Abstract
Anopheles melas is a brackish water-breeding member of the An. gambiae complex that is distributed along the coast of West Africa and is a major malaria vector within its range. Because little is known about the population structure of this species, we analyzed 15 microsatellite markers and 1,161 bp of mtDNA in 11 An. melas populations collected throughout its range. Compared to its sibling species An. gambiae, An. melas populations have a high level of genetic differentiation between them, representing its patchy distribution due to its fragmented larval habitat which is associated with mangroves and salt marsh grass. Populations clustered into three distinct groups representing Western Africa, Southern Africa, and Bioko Island populations that appear to be mostly isolated. Fixed differences in the mtDNA are present between all three clusters, and a Bayesian clustering analysis of the microsatellite data found no evidence for migration from mainland to Bioko Island populations, and little migration was evident between the Southern to the Western cluster. Surprisingly, mtDNA divergence between the three An. melas clusters is on par with levels of divergence between other species of the An. gambiae complex, and no support for monophyly was observed in a maximum likelihood phylogenetic analysis. Finally, an Approximate Bayesian Analysis of microsatellite data indicates that Bioko Island An. melas populations were connected to the mainland populations in the past, but became isolated, presumably when sea levels rose after the last glaciation period (≥10,000-11,000 years ago). This study has exposed species level genetic divergence within An. melas, and also has implications for control of this malaria vector.
Keywords: Anopheles melas, Anopheles gambiae, malaria, microsatellites, migration, population structure
Introduction
Anopheles melas is a brackish water-breeding member of the An. gambiae complex, which is comprised of at least seven largely morphologically indistinguishable species. This complex was thought to be a single species until crossing experiments between various “strains” revealed the presence of male hybrid sterility (Davidson 1962), White 1974, Hunt et al. 1998). All members of this complex are competent vectors of human malaria parasites, although they differ in host specificity ranging from highly anthropophillic (An. gambiae s.s.) to almost entirely zoophilic (An. quadriannulatus A), as well as in the extent of their distribution. For example, the two brackish water-breeding species, An. melas and An. merus are confined to the west and east coast of Africa, respectively. Hence the species in the complex vary considerably in their contribution to malaria transmission (Garret-Jones et al. 1980, Hunt et al. 1998, White et al. 1980), with only An. gambiae and An. arabiensis being considered primary malaria vectors.
Not surprisingly, research efforts have mainly focused on An. gambiae, Africa’s dominant malaria vector, which has turned out to have a remarkably complex population structure. Early on, the non-random distribution of polymorphic chromosomal inversions led to the description of the five so-called “chromosomal forms” (Touré et al. 1983, Coluzzi et al. 1985, Petrarca et al. 1987). These chromosomal forms are now thought to be local adaptations to various ecological conditions, but subsequent investigations of genetic differentiation among them revealed the presence of two molecular forms that are characterized by fixed differences in the ribosomal DNA (Favia et al. 1997). These have become known as the M and S molecular forms, and are now widely considered incipient species (della Torre et al. 2001, 2005). Fixed differences between these sympatric forms are primarily restricted to a few centromeric regions (Turner et al. 2005, White et al. 2010). The low recombination rate associated with these regions has been proposed as mechanism for the accumulation of genetic differentiation in the face of gene flow (Stump et al. 2005, Slotman et al. 2006).
An additional level of genetic complexity was identified within the M form. The M form in Mali, corresponding to a “Mopti” chromosomal form, shows a relatively high level of genetic differentiation from the “Forest” M form (Slotman et al. 2007). This suggests that the ecotypic adaptation associated with inversions has led to genetic differentiation within the M form, presumably due to geographic separation. In contrast, the S molecular form grades from a “Savanna” form in Mali, into a “Forest” S form in Cameroon, and with the exception of populations separated by the Rift Valley appears to have little genetic structure (Lehman et al. 2003). Such low levels of genetic differentiation across large distances also seem to be characteristic of the other major malaria vector in the complex, An. arabiensis (Besansky et al. 1997; Donnelly and Townson 2000).
This focus on the two primary vectors in the complex has resulted in a dearth of information about the genetic diversity and population structure of the other sibling species. Although generally considered a less anthropophilic mosquito than An. gambiae, An. melas will readily enter houses and feed on humans (Reddy et al. 2011). In The Gambia, up to 80.0% of An. melas sampled along The Gambia river had fed on humans (Caputo et al. 2008). Although data about malaria transmission by this species is limited, studies in Ghana (Tuno et al. 2010) and The Gambia (Bryan 1983, Bryan et al. 1987, Bøgh et al. 2007, Caputo et al. 2008) showed that this species is a dominant vector in locations close to its breeding habitat. For example, Bryan et al. (1987) found that populations of An. melas in The Gambia can comprise up to 100% of sampled An. gambiae complex mosquitoes, with a mean P. falciparum sporozoite infection rate of 1.5%. When found in sympatry with An. gambiae, the latter usually has a higher sporozoite rate (Akogbeto and Romano 1999), presumably because of a stronger preference for human hosts.
The small amount of available genetic data for An. melas comes from studies on paracentric inversion polymorphisms (Petrarca et al. 1983, Bafort and Petrarca 1983, Bryan et al. 1987, Akogbeto & Di Deco 1995, Calzetta et al. 2008) that are mainly aimed at investigating the distribution and the adaptive role of inversions. The range of this species is confined to the coast of West Africa, where it breeds primarily in mangrove swamps and tidal marshes (Sinka et al. 2010), and its population sizes fluctuate greatly with seasonal tides and rainfall. Because An. melas is confined to permanent saline water bodies (Bryan et al. 1987), it has a patchy distribution that may be reflected in its population genetic structure.
On Bioko Island, Equatorial Guinea, An. melas is also an important, and in some locations, dominant malaria vector (Sharp et al. 2007, Overgaard et al. 2012). Bioko Island is currently the focus of the Bioko Island Malaria Project (BIMCP), which implements indoor residual spraying (IRS) using carbamate insecticide, in conjunction with malaria screening and treatment. These interventions reduced mortality in children under the age of five by 64% in the first four years of the program (Kleinschmidt et al. 2009). Bioko Island is a potential candidate for a future malaria elimination campaign, and understanding the level of migration between mainland and Bioko Island vector populations is important for predicting the potential for the reintroduction of the malaria parasite and/or vector in the event of a successful elimination. The population genetic structure of An. gambiae has been examined in Equatorial Guinea using microsatellite markers (Moreno et al. 2007). This study indicated very high levels of migration of this species between the mainland and the island, both in the M and S molecular form. However, no information is available on An. melas, the other main vector on the island.
Considering the complex evolutionary history of the species complex, with varying levels of genetic isolation among and within species and forms, a basic knowledge of the genetic structure of other An. gambiae complex member species is vital for understanding their biology and the epidemiology of malaria. Understanding the degree of genetic homogeneity throughout the range of An. melas would allow us to predict whether insights into the biology of this species; e.g. indoor vs. outdoor feeding behavior (Reddy et al. 2011) or larval ecology (Walker and Lynch 2007), in one population may be applicable to other locations. In addition, information on the level and directionality of gene flow across the range of this species is important, as it can help us to predict the spread of insecticide resistance and inform better control of this malaria vector.
Using 15 species-specific microsatellite markers, and a fragment of the mitochondrial genes ND4 and ND5, we examined patterns of genetic variation and differentiation between 11 An. melas populations from The Gambia to Angola, including three populations on Bioko Island, Equatorial Guinea. Our aims were to assess patterns of population structure across the range of the species, and to determine the degree of genetic isolation between the mainland and island populations of Bioko.
Methods
Mosquito Collections
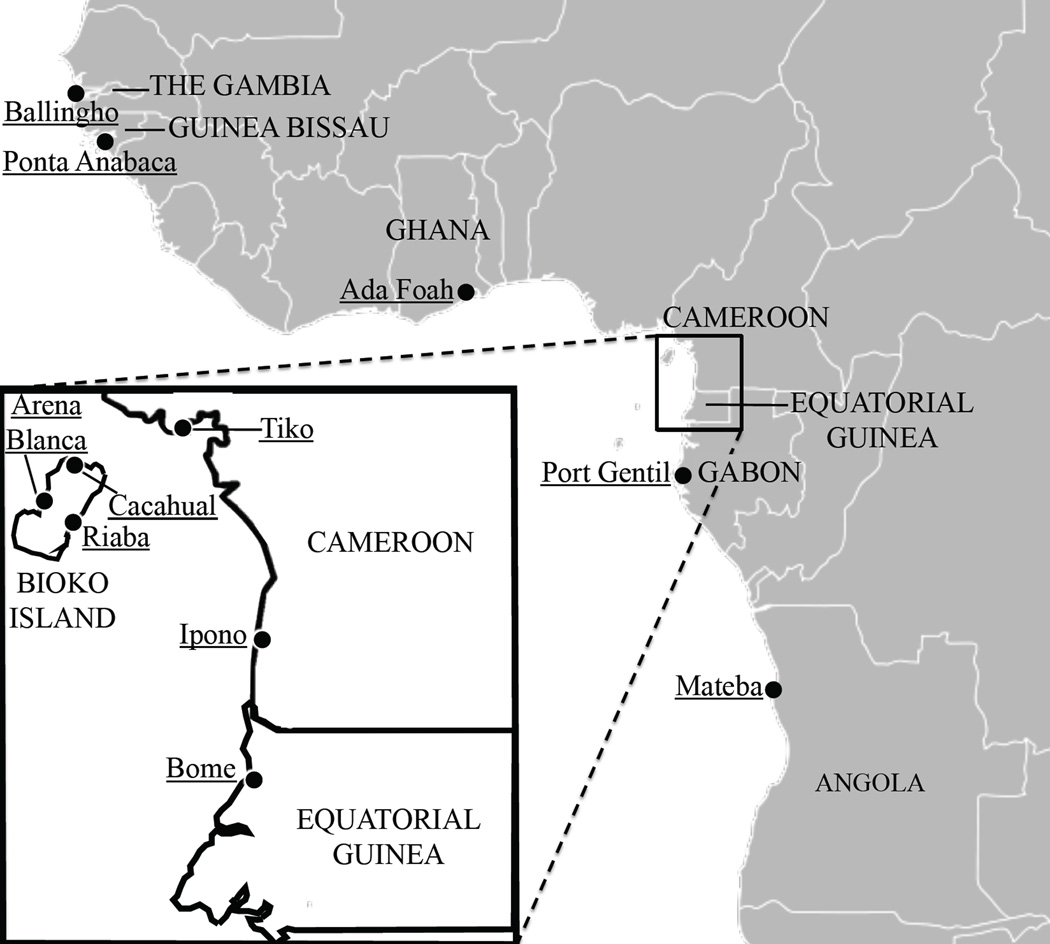
Adult female mosquitoes were collected using either CDC light traps, human landing catches, or through the aspiration resting females. Female An. melas from Equatorial Guinea were collected in Cacahual (Bioko Island) and Bomé (mainland), in October 2008, and Riaba and Arena Blanca (Bioko Island) in April 2009. Female mosquitoes were collected from Ipono, Cameroon in December 2005, from Tiko Cameroon in October 2010, from Ballingho, The Gambia in February 2010, from Ponta Abanaca, Guinea Bissau in December 2009, from Mateba, Angola in 2002 (Calzetta et al. 2008), and from Port Gentil, Gabon in 1999. Larval mosquitoes were collected in July 2010 in Ada Foah, Ghana from a roadside lagoon. Collections in Ghana and Tiko, Cameroon resulted in only 6 and 16 An. melas specimens collected respectively. Adult female An. gambiae s.s. (M form) mosquitoes were collected from Ukomba, a neighborhood of the city of Bata, Equatorial Guinea in March 2007 and from Mongola, Bioko Island in 2009.
Molecular Methods
Microsatellite DNA
Mosquito DNA was extracted from mosquito abdomens or whole mosquitoes using a Qiagen Biosprint 96 DNA extraction (Qiagen Inc., Valencia, CA). Species diagnostics were performed following Fanello et al. (2002). A polymerase chain reaction (PCR) was used to amplify 15 An. melas-specific polymorphic microsatellite loci (Deitz et al. 2012) in 6 to 96 individuals from each of the 11 An. melas populations, the average sample size being 56 (Table 1). Each microsatellite locus was amplified using a fluorescently labeled forward primer. PCR reactions contained 10–20 ng DNA template, with 1X PCR buffer (10 mM Tris-HCl pH 8.5, 50 mM KCl), 2.5 mM MgCl2, 200 µM of each dNTP, 2.0 µM of each forward (F) and reverse (R) primer, 0.03 U of Promega GoTaq DNA Polymerase (Promega Co., Madison, WI), and ddH2O to the final 20 µL reaction volume. PCRs were performed with an initial denaturing time of 2 min at 94°C followed by five cycles of 30 s at 94°C, 30 s at 50°C, 35 s at 72°C, 30 cycles of 30 s at 94°C, 30 s at 52°C, 35 s at 72°C, followed by a 15 min extension step at 72°C. PCR products were analyzed on a 3730xl DNA Genetic Analyzer (Life Technologies Corporation, Carlsbad, CA). Fragment lengths were determined using GeneMarker ver. 1.85 (SoftGenetics LLC., State College, PA).
Table 1.
An. melas and An. gambiae population collection location information. Microsatellite N defines the number of individuals genotyped for each of 15 microsatellite loci from each population. mtDNA N defines the number of individuals that was sequenced for the ND4-ND5 mtDNA locus.
| Species | Geographical Origin | Population Abbreviation | Microsatellite N | mtDNA N |
|---|---|---|---|---|
| An. melas | Arena Blanca, Equatorial Guinea (Bioko) | ARB | 96 | 61 |
| An. melas | Riaba, Equatorial Guinea (Bioko) | RIA | 94 | 16 |
| An. melas | Cacahual, Equatorial Guinea (Bioko) | CAC | 35 | 13 |
| An. melas | Tiko, Cameroon | TIK | 16 | 13 |
| An. melas | Ada Foah, Ghana | GHA | 6 | 6 |
| An. melas | Ballingho, The Gambia | GAM | 94 | 19 |
| An. melas | Ponta Anabaca, Guinea Bissau | GUI | 62 | 52 |
| An. melas | Ipono, Cameroon | IPO | 69 | 19 |
| An. melas | Bome, Equatorial Guinea | BOM | 69 | 11 |
| An. melas | Port Gentil, Gabon | GAB | 46 | 27 |
| An. melas | Mateba, Angola | ANG | 30 | 10 |
| An. gambiae (M Form) | Mongola, Equatorial Guinea (Bioko) | AGMG | − | 71 |
| An. gambiae (M form) | Ukomba, Equatorial Guinea | AGUK | − | 83 |
Mitochondrial DNA
Primers were designed to amplify a 1,578 bp region spanning part of the ND4 and ND5 genes of the mitochondrial genome based on the An. gambiae mitochondrial DNA (mtDNA) sequence (Beard et al. 1993) (FOR GGAGGATACGGTTTATTACGAA; REV CCTAATTGTCTTAAAGTTGATAAAGCA). PCR amplification was performed at a volume of 20 µL at the following reagent concentrations: 1X PCR buffer (10 mM Tris-HCl pH 8.5, 50mM KCl), 2.5 mM MgCl2, 200 µM of each dNTP, 2.0 µM of each primer, and 0.03 U of Promega GoTaq DNA Polymerase (Promega Co., Madison, WI). PCRs were run with an initial 10 min 94°C denaturation, followed by 35 cycles of 1 min at 94°C, 2 min at 53°C, 3 min at 72°C, then a 15 min extension at 72°C, and a hold step at 4°C. PCR products were purified using the PEG purification method (Lis 1980) and directly sequenced in both the forward and reverse directions using BigDye Terminator 3.1 Cycle Sequencing Kit (Life Technologies Corporation, Carlsbad, CA).
Analytical Methods
Microsatellite DNA Diversity and Divergence
Micro-Checker 2.2.3 (van Oosterhout et al. 2004) was used to test the microsatellite data set for the presence of null alleles using 10,000 permutations and a 99% confidence interval. Observed heterozygosity (HO), expected heterozygosity (HE), and other population genetic and diversity parameters were calculated in Arlequin ver. 3.5.1.2 (Excoffier and Lischer 2010). Tests for deviation from Hardy Weinberg Equilibrium (HWE) were run using 100,000 steps in the Markov chain, and linkage disequilibrium (LD) was calculated using 10,000 permutations. The program ADZE (Szpiech et al. 2008) was used to compute allelic richness (AR) using N=16 as the sample size. Both null allele corrected and uncorrected genotype frequencies were used to calculate population pair-wise FST and G”ST statistics (Meirmans and Hedrick 2011) in the program GenoDive (Meirmans and van Tienderen 2004). Population pair-wise FST values were calculated using 10,000 permutations. Pair-wise G”ST values were used to calculate an unrooted neighbour-joining tree in the program QuickTree (Howe et al. 2002). The tree was visualized in the program FigTree ver. 1.3.1 (Rambaut 2009).
A Bayesian assignment test was implemented in the program STRUCTURE ver. 2.3.3 (Pritchard et al. 2000) to determine the most likely number of populations (K) in our dataset, and to identify potential hybrids/migrants between populations. Simulations were replicated 20 times for each K, 1 through 10. Each Monte Carlo Markov Chain (MCMC, one per simulation) was run for 500,000 steps after a 100,00 step burn-in, under a population admixture model assuming independent allele frequencies, without using prior population information. Structure Harvester ver. 0.6.8 (Earl 2011), was used to examine the most likely number of populations (K). The individuals of each predicted cluster (K) were grouped in an Analysis of Molecular Variance (AMOVA) (Excoffier et al. 1992) which was calculated in Arlequin ver. 3.5.1.2 (Excoffier and Lischer 2010).
Mitochondrial DNA Diversity and Divergence
ND4-ND5 sequences were aligned manually in Sequencher ver. 4.9 (GeneCodes, Ann Arbor, USA) based on the translated An. gambiae amino acid sequence (Beard et al. 1993), and consensus sequences were trimmed to 1,161 bp. Chromatograms were visually inspected and only sequences with unambiguous basecalls were included in the analyses. Additional mtDNA sequences published by Besansky et al. (1994) (GenBank accession numbers U10123-U10133) were included for comparative evolutionary analysis between An. melas and other members of the An. gambiae complex. These included sequences spanning a portion of the ND4 and ND5 genes of two An. melas, three An. gambiae, two An. arabiensis, and two An. merus laboratory strains.
Estimates of within group, mean evolutionary distances were calculated for each sampled An. melas and An. gambiae population, each An. melas population cluster (see below), and within each species in the program MEGA ver. 5.05 (Tamura et al. 2011) using the method of Nei and Kumar (2000) under a Kimura 2-parameter substitution model (Kimura 1980) with a gamma distribution. Fixed nucleotide differences were calculated in the program DnaSP ver. 5.10.01 (Librado and Rozas 2009). Relationships between mtDNA haplotypes were visualized by computing minimum spanning trees (Kruskal 1956, Prim 1957) in the program Arlequin ver. 3.5.1.2 (Excoffier and Lischer 2010), which were visualized and manipulated in the program HapStar (Teacher and Griffiths 2011). Dxy values between major clusters, An. gambiae and An. arabiensis were calculated based on all available mtDNA sequences using DnaSP Ver. 5.10.01 (Librado and Rozas 2009).
A maximum likelihood approach was used to investigate evolutionary relationships between the mtDNA of members of the An. gambiae complex using the program RAxML ver. 7.0.0 (Stamatakis 2006), implemented in raxmlGUI (Silvestro and Michalak 2011). The program ModelTest ver. 0.1.1 (Posada 2008, Guindon and Gascuel 2003) was used to determine the most appropriate model of nucleotide evolution (GTR+ Gamma+ Invariant Sites) based on the Akaike Information Criterion (Posada and Buckley 2004). This mutation model was implemented in the maximum likelihood analysis in RAxML, which utilized the thorough bootstrap approach with 1,000 bootstrap replicates. The resulting tree with the highest bootstrap support values was visualized using FigTree ver. 1.3.1 (Rambaut 2009).
Next, we used a Bayesian analysis implemented in the program BEAST ver. 1.6.1 (Drummond and Rambaut 2007) to estimate divergence times between lineages of An. melas, and between An. melas, An. gambiae, and An. arabiensis. An initial starting tree was created in RAxML, which included the most sampled haplotypes from each of the An. melas population clusters, the Tiko haplotype most similar to Bioko Island haplotypes, and the most sampled haplotypes from the two An. gambiae populations. Additionally, ND4-ND5 sequences from An. gambiae, An. arabiensis, and An. melas laboratory strains published in Besansky et al. (1994) were included (Table S1). We performed a likelihood ratio test in MEGA 5 to examine if our nucleotide sequences evolved in a clock-like fashion based upon the topology of our starting tree. This hypothesis could not be rejected based upon a 95% confidence interval, therefore we used a strict molecular clock approach with a 2.3% My−1 insect mtDNA nucleotide substitution rate (Gaunt and Miles 2002) to estimate divergence dates. Nodes exceeding 50% bootstrap support in the starting tree were restricted to monophyly during the run, and the time to most recent common ancestor (tmrca) was estimated for each of these nodes. Four independent runs were performed in BEAST, each with a different random seed, and 500 million steps in the Monte Carlo Markov Chain. ND4-ND5 sequences were treated as a single partition and a GTR+ Gamma+ Invariant Sites mutation model was used. Log files of each run were analyzed in Tracer ver. 1.4 (Rambaut and Drummond 2007a) to assess convergence. LogCombiner ver. 1.4 (Rambaut and Drummond 2007b) was used to combine the log and tree file outputs of each run, and to remove the first 50 million (10%) of each as a burn-in. A maximum clade credibility tree was created in the program TreeAnnotator ver. 1.4 (Rambaut and Drummond 2007c) by excluding trees that did not have a posterior probability above the mean value. The maximum clade credibility tree was visualized in FigTree ver. 1.3.1 (Rambaut 2009).
Approximate Bayesian Computation
The microsatellite data set was used to test hypotheses regarding the origins of observed population clusters using Approximate Bayesian Computation (ABC) using 24 demographic scenarios (Table S2, S3) (Beaumont 2002). Our hypotheses regarding the history of An. melas populations addressed three questions: 1) Did isolation between two mainland population clusters arise through a vicariance (scenarios 1,5–7,11–13,17–19,23,24) or founder event (scenarios 2–4,8–10,14–16,20–22)? 2) Did the Bioko population originate through vicariance (disappearing land bridge) (scenarios 1,2,7,8,13,14,19,20) or a founder event (scenarios 3–6,9–12,15–18,21–24)? 3) Which mainland cluster was the source of the Bioko island populations (scenarios 1–12 vs. 13–24)? The data were analyzed as three populations: Bioko Island, Western and Southern. The 24 scenarios represented variations and reciprocal scenarios of the main hypothesis in the program DIYABC (Cornuet et al. 2008, 2010). Data were simulated by drawing values from a uniform prior distribution defined for each parameter - effective size (Ne) and time of divergence events (t generations before present (gbp)). As ABC is a coalescent modeling approach, simulations are encoded proceeding backwards in time from the most recent sample. Parameter estimates for both past and present effective population sizes (Ne) for each sampled population were estimated using a range of pre-defined priors. Estimates of the timing of past events (such as divergence, bottlenecks etc) are estimated as having taken place ‘t’ generations before present, also defined by a prior range of values. In our simulations, prior distributions are necessarily informed by existing estimates, but were also wide enough to encompass a wide range of potential values. As ABC utilizes summary statistics from sampled data, it is considered less vulnerable to assumption violations that are sometimes problematic in single-statistic estimation approaches. Initially 50,000 datasets were simulated for each scenario, and 13 scenarios with posterior probabilities equal to zero were discarded. The remaining pool of 11 scenarios was rerun for 50,000 simulations each, and another five scenarios with invariant zero posterior probabilities were excluded from further consideration. Subsequently, the remaining six scenarios were run for 100,000 simulations each, resulting in the exclusion of a further three scenarios with zero posterior probabilities. For each of the final three scenarios one million data sets were simulated. After computing the Euclidean distances between observed and simulated datasets, 1% of the closest datasets were retained for calculating the posterior probabilities. The scenario with the highest posterior probability was used to estimate population genetic parameters.
Results
Microsatellite DNA Diversity and Divergence
A total of 617 An. melas wild-caught specimens from 11 populations (Table 1, Figure 1) were genotyped at 15 microsatellite loci. The mean observed heterozygosity value (HO) ranged from 18% to 72%. HO deviated significantly (p<0.05) from the expected heterozygosity (HE) in 19 out of 165 tests before Bonferroni correction (Table S4). Homozygote excess was the cause of this deviation in 18 out of 19 instances, five of which occurred in Bioko Island, E.G. populations. However, HO deviated significantly from HE in only two out of 165 tests after Bonferroni correction (p<0.0003). Homozygote excess was the explanation for this deviation in both tests, and in one of these cases null alleles were also detected using Micro-checker (van Oosterhout et al. 2004). Micro-checker detected the presence of a small number of null alleles in four other instances, which were not associated with significant deviation from HWE after Bonferonni correction. These results are similar to other studies in the An. gambiae complex where some heterozygote deficiencies were observed (e.g. Donnelly and Townson 2000, Lehman et al. 2003). Perhaps not unexpectedly, the average HO (across all 15 loci) on Bioko Island is considerably less (38%), compared to mainland populations (66%). Overall rarefied allelic richness for individual loci ranged from 1.0 to 11.2. Mean rarefied allelic richness (across all 15 loci) was also much lower in Bioko Island populations than on the mainland (2.6 vs. 5.3).
Figure 1.
An. melas sample locations throughout West Africa, including Bioko Island, Equatorial Guinea and neighboring Cameroon and mainland Equatorial Guinea (inset).
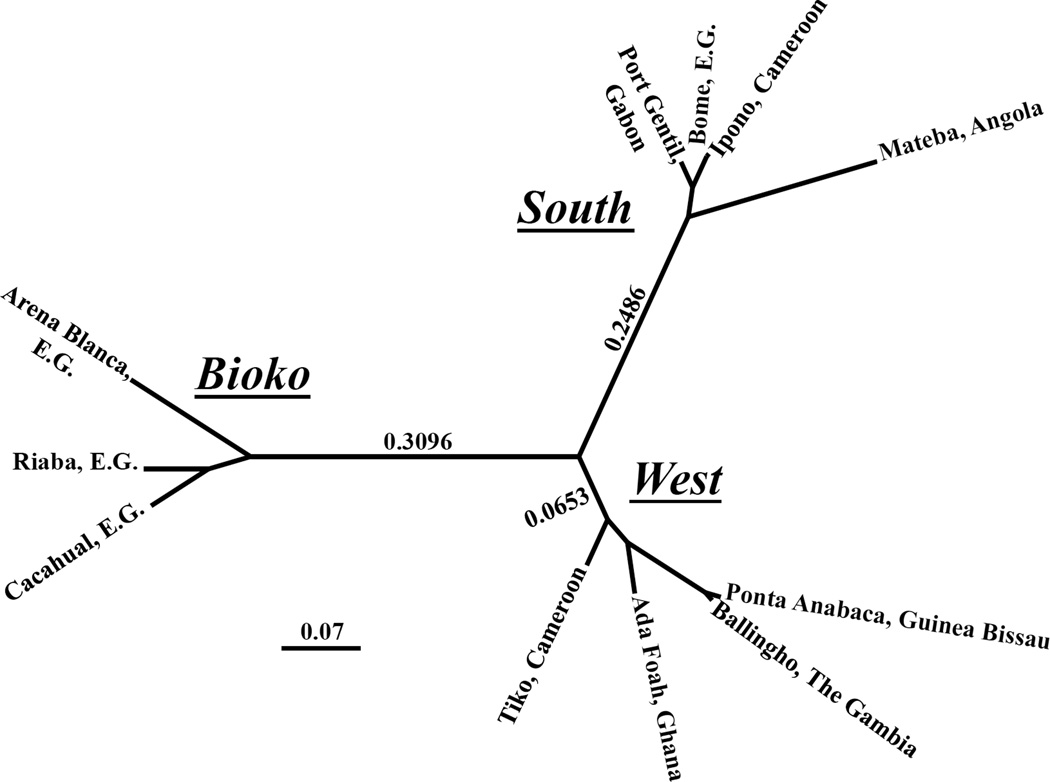
Population pair-wise FST and G”ST statistics are reported in Table 2. Significant genetic differentiation (FST, p<0.05) was observed in all but one population pair-wise comparison. Given the patchy distribution of this species this is not surprising. The one comparison for which no significant differentiation was observed between two populations only approximately 40 km apart, Ipono, Cameroon and Bome, Equatorial Guinea (FST=0.001). To visualize the pattern of genetic differentiation between An. melas populations, a neighbour-joining tree was constructed using G”ST values (Figure 2).
Table 2.
Pair-wise estimates of genetic divergence (G”ST and FST) values between 11 An. melas populations calculated using data from 15 microsatellite loci. Top diagonal: pair-wise G”ST values. Bottom diagonal: pair-wise FST values (significant values (p-val.<0.05) in bold). FST significance values were computed using 10,000 permutations. Population abbreviations correspond with those defined in Table 1.
| ARB | RIA | CAC | TIK | GHA | GAM | GUI | IPO | BOM | GAB | ANG | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ARB | − | 0.2620 | 0.2145 | 0.5546 | 0.6335 | 0.6143 | 0.6167 | 0.7513 | 0.7651 | 0.7497 | 0.8495 |
| RIA | 0.1548 | − | 0.1251 | 0.5097 | 0.5988 | 0.5891 | 0.5742 | 0.7173 | 0.7291 | 0.7125 | 0.8222 |
| CAC | 0.1285 | 0.0740 | − | 0.5053 | 0.5914 | 0.6014 | 0.5759 | 0.7385 | 0.7502 | 0.7304 | 0.8384 |
| TIK | 0.2803 | 0.2548 | 0.2408 | − | 0.1295 | 0.1628 | 0.1791 | 0.3859 | 0.4165 | 0.4332 | 0.5782 |
| GHA | 0.3468 | 0.3243 | 0.3260 | 0.0318 | − | 0.1512 | 0.1455 | 0.4155 | 0.4372 | 0.4227 | 0.5991 |
| GAM | 0.2722 | 0.2580 | 0.2475 | 0.0450 | 0.0457 | − | 0.0248 | 0.4787 | 0.5142 | 0.5147 | 0.6279 |
| GUI | 0.2885 | 0.2654 | 0.2490 | 0.0500 | 0.0449 | 0.0073 | − | 0.4883 | 0.5184 | 0.5229 | 0.6424 |
| IPO | 0.3476 | 0.3283 | 0.3205 | 0.1081 | 0.1252 | 0.1428 | 0.1480 | − | 0.0032 | 0.0713 | 0.2607 |
| BOM | 0.3589 | 0.3385 | 0.3309 | 0.1184 | 0.1336 | 0.1552 | 0.1592 | 0.0010 | − | 0.0487 | 0.2463 |
| GAB | 0.3696 | 0.3476 | 0.3408 | 0.1279 | 0.1361 | 0.1596 | 0.1654 | 0.0227 | 0.0157 | − | 0.2342 |
| ANG | 0.4635 | 0.4446 | 0.4519 | 0.2055 | 0.2390 | 0.2203 | 0.2325 | 0.0938 | 0.0897 | 0.0899 | − |
Figure 2.
Dendrogram of populations of An. melas based upon pair-wise G”ST values, constructed using a neighbour-joining cluster analysis. Branch lengths (average genetic distances) are indicated between clusters.
An. melas populations are clearly subdivided into three major clusters with a very high level of genetic differentiation between them. Perhaps not surprisingly, the Bioko Island populations constitute a single cluster. However, An. melas populations on the mainland have also diverged into two highly differentiated clusters. These clusters are allopatric and are hereinafter referred to as the Western and Southern cluster. The division between these two mainland clusters falls between two Cameroonian populations, which are only about 190 km apart along the coastline; Tiko and Ipono. None-the-less, these populations are highly differentiated (FST=0.108, G”ST=0.386). This is 3.7 times higher (G”ST) than that between Tiko and the most distant population within its cluster, The Gambia. It seems that the Tiko, Cameroon population lies at the center of divergence within An. melas, as it is also the closest mainland population to Bioko Island, both geographically and genetically.
The results of a global analysis of molecular variance (AMOVA), with populations assigned to the Bioko Island, Western, and Southern clusters, also indicate the high level of divergence within An. melas, since 22.3% of the genetic variation is explained by differences between the three clusters (Table 3). Differences between populations within these three groups account for only 4.3% of the observed genetic variation, with differences between individuals within populations explaining the remaining 73.4%.
Table 3.
Results of a global analysis of molecular variance (AMOVA), as a weighted average over 15 An. melas microsatellite loci. Groups were defined as Western, Southern, and Bioko Island population clusters.
| Source of Variation | Degrees of Freedom |
Sum of Squares |
Variance Compontents |
Percentage of Variation |
|---|---|---|---|---|
| Among Groups | 2 | 1092.189 | 1.23965 Va | 22.29 |
| Among Populations Within Groups | 8 | 223.697 | 0.23889 Vb | 4.30 |
| Within Populations | 1233 | 4992.281 | 4.08200 Vc | 73.41 |
| Total | 1233 | 6308.167 | 5.56053 | |
The Bayesian assignment test implemented in STRUCTURE strongly supports the presence of three populations, corresponding to the Bioko Island, Western and Southern population clusters (Figure 3). LnP(K) clearly flattened out at K=3 (Figure S1). This is indicative of the true K, as LnP(K) typically stops increasing once the real K is reached, and then plateaus at larger values (Pritchard and Wen 2003). Evanno et al. (2005) devised a method for detecting the break in this slope. Based on their simulation results, they reported that their method often provides a clear mode of their statistic ΔK at a single true value of K. This was not the case in our analyses. ΔK showed high values at both K=2 and K=3 (Figure S2). This is due to the larger increase in LnP(K) between K=1 and K=2 vs between K=2 and K=3.
Figure 3.
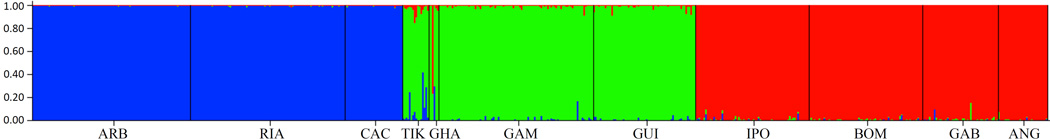
Results of the Bayesian assignment test for three putative populations (K=3) based upon microsatellite DNA data implemented in the program Structure (Pritchard et al. 2000). Each vertical bar corresponds to a single individual, and colors represent the proportion of the genome that is assigned to a particular cluster based upon the admixture model. Sample populations are annotated according to population abbreviations defined in Table 1. Bioko Island populations are represented in blue, mainland West populations are represented in green, and mainland South populations are represented in red.
Individuals with mixed genotypes from different clusters could possess immigrant ancestry, or could be evidence of remnant genetic similarity between recently diverged populations. The likelihood bar plot for K=3 (Figure 3, see Figure S3 for K=2) shows that Bioko Island represents a highly genetically distinct population, with no evidence of migrants from the mainland. The Western cluster has the highest proportion of migrants (Figure 3), which are found primarily in the two populations closest to the Southern cluster, Tiko and Ada Foah. In Tiko, several individuals show evidence of recent immigration from Bioko Island. These results indicate that migration between Bioko Island and the mainland has been from Bioko Island to the Western cluster, and migration between mainland populations is from Southern populations to Tiko and Ada Foah.
Mitochondrial DNA Diversity and Divergence
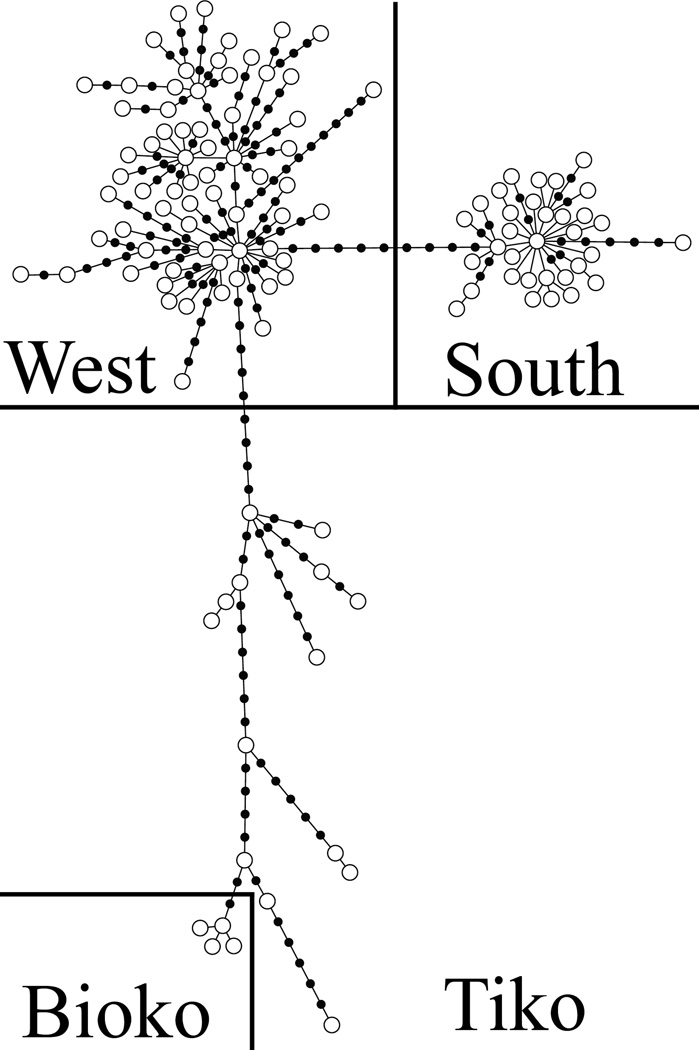
We obtained 1,161 bp of the ND4 and ND5 genes for a total of 247 An. melas specimens from all 11 populations, and from 154 An. gambiae specimens from Mongola, Bioko Island and Ukomba, mainland E.G. (Table 1). Patterns of mtDNA ND4-ND5 genetic diversity within, and between An. melas populations show a very similar pattern as that based on the microsatellite DNA. Bioko Island populations have extremely low levels of mtDNA diversity, with only four haplotypes observed in 90 samples (Table 4). Within-population genetic divergence on Bioko Island is between 2.5 to 26.8 times lower than genetic divergence in mainland populations. The three population clusters are evident in the mtDNA data as well (Figure 4). Haplotypes from each of the groups form distinct, highly differentiated clusters. The exception to this pattern is the Tiko population, which is intermediate between the Western and Bioko Island cluster. Three base-pair substitutions differentiate Bioko Island from the most closely related Tiko haplotype. No mtDNA haplotypes are shared between any of the three major clusters identified using microsatellite markers. The Western and Southern clusters have two fixed differences between them, but their haplotypes differ by a minimum of 10 base-pair substitutions. Interestingly, Tiko does not share any haplotypes with the Western cluster, and Tiko haplotypes are divergent from other West populations by a minimum of 10 base-pair substitutions. There are also two fixed nucleotide differences between the Western and Bioko Island groups, and no less than 14 between the Southern and Bioko Island clusters.
Table 4.
Estimates of mean mtDNA sequence divergence within sampled An. melas populations and clusters, and An. gambiae populations. The rate variation among sites was modeled with a gamma distribution (shape parameter = 6). Codon positions included were 1st+2nd+3rd+noncoding. There were a total of 1161 bp positions in the final dataset.
| Species / Population | Distance Estimate |
Standard Error |
|---|---|---|
| An. melas | ||
| Arena Blanca, Equatorial Guinea (Bioko) | 0.00021 | 0.00014 |
| Riaba, Equatorial Guinea (Bioko) | 0.00031 | 0.00021 |
| Cacaual, Equatorial Guinea (Bioko) | 0.00027 | 0.00019 |
| Tiko, Cameroon | 0.00706 | 0.00143 |
| Ada Foah, Ghana | 0.00509 | 0.00122 |
| Ballingho, The Gambia | 0.00579 | 0.00115 |
| Ponta Abanaca, Guinea Bissau | 0.00576 | 0.00101 |
| Ipono, Cameroon | 0.00317 | 0.00073 |
| Bome, Equatorial Guinea | 0.00261 | 0.00085 |
| Port Gentil, Gabon | 0.00259 | 0.00059 |
| Mateba, Angola | 0.00065 | 0.00039 |
| Western Cluster | 0.00786 | 0.00112 |
| Western Cluster (without Tiko, Cameroon) | 0.00588 | 0.00100 |
| Southern Cluster | 0.00255 | 0.00055 |
| Bioko Island Cluster | 0.00024 | 0.00014 |
| Within all An. melas | 0.01045 | 0.00173 |
| An gambiae | ||
| Mongola, Equatorial Guinea (Bioko) | 0.00312 | 0.00096 |
| Ukomba, Equatorial Guinea | 0.00366 | 0.00180 |
| Within all An. gambiae | 0.00389 | 0.00108 |
Figure 4.
An. melas minimum spanning tree (Kruskal 1956, Prim 1957) constructed in the program Arlequin ver. 3.5.1.2 (Excoffier and Lischer 2010), and visualized using HapStar (Teacher and Griffiths 2011). White circles represent sampled haplotypes and black intermediate circles represent ancestral or unsampled haplotypes. Population clusters are annotated according to An. melas genetic cluster.
This high level of mtDNA divergence between An. melas Bioko Island and mainland populations contrasts strongly with that between An. gambiae (M-form) populations located on Bioko Island (Mongola), and mainland Equatorial Guinea (Ukomba). These populations share the same haplotypes and unique haplotypes do not cluster by location (Figure S4). Although both of these have levels of within population genetic divergence on par with those from mainland An. melas populations (Table 4), no geographic structure was detected between mainland and Bioko Island An. gambiae populations. Both populations shared the two most sampled haplotypes, and haplotypes did not cluster by population (Figure S4).
A minimum spanning tree including mtDNA haplotypes from An. melas, An. gambiae, as well as An. arabiensis demonstrates that the most commonly sampled haplotypes from the Southern, Western (excluding Tiko), and Bioko Island population clusters are as divergent from each other (Dxy = 0.8% to 1.5%) as any of these An. melas clusters is from An. gambiae or An. arabiensis (Dxy = 0.5% to 1.6%) (Figure S5). This exemplifies the very high level of divergence observed within An. melas. The same patterns are observed in an unrooted Maximum Likelihood tree (Figure S6) that includes all unique haplotypes from An. melas and An. gambiae populations, as well as haplotypes from An. arabiensis, An. quadriannulatus, and An. merus (Besansky et al. 1994) (Table S1). The internal nodes of this Maximum Likelihood tree are not resolved as they have low bootstrap support. As expected, An. melas haplotypes once again form the previously identified monophyletic clusters, with Tiko, Cameroon haplotypes grouping with the Bioko Island haplotypes. A single exception was found in a specimen (GUIP905) from Guinea Bissau (Western cluster) that fell out more similar to the An. gambiae and An. arabiensis cluster. Importantly however, we did not find any support for An. melas being monophyletic within the An. gambiae complex (figure S6). This suggests that the An. melas clusters diverged from each other soon after the ancestral An. melas split from the other species of the complex. An alternative scenario of an independent origin of these clusters is less likely because of the similarity in larval ecology, shared fixed inversions (Coluzzi et al. 2002) and a diagnostic substitution in the rDNA.
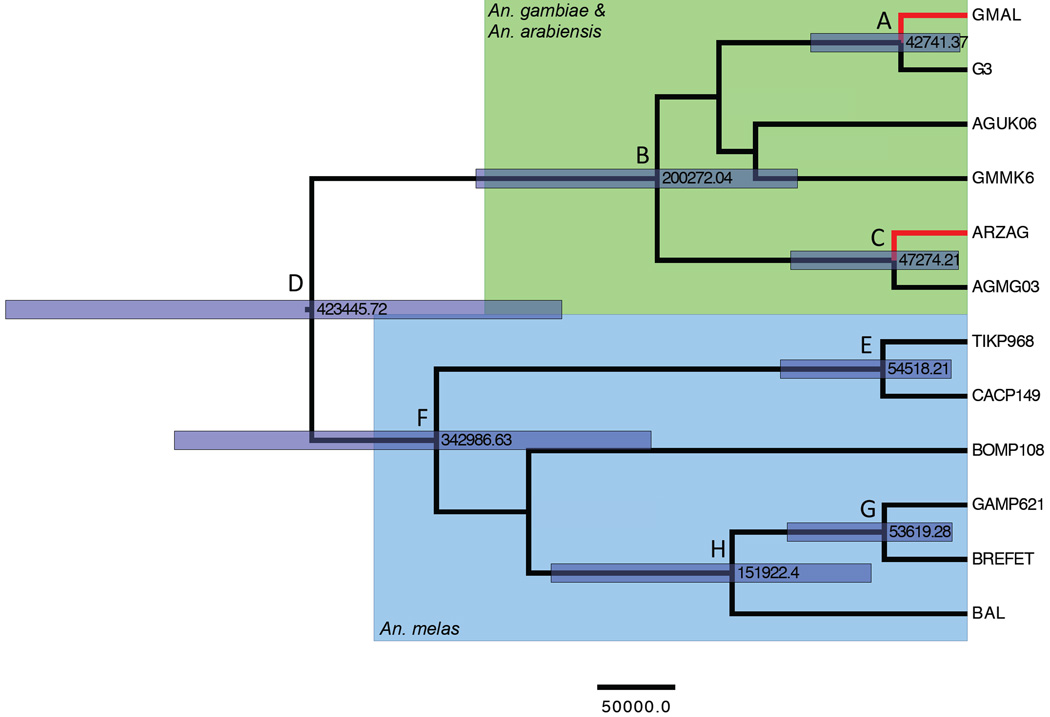
Using a Bayesian approach implemented in BEAST, we estimated divergence times between An. melas population clusters, as well as between An. gambiae, An. arabiensis, and An. melas (Figure 5 and Table 5). Bootstrap support in the starting tree was not sufficient between Western and Southern samples to calculate a divergence date between them. Median divergence estimates and 95% highest posterior density (HPD) statistics are reported in Table 5. This analysis dates the split of An. gambiae and An. melas lineages to 423,455 years before present (ybp) (95% HPD: 261,993 – 621,259), and the split between Bioko Island populations and Tiko, Cameroon to 54,518 ybp (95% HPD: 10,214 – 120,777)
Figure 5.
Unrooted phylogenetic tree including An. gambiae (G3 & GMMK6), An. arabiensis (GMAL & ARZAG), and An. melas (BREFET & BAL) (Besansky et al. 1994), An. gambiae from Mongola, Bioko Island (AGMG03) and Ukomba, Equatorial Guinea (AGUK06) as well as An. melas populations sampled for this study. Sampled An. melas individuals represent the most commonly sampled haplotypes from their respective population clusters: CACP149 is from Cacahual, Bioko Island, Equatorial Guinea, BOMP108 is from Bome, in the Southern cluster, and GAMP621 is from Ballingho, The Gambia in the Western cluster. TIKP968 is the Tiko haplotype most closely related to the Bioko Island population cluster. The green background indicates An. gambiae and An. arabiensis individuals, with An. arabiensis individuals indicated by red branches. Branches on a blue background indicate An. melas individuals. Violet bars represent the 95% HPD of divergence estimates, with the mean estimate annotated at the node in years before present.
Table 5.
Divergence date estimates between An. melas population clusters and An. gambiae and An. arabiensis individuals. Node names correspond with tree annotations in Figure 10. HPD, highest posterior density. BP, before present.
| Node | Median Divergence Date (years BP) |
Lower 95% HPD (years BP) |
Upper 95% HPD (years BP) |
|---|---|---|---|
| A | 42,741.37 | 4,801.66 | 101,272.76 |
| B | 200,272.04 | 109,769.74 | 317,389.87 |
| C | 47,274.21 | 5,784.25 | 114,177.44 |
| D | 423,445.72 | 261,993.99 | 621,259.42 |
| E | 54,518.21 | 10,214.47 | 120,777.02 |
| F | 342,986.63 | 204,171.86 | 512,187.90 |
| G | 53,619.28 | 9,744.75 | 116,168.92 |
| H | 151,922.40 | 62,089.65 | 268,870.96 |
Approximate Bayesian Computation
Among the 24 competing scenarios (Table S2), scenario 1, 2 and 7 were retained in the final pool of three scenarios for which 1 million simulations were conducted. This analysis resulted in scenario 1 having the highest posterior probability (0.66). This scenario describes a vicariance event (as opposed to a founder event) between the Southern and Western cluster. Additionally, this scenario also described a vicariance event between the Bioko population and the Western cluster. This is consistent with the isolation of the Bioko Island population being the result of the disappearance of a land bridge, rather than a founder event creating a population on an isolated island. Scenario 7 also had a relatively high posterior probability (0.317). This scenario describes an identical topography as scenario 1, except that in scenario 1 the Southern cluster is ancestral to the Western cluster and in scenario 7 this situation is reversed. This merely indicates that the ancestry between the Western and Southern is actually not well resolved. This is to be expected, as the issue of ancestry is largely irrelevant in a vicariance event.
Parameter estimates from the data sets simulated under scenario 1, indicate that the divergence of Bioko from the Western cluster was estimated to have occurred around 1,798,484 gbp (credibility interval (CI) 507,478–6,387,175) (Figure S7). Assuming 20 generations per year, this equates to approximately 90,000 years before present (ybp). The estimate of the divergence between the Western and Southern populations is less precise, as a wide range of values is almost equally likely (Figure S7), but the mode of the posterior density plot indicates that the most likely divergence time between the two clusters was approximately 7,215,619 gbp (CI 1,723,555–7,221,969). Again assuming 20 generation per year, this would equate to approximately 360,000 ybp. The Western and Southern populations have a very similar contemporary effective population size (Ne). The Ne of the Western cluster is estimated at 20,229 (CI 8,891–30,908), and that of the Southern cluster is estimated at 19,473 (CI 9,011–30,782) (Figure S9). Not surprisingly for an isolated island population, the Ne for Bioko Island, estimated at 1,010 (CI 450–5,735) (Figure S9), is much lower than either of the two mainland clusters.
Discussion
Our study of the population structure of An. melas has revealed yet another level of complexity within the Anopheles gambiae complex, by showing that this species is also subdivided into several genetically isolated groups, some of which may very well represent incipient species. This subdivision is evident in both the microsatellite and the mtDNA data. G”ST values based on microsatellite data are extraordinarily high, both between Bioko Island and the mainland, as well as between the two mainland groups. In addition, as far as we can tell, the Western and Southern clusters do not share mtDNA haplotypes and have fixed genetic differences between them.
This divergence process within An. melas, appears to be mostly allopatric. Populations on Bioko Island are separated from the mainland by approximately 40 km of the Gulf of Guinea, which poses an obvious barrier to gene flow. On the continent, the populations from the Western and Southern group that are closest (Tiko and Ipono, respectively) are approximately 190 km apart along the coastline. Although the Structure analyses indicated a small amount of introgression from the Southern into the Western group (primarily to Tiko), the high level of genetic isolation between the two clusters suggests that either little direct contact exists, or pre- and/or post-mating isolation mechanisms are in place that largely prevent the exchange of genes.
The results from our ABC analyses indicate that the two mainland clusters arose through a vicariance event, rather than a founder event. This indicates that a single ancestral An. melas population was divided in two by a geographic barrier. This barrier to gene flow arose approximately 360,000 ybp, separating this widespread species into the two current mainland clusters. Interestingly, the break between the Western and Southern clusters corresponds closely to the location of the Cameroon line, a chain of volcanoes that gave rise to Mount Cameroon and several islands in the Gulf of Guinea, including Bioko Island. Even though the Cameroon line is much older (> 30 million years (Burke 2001) than the vicariance event between the two mainland clusters, it is tempting to speculate that it played a role in separating the two clusters. Mount Cameroon is located on the coast, and in concert with a more recent rise in sea level could potentially have resulted in a geographic barrier, splitting the distribution of the ancestral An. melas in two, and preventing/limiting migration. Such a scenario should include the more recent separation of the Tiko population, which is located to the east of Mount Cameroon, from the Western cluster through a similar process. That Mount Cameroon may pose a barrier to gene flow between Tiko and the other Western populations, is supported by the mtDNA divergence between them.
Compared to a microsatellite study of An. gambiae populations across 1,700 km (Slotman et al. 2007), genetic differentiation (FST) between Western and Southern An. melas populations is remarkably high. It is between 1.5 to 10-fold the level of differentiation found between sympatric M and S forms of An. gambiae, which are widely considered incipient species (della Torre et al. 2001, Turner et al. 2005). It is also on par with levels of microsatellite differentiation found between An. gambiae and An. arabiensis. FST values between Bioko Island and Southern An. melas populations are actually about 1.5-fold higher than those observed between sympatric An. gambiae and An. arabiensis populations (Slotman et al. 2007).
The high level of divergence within An. melas is also evident from the fact that the mtDNA haplotypes did not form a monophyletic group in relation to the other species in the An. gambiae complex. This suggests that the An. melas clusters diverged soon after An. melas split from the other species in the complex, and our results raise the question whether these clusters represent incipient species within An. melas, or perhaps even previously unrecognized species. Although fixed genetic differences are a good indicator that these clusters may be independently evolving lineages (De Queiroz 2007), the presence of hybrid sterility would put the issue beyond discussion (Davidson 1962, Hunt et al. 1998).
The patchy distribution of An. melas throughout its range is clearly represented in its population structure. Even within the three major clusters, An. melas populations are far from panmictic. With a single exception, every population was highly significantly differentiated from all other populations in its cluster. In addition, pairwise FST values within clusters are high compared to its sister species An. gambiae, with microsatellite-based FST values being as much as 8.6 – fold higher than found between S form populations of An. gambiae more than 1,700 miles apart (Slotman et al. 2007). Furthermore, FST values similar to those observed within the Western cluster were the basis for concluding that M forms in Mali vs Cameroon are highly differentiated, indicating geographic isolation (Slotman et al. 2007). These comparisons indicate that the patchy distribution of An. melas due to its association with brackish-water mangrove coastal areas, has resulted in a deep population structure even on a smaller geographic scale, with limited current gene flow.
Coluzzi et al. (2002 and supporting online material) showed that allopatric populations of An. melas from The Gambia/Guinea Bissau, Benin/Togo and Congo/Angola were characterized by different inversion polymorphisms, leading authors to state “the nonrandom distribution of inversions … supports additional taxonomic splitting within … A. melas …”. The differences between the An. melas populations of Angola and the other populations have been recently confirmed (Calzetta et al. 2008 and Petrarca, personal communication). However, when inversions are used as genetic markers they can suggest population isolation where none exists, since the frequency of some chromosomal inversions in the gambiae complex is correlated with ecological conditions such as aridity (Coluzzi et al. 1979), indicating they are subject to selection. Whatever the case may be, the An. melas inversion distribution described by Coluzzi et al. (2002) appears to parallel the Western and Southern cluster.
Regardless of whether the two mainland clusters are in fact different species, the genetic distinctness of the three An. melas clusters has major implications for our understanding of the biology of this malaria vector. Very few studies of the biology of this species are available, but they do provide important information for malaria control. For example, Reddy et al. (2011) recently examined the host feeding behavior of An. melas on Bioko Island, and found that it readily feeds both indoors and outdoors. Such information is important for designing effective control strategies, as a partially outdoor feeding population would only be affected to some extent by the widely used indoor residual spraying (IRS) and insecticide treated nets (ITN). The high level of genetic differentiation within An. melas makes clear however that it cannot be assumed that information on the biology/ecology of the species gained from one location will be applicable throughout its range.
During this study, particular attention was paid to Bioko Island, where An. melas is a main malaria vector. Bioko Island lies on the continental shelf in the Gulf of Guinea. It is hypothesized that Bioko was connected to mainland Cameroon during the last glaciation and became isolated as sea levels rose. Therefore, the fauna of Bioko Island is species-rich and is closely associated with that of coastal Cameroon. Furthermore, endemism is low due to the island’s recent isolation (Jones 1994), which occurred approximately 10,000–11,000 years ago (Eisentraut 1965, Moreau 1966).
Based on our estimates of divergence times of the mtDNA, Bioko An. melas populations have been isolated from the mainland since between 10,214 to 120,777 ybp. This is a rather wide estimate, but it does indicate that An. melas on Bioko Island has been isolated from the mainland at least since the last glaciation. While molecular clock approaches are widely utilized to estimate times of divergence, they have also been criticized. Concerns include the incorrect estimation of calibration points (fossil dates) upon which mutation rates are calculated (Warnock et al. 2012), varied rates of molecular evolution between lineages, and variation of estimates among mutation models (Lanfear et al. 2010). As such, the results of this analysis should be interpreted with caution. None-the-less, the Approximate Bayesian Computation (ABC) analysis of the microsatellite data is consistent with this result, as it estimated the divergence time between Bioko and the Western cluster at approximately 90,000 ybp. The ABC analysis also showed that the An. melas populations on Bioko Island separated from the mainland through a vicariance event. Therefore, these populations were once connected, perhaps just prior to the last glaciation, but possibly earlier than that.
The alternative hypothesis of An. melas populations on Bioko Island originating from a founder event was not supported by the ABC analysis. Interestingly, genetic diversity is much lower on Bioko Island relative to the mainland, which is usually a typical result of a founder event. We know that recent malaria control activities under the BIMCP have drastically reduced the effective population size of this species on the island (Athrey et al., in review), which could also have resulted in a loss of genetic variation. However, this is unlikely to be the whole explanation as the three populations on the island are now nearly fixed for the same mtDNA haplotype, a situation that is unlikely to have resulted from recent vector control unless little haplotype diversity was present before the initiation of the control. Therefore, a small long-term Ne is likely to be at least partially responsible for the low level of genetic variation observed in An. melas populations on Bioko Island.
Surprisingly, the migration pattern of An. melas between Bioko Island and the mainland contrasts starkly with that of its sister species An. gambiae s.s, the other malaria vector on the island. MtDNA data of this latter species shows little or no evidence of genetic differentiation between the mainland and island populations, indicating recent/ongoing migration. Haplotypes are shared between the island and the mainland, and do not cluster by location. Additionally, previous work using microsatellites by Moreno et al. (2007) also found no substructure between An. gambiae Bioko Island and Equatorial Guinea mainland populations. Although we do not know the reason for the contrast in migration pattern between the two sibling species, perhaps this means that mosquito migration to the island is human-mediated. It is conceivable that An. gambiae, which is more closely associated with humans, would have greater opportunities for human-mediated migration.
Being an island with highly endemic malaria, Bioko Island is a potential candidate for a future malaria elimination campaign. Therefore, it is important to understand the probability of the re-introduction of eliminated vectors, or malaria parasites through infected mosquito migrants. Based on the present data, the probability of the reintroduction of An. melas is very low. An. gambiae s.s on the other hand, would have a high probability of being reintroduced into Bioko Island based on both this and other work (Moreno et al. 2007). Any elimination campaign would have to take measures to reduce this possibility. Malaria endemic islands are also of interest as potential sites for experimental releases of transgenic mosquitoes. If a high level of containment is desired for such an experiment, Bioko Island may not be an appropriate candidate.
Our study has unveiled an additional layer of complexity within the An. gambiae complex by identifying three highly isolated clusters within An. melas. Based on the observed level of divergence, these clusters may represent incipient species. Moreover, the patchy distribution of An. melas is reflected in the high levels of genetic differentiation between populations within each of the clusters. In addition to enhancing our understanding of the evolution of the An. gambiae complex, this work also has implications for malaria control programs in particular on Bioko Island, as well as other locations where this species is a dominant vector.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by an operational research grant to MAS by the Bioko Island Malaria Control Project. The BIMCP is funded by a consortium led by Marathon Oil Corporation (Houston, TX) and the Government of Equatorial Guinea.. Our thanks go out to Dr. Chris Schwabe, Dr. Luis Segura, and Ed Aldrich from Medical Care Development International, Dr. Gloria Nseng and Simon Abaga from the National Malaria Control Program and Jaime Kuklinski from One World Development Group for operations support in Equatorial Guinea. Vamsi Reddy and Vani Kulkarni provided technical support at Texas A&M University. We are thankful to Dr. Gregory Lanzaro for providing An. melas samples from Guinea Bissau and to Mr. Gian Carlo Carrara and representatives of the Angolan Ministry of Health for helping in the Angolan collections. Finally, we are also thankful to Dr Spencer Johnston, Dr Raul Medina, Dr Kostya V Krutovsky and Dr. Chris Schwabe for providing advise and/or comments on the manuscript. The partial support of the work was provided by NIH/NIAID grant R01AI085079 to MAS.
Footnotes
DATA ACCESSABILITY
Sequence data has been deposited in Genbank under accession numbers JX117425-JX117825. The microsatellite data has been deposited in Dryad doi:10.5061/dryad.d15d4
REFERENCES
- Akogbeto M, Di Deco MA. Répartition des membres du complexe Anopheles gambiae et de leurs variants chromosomiques au Bénin et au Togo, Afrique occidentale. Journal of African Zoology. 1995;109:443–454. [Google Scholar]
- Akogbeto M, Romano R. Infectivité d’Anopheles melas vis-à-vis du Plasmodium falcip arum dans le milieu côtier lagunaire du Benin. Bull. Soc. Pathol. Exot. 1999;92:3–5. [PubMed] [Google Scholar]
- Bafort JM, Petrarca V. Contribution to the knowledge of Anopheles melas and An. gambiae in West Africa. Annales de la Société Belge de Médecine Tropicale. 1983;63:167–170. [PubMed] [Google Scholar]
- Beard CB, Hamm DM, Collins FH. The mitochondrial genome of the mosquito Anopheles gambiae: DNA sequence, genome organization, and comparisons with mitochondrial sequences of other insects. Insect Molecular Biology. 1993;2:103–124. doi: 10.1111/j.1365-2583.1993.tb00131.x. [DOI] [PubMed] [Google Scholar]
- Beaumont MA, Zhang WY, Balding DJ. Approximate Bayesian computation in population genetics. Genetics. 2002;162:2025–2035. doi: 10.1093/genetics/162.4.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besansky NJ, Lehman T, Fahey GT, Fontenille D, Braack LEO, Hawley WA, Collins FH. Patterns of mitochondrial variation within and between African malaria vectors, Anopheles gambiae and An. arabiensis, suggest extensive gene flow. Genetics. 1997;147:1817–1828. doi: 10.1093/genetics/147.4.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besansky NJ, Powell JR, Caccone A, Hamm DM, Scott JA, Collins FH. Molecular phylogeny of the Anopheles gambiae complex suggests genetic introgression between principal malaria vectors. Proceedings of the National Academy of Sciences USA. 1994;91:6885–6888. doi: 10.1073/pnas.91.15.6885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bøgh C, Lindsay SW, Clarke SE, Dean A, Jawara M, Pinder M, Thomas CJ. High spatial resolution mapping of malaria transmission risk in The Gambia, West Africa, using landsat TM satellite imagery. American Journal of Tropical Medicine and Hygiene. 2007;76:875–881. [PubMed] [Google Scholar]
- Bryan JH. Anopheles gambiae and An. melas at Brefet, The Gambia, and their role in malaria transmission. Annals of Tropical Medicine and Parasitology. 1983;77:1–12. doi: 10.1080/00034983.1983.11811667. [DOI] [PubMed] [Google Scholar]
- Bryan JH, Petrarca V, Di Deco MA, Coluzzi M. Adult behavior of members of the Anopheles gambiae complex in the Gambia with special reference to An. melas and its chromosomal variants. Parassitologia. 1987;29:221–249. [PubMed] [Google Scholar]
- Burke K. Origin of the Cameroon Line of volcano-capped swells. The Journal of Geology. 2001;109:349–362. [Google Scholar]
- Calzetta M, Santolamazza F, Carrara GC, Cani PJ, Fortes F, Di Deco MA, della Torre A, Petrarca V. Distribution and chromosomal characterization of the Anopheles gambiae complex in Angola. American Journal of Tropical Medicine and Hygiene. 2008;78:169–175. [PubMed] [Google Scholar]
- Caputo B, Nwakanma D, Jawara M, Adiamoh M, Dia I, Konate L, Petrarca V, Conway DJ, della Torre A. Anopheles gambiae complex along The Gambia river, with particular reference to the molecular forms of An. gambiae s.s. Malaria Journal. 2008;7:182. doi: 10.1186/1475-2875-7-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coluzzi M, Petrarca V, Di Deco MA. Chromosomal inversion intergradation and incipient speciation in Anopheles gambiae . Bollenttino Di Zoologia. 1985;52:45–63. [Google Scholar]
- Coluzzi M, Sabatini A, della Torre A, Di Deco MA, Petrarca V. A polytene chromosome analysis of the Anopheles gambiae species complex. Science. 2002;298:1415–1418. doi: 10.1126/science.1077769. [DOI] [PubMed] [Google Scholar]
- Coluzzi M, Sabatini A, Petrarca V, Di Deco MA. Chromosomal differentiation and adaptation to human environments in the Anopheles gambiae complex. Transactions of the Royal Society of Tropical Medicine and Hygiene. 1979;73:483–497. doi: 10.1016/0035-9203(79)90036-1. [DOI] [PubMed] [Google Scholar]
- Cornuet JM, Ravigne V, Estoup A. Inference on population history and model checking using DNA sequence and microsatellite data with the software DIYABC (v1.0) BMC Bioinformatics. 2010;11:401. doi: 10.1186/1471-2105-11-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornuet JM, Santos F, Beaumont MA, Robert CP, Marin JM, et al. Inferring population history with DIY ABC: a user-friendly approach to approximate Bayesian computation. Bioinformatics. 2008;24:2713–2719. doi: 10.1093/bioinformatics/btn514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson G. The Anopheles gambiae complex. Nature. 1962;196:907. [Google Scholar]
- Deitz KC, Reddy VP, Reddy MR, Satyanarayanah N, Lindsey MW, Overgaard HJ, Jawara M, Caccone A, Slotman MA. Limited Usefulness of Microsatellite Markers from the Malaria Vector Anopheles gambiae when applied to the Closely Related Species An. melas . Journal of Heredity. 2012 doi: 10.1093/jhered/ess025. [DOI] [PubMed] [Google Scholar]
- della Torre A, Fanello C, Akogbeto M, Dossou-Yovo J, Favia G, Petrarca V, Coluzzi M. Molecular evidence of incipient speciation within Anopheles gambiae s.s. in West Africa. Insect Molecular Biology. 2001;20:9–18. doi: 10.1046/j.1365-2583.2001.00235.x. [DOI] [PubMed] [Google Scholar]
- della Torre A, Tu Z, Petrarca V. On the distribution and genetic differentiation of Anopheles gambiae s.s. molecular forms. Insect Biochemistry and Molecular Biology. 2005;35:755–769. doi: 10.1016/j.ibmb.2005.02.006. [DOI] [PubMed] [Google Scholar]
- de Queiroz K. Species concepts and species delimitation. Systematic Biology. 2007;56:879–886. doi: 10.1080/10635150701701083. [DOI] [PubMed] [Google Scholar]
- Donnelly MJ, Townson H. Evidence for extensive genetic differentiation among populations of the malaria vector Anopheles arabiensis in eastern Africa. Insect Molecular Biology. 2000;9:357–367. doi: 10.1046/j.1365-2583.2000.00197.x. [DOI] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evolutionary Biology. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earl DA. Structure Harvester ver 0.6.8. 2011. See http://users.soe.ucsc.edu/~dearl/software/structureHarvester/ [Google Scholar]
- Eisentraut M. Rassenbildung bei Säugetieren und Vögeln auf der Insel Fernando Poo. Zoologischer Anzeiger. 1965;174:37–53. [Google Scholar]
- Evanno G, Regnaut G, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Lischer HEL. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology Resources. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Smouse PE, Quattro JM. Analysis of Molecuar Variance Inferred from Metric Distances Among DNA Haplotypes: Application to Human Mitochondrial DNA Restriction Data. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanello C, Santolamazza F, della Torre A. Simultaneous identification of species and molecular forms of the Anopheles gambiae complex by PCR-RFLP. Medical and Veterinary Entomology. 2002;16:461–464. doi: 10.1046/j.1365-2915.2002.00393.x. [DOI] [PubMed] [Google Scholar]
- Favia G, della Torre A, Bagayoko M, Lanfrancotti A, Sagnon N, Touré YT, Coluzzi M. Molecular identification of sympatric chromosomal forms of Anopheles gambiae and further evidence of their reproductive isolation. Insect Molecular Biology. 1997;6:377–383. doi: 10.1046/j.1365-2583.1997.00189.x. [DOI] [PubMed] [Google Scholar]
- Garrett-Jones C, Borehamn PFL, Plant CP. Feeding habits of Anophelines (Diptera:Culicidae) in 1971–78, with reference to the human blood index: a review. Bulletin of Entomological Research. 1980;70:165–185. [Google Scholar]
- Gaunt MW, Miles MA. An insect molecular clock dates to the origin of the insects and accords with palaeontological and biogeographic landmarks. Molecular Biology and Evolution. 2002;19:748–761. doi: 10.1093/oxfordjournals.molbev.a004133. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast and accurate method to estimate large phylogenies by maximum-likelihood. Systematic Biology. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Howe K, Bateman A, Durbin R. QuickTree: building huge neighbour-joining trees of protein sequences. Bioinformatics. 2002;18:1546–1547. doi: 10.1093/bioinformatics/18.11.1546. [DOI] [PubMed] [Google Scholar]
- Hunt RH, Coetzee M, Fettene M. The Anopheles gambiae complex: a new species from Ethiopia. Transactions of The Royal Society of Tropical Medicine and Hygiene. 1998;92:231–235. doi: 10.1016/s0035-9203(98)90761-1. [DOI] [PubMed] [Google Scholar]
- Jones PJ. Biodiversity in the Gulf of Guinea: an overview. Biodiversity and Conservation. 1994;3:772–784. [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. Journal of Molecular Evolution. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Kleinschmidt I, Schwabe C, Benavente L, Torrez M, Ridl FC, et al. Marked increase in child survival after four years of intensive malaria control. American Journal of Tropical Medicine and Hygiene. 2009;80:882–888. [PMC free article] [PubMed] [Google Scholar]
- Kruskal JB. On the shortest spanning subtree of a graph and the travelling salesman problem. Proceedings of the American Mathematical Society. 1956;7:48–50. [Google Scholar]
- Lanfear R, Welch JJ, Bromham L. Watching the clock: Studying variation in rates of molecular evolution between species. Trends in Ecology and Evolution. 2010;25:495–503. doi: 10.1016/j.tree.2010.06.007. [DOI] [PubMed] [Google Scholar]
- Lehman T, Licht M, Elissa N, Maega BTA, Chimumbwa JM, Watsenga FT, Wondji CS, Simard F, Hawley WA. Population Structure of Anopheles gambiae in Africa. Journal of Heredity. 2003;94:133–147. doi: 10.1093/jhered/esg024. [DOI] [PubMed] [Google Scholar]
- Librado P, Rozas J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Lis JT. Fractionation of DNA fragments by polyethylene glycol induced precipitation. Methods in Enzymology. 1980;65:347–353. doi: 10.1016/s0076-6879(80)65044-7. [DOI] [PubMed] [Google Scholar]
- Meirmans PG, Hedrick PW. Assessing population structure: FST and related measures. Molecular Ecology Resources. 2011;11:5–18. doi: 10.1111/j.1755-0998.2010.02927.x. [DOI] [PubMed] [Google Scholar]
- Meirmans PG, van Tienderen GENOTYPE and GENODIVE: two programs for the analysis of genetic diversity of asexual organisms. Molecular Ecology Resources. 2004;4:792–794. [Google Scholar]
- Moreau RE. The Bird Faunas of Africa and its Islands. London: Academic Press; 1966. [Google Scholar]
- Moreno M, Salgueiro P, Vicente JL, Cano J, Berzosa PJ, de Lucio A, Simard F, Caccone A, Do Rosario VE, Pinto J, Benito A. Genetic population structure of Anopheles gambiae in Equatorial Guinea. Malaria Journal. 2007;6:137. doi: 10.1186/1475-2875-6-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Kumar S. Molecular Evolution and Phylogenetics. New York: Oxford University Press; 2000. [Google Scholar]
- Oliveira E, Salgueiro P, Palsson K, Vicente JL, Arez AP, Jaenson TG, Caccone A, Pinto J. High levels of hybridization between molecular forms of Anopheles gambiae from Guinea Bissau. Journal of Medical Entomology. 2008;45:1057–1063. doi: 10.1603/0022-2585(2008)45[1057:hlohbm]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Overgaard HJ, Sæbø S, Reddy MR, Reddy VP, Abaga S, Matias A, Slotman MA. Light traps fail to estimate reliable malaria mosquito biting rates on Bioko Island, Equatorial Guinea. Malaria Journal. 2012;11:56. doi: 10.1186/1475-2875-11-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrarca V, Carrara GC, Di Deco MA, Petrangeli G. Il complesso Anopheles gambiae in Guinea Bissau. Parassitologia. 1983;25:29–39. [PubMed] [Google Scholar]
- Petrarca V, Vercruysse J, Coluzzi M. Observations on the Anopheles gambiae complex in the Senegal River Basin, West Africa. Medical and Veterinary Entomology. 1987;1:303–312. doi: 10.1111/j.1365-2915.1987.tb00359.x. [DOI] [PubMed] [Google Scholar]
- Posada D. jModelTest: phylogenetic model averaging. Molecular Biology and Evolution. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- Posada D, Buckley TR. Model selection and model averaging in phylogenetics: advantages of Akaike information criterion and Bayesian approaches over likelihood ratio tests. Systematic Biology. 2004;53:793–808. doi: 10.1080/10635150490522304. [DOI] [PubMed] [Google Scholar]
- Prim RC. Shortest connection networks and some generalizations. Bell System Technical Journal. 1957;36:1389–1401. [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Wen W. Documentation for structure software: Version 2. 2003 Available from http://pritch.bsd.uchicago.edu.
- Rambaut A. FigTree: Tree figure drawing tool, ver 1.3.1. Institute of Evolutionary Biology, University of Edinburgh; 2009. http://tree.bio.ed.ac.uk/software/figtree/ [Google Scholar]
- Rambaut A, Drummond AJ. Tracer ver 1.4. Institute of Evolutionary Biology, University of Edinburgh; 2007a. See http://beast.bio.ed.ac.uk/Tracer/. [Google Scholar]
- Rambaut A, Drummond AJ. LogCombiner ver 1.4. Institute of Evolutionary Biology, University of Edinburgh; 2007b. See http://beast.bio.ed.ac.uk/LogCombiner/. [Google Scholar]
- Rambaut A, Drummond AJ. TreeAnnotator ver 1.4. Institute of Evolutionary Biology, University of Edinburgh; 2007c. See http://beast.bio.ed.ac.uk/TreeAnnotator/. [Google Scholar]
- Reddy MR, Overgaard HJ, Abaga S, Reddy VP, Caccone A, Kiszewski AE, Slotman MA. Outdoor host-seeking behavior of Anopheles gambiae mosquitoes following initiation of malaria vector control on Bioko Island, Equatorial Guinea. Malaria Journal. 2011;10:184. doi: 10.1186/1475-2875-10-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp BL, Ridl FC, Govender D, Kuklinski J, Kleinschmidt I. Malaria vector control by indoor residual insecticide spraying on the tropical island of Bioko, Equatorial Guinea. Malaria Journal. 2007;6:52. doi: 10.1186/1475-2875-6-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szpiech ZA, Jakobsson M, Rosenberg NA. ADZE: a rarefaction approach for counting alleles private to combinations of populations. Bioinformatics. 2008;24:2498–2504. doi: 10.1093/bioinformatics/btn478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvestro D, Michalak I. raxmlGUI: a graphical front-end for RAxML. Organisms Diversity and Evolution. 2011 [Google Scholar]
- Sinka ME, Bangs MJ, Manguin S, Coetzee M, Mbogo CM, Hemingway J, Patil AP, Temperley WH, Gething PW, Kabaria CW, et al. The dominant Anopheles vectors of human malaria in Africa, Europe, and the Middle East: occurrence data, distribution maps, and bionomic précis. Parasites and Vectors. 2010;3:117. doi: 10.1186/1756-3305-3-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotman MA, Reimer L, Thiemann T, Dolo G, Fondjo E, Lanzaro GC. Reduced recombination rate and genetic differentiation between the M and S forms of Anopheles gambiae . Genetics. 2006;174:2081–2093. doi: 10.1534/genetics.106.059949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotman MA, Tripet F, Cornel AJ, Meneses CR, Lee Y, Reimer LJ, Thiemann TC, Fondjo E, Fofana A, Traore SF, Lanzaro GC. Evidence for subdivision within the M molecular form of Anopheles gambiae . Molecular Ecology. 2007;16:639–649. doi: 10.1111/j.1365-294X.2006.03172.x. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Stump AD, Fitzpatrick MC, Lobo NF, Traoré S, Sagnon NF, Constantini C, Collins FH, Besansky NJ. Centromere-proximal differentiation and speciation in Anopheles gambiae . Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15930–15935. doi: 10.1073/pnas.0508161102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teacher AGF, Griffiths DJ. HapStar: automated haplotype network layout and visualization. Molecular Ecology Notes. 2011;11:151–153. doi: 10.1111/j.1755-0998.2010.02890.x. [DOI] [PubMed] [Google Scholar]
- Touré YT, Petrarca V, Coluzzi M. Nuove entità del complesso Anopheles gambiae in Mali. Parassitologia. 1983;25:367–370. [Google Scholar]
- Tuno N, Kjaerandsen J, Badu K, Kruppa T. Blood-feeding behavior of Anopheles gambiae and Anopheles melas in Ghana, western Africa. Journal of Medical Entomology. 2010;47:28–31. doi: 10.1603/033.047.0104. [DOI] [PubMed] [Google Scholar]
- Turner TL, Hahn MW, Nuzhdin SV. Genomic islands of speciation in Anopheles gambiae . PLoS Biology. 2005;3:e285. doi: 10.1371/journal.pbio.0030285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P. Micro-Checker: software for identifying and correcting genotyping error is microsatellite data. Molecular Ecology Notes. 2004;4:535–538. [Google Scholar]
- Walker K, Lynch M. Contributions of Anopheles larval control to malaria suppression in tropical Africa: review of achievements and potentials. Medical and Veterinary Entomology. 2007;21:2–21. doi: 10.1111/j.1365-2915.2007.00674.x. [DOI] [PubMed] [Google Scholar]
- Warnock RCM, Yang Z, Donoghue PCJ. Exploring uncertainty in the calibration of the molecular clock. Biology letters. 2012;8:156–159. doi: 10.1098/rsbl.2011.0710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White BJ, Cheng C, Simard F, Costantini C, Besansky NJ. Genetic association of physically unlinked islands of genomic divergence in incipient species of Anopheles gambiae. Molecular Ecology. 2010;19:925–939. doi: 10.1111/j.1365-294X.2010.04531.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White GB. The Anopheles gambiae complex and disease transmission in Africa. Transactions of the Royal Society of Tropical Medicine and Hygiene. 1974;68:278–298. doi: 10.1016/0035-9203(74)90035-2. [DOI] [PubMed] [Google Scholar]
- White GB, Tessafaye F, Boreham PFL, Lemma G. Malaria vector capacity of Anopheles arabiensis and An. quadriannulatus in Ethiopia: chromosomal interpretation after 6 years storage of field preparations. Transactions of the Royal Society of Tropical Medicine and Hygiene. 1980;74:683–684. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.