Abstract
BACKGROUND
Carbapenem-resistant Enterobacteriaceae (CRE) are rapidly emerging in hospitals in the United States and are posing a significant threat. To better understand the transmission dynamics and the acquisition of resistant strains, a thorough analysis of epidemiologic and molecular characteristics was performed.
METHODS
CRE isolated at Detroit Medical Center were analyzed from September 2008 to September 2009. blaKPC genes were investigated by polymerase chain reaction (PCR), and repetitive extragenic palindromic PCR (rep-PCR) was used to determine genetic similarity among strains. Epidemiologic and outcomes analyses were performed.
RESULTS
Ninety-two unique patient CRE isolates were recovered. Sixty-eight strains (74%) were Klebsiella pneumoniae, 7 were Klebsiella oxytoca, 15 were Enterobacter species, and 2 were Escherichia coli. Fifteen isolates (16%) were resistant to colistin, 14 (16%) were resistant to tigecycline, and 2 were resistant to all antimicrobials tested. The mean ± standard deviation age of patients was 63 ± 2 years. Sixty patients (68%) were admitted to the hospital from long-term care facilities. Only 70% of patients received effective antimicrobial therapy when infection was suspected, with a mean time to appropriate therapy of 120 ± 23 hours following sample culturing. The mean length of hospitalization after sample culturing was 18.6 ± 2.5 days. Of 57 inpatients, 18 (32%) died in the hospital. Independent predictors for mortality were intensive care unit stay (odds ratio [OR], 15.8; P =.003) and co-colonization with CRE and either Acinetobacter baumannii or Pseudomonas aeruginosa (OR, 17.2; P =.006). Among K. pneumoniae CRE, rep-PCR revealed 2 genetically related strains that comprised 70% and 20% of isolates, respectively.
CONCLUSIONS
In this large U.S. cohort of patients with CRE infection, which reflects the modern continuum of medical care, co-colonization with CRE and A. baumannii or P. aeruginosa was associated with increased mortality. Two predominant clones of K. pneumoniae accounted for the majority of cases of CRE infection.
Carbapenem-resistant Klebsiella pneumoniae (CRKP) was first reported in North Carolina in 2001.1 In 2004, outbreaks of CRKP infection were described in New York and its surrounding areas,2,3 and soon after CRKP spread and became endemic in various other parts of the United States and the world.4–11 In 2007, of the healthcare-associated infections reported to the Centers for Disease Control and Prevention, 8% of Klebsiella isolates were CRKP, compared with fewer than 1% in 2000.12 The incidence of other types of carbapenem-resistant Enterobacteriaceae (CRE)—mainly Escherichia coli and Enterobacter species—is also increasing.12 In southeast Michigan, CRKP and other CRE have become endemic in the past 2 years, causing outbreaks in various types of healthcare settings.13
CREs are resistant to most classes of antimicrobials; frequently, the only therapeutic options available are polymixin, tigecycline, and sometimes aminoglycosides. Since therapeutic options are scarce, infections due to CRE are associated with severe and adverse clinical outcomes. These poor outcomes are due primarily to a delay in the initiation of effective antimicrobial therapy.14
Risk factors reported for the isolation of CRE include advanced age, reduced functional status, invasive procedures, and recent use of antibiotics.4–6,8,10,11,15,16 The combination of a growing population of individuals at risk of infection with CRE and the frequent transfer of high-risk patients throughout different healthcare settings presents a major healthcare challenge. Gaining a greater understanding of the epidemiology and characteristics of patients with CRE infection will aid efforts to control the spread of and manage infections with CRE. We analyzed a large cohort of cases of CRE infection from an endemic location in the United States. Comprehensive and advanced epidemiological and molecular methods were utilized to achieve an improved understanding of the epidemiology of these pathogens and to describe the characteristics and manifestations of CRE infections.
METHODS
Study Settings and Design
The Detroit Medical Center (DMC) healthcare system consists of 8 hospitals, has more than 2,200 inpatient beds, and serves as a tertiary referral hospital for metropolitan Detroit and southeastern Michigan. Klebsiella pneumoniae carbapenemase (KPC)–producing Enterobacteriaceae that were isolated from September 1, 2008, to August 31, 2009, were analyzed and reviewed. Institutional review boards at Wayne State University and DMC approved the study before its initiation.
Patients and Clinical Variables
This retrospective cohort study included all patients who had a culture result that was positive for CRE, from all inpatient and outpatient facilities that submit specimens to the DMC clinical microbiology laboratory. Samples collected from all anatomic sites were included, and both infected and colonized patients were included.17 For patients who had more than 1 episode of CRE isolation during the study period, only the first episode was analyzed (eg, unique patient episodes). Parameters retrieved from patient charts included (1) patient demographics; (2) clinical characteristics and severity of illness indices at the time of hospital admission, including functional status, McCabe score,18 Charlson score,19 and immunosuppressive conditions; (3) recent healthcare-associated exposures; and (4) factors related to antimicrobial therapy, including empiric therapy (defined as therapy administered from 48 hours before to 72 hours following the date that the positive CRE culture result was obtained), the main “consolidative” antimicrobial regimen (ie, antibiotics provided greater than 72 hours following CRE isolation and up to 14 days following isolation), and the time to initiation of effective therapy (ie, therapy with an antimicrobial agent that demonstrated in vitro activity against the CRE isolate); (5) the incidence of co-colonization with CRE and a lactose-nonfermenting organism, such as Acinetobacter baumanni and/or Pseudomonas aeruginosa (co-colonization was defined as recovery of the lactose-nonfermenting pathogen during the period of 3 days before to 3 days after the date of first positive CRE culture result); and (6) patient outcomes, including mortality, length of hospital stay, functional status deterioration (defined as deterioration from admission to discharge in at least 1 activity of daily living according to Katz criteria20), and discharge to a long-term care facility (LTCF).
Microbiology
DMC has a single, centralized clinical microbiology laboratory, which processes ~500,000 samples annually. Multiple outpatient facilities in southeast Michigan use DMC’s laboratory services on a routine basis. Bacteria were identified to the species level, and susceptibilities to predefined antimicrobials were determined on the basis of an automated broth microdilution system (MicroScan) and in accordance with Clinical and Laboratory Standards Institute (CLSI) criteria.21 During the study period, minimum inhibitory concentrations (MICs) to colistin and tigecycline were determined with Etest strips (bioMérieux), using Mueller-Hinton media, and breakpoints to defined resistance was set to 4μg/dL, according to the manufacturer’s recommendation (established by the Food and Drug Administration). The level of resistance to carbapenems was determined to be “elevated” if the MIC of imipenem or that of meropenem exceeded 2mg/L.22 A positive phenotypic extended-spectrum β-lactamase (ESBL) test result was defined on the basis of the automated system and validated with a disc diffusion test; all analyses were conducted in accordance to CLSI criteria.21
All Enterobacteriaceae that were resistant to 1 or more extended-spectrum (or third-generation) cephalosporin and that had an MIC to ertapenem of at least 2 mg/L were screened for carbapenemase production with the modified Hodge test, which was conducted according to CLSI criteria.21 Subsequently, CREs were tested for the presence of blaKPC by using polymerase-chain reaction (PCR).4 Previously characterized KPC-producing K. pneumoniae isolates were used as controls.4,23
The association between the mucoid appearance of isolates and clinical outcomes was evaluated.24 Three independent investigators (eg, F.P., A.M.H., and R.A.B.), all “blinded” to clinical data, individually reviewed the appearances of the colonies and graded them from 0 (no mucoid appearance) to 3 (hypermucoid appearance). The mucoid appearance grade was then analyzed as both a continuous variable and a dichotomous variable (grade 0–1 vs. grade 2–3).
Repetitive extragenic palindromic PCR (rep-PCR) was conducted, using an automated system (DiversiLab). Band patterns were compared with the modified Kullback-Leibler method in order to generate a dendrogram, using the DiversiLab software. On the basis of previous work, isolates with band patterns of at least 95% similarity were considered to be genetically related.23 Results of rep-PCR typing were compared with those of previously characterized KPC-producing K. pneumoniae isolates.4,23 Analyses for K. pneumoniae and Enterobacter species were conducted separately.
Statistical Analysis
Some of the epidemiological parameters for outpatients could not be retrieved because of lack of records. Throughout the text, the percentages displayed are the “valid percent,” which is the percentage that excludes the data missing from the denominator. All bivariate and multivariate analyses of patient outcomes were conducted for inpatients only.
All analyses were performed with SPSS 18. Bivariate analyses were performed using the Fisher exact test or the χ2 test for categorical variables and the independent samples t test or the Wilcoxon rank-sum test for continuous variables. Multivariable models for outcomes were constructed using logistic regression. All variables with a P value less than 0.1 in the bivariate analyses were considered for inclusion in the multivariate analysis. A stepwise selection procedure was used to select variables for inclusion in the final model. The final selected model was tested for confounding. If a covariate affected the β coefficient of a variable in the model by more than 10%, the confounding variable was maintained in the multivariable model. All P values were 2 sided. In addition to examining statistical significance and confounding, effect modification between variables was evaluated by testing appropriate interaction terms for statistical significance. When effect modification was detected, subgroup analyses were performed.
RESULTS
Description of Cohort
Ninety-two unique-patient CRE isolates were recovered during the 1-year study period. Cultures were obtained from all 8 DMC hospitals (n = 57), from 1 long-term acute care (LTAC) facility located inside one of the study hospitals but owned by a different company (n = 24), and from outpatient clinics that used DMC’s laboratory services (n = 11).
The study cohort consisted primarily of elderly, debilitated, African American individuals who frequently resided in LTCFs (Table 1). Many of the patients were hospitalized in LTACs in the previous year. Most patients underwent 1 or more invasive procedures and were hospitalized in acute care hospitals in the 3 months before CRE isolation. Among the inpatient case patients (n = 57), 34 (60%) were from medical wards, 12 were from adult ICUs, 7 were from surgical wards, 2 were from pediatric ICUs, and 2 were from a rehabilitation institute. The study population had multiple comorbid conditions and high Charlson scores.19 The 10-year survival probability was only 17% for the entire cohort.19 The McCabe score indicated a high severity of acute illness and a poor expected prognosis (Table 1),18 with 14 (17%) of the patients expected to die in the 2 months following hospital admission. Many of the patients experienced prior cognitive impairments and an immunosuppressive state prior to hospitalization (Table 1).
TABLE 1.
Epidemiological Characteristics of 92 Patients with Carbapenem-Resistant Enterobacteriaceae (CRE) Infection, Detroit Medical Center, September 2008–September 2009
| Parameter | Value |
|---|---|
| Demographics | |
| Median (range) age | 65.5 yr (3 mo–94 yr) |
| Pediatrica | 3 (3) |
| Elderlyb | 53 (58) |
| Female sex | 47 (51) |
| African American | 71 (78) |
| Microbiology | |
| Co-colonization with a nonfermenterc | 33 (42) |
| ESBL positive | 61 (85) |
| blaKPC PCR positive | 69 (95) |
| Same species but carbapenem susceptible in previous month | 31 (37) |
| Recent isolation of multidrug-resistant pathogend | 59 (71) |
| Median imipenem MIC, μg/dL | 2 |
| Median meropenem MIC, μg/dL | 2 |
| “Elevated” level of carbapenem resistancee | 35 (42) |
| Colistin resistant | 15 (17) |
| Tigecycline resistant | 14 (16) |
| Amikacin resistant | 18 (38) |
| Tobramycin resistant | 66 (78) |
| Ciprofloxacin resistant | 65 (84) |
| TMP/SMX resistant | 58 (68) |
| Acute and chronic conditions on admission | |
| Dependent functional status | 68 (84) |
| Cognitive impairment | 39 (48) |
| McCabe score,18 mean ± SD | 2.05 ± 0.1 |
| Rapidly fatal McCabe score18 | 13 (16.7) |
| Diabetes mellitus | 58 (67) |
| Chronic renal failure | 47 (54) |
| Neurologic disease | 53 (61) |
| Hemiplegia | 32 (37) |
| Dementia | 27 (31) |
| Charlson weighted index comorbidity score,19 mean ± SD | 5.3 ± 0.3 |
| Charlson combined condition score,19 mean ± SD | 7.5 ± 0.4 |
| Charlson 10-year survival probability,19 mean % ± SD | 16.9 ± 3.3 |
| Chronic skin ulcers | 45 (52) |
| Immunosuppressive statef | 30 (34) |
| Exposure to healthcare settings and environments before CRE isolation | |
| Permanent residence at an LTCF | 60 (68) |
| LTAC exposure in past year | 48 (56) |
| Chronic hemodialysis | 15 (17) |
| Regular visits to outpatient clinic | 37 (42) |
| Hospitalized in past 3 months | 65 (76) |
| Surgery or invasive procedure in past 6 months | 74 (88) |
| ICU stay in past 3 months | 46 (64) |
| Permanent devicesg | 72 (87) |
| LOS before isolation during current hospitalization, mean days ± SD | 16 ± 3 |
| ICU stay during current hospitalization | 22 (39) |
| Severity of illness indices at time of CRE isolation | |
| Reduced consciousness | 36 (48) |
| Severe sepsis, septic shock, or multiorgan failure17 | 11 (18) |
| Received vasopressors | 7 (11) |
| Transferred to ICU | 9 (13) |
| Necessitated acute mechanical intubation or ventilation | 10 (17) |
| Necessitated insertion of central vascular catheter | 11 (26) |
| Necessitated insertion of urinary catheter | 11 (31) |
| Developed acute renal failure17 | 17 (20) |
| Developed acute liver injuryh | 2 (3) |
| Antimicrobial therapy | |
| Antibiotics in preceding 3 months | 71 (96) |
| Effective therapy administered | 29 (69) |
| Hours to effective therapy, mean ± SD | 120 ± 23 |
| Outcomes | |
| In-hospital mortality | 23 (32) |
| 3-month mortality | 28 (36) |
| Functional status deterioration | 24 (61) |
| Discharged to LTCF after being admitted from home | 14 (39) |
| Discharged to LTAC after being admitted from home | 16 (36) |
| Additional hospitalizations in 6 months following CRE isolation | 45 (56) |
| Surgeries or invasive procedures in 3 months following CRE isolation | 40 (53) |
| Total LOS, median days (IQR) | 19 (9–38.5) |
| LOS after isolation, median days (IQR) | 12 (6–26) |
| ICU LOS after isolation, median days (IQR) | 13 (7–35.5) |
NOTE. Unless otherwise indicated, data are no. (%), where the percentage is out of the patients for which data were available, for example, excluding the missing cases. For outpatient individuals, some of the clinical parameters could not be retrieved. ESBL, extended-spectrum β-lactamase; ICU, intensive care unit; IQR, interquartile range; LOS, length of stay; LTAC, long-term acute care facility; LTCF, long-term care facility; MIC, minimum inhibitory concentration; PCR, polymerase chain reaction; TMP/SMX, trimethoprim/sulfamethoxazole.
Defined as patients less than 16 years of age.
Defined as patients aged at least 65 years.
Isolation of Acinetobacter baumannii and/or Pseudomonas aeruginosa from 3 days before to 3 days after (including from the exact same culture) the index CRE isolation.
Isolation in previous 3 months of methicillin-resistant Staphylococcus aureus, heteroresistant vancomycin-intermediate S. aureus, vancomycin-intermediate S. aureus, vancomycin-resistant Enterococcus, an ESBL-producing organism, Acinetobacter baumannii, or Pseudomonas aeruginosa.
Imipenem or meropenem MICs of at least 4 μg/dL.
Includes 1 of the following: (1) neutropenia (<500 neutrophils) at time of culture, (2) glucocorticoid or steroid use in the past month, (3) chemotherapy in the past 3 months, (4) radiotherapy in the past 3 months, (5) human immunodeficiency virus infection, (6) having undergone transplantation, or (7) having received anti–tumor necrosis factor therapy in the past 3 months.
Chronic or permanent devices (eg, tracheotomies, central lines, urinary catheters, orthopedic external fixators) that were in place at least 48 hours before CRE isolation.
Defined as the presence of all of the following 3 conditions: (1) transaminases 2 times greater than baseline value, (2) international normalized ratio greater than 1.5 without the use of anticoagulants, and (3) total bilirubin 3 times greater than baseline.
The study strains included 74 Klebsiella pneumoniae strains, 1 Klebsiella oxytoca strain, 2 Escherichia coli strains, and 15 Enterobacter species strains (10 Enterobacter cloacae, 4 Enterobacter aerogenes, and 1 Enterobacter sakazakii). Fifteen (17%) were additionally resistant to colistin, and 14 were resistant (16%) to tigecycline. Two of the patients had CRE isolates that were resistant to all tested antibiotics, and both died during their hospital stay. Most CRE isolates were obtained from urine samples (37 [40%]) or the respiratory tract (24 [26%]), and the remaining isolates were obtained from samples of blood (16 [17%]), wounds or other soft tissues (13 [14%]), cerebrospinal fluid (1 [1%]), and rectal surveillance (1 [1%]). Twenty-one (29%) of the CRE isolates represented cases of colonization. The spectrum of infectious clinical syndromes among this cohort included urinary tract infections (n = 14 [19%]), pneumonia (n = 14 [19%]), skin and soft-tissue infections (n = 12 [17%]), catheter-related bloodstream infections (n = 8 [11%]), bacteremia without an identified focus (n = 2 [3%]), and a central nervous system infection (n = 1 [1%]).
Three epidemiological features of the cohort were inter-correlated and associated with unfavorable outcomes: (1) patients with recent (6 months) exposure to an LTAC (n = 48 [56% of the entire cohort]), (2) patients with CRE isolates with an “elevated” level of resistance to group-2 carbapenems (n = 35 [42% of the entire cohort]), and (3) patients who were co-colonized with CRE and Pseudomonas aeruginosa or Acinetobacter baumannii (n = 33 [42% of the entire cohort]). Ten of 15 patients with colistin-resistant CREs and 13 of 14 with tigecycline-resistant CREs had had recent LTAC exposure. In addition, 28 patients with recent LTAC exposure had elevated levels of carbapenem resistance and 24 were co-colonized with a nonfermenter. In addition, an elevated level of carbapenem resistance was significantly associated with co-resistance to other antimicrobials, including to colistin (P = .007), tigecycline (P < .001), amikacin (P = .006), tobramycin (P = .003), ciprofloxacin (P = .014), and trimethoprim/sulfamethoxazole (TMP/SMX; P = .04).
Antimicrobial Data
Of the 74 patients for whom information about earlier antibiotic exposure was obtained, 71 (96%) received systemic antibiotics in the 3 months preceding isolation of CRE. Antibiotics received included cephalosporins (61 [86%]), glycopeptides (47 [65%]), penicillins (36 [51%]), fluoroquinolones (22 [31%]), carbapenems (15 [21%]), tetracyclines including tigecycline (7 [10%]), and colistin (5 [7%]).
There were 41 inpatients with infections due to CRE. Only 28 (68%) received effective therapy at some point from 3 days prior to 14 days after their initial positive culture result for CRE. In 2 cases, effective therapy options were not available. Among the group of infected inpatients that received effective therapy, the median time to initiation of an effective drug was 100.5 hours (range, 0–336 hours; mean ± standard deviation [SD], 124 ± 132 hours).
Only 5 patients received empiric therapy that had in vitro activity against their CRE pathogen. In 2 cases the active agent was an aminoglycoside, and in 3 cases the active antimicrobial was colistin. Of the 28 cases of infected inpatients who received effective therapy, the main antibiotic classes that were used as consolidative therapies were colistin (13 [46%]), tigecycline (8 [29%]), and TMP/SMX (6 [21%]), with aminoglycosides and rifampin used as adjuncts in 9 (32%) and 7 (25%) of cases, respectively.
Outcomes
Mortality
Thirty-four (36%) of 92 patients died in the 3 months following the initial isolation of CRE. Of the 57 inpatients, 18 (32%) died during their index hospitalization. Several parameters were significantly associated with inhospital mortality (Table 2). Elderly patients with multiple comorbid conditions who were cared for in the ICU and who were co-colonized with a nonfermenter were prone to die during their hospital stay. All of the severity-of-illness indices measured at the time of CRE isolation (Table 1) were also associated with mortality. Recovery of CRE from the urine and susceptibility to TMP/SMX were associated with a decreased risk for in-hospital mortality. In multivariate analysis, co-colonization with a nonfermenting pathogen, age of greater than 60 years, and an ICU stay in the current hospitalization prior to CRE isolation were associated with an increased risk for in-hospital mortality (Table 3). In a separate analysis of 3-month mortality, the same predictors of mortality were identified.
TABLE 2.
Bivariable Analysis of Parameters Associated with In-Hospital Mortality among Hospitalized Patients (N =57)
| Parameter | Dead (n =18)a | Alive (n =39)a | OR (95% CI) | P |
|---|---|---|---|---|
| Demographics | ||||
| Age in years, mean ± SD | 70.6 ± 12.3 | 56.4 ± 23.3 | .004 | |
| Elderlyb | 14 (78) | 18 (46) | 4.1 (1.4–15) | .02 |
| Female sex | 10 (56) | 20 (51) | 1.25 (0.4–3.3) | .76 |
| African American | 13 (72) | 32 (84) | 0.5 (0.1–1.9) | .3 |
| Microbiology | ||||
| Co-colonization with a nonfermenterc | 13 (72) | 11 (28) | 6.6 (1.9–23) | .003 |
| Hypermucoid colony appearanced | 3 (23) | 5 (21) | 1.14 (0.2–5.8) | .9 |
| ESBL positive | 10 (83) | 24 (80) | 1.3 (0.2–7.3) | .8 |
| Isolation of same species but carbapenem-susceptible in previous month | 6 (33) | 11 (28) | 1.3 (0.4–4.2) | .7 |
| Recent isolation of multidrug-resistant pathogene | 14 (78) | 25 (64) | 2 (0.5–7) | .3 |
| Median imipenem MIC, μg/dL | 4.7 | 3.4 | .31 | |
| Median meropenem MIC, μg/dL | 8 | 5.2 | .25 | |
| “Elevated” level of carbapenem resistancef | 9 (53) | 13 (34) | 2.2 (0.7–6.9) | .19 |
| Colistin resistant | 3 (17) | 6 (16) | 1.03 (0.3–4.7) | .97 |
| Tigecycline resistant | 1 (6) | 4 (11) | 0.5 (0.05–4.9) | .5 |
| Amikacin resistant | 1 (33) | 9 (47) | 0.6 (0.04–7.2) | .65 |
| Tobramycin resistant | 10 (56) | 30 (77) | 0.4 (0.1–1.2) | .1 |
| Ciprofloxacin resistant | 9 (64) | 29 (83) | 0.37 (0.9–1.5) | .16 |
| TMP/SMX resistant | 6 (33) | 27 (69) | 0.2 (0.07–0.7) | .01 |
| Culture body site | ||||
| Blood | 4 (22) | 3 (8) | 2.9 (0.7–11.6) | .12 |
| Respiratory | 7 (39) | 8 (21) | 1.9 (0.8–4.4) | .14 |
| Urine | 3 (17) | 20 (51) | 0.3 (0.1–0.9) | .01 |
| Wound | 3 (17) | 8 (21) | 0.8 (0.2–2.7) | .7 |
| Infectious clinical syndrome | ||||
| Colonization only | 5 (28) | 11 (28) | 0.98 (0.2–4) | .97 |
| Catheter-related bloodstream infection | 3 (17) | 2 (5) | 3.7 (0.4–36) | .3 |
| Pneumonia | 5 (28) | 7 (18) | 1.8 (0.4–7.9) | .4 |
| Urinary tract infection | 1 (6) | 11 (28) | 0.15 (0.01–1.3) | .06 |
| Skin and soft tissue infection | 1 (6) | 8 (21) | 0.2 (0.01–2) | .2 |
| Acute and chronic conditions on admission | ||||
| Dependent functional status | 16 (89) | 30 (77) | 1.16 (0.9–1.5) | .3 |
| Cognitive impairment | 14 (78) | 13 (33) | 2.3 (1.4–3.9) | .005 |
| Rapidly fatal McCabe score18 | 6 (33) | 3 (8) | 6 (1–37) | .02 |
| Ischemic heart disease | 9 (50) | 8 (21) | 3.9 (1.2–13) | .03 |
| Congestive heart failure | 10 (56) | 11 (28) | 3.2 (1–10) | .05 |
| Diabetes mellitus | 17 (94) | 21 (54) | 14.6 (2–120) | .002 |
| Chronic renal failure | 10 (56) | 26 (67) | 0.6 (0.2–2.3) | .4 |
| Vascular neurologic disease | 10 (56) | 11 (28) | 3.2 (1–10.2) | .05 |
| Overall neurologic disease | 12 (67) | 19 (49) | 2.1 (0.7–6.7) | .2 |
| Hemiplegia | 5 (28) | 13 (33) | 0.8 (0.3–2.6) | .7 |
| Dementia | 8 (44) | 11 (28) | 2 (0.6–6.5) | .24 |
| Charlson weighted index comorbidity score,19 mean ± SD | 6.2 ± 2.5 | 5.2 ± 2.8 | .2 | |
| Charlson combined condition score,19 mean ± SD | 8.8 ± 3 | 7 ± 3.4 | .06 | |
| Charlson 10-year survival probability,19 mean % ± SD | 6.2 ± 17 | 23 ± 36 | .07 | |
| Chronic skin ulcers | 11 (61) | 22 (56) | 1.2 (0.4–3.8) | .7 |
| Immunosuppressive stateg | 6 (33) | 16 (41) | 0.7 (0.2–2.7) | .6 |
| Exposure to healthcare settings and environments before CRE isolation | ||||
| Permanent residence at an LTCF | 10 (56) | 23 (59) | 0.8 (0.3–2.7) | .8 |
| LTAC exposure in past year | 10 (56) | 14 (36) | 2.2 (0.7–7) | .16 |
| Chronic hemodialysis | 4 (22) | 9 (23) | 0.95 (0.3–3.6) | .94 |
| Regular visits to outpatient clinic | 5 (28) | 14 (36) | 0.7 (0.2–2.3) | .5 |
| Hospitalized in past 3 months | 12 (67) | 29 (74) | 0.7 (0.2–2.3) | .55 |
| Invasive procedure in past 6 months | 16 (89) | 33 (85) | 1.5 (0.3–8) | .67 |
| ICU stay in past 3 months | 13 (72) | 18 (46) | 3 (0.9–10) | .07 |
| Permanent devicesh | 15 (83) | 32 (82) | 1.1 (0.2–4.8) | .9 |
| LOS before isolation in current hospitalization, mean ± SD | 10.3 ± 10 | 11.4 ± 30 | .88 | |
| ICU stay in current hospitalization | 12 (67) | 10 (26) | 5.8 (1.7–20) | .004 |
| Antimicrobial therapy | ||||
| Any antibiotics in preceding 3 months | 17 (94) | 38 (97) | 0.4 (0.03–7.6) | .5 |
| Daptomycin in preceding 3 months | 2 (12) | 0 (0) | 3.4 (2.2–5.2) | .04 |
| Macrolide in preceding 3 months | 3 (18) | 1 (3) | 7.5 (0.7–78) | .09 |
| Rifampin in preceding 3 months | 2 (12) | 0 (0) | 3.4 (2.2–5.2) | .04 |
| Effective therapy was administeredi | 10 (77) | 18 (64) | 1.8 (0.4–8.3) | .4 |
| Hours to effective therapy, mean ± SD | 163 ± 207 | 103 ± 61 | .2 | |
| Colistin in main antibiotic course | 10 (56) | 7 (18) | 5.7 (1.7–20) | .006 |
| Rifampin in main antibiotic course | 6 (33) | 3 (8) | 6 (1.3–28) | .014 |
| Aztreonam in main antibiotic course | 2 (11) | 0 (0) | 3.4 (2.3–5.2) | .03 |
NOTE. CI, confidence interval; CRE, carbapene-resistant Enterobacteriaceae; ESBL, extended-spectrum β-lactamase; ICU, intensive care unit; LOS, length of stay; LTAC, long-term acute care facility; LTCF, long-term care facility; MIC, minimum inhibitory concentration; OR, odds ratio; TMP/SMX, trimethoprim/sulfamethoxazole. Bold type indicates significant values (P < .05).
Data are no. (%) of patients unless otherwise indicated. The percentages are out of the patients for which data were available, for example, excluding the missing cases.
Older than 65 years of age.
Isolation of Acinetobacter baumannii and/or Pseudomonas aeruginosa from 3 days before to 3 days after (including from the exact same culture) as the index CRE isolation.
Mucoid appearance were determined by 3 independent coinvestigators blinded to clinical data, who graded the colony from 0 (no mucoid) to 3 (very mucoid). “Hypermucoid” was defined as grades of 2 and 3 combined.
Isolation in previous 3 months of methicillin-resistant Staphylococcus aureus, heteroresistant vancomycin-intermidiate S. aureus, vancomycin-intermidiate S. aureus, vancomycin-resistant Enterococcus, an ESBL-producing organism, A. baumannii, or P. aeruginosa.
Imipenem or meropenem MICs of at least 4 μg/dL.
Includes 1 of the following: (1) neutropenia (<500 neutrophils) at time of culture, (2) glucocorticoid or steroid use in the past month, (3) chemotherapy in the past 3 months, (4) radiotherapy in the past 3 months, (5) human immunodeficiency virus infection, (6) having undergone transplantation, or (7) having received anti–tumor necrosis factor therapy in the past 3 months.
Chronic or permanent devices (eg, tracheotomies, central lines, urinary catheters, orthopedic external fixators) that were in place at least 48 hours prior to CRE isolation.
An effective antibiotic per in vitro susceptibility results was administered in the 14 days following the positive CRE culture result.
TABLE 3.
Multivariable Analysis of Parameters Associated with In-Hospital Mortality among Hospitalized Patients (N =57)
| Variable | OR (95% CI) | P |
|---|---|---|
| Co-colonization with a nonfermenter | 10.5 (2.1–53.7) | .005 |
| Elderlya | 5.9 (1.1–32) | .04 |
| ICU stay during current hospitalization, before isolation | 6.6 (1.3–34) | .02 |
| Rapidly fatal McCabe score18 | 3 (0.4–24) | .3 |
NOTE. CI, confidence interval; ICU, intensive care unit; OR, odds ratio.
Older than 65 years of age.
Length of hospital stay
The median length of stay for the inpatient cohort (n = 57) was 19 days (Table 1). After excluding patients who died in the hospital, the median length of stay from culture date to discharge was 10 days (range, 1–156 days). The median length of stay in the ICU from culture date to ICU discharge is presented in Table 1. If effective therapy was administered in the first 14 days after culture date, there was a trend toward decreased length of stay (P = .09).
Additional outcomes
Among the inpatients who survived hospitalization (n = 39), (1) 24 (62%) experienced deterioration in their functional status, according to Katz criteria, during their hospitalization;20 (2) 31 (79%) were discharged to an LTCF or LTAC (of these, 18 had been admitted to the hospital from home); (3) 32 (82%) were readmitted within 6 months of their CRE isolation; and (4) 23 (59%) underwent an invasive procedure (including surgeries) in the 6 months following CRE isolation. Separate multivariate models were constructed for each of these outcome parameters. Co-colonization with nonfermenting gram-negative bacteria was independently associated with readmission to the hospital within 6 months (OR, 3.04 [95% confidence interval, 1–10.9]; P = .05).
Microbiologic and molecular analyses
Sixty-three K. pneumoniae isolates were examined for their mucoid appearance. Nineteen of these strains were hypermucoid, and all but 1 of these 19 belonged to clone 1 (Figure 1). No significant associations between hypermucoid appearance and South Asian ethnicity, liver disease, intravenous drug abuse, or clinical outcomes were identified.
FIGURE 1.
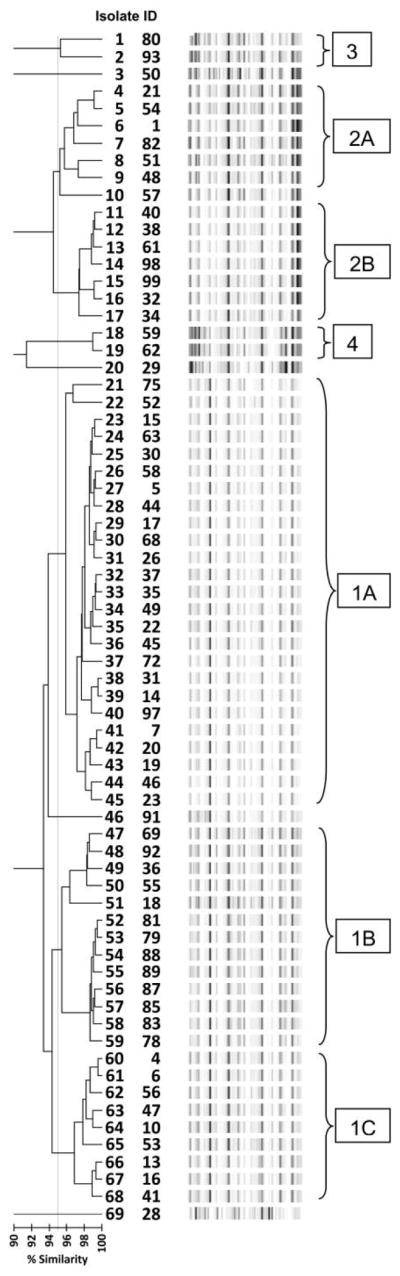
Repetitive extragenic palindromic polymerase chain reaction of Klebsiella pneumonia isolates, Detroit Medical Center, September 2008–September 2009.
Sixty-eight of 74 K. pneumonia isolates were available for genotyping by rep-PCR. Forty-eight were defined as clone 1, 14 were clone 2, 3 were clone 3, and 3 were clone 4 (Figure 1). A majority (30 of 43 individuals) of the members of the cohort who had a history of residence in an LTAC had a clone-1 K. pneumonia strain (P = .4). Conversely, clone-2 isolates were associated with antecedent exposure to non-LTAC LTCFs (P = .05) and were more likely to be colistin resistant (P = .001) and display elevated levels of resistance to group-2 carbapenems (P = .014). Clone 4 was more susceptible to tobramycin (P < .001), TMP/SMX (P = .012), and ciprofloxacin (P < .001). No association between specific clone type and clinical outcomes was identified.
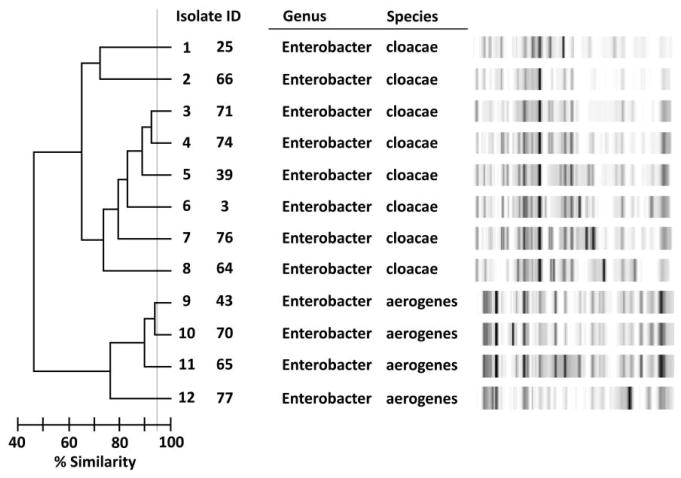
The results of the Enterobacter species rep-PCR are presented in Figure 2. Twelve of the 15 isolates were available for genotyping. The isolates were not closely genetically related, and each clone complex included only 1–2 strains.
FIGURE 2.
Repetitive extragenic palindromic polymerase chain reaction of Enterobacter species isolates, Detroit Medical Center, September 2008–September 2009.
DISCUSSION
This report identified unique epidemiological characteristics and outcomes associated with CRE. A unique manifestation of this cohort is that more than one-third of the isolates were from outpatients, many of whom resided permanently in LTCFs, underscoring the importance of LTCFs as reservoirs for multidrug-resistant (MDR) organisms, particularly gram-negative bacteria. An additional important feature of this cohort is the high rate of colistin resistance among the studied CREs.
The role of LTCFs as reservoirs for MDR gram-negative rods has been reported by others,4,6 but our cohort is the largest examining CRE to detail LTCF exposures among patients admitted to the hospital and, as far as we know, represents the largest collection of colistin-resistant CRE examined (despite the known discrepancies between methodologies in MIC determination to colistin or tigecycline).25 LTCFs often have less rigorous infection control and antimicrobial stewardship programs, with fewer resources, compared with acute care hospitals.26 Because of the advanced and invasive care that some patients receive in LTCFs (particularly in LTACs) and the frequent transfer of patients between LTCF and hospital settings,4,26 the spread of MDR gram-negative rods has become a regional issue in some parts of the country. Acute care hospitals can no longer be considered as “isolated” environments. When implementing programs that address CRE, it is now important to consider all reservoirs, including LTCFs.
Patients admitted from LTACs and those who had exposure to an LTAC in the 6 months before CRE isolation were the most morbid patients in the cohort and suffered the poorest clinical outcomes. While this association is not surprising, it highlights the clinical challenges that patients of LTACs present to acute care hospitals.
Overall outcomes were extremely poor in our CRE cohort. While others have reported on mortality and length of stay among patients with CRE infection,5,7–9,27 this study is the first to report on outcomes such as hospital readmissions, functional status deterioration, discharge to nonhospital institutional settings, and invasive procedures after isolation. We observed a mortality rate of 36% in our cohort, which is similar to the mortality reported in other CRE studies.5,27 The subjects in our CRE cohort were generally frail, with impaired functional status. They represent some of the most vulnerable and dependent patients cared for in hospitals. The association between functional status and CRE, combined with the magnitude of functional status deterioration following CRE isolation, has not been published previously.28
One particularly interesting finding was the association between co-colonization with a nonfermenting gram-negative organism and poor outcomes: after adjustment, co-colonization was associated with a greater than 10-fold increase in hospital mortality risk. This association might relate to the severity of illness of patients who are co-colonized. However, the possibility of transfer of mobile genetic elements between these types of pathogens is of concern.29 If co-colonization between a nonfermenting gram-negative pathogen and CRE increases mortality risk, then preventive infection-control practices might need to be adjusted in an effort to keep patients infected with CRE separated from patients infected with nonfermenting gram-negative bacteria. Co-colonization was, in addition, a significant risk factor for isolation of colistin-resistant CRE, and whether the lactose-nonfermenting gram-negative bacillus can facilitate transfer of genetic elements conferring resistance to colistin to Enterobacteriaceae is an intriguing concept that should be subjected to further investigation.
Previously, an association between the mucoid appearance of colonies of Klebsiella pneumoniae strains and increased virulence was reported.24 All but 1 of the hypermucoid strains belonged to 1 clone (clone 1; Figure 1), but in this cohort we did not find associations between mucoid or hypermucoid appearance and clinical outcomes or decreased susceptibility to carbapenems.
Molecular results demonstrated that most K. pneumoniae CRE strains in our cohort belonged to 4 major genotypes, and those different genotypes were common in different populations. For example, CRE of clone type 1 were frequently recovered from subjects with LTAC exposure, and patients admitted from LTCFs other than LTACs frequently had CRE of clone type 2. Thus, although polyclonality of CRE strains was evident in the study cohort, there probably was patient-to-patient spread within particular groups and within institutional settings outside of the hospital setting. These findings further demonstrate the need for regional approaches to the control and management of CREs. There might have been additional patient-to-patient transmission of resistance determinants (as opposed to bacterial strains), but such an investigation was beyond the scope of this study.
This is the first study, to our knowledge, to examine CREs that had been isolated from samples obtained from patients in different types of healthcare settings within the same region, including acute care hospitals, rehabilitation institutions, LTCFs, and outpatient clinics. Our comprehensive and advanced epidemiological analysis of a large CRE-infected or -colonized cohort from an endemic location in the United States stands as an important benchmark for future work. This article highlights several novel and meaningful analyses, including evaluation of various outcomes in addition to mortality; evaluation of strains with an elevated level of resistance to group-2 carbapenems, which might serve as a marker for the presence of resistance mechanisms in addition to blaKPC;22 and the association between mucoid strain appearance and virulence.
Carbapenem-resistant Enterobacteriaceae (CRE) became endemic in many parts of the United States in a short period of time. In less than a decade, the prevalence of blaKPC-producing K. pneumoniae causing hospital-acquired infections that were reported to the Centers for Disease Control and Prevention escalated, from less than 1% in 2000 to 8% in 2007. The prevalence today is probably much higher. The reasons for the rapid spread of CRE probably relate in part to the difficulties in eradicating the organism once a patient is colonized and to the limited antimicrobial options available. This study clearly demonstrates that within metropolitan Detroit, CREs have become firmly established in both hospital and nonhospital settings, and resistance to colistin and tigecycline, 2 of the only remaining antimicrobials with activity against CRE, is emerging. Regional approaches are necessary to control the spread of CRE from these environments to the community, and more information is needed regarding optimal dosing of colistin and mechanisms of resistance to colistin and tigecycline.
Acknowledgments
Financial support. The molecular investigation was supported in part by the Veterans Affairs Merit Review Program; Geriatric Research, Education, and Clinical Centers VISN 10; National Institutes of Health grant RO1 AI072219 (to R.A.B.); and the Steris Corporation (to F.P.).
Footnotes
Potential conflicts of interests. J.M.P. has board membership at Astellas and Forest, has served as a consultant for Cubist, and has received payment for lectures, including service on speakers bureaus for Merck and Pfizer. R.A.B. served as a consultant for Pfizer. K.S.K. served as a consultant for Merck, Pfizer, OrthoMcneil, and Forest and received payment for lectures, including service on speakers’ bureaus for Merck, Pfizer, Cubist, and OrthoMcneil. All other authors report no conflicts of interest relevant to this article.
References
- 1.Yigit H, Queenan AM, Anderson GJ, et al. Novel carbapenem-hydrolyzing beta-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob Agents Chemother. 2001;45(4):1151–1161. doi: 10.1128/AAC.45.4.1151-1161.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Patel G, Huprikar S, Factor SH, Jenkins SG, Calfee DP. Outcomes of carbapenem-resistant Klebsiella pneumoniae infection and the impact of antimicrobial and adjunctive therapies. Infect Control Hosp Epidemiol. 2008;29(12):1099–1106. doi: 10.1086/592412. [DOI] [PubMed] [Google Scholar]
- 3.Bradford PA, Bratu S, Urban C, et al. Emergence of carbapenem-resistant Klebsiella species possessing the class A carbapenem-hydrolyzing KPC-2 and inhibitor-resistant TEM-30 beta-lactamases in New York City. Clin Infect Dis. 2004;39(1):55–60. doi: 10.1086/421495. [DOI] [PubMed] [Google Scholar]
- 4.Endimiani A, Depasquale JM, Forero S, et al. Emergence of blaKPC-containing Klebsiella pneumoniae in a long-term acute care hospital: a new challenge to our healthcare system. J Antimicrob Chemother. 2009;64(5):1102–1110. doi: 10.1093/jac/dkp327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gasink LB, Edelstein PH, Lautenbach E, Synnestvedt M, Fishman NO. Risk factors and clinical impact of Klebsiella pneumoniae carbapenemase-producing K. pneumoniae. Infect Control Hosp Epidemiol. 2009;30(12):1180–1185. doi: 10.1086/648451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Munoz-Price LS, Hayden MK, Lolans K, et al. Successful control of an outbreak of Klebsiella pneumoniae carbapenemase-producing K. pneumoniae at a long-term acute care hospital. Infect Control Hosp Epidemiol. 2010;31(4):341–347. doi: 10.1086/651097. [DOI] [PubMed] [Google Scholar]
- 7.Hirsch EB, Tam VH. Detection and treatment options for Klebsiella pneumoniae carbapenemases (KPCs): an emerging cause of multidrug-resistant infection. J Antimicrob Chemother. 2010;65(6):1119–1125. doi: 10.1093/jac/dkq108. [DOI] [PubMed] [Google Scholar]
- 8.Leavitt A, Navon-Venezia S, Chmelnitsky I, Schwaber MJ, Carmeli Y. Emergence of KPC-2 and KPC-3 in carbapenem-resistant Klebsiella pneumoniae strains in an Israeli hospital. Antimicrob Agents Chemother. 2007;51(8):3026–3029. doi: 10.1128/AAC.00299-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Samra Z, Ofir O, Lishtzinsky Y, Madar-Shapiro L, Bishara J. Outbreak of carbapenem-resistant Klebsiella pneumoniae producing KPC-3 in a tertiary medical centre in Israel. Int J Antimicrob Agents. 2007;30(6):525–529. doi: 10.1016/j.ijantimicag.2007.07.024. [DOI] [PubMed] [Google Scholar]
- 10.Schwaber MJ, Carmeli Y. Carbapenem-resistant Enterobacteriaceae: a potential threat. JAMA. 2008;300(24):2911–2913. doi: 10.1001/jama.2008.896. [DOI] [PubMed] [Google Scholar]
- 11.Souli M, Galani I, Antoniadou A, et al. An outbreak of infection due to beta-lactamase Klebsiella pneumoniae carbapenemase 2–producing K. pneumoniae in a Greek university hospital: molecular characterization, epidemiology, and outcomes. Clin Infect Dis. 2010;50(3):364–373. doi: 10.1086/649865. [DOI] [PubMed] [Google Scholar]
- 12.Centers for Disease Control and Prevention. Guidance for control of infections with carbapenem-resistant or carbapenemase-producing Enterobacteriaceae in acute care facilities. MMWR Morb Mortal Wkly Rep. 2009;58(10):256–260. [PubMed] [Google Scholar]
- 13.Marchaim D, Chopra T, Pogue JM, et al. Outbreak of colistin-resistant, carbapenem-resistant Klebsiella pneumoniae in metropolitan Detroit, Michigan. Antimicrob Agents Chemother. 2011;55(2):593–599. doi: 10.1128/AAC.01020-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paul M, Shani V, Muchtar E, Kariv G, Robenshtok E, Leibovici L. Systematic review and meta-analysis of the efficacy of appropriate empiric antibiotic therapy for sepsis. Antimicrob Agents Chemother. 2010;54(11):4851–4863. doi: 10.1128/AAC.00627-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maltezou HC, Giakkoupi P, Maragos A, et al. Outbreak of infections due to KPC-2-producing Klebsiella pneumoniae in a hospital in Crete (Greece) J Infect. 2009;58(3):213–219. doi: 10.1016/j.jinf.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 16.Zarkotou O, Pournaras S, Voulgari E, et al. Risk factors and outcomes associated with acquisition of colistin-resistant KPC-producing Klebsiella pneumoniae: a matched case-control study. J Clin Microbiol. 2010;48(6):2271–2274. doi: 10.1128/JCM.02301-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dellinger RP, Levy MM, Carlet JM, et al. Surviving sepsis campaign: international guidelines for management of severe sepsis and septic shock: 2008. Crit Care Med. 2008;36(1):296–327. doi: 10.1097/01.CCM.0000298158.12101.41. [DOI] [PubMed] [Google Scholar]
- 18.Bion JF, Edlin SA, Ramsay G, McCabe S, Ledingham IM. Validation of a prognostic score in critically ill patients undergoing transport. Br Med J (Clin Res Ed) 1985;291(6493):432–434. doi: 10.1136/bmj.291.6493.432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Charlson ME, Pompei P, Ales KL, MacKenzie CR. A new method of classifying prognostic comorbidity in longitudinal studies: development and validation. J Chronic Dis. 1987;40(5):373–383. doi: 10.1016/0021-9681(87)90171-8. [DOI] [PubMed] [Google Scholar]
- 20.Katz S, Ford AB, Moskowitz RW, Jackson BA, Jaffe MW. Studies of illness in the aged: the index of ADL: a standardized measure of biological and psychosocial function. JAMA. 1963;185:914–919. doi: 10.1001/jama.1963.03060120024016. [DOI] [PubMed] [Google Scholar]
- 21.Clinical and Laboratory Standards Institute (CLSI) Performance Standards for Antimicrobial Susceptibility Testing: 19th Informational Supplement. CLSI approved standard M100-S19. Wayne, PA: CLSI; 2009. [Google Scholar]
- 22.Kitchel B, Rasheed JK, Endimiani A, et al. Genetic factors associated with elevated carbapenem resistance in KPC-producing Klebsiella pneumoniae. Antimicrob Agents Chemother. 2010;54(10):4201–4207. doi: 10.1128/AAC.00008-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Endimiani A, Hujer AM, Perez F, et al. Characterization of blaKPC-containing Klebsiella pneumoniae isolates detected in different institutions in the eastern USA. J Antimicrob Chemother. 2009;63(3):427–437. doi: 10.1093/jac/dkn547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu VL, Hansen DS, Ko WC, et al. Virulence characteristics of Klebsiella and clinical manifestations of K. pneumoniae bloodstream infections. Emerg Infect Dis. 2007;13(7):986–993. doi: 10.3201/eid1307.070187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lat A, Clock SA, Wu F, et al. Comparison of polymyxin B, tigecycline, cefepime, and meropenem MICs against KPC-producing Klebsiella pneumoniae by broth microdilution, Vitek 2, and Etest. J Clin Microbiol. 2011;49(5):1795–1798. doi: 10.1128/JCM.02534-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Munoz-Price LS. Long-term acute care hospitals. Clin Infect Dis. 2009;49(3):438–443. doi: 10.1086/600391. [DOI] [PubMed] [Google Scholar]
- 27.Schwaber MJ, Klarfeld-Lidji S, Navon-Venezia S, Schwartz D, Leavitt A, Carmeli Y. Predictors of carbapenem-resistant Klebsiella pneumoniae acquisition among hospitalized adults and effect of acquisition on mortality. Antimicrob Agents Chemother. 2008;52(3):1028–1033. doi: 10.1128/AAC.01020-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perez F, Endimiani A, Ray AJ, et al. Carbapenem-resistant Acinetobacter baumannii and Klebsiella pneumoniae across a hospital system: impact of post-acute care facilities on dissemination. J Antimicrob Chemother. 2010;65(8):1807–1818. doi: 10.1093/jac/dkq191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sidjabat HE, Silveira FP, Potoski BA, et al. Interspecies spread of Klebsiella pneumoniae carbapenemase gene in a single patient. Clin Infect Dis. 2009;49(11):1736–1738. doi: 10.1086/648077. [DOI] [PubMed] [Google Scholar]