Abstract
When inhibitors of enzymes that utilize isoprenoid pyrophosphates are based on the natural substrates, a significant challenge can be to achieve selective inhibition of a specific enzyme. One element in the design process is the stereochemistry of the isoprenoid olefins. We recently reported preparation of a series of isoprenoid triazoles as potential inhibitors of geranylgeranyl transferase II but these compounds were obtained as a mixture of olefin isomers. We now have accomplished the stereoselective synthesis of these triazoles through the use of epoxy azides for the cycloaddition reaction followed by regeneration of the desired olefin. Both geranyl and neryl derivatives have been prepared as single olefin isomers through parallel reaction sequences. The products were assayed against multiple enzymes as well as in cell culture studies and surprisingly a Z-olefin isomer was found to be a potent and selective inhibitor of geranylgeranyl diphosphate synthase.
Keywords: Isoprenoid biosynthesis, inhibition, GGDP synthase, olefin stereochemistry
1. Introduction
Biosynthesis of isoprenoids in mammals and higher plants proceeds through a series of enzymes that converts 3-hydroxy-3-methylglutaryl-coenzyme A (HMG-CoA) to products with larger hydrocarbon chains including farnesyl pyrophosphate (FPP) and geranylgeranyl pyrophosphate (GGPP). These key compounds are elaborated to a constellation of more complex derivatives, including sesquiterpenes, diterpenes, steroids, and dolichols among many others.1 Furthermore, both FPP and GGPP also are employed in post-translational prenylation reactions mediated by farnesyl transferase (FTase) and geranylgeranyl transferase (GGTase) I and II. This processing is particularly important to proteins of the Ras superfamily, because their function requires membrane localization that is brought about by incorporation of one or more hydrophobic isoprenoid chains.
Given the importance of isoprenoid biosynthesis to metabolism, it is not surprising that these enzymes are the proximate targets of therapeutic agents employed for treatment of a number of important diseases. For example, hypercholesterolemia often is treated by administration of statins that inhibit HMG-CoA reductase, and osteoporosis commonly is treated with bisphosphonates that inhibit farnesyl diphosphate synthase (FDPS). There has been increasing interest in geranylgeranyl diphosphate synthase (GGDPS) inhibitors because of their anti-proliferative effects and as potential anti-malarial agents.2–4 Inhibitors of FTase have been investigated extensively as potential chemotherapeutic agents by virtue of their anti-Ras activity.5
While there have been a select number of studies of FTase inhibitors that are modeled after the isoprenoid substrates,6–8 the FTase inhibitors which have been clinically investigated have been derived through strategies such as peptide mimics or bisubstrate analogues.9 The latter approaches, while feasible for FTase and GGTase I, have not been possible for GGTase II, as this enzyme does not directly recognize its protein substrate. Instead GGTase II “outsources” its substrate specificity to the Rab escort protein (REP) which delivers Rabs to the active site of the transferase.10
We have been interested in the strategy of inhibiting GGTase II in multiple myeloma cells to disrupt monoclonal protein secretion and induce apoptosis via the unfolded protein response pathway.11 There has also been increasing interest in Rabs in other malignancies, particularly with respect to the role that Rabs play in mediating tumor cell migration and invasion.12 We recently reported the preparation of a small set of triazole bisphosphonate salts represented by compound 1 (Figure 1), and found that incorporation of a longer isoprenoid chain on this heterocyclic template resulted in modest inhibitors of GGTase II.13 The synthesis was based on “click” chemistry of acetylene 214 with an isoprenoid derived azide. That approach allowed rapid assembly of the target compounds but it was limited in that a [3,3]-sigmatropic rearrangement of the allylic azides allows them to equilibrate and the cycloaddition reaction then afforded products that were mixtures of olefin isomers. For example, whether geranyl azide (3) or neryl azide (4) was employed, a 2:1 mixture of E- and Z-olefin isomers was obtained and these isomers were not readily separable.13 To support more detailed biological studies of isoprenoid triazoles we sought a synthetic route that would avoid the issue of olefin isomerization. In this manuscript we report development of a successful strategy through studies of geranyl and neryl epoxides, as well as the surprising biological results discovered with one of the new compounds.
Figure 1.
Synthesis of isoprenoid triazoles via click chemistry.
2. Synthesis
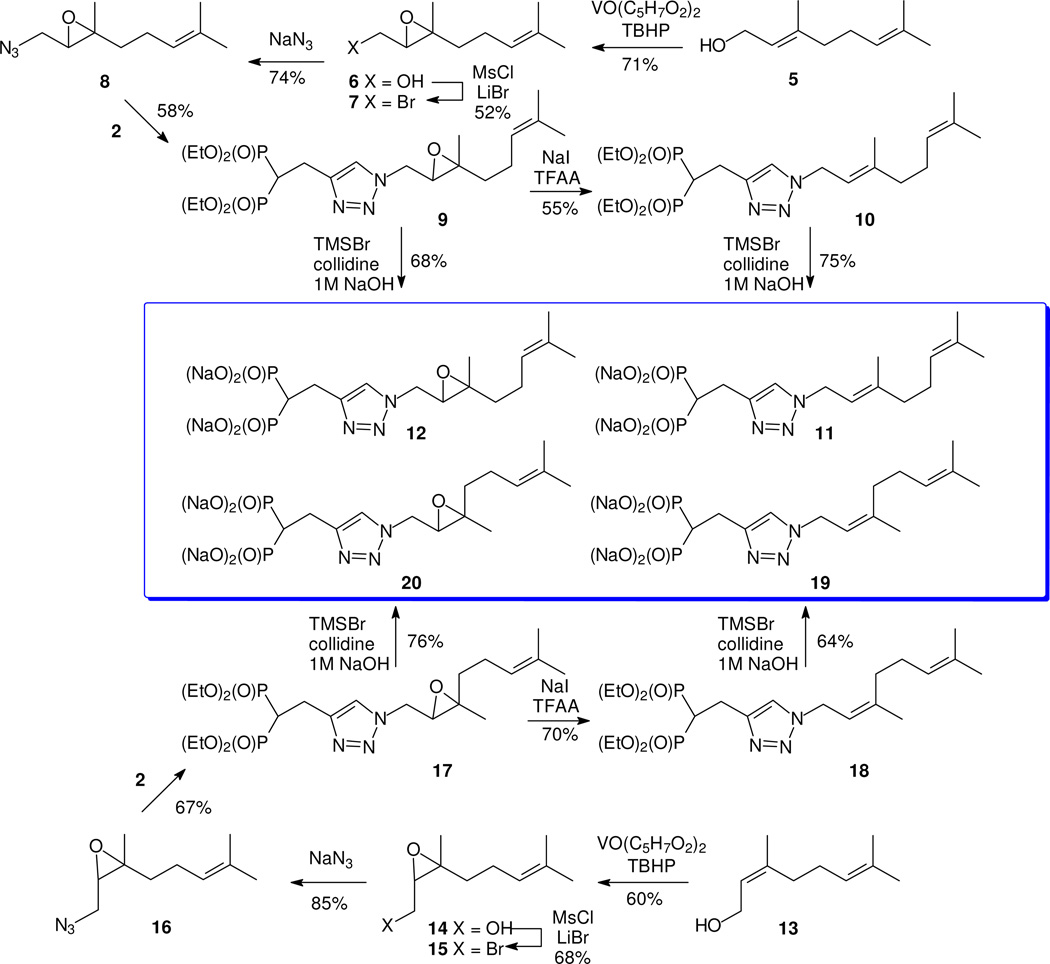
Given that the equilibration of geranyl (3) and neryl azides (4) is a sigmatropic process, an apparent way to circumvent the rearrangement is to protect the C-2 olefin in a way that allows stereocontrolled regeneration after formation of the azide and the cycloaddition. While there may be a number of viable strategies, we were intrigued by the possibility of epoxidation. Regiospecific introduction of the epoxide is well established.15 The only concern might be regeneration of the olefin with stereocontrol, especially in the presence of the triazole ring and the bisphosphonate moiety.
To begin exploration of this strategy, geraniol (5) was converted regioselectively to the corresponding epoxide 6 (Figure 2).15, 16 While material prepared in this way is racemic, because this intermediate was viewed primarily as a protected olefin control of the absolute stereochemistry was of no lasting concern. More importantly, only one regioisomer of this epoxide was detectable. After formation of bromide 716,17 via the mesylate of alcohol 6, reaction with NaN3 gave the expected product 8. Out of concern for the stability of this azide, after minimal purification it was used immediately in a cycloaddition with acetylene 2. This click reaction proceeded smoothly to afford the expected triazole 9. Attempted reaction of this triazole epoxide with TMSCl and NaI under the literature conditions18 gave no sign of an olefin product and instead returned only unreacted starting material. However when treated with NaI and trifluoroacetic anhydride (TFAA) under the conditions of Sonnet,19 which is believed to result in formation of trifluoroacetyl iodide in situ, epoxide 9 was converted to a single olefin. This product was assigned as the E-isomer 10 on the basis of its NMR data, and those spectral data matched that of the major isomer formed through reaction of geranyl (3) or neryl (4) azide with acetylene 2.13 Perhaps the most diagnostic signal was that observed at 39.6 ppm in the 13C NMR spectrum, corresponding to the methylene group adjacent to the more substituted carbon of newly formed olefin. This is very close to the value reported for geraniol (39.7), and quite different from that reported for nerol (32.2).20
Figure 2.
Synthesis of geranyl and neryl triazole bisphosphonates.
Once bisphosphonate ester 10 was in hand, hydrolysis under standard conditions afforded the salt 11. While this salt was the primary objective of these efforts, intermediate 9 also was viewed as a potential source of biological activity if the epoxide could be preserved through the course of phosphonate ester hydrolysis. Standard conditions for ester hydrolysis appeared to result in diol formation, but brief treatment with NaOH after reaction with TMSBr and collidine gave the new epoxide triazole salt 12. Assay of this second bisphosphonate allows a direct comparison with the activity of the corresponding olefin 11.
After preparation of the geranyl derivatives 11 and 12, it was of interest to attempt a parallel series of reactions starting with nerol, to allow determination of activity as a function of the olefin or epoxide stereochemistry. Therefore nerol (13) was converted to the azido expoxide 16 through the intermediate epoxides 1416 and 15.17 The click reaction of azide 16 and acetylene 2 proceeded smoothly to give triazole 17. Treatment of this epoxide isomer with NaI and TFAA also gave a single olefin product, identified as the Z-isomer 18 by comparison with the pure E-isomer 10 or the minor component of the isomer mixture prepared earlier,13 which confirms that the reduction itself is a stereospecific process with these isoprenoid olefins. Again, a diagnostic signal was observed in the 13C NMR spectrum at 32.2 ppm representing the methylene group adjacent to the newly formed olefin. This is equal to the value reported for that carbon in nerol (32.2), and quite different from that reported for geraniol (39.7).20 Finally both esters 18 and 17 were converted to the corresponding bisphosphonate salts (19 and 20, respectively).
3. Biological results and discussion
Our preliminary studies with compound 1 revealed that it disrupts protein geranylgeranylation in intact cells (unpublished data). To determine whether this activity is a consequence of the geranyl and/or neryl stereochemistry, and to begin to determine the underlying mechanism of action, a series of western blot experiments was performed. Rap1a is a substrate of GGTase I while Rab6 is a substrate of GGTase II. For detection of Rap1a, whole cell lysate was prepared and an antibody that detects only unmodified Rap1a was utilized. For Rab6, a Triton X-114 lysis was employed to generate aqueous and detergent fractions. Under control conditions, the majority of Rab6 is found in the detergent (membrane) fraction, while in the setting of an agent which disrupts Rab geranylgeranylation, Rab6 becomes localized to the aqueous (cytosolic) fraction. The known GGDPS inhibitor21 digeranyl bisphosphonate (DGBP)22 was used as a positive control. As shown in Figure 3A, treatment with DGBP results in the accumulation of unmodified Rap1a and Rab6. A similar pattern was displayed upon treatment with compound 1 which is a mixture of olefin isomers. Evaluation of the individual isomers revealed that the geranyl isomer 11 only weakly disrupted Rap1a geranylgeranylation while the neryl isomer 19 effectively disrupted both Rap1a and Rab6 geranylation. Interestingly, neither of the epoxide derivatives (12 and 20) displayed activity in this assay, and none of the tested compounds disrupted Ras farnesylation (data not shown).
Figure 3.
Neryl triazole 19 potently disrupts protein geranylgeranylation in human myeloma cells. RPMI-8226 cells were incubated for 48 hours in the presence or absence of test compounds. Cells were lysed using RIPA buffer to generate whole cell lysate or with Triton X-114 to generate aqueous and detergent fractions. Immunoblot analysis was performed. The Rap1a antibody detects only unmodified protein. β-tubulin was used as a loading control for whole cell lysate and aqueous fractions while calnexin was used the loading control for the detergent fraction. The gels are representative of two independent experiments. A) Cells were incubated in the presence or absence of 10 µM DGBP (positive control) or test compound. B) Cells were incubated in the presence or absence of increasing concentration (0.1, 1, 10 µM) of either DGBP, 11, or 19. C) Addition of GGPP, but not mevalonate (Mev) or FPP, prevents the effects of 19 on Rap1a geranylgeranylation. Cells were incubated in the presence or absence of inhibitor (10 µM lovastatin (Lov) or 10 µM 19) and/or isoprenoid (1 mM Mev, 10 µM FPP, or 10 µM GGPP).
To evaluate further the differences in potency between the two isomers, the geranyl and neryl triazoles were tested at varying concentrations and compared to the known GGDPS inhibitor DGBP. As shown in Figure 3B, accumulation of unmodified Rap1a was observed with 10 µM DGBP but not at lower concentrations. The geranyl isomer 11 only weakly disrupted Rap1a geranylgeranylation at 10 µM while unmodified Rap1a is detected following treatment with 1 µM of compound 19, indicating that the neryl stereochemistry confers a potency which is at least an order of magnitude greater than the geranyl isomer.
Add-back experiments in which cells were incubated in the presence of both compound 19 and either mevalonate, FPP, or GGPP, demonstrated that only GGPP prevents the accumulation of unmodified Rap1a that is induced by compound 19 (Figure 3C). This is in contrast to the results observed when cells were treated with the HMG-CoA reductase inhibitor lovastatin, where either mevalonate or GGPP could prevent lovastatin’s effects. In aggregate, these western blot studies are consistent with the hypothesis that the neryl isomer 19 disrupts protein geranylgeranylation via inhibition of GGDPS.
To address this hypothesis further, the effects of the geranyl and neryl triazoles on intracellular FPP and GGPP levels were examined. As shown in Figure 4, the isomer mixture represented by structure 1 (~2:1 E to Z) decreased GGPP levels and increased FPP levels, consistent with inhibition of GGDPS. This activity was a consequence of the neryl component (19) which even more potently decreased GGPP levels and increased FPP levels while the geranyl isomer did not significantly alter GGPP levels.
Figure 4.
Neryl triazole 19 depletes cells of GGPP and increases intracellular FPP levels. RPMI-8226 cells were incubated for 24 hours in the presence or absence of 1 µM test compound. DGBP was included as a positive control. Cells were counted and intracellular FPP/GGPP levels were measured. Data are expressed as percentage of control (mean ± SD, n=2). The * denotes p<0.05 per unpaired two-tailed t-test and compares treated cells to untreated control cells.
The group of triazole compounds was tested for the ability to directly inhibit GGDPS in an in vitro enzyme assay. As shown in Table 1, the neryl triazole 19 is the most potent inhibitor with an IC50 of 375 nM while the geranyl triazole 11 is approximately 40-fold less potent. To determine whether there was any cross-reactivity against FDPS or the prenyl transferase enzymes, the triazole phosphonates were tested against FDPS, FTase, GGTase I, and GGTase II. The FDPS studies revealed that compound 19 is over 200-fold less potent as an inhibitor against FDPS than it is against GGDPS. Consistent with our prior studies demonstrating that compound 1 does not inhibit GGTase II, neither of the individual olefin isomers (11 or 19) nor the related epoxides (12 or 20) displayed inhibitory activity against this enzyme at concentrations up to 1 mM. The triazole phosphonates displayed weak activity against both FTase and GGTase I, with IC50 values in the 300–500 µM range, which is unlikely to be relevant to the observed activity of these agents in cell culture.
Table 1.
Evaluation of triazole phosphonates against GGDPS and related enzymes.
| Compound | GGDPS IC50 (µM) |
FDPS IC50 (µM) |
FTase IC50 (µM) |
GGTase I IC50 (µM) |
GGTase II IC50 (µM) |
|---|---|---|---|---|---|
| 1 | 2.2 | 84 | 500 | 200 | >1000 |
| 11 | 17 | 57 | >500 | 340 | >1000 |
| 19 | 0.38 | 79 | 400 | 500 | >1000 |
| 12 | 23 | 33 | 400 | 500 | >1000 |
| 20 | 17 | 47 | 340 | 300 | >1000 |
Finally, the ability of these agents to disrupt a key cellular process in myeloma cells was examined. RPMI-8226 cells were incubated with the test compounds and the effects on monoclonal protein trafficking were determined via lambda light chain ELISA performed on whole cell lysate. We already have demonstrated that agents which impair Rab geranylgeranylation disrupt monoclonal protein trafficking in myeloma cells, resulting in decreased secretion and increased intracellular levels of light chain.11 As shown in Figure 5, the olefin mixture 1 induces an accumulation of intracellular lambda light chain equivalent to that induced by the positive control DGBP. This activity is a consequence of the neryl triazole 19 as the geranyl triazole 11 has no effect on intracellular light chain levels.
Figure 5.
Neryl triazole 19 disrupts monoclonal protein trafficking in human myeloma cells. RPMI- 8226 cells were incubated for 48 hours in the presence or absence of 10 µM test compound or DGBP. Intracellular lambda light chain concentrations were determined via ELISA. Data are expressed as percentage of control (mean ± SD, n=3). The * denotes p<0.05 per unpaired two-tailed t-test and compares treated cells to untreated control cells.
We have previously prepared a series of mono- and dialkyl bisphosphonates with isoprenoid chains that differed in both chain length and olefin stereochemistry.23 The most potent GGDPS inhibitors were digeranyl bisphosphonate (DGBP) and 2E,6E-farnesyl bisphosphonate (21). The geranyl/neryl and dineryl bisphosphonate versions were approximately 14-fold less potent than DGBP while the 2E,6Z- (22), 2Z,6E- and 2Z,6Z-isomers were approximately 6-, 400-, and 700-fold less potent than the 2E,6E-isomer. In addition, in the monoalkyl series the geranyl bisphosphonate6 was at least one order of magnitude more potent than the corresponding neryl bisphosphonate.24 In our present study, we discovered that the neryl triazole bisphosphonate 19 is a significantly more potent inhibitor of GGDPS than the corresponding geranyl triazole bisphosphonate 11.
The incorporation of a triazole with a 10-carbon substituent results in a total chain length which is equivalent to the previously described farnesyl bisphosphonates. As shown in Figure 6, from one perspective the triazole ring system can be viewed as a nearly isosteric replacement for an isoprenoid unit in an E-configuration, although the C-3 to C-5 bond lengths and angles in compounds 21 and 22 differ slightly from the C-3 to N-5 array in compounds 11 and 19. Even so, the geranyl isomer 11 clearly would be viewed as most closely parallel to 2E,6E-farnesylbisphosphonate (21) while the neryl isomer 19 would more closely approximate 2E,6Z-farnesylbisphosphonate (22). Because earlier studies have shown that the 2E,6E-isomer 21 is about 6-fold more potent than the 2E,6Z-isomer 22, it would be reasonable to predict that the geranyl derivative 11 would be more active than the neryl isomer 19. Nevertheless, in the actual experiments the potency of the neryl compound 19 is actually more than 40-fold greater than the geranyl isomer 11. This experimental data suggests that the triazole moiety itself may aid in binding to GGDPS. If so, this affords a new design element that might be exploited in further development of GGDPS inhibitors. Finally, the importance of the first isoprenoid olefin in this compound series also is clear as the incorporation of an epoxide in the cis-configuration (20) reduces the activity against GGDPS to a level comparable to the geranyl triazole.
Figure 6.
Comparison of farnesyl and triazole bisphosphonates.
Of note, the neryl triazole 19 is approximately an order of magnitude more potent in disrupting protein geranylgeranylation in cell culture studies than DGBP, but is slightly less potent in the in vitro enzyme assay where DGBP has a reported IC50 of 0.2 µM.21 This may be a consequence of enhanced cellular uptake of compound 19 as compared to DGBP by virtue of the presence of one isoprenoid chain as opposed to two. In addition, its effectiveness might be further enhanced through prodrug strategies,24 although this has yet to be examined with these triazoles. DGBP has been demonstrated to be competitive with respect to FPP as opposed to IPP.25 Whether compound 19 inhibits GGDPS via a similar mechanism also remains to be determined.
4. Conclusions
In conclusion, we have developed a new synthetic route which affords the preparation of individual olefin isomers of isoprenoid triazole bisphosphonates. This synthetic approach should be applicable to the preparation of the isomers of the longer chained triazole bisphosphonates which are of interest because of their potential activity as GGTase II inhibitors. We have demonstrated that the neryl triazole 19 is a potent and specific inhibitor of GGDPS in both enzymatic and cellular assays. This potent inhibitor has important therapeutic implications because of the integral roles that geranylgeranylated proteins play in diverse cellular activities.
5. Experimental procedures and methods
5.1 General experimental conditions
Tetrahydrofuran was freshly distilled from sodium/benzophenone, while methylene chloride was distilled from calcium hydride prior to use. All other reagents and solvents were purchased from commercial sources and used without further purification. All reactions in nonaqueous solvents were conducted in flame-dried glassware under a positive pressure of argon and with magnetic stirring. All NMR spectra were obtained at 300, 400, or 500 MHz for 1H, and 75, 100, or 125 MHz for 13C, with internal standards of (CH3)4Si (1H, 0.00) or CDCl3 (1H, 7.27; 13C, 77.2 ppm) for non-aqueous samples or D2O (1H, 4.80) and 1,4-dioxane (13C, 66.7 ppm) for aqueous samples. The 13P chemical shifts were reported in ppm relative to 85% H3PO4 (external standard). High resolution mass spectra were obtained at the University of Iowa Mass Spectrometry Facility. Silica gel (60 Å, 0.040–0.063 mm) was used for flash chromatography.
5.2 Tetraethyl (E)-(2-(1-((3-methyl-3-(4-methylpent-3-en-1-yl)oxiran-2-yl)methyl)-1H-1,2,3-triazol-4-yl)ethane-1,1-diyl)bis(phosphonate) (9)
Solid NaN3 (171 mg, 2.63 mmol) was added to a solution of bromide 716,17 (403 mg, 1.73 mmol) in DMF (7 mL) and the reaction was allowed to stir overnight at rt while protected from light in a flask wrapped with aluminum foil.26 Once the reaction was complete, it was diluted with ether. After the filtrate was washed with water (five times) and then with brine to remove any remaining DMF, the organic layer was dried (Na2SO4) and concentrated in vacuo to obtain the azide as a yellow-orange oil (250 mg, 74%). (1H NMR (300 MHz, CDCl3) δ 5.08 (t, J = 7.1 Hz, 1H), 3.47–3.41 (m, 2H), 2.98 (t, J = 6.0 Hz, 1H), 2.15–2.05 (m, 2H), 1.76–1.42 (m, 2H), 1.70 (s, 3H), 1.62 (s, 3H), 1.32 (s, 3H)). Without further purification, a portion of the azide 8 (140 mg, 0.72 mmol) and tetraethyl-(3-butyn- 1-ylidene)-1,1-bisphosphonate14 (2, 180 mg, 0.55 mmol) were dissolved in a solution of water in t-BuOH (1:4, 3 mL), followed by addition of CuSO4 (5 M, 0.01 mL) and sodium ascorbate (33 mg, 0.17 mmol) in sequence. After the reaction was allowed to stir overnight at rt, the solvent was removed under vacuum. The resulting residue was dissolved in brine and extracted with EtOAc. The combined organic layer was washed with 5% NH4OH, dried (Na2SO4), and concentrated in vacuo. The resulting oil was purified via flash chromatography (10% EtOH in hexanes) to provide the desired triazole 9 as a colorless oil (170 mg, 58%): 1H NMR (400 MHz, CDCl3) δ 7.63 (s, 1H), 5.04 (t, J = 7.1 Hz, 1H), 4.69 (dd, J = 14.4 Hz, 4.1 Hz, 1H), 4.26 (dd, J = 14.8 Hz, 7.2 Hz, 1H), 4.22–4.09 (m, 8H), 3.35 (td, JHP = 16.0 Hz, J = 6.2 Hz, 2H), 3.10 (dd, J = 7.6 Hz, 4.4 Hz, 1H), 2.98 (tt, JHP = 23.5 Hz, J = 6.4 Hz, 1H), 2.07 (td, J = 8.2 Hz, 7.6 Hz, 2H), 1.73–1.63 (m, 1H), 1.67 (s, 3H), 1.60 (s, 3H), 1.54–1.45 (m, 1H), 1.42 (s, 3H), 1.33–1.27 (m, 12H); 13C NMR (75 MHz, CDCl13) δ 145.5 (t, JCP = 7.8 Hz), 132.5, 122.9, 122.7, 62.8 (d, JCP = 6.5 Hz, 2C), 62.6 (d, JCP = 6.6 Hz, 2C), 61.4, 60.4, 49.9, 38.1, 36.6 (t, JCP = 132.5 Hz), 25.7, 23.5, 22.1 (t, JCP = 4.0 Hz), 17.7, 17.0, 16.4 (d, JCP = 3.1 Hz, 2C), 16.3 (d, JCP = 2.3 Hz, 2C); 31P NMR (121 MHz, CDCl3) δ 22.3; HRMS (ES−, m/z) calcd for (M-H)− C14H24N3O7P2: 408.1090; found: 408.1114.
5.3 Tetraethyl (E)-(2-(1-(3,7-dimethylocta-2,6-dien-1-yl)-1H-1,2,3-triazol-4-yl)ethane-1,1-diyl)bis(phosphonate) (10)
Sodium iodide (60 mg, 0.4 mmol) was weighed in a round bottom flask and dried in an oven overnight. After the salt was dissolved in acetonitrile/THF (1:1, 1 mL), trifluoroacetic anhydride (0.014 mL, 0.1 mmol) was added. After 5 minutes, when the solution had turned to a deep yellow color, it was cooled in an ice bath. The starting material 9 (50 mg, 0.1 mmol) was then added to the reaction vessel as a neat oil. After an additional 5 minutes, the ice bath was removed and the reaction mixture was allowed to stir overnight at rt. Once the reaction was complete based on TLC analysis (5% MeOH/EtO2), it was diluted with saturated NaHSO3. The aqueous layer was extracted with Et2O, the organic extracts were combined, dried (Na2SO4), and concentrated in vacuo. Final purification by column chromatography (10% MeOH/EtO2) afforded the desired product 10 as a colorless oil (28 mg, 55%): 1H NMR (500 MHz, CDCl3) δ 7.44 (s, 1H), 5.40 (tq, J = 7.1 Hz, 1.0 Hz, 1H), 5.06 (t, J = 5.7 Hz, 1H), 4.92 (d, J = 6.9 Hz, 2H), 4.22–4.07 (m, 8H), 3.32 (td, JHP = 15.6 Hz, J = 6.3 Hz, 2H), 3.03 (tt, JHP = 23.6 Hz, J = 6.3 Hz, 1H), 2.14–2.04 (m, 4H), 1.78 (s, 3H), 1.68 (s, 3H), 1.60 (s, 3H), 1.28 (t, J = 7.0 Hz, 6H), 1.28 (t, J = 7.1 Hz, 6H); 13C NMR (125 MHz, CDCl13) δ 145.2 (t, JCP = 7.8 Hz), 143.1, 132.3, 123.6, 121.7, 117.3, 63.1, 63.0, 62.7, 62.7, 48.0, 39.6, 36.7 (t, JCP = 132.6 Hz), 30.5, 26.3, 25.9, 22.3 (t, JCP = 4.4 Hz), 17.9, 16.5, 16.4, 16.4, 16.4; 31P NMR (202 MHz, CDCl3) δ 22.5; HRMS (ES+, m/z) calcd for (M+H)+ C22H42N3O6P2: 506.2549; found: 506.2547.
5.4 Sodium (E)-(2-(1-(3,7-dimethylocta-2,6-dien-1-yl)-1H-1,2,3-triazol-4-yl)ethane-1,1-diyl)bis(phosphonate) (11)
Collidine (0.21 mL, 1.57 mmol) was added into an ice cold solution of compound 10 (79 mg, 0.16 mmol) in CH2Cl2 (3.5 mL) followed by addition of TMSBr (0.25 mL, 1.88 mmol) and the reaction was allowed to stir overnight at rt. Once the reaction was complete, based on analysis of the 31P NMR spectrum of the reaction mixture, it was diluted with toluene. After the solvent was removed in vacuo, the residue was washed with toluene five more times and dried to remove any remaining TMSBr. It was then treated with 1N NaOH (1.0 mL, 1 mmol) overnight. The reaction mixture was then dried on a lyophilizer to obtain the salt, which was then dissolved in a small amount of water, precipitated by addition of acetone, isolated by filtration, and dried. This material was further dissolved in water and lyophilized to produce the pure white salt 11 (59 mg, 75%): 1H NMR (500 MHz, D2O) δ 7.81 (s, 1H), 5.45 (t, J = 6.6 Hz, 1H), 5.12 (s, 1H), 4.97 (d, J = 7.4 Hz, 2H), 3.22 (td, JHP = 14.7 Hz, J = 6.2 Hz, 2H), 2.42 (tt, JCP = 21.2 Hz, J = 6.4 Hz, 1H), 2.17–2.05 (m, 4H), 1.78 (s, 3H), 1.64 (s, 3H), 1.56 (s, 3H), 13C NMR (125 MHz, D2O) δ 147.0 (t, JCP = 3.7 Hz), 143.7, 133.5, 123.9, 123.6, 117.1, 47.9, 39.6 (t, JCP = 118.6 Hz), 38.7, 25.6, 25.0, 21.8 (t, JCP = 3.4 Hz), 17.1, 15.7; 31P NMR (202 MHz, D2O) δ 18.8; HRMS (ES+, m/z) calcd for (M+H)+ C14H22N3O6P2Na4: 482.0575; found: 482.0570.
5.5 Sodium (E)-(2-(1-((3-methyl-3-(4-methylpent-3-en-1-yl)oxiran-2-yl)methyl)-1H-1,2,3-triazol-4-yl)ethane-1,1-diyl)bis(phosphonate) (12)
An ice cold solution of epoxide 9 (76 mg, 0.15 mmol) in dichloromethane (4 mL) was treated with collidine (0.20 mL, 1.5 mmol) and TMSBr (0.23 mL, 1.73 mmol) in sequence. After it was allowed to react overnight at rt, and the reaction was complete based on analysis of the 31P NMR spectrum, the reaction solvent was removed in vacuo. The resulting residue was then dissolved in toluene and dried under vacuo, and this process was repeated three times. The white solid that formed was allowed to stir with 1N NaOH (0.75 mL, 0.75 mmol) for 2 minutes at rt. This material was precipitated by addition of acetone followed by removal of water on a lyophilizer to obtain the initial salt. The salt was dissolved in water and allowed to stir in 1N NaOH (0.12 mL, 0.12 mmol) for 2 minutes at rt to remove residual collidine observed in 1H NMR spectrum. The desired product then was precipitated by addition of acetone followed by removal of water on a lyophilizer to produce the final product 12 as a white solid (49 mg, 68%): 1H NMR (500 MHz, D2O) δ 7.93 (s, 1H), 5.15–5.09 (m, 1H), 4.76 (dd, J = 14.9 Hz, 4.3 Hz, 1H), 4.51 (dd, J = 14.7 Hz, 7.4 Hz, 1H), 3.43 (dd, J = 7.4 Hz, 4.6 Hz, 1H), 3.21 (td, JHP = 14.9 Hz, J = 7.1 Hz, 2H), 2.21 (tt, JHP = 21.1 Hz, J = 6.7 Hz, 1H), 2.18–2.11 (m, 2H), 1.84–1.74 (m, 1H), 1.74–1.70 (m, 1H), 1.66 (s, 3H), 1.62 (s, 3H), 1.57–1.50 (m, 1H), 1.49 (s, 3H); 13C NMR (125 MHz, D2O) δ 148.5, 134.0, 124.3, 123.1, 64.2, 62.0, 49.4, 40.0 (t, JCP = 112.5 Hz), 37.3, 24.9, 23.1, 22.1, 16.9, 15.6; 31P NMR (202 MHz, D2O) δ 18.7; HRMS (ES−, m/z) calcd for (M-H)− C14H24N3O7P2: 408.1090; found: 408.1101.
5.6 Tetraethyl (Z)-(2-(1-((3-methyl-3-(4-methylpent-3-en-1-yl)oxiran-2-yl)methyl)-1H-1,2,3-triazol-4-yl)ethane-1,1-diyl)bis(phosphonate) (17)
According to the procedure described for preparation of compound 9, bromide 1517 (400 mg, 1.7 mmol) was treated with NaN3 (170 mg, 2.6 mmol). The resulting intermediate organic azide (16, 260 mg, 1.33 mmol) was then isolated and treated with bisphosphonate 2 (330 mg, 1.02 mmol) to afford the desired triazole 17 (354 mg, 67%) as a colorless oil after purification by flash chromatography (10% EtOH in hexanes): 1H NMR (300 MHz, CDCl3) δ 7.68 (s, 1H), 5.14 (t, J = 7.6 Hz, 1H), 4.76 (dd, J = 14.1 Hz, 3.3 Hz, 1H), 4.28–4.05 (m, 9H), 3.34 (td, JHP = 16.5 Hz, J = 6.3 Hz, 2H), 3.10 (dd, J = 7.4 Hz, 3.7 Hz, 1H), 3.00 (tt, JCP = 23.4 Hz, J = 6.3 Hz, 1H), 2.25–2.13 (m, 2H), 1.82–1.56 (m, 2H), 1.71 (s, 3H), 1.65 (s, 3H), 1.36 (s, 3H), 1.30 (t, J = 7.1 Hz, 12H); 13C NMR (75 MHz, CDCl3) δ 145.0 (t, JCP = 7.6 Hz), 132.2, 122.6, 122.3, 62.4, 62.3, 62.1, 62.0, 61.3, 61.2, 49.4, 36.2 (t, JCP = 132.6 Hz), 32.7, 25.3, 23.6, 21.7 (t, JCP = 3.8 Hz), 21.5, 17.3, 16.0, 15.9, 15.9, 15.8; 31P NMR (121 MHz, CDCl3) δ 22.2; HRMS (ES+, m/z) calcd for (M+H)+ C22H42N3O7P2: 522.2498; found: 522.2507.
5.7 Tetraethyl (Z)-(2-(1-(3,7-dimethylocta-2,6-dien-1-yl)-1H-1,2,3-triazol-4-yl)ethane-1,1-diyl)bis(phosphonate) (18)
Bisphosphonate ester 18 was synthesized according to the procedure employed for the preparation of compound 10 with some modifications. Sodium iodide (80 mg, 0.53 mmol) was weighed in a round bottom flask and dried in an oven overnight. After the salt was dissolved in acetonitrile/THF (1:1, 1.14 mL), trifluoroacetic anhydride (0.019 mL, 0.14 mmol) was added. After 5 minutes, the solution was cooled to 0 °C, and epoxide 17 (69 mg, 0.13 mmol) in acetonitrile/THF (1:1, 0.5 mL) was added. After an additional 5 minutes, the ice bath was removed and the reaction was allowed to stir overnight at rt. Once the reaction was complete based on TLC analysis (25% EtOH/hexane), it was diluted with saturated NaHSO3. The aqueous layer was extracted with Et2O, the organic extracts were combined, dried (Na2SO4), and concentrated in vacuo to afford the desired product 18 (47 mg, 70%) as a yellow oil which was used without further purification: 1H NMR (400 MHz, CDCl3) δ 7.45 (s, 1H), 5.40 (t, J = 7.5 Hz, 1H), 5.13–5.06 (m, 1H), 4.90 (d, J = 7.5 Hz, 2H), 4.21– 4.06 (m, 8H), 3.34 (td, JHP = 16.2 Hz, J = 6.5 Hz, 2H), 3.04 (tt, JHP = 23.6 Hz, J = 6.2 Hz, 1H), 2.22–2.09 (m, 4H), 1.79 (s, 3H), 1.69 (s, 3H), 1.62 (s, 3H), 1.28 (t, J = 6.9 Hz, 12H); 31C NMR (100 MHz, CDCl3) δ 145.0 (t, JCP = 11.5 Hz), 142.8, 132.7, 123.3, 121.7, 118.1, 63.0, 62.9, 62.7, 62.6, 47.7, 36.6 (t, JCP = 132.6 Hz), 32.2, 26.4, 25.8, 23.5, 22.2 (t, JCP = 4.0 Hz), 17.8, 16.4, 16.4, 16.3, 16.3; 31P NMR (121 MHz, CDCl3) δ 22.5; HRMS (ES+, m/z) calcd for (M+H)+ C22H42N3O6P2: 506.2549; found: 506.2553.
5.8 Sodium (Z)-(2-(1-(3,7-dimethylocta-2,6-dien-1-yl)-1H-1,2,3-triazol-4-yl)ethane-1,1-diyl)bis(phosphonate) (19)
According to the procedure employed for preparation of compound 11, bisphosphonate 18 (66 mg, 0.13 mmol) was treated with collidine (0.17 mL, 1.31 mmol), TMSBr (0.21 mL, 1.57 mmol) and then 1N NaOH (0.75 mL, 0.75 mmol). A parallel work-up and final purification by precipitation gave the desired product 19 (40 mg, 64%) as a white solid: 1H NMR (500 MHz, D2O) δ 7.81 (s, 1H), 5.52 (t, J = 7.0 Hz, 1H), 5.19 (t, J = 7.1, 1H), 4.95 (d, J = 7.4 Hz, 2H), 3.14 (td, JHP = 14.9 Hz, J = 5.5 Hz, 2H), 2.29–2.13 (m, 4H), 2.05 (tt, JCP = 21.7 Hz, J = 5.6 Hz, 1H), 1.80 (s, 3H), 1.67 (s, 3H), 1.62 (s, 3H), 13C NMR (125 MHz, D2O) δ 150.7, 143.7, 134.2, 123.7, 123.5, 117.8, 47.8, 41.5 (t, JCP = 118.0 Hz), 31.4, 25.8, 25.0, 24.4, 22.6, 17.0; 31P NMR (202 MHz, D2O) δ 19.8; HRMS (ES−, m/z) calcd for (M-H)− C14H24N3O6P2: 392.1140; found: 392.1135.
5.9 Sodium (Z)-(2-(1-((3-methyl-3-(4-methylpent-3-en-1-yl)oxiran-2-yl)methyl)-1H-1,2,3-triazol-4-yl)ethane-1,1-diyl)bis(phosphonate) (20)
To a stirred solution of bisphosphonate 17 (168 mg, 0.32 mmol) in CH2Cl2 (8.7 mL) at 0 °C, collidine (0.43 mL, 3.2 mmol) and TMSBr (97%, 0.52 mL, 3.9 mmol) were each added dropwise in succession. The reaction was allowed to stir overnight while it warmed to room temperature. Once the reaction was complete, which was determined by analysis of the 31P NMR spectrum of the reaction mixture, the solvent was removed in vacuo. The residue then was diluted with toluene (10 mL) and concentrated in vacuo to remove excess TMSBr (3×). The resulting residue was dried on a vacuum line overnight. It then was treated with 1N NaOH (1.61 mL, 1.61 mmol), allowed to stir at room temperature for 2 minutes, and immediately precipitated from anhydrous acetone. The resulting solid was removed by filtration, dissolved in water, and freeze dried to provide compound 20 (122 mg, 76%) as a white powder: 1H NMR (500 MHz, D2O) δ 7.94 (s, 1H), 5.26–5.23 (m, 1H), 4.87–4.84 (m, 1H), 4.43–4.38 (dd, J = 15.0, 8.5 Hz, 1H), 3.45–3.44 (m, 1H), 3.22 (td, JHP = 15.0 Hz, J = 7.0 Hz, 2H), 2.31–2.18 (m, 3H), 1.86–1.80 (m, 1H) 1.74 (s, 3H), 1.71–1.70 (m, 1H) 1.68 (s, 3H), 1.42 (s, 3H); 13C NMR (125 MHz, D2O) δ 148.3, 133.8, 124.1, 123.1, 64.0, 62.9, 49.6, 39.9 (t, JCP = 111.0 Hz), 32.2, 25.1, 23.5, 22.0, 21.0, 17.1; 31P NMR δ 19.0; HRMS (ES−, m/z) calcd for (M-H)− C14H24N3O6P2: 392.1140; found: 392.1135.
5.10 Immunoblot analysis
RPMI-8226 cells were incubated with drugs for 48 hrs. Whole cell lysate was obtained using RIPA buffer (0.15M NaCl, 1% sodium deoxycholate, 0.1% SDS, 1% Triton (v/v) X-100, 0.05 M Tris HCl) containing protease and phosphatase inhibitors. Aqueous and detergent fractionations were obtained using Triton X-114 as previously described.27,28 Protein content was determined using the bicinchoninic acid (BCA) method (Pierce Chemical, Rockford, IL). Equivalent amounts of cell lysate were resolved by SDS-PAGE, transferred to polyvinylidene difluoride membrane, probed with the appropriate primary antibodies, and detected using HRP-linked secondary antibodies and Amersham Pharmacia Biotech ECL Western blotting reagents per manufacturer’s protocols.
5.11 Measurement of intracellular FPP and GGPP levels
Intracellular FPP and GGPP levels were measured using the previously reported reversed phase HPLC methodology.29 Briefly, following incubation with drugs, cells were collected and counted using Trypan blue staining and a Bio-Rad TC10 automated cell counter. Isoprenoid pyrophosphates were extracted from cell pellets with extraction solvent (butanol/75mM ammonium hydroxide/ethanol 1:1.25:2.75). After drying down by nitrogen gas, the FPP and GGPP in the residue were incorporated into fluorescent GCVLS or GCVLL peptides by FTase or GGTase I. The prenylated fluorescent peptides were separated and quantified by reversed phase HPLC with fluorescence detection.
5.12 FDPS and GGDPS enzyme assay
Recombinant FDPS and GGDPS were kindly provided by Dr. Raymond Hohl, University of Iowa. Recombinant enzyme (10 nM FDPS, 20 nM GGDPS) was incubated with assay buffer (50 mM Tris-HCl, pH 7.7, 20 mM MgCl2, 5 mM TCEP, 5 µg/mL BSA) and test compounds for 10 minutes at room temperature. The reaction was initiated by the addition of 10 µM GPP (for FDPS) or 10 µM FPP (for GGDPS) and 10 µM [14C]-IPP and was carried out at 37 °C for 30 minutes. The reaction was stopped by the addition of saturated NaCl. Radiolabeled GGPP was extracted with n-butanol and counted via liquid scintillation counting.
5.13 FTase and GGTase I enzyme assay
FTase and GGTase I activity was determined using the methods of Cassidy et al.30 and Pickett et al.31 with some modifications. For FTase, reaction mixtures were initiated by the addition of 3 nM recombinant FTase (Jena Biosciences) to reaction buffer (50 mM Tris-HCl, pH 7.5, 5 mM DTT, 10 mM MgCl2, 10 µM ZnCl2, 0.04% n-dodecyl β-d-maltoside (DDM)) containing 10 µM FPP and 5 µM dansyl-GCVLS (Biosynthesis, Inc.). For GGTase I, 12 nM of recombinant GGTase I (Jena Biosciences) was added to reaction buffer (50 mM Tris-HCl, pH 7.5, 5 mM DTT, 5 mM MgCl2, 50 µM ZnCl2, 10 mM KCl, 0.04% DDM) containing 10 µM GGPP and 7 µM dansyl-GCVLS (Biosynthesis, Inc.). Reactions were carried out at 30 °C in the presence or absence of test compounds. After one hour, reaction mixtures were placed on ice, and the reaction was stopped by the addition of acetonitrile containing 5% HCl. The mixture was briefly vortexed and then spun at 14000 rpm for 5 minutes. 100 µL of the supernatant was injected into an HPLC with fluorescence detection and the prenylated fluorescent peptides were quantified.
5.14 GGTase II enzyme assay
GGTase II activity was determined using the method of Seabra and James with some modifications as previously described.13,32 Briefly, reactions were initiated with the addition of 5 µM [3H]-GGPP to reaction buffer (50 mM sodium HEPES, 5 mM MgCl2, 1 mM dithiothreitol, 1 mM NP-40) containing 30 nM recombinant GGTase II (Jena Biosciences), 2 µM recombinant Rab1A, and 1 µM recombinant REP-2 (Jena Biosciences) with or without the test compounds. After a 20 minute incubation at 37 °C, reactions were terminated by the addition of 200 µL 10% HCl/EtOH. Samples were filtered through P-81 filters (Whatman) by a Brandel harvester. Filters were then washed in EtOH, dried, and counted using liquid scintillation counting. Recombinant Rab1A was prepared by transforming BL21 Star DE3 competent cells with a plasmid containing N-His-tagged Rab1A (GeneCopoeia). A Qiagen Ni-NTA Fast Spin kit was used to purify the recombinant protein. Protein content was determined using the BCA method.
5.15 Monoclonal protein quantitation
Cells were incubated in the presence or absence of test compounds for specified periods of time. The cells were lysed in RIPA buffer containing protease and phosphatase inhibitors. Protein content was determined using the BCA method. Human lambda light chain kit (Bethyl Laboratories, Montgomery, TX) was used to quantify intracellular monoclonal protein levels.
5.16 Statistics
Two-tailed t-testing was used to calculate statistical significance. An α of 0.05 was set as the level of significance. Concentration response curves for the enzyme assays were analyzed via CalcuSyn software (Biosoft, Cambridge, UK) to determine IC50 values.
Supplementary Material
Acknowledgements
Financial support from the University of Iowa Graduate College in the form of an Iowa AGEP/Dean’s Graduate Fellowship (to VSW), the Center for Biocatalysis and Bioprocessing in the form of an NIH Predoctoral Fellowship in Biotechnology (to VSW), the Roy J. Carver Charitable Trust, and the NIH (R01CA-172070) is gratefully acknowledged.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary data
Supplementary data associated with this article, including NMR spectra, can be found in the online version, at
References
- 1.Holstein SA, Hohl RJ. Lipids. 2004;39:293–309. doi: 10.1007/s11745-004-1233-3. [DOI] [PubMed] [Google Scholar]
- 2.Wiemer AJ, Wiemer DF, Hohl RJ. Clin. Pharmacol. Ther. 2011;90:804–812. doi: 10.1038/clpt.2011.215. [DOI] [PubMed] [Google Scholar]
- 3.Dudakovic A, Wiemer AJ, Lamb KM, Vonnahme LA, Dietz SE, Hohl RJ. J. Pharmacol. Exp. Ther. 2008;324:1028–1036. doi: 10.1124/jpet.107.132217. [DOI] [PubMed] [Google Scholar]
- 4.Zhang YH, Zhu W, Liu YL, Wang H, Wang K, Li K, No JH, Ayong L, Gulati A, Pang R, Freitas-Junior L, Morita CT, Oldfield E. ACS Med. Chem. Lett. 2013;4:423–427. doi: 10.1021/ml4000436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holstein SA, Hohl RJ. Curr. Opin. Pharmacol. 2012;12:704–709. doi: 10.1016/j.coph.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 6.Holstein SA, Cermak DM, Wiemer DF, Lewis K, Hohl RJ. Bioorg. Med. Chem. 1998;6:687–694. doi: 10.1016/s0968-0896(98)00034-0. [DOI] [PubMed] [Google Scholar]
- 7.Hohl RJ, Lewis KA, Cermak DM, Wiemer DF. Lipids. 1998;33:39–46. doi: 10.1007/s11745-998-0178-x. [DOI] [PubMed] [Google Scholar]
- 8.Gibbs BS, Zahn TJ, Mu YQ, Sebolt-Leopold JS, Gibbs RA. J. Med. Chem. 1999;42:3800–3808. doi: 10.1021/jm9902786. [DOI] [PubMed] [Google Scholar]
- 9.Sousa SF, Fernandes PA, Ramos MJ. Curr. Med. Chem. 2008;15:1478–1492. doi: 10.2174/092986708784638825. [DOI] [PubMed] [Google Scholar]
- 10.Guo Z, Wu YW, Das D, Delon C, Cramer J, Yu S, Thuns S, Lupilova N, Waldmann H, Brunsveld L, Goody RS, Alexandrov K, Blankenfeldt W. EMBO J. 2008;27:2444–2456. doi: 10.1038/emboj.2008.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holstein SA, Hohl RJ. Leuk. Res. 2011;35:551–559. doi: 10.1016/j.leukres.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Recchi C, Seabra MC. Biochem. Soc. Trans. 2012;40:1398–1403. doi: 10.1042/BST20120199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou X, Hartman SV, Born EJ, Smits JP, Holstein SA, Wiemer DF. Bioorg. Med. Chem. Lett. 2013;23:764–766. doi: 10.1016/j.bmcl.2012.11.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Skarpos H, Osipov SN, Vorob'eva DV, Odinets IL, Lork E, Roschenthaler GV. Org. Biomol. Chem. 2007;5:2361–2367. doi: 10.1039/b705510b. [DOI] [PubMed] [Google Scholar]
- 15.Sharpless KB, Michaels RC. J. Am. Chem. Soc. 1973;95:6136–6137. [Google Scholar]
- 16.Lempers HEB, Garcia ARI, Sheldon RA. J. Org. Chem. 1998;63:1408–1413. [Google Scholar]
- 17.Gash RC, MacCorquodale F, Walton JC. Tetrahedron. 1989;45:5531–5538. [Google Scholar]
- 18.Caputo R, Mangoni L, Neri O, Palumbo G. Tetrahedron Lett. 1981;22:3551–3552. [Google Scholar]
- 19.Sonnet PE. J. Org. Chem. 1978;43:1841–1842. [Google Scholar]
- 20.Bohlmann F, Zeisberg R, Klein E. Organic Magnetic Resonance. 1975;7:426–432. [Google Scholar]
- 21.Wiemer AJ, Tong H, Swanson KM, Hohl RJ. Biochem. Biophys. Res. Commun. 2007;353:921–925. doi: 10.1016/j.bbrc.2006.12.094. [DOI] [PubMed] [Google Scholar]
- 22.Shull LW, Wiemer AJ, Hohl RJ, Wiemer DF. Bioorg. Med. Chem. 2006;14:4130–4136. doi: 10.1016/j.bmc.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 23.Wiemer AJ, Yu JS, Lamb KM, Hohl RJ, Wiemer DF. Bioorg. Med. Chem. 2008;16:390–399. doi: 10.1016/j.bmc.2007.09.029. [DOI] [PubMed] [Google Scholar]
- 24.Wiemer AJ, Yu JS, Shull LW, Barney RJ, Wasko BM, Lamb KM, Hohl RJ, Wiemer DF. Bioorg. Med. Chem. 2008;16:3652–3660. doi: 10.1016/j.bmc.2008.02.016. [DOI] [PubMed] [Google Scholar]
- 25.Holstein SA, Hohl RJ. The Enzymes. Vol. 30. Elsevier Inc; 2011. pp. 301–319. [Google Scholar]
- 26.Limberg G, Lundt I, Zavilla J. Synthesis. 1999:178–183. [Google Scholar]
- 27.Bordier C. J. Biol. Chem. 1981;256:1604–1607. [PubMed] [Google Scholar]
- 28.Wasko BM, Dudakovic A, Hohl RJ. J. Pharmacol. Exp. Ther. 2011;337:540–546. doi: 10.1124/jpet.110.175521. [DOI] [PubMed] [Google Scholar]
- 29.Tong H, Holstein SA, Hohl RJ. Anal. Biochem. 2005;336:51–59. doi: 10.1016/j.ab.2004.09.024. [DOI] [PubMed] [Google Scholar]
- 30.Cassidy PB, Dolence JM, Poulter CD. Methods in enzymology. 1995;250:30–43. doi: 10.1016/0076-6879(95)50060-x. [DOI] [PubMed] [Google Scholar]
- 31.Pickett WC, Zhang FL, Silverstrim C, Schow SR, Wick MM, Kerwar SS. Anal. Biochem. 1995;225:60–63. doi: 10.1006/abio.1995.1108. [DOI] [PubMed] [Google Scholar]
- 32.Seabra MC, James GL. Methods Mol Biol. 1998;84:251–260. doi: 10.1385/0-89603-488-7:251. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.