Abstract
The hypothalamus is a central regulator of many behaviors that are essential for survival, such as temperature regulation, food intake and circadian rhythms. However, the molecular pathways that mediate hypothalamic development are largely unknown. To identify genes expressed in developing mouse hypothalamus, we performed microarray analysis at 12 different developmental time points. We then conducted developmental in situ hybridization for 1,045 genes that were dynamically expressed over the course of hypothalamic neurogenesis. We identified markers that stably labeled each major hypothalamic nucleus over the entire course of neurogenesis and constructed a detailed molecular atlas of the developing hypothalamus. As a proof of concept of the utility of these data, we used these markers to analyze the phenotype of mice in which Sonic Hedgehog (Shh) was selectively deleted from hypothalamic neuroepithelium and found that Shh is essential for anterior hypothalamic patterning. Our results serve as a resource for functional investigations of hypothalamic development, connectivity, physiology and dysfunction.
The vertebrate hypothalamus is a central homeostatic regulator of the neuroendocrine system and numerous other behavioral and physiological processes that are essential for survival. Developmental defects of the hypothalamus are proposed to disrupt the control of hypothalamic neuropeptide release and may serve as the etiological underpinnings of multiple diseases, potentially including obesity, mood disorders and autism1–4. A detailed understanding of the gene expression pattern of the developing hypothalamus may help to identify genes whose altered expression underlies these disorders.
Previous work has found that morphogens such as the Shh and Wnt proteins pattern the hypothalamic neuroepithelium along the dorsoventral and anteroposterior axes and are critical later in hypothalamic development5–8. Hypothalamic neurogenesis occurs between embryonic day 10 (E10) and E16 in the mouse, with individual hypothalamic nuclei typically being generated in an inside-out sequence9. Previous studies have identified a number of transcription factors that regulate the development of specific hypothalamic nuclei and neuronal subtypes10–16. However, even though several studies have examined the expression pattern of many genes in the developing mouse brain17,18, the hypothalamic expression patterns of these genes is difficult to interpret, given the complex structure of the developing hypothalamus and the lack of well-defined marker genes.
Furthermore, although the anatomical structure of the developed mammalian hypothalamus is quite well characterized, the precise contribution of individual hypothalamic neuronal subtypes to the regulation of appetitive and emotional behaviors is often unclear. Historically, bilateral stereotactic lesions have been the method of choice used to link specific hypothalamic nuclei to the regulation of behavior, and such manipulations typically destroy a diverse assortment of neuronal subtypes and often also disrupt the axons of neurons whose cell bodies are located outside the targeted region. More recently, genetic studies have examined the consequences of eliminating the expression of hypothalamic neuropeptides or of ablating the cells that express these transmitters altogether19–23. Although these approaches often produce marked behavioral phenotypes, they have limitations. Hypothalamic anatomy is highly complex and it is rarely possible to produce selective surgical lesions that do not affect multiple neuronal subtypes or axonal projections that regulate a specific physiological process. Similarly, genetic approaches are complicated by the fact that relatively few neuropeptides are expressed exclusively in the hypothalamus. A detailed understanding of the gene expression pattern of the developing hypothalamus may help to elucidate which specific neuronal subtypes regulate which specific physiological processes. To identify markers that would serve as a resource for functional investigations of hypothalamic development, connectivity, physiology and dysfunction, we began construction of a detailed molecular atlas of the developing mouse hypothalamus. We employed genome-scale microarray and in situ hybridization (ISH) analysis to systematically analyze changes in gene expression over the course of hypothalamic neurogenesis. Using these data, we generated a model for the organization of the developing hypothalamic neuroepithelium and scored expression levels for each individual transcript tested. We anticipate that these findings will substantially accelerate mechanistic analysis of nucleogenesis and cell-fate specification in the developing mammalian hypothalamus.
RESULTS
Microarray analysis of developing hypothalamus
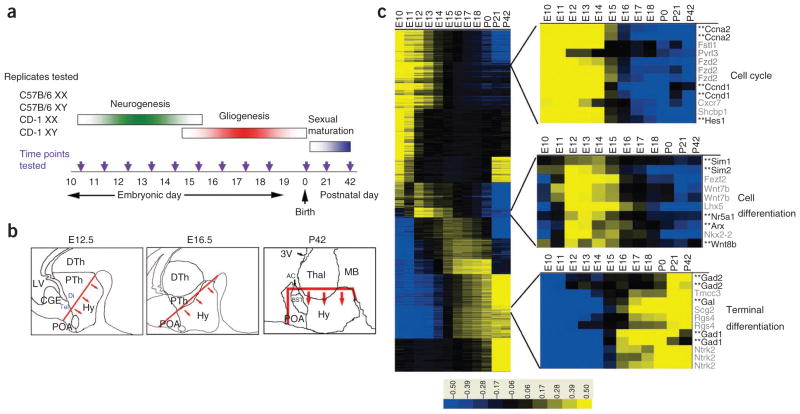
We used a genome-scale microarray-based analysis to profile gene expression over the course of development of the hypothalamus and preoptic area (Fig. 1a). These structures regulate sexually dimorphic behaviors and sex-dependent differences in gene expression are seen in both structures during development24,25. Furthermore, strain-dependent differences in hypothalamic function have been reported in mice26,27. As our goal was to identify genes that regulate hypothalamic development across a broad range of genotypes, we sought to clarify the effects of sex and strain on gene expression. To this end, we used four separate biological replicates, consisting of microdissected tissue obtained from male and female C57Bl/6 and CD-1 mice, for expression profiling (Fig. 1a). We obtained RNA from E10.5 hypothalamic neuroepithelium and every day thereafter until E18.5. Samples from postnatal day 0 (P0), P21 and P42 were also analyzed (Fig. 1b). To detect sexually dimorphic patterns of gene expression that occur transiently during development, we grouped three successive developmental time points for statistical analysis (Supplementary Fig. 1). We identified 37 probe sets by applying a low-stringency threshold of B-statistic 0 log-odds ratio, which means that there is a 50% probability that these probe sets are differentially expressed between male and female hypothalamus. Most represented either Y-chromosomal transcripts, noncoding transcripts involved in mediating X inactivation or X-chromosomal genes that escape X inactivation (Supplementary Fig. 1), all of which are differentially expressed in both developing and mature mouse brain28. Several other probe sets matching autosomal genes showed significantly higher expression (≤0 log-odds and ≥twofold change) in male embryos in the E11–13 or E12–14 window. Most of these corresponded to uncharacterized intronic transcripts, with only seven matching exonic sequences, and none of these transcripts had more than a twofold difference in expression at any of the time points tested. Comparison of C57Bl/6 versus CD-1 mice yielded a larger number of differentially expressed probe sets, and we applied a twofold change cut-off and 0 log-odds ratio threshold to the data. A total of 221 probe sets were significantly different (≤0 log-odds and ≥twofold change) after these two filters were applied (Supplementary Fig. 2). A more careful examination of these differences revealed that the majority reflected consistent differences in transcript abundance at all developmental stages that did not result in differences in the overall temporal pattern of transcript expression between strains (Supplementary Table 1). We conclude that only a relatively small fraction, about 1%, of all dynamically expressed transcripts differ in expression by sex or strain in developing hypothalamus.
Figure 1.
Microarray-based identification of developmentally dynamic transcripts in mouse hypothalamus. (a) Schema for microarray analysis in developing mouse hypothalamus. Hypothalamic tissue from a minimum of six individuals was collected for each of the four indicated genotypes at each of the time points indicated. (b) Anatomical landmarks used for dissection of hypothalamic and preoptic tissue at each indicated time point are shown. Arrows indicate tissue that was collected for RNA extraction. 3V, third ventricle; AC, anterior commissure; BST, bed nucleus of stria terminalis; CGE, caudal ganglionic eminence; Di, diencephalon; DTh, dorsal thalamus; Hy, hypothalamus; LV, lateral ventricle; MB, midbrain; POA, preoptic area; PTh, prethalamus; Tel, telencephalon; Thal, thalamus. (c) Self-organizing map–based cluster analysis of the 25,471 probe sets that showed significantly different expression levels (<1% false discovery rate and >twofold change) over the course of development is shown. The signal intensity indicates the difference between the median log2-scaled signal intensity at the indicated time point and the median log2-scaled signal intensity observed for that probe set across all time points tested. Magnified images of clusters enriched for genes involved in neuronal progenitor proliferation, hypothalamic neuronal differentiation and terminal markers of hypothalamic neuronal fate are shown. Genes whose expression in developing hypothalamus has been previously examined are shown in black and marked by a double asterisk, whereas transcripts showing similar expression, but which have not been previously functionally analyzed in hypothalamus are shown in gray.
We analyzed the much larger set of transcripts that were dynamically expressed in developing hypothalamus in both sexes and strains. Of these, 25,471 probe sets were significantly different (one-way ANOVA, false discovery rate 1%) from the median signal intensity at one or more time point (Fig. 1c and Supplementary Table 2). We were able to readily identify genes whose expression was enriched during the period of neurogenesis, which occurs from E10 to E16 in the mouse, with the bulk of the neurons being born in the interval from E12 to E14 (ref. 9). Genes that are broadly expressed in neuronal progenitor cells, such as Ccnd1, Cdk4, Hes1 and Nes, were highly expressed early in the course of neurogenesis, with expression levels declining steadily in the postnatal period (Fig. 1c and data not shown). Genes that regulate regional patterning of hypothalamic progenitor cells, such as Sim1, Sim2, Arx and Nr5a1, reached their highest level of expression between E12 and E14, coincident with the peak interval of hypothalamic neurogenesis (Fig. 1c). Finally, markers of terminal differentiation, such as Gad67, Gal (galanin), Pmch (pro-melanin concentrating hormone) and Hcrt (hypocretin), peaked postnatally (Fig. 1c and data not shown). These data suggest that our microarray analysis accurately detects changes in gene expression in the developing mouse hypothalamus.
ISH validation of genes in the developing hypothalamus
To characterize the cellular expression patterns of transcripts identified as being developmentally dynamically expressed by our microarray analysis, we conducted high-throughput ISH analysis using a similar approach to that used in our previous analysis of gene expression in developing retina29. Transcripts that were dynamically expressed during the course of hypothalamic neurogenesis on the basis of our microarray analysis (Fig. 1c) were then selected for ISH analysis. We examined 1,166 unique ISH probes, representing a total of 1,045 genes, on fresh-frozen coronal sections of mouse hypothalamus at E10.5, E12.5, E14.5, E16.5, P0 and P42. Of these, 881 probes showed cell-, region- and/or stage-specific expression in at least one region of the developing brain and 352 probes showed strong and/or selective expression in the hypothalamic neuroepithelium (Supplementary Table 3).
We conducted a second round of high-quality two-color ISH analysis to reliably map the expression of genes of interest. We selected Shh as the second color for these studies, as it is robustly and selectively expressed in a number of different regions in the developing fore-brain, including ventral telencephalic neuroepithelium, the basal plate of the developing hypothalamus and the zona limitans intrathalamica (Zli)30,31. Thus, any gene expression pattern can be characterized in relation to these regions. For this second round of ISH studies, we analyzed a total of 352 test probes using a previously described protocol32, examining 40-μm coronal sections spaced 40 μm apart from E11.5, E12.5, E14.5 and E16.5 developing hypothalamus. For many genes, sagittal sections were also analyzed for these time points. We obtained a reliable signal for both the test transcript and Shh in coronal sections with 197 of the probes and a specific signal in sagittal sections with another 92 of the probes (Supplementary Table 4). Images of all two-color ISH data are available at http://blackshaw.bs.jhmi.edu.
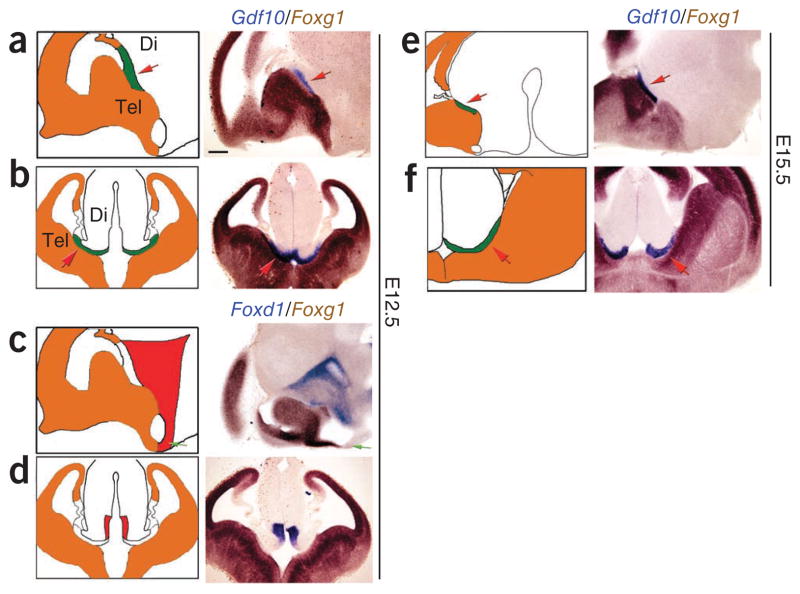
Using these data, we sought to identify molecular markers delineating specific regions of the developing hypothalamic neuroepithelium. We first determined the borders of the developing hypothalamus (Fig. 2). At E12.5, our expression analysis identified the BMP family member Gdf10 as a marker of the dorsal diencephalic border of the eminentia thalami (EmThal), which was confirmed by a lack of overlap with the telencephalic and cortical hem33 markers Foxg1, Wnt2b or Ttr (Fig. 2a,b and Supplementary Fig. 3). This expression pattern persisted at E15.5 (Fig. 2e,f and Supplementary Fig. 3), indicating that Gdf10 delineates the anterior border of the dorsal diencephalon At E12.5, Foxd1 was broadly expressed in progenitors of both prethalamus and anterior hypothalamus and was adjacent to the telencephalic marker Foxg1 (Fig. 2c,d), consistent with previous reports34,35. Therefore, Foxd1 and Gdf10 define the anterior border of the diencephalon at early stages of hypothalamic neurogenesis.
Figure 2.
Marker genes that delineate the telencephalic-diencephalic border. (a,b) Sagittal and coronal views of two-color ISH analysis of Gdf10 (blue) and Foxg1 (brown) at E12.5. Gdf10 demarcated the anterior border of prethalamus (red arrow). (c,d) Sagittal and coronal views of two-color ISH analysis of Foxd1 (blue) and Foxg1 (brown) at E12.5. Foxg1 and Foxd1 were expressed in adjacent, but nonoverlapping, zones in telencephalon and anterior diencephalon, respectively. The green arrows indicate the ventral telencephalic-diencephalic boundary. (e,f) Sagittal and coronal views of two-color ISH analysis of Gdf10 (blue) and Foxg1 (brown) at E15.5. Gdf10 expression continued to demarcate the anterior border of prethalamus at this later stage of development. Schematic figures indicate zones of gene expression at all of the stages shown. Di, diencephalon; Tel, telencephalon. Scale bar represents 0.2 mm.
The anterior diencephalon is divided into multiple regions
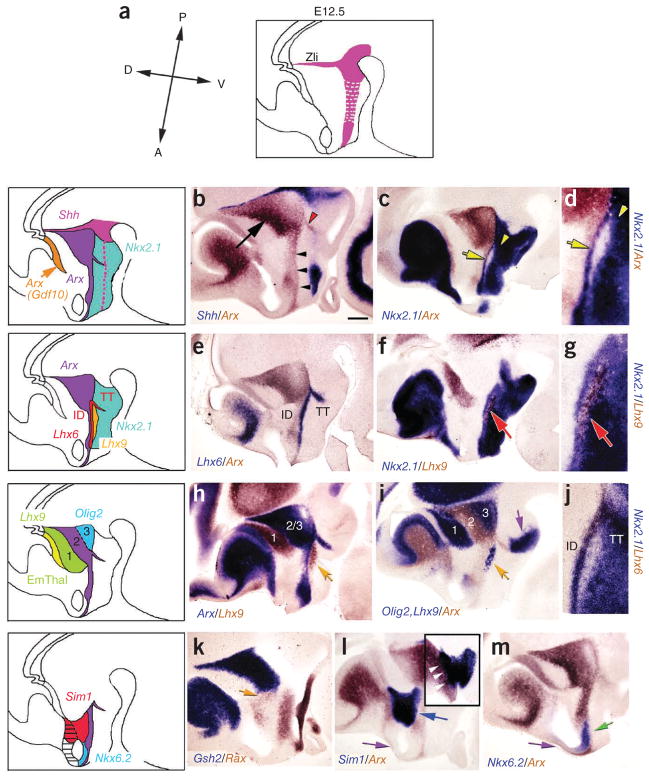
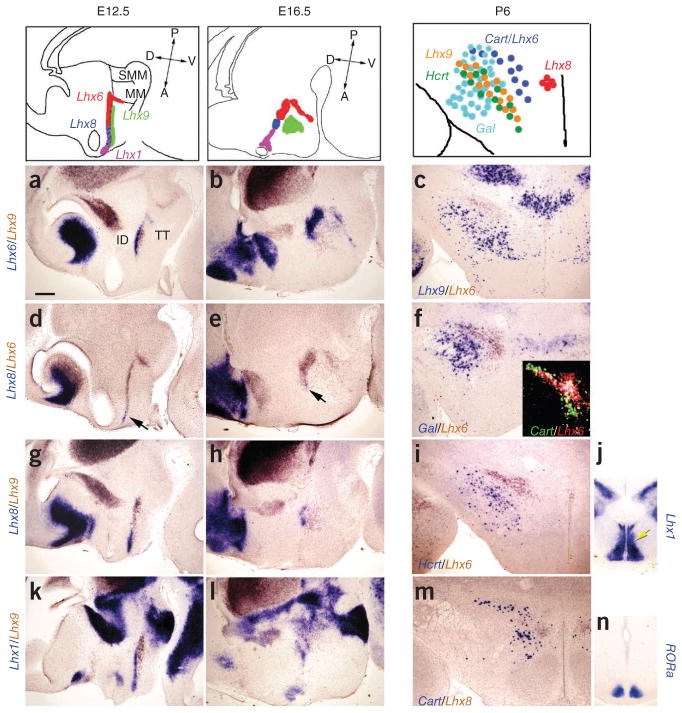
We next attempted to characterize the border of the prethalamus and the hypothalamus using Shh and other marker genes to orient ourselves to the neuraxis (Fig. 3). We performed two-color ISH with the previously described posteroventral hypothalamic marker Nkx2.1 (ref. 36) and the prethalamic marker Arx37. In addition to Arx expression in presumptive prethalamic region (Fig. 3b), we found two linear domains of Arx expression, one that extended toward the ventral hypothalamic surface (Fig. 3b) and another that ran parallel to the basal domain of Shh expression toward the optic recess (Fig. 3b). Careful analysis revealed two previously unreported domains with overlapping Arx and Nkx2.1 expression (Fig. 3c,d). We also found that the posterior domain of Arx and Nkx2.1 expression also expressed Lhx6 (Fig. 3e,j). We termed this domain the tuberomamillary terminal, as it demarcates the border between mamillary and premamillary neuroepithelium (Fig. 3e,j and Fig. 4). The second domain of overlapping Lhx6, Nkx2.1 and Arx expression ran parallel to the neuraxis in the hypothalamus and extended anterior in the direction of the optic recess. This domain of overlap bisected the hypothalamus into anterodorsal and anteroventral domains, and we therefore referred to it as the intrahypothalamic diagonal (Fig. 3e,j). Expression of another LIM homeodomain family member Lhx9 demarcated a Nkx2.1-negative ventral hypothalamic region (Fig. 3f,g).
Figure 3.
Regional patterning of prethalamus and anterior hypothalamus is revealed by analysis of marker gene expression. (a) Schematic indicating Shh expression in the developing diencephalon at E12.5. (b) Two-color ISH analysis of Shh (blue) and Arx (brown) indicated that Arx was expressed in prethalamus (black arrow) and two distinctive regions in hypothalamic neuroepithelium (black arrowheads and red arrowhead). (c,d) Nkx2.1 (blue) expression partially overlapped with Arx (brown) expression in the hypothalamic zone running parallel to the basal plate (yellow arrow) and in posterior hypothalamus (yellow arrowhead). (e) Lhx6 expression (purple) demarcated the intrahypothalamic diagonal and the tuberomamillary terminal, overlapping with Arx (brown). (f,g) Lhx9 expression (brown) marked a region ventral to the intrahypothalamic diagonal (red arrow) that was devoid of Nkx2.1 (purple). (h) Arx (2/3, blue) and Lhx9 (EmThal (1), brown) defined nonoverlapping domains of gene expression in prethalamus and hypothalamus. Lhx9 was expressed immediately ventral to the Arx-positive zone in the intrahypothalamic diagonal (yellow arrow). (i) Olig2 expression (blue, in combination with Lhx9) selectively marked the most ventral portion of the prethalamus (3). (j) Lhx6 expression (brown) overlapped with Nkx2.1 (blue) in the intrahypothalamic diagonal and tuberomamillary terminal. (k) Gsh2 (blue) and Rax (brown) were expressed on the border of prethalamus and hypothalamus (orange arrow). (l) Sim1 (blue) was expressed immediately anterodorsal to the zone of Arx expression (brown) in intrahypothalamic diagonal (blue arrowhead). (m) A domain of ventral anterior hypothalamic neuroepithelium was labeled by Nkx6.2 (green arrow). Scale bar represents 0.2 mm (b, c, e, f, h, i and k–m) and 0.10 mm for (d, g and j). ID, intrahypothalamic diagonal; TT, tuberomamillary terminal.
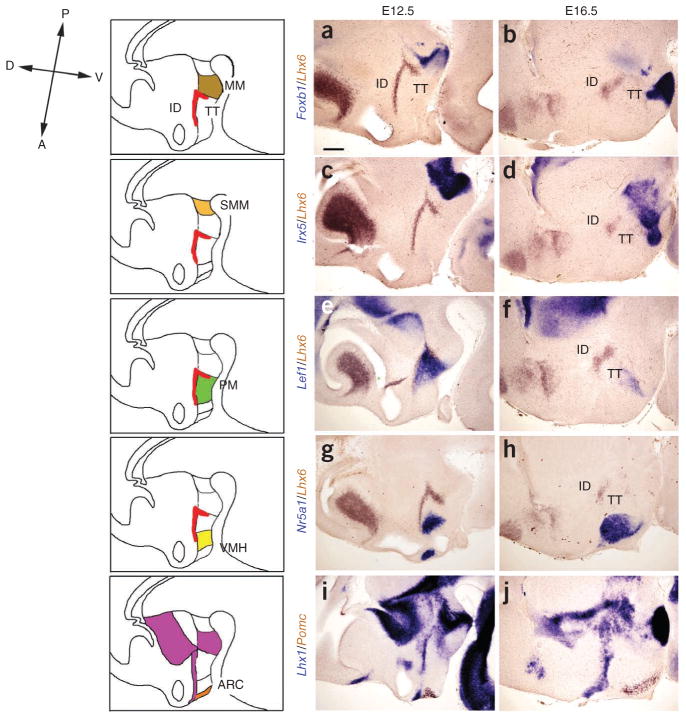
Figure 4.
Characterization of molecular markers of regional identity in posteroventral hypothalamus. Selective and nucleus-specific expression of markers is indicated at both E12.5 and E16.5. (a,b) Foxb1 (blue) marked the mamillary nucleus (MM), which was next to the Lhx6-expressing tuberomamillary terminal (brown) at E12.5 (a). Their spatial relationship was conserved at E16.5 (b). (c,d) Irx5 expression demarcated the supramamillary nucleus (SMM), which is located posterior to the tuberomamillary terminal marked by Lhx6 (brown) at E12.5 (c). A possible cell migration made the Irx5-expressing domain crossed the Lhx6-expressing tuberomamillary terminal at E16.5 (d). (e,f) Lef1 (blue) expression marked the premamillary (PM) region at E12.5 and E16.5, demonstrating a conserved domain of expression relative to the domain of Lhx6 expression (brown). (g,h) Nr5a1 was expressed in the ventromedial hypothalamic nucleus (VMH) at E12.5 and E16.5. (i,j) Pomc was expressed in the arcuate nucleus (ARC) at E12.5 and E16.5. To aid in the identification of these structures, we used Lhx6 as a second ISH color (brown) in a–h and Lhx1 (blue) for i,j. Scale bar represents 0.2 mm. ID, intrahypothalamic diagonal; TT, tuberomamillary terminal.
To more precisely delineate the prethalamic-hypothalamic border, we identified additional markers that selectively labeled the prethalamus. We found that prethalamic neuroepithelium could be further delineated by the markers Lhx9, which labels the ventral half of EmThal, and Olig2, which labels a posteroventral domain of prethalamus (Fig. 3h,i). These markers, in combination with Arx and Gdf10, can be used to fully mark prethalamic neuroepithelium and to demarcate subdivisions of the developing prethalamus along the neuraxis. In addition, we found that prethalamic progenitor cells and hypothalamic progenitor cells could be distinguished by two-color ISH of the transcription factors Gsh2 and Rax (Fig. 3k). We found that Sim1, which has previously been shown to selectively mark developing paraventricular hypothalamic nucleus (PVH)10, was expressed in a domain residing between prethalamus and optic recess (Fig. 3l) and straddling the border of telencephalon and diencephalon, as defined by Foxg1 expression (Fig. 3l). Immediately anteroventral to the Sim1-expressing domain, we detected a domain of diencephalic neuroepithelium that was selectively marked by Nkx6.2 in the Arx-negative region (Fig. 3m). We did not identify any markers that selectively labeled ventral telencephalic preoptic neuroepithelium, but this domain can nonetheless be defined as expressing Foxg1 (Fig. 2e), but not Sim1 and Nkx6.2 (Fig. 3l,m).
Molecular markers for developing ventral hypothalamus
To unambiguously identify the position of tuberomamillary terminal and intrahypothalamic diagonal relative to ventral hypothalamic nuclei, we conducted two-color ISH analysis of Lhx6 with additional markers (Fig. 4). We first analyzed the expression of Lhx6 and Foxb1 at E12.5, which selectively marks mamillary neuroepithelium12,13 and found that the tuberomamillary terminal was anterior to this zone (Fig. 4a). The relative position of these two markers was maintained at later stages of development after neurogenesis was complete (E16.5) (Fig. 4b and Supplementary Fig. 4). Irx5 expression similarly demarcated the supramamillary nucleus (SMM), which is located posterior to the mamillary neuroepithelium (Fig. 4c,d). Lef1, on the other hand, selectively labeled premamillary neuroepithelium, which lies immediately anterior to mamillary neuroepithelium, at E12.5 and also labeled premamillary neuroepithelium neurons after the completion of hypothalamic neurogenesis (Fig. 4e,f). Lhx6 expression overlapped with Lef1 expression in the tuberomamillary terminal, and Lhx6 expression in intrahypothalamic diagonal marked the dorsal domain of Lef1-positive premamillary neuroepithelium (data not shown). Finally, we compared the expression of Nr5a1 and pro-opiomelanocortin (Pomc), which delineate ventromedial hypothalamic nucleus (VMH)14 and arcuate nucleus (ARC)38, respectively. Neither marker overlapped with the intrahypothalamic diagonal or tuberomamillary terminal (Fig. 4g and data not shown) and Pomc, which is expressed directly ventroposterior to the zone of Lhx1 expression at both E12.5 and E16.5 (Fig. 4i,j). These data provide molecular landmarks that can be used to readily distinguish between functionally distinct nuclei during early development of the ventrobasal hypothalamus.
To further characterize the cell types of the intrahypothalamic diagonal and tuberomamillary terminal, we conducted two-color ISH with Lhx6 and the inhibitory interneuron marker Gad67. We found that Lhx6 and Gad67 expression fully overlapped at all of the developmental stages that we examined (data not shown). Several other Lhx family members were also expressed in distinct domains in the intrahypothalamic diagonal, as defined by Gad67 and Lhx6 expression (Fig. 5). Because Lhx9 was expressed ventral to Arx (Fig. 3i), we compared its expression with Lhx6 and found that Lhx9 was expressed immediately ventral to the Lhx6-positive zone of the intrahypothalamic diagonal (Fig. 5a). We next determined that Lhx8 was expressed immediately anterior to the Lhx6-positive region of the intrahypothalamic diagonal (Fig. 5d). However, although the size of each gene expression domain was changed, the relative positions of each gene expression domain were maintained, even at E16.5 (Fig. 5b,e). Lhx8 was expressed in a zone anterodorsal to Lhx9 at E12.5 (Fig. 5g) and its expression anterior to Lhx6 was preserved at E16.5 (Fig. 5h). Lhx1 expression overlapped that of Lhx8 at E12.5 and extended anterior from the zone of Lhx6 expression (Fig. 5k). These Lhx-expressing regions therefore appear to define a series of nuclei residing both between and lateral to the paraventricular nucleus and VMH. To further characterize these Lhx-positive nuclei, we confirmed that the anterior expression domain of Lhx1 delineated the suprachiasmatic nucleus (SCN), as indicated by Rorα expression39. At E16.5, Lhx1 was clearly expressed in both suprachiasmatic nucleus and in a region immediately posterodorsal to the SCN that probably represents the ventral subparaventricular zone40 (Fig. 5j,n).
Figure 5.
Lhx family members delineate discrete regions of the developing hypothalamus. Schematic figures indicate the patterns of Lhx gene expression at E12.5 and E16.5. (a,b) Lhx6 (blue) was expressed in the posterior portion of the intrahypothalamic diagonal immediately dorsal to the zone of Lhx9 expression (brown) (a). This pattern of Lhx6 and Lhx9 expression was preserved at E16.5 (b). (d,e) Lhx8 (blue) was expressed anterior to the Lhx6-positive region of the intrahypothalamic diagonal (brown). (g,h) Lhx8 (blue) was expressed anterior and dorsal to Lhx9 expression (brown). (k,l) Lhx1 (blue) was expressed anterior and dorsal to Lhx9 expression (brown), but overlapped with Lhx8 and the anterior domain of Lhx6 expression (data not shown). (c,f,i,m) Postnatal expression patterns of intrahypothalamic diagonal–expressed Lhx6 overlapped with Cart. A coronal section is shown in c, indicating that there is a conserved spatial relationship in lateral hypothalamic expression of Lhx9 (purple) and Lhx6 (brown) at P6. Lhx6-positive cells (brown) were found medial and dorsal to galanin (Gal)-positive neurons (purple) (f). Lhx6-positive cells (brown) were found medial and dorsal to hypocretin (Hcrt)-positive neurons (blue) (i). Lhx8 (brown) was expressed medial to Cart (blue) in dorsomedial hypothalamic nucleus (m). Schematic drawings indicating expression of neuropeptides and Lhx genes in postnatal hypothalamus, along with regions of overlap, are shown at the top of each column. (j,n) The anterior terminus of the zone of Lhx1 expression corresponded to the Rorα-positive SCN at E16.5. However, expression also extended posterodorsal from the SCN into the ventral subparaventricular zone (yellow arrow). Scale bar represents 0.2 mm (a, b, d, e, g, h, k and l) and 0.3 mm (c, f, i, j, m and n).
We examined the expression of markers of anterior, dorsomedial and lateral hypothalamic neuronal subtypes in conjunction with each Lhx gene at P6. Lhx6 was coexpressed with Cart-expressing cells in the lateral and dorsomedial hypothalamus (Fig. 5c,m,j). Lhx9, on the other hand, was coexpressed with a subset of hypocretin-positive cells and galanin-positive cells in the lateral hypothalamus (Fig. 5c,f,i), but was absent from the more laterally located Pmch-positive cells in this region (data not shown). This expression pattern persisted at E16.5 and P6. Lhx8 was expressed in the central region of the dorsomedial hypothalamic nucleus, a region that has been reported to be important for feeding-dependent entrainment of circadian rhythms41.
To further confirm that the marker genes that we identified are selectively and persistently expressed over the full course of hypothalamic neurogenesis, we examined the expression of all of the nucleus-specific markers described above at E11.5, E14.5 and E16.5 using two-color ISH for the marker in question and Shh. We found the same gene expression patterns seen for these markers at E12.5 at E11.5, E14.5 and E16.5 (Supplementary Figs. 4 and 5). We conclude that use of these markers can unambiguously identify functionally distinct subregions of hypothalamus throughout the course of neurogenesis.
Basal hypothalamic Shh patterns the anterior hypothalamus
Although Shh expression is confined to the ventral portion of the neural tube along most of the neuraxis, we found a more dorsally located stripe of Shh expression in the hypothalamus by E9.5 (ref. 30) (Fig. 3a). Several previous studies have addressed the role of Shh in regulating hypothalamic development7,42. However, these studies did not dissociate the contributions of basal plate– and Zli-derived Shh to the development of the prethalamus and hypothalamus, as Shh is expressed in both Zli and hypothalamus. We asked whether the markers that we identified could aid in interpreting the phenotype seen following the selective deletion of Shh from the basal plate domain of developing hypothalamus.
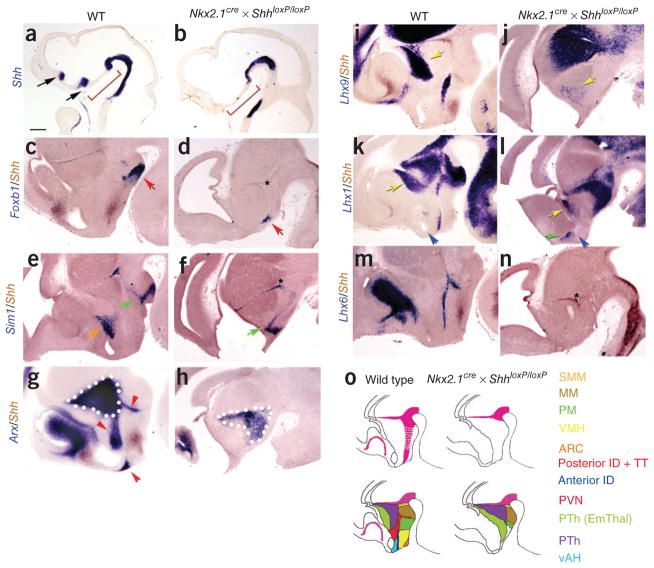
To selectively remove Shh from the hypothalamic basal plate, we generated Nkx2.1-cre × ShhloxP/loxP mice43,44. These mice showed complete deletion of Shh from the basal regions of both hypothalamus and telencephalon by E10.5, while Shh expression in more posterior regions and the Zli was unaffected (Fig. 6a,b and Supplementary Fig. 6). These mice show a notable thinning of both anteriotuberal hypothalamic and ventral telencephalic neuroepithelium by E12.5, along with a corresponding expansion of the third ventricle. Using the molecular markers that we characterized, we determined that these mice maintained expression of the mamillary neuroepithelium markers Foxb1 and Sim1 (Fig. 6c–f) and the SMM marker Irx5 (Supplementary Fig. 6). Furthermore, the prethalamic domains of expression of Arx, Lhx1, Lhx9 and Gsh2 are also preserved (Fig. 6g–l and Supplementary Fig. 6). This stands in stark contrast with previous studies, in which Shh deletion in the Zli led to a complete loss of prethalamic markers31,45,46. However, markers of tuberal and anterior hypothalamic nuclei were essentially absent. Lhx1, Lhx9 and Lhx6 were not detected in the intrahypothalamic diagonal and Lhx6 was not expressed in tuberomamillary terminal (Fig. 6i–n). Lef1 expression, which marks the premamillary neuroepithelium region, was markedly reduced (Supplementary Fig. 6). Nr5a1, Pomc and Nkx6.2 were not expressed (Supplementary Fig. 6). This does not, however, represent a change in the regional fate of anteriotuberal hypothalamic progenitors, as the telencephalic marker Foxg1 was not expanded into diencephalon and Rax expression was maintained, implying that these cells still retain their hypothalamic identity, but fail to undergo later stages of differentiation (Supplementary Fig. 6). Although preservation of posterior hypothalamic markers has previously been reported following Shh loss of function7,42,47, the precise source of the Shh signal that mediated this effect remains unclear. Furthermore, these studies reported different effects of Shh on the development of anterior and tuberal hypothalamic neurons, which may reflect a persistence of Shh expression in the ventral telencephalon in these studies7,42. Our data indicate that Shh derived from ventral forebrain neuroepithelium is essential for differentiation of the anterior and tuberal hypothalamus (Fig. 6o).
Figure 6.
Diencephalic phenotype of Nkx2.1-Cre × Shh loxP/loxP mice at E10.5 and E12.5, as indicated by analysis of region-specific marker genes. (a,b) Shh was not expressed in the basal hypothalamus at E10.5 in Nkx2.1-Cre × Shh loxP/loxP mice, as visualized by Shh probe (red bracket). (c,d) Expression of Foxb1 (blue) in mamillary neuroepithelium (red arrow) was preserved in Nkx2.1-Cre × Shh lox/lox mice. (e,f) Sim1 (blue) was not expressed in mamillary neuroepithelium (green arrow), but was not expressed in paraventricular nucleus (PVN) in mutant mice (orange arrow). (g,h) Arx was not expressed in the intrahypothalamic diagonal and tuberomamillary terminal (blue) in Nkx2.1-Cre × Shh lox/lox mice, but was expressed in the posterior prethalamic region. (i,j) Lhx9 (blue) was not expressed in ventral hypothalamus, but its expression in anterior prethalamus was expanded anteriorly (yellow arrow). (k,l) Lhx1 was not expressed in the intrahypothalamic diagonal, but it was expressed in anterior and posterior prethalamus, as well as in the mamillary hypothalamus. Lhx1 was expressed in a prominent anterior-located region (green arrow), but this region was dorsal to the optic recess (blue arrow) and resided in preoptic neuroepithelium. (m,n) Hypothalamic Lhx6 expression (blue) was absent in Nkx2.1-Cre × Shh lox/lox mice. Shh (brown) was expressed in Zli (asterix) and posterior to the diencephalon in mutant mice (d,f,n), but not in ventral diencephalon and telencephalon. (o) Schematic drawings of gene expression in wild-type and mutant mice with color combination. Scale bar represents 0.2 mm.
Other gene expression patterns in developing hippocampus
Many other genes were selectively expressed in one or more hypothalamic nuclei over the course of neurogenesis (see Supplementary Table 4 for a full scoring of all two-color ISH data and http://blackshaw.bs.jhmi.edu for all two-color ISH data; an example is shown for the developing SCN in Supplementary Fig. 7). After Lhx1 expression was initiated, multiple genes known to regulate neuronal development were sequentially expressed. Igfbp5 was expressed in progenitors throughout the anteroventral hypothalamus at this stage, becoming focally expressed in SCN precursors by E14.5, along with the transcription factors Id4 and Rorβ (Supplementary Fig. 7). The transcription factors Rorα and Nr1d1 and the axon guidance factor Sema6a were not detectably expressed until E16.5 (Supplementary Fig. 7 and data not shown).
The expression of other candidate developmental regulators did not follow obvious nuclear borders, but instead defined discrete domains of the developing hypothalamus and prethalamus that run parallel to the basal domain of Shh expression (Supplementary Fig. 8). For instance, at E11.5, Fgf15 and Dkk3 occupied a dorsal domain, which encompasses EmThal, prethalamus and PVH, whereas Nrg1 occupied a more ventral domain closer to the basal Shh domain. Wnt7a and Wnt7b were expressed throughout the prethalamus and PVH and in intrahypothalamic diagonal and tuberomamillary terminal, ventral to the basal Shh domain. Finally, Igfbp5 and Fst were expressed in intrahypothalamic diagonal, with their expression also partially overlapping the Shh-expressing basal plate.
We also identified genes that were broadly expressed only in early-stage hypothalamic progenitor cells. Such stage-specific expression patterns may be associated with conferring competence to generate early-born cell fates and with changes in cell cycle length over the course of neurogenesis, or with the probability of progenitors undergoing asymmetric division. When we examined the temporal expression pattern of these genes, we observed a much more rapid decrease in the level of these transcripts than we observed for more generic neuronal progenitor markers, such as Ccnd1 and the S phase–specific histone Hist3h2a (Supplementary Fig. 9). ISH analysis revealed that Foxd1, Igf2bp1, Rbpms2 and Shcbp1 mRNA were robustly expressed in the hypothalamic and prethalamic ventricular zone at E11.5, but that expression declined rapidly and was virtually absent by E14.5, at which point the expression of more generic progenitor markers was still readily detectable. Finally, we found that both Rax and Gpr50 were broadly and essentially exclusively expressed in hypothalamic progenitors at E14.5 (Supplementary Fig. 10), suggesting that either of these genes could be used to drive the expression of inducible transgenes for both widespread and selective disruption of gene function in the developing hypothalamus.
DISCUSSION
To the best of our knowledge, this is the first study to systematically characterize the gene expression pattern of the developing mammalian hypothalamus at the genomic level. We identified molecular markers that allowed us to create a molecular atlas of the developing hypothalamus. This gene expression atlas selectively delineated domains with hypothalamic neuroepithelium that correspond to specific nuclei in postnatal hypothalamus. Our data confirmed the previously proposed division between Sim1-positive anterodorsal hypothalamic neuroepithelium, which contain the primordial of the paraventricular nucleus, and the Nkx2.1-positive posteroventral hypothalamic neuroepithelium, which contains the primordial of VMH, ARC, premamillary neuroepithelium and mamillary neuroepithelium. Notably, our study also revealed that the preoptic area which was previously believed to be derived from ventral diencephalon, may be actually be derived from Foxg1-positive telencephalic neuroepithelium. However, from E14.5 on, scattered Foxg1-expressing cells are observed in anterior hypothalamus (data not shown). This suggests that cell migration may occur between the ventral telencephalic and diencephalic domains during later stages of development, as previous studies have indicated48.
Our study also identified a domain between these two regions that appeared to give rise to multiple nuclei in the dorsal and anterior hypothalamus. This region runs parallel to the hypothalamic zone of Shh expression and also overlaps with the previously described hypothalamic zone of Dlx2 expression49. In this region was a zone of Arx-positive, Gad67-positive cells that we term the intrahypothalamic diagonal and it was also defined by expression of Lhx6, Lhx8 and Lhx1. This domain appeared to later give rise to the inhibitory interneurons of the SCN, dorsomedial hypothalamic nucleus and posterior hypothalamus. Expression of Lhx6 overlapped with that of Cart-positive neurons of the dorsolateral postnatal hypothalamus. Immediately ventral to the posterior domain of the intrahypothalamic diagonal was another zone marked by selective expression of Lhx9, which, at least in part, gave rise to the Gal- and Hcrt-expressing cells of the lateral hypothalamus. In addition, Lhx6 expression extended ventral from the intrahypothalamic diagonal immediately anterior to mamillary neuroepithelium. We termed this domain, which also robustly expressed the Dlx genes, Arx and Gad67, the tuberomamillary terminal (Figs. 3 and 4 and data not shown). A dispersal of Arx- and Gad67-expressing cells from tuberomamillary terminal into developing premamillary and arcuate regions was observed during the course of neurogenesis (Supplementary Table 4 and data not shown) and it may be a point of origin for inhibitory interneurons in the ventrobasal hypothalamus.
Some of these genes guide the development of hypothalamic nuclei and serve as molecular markers that stably and selectively demarcate the major nuclei of the developing hypothalamus10,12,14. We expect that other such markers that we characterized, such as the Lhx genes, which delineate the intrahypothalamic diagonal and tuberomamillary terminal, may also have a role in patterning these structures. In addition to this set of well-characterized marker genes, we identified a large cohort of other genes that may also be important in hypothalamic patterning. We identified transcripts that were selectively activated at multiple different stages of development of every major hypothalamic nucleus (Fig. 4, Supplementary Fig. 4 and Supplementary Table 4). We identified multiple growth and differentiation factors that were expressed in discrete regions of hypothalamic neuroepithelium and genes that were selectively expressed in early hypothalamic progenitor cells, which may regulate developmental competence or proliferation potential. Finally, we identified genes that were broadly and selectively expressed in hypothalamic progenitors and that may prove useful for designing tools to manipulate hypothalamic gene function.
These data will provide tools for spatially and temporally restricted gain- and loss-of-function analysis of these candidate molecules in discrete domains of hypothalamic neuroepithelium and provide insight into the molecular pathways that guide development of individual hypothalamic neuronal subtypes. These studies may ultimately allow selective manipulation of the function of individual subtypes of hypothalamic neurons and a detailed analysis of their contribution to mammalian physiology and behavior.
ONLINE METHODS
Mice
C57Bl/6 and CD-1 timed pregnant mice were obtained from Charles River and Japan SLC, respectively. Midday of the day of discovery of a vaginal plug was considered to be E0.5. All experimental procedures using mice were approved by the RIKEN Institutional Animal Care Committee and the Johns Hopkins University Institutional Animal Care and Use Committee.
Tissue dissection, embryo genotypic and RNA purification
Timed pregnant mice and postnatal mice were killed via CO2 gas followed by decapitation. Embryos were decapitated and individually dissected in phosphate-buffered saline. For embryos, the neural tube was initially opened along the dorsal mid-line and heads were divided into individual halves. Tissue from hypothalamus and preoptic area was separated from the surrounding skull by microdissection following the anatomical landmarks shown in Figure 1c. Dissected tissue from individual mice was stored in RNAlater (Ambion). For embryonic samples, tissue was also isolated from the leg or tail region, genomic DNA was isolated, and sex was determined by PCR-based genotyping using a single primer pair (5′-GAG GCC CAG ACM AGA GKA AA-3′ and 5′-TAA CAC YAC CTY CTT CCA CC-3′) that amplified both the X chromosomal gene Kdm5c, producing a 417 base pair product, and the Y chromosomal gene Kdm5d, producing a 380 base pair product. The sex of postnatal mice was determined by genital examination. Following determination of sex, dissected tissue from a minimum of six individuals from each sex and genotype was pooled for RNA isolation using RNEasy (Qiagen).
Analysis of microarray data
To identify sexually dimorphic pattern and strain-specific difference of gene expression that occur during the developmental time course, we performed microarray experiments using mouse 430_2.0 Genome Genechip arrays (Affymetrix). We collected samples from 12 developmental stages: E10.5, E11.5, E12.5, E13.5, E14.5, E15.5, E16.5, E17.5, E18.5, P0.5, P21 and P42. At each stage, samples were collected from CD-1 and C57Bl/6 mice, with a single sample from male and female mice of each strain analyzed for each time point. This represents a total of 48 microarray hybridizations, which were performed at the Johns Hopkins Microarray Core Facility, using standard protocols.
Hybridization intensity raw data (*.CEL files) were uploaded to the R-Project Bioconductor statistical tools package. Normalized gene expression values were generated for each array chip by RMA method in Bioconductor Affymetrix package. RMA method consists of three steps: background correction, quantile normalization and final summarization. We then applied Bioconductor ‘limma’ package software, which utilizes linear modeling to calculate moderated t statistics for computing fold changes and adjusted P values for each gene and each comparison between groups. The false discovery rate was controlled as previously described50. Comparing two different strains yielded larger numbers of differentially expressed transcripts; therefore, we applied a twofold cutoff ratio to the data as an additional filter. We then applied K-mean (n = 5) method for clustering analysis to both sex- and strain-specific probe sets.
To identify classes of coexpressed genes that are enriched for biologically relevant categories of hypothalamic genes, we performed a time course analysis of the developmental data in which all four replicates are included and sex/strain-specific differences excluded. We first applied ‘limma’ package for sliding time window comparisons to identify the developmental timing group. All differential expressed gene lists were generated at 1% false discover rate and twofold change. Comparing these gene lists between different time frame groups, we used E10–11, E12–14, E15–16, E17–18 to P0 and P21–42 as time frame combinations. We then applied moderated F statistics to select the differential expressed genes during these five time points.
ISH
The first-pass single-color ISH analysis was performed essentially as described previously29. The accession numbers of the DNA templates used for probe generation and the genomic sequence (mm9 assembly) that was amplified by PCR for template generation are listed in Supplementary Table 3. For the two-color section ISH analysis, maternal animals were anesthetized and embryos were immediately dissected out. The brains were removed, fixed overnight in 30% sucrose/4% paraformaldehyde (vol/vol) and sectioned in the coronal plane on a Leica sledge microtome at 40 μm. Sections were individually mounted on slides and processed for ISH to visualize expression of various genes. Single- or two-color nonradioactive ISH was performed using previously described method with some modifications32. Glass slides with sections were fixed by 4% paraformaldehyde and treated with proteinase K (Roche). Sections were then fixed again and hybridized with digoxigenin-labeled or fluorescein-labeled probes (Roche) at 70 °C overnight. Excess probes were washed out and sections were blocked with lamb serum and incubated with solution containing alkaline phosphatase–conjugated with digoxigenin-labeled or fluorescein-labeled antibodies (Roche). Color was developed with combinations of the chromagens nitroblue tetrazolium (Nacalai, 350 mg ml−1) and 5-bromo, 4-chloro, 3-indolylphosphate for blue staining or tetranitroblue tetrazolium (Research Organics, 350 mg ml−1) and 5-bromo, 4-chloro, 3-indolylphosphate for brown staining. The probes used for this analysis that gave good signal using two-color ISH are listed in Supplementary Table 4, whereas probes that were tested, but did not give good signal, are included in Supplementary Table 3. All two-color ISH data using candidate genes and Shh can be accessed at http://blackshaw.bs.jhmi.edu and in the Mouse Gene Expression Database at Jackson Labs (http://www.informatics.jax.org/expression.shtml).
Supplementary Material
Acknowledgments
We would like to thank C.-M. Fan for guidance concerning dissection of embryonic hypothalamus and S. Hattar, A. Moore and B. Nelson for comments on the manuscript. This work was supported by the RIKEN Brain Science Institute and Human Frontier Science Project (T.S.), a National Science Foundation Predoctoral Fellowship (D.A.L.), a Basil O’Connor Starter Scholar Award from the March of Dimes (S.B.) and an award from the Klingenstein Fund (S.B.). S.B. is a W.M. Keck Distinguished Young Investigator in Medical Science.
Footnotes
AUTHOR CONTRIBUTIONS
S.B. conceived the study. L.Q. and L.J. dissected tissue, purified RNA and conducted microarray analysis. Y.Y. and J.Q. analyzed microarray data. D.A.L., A.M.-A., L.J., H.W. and M.A. carried out first-pass single-color ISH analysis. A.K., A.C.Y., H.M., L.J. and H.W. carried out the two-color ISH analysis. T.S. and S.B. contributed to the experimental design, supervised experiments, discussed the results and wrote the paper.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Accession codes. All microarray data reported here has been deposited in GEO and is available as GEO accession no. GSE21278. All two-color in situ hybridization data are deposited in the Jackson Labs Mouse Gene Expression Database and are available at http://informatics.jax.org/ as accession ID J:157819. This data is also available for download at http://blackshaw.bs.jhmi.edu.
Reprints and permissions information is available online at http://www.nature.com/reprintsandpermissions/.
Note: Supplementary information is available on the Nature Neuroscience website.
References
- 1.O’Rahilly S. Human genetics illuminates the paths to metabolic disease. Nature. 2009;462:307–314. doi: 10.1038/nature08532. [DOI] [PubMed] [Google Scholar]
- 2.Caqueret A, Yang C, Duplan S, Boucher F, Michaud JL. Looking for trouble: a search for developmental defects of the hypothalamus. Horm Res. 2005;64:222–230. doi: 10.1159/000088977. [DOI] [PubMed] [Google Scholar]
- 3.Fyffe SL, et al. Deletion of Mecp2 in Sim1-expressing neurons reveals a critical role for MeCP2 in feeding behavior, aggression and the response to stress. Neuron. 2008;59:947–958. doi: 10.1016/j.neuron.2008.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McGill BE, et al. Enhanced anxiety and stress-induced corticosterone release are associated with increased Crh expression in a mouse model of Rett syndrome. Proc Natl Acad Sci USA. 2006;103:18267–18272. doi: 10.1073/pnas.0608702103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee JE, Wu SF, Goering LM, Dorsky RI. Canonical Wnt signaling through Lef1 is required for hypothalamic neurogenesis. Development. 2006;133:4451–4461. doi: 10.1242/dev.02613. [DOI] [PubMed] [Google Scholar]
- 6.Dale JK, et al. Cooperation of BMP7 and SHH in the induction of forebrain ventral midline cells by prechordal mesoderm. Cell. 1997;90:257–269. doi: 10.1016/s0092-8674(00)80334-7. [DOI] [PubMed] [Google Scholar]
- 7.Manning L, et al. Regional morphogenesis in the hypothalamus: a BMP-Tbx2 pathway coordinates fate and proliferation through Shh downregulation. Dev Cell. 2006;11:873–885. doi: 10.1016/j.devcel.2006.09.021. [DOI] [PubMed] [Google Scholar]
- 8.Ohyama K, Das R, Placzek M. Temporal progression of hypothalamic patterning by a dual action of BMP. Development. 2008;135:3325–3331. doi: 10.1242/dev.027078. [DOI] [PubMed] [Google Scholar]
- 9.Shimada M, Nakamura T. Time of neuron origin in mouse hypothalamic nuclei. Exp Neurol. 1973;41:163–173. doi: 10.1016/0014-4886(73)90187-8. [DOI] [PubMed] [Google Scholar]
- 10.Michaud JL, Rosenquist T, May NR, Fan CM. Development of neuroendocrine lineages requires the bHLH-PAS transcription factor SIM1. Genes Dev. 1998;12:3264–3275. doi: 10.1101/gad.12.20.3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schonemann MD, et al. Development and survival of the endocrine hypothalamus and posterior pituitary gland requires the neuronal POU domain factor Brn-2. Genes Dev. 1995;9:3122–3135. doi: 10.1101/gad.9.24.3122. [DOI] [PubMed] [Google Scholar]
- 12.Labosky PA, et al. The winged helix gene, Mf3, is required for normal development of the diencephalon and midbrain, postnatal growth and the milk-ejection reflex. Development. 1997;124:1263–1274. doi: 10.1242/dev.124.7.1263. [DOI] [PubMed] [Google Scholar]
- 13.Wehr R, Mansouri A, de Maeyer T, Gruss P. Fkh5-deficient mice show dysgenesis in the caudal midbrain and hypothalamic mammillary body. Development. 1997;124:4447–4456. doi: 10.1242/dev.124.22.4447. [DOI] [PubMed] [Google Scholar]
- 14.Davis AM, et al. Loss of steroidogenic factor 1 alters cellular topography in the mouse ventromedial nucleus of the hypothalamus. J Neurobiol. 2004;60:424–436. doi: 10.1002/neu.20030. [DOI] [PubMed] [Google Scholar]
- 15.Acampora D, et al. Progressive impairment of developing neuroendocrine cell lineages in the hypothalamus of mice lacking the Orthopedia gene. Genes Dev. 1999;13:2787–2800. doi: 10.1101/gad.13.21.2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goshu E, et al. Sim2 contributes to neuroendocrine hormone gene expression in the anterior hypothalamus. Mol Endocrinol. 2004;18:1251–1262. doi: 10.1210/me.2003-0372. [DOI] [PubMed] [Google Scholar]
- 17.Gray PA, et al. Mouse brain organization revealed through direct genome-scale TF expression analysis. Science. 2004;306:2255–2257. doi: 10.1126/science.1104935. [DOI] [PubMed] [Google Scholar]
- 18.Alvarez-Bolado G, Eichele G. Analysing the developing brain transcriptome with the GenePaint platform. J Physiol (Lond ) 2006;575:347–352. doi: 10.1113/jphysiol.2006.112763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elmquist JK, Elias CF, Saper CB. From lesions to leptin: hypothalamic control of food intake and body weight. Neuron. 1999;22:221–232. doi: 10.1016/s0896-6273(00)81084-3. [DOI] [PubMed] [Google Scholar]
- 20.Charlton H. Hypothalamic control of anterior pituitary function: a history. J Neuroendocrinol. 2008;20:641–646. doi: 10.1111/j.1365-2826.2008.01718.x. [DOI] [PubMed] [Google Scholar]
- 21.Kruk MR, et al. The hypothalamus: cross-roads of endocrine and behavioural regulation in grooming and aggression. Neurosci Biobehav Rev. 1998;23:163–177. doi: 10.1016/s0149-7634(98)00018-9. [DOI] [PubMed] [Google Scholar]
- 22.Saper CB, Scammell TE, Lu J. Hypothalamic regulation of sleep and circadian rhythms. Nature. 2005;437:1257–1263. doi: 10.1038/nature04284. [DOI] [PubMed] [Google Scholar]
- 23.Ulrich-Lai YM, Herman JP. Neural regulation of endocrine and autonomic stress responses. Nat Rev Neurosci. 2009;10:397–409. doi: 10.1038/nrn2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Büdefeld T, Grgurevic N, Tobet SA, Majdic G. Sex differences in brain developing in the presence or absence of gonads. Dev Neurobiol. 2008;68:981–995. doi: 10.1002/dneu.20638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tobet S, et al. Brain sex differences and hormone influences: a moving experience? J Neuroendocrinol. 2009;21:387–392. doi: 10.1111/j.1365-2826.2009.01834.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brown AE, Mani S, Tobet SA. The preoptic area/anterior hypothalamus of different strains of mice: sex differences and development. Brain Res Dev Brain Res. 1999;115:171–182. doi: 10.1016/s0165-3806(99)00061-9. [DOI] [PubMed] [Google Scholar]
- 27.Peinado JR, et al. Strain-dependent influences on the hypothalamopituitary-adrenal axis profoundly affect the 7B2 and PC2 null phenotypes. Endocrinology. 2005;146:3438–3444. doi: 10.1210/en.2004-1289. [DOI] [PubMed] [Google Scholar]
- 28.Rinn JL, Snyder M. Sexual dimorphism in mammalian gene expression. Trends Genet. 2005;21:298–305. doi: 10.1016/j.tig.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 29.Blackshaw S, et al. Genomic analysis of mouse retinal development. PLoS Biol. 2004;2:e247. doi: 10.1371/journal.pbio.0020247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chiang C, et al. Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function. Nature. 1996;383:407–413. doi: 10.1038/383407a0. [DOI] [PubMed] [Google Scholar]
- 31.Kiecker C, Lumsden A. Hedgehog signaling from the ZLI regulates diencephalic regional identity. Nat Neurosci. 2004;7:1242–1249. doi: 10.1038/nn1338. [DOI] [PubMed] [Google Scholar]
- 32.Kataoka A, Shimogori T. Fgf8 controls regional identity in the developing thalamus. Development. 2008;135:2873–2881. doi: 10.1242/dev.021618. [DOI] [PubMed] [Google Scholar]
- 33.Grove EA, Tole S, Limon J, Yip L, Ragsdale CW. The hem of the embryonic cerebral cortex is defined by the expression of multiple Wnt genes and is compromised in Gli3-deficient mice. Development. 1998;125:2315–2325. doi: 10.1242/dev.125.12.2315. [DOI] [PubMed] [Google Scholar]
- 34.Herrera E, et al. Foxd1 is required for proper formation of the optic chiasm. Development. 2004;131:5727–5739. doi: 10.1242/dev.01431. [DOI] [PubMed] [Google Scholar]
- 35.Hatini V, Tao W, Lai E. Expression of winged helix genes, BF-1 and BF-2, define adjacent domains within the developing forebrain and retina. J Neurobiol. 1994;25:1293–1309. doi: 10.1002/neu.480251010. [DOI] [PubMed] [Google Scholar]
- 36.Pera EM, Kessel M. Demarcation of ventral territories by the homeobox gene NKX2.1 during early chick development. Dev Genes Evol. 1998;208:168–171. doi: 10.1007/s004270050170. [DOI] [PubMed] [Google Scholar]
- 37.Miura H, Yanazawa M, Kato K, Kitamura K. Expression of a novel aristaless related homeobox gene Arx in the vertebrate telencephalon, diencephalon and floor plate. Mech Dev. 1997;65:99–109. doi: 10.1016/s0925-4773(97)00062-2. [DOI] [PubMed] [Google Scholar]
- 38.Watson SJ, Barchas JD, Li CH. beta-Lipotropin: localization of cells and axons in rat brain by immunocytochemistry. Proc Natl Acad Sci USA. 1977;74:5155–5158. doi: 10.1073/pnas.74.11.5155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ino H. Immunohistochemical characterization of the orphan nuclear receptor ROR alpha in the mouse nervous system. J Histochem Cytochem. 2004;52:311–323. doi: 10.1177/002215540405200302. [DOI] [PubMed] [Google Scholar]
- 40.Moga MM, Moore RY. Organization of neural inputs to the suprachiasmatic nucleus in the rat. J Comp Neurol. 1997;389:508–534. doi: 10.1002/(sici)1096-9861(19971222)389:3<508::aid-cne11>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 41.Fuller PM, Lu J, Saper CB. Differential rescue of light- and food-entrainable circadian rhythms. Science. 2008;320:1074–1077. doi: 10.1126/science.1153277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Szabó NE, et al. Role of neuroepithelial Sonic hedgehog in hypothalamic patterning. J Neurosci. 2009;29:6989–7002. doi: 10.1523/JNEUROSCI.1089-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dassule HR, Lewis P, Bei M, Maas R, McMahon AP. Sonic hedgehog regulates growth and morphogenesis of the tooth. Development. 2000;127:4775–4785. doi: 10.1242/dev.127.22.4775. [DOI] [PubMed] [Google Scholar]
- 44.Xu Q, Tam M, Anderson SA. Fate mapping Nkx2.1-lineage cells in the mouse telencephalon. J Comp Neurol. 2008;506:16–29. doi: 10.1002/cne.21529. [DOI] [PubMed] [Google Scholar]
- 45.Scholpp S, Wolf O, Brand M, Lumsden A. Hedgehog signaling from the zona limitans intrathalamica orchestrates patterning of the zebrafish diencephalon. Development. 2006;133:855–864. doi: 10.1242/dev.02248. [DOI] [PubMed] [Google Scholar]
- 46.Szabó NE, Zhao T, Zhou X, Alvarez-Bolado G. The role of Sonic hedgehog of neural origin in thalamic differentiation in the mouse. J Neurosci. 2009;29:2453–2466. doi: 10.1523/JNEUROSCI.4524-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mathieu J, Barth A, Rosa FM, Wilson SW, Peyrieras N. Distinct and cooperative roles for Nodal and Hedgehog signals during hypothalamic development. Development. 2002;129:3055–3065. doi: 10.1242/dev.129.13.3055. [DOI] [PubMed] [Google Scholar]
- 48.Tobet SA, Paredes RG, Chickering TW, Baum MJ. Telencephalic and diencephalic origin of radial glial processes in the developing preoptic area/anterior hypothalamus. J Neurobiol. 1995;26:75–86. doi: 10.1002/neu.480260107. [DOI] [PubMed] [Google Scholar]
- 49.Porteus MH, et al. DLX-2, MASH-1, and MAP-2 expression and bromodeoxyuridine incorporation define molecularly distinct cell populations in the embryonic mouse forebrain. J Neurosci. 1994;14:6370–6383. doi: 10.1523/JNEUROSCI.14-11-06370.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hochberg Y, Benjamini Y. More powerful procedures for multiple significance testing. Stat Med. 1990;9:811–818. doi: 10.1002/sim.4780090710. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.