gamma chains linked by disulphide bonds. The three genes encoding its subunits (FGA, FGB and FGG) contain variants that associate with fibrinogen levels (5;6). Other variants associating with fibrinogen levels are located in the APG4C region (7), the IL6R region (5), the CPS1 region (5), IRF1, PCCB, NLRP3 (6), NR3C2, GRIK2, PCSK2 (8).
Fibrinogen affects plasma viscosity (along with immunoglobulins), but the latter has not been extensively analysed for genetic association. However, there is some evidence of associations with intergenic SNPs rs10490258 on chromosome 2 and rs10485968 on chromosome 7, and intergenic SNPs near IBRDC2 and MAMDC1 (7) with plasma viscosity. In contrast, for platelet count genome-wide association studies have identified a number of loci, including SH2B3, RCL1, THPO, GP1BA (9), ATXN2, PTPN11, AK3 (10), BAK1 (9;10), PIK3CG (11), HBS1L/MYB (9;12), GCKR and others (13). Recently Gieger et al reported a comprehensive meta-analysis of GWAS for platelet count which identified a total of 56 variants (13).
Current evidence concerning SNPs associated with clotting phenotypes has some important limitations. Despite genome-wide association studies on these traits, studies have not reported on intra-system pleiotropy with coagulation traits. Levels of a clotting factor may reflect the direct effect of variation on the gene for that factor, or the indirect effect of variation on another gene acting via a particular biochemical pathway. The latter offers scope for a single genetic variant to influence multiple coagulation traits downstream of that biochemical pathway. One such pathway is triglyceride metabolism: triglyceride levels associate with factor VII and other vitamin K-dependent clotting factor levels (14), and with elevated plasma viscosity(15). A genetic analysis of the pleiotropic role of triglyceride-associated SNPs in coagulation phenotypes therefore offers scope to explore the potential “causal” role of triglycerides in this system.
To address these limitations, we present a genetic analysis of four phenotypes, factor VII levels, fibrinogen levels, plasma viscosity and platelet count, measured in two large cohort studies with genotype data from the HumanCVD BeadArray (16). Associated SNPs were also examined for pleiotropic effects between the phenotypes studied and triglyceride-associated SNPs were tested for elevated association with clotting traits. Though not providing genome wide coverage, the HumanCVD BeadChip specifically targets rare SNPs and so may have an advantage over standard GWAS platforms to detect association with rare variants.
Materials and Methods
Cohorts
Our analysis was based on the British Women’s Heart and Health Study (BWHHS, n=3445) and the Whitehall II study (WHII, n=5059) (supplementary table 1). In the BWHHS, a prospective study of heart disease in British women, over 4000 participants were recruited from general practice registers in 23 UK towns (17;18). Baseline measurements were taken in 1999-2001 (participant ages 60-79), with subsequent follow-ups in 2003, 2007 and 2010. Blood samples were collected at baseline and DNA extracted from whole blood using a salting-out procedure (19). Clotting traits were measured on baseline blood samples. Fibrinogen and factor VII were measured in an automated coagulometer (MDA, Organon Teknika, Cambridge, UK) using the manufacturer’s reagents and standards. Plasma viscosity was measured at 37°C in a semi-automated capillary viscometer (Coulter, Luton, Beds). Platelet count was performed using laser light scatter flow cytometry on an Advia 120 haematology system (Bayer Healthcare, Newbury, UK).
The WHII study recruited 10,308 participants (70% men) between 1985 and 1989 from 20 London based Civil service departments (20;21). Blood samples were collected in 2002-4 and DNA was extracted from 6,156 whole blood samples using magnetic bead technology (Medical Solutions, Nottingham, UK) and normalised to a concentration of 50ng/μl. Clotting traits were measured in 1991-1993. Fibrinogen and factor VII were measured in an automated coagulometer (MDA, Organon Teknika, Cambridge, UK) using the manufacturer’s reagents and standards. Plasma viscosity was measured at 37°C in a semi-automated capillary viscometer (Coulter, Luton, Beds), and platelet count used a Coulter counter (Coulter, Luton, Beds).
Genotyping
HumanCVD BeadChip (Illumina Inc) content was selected by a consortium comprising the Institute of Translational Medicine and Therapeutics (ITMAT), the Broad Institute and the National Heart Lung and Blood Institute (NHLBI) supported Candidate-gene Association Resource (CARe) Consortium (16). This BeadChip tests nearly 50,000 SNPs in approximately 2000 genes selected for functional candidacy in cardiovascular disease and associated traits.
For BWHHS, genotyping was successfully performed on 3445 samples using the HumanCVD BeadChip (Illumina). The following “hard” limits for Illumina BeadStudio parameters were applied to the data: cluster separation < 0.3, call frequency < 0.95, AB R mean < 0.3, AB T mean < 0.2 or > 0.8, heterozygote excess < −0.3 or > 0.1. SNPs at the borderline of these limits for each parameter were then checked and either re-clustered or discarded as necessary. Principal components analysis confirmed self-reported ethnicity and 32 individuals were excluded on the basis of non-European ancestry, leaving 3413 samples for analysis.
For WHII, full details of the genotyping and data cleaning have been published previously (22). After quality control (filtering for duplicates, cryptic relatedness, ambiguous sex, self-reported non-Caucasians, outliers based on the genome-wide identity-by-state analysis implemented in PLINK, sample call rate <80% and SNP call rate <98%), 5059 HumanCVD genotyped samples were available for the analysis.
Statistical analyses
Preliminary association analyses for each trait were carried out separately in each study using PLINK (23) (http://pngu.mgh.harvard.edu/~purcell/plink/epi.shtml) to perform a linear regression (per-allele test) for association between each SNP and the trait with and without age and sex adjustment, and excluding participants on warfarin or aspirin therapy (whilst aspirin acts primarily on platelet function, there is some uncertainty about other antithrombotic properties (24)). Data for BWHHS and WHII were then meta-analysed using METAL (25). Results were ranked in ascending order of p-value for the purposes of interpretation, and a p-value threshold of 1×10−5 was selected to balance the risk of false positives against the higher prior probability of true positives expected with a candidate gene array (16). These analyses were repeated for both plasma viscosity and factor VII in a post-hoc analysis adjusted for triglyceride levels to determine for which genotypes triglyceride levels are likely to be in the causal chain.
Variable selection was applied to identify the genetic variants with strongest predictive effect at each locus. Step-wise selection was implemented using Akaike’s Information Criterion (26) separately for each chromosome. The selected variants were then analysed for proportion of the total variance explained (r2) using a regression of the SNP on phenotype after adjustment for age.
Analysis of pleiotropy
As the phenotypes examined are part of an interlinked biological process we examined the hypothesis that genetic variants associated with one phenotype might also affect others. Potential pleiotropic effects were examined for all SNPs associated at p < 1 × 10−6 (63 unique SNPs) with any of the four traits. Each of these SNPs was examined for its association (analysed as above) with the three traits other than the one by which it had originally been selected within the meta-analysis data for BWHHS and WHII. These data were plotted to give a graphical representation of potential pleiotropy.
In addition to this analysis, we cross-referenced our top hits from table 1 (and any tag SNP with r2 > 0.8 linkage disequilibrium from 1000 genomes CEU) with the NHGRI GWAS Catalog (27) to identify pleiotropic effects on other phenotypes (coagulation, other cardiovascular or any other reported).
Table 1. Most significantly associated SNP at each locus associated with four coagulation phenotypes in a meta-analysis between BWHHS and WHII at p<1×10−5.
| SNP ID | Allele | Dir | P | Z-score | N | Gene/region | Chr | Pos |
|---|---|---|---|---|---|---|---|---|
| Platelet Count | ||||||||
| rs17696736 | G | ++ | 1.24×10−06 | 4.850 | 4385 | NAA25-SH2B3 | 12 | 110,971,201 |
| rs333948 | A | ++ | 8.88×10−06 | 4.443 | 4396 | CSF1 | 1 | 110,272,115 |
| Plasma Viscosity | ||||||||
| rs10128498 | G | −− | 1.30×10−06 | −4.840 | 6728 | GRK5 | 10 | 121,042,898 |
| rs1260326 | A | ++ | 1.63×10−06 | 4.795 | 6718 | GCKR | 2 | 27,584,444 |
| rs651821 | G | ++ | 7.17×10−06 | 4.489 | 6745 | ZNF259-APOA5 | 11 | 116,167,789 |
| Fibrinogen Levels | ||||||||
| rs4508864 | A | ++ | 5.59×10−10 | 6.202 | 7258 | FGB | 4 | 155,700,739 |
| rs743562 | A | −− | 4.23×10−06 | −4.600 | 7249 | IL5 | 5 | 131,900,282 |
| Factor VII Levels | ||||||||
| rs6041 | A | −− | 2.22×10−307 | −37.477 | 7218 | F10-F7 | 13 | 112,820,708 |
| rs8119351 | A | ++ | 1.02×10−22 | 9.810 | 7232 | PROCR | 20 | 33,218,066 |
| rs1260326 | A | ++ | 2.16×10−07 | 5.185 | 7207 | GCKR | 2 | 27,584,444 |
Dir = direction of effect in the two studies (where ++ indicates both studies show an increase in trait per allele and – indicates a decrease in trait per allele).
Analysis of triglyceride-associated SNP enrichment in factor VII and plasma viscosity results
SNPs reported as associated with triglycerides in the Teslovich et al “95 lipid loci” paper and also present on the HumanCVD array (n=14, listed below) (28) were tested for their potential association with factor VII and plasma viscosity in BWHHS using a linear regression (per-allele test) for association between SNP and trait in PLINK (23). Phenotype association results (Z-scores) for all SNPs from the intersection of this set with the HumanCVD were extracted and compared to the HumanCVD set as a whole using a quantile-quantile comparison of Z-score distribution. Significantly associated SNPs for each of plasma viscosity and factor VII were analysed with adjustment for triglyceride levels in both cohorts to determine whether they show any effect independent of triglycerides. This was done by including triglyceride levels as a covariate in the linear regression. Finally, a triglyceride “allele score” was constructed by summing the number of risk alleles for each individual at the following loci: rs1260326, rs17145713, rs285, rs331, rs3289, rs10503669, rs17321515, rs17108993, rs6589565, rs12286037, rs651821, rs10750097, rs5072 and rs2304128. This allele score was then tested for association with plasma viscosity and factor VII in BWHHS and WHII using a regression of phenotype on allele score, then meta-analysed using the rmeta package in R.
Results
Association analyses
Table 1 presents summary results (the most significantly associated SNP at each genomic locus) from a HumanCVD-wide meta-analysis of 36,033 SNPs against each of the four coagulation traits (for summary Manhattan plots see supplementary figure 1 and for full details of all associated SNPs see supplementary table 2). These analyses were unadjusted for age and sex (adjustment made little difference to the results), but users of warfarin or aspirin were excluded from the analyses.
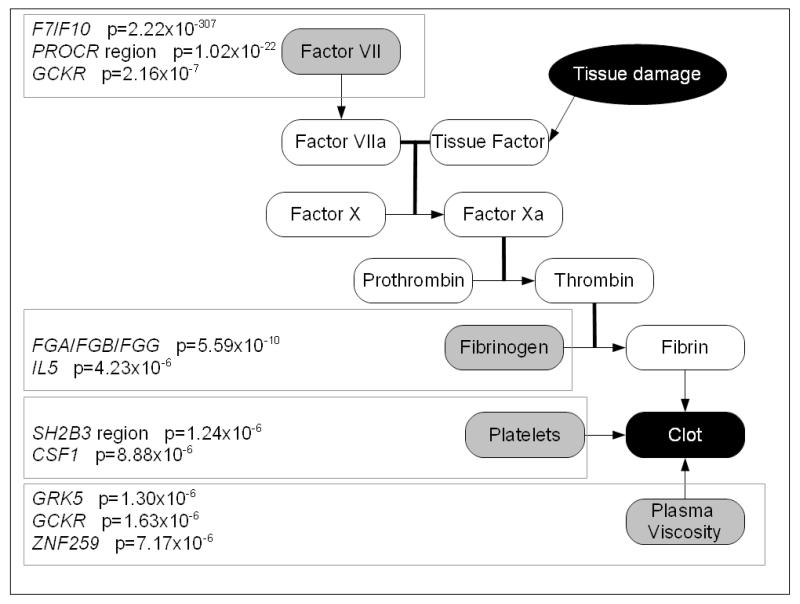
The associated genes and the p-value for the most-significantly associated SNP at each gene locus are presented on a schematic of the coagulation cascade in Figure 1. Novel associations with plasma viscosity were observed at the GRK5 (rs10128498, p=1.30×10−6), GCKR (rs1260326, 1.63×10−6) and ZNF259-APOA5 (rs651821, p=7.17×10−6) loci, and with platelet count at the CSF1 locus (rs333948, p=8.88×10−6). Previously reported loci replicated in our analyses included: the SH2B3 region and platelet count (rs17696736, p=1.24×10−6), and the F7/F10 (rs6041, p=2.22×10−307), PROCR (rs8119351, p=1.02×10−22), GCKR (rs1260326, p=2.16×10−07) loci and factor VII levels, and the FGA/FGB/FGG locus (rs4508864, p=5.59×10−10) and IL5 locus (rs743562, p=4.23×10−6) and fibrinogen levels. The association of the IL5 SNP with fibrinogen was not replicated in a “look-up” in existing data from Northwick Park Heart Study II (NPHSII, n=2605, data not shown), but was replicated in a large GWAS of fibrinogen levels in 22,096 Europeans (6).
Figure 1.
Genes containing SNPs associated with parts of the coagulation system in the meta-analysis
The proportion of trait variance for each locus (r2) was tested by using the SNPs surviving a variable selection (step-wise regression) in a multiple regression model. In BWHHS, variation in the F7 region accounts for 15.6% of variance in factor VII levels, with 1.6% attributable to the chromosome 20 PROCR region, while GCKR accounted for around 0.5%. Variation in the FBA/B/G region accounted for ~ 3% of the variance in fibrinogen levels. For plasma viscosity, the GCKR region accounted for 1.5% variance, GRK5 for 1% and ZNF259-APOA5 for 1.2%. Platelet count SNPs in our top set accounted for around 0.2% variance in each region.
Analysis of pleiotropy
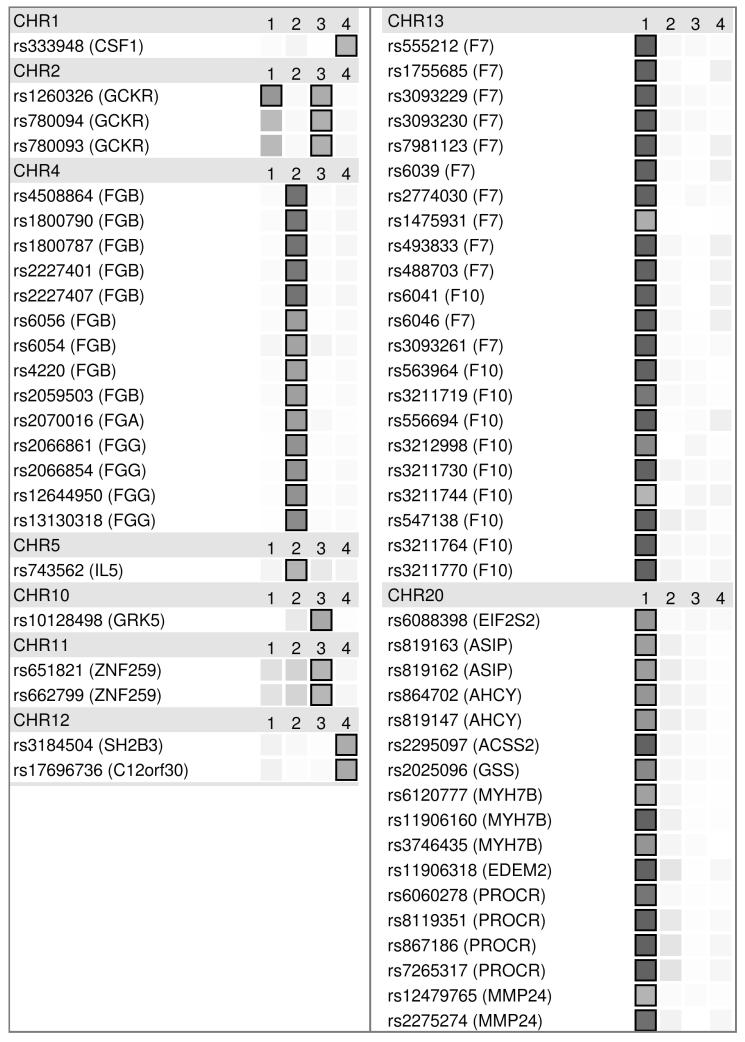
Figure 2 presents the results from the analysis of pleiotropy. For each SNP, the strength of evidence against the null hypothesis of no association is plotted as intensity (all associations exceeding our threshold of p = 1 × 10−5 are boxed, p < 1 × 10−10 are represented as saturated). None of the SNPs associated with platelet count were associated with any of the other traits, but for each of the other traits there were some SNPs that showed some evidence of pleiotropic effects on other traits. Of these, the only one passing a p = 1 × 10−5 threshold was GCKR SNP rs1260326, which associated with both plasma viscosity (p = 1.63 × 10−6) and factor VII (p = 2.16 × 10−7). The only others showing some evidence (but not exceeding our HumanCVD-wide threshold) for two or more traits included ZNF259-APOA5 SNPs rs651821 and rs662799 (both SNPs p < 1.00 × 10−2 for factor VII, p < 1.00 × 10−3 for fibrinogen and p < 1.00 × 10−5 for plasma viscosity).
Figure 2.
Pleiotropy analysis of most significant SNPs for each trait (from table 1). Traits are in columns labelled: [1] Factor VII, [2] Fibrinogen, [3] Plasma Viscosity and [4] Platelet Count. Results are ordered by chromosome then position (in one section per chromosome), with dbSNP accession number and mapped gene name given for each. Significance is indicated by grey intensity, max grey threshold at 1×10−10 and boxed at 1×10−5 (our HumanCVD-wide significance threshold).
Patterns of pleiotropy (in terms of the combination of phenotypes for which there was some evidence of association) were consistent across associated SNPs within a particular locus, most likely reflecting high linkage disequilibrium between associated SNPs at that locus. In contrast, there were marked differences in patterns between some loci (eg comparing the pleiotropic pattern with GCKR SNPs versus the trait-specific pattern observed with F7 SNPs).
Supplementary table 3 shows the results of cross-referencing our results from table 1 (or tag SNPs with r2 > 0.8) with the NHGRI GWAS catalog (27). Unsurprisingly SNPs associated with Factor VII levels in our analysis are reported as associated with coagulation phenotypes in several GWAS (rs6041 and rs8119351). However, a range of other associations are seen, including with a range of metabolic, cardiovascular and inflammatory phenotypes and outcomes, supporting a pleiotropic role of these genes in disease phenotypes.
Analysis of triglyceride-associated SNP enrichment in factor VII and plasma viscosity
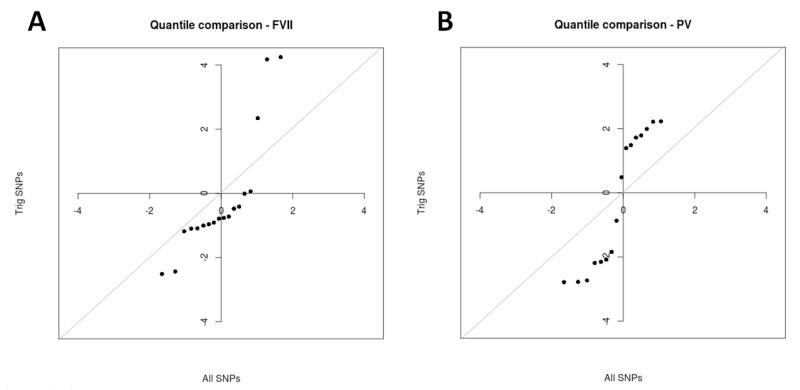
The hypothesis that triglyceride SNPs are likely to associate with factor VII levels and plasma viscosity was tested by performing a quantile-quantile comparison of z-score distribution for all SNPs in HumanCVD vs. just those SNPs reported as associated with triglycerides in the 95 lipid loci paper (28) (Figure 3). These results suggest that SNPs associated with triglycerides are more likely to be associated with factor VII levels or plasma viscosity than average.
Figure 3.
Enrichment of triglyceride SNPs amongst (A) factor VII and (B) plasma viscosity results. Plots show QQ comparison of Z-score distribution for all SNPs in HumanCVD vs just those SNPs reported as associated with triglycerides in a large GWAS of lipid levels (28).
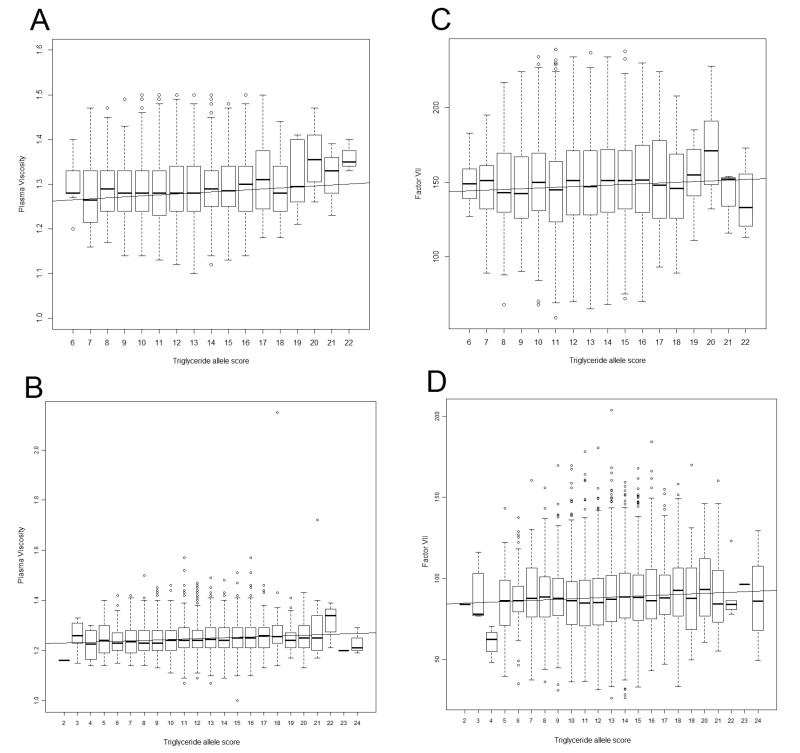
Supplementary tables 4 and 5 show association analyses of our top hits adjusted for triglyceride levels, illustrating that effects in GCKR and ZNF259-APOA5 are substantially due to genotypic effects acting through triglyceride levels: the original associations attenuated substantially following the adjustment for triglycerides. Figure 4 shows the relationship between a triglyceride “allele score” (i.e. sum of triglyceride-raising alleles across multiple loci) and plasma viscosity and factor VII. A linear model fitted to these data and a meta-analysis across BWHHS and WHII gives a regression coefficient of 0.0019 milliPascal seconds per triglyceride-raising allele for plasma viscosity (95% CI 0.0013 to 0.0025, p=9.8×10−10). For factor VII, meta-analysis gives a regression coefficient of 0.37 international units/decilitre per triglyceride-raising allele (95% CI 0.16 to 0.57, p=0.0004). This analysis is consistent with the observed attenuation of effects of GCKR and ZNF259-APOA5 when triglycerides are considered, and illustrate a general relationship between triglyceride-raising genetic variation and both of these phenotypes.
Figure 4.
(A) Relationship between triglyceride allele score and plasma viscosity in BWHHS, (B) Relationship between triglyceride allele score and plasma viscosity in WHII, (C) Relationship between triglyceride allele score and factor VII levels in BWHHS, (D) Relationship between triglyceride allele score and factor VII levels in WHII.
Discussion
Using the HumanCVD array to examine four coagulation traits we confirmed a number of previously reported associations (SH2B3, F7/F10, PROCR, GCKR, FGA/FGB/FGG, and IL5), identified several previously unreported associations and found that SNPs in GCKR and ZNF259-APOA5 appear to have a pleiotropic effect on coagulation phenotypes. Loci newly identified as associated with plasma viscosity were GRK5 (rs10128498, p=1.30×10−6), GCKR (rs1260326, 1.63×10−6) and ZNF259-APOA5 (rs651821, p=7.17×10−6); and with platelet count was CSF1 (rs333948, p=8.88×10−6). G protein-coupled receptor kinase 5 (encoded by GRK5) is involved in deactivation of G protein-coupled receptors, and is believed to play a role in regulating neutrophil motility (29). Glucokinase (hexokinase 4) regulator (encoded by GCKR) inactivates glucokinase in liver and pancreatic islets by binding to the enzyme. This particular SNP (rs1260326, Pro446Leu) is associated with impaired glucose tolerance (IGT) (30) and the T (Leu) allele associates with increased triglyceride levels (31-34).
The ZNF259-APOA5 locus contains APOA5, APOA4, APOC3 and APOA1, and has been robustly demonstrated to be associated with triglyceride levels (35-37). The relationship between triglyceride levels and coagulation measures is well established, with associations between hyperlipidaemia and hypercoagulability demonstrated in both animal models and humans (38-40). The associations of ZNF259-APOA5 and GCKR with plasma viscosity (and factor VII for GCKR) may therefore be caused by the influence of genetic variants in these regions on triglyceride levels. An analysis of SNPs associated with triglyceride levels showed that these were indeed enriched in our results for plasma viscosity, supporting an important role of triglyceride levels in this phenotype. A specific check in BWHHS of our ZNF259-APOA5 top SNPs associated with plasma viscosity also demonstrated association with triglycerides (p < 0.01, data not shown). As triglycerides are hypothesised to be on the causal pathway between genotype and both factor VII levels(14) and plasma viscosity(15), we re-analysed the most significantly associated SNPs for these traits adjusted for triglyceride levels (supplementary tables 4 & 5). A strong attenuation of signal for both GCKR and ZNF259-APOA5 in these analyses supports the hypotheses that these SNPs act primarily through triglyceride levels. Interestingly, these are the two loci that show the strongest evidence for potential pleiotropic effects (figure 2, discussed below), which is consistent with an indirect causal pathway via triglyceride levels. In contrast, the GRK5 plasma viscosity association was relatively unchanged (as were all the other main factor VII associations), suggesting that triglyceride levels are unimportant in the functional mechanisms underlying this association. A gene score approach (using sum of triglyceride-raising alleles) supports the hypothesis that genetic effects on triglycerides have downstream consequences on plasma viscosity and levels of factor VII (figure 4). Actually, the association between triglyceride-associated genes and plasma viscosity is likely to be mediated by triglyceride-associated lipoproteins which have direct effects on viscosity (whereas lipids themselves do not) (15;41).
In a system of interacting components such as the coagulation cascade, it is important to consider the possibility of pleiotropy. In addition to a selection of pleiotropic associations not surviving a Bonferroni correction, the GCKR SNP rs1260326, which associated with plasma viscosity (p = 1.63 × 10−6), was also observed to associate with factor VII (p = 2.16 × 10−7). This gene is likely to be acting through an effect on triglyceride levels, which in turn affects plasma viscosity and factor VII levels. There may be potential for functional classification of genes using the pleiotropic association profile observed for SNPs within that gene. For example, GCKR SNPs show effects on both plasma viscosity and factor VII levels, whilst PROCR SNPs only show evidence of association with factor VII levels. This difference in pattern is consistent with the likely different functional mechanisms of these two genes, with GCKR acting in part through triglyceride levels, with PROCR acting on activation of protein C. A GWAS of plasma levels of protein C reports that variants in both genes associate with protein C levels (42), suggesting that the GCKR may act on factor VII levels via protein C and on plasma viscosity via triglycerides. Our results suggest a biologically important inference: if triglyceride-associated SNPs associate with coagulation traits, this indicates that triglycerides are potentially functionally important in the mechanisms of coagulation.
To investigate whether any of the observed genetic associations with quantitative coagulation traits showed evidence of disease implications, we explored the relationship between our most significantly associated SNPs and deep vein thrombosis (DVT), and D-dimer levels. Our analysis showed no strong evidence of association with either DVT outcome or D-dimer levels (data not shown), but limited statistical power means we can’t exclude the possibility of a small effect.
A limitation of this study is that only four clotting-related phenotypes were available in both of the cohorts, which limited the extent to which we could analyse the coagulation system as a whole. A second limitation is that the associations described explained only a small proportion of trait variance. This is in common with many large-scale genetic association studies. However, the results still provide potentially important functional information about the molecular basis of coagulation that could be important in the contexts of drug development or rare variant effect prediction. In either of these cases a larger effect through the same molecular mechanism could be clinically important, hence identifying the mechanism adds important information to the field. Finally, we opted to perform a meta-analysis rather than separate discovery/replication analyses. This ensured we maximised statistical power, but could be susceptible to “winner’s curse”.
In conclusion, we confirm a number of previously reported genetic associations with four coagulation phenotypes and report some novel associations. Whilst informative, none of the SNPs identified appears to have an individual effect likely to be of clinical importance. However, an analysis of triglyceride-associated SNPs with pleiotropic effects on clotting phenotypes suggests a possible causal role for triglycerides in coagulation (consistent with observed associations (38-40)) that merits further investigation.
Supplementary Material
| What is known on this topic | What this paper adds |
|---|---|
|
|
Acknowledgements
We thank all of the participants and the general practitioners, research nurses and data management staff who supported data collection and preparation.
Funding
BWHHS: The British Women’s Heart and Health Study has been supported by funding from the BHF (PG/07/131/24254) and the Department of Health Policy Research Programme (England). The BWHHS is co-ordinated by Shah Ebrahim (PI), Debbie Lawlor and Juan-Pablo Casas, with cardiochip work funded by the BHF (PG/07/131/24254, PI Tom Gaunt, co-Is Ian Day, Debbie Lawlor, Shah Ebrahim, George Davey Smith, Yoav Ben-Shlomo, Santiago Rodriguez). CL is supported by an MRC project grant (G1000427, PI TRG).
WHII: Prof Humphries is a British Heart Foundation (BHF) Chairholder (PG08/008). The UCL Genetics Institute supports DZ, and SS. The work on WH-II was supported by the BHF PG/07/133/24260, RG/08/008, SP/07/007/23671 and a Senior Fellowship to Professor Hingorani (FS/2005/125). Dr Kumari’s and Prof. Kivimaki’s time on this manuscript was partially supported by the National Heart Lung and Blood Institute (NHLBI: HL36310). The WH-II study has been supported by grants from the Medical Research Council; Economic and Social Research Council; British Heart Foundation; Health and Safety Executive; Department of Health; National Institute on Aging (AG13196), US, NIH; Agency for Health Care Policy Research (HS06516); and the John D and Catherine T MacArthur Foundation Research Networks on Successful Midlife Development and Socio-economic Status and Health.
Footnotes
Disclosures JW is employed by GlaxoSmithKline and owns GlaxoSmithKline shares. ADH has served on an advisory board for Roche (no personal remuneration).
References
- (1).MacCallum PK, Meade TW. Haemostatic function, arterial disease and the prevention of arterial thrombosis. Baillieres Best Pract Res Clin Haematol. 1999;12:577–99. doi: 10.1053/beha.1999.0041. [DOI] [PubMed] [Google Scholar]
- (2).Davie EW, Fujikawa K, Kisiel W. The coagulation cascade: initiation, maintenance, and regulation. Biochemistry. 1991;30:10363–70. doi: 10.1021/bi00107a001. [DOI] [PubMed] [Google Scholar]
- (3).Smith NL, Chen MH, Dehghan A, Strachan DP, Basu S, Soranzo N, et al. Novel Associations of Multiple Genetic Loci With Plasma Levels of Factor VII, Factor VIII, and von Willebrand Factor. Circulation. 2010;121:1382–92. doi: 10.1161/CIRCULATIONAHA.109.869156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Taylor KC, Lange LA, Zabaneh D, Lange E, Keating BJ, Tang W, et al. A gene-centric association scan for Coagulation Factor VII levels in European and African Americans: the Candidate Gene Association Resource (CARe) Consortium. Human Molecular Genetics. 2011;20:3525–34. doi: 10.1093/hmg/ddr264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Danik JS, Paré G, Chasman DI, Zee RYL, Kwiatkowski DJ, Parker A, et al. Novel Loci, Including Those Related to Crohn Disease, Psoriasis, and Inflammation, Identified in a Genome-Wide Association Study of Fibrinogen in 17 686 Women / CLINICAL PERSPECTIVE. Circulation: Cardiovascular Genetics. 2009;2:134–41. doi: 10.1161/CIRCGENETICS.108.825273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Dehghan A, Yang Q, Peters A, Basu S, Bis JC, Rudnicka AR, et al. Association of Novel Genetic Loci With Circulating Fibrinogen Levels / CLINICAL PERSPECTIVE. Circulation: Cardiovascular Genetics. 2009;2:125–33. doi: 10.1161/CIRCGENETICS.108.825224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Yang Q, Kathiresan S, Lin JP, Tofler GH, O’Donnell CJ. Genome-wide association and linkage analyses of hemostatic factors and hematological phenotypes in the Framingham Heart Study. BMC Med Genet. 2007;8(Suppl 1):S12. doi: 10.1186/1471-2350-8-S1-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Zemunik T, Boban M, Lauc G, Jankovic S, Rotim K, Vatavuk Z, et al. Genome-wide association study of biochemical traits in Korcula Island, Croatia. Croat Med J. 2009;50:23–33. doi: 10.3325/cmj.2009.50.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Kamatani Y, Matsuda K, Okada Y, Kubo M, Hosono N, Daigo Y, et al. Genome-wide association study of hematological and biochemical traits in a Japanese population. Nat Genet. 2010;42:210–5. doi: 10.1038/ng.531. [DOI] [PubMed] [Google Scholar]
- (10).Soranzo N, Spector TD, Mangino M, K++hnel B, Rendon A, Teumer A, et al. A genome-wide meta-analysis identifies 22 loci associated with eight hematological parameters in the HaemGen consortium. Nat Genet. 2009;41:1182–90. doi: 10.1038/ng.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Soranzo N, Rendon A, Gieger C, Jones CI, Watkins NA, Menzel S, et al. A novel variant on chromosome 7q22.3 associated with mean platelet volume, counts, and function. Blood. 2009;113:3831–7. doi: 10.1182/blood-2008-10-184234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Ferreira MAR, Hottenga JJ, Warrington NM, Medland SE, Willemsen G, Lawrence RW, et al. Sequence Variants in Three Loci Influence Monocyte Counts and Erythrocyte Volume. The American Journal of Human Genetics. 2009;85:745–9. doi: 10.1016/j.ajhg.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Gieger C, Radhakrishnan A, Cvejic A, Tang W, Porcu E, Pistis G, et al. New gene functions in megakaryopoiesis and platelet formation. Nature. 2011;480:201–8. doi: 10.1038/nature10659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Constantino M, Merskey C, Kudzma DJ, Zucker MB. Increased activity of vitamin K-dependent clotting factors in human hyperlipoproteinaemia - association with cholesterol and triglyceride levels. Thromb Haemost. 1977;38:465–74. [PubMed] [Google Scholar]
- (15).Rosenson RS, Shott S, Tangney CC. Hypertriglyceridemia is associated with an elevated blood viscosity Rosenson: triglycerides and blood viscosity. Atherosclerosis. 2002;161:433–9. doi: 10.1016/s0021-9150(01)00656-6. [DOI] [PubMed] [Google Scholar]
- (16).Keating BJ, Tischfield S, Murray SS, Bhangale T, Price TS, Glessner JT, et al. Concept, Design and Implementation of a Cardiovascular Gene-Centric 50 K SNP Array for Large-Scale Genomic Association Studies. PLoS ONE. 2008;3:e3583. doi: 10.1371/journal.pone.0003583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Lawlor DA, Bedford C, Taylor M, Ebrahim S. Aspirin use for the prevention of cardiovascular disease: the British Womens Heart and Health Study. British Journal of General Practice. 2001;51(470):743–745. Ref Type: Abstract. [PMC free article] [PubMed] [Google Scholar]
- (18).Lawlor DL, Ebrahim SE, Davey Smith GDS. The association between components of adult height and Type II diabetes and insulin resistance: British Women’s Heart and Health Study. Diabetologia. 2002;45:1097–106. doi: 10.1007/s00125-002-0887-5. [DOI] [PubMed] [Google Scholar]
- (19).Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Research. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Marmot MG, Stansfeld S, Patel C, North F, Head J, White I, et al. Health inequalities among British civil servants: the Whitehall II study. The Lancet. 1991;337:1387–93. doi: 10.1016/0140-6736(91)93068-k. [DOI] [PubMed] [Google Scholar]
- (21).Marmot M, Brunner E. Cohort Profile: The Whitehall II study. Int J Epidemiol. 2005;34:251–6. doi: 10.1093/ije/dyh372. [DOI] [PubMed] [Google Scholar]
- (22).Talmud PJ, Drenos F, Shah S, Shah T, Palmen J, Verzilli C, et al. Gene-centric Association Signals for Lipids and Apolipoproteins Identified via the HumanCVD BeadChip. The American Journal of Human Genetics. 2009;85:628–42. doi: 10.1016/j.ajhg.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. The American Journal of Human Genetics. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Undas A, Brummel-Ziedins KE, Mann KG. Antithrombotic properties of aspirin and resistance to aspirin: beyond strictly antiplatelet actions. Blood. 2007;109:2285–92. doi: 10.1182/blood-2006-01-010645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–1. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Akaike H. New Look at Statistical-Model Identification. Ieee Transactions on Automatic Control. 1974;AC19:716–23. [Google Scholar]
- (27).Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106:9362–7. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–13. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Arraes SM, Freitas MS, da Silva SV, de Paula Neto HA, Alves-Filho JC, Martins MA, et al. Impaired neutrophil chemotaxis in sepsis associates with GRK expression and inhibition of actin assembly and tyrosine phosphorylation. Blood. 2006;108:2906–13. doi: 10.1182/blood-2006-05-024638. [DOI] [PubMed] [Google Scholar]
- (30).Saxena R, Hivert MF, Langenberg C, Tanaka T, Pankow JS, Vollenweider P, et al. Genetic variation in GIPR influences the glucose and insulin responses to an oral glucose challenge. Nat Genet. 2010;42:142–8. doi: 10.1038/ng.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Chambers JC, Zhang W, Sehmi J, Li X, Wass MN, Van der Harst P, et al. Genome-wide association study identifies loci influencing concentrations of liver enzymes in plasma. Nat Genet. 2011;43:1131–8. doi: 10.1038/ng.970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Diabetes Genetics Initiative of Broad Institute of Harvard and MIT LUaNIoBR. Saxena R, Voight BF, Lyssenko V, Burtt NlP, de Bakker PIW, et al. Genome-Wide Association Analysis Identifies Loci for Type 2 Diabetes and Triglyceride Levels. Science. 2007;316:1331–6. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- (33).Kathiresan S, Melander O, Guiducci C, Surti A, Burtt NP, Rieder MJ, et al. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat Genet. 2008;40:189–97. doi: 10.1038/ng.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Johansen CT, Wang J, Lanktree MB, Cao H, McIntyre AD, Ban MR, et al. Excess of rare variants in genes identified by genome-wide association study of hypertriglyceridemia. Nat Genet. 2010;42:684–7. doi: 10.1038/ng.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Waterworth DM, Ricketts SL, Song K, Chen L, Zhao JH, Ripatti S, et al. Genetic Variants Influencing Circulating Lipid Levels and Risk of Coronary Artery Disease. Arteriosclerosis, Thrombosis, and Vascular Biology. 2010;30:2264–76. doi: 10.1161/ATVBAHA.109.201020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Pollin TI, Damcott CM, Shen H, Ott SH, Shelton J, Horenstein RB, et al. A Null Mutation in Human APOC3 Confers a Favorable Plasma Lipid Profile and Apparent Cardioprotection. Science. 2008;322:1702–5. doi: 10.1126/science.1161524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Aulchenko YS, Ripatti S, Lindqvist I, Boomsma D, Heid IM, Pramstaller PP, et al. Loci influencing lipid levels and coronary heart disease risk in 16 European population cohorts. Nat Genet. 2008;41:47. doi: 10.1038/ng.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Kim WM, Merskey C, Deming QB, Adel HN, Wolinsky H, Clarkson TB, et al. Hyperlipidemia, hypercoagulability, and accelerated thrombosis: studies in congenitally hyperlipidemic rats and in rats and monkeys with induced hyperlipidemia. Blood. 1976;47:275–86. [PubMed] [Google Scholar]
- (39).Andersen P. Hypercoagulability and reduced fibrinolysis in hyperlipidemia: relationship to the metabolic cardiovascular syndrome. J Cardiovasc Pharmacol. 1992;20(Suppl 8):S29–S31. doi: 10.1097/00005344-199200208-00007. [DOI] [PubMed] [Google Scholar]
- (40).Simpson HC, Mann JI, Meade TW, Chakrabarti R, Stirling Y, Woolf L. Hypertriglyceridaemia and hypercoagulability. Lancet. 1983;1:786–90. doi: 10.1016/s0140-6736(83)91849-4. [DOI] [PubMed] [Google Scholar]
- (41).Rosenson RS, Lowe GD. Effects of lipids and lipoproteins on thrombosis and rheology. Atherosclerosis. 1998;140:271–80. doi: 10.1016/s0021-9150(98)00144-0. [DOI] [PubMed] [Google Scholar]
- (42).Tang W, Basu S, Kong X, Pankow JS, Aleksic N, Tan A, et al. Genome-wide association study identifies novel loci for plasma levels of protein C: the ARIC study. Blood. 2010;116:5032–6. doi: 10.1182/blood-2010-05-283739. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.