Abstract
Humans and viruses have a long co-evolutionary history. Viral illnesses have and will continue to shape human history: from smallpox, to influenza, to HIV, and beyond. Animal models of human viral illnesses are needed in order to generate safe and effective antiviral medicines, adjuvant therapies, and vaccines. These animal models must support the replication of human viruses, recapitulate aspects of human viral illnesses, and respond with conserved immune signaling cascades. The zebrafish is perhaps the simplest, most commonly used laboratory model organism in which innate and/or adaptive immunity can be studied. Herein, we will discuss the current zebrafish models of human viral illnesses and the insights they have provided. We will highlight advantages of early life stage zebrafish and the importance of innate immunity in human viral illnesses. We will also discuss viral characteristics to consider before infecting zebrafish with human viruses as well as predict other human viruses that may be able to infect zebrafish.
Keywords: Immunity, zebrafish, virus, human, infection
1. Introduction
It has been estimated that 90% of the infectious diseases that afflict human beings are caused by viruses (Norkin, 2010), and over 200 different viruses have been isolated from the human upper respiratory tract alone (Mackie, 2003). Many viral infections are asymptomatic and may go unnoticed, but others cause severe or even life threatening diseases in humans. The immune system has evolved in many species, including humans, to recognize and clear foreign infectious agents from the body. In vertebrates, there are two branches of the immune system that work in concert: innate and adaptive. Innate immunity is more evolutionarily ancient and functions to initially recognize the threat and limit the spread of infection. In jawed vertebrates, adaptive immunity triggers cell-mediated and antibody-mediated responses that can curtail existing infections and provide the host with long-term immunological memory. The adaptive immune response has been exploited in the development of vaccines. Through immunizations, some viral illnesses have been essentially eradicated from the human population. Due to the success of some vaccination programs, the importance of adaptive immunity in virus infections is widely recognized; however, the important role of the innate immune response in viral diseases is only now being appreciated.
Viruses are recognized by the innate immune system upon binding to pattern recognition receptors (PRRs) on or within host cells. PRRs that recognize viruses include Toll-like receptors, retinoic acid-inducible gene I-like receptors, nucleotide oligomerization domain-like receptors, and receptors that detect DNA in the cytosol of cells. Several reviews provide detailed information on viral detection by PRRs and the signaling pathways elicited (Kawai and Akira, 2006; Pichlmair and Reis e Sousa, 2007; Saito and Gale, 2007; McCartney and Colonna, 2009; Shayakhmetov et al., 2009; Takeuchi and Akira, 2009; Pedraza et al., 2010; Drutskaya et al., 2011; Thompson et al., 2011). Signaling by PRRs through distinct or overlapping signal transduction pathways culminates in the production of cytokines, such as interferon (IFN), and chemokines. Through paracrine signaling, IFN is secreted by infected cells and binds to receptors on nearby uninfected cells in an attempt to wall off the spread of infection by instructing cells to suspend transcription and translation. Other cytokines and chemokines are involved in pro-inflammatory signaling cascades that recruit phagocytes to sites of infection. A better understanding of the innate immune response to viral infections will be beneficial in developing preventative and adjuvant therapies for viral illnesses because activation of innate immune signaling pathways and phagocytic cells is common to all viral infections, potentiates a robust adaptive immune response, has both positive and negative effects on the health of the host, and can be thwarted by certain viral adaptations.
The dynamics between host immune responses and human viral pathogens can only be studied in animal models where tissue interactions are intact and where genetics, cell types, and signaling cascades are homologous to those in humans. Commonly used animal models of human viral infections include small mammals and non-human primates. In an effort to reduce, replace, and refine the animal models used in scientific research, the zebrafish infectious disease model has become an attractive option. Zebrafish are vertebrates that possess both innate and adaptive immune systems. The conservation of innate and adaptive immunity between zebrafish and humans has been well-characterized (reviewed in Traver et al., 2003; Trede et al., 2004; van der Sar et al., 2004; Phelps and Neely, 2005; Jakovlic et al., 2006; Sullivan and Kim, 2008; Cui et al., 2011; Meijer and Spaink, 2011; Novoa and Figueras, 2011; Crim and Riley, 2012; van der Vaart et al., 2012). This review will focus on the use of zebrafish to study human viral illnesses and the host’s innate immune response, largely because this represents the bulk of current research. Zebrafish phagocytic macrophages and neutrophils are similar to those in mammals in terms of their morphology, molecular signatures, and functionality (Bennett et al., 2001). Certain other innate immune cell types found in mammals, including monocytes, NK cells, dendritic cells, eosinophils, and basophils, have been identified in zebrafish and are beginning to be characterized (Dobson et al., 2008; Moss et al., 2009; Balla et al., 2010; Lugo-Villarino et al., 2010; Da’as et al., 2011). In addition, innate immune signal transduction pathways downstream from pathogen receptors, such as IFN signaling, are well-conserved between mammals and zebrafish (Stein et al., 2007; Langevin et al., 2013). Thus, the zebrafish is a useful model for the study of the innate immune response to human infectious diseases. Moreover, adult zebrafish have adaptive immunity and could be used to study the adaptive immune response to human viruses that are able to infect adult zebrafish. In this review we will discuss the studies utilizing the four known zebrafish models of human viral illnesses. Roles for zebrafish neutrophils, macrophages, and IFN signaling in response to these human virus infections will be described. Given the involvement of other innate immune cell types (e.g. NK cells, dendritic cells) in these four specific virus infections in mammals and the recent identification of similar cell types in zebrafish, the zebrafish infection model will likely contribute additional important insights regarding roles for these cell types in in vivo infections in the near future. We will also propose additional human viral pathogens that may be able to infect zebrafish and describe the insights that the zebrafish infectious disease model can provide due to the unique research opportunities possible in the zebrafish system.
2. Zebrafish models of human viral illnesses
The zebrafish is rapidly gaining in popularity as an infectious disease model. Zebrafish have been used to study fish-specific infectious diseases that afflict economically important fish species (reviewed in Trede et al., 2004; van der Sar et al., 2004; Phelps and Neely, 2005; Sullivan and Kim, 2008; Meijer and Spaink, 2011; Milligan-Myhre et al., 2011; Novoa and Figueras, 2011; Crim and Riley, 2012). It has recently been shown that zebrafish can be good models in which to study human infectious diseases as well. It has been suggested that to be able to preserve and study the complexities of host-pathogen co-evolution when using animals to model human infectious diseases, it is important to employ the most closely related pathogen that naturally infects the model species (Baker, 1998; Crim and Riley, 2012; Keebaugh and Schlenke, 2014). However, the natural pathogens of zebrafish are currently unknown (Crim and Riley, 2012). A different approach, that potentially has more direct translational impact, is to use an animal model with an immune response similar to humans that can be infected by human isolates of a pathogen. The first reported human pathogens that could infect and cause disease in zebrafish were bacteria (reviewed in Trede et al., 2004; van der Sar et al., 2004; Phelps and Neely, 2005; Sullivan and Kim, 2008; Meijer and Spaink, 2011; Milligan-Myhre et al., 2011; Novoa and Figueras, 2011). There are now reports of zebrafish models of human fungal (Chao et al., 2010; Brothers et al., 2011; Brothers et al., 2013; Chen et al., 2013; Gratacap et al., 2013; Kuo et al., 2013; Y.-C. Wang et al., 2013) and human viral pathogen infections (Burgos et al., 2008; Ding et al., 2011; Antoine et al., 2013; Palha et al., 2013; K. A. Gabor and C. H. Kim, personal communication). We will describe the human viral illnesses for which there are currently zebrafish infection models and then discuss the findings and insights obtained thus far from these zebrafish models of human viral infections.
2.1. Human viral illnesses for which there are zebrafish infection models
To date, there are publications of four human viral illnesses that can be modeled in zebrafish, but we believe that many more zebrafish models of human viral diseases can and will be developed. The following are descriptions of the diseases that occur in humans infected with the four human viruses that have been shown to also infect zebrafish.
Herpes simplex virus (HSV)-1 is a DNA virus belonging to the Herpesviridae family of human herpesviruses that also includes the closely related HSV-2 and varicella zoster virus. HSV-1 is distributed ubiquitously worldwide throughout human populations, with infection rates approaching 90%. In the U.S., infection rates are lower but still overwhelming, with reported rates ranging from 57-65% (Koelle and Corey, 2008; Nicoll et al., 2012). HSV-1 infections can be spread by horizontal transmission via contact with infected individuals during an active infection state, or by vertical transmission from mother to neonate (Corey and Wald, 2009). Primary infections typically present as skin blisters on the mouth or genitals. Recurrent outbreaks of active infection are caused by emergence of the virus from a latent phase, often in the trigeminal sensory ganglia (Arduino and Porter, 2006). HSV-1 and -2 infections in neonates can have serious consequences. Approximately one-quarter of infected neonates will present with disseminated infection involving multiple organs; approximately one-third will present with central nervous system involvement leading to seizures, lethargy, tremors, and lack of feeding; and approximately 45% will have manifestations localized to the skin, ears, and mouth (Kimberlin, 2005). With antiviral therapies, mortalities have been reduced. However, the development of antiviral therapies, especially vaccines, has been hampered by the considerable genetic diversity exhibited by HSV-1 (Szpara et al., 2013).
Hepatitis C virus (HCV) is a positive-sense, single-stranded RNA virus belonging to the Hepacivirus genus of the Flaviviridae family. HCV infects 130-185 million people worldwide, with an estimated 5 million people infected in the U.S (Edlin, 2011; Rehermann, 2013; Scheel and Rice, 2013; Simmonds, 2013; Thomas, 2013). HCV is transmitted between individuals through parenteral routes such as blood transfusion, contaminated needles and medical equipment, and tattoos (Rehermann, 2013; Simmonds, 2013). There are also instances of transmission through sexual contact and by perinatal exposure. The acute phase of HCV infection is usually mild to asymptomatic, with up to one-third of patients spontaneously clearing the infection (Thomas, 2013). During the acute phase, a small percentage of patients (approximately 15%) demonstrate symptoms: such as jaundice, influenza-like illness, dark urine, clay-colored stools, and pain in the upper right abdominal quadrant (Maheshwari et al., 2008). HCV infects individuals by targeting hepatocytes and can often establish a chronic liver infection without triggering a robust host antiviral innate immune response. Chronic infection often leads to liver cirrhosis and/or cancer and is the leading cause of disease requiring liver transplantation (Horner and Gale, 2013). Chronic infection can also lead to fibrosis, steatosis, insulin resistance and type 2 diabetes, hepatocellular carcinoma, and extrahepatic diseases including non-Hodgkin lymphoma, mixed cryoglobulinemia, and glomerulonephritis (Yamane et al., 2013). There are some indications that HCV can trigger pathology in the central nervous system (CNS) through direct infection of CNS tissues. In some individuals, HCV infection can be cleared with antiviral and IFN therapy; but in other cases, infection persists.
The chikungunya virus (CHIKV) is an enveloped, positive-sense, single-stranded RNA virus in the Alphavirus genus of the Togaviridae family. CHIKV is an arbovirus spread by Aedes mosquitoes (A. albopictus and A. aegypti) (Schwartz and Albert, 2010; Tsetsarkin et al., 2011; Burt et al., 2012). In the Makonde language, native to parts of Tanzania and Mozambique, chikungunya means “that which bends up,” and this refers to the contorted postures of affected individuals who are experiencing the extreme arthralgia and myalgia from this disease that can arise even months or years after initial infection. CHIKV has been detected in nearly 40 countries, with epidemics in Africa and Asia and documented cases in Australia and western Europe (Schwartz and Albert, 2010). The most recent epidemic occurred in 2006 in La Réunion (east of Madagascar) where there were 300,000 cases in a population of 785,000 people, with 237 deaths. CHIKV disease has acute and chronic phases (Burt et al., 2012; Dupuis-Maguiraga et al., 2012). The acute phase is typical of viral infections and marked by robust type I IFN responses. Patients typically present with fever, arthralgia, myalgia, cephalgia, rash, erythema, and fatigue. In the 2006 outbreak in La Réunion, a small number of affected individuals with severe, acute CHIKV disease also presented with encephalitis and hemorrhage. In children, additional symptoms included diarrhea, vomiting, and convulsions, among others. Symptoms usually last for approximately two weeks. Interestingly, some infected individuals are asymptomatic for decades. In its chronic phases, patients may suffer from crippling arthralgia, myalgia, and arthritis. At this time, no vaccines exist.
Influenza A virus (IAV) belongs to the Orthomyxoviridae family. It has a negative-sense, single-stranded, and segmented RNA genome. IAV infects human populations on a yearly basis during seasonal outbreaks and epidemics. Due to antigenic drift, IAV is continually evolving to cope with antibody-mediated immune selective pressures applied by its hosts (Medina and García-Sastre, 2011; Palese and T. T. Wang, 2011). On occasion, an antigenic shift that transforms the virus into a novel subtype with altogether unique antigenic profiles may occur and this can lead to serious pandemics. At least four such pandemics have occurred in the past 100 years. In 1918, the Spanish influenza pandemic caused by the H1N1 subtype killed an estimated 50-100 million people around the world. Additional pandemics occurred in 1957, 1968, and 2009. Individuals infected with IAV typically present with fever, myalgia, non-productive cough, sore throat, and fatigue (Hamborsky et al., 2007). Fever and other symptoms typically subside within 2-3 days. The elderly and young children are at particular risk for incurring serious complications from IAV infections. An average of approximately 24,000 individuals died each year from influenza complications between 1984 and 2007. Complications include viral or secondary bacterial pneumonia, Reye syndrome (from inappropriate administration of aspirin to sick children), and cardiopulmonary events. Individuals suffering from pre-existing ailments including chronic cardiovascular or pulmonary diseases are at particular risk. Because of antigenic drift and shift, vaccines provide limited protection against IAV and strains of the virus can rapidly evolve antiviral resistance.
The human health and economic burdens posed by these viruses are readily apparent. A common thread in these four viral illnesses is treatment and prevention difficulties due to virus genetic diversity and immune evasion strategies. Zebrafish models of these human viral illnesses will complement other animal models by providing the opportunity to visualize virus-host dynamics and to conduct genetic and chemical screens, which will facilitate the development and testing of new antiviral strategies.
2.2. Virological and immunological assays and insights from zebrafish models of human viral infections
In this section, we will discuss the virological and immunological assays that have been conducted in zebrafish infected with human viral pathogens. We will highlight those that are ideally suited to the zebrafish infection model (see Table 1) and discuss the insights that have been garnered from these experiments.
Table 1.
The advantages of using zebrafish to model human viral infections
| Research Areas | Tools/Techniques | Insights | Example References |
|---|---|---|---|
| Virology | Fluorescent human viral pathogens |
In vivo infection kinetics, cell tropism, and viral burden |
Ding 2011; Palha 2013; K. A. Gabor and C. H. Kim, personal communication |
| Immunology | Transgenic lines | Phagocyte behavior, spatiotemporal induction of antiviral signaling |
This review; Palha 2013 |
| Morpholino knockdown | Gene products regulating infection |
Palha 2013 | |
| Chemical treatment | Druggable signaling pathways regulating infection |
Ding 2011; Palha 2013; K. A. Gabor and C. H. Kim, personal communication |
2.2.1. Virology
Infection kinetics, such as viral gene expression and viral burden, can be measured in zebrafish and other animal models of viral infections and compared to human viral infections. Viral DNA or RNA can be quantified by PCR and spatiotemporal expression can be observed via in situ hybridization (ISH). Amounts of viral protein can be quantified by western blot or localization can be observed via immunohistochemistry (IHC). As opposed to fish-specific viruses, many antibodies already exist for the detection of human viral proteins via IHC. However, it is the unique opportunity to visualize infection kinetics in vivo using fluorescent human viruses that distinguishes the zebrafish from other animal models of infectious diseases.
2.2.1.1. Viral gene expression
The expression of viral DNA or RNA has been quantified to show replication of human viral pathogens in zebrafish. In adult zebrafish infected with HSV-1 via intraperitoneal injection, DNA was isolated from different areas of the body to show viral replication, clearance, or persistence and the spread of virus to different tissues over time. Isolating DNA from different tissues is easier in adult zebrafish due to the size of the organism, but it is still possible in embryonic/larval zebrafish. Oligonucleotide primers to amplify thymidine kinase were used to detect viral DNA and alpha-actin was amplified to detect zebrafish genomic DNA in HSV-1-infected zebrafish. DNA was isolated from the ventral body (the site of injection/initial infection), the dorsal body (site of the spinal column), and the head (containing the brain) at different infection doses and multiple time points. This experiment revealed a peak in HSV-1 viral genomes in the ventral body, followed by a peak in viral genomes in the dorsal body, followed by the appearance and persistence of HSV-1 viral genomes in the brain at the highest viral concentration tested (Burgos et al., 2008). Anti-HSV-1 antibody staining illustrated CNS tissue tropism and showed that viral proteins were localized to the cytoplasm or microtubules in spinal cord neurons and axons, encephalon neurons, and pituitary gland cells (Burgos et al., 2008). These results demonstrated replication of the human viral pathogen HSV-1 in zebrafish, confirmed CNS tissue tropism for HSV-1 in zebrafish, and showed that the adult zebrafish brain is a potential viral reservoir.
PCR quantification of DNA from DNA viruses is a more straightforward indicator of viral number and replication than is RT-PCR for RNA viruses. Care should be taken when designing primers to amplify cDNA reverse-transcribed from RNA viruses to determine whether viral genomic RNA, subgenomic mRNA transcripts, or both will be amplified. However, the expression profile of viral cDNA from RNA viruses can still provide information about viral infection kinetics. In CHIKV-infected zebrafish larvae, primers were designed to detect expression of both genomic RNA and subgenomic mRNA for the E1 viral spike gene. Viral transcript levels were normalized to host ef1alpha transcripts. Quantitative PCR results revealed a rapid increase in viral E1 expression up to 24 hpi followed by a slow, steady decline in viral E1 expression (Palha et al., 2013). The peak timing of CHIKV virus replication seen via E1 expression was confirmed with viral burden assays as discussed below.
In zebrafish injected at the one cell stage with a HCV sub-replicon construct, amplification of the viral sub-replicon was demonstrated by PCR and detection of viral gene products was confirmed by ISH and western blot. PCR showed that viral core cDNA expression increased between 4 and 10 dpi and a western blot with anti-HCV core antibody confirmed the presence of viral core protein in zebrafish. ISH also detected HCV core transcripts and revealed that the expression of HCV sub-replicon genes was spatially restricted to the zebrafish liver and intestine (Ding et al., 2011). These assays ((q)PCR, ISH, IHC, and western blot) can all be used to determine whether human viral pathogens are capable of replication in zebrafish and thus whether the zebrafish may be a good model organism in which to study certain human viral infections.
2.2.1.2. Viral burden
As a complement to quantification of viral gene/gene product expression, viral burden indicates the functionality of replicated viruses as infectious agents. Homogenates from HSV-1-infected adult zebrafish brains were shown to infect cultured human neuroblastoma cells; however, viral titer was not quantified or examined over time (Burgos et al., 2008). The dilution of virus-containing sample that infects 50% of tissue culture samples (TCID50) provides a measure of viral titer, which can be used to both report the concentration of virus used for infection and to assess the viral load generated during the course of infection. Homogenates from CHIKV-infected larvae were added to cultured Vero cells to quantify viral titer. It was determined that injection of approximately 102 TCID50 CHIKV into the circulating blood of 3 day old zebrafish larvae resulted in disease symptoms. TCID50/larva was also determined over the course of infection and the peak production of infectious agents was found to occur between 24 and 48 hpi, which correlated with the CHIKV viral E1 expression profile observed via qPCR (Palha et al., 2013).
When viruses, such as IAV, are propagated in fertilized chicken eggs, viral titer can be quantified by determining the dose that infects 50% of exposed embryos (EID50). Approximately 1×104 EID50 IAV injected into the circulation of 2 dpf zebrafish embryos achieved systemic infection. TCID50 assays were performed by adding homogenized IAV-infected zebrafish samples to cultured MDCK cells. Viral load at 0 hpi revealed 2.55-2.80×102 TCID50/mL IAV in infected zebrafish. The viral titer of either IAV strain in infected zebrafish increased by over 10 fold during the 96 hours it was monitored (K. A. Gabor and C. H. Kim, personal communication). TCID50 assays provide additional demonstration of human viral pathogen replication in zebrafish and show whether increased viral gene expression leads to the production of more functional (i.e. infectious) virions.
2.2.1.3. Visualization of human viral infection in zebrafish
The virological assays and results discussed thus far could be obtained from any animal model of an infectious disease. The zebrafish infectious disease model is an ideal system in which to visualize fluorescent viruses or viral sub-replicons in vivo. Fluorescence can be visualized and imaged effectively in early life stage zebrafish because they are transparent; however, chemical and genetic strategies for reducing pigmentation in older zebrafish are available (e.g. use of zebrafish pigmentation mutant lines, 1-phenyl-2-thiourea treatment). Infection of a transparent host with a fluorescent virus allows for direct observation of viral cell tropism, infection kinetics, and viral burden in living animals. A strain of CHIKV was engineered to express GFP. Infection of zebrafish larvae with CHIKV-GFP revealed a wide tissue tropism, including fluorescence observed in liver, jaw, gills, vascular endothelium, eyes, fins, red blood cells, muscle fibers, brain, spinal cord, swim bladder, and the yolk syncytial layer. The range of zebrafish tissues infected by CHIKV is similar to the wide cellular tropism observed in vitro. CHIKV-GFP fluorescence, corresponding to the emergence of newly infected cells of all cell types, reached a peak at about 14 hpi. This was followed by a reduction in fluorescence in certain cell types at different times, depending on the degree of apoptosis brought about by CHIKV. Liver cells were found to undergo apoptosis upon CHIKV infection with the highest frequency, whereas brain cells were found to be the most refractory. CHIKV-GFP fluorescence was still detectable in the brain of infected zebrafish at 7 dpi. A CHIKV capsid-specific antibody was used to confirm that cells expressing GFP co-localized with CHIKV viral proteins detected via IHC and GFP was determined to be a reliable read out of the presence of virus (Palha et al., 2013). Certain symptoms of CHIKV infection can persist for months to years in humans, leading to the speculation that a viral reservoir exists. These studies, using zebrafish as a model for CHIKV infection, suggest that brain tissue might be a potential viral reservoir. This result, combined with the fact that more than half of CHIKV-infected human neonates develop encephalopathy and infected adults do not, suggests that early life stage human and zebrafish brain tissue can be infected by CHIKV and adult brain tissue cannot. Alternatively, the brain of adult humans and zebrafish larvae may harbor a latent reservoir of CHIKV, whereas CHIKV in human neonate brain tissue causes an immune response. These possibilities can be further studied in the zebrafish CHIKV infection model by attempting to infect adult zebrafish brain tissue with CHIKV or by manipulating host or viral factors to try to cause encephalopathy in early life stage zebrafish.
A strain of human IAV that results in translation of GFP in infected cells has been generated (NS1-GFP, (Manicassamy et al., 2010)). This virus was developed to visualize IAV infection in mouse lungs in vivo but, due to background autofluorescence, only ex vivo imaging was possible. In mice, NS1-GFP virus was found to be significantly attenuated compared to wild-type IAV (approximately 100-fold less infectious); yet infection with a high titer of NS1-GFP was still lethal (Manicassamy et al., 2010). Zebrafish were infected with this fluorescent reporter strain of IAV to observe in vivo infection kinetics and tissue tropism. In NS1-GFP-infected zebrafish larvae, fluorescence was first observed at 6 hpi and then declined after reaching a peak at approximately 24 hpi (unpublished observation). Fluorescence was observed in the yolk syncytial layer, heart, skeletal muscle, blood vessels, and swimbladder (K. A. Gabor and C. H. Kim, personal communication). The IAV infected tissues observed in zebrafish paralleled the epithelial and endothelial cellular tropism of IAV observed in vitro.
The HCV sub-replicon constructs injected into zebrafish were engineered in such a way that translation of the virally encoded RNA-dependent RNA polymerase generated red fluorescence and translation of the viral core gene product generated green fluorescence. These constructs were injected separately and in combination. Red fluorescence (viral RNA-dependent RNA polymerase) was observed when this construct was injected alone; however, given the requirement for the viral RNA-dependent RNA polymerase in the production of HCV viral gene products, green fluorescence was only observed with red fluorescence if the constructs were co-injected. Green and red fluorescence were observed at 8 and 12 dpi in the zebrafish liver (Ding et al., 2011), suggesting that these sub-replicon constructs show a similar tissue tropism to HCV in humans. These reports illustrate some of the virological parameters that can be observed or quantified through visualization of a fluorescent human viral pathogen in an animal host. In addition to infecting zebrafish with fluorescent human viral pathogens and visualizing in vivo infection kinetics and tissue tropism, fluorescence could also be used to compare the relative viral burden after experimental manipulations.
2.2.2. Host response
There are always two opposing forces during viral infections: viral exploitation of the host and the host’s response. Studying viral infections in animal models allows observation and manipulation of the dynamic interplay between virus and host, as both pursue their mostly conflicting agendas. Both the virus and the host’s immune response can contribute to the symptoms and the severity of viral illnesses. While the immune response protects the host by recognizing pathogens, limiting their spread, and ultimately killing them, the mechanisms employed by the immune system can result in host tissue damage. Understanding the beneficial and the deleterious effects of the immune response in each viral illness will inform new ways to improve treatment of viral diseases and to mitigate unwanted effects. We have previously described assays to measure the success of viruses in terms of replication and spread and now we will discuss assays to examine the success of the hosts. We will summarize what has been done in this regard in zebrafish infected with human viral pathogens and focus on the opportunities unique to the zebrafish infection model to learn about host immunology.
2.2.2.1. Mortality, gross pathology, and histology
One measure of defense against a viral pathogen is survival of the host. Mortality was monitored in zebrafish infected with HSV-1, CHIKV, or IAV. In adult zebrafish infected via intraperitoneal injection with 106 pfu HSV-1, 4.3% of the zebrafish died within 4 dpi whereas none of the controls died during this same period (Burgos et al., 2008). Less than 10% of the zebrafish larvae infected via tail vein injection with 102 TCID50 CHIKV died. Greater than 90% of CHIKV-infected zebrafish larvae that survived had recovered by 5 dpi (Palha et al., 2013). Mortality was observed within the first 24 hpi in zebrafish larvae injected in the Duct of Cuvier with 1×104 EID50 IAV. Cumulatively, after 5 dpi, infection with H1N1 or H3N2 strains of IAV resulted in 54% and 59% mortality, respectively; whereas over 90% of the mock-infected zebrafish larvae survived (K. A. Gabor and C. H. Kim, personal communication). These studies show how infection dose, site of infection, and the life stage of the zebrafish at time of infection could be impacting mortality. They also suggest that the relative pathogenicity of human viruses may be maintained in the zebrafish infection model (i.e. zebrafish recover from viral illnesses from which humans are likely to recover and zebrafish die from viral illnesses that are more likely to be lethal to humans).
Other observations that can be made regarding the physical condition of virus-infected hosts include gross pathology and histopathology. Adult HSV-1-infected zebrafish showed changes in pigmentation, swimming, and opercular ventilation. HSV-1 infection caused hemorrhaging under the dermis in muscle, infiltration of lymphoid cells, and degeneration of secondary oocytes (Burgos et al., 2008). There is some controversy as to whether HSV-1 infection impacts human fertility. Studies have found a correlation between HSV-1 infection and sperm abnormalities, such as reduced sperm count (Monavari et al., 2013). Histopathological analyses of HSV-1-infected zebrafish suggest female infertility may be compromised and that zebrafish could be used to further study whether a causal link exists between HSV-1 and infertility. In CHIKV-infected zebrafish larvae, opacification of the yolk was observed along with delayed swim bladder inflation, abnormal heartbeat and blood flow, edema, loss of equilibrium, and impaired swimming (Palha et al., 2013). Zebrafish larvae infected with IAV were lethargic and displayed edema that worsened over time. IAV-infected larvae also had pigmentation and craniofacial abnormalities. Histology revealed pericardial edema and necrosis in the liver, gills, and hematopoietic tissue of the head kidney in IAV-infected zebrafish (K. A. Gabor and C. H. Kim, personal communication). Comparative pathology and histology between humans (or other animal models) and zebrafish infected with human viral pathogens will help to determine the translational validity of the model. Analysis of histopathological changes in the entire body of virus-infected model organisms may also reveal changes in tissues not previously known to be affected in humans during certain viral infections.
2.2.2.2. Visualizing phagocyte behavior in zebrafish
Similar to exploiting the transparency of zebrafish and fluorescent viruses to study viral infection kinetics, cell tropism, and viral burden in vivo, there are opportunities ideally suited to the zebrafish model to generate fluorescent tools to learn about immunology and the host response to viral infection. The ability to visualize, track, and image fluorescently labeled components of the host immune response in the zebrafish infection model renders this system ideal for immunological research. Transgenic zebrafish lines have been generated to express fluorescent proteins under the control of innate immunity cell-specific promoters or as a read out of immune cell signaling pathway activation.
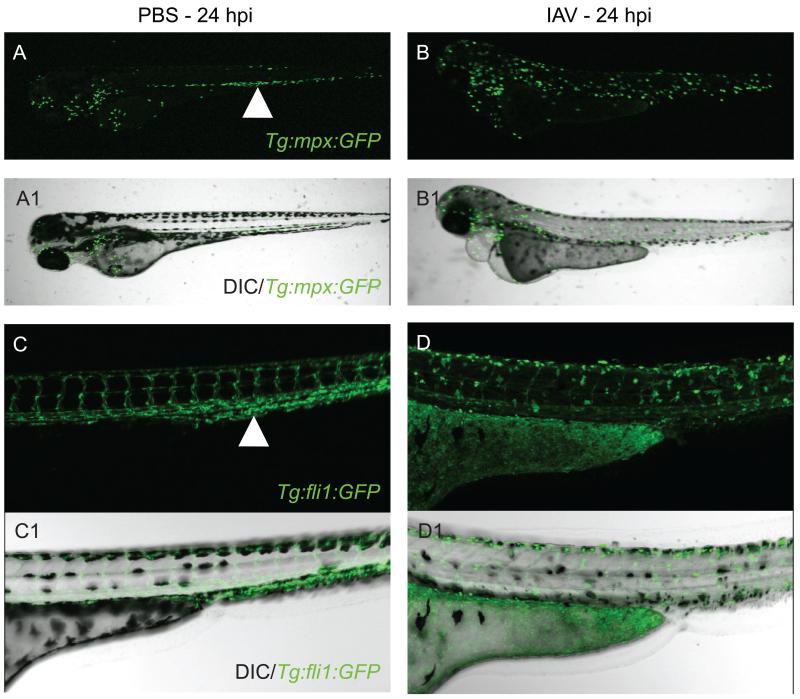
Neutrophils and macrophages involved in the innate immune response are known to cause both beneficial and undesirable effects during the course of human viral infections. Deregulated neutrophil behavior was recently reported to be predictive of host mortality in IAV-infected mice (Brandes et al., 2013). Zebrafish lines with fluorescent neutrophils and/or macrophages have been engineered to allow direct visualization of phagocyte behavior under normal or infection conditions in vivo (Lawson and Weinstein, 2002; Mathias et al., 2006; Renshaw et al., 2006). Using the Tg(mpx:GFP) or Tg(fli1:GFP) zebrafish lines to visualize neutrophils or macrophages, respectively, we observed that the localization of neutrophils and macrophages changed dramatically in IAV-infected zebrafish larvae by 24 hpi/72 hpf (Fig. 1A-D1). In uninfected Tg(mpx:GFP) zebrafish, neutrophils congregated in the caudal hematopoietic tissue (Fig. 1A, white arrowhead). In IAV-infected mpx transgenic zebrafish, neutrophils were dispersed throughout the body by 24 hpi (Fig. 1B). The fli1 promoter is expressed in zebrafish macrophages as well as endothelial cells lining blood vessels. In uninfected fli1 transgenic zebrafish, the vasculature was readily observed and macrophages were difficult to distinguish, but likely residing in the caudal hematopoietic tissue (Fig. 1C, white arrowhead). In IAV-infected Tg(fli1:GFP) zebrafish at 24 hpi, vascular architecture appeared severely disrupted and macrophages (amorphous GFP+ cells in Fig. 1D) were observed throughout the body. In zebrafish, at 24 hpi, it appeared that IAV had a greater impact on neutrophil localization than on total cell population; however, total numbers of neutrophils per larva could be quantified by flow cytometry. It will be important to observe and quantify neutrophils at other time points during IAV infection in zebrafish because total neutrophil numbers were found to reach a peak at 72 hpi in CHIKV-infected zebrafish (Palha et al., 2013).
Figure 1.
Visualizing the phagocyte response to human IAV infection using transgenic zebrafish lines. (A-D1) Side oriented, anterior left, dorsal top, live, 72 hpf transgenic zebrafish larvae. (A-B1) Tg(mpx:GFP) line in which neutrophils are fluorescent. (C-D1) Tg(fli1:GFP) line with fluorescent endothelial cells (vascular endothelial cells and macrophages). (A) Fluorescent micrograph of mock-infected Tg(mpx:GFP) zebrafish at 24 hpi. White arrowhead points to neutrophils localized to the caudal hematopoietic tissue. (A1) Merge of fluorescent and DIC images of mock-infected Tg(mpx:GFP) zebrafish. (B) Fluorescent micrograph of IAV-infected Tg(mpx:GFP) zebrafish at 24 hpi. Note the altered distribution of neutrophils throughout the body in response to IAV infection. (B1) Merge of fluorescent and DIC panels of IAV-infected Tg(mpx:GFP) zebrafish. (C) Fluorescent micrograph of mock-infected Tg(fli1:GFP) zebrafish at 24 hpi. The architecture of the vasculature can be clearly observed. White arrowhead again denotes the caudal hematopoietic region. (C1) Merge of fluorescent and DIC images of mock-infected Tg(fli1:GFP) zebrafish. (D) Fluorescent micrograph of IAV-infected Tg(fli1:GFP) zebrafish at 24 hpi. Note that fluorescence in the vasculature is faint and fluorescent macrophages are easily visualized and have a different distribution in response to IAV infection. (D1) Merge of fluorescent and DIC images of IAV-infected Tg(fli1:GFP) zebrafish.
The increase in total neutrophil number due to an infection (infection-induced granulopoiesis) is known to be regulated by granulocyte colony stimulating factor (GCSF)-dependent and –independent mechanisms (Basu et al., 2000; Panopoulos and Watowich, 2008). In zebrafish, it has been shown that an inducible nitric oxide synthase gene (nos2a) and IFN are required for infection-induced granulopoiesis (Hall et al., 2012; Palha et al., 2013). Determining how the expression and/or activity of these genes differs between IAV- or CHIKV-infected zebrafish could shed light on potential differences observed in infection-induced granulopoiesis in zebrafish models of these viral diseases. Interestingly, while the total number of neutrophils was reported to increase in CHIKV-infected zebrafish, their spatial distribution was found to be unaltered (Palha et al., 2013). This observation is in contrast to what is shown during IAV infection in zebrafish in Figure 1A-B1. Thus, regulation of neutrophil motility during these human viral infections could be studied in zebrafish and may contribute to differing disease symptoms and severity. Transgenic zebrafish with fluorescent phagocytes will be an important tool for determining how phagocytes respond to infection with a range of human viral pathogens and how their differential responses impact viral illnesses.
2.2.2.3. Measuring and visualizing IFN signal transduction in zebrafish
In addition to their use in visualizing and tracking phagocyte behavior in response to viral infection, transgenic zebrafish can also be generated to show spatiotemporal induction of immune cell signaling pathways. A component of innate immune antiviral signaling, IFN, is known to play a major role in controlling CHIKV infection in mammals (Schwartz and Albert, 2010) and viral infection-inducible IFN genes have been identified in zebrafish (Altmann et al., 2003; Aggad et al., 2009; Sieger et al., 2009). For a detailed discussion of IFN signaling in fish and mammals, we refer readers to reviews on the topic such as Langevin et al., 2013. Given the importance of antiviral IFN signaling in controlling mammalian CHIKV infection, an Tg(IFNphi1:mCherry) transgenic zebrafish line was generated to identify the cellular source of IFN during CHIKV infection. At 2 dpi, mCherry fluorescence was observed in hepatocytes and motile leukocytes. The authors compared IFN expression observed in their transgenic line to ISH detection of IFN expression and found the transgenic line to be a faithful, although slightly delayed, reporter of IFN expression. An experiment was conducted in which CHIKV-GFP virus was injected into Tg(IFNphi1:mCherry) zebrafish, and it was found that IFN was produced in non-CHIKV-infected cells (Palha et al., 2013). Along with visualizing IFN signal transduction in zebrafish infected with human viruses, the expression of genes involved in IFN signal transduction can be quantified by PCR. The expression of IFN phi 1 and a downstream target (viperin) were induced nearly 100-fold at 24 hpi in CHIKV-infected zebrafish larvae (Palha et al., 2013). In IAV-infected zebrafish larvae, IFN phi 1 expression and the expression of a downstream target (MxA) increased approximately 10- and 5-fold, respectively, and peaked at approximately 48 hpi (K. A. Gabor and C. H. Kim, personal communication). The difference in the degree of IFN phi 1 induction between CHIKV- and IAV-infected zebrafish larvae could reflect a lesser involvement of IFN in controlling IAV infection or the ability of certain IAV viral gene products to inhibit IFN expression and activity (Hale et al., 2008). The latter is thought to be one of the major reasons why IAV infections can cause such serious illness. The zebrafish will likely be a good model for dissecting out the different roles of IFN in human viral illnesses and the application of current and future transgenic zebrafish tools to visualize innate immune signal transduction will enable roles for IFN and other signal transduction components in human viral illnesses to be illuminated.
2.2.2.4. Manipulating IFN signal transduction or phagocytes with genetic or chemical treatments in zebrafish
Along with transgenesis, the zebrafish model is amenable to reverse genetic experimental approaches and chemical exposures. Injection of specifically designed morpholino oligonucleotides into 1-4 cell stage zebrafish embryos can block translation or splicing of specific transcripts, which results in temporary knockdown of a protein of interest. Vivo morpholinos contain a moiety that enables them to enter cells after the earliest stages of development and thus can be used for protein knockdown in older life stage zebrafish. Targeted mutagenesis strategies have been shown to be effective in the zebrafish model and are now being improved upon and their use is becoming more feasible in this system (Meng et al., 2008; Huang et al., 2011; Hwang et al., 2013). Morpholinos can cause non-specific side effects and must be validated before being used for experimentation. Even so, morpholinos can and have been used in the context of zebrafish infection studies to elucidate roles of certain gene products and cell types during the course of infection. The importance of the role of IFN phi 1 in CHIKV-infected zebrafish, suggested by drastically increased expression, was definitively demonstrated in zebrafish upon morpholino-mediated knockdown of MAVS (a signaling component upstream of IFN) or CRFB1 and CRFB2 (IFN receptor subunits, downstream of IFN). These morpholinos blocked either IFN production and downstream target expression or downstream target expression alone, respectively. Both manipulations significantly increased the number of viral genomes/transcripts and the percent mortality in CHIKV-infected zebrafish (Palha et al., 2013).
Since IFN was found to be necessary to control CHIKV infection and neutrophils, macrophages, and hepatocytes were determined to be the main cell types producing IFN in zebrafish, the authors chemically or genetically depleted each cell type individually and assessed the impact on CHIKV-infected zebrafish. The nitro-reductase system has proven to be a powerful tool for conditional, chemical ablation of certain cell types in zebrafish (Pisharath and Parsons, 2009). A transgenic zebrafish strain for macrophage-specific ablation was generated using this strategy (Davison et al., 2007; Palha et al., 2013). When zebrafish with ablated macrophages were infected with CHIKV, disease was slightly worse but mortality did not occur (Palha et al., 2013). Therefore, macrophages are not crucial to host protection against CHIKV infection. This tool could easily be used in the context of other human viral infections to determine whether the contributions of macrophages result in host protection. The development of zebrafish lines in which other immune cell types can be ablated will be extremely useful. Genetic depletion of certain cell types can also be achieved in zebrafish using morpholinos. Tom22 knockdown, which results in hepatocyte depletion, led to the production of more CHIKV viral transcripts but did not increase mortality in zebrafish. Depletion of predominately neutrophils with Csf3r morpholinos or blocking the CHIKV infection-induced increase in neutrophil numbers in zebrafish with Nos2a morpholinos resulted in more severe disease phenotypes, increased viral transcripts, and high mortality (Palha et al., 2013). CHIKV infection has previously been shown to induce neutrophilia in humans (Chow et al., 2010) but this zebrafish infection study was the first to report that increased numbers of neutrophils protect the host against CHIKV infection and mortality, possibly through increased IFN production. The association between increased IFN and neutrophilia in CHIKV infection is interesting because IFN expression can be associated with neutropenia (Navarini et al., 2006; Rønneseth et al., 2006). Increased numbers of neutrophils have been hypothesized to be involved in the extreme joint pain associated with CHIKV infection in humans (Chow et al., 2010), as the presence of neutrophils also seems to be correlated with joint pain and damage in rheumatoid arthritis (Pillinger and Abramson, 1995). These results together suggest that increased neutrophils protect the host during CHIKV infection, likely through increasing IFN production, but that joint pain may be an undesired consequence of CHIKV-induced neutrophilia. Therefore, future studies aimed at maximizing the benefits and minimizing the side effects of neutrophilia could improve treatments for CHIKV infection and other diseases. The relative ease with which neutrophils or macrophages or other cell types can be chemically or genetically depleted in zebrafish will make it possible to discern positive from negative roles for individual cell types in the immune response.
2.2.2.5. Using zebrafish models of human viral infections to screen for novel antiviral compounds
In addition to using chemical treatments to ablate cell types, chemical treatments can be used to activate or inhibit signaling pathways or can be tested for antiviral activity. IAV infections can be treated with certain antiviral compounds; however, antiviral-resistant IAV strains are rapidly emerging and necessitate the discovery of new antiviral therapies. Zebrafish are an ideal model system in which to conduct large-scale chemical screens to test the efficacy and safety of water-soluble compounds. Amantadine HCl is used to ameliorate H1N1, H2N2, and H3N2 infections but has little or no activity against influenza B virus isolates. As a proof of concept that the zebrafish model of human IAV infection can be used to screen for chemicals with anti-IAV activity, IAV-infected zebrafish were immersed in Amantadine HCl at 24 hpi. Amantadine HCl treatment of IAV-infected zebrafish significantly reduced viral burden and mortality (K. A. Gabor and C. H. Kim, personal communication). A similar experiment was performed in HCV sub-replicon injected zebrafish. Two anti-HCV medications, ribavirin and oxymatrine, were shown to reduce HCV core RNA and protein levels in zebrafish injected with the HCV sub-replicon (Ding et al., 2011). Immersion can be an effective method for exposing early life stage zebrafish to a range of chemical agents, but these agents may have to be injected into adult zebrafish. Adult zebrafish were co-injected with HSV-1 and either an antiviral (acyclovir) or an immunosuppressant (cyclophosphamide) compound. Co-injection with the antiviral compound significantly reduced the number of viral genomes in all of the parts of the body that were analyzed, but did not reduce mortality. HSV-1 infection in combination with immunosuppression resulted in a dose-dependent increase in mortality and an increase in viral genomes in the ventral trunk region of the body (Burgos et al., 2008). The assays done thus far using zebrafish to test the efficacy of chemicals against human viral pathogens have been largely proof-of-concept experiments. The success of these experiments has laid the foundation for the exciting possibility of using fluorescent virus-infected zebrafish to conduct automated, large-scale chemical screens to rapidly detect novel compounds with antiviral activity.
3. Predictions and considerations regarding zebrafish infection with human viral pathogens
We have discussed how zebrafish models of human viral infections complement other animal models by providing the unique opportunity to visualize virus-host interactions in a small, simple host with a relatively well-conserved immune system. However, given the millions of years of evolution separating zebrafish and humans and the aspect of human-virus co-evolution that cannot be recapitulated in this model system, zebrafish will not be able to host or provide these insights for all human viral infections. In an effort to predict which human viral pathogens can infect zebrafish, we stress the necessity for considering viral incubation temperature, breadth of host range, and the expression of zebrafish orthologs of known viral receptors. To help determine whether zebrafish may be a good infection model for a certain human viral pathogens, a flow chart of essential questions to ask about the human viral pathogen is provided in Figure 2. Accounting for the constraints imposed by temperature, host range, and conservation of receptors, we believe zebrafish are most likely to be able to host certain viruses that infect the human upper respiratory tract, examples of which are listed in Table 2.
Figure 2.
A flow chart of questions to ask when predicting whether a human viral pathogen can infect zebrafish. Human viral pathogens of the upper respiratory tract with broad host ranges and receptors that are conserved and highly expressed in zebrafish are good candidates to test in the zebrafish infectious disease model.
Table 2.
Selected viruses found in the upper respiratory tract of humans and predictions whether they can infect zebrafish.
| Human Virus | Viral Receptor/ Binding Moiety |
Ensembl Prediction | Zebrafish Expression Data |
|---|---|---|---|
| Human influenza B virus |
N-linked α2,6 sialic acid |
N/A | Present at 48 hpf (K. A. Gabor and C. H. Kim, personal communication) |
| Reovirus serotype 3 | N-linked α2,6 sialic acid |
N/A | Present at 48 hpf (K. A. Gabor and C. H. Kim, personal communication) |
| JC virus | N-linked α2,6 sialic acid |
N/A | Present at 48 hpf (K. A. Gabor and C. H. Kim, personal communication) |
| Human parainfluenza virus 3 |
N-linked α2,6 sialic acid |
N/A | Present at 48 hpf ( K. A. Gabor and C. H. Kim, personal communication) |
| SARS, Human coronoavirus NL63 |
ACE2 | One-to-one orthologous relationship with zebrafish ace2 gene |
Uncharacterized |
| Coxsackieviruses and adenoviruses |
CXADR | One-to-one orthologous relationship with zebrafish cxadr gene |
Embryonic central nervous system (Thisse et al., 2001) |
| Respiratory syncytial virus |
NCL | One-to-one orthologous relationship with zebrafish ncl gene |
Identified as zebrafish embryonic-essential gene (Amsterdam et al., 2004). Mutant phenotype in central nervous system, head, eye, heart, liver, and gut. |
| Poliovirus | PVR/CD155 | Many-to-many orthologous relationship with zebrafish pvr genes |
Isoforms expressed in embryonic/larval central nervous system and eye (Helvik et al., 2009; Thisse et al., 2008). |
| Echovirus | CD55 | Many-to-many orthologous relationship with zebrafish cfh gene |
Unspecified embryonic expression (Thisse et al., 2004). Expressed in adult eye, fin, gill, heart, intestine, liver, and muscle (Sun et al., 2010). |
| Rhinoviruses | ICAM-1 | No zebrafish ortholog | N/A |
| Epstein Barr virus | CR2 | No zebrafish ortholog | N/A |
| Human coronoaviruses HKU1 |
HLA-C | No zebrafish ortholog | N/A |
| Human coronoaviruses 229E |
ANPEP | One-to-many orthologous relationship with zebrafish anpep genes and others |
Uncharacterized |
3.1. Incubation temperature
Viruses are typically able to replicate only in defined temperature ranges. This is a much more limiting factor for some viruses than others. An issue that arises when attempting to use zebrafish to host human viral pathogen infections is that human body temperature is warmer than the ideal temperature for rearing zebrafish. The core body temperature of humans is approximately 37 °C. Therefore, human isolates of viral pathogens are likely to be adapted to thrive at this temperature. The optimal temperature for rearing zebrafish is 28.5 °C. However, zebrafish are poikilotherms so their body temperature will vary with the temperature of their surroundings. Zebrafish embryos/larvae can be reared between 25-33 °C without temperature-induced abnormalities (Kimmel et al., 1995) and adult zebrafish can be slowly acclimated to an even wider temperature range. While the core body temperature of humans is 37 °C, the respiratory tract, which is exposed to the outside environment, can be closer to 33 °C (Massin et al., 2001). Thus, there is overlap between the temperature of the human respiratory tract and the temperature range over which zebrafish can be raised. Propagation of human IAV was shown to be more successful at 33 °C than 37 °C (Stern and Tippett, 1963). Human viruses, such as IAV, that can replicate at the cooler end of the human body temperature spectrum are good candidates for zebrafish infection.
3.2. Host range
The species that viral pathogens are able to infect depends on a variety of factors; including temperature, which must be in a range that is permissible for viral replication. The broader the host range of a viral pathogen, the more likely it may be able to infect zebrafish. CHIKV has a broad host range, spanning insects to humans. IAV has a narrower host range than CHIKV, infecting birds and mammals. HSV-1 infections are limited to humans; however, other herpesviruses can infect mollusks, fish, reptiles, birds, and mammals. HCV specifically infects humans and chimpanzees. Of the four human viruses attempted thus far, HCV was the only virus that required generation of sub-replicon constructs for viral gene expression in zebrafish. This alternative approach may be effective when zebrafish cannot be infected with certain human pathogens. The generation and injection of fluorescent sub-replicon constructs still allows the advantages of the zebrafish infection model to be exploited. Analysis of the HCV sub-replicons in zebrafish revealed viral tissue tropism and viral burden in response to chemical treatments. Viral sub-replicon constructs and the zebrafish infection model have the potential to provide valuable insights into infectious diseases even when the human viral pathogen is unable to replicate in zebrafish.
3.3. Viral receptors
An important attribute to consider before attempting to infect zebrafish with human viruses is whether the human viral pathogen’s receptor is conserved and expressed in zebrafish. Viruses are obligate intracellular parasites that require entry into host cells and access to host cell machinery for replication and spread of infection. The necessary viral receptors must be present on zebrafish cells in order for human viral pathogens to be able to infect zebrafish. For some human viral pathogens, the receptors may be unknown, their identity debated, or they may be only partially characterized. A cellular receptor for HSV-1 is a modified heparin sulfate proteoglycan. The zebrafish enzyme responsible for this specific modification is 3-O-sulfotransferase-3. Expression of this enzyme was shown to be necessary for HSV-1 infection of zebrafish fibroblast cells and sufficient for HSV-1 entry into CHO-K1 cells (Hubbard et al., 2013). This enzyme is known to be expressed in the CNS and other tissues in embryonic zebrafish (B. Thisse and C. Thisse, 2004), which supports the observation that HSV-1 can infect zebrafish embryos in addition to adult zebrafish (Burgos et al., 2008). Host proteins mediating CHIKV binding were unknown until Prohibitin (PHB) was recently identified (Wintachai et al., 2012). Zebrafish have an ortholog of the human PHB gene and a possible ortholog of the human PHB2 gene. Both of these genes are highly expressed and ubiquitous in zebrafish embryos/larvae (B. Thisse and C. Thisse, 2004). The zebrafish expression data support the idea that PHB is involved in CHIKV infection of zebrafish larvae, a hypothesis that can easily be tested in the zebrafish infection model with PHB morpholinos. Human isolates of IAV and several other human viral pathogens bind to host proteins with terminal alpha2,6-linked sialic acids. These specific sialic acid linkages were identified in 48 hpf zebrafish embryos (K. A. Gabor and C. H. Kim, personal communication), suggesting that the subset of human viral pathogens that bind this moiety may also be able to infect 48 hpf zebrafish embryos. Many human viruses bind host proteins with alpha2,3 sialic acid linkages. Sialic acids linked in the alpha2,3 conformation were not detected in 48 hpf zebrafish embryos (K. A. Gabor and C. H. Kim, personal communication). However, 48 hpf zebrafish express sialyltransferase enzymes to attach sialic acids in the alpha2,3 confirmation, so it may still be possible to infect early life stage zebrafish with human viral pathogens that utilize alpha2,3 linked sialic acid receptors. Knowing the spatiotemporal expression patterns of viral receptors will inform the life stage at which zebrafish can most likely be infected as well as the viral tissue tropism expected. If the receptor for a human viral pathogen is not conserved or expressed in zebrafish, this obstacle could potentially be circumvented with the use of the viral sub-replicon strategy.
4. Conclusions and future perspectives
We believe that the zebrafish infectious disease model is at an exciting time in its history. The idea of exploiting certain zebrafish characteristics to learn about human pathogen infections is gaining widespread acceptance, the toolkit for the genetic manipulation of zebrafish is largely in place and expanding, and proof-of-concept experiments have already provided results indicating that zebrafish infectious disease models are likely to have important translational impacts. The zebrafish system as a host for human pathogens is poised to quickly make meaningful contributions to the better understanding of and improved therapies for human infectious diseases.
Highlights.
There are unique opportunities to learn about virus-host interactions in zebrafish.
Innate immunity can be both beneficial and detrimental in human viral illnesses.
Temperature, host range, and receptors are human virus characteristics to consider.
Acknowledgements
The authors would like to thank past and present members of the Kim laboratory and Drs. Remi Gratacap and Paul Millard for helpful discussions and ideas.
Funding
This work was supported by the National Institutes of Health grant RO1GM087308 and the Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health grant P20GM12345.
Abbreviations
- HSV-1
Herpes simplex virus type 1
- CHIKV
Chikungunya virus
- IAV
Influenza A virus
- HCV
Hepatitis C virus
- (q)PCR
(quantitative) polymerase chain reaction
- ISH
in situ hybridization
- IHC
immunohistochemistry
- CNS
central nervous system
- TCID50
50% tissue culture infectious dose
- EID50
50% embryo infectious dose
- MDCK
Madin-Darby canine kidney
- IFN
interferon
- Hpi/dpi
hours post-infection/days post-infection
- Hpf/dpf
hours post-fertilization/days post-fertilization
- MAVS
mitochondrial antiviral signaling protein
- CRFB1/CRFB2
cytokine receptor family member b1/b2
- PRR
pattern recognition receptor
- PHB
prohibitin
- PFU
plaque forming units
- NS1-GFP
strain of human IAV engineered to express GFP fused to the non-structural NS1 gene product
- mpx
myeloid-specific peroxidase
- mpeg1
macrophage expressed 1
- fli1
friend leukemia integration 1
- mxa
myxovirus (influenza) resistance a
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errorsmaybe discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aggad D, Mazel M, Boudinot P, Mogensen KE, Hamming OJ, Hartmann R, Kotenko S, Herbomel P, Lutfalla G, Levraud JP. The Two Groups of Zebrafish Virus-Induced Interferons Signal via Distinct Receptors with Specific and Shared Chains. J Immunol. 2009;183:3924–3931. doi: 10.4049/jimmunol.0901495. [DOI] [PubMed] [Google Scholar]
- Altmann SM, Mellon MT, Distel DL, Kim CH. Molecular and functional analysis of an interferon gene from the zebrafish, Danio rerio. J Virol. 2003;77:1992–2002. doi: 10.1128/JVI.77.3.1992-2002.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoine TE, Jones KS, Dale RM, Shukla D, Tiwari V. Zebrafish: Modeling for Herpes Simplex Virus Infections. Zebrafish. 2013;11:17–25. doi: 10.1089/zeb.2013.0920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arduino PG, Porter SR. Oral and perioral herpes simplex virus type 1 (HSV-1) infection: review of its management. Oral Dis. 2006;12:254–270. doi: 10.1111/j.1601-0825.2006.01202.x. [DOI] [PubMed] [Google Scholar]
- Baker DG. Natural Pathogens of Laboratory Mice, Rats, and Rabbits and Their Effects on Research. Clinical Microbiology Reviews. 1998;11:231–266. doi: 10.1128/cmr.11.2.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balla KM, Lugo-Villarino G, Spitsbergen JM, Stachura DL, Hu Y, Banuelos K, Romo-Fewell O, Aroian R, Traver D. Eosinophils in the zebrafish: prospective isolation, characterization, and eosinophilia induction by helminth determinants. Blood. 2010;116:3944–3954. doi: 10.1182/blood-2010-03-267419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu S, Hodgson G, Zhang H-H, Katz M, Quilici C, Dunn AR. “Emergency” granulopoiesis in G-CSF-deficient mice in response to Candida albicans infection. Blood. 2000;95:3725–3733. [PubMed] [Google Scholar]
- Bennett CM, Kanki JP, Rhodes J, Liu TX, Paw BH, Kieran MW, Langenau DM, Delahaye-Brown A, Zon LI, Fleming MD, Look AT. Myelopoiesis in the zebrafish, Danio rerio. Blood. 2001;98:643–651. doi: 10.1182/blood.v98.3.643. [DOI] [PubMed] [Google Scholar]
- Brandes M, Klauschen F, Kuchen S, Germain RN. A systems analysis identifies a feedforward inflammatory circuit leading to lethal influenza infection. Cell. 2013;154:197–212. doi: 10.1016/j.cell.2013.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brothers K, Newman Z. Live imaging of disseminated candidiasis in zebrafish reveals role of phagocyte oxidase in limiting filamentous growth. Eukaryotic Cell. 2011;10:932–944. doi: 10.1128/EC.05005-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brothers KM, Gratacap RL, Barker SE, Newman ZR, Norum A, Wheeler RT. NADPH Oxidase-Driven Phagocyte Recruitment Controls Candida albicans Filamentous Growth and Prevents Mortality. PLoS Pathog. 2013;9:e1003634. doi: 10.1371/journal.ppat.1003634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgos JS, Ripoll-Gomez J, Alfaro JM, Sastre I, Valdivieso F. Zebrafish as a New Model for Herpes Simplex Virus Type 1 Infection. Zebrafish. 2008;5:323–333. doi: 10.1089/zeb.2008.0552. [DOI] [PubMed] [Google Scholar]
- Burt FJ, Rolph MS, Rulli NE, Mahalingam S, Heise MT. Chikungunya: a re-emerging virus. The Lancet. 2012;379:662–671. doi: 10.1016/S0140-6736(11)60281-X. [DOI] [PubMed] [Google Scholar]
- Chao CC, Hsu PC, Jen CF, Chen IH, Wang CH, Chan HC, Tsai PW, Tung KC, Wang CH, Lan CY, Chuang YJ. Zebrafish as a Model Host for Candida albicans Infection. Infect Immun. 2010;78:2512–2521. doi: 10.1128/IAI.01293-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YY, Chao C-C, Liu F-C, Hsu P-C, Chen H-F, Peng S-C, Chuang Y-J, Lan C-Y, Hsieh W-P, Wong DSH. Dynamic Transcript Profiling of Candida albicans Infection in Zebrafish: A Pathogen-Host Interaction Study. PLoS ONE. 2013;8:e72483. doi: 10.1371/journal.pone.0072483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow A, Her Z, Ong EKS, Chen JM, Dimatatac F, Kwek DJC, Barkham T, Yang H, Renia L, Leo YS, Ng LFP. Persistent Arthralgia Induced by Chikungunya Virus Infection is Associated with Interleukin-6 and Granulocyte Macrophage Colony-Stimulating Factor. Journal of Infectious Diseases. 2010;203:149–157. doi: 10.1093/infdis/jiq042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corey L, Wald A. Maternal and neonatal herpes simplex virus infections. N Engl J Med. 2009;361:1376–1385. doi: 10.1056/NEJMra0807633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crim MJ, Riley LK. Viral Diseases in Zebrafish: What Is Known and Unknown. ILAR Journal. 2012;53:135–143. doi: 10.1093/ilar.53.2.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui C, Benard EL, Kanwal Z, Stockhammer OW, van der Vaart M, Zakrzewska A, Spaink HP, Meijer AH. Infectious Disease Modeling and Innate Immune Function in Zebrafish Embryos. Methods in Cell Biology. 2011;105:273–308. doi: 10.1016/B978-0-12-381320-6.00012-6. [DOI] [PubMed] [Google Scholar]
- Davison JM, Akitake CM, Goll MG, Rhee JM, Gosse N, Baier H, Halpern ME, Leach SD, Parsons MJ. Transactivation from Gal4-VP16 transgenic insertions for tissue-specific cell labeling and ablation in zebrafish. Dev Biol. 2007;304:811–824. doi: 10.1016/j.ydbio.2007.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da’as S, Teh EM, Dobson JT, Nasrallah GK, McBride ER, Wang H, Neuberg DS, Marshall JS, Lin T-J, Berman JN. Zebrafish mast cells possess an FcεRI-like receptor and participate in innate and adaptive immune responses. Dev Comp Immunol. 2011;35:125–134. doi: 10.1016/j.dci.2010.09.001. [DOI] [PubMed] [Google Scholar]
- Ding C-B, Zhang J-P, Zhao Y, Peng Z-G, Song D-Q, Jiang J-D. Zebrafish as a Potential Model Organism for Drug Test Against Hepatitis C Virus. PLoS ONE. 2011;6:e22921. doi: 10.1371/journal.pone.0022921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson JT, Seibert J, Teh EM, Da’as S, Fraser RB, Paw BH, Lin T-J, Berman JN. Carboxypeptidase A5 identifies a novel mast cell lineage in the zebrafish providing new insight into mast cell fate determination. Blood. 2008;112:2969–2972. doi: 10.1182/blood-2008-03-145011. [DOI] [PubMed] [Google Scholar]
- Drutskaya MS, Belousov PV, Nedospasov SA. Innate mechanisms of viral recognition. Mol Biol. 2011;45:5–15. [Google Scholar]
- Dupuis-Maguiraga L, Noret M, Brun S. Chikungunya disease: infection-associated markers from the acute to the chronic phase of arbovirus-induced arthralgia. PLoS Neglected Tropical Diseases. 2012;6:e1446. doi: 10.1371/journal.pntd.0001446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edlin BR. Perspective: test and treat this silent killer. Nature. 2011;474:S18–9. doi: 10.1038/474S18a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratacap RL, Rawls JF, Wheeler RT. Mucosal candidiasis elicits NF-kappaB activation, proinflammatory gene expression and localized neutrophilia in zebrafish. Dis Model Mech. 2013;6:1260–1270. doi: 10.1242/dmm.012039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale BG, Randall RE, Ortin J, Jackson D. The multifunctional NS1 protein of influenza A viruses. Journal of General Virology. 2008;89:2359–2376. doi: 10.1099/vir.0.2008/004606-0. [DOI] [PubMed] [Google Scholar]
- Hall CJ, Flores MV, Oehlers SH, Sanderson LE, Lam EY, Crosier KE, Crosier PS. Infection-responsive expansion of the hematopoietic stem and progenitor cell compartment in zebrafish is dependent upon inducible nitric oxide. Cell Stem Cell. 2012;10:198–209. doi: 10.1016/j.stem.2012.01.007. [DOI] [PubMed] [Google Scholar]
- Hamborsky J, McIntyre L, Atkinson CWW. Epidemiology and Prevention of Vaccine-Preventable Diseases. 10 ed. CDC, Department of Health and Human Services; 2007. [Google Scholar]
- Horner SM, Gale M. Regulation of hepatic innate immunity by hepatitis C virus. Nature Medicine. 2013;19:879–888. doi: 10.1038/nm.3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P, Xiao A, Zhou M, Zhu Z, Lin S, Zhang B. Heritable gene targeting in zebrafish using customized TALENs. Nature biotechnology. 2011;29:699–700. doi: 10.1038/nbt.1939. [DOI] [PubMed] [Google Scholar]
- Hubbard S, Darmani NA, Thrush GR, Dey D, Burnham L, Thompson JM, Jones K, Tiwari V. Zebrafish-Encoded 3-O-Sulfotransferase-3 Isoform Mediates Herpes Simplex Virus Type 1 Entry and Spread. Zebrafish. 2013;7:181–187. doi: 10.1089/zeb.2009.0621. [DOI] [PubMed] [Google Scholar]
- Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nature. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakovlic I, Zhang YB, Wu QJ, Gui JF. Key Players in Innate Immune Recognition of Virus Infection in Mammals and Fish. Ribarstvo. 2006;64:65–74. [Google Scholar]
- Kawai T, Akira S. Innate immune recognition of viral infection. Nat Immunol. 2006;7:131–137. doi: 10.1038/ni1303. [DOI] [PubMed] [Google Scholar]
- Keebaugh ES, Schlenke TA. Insights from natural host-parasite interactions: The Drosophila model. Dev Comp Immunol. 2014;42:111–123. doi: 10.1016/j.dci.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimberlin DW. Herpes simplex virus infections in neonates and early childhood. Semin Pediatr Infect Dis. 2005;16:271–281. doi: 10.1053/j.spid.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Koelle DM, Corey L. Herpes simplex: insights on pathogenesis and possible vaccines. Annu Rev Med. 2008;59:381–395. doi: 10.1146/annurev.med.59.061606.095540. [DOI] [PubMed] [Google Scholar]
- Kuo Z-Y, Chuang Y-J, Chao C-C, Liu F-C, Lan C-Y, Chen B-S. Identification of Infection- and Defense-Related Genes via a Dynamic Host-Pathogen Interaction Network Using a Candida Albicans-Zebrafish Infection Model. J Innate Immun. 2013;5:137–152. doi: 10.1159/000347104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langevin C, Aleksejeva E, Passoni G, Palha N, Levraud J-P, Boudinot P. The Antiviral Innate Immune Response in Fish: Evolution and Conservation of the IFN System. Journal of molecular biology. 2013;425:4904–4920. doi: 10.1016/j.jmb.2013.09.033. [DOI] [PubMed] [Google Scholar]
- Lawson ND, Weinstein BM. In Vivo Imaging of Embryonic Vascular Development Using Transgenic Zebrafish. Dev Biol. 2002;248:307–318. doi: 10.1006/dbio.2002.0711. [DOI] [PubMed] [Google Scholar]
- Lugo-Villarino G, Balla KM, Stachura DL, Bañuelos K, Werneck MBF, Traver D. Identification of dendritic antigen-presenting cells in the zebrafish. PNAS. 2010;107:15850–15855. doi: 10.1073/pnas.1000494107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackie PL. The classification of viruses infecting the respiratory tract. Paediatr Respir Rev. 2003;4:84–90. doi: 10.1016/S1526-0542(03)00031-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maheshwari A, Ray S, Thuluvath PJ. Acute hepatitis C. The Lancet. 2008;372:321–332. doi: 10.1016/S0140-6736(08)61116-2. [DOI] [PubMed] [Google Scholar]
- Manicassamy B, Manicassamy S, Belicha-Villanueva A, Pisanelli G, Pulendran B, Garcia-Sastre A. Analysis of in vivo dynamics of influenza virus infection in mice using a GFP reporter virus. PNAS. 2010;107:11531–11536. doi: 10.1073/pnas.0914994107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massin P, van der Werf S, Naffakh N. Residue 627 of PB2 Is a Determinant of Cold Sensitivity in RNA Replication of Avian Influenza Viruses. J. Virol. 2001;75:5398–5404. doi: 10.1128/JVI.75.11.5398-5404.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathias JR, Perrin BJ, Liu T-X, Kanki J, Look AT, Huttenlocher A. Resolution of inflammation by retrograde chemotaxis of neutrophils in transgenic zebrafish. Journal of leukocyte biology. 2006;80:1281–1288. doi: 10.1189/jlb.0506346. [DOI] [PubMed] [Google Scholar]
- McCartney SA, Colonna M. Viral sensors: diversity in pathogen recognition. Immunological Reviews. 2009;227:87–94. doi: 10.1111/j.1600-065X.2008.00726.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina RA, García-Sastre A. Influenza A viruses: new research developments. Nat Rev Micro. 2011;9:590–603. doi: 10.1038/nrmicro2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer AH, Spaink HP. Host-Pathogen Interactions Made Transparent with the Zebrafish Model. Current Drug Targets. 2011;12:1000–1017. doi: 10.2174/138945011795677809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Noyes MB, Zhu LJ, Lawson ND, Wolfe SA. Targeted gene inactivation in zebrafish using engineered zinc-finger nucleases. Nature Biotechnology. 2008;26:695–701. doi: 10.1038/nbt1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan-Myhre K, Charette JR, Phennicie RT. Study of host-microbe interactions in zebrafish. Methods in Cell Biology. 2011;105:87–116. doi: 10.1016/B978-0-12-381320-6.00004-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monavari SH, Vaziri MS, Khalili M, Shamsi-Shahrabadi M, Heyvani H, Mollaei H, Fazlalipour M. Asymptomatic seminal infection of herpes simplex virus: impact on male infertility. Journal of Biomedical Research. 2013;27:56–61. doi: 10.7555/JBR.27.20110139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss LD, Monette MM, Jaso-Friedmann L, Leary JH, Dougan ST, Krunkosky T, Evans DL. Identification of phagocytic cells, NK-like cytotoxic cell activity and the production of cellular exudates in the coelomic cavity of adult zebrafish. Developmental & Comparative Immunology. 2009;33:1077–1087. doi: 10.1016/j.dci.2009.05.009. [DOI] [PubMed] [Google Scholar]
- Navarini AA, Recher M, Lang KS. Increased susceptibility to bacterial superinfection as a consequence of innate antiviral responses. PNAS. 2006;103(42):15535–15539. doi: 10.1073/pnas.0607325103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicoll MP, Proença JT, Efstathiou S. The molecular basis of herpes simplex virus latency. FEMS Microbiol Rev. 2012;36:684–705. doi: 10.1111/j.1574-6976.2011.00320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norkin LC. Virology: Molecular Biology and Pathogenesis. first ed. ASM Press; Washington, DC: 2010. [Google Scholar]
- Novoa B, Figueras A. Zebrafish: Model for the Study of Inflammation and the Innate Immune Response to Infectious Diseases. Adv Exp Med Biol. 2012;946:253–275. doi: 10.1007/978-1-4614-0106-3_15. [DOI] [PubMed] [Google Scholar]
- Palese P, Wang TT. Why do influenza virus subtypes die out? A hypothesis. MBio. 2011;2:e00150–11. doi: 10.1128/mBio.00150-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palha N, Guivel-Benhassine F, Briolat V, Lutfalla G, Sourisseau M, Ellett F, Wang C-H, Lieschke GJ, Herbomel P, Schwartz O, Levraud J-P. Real-Time Whole-Body Visualization of Chikungunya Virus Infection and Host Interferon Response in Zebrafish. PLoS Pathog. 2013;9:e1003619. doi: 10.1371/journal.ppat.1003619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panopoulos AD, Watowich SS. Granulocyte colony-stimulating factor: Molecular mechanisms of action during steady state and “emergency” hematopoiesis. Cytokine. 2008;42:277–288. doi: 10.1016/j.cyto.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedraza ST, Betancur JG, Urcuqui-Inchima S. Viral recognition by the innate immune system: the role of pattern recognition receptors. Colombia Medica. 2010;41:377–387. [Google Scholar]
- Phelps HA, Neely MN. Evolution of the Zebrafish Model: From Development to Immunity and Infectious Disease. Zebrafish. 2005;2:87–103. doi: 10.1089/zeb.2005.2.87. [DOI] [PubMed] [Google Scholar]
- Pichlmair A, Reis e Sousa C. Innate Recognition of Viruses. Immunity. 2007;27:370–383. doi: 10.1016/j.immuni.2007.08.012. [DOI] [PubMed] [Google Scholar]
- Pillinger MH, Abramson SB. The neutrophil in rheumatoid arthritis. Rheum Dis Clin North Am. 1995;21:691–714. [PubMed] [Google Scholar]
- Pisharath H, Parsons MJ. Nitroreductase-Mediated Cell Ablation in Transgenic Zebrafish Embryos. Methods in Molecular Biology. 2009;546:133–143. doi: 10.1007/978-1-60327-977-2_9. [DOI] [PubMed] [Google Scholar]
- Rehermann B. Pathogenesis of chronic viral hepatitis: differential roles of T cells and NK cells. Nature Medicine. 2013;19:859–868. doi: 10.1038/nm.3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renshaw SA, Loynes CA, Trushell DMI, Elworthy S, Ingham PW, Whyte MKB. A transgenic zebrafish model of neutrophilic inflammation. Blood. 2006;108:3976–3978. doi: 10.1182/blood-2006-05-024075. [DOI] [PubMed] [Google Scholar]
- Rønneseth A, Pettersen EF, Wergeland HI. Neutrophils and B-cells in blood and head kidney of Atlantic salmon (Salmo salar L.) challenged with infectious pancreatic necrosis virus (IPNV) Fish Shellfish Immunol. 2006;20:610–620. doi: 10.1016/j.fsi.2005.08.004. [DOI] [PubMed] [Google Scholar]
- Saito T, Gale M., Jr Principles of intracellular viral recognition. Current Opinion in Immunology. 2007;19:17–23. doi: 10.1016/j.coi.2006.11.003. [DOI] [PubMed] [Google Scholar]
- Scheel TKH, Rice CM. Understanding the hepatitis C virus life cycle paves the way for highly effective therapies. Nature Medicine. 2013;19:837–849. doi: 10.1038/nm.3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz O, Albert ML. Biology and pathogenesis of chikungunya virus. Nat Rev Micro. 2010;8:491–500. doi: 10.1038/nrmicro2368. [DOI] [PubMed] [Google Scholar]
- Shayakhmetov DM, Di Paolo NC, Mossman KL. Recognition of Virus Infection and Innate Host Responses to Viral Gene Therapy Vectors. Mol Ther. 2009;18:1422–1429. doi: 10.1038/mt.2010.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieger D, Stein C, Neifer D, van der Sar AM, Leptin M. The role of gamma interferon in innate immunity in the zebrafish embryo. Dis Model Mech. 2009;2:571–581. doi: 10.1242/dmm.003509. [DOI] [PubMed] [Google Scholar]
- Simmonds P. The origin of hepatitis C virus. Hepatitis C Virus: From Molecular Virology to Antiviral Therapy. Current Opinion in Microbiology and Immunology. 2013;369:1–15. doi: 10.1007/978-3-642-27340-7_1. [DOI] [PubMed] [Google Scholar]
- Stein C, Caccamo M, Laird G, Leptin M. Conservation and divergence of gene families encoding components of innate immune response systems in zebrafish. Genome Biol. 2007;8:R251. doi: 10.1186/gb-2007-8-11-r251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern H, Tippett KC. Primary isolation of influenza viruses at 33 degrees C. The Lancet. 1963;1:1301–1302. doi: 10.1016/s0140-6736(63)91992-5. [DOI] [PubMed] [Google Scholar]
- Sullivan C, Kim CH. Zebrafish as a model for infectious disease and immune function. Fish Shellfish Immunol. 2008;25:341–350. doi: 10.1016/j.fsi.2008.05.005. [DOI] [PubMed] [Google Scholar]
- Szpara ML, Gatherer D, Ochoa A, Greenbaum B, Dolan A, Bowden RJ, Enquist LW, Legendre M, Davison AJ. Evolution and diversity in human herpes simplex virus genomes. J Virol. 2014;88:1209–1227. doi: 10.1128/JVI.01987-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi O, Akira S. Innate immunity to virus infection. Immunological Reviews. 2009;227:75–86. doi: 10.1111/j.1600-065X.2008.00737.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse B, Heyer V, Lux A, Alunni A, Degrave A, Seiliez I, Kirchner J, Parkhill J-P, Thisse C. Spatial and Temporal Expression of the Zebrafish Genome by Large-Scale In Situ Hybridization Screening. Meth Cell Biol. 2004;77:505–519. doi: 10.1016/s0091-679x(04)77027-2. [DOI] [PubMed] [Google Scholar]
- Thomas DL. Global control of hepatitis C: where challenge meets opportunity. Nature Medicine. 2013;19:850–858. doi: 10.1038/nm.3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson MR, Kaminski JJ, Kurt-Jones EA, Fitzgerald KA. Pattern Recognition Receptors and the Innate Immune Response to Viral Infection. Viruses. 2011;3:920–940. doi: 10.3390/v3060920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traver D, Herbomel P, Patton EE, Murphey RD, Yoder JA, Litman GW, Catic A, Amemiya CT, Zon LI, Trede NS. The zebrafish as a model organism to study development of the immune system. Adv Immunol. 2003;81:253–330. [PubMed] [Google Scholar]
- Trede N, Langenau D, Traver D, Look A. The use of zebrafish to understand immunity. Immunity. 2004;20:367–379. doi: 10.1016/s1074-7613(04)00084-6. [DOI] [PubMed] [Google Scholar]
- Tsetsarkin KA, Chen R, Sherman MB, Weaver SC. Chikungunya virus: evolution and genetic determinants of emergence. Curr Opin Virol. 2011;1:310–317. doi: 10.1016/j.coviro.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Sar AM, Appelmelk BJ, Vandenbroucke-Grauls CMJE, Bitter W. A star with stripes: zebrafish as an infection model. Trends in Microbiology. 2004;12:451–457. doi: 10.1016/j.tim.2004.08.001. [DOI] [PubMed] [Google Scholar]
- van der Vaart M, Spaink HP, Meijer AH. Pathogen Recognition and Activation of the Innate Immune Response in Zebrafish. Advances in Hematology. 2012;2012:1–19. doi: 10.1155/2012/159807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y-C, Lin C, Chuang M-T, Hsieh W-P, Lan C-Y, Chuang Y-J, Chen B-S. Interspecies protein-protein interaction network construction for characterization of host-pathogen interactions: a Candida albicans-zebrafish interaction study. BMC Systems Biology. 2013;7:1–1. doi: 10.1186/1752-0509-7-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wintachai P, Wikan N, Kuadkitkan A, Jaimipuk T, Ubol S, Pulmanausahakul R, Auewarakul P, Kasinrerk W, Weng W-Y, Panyasrivanit M, Paemanee A, Kittisenachai S, Roytrakul S, Smith DR. Identification of prohibitin as a Chikungunya virus receptor protein. J Med Virol. 2012;84:1757–1770. doi: 10.1002/jmv.23403. [DOI] [PubMed] [Google Scholar]
- Yamane D, McGivern DR, Masaki T. Liver Injury and Disease Pathogenesis in Chronic Hepatitis C. Hepatitis C Virus: From Molecular Virology to Antiviral Therapy. Current Opinion in Microbiology and Immunology. 2013;369:263–288. doi: 10.1007/978-3-642-27340-7_11. [DOI] [PubMed] [Google Scholar]