I. Abstract
Membrane traffic between organelles is essential for a multitude of processes that maintain cell homeostasis. Many steps in these tightly regulated trafficking pathways take place in microdomains on the membranes of organelles, which require analysis at nanometer resolution. Electron Microscopy (EM) can visualize these processes in detail and is mainly responsible for our current view of morphology on the subcellular level. This review discusses how EM can be applied to solve many questions of intracellular membrane traffic, with a focus on the endosomal system. We describe the expansion of the technique from purely morphological analysis to cryo-immuno-EM, Correlative Light Electron Microscopy (CLEM) and 3D electron tomography. In this review we go into some technical details of these various techniques. Furthermore, we provide a full protocol for immunolabeling on Lowicryl sections of high-pressure frozen cells as well as a detailed description of a simple CLEM method that can be applied to answer many membrane trafficking questions. We believe that these EM-based techniques are important tools to expand our understanding of the molecular details of endosomal sorting and intracellular membrane traffic in general.
Keywords: cargo sorting, endosome, electron microscopy, high-pressure freezing, freeze substitution, Tokuyasu, tomography
II. Introduction
II.a. Intracellular membrane traffic
Membrane-mediated transport is one of the major fields of focus in cell biology research. It is essential for delivering newly synthesized proteins to the location where they can perform their function, maintaining lipid bilayers, storing signal molecules, recycling or degrading used proteins, and providing membrane expansion in cell division. Membrane carriers are tubular- or vesicular-shaped structures that are derived from the limiting membrane of the donor organelle where the cargo is initially located. To produce such a carrier, the donor bilayer is deformed into a budding profile (Fig. 1A). This process is regulated by proteins that can reshape membranes by forming a scaffold on the membrane of a specific curvature and/or inserting domains into the lipid bilayer that introduce membrane curvature (reviewed by (McMahon and Gallop, 2005)). Budding of the donor membrane coincides with sorting of specific cargo. Once the carrier is formed and loaded with cargo, the tubular/vesicular carrier detaches from the donor membrane in the fission step. This process is regulated by several proteins that narrow the carrier neck, e.g. dynamin, in combination with longitudinal force that is applied by motor proteins that pull the carrier along the cytoskeleton. After transport to the target organelle, the carrier is recognized by tethering proteins. The transport vesicle subsequently docks to the target membrane, involving proteins from the SNARE family. Finally the carrier fuses with the target organelle to deliver its cargo.
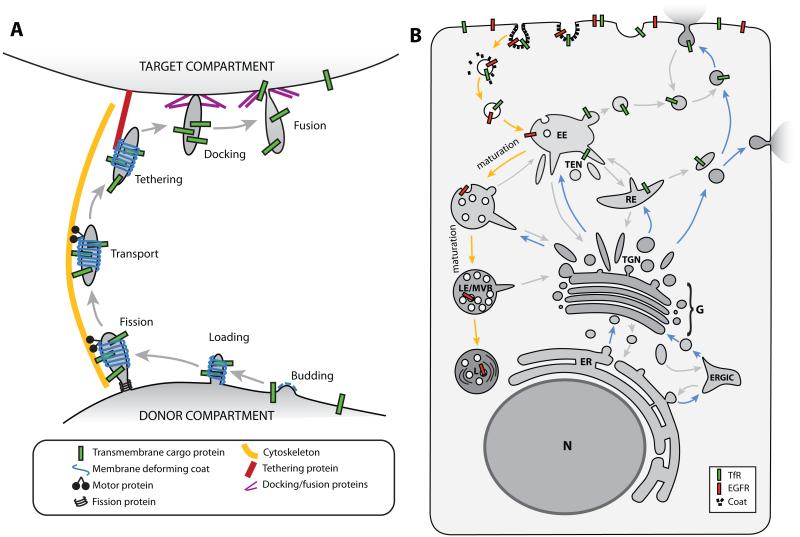
Figure 1. Steps and routes of intracellular membrane transport.
A: Schematic diagram of the different steps in which membrane transport is generally divided. A bud is formed on the donor membrane, which is further shaped into a tubular/vesicular membrane carrier and loaded with the cargo. The membrane carrier pinches off from the donor compartment (fission) and is transported to the target compartment along the cytoskeleton. At the target compartment, the membrane carrier is tethered and docked before it fuses with the target membrane to deliver the cargo. B: Cartoon of a cell depicting the major membrane trafficking routes. Blue arrows mark the biosynthetic pathway, orange arrows show the degradative route, and grey arrows mark the many recycling routes. See section I.a. for details. (ER) Endoplasmic Reticulum, (G) Golgi complex, (ERGIC) ER-Golgi Intermediate Compartment, (TGN) trans-Golgi network, (EE) early endosome, (TEN) tubular endosomal network, (RE) recycling endosome, (LE) late endosome, (MVB) multi-vesicular body, (L) lysosome, (TfR) Transferrin Receptor, (EGFR) Epidermal Growth Factor Receptor.
The cell contains many different trafficking routes (Fig. 1B). Most of the steps in the trafficking processes are therefore highly regulated to obtain the required specificity. In general, there are two main routes: the biosynthetic pathway and the degradative route. The biosynthetic pathway (Fig. 1B, blue arrows) starts with protein production in the Endoplasmic Reticulum (ER). Proteins are transported to the Golgi complex for modification either directly or through an intermediate compartment. In the trans-Golgi network (TGN) the proteins are sorted for transport to different locations in the cell where each protein performs its function, for instance in the plasma membrane or in the endosomal system.
The degradative route (Fig. 1B, orange arrows) starts with the internalization or endocytosis of material from the plasma membrane. The endocytosed vesicles fuse with the early endosome where the cargo is sorted in a tubular endosomal network. Many proteins are recycled back to their steady-state compartments, including the plasma membrane and the TGN. There are many points of crosstalk between both major transport pathways but it is beyond the scope of this review to highlight them all. Proteins destined for degradation are sorted into intraluminal vesicles of the endosome and remain there while the endosomal vacuole matures into a late endosome also known as multi-vesicular body (MVB). The MVB fuses with the lysosome where their content is finally degraded.
II.b. Visualization of membrane transport pathways
The use of fluorescently labeled proteins and advanced live-cell imaging techniques has identified many proteins and protein complexes that fulfill essential roles in the dynamic process of membrane traffic. However, the resolution of any microscope is restricted by the wavelength of the source of imaging radiation; in the case of conventional light microscopes this results in a limit in resolution of around 200 nm. This resolution can be improved by various super resolution techniques (Schönle et al., 2000; Gustafsson, 2005; Rust et al., 2006; Betzig et al., 2006), but even these techniques are not sufficient to visualize processes that take place at the nanometer scale. Furthermore, in fluorescence microscopy only labeled proteins are visible, not their cellular environment (so-called reference space). Finally, it is difficult to reliably co-localize non-interacting proteins to specific subdomains on membrane organelles using light microscopy due to insufficient resolution and the lack of reference space information (compare (Rink et al., 2005) and (Hughes et al., 2009)). The only technique so far that can localize proteins at the nanometer resolution is EM. Immuno-EM methods have proven to be very powerful tools for cell biology and have made many important contributions to the field. Immuno-EM as we know it today started with the presentation of the first electron-dense marker, Ferritin, to label antibodies for EM (Singer, 1959). Over the years, the technique has been improved and expanded to a range of EM-based methods, such as CLEM and electron tomography. These techniques can allow cell biologists to gain novel insights in the process of membrane traffic.
II.c. Development of ultrastructural labeling
Membrane carriers of different trafficking routes often appear quite similar in Transmission EM (TEM) and can only be identified by the presence or absence of specific marker proteins. Several labeling techniques can be applied in TEM to visualize the different trafficking routes and localize specific proteins at the ultrastructural level. We will discuss a couple of these labeling techniques as applied in the endosomal system.
The first method of ultrastructural labeling was performed by internalization of a fluid-phase EM marker by living cells and following the fate of that marker by fixing the cells at different time intervals. Using this method, Heuser and Reese analyzed synaptic vesicle endocytosis in the neuromuscular junction during different stimulation protocols, by which different numbers of synaptic vesicles fuse with the membrane to release their neurotransmitter (Heuser and Reese, 1973). The nerve terminal was incubated with medium containing horseradish peroxidase (HRP). HRP itself is not visible in the EM but it catalyzes the precipitation of diaminobenzidine (DAB) in the presence of hydrogen peroxide, which appears as electron-dense material after osmium staining (Seligman et al., 1968). Heuser and Reese observed that synaptic vesicles were being recycled locally in the nerve terminal by fusing with endosomes and proposed a model for the presynaptic vesicle cycle (Heuser and Reese, 1973). In the same issue of that journal, Ceccarelli and co-workers loaded the nerve terminals with HRP or dextran and, combined with electrophysiological experiments, came to the conclusion that synaptic vesicles are not fully retrieved to an endosomal compartment as suggested by Heuser and Reese, but were refilled at the site of release (Ceccarelli et al., 1973). The relative importance of these different types of synaptic vesicle recycling is one of the most studied processes in neuronal cell biology and is still actively debated (e.g. (Granseth et al., 2006; Balaji and Ryan, 2007; Zhang et al., 2009)).
As an alternative to fluid phase uptake, endocytosis of specific receptors can be followed by labeling their substrate. This technique revealed different sorting routes in the endocytic network and characterized the different organelles in the pleiomorphic endosomal system. The iron transporter Transferrin (Tf) is still one of the most used endosomal markers. The receptor for Transferrin (TfR) is constitutively internalized and recycled back to the plasma membrane. The Tf-iron complex binds the TfR at the cell surface and is transported into the cell by endocytosis (Fig. 1B). In the early endosome, the Tf-TfR complex releases the iron ions and is recycled back to the cell surface. This recycling pathway can take either a direct route from the early endosome to the plasma membrane or a slow recycling route through the recycling endosome compartment (Hopkins et al., 1994) (Fig. 1B). Tf coupled to a reporter molecule (e.g. fluorophores, gold particles, or HRP) is applied to study processes of endocytosis and early endosomal sorting. Due to the efficient recycling of the Tf-TfR complex, late endosomes no longer contain Tf. Therefore Tf can be used as a tool to study the maturation process from early to late endosomes. Mari and co-authors showed in an elegant quantitative TEM study the maturation of early to late endosomes correlates with an increase of intraluminal vesicles in the endosomal vacuole (Mari et al., 2008). The accumulation of intraluminal vesicles in late endosomes gave them the name of multi-vesicular bodies (MVBs). However, the function of this organelle was unclear until labeled Epidermal Growth Factor (EGF) identified MVBs as part of the endosomal-lysosomal degradation pathway (Gorden et al., 1978; Haigler et al., 1979). When EGF associates with its receptor, it stimulates cell proliferation upon which the ligand-receptor complex is internalized. The complex is then targeted to the intraluminal vesicles of MVBs for breakdown to prevent uncontrolled growth (Stoscheck and Carpenter, 1984). This makes EGF and the EGF receptor (EGFR) a popular tool to study the degradative pathway.
Lysosomes were characterized at the ultrastructural level by a slightly different approach. Essner and Novikoff used the activity of an endogenous acid phosphatase present in lysosomes to form a lead-phosphate precipitate on ultrathin sections (Essner and Novikoff, 1961). They found that the electron-dense precipitate was contained in “dense bodies”; organelles with high protein content and multilaminar internal membranes. These ultrastructural hallmarks are still used to recognize lysosomes in TEM.
Although the above mentioned labeling techniques provided many new insights and are still used today, they are limited by the accessibility of the compartment from the outside of the cell or the presence of unique enzymes that catalyze a suitable reaction that is detectable in the EM. The application of immuno-histochemistry, i.e. labeling proteins using specific antibodies, should have overcome these limitations but it has proven to be complicated to combine with good ultrastructural preservation until the introduction of the Tokuyasu cryosectioning technique in 1973 (Tokuyasu, 1973). This simple method made immuno-EM widely accessible and still sets the standard for protein localization in the membrane traffic field at the nanometer resolution.
III. Methods
III.1 The Tokuyasu cryosectioning technique
III.1.a. Principle of the Tokuyasu method
The key to a successful immuno-EM experiment is to retain both ultrastructural detail and antigenicity of the molecules in the ultrathin section. As biological material is too soft to be used for ultrathin sectioning straight away, the material is generally embedded in a resin to provide sufficient support. However, these resins cover the epitopes in the section and prevent/decrease antibody recognition. Some resins preserve the antigenicity of the proteins just at the section surface (methacrylic resins such as Lowicryl HM20, further described in section III.2.b.). Alternatively, biological material can be frozen to gain sufficient rigidity for ultrathin sectioning and still retain antigenicity. However, simple freezing of biological material will produce ice of a crystalline nature. These ice crystals take up more space than liquid water, thereby severely damaging the ultrastructure of the cell (McDonald, 2007; Brown et al., 2009). Dr. Kyoteru Tokuyasu introduced a method to prevent ice crystal formation by infusing chemically fixed cells with high concentrations of sucrose (Tokuyasu, 1973). Sucrose is highly hydrophilic, which at high concentrations binds sufficient water molecules to prevent water from adopting its crystalline organization when cooled down. The non-crystalline ice (so-called vitreous ice) that is formed occupies the same amount of space as water and has minimal impact on the ultrastructure. This development allowed ultrathin sectioning of frozen cells that have preserved most of their ultrastructural information as well as antigenicity of their molecules.
III.1.b. The Tokuyasu approach
Since its introduction, the Tokuyasu method has been improved and perfected (for a detailed protocol, see e.g. (Slot and Geuze, 2007)). Also the introduction of diamond cryo-knives as well as major improvements in the stability of the ultra-microtomes have made the technique much easier and accessible as a “standard” EM technique. The flowchart (Fig. 2A) shows the workflow of a Tokuyasu cryo-immuno-EM experiment. In brief, cells and tissues can be fixed by conventional fixatives, such as paraformaldehyde (PFA) and glutaraldehyde (GA) in a sufficiently strong buffer solution (e.g. Sorensen’s phosphate buffer, buffering capacity of PBS is not strong enough) to scavenge the free protons produced during the fixation process. GA is preferable to PFA for preservation of ultrastructural detail, but its disadvantage is that more epitopes are distorted, thereby reducing the immunolabeling efficiency. Therefore, combinations of PFA and GA fixative are a popular choice for biological samples in order to obtain the optimal balance between ultrastructural preservation and antigenicity of the material. It is advisable to test different concentrations and/or combinations of fixative when starting an immuno-EM experiment on new cell-types/tissues or using new antibodies. Generally, successful labeling with an antibody/fixative combination in immuno fluorescence light microscopy is a good indicator for a successful immuno-EM experiment.
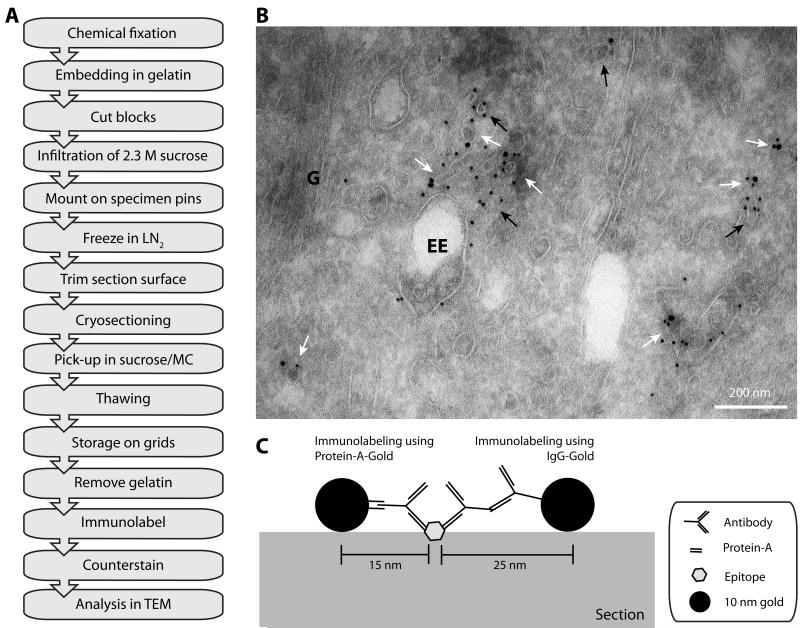
Figure 2. Immunolabeling the endosomal compartment using the Tokuyasu method.
A: Flow diagram of a typical immuno-EM experiment using the Tokuyasu method (see section III.1.b. for details). (LN2) liquid nitrogen, (MC) methylcellulose, (TEM) transmission electron microscope. B: Electron micrograph of immunogold labeling for GFP and RFP of HeLa cells, expressing GFP-SNX1 and RFP-Rab5, processed according to the Tokuyasu method. Ultrathin sections were labeled by GFP and RFP antibodies and PAG (10 and 15 nm, respectively). White arrows indicate tubular/vesicular profiles that are labeled by both sizes of gold, black arrows indicate membrane structures only labeled for one protein. (G) Golgi complex, (EE) early endosomal vacuole. Scale bar represents 200 nm. C: Cartoon of immunogold labeling of an epitope on the surface of the ultrathin section. Protein-A-Gold markers have a higher resolution (within 15 nm distance of the epitope) compared to IgG-Gold (within 25 nm distance of the epitope) due to the larger size of the secondary antibody. Adapted from (Verkade et al., 1997).
After the material is fixed, the extracellular space is stabilized by a gelatin matrix. Monolayers of cells can be scraped, pelleted, and embedded in gelatin or embedded as an intact layer (“flat embedding”; for details see (Oorschot et al., 2002)). These gelatin-encapsulated blocks of cells or tissue are infiltrated with a 2.3 M sucrose solution, mounted on specimen holders and frozen in liquid nitrogen (LN2) for cryosectioning. As the material is now cryo-protected by sucrose, blocks can be thawed, remounted and refrozen without damaging the ultrastructure.
The cryo-ultramicrotome is the only piece of specialized equipment required for the Tokuyasu technique, but most conventional ultramicrotomes can be used as such by attaching a cryo-chamber. Ultrathin sections are collected at low temperatures, generally −100 °C to −120 °C. Other temperatures can be chosen to obtain the preferred rigidity of the block, for instance filter-grown cells are sectioned at −90 °C in our lab. Ultrathin cryosections are captured in sucrose solution, usually supplemented with methylcellulose to provide additional support (Liou et al., 1996). The sections are thawed and stored, still covered by the pick-up solution, on carbon/formvar-coated grids at 4 °C.
After removal of the gelatin, the ultrathin sections can be immuno-stained in a manner that is very similar to widely used immuno fluorescence labeling of cells for confocal microscopy. The quality of the antibody (in combination with the applied fixative) is the main determinant of a successful immunolabeling experiment. New antibodies should be tested in different concentrations for optimal labeling. The antibodies can be visualized using electron-dense markers, such as colloidal gold particles of different sizes, which allows co-localization studies of multiple proteins in the same section (Fig. 2B). Gold particles can be directly linked to the primary antibody, attached to a secondary antibody raised against a specific species or coated in Protein-A/G that bind antibodies with high affinity (Griffiths, 1993). Finally, the tissue in the section can be stained with uranyl acetate to improve contrast in TEM. It should be noted that in the Tokuyasu procedure uranyl acetate acts as a negative stain and deposits along the membranes rather than staining the membranes itself (Fig. 2B). The full procedure from fixing the cells up to analysis in the TEM can be completed in two to three days, which makes the Tokuyasu technique a popular technique to determine the nature of the many different membrane transport carriers in the cell and localize their proteins of interest at the ultrastructural level.
III.1.c. Remarks
As mentioned above, antibodies can be labeled indirectly by either secondary antibodies (IgG) coupled to gold (IgG-G) or by Protein-A-Gold (PAG). PAG binds several species of antibodies (not all, to check compatibility see e.g. (Harlow and Lane, 1988)) at the Fc part of the primary antibody. This results in a one-to-one labeling pattern, i.e. each gold particle labels one primary antibody. IgG-G on the other hand, recognizes multiple epitopes on a single primary antibody. This results in a labeling pattern by which multiple (generally 2 to 3) gold particles label a single primary antibody. This difference makes PAG-based labeling more suitable for quantification, while the IgG-G technique can amplify labeling when the number of epitopes is low. Another advantage of the PAG method is that it improves the resolution of the immunolabeling. The length of an IgG molecule can be estimated to be approximately 16 nm in its crystal structure. A secondary IgG antibody and gold particles will bind to the middle of the Fc region resulting in a distance of 8 to 12 nm for one antibody (Verkade et al., 1997). Therefore, the total distance from the detected molecule to the centre of a 10 nm gold particle linked to a secondary antibody can span up to 25 nm (Fig. 2C). With a molecular weight of 40 – 60 kDa Protein-A is significantly smaller than the 150 kDa of a complete IgG molecule. Therefore, the gold particles will be located closer to the epitope in PAG labeled sections (within ~15 nm) compared to IgG-G labeling. Other strategies for improving the labeling resolution are using Fab fragments (~50 kDa) of antibodies instead of the complete antibody or directly linking the primary antibody to gold particles.
Both the PAG and IgG-G labeling technique can be used to study multiple proteins at the same time. For IgG-G, this can be achieved by using primary antibodies of different species that can be selectively recognized by IgG-G linked to different sizes of gold. For PAG, epitopes can be labeled by different sizes of gold consecutively by including a 5-minute GA fixation step after each labeling step. This fixation step distorts the Fc part of the primary antibody; therefore the same species of primary antibody can be used in multiple rounds. It is important to keep in mind that double labeling can introduce two artifacts: co-labeling, the different sized gold particles label the same primary antibody, and steric hindrance, gold particles obscuring epitopes in the consecutive labeling reaction. Therefore it is advisable to always include single labeled control conditions for each primary antibody to check that the labeling pattern is not affected in the double labeling condition.
Although the Tokuyasu cryo-immuno method currently is the best method for high antibody labeling in EM, it should be noted that the labeling efficiency of such experiments is estimated to be around 10 to 15% (Griffiths and Hoppeler, 1986; D’Amico and Skarmoutsou, 2008).
III.2. Cryofixation and High-Pressure Freezing (HPF)/Freeze-substitution(FS)
III.2.a. Principle of cryofixation
Although the Tokuyasu cryosectioning method was a great step forward in the applications of EM, the method does have disadvantages. The major disadvantage is that the material has to be chemically fixed due to the osmotic pressure of the 2.3M sucrose solution. Chemical fixation introduces several artifacts, which can have a great impact on the data (Murk et al., 2003; McDonald, 2007; Brown et al., 2009). Firstly, chemical fixation is relatively slow (in the order of seconds to minutes depending on the tissue and fixative) which can be too slow if you are studying a process that is significantly faster (see e.g. (Rosenmund and Stevens, 1997)). Secondly, chemical fixation of membrane proteins creates pores in the plasma membrane, which allows exchange of ions and small molecules between the extracellular medium and the cytosol during the fixation (Penttila et al., 1974). Thirdly, many cytoskeletal elements are destabilized during the chemical fixation (Small, 1981) and highly dynamic structures like sorting tubules can collapse upon chemical fixation (JRTvW & PJC, unpublished observation). Fourthly, chemical fixation can dehydrate the specimen due to osmotic difference between the fixative and the cell (Kellenberger et al., 1992; Murk et al., 2003). Finally, chemical fixation can distort the epitope on the protein of interest, thereby reducing efficient immunolabeling. Although these artifacts of chemical fixation may have no major consequences on the outcome of the experiment, in some cases it might be crucial to circumvent them by using cryofixation.
The objective of cryofixation is to physically immobilize the specimen by freezing without ice crystal formation. As discussed above, vitreous ice can be obtained by using high concentrations of sucrose. Alternatively, water in living specimens can be vitrified by cooling down the samples so rapidly that water is not organized in ice crystals (Steinbrecht and Zierold, 1984). The cooling rate required for such vitreous freezing of water is about 105 K/s. This cooling rate can be achieved in very thin specimens (such as liposomes or platelets) by “plunge freezing” these samples in liquid ethane. Thicker specimens can be vitrified by High-Pressure Freezing (HPF). At 2000 bar the cooling rate for vitreous ice is reduced to 103 K/s that allows vitreous freezing of specimens up to a thickness of 200 μm (Moor, 1987; Studer et al., 2008). HPF of biological specimens introduces minimal artifacts when well frozen and allows analysis of the ultrastructure of cells in their most natural state.
III.2.b. HPF post processing
The optimal method to observe a HPF specimen is directly in its frozen-hydrated state at temperatures close to that of liquid nitrogen or below (cryo-EM). Plunge-frozen, isolated material only contains a very thin layer of frozen water and can be directly observed in a cryo-EM without any additional fixation or staining. Thicker specimens can be cryosectioned directly after HPF and observed in cryo-EM. This technique is called CEMOVIS (Cryo EM Of Vitreous Sections) and is discussed in detail elsewhere in this volume by Andy Hoenger and Günter Resch. Since no post-freezing labeling and/or staining is used in CEMOVIS, proteins can only be labeled by live-labeling before the specimen is frozen. When labeling and/or staining of the material is required, the cryofixation needs to be replaced by chemical fixation to allow the sample to be handled at higher temperatures. This process is called freeze-substitution.
Freeze substitution is done by warming up the sample in a very controlled manner while fixing and dehydrating the sample at minimal temperatures. The dehydration step is done with an organic solvent, such as acetone, that is liquid at the starting temperature for a freeze substitution experiment (often −90 °C). The dehydration and chemical fixation processes at these temperatures are slow, but are thought not to introduce as many artifacts as fixation and dehydration at room temperature. However, shrinkage and reduced antigenicity can still occur. This critically depends on the organic solvent in which the freeze substitution is performed. Methanol, for instance, extracts more water and cytoplasmic material compared to acetone (Weibull et al., 1984) and is therefore hardly used in FS anymore. Addition of chemical compounds such as osmium tetroxide, uranyl acetate, and glutaraldehyde mainly determine whether antigenicity is retained. Ultimately, it is the resin that further determines both the ultrastructure and immuno-reactivity. In Figure 3A we provide an overview of the process. For the best ultrastructural preservation, we use a relatively short freeze substitution protocol that only lasts 23 hours after which the sample is infiltrated with Epon and hardened in an oven. The freeze substitution cocktail contains 1 % osmium tetroxide and 0.1 % uranyl acetate in acetone (see e.g. (McDonald et al., 2007; Verkade, 2008; Brown et al., 2009)). This protocol is unsuitable for subsequent immunolabeling because the osmium has destroyed most antigenic sites and Epon sections expose very few antigens. It is thought that the surface of Epon is much smoother compared to other resins such as Lowicryls (Hayat, 2000). A rougher surface results in a larger surface area of the section and therefore more epitopes will be exposed. In addition, Lowicryls are liquid at relatively low temperatures (from −35 to −80 °C) and they can be polymerized at these temperatures using UV light. This makes Lowicryls very suitable as embedding medium after HPF. Lowicryls come in a polar/hydrophilic (K4M and K11M) and non-polar/hydrophobic (HM20 and HM23) form. We prefer Lowicryl HM20 for our studies as it gives both a good ultrastructure and antigenicity, albeit that the labeling efficiency is still lower compared to the Tokuyasu method (see section III.1.a and compare Fig. 2B and 3B). To utilize the higher labeling efficiency of Tokuyasu sections, HPF samples can be processed with a FS protocol compatible with the Tokuyasu technique (van Donselaar et al., 2007; Ripper et al., 2008). After the fixation and dehydration have been performed in FS, the sample is re-hydrated and further processed using the Tokuyasu method (see for details (van Donselaar et al., 2007)). This procedure is very time-consuming and has not been applied to intracellular membrane studies although it could offer a method to study sensitive membrane structures such as sorting tubules with the immunolabeling efficiency of the Tokuyasu method.
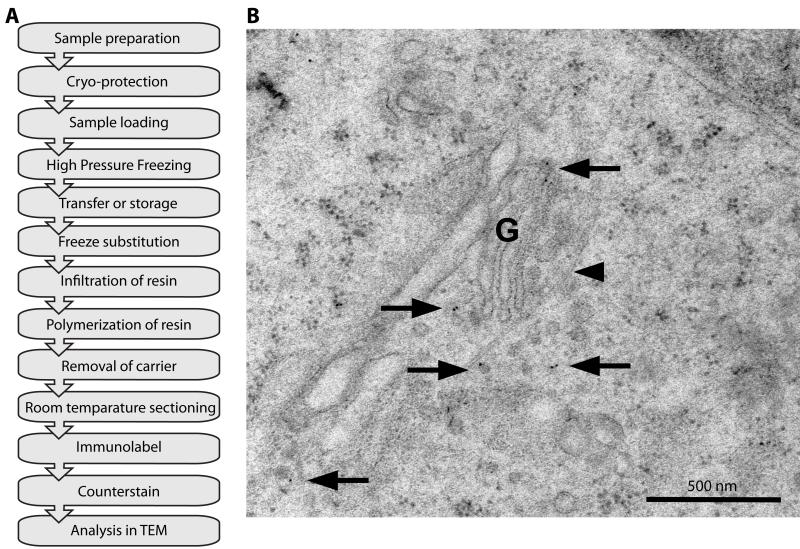
Figure 3. Immuno-labeling of VSVG using HPF/FS and Lowicryl embedding.
A: Flow diagram of a typical HPF/FS experiment using Lowicryl HM20 embedding and immunolabeling (see section III.2.c. for details). B: Electron micrograph of immunogold labeling for GFP in a HeLa cell after HPF/FS and Lowicryl HM20 embedding. HeLa cells grown on sapphire discs were transfected with a construct expressing a temperature-sensitive VSVG-GFP. With the temperature block present the protein accumulates in the Golgi complex (G). 30 min after release of the temperature block the cells were high-pressure frozen and processed. Most of the protein has reached the cell surface but there is still protein present inside the Golgi and in vesicular profiles near the Golgi (arrows). This procedure preserves antigenicity as well as structural elements such as microtubules (arrowhead), which are very sensitive to chemical fixation. Scale bar represents 500 nm.
III.2.c. Protocol for immunolabeling using HPF and Lowicryl embedding
High-pressure freezing
For our high-pressure freezing experiments we use the Leica EM PACT2 + Rapid Transfer System (RTS) (see material section for details) (Verkade, 2008). For detailed protocols of sample preparation for HPF of cell culture grown on sapphire discs, we refer to previous publications (McDonald et al., 2007; Verkade, 2008). A few notes on the cell culture and HPF:
-
1.
Sapphire discs are used as opposed to glass due to better conductive properties necessary for the rapid freezing process. However, sapphire discs are expensive and can be replaced by relatively cheap aclar film as cell support (Jiménez et al., 2006). Aclar films are bought as sheets and 1.5 mm circles have to be punched out. In our hands cell growth is similar on aclar compared to glass or plastic and the HPF procedure works equally well.
-
2.
Grow cells to full confluency. This is important, because then they will detach easily from the growth support.
-
3.
Cells in suspension are also very suitable for HPF. We leave the cell suspension to sediment (do not spin), and pipette 1 μl of the thick cell suspension into the well of a 100 μm deep HPF membrane carrier (Leica Microsystems), which is frozen in the Leica EM PACT2.
-
4.
We generally add a cryo-protectant to the growth medium, but this is not always required (Verkade, 2008). We prefer BSA as it improves the sectioning properties of the block.
Freeze substitution
A short freeze-substitution protocol for pure ultrastructural analysis (i.e. embedding in Epon/Araldite) has been described previously (Verkade, 2008). Here, we will focus on a freeze-substitution protocol for embedding in Lowicryl (HM20) that provides both good ultrastructure and immunolabeling. We make use of the Automated Freeze Substitution device (AFS2, Leica Microsystems) equipped with the attachment for automated exchange of the reagents (Freeze Substitution Processor, FSP, Leica Microsystems). We particularly value the use of the FSP in these embedding experiments as it brings the researcher in minimal contact with the toxic Lowicryl resin. The FSP exchanges the media through a syringe that is connected with the sample chambers via flow-through baskets.
-
1.Bring the AFS2 to −90 °C and fill the flow-through baskets with the freeze-substitution ‘cocktail’. In the example shown (Fig. 3B) we have used 0.1% uranyl acetate in acetone but we have also had good experience with adding an additional 0.01% osmium tetroxide or just incubation in pure acetone (see (Brown et al., 2009)).
- Uranyl acetate hardly dissolves in acetone. A 10% stock solution is made in methanol and frozen in LN2. The stock solution is diluted 1:100 in acetone.
-
2.
Perform freeze-substitution using the FSP.
24 hours at −90 °C in the freeze substitution cocktail.
Raise the temperature to −50 °C with steps of 5 °C/hour.
24 hours at −50 °C in the freeze substitution cocktail.
Three washes with pure acetone, 1 hour each.
Gradual infiltration with Lowicryl HM20, mixed with acetone, 25, 50, 75, 100%, 2 hours each step.
24 hours at −50 °C with UV treatment.
Raise the temperature to 20 °C, 5 °C /hour with UV light switched on.
24 - 48 hours at 20 °C with UV light switched on. The length of this step is not critical, but ascertain that the Lowicryl is fully polymerized. With the current use of LEDs instead of the older UV light sources the polymerization has become more reliable.
We have recently shortened this Lowicryl embedding procedure based on successful experiments described elsewhere (Hawes et al., 2007). The initial step at −90 °C is shortened to only 5 hours and all warm-up steps are done with an increase of 20 °C/hour. The ultrastructure of these samples looks very similar to samples processed using the longer procedure (not shown).
-
3.
After the polymerization, the Lowicryl blocks are removed from the flow through baskets before sectioning (see (Verkade, 2008)). The sapphire discs are removed from the resin by repeated submergence in boiling water followed by liquid nitrogen. Lowicryl is harder than Epon, but removal of the sapphire disc is actually much easier. This procedure leaves the cells directly exposed at the surface of the Lowicryl.
Sectioning and immunolabeling
-
4.
Sectioning of Lowicryl HM20 blocks is a bit different from Epon since the hydrophobic resin sections do not easily float on water. To reduce compression or even complete loss of the section, the water level is lowered in the knife trough so the initial gliding is done over the diamond surface.
-
5.
Immunolabeling the Lowicryl sections is similar to labeling Tokuyasu sections, however the quenching step using glycine or sodium borohydrate can be skipped since no aldehydes were used in the procedure. The sections are taken through the following steps at room temperature:
-
6.
Incubate 15 minutes 0.1% BSA in PBS (blocking solution).
-
7.
Label 1 hour with the primary antibody in blocking solution (10 μl single grid or 20 μl for two grids/drop and so on). Note that antibodies need to be tested for Lowicryl labeling. For example, we use a goat anti-GFP (Rockland, 600-101-215) that works well on Lowicryl HM20 embedded samples processed following the above-described method (see Fig. 3B).
-
8.
Wash 3 times with blocking solution.
-
9.
Label 30-60 minutes with secondary antibody in blocking solution.
-
10.
Wash 3 times with blocking solution.
-
11.
Wash 6 times with distilled water.
-
12.
Contrast with uranyl acetate and lead citrate.
III.2.d. Remarks
In general, immunolabeling efficiency after HPF/FS and plastic embedding in Lowicryl is considered to be lower than with the Tokuyasu immuno-EM method, although a direct comparison has not been described. One of the main advantages of using the HPF method is that cytoskeletal elements, which are an integral part of the membrane trafficking pathways, are well preserved (compare Figs. 2B and 3B).
III.3. Correlative Light Electron Microscopy (CLEM)
III.3.a. Introduction to CLEM
EM has given us many insights into cell biology, but its static nature makes it difficult if not impossible to reconstruct a highly dynamic process such as membrane trafficking. Combining the advantages of ultra-fast fluorescent live-cell imaging with the potential of EM for high-resolution imaging is one of the major objectives of CLEM (Verkade, 2008; Brown et al., 2009; McDonald, 2009). Only advanced CLEM techniques can allow us to reliably visualize and characterize intracellular trafficking events at high resolution, e.g. only with the accompanying live-cell data is it possible, for example, to discriminate between fusion and budding of a membrane carrier in EM (Verkade, 2008). Trying to achieve this, however, introduces many technical challenges that have led to a wide variety of CLEM procedures. Some approaches focus on the retracing aspect where light microscopy is mainly used to locate a region of interest, e.g. the fluorescence indicates the presence of a cell (Plitzko et al., 2009). Others highlight the live-cell imaging (Polishchuk et al., 2000) or fixation aspect (Verkade, 2008). The choice of the CLEM method ultimately depends on the research question. For intracellular membrane transport studies, the LM part of a CLEM experiment should include a live-cell imaging component to reconstruct the dynamic process.
The need for CLEM can be illustrated by the fact that there still remains confusion over issues, such as the co-localization of Tf and EGF receptors throughout the endocytic pathway (Leonard et al., 2008). Light microscopy alone (even when performed with a higher resolution LM technique, such as Total Internal Reflection Fluorescence microscopy (TIRF-M)) lacks sufficient resolution to conclude whether these receptors are found on the same organelle. Furthermore, the reference space provides essential structural information on the subdomains within one organelle where these receptors are present (Fig. 4B).
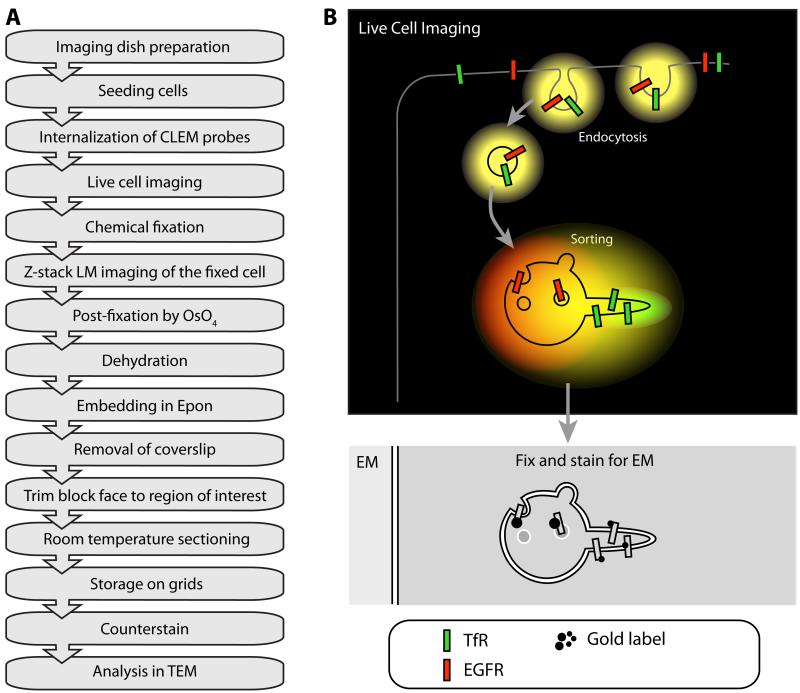
Figure 4. The principle of CLEM.
A: Flow diagram of a typical CLEM experiment using the “simple” approach described in III.3.c consisting of live labeling, chemical fixation and Epon embedding. (OsO4) osmium tetroxide B: Cartoon of a CLEM experiment on RFP-EGFR and GFP-TfR trafficking. Both types of cargo are internalized by endocytosis, showing up as yellow fluorescent compartments in live cell imaging due to the co-localization of GFP and RFP. In the early endosome, GFP-TfR is sorted into tubular/vesicular carriers that retrieve the receptor to plasma membrane. In the fluorescence microscope, these carriers appear green due to the high concentration of GFP-TfR. The RFP-EGFR is collected in intraluminal vesicles in the endosomal vacuole, which shows up as predominately red. Although this sorting process is visible at the LM level, the endosome itself or its subdomains are not visible. By capturing the event by HPF and analyzing the sample by EM, it is possible to define the localization of GFP-TfR and RFP-EGFR in their different subdomains within the early endosome compartment.
In a landmark paper, introducing the term CLEM, the morphology of carriers operating between the Golgi apparatus and the plasma membrane was established (Polishchuk et al., 2000). These carriers were followed using VSVG-GFP as a biosynthetic reporter protein and chemically fixed at time-specific time points. Detection of previously followed carriers in the EM was performed by immunolabeling of GFP with secondary antibodies attached to either gold or HRP to precipitate DAB. This approach allowed the authors to study the fine ultrastructure of the organelles involved in Golgi-to-plasma-membrane traffic and relate these structures back to their stage in this membrane trafficking pathway.
III.3.b. CLEM markers
One of the most important decisions to make when performing a CLEM experiment is the choice of the marker or probe. A CLEM probe must be fluorescent in the LM and electron-dense in the EM. Different experimental aims also require different probes. Thus far, no genetically encoded CLEM probe has been described that can be expressed in the cytoplasm of living cells. The current CLEM markers can only enter the cell by live labeling through endocytosis or microinjection (Verkade, 2008).
FluoroNanoGold is composed of a fluorescent moiety, such as fluorescein or rhodamine, and an ultrasmall gold particle (0.8 to 1.4 nm) attached to either an antibody or to streptavidin (Robinson and Takizawa, 2009). Since ultrasmall gold is not directly visible in a standard EM, it has to be silver-enhanced for reliable imaging (Tchelidze et al., 2006). The range of colors and sizes available, as well as the high contrast achieved with gold particles have resulted in widespread use of these fluoro-gold probes throughout the CLEM community. Similarly, quantum dots (QDs) can be used to study endocytic events. QDs are brightly fluorescent nanocrystals with an electron-dense core usually comprising of either cadmium selenide or cadmium telluride (Michalet et al., 2001; Giepmans et al., 2005). QDs can be conjugated to antibodies or other targeting molecules, such as Protein-A or streptavidin (Giepmans et al., 2006). Although QDs are available in colors that span the whole visible spectrum (Jaiswal and Simon, 2004), there are only three that have been reported to be reliably differentiated in the EM; spherical-blue QD525, spherical-green QD565 and cylindrical-red QD655 (Giepmans et al., 2005). Because the cadmium in the quantum dot is less electron-dense than colloidal gold particles these particles can appear grey in the TEM ((Giepmans et al., 2005), but see Fig. 5K and 5N). As a probe for CLEM, QDs suffer from the same drawbacks as other exogenous labels: they can only be utilized for live cell imaging after live labeling.
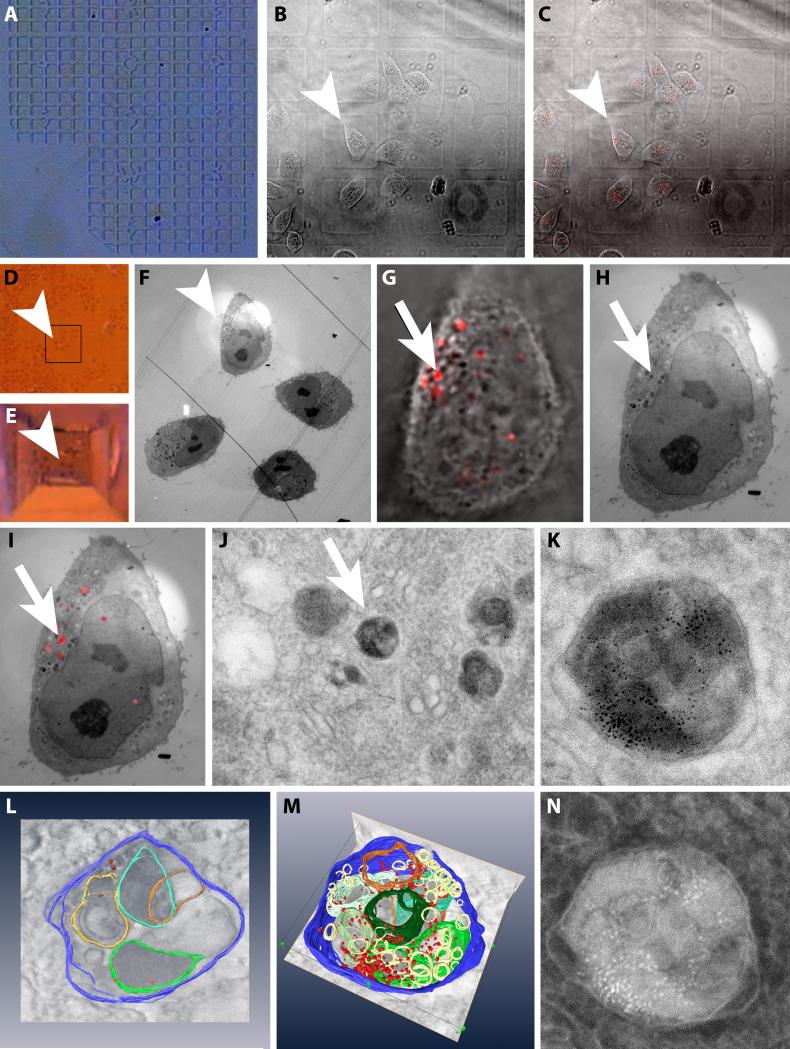
Figure 5. A simple CLEM experiment including 3-D reconstruction by electron tomography.
Example images of a CLEM experiment using the simple method described in section III.3.a A: The finder pattern of the embossed coverslip. B: DIC image of HeLa cells grown on the embossed coverslip. Arrowhead points to the cell of interest (images B-F). C: Overlay of B and a fluorescence image of the cells that have internalized EGF-biotin coupled to streptavidin-QD 655. D: Stereo microscopic image of the region of the cells of interest after fixation and embedding in the Epon resin. E: As D after trimming the block face. F: EM overview image highlighting the area containing the cell of interest. G: Zoomed image of the overlay of the DIC and fluorescence of the cell of interest. Arrow points to the organelle containing EGF-biotin coupled to streptavidin-QD 655 (image G to J) H: EM image of the same region as G. I: Overlay of the fluorescence image and the electron micrograph of the same region as G. J: Electron micrograph of the subcellular area containing the organelle of interest. K: Standard high magnification bright field EM image of the organelle of interest. L: Screen shot of the modeling part of an electron tomography experiment where structures of interest have to be manually segmented. M: Screen shot of the modeling of the structure of interest. N: HAADF image of the same region as K.
Live labeling can be circumvented by genetic manipulation of the protein of interest to adjoin either a fluorescent moiety such as GFP, or a targeting motif for the biarsenical molecules FlAsH and ReAsH. Fluorescein bi-Arsenical Hairpin-binding (FlAsH) and the Resorufin red variant (ReAsH) are small, membrane-permeable fluorescent molecules that selectively label a tetracysteine motif (CCxxCC) appended to the protein of interest (Griffin et al., 1998). Although well suited to imaging in the LM, as well as for more specialized techniques such as pulse-chase experiments (Gaietta et al., 2002), neither FlAsH nor ReAsH are directly visible in the EM. However, ReAsH can photo-polymerize DAB in the same manner as HRP to form an osmiophilic precipitate (Gaietta et al., 2002; Adams et al., 2002). The drawbacks to using DAB include a diffuse staining and the possibility that DAB will drift from the site of production and fill the organelle/region surrounding the protein of interest. DAB is therefore a relatively low-resolution technique that should be used when the protein is encapsulated within an organelle.
Genetically encoded fluorescent proteins (Tsien, 2009), such as GFP and mRFP, are the most reliable targeting method for LM due to the one-to-one labeling ratio; if the protein is there it will be fluorescent and vice versa. Advantages include fluorescence, a wide variety of colors, and with the advent of photo-switchable and photo-convertible proteins the ability for super-resolution and pulse-chase experiments. The major limitation of GFP as a probe for CLEM is the total lack of visibility in the EM; proteins are not electron-dense and fluorescence is usually quenched by the fixation and embedding methods. Commercial antibodies to GFP are available but can only be applied after fixation and do not show one-to-one labeling efficiency. One method of visualization involves photo-conversion of DAB by production of singlet oxygen from fluorescing GFP (Monosov et al., 1996). Known as GFP-recognition after photo-bleaching (GRAB) the disadvantages of using DAB remain, although successful electron tomography has been demonstrated (Grabenbauer et al., 2005; Meiblitzer-Ruppitsch et al., 2008).
We have summarized and commented on the properties of the probes discussed above (Table 1). In the next sections we will present applications of these probes for three different CLEM approaches: i) a simple CLEM approach ii) CLEM using Tokuyasu, iii) CLEM using HPF.
Table 1. Summary of various techniques and probes used for CLEM.
Different techniques are scored from – (not suitable) to +++ (highly suited) based on the research reviewed above and on our own judgments. ‘Membrane penetration’ indicates the ability of the probe to cross the plasma membrane non-invasively to label the intracellular target. ‘Penetration after fixation’ refers to the depth of labeling in the sample that the probe can achieve after fixation with glutaraldehyde; HRP can be genetically attached to an intracellular target, in which case DAB is the penetrating reagent. ‘Multiprotein labeling’ relies on different colors and sizes of the probe being distinguished in both LM and EM; for example, whilst there are two colors of FlAsH/ReAsH they both bind to the same motif and therefore label the same protein. ‘Endogenous target proteins’ are those with no genetic modification, so although labeling with antibodies enables endogenous protein targeting, both Fluorescent Proteins (FPs) and FlAsH/ReAsH require genetic modification. ‘Pulse-chase’ is a live cell imaging technique that allows the researcher to image the same protein at different time-points in different colors, i.e. newer proteins appear green whereas old ones are red. ‘Specificity’ is a measurement of the fidelity of labeling; FPs label one-to-one due to the nature of genetic covalent modifications, whereas antibodies can label one protein more than once or not at all, FlAsH/ReAsH are comparable to antibodies. Longevity is the resistance of the probe to photobleaching, whereas phototoxicity is the ability of the probe to perturb the cell. NA = not applicable.
| FPs | FlAsH/ReAsH | QDs | Fluoronanogold | Immunogold | HRP | |
|---|---|---|---|---|---|---|
|
|
||||||
| Live cell LM | ++ | ++ | ++1 | +1 | - | − |
| EM | − | − | + | ++ | ++ | +2 |
| CLEM | −/+2 | −/+2 | ++ | +++ | ++ | +2 |
| Membrane penetration in live cell | NA | + | − | − | − | − |
| Penetration after fixation for EM | NA | NA | + | + | + | + |
| Multiprotein labeling | + | − | +++ | ++ | + | − |
| Endogenous target protein | − | − | + | + | + | + |
| Pulse-chase | +3 | ++ | −/+1 | −/+1 | − | − |
| Specificity | +++ | ++ | + | + | + | + |
| Time delay for fluorescence | mins-hours | <20 min | 01 | 01 | NA | NA |
Specifically for endocytosis events or direct microinjection of probes
Requires photo-polymerisation of DAB and Osmium staining
Only for photoconvertible FPs, such as Kaede
III.3.c. A simple CLEM approach
CLEM techniques can be very technically challenging, aiming to combine ultra fast live-cell imaging with high immunolabeling efficiency and/or HPF for maximal ultrastructural preservation in EM (see below, sections III.3.d and III.3.e). For many questions, it might not be necessary to use such high-end techniques. When optimal preservation of antigenicity, instantaneous fixation and/or maximal ultrastructural detail are not essential for the experiment, live cell imaging followed by a simple chemical fixation of cells on an embossed coverslip may be sufficient (Fig. 4A).
Such a technique is significantly easier and has a much higher success rate than the other two methods discussed below (sections III.3.d and III.3.e). Here we describe a detailed protocol for a simple CLEM approach, where we illustrate the degradative pathway taken by EGF receptors after internalization (Fig. 5).
Preparing live cell imaging dish
-
1.
Drill a 5mm hole in the centre of a standard 35mm cell culture dish.
-
2.
Attach an embossed coverslip using vacuum grease to the underside of the dish so that the raised finder pattern (Fig. 5A) is facing upwards and positioned within the hole.
Internalization of endocytic markers
-
3.
Seed cells onto the coverslip and leave to grow to 30-70% confluence.
-
4.
Prepare the CLEM marker. In our experiment, we pre-bind the EGF-biotin with Streptavidin-QD 655, 30 min at 37°C.
-
5.
Remove growth media and replace with the CLEM marker solution (EGF-QD-655 in Fig. 5).
-
6.
After a suitable incubation period remove the excess CLEM marker.
-
7.
Wash the cells with warm PBS to remove any unbound ligands and add approximately 100 μl of imaging medium (Phenol red-free imaging medium results in better quality LM images).
Live-cell imaging
-
8.
Record the location of the cell of interest in relation to its surrounding by taking low-magnification bright field images (Fig. 5B).
-
9.
Image fluorescence marker. Live cell imaging through the embossed coverslip is compatible with almost any fluorescence microscope setup, with the exception of TIRF-M due to the varying thickness of glass coverslip.
Fixation and embedding
-
10.
Fix the cells by adding 1 ml of 2.5% glutaraldehyde in 0.1M cacodylate buffer (pH 7.4) to the dish.
-
11.
Record a z-stack of fluorescence images with ~0.2 μm steps before autofluorescence from fixation masks the fluorescent signal.
-
12.
Remove the glutaraldehyde after 30 minutes of fixation and replace with cacodylate buffer.
-
13.
Stain the cell by adding 1% osmium tetroxide in cacodylate buffer for 30 minutes.
-
14.
Remove the osmium and wash with cacodylate buffer.
-
15.
Replace cacodylate buffer with deionized water.
-
16.
Dehydrate the sample in a graded series of ethanol going from 70% to 100% ethanol.
-
17.
Embed in Epon and polymerize resin at 60 °C for 24 hours.
Relocation and Sectioning
-
18.
Remove the coverslip by submerging the dish in LN2 and immediate subsequent plunging into boiling water.
-
19.
Carefully lift off the coverslip from the Epon block and cut away the cell culture dish. If the entire coverslip does not detach from the Epon block, repeat step 18.
-
20.
Trace back the region of interest using the bright field images and a stereomicroscope. Trim region down to a suitable size (Fig. 5D and 5E).
-
21.
Acquire serial sections of the trimmed block at the desired thickness (~70 nm for standard TEM, ~300 nm for tomography).
-
22.
Contrast cells with uranyl acetate and lead citrate following standard protocols.
-
23.
Relocate cells of interest (Fig. 5F). The structure of interest within a selected cell can be found with the aid of an overlay of the fluorescence onto the EM image (Fig. 5H-J). The structure of interest is now available for further detailed analysis, using standard bright-field TEM (Fig. 5K), electron tomography (Fig. 5L and 5M) and Scanning TEM (STEM) imaging (Fig. 5N, below section III.4.).
III.3.d. CLEM using Tokuyasu
CLEM using the Tokuyasu method aims to combine dynamic live cell imaging data with the immunolabeling efficiency of Tokuyasu sections (see section III.1.a.). This method is technically challenging; a detailed description of the procedure is published elsewhere (van Rijnsoever et al., 2008). Here, we will give a brief overview of the procedure.
An embossed coverslip is coated with Formvar followed by gelatin. This coverslip is mounted onto a culture dish in which cells expressing the fluorescent protein of interest are seeded. After live imaging, a PFA-based chemical fixation (e.g. 4% PFA and 0.05% GA) is applied to preserve antigenicity. Chemical fixation is intrinsically slow, therefore this CLEM approach is not recommended for studying highly dynamic processes or studying structures that are not well preserved during the fixation process. An advantage of this fixation method is that one can record a z-stack of fluorescent images of the fixed material for later relocation of the same structure in EM, while this is not possible when using cryofixation (see below III.3.e.). DIC images are also taken to map the location of the cell on the grid. After post fixation overnight in 2% PFA and 0.2% GA, the cell is relocated and marked with nail varnish. Cells are subsequently embedded in 6% gelatin (not 12%, which is used in standard Tokuyasu cryo-immuno-EM protocol, since the gelatin will be fixed later on). A layer of erythrocytes in 12% gelatin is added and solidified to facilitate orientation of the block. The sample is further fixed with 0.5% PFA and incubated with cold sucrose for 3 days on a rocker. A block containing the region of interest is cut out and mounted on a specimen pin. The specimen is then frozen in liquid nitrogen, serial cryosectioned at −100 °C and sections are collected in droplets of methylcellulose/sucrose. Every fifth grid is immunolabeled and viewed in EM. Then, using the z-stack from the LM, the sections containing the structures of interest can be narrowed down and immunolabeled for detailed analysis.
III.3.e. CLEM using HPF
For optimal preservation of ultrastructure, the most suitable CLEM experiment is HPF with freeze substitution. This technique usually focuses on the trafficking of internalized CLEM markers rather then immunolabeling of fluorescent proteins. Surface immunolabeling of the section for endogenous proteins is possible when combined with freeze substitution to Lowicryl HM20, although this remains technically challenging. HPF-CLEM can combine live-cell imaging with the superior fixation quality of high-pressure freezing (Verkade, 2008). A disadvantage compared with Tokuyasu CLEM is that the reference z-stack for serial sectioning cannot be taken after cryofixation since there is currently no device that can integrate cryo-immobilization with light microscopy (See also Buser et al. this volume). Therefore, it is necessary to record the z-stack before physically transferring the sample between the light microscope and the HPF apparatus. When studying processes such as membrane trafficking, the speed of this transfer is critical. To maximally reduce the time interval between imaging and HPF, we perform live-cell imaging with the sample already held in the HPF rapid loader (Verkade, 2008). An unavoidable consequence of this setup is that it creates a ‘distance’ between the cell and objective. With the modifications described previously (Brown et al., 2009) this distance has been reduced and the system of cell retracing improved. Briefly, the technique involves imaging cells grown on sapphire discs with a carbon coated finder pattern. The sapphire discs are held in modified HPF carriers and imaging is performed in an imaging dish containing medium with 20% BSA to act as a filler and cryo-protectant during HPF. Another modified rapid loader has recently been described that allows imaging in an intact imaging dish (McDonald, 2009). The time between recording the last image on the light microscope and having a fixed specimen can be as little as 4 seconds (Verkade, 2008).
Frozen samples have to undergo a freeze substitution protocol and resin embedding with either Epon or Lowicryl as described above (Section III.2.b.). Cells are relocated with the aid of the carbon finder pattern, which transfers to the surface of the polymerized resin block. After trimming down to the area of interest, the block is serially sectioned with a diamond knife using a standard room temperature ultramicrotome. Achieving serial sections under room temperature conditions is significantly less challenging as compared to serial cryosectioning for the Tokuyasu approach.
III.3.f. Remarks
CLEM techniques are valuable tools that allow the cell biologist to meet the two main criteria for answering intracellular membrane trafficking questions: combining high spatial resolution with dynamic information of the trafficking pathway of interest. In Table 2, we compare the three CLEM techniques discussed above and highlight the strengths and weaknesses of each. Since CLEM can be time-consuming and technically challenging, it is important to choose the appropriate method that can provide an answer to your specific research question.
Table 2. Comparison of three different CLEM techniques, highlighting some properties of each technique and the advantages and disadvantages.
For further comments see section III.3.
| Tokuyasu | HPF | Simple | |
|---|---|---|---|
|
|
|||
| Quality of live cell imaging | Good | Not optimal due to distance between cells and objective. | Good |
| Labels | Internalised markers and high efficiency Immunolabelling | Internalised markers and surface Immunolabelling with HM20 | Only internalised markers |
| Time resolution | Seconds to minutes | 4 seconds | Seconds to minutes |
| Processing | Technically challenging | Automated if using an AFS with FSP. | Simple |
| Retracing cells | Embossed coverslip | Finder grids or carbon coated finder pattern | Embossed coverslip |
| Sectioning | Cryo sectioning. Difficult to achieve serial sections | Standard diamond knife sectioning of resin block | Standard diamond knife sectioning of resin block |
| Preservation of ultrastructure | Good | Excellent | Good |
| Time to complete experiment | 4-5 days | 4-7 days | 2-3 days |
| Pitfalls | Formvar film too thick, cryosectioning, immunolabelling | Making the live cell carrier, achieving a well frozen sample | Removal of the coverslip |
III.4. Electron tomography
III.4.a. Principles and use of electron tomography
The geometry and size of the membrane carriers is thought to play an important part in efficient sorting and transport of specific cargo (Carlton and Cullen, 2005; Traer et al., 2007). TEM usually produces a 2-dimensional projection of a 3-dimensional sample, which makes the geometry of organelles in the sample hard to interpret. For example, cellular features that are seen as a single entity may actually be small features in different planes within the section. Furthermore, organelles and other cellular structures may only be partially present in the standard 70 nm sections, which can make interpretation difficult. For instance, vesicles and perpendicular-sectioned tubules both appear as circular projections in TEM. In line with this, structures might be bent, coming in-and-out of the section plane, and appear as two separate structures in the TEM projection. We can solve these difficulties by either combining serial thin sections or even better by viewing a semi-thick sample from different tilt angles (McIntosh et al., 2005; Frey et al., 2006). If a whole series of tilted images is collected, it is possible to computationally reconstruct a three-dimensional view or “tomogram”. Unless samples are specially shaped into a ‘rod’ using a Focused Ion Beam (FIB) instrument, we are unable to collect data through a full 360° rotation which would yield perfect reconstructions. Resolution in electron tomography is limited by the sample holder geometry. Since the FIB preparation of samples for tomography is not routinely established yet (Marko et al., 2007), reconstructions usually contain a “missing wedge” of information.
To create a tomogram, the sample is generally tilted from −65° to +65°, typically with 1° increments (see also chapters by O’Toole et al., Giddings et al., Hoenger et al. in this volume). Reconstructions can be improved by collecting so-called dual axes series, where a second set of data is collected from the same sample area at 90° to the first and the resulting tomograms are merged. Better results are achieved with smaller tilt increments, but a balance must then be found between acquisition time and electron dose to the sample to obtain the optimal image quality.
The images in a tilt series need to be further aligned and then the tomogram can be mathematically reconstructed using Fourier transform back projection algorithms (e.g. (Frank, 2006). The alignment can be improved by the use of “fiducials” or large gold markers applied to the top and bottom surfaces of the sample. These are then used in feature tracking software to better align images in the series. Many different software packages and procedures are available for performing reconstructions and helping with visualizations, including freeware packages such as IMOD (Kremer et al., 1996; Mastronarde, 1997). Finally, these structures can be rendered as three-dimensional models (Fig. 5L and 5M and supplementary movie 1). It should be noted that modeling of the tomogram is time consuming and can introduce subjectivity.
The thickness of a specimen that can be imaged depends on the microscope performance (mostly the acceleration voltage). Examples where electron tomography has been used include visualization of the complex structure of the Golgi apparatus (Ladinsky et al., 1994; Ladinsky et al., 1999), structural studies of mitochondria (Perkins et al., 1997; Perkins et al., 2009), and the microtubule cytoskeleton and centrosomes (Moritz et al., 1995; O’Toole et al., 2003). Ladinsky and co-workers combined serial tomographic reconstructions to create a 1 μm3 volume containing the Golgi complex from several 300 nm tomographic sections (Ladinsky et al., 1999). In our own lab, we have recently combined five serial 300 nm sections to reconstruct a multi-vesicular body labeled with different markers (Brown et al., 2009).
Nowadays, sections with thicknesses of 200-300 nm can also be analyzed routinely by tomography using conventional 120 kV TEM mainly due to the implementation of computer assisted acquisition available on modern microscopes. With the advance of intermediate-voltage electron microscopes with Field Emission guns and the combination with zero-loss Electron Energy Loss Spectroscopy (EELS) or STEM techniques, single sections of up to 1 to 2 μm thickness have been reconstructed (Beorchia et al., 1993; Sousa et al., 2009) (see also Walther et al., this volume). The so-called High-Angle Annular Dark Field (HAADF) STEM detector collects electrons scattered to high diffraction angles by the sample. The signal is proportional to the square of the atomic number of the atoms in the sample. This opens up the possibility of discriminating similar sized particles of different atomic compositions (e.g. silver vs. gold) (Sousa et al., 2007). HAADF images can be superior for reconstructions due to a much better signal-to-noise ratio (Yakushevska et al., 2007). As an example, whereas QDs are not easily visualized in standard bright field EM mode (Fig. 5K), the quantum dots ‘light up’ using HAADF STEM (Fig. 5N; see also (Brown et al., 2009)). However, it takes significantly longer to collect the data in the STEM mode compared to bright field TEM.
In electron tomography we should be aware of sample shrinkage. Due to locally produced heat leading to evaporation, the section will shrink during repetitive beam exposure. In order to facilitate alignment of the collected images the sample has to be pre-irradiated (i.e. pre-illuminated) before starting the collection of tilted views. This shrinkage of the section is especially an issue when tomography is applied to Tokuyasu and native cryosections, which are considerably more ‘vulnerable’ to beam damage than resin sections. Electron tomography has been successfully applied on Tokuyasu cryosections, especially using nanogold markers to improve the labeling efficiency of the thick section (Tchelidze et al., 2006; Zeuschner et al., 2006). In our own lab, we recently used thick immunogold labeled Tokuyasu cryosections to demonstrate the localization of different proteins of the COPII complex on budding vesicles of the transitional endoplasmic reticulum (Hughes et al., 2009). Observing the sample at low temperature using a cryo-specimen holder, where the sample is held at liquid nitrogen temperatures, may help to reduce shrinkage but adds an extra degree of complexity to the experiment. This holder can also be used to perform tomography on frozen samples, cryo electron tomography (cryo-ET), which is described in more depth by Kimse and Hoenger in this volume. A beautiful example of cryo-ET applied in the field of membrane traffic is the imaging of clathrin-coated transport vesicles (Cheng et al., 2007). The technique can, in principle, be used to freeze whole cells and analyze the structure of protein complexes in their natural environment in nanometer resolution (reviewed by (Baumeister et al., 1999) and demonstrated by (Medalia et al., 2002)). This detail is essential to answer some of the key questions on the molecular mechanism of membrane traffic and cargo sorting.
IV. Outlook
Although intracellular membrane transport is a highly dynamic process, the importance of ‘still’ images at high resolution as provided by EM is still invaluable. The introduction of new EM techniques, such as CLEM and electron tomography, especially in combination with immunolabeling techniques, will open up new avenues of research to gain even deeper insights into the mechanisms of transport. In this paper we have presented an overview of the most important techniques currently available for transport studies. We are convinced the use of live-cell imaging techniques in combination with high-end EM techniques, such as CLEM and tomography, will soon set the standard for membrane transport studies.
V. Materials and Equipment
Equipment
AFS2 + FSP (Leica Microsystems, Vienna, Austria).
Cryo-ultramicrotome EM-UC6 + FC6 (Leica Microsystems, Vienna, Austria).
Diamond knife (Diatome, Biel, Switzerland),
EM PACT2 + RTS (Leica Microsystems, Vienna, Austria).
Forceps (Manufactures des Outils Dumont, Switzerland)
Materials
Acetone (EMS, Hatfield, USA),
BSA, fraction V (Sigma-Aldrich Co., Poole, UK)
Cell culture dish, 35mm (Corning Inc., Corning, USA)
CO2 independent medium (GIBCO/Invitogen Life Technologies, Paisley, UK)
EGF-Biotin (Invitogen Life Technologies, Paisley, UK)
Embossed coverslip (MatTek Inc., Ashland, USA)
Epon812 (TAAB, Aldermaston, UK)
Ethanol (Fisher, Loughborough, England)
GFP antibody, 600-101-215 Goat (Rockland Inc, Gilbertsville, USA)
GFP antibody, A11122 Rabbit (Invitogen Life Technologies, Paisley, UK)
Glutaraldehyde, 25% EM grade (Agar Scientific, Stansted, UK)
Lowicryl HM20 Mono Step (Agar Scientific, Stansted, UK)
Membrane carrier, 100 μm deep (Leica Microsystems, Vienna, Austria)
Methanol (Fisher, Loughborough, UK)
Osmium tetroxide (EMS, Hatfield, USA)
Phenol red free medium (GIBCO/Invitogen Life Technologies, Paisley, UK)
Sapphire discs, 1.5 mm (Leica Microsystems, Vienna, Austria)
Sodium cacodylate buffer (EMS, Hatfield, USA)
Streptavidin-Quatum dot 655 (Invitogen Life Technologies, Paisley, UK)
Uranyl acetate (Agar Scientific, Stansted, UK)
Supplementary Material
Acknowledgements
The Wellcome Trust Fund supports JRTvW and JM, EB is supported by BBSRC, and THS is supported by the EPSRC.
References
- Adams SR, Campbell RE, Gross LA, Martin BR, Walkup GK, Yao Y, Llopis J, Tsien RY. New biarsenical ligands and tetracysteine motifs for protein labeling in vitro and in vivo: synthesis and biological applications. J Am Chem Soc. 2002;124:6063–6076. doi: 10.1021/ja017687n. [DOI] [PubMed] [Google Scholar]
- Balaji J, Ryan TA. Single-vesicle imaging reveals that synaptic vesicle exocytosis and endocytosis are coupled by a single stochastic mode. Proc Natl Acad Sci U S A. 2007;104:20576–20581. doi: 10.1073/pnas.0707574105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumeister W, Grimm R, Walz J. Electron tomography of molecules and cells. Trends Cell Biol. 1999;9:81–85. doi: 10.1016/s0962-8924(98)01423-8. [DOI] [PubMed] [Google Scholar]
- Beorchia A, Heliot, Menager M, Kaplan H, Ploton D. Applications of medium-voltage STEM for the 3-D study of organelles within very thick sections. J Microsc. 1993;170:247–258. doi: 10.1111/j.1365-2818.1993.tb03348.x. [DOI] [PubMed] [Google Scholar]
- Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, Hess HF. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- Brown E, Mantell J, Carter DA, Tilly G, Verkade P. Studying intracellular transport using high-pressure freezing and Correlative Light Electron Microscopy. Semin Cell Dev Biol. 2009;20:910–919. doi: 10.1016/j.semcdb.2009.07.006. [DOI] [PubMed] [Google Scholar]
- Carlton JG, Cullen PJ. Coincidence detection in phosphoinositide signaling. Trends Cell Biol. 2005;15:540–547. doi: 10.1016/j.tcb.2005.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccarelli B, Hurlbut WP, Mauro A. Turnover of transmitter and synaptic vesicles at the frog neuromuscular junction. J Cell Biol. 1973;57:499–524. doi: 10.1083/jcb.57.2.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Boll W, Kirchhausen T, Harrison SC, Walz T. Cryo-electron tomography of clathrin-coated vesicles: structural implications for coat assembly. J Mol Biol. 2007;365:892–899. doi: 10.1016/j.jmb.2006.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Amico F, Skarmoutsou E. Quantifying immunogold labelling in transmission electron microscopy. J Microsc. 2008;230:9–15. doi: 10.1111/j.1365-2818.2008.01949.x. [DOI] [PubMed] [Google Scholar]
- Essner E, Novikoff AB. Localization of acid phosphatase activity in hepatic lysosomes by means of electron microscopy. The Journal of biophysical and biochemical cytology. 1961;9:773–784. doi: 10.1083/jcb.9.4.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J. Electron Tomography: Methods for Three-Dimensional Visualization of Structures in the Cell. Springer Science + Business Media; New York, NY, USA: 2006. [Google Scholar]
- Frey TG, Perkins GA, Ellisman MH. Electron tomography of membrane-bound cellular organelles. Annual review of biophysics and biomolecular structure. 2006;35:199–224. doi: 10.1146/annurev.biophys.35.040405.102039. [DOI] [PubMed] [Google Scholar]
- Gaietta G, Deerinck TJ, Adams SR, Bouwer J, Tour O, Laird DW, Sosinsky GE, Tsien RY, Ellisman MH. Multicolor and electron microscopic imaging of connexin trafficking. Science. 2002;296:503–507. doi: 10.1126/science.1068793. [DOI] [PubMed] [Google Scholar]
- Giepmans BN, Deerinck TJ, Smarr BL, Jones YZ, Ellisman MH. Correlated light and electron microscopic imaging of multiple endogenous proteins using Quantum dots. Nature Methods. 2005;2:743–749. doi: 10.1038/nmeth791. [DOI] [PubMed] [Google Scholar]
- Giepmans BN, Adams SR, Ellisman MH, Tsien RY. The fluorescent toolbox for assessing protein location and function. Science. 2006;312:217–224. doi: 10.1126/science.1124618. [DOI] [PubMed] [Google Scholar]
- Gorden P, Carpentier JL, Cohen S, Orci L. Epidermal growth factor: morphological demonstration of binding, internalization, and lysosomal association in human fibroblasts. Proc Natl Acad Sci USA. 1978;75:5025–5029. doi: 10.1073/pnas.75.10.5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabenbauer M, Geerts WJ, Fernadez-Rodriguez J, Hoenger A, Koster AJ, Nilsson T. Correlative microscopy and electron tomography of GFP through photooxidation. Nat Methods. 2005;2:857–862. doi: 10.1038/nmeth806. [DOI] [PubMed] [Google Scholar]
- Granseth B, Odermatt B, Royle SJ, Lagnado L. Clathrin-mediated endocytosis is the dominant mechanism of vesicle retrieval at hippocampal synapses. Neuron. 2006;51:773–786. doi: 10.1016/j.neuron.2006.08.029. [DOI] [PubMed] [Google Scholar]
- Griffin BA, Adams SR, Tsien RY. Specific covalent labeling of recombinant protein molecules inside live cells. Science. 1998;281:269–272. doi: 10.1126/science.281.5374.269. [DOI] [PubMed] [Google Scholar]
- Griffiths G. Fine Structure Immunocytochemistry. Springer-Verlag; Berlin Heidelberg: 1993. [Google Scholar]
- Griffiths G, Hoppeler H. Quantitation in immunocytochemistry: correlation of immunogold labeling to absolute number of membrane antigens. J Histochem Cytochem. 1986;34:1389–1398. doi: 10.1177/34.11.3534077. [DOI] [PubMed] [Google Scholar]
- Gustafsson MG. Nonlinear structured-illumination microscopy: wide-field fluorescence imaging with theoretically unlimited resolution. Proc Natl Acad Sci USA. 2005;102:13081–13086. doi: 10.1073/pnas.0406877102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigler HT, McKanna JA, Cohen S. Direct visualization of the binding and internalization of a ferritin conjugate of epidermal growth factor in human carcinoma cells A-431. J Cell Biol. 1979;81:382–395. doi: 10.1083/jcb.81.2.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- Hawes P, Netherton CL, Mueller M, Wileman T, Monaghan P. Rapid freeze-substitution preserves membranes in high-pressure frozen tissue culture cells. J Microsc. 2007;226:182–189. doi: 10.1111/j.1365-2818.2007.01767.x. [DOI] [PubMed] [Google Scholar]
- Hayat MA. Principles and techniques of electron microscopy: biological applications. Cambridge University Press; Cambridge, UK: 2000. [Google Scholar]
- Heuser JE, Reese TS. Evidence for recycling of synaptic vesicle membrane during transmitter release at the frog neuromuscular junction. J Cell Biol. 1973;57:315–344. doi: 10.1083/jcb.57.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins CR, Gibson A, Shipman M, Strickland DK, Trowbridge IS. In migrating fibroblasts, recycling receptors are concentrated in narrow tubules in the pericentriolar area, and then routed to the plasma membrane of the leading lamella. J Cell Biol. 1994;125:1265–1274. doi: 10.1083/jcb.125.6.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes H, Budnik A, Schmidt K, Palmer KJ, Mantell J, Noakes C, Johnson A, Carter DA, Verkade P, Watson P, Stephens DJ. Organisation of human ER-exit sites: requirements for the localisation of Sec16 to transitional ER. J Cell Sci. 2009;122:2924–2934. doi: 10.1242/jcs.044032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal JK, Simon SM. Potentials and pitfalls of fluorescent quantum dots for biological imaging. Trends Cell Biol. 2004;14:497–504. doi: 10.1016/j.tcb.2004.07.012. [DOI] [PubMed] [Google Scholar]
- Jiménez N, Humbel BM, van Donselaar E, Verkleij AJ, Burger KN. Aclar discs: a versatile substrate for routine high-pressure freezing of mammalian cell monolayers. J Microsc. 2006;221:216–223. doi: 10.1111/j.1365-2818.2006.01558.x. [DOI] [PubMed] [Google Scholar]
- Kellenberger E, Johansen R, Maeder M, Bohrmann B, Stauffer E, Villiger W. Artefacts and morphological changes during chemical fixation. J Microsc. 1992;168:181–201. doi: 10.1111/j.1365-2818.1992.tb03260.x. [DOI] [PubMed] [Google Scholar]
- Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- Ladinsky MS, Kremer JR, Furcinitti PS, McIntosh JR, Howell KE. HVEM tomography of the trans-Golgi network: structural insights and identification of a lace-like vesicle coat. J Cell Biol. 1994;127:29–38. doi: 10.1083/jcb.127.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladinsky MS, Mastronarde DN, McIntosh JR, Howell KE, Staehelin LA. Golgi structure in three dimensions: functional insights from the normal rat kidney cell. J Cell Biol. 1999;144:1135–1149. doi: 10.1083/jcb.144.6.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard D, Hayakawa A, Lawe D, Lambright D, Bellve KD, Standley C, Lifshitz LM, Fogarty KE, Corvera S. Sorting of EGF and transferrin at the plasma membrane and by cargo-specific signaling to EEA1-enriched endosomes. J Cell Sci. 2008;121:3445–3458. doi: 10.1242/jcs.031484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou W, Geuze HJ, Slot JW. Improving structural integrity of cryosections for immunogold labeling. Histochem Cell Biol. 1996;106:41–58. doi: 10.1007/BF02473201. [DOI] [PubMed] [Google Scholar]
- Mari M, Bujny M, Zeuschner D, Geerts W, Griffith J, Petersen C, Cullen PJ, Klumperman J, Geuze H. SNX1 defines an early endosomal recycling exit for sortilin and mannose 6-phosphate receptors. Traffic. 2008;9:380–393. doi: 10.1111/j.1600-0854.2007.00686.x. [DOI] [PubMed] [Google Scholar]
- Marko M, Hsieh C, Schalek R, Frank J, Mannella C. Focused-ion-beam thinning of frozen-hydrated biological specimens for cryo-electron microscopy. Nat Methods. 2007;4:215–217. doi: 10.1038/nmeth1014. [DOI] [PubMed] [Google Scholar]
- Mastronarde DN. Dual-axis tomography: an approach with alignment methods that preserve resolution. J Struct Biol. 1997;120:343–352. doi: 10.1006/jsbi.1997.3919. [DOI] [PubMed] [Google Scholar]
- McDonald KL. Cryopreparation methods for electron microscopy of selected model systems. Methods Cell Biol. 2007;79:23–56. doi: 10.1016/S0091-679X(06)79002-1. [DOI] [PubMed] [Google Scholar]
- McDonald KL, Morphew M, Verkade P, Muller-Reichert T. Recent advances in high-pressure freezing: equipment- and specimen-loading methods. Methods Mol Biol. 2007;369:143–173. doi: 10.1007/978-1-59745-294-6_8. [DOI] [PubMed] [Google Scholar]
- McDonald KL. A review of high-pressure freezing preparation techniques for correlative light and electron microscopy of the same cells and tissues. J Microsc. 2009;235:273–281. doi: 10.1111/j.1365-2818.2009.03218.x. [DOI] [PubMed] [Google Scholar]
- McIntosh R, Nicastro D, Mastronarde D. New views of cells in 3D: an introduction to electron tomography. Trends Cell Biol. 2005;15:43–51. doi: 10.1016/j.tcb.2004.11.009. [DOI] [PubMed] [Google Scholar]
- McMahon HT, Gallop JL. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature. 2005;438:590–596. doi: 10.1038/nature04396. [DOI] [PubMed] [Google Scholar]
- Medalia O, Weber I, Frangakis AS, Nicastro D, Gerisch G, Baumeister W. Macromolecular architecture in eukaryotic cells visualized by cryoelectron tomography. Science. 2002;298:1209–1213. doi: 10.1126/science.1076184. [DOI] [PubMed] [Google Scholar]
- Meiblitzer-Ruppitsch C, Vetterlein M, Stangl H, Maier S, Neumüller J, Freissmuth M, Pavelka M, Ellinger A. Electron microscopic visualization of fluorescent signals in cellular compartments and organelles by means of DAB-photoconversion. Histochem Cell Biol. 2008;130:407–419. doi: 10.1007/s00418-008-0429-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalet X, Pinaud F, Lacoste TD, Dahan M. Properties of Fluorescent Semiconductor Nanocrystals and their Application to Biological Labeling. Single Molecules. 2001;2:261–276. [Google Scholar]
- Monosov EZ, Wenzel TJ, Luers GH, Heyman JA, Subramani S. Labeling of peroxisomes with green fluorescent protein in living P. pastoris cells. J Histochem Cytochem. 1996;44:581–589. doi: 10.1177/44.6.8666743. [DOI] [PubMed] [Google Scholar]
- Moor H. Theory and practice of high pressure freezing. In: Steinbrecht RA, Zierold K, editors. Cryo-Techniques in Biological Electron Microscopy. Springer-Verlag; Berlin Heidelberg: 1987. pp. 175–191. [Google Scholar]
- Moritz M, Braunfeld MB, Sedat JW, Alberts B, Agard DA. Microtubule nucleation by gamma-tubulin-containing rings in the centrosome. Nature. 1995;378:638–640. doi: 10.1038/378638a0. [DOI] [PubMed] [Google Scholar]
- Murk JLAN, Posthuma G, Koster AJ, Geuze HJ, Verkleij AJ, Kleijmeer MJ, Humbel BM. Influence of aldehyde fixation on the morphology of endosomes and lysosomes: quantitative analysis and electron tomography. J Microsc. 2003;212:81–90. doi: 10.1046/j.1365-2818.2003.01238.x. [DOI] [PubMed] [Google Scholar]
- O’Toole ET, McDonald KL, Mäntler J, McIntosh JR, Hyman AA, Müller-Reichert T. Morphologically distinct microtubule ends in the mitotic centrosome of Caenorhabditis elegans. J Cell Biol. 2003;163:451–456. doi: 10.1083/jcb.200304035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oorschot V, de Wit H, Annaert W, Klumperman J. A novel flat-embedding method to prepare ultrathin cryosections from cultured cells in their in situ orientation. J Histochem Cytochem. 2002;50:1067–1080. doi: 10.1177/002215540205000809. [DOI] [PubMed] [Google Scholar]
- Penttila A, Kalimo H, Trump BF. Influence of glutaraldehyde and-or osmium tetroxide on cell volume, ion content, mechanical stability, and membrane permeability of Ehrlich ascites tumor cells. J Cell Biol. 1974;63:197–214. doi: 10.1083/jcb.63.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins GA, Renken C, Martone ME, Young SJ, Ellisman MH, Frey TG. Electron tomography of neuronal mitochondria: three-dimensional structure and organization of cristae and membrane contacts. J Struct Biol. 1997;119:260–272. doi: 10.1006/jsbi.1997.3885. [DOI] [PubMed] [Google Scholar]
- Perkins GA, Sun MG, Frey TG. Chapter 2 Correlated light and electron microscopy/electron tomography of mitochondria in situ. Meth Enzymol. 2009;456:29–52. doi: 10.1016/S0076-6879(08)04402-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plitzko JM, Rigort A, Leis A. Correlative cryo-light microscopy and cryo-electron tomography: from cellular territories to molecular landscapes. Curr Opin Biotechnol. 2009;20:83–89. doi: 10.1016/j.copbio.2009.03.008. [DOI] [PubMed] [Google Scholar]
- Polishchuk RS, Polishchuk EV, Marra P, Alberti S, Buccione R, Luini A, Mironov AA. Correlative light-electron microscopy reveals the tubular-saccular ultrastructure of carriers operating between Golgi apparatus and plasma membrane. J Cell Biol. 2000;148:45–58. doi: 10.1083/jcb.148.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rink J, Ghigo E, Kalaidzidis Y, Zerial M. Rab conversion as a mechanism of progression from early to late endosomes. Cell. 2005;122:735–749. doi: 10.1016/j.cell.2005.06.043. [DOI] [PubMed] [Google Scholar]
- Ripper D, Schwarz H, Stierhof YD. Cryo-section immunolabelling of difficult to preserve specimens: advantages of cryofixation, freeze-substitution and rehydration. Biol Cell. 2008;100:109–123. doi: 10.1042/BC20070106. [DOI] [PubMed] [Google Scholar]
- Robinson JM, Takizawa T. Correlative fluorescence and electron microscopy in tissues: immunocytochemistry. J Microsc. 2009;235:259–272. doi: 10.1111/j.1365-2818.2009.03221.x. [DOI] [PubMed] [Google Scholar]
- Rosenmund C, Stevens CF. The rate of aldehyde fixation of the exocytotic machinery in cultured hippocampal synapses. J Neurosci Methods. 1997;76:1–5. doi: 10.1016/s0165-0270(97)00061-7. [DOI] [PubMed] [Google Scholar]
- Rust MJ, Bates M, Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nat Methods. 2006;3:793–795. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schönle A, Glatz M, Hell SW. Four-dimensional multiphoton microscopy with time-correlated single-photon counting. Applied optics. 2000;39:6306–6311. doi: 10.1364/ao.39.006306. [DOI] [PubMed] [Google Scholar]
- Seligman AM, Karnovsky MJ, Wasserkrug HL, Hanker JS. Nondroplet ultrastructural demonstration of cytochrome oxidase activity with a polymerizing osmiophilic reagent, diaminobenzidine (DAB) J Cell Biol. 1968;38:1–14. doi: 10.1083/jcb.38.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer SJ. Preparation of an electron-dense antibody conjugate. Nature. 1959;183:1523–1524. doi: 10.1038/1831523a0. [DOI] [PubMed] [Google Scholar]
- Slot JW, Geuze HJ. Cryosectioning and immunolabeling. Nat Protoc. 2007;2:2480–2491. doi: 10.1038/nprot.2007.365. [DOI] [PubMed] [Google Scholar]
- Small JV. Organization of actin in the leading edge of cultured cells: influence of osmium tetroxide and dehydration on the ultrastructure of actin meshworks. J Cell Biol. 1981;91:695–705. doi: 10.1083/jcb.91.3.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa AA, Aronova MA, Kim YC, Dorward LM, Zhang G, Leapman RD. On the feasibility of visualizing ultrasmall gold labels in biological specimens by STEM tomography. J Struct Biol. 2007;159:507–522. doi: 10.1016/j.jsb.2007.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa AA, Hohmann-Marriott MF, Zhang G, Leapman RD. Monte Carlo electron-trajectory simulations in bright-field and dark-field STEM: implications for tomography of thick biological sections. Ultramicroscopy. 2009;109:213–221. doi: 10.1016/j.ultramic.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbrecht RA, Zierold K. A cryoembedding method for cutting ultrathin cryosections from small frozen specimens. J Microsc. 1984;136:69–75. doi: 10.1111/j.1365-2818.1984.tb02546.x. [DOI] [PubMed] [Google Scholar]
- Stoscheck CM, Carpenter G. Down regulation of epidermal growth factor receptors: direct demonstration of receptor degradation in human fibroblasts. J Cell Biol. 1984;98:1048–1053. doi: 10.1083/jcb.98.3.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studer D, Humbel BM, Chiquet M. Electron microscopy of high pressure frozen samples: bridging the gap between cellular ultrastructure and atomic resolution. Histochem Cell Biol. 2008;130:877–889. doi: 10.1007/s00418-008-0500-1. [DOI] [PubMed] [Google Scholar]
- Tchelidze P, Sauvage C, Bonnet N, Kilian L, Beorchia A, O’Donohue MF, Ploton D, Kaplan H. Electron tomography of amplified nanogold immunolabelling: Improvement of quality based on alignment of projections with sinograms and use of post-reconstruction deconvolution. J Struct Biol. 2006;156:421–431. doi: 10.1016/j.jsb.2006.07.004. [DOI] [PubMed] [Google Scholar]
- Tokuyasu KT. A technique for ultracryotomy of cell suspensions and tissues. J Cell Biol. 1973;57:551–565. doi: 10.1083/jcb.57.2.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traer C, Rutherford A, Palmer K, Wassmer T, Oakley J, Attar N, Carlton J, Kremerskothen J, Stephens D, Cullen PJ. SNX4 coordinates endosomal sorting of TfnR with dynein-mediated transport into the endocytic recycling compartment. Nat Cell Biol. 2007;9:1370–1380. doi: 10.1038/ncb1656. [DOI] [PubMed] [Google Scholar]
- Tsien RY. Constructing and exploiting the fluorescent protein paintbox (Nobel Lecture) Angew Chem Int Ed Engl. 2009;48:5612–5626. doi: 10.1002/anie.200901916. [DOI] [PubMed] [Google Scholar]
- van Donselaar E, Posthuma G, Zeuschner D, Humbel B, Slot J. Immunogold labeling of cryosections from high-pressure frozen cells. Traffic. 2007;8:471–485. doi: 10.1111/j.1600-0854.2007.00552.x. [DOI] [PubMed] [Google Scholar]
- van Rijnsoever C, Oorschot V, Klumperman J. Correlative light-electron microscopy (CLEM) combining live-cell imaging and immunolabeling of ultrathin cryosections. Nat Methods. 2008;5:973–980. doi: 10.1038/nmeth.1263. [DOI] [PubMed] [Google Scholar]
- Verkade P, Schrama LH, Verkleij AJ, Gispen WH, Oestreicher AB. Ultrastructural co-localization of calmodulin and B-50/growth-associated protein-43 at the plasma membrane of proximal unmyelinated axon shafts studied in the model of the regenerating rat sciatic nerve. Neuroscience. 1997;79:1207–1218. doi: 10.1016/s0306-4522(97)00041-9. [DOI] [PubMed] [Google Scholar]
- Verkade P. Moving EM: the Rapid Transfer System as a new tool for correlative light and electron microscopy and high throughput for high-pressure freezing. J Microsc. 2008;230:317–328. doi: 10.1111/j.1365-2818.2008.01989.x. [DOI] [PubMed] [Google Scholar]
- Weibull C, Villiger W, Carlemalm E. Extraction of lipids during freeze-substitution of Acholeplasma laidlawii-cells for electron microscopy. J Microsc. 1984;134:213–216. doi: 10.1111/j.1365-2818.1984.tb02513.x. [DOI] [PubMed] [Google Scholar]
- Yakushevska AE, Lebbink MN, Geerts WJ, Spek L, Donselaar EG, Jansen KA, Humbel BM, Post JA, Verkleij, Koster AJ. STEM tomography in cell biology. J Struct Biol. 2007;159:381–391. doi: 10.1016/j.jsb.2007.04.006. [DOI] [PubMed] [Google Scholar]
- Zeuschner D, Geerts W, van Donselaar E, Humbel B, Slot J, Koster AJ, Klumperman J. Immuno-electron tomography of ER exit sites reveals the existence of free COPII-coated transport carriers. Nat Cell Biol. 2006;8:377–383. doi: 10.1038/ncb1371. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Li Y, Tsien RW. The dynamic control of kiss-and-run and vesicular reuse probed with single nanoparticles. Science. 2009;323:1448–1453. doi: 10.1126/science.1167373. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.